Abstract
Direct detection of SARS-CoV-2 viral antigens could replace RT-PCR, provided that its clinical performance is validated in different epidemiological settings. Here, we evaluated the performance of the VITROS Antigen test, an enzyme immunoassay detecting a SARS-CoV-2 antigen, in NPSs from 3 cohorts of patients.
Methods
: Three cohorts including SARS-CoV-2 RNA-positive samples collected during the first and second wave of the French epidemic between March 2020 and February 2021 (including variant B.1.1.7/α and variant B.1.351/β).
Results
: Among the 1763 prospectively tested subjects, 8.2% (145/1763) were SARS-CoV-2 RNA-positive by RT-PCR. Using Ct ≤ 30 and Ct ≤ 35 as thresholds, the sensitivities of the antigen assay were 98.8% (93.6–100%) and 93.5% (87.0–97.3%), respectively. The overall specificity of the assay was 100% (1614/1614; 99.8–100%). In a retrospective cohort of subjects infected with variants of concern, 90.4% (47/52) of NPSs containing B. B.1.1.7/α (Ct ≤ 35) and 100% (7/7) of those containing B.1.351/β were positive with the VITROS EIA SARS-CoV-2 Antigen test.
Conclusion
: The excellent performance of the EIA Antigen test reported here, including in patients infected with viral “variants of concern”, support the use of high-throughput, EIA-based SARS-CoV-2 antigen assays as an alternative or complement to nucleic acid testing in order to scale-up laboratory screening and diagnostic capacities.
Keywords: SARS CoV-2, EIA, Antigen test, Sensitivity, Specificity, Variants of concern
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the etiologic agent responsible for Coronavirus Disease 2019 (COVID-19). Various diagnostic tests are available to diagnose and screen SARS-CoV-2 infection. The reference method is the detection of viral RNA in nasopharyngeal swabs (NPS) by means of a nucleic acid amplification tests (NAAT), including reverse transcription-polymerase chain reaction (RT-PCR), transcription-mediated amplification (TMA) or loop-mediated isothermal amplification (LAMP). NAAT methods are both specific and highly sensitive, being able to detect small amounts of viral RNA, i.e. with low Ct value positivity, which do not always correlate with infectious viruses, as <3% of SARS CoV-2 can infect cell cultures in samples with low viral levels (Ct >35). NAAT tests are costly and must be performed in certified biology laboratories. They require technical skills and expensive equipment. At least 4 to 6 h are generally required to complete the analyses. Intermittent worldwide or local reagent and materials shortages have seriously hampered COVID-19 diagnostic capacities, especially during the two principal waves of the pandemic. This emphasizes the need for alternative, more flexible viral detection methods making it possible to rapidly scale-up the capacity of virology laboratories in case of epidemic outbreaks.
Direct detection of SARS-CoV-2 viral antigens in NPSs by means of lateral flow immunoassays (rapid diagnostic tests [RDT]) has been widely used as a fast and cheap approach to identify infected individuals within the framework of large-scale testing for subsequent tracing and isolation. Nevertheless, the reported sensitivities and specificities of SARS-CoV-2 antigen RDTs were generally reduced compared to those of NAATs [1]. Therefore, the use of SARS-CoV-2 antigen RDTs to diagnose the infection in clinical practice has been controversial and limited to specific indications (large-scale screening, early diagnosis of symptomatic infection). More recently, chemiluminescent enzyme immunoassays (EIA), run on high-throughput, automated, integrated platforms, have been developed for the detection of SARS-CoV-2 antigens. Such assays would be extremely helpful to complement NAAT testing in biology laboratories.
