Abstract
ADP ribosylation factors (ARFs) are ∼20-kDa guanine nucleotide-binding proteins that activate cholera toxin and phospholipase D and are critical components of vesicular trafficking pathways. ARF domain protein 1 (ARD1), a member of the ARF superfamily, contains a 46-kDa amino-terminal extension, which acts as a GTPase-activating protein (GAP) with activity towards its ARF domain. When overexpressed, ARD1 was associated with lysosomes and the Golgi apparatus. In agreement with this finding, lysosomal and Golgi membranes isolated from human liver by immunoaffinity contained native ARD1. ARD1, expressed as a green fluorescent fusion protein, was initially associated with the Golgi network and subsequently appeared on lysosomes, suggesting that ARD1 might undergo vectorial transport between the two organelles. Here we show by microscopic colocalization that GAP and ARF domains determine lysosomal and Golgi localization, respectively, consistent with the presence of more than one signal motif. Using truncated ARD1 molecules, expressed as green fluorescent fusion proteins, it was found that the signal for lysosomal localization was present in residues 301 to 402 of the GAP domain. Site-specific mutagenesis demonstrated that the sequence 369KXXXQ373 in the GAP domain was responsible for lysosomal localization. Association of ARD1 with the Golgi apparatus required tyrosine-based motifs. A green fluorescent fusion protein containing the QKQQQQF motif was partially associated with lysosomes, suggesting that this motif contains the information sufficient for lysosomal targeting. These results suggest that ARD1 is a multidomain protein with ARF and GAP regions, which contain Golgi and lysosomal localization signals, respectively, that could function in vesicular trafficking.
The interaction of signal motifs in the cytoplasmic tails of proteins en route to specific organelles with membrane coat proteins is now regarded as a general mechanism of protein sorting at several stages of the endocytic and secretory pathways. Among the best-characterized sorting signals are sequences of four to six amino acids that include a critical tyrosine and hydrophobic residues (4) and di-leucine-based sorting signals (32). Di-leucine- and tyrosine-based signal elements bind to distinct sites on adapter proteins (APs) (30), and it is likely that different tyrosine-based motifs interact preferentially with each of the APs. These selective associations might be the basis of sorting processes in which specific signals are involved (25).
ADP ribosylation factors (ARFs), a family of ∼20-kDa guanine nucleotide-binding proteins, originally identified as activators of the ADP ribosylation of Gαs by cholera toxin, are believed to play a critical role in vesicular trafficking. Members of the family include the ARF proteins, the ARF-like proteins, and the related much larger ARF domain protein 1 (ARD1) (23). Six mammalian ARFs have been classified into three groups according to size and amino acid sequence, phylogenetic analysis, and gene structure. ARF1, ARF2, and ARF3 form class I; ARF4 and ARF5 class II; and ARF6 class III (37, 45).
Like other monomeric GTPases, ARFs bind and hydrolyze GTP very slowly. The ratio of inactive ARF-GDP and active ARF-GTP is regulated by guanine nucleotide-exchange proteins (GEPs) and GTPase-activating proteins (GAPs). Several ARF GAPs and ARF GEPs have been purified and cloned. The latter fall into two families, ∼200-kDa, brefeldin A-sensitive GEPs and ∼50-kDa, brefeldin A-insensitive GEPs (23). ARF GAPs differ in their phospholipid sensitivity and ARF specificity (9, 11, 27).
ARD1 is a 64-kDa protein with an 18-kDa carboxy-terminal ARF domain linked to a 46-kDa amino-terminal extension (20). Like ARFs, the 18-kDa ARF domain of ARD1 specifically binds GDP and GTP and lacks detectable GTPase activity (41). Using recombinant proteins, it was shown that the 46-kDa amino-terminal domain of ARD1 physically binds to the ARF domain and stimulates hydrolysis of bound GTP; i.e., it possesses GAP activity (39). The amino-terminal domain interacts specifically with the ARF domain of ARD1 as it did not increase GTP hydrolysis by other members of the ARF and ARL families (11). Two negatively charged amino acids (Asp427 and Glu428), which are located in the effector region of the ARF domain, interact with two positively charged amino acids (Arg249 and Lys250) in the amino-terminal domain and are required for both functional and physical interactions between the GTP-binding and GAP domains (41, 42). By site-directed mutagenesis, it was further demonstrated that, in the amino-terminal GAP domain, an intact zinc finger motif, two arginines, and a sequence that resembles a consensus motif present in Rho or Rac GAPs are required for GAP activity (42).
Native ARD1 was found associated with lysosomal and Golgi membranes isolated from human liver by immunoaffinity (43). When overexpressed in NIH 3T3, COS 7, and HeLa cells, ARD1 had a subcellular localization typical of the Golgi apparatus and lysosomes (43) and distinct from those of other ARFs (12). We report here that the GAP domain of ARD1 contains a structure responsible for its lysosomal localization, whereas the ARF domain is responsible for the association with Golgi membranes. By site-specific mutagenesis, we have identified two tyrosine-based motifs in the ARF region and the sequence 369KXXXQ373 in the GAP domain as elements critical for Golgi and lysosomal localization, respectively.
MATERIALS AND METHODS
Materials.
Restriction enzymes were purchased from Boehringer Mannheim; brefeldin A was purchased from Epicentre Technologies; Dulbecco's modified Eagle's medium, fetal bovine serum, penicillin-streptomycin solution, and glutamine were purchased from Life Technologies, Inc.; and cells were purchased from the American Type Culture Collection (Manassas, Va.). Sources of other materials have been published (42, 43).
Construction of a eucaryotic expression vector containing ARD1.
ARD1 cDNA with its original Kozak sequence was amplified from a human liver library (Origene Technologies Inc., Rockville, Md.) by PCR in the presence of Pfu (Stratagene), essentially as published (43). The PCR product was extracted from low-melting-point agarose gel, purified with a Wizard PCR purification kit (Promega), and ligated in frame to the XhoI- and BamHI-digested pcDNA3.1/Zeo(−) expression vector (Invitrogen). Ultracompetent cells (Stratagene) were transformed with the resulting plasmid, pcDNA3.1/Zeo(ARD1), which had been purified with a plasmid Maxiprep kit (Qiagen). The entire sequence of the ARD1 construct was confirmed by automated sequencing (373 DNA sequencer; Applied Biosystems) using the primers 5′-TTATACGACTCACTATAGGG-3′, 5′-AGCTGCAGAAGAATCCATT-3′, 5′-ATCAATTTTAGATATGGCT-3′, 5′-ATGATTGTAGAGTTGTCTT-3′, 5′-TTATTACCTCAATACTCAA-3′, and 5′-GCTAGTTATTGCTCAGCGG-3′.
Construction of eucaryotic expression vectors containing GAP or ARF domains of ARD1.
The GAP fragment of ARD1 was amplified by PCR from pcDNA3.1/Zeo(ARD1) using Pfu (Stratagene) with the forward primer 5′-TCCCCTCTCGAGATGGCTACCCTGGTTGTAAAC-3′ (italicized sequence is an XhoI site) and the reverse primer 5′-GAATGGATCCTCATTTTGGTCCAATGTG-3′ (italicized sequence is a BamHI site). The reverse primer introduced a stop signal at the 3′ end of the GAP domain (boldface sequence). The ARF fragment of ARD1 was amplified by PCR from pcDNA3.1/Zeo(ARD1) using Pfu with the forward primer 5′-CACATTGACTCGAGGATGGAAATTCGGGTCGTTACG-3′ and the reverse primer 5′-TAGAAGGCACAGTCGAGG-3′ (differences from pcDNA3.1/Zeo(ARD1) are underlined). The forward primer introduced an XhoI restriction site (italicized) and a Kozak sequence (boldface letters). The PCR products, after digestion with XhoI and BamHI, were extracted from low-melting-point agarose gel, purified with a Wizard PCR purification kit (Promega), and ligated in frame to the XhoI- and BamHI-digested pcDNA3.1/Zeo(−) expression vector. DH5α cells (GIBCO-BRL) were transformed with the resulting plasmid pcDNA3.1/Zeo(GAP domain) and pcDNA3.1/Zeo(ARF domain) according to the manufacturer's instruction. Plasmids were purified with a plasmid Maxiprep kit (Qiagen) and entire sequences confirmed as described above.
Construction of GFP fusion vector containing ARD1 fragments.
