Abstract
Heparan sulfate 3-O-sulfotransferases generate highly sulfated but rare 3-O-sulfated heparan sulfate (HS) epitopes on cell surfaces and in the extracellular matrix. Previous ex vivo experiments suggested functional redundancy exists among the family of seven enzymes but that Hs3st3a1 and Hs3st3b1 sulfated HS increases epithelial FGFR signaling and morphogenesis. Single-cell RNAseq analysis of control SMGs identifies increased expression of Hs3st3a1 and Hs3st3b1 in endbud and myoepithelial cells, both of which are progenitor cells during development and regeneration. To analyze their in vivo functions, we generated both Hs3st3a1−/− and Hs3st3b1−/− single knockout mice, which are viable and fertile. Salivary glands from both mice have impaired fetal epithelial morphogenesis when cultured with FGF10. Hs3st3b1−/− mice have reduced intact SMG branching morphogenesis and reduced 3-O-sulfated HS in the basement membrane. Analysis of HS biosynthetic enzyme transcription highlighted some compensatory changes in sulfotransferases expression early in development. The overall glycosaminoglycan composition of adult control and KO mice were similar, although HS disaccharide analysis showed increased N- and non-sulfated disaccharides in Hs3st3a1−/− HS. Analysis of adult KO gland function revealed normal secretory innervation, but without stimulation there was an increase in frequency of drinking behavior in both KO mice, suggesting basal salivary hypofunction, possibly due to myoepithelial dysfunction. Understanding how 3-O-sulfation regulates myoepithelial progenitor function will be important to manipulate HS-binding growth factors to enhance tissue function and regeneration.
Keywords: Heparan sulfate, HS-3-O-sulfotransferase, proteoglycan, basement membrane, FGF10, salivary gland development
Introduction
Heparan sulfate (HS) proteoglycans, with linear polysulfated HS chains covalently attached to their core, are present on all cell surfaces and in extracellular matrices. Their high negative charge due to HS sulfation, influence the activity of many growth factors, affecting processes such as cell proliferation, differentiation, developmental patterning, regeneration, hemostasis, lipid metabolism, immunity, and inflammation [1]. Understanding how the sulfation code is regulated during development is critical to investigating the mechanisms by which HS regulates its broad biological functions. The complex fine structure of HS is generated during biosynthesis, and includes N-deacetylation/N-sulfation, epimerization, and 2-, 3- and 6-O-sulfation, through the coordinated action of several sulfotransferase enzymes [2]. The addition of 3-O-sulfation is a terminal step that generates the least abundant sulfated epitope comprising of less than 0.5 % of the total sulfation [3, 4]. However, the 3-O-sulfotransferases (Hs3sts) are the largest HS enzyme family with seven isoforms in mammals. Biochemical studies show that Hs3st3s can generate the most highly sulfated disaccharide epitope in HS [5–7]. The presence of such a large family of enzymes also suggests that they may have overlapping or compensatory enzymatic functions, which adds complexity of analyzing their in vivo function using murine genetic approaches.
Research investigating the function of 3-O-sulfated HS shows that many receptors and ligands prefer 3-O-sulfated HS, including the ectodomain of FGF receptor (FGFR)-1, FGF1, cyclophilin, stabilin, neuropilin, FGF10-FGFR2b complex and Tau [8–13]. Studies in zebrafish suggest that BMP4 and FGF8 also prefer 3-O-sulfated HS [14, 15]. The 3-O-sulfotransferases are implicated in binding FGF and FGF receptors, epithelial-mesenchymal transition, neurite branching [16], left-right patterning [14], BMP-dependent myocardium contraction [15], regulation of intestinal homeostasis [17], kidney fibrosis correlated with reduced HS3ST1 expression [18] and regulation of neural activity and synaptogenesis [19]. Together, these data suggest that specific interactions of ligands with highly negatively charged 3-O-sulfated HS epitopes modulate growth factor functions in many biological contexts.
We previously showed that Hs3st3a1 and Hs3st3b1 are expressed in the endbuds of E13 fetal submandibular glands (SMG) and 3-O-sulfated HS stabilizes the FGF10/FGFR2b complex, promoting MAPK signaling and the expansion of progenitors within the endbuds [10]. Our previous studies used exogenous 3-O-sulfated HS in epithelial organ culture to investigate FGF-dependent epithelial progenitor growth and morphogenesis. We used siRNA knockdown of sulfotransferases, showing that knockdown of multiple sulfotransferase isoforms was required to reduce epithelial morphogenesis ex vivo. These studies led us to predict that functional compensation of 3-O-sulfotransferases may occur in vivo in the absence of one isoform at either the transcriptional or enzymatic level.
Here we investigate the functions of 3-O-sulfated HS by interrogating HS biosynthetic enzyme expression using single-cell RNAseq (scRNAseq) data and generating knockout (KO) mice of two of the seven 3-O-sulfotransferase enzymes detected in SMG epithelium. We compare these KO mice as we investigate fetal epithelial morphogenesis, transcription of the HS biosynthetic machinery, glycosaminoglycan composition and HS disaccharide analysis and discover defects in adult gland function. We propose that reduced 3-O-sulfation of the myoepithelial basement membrane compromises salivary gland function. Our studies highlight the complexity of the regulation of 3-O-sulfation of HS and its biological functions.
