Abstract
Potyviruses are among the most important pathogens of dicotyledonous and monocotyledonous ornamentals and crop plants. In this study, leaf samples were collected from symptomatic narcissus plants and weeds in Fars and Tehran provinces of Iran. Enzyme-linked immunosorbent assay using broad-spectrum potyvirus antibodies gave a positive reaction with 38 out of 61 narcissus samples tested (62.3%); the results were confirmed by reverse-transcription polymerase chain reaction using universal NIb primers, and for thirty samples, by sequencing and phylogenetic studies. The results suggested the infection of almost all positive samples with narcissus yellow stripe virus (NYSV); only one sample seemed to be infected with narcissus late season yellows virus (NLSYV). The 3'-end of the genome of the NLSYV isolate and six NYSV isolates, encompassing the complete coat protein gene, was amplified and sequenced using species-specific and universal potyvirus primers. Sequence analysis indicated the presence of NLSYV and NYSV, not previously identified from Western Asia. No evidence of recombination was found in Iranian isolates. Based on phylogenetic analyses, isolates of NLSYV and NYSV clustered into five and three phylogroups, respectively, where all the Iranian isolates fell into distinct subpopulations in groups NLSYV-I and NYSV-II. Multiple sequence alignments showed some phylogroup-specific amino acid substitutions for both viruses. Phylogroup IV and II populations had higher nucleotide diversities as compared with other populations of NLSYV and NYSV, respectively. Our findings revealed the presence of negative selection in the populations of both viruses. Almost no statistically significant gene flow was found between populations of these viruses.
Supplementary information
The online version contains supplementary material available at 10.1007/s42161-021-00985-0.
Keywords: West Asia, Iran, Narcissus viruses, Potyvirus, Evolutionary analyses
Introduction
The genus Potyvirus (family Potyviridae) is one of the largest genera of viruses with 183 species (Lefkowitz et al. 2018; ICTV; https://talk.ictvonline.org/taxonomy/). Members of this genus are among the most important pathogens of ornamentals and crop plants all through the world. They infect both mono- and dicotyledonous plants and are found infecting wild plants as well (Gibbs and Ohshima 2010; Roossinck 2012). They have flexuous filamentous particles, 700–750 nm long, containing a monopartite linear positive sense single-stranded RNA genome of approximately 10 kb. Members of this genus have one major open reading frame (ORF) in their genome that is translated into a large polyprotein, which is processed proteolytically into at least ten different functional proteins (Wylie et al. 2017). Potyviruses are transmitted by aphid vectors in a non-persistent manner to a wide range of plant species. They also can spread through seeds and/or infected vegetative organs (Gibbs and Ohshima 2010). Previous studies have shown that the genus Potyvirus emerged from viruses infecting wild monocotyledonous plants about 7,250 years ago in southwestern Eurasia (i.e., Europe, Asia Minor and the Middle East) and/or North Africa (Gibbs and Ohshima 2010; Moury and Desbiez 2020). Interestingly, members of the two earliest lineages of potyviruses were first isolated from monocotyledonous bulb and grass species of the families Liliaceae or Poaceae which were first domesticated in the same region. This resembles those recorded for Rymovirus species, the close sister genus of Potyvirus (Nguyen et al. 2013a, b). A variety of potyviruses have been reported from West Asia, including the Middle East, and the rest of southwestern Eurasian region (e.g., Brunt et al. 1996; Farzadfar et al. 2002).
The genus Narcissus, belonging to the family Amaryllidaceae, consists of 63 species and many natural hybrids propagated vegetatively from bulbs (Sochacki and Chojnowska 2016). Narcissus species were primarily found in the western Mediterranean regions with a center of origin in the Iberian Peninsula (Spain and Portugal) and then spread into Far East prior to the tenth century (Wylie et al. 2014; Kamenetsky and Okubo 2016), with the exception of Narcissus tazetta, which is found across China, Japan and West Asia (Hanks 2002). Narcissus is one of the important ornamental plants grown for their beautiful flowers (Hanks 2002) and is a source of different alkaloids; therefore, the plant is exploited for essential oils and medical purposes as well (Masi et al. 2015). Narcissus is susceptible to a number of diseases caused by fungi, bacteria and viruses resulting in drastic reduction in the quality and quantity of bulbs and flowers (Brunt 1995). Viruses are one of the most important problems of plants propagated by vegetative organs like bulbs (Gera and Zeidan 2006). As reported, most bulb collections of narcissus around the world are heavily infested with viruses (Hanks 2002). Several species of the genus Potyvirus are reported to infect narcissus plants (Brunt et al. 1996; Chen et al. 2003; Wylie et al. 2010; Wylie and Jones 2012; Ohshima et al. 2016). Mixed infections with different Potyviridae species/strains have been frequently observed in these plants. While the level of genetic diversity of the coat protein (cp) gene region differs between potyviruses infecting narcissus, their evolutionary mutation rate is similar (Ohshima et al. 2016). Studies of narcissus plants with mixed infections also have resulted in finding new potyvirus species in the turnip mosaic virus (TuMV) group and natural recombination events between different potyviruses. Narcissus potyviruses in this group were then suggested to be members of a single mega-species, i.e., narcissus virus (Ohshima et al. 2018).
Iran has a great potential for cultivation of various ornamental plants, but this has recently been threatened by various plant diseases, including those caused by viruses. In this country, Narcissus is extensively cultivated in greenhouse and open-farm systems. In some regions, e.g., in Khuzestan and Fars provinces (southwest), and Khorasan-e-Jonoobi (southeast), Narcissus is grown as semi-wild plants under dry farming conditions. Kazeroon County (Fars province) is the main center of narcissus production in Iran; Jareh and Baladeh District, where the plant has been grown for tens of years and has the largest semi-wild narcissus farms in Iran. Long-term and extensive cultivation as well as vegetative propagation of the plant favor the occurrence of diverse viruses in Iranian narcissi. To our knowledge, only one study is available on potyviruses infecting Narcissus in Iran, i.e., narcissus degenerate virus (Koohi Habibi et al. 2010). Preliminary studies have shown that different potyvirus species/strains are occurring in this plant in the country (Golnaraghi et al. 2015; Khosroshahi et al. 2015). In this study, we attempted to identify potyviruses infecting narcissus plants and compare them with others using gene sequences from different geographical regions to provide the first detailed information of evolutionary characterization of potyviruses infecting narcissus plants in Iran.
Material and methods
Plant sampling
During springs of 2013 and 2014, surveys were conducted in two main provinces for narcissus production to identify potyviruses infecting this plant in Iran. Totally, 61 narcissus leaf samples showing virus-like symptoms of leaf chlorosis, streak mosaic and stunting were collected from field-grown (semi-wild) plants in Jareh and Baladeh District in Kazeroon County (Fars province) or greenhouse-grown plants in the city of Tehran (Tehran province). Moreover, twelve leaf samples from different weed plants, including Amaranthus sp., Malva sp., Taraxacum officinale and four unknown weeds, showing mottling, yellowing or chlorosis symptoms, were collected from the fields visited (Table S1 and Fig. S1). Samples were kept at 4 °C until tested serologically using monoclonal antibodies for viruses of the genus Potyvirus. Leaf samples also were silica gel/freeze dried and stored at –20 °C until required. Virus infection rate in each field/greenhouse visited was calculated according to the reaction of leaf sample extracts in diagnostic assays, as described below; virus disease incidence also was roughly estimated based on virus-like symptoms observed in ca. 20–50 randomly selected plants in each field/greenhouse.
Serological assays using species and genus specific antibodies
Potyvirus infections of the collected leaf samples were tested by plate-trapped antigen enzyme linked immunosorbent assay (PTA-ELISA) using monoclonal antibodies for viruses of the genus Potyvirus (Bioreba; Reinach, Switzerland). The samples were also tested for cucumber mosaic virus (CMV) using double-antibody sandwich ELISA (DAS-ELISA) and virus specific antibodies (Bioreba, Switzerland). The OD values were measured using a microtitre plate reader (BioTek ELx 800, USA). A reaction was considered positive when the absorbance value at 405 nm was equal to or more than three times of the mean value of the healthy controls. ELISA tests were carried out in two replicates using buffers as well as positive and negative controls provided by Bioreba (Switzerland), and performed according to the manufacturer’s instructions. Reverse-transcription followed by polymerase chain reaction (RT-PCR) tests and sequencing were subsequently utilized to confirm ELISA results.