Fragments of ARD1 were amplified by PCR from the cDNA pEGFP-C2(ARD1) (53), using Pfu with the forward primers (primers 1 to 7) and reverse primers (primers 8 to 14) as indicated in Table 1. Primer sequences were as follows: 1, 5′-CCGGCCGCACTCAGATCTCTATG-3′; 2, 5′-GCTTTATTGGAGCAGATCTCACAGAATGGGCC-3′; 3, 5′-GAAGGTTGTCAAACTAAGATCTTCATGTGCTGTGTCTGC-3′; 4, 5′-TATGATCTACATGAAAAGATCTGTCGTCAAGAAGAAATGGCT-3′; 5, 5′-CGAGTTCACATTGAGATCTCAATGGAAATTCGGGTC-3′; 6, 5′-AGCGAACTTGCAAAGTTGAGATCTACGATGAAAG-3′; 7, 5′-CTCAAGCTGTTGTGTTTGAGATCTATATGAGCAGTC-3′; 8, 5′-CAATTAGGCCCATTGTCGACTCATTCCAAAAGCTCC-3′; 9, 5′-TCCATATTCTTTGCAGCAGACGTCGACTCAGAGTGGGCTAGTTTGACA-3′; 10, 5′-AACACTTAGAGCCATGTCGACTCAACACAGAGTTTCATGTGA-3′; 11, 5′-TCCTAA CGTAACGACCCGGTCGACTCATTTTGGTCCAATGTGAACTCG-3′; 12, 5′-TTCTGAGATGAGTTGTCGACCTCAAGCAACATCCAATACTCCAGC-3′; 13, 5′-GCATCTCGGTCGACTCATTCCGTTAACAACTTTGCAAG-3′; 14, 5′-CTAATTCTGTCTCTGTCGACTCAATCTACAACAAACACAACAGC-3′. The forward and reverse primers introduced BglII and SalI restriction sites (italicized sequences) and initiation and stop codons (boldface sequences), respectively. PCR products were digested with BglII and SalI restriction sites and were purified as described above. They were ligated in frame to the BglII- and SalI-digested pEGFP-C2 expression vector (CLONTECH). Sequences were confirmed by automated sequencing.
TABLE 1.
Intracellular localization of ARD1 fragments expressed as GFP fusion proteins
| GFP and ARD1 fragmentsa | Primersb
|
Fragment localizationc
|
Colocalization | ||
|---|---|---|---|---|---|
| F | R | Nontreated | Plus AcR | ||
| GFP | Cytosolic | Cytosolic | Noned | ||
| GFP–1–574 (ARD1) | Perinuclear | Dispersed | Golgi/Lysog | ||
| GFP–1–100 | 1 | 8 | Cytosolice | Cytosolice | None |
| GFP–1–200 | 1 | 9 | Cytosolice | Cytosolice | None |
| GFP–1–300 | 1 | 10 | Cytosolice | Cytosolice | None |
| GFP–1–402 (GAP) | 1 | 11 | Vesicularf | Dispersed | Lyso |
| GFP–101–574 | 2 | 12 | Perinuclear | Dispersed | Golgi/Lyso |
| GFP–201–574 | 3 | 12 | Perinuclear | Dispersed | Golgi/Lyso |
| GFP–301–574 | 4 | 12 | Perinuclear | Dispersed | Golgi/Lyso |
| GFP–403–574 (ARF) | 5 | 12 | Perinuclear | Perinuclear | Golgi |
| GFP–403–500 | 5 | 13 | Cytosolic | Cytosolic | None |
| GFP–501–574 | 6 | 12 | Cytosolic | Cytosolic | None |
| GFP–101–500 | 2 | 13 | Vesicularf | Dispersed | Lyso |
| GFP–201–500 | 3 | 13 | Vesicularf | Dispersed | Lyso |
| GFP–301–500 | 4 | 13 | Vesicularf | Dispersed | Lyso |
| GFP–101–402 | 2 | 11 | Vesicularf | Dispersed | Lyso |
| GFP–101–300 | 2 | 10 | Cytosolic | Cytosolic | None |
| GFP–101–200 | 2 | 9 | Cytosolic | Cytosolic | None |
| GFP–201–402 | 3 | 11 | Vesicularf | Dispersed | Lyso |
| GFP–301–402 | 4 | 11 | Vesicularf | Dispersed | Lyso |
| GFP–201–300 | 3 | 10 | Cytosolic | Cytosolic | None |
| GFP–403–480 | 5 | 14 | Cytosolic | Cytosolic | None |
| GFP–481–574 | 7 | 12 | Cytosolic | Cytosolic | None |
Constructs used to transfect NIH 3T3 cells.
Forward (F) and reverse (R) primers were used, as described in Materials and Methods, to clone ARD1 fragments into pEGFP-C2. Primer sequences are given in Materials and Methods.
At 48 h after transfection, cells were incubated for 15 min at 37°C with PBS (nontreated) or with AcR, and coverslips were briefly washed with PBS before cells were fixed and mounted for microscopic observation. Identical results were obtained with three different cell cultures. For colocalization of GFP proteins and marker proteins, cells were stained with anti-p58 and anti-LAMP-1 antibodies, specific markers for the Golgi apparatus and lysosomes, respectively, followed by rhodamine-conjugated secondary antibody. Preparations were examined by confocal microscopy.
None, absence of colocalization with either p58 or LAMP-1.
In addition to strong cytosolic staining, the nucleus appeared fluorescent.
Fluorescent vesicular structures were not restricted to the perinuclear region and Golgi staining was not detected.
Golgi/Lyso, colocalization with both Golgi and lysosome markers.
Construction of GFP fusion vector containing potential targeting motifs.
The oligonucleotides 5′-ACTCAGATCTCGCAGAAACAGCAGCAGCAGTTTAGTCGACGGTAC-3′, 5′-ACTCAGATCTCGGAATATAAAAATCTAAAAAGTCGACGGTAC-3′, and 5′-ACTCAGATCTCGCTGTATGAAGGGTTGGACAGTCGACGGTAC-3′ (italicized sequences are BglII and SalI restriction sites, respectively; boldface sequences encode three potential targeting motifs), and their complementary sequences were used to introduce, at the carboxyl-terminal end of the enhanced-GFP protein, QKQQQQF, EYKNLK, and LYEGLD peptide motifs, respectively. One microgram of each oligonucleotide was incubated at 95°C for 5 min and then annealed with its complementary sequence for 10 min at 60°C. Double-stranded DNA products were digested with BglII and SalI restriction enzymes, purified on NAP-5 columns (Pharmacia), and ligated in-frame to the BglII- and SalI-digested pEGFP-C2 expression vector (CLONTECH). Similarly, the construct containing the EYKNLK peptide motif was used with the oligonucleotide 5′-ACTCGTCGACCGCTGTATGAAGGGTTGGACAGTCGACGGTAC-3′ (italicized sequence is a SalI restriction site) and its complementary sequence to generate a protein containing both EYKNLK and LYEGLD peptide motifs. Introduction of sequences to pEGFP-C2 was confirmed by automated sequencing. Constructs have a linker peptide sequence (SGTQIS) between enhanced green fluorescent protein (GFP) and ARD1 peptide sequences and a carboxyl-terminal SRRYRGPGIHRI extension, both contributed by the pEGFP-C2 vector.
Site-directed mutagenesis.
Point mutations were created in pcDNA3.1/Zeo(ARD1) with a Quickchange site-directed mutagenesis kit from Stratagene according to the manufacturer's instruction. Briefly, complementary oligonucleotides containing the desired changes, flanked by sequences of 20 unmodified nucleotides, were synthesized and purified. Forty nanograms (each) of pcDNA3.1/Zeo(ARD1), pcDNA3.1/Zeo(ARF domain), and pcDNA3.1/Zeo(GAP domain) was amplified by PCR with Pfu and 125 ng of complementary oligonucleotides with incubation at 96°C for 1 min followed by 18 cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 1 min. Original pcDNA3.1/Zeo(ARD1) was digested in the PCR mixture with 20 U of DpnI for 1 h at 37°C. The pcDNA3.1/Zeo plasmids produced by PCR encoding GAP domain(344ATLQA348), GAP domain(369AQQQA373), ARD1(344ATLQA348), ARD1(369AQQQA373), ARF domain(445AKNA448), ARF domain(555AEGA558), ARD1(445AKNA448), and ARD1(555AEGA558) were used to transform XL-1-Blue supercompetent cells for amplification. Plasmids were purified with a plasmid Maxiprep kit (Qiagen). The double-mutant ARF domain(445AKNA448-555AEGA558) and ARD1(445AKNA448-555AEGA558) were synthesized using the same procedure with, respectively, pcDNA/Zeo-ARF domain(445AKNA448) and pcDNA/Zeo-ARD1(445AKNA448) as templates. Mutations and sequences of the entire clones were confirmed by automated sequencing.
Preparation of recombinant proteins.
Glutathione S-transferase (GST) fusion proteins synthesized using a ligation-independent cloning method were purified on glutathione-Sepharose beads (Pharmacia) as described (39). After cleavage by bovine thrombin, GST was removed with glutathione-Sepharose beads and thrombin was removed with benzamidine-Sepharose 6B (39). The proteins ARF domain, ARF domain(445AKNA448), ARF domain(555AEGA558), and ARF domain(445AKNA448-555AEGA558) were further purified by gel filtration through Ultrogel AcA 54. Purity estimated by silver staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis was >90%. Amounts of purified proteins were estimated by a dye-binding assay and by sodium dodecyl sulfate-polyacrylamide gel electrophoresis using bovine serum albumin (BSA) as a standard.
