Abstract
Alternative splicing of exon 7B in the hnRNP A1 pre-mRNA produces mRNAs encoding two proteins: hnRNP A1 and the less abundant A1B. We have reported the identification of several intron elements that contribute to exon 7B skipping. In this study, we report the activity of a novel element, conserved element 9 (CE9), located in the intron downstream of exon 7B. We show that multiple copies of CE9 inhibit exon 7B-exon 8 splicing in vitro. When CE9 is inserted between two competing 3′ splice sites, a single copy of CE9 decreases splicing to the distal 3′ splice site. Our in vivo results also support the conclusion that CE9 is a splicing modulator. First, inserting multiple copies of CE9 into an A1 minigene compromises the production of fully spliced products. Second, one copy of CE9 stimulates the inclusion of a short internal exon in a derivative of the human β-globin gene. In this case, in vitro splicing assays suggest that CE9 decreases splicing of intron 1, an event that improves splicing of intron 2 and decreases skipping of the short internal exon. The ability of CE9 to act on heterologous substrates, combined with the results of a competition assay, suggest that the activity of CE9 is mediated by a trans-acting factor. Our results indicate that CE9 represses the use of the common 3′ splice site in the hnRNP A1 alternative splicing unit.
Alternative RNA splicing is a crucial event in the expression of many eucaryotic genes transcribed by RNA polymerase II. Regulation of alternative splicing plays a key role in the production of distinct protein isoforms during development and in different cell types. Although the mechanisms that control splice site selection in mammalian cells remain poorly understood, recent progress indicates that a variety of sequence elements within precursor mRNAs can have positive or negative effects on splice site recognition and pairing (reviewed in references 5, 13, and 43). A class of elements called splicing enhancers stimulate the use of splice sites. Many of these sequences are bound by proteins that are part of the SR family of splicing factors (45). The binding of SR proteins to exonic enhancer elements can increase U2AF65 or U2 snRNP binding to an upstream 3′ splice site region (38, 42, 57, 60). Because SR proteins can also improve the binding of U1 snRNP to 5′ splice sites (29, 40), it is assumed that enhancer elements located near a 5′ splice site may also facilitate the stable recruitment of U1 snRNP (9, 20, 28, 36). When located directly in a 3′ splice site region, a binding site for SR protein can impair U2 snRNP binding (39), indicating that the position of the SR protein binding site is crucial to determining whether the site will act as an activator or a repressor.
Elements that repress splice site utilization have also been uncovered in mammalian pre-mRNAs. Some silencer elements act by forming a duplex structure that impairs splice site recognition (7, 17, 19, 30, 37). Other negative elements require the contribution of trans-acting factors. While the binding of the polypyrimidine tract-binding protein has been linked to enhancer function in one case (44), polypyrimidine tract-binding protein binding to intronic elements has generally been associated with splicing inhibition (58, 63). Recently, the hnRNP A1 protein has been implicated in the activity of silencer elements located in the alternative exon of fibroblast growth factor receptor 2 (24) and the human immunodeficiency virus tat exon 2 (11). While additional silencer elements have been uncovered in other mammalian pre-mRNAs, the mechanisms by which they inhibit splicing are not understood (8, 50, 51, 56). Interestingly, the binding of hnRNP A1 proteins to intron elements can modulate 5′ splice site utilization without affecting 5′ splice site recognition (8, 15). In this case, an interaction between bound A1 proteins has been proposed to bring into close proximity distant splicing partners (8).
While it is clear that several regulatory elements can affect the recognition of splicing signals, several studies now suggest that proper control of a single alternative splicing event requires coordination between distinct elements and factors (1, 3, 8, 10, 12, 19, 20, 22, 23, 31, 33, 35, 41, 48, 53, 56, 59). The convergence of many elements acting on a single splicing event may be essential to modulate pre-mRNA splicing in response to a large variety of tissue-specific effectors and developmental cues.
We are using the hnRNP A1 gene as a model system to study the control of alternative splicing. This gene produces two different mRNAs, encoding the A1 (34 kDa) and A1B (38 kDa) proteins. These two mRNAs are produced by the exclusion and inclusion of exon 7B, respectively. Sequence alignment between the mouse and human hnRNP A1 genes has revealed several conserved regions in the introns flanking exon 7B. Previous results have shown that at least four elements influence the alternative splicing of exon 7B (7, 8, 15). In this study, we report the activity of an intron sequence called conserved element 9 (CE9). Our results show that CE9 is a silencer element that can repress the use of a variety of downstream 3′ splice sites, including the 3′ splice site of hnRNP A1 exon 8. We discuss the role of this novel element in the control of hnRNP A1 pre-mRNA splicing.
MATERIALS AND METHODS
Plasmid constructs.
To insert multiple copies of CE9 we used an oligonucleotide with a mutated HindIII site, the CE9 sequence, a HindIII site, and an EcoRV site. A partial duplex form of this oligonucleotide was inserted in between the HindIII and EcoRV sites of pBluescript II KS(+). Thereafter, additional CE9 oligonucleotides were inserted to produce pK9-2x and pK9-3x. pA was constructed by inserting a HincII-EcoRV fragment of pK9-2x and pK9-3x into the StuI site of pSPAdStu (42). To generate pBΔ, pK78A1 was digested with StuI and HincII and self-ligated. To construct pB derivatives, the HincII-EcoRV fragments of pK9 derivatives were inserted between the StuI and HincII sites of pK78A1 (14). pC3′ −/9 was constructed by reinserting the reannealed CE9 oligonucleotides (38 bp) into the EcoRV site of pC3′ −/− (6). To produce pmA1Δ9, the StuI-BbsI fragment of STE (7) was inserted into pmA1ΔSTE, which had been previously cut with StuI and HincII. pmA1-3x and pmA1-3xα were generated by insertion of a HincII and EcoRV fragment, taken from pK9-3x, into a pmA1 derivative lacking CE9 and previously digested with MscI. To produce DUP derivatives, oligonucleotides were inserted into DUP4-1 and DUP5-1 (kindly provided by E. Modafferi and D. Black [49]), which had been previously digested with ApaI or BglII and treated with T4 DNA polymerase or Klenow, respectively. Derivatives used for in vitro splicing studies were made by inserting the BamHI-SacI fragment from DUP constructs into pBluescript II KS(+) previously cut with BamHI and SacI. To generate pKCE9 and derivatives, pBluescript II KS(+) was cut with HincII, and reannealed oligonucleotides were inserted at this site. All constructs were verified by extensive restriction enzyme digestion analysis and DNA sequencing when appropriate.
Transfection assays.
Transfection of HeLa cells with different constructs of DUP was accomplished using 20 μg of Dosper liposomal transfection reagent (Boehringer Mannheim). For pmA1 derivatives, transfection in HeLa cells was performed using the standard calcium phosphate coprecipitation procedure. At 48 h posttransfection, total RNA was extracted using the guanidine hydrochloride procedure (14).
RNA analysis.