Results
Spatial and temporal localization of the Hs3st enzyme transcripts during SMG development
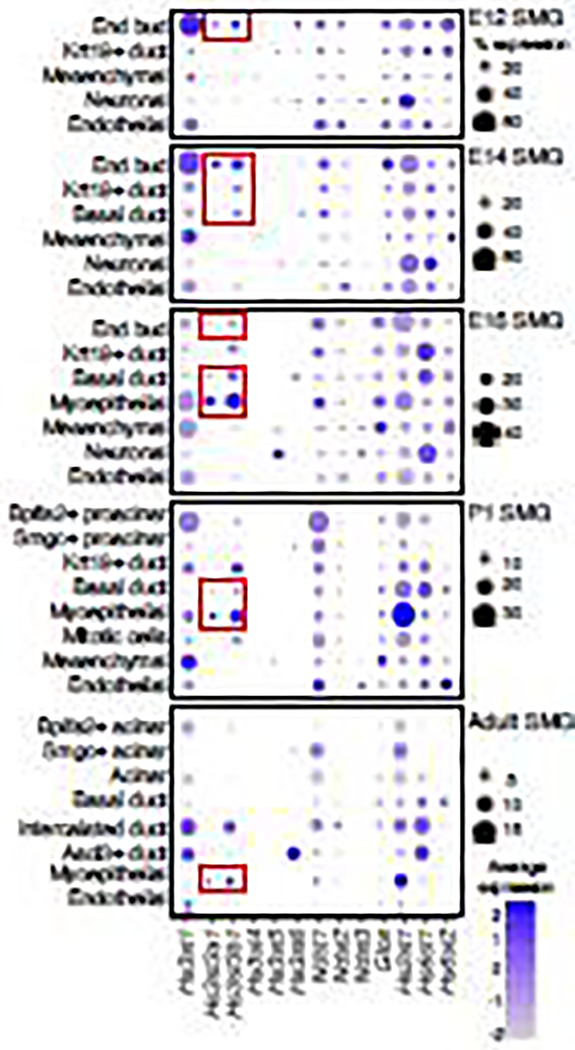
We mined our scRNAseq atlas of murine SMGs development [20] to interrogate the cellular expression of the Hs3st and other HS biosynthetic gene transcripts at embryonic day 12 (E12), E14, E16, postnatal day 1 (P1) and adult stages (Fig. 1). Gene expression was visualized using DotPlots showing both the level of expression and the percentage of cells expressing each gene in cell clusters annotated with known cell-type markers using uniform manifold approximation and projection (UMAP) [20]. At E12, when SMG development begins and the endbud forms, Hs3st1, Hs3st3a1, Hs3st3b1, and Hs3st6 were mainly expressed in the endbud epithelial cells. Hs3st1, the most abundant isoform, was broadly expressed in all cell clusters. At E14 SMG, when branching morphogenesis occurs, Hs3st3a1, and Hs3st3b1 were expressed in the endbud and basal and Krt19+ duct clusters, whereas Hs3st1 was broadly expressed in all cell clusters. Hs3st6 was expressed in a few endbud and basal duct cells. At E16, the onset of cell differentiation, Hs3st3a1 and Hs3st3b1 were more highly expressed in developing myoepithelial cells, as well as some endbud and basal duct cells. Hs3st3b1 was also expressed in Krt19+ differentiated duct cells. Hs3st1 was expressed in endbud, myoepithelial cells, mesenchymal and endothelial cells. Hs3st6 was detected in basal ducts. At P1, when the gland starts functioning, Hs3st3a1, Hs3st3b1 and Hs3st1 continue to be expressed in myoepithelial cells. Expression of both Hs3st1 and Hs3st3b1 was detected in mitotic, Krt19+ duct, and Bpifa2+ proacinar cells. In addition, Hs3st1 was expressed in the mesenchymal cells whereas Hs3st6 was only detected in Smgc+ proacinar cells. In the adult SMG, Hs3st3a1 and Hs3st3b1 were both expressed in myoepithelial cells, Hs3st3b1 was expressed in intercalated ducts and Hs3st1 was broadly expressed in the intercalated duct, endothelial, Bpifa2+ proacinar, and Ascl3+ duct cells. Hs3st6 was localized in the Ascl3+ duct cells (Fig. 1).
Figure 1. DotPlot visualization of HS biosynthetic gene expression from scRNA-seq analysis of SMG development.
Embryo day(E)12 SMG, E14 SMG, E16 SMG, Postnatal day (P)1 SMG and adult SMGs. Cell identities listed on y-axis, showing unbiased gene expression per cell cluster identified by log Fold Change; HS biosynthetic genes (features) are listed along the x-axis. Dot size reflects the percentage of cells in a cluster expressing each gene; dot color reflects expression level. Epithelial clusters expressing both Hs3st3a1 and Hs3st3b1 are highlighted with a red box.
Other HS biosynthetic enzymes were also expressed in both the developing and adult SMG. Unlike Hs3sts, which showed spatial expression at various SMG stages, the majority of the other HS biosynthetic genes such as Ndst1, Ndst2, Glce, Hs2st1 and Hs6st1 were expressed in all cell clusters throughout development (Fig. 1). Ndst1 and Hs2st1 were the most abundantly expressed of all HS enzyme transcripts. Interestingly, Hs2st1 showed the highest expression of all transcripts in the myoepithelial cells at P1 and adult stages when the gland begins to secrete saliva.
Overall, the scRNAseq expression data shows that Hs3st3b1was more highly expressed than Hs3st3a1 throughout development, and both were expressed in the endbud and myoepithelial cells. Endbud cells are progenitor cells during development and myoepithelial cells are involved in acinar secretion and are progenitors during gland regeneration [21].