Extraction of total RNA and RT-PCR
Total RNA was extracted from 0.02–0.03 g of dried leaf tissues using innuSOLV RNA solution (AJ Innuscreen GmbH, Berlin, Germany), according to the manufacturer’s instruction. Extracted RNA preparations were finally precipitated and resuspended in RNase/DNase free water. Their quality was then checked by gel electrophoresis; each RNA extract was then aliquoted into two microtubes and stored at –20 °C (for the following tests) and –70 °C (for long-term storage). First-strand cDNA synthesis was performed using 1 μg of total RNA, 1 µl (10 µM) of reverse primer (NIb3R or Tu3T9M) and moloney murine leukemia virus (M-MuLV) reverse transcriptase (CinnaGen, Tehran, Iran), according to the instructions provided by the supplier. PCR amplifications were carried out using SmarTaq DNA polymerase (CinnaGen, Iran), and thermocycling was done in a MyCycler thermal cycler (Biorad, CA, USA). All primers used in this work were synthesized by Metabion (Martinsried, Germany) or CinnaGen (Iran), and are listed in Table S2.
As a first step, 38 serologically positive leaf samples were tested by RT-PCR using universal NIb primers (NIb2F and NIb3R) to amplify partial nuclear inclusion body protein (NIb) gene of potyviruses to confirm the potyvirus infection of the samples. In the second step, seven isolates were selected, and RT-PCR tests were performed using different potyvirus universal primer pairs to amplify the 3'-end of the genome of the isolates; however they failed to produce expected DNA fragments in the tests. Therefore, two new forward primers NY-NIb1 and NL-NIb1 were designed based on the partial NIb gene sequences of the isolates with the highest identities to narcissus yellow stripe virus (NYSV) (six isolates) and narcissus late season yellows virus (NLSYV) (one isolate), respectively, and used in combination with primer Tu3T9M (Fig. S2). The PCR cycles were 94 °C for 4 min, and 38 cycles of 94 °C for 1 min, 50 °C for 45 s, 72 °C for 2 min, and final extension of 72 °C for 10 min. PCR products and DNA ladder (Thermo Scientific, CA, USA) were separated by agarose gel electrophoresis, visualized using GelRed staining (Biotium Inc., CA, USA), and photographed with ultraviolet-illumination.
Sequencing
To determine the nucleotide sequence of PCR products, all amplified fragments with the expected sizes were extracted and purified from agarose gels by the sequencing service of Bioneer (Daejeon, Korea). Sanger sequencing was carried out by primer walking in both directions using the aforementioned amplification primers, or newly designed primers NL-Seq and NY-Seq, and Narci (the first two primers for NLSYV- and NYSV-like isolates, and the latter for all the isolates) (Table S2 and Fig. S2). Sequences were determined using three independent PCR products. The sequences of adjacent regions were overlapped by about 100 bp to ensure that they were from the same genome and were not from different components of a genome mixture (Farzadfar et al. 2009). Bio-Edit 7.2.5 software (Hall 1999) was utilized for trimming and assembling of nucleotide sequence data. Basic Local Alignment Search Tool (BLAST) was then used to search GenBank databases (https://www.ncbi.nlm.nih.gov/) to find the sequences that most closely matched with the nucleotide and deduced amino acid sequences.
Potyvirus species phylogenetic analysis
These analyses were performed using different applications implemented in MEGA 7.0 software (Kumar et al. 2016). Multiple nucleotide sequence alignment of the partial NIb gene of 34 Iranian isolates (Table S3) and other 34 publicly available sequences of monocot infecting species from the genus Potyvirus (Table S4) was performed using CLUSTAL W. To ensure that the alignment was in frame, codon-based alignment of the corresponding amino acid sequences was carried out, and the gaps were subsequently deleted manually (Farzadfar et al. 2009). In this study, a homologous sequence from the sister genus Rymovirus, i.e., ryegrass mosaic virus (accession No. NC_001814), was used as the outgroup (Zheng et al. 2010). The alignment was then used to construct maximum-likelihood (ML) phylogenetic trees. General time-reversible model with a proportion of invariant sites and a gamma distribution of site-rate variants across four categories (GTR + G4 + I) was used as the best nucleotide substitution model with the lowest values for Bayesian information criterion (BIC) score. The trees were constructed using 1,000 bootstraps to evaluate node significances. Nodes with less than 50% bootstrap support were collapsed and the joining branches of the collapsed node were adjusted to include the length of the collapsed branch and maintain most patristic distances (Zheng et al. 2010).
Recombination analysis of narcissus potyviruses
To investigate the possible occurrence of recombination events in the sequences obtained for the Iranian isolates of NLSYV and NYSV and those published previously (Table S5), network trees were first generated using the NN method in SPLITSTREE 4.11.3 (Huson and Bryant 2006). The sequences were subsequently analyzed for possible recombination events by using different recombination detection programs implemented in RDP 4.39 software (Martin et al. 2015). A recombination event was credibly accepted when detected by three or more methods with p-values < 1.0 × 10–6. The analyses were performed using default settings for different detection programs, according to the earlier recommendations (Posada 2002; Farzadfar et al. 2009). The sequences were first checked using Bonferroni corrected p-value cut-off of 0.05; the cut-off value of 0.01 was then applied to confirm a recombination event.
To ensure that all possible recombination events would be detected, degapped alignments of coding regions, including partial NIb and complete cp sequences were first generated separately by using amino acid sequences alignment, as described above. A degapped alignment of 3′UTR sequences was also made. Recombination analyses were then performed using each of these regions individually as well as concatenated sequences of NIb-CP, NIb-3′UTR, CP-3′UTR and NIb-CP-3'UTR. No outgroup was utilized in these alignments. In this procedure, the length of the aligned and degapped partial NIb, complete cp and 3'UTR sequences was 633 nt, 822 nt and 203 nt, respectively.
Phylogenetic analysis of narcissus potyviruses
The phylogenetic relationships of nucleotide sequences obtained for NLSYV and NYSV isolates under study with those of other isolates from different countries were investigated by ML method using MEGA 7.0 software. Corresponding sequences of turnip mosaic virus (TuMV; accession No. AB989659), wild onion symptomless virus (WoSV; LC159495) and scallion mosaic virus (ScaMV; LC064113) were applied as outgroups because BLAST searches in the international sequence databases identified them as the most closely related species to those viruses, similar to the earlier recommendations (Ohshima et al. 2016, 2018). A degapped alignment was then made, as above, using the NLSYV and NYSV sequences and the outgroups. The nucleotide sequences of degapped partial NIb and complete CP coding regions were 633 and 822 nucleotides, respectively. For ML analyses, Tamura-Nei model with a gamma distribution of site-rate variants across four categories (T93 + G4) and Tamura 3-parameter model with a proportion of invariant sites and a gamma distribution of site-rate variants across four categories (T92 + G4 + I) were determined as the best nucleotide substitution models for partial NIb and complete cp genes. Both phylogenetic trees were constructed using 1,000 bootstraps to evaluate node support. Finally, nodes with less than 50% bootstrap support were collapsed. The pairwise identity of the nucleotide sequences of cp and NIb genes were determined using SDT 1.2 (Muhire et al. 2013).
Genetic diversity and population differentiation
Population genetics for NYSV and NLSYV cp and partial NIb genes were estimated within and between different phylogenetic groups, including three and four groups for NYSV and NLSYV, respectively. Recombinant isolates were removed prior to population and selection analysis. Due to the low number of isolates within NLSYV group II (only one isolate), this group was eliminated from these analyses. The parameters calculated were statistic Watterson’s estimator of θ (θw), number of segregating sites (S), nucleotide diversity π (Pi), the average number of nucleotide differences per site between two sequences, number of haplotypes and haplotype diversity. These parameters were calculated by the DnaSP 5.10.01 program (Librado and Rozas 2009).
