Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic of coronavirus disease 2019 (COVID-19) since December 2019 that has led to more than 160 million confirmed cases, including 3.3 million deaths. To understand the mechanism by which SARS-CoV-2 invades human cells and reveal organ-specific susceptible cell types for COVID-19, we conducted comprehensive bioinformatic analysis using public single-cell RNA sequencing datasets. Utilizing the expression information of six confirmed COVID-19 receptors (ACE2, TMPRSS2, NRP1, AXL, FURIN and CTSL), we demonstrated that macrophages are the most likely cells that may be associated with SARS-CoV-2 pathogenesis in lung. Besides the widely reported ‘chemokine storm’, we identified ribosome related pathways that may also be potential therapeutic target for COVID-19 lung infection patients. Moreover, cell-cell communication analysis and trajectory analysis revealed that M1-like macrophages showed the highest relation to severe COVID-19 patients. And we also demonstrated that up-regulation of chemokine pathways generally lead to severe symptoms, while down-regulation of ribosome and RNA activity related pathways are more likely to be mild. Other organ-specific susceptible cell type analyses could also provide potential targets for COVID-19 therapy. This work can provide clues for understanding the pathogenesis of COVID-19 and contribute to understanding the mechanism by which SARS-CoV-2 invades human cells.
Keywords: COVID-19, Single-cell RNA sequencing, Bioinformatics, Macrophage, Data mining
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic of coronavirus disease 2019 (COVID-19) since December 2019 [[1], [2], [3]]. Similar to the other two members of the β coronavirus genus, i.e., severe acute respiratory syndrome coronavirus (SARS-CoV [4]) and Middle East respiratory syndrome coronavirus (MERS-CoV [5]), SARS-CoV-2 can lead to not only serious respiratory tract diseases but also damage to many other human organs [[6], [7], [8]]. As of May 2021, no effective antiviral for COVID-19 is available, and COVID-19 has caused more than 3.3 million global deaths according to the World Health Organization (WHO). Hence, there is an urgent need to understand the physiological and pathological mechanisms by which SARS-CoV-2 infects humans. Various studies have demonstrated that a glycosylated spike (S) protein of coronavirus plays a key role in the process of cell entry [9,10]. As a cell entry receptor of SARS-CoV [11], angiotensin-converting enzyme 2 (ACE2) has also been proven to be an important entry receptor for SARS-CoV-2 [1]. Previous studies have elucidated that the spike protein can bind with the ACE2 receptor in the receptor binding domain (RBD), and the spike protein is primed by the serine protease TMPRSS2 [12]. Utilizing the expression value of ACE2 in single cell data, Dey et al. identified several cell types susceptible to the virus and PPAR signaling pathway as key regulator during infection [13].
However, single-cell RNA sequencing data have revealed that the expression of ACE2 is very low in most human tissues [14]. This indicates that other receptors may exist for target cell entry due to the rapid and multiorgan infection of COVID-19. Recently, many studies have reported several candidate receptors that may facilitate SARS-CoV-2 entry. Cantuti-Castelvetri et al. reported that neuropilin-1 (NRP1) could facilitate SARS-CoV-2 cell entry and may serve as a potential target for drug development [15]. Wu et al. demonstrated that a redundant furin cut site is responsible for the stronger infection of SARS-CoV-2 [16]. Zhao et al. reported that cathepsin L (CTCL) plays a crucial role during the process of SARS-CoV-2 infection [17]. Wei et al. found that scavenger receptor B type 1 (SR-B1) can act as a host factor and promote SARS-CoV-2 entry [18]. Wang et al. revealed that tyrosine-protein kinase receptor UFO (AXL) interacts with SARS-CoV-2 [19].
Although various studies have elucidated the cell entry mechanism of ACE2 and TMPRSS2, little is known about the other newly discovered receptors. Since single-cell RNA sequencing technology has shown great advantages in biological studies [[20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31]], in this study, we utilized five single-cell RNA sequencing public datasets, each for a tissue vulnerable to COVID-19 infection (specifically, lung, heart, kidney, liver and bladder) [32,33]. Organ-specific susceptible cell types to SARS-CoV-2 were identified, and in lung tissue, we demonstrated that up-regulation of chemokine pathways generally lead to severe symptoms, while down-regulation of ribosome and RNA activity related pathways is more likely to be mild [34]. This work can provide clues for understanding the pathogenesis of COVID-19 and contribute to understanding the mechanism by which SARS-CoV-2 invades human cells [35,36].
2. Material and methods
2.1. Data resources
Public single-cell RNA sequencing datasets were downloaded from the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). The lung dataset used in this study was obtained as NO. GSE145926. This dataset contained the bronchoalveolar lavage fluid (BALF) of nine COVID-19 patients and three healthy controls. To be consistent with the authors [37], we also obtained other healthy lung scRNA-seq data from NO. GSM3660650 [38]. To explore the expression levels of these candidate receptors, we then utilized scRNA-seq datasets of four other healthy tissues to discover the highly expressed cell types. Specifically, healthy heart samples were acquired from NO. GSE109816 [39]. Five heathy liver samples of scRNA-seq data were acquired from NO. GSE115469 [40]. Three scRNA-seq samples of healthy human kidney were obtained from NO. GSE131685 [41]. Finally, we utilized three healthy bladder scRNA-seq datasets from NO. GSE129845 [42]. The details of all single-cell datasets used in this study was shown in Table 1 .
Table 1.
Summary of the datasets used in this study.
| Dataset GEO number | Organ | DOI | URL |
|---|---|---|---|
|
GSE145926 GSM3660650 |
Lung | 10.1038/s41591-020-0901-9 | https://www.nature.com/articles/s41591-020-0901-9 |
| GSE109816 | Heart | 10.1038/s41556-019-0446-7 | https://www.nature.com/articles/s41556-019-0446-7 |
| GSE115469 | Liver | 10.1038/s41467-018-06318-7 | https://www.nature.com/articles/s41467-018-06318-7 |
| GSE131685 | Kidney | 10.1038/s41597-019-0351-8 | https://www.nature.com/articles/s41597-019-0351-8 |
| GSE129845 | Bladder | 10.1681/ASN.2019040335 | https://pubmed.ncbi.nlm.nih.gov/31462402/ |
2.2. Dataset processing and analysis
Seurat 3.0 was used to process and analyse the lung dataset [43]. Following the authors’ description, we removed low-quality cells by selecting feature numbers between 200 and 6000. Moreover, cells with a mitochondrial percentage >10% and library size <1000 were also identified. The data were normalized by the “NormalizeData” function with the “LogNormalize” method. A total of 2000 highly variable genes were obtained for each sample using the “vst” selection method in the “FindVariableFeatures” function. To correct the batch effect, we utilized the “FindIntegrationAnchors” function and integrated the COVID-19 patients and healthy human samples together for further analysis. Principal component analysis (PCA) was utilized to find the k-nearest neighbours. The “FindClusters” function, which employs the “Louvain” algorithm, showed 31 clusters with a resolution parameter setting of 1.2 [44]. The dimension reduction method UMAP (Uniform Manifold Approximation and Projection) was used for data visualization [45]. Based on these clusters, cell type annotation was carried out with canonical marker genes.
Downstream analysis started by finding the top 100 differentially expressed genes (DEGs) of the COVID-19 group and healthy group with the Seurat function “FindMarkers”. Differentially expressed genes (DEGs) were filtered by (p value <= 0.05 and q value <= 0.2) for enrichment analysis. The DEGs were further utilized for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment to identify biological pathways. The R package clusterProfiler (version 3.16.1) was used for enrichment analysis [46]. CellChat (version 1.1.0) was utilized for cell-cell communication analysis [47]. Gene set variation analysis (GSVA) analysis was conducted using R package GSVA (version 1.42.0), 50 hallmark gene sets in the Molecular Signatures Database (MSigDB) were utilized for enrichment [48]. We utilized Monocle 2 (version 2.18.0) for trajectory inference and GeneSwitches was used for identifying functional changes pathways [49,50].