Generation of the Hs3st3a1 and Hs3st3b1 single knockout mouse
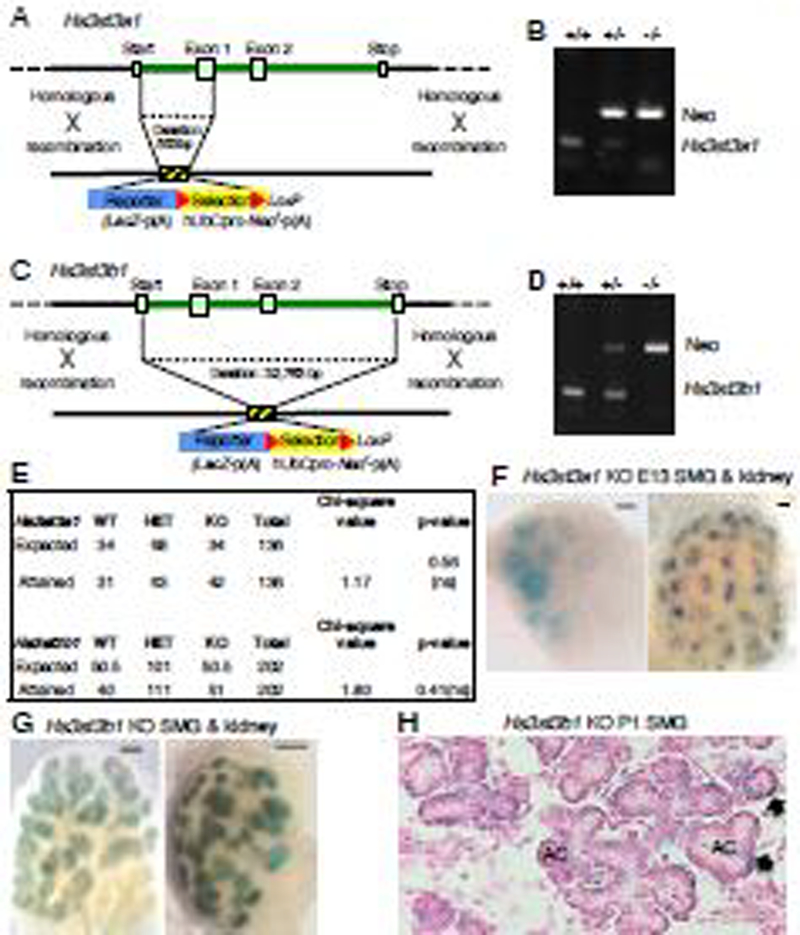
To investigate the function of Hs3st3a1 and Hs3st3b1 in vivo during SMG development, we used ES cells obtained from the KOMP Repository that used homologous recombination to partially delete these genes. The targeting vector contained a lacZ reporter followed by a floxed neomycin selection marker to replace part of the coding sequence. For Hs3st3a1, the insertion of a lacZ neo cassette created a 553bp deletion between positions 64,436,622 and 64,436,070 of chromosome 11 (Fig. 2A–B). For Hs3st3b1, the insertion generated a deletion of 32,763bp between positions 63702632 and 63735394 of chromosome 11 (Fig. 2C–D). Loss of allele analysis was performed by KOMP to confirm the mutants. Male chimeras from two independent ES cell clones were crossed with C57BL/6 females to establish heterozygous (HET) pups. Intercrossing of these heterozygous mice produced homozygous knockout (KO) offspring. Homologous recombination in offspring was confirmed by PCR based on the detection of either Hs3st3a1 or Hs3st3b1 gene and the detection of lacZ neo in the targeting cassette (Fig. 2C and D). Both Hs3st3a1 KO and Hs3st3b1 KO mice are viable, fertile, do not show any gross abnormalities and were obtained at the expected Mendelian ratio (Fig. 2E).
Figure 2. Generation of both Hs3st3a1 and Hs3st3b1 knockout mice.
Schematic and genotyping of the wild-type Hs3st3a1 (A-B) or Hs3st3b1 (C-D) locus, targeting vector and genotyping. A lacZ reporter with a floxed neomycin selection marker replaces most of the coding sequence of the gene. (B and D) Hs3st3a1 and Hs3st3b1 knockout mice genotyping. PCR amplified the WT allele (lower band) and detected the targeting vector (Neomycin cassette- upper band). (E) Data shown are the number of mice of a given genotype for each strain. Chi square analysis shows no significant difference from the expected numbers (F-G) Whole mount X-gal staining in SMG and kidney of E13 Hs3st3a1 KO (F) and Hs3st3b1 KO (G) embryos. Whole-mount X-gal staining was performed on E14 embryos showing SMG and kidneys with positive beta galactosidase staining. Scale bar; 100 μm. (H) Section of X-gal staining of Hs3st3b1 KO SMG in P1 SMG showing positive beta galactosidase staining in cells surrounding the acinar cells (AC). Black arrows point to myoepithelial cells.
When SMG from E13 and E14 embryos of Hs3st3a1 KO and Hs3st3b1 KO mice were stained with beta galactosidase, staining was detected in endbud cells of the SMG epithelium (Fig. 2F and G), consistent with the scRNAseq data. The kidneys from these embryos showed that the beta galactosidase was localized in the branching ureteric bud tips in the kidney. Similar pattern of localization for Hs3st3a1 and Hs3s3b1 mRNA has been shown previously by in situ hybridization in embryonic kidneys [22]. In sections of P1 Hs3st3b1 KO SMGs, beta galactosidase expression was localized in myoepithelial cells that surround the acinar structures (Fig. 2H), which is consistent with the scRNAseq showing expression of Hs3st3b1 in myoepithelial cells (Fig. 1). However, beta galactosidase expression was not detectable above background in P1 Hs3st3a1 KO SMGs, due to the high endogenous beta galactosidase background in SMGs.