Primer extension analysis was performed on total RNA essentially as described by Modafferi and Black (49). The reactions were run on a 5% denaturing gel (38:2 acrylamide-bisacrylamide, 8 M urea, 1× Tris-borate-EDTA) in 1× Tris-borate-EDTA buffer. For pmA1 derivatives, reverse transcription (RT)-PCR was performed as described previously (61). The oligonucleotides used in this assay were CMV-1 (15), A1E9 (8), and A1E7 (TGCCAAATCCATTATAGCCA). RT-PCR assays on the endogenous β-actin mRNA were performed by using oligonucleotides AC-1 (GGAGCATTTGCGGTGGACGAT) and AC-2 (ACCACCATGTACCCTGGCATT).
In vitro transcription and splicing assays.
All derived DUP substrates were produced from pBluescript-based plasmids linearized with BamHI and transcribed with T7 RNA polymerase (Amersham Pharmacia Biotech) in the presence of cap analog and [α-32P]UTP (Amersham Pharmacia Biotech). The A RNA and derivatives were obtained from plasmids linearized with HincII. C3′ −/− and C3′ −/9 RNAs were obtained from plasmids linearized with ScaI. B RNA and derivatives were obtained from plasmids linearized with BamHI. Transcription was accomplished with T3 or SP6 RNA polymerase (Amersham Pharmacia Biotech). CE9 and K(+) RNAs were produced from plasmids linearized with ClaI and transcribed with T3 RNA polymerase. Cold RNAs were produced as described above except that the relative amount of [α-32P]UTP was reduced 2,000-fold. The purification of all RNA molecules was performed as described by Chabot (14).
HeLa nuclear extracts were prepared (25) and used in splicing reactions as previously described (15). Identification of lariat molecules and other splicing products was confirmed by performing debranching reactions in S100 extracts (20) followed by gel migration relative to molecular weight standards. The competition assay was performed by preincubating the splicing mixture with cold competitor RNAs for 10 min at 30°C prior to addition of the radiolabeled RNA substrate.
Gel shift assays.
For complex formation, we used the procedure described by Das and Reed (21). Some of the extracts used for splicing complex formation were treated with RNase H and oligonucleotides as described by Black et al. (6). The oligonucleotides used for these experiments were U2A (GGCCGAGAAGCGAT) and U4A (CCACTGCGCAAAGCT).
RESULTS
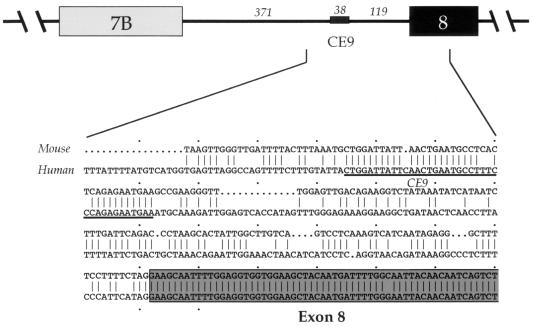
To address the molecular mechanisms controlling the alternative splicing of the hnRNP A1 pre-mRNA, we are investigating the contribution of sequence elements located in the introns flanking alternative exon 7B. The rationale for selecting intron elements is based on their high degree of sequence conservation between the mouse and human hnRNP A1 genes. Each of the four intron elements that have been analyzed so far have demonstrated an effect on the alternative splicing of exon 7B: CE6 base-pairs with the 5′ splice site region of exon 7B to decrease its use (7), CE4m represses the 3′ splice site of exon 7B (8), and hnRNP A1 binding sites located in each of the introns flanking exon 7B promote exon 7B skipping (8, 15). Here, we focus on CE9, which is located in the intron downstream of exon 7B. The 38-nucleotide (nt)-long CE9 is located 119 nt upstream from the 3′ splice site of exon 8 in the hnRNP A1 pre-mRNA (Fig. 1).
FIG. 1.
Schematic representation of the downstream portion of the hnRNP A1 alternative splicing unit, with the length indicated in nucleotides. An alignment between the mouse and the human sequences from the middle of the intron to a portion of exon 8 is shown. The sequence of CE9 is underlined.
CE9 represses a downstream 3′ splice site in vitro.
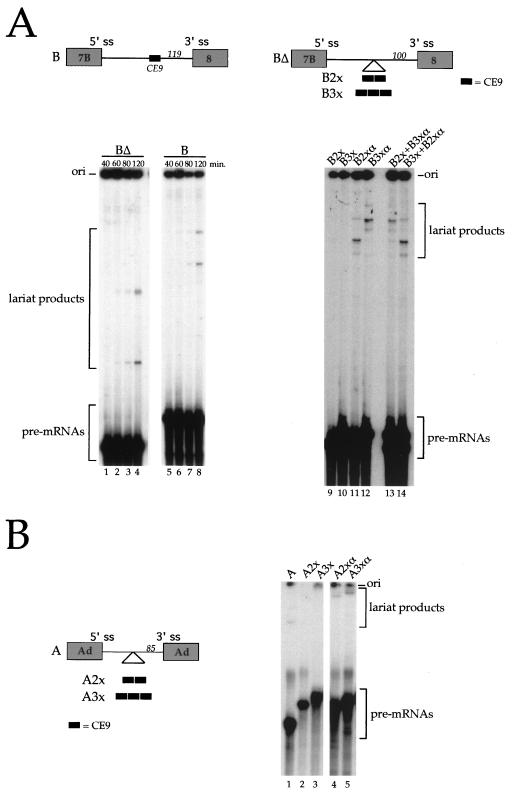
To determine whether CE9 can affect splicing, we tested the effect of deleting CE9 from the intron of a simple exon 7B-exon 8 pre-mRNA. Compared to the pre-mRNA containing CE9, no significant difference in splicing efficiency was observed following a time course incubation in a HeLa nuclear extract (Fig. 2A, lanes 1 through 8). Because the effect of weak elements can be difficult to detect in vitro, we tested the effect of inserting several copies of the element in the single intron construct. This approach often reveals the activity of weak elements in vitro and in vivo (for examples, see references 4, 34, 46, 48, and 49). Compared to the insertion of complementary sequences in the pre-mRNA lacking CE9 (B2xα and B3xα) (Fig. 2A, lanes 11 and 12), two or three copies of CE9 completely inhibited splicing (B2x and B3x) (Fig. 2A, lanes 9 and 10). Lack of splicing was not due to the presence of a nonspecific inhibitor in the B2x and B3x RNA preparations, since coincubation with B2xα and B3xα RNAs did not compromise splicing activity (Fig. 2A, lanes 13 and 14). Copies of CE9 were also inserted in an adenovirus model pre-mRNA (Fig. 2B). The presence of two or three copies of CE9 inhibited in vitro splicing (Fig. 2B, lanes 2 and 3), whereas the insertion of two or three copies of complementary sequences was not inhibitory (Fig. 2B, lanes 4 and 5). Thus, the presence of at least two copies of CE9 can inhibit the in vitro splicing of the intron normally separating exon 7B from exon 8. Moreover, multiple copies of CE9 can inhibit the splicing of a heterologous intron.
FIG. 2.