The other four scRNA-seq datasets were also analysed similar to the process for the lung dataset. We have uploaded all of the code used in this study at GitHub (https://github.com/ZilongZhang44/COVID-19).
3. Results
3.1. Lung dataset
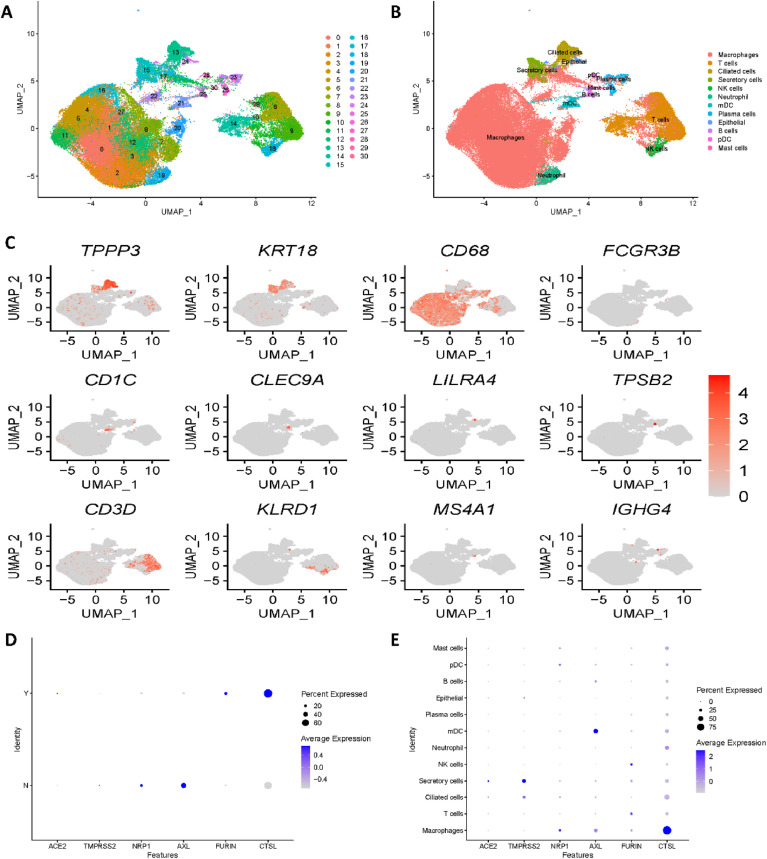
Unsupervised clustering of the single-cell RNA sequencing lung dataset showed 31 clusters (Fig. 1 A). Canonical marker genes provided by the authors were utilized for cell type annotation; subsequently, twelve cell types were identified, as shown in Fig. 1B. Fig. 1C shows the gene expression of the most important canonical marker genes, specifically TPPP3, KRT18, CD68, FCGR3B, CD1C, CLEC9A, LILRA4, TPSB2, CD3D, KLRD1, MS4A1 and IGHG4. Clusters 0, 1, 2, 3, 4, 5, 7, 8, 10, 11, 12, and 16 were annotated as macrophages with the canonical marker gene CD68. With CD3D, T cells were identified corresponding to clusters 6, 9, 14 and 29. With TPP3 and KRT18, ciliated cells and secretory cells were annotated corresponding to cluster 13 and cluster 15, respectively. Cluster 18 was identified as NK cells with KLRD1, while cluster 19 was identified as neutrophil cells with FCGR3B. Cluster 21 was annotated to myeloid dendritic cells with marker genes CD1C and CLEC9A. Subsequently, B cells and mast cells were annotated with MS4A1 and TPSB2, which corresponded to cluster 25 and cluster 30, respectively. With IFHG4, clusters 23 and 26 were identified as plasma cells. Cluster 24 was annotated as epithelial cells with marker genes TPPP3 and KRT18. Finally, plasmacytoid dendritic cells were identified as corresponding to cluster 28 with the marker gene LILRA4.
Fig. 1.
Single-cell RNA sequencing analysis of the lung dataset. (A) Unsupervised clustering revealed 31 clusters in integrated lung dataset. (B) Cell type annotation was conducted and 12 cell types were identified. (C) Expression of canonical marker genes used for cell type annotation. (D) Expression differences in all 6 reported receptors between COVID-19 patients and healthy controls. Y stands for COVID-19 patients, N stands for healthy controls. (E) Dot plot shows the expression of six receptors in all identified cell types.
Moreover, we divided the lung dataset into a COVID-19 group and a healthy group and compared the expression differences of all potential receptors mentioned above: ACE2, TMPRSS2, NRP1, AXL, FURIN and CTSL (SR-B1 gene was not detected). Fig. 1D illustrates significant expression differences in all 6 reported receptors, which is consistent with the original paper. As ACE2 and TMPRSS2 have already been investigated in many studies, we mainly focused on the expression of the most recently reported genes NRP1, AXL, FURIN and CTSL. As shown in Fig. 1D, in the COVID-19 group, NRP1 and AXL were downregulated, while FURIN and CTSL were upregulated. To explore specific cell types vulnerable to SARS-CoV-2 infections in the lung, we then revealed the expression of receptors by cell type. Fig. 1E illustrates that all four of the latest reported receptors showed high expression in macrophages. Hence, we concentrated on macrophage cell types for further analysis.
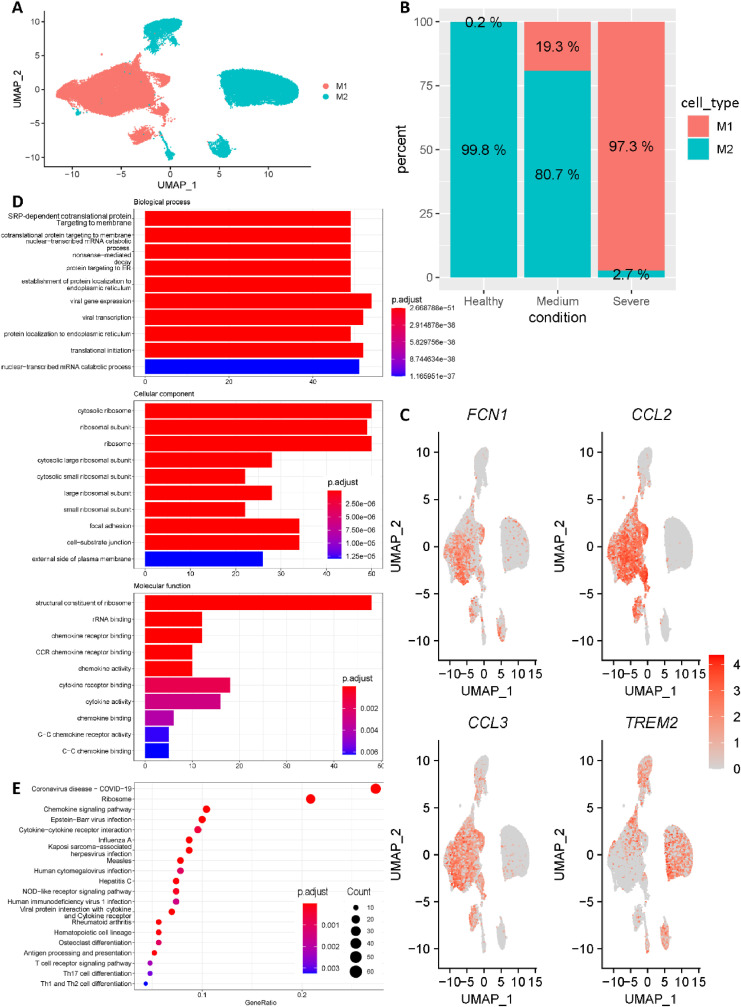
To reveal the heterogeneity among macrophages, the macrophages cells were further clustered into M1-like macrophages and M2-like macrophages based on the canonical marker genes FCN1, CCL2, CCL3 and TREM2 (Fig. 2 A, C). Interestingly, we noticed that percentage of M1-like macrophages was highly correlated with COVID-19 progression. Specifically, M1-like macrophages only accounted for 0.2% of macrophages in healthy control group, while increasing to 19.3% and 97.3% in medium COVID-19 cases and severe COVID-19 cases, respectively (Fig. 2 B). In addition, all M1-like macrophages were divided into a COVID-19 group and a healthy group, and differential gene expression analysis was applied between these two groups. Differentially expressed genes (DEGs) were filtered by (p value <= 0.05 and q value <= 0.2) for enrichment analysis. Fig. 2D showed the top 10 enriched Gene Ontology (GO) terms for each of three ontologies. Interestingly, besides the widely reported ‘chemokine storm’, ribosome related terms were strongly enriched. Strikingly, as shown in Fig. 2E, ribosome pathway was the greatest enrichment besides COVID-19 pathway. Collectively, ribosome pathway may be possible therapeutic target for COVID-19 lung infection patients.