GTPγS binding assay.
GTPγS binding to purified recombinant ARF domain proteins was assessed using a rapid filtration technique. Samples were incubated for 30 min at 30°C in 20 mM Tris (pH 8.0)–10 mM dithiothreitol (DTT)–2.5 mM EDTA with BSA, 0.3 mg/ml, and cardiolipin, 1 mg/ml, and then for 40 min at 30°C in the same medium plus 10 mM MgCl2 and 3 μM [35S]GTPγS (∼106 cpm; total volume, 150 μl). Samples (70 μl) were then transferred to nitrocellulose filters in a manifold (Millipore) for rapid filtration, followed by washing five times each with 1 ml of ice-cold buffer (25 mM Tris-HCl [pH 8.0]–100 mM NaCl–1 mM DTT–1 mM EDTA–5 mM MgCl2). Dried filters were dissolved in scintillation fluid for radioassay.
Assay of CTA-catalyzed ADP-ribosylagmatine formation.
Recombinant ARF domain proteins were incubated for 30 min at 30°C in 40 μl of 20 mM Tris (pH 8.0)–10 mM DTT–2.5 mM EDTA with BSA, 0.3 mg/ml, and cardiolipin, 1 mg/ml, before addition of 20 μl of solution to yield final concentrations of 100 μM GTPγS or GTP and 10 mM MgCl2. Components needed to quantify ARF stimulation of cholera toxin (CTA)-catalyzed ADP-ribosylagmatine formation were then added in 70 μl to yield final concentrations of 50 mM potassium phosphate (pH 7.5), 6 mM MgCl2, 20 mM DTT, ovalbumin (0.3 mg/ml), 0.2 mM [adenine-14C]NAD (0.05 μCi), 20 mM agmatine, cardiolipin (1 mg/ml), and 100 μM GTPγS or GTP with 0.5 μg of cholera toxin (41). After incubation at 30°C for 1 h, samples (70 μl) were transferred to columns of AG1-X2 equilibrated with water and eluted with five 1-ml volumes of water. The eluate, containing [14C]ADP-ribosylagmatine, was collected for radioassay.
Antibodies.
High-performance liquid chromatography-purified (>95% pure) synthetic peptides representing N- and C-terminal sequences of ARD1 (N-terminal ARD1 [Nt-ARD1] ATLVVNKLGAG; C-terminal ARD1 [Ct-ARD1], QLVAAGVLDVA) were purchased from Bio-Synthesis, Inc. Mass spectroscopy, amino acid analysis, and sequencing were performed on each peptide. Peptides were dissolved in water at a final concentration of 10 mg/ml. Rabbits were immunized with either Nt-ARD1 or Ct-ARD1 peptide coupled to hemocyanin as published (38). After (NH4)2SO4 precipitation and dialysis, immunoglobulin G (IgG) from antisera against the Nt- and Ct-ARD1 peptides was purified on protein A/G-Sepharose (Pierce) and then affinity purified on Affi-Gel 15 and Affi-Gel 10 (Bio-Rad) coupled to Nt-ARD1 and Ct-ARD1, respectively. Specific antibodies were eluted in 12 ml of 0.2 M glycine (pH 2.7)–10% ethylene glycol, and the pH was immediately adjusted to 7.5 with 1 N NaOH. After dialysis against phosphate-buffered saline (PBS), antibodies were concentrated (Centricon 50) and stored at −80°C in 30% glycerol. Polyclonal antibodies against ARD1 were prepared by injecting purified recombinant fusion proteins (GST-ARD1) into rabbits as described (42). The ARD1 antibody was affinity-purified on His6-ARD1-Ni2+ columns (Novagen) and stored as described above. Specificity of NH2- and COOH-terminal and ARD1 antibodies was demonstrated by Western blotting and immunofluorescence (42, 43). Antibodies against β-COP, p58, AP-1, and the secondary antibodies used for immunofluorescence studies were from Sigma.
Cell culture and eucaryotic expression.
NIH 3T3 fibroblasts were grown in Dulbecco's modified Eagle's medium containing 25 mM glucose, 10% fetal bovine serum, penicillin and streptomycin (each at 10 U/ml), and 200 mM glutamine. Absence of mycoplasma in culture was confirmed by PCR using the mycoplasma PCR primer set from Stratagene. Expression plasmids with DNA encoding ARD1, ARD1 fragments, or mutant proteins were introduced into NIH 3T3 cells (100-mm-diameter dishes; 80% confluent) using Transfectam (Promega) as described (43), or Lipofectamine Plus (GIBCO BRL) according to the manufacturer's instruction. After 2 to 3 h of incubation at 37°C, 10 ml of culture medium with fetal bovine serum and antibiotics were added. Expression of ARD1, ARD1 fragments, or mutant proteins was assessed after 48 h by immunofluorescence. Transfection efficiency, estimated after each transfection by counting 500 cells in planar sections randomly selected, ranged from 4 to 17%.
Immunocytochemistry.
Cells were fixed for 20 min with 4% paraformaldehyde in 0.12 M sodium–potassium phosphate buffer, pH 7.0 (7). After several rinses with PBS, cells were permeabilized for 4 min in PBS containing 0.1% Triton X-100 and incubated for 1 h with 3% BSA and 10% normal goat serum in PBS to reduce nonspecific reaction. Cells were then incubated for 1 h with the primary antibody diluted in PBS containing 3% BSA in a moist chamber, washed, and subsequently incubated with the appropriate secondary antibodies diluted 1:400. Coverslips were extensively washed with PBS, rinsed with water, and mounted in Mowiol 4-88 (Hoechst). For evaluation of immunofluorescence, samples were inspected with a PlanApo oil immersion objective (60×) on a Nikon ELWD0.3 microscope. Acquisition of labeled cell images was accomplished with a Leica laser-scanning confocal microscope as described earlier (7, 40). No staining was observed with the secondary antibody alone, when the cells were not permeabilized, or after mock transfection. All experiments were repeated at least once. Photomicrographs are representative of at least 90% of transfected cells, except as indicated in Fig. 4 and 5.
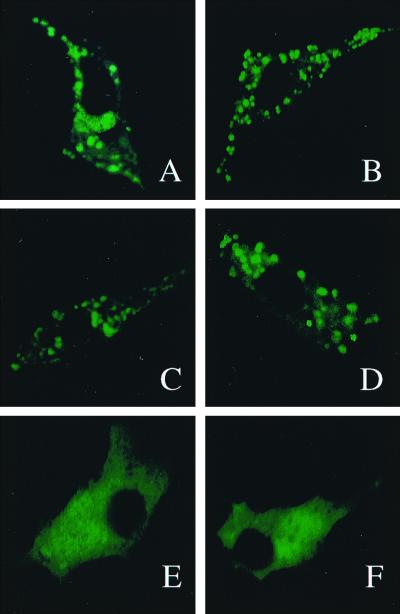
FIG. 4.
At 48 h after transfection, NIH 3T3 cells expressing ARD1, ARD1(301–402) or ARD1(201–300) were incubated for 15 min at 37°C with PBS (A, C, and E) or with AcR (B, D, and F), and coverslips were briefly washed with PBS before cells were fixed and mounted for microscopic observation. Identical results were obtained with three different cell cultures.
FIG. 5.
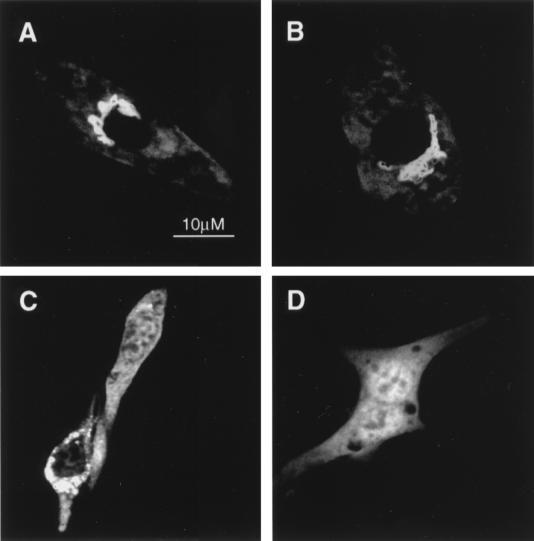
Effect of site-specific mutation of KXXXQ motifs on localization of the GAP domain of ARD1. NIH 3T3 cells were transfected with pcDNA3.1(GAP domain) (A), pcDNA3.1(344ATLQA348GAP domain) (B), or pcDNA3.1(369AQQQA373GAP domain) (C). (Mutated amino acids are underlined.) Immunofluorescence images obtained after incubation of the cells with anti-Nt-ARD1 (1:10,000) antibodies are shown. Similar results were obtained with two different cell preparations.