There is reduced FGF10-dependent morphogenesis of isolated fetal SMG epithelia from both the Hs3st3a1 and Hs3st3b1 KO embryos
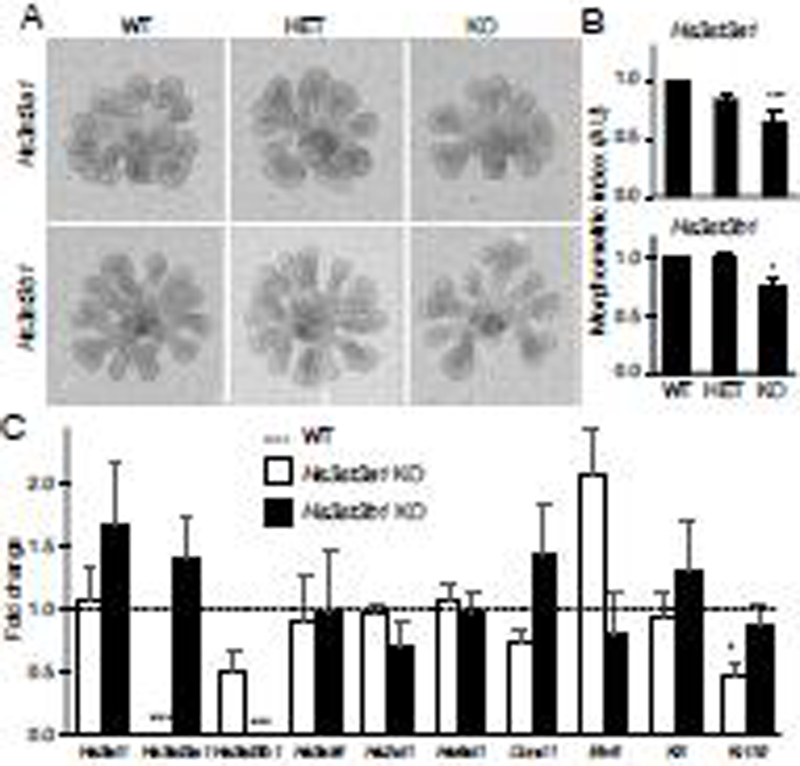
We had previously shown that addition of exogenous 3-O-sulfated HS increases FGF10-dependent growth of isolated fetal epithelia by increasing proliferation and FGFR2b-dependent downstream gene expression [10]. Therefore, epithelial rudiments were isolated from fetal E13 KO SMGs and cultured in laminin-111 matrix with FGF10 in a minimal media for 24-hours to test whether loss of Hs3st3 enzymes would affect FGF10-dependent signaling and epithelial morphogenesis (Fig. 3A). Morphogenesis was calculated as a morphogenic index, which is a product of the endbud number X width of endbuds X duct length in arbitrary units. A decrease in epithelial morphogenesis was observed with loss of either Hs3st3a1 or Hs3st3b1 compared to HET and WT epithelium (Fig. 3B).
Figure 3. There is reduced FGF10-dependent epithelial morphogenesis in both Hs3st3a1 and Hs3st3b1 KO SMG epithelia.
(A) Images of isolated epithelia from E13 SMGs treated with 400 ng/ml FGF10 for 24 hr. (B) Morphogenic index (number of endbuds × duct length × endbud width, in AU) decreases in Hs3st3a1 and Hs3st3b1 KO epithelia compared to the WT. Error bars represent ±SEM. ANOVA; * p<0.05 and *** p < 0.001, compared to WT.
(C) Deletion of Hs3st3a1 decreases expression of Krt19. Deletion of Hs3st3b1 does not affect expression of other genes tested. Gene expression was measured by qPCR and normalized to Rps29 and WT epithelia (dotted line). Error bars: SEM. Unpaired t-test; * p<0.05 and *** p < 0.001.
Analysis of gene expression in Hs3st3b1 KO isolated epithelia, showed reduced Hs3st3b1 as expected and no other changes in gene expression, although there was a trend of increased Hs3st1 and Hs3st3a1 expression (Fig. 3C), suggesting these isoforms may compensate for the loss of Hs3st3b1, and that this was only detectable when analyzing isolated epithelium. In contrast, in epithelial cultures of Hs3st3a1 KO, Hs3st3b1 expression was reduced. This may explain why there was a more significant reduction in epithelial morphogenesis than the Hs3st3b1 KO epithelia. (Fig. 3B). This may also suggest Hs3st3a1 regulates Hs3st3b1 expression, which remains to be explored. There was also no change in expression of Hs2st1 and Hs6st1, in either of the KO SMG epithelia. There was also a trend of increased Etv5 expression with Hs3st3a1 KO which suggests increased downstream FGFR signaling. Interestingly, there was reduced Krt19 expression, suggesting a relative reduction in duct differentiation (Fig. 3C). Overall, these data confirm that in an isolated epithelial culture assay, the loss of either Hs3st3a1 or Hs3st3b1 reduces FGF10-dependent growth.