CE9 inhibits in vitro splicing. (A) The structures of pre-mRNA B and BΔ, as well as derivatives containing multiple copies of CE9 or complementary sequences, are shown at the top. The distance of the insertion point to the 3′ splice site (ss) is indicated in nucleotides. Note that the CE6 element is absent from all pre-mRNAs. Labeled pre-mRNAs were incubated in HeLa nuclear extracts for the times indicated for BΔ and B RNAs or for 2 h for multiple inserts. Splicing products were run on an 11% acrylamide–8 M urea gel. Mixtures containing two different RNAs were analyzed to rule out the presence of a nonspecific inhibitor in the B2x and B3x RNA preparations (lanes 13 and 14). (B) Copies of CE9 or complementary sequences were inserted into a pre-mRNA substrate derived from the adenovirus (Ad) major late transcription unit (A RNA). The labeled splicing products were resolved on a 7% acrylamide–8 M urea gel. Because mRNA products were obscured by the degradation of the pre-mRNAs, only the portion of the gels that indicates the pre-mRNAs and lariat products is shown.
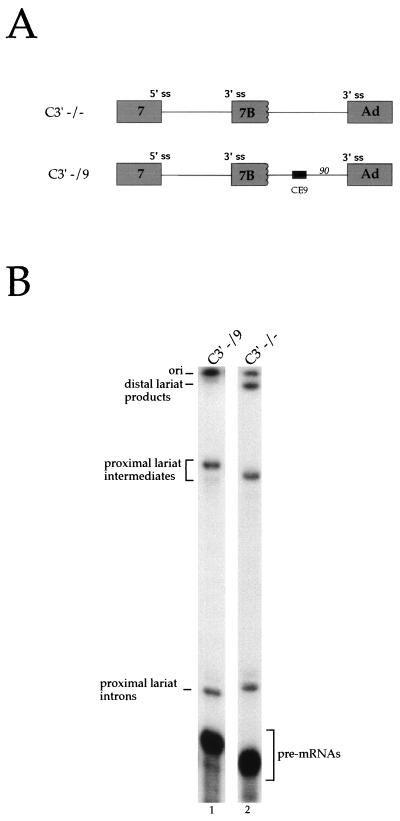
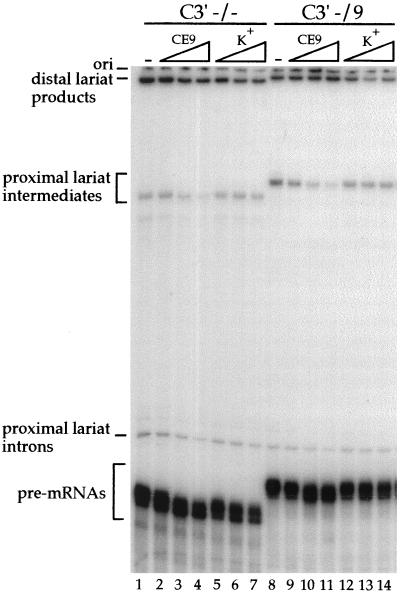
An approach that is more sensitive to the activity of weak elements relies on the use of pre-mRNAs containing alternative splice sites (for examples, see references 8, 15, and 52). Moreover, this approach has the advantage of simultaneously addressing the ability of the element to modulate splice site selection. To analyze this activity for CE9, we used a model pre-mRNA carrying the two 3′ splice sites competing for a single 5′ splice site (C3′ −/− RNA) (8). This pre-mRNA is spliced to each 3′ splice site with approximately equivalent efficiency (Fig. 3B, lane 2). Inserting one copy of CE9 between the two 3′ splice sites repressed the use of the distal site (C3′ −/9 RNA [Fig. 3B, lane 1]). Adding two or three copies of CE9 gave the same effect, with no reduction in total splicing efficiency (data not shown). Insertion of unrelated sequences of the same length had no effect on 3′ splice site selection, ruling out a distance effect (data not shown, but see reference 8). CE9 did not influence 5′ splice site selection when positioned between the 5′ splice sites of exon 7 and exon 7B (data not shown). Given that multiple copies of CE9 inhibit the splicing of one-intron pre-mRNAs (Fig. 2), these results suggest that CE9 is a silencer element and that one copy of CE9 can repress the use of a downstream 3′ splice site in a model pre-mRNA carrying competing 3′ splice sites.
FIG. 3.
CE9 represses the utilization of a downstream 3′ splice site (ss). (A) Schematic representation of pre-mRNAs containing competing 3′ splice sites. The position of CE9 is indicated, as is its distance from the 3′ splice site, in nucleotides. Ad, adenovirus. (B) Labeled pre-mRNAs were incubated in a HeLa nuclear extract for 2 h. Splicing products were fractionated on an 11% acrylamide–8 M urea gel. The positions of the lariat products generated from the use of the distal (Ad) or the proximal (7B) 3′ splice site are indicated.
CE9 affects splicing in vivo.
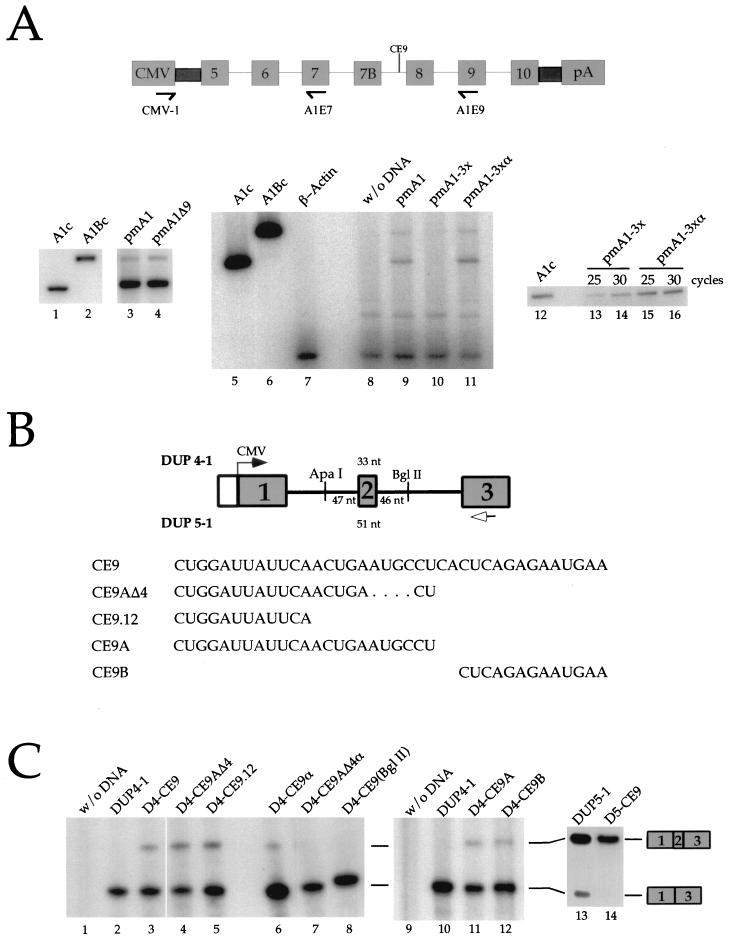
To determine whether CE9 has the same activity in vivo, we tested the effect of deleting CE9 or inserting several copies of CE9 into an hnRNP A1 minigene. The wild-type A1 minigene was spliced to yield predominantly the skipped A1 isoform, as judged by an RT-PCR assay (Fig. 4A, lanes 3 and 8) (7, 8, 15). The deletion of CE9 did not affect the frequency of exon 7B inclusion (Fig. 4A, lane 4). We then tested the effect of adding several copies of CE9 into pmA1Δ9. In comparison to a control construct carrying three copies of the complementary sequence of CE9 (pmA1-3xα) (Fig. 4A, lane 11), three copies of CE9 (pmA1-3x) promoted a large decrease in the accumulation of spliced products (Fig. 4A, lane 10). This effect was seen in three independent transfection assays (data not shown). The lack of signal was not due to RNA degradation or to a nonspecific inhibitor of RT-PCR, since the signal corresponding to the actin mRNA was obtained by coamplification in all samples (Fig. 4A, lanes 8 to 11). Moreover, a separate RT-PCR assay performed with the CMV-1 and A1E7 oligonucleotides indicated that splicing had occurred normally between exon 5 and exon 7 (Fig. 4A, lanes 12 to 16), ruling out a general problem of expression with pmA1-3x. The absence of products with the CMV-1–A1E9 pair of oligonucleotides may mean that an RNA missing exon 9 is produced. Alternatively, if the intron separating exon 7B from exon 8 is retained, the formation of a very stable secondary structure between the CE6 element and the 5′ splice site of exon 7B (6) may prevent progression by reverse transcriptase and hence affect the accumulation of amplified products. In any case, the results indicate that the CE9 elements have locally perturbed splicing, consistent with the predicted outcome for a splicing repressor element.