RESULTS
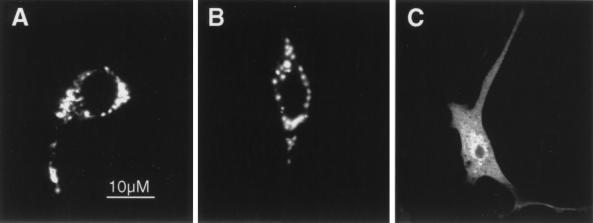
Confirming our previous observations (43), ARD1 overexpressed in NIH 3T3 cells was localized in the perinuclear region based on immunoreactivity with antibodies raised against recombinant ARD1 (Fig. 1A). Some vesicular structures scattered throughout the cytosol and in cellular protrusions were also labeled (Fig. 1A). Identical results had been observed with antibodies raised against undecapeptides corresponding to the N and the C termini of ARD1 (43). In NIH 3T3 cells overexpressing the ARF domain of ARD1, anti-Ct-ARD1 antibodies stained the juxtanuclear region in a pattern similar to that of a Golgi marker (Fig. 1B), whereas no staining was detected with an anti-Nt-ARD1 antibody (data not shown). Conversely, in cells overexpressing the GAP domain of ARD1, an anti-Nt-ARD1 antibody stained vesicular structures throughout the cells and less strongly stained the perinuclear region, a pattern reminiscent of the endosomal-lysosomal complex (Fig. 1C). As expected, an anti-Ct-ARD1 antibody did not label any structures in cells expressing the GAP domain of ARD1 (data not shown). These results show that, when expressed separately, the ARF and GAP domains of ARD1 have distinct subcellular distributions.
FIG. 1.
Localization of ARD1 and of the ARF and GAP domains of ARD1. NIH 3T3 cells were transfected with pcDNA3.1(ARD1) (A), pcDNA3.1(ARF domain) (B), pcDNA3.1(GAP domain) (C). Immunofluorescence images obtained after incubation of the cells with anti-ARD1 (1:1,000), anti-Ct-ARD1 (1:10,000), and anti-Nt-ARD1 ARD1 (1:10,000) antibodies are shown. Identical results were obtained with four different NIH 3T3 cell preparations and with COS 7 cells.
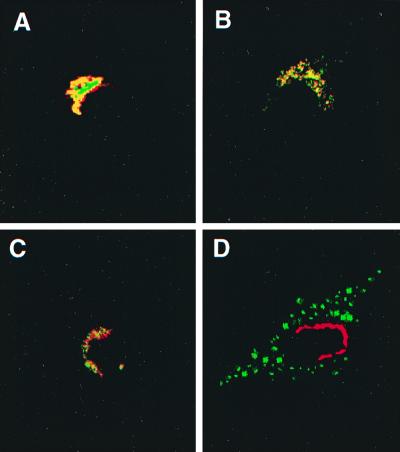
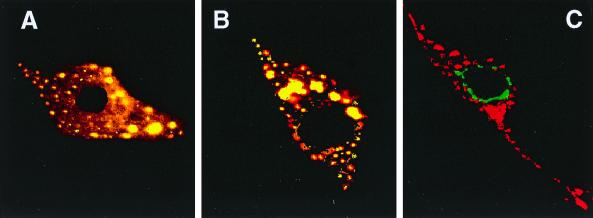
In cells expressing the ARF domain of ARD1, we compared the patterns of immunofluorescence obtained with anti-Ct-ARD1 and those with anti-p58, anti-β-COP, or anti-AP-1 (Golgi markers) antibodies (Fig. 2). Most, but not all, of the p58-labeled structures were labeled by the anti-Ct-ARD1 antibody, and vice versa (Fig. 2A). The AP-1 and ARD1 were colocalized more completely (Fig. 2B), suggesting that the ARF domain of ARD1 was present in the trans-Golgi network (TGN). Although there was partial colocalization of ARD1 and the cis-Golgi β-COP (Fig. 2C), it was less than that with the other Golgi markers. In contrast, distributions of the ARF domain and the lysosomal marker lysosome-associated membrane protein 1 (LAMP-1) (Fig. 2D) were completely different. The data are consistent with the hypothesis that the overexpressed ARF domain was concentrated in the Golgi apparatus.
FIG. 2.
Subcellular localization of the ARF domain of ARD1. NIH 3T3 cells transfected with pcDNA3.1(ARF domain) were incubated simultaneously with rabbit anti-Ct-ARD1 (1:10,000) (A to D) and mouse anti-p58 (1:100) (A), mouse anti-AP-1 (1:50) (B), mouse anti-β-COP (1:20) (C), or mouse anti-LAMP-1 (1:1,000) (D) antibodies, followed by the secondary anti-rabbit-IgG-fluorescein isothiocyanate and anti-mouse-IgG-rhodamine antibodies (A to C) or anti-rabbit-IgG-rhodamine and anti-mouse-IgG-fluorescein isothiocyanate antibodies (D). Preparations were examined by confocal microscopy. Areas where rhodamine and fluorescein are colocalized are yellow. The experiment was repeated once with a different cell preparation and similar results were obtained.
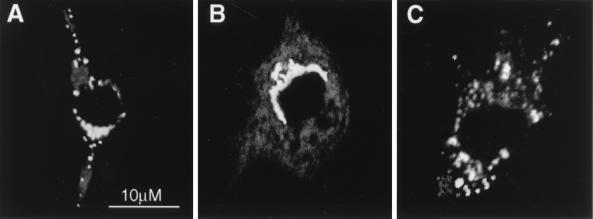
To demonstrate association of the overexpressed GAP domain of ARD1 with lysosomes in transfected NIH 3T3 cells, double immunofluorescence experiments were performed with anti-LAMP-1 antibodies and the fluorescent probe Lysotracker, both markers for lysosomes, and the anti-Nt-ARD1 antibodies. Confocal microscopy indicated extensive colocalization of the GAP domain of ARD1 with LAMP-1 (Fig. 3A) and Lysotracker (Fig. 3B), but not with the Golgi marker p58 (Fig. 3C). Data from the dual staining experiments suggested that ARD1 contains different signal elements in the ARF and GAP domains, which are determinants of Golgi and lysosomal localization, respectively.
FIG. 3.
Subcellular localization of the GAP domain of ARD1. NIH 3T3 cells transfected with pcDNA3.1(GAP domain) were incubated at 37°C with 4 ml of DMEM without (A and C) or with (B) 60 nM Lysotracker (lysosomal marker) for 60 min before fixation and permeabilization for 10 min in PBS containing 10 μM digitonin. Cells were then incubated simultaneously with rabbit anti-Nt-ARD1 (1:10,000) (A to C) and mouse anti-LAMP-1 (1:1,000) (A) or the mouse anti-p58 (1:100) (C) antibodies, followed by the secondary anti-rabbit-IgG-fluorescein isothiocyanate and anti-mouse-IgG-rhodamine antibodies (A), the anti-rabbit-IgG-fluorescein isothiocyanate antibody (B), or anti-rabbit-IgG-rhodamine and anti-mouse-IgG-fluorescein isothiocyanate antibodies (C). As in Fig. 2, preparations were examined by confocal microscopy. The yellow staining corresponds to areas where rhodamine and fluorescein colocalize. Similar results were obtained with two different cell preparations.
A membrane fraction of NIH 3T3 cells expressing the GAP domain of ARD1 was probed by Western blotting with anti-ARD1 and anti-Nt-ARD1 antibodies. No effect of serum starvation or NH4Cl addition on the amounts of the single immunoreactive band of ∼46-kDa, which comigrated with recombinant ARD1 GAP domain, was detected (data not shown), suggesting that the overexpressed protein was not degraded in lysosomes.