To investigate the role of natural selection at the molecular level, the rate of synonymous substitutions per synonymous sites (dS) and the rate of nonsynonymous substitutions per nonsynonymous sites (dN) were estimated by DnaSP 5.10.01 program; the dN/dS ratios was then calculated. Each coding region of NYSV and NLSYV sequences was also separately analyzed for evidence of selection acting on individual codon sites using mixed effects model evolution (MEME), random effect likelihood (REL) and fixed effects likelihood (FEL) methods implemented in the HyPhy package via the online DATAMONKEY server (http://www.datamonkey.org/) (Delport et al. 2010). To classify a site as positively or negatively selected, the cut-off p-value was selected as recommended. Moreover, Tajima's D, Fu and Li's D*, and Fu and Li's F* statistical tests were also applied using DnaSP 5.10.01 program to assess any evidence of selection.
Finally, three permutation-based statistical tests, including Ks*, Z* and Snn were used to evaluate genetic differentiation between populations. These statistical tests also were carried out using DnaSP 5.10.01. Moreover, the absolute value of the standardized variance in allele frequencies across populations (FST) was utilized to estimate genetic differentiation and the gene flow level between populations. These values were measured as Wright’s FST values and estimated by Arlequin 3.11 software (Excoffier et al. 2005); the significance between each comparison pair was evaluated through 1,000 permutation procedures.
Results
Samplings and serological assays
Based on symptoms, about 40.3% of the plants visited probably had a viral disease infection; the rate was higher in the fields (52.5%) than that in the greenhouse (20.6%) (Table S1); the most common symptom observed on narcissus plants was (mild) streak mosaic (mottling), which was found in almost all symptomatic plants collected (Fig. S1). Among the samples tested, 38 out of 61 narcissus samples (62.3%) positively reacted with the potyvirus-monoclonal antibodies. None of the samples reacted with CMV antibodies. Moreover, weed samples collected from the area surveyed were found uninfected with potyviruses. Leaf chlorosis, streak mosaic and stunting were associated with the potyvirus infections (Fig. S1). Mild (mild mosaic) to severe (stunting) symptoms were observed in samples with the virus infections. Moreover, almost all samples with severe symptoms (severe mosaic and stunting) strongly reacted with Potyvirus genus-specific antibodies.
RT-PCR, sequencing and sequence analysis
Among 38 serologically positive samples, DNA fragments of ca. 0.3 kb were amplified for 33 isolates using NIb primers, confirming infection with potyviruses (Zheng et al. 2010). For virus identification, the amplified fragments of 30 isolates (different for field/greenhouse, region and symptoms) with high quantities of DNA amplicons and sharp bands in gel electrophoresis were then sequenced (three of the isolates with low quantities of DNA amplicons failed in sequencing). BLASTn analyses of these sequences showed 98.6% and 90.3–94.8% pairwise identity with NLSYV (n = 1) and NYSV (n = 29), respectively, as the most closely matched species. Phylogenetic analysis also confirmed the results of the database searches and these sequences clustered with those of other Iranian NLSYV and NYSV isolates (Fig. S3).
Also, DNA fragments of about 2.0 kb were amplified using primers Tu3T9M, and NY-NIb1 or NL-NIb1 designed to amplify the 3'-end region of seven potyvirus isolates: one isolate of NLSYV (Th-Nar72) and six isolates of NYSV (Fa-Nar3, Fa-Nar16, Fa-Nar23, Fa-Nar24, Fa-Nar34 and Fa-Nar43). Except for Fa-Nar24, which cp gene was sequenced partially and not utilized for further analysis, other sequences were 1929–1932 nt long, encompassing partial NIb (895–902 nt), complete cp (825 nt) and 3'UTR (206–210 nt). All sequences obtained in this study were deposited in the GenBank and received accession numbers MT345535-MT345549 and MT348713-MT348727 (Table S3).
Recombination analysis of narcissus potyviruses
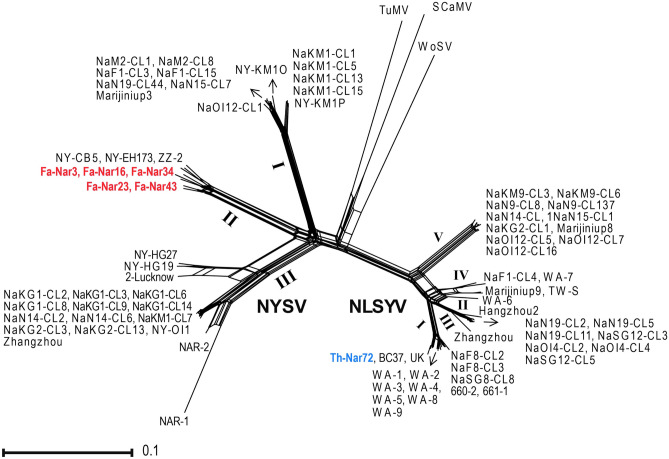
Phylogenetic network trees were constructed to examine the possibility of the presence of recombinant sequences in partial NIb and CP regions. Reticulated nodes were observed in the tree generated using a dataset of NYSV cp gene sequences, indicating the possibility of the occurrence of recombination events in CP coding region (Fig. 1); such nodes were not found for a dataset of partial NIb gene sequences (Fig. S4). The pairwise homoplasty index (PHI) also confirmed the significant evidence of recombination in cp gene sequences of NYSV (P = 0.0094). To assess the evidence of intergenic and inter-species recombination, multiple sequence alignments of NIb-CP, NIb-3'UTR, CP-3'UTR and NIb-CP-3'UTR were analyzed by RDP 4.39 software. The analysis showed that NYSV NAR-2 from India had evidence of recombination (p = 4.16 × 10–35- 4.41 × 10–9), as previously reported (Ohshima et al. 2018). Therefore, this isolate was removed from further phylogenetic and evolutionary analysis. The analyses showed no evidence of recombination for Iranian NLSYV and NYSV sequences.
Fig. 1.
Phylogenetic network tree of the coat protein (cp) gene of narcissus late season yellows virus (NLSYV) and narcissus yellow stripe virus (NYSV) isolates. The tree was rooted with sequences of turnip mosaic virus (TuMV), scallion mosaic virus (SCaMV) and wild onion symptomless virus (WoSV). NLSYV and NYSV isolates from this study are shown in blue and red, respectively. Detailed information of isolates used to construct the tree is listed in Tables S3 and S5. Latin numbers refer to phylogenetic groups which are shown in Fig. 2. Scale bar is 0.1 substitutions/site
Phylogenetic analysis and identity matrixes of narcissus potyviruses
All non-recombinant sequences and the outgroups were utilized for phylogenetic analysis using ML algorithm (Fig. 2). The trees obtained from ML method placed the NLSYV cp sequences into five distinct phylogroups I, II, III, IV and V; the Iranian isolate Th-Nar72 clustered together with isolates from Australia, Hungary, Japan and United Kingdom into group I, and categorized into a distinct subgroup with one isolate from United Kingdom and one isolate from Australia. Moreover, NYSV cp sequences fell into three phylogroups I, II and III, and the Iranian isolates (Fa-Nar3, Fa-Nar16, Fa-Nar23, Fa-Nar34 and Fa-Nar43) grouped with isolates from Japan and China, and formed distinct subgroups in the group II. Phylogenetic grouping of the isolates using partial NIb sequences of NLSYV and NYSV was similar to that formed using cp sequences, except that NLSYV group II did not exist due to the lack of NIb sequence for WA-6 isolate (Fig. S5). The grouping of the isolates for both CP and NIb regions was supported by high bootstrap values. Similar trees were obtained using deduced amino acid sequences of both regions for the two viruses. No correlation was found between phylogenetic grouping and geographic location of the isolates for both NLSYV and NYSV.
Fig. 2.