Fig. 2.
Macrophages subgroup classification. (A) All macrophages were further clustered into M1-like macrophages and M2-like macrophages. (B) The fraction of cells originated from different groups (Healthy controls, Medium patients and Severe patients). (C) Expression of canonical marker genes used for cell subtype annotation. (D) Enriched terms for each of three ontologies revealed by Gene Ontology (GO) enrichment results of M1-like macrophages. (E) Most enriched KEGG pathways revealed by differentially expressed genes of M1-like macrophages.
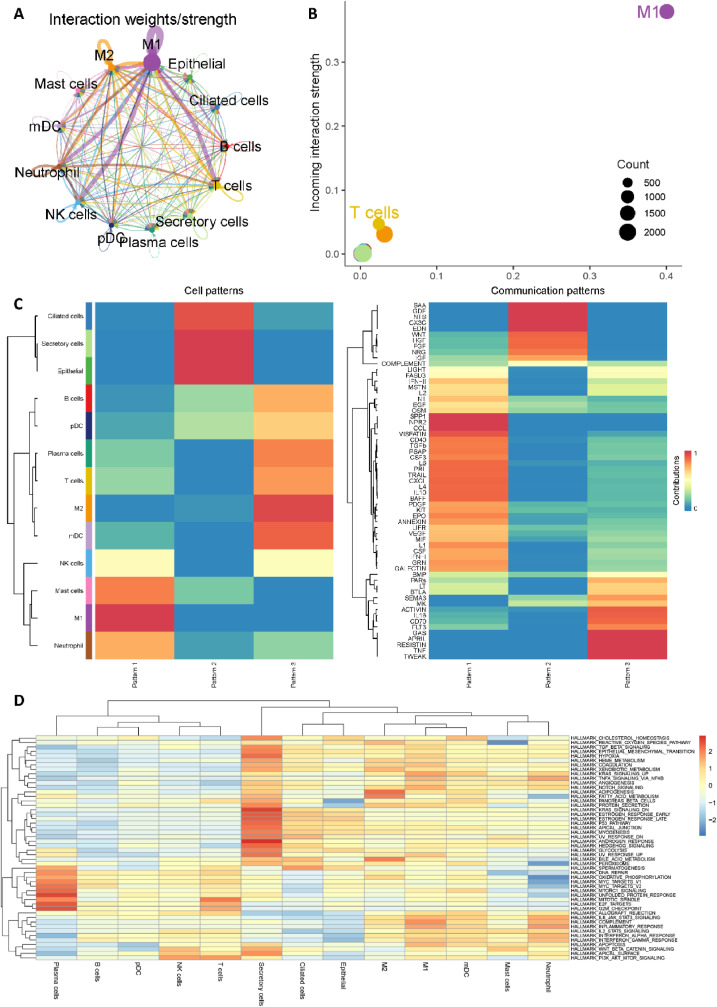
We then utilized CellChat to investigate cell-cell communication in COVID-19 patients [47]. Notably, M1-like macrophages showed significantly interactions with T cells, Neutrophil and NK cells (Fig. 3 A), and Fig. 3B revealed the dominant role of M1-like macrophages in COVID-19 patients. These findings have also been verified by Lv et al. that M1-like macrophages could facilitate SARS-CoV-2 infection of the lungs by allowing the entry of SARS-CoV-2 RNA from the endosomes into the cytoplasm [51]. In addition, as shown in Fig. 3C, CellChat revealed three communication outgoing patterns. M1-like macrophages were grouped into pattern 1 and SPP1 (secreted phosphoprotein 1), NPR2 (Natriuretic Peptide Receptor 2) and CCL (Chemokine ligands) may be possible significant pathways. Moreover, GSVA software was utilized to identify differences in hallmark gene sets of Molecular Signatures Database (MSigDB) across different cell types [48,[52], [53], [54], [55]]. As shown in Fig. 3D, complement pathway, KRAS_singaling pathway and Inflammatory response pathway are significantly enriched in M1-like macrophages, which may be considered as possible therapeutic targets for further study.
Fig. 3.
Cell-cell communication analysis of macrophages. (A) Aggregated cell-cell communication network of total interaction strength (weights) between any two cell groups. (B) Signaling role analysis on the aggregated cell-cell communication network from all signaling pathways. (C) Global communication pattern of multiple cell types. (D) Gene set variation analysis (GSVA) for 50 hallmark gene sets in the Molecular Signatures Database (MSigDB).