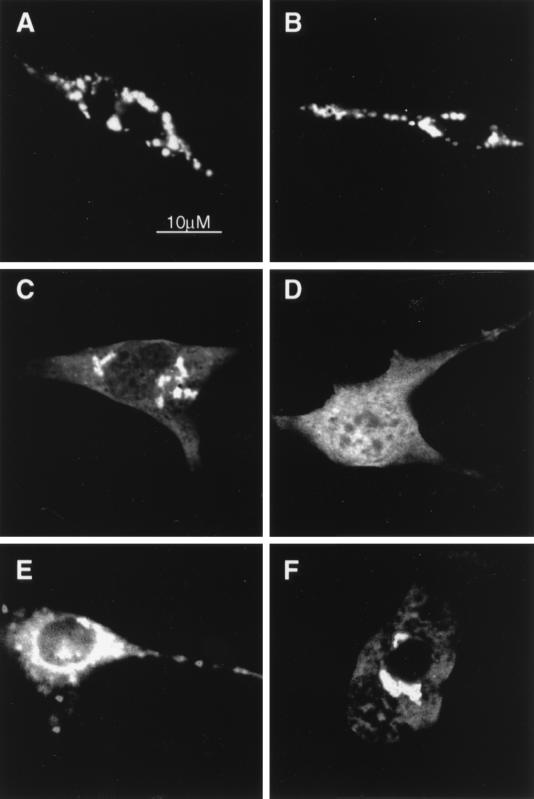
Green fluorescent fusion proteins have been widely used to study the subcellular distribution of proteins (reviewed in reference 36). Confirming our earlier report (43), 48 h after transfection, GFP-ARD1 was mainly concentrated in the perinuclear region (Table 1). Incubation of cells in an acidic acetate Ringer's medium (AcR), which is known to cause dispersion of lysosomes from the perinuclear area (15), resulted in dispersion of the GFP-ARD1 fluorescence (Fig. 4A and B; Table 1). Fragments corresponding to the N-terminal first 100, 200, or 300 amino acids were mainly found in the cytoplasm, and no effect of AcR treatment was observed (Table 1). Similarly, removal of 100, 200, or 300 amino acids from the N terminus of ARD1 did not affect localization before or after AcR treatment (Table 1), suggesting that signal elements involved in targeting ARD1 to the Golgi area or to lysosomes are not present in its N terminus. When expressed as a GFP fusion protein, however, the GAP domain of ARD1 (first 402 residues) was detected in vesicular structures concentrated mainly in the perinuclear region and was also present in cellular protrusions, presumably associated with lysosomes (Table 1). AcR treatment induced dispersion of the latter structures over the entire cell body, as expected of lysosomes (Table 1).
In cells expressing a GFP fusion protein corresponding to 172 residues from the C terminus of ARD1 (i.e., the ARF domain), fluorescence was detected only in the perinuclear region (Table 1). After AcR treatment, fluorescence was still restricted to the perinuclear area, presumably associated with Golgi membranes (Table 1). Expression of other fragments of ARD1 confirmed that proteins containing amino acids 301 to 402 always exhibited a lysosomal-type pattern of fluorescence, whereas fragments without this segment were not found in lysosomes (Fig. 4C to F; Table 1). Colocalization of ARD1 GFP fusion fragments with a Golgi marker (p58) and a lysosomal marker (LAMP-1) confirmed these observations (Table 1). These data are consistent with the conclusion that the signal element responsible for lysosomal localization of ARD1 is present in amino acids 301 to 402 of the GAP domain. Fragments containing residues 403 to 500, 403 to 480, 500 to 574, and 481 to 574 were detected only in the cytosol, suggesting that perinuclear localization of the ARD1-ARF domain requires a largely intact ARF domain or multiple signal elements present in it.
At least two different types of signal elements have been implicated in targeting proteins to lysosomes. Well-known tyrosine-based motifs have been described as critical for localization of LAMPs and other proteins to lysosomes (reviewed in reference 35). The pentapeptide KFERQ and sequence immunologically related to it were reported by Dice and coworkers to be the signal motif recognized by members of the 70-kDa heat shock protein family that are involved in a specific lysosomal proteolytic pathway (10). Two KXXXQ motifs are present in the sequence of ARD1 between amino acids 301 and 402, a region that contains no tyrosine residues. The potential involvement of 344KTLQQ348 and 369KQQQQ373 sequences in lysosomal targeting of the GAP domain of ARD1 was assessed by site-specific mutagenesis. When 344KTLQQ348 was mutated to 344ATLQA348, the mutant GAP domain had a distribution pattern similar to that of the nonmutant protein (Fig. 5A and B). Localization of the mutant GAP domain containing 369AQQQA373, however, was dramatically different. The protein was detected mainly in the cytoplasm and accumulated in the perinuclear region (Fig. 5C), suggesting that the sequence 369KQQQQ373 in the GAP domain of ARD1 is critical for its lysosomal localization. In agreement with the results obtained from overexpression of mutants of the GAP domain, the subcellular distribution of ARD1(344ATLQA348) was indistinguishable from that of ARD1 (Fig. 6A and E), whereas ARD1(369AQQQA373) was restricted to the perinuclear region (Fig. 6F). ARD1(369AQQQA373) colocalized with the Golgi marker p58 (data not shown), confirming a critical role for the sequence 369KQQQQ373 in the lysosomal localization of ARD1.
FIG. 6.
Effect of site-specific mutation of tyrosine-based and KXXXQ motifs on localization of ARD1. NIH 3T3 cells were transfected with pcDNA3.1(ARD1) (A), pcDNA3.1(445AKNA448ARD1) (B), pcDNA3.1(555AEGA558ARD1) (C), pcDNA3.1(445AKNA448-555AEGA558ARD1) (D), pcDNA3.1(344ATLQA348ARD1) (E), or pcDNA3.1(369AQQQA373ARD1) (F). (Mutated amino acids are underlined.) Immunofluorescence images obtained after incubation of the cells with anti-ARD1 (1:1,000) antibodies are shown. Similar results were obtained with at least three different cell preparations.
The region 301 to 402 in ARD1 also contains two di-leucine motifs (20), which in other proteins have been shown to play a role in targeting of proteins to organelles (32). Single replacement of Leu331, Leu332, Leu363, or Leu364 with glycine did not alter localization of ARD1, nor did the double replacements Leu331-Leu332 or Leu363-Leu364 (data not shown). Identical results were obtained when the equivalent mutations were made in the GAP domain of ARD1 (data not shown). Although an interaction of this di-leucine motif with some APs cannot be completely excluded, it appears that these leucines do not have a critical function in the subcellular localization of ARD1.
The ARF domain of ARD1 has two YXXL motifs (20). The role of the 445YKNL448 and 555YEGL558 sequences in Golgi localization of the ARF domain was investigated by site-specific mutagenesis. Distribution of the mutant ARF domain containing 445AKNA448 was not apparently different from that of the nonmutant (Fig. 7A and B). Localization of the ARF domain containing 555AEGA558 was, however, more complex. Of 320 transfected cells, 64% contained the mutant ARF domain in both the cytosol and the nucleus (Fig. 7C, upper cell), whereas in 36% it was found in both the cytosol and the perinuclear region (Fig. 7C, lower cell). The perinuclear fluorescence coincided with that for p58 (data not shown). The ARF domain containing both sets of mutations (Fig. 7D) was present in both cytosol and nucleus, without obvious perinuclear concentration.
FIG. 7.
Effect of site-specific mutation of tyrosine-based motifs on localization of the ARF domain of ARD1. NIH 3T3 cells were transfected with pcDNA3.1(ARF domain) (A), pcDNA3.1(445AKNA448ARF domain) (B), pcDNA3.1(555AEGA558ARF domain) (C), or pcDNA3.1(445AKNA448-555AEGA558ARF domain) (D). (Mutated amino acids are underlined.) Immunofluorescence images obtained after incubation of the cells with anti-Ct-ARD1 (1:10,000) antibodies are shown. Similar results were obtained with two different cell cultures.
We used functional assays to monitor conformational integrity of the mutated ARF domain proteins. Binding of GTPγS to ARF requires a strict positioning of residues involved in the nucleotide-binding pocket and is responsible for the conformational switch that activates ARFs. No significant differences in GTPγS binding among ARF domain, ARD1(445AKNA448), ARD1(555AEGA558), and ARD1(445AKNA448-555AEGA558) were observed (Table 2), consistent with no difference in architecture of the guanine nucleotide-binding site of these recombinant proteins. Activation of cholera toxin-catalyzed ADP-ribosylagmatine formation by ARFs requires binding of GTP followed by an interaction with the bacterial toxin that induces a change in its catalytic activity. Therefore, the ability of the ARF domain mutants to activate CTA should be a good indicator of conformational integrity. No significant difference among the recombinant proteins in their abilities to activate CTA was observed (Table 2), confirming that the three mutant proteins were probably folded correctly. Together, these results suggest that the 445YKNL448 sequence has a critical role in Golgi localization of the ARF domain of ARD1 and that the 555YEGL558 motif may function coordinately with it.
TABLE 2.
GTPγS binding and ARF activity of the ARF domain of ARD1 and mutated ARF domains of ARD1a
| Protein | GTPγS binding (pmol) | ARF activity (nmol/h) |
|---|---|---|
| ARF domain | 1.07 ± 0.055 | 0.51 ± 0.03 |
| 445AKNA448 | 0.97 ± 0.090 | 0.49 ± 0.04 |
| 555AEGA558 | 1.09 ± 0.064 | 0.47 ± 0.05 |
| 445AKNA448-555AEGA558 | 1.11 ± 0.087 | 0.51 ± 0.03 |
[35S]GTPγS binding to the indicated recombinant ARF domain protein (70 pmol) was assessed by a rapid filtration technique in the presence of 3 μM GTPγS and of cardiolipin (1 mg/ml). In the absence of cardiolipin, GTPγS binding was very low (≈0.035 ± 0.006 pmol) and did not differ significantly among the proteins. Binding in the absence of protein has been subtracted. After the recombinant proteins were incubated with 100 μM of GTPγS and cardiolipin (1 mg/ml), ARF stimulation of cholera toxin-catalyzed ADP-ribosylagmatine formation was assessed for 60 min at 30°C. ARF activity is the difference between the cholera toxin-catalyzed formation of [14C]ADP-ribosylagmatine without and with ARF domain protein. In the absence of cardiolipin, ARF activity was less than 0.005 nmol/h.