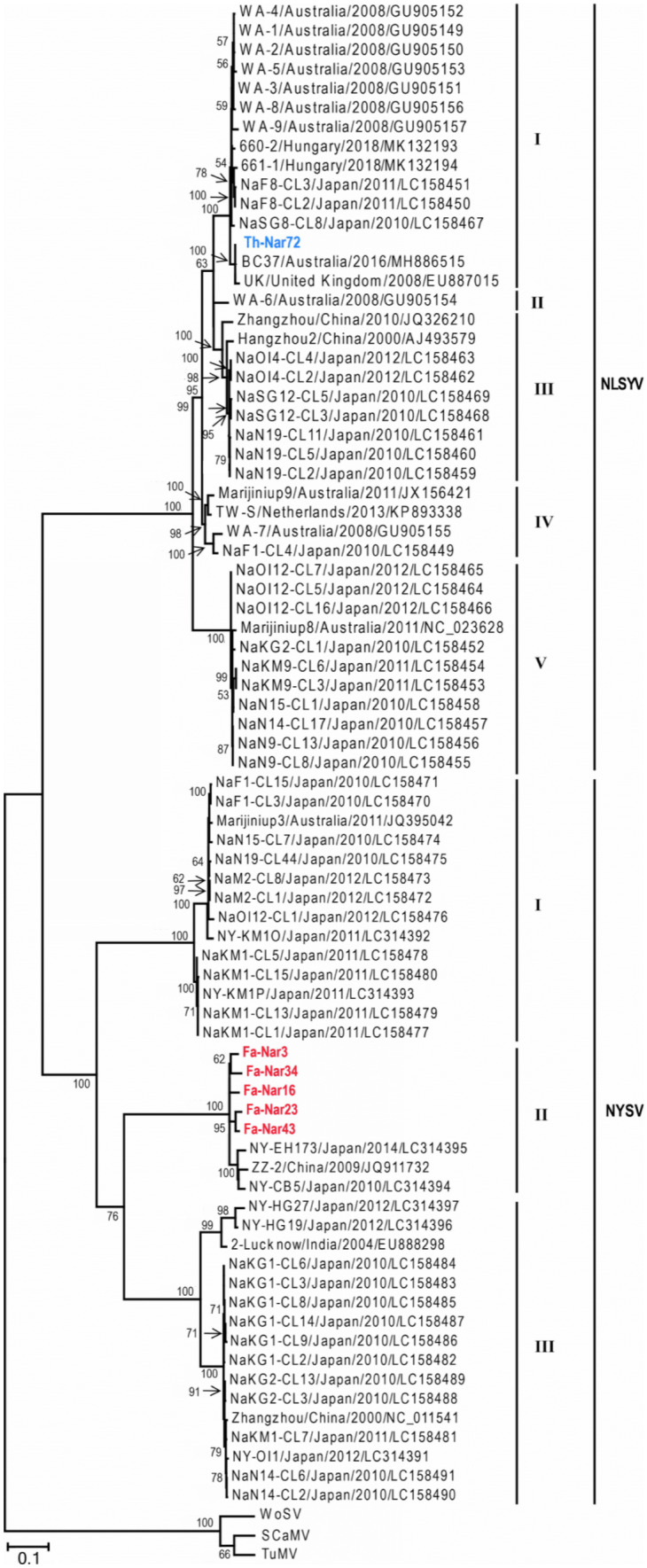
A maximum-likelihood (ML) phylogenetic tree of coat protein (cp) gene sequences of narcissus late season yellows virus (NLSYV) and narcissus yellow stripe virus (NYSV) isolates. Bootstrap values higher than 50 are shown at each node. The tree was rooted with sequences of turnip mosaic virus (TuMV), scallion mosaic virus (SCaMV) and wild onion symptomless virus (WoSV). NLSYV and NYSV isolates from this study are shown in blue and red, respectively. Scale bar is 0.1 substitutions/site
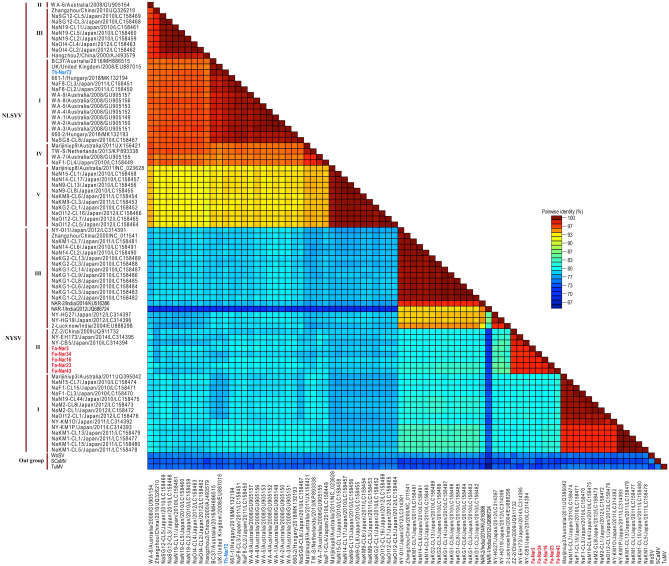
While pairwise analysis of NLSYV isolates between different phylogenetic groups indicated nucleotide sequence identities of 86.3–93.9% and 81.8–92.4% for CP and NIb regions, respectively, these identities for NYSV isolates were 77.1–79.8% and 70.6–76.4%. The nucleotide sequence identities of NLSYV and NYSV isolates in each group with respect to the outgroups were 70.7–74.3% and 69.5–74.5% (for CP), and 64.6–71.3% and 63.9–70.2% (for NIb), respectively. The pairwise nucleotide identity matrixes for CP and partial NIb regions of the isolates are shown (Figs. 3 and S6). Moreover, multiple sequence alignment showed high similarities for deduced amino acid sequence of cp and NIb genes of Iranian and other isolates of NLSYV (cp: 94.1–99.6%, NIb: 95.2–99.5%) and NYSV (cp: 82.7–99.2%, NIb: 79.1–96.6%). Although numerous conserved amino acid sequences were found among the isolates, especially among those belonging to the same phylogenetic group, a number of amino acid changes were observed in the isolates belonging to different groups. The frequency of amino acid substitutions among NYSV groups was more than among NLSYV groups (Figs. S7 and S8).
Fig. 3.
Nucleotide pairwise identity color matrix of coat protein (cp) gene sequences of narcissus late season yellows virus (NLSYV) and narcissus yellow stripe virus (NYSV) isolates calculated by SDT 1.2 program (Muhire et al. 2013). Corresponding regions of turnip mosaic virus (TuMV), scallion mosaic virus (SCaMV) and wild onion symptomless virus (WoSV) were included for comparison. NLSYV and NYSV isolates from this study are shown in blue and red, respectively
Genetic diversity and structure analysis
To assess the genetic variability within and between NLSYV and NYSV populations, each phylogenetic group was assigned as a subpopulation (four subpopulations for NLSYV and three for NYSV) and associated population genetic parameters were subsequently estimated. Due to the low number of isolates in NLSYV-II, this subpopulation was ignored from these analyses. Based on the results for CP and NIb regions, NYSV populations have more genetic diversity than NLSYV. For CP region, comparison of genetic diversity within each subpopulation indicated a higher diversity of NYSV group II (π = 0.0474 ± 0.0038; θw = 0.0502 ± 0.0048) as compared with the group III (π = 0.0408 ± 0.0136; θw = 0.0454 ± 0.004) and the group I (π = 0.0293 ± 0.003; θw = 0.0267 ± 0.00001). However, the standard deviation of π for group III was considerably higher than that obtained for the other groups. For NLSYV cp, results revealed that the groups V and IV had the lowest (π = 0.0058 ± 0.0009; θw = 0.0074 ± 0.000003) and the highest (π = 0.0377 ± 0.0079; θw = 0.0358 ± 0.00002) genetic diversities, respectively. Similar results were obtained for NIb regions. The haplotype diversity in all analyzed subpopulations was > 0.87. To understand the evolutionary constraints imposed on different populations, the ratio of dN/dS (Ѡ) for all populations was calculated. The Ѡ values in all cases were < 1, indicating that both CP and NIb regions were subjected to negative selection and were strongly conserved, however, purifying selection for NLSYV (0.077 ± 0.002 for CP; 0.062 ± 0.001 for NIb) was stronger than NYSV (0.139 ± 0.005 for CP; 0.142 ± 0.001 for NIb). The results for CP and NIb regions are presented in detail in Table 1.