Ex vivo branching of intact SMGs is only reduced in Hs3st3b1 KO mice
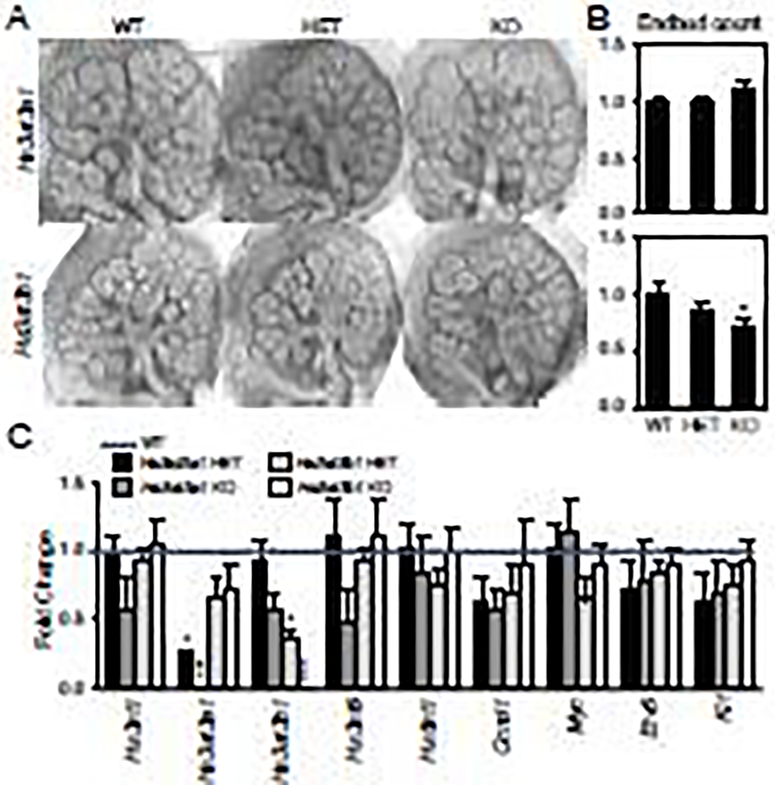
Isolated epithelial culture is dependent on exogenous FGF10 and a laminin 3D matrix, so we next investigated whether KO of Hs3st3a1 or Hs3st3b1 would affect SMG branching morphogenesis when the intact endogenous mesenchyme and basement membrane were present. The culture of fetal SMGs is dependent on HS sulfation and ex vivo culture in minimal media may exacerbate subtle phenotypes that are compensated for in vivo. E13 SMGs from Hs3st3a1 and Hs3st3b1 KO embryos obtained from heterozygous crosses were cultured for 48 hours and branching morphogenesis was calculated as the number of endbuds at 48 hours/number of endbuds at 1 hour (Fig. 4A and B). Freshly dissected SMGs showed no differences in endbud number between the Hs3st3a1 wildtype (WT), HET and KO glands, whereas SMGs from Hs3st3b1 KO embryos had fewer endbuds than WT. Furthermore, ex vivo culture of Hs3st3a1 SMGs for 48 hours did not show any differences in the bud count ratio among the three genotypes (Fig. 4A–B). However, branching morphogenesis was reduced in Hs3st3b1 KO compared to the WT SMGs, whereas the HET glands were not affected (Fig. 4A–B). This data suggests that deletion of Hs3st3b1, which is more highly expressed in the endbud than Hs3st3a1, reduces branching morphogenesis of E13 SMGs in ex vivo culture.
Figure 4. Branching morphogenesis of embryonic E13 SMGs from Hs3st3b1, but not Hs3st3a1, KO mice is reduced in ex vivo culture.
(A) Light micrographs of SMGs isolated from E13 embryos and cultured ex vivo for 48 hours. (B) Quantification of the number of endbuds (expressed as a ratio of the number at 48h/the number at 1h) of WT, HET and KO SMGs. Error bars; SEM. One-way ANOVA compared to WT, * p < 0.05, ** p < 0.01. (C) Gene expression changes was normalized to WT control (dotted line) and Rps29. Error bars: SEM. One-way ANOVA compared to WT, * p < 0.05, ** p < 0.01 and *** p < 0.001.
Analysis of gene expression by qPCR was used to determine if deletion of either gene resulted in compensation by expression of other Hs3sts. As expected, Hs3st3a1 expression was reduced in the HET and not detected in the KO, and no transcriptional changes for the other Hs3sts were detected (Fig. 4C). In addition, a reduction in Hs3st3b1 expression was detected in the HET and lack of detection in the KO, and the expression of other Hs3sts was not affected (Fig. 4C).
We also analyzed expression of markers associated with FGFR2 signaling (Etv5), proliferation (Ccnd1 and Myc) and a progenitor cell marker (Kit), which we previously showed to be involved in 3-O-HS/FGF10/FGFR2b mediated gene expression, but these were not affected significantly in the SMG cultures (Fig. 4C). Together, these data show that loss of Hs3st3b1, but not Hs3st3a1, reduces branching morphogenesis in ex vivo culture, without affecting gene expression of other sulfotransferase or genes related to FGFR2b signaling and proliferation. Since no significant differences were observed in the heterozygous SMGs compared to WT, we continued to perform subsequent experiments with only the KOs.
Expression of 3-O-sulfated HS epitopes is reduced in Hs3st3b1 KO SMGs
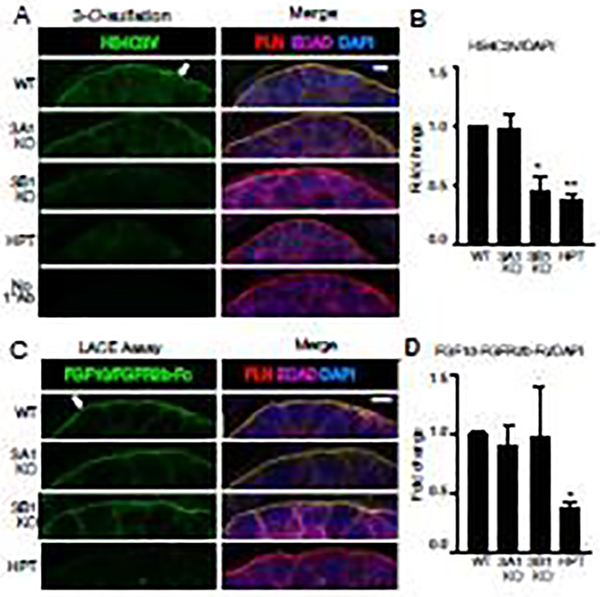
Since the Hs3st3a1 and Hs3s3b1 enzyme add 3-O-sulfated epitopes to HS, we next determined whether these epitopes were reduced in the fetal Hs3st3a1 and Hs3st3b1 KO SMGs. Due to lack of specific antibodies to detect the Hs3st enzymes or the complex range of 3-O-sulfated epitopes, we used a single-phage display anti-HS antibody, HS4C3V, which detects a 3-O-sulfated HS epitope in kidney, lung and liver [23]. We co-stained with antibodies to perlecan (PLN), which detects the basement membrane, and E-cadherin (ECAD), an epithelial cell-cell junction marker, and quantitated the fluorescence. Staining with the HS4C3V antibody in the fetal WT SMGs was localized to the basement membrane (shown by the arrow) and at epithelial cell-cell surfaces (Fig. 5A). The specificity of the antibody staining was validated by treating the SMGs with heparinase III enzyme, which removes HS and abolishes the staining to the basement membrane and the epithelial cell surfaces (Fig. 5A and B). In addition, SMGs stained with only the secondary antibody were devoid of fluorescence. HS4C3V staining was quantified and found to be markedly reduced in the Hs3st3b1 KO SMGs both at the basement membrane and cell surface (Fig. 5A and B) suggesting Hs3st3b1 is involved in generating the epitope recognized by HS4C3V. Interestingly, we could not measure a reduction in the epitope in the Hs3st3a1 KO SMGs, although it is possible that a subtle reduction in an epitope is not detectable or that Hs3st3a1 is expressed at a low level and its loss is compensated for by Hs3st3b1 or another enzyme.