The role of these motifs in the dual localization of intact ARD1 was also evaluated. Immunofluorescence microscopy revealed that ARD1(445AKNA448), like ARD1, was present in the perinuclear region as well as in vesicular structures in cytoplasmic processes (Fig. 6A and B). The same mutation, likewise, had no apparent effect on localization of the ARF domain, which was entirely perinuclear (Fig. 7A and B). The mutant ARD1(555AEGA558), however, was distributed in a pattern very similar to that of the equivalent mutant of the ARF domain (compare Fig. 6C and 7C). Of 210 transfected cells, the ARD1 mutant (Fig. 6C) was present in the cytosol in 73% of the cells and in the perinuclear region as well as in the cytosol in 27% of the cells. Dual staining confirmed that the immunoreactive material in perinuclear structures, in cells expressing ARD1(555AEGA558), colocalized with p58 (data not shown). After mutation of both tyrosine-based motifs, ARD1 was found in the cytosol and nucleus, but not lysosomes (Fig. 6D). These data are consistent with the view that each of the tyrosine-based motifs in the ARF region of ARD1 may contribute to its localization in the Golgi complex and that passage through the Golgi is required to reach lysosomes.
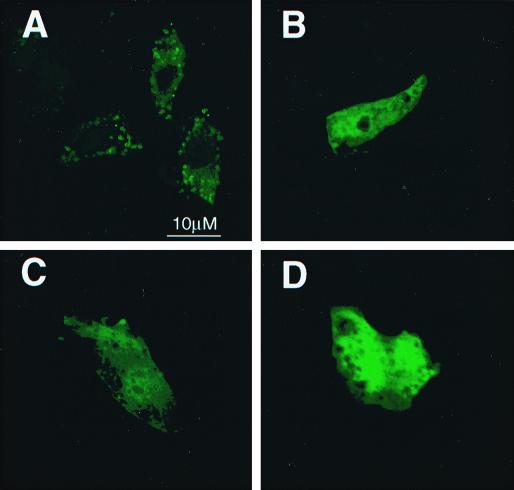
GFP fusion proteins containing the individual localization motifs identified in ARD1 were used to determine whether any of them would be sufficient to localize GFP in the Golgi apparatus or lysosomes rather than cytosol. A GFP-QKQQQQF fusion protein appeared partially cytosolic but was also associated with lysosomes (Fig. 8A). On the other hand, GFP fusion proteins containing either or both of the tyrosine-based motifs were present in cytoplasm and nucleus (Fig. 8B to D). Thus, although the QKQQQQF motif was adequate for lysosomal localization, in a non-ARF context, the tyrosine-based motifs were insufficient to achieve localization in the Golgi system.
FIG. 8.
Localization of GFP fusion protein containing peptide localization motifs. NIH 3T3 cells were transfected with a plasmid encoding GFP-QKQQQQF (A), GFP-EYKNLK (B), GFP-LYEGLD (C), and GFP-EYKNL(K/L)YEGLD (D). Immunofluorescence images were obtained 12 h after transfection. Similar results were obtained with at least three different cell preparations.
DISCUSSION
The experiments described here were undertaken to characterize better the subcellular localization of ARD1 and test directly the involvement of potential signal motifs in delivery of this protein to lysosomal and Golgi structures. Two tyrosine-based motifs were identified that are important in association of the ARF domain of ARD1 with the Golgi apparatus, as well as a KXXXQ sequence that has a critical role in localization of the GAP domain to lysosomes.
Previous studies have implicated the heat shock cognate protein of 73 kDa (hsc73) in stimulating a lysosomal pathway of proteolysis that is selective for particular cytosolic proteins (8). Interestingly, hsc70 also was implicated in the uncoating of clathrin-coated endosomes (31). The KFERQ pentapeptide sequence is an important determinant of cytosolic protein binding by hsc70 (34). Using GFP fusions with truncated ARD1 sequences, we observed that residues 301 to 402 in the GAP domain of ARD1 contain a signal for lysosomal localization (Table 1). Proteins containing the first 300 amino-terminal residues, but not amino acids 301 to 402, were distributed in the cytosol and the nucleus. It is possible that the sequence CX2C-X16CXHX2CHXCX12CX2, analogous to a zinc-binding domain, which is found in the N-terminal part of ARD1 (42), could play a role in the nuclear localization of these fragments. The fact that mutation of the sequence 369KQQQQ373 in the GAP domain (or in ARD1) prevented lysosomal localization might be consistent with a relationship between the latter protein and the hsc70 pathway. Addition of the QKQQQQF motif to GFP resulted in appearance of the fusion protein with lysosomes.
Based on our earlier findings that native ARD1 was associated with lysosomes and that distribution was not affected by treatment of cells with chloroquine or ammonium chloride (43), which are known to inhibit the hsc70 proteolytic pathway (1), we believe that ARD1 had not been routed to lysosomes for degradation. Accordingly, we detected no degradation products of the GAP domain of ARD1 in a membrane fraction of transfected cells, even when the hsp70 pathway had been activated by serum starvation (24). Rather, we speculate that similar to other members of the ARF family, ARD1 might have a direct role in the process of vesicular trafficking from the TGN to lysosomes. Therefore, unlike cytosolic proteins containing KFERQ motifs, which are routed to lysosomes by hsc70 for degradation, ARD1 could interact with the latter to regulate its activity, hypothetically in the uncoating of clathrin-coated endosomes. Accordingly, it was postulated that hsp70 proteins act in the lysosomal synthetic pathway or in the uncoating of clathrin-coated vesicles under normal conditions and that serum starvation triggers a specific degradation pathway by promoting interaction with cytosolic proteins containing KFERQ motifs (1). Whether ARD1 participates in the hsc70 degradation pathway remains to be determined.
A recent study has shown that although the catalytic site of ARF GAP1 (a GAP protein for ARF1) is located in the amino-terminal part of the protein (9), the carboxyl terminus is also required for function in cells (16). It was shown that the carboxyl terminus of ARF GAP1 could be involved in targeting the protein to the Golgi apparatus, presumably through interaction with another Golgi-associated protein (16). Accordingly, we demonstrated that, in vitro, the polypeptide containing amino acids 100 to 300 of the GAP domain of ARD1 was a bona fide GAP protein for its ARF domain (42). Our results now suggest that as in ARF GAP1, some targeting information is located carboxy-terminal to the catalytic site of the built-in GAP domain of ARD1.
Proteins resident in the TGN include TGN38 and TGN41, isoforms of a highly glycosylated type I membrane protein of unknown function (18, 29), and furin, which belongs to a subfamily of the subtilisin-related mammalian endoproteases (3). Although it is clear that furin and TGN38 and TGN41 cycle between the TGN and the plasma membrane (5, 21), the molecular mechanisms responsible for retention of the bulk of these molecules in the TGN at steady state remains poorly understood. Intracellular trafficking of furin requires a tyrosine-based motif and an acidic region (44), whereas that of TGN38 relies on a tyrosine-based motif and the transmembrane region (5). Intracellular trafficking of the varicella-zoster virus glycoprotein I is regulated by a tyrosine-based motif and a casein kinase II phosphorylation site (2). As reported here, ARD1, like those proteins, uses distinct small peptide motifs to accomplish its association with different subcellular compartments. ARD1, however, appears to be the first example of nonintegral membrane protein that uses tyrosine-based motifs for targeting.
Although a YEGL sequence is present in all six mammalian ARFs, there are slight variations in the other tyrosine-based motif, which in ARD1 is YKNL. ARFs 1 to 5 contain the sequence YKNI and ARF6 contains YKNV. ARD1 has a subcellular distribution, seemingly distinct from those of the ARFs (43). Class I ARFs 1 to 3 appear to be cytosolic proteins that associate specifically and reversibly with the Golgi apparatus. The much more limited data on ARF4 and -5 are probably consistent with a similar distribution (reviewed in reference 22). ARF6, however, was found predominantly associated with membrane compartments such as endosomes (13), plasma membrane (6, 26, 33), and secretory granules (14), although cytosolic ARF6 was also reported in some cells (46). A recent study employing chimeric ARF1-ARF6 proteins suggested that important localization elements are present in both the amino- and carboxyl-terminal parts of ARF6 and ARF1 and that amino acids 53 to 58 of ARF6, equivalent to amino acids 444 to 449 in ARD1, were critical for localization (1a).