Our study evaluates the performance of a SARS-CoV-2 antigen EIA, VITROS SARS-CoV-2 Antigen (Ortho Clinical Diagnostics, Tarrytown, New Jersey), using the VITROS 3600 high-throughput automated integrated platform. The VITROS SARS-CoV-2 Antigen test is a chemiluminescent immunoassay that uses a capture antibody recognizing an epitope in the N-terminal domain (NTD) of the viral nucleoprotein. The study aimed to evaluate the performance (sensitivity, specificity, positive and negative predictive values) of the VITROS SARS-CoV-2 Antigen test in NPSs collected from a large series of patients and to describe the potential integration of this assay into diagnostic algorithms in biology laboratories. The study included 3 parts: (i) a retrospective analysis performed on frozen samples from symptomatic subjects infected during the first French epidemic wave (March to April 2020) with confirmed presence of SARS-CoV-2 RNA by means of RT-PCR; (ii) a retrospective analysis performed on frozen samples from symptomatic subjects diagnosed in February 2021 with positive SARS-CoV-2 RNA detection due to a known “variant of concern”, either B.1.1.7/α or B.1.351/β; (iii) a prospective study including all subjects consecutively tested for the presence of SARS-CoV-2 RNA by means of RT-PCR in our virology laboratory between November 18 and December 3, 2020 (excluding weekends and Mondays, for internal organizational purposes).
2. Materials and methods
The performance of the VITROS SARS-CoV-2 Antigen EIA test has been assessed in 3 cohorts of individuals tested for the presence of SARS-CoV-2 RNA, including hospitalized patients, outpatients and healthcare workers from the Henri Mondor university hospital. The first, retrospective cohort included 147 samples found to be positive for SARS-CoV-2 RNA during the first epidemic wave between March and April 2020. The second, retrospective cohort included frozen samples from symptomatic patients diagnosed in February 2021 as infected with variants of concern by means of full-length genome sequence analysis (54 individuals infected with variant B.1.1.7/α and 7 with variant B.1.351/β). The third, prospective cohort included 1763 unselected, fresh NPSs consecutively collected between November and December 2020, as part of the management of cases suspected of COVID-19 in our hospital. This anonymous retrospective study protocol followed the ethical guidelines of the declaration of Helsinki and was approved by our institutional review board.
2.1. Retrospective cohort collected during the first epidemic wave
For each participant, an NPS was collected in a viral transport medium (VTM) containing 0.9% NaCl for nucleic acid extraction. Part of the suspension had been used for SARS-CoV-2 RNA determination. The remaining part was stored at −70 °C until use in the present study. SARS-CoV-2 RNA had been sought by means of an “in-house“ assay based on the Charité protocol targeting the E gene or the RNA-dependent RNA polymerase gene [2] or of a commercially available RT-PCR assay targeting the E or S genes (RealStar® SARS-CoV-2 RT PCR Kit 1.0, Altona Diagnostics, Hamburg, Germany) [3]. SARS-CoV-2 RNA-positive NPSs were stratified according to the viral load, estimated by the cycle threshold value (Ct).
2.2. Retrospective cohort including SARS-CoV-2 “variants of concern” collected in February 2021
For each participant, an NPS was collected in a viral transport medium (Ozyme, Saint-Cyr-l’École, France). Part of the suspension had been used for SARS-CoV-2 RNA determination. The remaining part was stored at −70 °C until use in the present study. SARS-CoV-2 RNA had been sought by means of TaqPath COVID19 RT-PCR assay (ThermoFisher Scientific, Waltham, Massachusetts, USA), according to the manufacturer's instructions. Molecular characterization of the “variants of concern” was based on next-generation sequencing, by means of the COVIDSeq Test (Illumina, San Diego, California), that uses 98-target multiplex amplifications along the full SARS-CoV-2 genome. The libraries were sequenced with NextSeq 500/550 High Output Kit v2.5 on a NextSeq 500 device. The sequences were demultiplexed and assembled as full-length genomes by means of the DRAGEN COVIDSeq Test Pipeline on a DRAGEN server. Lineages and clades were interpreted using Pangolin and NextClade.
