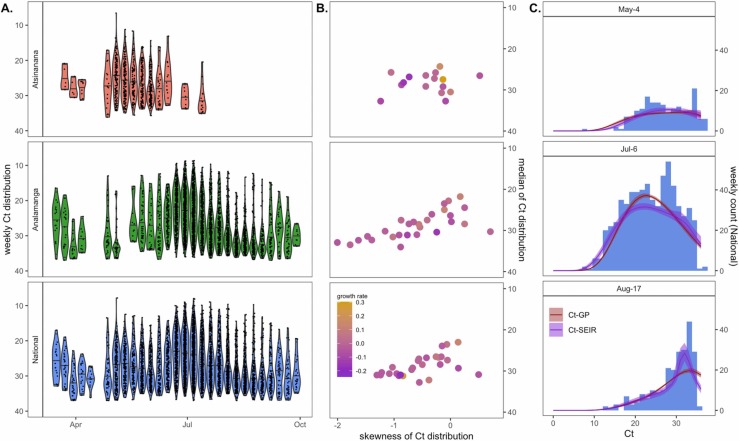
Fig. 3.
Population-level Ct distribution reflects epidemic dynamics of the first wave of COVID-19 across three Madagascar regions. (A.) Simulated weekly Ct distributions by Madagascar region, derived from population-level longitudinal GAMs (Fig. 2A), excluding random effects of test and target. (B.) Higher skew and lower median Ct from each cross-sectional Ct distribution in (A.) were loosely associated with higher epidemic growth rates from the corresponding week, here derived from EpiNow2 estimation from IPM case count data (Fig. 1B.) (C.) Cross-sectional Ct distributions from Analamanga time series in (A.) were fit via Gaussian process (GP) and SEIR mechanistic models incorporating a within-host viral kinetics model. Modeled Ct distributions are shown as solid lines (GP = red; SEIR = purple), with 95% quantiles in surrounding sheer shading. Both models effectively recapture the shape of the Ct histogram as it changes (skews left) across the duration of the first epidemic wave. Model fits to the full time series of Ct histograms across all three regions are visualized in Figs. S7–S9. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)