FIG. 4.
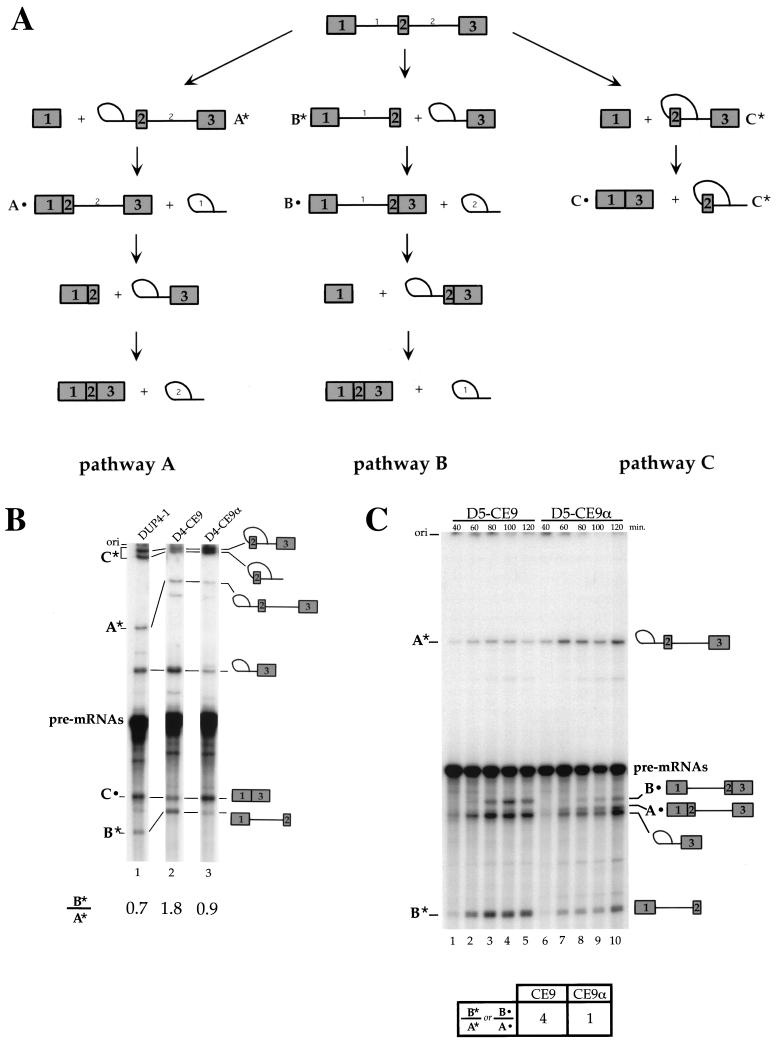
CE9 affects splicing in vivo. (A) CE9 prevents the production of fully spliced mRNAs. A genomic portion of the murine A1 gene was expressed in HeLa cells using the CMV-1 promoter. The relative position of CE9 is indicated, as well as the position of the oligonucleotides used for the RT-PCR assays performed on total RNA isolated 48 h posttransfection. A derivative carrying a deletion of CE9 was used (pmA1Δ9 [lane 4]). Three copies of CE9 or three copies of the complementary sequence of CE9 were inserted into pmA1Δ9 (pmA1-3x or pmA1-3xα, respectively). Reconstructed A1 or A1B cDNAs were used as controls in PCR assays (lanes 1, 2, 5, 6, and 12). RT-PCR amplification of the endogenous β-actin mRNA was performed independently on a mock transfection (lane 7) or simultaneously with the A1 minigene analysis with oligonucleotides CMV-1 and A1E9 (lanes 8 to 11). A separate RT-PCR assay was carried out with the CMV-1 and A1E7 oligonucleotides (lanes 12 to 16). The number of cycles used in the amplification rounds is indicated above the lanes. (B) Schematic representation of the DUP constructs and derivatives. The length of the central exon for DUP 4-1 and DUP 5-1 is indicated. The transcription start site is indicated by an arrow. The distance between the different cloning sites and the central exon is indicated. The arrow below exon 3 represents the oligonucleotide used for primer extension analysis. The RNA versions of the different oligonucleotides cloned into the DUP plasmids are shown at the bottom. (C) CE9 promotes the inclusion of artificial globin exon in vivo. DUP expression was analyzed by primer extension. Plasmid names are indicated above each lane. Each DUP 4-1, DUP 5-1, or derivative was generated by inserting oligonucleotides at the ApaI site, except for D4-CE9(BglII), for which BglII in the downstream intron was used (lane 8). Extension products were loaded onto a 5% acrylamide–8 M urea. The slightly abnormal migration of the 1-3 product in lane 8 is a gel artifact.