2.3. Prospective cohort
For each participant, an NPS was collected in a viral transport medium (Greiner Bio One® or Labo Moderne LMR®, Ozyme®). Each fresh sample was subsequently split into two aliquots: one 400-µL aliquot for antigen testing by EIA and another aliquot for concomitant SARS-CoV-2 RNA detection by means of a commercially available NAAT (TMA-based Aptima™ SARS CoV-2 Assay, Hologic, San Diego, California; or PCR-based Alinity m® SARS CoV-2 Assay Abbott, Germany). Samples found to be RNA-positive in TMA were retested by RT-PCR (ARGENE® SARS-COV-2 R-GENE, bioMérieux, France, or RealStar® SARS-CoV-2 RT-PCR Kit 1.0) for the determination of Ct values.
2.4. SARS-CoV-2 antigen detection by means of VITROS SARS-CoV-2 antigen test
Briefly, 400 μL of viral transport medium was mixed with 100 μL of the extraction buffer and then processed with random access according to the manufacturer's instructions. The samples were loaded on the VITROS 3600 platform which has a capacity of 130 samples per hour and a time to first result of 48 min. The results were interpreted as “positive“ or “negative“ according to the manufacturer's instructions. Discordant results (positive RT-PCR test with a Ct <30 and negative antigen test) were systematically retested by NAAT. In this setting, because freeze/thaw procedures could alter the RNA quality, the Ct value of the latest test was used for analysis.
3. Statistical analysis
Quantitative variables were expressed as median, interquartile range (IQR) and range (minimum-maximum). Qualitative data were expressed as raw numbers in percentages. The diagnostic performance analysis of VITROS SARS-CoV-2 Antigen test was conducted considering RT-PCR results as the reference, computing sensitivity and specificity along with their 95% confidence intervals calculated using the exact method. To illustrate the clinical significance of the results in a real-life setting, positive and negative predictive values were calculated for a range of hypothetical prevalence values. All statistical analyses were performed using Stata software version v16.1 (StataCorp LP, College Station, Texas).
4. Results
4.1. Characteristics of the study population
First-wave retrospective cohort. One hundred and forty-seven samples from patients with confirmed COVID-19 based on SARS-CoV-2 RNA detection were tested with the antigen EIA (Table 1 ). Their median age was 65 years (IQR: 51–83 years), and 53.7% of them were males. When the information was available (n = 71), 66.2% of samples had been collected within 7 days after symptom onset, and 35.2% had been collected within the first 3 days after symptom onset. Ct values ranged from 13 to 39 (median: 27) with the E gene target.
Table 1.
Characteristics of the SARS-CoV-2 RNA-positive study population, including a retrospective cohort of 147 patients sampled during the first French epidemic wave (between March and April 2020) and 145 patients prospectively tested during the second French epidemic wave (between November and December 2020).
| Retrospective cohort of SARS-CoV-2 RNA-positive patients(N = 147) | Prospective cohort of SARS-CoV-2 RNA-positive patients(N = 145) | |
|---|---|---|
| Median age (min-max), year | 65 (19–95) | 60 (17–100) |
| % male gender (n/N) | 53.7% (79/147) | 36.6% (53/145) |
| Wards | ||
| Intensive care units [% (n/N)] | 6.8% (10/147) | 3.4% (5/145) |
| Geriatric wards [% (n/N)] | 34.7% (51/147) | 43.4% (63/145) |
| Medical wards [% (n/N)] | 8.2% (12/147) | 11.0% (16/145) |
| Outpatients [% (n/N)] | 29.3% (43/147) | 13.8% (20/145) |
| Healthcare workers [% (n/N)] | 13.6% (20/147) | 24.8% (36/145) |
| Other [% (n/N)] | 7.5% (11/147) | 3.4% (5/145) |
“Variant of concern retrospective cohort. The 59 samples infected with either of the two “variants of concern” studied were collected within the framework of the French national SARS-CoV-2 sequencing surveillance program. Ct values ranged from 10 to 40 in samples containing the B.1.1.7/α variant, 19 to 26 in those containing the B.1.351/β variant with the N gene target.