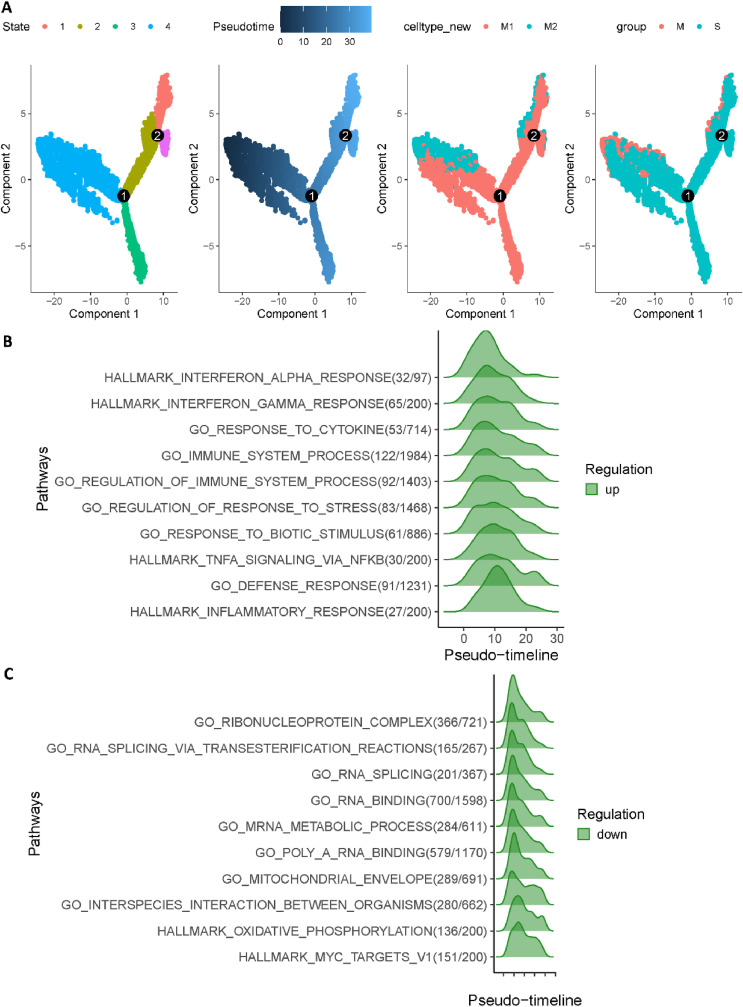
Finally, we used Monocle 2 to investigate cell trajectory of macrophages in COVID-19 patients [49]. Fig. 4 A revealed two major cell trajectories (i.e., path1: from state 4 to state 3, and path2: from state 4 through state 2 to state 1). Notably, we noticed that cell fates belong to path1 (states 4, 3) had a much higher severe rate than path2 (states 4, 2, 1). Subsequently, we utilized GeneSwitches to discover functional events during cell state transitions [50]. Moreover, as shown in Fig. 4B, the top ten significantly changed pathways for path1 are strongly related to previous studies about COVID-19 severe patients, which included interferon alpha, cytokine and immune system. Strikingly, Fig. 4C revealed that the highest functional change pathways in path2 (states 4, 2, 1) are related to ribonucleoprotein and RNA activity, which is consistent with enrichment analysis. Collectively, we demonstrated that up-regulation of Chemokine pathways generally lead to severe symptoms, while down-regulation of ribosome and RNA activity related pathways is more likely to be mild.
Fig. 4.
Trajectory inference of macrophages in COVID-19 patients. (A) Trajectory analysis results of macrophages in COVID-19 patients. (From left to right, cells were coloured by states, cell type, pseudotime and patient group, respectively.) (B) Top 10 significantly changed pathways ordered by the switching time of path1 (states 4, 3). (C) Top 10 significantly changed pathways ordered by the switching time of path2 (states 4, 2, 1).
3.2. Heart dataset
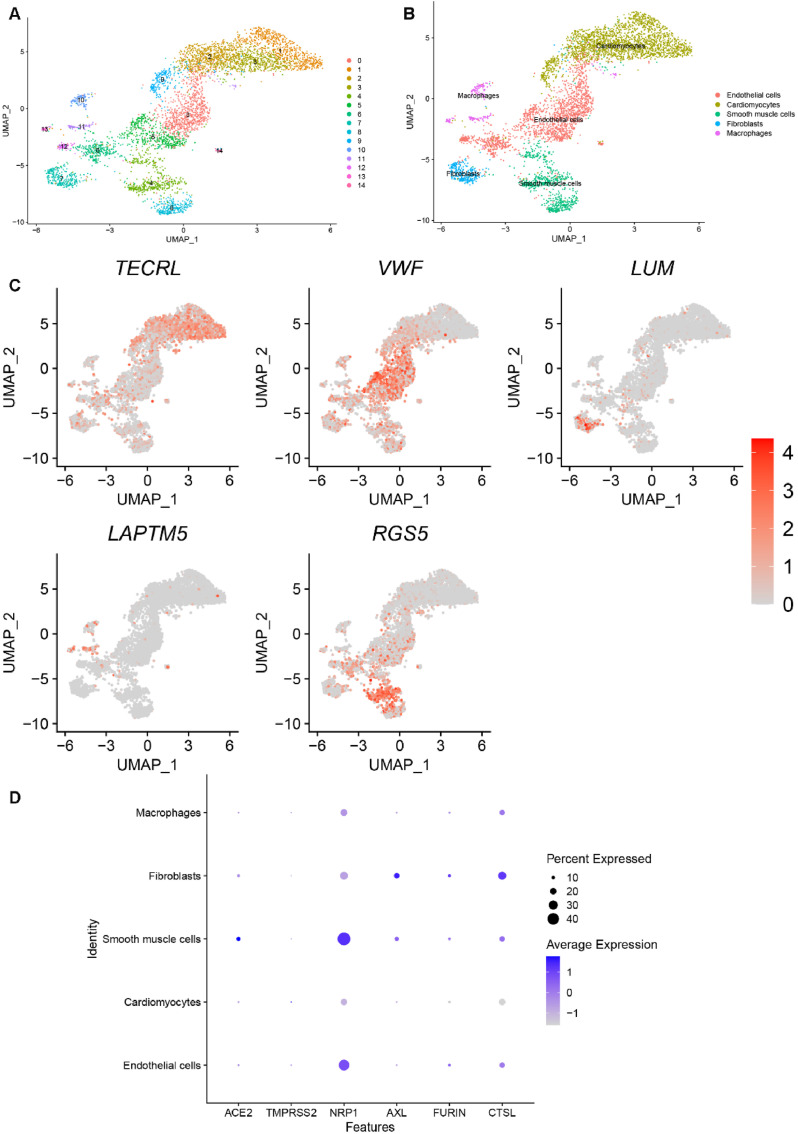
Cells in the heart dataset were grouped into 15 clusters, as shown in Fig. 5 A. We then chose canonical marker genes (TECRL, VWF, LUM, LAPTM5 and RGS5) for biological annotation. The annotation processes and results are shown in Fig. 5B–C. Clusters 0, 5, 6 and 14 were identified as endothelial cells by the marker gene VWF. With TECRL, clusters 1, 2, 3, and 9 were identified as cardiomyocytes. Cluster 7 was annotated as fibroblasts by the marker gene LUM. With LAPTM5, clusters 10, 11 and 13 were annotated as macrophages. Finally, clusters 4 and 8 were identified as smooth muscle cells using RGS5. Fig. 5D depicts possible heart-specific cell types vulnerable to COVID-19 by showing the gene expression conditions of potential receptors. We found that fibroblasts and smooth muscle cells had higher expression of almost all receptors, which indicates a high risk for COVID-19 infection.
Fig. 5.
Single-cell RNA sequencing analysis of the heart dataset. (A) Unsupervised clustering revealed 15 clusters in heart dataset. (B) Cell type annotation was conducted and 5 cell types were identified. (C) Expression of canonical marker genes used for cell type annotation. (D) Dot plot shows the expression of six receptors in all identified cell types.