Table 1.
Population genetic parameters estimated for narcissus late season yellows virus (NLSYV) and narcissus yellow stripe virus (NYSV)a
|
Virus/ region |
Sub-populationb | Nc | Parameterd | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S | H | Hd | Pi | θw | dN | dS | dN/dS | |||
|
NLSYV CP |
All | 40 | 206 | 33 | 0.98 | 0.080 ± 0.003 | 0.058 ± 0.0041 | 0.021 | 0.279 | 0.077 ± 0.002 |
| I | 15 | 53 | 15 | 1 | 0.016 ± 0.002 | 0.019 ± 0.0027 | 0.005 | 0.053 | 0.104 ± 0.002 | |
| III | 9 | 52 | 7 | 0.94 | 0.016 ± 0.005 | 0.023 ± 0.00001 | 0.006 | 0.053 | 0.114 ± 0.002 | |
| IV | 4 | 54 | 4 | 1 | 0.037 ± 0.007 | 0.035 ± 0.00002 | 0.017 | 0.107 | 0.160 ± 0.002 | |
| V | 11 | 18 | 6 | 0.87 | 0.005 ± 0.0001 | 0.007 ± 0.000003 | 0.001 | 0.021 | 0.055 ± 0.001 | |
| Partial NIb | All | 29 | 196 | 24 | 0.98 | 0.113 ± 0.004 | 0.078 ± 0.00003 | 0.0257 | 0.413 | 0.062 ± 0.001 |
| I | 7 | 39 | 6 | 0.95 | 0.024 ± 0.002 | 0.025 ± 0.00001 | 0.0066 | 0.085 | 0.077 ± 0.001 | |
| III | 9 | 55 | 6 | 0.88 | 0.024 ± 0.010 | 0.033 ± 0.00001 | 0.0057 | 0.090 | 0.063 ± 0.011 | |
| IV | 2 | 35 | 2 | 1 | 0.055 ± 0.025 | 0.055 ± 0.00008 | 0.0129 | 0.197 | 0.065 ± 0.032 | |
| V | 11 | 31 | 10 | 0.98 | 0.013 ± 0.002 | 0.016 ± 0.000009 | 0.0037 | 0.047 | 0.078 ± 0.008 | |
|
NYSV CP |
All | 38 | 298 | 34 | 0. 99 | 0.158 ± 0.006 | 0.086 ± 0.005 | 0.066 | 0.478 | 0.139 ± 0.005 |
| I | 14 | 70 | 12 | 0.97 | 0.029 ± 0.003 | 0.026 ± 0.00001 | 0.013 | 0.084 | 0.163 ± 0.003 | |
| II | 8 | 107 | 8 | 1 | 0.047 ± 0.003 | 0.050 ± 0.004 | 0.006 | 0.186 | 0.033 ± 0.001 | |
| III | 16 | 124 | 14 | 0.97 | 0.040 ± 0.013 | 0.045 ± 0.004 | 0.006 | 0.159 | 0.040 ± 0.001 | |
| Partial NIb | All | 39 | 283 | 33 | 0.98 | 0.189 ± 0.008 | 0.105 ± 0.00003 | 0.080 | 0.565 | 0.142 ± 0.001 |
| I | 14 | 65 | 10 | 0.92 | 0.038 ± 0.006 | 0.032 ± 0.00001 | 0.007 | 0.145 | 0.051 ± 0.007 | |
| II | 9 | 135 | 9 | 1 | 0.074 ± 0.006 | 0.078 ± 0.00004 | 0.013 | 0.288 | 0.045 ± 0.005 | |
| III | 16 | 131 | 14 | 0.98 | 0.057 ± 0.020 | 0.062 ± 0.00002 | 0.016 | 0.202 | 0.079 ± 0.008 | |
aPopulation genetic parameters were calculated using DnaSP 5.10.01 program (Librado and Rozas 2009). The results were estimated for coat protein (CP) and partial nuclear inclusion b protein (NIb) regions of NLSYV and NYSV
bNon-recombinant sequences from Iran and other parts of the world, listed in Tables S3, S5 and Figs. 2, S5, were used
cNumber of sequences (population size of each subpopulation)
dThe parameters calculated were the number of segregating sites (S), number of haplotypes (H) and haplotype diversity (Hd), nucleotide diversity π (Pi), statistic Watterson’s estimator of θ (θw), synonymous substitutions per synonymous site (dS), nonsynonymous substitutions per nonsynonymous sites (dN), and dN/dS ratios
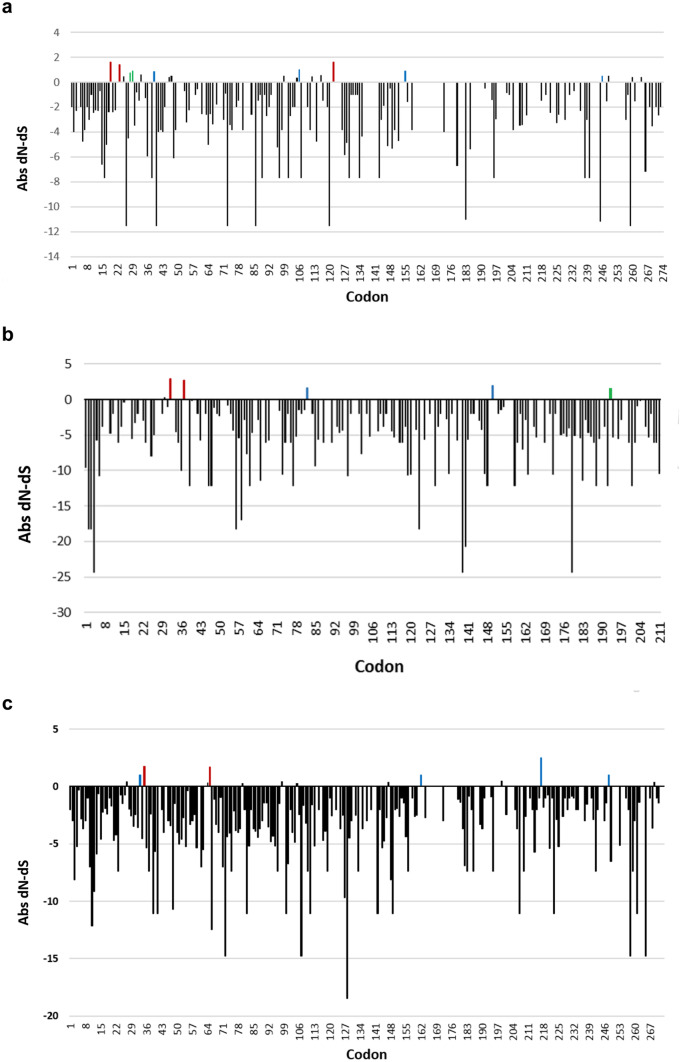
Investigations on the selection pressures acting on codon sites within the CP and NIb coding regions of NLSYV and NYSV were also performed. To identify signals of selection, three different selection detection models, including MEME, FEL and REL were used. As expected, most of the codons throughout the CP and NIb regions for both viruses were under negative selection. Few sites in each gene were detected significantly to be evolving under pervasive (FEL and REL methods), episodic (MEME method) or both pervasive and episodic diversifying selection. In NLSYV, more sites were found under positive selection (CP = 9: codons 19, 23, 28, 29, 39, 106, 122, 155, 246; and NIb = 5: codons 32, 37, 82, 150, 193) than those found in NYSV (CP = 6: codons 33, 35, 65, 162, 217, 248; and NIb = no positive site) (Fig. 4).
Fig. 4.