Figure 5. 3-O-sulfation of HS in the basement membrane of embryonic SMGs is reduced in Hs3st3b1 KO but FGF10/FGFR2b complex binding is not altered.
(A) Representative single confocal images of HS4C3V (green), perlecan (red), E-cadherin (magenta) and nuclei in blue. Scale bar: 5 μm. (B) Quantification of fluorescence intensity normalized to total nuclei staining and expressed as a fold change compared to WT. At least five SMGs from three independent experiments were imaged and used for quantification. Error bars: SEM. One-way ANOVA compared to WT, * p < 0.05, ** p < 0.01 and *** p < 0.001. (C) Representative images of single confocal sections showing FGF10-FGFR2b-Fc complex (green), perlecan (red), E-cadherin (magenta) and nuclei (blue). Scale bar: 5 μm. (D) Quantification of fluorescence intensity normalized to total nuclei staining and expressed as a fold change compared to WT. At least five SMGs from three independent experiments were imaged and used for quantification. Error bars: SEM. One-way ANOVA compared to WT, * p < 0.05, ** p < 0.01 and *** p < 0.001. White arrows point to basement membrane.
Binding of FGF10/FGFR2b complex to endogenous HS is not altered in Hs3st3a1 or Hs3st3b1 KO SMG epithelium
Another method to analyze changes in endogenous HS structures, is the ligand and carbohydrate engagement (LACE) assay, which maps the binding of an exogenous growth factor-growth factor receptor complex to the endogenous HS in a tissue section. We used recombinant FGF10 and FGFR2b-Fc for the LACE assay. We have previously shown that 3-O-sulfated HS enhanced their complex formation in a pulldown assay [10]. The LACE binding was detected with an anti-human Fc antibody. FGF10/FGFR2b-Fc binding was detected on the basement membrane (shown by the arrow) that surrounds the epithelial buds and on the epithelial cell surface. We co-stained with antibodies to PLN and ECAD, and quantitated the fluorescence. Pretreatment of SMGs with heparinase III enzyme, which degrades HS, reduced FGF10/FGFR2b-Fc complex binding, but did not affect PLN and ECAD staining, confirming the specificity of the complex binding to HS (Fig. 5C and D). However, we could not measure differences in binding of FGF10/FGFR2b complex in either Hs3st3a1 KO or Hs3st3b1 KO SMGs compared to WT (Fig. 5C and D). Taken together, these data suggest that although there was a reduction of 3-O-sulfated epitopes (HS4C3 staining) in the Hs3st3b1 KO SMG the changes in 3-O-sulfation did not alter FGF10/FGFR2b-Fc binding to the endogenous HS. This may also suggest that the HS-epitope required for FGF10-FGFR2b binding is not the most highly 3-O-sulfated, or that it is generated by other enzymes with the loss of a single Hs3st3 isoform.
Both Hs3st3a1 and Hs3st3b1 KO adult SMGs appear similar to WT.
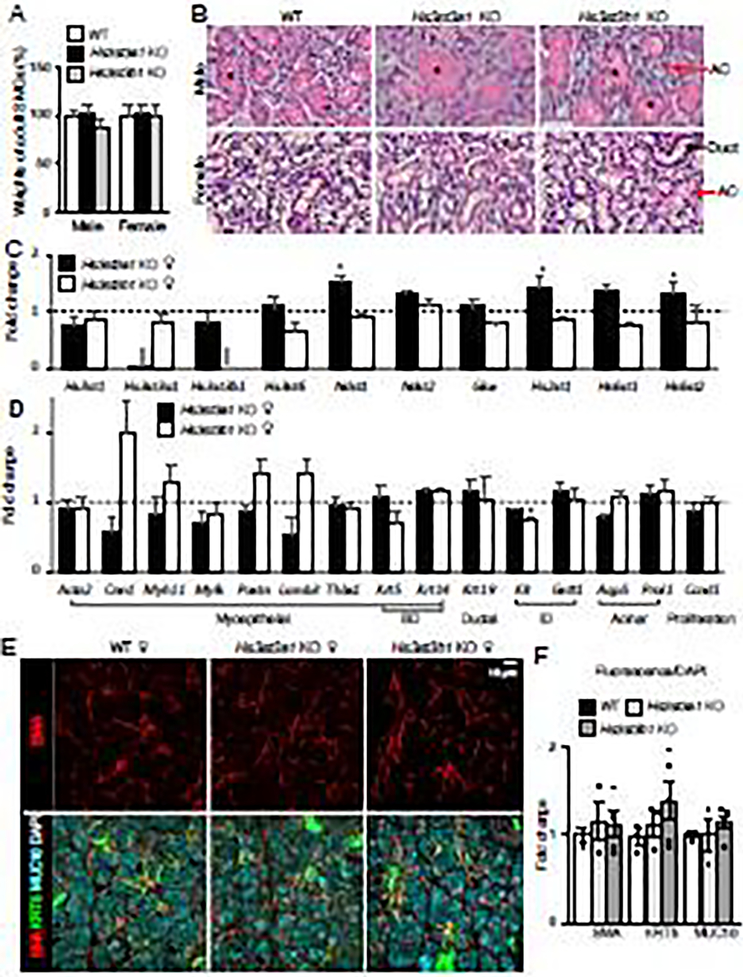
We next analyzed the histology of adult salivary glands from the KO mice and compared to the WT littermates. The SMGs of mice are sexually dimorphic with the male glands containing prominent granular ducts (indicated by the asterisks). The weights of the adult SMGs and the gross histological appearance of the SMGs from the KO mice were comparable to the WT in both males and females (Fig. 6A and B). Gene expression analysis was performed on the adult SMGs to determine if there were differences in expression of the epithelial Hs3sts and if there was any transcriptional compensation. As expected, deletion of either Hs3st3a1 or Hs3st3b1 gene resulted in the loss of the transcripts in all the KOs (Fig. 6C and Supplementary Fig.1A). Interestingly, expression of Hs3st3b1 was reduced in Hs3st3a1 KO male glands (Supplementary Fig.1A), but