3.3. Kidney dataset
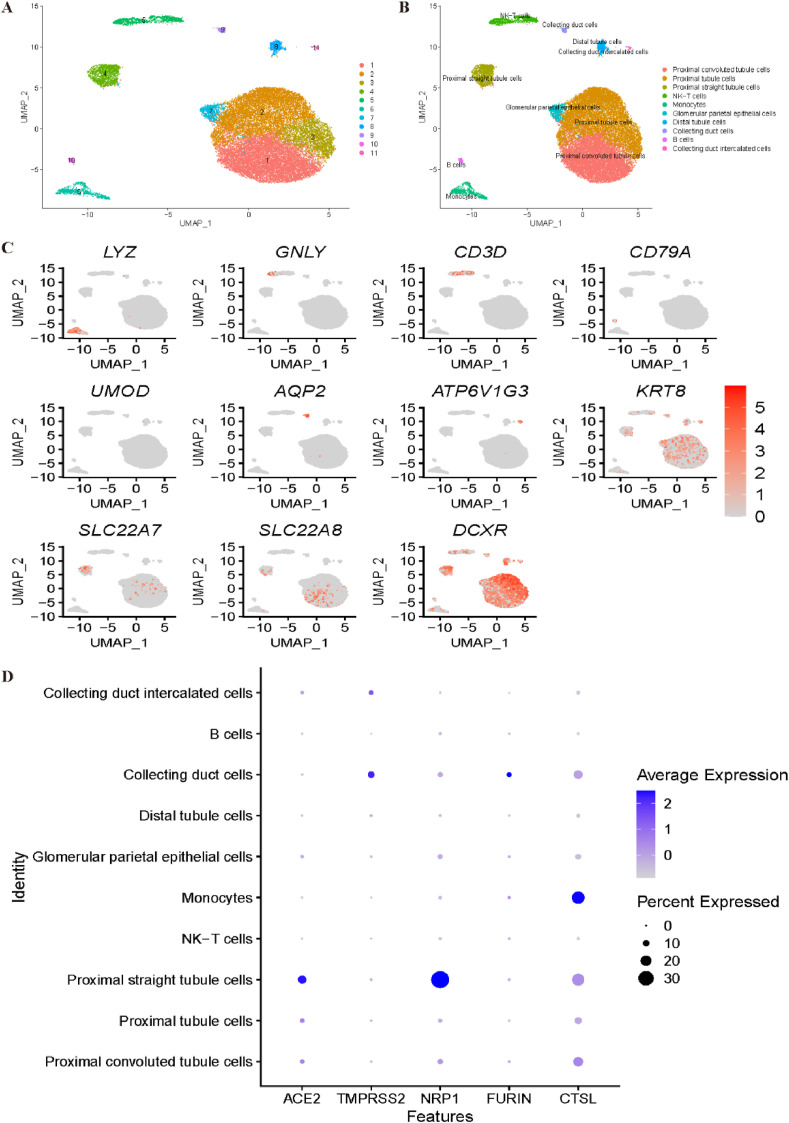
We then reanalysed the expression profiles of COVID-19 receptors in a kidney dataset. Unsupervised clustering by Seurat illustrated 11 clusters, as shown in Fig. 6 A. The annotation step for the kidney dataset is shown in Fig. 6B–C. With the canonical marker genes LYZ and CD14, cluster 6 was annotated as monocytes. NK cells and T cells were identified with GNLY, NKG7 and CD3D, which corresponded to cluster 5. Cluster 10 was identified as B cells by the CD79A and CD79B genes, while cluster 8 was annotated as distal tubule cells by the UMOD and DEFB1 marker genes. With AQP2 and CLDN8, cluster 9 was identified as collecting duct cells. Cluster 11 was identified as collecting duct intercalated cells by the ATP6V1G3 and ATP6V0D2 genes. The expression profiles of KRT8 and KRT18 indicated that cluster 7 corresponded to glomerular parietal epithelial cells. SLC22A8 and SLC22A7 were utilized to annotate cluster 4 and cluster 1 as proximal convoluted tubule cells and proximal straight tubule cells, respectively. Finally, clusters 2 and 3 were identified as proximal tubule cells using marker genes DCXR and GPX3. Fig. 6D demonstrates that collecting duct cells and proximal tubule cells may be most vulnerable to COVID-19 infection.
Fig. 6.
Single-cell RNA sequencing analysis of the kidney dataset. (A) Unsupervised clustering revealed 11 clusters in kidney dataset. (B) Cell type annotation was conducted and 10 cell types were identified. (C) Expression of canonical marker genes used for cell type annotation. (D) Dot plot shows the expression of six receptors in all identified cell types.
3.4. Liver dataset
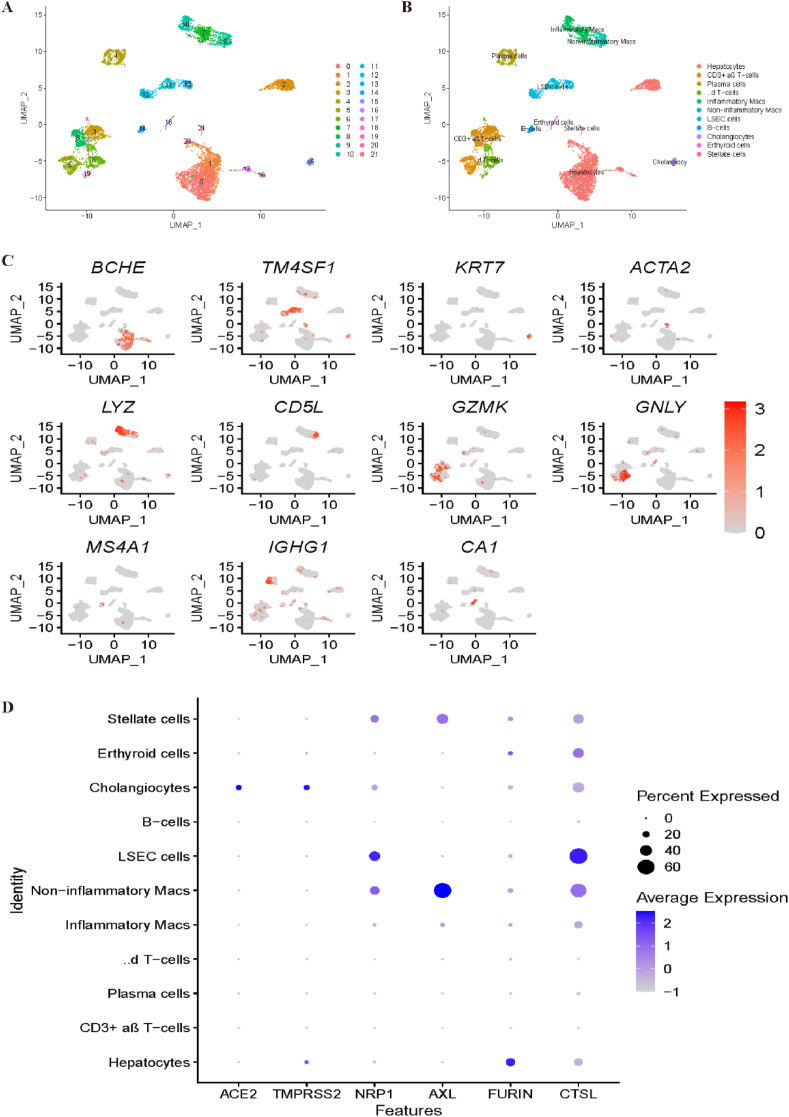
Similarly, we reanalysed liver scRNA-seq data. As shown in Fig. 7 A, all cells were grouped into 22 clusters. With the canonical marker genes BCHE and G6PC, clusters 0, 1, 2, 17, 18 and 20 were identified as hepatocytes. Clusters 11, 12, and 13 were annotated as limbal epithelial stem cells (LESCs) by TM4SF1 and CCL14. With the marker genes KRT7 and KRT19, cluster 15 was identified as cholangiocytes. Cluster 21 was identified as stellate cells by ACTA2 and RBP1, while cluster 7 was identified as inflammatory macs based on the expression profile of LYZ and HLA-DPB1. With CD5L, MARCO and VSIG4, noninflammatory macs were annotated corresponding to cluster 9. Clusters 6, 8 and 19 were all annotated as T cells; specifically, clusters 6 and 19 were identified as γδ T-cells by the marker genes GNLY and STMN1, while cluster 8 was identified as CD3+ αβ T-cells by GZMK. B cells were identified corresponding to cluster 14 by MS4A1 and CD37. Finally, plasma cells and erythroid cells were annotated corresponding to clusters 4 and 16 by IGHG1 and CA1, respectively (Fig. 7B–C). Fig. 7D demonstrates that overall, the most specific cell type that is vulnerable to COVID-19 is cholangiocyte, while stellate cells and noninflammatory macs cells may also be potential virus targets based on these four newly reported receptors.