The lack of an effect associated with the deletion of CE9 may be due to the presence of redundant elements in the A1 pre-mRNA. Redundancy in sequence elements that affect the alternative splicing of the neurospecific c-src exon has also complicated the analysis of individual elements in their natural context (48, 49). A strategy to assess the potential effect of individual elements in vivo is to insert them into a heterologous alternative splicing unit. An artificial human β-globin minigene has been used for this purpose (DUP4-1) (Fig. 4B) (48, 49). This reporter gene contains an internal exon of 33 nt which is skipped at a frequency of >95% following expression in HeLa cells (27). Whereas exclusion of the internal exon is attributed to the proximity of the abutting 3′ and 5′ splice sites, it remains unclear whether this proximity prevents exon definition or hinders the simultaneous assembly of spliceosomes on flanking introns (26, 27). Exon inclusion is assessed by performing a primer extension assay on total RNA isolated 48 h posttransfection. As shown in Fig. 4C, insertion of one copy of CE9 in the upstream intron promoted inclusion of the internal exon (20% inclusion [lane 3]). Insertion of the complementary sequence of CE9 at the same position had a modest effect (9% inclusion [lane 6]), and insertion of CE9 in the downstream intron had no effect (<5% inclusion [lane 8]). The insertion of CE9 in a similar construct containing an internal exon of 51 nt also stimulated exon inclusion in vivo (DUP5-1 [lanes 13 and 14]).
To better characterize the sequences within CE9 that are responsible for this activity, we tested the activity of the first 23 nt and the last 13 nt of CE9 (CE9A and CE9B, respectively). CE9A promoted exon inclusion of the DUP4-1 exon almost as efficiently as the complete CE9 element, whereas CE9B was slightly less efficient at promoting inclusion of the internal exon (17 and 13% inclusion [Fig. 4B, lanes 11 and 12, respectively]). A derivative of CE9A, CE9AΔ4, which lacks 4 nt near the 3′ end of CE9A, was at least as efficient as CE9 at promoting inclusion of the internal exon (30% inclusion [Fig. 4C, lane 4]). Finally, the first 12 nt of CE9 retained the ability to promote inclusion of the globin internal exon (25% inclusion [lane 5]). An insert carrying a portion of the complementary sequence of CE9 did not promote exon inclusion (<5% inclusion [lane 7]). Our in vivo results indicate that at least two regions of CE9 can affect the alternative splicing of DUP4-1 (CE9A and CE9B). However, the first 12 nt of CE9A possess the ability to stimulate the inclusion of the internal globin exon as efficiently as the complete CE9 element.
CE9 can relieve interference of closely positioned splice sites.
If exclusion of the internal globin exon is caused by poor exon definition, improving splice site recognition should reduce exon skipping, consistent with the observations of Dominski and Kole (26). On the other hand, if exon skipping is due to steric interference between closely positioned splice sites, reducing the rate of spliceosome assembly on the first intron may improve spliceosome assembly on the second intron and should also lead to more efficient inclusion of the internal exon in vivo. Thus, the more frequent inclusion of the short globin exon could be due to either an enhancer or a silencer element. To verify whether more efficient inclusion of the central globin exon was due to the silencer activity of CE9, we monitored the in vitro splicing of labeled pre-mRNAs produced from DUP4-1 and derivatives (D4-CE9 and D4-CE9α). The identity of each band in Fig. 5B was confirmed by gel purifying each RNA species, submitting it to a debranching reaction in a HeLa 100 extract, and assessing its size relative to length markers (data not shown). The splicing profile of the DUP4-1 pre-mRNA matched the profile observed previously by Dominski and Kole (26, 27) for a related pre-mRNA (DUP33) in HeLa extracts. To facilitate the presentation, we compared products that are unique to each of the three splicing pathways. As shown in Fig. 5A, A* represents an RNA splicing intermediate specific for the pathway where intron 1 is removed first (pathway A), while B* is a splicing intermediate specific for the pathway where intron 2 is removed first (pathway B). C* is a doublet band that contains a splicing intermediate and a splicing product diagnostic of exon skipping (pathway C). Incubation of DUP4-1 pre-mRNA generated products corresponding to the three splicing pathways (Fig. 5B, lane 1), in agreement with previous results obtained with DUP33 (26, 27). The preference in the order of intron removal can be estimated by measuring the ratio of B* to A* products, which is approximately 0.7 with DUP4-1 pre-mRNA. In addition, exon 2 skipping (C* and C• products) is the most efficient pathway by which DUP4-1 is spliced. The insertion of CE9 strongly decreased the appearance of skipped products (C* and C•) and increased the ratio of B* to A* products to 1.8 (Fig. 5B, lane 2). In contrast, a derivative carrying the complementary sequence of CE9 allowed relatively efficient exon 2 skipping (C* and C• products) and a ratio of B* to A* products that was similar to that of DUP4-1 (Fig. 5B, lane 3).
FIG. 5.
Effect of CE9 on DUP 4-1 and DUP 5-1 splicing in vitro. (A) Representation of different pathways used for the splicing of DUP pre-mRNAs. Pathway A occurs when intron 1 is the first intron to be removed, while pathway B takes place when intron 2 is spliced first. Pathway C corresponds to skipping of the internal exon. The splicing products, A*, A•, B*, B•, C*, and C•, indicate molecules that are unique to each pathway and correspond to the bands shown in the splicing gels (panels B and C). (B) CE9 shifts splicing in favor of pathway B. Labeled pre-mRNAs were incubated in HeLa extracts for 1 h. Splicing products were fractionated on an 11% acrylamide–8 M urea gel. Selected products are indicated. (C) CE9 also improves pathway B in DUP 5-1. Products specific to pathway A or B are indicated. Note that the band immediately below product A• corresponds to the intron 2-exon 3 lariat intermediate, a product common to pathways A and B. Pre-mRNAs were incubated under splicing conditions for the times indicated. Splicing products were loaded onto an 8% acrylamide–8 M urea gel. The ratio of product B* to product A* (or B•/A•) is indicated and is based on the 2-h time point. The structure of DUP pre-mRNA and derivatives is as shown in Fig. 4A, except that the labeled pre-mRNAs were synthesized in vitro using T7 RNA polymerase.
The effect of CE9 was confirmed using a DUP pre-mRNA produced from DUP5-1, which contains a slightly larger internal exon (51 nt). In our laboratory, no exon 2 skipping was observed with the DUP5-1 pre-mRNA in vitro (data not shown). By monitoring the appearance of intermediates and products that are unique to pathways A and B, we observed that the ratio of B* to A* products is approximately 1 for the control pre-mRNA that contains the complementary sequence of CE9 (Fig. 5C, lane 10). When CE9 is present in intron 1, this ratio changes to 4 (Fig. 5C, lane 5). These values are confirmed by assessing the intensity of another set of diagnostic products that are specific for pathways A and B (Fig. 5A). Thus, for both DUP4-1 and DUP5-1 pre-mRNAs, the presence of CE9 decreased the splicing efficiency of the first intron, consistent with the silencer effect of CE9. This decrease was accompanied by an improvement in the splicing efficiency of the second intron. Better splicing of intron 2 likely explains why exon 2 skipping is less efficient in the D4-CE9 pre-mRNA. Thus, the silencing effect of CE9 would indirectly promote more efficient splicing of the second intron possibly because of reduced interference in spliceosome assembly. This would reduce the frequency of exon 2 skipping and would lead to more frequent inclusion of the internal exon. Although CE9 reduces splicing of the first intron in vitro, splicing of this intron may ultimately take place by default in vivo.