Although the subcellular distribution of ARD1 has not been studied as extensively as that of ARF1 and ARF6, overexpressed epitope-tagged ARD1 was found associated with Golgi and lysosomal membranes and was not detected in cytosol. The endogenous protein was also identified in membranes from these organelles (43). In the experiments reported here, we detected no cytosolic epitope-tagged ARF6 or ARD1 in transfected NIH 3T3 cells (data not shown), although it is possible, of course, that cytosolic ARD1 is detectable in other types of cells. It may be relevant that ARD1 does not possess an amino-terminal glycine that is myristoylated in all of the ARF proteins and believed to be important in their interaction with cellular membranes (20). It is tempting to speculate that tyrosine-based motifs contribute to the predominant membrane association of ARF6 and ARD1 and that in the YKNΘ motifs (where Θ represents a hydrophobic amino acid), the hydrophobic residue contributes to the specific association with subcellular organelles through interaction with distinct adapter proteins (19, 25).
We found that mutation of the two tyrosine-based motifs in ARD1 resulted in a cytoplasmic localization, whereas the GAP domain, which lacks these sequences, was associated with lysosomes when expressed separately (Fig. 1C and 3A to C). These results clearly indicate that the GAP domain contains some part of a targeting signal for lysosomal localization, but in the intact protein structure, tyrosine-based motifs in the ARF domain also play a critical role. In agreement with these observations, the mutant ARD1(555AEGA558) was found in the cytosol and Golgi structures. Our previous observation that newly synthesized GFP-ARD1 protein appeared first associated with the Golgi apparatus and subsequently was accumulated in lysosomes, could suggest that the interaction of ARD1 with a component of the Golgi complex is a requirement for transport to lysosomes. The partial colocalization of ARD1 (43) or its ARF domain with markers of distinct portions of the Golgi apparatus also suggests that ARD1 (or the ARF domain) can move throughout the Golgi complex and is not restricted to one specific subcompartment. The presence of the amino-terminal zinc finger motif may account for localization of some of ARD1 in the nucleus after mutation of both tyrosine-based signal sequences.
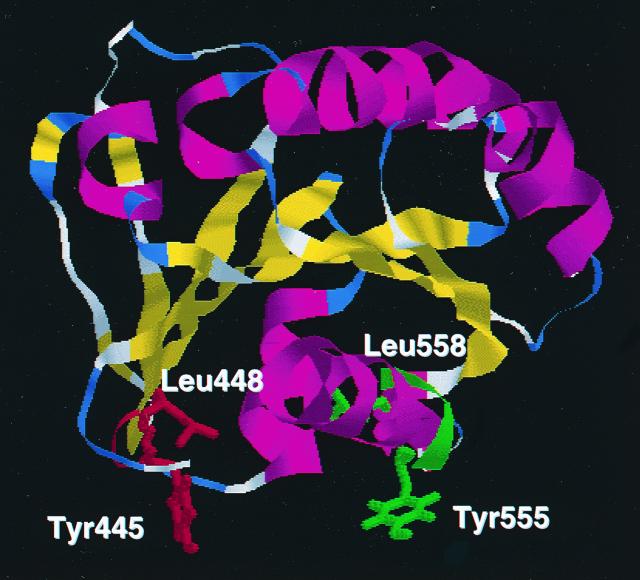
Multiple tyrosine-based motifs appear to play a role in shuttling of the TGN51 protein between the cell surface and the TGN (17). Because mutation of the two tyrosine-based signals was required to induce a completely cytosolic localization of the ARF domain of ARD1 in transfected cells, it seems that both tyrosine-based elements may participate in the association with Golgi membranes. The data show, however, that mutation of 555YEGL558 in ARD1, or its ARF domain, produced the more dramatic effect on localization. It is perhaps important that the YEGL motif is located near the carboxy terminus of ARD1, because it has been demonstrated that the spacing of tyrosine-based motifs relative to the membrane is critical for localization (30). A three-dimensional structure of the ARF domain of ARD1 (prepared by computer modeling obtained from SwissProt at http://www.expasy.ch/swissmod /SM_3DCrunch.htm) based on information derived from the crystal structure of ARF1, is very similar to that of ARF1 and has both tyrosine-based motifs on the same face of the molecule (Fig. 9). The tyrosine residues surround the carboxy-terminal α-helix and an α-helix analogous to the amino terminus of ARFs. In ARFs, both α-helices are believed to be critical for the membrane interaction, as they may be also in ARD1. Orientation of the two tyrosine-based motifs appended to the GFP was almost certainly different from that in Fig. 9, perhaps the reason for their failure to alter its distribution.
FIG. 9.
Tyrosine-based signals in the ARF domain of ARD1. A ribbon model of the structure of the ARF domain of ARD1 was generated using the RasMol software based on information derived from the crystal structure of ARF1. The tyrosine-based motif 445 to 448 is shown in red and the motif 555 to 558 is shown in green, with the tyrosine and leucine residues shown as sticks.
ACKNOWLEDGMENTS
We thank W. A. Patton for stimulating discussions and help obtaining the three-dimensional model of the ARF domain of ARD1. We also thank W. Riemenschneider for his help with the confocal microscopy and J. G. Donaldson and J. S. Bonifacino for interesting discussions.
REFERENCES
- 1.Agarraberes F A, Terlecky S R, Dice J F. An intralysosomal hsc70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1a.Al-Awar O, Rodhakrishna H, Powell N N, Donaldson J B. Separation of membrane trafficking and actin remodeling functions of ARF6 with an effective domain mutant. Mol Cell Biol. 2000;20:5998–6007. doi: 10.1128/mcb.20.16.5998-6007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alconada A, Bauer U, Hoflack B. A tyrosine-based motif and a casein kinase II phosphorylation site regulate the intracellular trafficking of the varicella-zoster virus glycoprotein I, a protein localized in the trans Golgi network. EMBO J. 1996;15:6096–6110. [PMC free article] [PubMed] [Google Scholar]
- 3.Barr P J, Mason O B, Landsberg P A, Wong M C, Kiefer M C, Brake A J. cDNA and gene structure for a human subtilisin-like protease with cleavage specificity for paired basic amino acid residues. DNA Cell Biol. 1991;10:319–328. doi: 10.1089/dna.1991.10.319. [DOI] [PubMed] [Google Scholar]
- 4.Bonifacino J S, Marks M S, Ohno H, Kirchhausen T. Mechanisms of signal-mediated protein sorting in the endocytic and secretory pathways. Proc Assoc Am Physician. 1996;108:285–295. [PubMed] [Google Scholar]
- 5.Bos K, Wraight C, Stanley K. TGN38 is maintained in the trans-Golgi network by a tyrosine-containing motif in the cytoplasmic domain. EMBO J. 1993;12:2219–2228. doi: 10.1002/j.1460-2075.1993.tb05870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caumont A-S, Galas M-C, Vitale N, Aunis D, Bader M-F. Regulated exocytosis in chromaffin cells. Translocation of ARF6 stimulates a plasma membrane-associated phospholipase D. J Biol Chem. 1998;273:1373–1379. doi: 10.1074/jbc.273.3.1373. [DOI] [PubMed] [Google Scholar]
- 7.Chasserot-Golaz S, Vitale N, Sagot I, Delouche B, Dirrig S, Pradel L-A, Henry J-P, Aunis D, Bader M-F. Annexin II in exocytosis: catecholamine secretion requires the translocation of p36 to the subplasmalemmal region in chromaffin cells. J Cell Biol. 1996;133:1217–1236. doi: 10.1083/jcb.133.6.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang H L, Dice J F. Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J Biol Chem. 1989;263:6797–6805. [PubMed] [Google Scholar]
- 9.Cukierman E, Huber I, Rotman M, Cassel D. The ARF1-GTPase-activating protein: zinc finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- 10.Dice J F, Chiang H-L, Spencer E P, Backer J M. Regulation of catabolism of microinjected ribonuclease A. J Biol Chem. 1986;261:6853–6859. [PubMed] [Google Scholar]
- 11.Ding M, Vitale N, Tsai S-C, Moss J, Vaughan M. Characterization of a GTPase-activating protein that stimulates GTP hydrolysis by both ADP-ribosylation factor (ARF) and ARF-like proteins. J Biol Chem. 1996;271:24005–24009. doi: 10.1074/jbc.271.39.24005. [DOI] [PubMed] [Google Scholar]
- 12.Donaldson J G, Radhakrishna H, Peters P J. The ARF GTPases: defining roles in membrane traffic and organelle structure. Cold Spring Harb Symp Quant Biol. 1995;60:229–234. doi: 10.1101/sqb.1995.060.01.026. [DOI] [PubMed] [Google Scholar]
- 13.D'Souza-Schorey C, Li G, Colombo M I, Stahl P D. A regulatory role for ARF6 in receptor-mediated endocytosis. Science. 1995;267:1175–1178. doi: 10.1126/science.7855600. [DOI] [PubMed] [Google Scholar]