Prospective cohort. In this part of the study, 1763 subjects tested for a suspicion of COVID-19 in our hospital between November 18 and December 3, 2020, were included. SARS CoV-2 RNA detection by RT-PCR or TMA was performed in all samples, that were subsequently classified into 2 categories: 1614 patients with negative SARS-CoV-2 RNA detection, and 145 patients with detectable SARS-CoV-2 RNA. Four patients were excluded from the study because molecular testing was not conclusive. All of the 1763 fresh NPSs were tested in parallel with the VITROS EIA SARS-CoV-2 Antigen test.
The 145 prospectively collected samples found to be SARS-CoV-2 RNA-positive were used to characterize the sensitivity of the antigen EIA assay (Table 1). Their median age was 60 years (IQR: 43–84 years) and 36.6% of them were males. When the information was available (n = 119), 36.1% (43/119) of patients were asymptomatic, 47.0% (56/119) of samples had been collected within 7 days after symptom onset, and 40.3% (48/119) had been collected within the 3 first days after symptom onset. Ct values ranged from 15 to 39 (median: 25) using the mean value of the N and RNA-dependent RNA polymerase gene targets (ARGENE® SARS-COV-2 R-GENE).
The samples from the 1614 SARS-CoV-2 RNA-negative individuals were tested to determine the specificity of the antigen EIA. Their median age was 57 years (IQR: 39–80 years), and 37.1% of them were males.
4.2. Performance of the VITROS EIA SARS-CoV-2 antigen assay
The evaluable assay rate of the VITROS EIA SARS-CoV-2 Antigen test was high (99.8%). The 3 specimens with indeterminate results were all in the SARS-CoV-2 RNA-negative group.
Specificity. All of the 1614 SARS CoV-2 RNA-negative samples, collected prospectively between November and December 2020, tested negative for the detection of SARS-CoV-2 antigen by EIA (100% specificity, 95% confidence interval [CI]: 99.8–100%). Notably, specificity could not be tested against other respiratory viral pathogens, as circulation of such viruses was rarely observed during the period of sampling.
Sensitivity. The sensitivity of the VITROS EIA SARS-CoV-2 Antigen test is shown in Table 2 , using SARS-CoV-2 RNA detection as the reference. As mentioned by the manufacturer, the test is strictly qualitative and there was no correlation between the quantitative S/C signal by the EIA Ag test and Ct values. Sensitivity correlated with the delay of sampling after symptom onset and the viral load measured by the Ct value in RT-PCR. In patients with Ct values ≤25, sensitivities for the detection of SARS-CoV-2 antigen were 98.5% (95%CI: 92.0%−100%) and 98.4% (95%CI: 91.2%−100%) in the first-wave retrospective and in the prospective cohorts, respectively. The detection rates of the antigen test remained high (98.8%, 95.0%, 93.5%) in the prospective cohort for Ct values ≤30, ≤33 and ≤35, respectively (Table 2). In the first-wave retrospective cohort, the detection rates of the test decreased only for Ct values >33 (28.6%), suggesting a possible deleterious effect of freezing/thawing cycles on the sensitivity of antigen testing for these very low viral levels.
Table 2.
Sensitivity (95%CI) of the VITROS SARS-CoV-2 Antigen test in SARS-CoV-2 RNA-positive samples from the retrospective and prospective cohorts, according to the number of days after symptom onset and the viral load assessed by the cycle threshold value (Ct) in RT-PCR.