Plot representing significant signals of natural selection acting on individual codon sites within: (a) narcissus late season yellows virus (NLSYV) coat protein (CP) region, (b) NLSYV partial nuclear inclusion b protein (NIb) region, and (c) narcissus yellow stripe virus (NYSV) CP region. The Abs (Absolut) dN-dS values are plotted for negative (black) and positive (red, blue and green) selection signals. Sites at which episodic diversifying selection was detected using mixed effects model evolution (MEME) method are represented by blue bars, sites at which pervasive diversifying selection was detected using fixed effects likelihood (FEL) and random effect likelihood (REL) methods are shown in green, and those detected to be under both pervasive and episodic selection are shown in red. Codon 1 is the beginning of each cistron and codons 274, 211 and 267 are the termination of NLSYV CP, NLSYV NIb and NYSV CP, respectively
Tajima's D, Fu and Li's D*, and Fu and Li's F* statistical tests were also utilized using DnaSP 5.10.01 program to assess any evidence of selection. We expect that the three statistical tests produce negative values resulting from background selection, genetic hitchhiking or demographic expansion. We also expect that the statistical tests generate positive values resulting from balancing selection and a reduction in the size of the population. Although most of the values obtained were negative, none of them were statistically significant (P value > 0.05) (Table S6).
Genetic differentiation analysis, Ks*, Z* and Snn parameters, was performed to evaluate if virus isolates from different phylogenetic groups (Table 2) or geographical origins (Table S7) were genetically differentiated. Results indicated that isolates belonging to different phylogenetic groups are partially differentiated populations, however, subpopulations belonging to the same phylogenetic group but different geographical origin (i.e., China, Iran, Japan, Australia and Hungary) were mostly not genetically differentiated populations. To evaluate the extent of genetic differentiation or the gene flow between subpopulations within viruses, FST values were calculated. Calculation of the coefficient showed that all the values of FST among phylogenetic populations were < 0.33, and in most cases, the values obtained were statistically significant (P < 0.001) (Table 2). This value for most of geographical subpopulations were < 0.33; all the values were not statistically significant (P value > 0.05) or had low significance levels (0.01 < P value < 0.05) (Table S7).
Table 2.
Gene flow and genetic differentiation of narcissus late season yellows virus (NLSYV) and narcissus yellow stripe virus (NYSV), considering their phylogenetic groupa
| Virus/region/subpopulationb | Parameterc | ||||
|---|---|---|---|---|---|
| Ks* (P value)d | Z* (P value) | Snn (P value) | FST (P value) | Nm | |
| NLSYV cp | |||||
| I (n = 15)e vs. III (n = 9) | 2.441 (0.000***) | 3.928 (0.000***) | 1.000 (0.000***) | 0.777 (0.000***) | 0.07 |
| I (n = 15) vs. IV (n = 4) | 2.665 (0.000***) | 3.813 (0.000***) | 1.000 (0.000***) | 0.743 (0.000***) | 0.12 |
| I (n = 15) vs. V (n = 11) | 2.166 (0.000***) | 4.037 (0.000***) | 1.000 (0.000***) | 0.903 (0.000***) | 0.02 |
| III (n = 9) vs. IV (n = 4) | 2.499 (0.003**) | 2.877 (0.003**) | 1.000 (0.007**) | 0.698 (0.000***) | 0.13 |
| III (n = 9) vs. V (n = 11) | 1.885 (0.000***) | 3.585 (0.000***) | 1.000 (0.000***) | 0.919 (0.000***) | 0.02 |
| IV (n = 4) vs. V (n = 11) | 1.937 (0.000***) | 3.269 (0.000***) | 1.000 (0.000***) | 0.873 (0.000***) | 0.06 |
| NIb | |||||
| I (n = 7) vs. III (n = 9) | 2.348 (0.000***) | 3.130 (0.000***) | 1.000 (0.001**) | 0.730 (0.000***) | 0.09 |
| I (n = 7) vs. IV (n = 2) | 2.676 (0.340*) | 2.162 (0.0340*) | 1.000 (0.1050 ns) | 0.709 (0.0180*) | 0.16 |
| I (n = 7) vs. V (n = 11) | 2.318 (0.000***) | 3.431 (0.000***) | 1.000 (0.001**) | 0.898 (0.000***) | 0.03 |
| III (n = 9) vs. IV (n = 2) | 2.114 (0.005**) | 2.689 (0.008**) | 1.000 (0.039*) | 0.728 (0.054 ns) | 0.15 |
| III (n = 9) vs. V (n = 11) | 2.117 (0.000***) | 3.543 (0.000***) | 1.000 (0.000***) | 0.893 (0.000***) | 0.03 |
| IV (n = 2) vs. V (n = 11) | 2.120 (0.017*) | 3.065 (0.009**) | 1.000 (0.027*) | 0.893(0.0180*) | 0.06 |
| NYSV CP | |||||
| I (n = 14) vs. II (n = 8) | 3.111 (0.000***) | 3.878 (0.000***) | 1.000 (0.000***) | 0.845 (0.000***) | 0.05 |
| I (n = 14) vs. III (n = 16) | 2.756 (0.000***) | 4.382 (0.0000***) | 1.000 (0.000***) | 0.845 (0.000***) | 0.05 |
| II (n = 8) vs. III (n = 16) | 2.980 (0.000***) | 4.096 (0.000***) | 1.000 (0.000***) | 0.801 (0.000***) | 0.06 |
| NIb | |||||
| I (n = 14) vs. II (n = 9) | 3.203 (0.000***) | 3.958 (0.000***) | 1.000 (0.000***) | 0.798 (0.000***) | 0.07 |
| I (n = 14) vs. III (n = 16) | 2.711 (0.000***) | 4.384 (0.000***) | 1.000 (0.000***) | 0.806 (0.000***) | 0.06 |
| II (n = 9) vs. III (n = 16) | 3.028 (0.000***) | 4.145 (0.000***) | 1.000 (0.000***) | 0.751 (0.000***) | 0.09 |
aGenetic differentiation and gene flow level between populations were assessed by four statistical tests in DnaSP version 5.10.01 (Librado and Rozas 2009). FST values were estimated by Arlequin 3.11 software (Excoffier et al. 2005). The results were estimated for coat protein (CP) and partial nuclear inclusion b protein (NIb) regions of NLSYV and NYSV
bNon-recombinant sequences from Iran and other parts of the world, listed in Tables S3, S5 and Figs. 2, S5, were used
cKs* and Z* are the sequence-based statistics; Snn is the nearest-neighbor statistic; FST is the inter-population component of genetic variation of the standardized variance in allele frequencies across populations; and Nm is the migration fraction per generation
dP-values were determined by permutation tests with 1000 replicates (ns: not significant, * 0.01 < P < 0.05, ** 0.001 < P < 0.01 and *** P < 0.001)
eNumber of sequences (population size of each subpopulation)
Discussion
Potyviruses represent one-fourth of known plant RNA viruses (Gibbs and Ohshima 2010). Our results indicated the prevalence of (poty) virus-like symptoms in the fields visited and showed the occurrence of potyviruses in more than 60% of the symptomatic narcissus samples tested (Table S1). Failure in potyvirus detection in some collected samples with symptoms has been previously described earlier that might be due to other virus infections, e.g., carlaviruses which cause the same symptoms (Chen et al. 2003; Wylie and Jones 2012). However, almost all the plants with severe symptoms strongly reacted to potyvirus antibodies. Most of potyvirus degenerate primer pairs used in this study failed to amplify expected DNA fragments in RT-PCR tests. Moreover, potyvirus infections of some ELISA positive samples with good quantities and qualities of RNA extracts failed to be detected by the primers used. This may be due to the presence of nucleotide variants in the genome of these isolates that are not complementary to the primer sequences, or that the potyvirus antibodies non-specifically react with other viruses (Valouzi et al. 2019). This study also confirmed the capacity of NIb and Tu3T9M universal primers to amplify Iranian potyvirus genomes (Golnaraghi et al. 2015; Valouzi et al. 2017, 2019). We found NYSV and NLSYV in Iranian field- and greenhouse-grown narcissus plants, respectively. Both viruses are among important narcissus viruses in the world which affect flower quality and reduce bulb yield (Brunt 1995). NYSV was prevalent among the sampled symptomatic narcissus plants from fields visited, but we could not detect NLSYV and other potyviruses in these fields. Some narcissus potyviruses induce mild or latent symptoms, and in some cases, e.g., NLSYV, the symptoms develop late in the growing season, i.e., after flowering stage when the virus concentration is high enough to be detected by serological assays (Mowat et al. 1988; Brunt et al. 1996). Therefore, extended surveys of other narcissus growing areas in different growing stages and application of various virus detection approaches are needed to understand more about narcissus (poty) viruses in West Asian region of Iran.