Fig. 7.
Single-cell RNA sequencing analysis of the liver dataset. (A) Unsupervised clustering revealed 22 clusters in liver dataset. (B) Cell type annotation was conducted and 11 cell types were identified. (C) Expression of canonical marker genes used for cell type annotation. (D) Dot plot shows the expression of six receptors in all identified cell types.
3.5. Bladder dataset
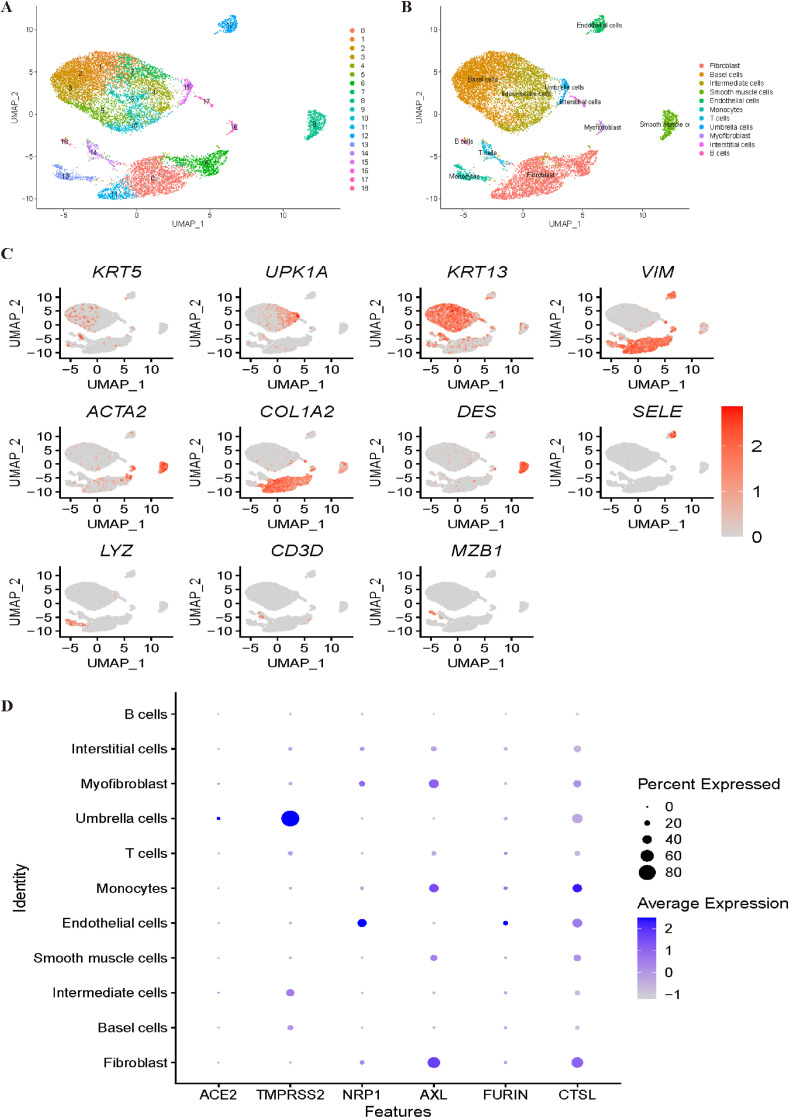
For the bladder dataset, all cells were grouped into 19 clusters (Fig. 8 A). The annotation processes are shown in Fig. 8B and C. Clusters 1, 2, 3, and 5 were identified as base cells with genes KRT5 and KRT17. Intermediate cells were annotated with KRT13 corresponding to cluster 4. With the canonical marker genes UPK1A and UPK1B, cluster 15 was annotated as umbrella cells. Interstitial cells were identified corresponding to cluster 17 by VIM. With ACTA2, cluster 15 was annotated as myofibroblasts. Cluster 8 was annotated as smooth muscle cells with the DES gene. With SELE and PECAM1, cluster 12 was identified as endothelial cells. Subsequently, T cells and B cells were identified corresponding to cluster 14 and cluster 18, respectively, by marker genes CD3D and MZB1. With the marker genes LYZ and MS4A7, cluster 13 was annotated as monocytes. Last, cluster 11 was annotated as fibroblasts with COL1A2. However, in Fig. 8D, it was difficult to determine the most vulnerable cell types to COVID-19, and further investigation is needed to obtain a convincing conclusion.
Fig. 8.
Single-cell RNA sequencing analysis of the bladder dataset. (A) Unsupervised clustering revealed 19 clusters in bladder dataset. (B) Cell type annotation was conducted and 11 cell types were identified. (C) Expression of canonical marker genes used for cell type annotation. (D) Dot plot shows the expression of six receptors in all identified cell types.
4. Discussion
As of May 2021, the COVID-19 global pandemic has caused more than 160 million confirmed cases, including 3.3 million deaths. Hence, there is an urgent need to understand the physiological and pathological mechanisms by which SARS-CoV-2 infects humans. In this study, we focused on the expression profiles of several reported receptors of SARS-CoV-2 (specifically, ACE2, TMPRSS2, NRP1, AXL, FURIN and CTSL) in different organs.
Since lung abnormalities are one of the most common symptoms for COVID-19 patients, we first explored the different expression profiles of potential receptors between COVID-19 patients and healthy donors [56]. As expected, all six receptors showed significant transcription differences upon infection with SARS-CoV-2. The mRNA expression levels of ACE2 and TMPRSS2 are highest in secretory cells and ciliated cells, which is consistent with other previous studies [14,57,58]. Interestingly, after focusing on observing the expression profiles of the other four newly confirmed receptors, we found that macrophages showed high expression levels of all of these receptors. Cell-cell communication analysis and trajectory inference were conducted to COVID-19 patients scRNA-seq data, which showed that M1-like macrophages were significantly associated with SARS-CoV-2 pathogenesis in lung. These results were consistent with a recent study which revealed M1 alveolar macrophages facilitate SARS-CoV-2 infection of the lungs [51]. GO and KEGG enrichment were conducted to M1-like macrophages, as expected, the GO enrichment results showed many virus-related pathways. Moreover, various GO terms related to cytokines, such as “cytokine activity” and “chemokine activity”, were also significantly enriched, which indicates the important role of the presence of so-called ‘cytokine storm’ [28,34,59]. Recent study by Jaggi et al. has also proven that M1 macrophages played an essential role in blocking cytokine storm [60]. As we know, the “classically” activated M1-like macrophages are generally pro-inflammatory. Large proportion of M1-like macrophages in the lung could stimulate the SARS-CoV-2 associated cytokine storm, which may be the reason for the serious condition of severity patients. Excitingly, we also noticed that ribosome related pathways were also strongly enriched in both GO and KEGG enrichment, which may be potential therapeutic target for COVID-19 lung infection patients.
Notably, we noticed that up-regulation of chemokine pathways generally lead to severe symptoms, while down-regulation of ribosome and RNA activity related pathways is more likely to be mild. Various studies have already revealed that due to the up-regulation of chemokine, which caused over-production of soluble markers of inflammation [28]. However, down-regulated ribosome pathways have been rarely discussed. As we know, ribosomes are complex molecular inside the living cells for producing proteins. Down-regulated of ribosomes may hinder the SARS-CoV-2 replication and inhibit the progression of the disease. Hence, ribosomes activity may be used as an indicator for COVID-19 disease assessment.