| Retrospective cohort(N = 147) | Prospective cohort(N = 145) | |||
|---|---|---|---|---|
| Days from symptom onset | ||||
| N Ag+/ N PCR+ | Sensitivity (95% CI) | N Ag+/ N PCR+ | Sensitivity (95% CI) | |
| Days ≤3 | 18/25 | 72.0% (50.6–87.9) | 40/48 | 83.3% (69.8–92.5) |
| Days 4–7 | 17/22 | 77.3% (54.6–92.2) | 8/8 | 100% (63.1–100) |
| Days 8–11 | 12/20 | 60.0% (36.1–80.9) | 6/8 | 75.0% (34.9–96.8) |
| Days ≥12 | 2/4 | 50.0% (6.8–86.1) | 3/12 | 25.0% (5.5–57.2) |
| Asymptomatic | 0/1 | 0.0% (0.0 −97.5) | 30/43 | 69.8% (53.9–82.8) |
| Unknown | 64/75 | 85.3% (75.3–92.4) | 15/25 | 60% (38.7–79.8) |
| Ct value category | ||||
| Ct ≤20 | 20/21 | 95.2% (76.2–99.9) | 23/23 | 100% (85.2–100) |
| Ct 21–25 | 46/46 | 100% (92.3–100.0) | 37/38 | 97.4% (86.2–99.9) |
| Ct 26–30 | 30/32 | 93.8% (79.2–99.2) | 24/24 | 100% (85.8–100) |
| Ct 31–35 | 15/35 | 42.9% (26.3–60.6) | 16/22 | 72.7% (49.8–89.3) |
| Ct 36–39 | 2/13 | 15.4% (1.9–45.4) | 1/6 | 16.7% (0.4–64.1) |
| Ct ≤30 | 96/99 | 97.0% (91.4–99.4) | 84/85 | 98.8% (93.6–100.0) |
| Ct ≤33 | 105/113 | 92.9% (86.5–96.9) | 95/100 | 95.0% (88.7–98.4) |
| Ct ≤35 | 111/134 | 82.8% (75.9–88.8) | 100/107 | 93.5% (87.0–97.3) |
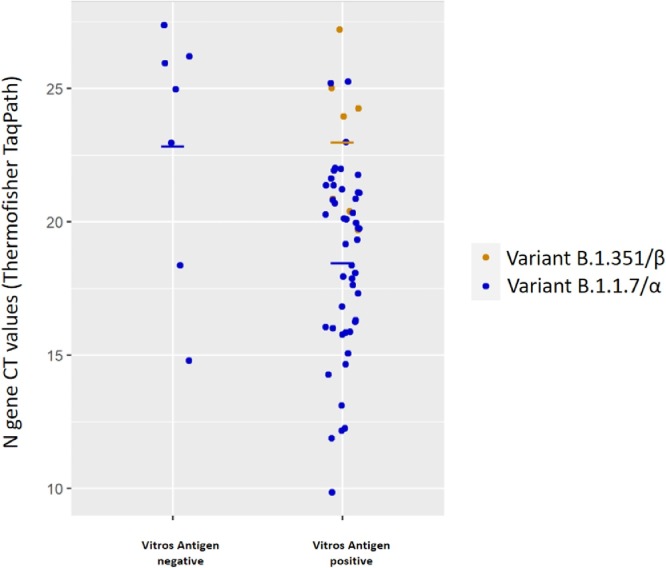
We tested the sensitivity of antigen detection of the assay in samples containing two recently emerged SARS-CoV-2 “variants of concern”. Fig. 1 shows the EIA assay results according to Ct values measured by TaqPath COVID19 RT-PCR kit. Among the 52 samples containing the B.1.1.7/α variant, 47 tested positive with VITROS EIA SARS-CoV-2 Antigen (sensitivity: 90.4%; for Ct values Ct ≤35). All of the 7 samples containing the B.1.351/β variant were positive with the antigen EIA (sensitivity: 100%; 95%CI: 57.1%−100% for Ct values ranging from 19 to 26).
Fig. 1.
Sensitivity of the VITROS EIA SARS-CoV-2 Antigen test to detect two recently emerged SARS-CoV-2 “variants of concern”. The antigen test results are presented according to Ct values measured by TaqPath COVID19 RT-PCR kit. Among the 52 samples containing the B.1.1.7/α variant (with Ct ≤35), 47 tested positive with VITROS EIA SARS-CoV-2 Antigen (sensitivity: 90.4%). All of the 7 samples containing the B.1.351/β variant were positive with the antigen EIA (sensitivity: 100%; 95%CI: 57.1%−100% for Ct values ranging from 19 to 26).