The effect of CE9 is mediated by a trans-acting factor.
The ability of CE9 to influence the splicing of heterologous pre-mRNAs suggests that a trans-acting factor(s) mediates the activity of CE9. To confirm that a cellular factor is required for the activity of CE9, we performed splicing assays in the presence of an excess of cold competitor RNA. When a short RNA containing the complete CE9 sequence was preincubated in a HeLa extract, the effect of the cis-acting CE9 element was abrogated, and splicing to the distal 3′ splice site of the C3′ −/9 pre-mRNA was improved (Fig. 6, lanes 9 to 11). No effect on 3′ splice site selection was seen when the competitor RNA contained plasmid-derived sequences only (lanes 12 to 14). These results suggest that titratable factors bind to the CE9 element to mediate its effect. Notably, an excess of the CE9 competitor RNA also affected 3′ splice site selection on a pre-mRNA lacking CE9 (C3′ −/−) (Fig. 6, lanes 2 to 4). This result suggests that the factor binding to CE9 may play a general role in 3′ splice site selection.
FIG. 6.
A trans-acting factor mediates the repression by CE9 on a downstream 3′ splice site. Splicing was performed in a HeLa nuclear extract preincubated for 10 min with increasing amounts of the CE9 RNA as a competitor (lanes 2 to 4 and 9 to 11) or with a control RNA derived from pBluescript K(+) (lanes 5 to 7 and 12 to 14). Each set of the competition was performed with 0.5 fmol of pre-mRNA and 50, 250, or 500 fmol of unlabeled competitor RNA.
The CE9 repressor element does not prevent splicing complex formation.
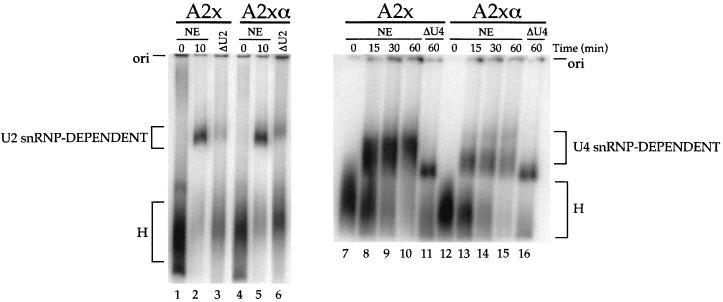
To address the mechanism by which CE9 inhibits splicing, we analyzed splicing complex assembly. As shown in Fig. 7, early complex formation was as efficient with the adenovirus pre-mRNA carrying two copies of CE9 as with the pre-mRNA carrying two copies of the complementary sequences (compare lane 2 with lane 5). The major complex likely corresponds to complex A, since its assembly was strongly reduced in an extract that had been pretreated with RNase H and an oligonucleotide complementary to the 5′ end of U2 snRNA (lanes 3 and 6). Using longer incubation periods, we could monitor more advanced stages of spliceosome assembly, although the resolution was not sufficient to distinguish complex B from complex C (lanes 7 to 16). When pre-mRNAs were incubated in an extract that had been pretreated with RNase H and an oligonucleotide complementary to U4 snRNA, slower-migrating complexes were converted into a complex probably equivalent to complex A (Fig. 7, lanes 11 and 16). The pre-mRNA containing two copies of CE9 (A2x) underwent complex formation at least as efficiently as that of the control A2xα version (compare lanes 8 to 10 with lanes 13 to 15). Thus, although two copies of CE9 blocked splicing of the A2x pre-mRNA, this substrate was efficiently assembled into snRNP-containing complexes.
FIG. 7.
CE9 does not block the assembly of snRNP-containing complexes. The time course of splicing complex assembly was determined by using adenovirus pre-mRNAs containing two copies of CE9 or two copies of the complementary sequences (A2x or A2xα, respectively) (see Fig. 2B for a schematic diagram of the pre-mRNAs). Nuclear extracts were pretreated for 1 h at 30°C in the presence of RNase H alone (NE) or RNase H and oligonucleotides complementary to U2 snRNA (ΔU2) or U4 snRNA (ΔU4). The reaction mixtures were loaded on a 2% low-melting-point agarose gel. The origins of the gels and the identities of the complexes are indicated.
DISCUSSION
We have shown that the intron element CE9 can modulate in vivo and in vitro splicing. Several observations are consistent with the conclusion that CE9 is a silencer element that represses the use of a downstream 3′ splice site. First, the insertion of multiple copies of CE9 in the intron of simple pre-mRNAs abrogates in vitro splicing. This effect was seen for an adenovirus model pre-mRNA and, more importantly, for an A1 pre-mRNA carrying the 5′ splice site of exon 7B and the 3′ splice site of exon 8. In vivo, the insertion of several copies of CE9 in an A1 minigene also compromises the accumulation of fully spliced products. Second, the insertion of a single CE9 element between two competing 3′ splice sites reduces splicing to the downstream 3′ splice site. Third, the presence of CE9 in the first intron of an artificial globin pre-mRNA represses splicing of this intron in vitro. Because this effect is accompanied by an improvement in splicing of the second intron, the repressing activity of CE9 apparently relieves the interference created by closely positioned splice sites on this short exon. Assuming that splicing of the first intron ultimately occurs by default, this would explain why CE9 improves the inclusion of the short internal globin exon in vivo. Overall, these results are consistent with the notion that the normal function of CE9 is to reduce the use of the 3′ splice site of exon 8.
In the hnRNP A1 pre-mRNA, the frequency of inclusion of exon 7B is determined to a large extent by a competition between the 3′ splice sites of exon 7B and exon 8. This is because a duplex structure impairs the use of the 5′ splice site of exon 7B (7). At least two types of elements favor selection of the 3′ splice site of exon 8 over that of exon 7B in HeLa cells: the CE4m element represses the 3′ splice site of exon 7B, and A1 binding elements have been proposed to facilitate pairing between the 5′ splice site of exon 7 and the 3′ splice site of exon 8 (8, 15). This combination of elements may neutralize the activity of a weak element like CE9 and explain why deleting CE9 from the A1 minigene has little impact in vivo. However, in certain types of cells or in some situations, repressing the 3′ splice site of exon 8 may be important to favor selection of the 3′ splice site of exon 7B.
The interaction of CE9 with a cellular factor appears to be important for its modulating activity, since an excess of competitor RNA containing the CE9 sequences can stimulate splicing to a distal 3′ splice site. The ability of CE9 to function in a variety of pre-mRNA substrates also supports the notion that a cellular factor is required for the activity of CE9. Further work will be required to identify the factor that binds to CE9. It is intriguing that the first half of CE9, which displays most of the activity of CE9, contains a sequence that resembles the CE4m repressor element downstream of exon 7B (8) and the human immunodeficiency virus repressor element in exon 3 of tat-rev (2, 56) (consensus sequence = CU[A/G]GA[C/U]UA). Despite this similarity between CE4m and CE9, the mechanisms of inhibition appear to be different, since CE4m and CE9 repress the upstream and the downstream 3′ splice sites, respectively, in C3′ −/− pre-mRNA. In contrast to silencer elements that prevent splicing complex formation (16, 38, 54), the inhibition of pre-mRNA splicing by CE9 was not associated with a block in the assembly of snRNP-containing complexes. Some silencer elements do not prevent snRNP binding (51), and others are bound by snRNPs (18, 32, 38, 47, 55). Thus, CE9 apparently belongs to the latter category of silencer elements and may block a late step of spliceosome assembly or promote the formation of aberrant splicing complexes.