As COVID-19 has been proven to cause many symptoms in tissues other than the lung, we then collected scRNA-seq data from the heart, kidney, liver and bladder in healthy donors [61]. We mainly focused on revealing the organ-specific susceptible cell types for SARS-CoV-2. In the heart dataset, fibroblasts and smooth muscle cells had high expression of all six confirmed receptors, which indicates that they may be potentially associated with SARS-CoV-2 pathogenesis. Similarly, collecting duct cells and proximal tubule cells may be associated with SARS-CoV-2 infection in the kidney. For the liver, stellate cells and noninflammatory macs cells show the highest risk for SARS-CoV-2 infection. However, for bladder single-cell data, it was difficult to identify the possible cell types, which need further investigation.
5. Conclusion
In summary, we reanalysed the public single-cell RNA sequencing dataset to understand the possible tropism of SARS-CoV-2. By comprehensive bioinformatics analysis of a lung single-cell RNA sequencing dataset of COVID-19 patients and healthy donors, macrophages were identified as the most likely cells that may be associated with SARS-CoV-2 pathogenesis. Further analysis of macrophages in lung demonstrates that up-regulation of chemokine pathways generally lead to severe symptoms, while down-regulation of ribosome and RNA activity related pathways is more likely to be mild. Other organ-specific susceptible cell type analyses could also provide a potential target for COVID-19 therapy [62,63]. Of course, we admit that there are certain limitations, such as no experimental validation and limited receptor genes [64,65]. Nonetheless, our studies may provide potential clues for further investigation of the pathogenesis of COVID-19.
Availability
Code used in this study is available from https://github.com/ZilongZhang44/COVID-19.
Funding
The work was supported by the National Natural Science Foundation of China (No. 61922020, No. 61771331) and the Special Science Foundation of Quzhou (2021D004).
Declaration of competing interest
None declared.
Biographies
Zilong Zhang is currently working as a postdoctoral researcher in the University of Electronic Science and Technology of China. He received his Ph.D. degree from the University of Tokyo, Japan in 2020. His research interests include single-cell sequencing data analysis, bioinformatics and machine learning.
Feifei Cui received her Ph.D. degree from the University of Tokyo, Japan. She is currently a postdoctoral researcher at the University of Electronic Science and Technology of China. Her research interests include bioinformatics, deep learning and biological data mining.
Chen Cao is currently working as an associate professor in the Yangtze Delta Region Institute (Quzhou), University of Electronic Science and Technology of China. He received his Ph.D. degree from the Jilin University in 2016. His research interests include bioinformatics, machine learning.
Qingsuo Wang is currently working in the Beidahuang Industry Group General Hospital, Harbin 150001, China. His research interests include data mining and bioinformatics.
Quan Zou is a professor at the Institute of Fundamental and Frontier Sciences, University of Electronic Science and Technology of China. He received his Ph.D. from Harbin Institute of Technology, P.R.China in 2009. He is a senior member of IEEE and ACM. His research is in the areas of bioinformatics, machine learning and parallel computing.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng L., Han X., Zhu Z., Qi C., Wang P., Zhang X. Functional alterations caused by mutations reflect evolutionary trends of SARS-CoV-2. Briefings Bioinf. 2021 doi: 10.1093/bib/bbab042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Y., Li F., Zhang Y., Zhou Y., Tan Y., Chen Y., Zhu F. Databases for the targeted COVID-19 therapeutics. Br. J. Pharmacol. 2020;177(21):4999–5001. doi: 10.1111/bph.15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348(20):1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 5.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D., Fouchier R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.Zou L., Ruan F., Huang M., Liang L., Huang H., Hong Z., Yu J., Kang M., Song Y., Xia J., et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamers M.M., Beumer J., van der Vaart J., Knoops K., Puschhof J., Breugem T.I., Ravelli R.B.G., Paul van Schayck J., Mykytyn A.Z., Duimel H.Q., et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369(6499):50–54. doi: 10.1126/science.abc1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Y., Zhang S., Li F., Zhou Y., Zhang Y., Wang Z., Zhang R., Zhu J., Ren Y., Tan Y., et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020;48(D1):D1031–d1041. doi: 10.1093/nar/gkz981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 10.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dey A., Sen S., Maulik U. Unveiling COVID-19-associated organ-specific cell types and cell-specific pathway cascade. Briefings Bioinf. 2021;22(2):914–923. doi: 10.1093/bib/bbaa214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F., et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26(5):681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu C., Zheng M., Yang Y., Gu X., Yang K., Li M., Liu Y., Zhang Q., Zhang P., Wang Y., et al. Furin: a potential therapeutic target for COVID-19. iScience. 2020;23(10):101642. doi: 10.1016/j.isci.2020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao M.-M., Yang W.-L., Yang F.-Y., Zhang L., Huang W., Hou W., Fan C., Jin R., Feng Y., Wang Y., et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. medRxiv. 2020 doi: 10.1038/s41392-021-00558-8. 2020.2010.2025.20218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei C., Wan L., Yan Q., Wang X., Zhang J., Yang X., Zhang Y., Fan C., Li D., Deng Y., et al. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. metab. 2020;2(12):1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- 19.Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., Wu P., Xie S., Bian W., Zhang C., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021:1–15. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu X., Li H.-D., Guo L., Wu F.-X., Wang J. Analysis of single-cell RNA-seq data by clustering approaches. Curr. Bioinf. 2019;14(4):314–322. [Google Scholar]
- 21.Qi R., Ma A., Ma Q., Zou Q. Clustering and classification methods for single-cell RNA-sequencing data. Briefings Bioinf. 2020;21(4):1196–1208. doi: 10.1093/bib/bbz062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Z., Ding H., Zou Q. Identifying cell types to interpret scRNA-seq data: how, why and more possibilities. Brief. Funct. Geno. 2020;19(4):286–291. doi: 10.1093/bfgp/elaa003. [DOI] [PubMed] [Google Scholar]
- 23.Jin S., Zeng X., Xia F., Huang W., Liu XJBiB. Application of deep learning methods in biological networks. Briefings Bioinf. 2020 doi: 10.1093/bib/bbaa043. [DOI] [PubMed] [Google Scholar]
- 24.Liu X., Hong Z., Liu J., Lin Y., Rodríguez-Patón A., Zou Q., Zeng XJBib. Computational methods for identifying the critical nodes in biological networks. Briefings Bioinf. 2020;21(2):486–497. doi: 10.1093/bib/bbz011. [DOI] [PubMed] [Google Scholar]
- 25.IRIS3: Integrated Cell-type-specific Regulon Inference Server from Single-Cell RNA-Seq. [DOI] [PMC free article] [PubMed]