not in the female (Fig. 6C). Hs3st3a1 female KO mice showed upregulation in expression of both Ndst1, Hs2st1 and Hs6st2, which are HS enzymes that modify N, 2-O- and 6-O-sulfation on HS, respectively (Fig. 6C). However, this finding was not observed in the male Hs3st3a1 KO SMGs. Genes associated with cell clusters that expressed Hs3st3a1 and Hs3st3b1 transcripts in the adult SMG such as myoepithelial cells (Acta2, Cnn1, Myh11, Mylk, Postn, Lamb3, Thbs1, Krt5 and Krt14), basal duct cells (Krt5 and Krt14), ductal cells (Krt19), intercalated duct (ID) cells (Kit, and Gstt1) and acinar cells (Aqp5 and Prol1) were also screened. No changes in expression of these genes were detected except Kit was decreased in the Hs3st3b1 KO adult female SMG (Fig. 6D) and the myoepithelial marker, Cnn1 was decreased in Hs3st3b1 KO adult male SMG (Supplementary Fig. 1D) when compared to the control. Although not significant, decreasing trends of Kit were detected in the other KO SMGs. Interestingly, both adult myoepithelial and intercalated duct cells also express higher levels of Hs3st3b1 (Fig. 1) suggesting the loss may have subtle effects on cell differentiation. Together, this indicates that even though gross histology is similar to WT, there might be subtle changes in specific cell populations due to loss of either Hs3st3a1 or Hs3s3b1. The data also suggest that there may be sex-dependent differences in cellular responses to modifying HS in sulfotransferase KOs, although a more detailed analysis of HS differences in specific cell types would be needed.
Figure 6. Adult Hs3st3a1 and Hs3st3b1 KO SMGs are similar in size and histology as the WT but show differences in HS biosynthetic enzyme transcripts.
(A) Comparison of the weights of the adult SMGs from males and females show no difference. Weights of SMGs are normalized to the weight of the mouse and then normalized to that of the wildtype and expressed as a %. (B) H&E staining of the adult SMG show no gross histological differences between the WT, Hs3st3a1 KO and Hs3st3b1 KO in both male and female. Black asterisks label granular ducts, black arrows point to ducts, and red arrows point to acinar cells. (C-D) Gene expression of HS biosynthetic enzymes and markers of different cell types in adult female SMGs. Gene expression was normalized to wildtype control (dotted line) and Rps29. Error bars: SEM. Unpaired t-test compared to WT, * p < 0.05, ** p < 0.01 and *** p < 0.001. (E) Representative maximum projection images of confocal sections from WT, Hs3st3a1 KO and Hs3st3b1 KO female SMGs showing KRT5 (green), SMA (red), MUC10 (cyan) and nuclei (gray). Scale bar: 10 μm. (F) Quantification of fluorescence intensity normalized to total nuclei staining and expressed as a fold change compared to WT. At least three SMGs with a minimum of 5 different positions were imaged and used for quantification. Error bars: SEM. One-way ANOVA compared to WT, not significant.
Next, immunostaining was performed to determine if changes were observed in myoepithelial cells, where both Hs3st3a1 and Hs3st3b1 were expressed. SMG sections were stained for detection of smooth muscle actin (SMA), a marker of myoepithelial cells transcribed by Acta2, mucin10 (MUC10), an acinar marker transcribed by Prol1, and keratin 5 (KRT5), which marks both myoepithelial and basal duct cells. No obvious differences in protein expression or localization were detected between the WT and Hs3st3b1 KO SMGs (Fig. 6E and 6F and Supplementary Fig. 1C and 1D). The trend in increased SMA and KRT5 staining in KO myoepithelial cells may reflect a subtle change in differentiation state, although this remains to be explored.