The mechanisms that control the alternative splicing of cassette exons have implicated elements that affect the recognition or use of splice sites directly flanking the alternative exon. However, modulating the use of common splice sites in alternative splicing units may be equally important for producing the proper ratio of mRNA isoforms. Although the common splice sites of alternative splicing units are often suboptimal, less is known about elements that affect their use. Enhancers elements in 3′ and 5′ common exons have been identified in the fibronectin ED1 and calcitonin/calcitonin gene-related peptide alternative splicing units, respectively (42, 62). In these cases, it remains unclear whether these elements affect alternative splicing in their natural setting. However, in the neural cell adhesion molecule splicing unit, an exon enhancer in the 5′ common exon alters the inclusion rate of a downstream alternative exon (20). To our knowledge, CE9 is the first element documented to repress splicing to a 3′ common exon. We expect that the current efforts aimed at identifying elements that modulate splice site selection will reinforce the notion that alternative splicing requires controlling the use of common as well as alternative splice sites.
ACKNOWLEDGMENTS
We thank D. Black and E. Modaferri for kindly providing DUP4-1 and DUP5-1 plasmids. We thank Johanne Toutant for performing transfections and preparing nuclear extracts, and we thank M. Blanchette and S. Hutchison for comments on the manuscript.
M.J.S. is the recipient of a studentship from the FCAR/FRSQ. This work was supported by a grant from the Medical Research Council of Canada. B.C. is a Chercheur-Boursier Senior from the FRSQ and is a member of the Sherbrooke RNA/RNP group supported by the FCAR.
REFERENCES
- 1.Amendt B A, Hesslein D, Chang L-J, Stoltzfus C M. Presence of negative and positive cis-acting RNA splicing elements within and flanking the first tat coding exon of human immunodeficiency virus type 1. Mol Cell Biol. 1994;14:3960–3970. doi: 10.1128/mcb.14.6.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amendt B A, Si Z-H, Stoltzfus C M. Presence of exon splicing silencers within human immunodeficiency virus type 1 tat exon 2 and tat-rev exon 3: evidence for inhibition mediated by cellular factors. Mol Cell Biol. 1995;15:4606–4615. doi: 10.1128/mcb.15.8.4606. . (Erratum, 15:6480.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashiya M, Grabowski P J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 4.Black D L. Activation of c-src neuron-specific splicing by an unusual RNA element in vivo and in vitro. Cell. 1992;69:795–807. doi: 10.1016/0092-8674(92)90291-j. [DOI] [PubMed] [Google Scholar]
- 5.Black D L. Finding splice sites within a wilderness of RNA. RNA. 1995;1:763–771. [PMC free article] [PubMed] [Google Scholar]
- 6.Black D L, Chabot B, Steitz J A. U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing. Cell. 1985;42:737–750. doi: 10.1016/0092-8674(85)90270-3. [DOI] [PubMed] [Google Scholar]
- 7.Blanchette M, Chabot B. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA. 1997;3:405–419. [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchette M, Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18:1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bourgeois C F, Popielarz M, Hildwein G, Stevenin J. Identification of a bidirectional splicing enhancer: differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol Cell Biol. 1999;19:7347–7356. doi: 10.1128/mcb.19.11.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo C A, Baralle F E. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caputi M, Mayeda A, Krainer A R, Zahler A M. hnRNP A/B proteins are required for inhibition of HIV-1 pre-mRNA splicing. EMBO J. 1999;18:4060–4067. doi: 10.1093/emboj/18.14.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carstens R P, McKeehan W L, Garcia-Blanco M A. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol Cell Biol. 1998;18:2205–2217. doi: 10.1128/mcb.18.4.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chabot B. Directing alternative splicing: cast and scenarios. Trends Genet. 1996;12:472–478. doi: 10.1016/0168-9525(96)10037-8. [DOI] [PubMed] [Google Scholar]
- 14.Chabot B. Synthesis and purification of RNA substrates. In: Hames D, Higgins S, editors. RNA processing. Vol. 1. Oxford, England: Oxford University Press; 1994. pp. 1–29. [Google Scholar]
- 15.Chabot B, Blanchette M, Lapierre I, La Branche H. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol. 1997;17:1776–1786. doi: 10.1128/mcb.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chew S L, Baginsky L, Eperon I C. An exonic splicing silencer in the testes-specific DNA ligase III beta exon. Nucleic Acids Res. 2000;28:402–410. doi: 10.1093/nar/28.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clouet d'Orval B, d'Aubenton Carafa Y, Sirand-Pugnet P, Gallego M, Brody E, Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991;252:1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- 18.Cook C R, McNally M T. SR protein and snRNP requirements for assembly of the Rous sarcoma virus negative regulator of splicing complex in vitro. Virology. 1998;242:211–220. doi: 10.1006/viro.1997.8983. [DOI] [PubMed] [Google Scholar]
- 19.Côté J, Chabot B. Natural base-pairing interactions between 5′ splice site and branch site sequences affect mammalian 5′ splice site selection. RNA. 1997;3:1248–1261. [PMC free article] [PubMed] [Google Scholar]
- 20.Côté J, Simard M J, Chabot B. An element in the 5′ common exon of the NCAM alternative splicing unit interacts with SR proteins and modulates 5′ splice site selection. Nucleic Acids Res. 1999;27:2529–2537. doi: 10.1093/nar/27.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das R, Reed R. Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA. 1999;5:1504–1508. doi: 10.1017/s1355838299991501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Gatto F, Breathnach R. Exon and intron sequences, respectively, repress and activate splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1995;15:4825–4834. doi: 10.1128/mcb.15.9.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Del Gatto F, Plet A, Gesnel M-C, Fort C, Breathnach R. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1997;17:5106–5116. doi: 10.1128/mcb.17.9.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del Gatto-Konczak F, Olive M, Gesnel M-C, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dominski Z, Kole R. Cooperation of pre-mRNA sequence elements in splice site selection. Mol Cell Biol. 1992;12:2108–2114. doi: 10.1128/mcb.12.5.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dominski Z, Kole R. Selection of splice sites in pre-mRNAs with short internal exons. Mol Cell Biol. 1991;11:6075–6083. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elrick L L, Humphrey M B, Cooper T A, Berget S M. A short sequence within two purine-rich enhancers determines 5′ splice site specificity. Mol Cell Biol. 1998;18:343–352. doi: 10.1128/mcb.18.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estes P A, Cooke N E, Liebhaber S A. A native RNA secondary structure controls alternative splice-site selection and generates two human growth hormone isoforms. J Biol Chem. 1992;267:14902–14908. [PubMed] [Google Scholar]