- 26.Xie J., Ma A., Zhang Y., Liu B., Cao S., Wang C., Xu J., Zhang C., Ma Q. QUBIC2: a novel and robust biclustering algorithm for analyses and interpretation of large-scale RNA-Seq data. Bioinformatics. 2020;36(4):1143–1149. doi: 10.1093/bioinformatics/btz692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Ma A., Mathé E.A., Li L., Liu B., Ma Q. Elucidation of biological networks across complex diseases using single-cell omics. Trends Genet. 2020;36(12):951–966. doi: 10.1016/j.tig.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R. The COVID-19 cytokine storm; what we know so far. Front. Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang J., Ma A., Chang Y., Gong J., Jiang Y., Fu H., Wang C., Qi R., Ma Q., Xu D. scGNN: a novel graph neural network framework for single-cell RNA-Seq analyses. bioRxiv. 2020 doi: 10.1038/s41467-021-22197-x. 2020.2008.2002.233569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang Y., Allen C., Wan C., Chung D., Zhang C., Li Z., Ma Q. IRIS-FGM: an integrative single-cell RNA-Seq interpretation system for functional gene module analysis. bioRxiv. 2020 doi: 10.1093/bioinformatics/btab108. 2020.2011.2004.369108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z., Cui F., Lin C., Zhao L., Wang C., Zou Q. Critical downstream analysis steps for single-cell RNA sequencing data. Briefings Bioinf. 2021 doi: 10.1093/bib/bbab105. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z., Cui F., Wang C., Zhao L., Zou Q. Goals and approaches for each processing step for single-cell RNA sequencing data. Briefings Bioinf. 2020 doi: 10.1093/bib/bbaa314. [DOI] [PubMed] [Google Scholar]
- 33.Zhu T., Guan J., Liu H., Zhou S. RMDB: an integrated database of single-cytosine-resolution DNA methylation in oryza sativa. Curr. Bioinf. 2019;14(6):524–531. [Google Scholar]
- 34.Fajgenbaum D.C., June C.H., Storm Cytokine. N. Engl. J. Med. 2020;383(23):2255–2273. doi: 10.1056/NEJMra2026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P., Jin X., Zhou W., Luo M., Xu Z., Xu C., Li Y., Ma K., Cao H., Huang Y., et al. Comprehensive analysis of TCR repertoire in COVID-19 using single cell sequencing. Genomics. 2020;113(2):456–462. doi: 10.1016/j.ygeno.2020.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li F., Luo M., Zhou W., Li J., Jin X., Xu Z., Juan L., Zhang Z., Li Y., Liu R., et al. Single cell RNA and immune repertoire profiling of COVID-19 patients reveal novel neutralizing antibody. Protein Cell. 2020 doi: 10.1007/s13238-020-00807-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26(6):842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 38.Morse C., Tabib T., Sembrat J., Buschur K.L., Bittar H.T., Valenzi E., Jiang Y., Kass D.J., Gibson K., Chen W., et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 2019;54(2) doi: 10.1183/13993003.02441-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Yu P., Zhou B., Song J., Li Z., Zhang M., Guo G., Wang Y., Chen X., Han L., et al. Single-cell reconstruction of the adult human heart during heart failure and recovery reveals the cellular landscape underlying cardiac function. Nat. Cell Biol. 2020;22(1):108–119. doi: 10.1038/s41556-019-0446-7. [DOI] [PubMed] [Google Scholar]
- 40.MacParland S.A., Liu J.C., Ma X.Z., Innes B.T., Bartczak A.M., Gage B.K., Manuel J., Khuu N., Echeverri J., Linares I., et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018;9(1):4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao J., Yu Z., Chen Y., Bao M., Zou C., Zhang H., Liu D., Li T., Zhang Q., Li J., et al. Single-cell RNA sequencing of human kidney. Sci Data. 2020;7(1):4. doi: 10.1038/s41597-019-0351-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Z., Liao J., Chen Y., Zou C., Zhang H., Cheng J., Liu D., Li T., Zhang Q., Li J., et al. Single-cell transcriptomic map of the human and mouse bladders. J. Am. Soc. Nephrol. 2019;30(11):2159–2176. doi: 10.1681/ASN.2019040335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E. Fast unfolding of communities in large networks. J. Stat. Mech. Theor. Exp. 2008;2008(10):P10008. [Google Scholar]
- 45.Leland McInnes J.H., James Melville. 2018. UMAP: uniform Manifold approximation and projection for dimension reduction. arXiv:180203426. [Google Scholar]
- 46.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.H., Myung P., Plikus M.V., Nie Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu X., Hill A., Packer J., Lin D., Ma Y.A., Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods. 2017;14(3):309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao E.Y., Ouyang J.F., Rackham O.J.L. GeneSwitches: ordering gene expression and functional events in single-cell experiments. Bioinformatics. 2020;36(10):3273–3275. doi: 10.1093/bioinformatics/btaa099. [DOI] [PubMed] [Google Scholar]
- 51.Lv J., Wang Z., Qu Y., Zhu H., Zhu Q., Tong W., Bao L., Lv Q., Cong J., Li D., et al. Distinct uptake, amplification, and release of SARS-CoV-2 by M1 and M2 alveolar macrophages. Cell discovery. 2021;7(1):24. doi: 10.1038/s41421-021-00258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu Y., Sun J.Y., Zhang Y., Zhang H., Gao S., Wang T., Han Z., Wang L., Sun B.L., Liu G. rs1990622 variant associates with Alzheimer's disease and regulates TMEM106B expression in human brain tissues. BMC Med. 2021;19(1):11. doi: 10.1186/s12916-020-01883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu Y., Qiu S., Cheng L. Integration of multiple-omics data to analyze the population-specific differences for coronary artery disease. Comput. Math. Meth. Med. 2021;2021:7036592. doi: 10.1155/2021/7036592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu Y., Zhang H., Liu B., Gao S., Wang T., Han Z., International Genomics of Alzheimer's P., Ji X., Liu G. rs34331204 regulates TSPAN13 expression and contributes to Alzheimer's disease with sex differences. Brain. 2020;143(11):e95. doi: 10.1093/brain/awaa302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. Jama. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 57.Yang Q., Li B., Tang J., Cui X., Wang Y., Li X., Hu J., Chen Y., Xue W., Lou Y., et al. Consistent gene signature of schizophrenia identified by a novel feature selection strategy from comprehensive sets of transcriptomic data. Briefings Bioinf. 2020;21(3):1058–1068. doi: 10.1093/bib/bbz049. [DOI] [PubMed] [Google Scholar]
- 58.Jia H.P., Look D.C., Shi L., Hickey M., Pewe L., Netland J., Farzan M., Wohlford-Lenane C., Perlman S., McCray P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005;79(23):14614–14621. doi: 10.1128/JVI.79.23.14614-14621.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang Y., Liu J., Zhang D., Xu Z., Ji J., Wen C. Cytokine storm in COVID-19: the current evidence and treatment strategies. Front. Immunol. 2020;11:1708. doi: 10.3389/fimmu.2020.01708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jaggi U., Matundan H.H., Yu J., Hirose S., Mueller M., Wormley F.L., Jr., Ghiasi H. Essential role of M1 macrophages in blocking cytokine storm and pathology associated with murine HSV-1 infection. PLoS Pathog. 2021;17(10) doi: 10.1371/journal.ppat.1009999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berlin D.A., Gulick R.M., Martinez F.J. Severe covid-19. N. Engl. J. Med. 2020;383(25):2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- 62.Zeng X., Song X., Ma T., Pan X., Zhou Y., Hou Y., Zhang Z., Li K., Karypis G., Cheng FJJopr. Repurpose open data to discover therapeutics for COVID-19 using deep learning. J. Proteome Res. 2020;19(11):4624–4636. doi: 10.1021/acs.jproteome.0c00316. [DOI] [PubMed] [Google Scholar]
- 63.Zeng X., Zhu S., Lu W., Liu Z., Huang J., Zhou Y., Fang J., Huang Y., Guo H., Li L.J.C.S. Target identification among known drugs by deep learning from heterogeneous networks. Chem. Sci. 2020;11(7):1775–1797. doi: 10.1039/c9sc04336e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeng X., Lin Y., He Y., Lv L., Min X. Deep collaborative filtering for prediction of disease genes. IEEE ACM Trans. Comput. Biol. Bioinf. 2020;17(5):1639–1647. doi: 10.1109/TCBB.2019.2907536. [DOI] [PubMed] [Google Scholar]
- 65.Zhang X., Zou Q., Rodriguez-Paton A., Zeng X., Bioinformatics Meta-path methods for prioritizing candidate disease miRNAs. IEEE ACM Trans. Comput. Biol. Bioinf. 2017;16(1):283–291. doi: 10.1109/TCBB.2017.2776280. [DOI] [PubMed] [Google Scholar]