Negative and positive predictive values.Table 3 shows the calculated negative and positive predictive values (NPV and PPV) of the antigen EIA assay under varying hypothetical prevalence rates of infection in the tested population. Simulation was based on sensitivities and specificities estimated in symptomatic patients in the prospective cohort during the first 7 days following symptom onset. Globally, the PPV (100%) and NPV values (94.2% to 99.9% for a prevalence of 1 to 30%) were very high.
Table 3.
Simulations of the calculated negative and positive predictive values (NPV and PPV) of the VITROS SARS-CoV-2 Antigen test according to the prevalence of infection in the tested population. The simulation is based on sensitivities and specificities estimated in symptomatic patients in the prospective cohort during the first 7 days following symptom onset.
| Prevalence (%) | Case (n) | Non-case (n) | True positive(n) | False negative (n) | True negative(n) | False positive (n) | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|---|
| 1.0% | 10 | 990 | 9 | 1 | 990 | <1 | 100% | 99.9% |
| 2.0% | 20 | 980 | 17 | 3 | 980 | <1 | 100% | 99.7% |
| 5.0% | 50 | 950 | 43 | 7 | 950 | <1 | 100% | 99.3% |
| 10.0% | 100 | 900 | 86 | 14 | 900 | <1 | 100% | 98.4% |
| 15.0% | 150 | 850 | 129 | 21 | 850 | <1 | 100% | 97.5% |
| 20.0% | 200 | 800 | 171 | 29 | 800 | <1 | 100% | 96.6% |
| 30.0% | 300 | 700 | 257 | 43 | 700 | <1 | 100% | 94.2% |
| Simulation based on the sensitivity values found in symptomatic individuals during the 7 days following symptom onset. Sensitivity = 85.7%, specificity = 100.0% | ||||||||
5. Discussion
In the present study, we used a large collection of retrospectively and prospectively collected NPSs to evaluate the performance (sensitivity, specificity, positive and negative predictive values) of the VITROS EIA SARS-CoV-2 Antigen test, an EIA assay detecting SARS-CoV-2 antigen. The excellent specificity we observed without any false-positive result, based on over 1600 SARS-CoV-2 negative samples, implies that confirmation of positive EIA results by means of nucleic acid testing is not required, even when testing populations with a low prevalence of infection, such as groups of individuals targeted by large-scale screening campaigns.
Although SARS-CoV-2 antigen testing is generally thought to be less sensitive for the diagnosis of infection than viral RNA detection, we show here that the VITROS antigen test is highly sensitive for the entire range of viral loads associated with viral infectivity (Ct ≤35). Indeed, the test achieved over 90% sensitivity for prospectively collected samples with a Ct ≤35 in RT-PCR. This performance largely exceeds the requirements from the ECDC and WHO for the diagnosis of infection and large-scale screening [1,4]. Several groups worldwide routinely use a cut-off Ct of 35 to report a positive result of RT-PCR, because only <3% of cell cultures can be infected by samples with low viral levels (Ct >35) (5). Overall, the sensitivity of the VITROS EIA SARS-CoV-2 Antigen test was close to that of RT-PCR for prospectively tested samples with Ct values compatible with an actual infection. Our data are in line with other evaluations of SARS-CoV-2 antigen tests based on EIA (6,7,8). Thus, EIA assays can be used as an alternative to RT-PCR or in complement to it to scale-up COVID-19 diagnostic capacities in biology laboratories. This may prove particularly useful in the context of new epidemic waves, with the ability of each platform to run 130 samples per hour and to provide “random access” for emergency testing.