Recombination is an important source of genetic variation of different plant viruses, including members of the family Potyviridae (Yasaka et al. 2017). No evidence of recombination was found in Iranian isolates. Our study along with previous studies (Ohshima et al. 2018) showed that among 79 isolates of NYSV and NLSYV, only one isolate, i.e., NYSV NAR-2, was recombinant. This seems to indicate a low recombination frequency between these viruses isolates that might be due to the narrow host range of narcissus potyviruses, which are limited to Narcissus in nature (Ohshima et al. 2016). However, only the 3'-end of the genome of these viruses was checked for the presence of recombination events, and so, more studies are necessary to confirm this hypothesis.
Phylogenetic studies of cp sequences of NLSYV and NYSV indicated a grouping of the isolates into five and three distinct phylogroups, respectively; previous studies reported five groups for NLSYV, but four groups for NYSV (Ohshima et al. 2016). Moreover, similar trees were obtained using NIb sequences as well as the deduced amino acid sequences of both regions for the two viruses. No correlation was found between phylogenetic grouping and geographic location of the isolates for both NLSYV and NYSV. Our findings seem to reveal that Iranian NYSV and NLSYV isolates might have different geographical origins, as they were clustered with geographically different isolates. Analysis of genetic diversity of CP cistron showed that NYSV populations were genetically more diverse than NLSYV (Table 1), which is consistent with earlier studies and shows that the virus has a higher genetic diversity as compared with other narcissus potyviruses studied (Ohshima et al. 2016). Both viruses infect a similar narrow host range and are transmitted through similar manners, i.e., vegetative propagation and aphid vectors (Brunt et al. 1996). So, the higher diversity of NYSV as compared with NLSYV probably seems to show that different driving evolutionary forces or unidentified diversity sources may be involved in the evolution of the two viruses, or these viruses differ in the time of origin. Similar results have been reported for several viruses, e.g., TuMV (Yasaka et al. 2017). Many amino acid substitutions were also found in cp sequences of NYSV, most of them were phylogroup specific substitutions (Fig. S7) that can be easily used to design phylogroup-specific primers, similar to those reported earlier for other viruses (Golnaraghi et al. 2018; Valouzi et al. 2020). Our results also revealed that nucleotide identities of cp sequences between different phylogenetic groups of the virus (Fig. 3) were at or close to the species demarcation limit (< 76%) (Wylie et al. 2017), supporting the idea that different NYSV phylogenetic groups should be considered as new species of NYSV-1, NYSV-2 and NYSV-3 (Ohshima et al. 2018). Therefore, Iranian NYSV isolates studied in this work belong to NYSV-2. Also, comparison of genetic diversities among CP and NIb genomic regions showed that NIb had a higher nucleotide diversity than cp in both NLSYV and NYSV (Table 1), similar to those reported for other potyviruses (Nguyen et al. 2013a, b); however, NIb sequences obtained in our study were partial.
The majority of cistrons in potyviral genomes are under negative selection, and so, they are remarkably stable (Nigam et al. 2019). Investigation on selection pressure on each genomic region of both NLSYV and NYSV revealed that dN/dS ratios were significantly less than one, indicating a purifying selection pressure in which NLSYV is under higher (lower rates of dN/dS) negative selection than NYSV (higher rates of dN/dS) (Table 1). Identification of selection pressure revealed the fact that cp had a higher number of codons evolved under positive selection as compared with NIb (Fig. 4). Positive selection in cp allows generation of the diversity required for variants with a competitive advantage to be selected and promote virus adaptation (Nigam et al. 2019). Our results for cp also showed that sites under positive selection were mostly located within the first 65 amino acids at the N terminal part of the CP (Fig. 4; Nigam et al. 2019). This part of the CP is involved in aphid transmission because of the presence of DAG motif, binds the subunits of virions and effect on the RNA stability and host specificity determinant (Nigam et al. 2019). The results also indicated partial genetic differentiation between phylogenetic, but almost not geographical, groups of NLSYV and NYSV (Tables 2 and S7). The high values of FST revealed rare gene flow between phylogenetic groups and were in agreement with phylogenetic analyses. As the bulbs are believed to be the main source for the spread of narcissus plants and their viruses worldwide, we expected to see frequent gene flow between geographical subpopulations of the two viruses. Although low values of FST were calculated for almost all the geographical subpopulations, the values were not statistically significant; this may be due to the low number of geographically different isolates in each phylogroup. In other word, most of the NLSYV and NYSV sequences in the GenBank are obtained from isolates in Japan (East Asia) and Australia (Oceania) as well as few numbers from Europe and West Asia (this study).
Narcissus is propagated vegetatively through its bulbs and therefore, virus infection could be continuously passed from generation to generation through propagations of infected mother stocks (Brunt 1995). Hence, reliable and early diagnosis and identification of virus(es) is essential for designing an efficient disease management. Worldwide studies on narcissus viruses have been mainly focused on the host plant as a source of viruses. This is somehow reasonable as most of narcissus viruses can be easily transmitted through vegetative propagation. However, future investigations on vectorome, viruses present in a vector which can be transmitted to a non-homologous host, e.g., from aphids to plant hosts (Valouzi et al. 2020), in narcissus fields may help us understand dynamics of narcissus viruses and find new viruses capable of infecting this plant. Covid-19 pandemic and Coronaceous period (Nazari et al. 2021) have remarkably increased human attention to the importance of biodiversity and viruses. Therefore, we expect to see further efforts in the near future to know more about viruses and their ecological roles in different ecosystems, including plant ecosystems.
Supplementary information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors wish to thank Dr. Yasuhiro Tomitaka (Kyushu Okinawa Agricultural Research Center, National Agriculture and Food Research Organization, Kumamoto, Japan) for his help and advice.
Funding
This work was supported by grant no. 98001001 from the Iranian Group for the Promotion of Science (IGPS).
Availability of data and material
The data that support the findings of this study are available from corresponding author upon response request.