- 31.Gallego M E, Balvay L, Brody E. cis-acting sequences involved in exon selection in the chicken β-tropomyosin gene. Mol Cell Biol. 1992;12:5415–5425. doi: 10.1128/mcb.12.12.5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gontarek R R, McNally M T, Beemon K. Mutation of an RSV intronic element abolishes both U11/U12 snRNP binding and negative regulation of splicing. Genes Dev. 1993;7:1926–1936. doi: 10.1101/gad.7.10.1926. [DOI] [PubMed] [Google Scholar]
- 33.Gooding C, Roberts G C, Smith C W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated alpha-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- 34.Hertel K J, Maniatis T. The function of multisite splicing enhancers. Mol Cell. 1998;1:449–455. doi: 10.1016/s1097-2765(00)80045-3. [DOI] [PubMed] [Google Scholar]
- 35.Huh G S, Hynes R O. Elements regulating an alternatively spliced exon of the rat fibronectin gene. Mol Cell Biol. 1993;13:5301–5314. doi: 10.1128/mcb.13.9.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Humphrey M B, Bryan J, Cooper T A, Berget S M. A 32-nucleotide exon-splicing enhancer regulates usage of competing 5′ splice sites in a differential internal exon. Mol Cell Biol. 1995;15:3979–3988. doi: 10.1128/mcb.15.8.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutton M, Lendon C L, Rizzu P, Baker M, Froelich S, Houlden H, Pickering-Brown S, Chakraverty S, Isaacs A, Grover A, Hackett J, Adamson J, Lincoln S, Dickson D, Davies P, Petersen R C, Stevens M, de Graaff E, Wauters E, van Baren J, Hillebrand M, Joosse M, Kwon J M, Nowotny P, Heutink P, et al. Association of missense and 5′-splice-site mutations in tau with the inherited dementia FTDP-17. Nature. 1998;393:702–705. doi: 10.1038/31508. [DOI] [PubMed] [Google Scholar]
- 38.Kan J L, Green M R. Pre-mRNA splicing of IgM exons M1 and M2 is directed by a juxtaposed splicing enhancer and inhibitor. Genes Dev. 1999;13:462–471. doi: 10.1101/gad.13.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanopka A, Mühlemann O, Aküsjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381:535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 40.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 41.Kuo B A, Norton P A. Accurate selection of a 5′ splice site requires sequences within fibronectin alternative exon B. Nucleic Acids Res. 1999;27:3945–3952. doi: 10.1093/nar/27.19.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 43.Lopez A J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet. 1998;32:279–305. doi: 10.1146/annurev.genet.32.1.279. [DOI] [PubMed] [Google Scholar]
- 44.Lou H, Helfman D M, Gagel R F, Berget S M. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol Cell Biol. 1999;19:78–85. doi: 10.1128/mcb.19.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manley J L, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- 46.McCullough A J, Berget S M. G triplets located throughout a class of small vertebrate introns enforce intron borders and regulate splice site selection. Mol Cell Biol. 1997;17:4562–4571. doi: 10.1128/mcb.17.8.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McNally L M, McNally M T. An RNA splicing enhancer-like sequence is a component of a splicing inhibitor element from Rous sarcoma virus. Mol Cell Biol. 1998;18:3103–3111. doi: 10.1128/mcb.18.6.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Modafferi E F, Black D L. Combinatorial control of a neuron-specific exon. RNA. 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Modafferi E F, Black D L. A complex intronic splicing enhancer from the c-src pre-mRNA activates inclusion of a heterologous exon. Mol Cell Biol. 1997;17:6537–6545. doi: 10.1128/mcb.17.11.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muro A F, Caputi M, Pariyarath R, Pagani F, Buratti E, Baralle F E. Regulation of fibronectin EDA exon alternative splicing: possible role of RNA secondary structure for enhancer display. Mol Cell Biol. 1999;19:2657–2671. doi: 10.1128/mcb.19.4.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nemeroff M E, Utans U, Krämer A, Krug R M. Identification of cis-acting intron and exon regions in influenza virus NS1 mRNA that inhibit splicing and cause the formation of aberrantly sedimenting presplicing complexes. Mol Cell Biol. 1992;12:962–970. doi: 10.1128/mcb.12.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed R, Maniatis T. Intron sequences involved in lariat formation during pre-mRNA splicing. Cell. 1985;41:95–105. doi: 10.1016/0092-8674(85)90064-9. [DOI] [PubMed] [Google Scholar]
- 53.Ryan K J, Cooper T A. Muscle-specific splicing enhancers regulate inclusion of the cardiac troponin T alternative exon in embryonic skeletal muscle. Mol Cell Biol. 1996;16:4014–4023. doi: 10.1128/mcb.16.8.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Si Z-H, Rauch D, Stoltzfus C M. The exon splicing silencer in human immunodeficiency virus type 1 tat exon 3 is bipartite and acts early in spliceosome assembly. Mol Cell Biol. 1998;18:5404–5413. doi: 10.1128/mcb.18.9.5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Siebel C W, Fresco L D, Rio D C. The mechanism of somatic inhibition of Drosophila P-element pre-mRNA splicing: multiprotein complexes at an exon pseudo-5′ splice site control U1 snRNP binding. Genes Dev. 1992;6:1386–1401. doi: 10.1101/gad.6.8.1386. [DOI] [PubMed] [Google Scholar]
- 56.Staffa A, Cochrane A. Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol Cell Biol. 1995;15:4597–4605. doi: 10.1128/mcb.15.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valcárcel J, Gebauer F. Post-transcriptional regulation: the dawn of PTB. Curr Biol. 1997;7:R705–R708. doi: 10.1016/s0960-9822(06)00361-7. [DOI] [PubMed] [Google Scholar]
- 59.van Oers C C M, Adema G J, Zandberg H, Moen T C, Bass P D. Two different sequence elements within exon 4 are necessary for calcitonin-specific splicing of the human calcitonin/calcitonin gene-related peptide I pre-mRNA. Mol Cell Biol. 1994;14:951–960. doi: 10.1128/mcb.14.2.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Z, Hoffmann H M, Grabowski P J. Intrinsic U2AF binding is modulated by exon enhancer signals in parallel with changes in splicing activity. RNA. 1995;1:21–35. [PMC free article] [PubMed] [Google Scholar]
- 61.Yang X, Bani M R, Lu S J, Rowan S, Ben-David Y, Chabot B. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc Natl Acad Sci USA. 1994;91:6924–6928. doi: 10.1073/pnas.91.15.6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yeakley J M, Hedjran F, Morfin J-P, Merillat N, Rosenfeld M G, Emeson R B. Control of calcitonin/calcitonin gene-related peptide pre-mRNA processing by constitutive intron and exon elements. Mol Cell Biol. 1993;13:5999–6011. doi: 10.1128/mcb.13.10.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L, Liu W, Grabowski P J. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]