- 14.Galas M-C, Helms J B, Vitale N, Thiersé D, Aunis D, Bader M-F. Regulated exocytosis in chromaffin cells: a potential role for a secretory granule-associated ARF6 protein. J Biol Chem. 1997;272:2788–2793. doi: 10.1074/jbc.272.5.2788. [DOI] [PubMed] [Google Scholar]
- 15.Heuser J. Changes in lysosome shape and distribution correlated with changes in cytoplasmic pH. J Cell Biol. 1989;108:855–864. doi: 10.1083/jcb.108.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber I, Cukierman E, Rotman M, Aoe T, Hsu V W, Cassel D. Requirement for both the amino-terminal catalytic domain and a noncatalytic domain for in vivo activity of ADP-ribosylation factor GTPase-activating protein. J Biol Chem. 1998;273:24786–24791. doi: 10.1074/jbc.273.38.24786. [DOI] [PubMed] [Google Scholar]
- 17.Kain R, Angata K, Kerjaschki D, Fukuda M. Molecular cloning and expression of a novel human trans-Golgi network glycoprotein, TGN51, that contains multiple tyrosine-containing motifs. J Biol Chem. 1998;273:981–988. doi: 10.1074/jbc.273.2.981. [DOI] [PubMed] [Google Scholar]
- 18.Luzio J P, Brake B, Banting G, Howell K E, Braghetta P, Stanley K K. Identification, sequencing and expression of an integral protein of the trans-Golgi network. Biochem J. 1990;270:97–102. doi: 10.1042/bj2700097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mellman I. Endocytosis and molecular sorting. Annu Rev Cell Dev Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 20.Mishima K, Tsuchiya M, Nightingale M S, Moss J, Vaughan M. ARD1, a 64-kDa guanine nucleotide-binding protein with a carboxyl-terminal ADP-ribosylation factor domain. J Biol Chem. 1993;268:8801–8807. [PubMed] [Google Scholar]
- 21.Molloy S S, Thomas L, VanSlyke J K, Stenberg P E, Thomas G. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 1994;13:18–33. doi: 10.1002/j.1460-2075.1994.tb06231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moss J, Vaughan M. Structure and function of ARF proteins: activators of cholera toxin and critical components of intracellular vesicular transport processes. J Biol Chem. 1995;270:12327–12330. doi: 10.1074/jbc.270.21.12327. [DOI] [PubMed] [Google Scholar]
- 23.Moss J, Vaughan M. Molecules in the ARF orbit. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 24.Neff N T, Bourret L, Maio P, Dice J F. Degradation of proteins microinjected into IMR-90 human diploid fibroblasts. J Cell Biol. 1981;91:184–194. doi: 10.1083/jcb.91.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohno H, Stewart J, Fournier M C, Bosshart H, Rhee I, Miyatake S, Saito T, Gallusser A, Kirchhausen T, Bonifacino J S. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 1995;269:1872–1875. doi: 10.1126/science.7569928. [DOI] [PubMed] [Google Scholar]
- 26.Radhakrishna H, Donaldson J G. ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J Cell Biol. 1997;139:49–61. doi: 10.1083/jcb.139.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Randazzo P A. Functional interaction of ADP-ribosylation factor 1 with phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 1997;272:7688–7692. [PubMed] [Google Scholar]
- 28.Rapoport I, Chen Y C, Cuppers P, Shoelson S E, Kirchhausen T. Dileucine-based sorting signals bind to the beta chain of AP-1 at a site distinct and regulated differently from the tyrosine-based motif-binding site. EMBO J. 1998;8:2148–2155. doi: 10.1093/emboj/17.8.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reaves B, Wilde A, Banting G. Identification, molecular characterization and immunolocalization of an isoform of the trans-Golgi network (TGN)-specific integral membrane protein. Biochem J. 1992;283:313–316. doi: 10.1042/bj2830313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rohrer J, Schweizer A, Russell D, Kornfeld S. The targeting of LAMP-1 to lysosomes is dependent on the spacing of its cytoplasmic tail tyrosine sorting motif relative to the membrane. J Cell Biol. 1996;132:565–576. doi: 10.1083/jcb.132.4.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothman J E, Schmidt S L. Enzymatic recycling of clathrin from coated vesicles. Cell. 1986;46:5–9. doi: 10.1016/0092-8674(86)90852-4. [DOI] [PubMed] [Google Scholar]
- 32.Sandoval I V, Bakke O. Targetting of membrane proteins to endosomes and lysosomes. Trends Cell Biol. 1994;4:292–297. doi: 10.1016/0962-8924(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 33.Song J, Khachikian S J, Radhakrishna H, Donaldson J G. Localization of endogenous ARF6 to the sites of cortical actin rearrangement and involvement of ARF6 in cell spreading. J Cell Sci. 1998;111:2257–2267. doi: 10.1242/jcs.111.15.2257. [DOI] [PubMed] [Google Scholar]
- 34.Terlecky S R, Chiang H-L, Olson T S, Dice J F. Protein and peptide binding and stimulation of in vitro lysosomal proteolysis by the 73-kDa heat shock cognate protein. J Biol Chem. 1992;267:9202–9209. [PubMed] [Google Scholar]
- 35.Trowbridge I S, Collawn J F, Hopkins C R. Signal-dependent membrane protein trafficking in the endocytotic pathway. Annu Rev Cell Biol. 1993;9:129–161. doi: 10.1146/annurev.cb.09.110193.001021. [DOI] [PubMed] [Google Scholar]
- 36.Tsien R Y. The green fluorescent protein. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 37.Tsuchiya M, Price S R, Tsai S-C, Moss J, Vaughan M. Molecular identification of ADP-ribosylation factor mRNAs and their expression in mammalian cells. J Biol Chem. 1991;266:2772–2777. [PubMed] [Google Scholar]
- 38.Vitale N, Mukai H, Rouot B, Thiersé D, Aunis D, Bader M-F. Exocytosis in chromaffin cells: possible involvement of the heterotrimeric GTP-binding Go. J Biol Chem. 1993;268:14715–14723. [PubMed] [Google Scholar]
- 39.Vitale N, Moss J, Vaughan M. ARD1, a 64-kDa bifunctional protein containing an 18-kDa GTP-binding ADP-ribosylation factor domain and a 46-kDa GTPase-activating domain. Proc Natl Acad Sci USA. 1996;93:1941–1944. doi: 10.1073/pnas.93.5.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vitale N, Gensse M, Chasserot-Golaz S, Aunis D, Bader M-F. Trimeric G proteins control regulated exocytosis in bovine chromaffin cells: sequential involvement of Go associated with secretory granules and Gi3 bound to the plasma membrane. Eur J Neurosci. 1996;8:1275–1285. doi: 10.1111/j.1460-9568.1996.tb01296.x. [DOI] [PubMed] [Google Scholar]
- 41.Vitale N, Moss J, Vaughan M. Interaction of the GTP-binding and GTPase-activating domains of ARD1 involves the effector region of the ADP-ribosylation factor domain. J Biol Chem. 1997;272:3897–3904. doi: 10.1074/jbc.272.7.3897. [DOI] [PubMed] [Google Scholar]
- 42.Vitale N, Moss J, Vaughan M. Molecular characterization of the GTPase-activating domain of ADP-ribosylation factor domain protein 1 (ARD1) J Biol Chem. 1998;273:2553–2560. doi: 10.1074/jbc.273.5.2553. [DOI] [PubMed] [Google Scholar]
- 43.Vitale N, Horiba K, Ferrans V J, Moss J, Vaughan M. Localization of the ADP-ribosylation factor domain protein 1 (ARD1) in lysosomes and Golgi apparatus. Proc Natl Acad Sci USA. 1998;98:8613–8618. doi: 10.1073/pnas.95.15.8613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Voorhoes P, Deignan E, van Donselaar E, Humphrey J, Marks M, Peters P J, Bonifacino J S. An acidic sequence within the cytoplasmic domain of furin functions as a determinant of trans-Golgi network localization and internalization from the cell surface. EMBO J. 1995;14:4961–4975. doi: 10.1002/j.1460-2075.1995.tb00179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welsh C F, Moss J, Vaughan M. ADP-ribosylation factors: a family of approximately 20-kDa guanine nucleotide-binding proteins that activate cholera toxin. Mol Cell Biochem. 1994;139:157–166. doi: 10.1007/BF00928458. [DOI] [PubMed] [Google Scholar]
- 46.Yang C Z, Heimberg H, D'Souza-Schorey C, Mueckler M M, Stahl P D. Subcellular distribution and differential expression of endogenous ADP-ribosylation factor 6 in mammalian cells. J Biol Chem. 1998;273:4006–4011. doi: 10.1074/jbc.273.7.4006. [DOI] [PubMed] [Google Scholar]