Declarations
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Conflicts of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Brunt AA (1995) Narcissus. In: Loebenstein G, Lawson RH, Brunt AA (eds) Virus and virus-like diseases of bulb and flower crops. John Wiley & Sons Ltd, Chichester, UK, pp 322–334
- Brunt AA, Crabtree K, Dallwitz MJ, Gibbs AJ, Watson L, Zurcher EJ (eds) (1996) Plant viruses online: descriptions and lists from the VIDE database. Version: 20th August 1996
- Chen J, Chen JP, Langeveld SA, Derks AFLM, Adams MJ. Molecular characterization of carla- and potyviruses from Narcissus in China. J Phytopathol. 2003;151:26–29. doi: 10.1046/j.1439-0434.2003.00674.x. [DOI] [Google Scholar]
- Delport W, Poonm AFY, Frost SDW, Kosakovsky-Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26:2455–2457. doi: 10.1093/bioinformatics/btq429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (Version 3.0): An integrated software package for population genetic data analysis. Evol Bioinform Online. 2005;1:47–50. doi: 10.1177/117693430500100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzadfar S, Golnaraghi AR, Pourrahim R. Plant viruses of Iran (in English) Tehran, Iran: Saman Co.; 2002. [Google Scholar]
- Farzadfar S, Tomitaka Y, Ikematsu M, Golnaraghi AR, Pourrahim R, Ohshima K. Molecular characterization of Turnip mosaic virus isolates from Brassicaceae weeds. Eur J Plant Pathol. 2009;124:45–55. doi: 10.1007/s10658-008-9390-2. [DOI] [Google Scholar]
- Gera A, Zeidan M (2006) New and emerging virus diseases in ornamental crops. Acta Hortic 722:175–180. 10.17660/ActaHortic.2006.722.21
- Gibbs A, Ohshima K. Potyviruses and the digital revolution. Annu Rev Phytopathol. 2010;48:205–223. doi: 10.1146/annurev-phyto-073009-114404. [DOI] [PubMed] [Google Scholar]
- Golnaraghi A, Shahraeen N, Nguyen HD. Characterization and genetic structure of a Tospovirus causing chlorotic ring spots and chlorosis disease on peanut; comparison with Iranian and Polish populations of Tomato yellow fruit ring virus. Plant Dis. 2018;102:1509–1519. doi: 10.1094/PDIS-09-17-1350-RE. [DOI] [PubMed] [Google Scholar]
- Golnaraghi A, Valouzi H, Khosroshahi TS, Yazdani-Khameneh S, Hashemi S, Diyanat M (2015) A pair of universal primers facilitates the detection of potyviruses occurring in weeds and wild plants in Iran. In: International Plant Protection Congress. Berlin, Germany
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hanks GR (2002) Narcissus and daffodil. In: Hardman R (ed) Medicinal and Aromatic Plants – Industrial Profiles. Taylor & Francis Inc., London, UK
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Kamenetsky R, Okubo H. Ornamental geophytes : from basic science to sustainable production. Florida, USA: CRC Press; 2016. [Google Scholar]
- Khosroshahi TS, Golnaraghi A, Diyanat M, Rakhshandehroo F (2015) Detection of NLSYV infecting Narcissus tazetta in Tehran province. In: 1st International & 9th Iranian Biotechnology Congress. Tehran, Iran
- Koohi Habibi M, Hamzeh N, Ghazanfari K (2010) Report of narcissus degeneration virus on narcissus in Iran. In: 19th Iranian Plant Protection Congress. Tehran, Iran
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV) Nucleic Acids Res. 2018;46:D708–D717. doi: 10.1093/nar/gkx932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol 1:vev003. 10.1093/ve/vev003 [DOI] [PMC free article] [PubMed]
- Masi M, Frolova LV, Yu X, Mathieu V, Cimmino A, De Carvalho A, Kiss R, Rogelj S, Pertsemlidis A, Kornienko A, Evidente A. Jonquailine, a new pretazettine-type alkaloid isolated from Narcissus jonquilla quail, with activity against drug-resistant cancer. Fitoterapia. 2015;102:41–48. doi: 10.1016/j.fitote.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moury B, Desbiez C. Host range evolution of potyviruses: A global phylogenetic analysis. Viruses. 2020;12:111. doi: 10.3390/v12010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat WP, Duncan GH, Dawson S. Narcissus late season yellows potyvirus: symptoms, properties and serological detection. Ann App Biol. 1988;113:531–544. doi: 10.1111/j.1744-7348.1988.tb03330.x. [DOI] [Google Scholar]
- Muhire B, Martin DP, Brown JK, Navas-Castillo J, Moriones E, Zerbini MF, Rivera-Bustamante R, Malathi VG, Briddon RW, Varsani A. A genome-wide pairwise identity-based proposal for the classification of viruses in the genus Mastrevirus (family Geminiviridae) Arch Virol. 2013;158:1411–1424. doi: 10.1007/s00705-012-1601-7. [DOI] [PubMed] [Google Scholar]
- Nazari K, Sheykhrezaee Z, Golnaraghi Z (2021) Man in the Coronaceous period (1): how long will Corona vaccination last in the world? Iranian Students' News Agency, Report No. 1400020503193
- Nigam D, La Tourrette K, Souza PFN, Garcia-Ruiz H. Genome-wide variation in potyviruses. Front Plant Sci. 2019;10:1439. doi: 10.1007/s00705-012-1601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HD, Tomitaka Y, Ho SYW, Duchene S, Vetten HJ, Lesemann D, Ohshima K. Turnip mosaic potyvirus probably first spread to Eurasian brassica crops from wild orchids about 1000 years ago. PLoS ONE. 2013;8:e55336. doi: 10.1371/journal.pone.0055336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HD, Tran HTN, Ohshima K. Genetic variation of the Turnip mosaic virus population of Vietnam: A case study of founder, regional and local influences. Virus Res. 2013;171:138–149. doi: 10.1016/j.virusres.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Ohshima K, Mitoma S, Gibbs AJ (2018) The genetic diversity of narcissus viruses related to turnip mosaic virus blur arbitrary boundaries used to discriminate potyvirus species. PLoS One 13:e0190511. 10.1371/journal.pone.0190511 [DOI] [PMC free article] [PubMed]
- Ohshima K, Nomiyama R, Mitoma S, Honda Y, Yasaka R, Tomimura K. Evolutionary rates and genetic diversities of mixed potyviruses in Narcissus. Infect Genet Evol. 2016;45:213–223. doi: 10.1016/j.meegid.2016.08.036. [DOI] [PubMed] [Google Scholar]
- Posada D. Evaluation of methods for detecting recombination from DNA sequences: empirical data. Mol Biol Evol. 2002;19:708–717. doi: 10.1093/oxfordjournals.molbev.a004129. [DOI] [PubMed] [Google Scholar]
- Roossinck MJ. Plant virus metagenomics: biodiversity and ecology. Annu Rev Genet. 2012;46:359–369. doi: 10.1146/annurev-genet-110711-155600. [DOI] [PubMed] [Google Scholar]
- Sochacki D, Chojnowska E. The frequency of viral infections on two narcissus plantations in Central Poland. J Hortic Res. 2016;24:19–24. doi: 10.1515/johr-2016-0016. [DOI] [Google Scholar]
- Valouzi H, Golnaraghi A, Abedini-Aminabad L, Diyanat M. Serological and molecular identification of Turnip mosaic virus in some wild plants in Iran. Australas Plant Dis Notes. 2017;12:3. doi: 10.1007/s13314-016-0225-2. [DOI] [Google Scholar]
- Valouzi H, Golnaraghi A, Hashemi S, Abedini-Aminabad L, Yazdani-Khameneh S. Detection of potyviruses infecting Iranian common beans using broad-spectrum antibodies and universal primer pairs. Can J Plant Pathol. 2019;41:535–543. doi: 10.1080/07060661.2019.1579756. [DOI] [Google Scholar]
- Valouzi H, Hashemi S, Wylie S, Ahadiyat A, Golnaraghi A. Wisteria vein mosaic virus detected for the first time in Iran from an unknown host by analysis of aphid vectors. Plant Pathol J. 2020;36:87–97. doi: 10.5423/PPJ.OA.10.2019.0268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie SJ, Adams M, Chalam C, Kreuze J, López-Moya JJ, Ohshima K, Praveen S, Rabenstein F, Stenger D, Wang A, Zerbini FM. ICTV Virus Taxonomy Profile: Potyviridae. J Gen Virol. 2017;98:352–354. doi: 10.1099/jgv.0.000740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylie SJ, Jones MGK. Complete genome sequences of seven carlavirus and potyvirus isolates on Narcissus and Hippeastrum plants in Australia, and proposals to clarify their naming. Arch Virol. 2012;157:1471–1480. doi: 10.1007/s00705-012-1319-6. [DOI] [PubMed] [Google Scholar]
- Wylie SJ, Li H, Sivasithamparam K, Jones MGK. Complete genome analysis of three isolates of narcissus late season yellows virus and two of narcissus yellow stripe virus: Three species or one? Arch Virol. 2014;159:1521–1525. doi: 10.1007/s00705-013-1969-z. [DOI] [PubMed] [Google Scholar]
- Wylie SJ, Nouri S, Coutts BA, Jones MGK. Narcissus late season yellows virus and Vallota speciosa virus found infecting domestic and wild populations of Narcissus species in Australia. Arch Virol. 2010;155:1171–1174. doi: 10.1007/s00705-010-0682-4. [DOI] [PubMed] [Google Scholar]
- Yasaka R, Fukugawa H, Ikematsu M, Soda H, Korkmaz S, Golnaraghi A, Katis N, Ho SYW, Gibbs AJ, Ohshima K. The timescale of emergence and spread of Turnip mosaic potyvirus. Sci Rep. 2017;7:4240. doi: 10.1038/s41598-017-01934-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Rodoni BC, Gibbs MJ, Gibbs AJ. A novel pair of universal primers for the detection of potyviruses. Plant Pathol. 2010;59:211–220. doi: 10.1111/j.1365-3059.2009.02201.x. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from corresponding author upon response request.