The world is currently facing a rapid increase in COVID-19 case rates, potentially associated with the emergence of new SARS-CoV-2 variants of concern, including B.1.1.7/α and B.1.351/β. Most of the amino acid changes observed in these variants occur in the spike protein. As is the case of many other antigen tests, VITROS EIA SARS-CoV-2 Antigen test targets the viral nucleocapsid protein. We confirm here that its performance is not affected by changes carried by these two variants of concern. Nevertheless, the emergence of variants carrying mutations in the nucleoprotein is possible and epidemiological surveillance and a regular assessment of the consequences of such changes on antigen test performance will be needed.
In conclusion, the VITROS EIA SARS-CoV-2 Antigen test, a high-throughput, automated EIA assay for the detection of SARS-CoV-2 antigen, has excellent specificity. Its sensitivity is close to that of RT-PCR in patients with viral loads indicating the presence of infectious viruses. Based on these results and others from the literature (6-8), the place of antigen testing in diagnostic strategies should be revisited and the use of EIA platforms, as alternatives or complements to RT-PCR should be encouraged, in order to increase the diagnostic capabilities of biology laboratories. Other EIA-based antigen tests are being developed and could help in this endeavor, provided that structured evaluations are performed and the assay performance is considered as acceptable. Overall, the performance, ease of use, simplicity, low cost, high-throughput capacity and rapidity of results of automated EIA-based antigen tests make them a technique of choice for the laboratory diagnosis of SARS-CoV-2 infections.
Funding sources
None
Declaration of Competing Interest
S.F. has served as a speaker for Abbvie and Abbott diagnostics. C.R. has served as an advisor, and/or speaker for Illumina, and Vela Diagnostics. SC has served as a speaker for Gilead, Abbvie and Abbott diagnostics. J.-M.P. has served as an advisor, and/or speaker for Abbvie, Gilead, Assembly Biosciences, Arbutus, Merck, Regulus, and Memo Therapeutics. The remaining authors have no conflict of interest to disclose.
References
- 1.www.ecdc.europa.eu/en/publications-data/options-use-rapid-antigen-tests-covid-19-eueea-and-uk., 2019.
- 2.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.-.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Visseaux B., Le Hingrat Q., Collin G., Ferré V., Storto A., Ichou H., Bouzid D., Poey N., de Montmollin E., Descamps D., Houhou-Fidouh N. Evaluation of the RealStar® SARS-CoV-2 RT-PCR kit RUO performances and limit of detection. J. Clin. Virol. 2020;129 doi: 10.1016/j.jcv.2020.104520. https://www.who.int/publications/i/item/WHO-2019-nCoV-lab-testing-2021.1-eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.https://www.who.int/publications/m/item/covid-19-target-product-profiles-for-priority-diagnostics-to-support-response-to-the-covid-19-pandemic-v.0.1.
- 5.Jaafar R., Aherfi S., Wurtz N., Grimaldier C., Hoang V.T., Colson P., et al. Correlation between 3790 qPCR positives samples and positive cell cultures including 1941 SARS-CoV-2 isolates. Clin. Infect. Dis. 2020 [Google Scholar]
- 6.Aoki K., Nagasawa T., Ishii Y., Yagi S., Okuma S., Kashiwagi K., Maeda T., Miyazaki T., Yoshizawa S., Tateda K. Clinical validation of quantitative SARS-CoV-2 antigen assays to estimate SARS-CoV-2 viral loads in nasopharyngeal swabs. J. Infect. Chemother. 2021;27:613–616. doi: 10.1016/j.jiac.2020.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., Kakizaki Y., Miyashita Y., Yagi S., Kojima S., Omata M. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int. J. Infect. Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levett P.N., Cheung B., Kustra J., Pidduck T., Mak A., Tsang F., Petric M., Krajden M. Evaluation of a high volume antigen test for detection of SARS-CoV-2. J. Clin. Virol. 2021;142 doi: 10.1016/j.jcv.2021.104938. [DOI] [PMC free article] [PubMed] [Google Scholar]