Hs3st3a1 KO SMGs have decreased N-sulfated but increased non-sulfated HS disaccharides, whereas HS disaccharides are not altered in Hs3st3b1 KO SMGs
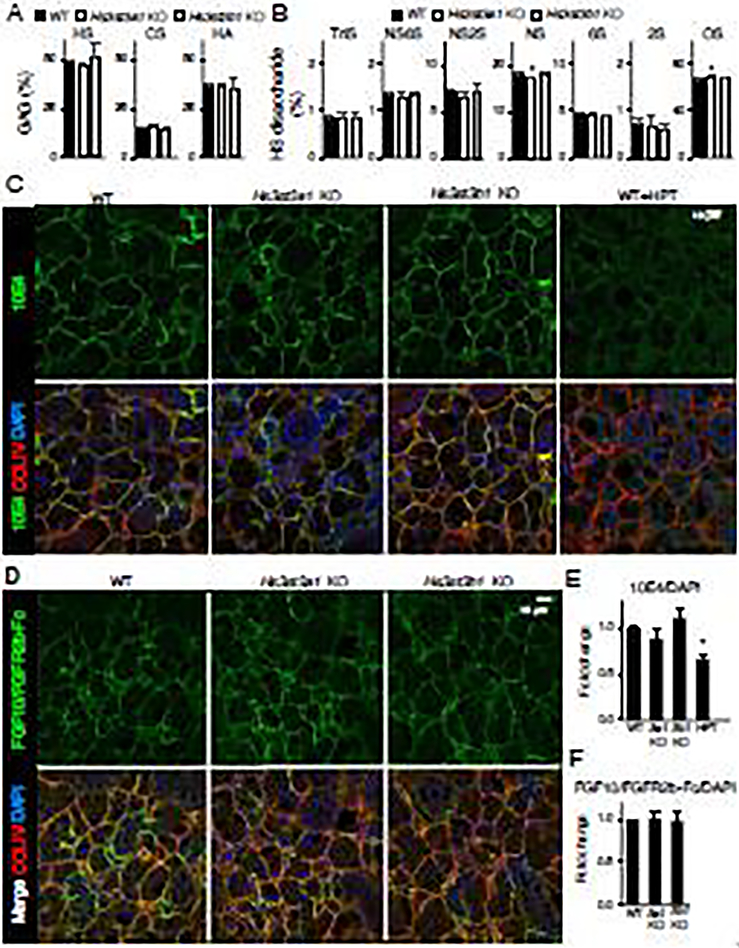
We next determined if there were changes in the composition of total HS and others glycosaminoglycans (GAGs) such as chondroitin sulfate (CS), and hyaluronic acid (HA) in the SMGs of adult Hs3st3a1 or Hs3st3b1 KOs compared to WT. Analysis shows that all the SMGs from the WT, Hs3st3a1 KO and Hs3st3b1 KO contain very similar composition of total GAGs. No differences were detected in the total levels of HS, CS and HA between the KOs and WT. HS was the most abundant of the three GAGs analyzed accounting for ~50 % of the total GAGs while the HA and CS accounted for ~35 % and ~15 %, respectively (Fig. 7A).
Figure 7. Composition of total GAGs, HS disaccharides and HS staining in adult SMGs.
Analysis of total GAG (A) and HS disaccharide (B) composition between WT, Hs3st3a1 KO and Hs3st3b1 KO SMGs. All data were averaged. Error bars: SD. One-way ANOVA compared to WT, * p < 0.05. N = 4 for each group. (C) Representative images of single confocal sections from WT, Hs3st3a1 KO and Hs3st3b1 KO SMGs showing 10E4 (green), ColIV (red), and nuclei (blue). (D) Representative images of single confocal sections from WT, Hs3st3a1 KO and Hs3st3b1 KO SMGs showing FGF10-FGFR2b-Fc complex (green), ColIV (red), and nuclei (blue). (E-F). Scale bar: 10 μm. Quantification of 10E4 (E) and FGF10/FGFR2b-Fc (F) fluorescence intensity normalized to total nuclei staining and expressed as a fold change compared to WT. Three SMGs were imaged and used for quantification. Error bars: SEM. One-way ANOVA compared to WT, * p < 0.05 or not significant.
To further characterize the HS changes in the SMGs, disaccharide composition analysis was conducted using liquid chromatography mass spectrometry (LCMS) following treatment of the GAGs with heparin lyases. It is important to note that current LCMS analysis is not able to resolve 3-O-sulfated disaccharides, therefore this analysis is an attempt to see if loss of either enzyme has an effect on the percentages of disaccharides within the HS. The WT and Hs3st3b1 KO exhibited similar HS disaccharide composition. However, Hs3st3a1 KO HS had a decrease in N-sulfated (NS) and an increase in non-sulfated (OS) disaccharides (Figure 7B). This suggests that deletion of Hs3st3a1 affects the sulfated composition of the HS. This is interesting because analysis of HS biosynthetic enzyme expression also suggested that in the Hs3st3a1 KO there were transcriptional increases in other sulfotransferase enzymes (Ndst1, Hs2st1 and Hs6st2). It is not clear how transcriptional changes in sulfotransferases directly correlate with HS disaccharide composition. Additionally, analysis of the CS disaccharides was also performed, and no differences were observed between the WT and the knockout SMGs suggesting there is no compensatory change in CS when a either Hs3st3a1 or Hs3st3b1 are deleted (Supplementary Fig. 2).
In order to further determine whether the distribution of HS was altered in the knockout mice, antibody staining was performed using 10E4 antibody, which reacts with an epitope containing N-sulfated glucosamine residues present in HS [24]. The antibody bound to the basement membrane, as shown by collagen IV staining, in the adult SMG and this binding was abolished following heparinase III (heparitinase I) treatment (Fig. 7C and E). No gross differences were observed in the 10E4 HS antibody staining between the WT, Hs3st3a1 and Hs3st3b1 KO SMGs (Fig. 7D). Further confirming there was no reduction in HS in the KO SMGs.
We next used the LACE assay to analyze the binding of FGF10/FGFR2b complex to endogenous HS in the adult tissue. In the adult SMG, binding of FGF10/FGFR2b complex was localized to the basement membrane surrounding the acinar and ductal structures as the binding colocalized with collagen IV (Fig. 7E). However, no differences were observed in the binding of the complex to endogenous HS between WT, Hs3st3a1 and Hs3st3b1 KO SMGs (Fig. 7F).
Together, these biochemical and immunostaining data suggests that loss of Hs3st3a1 or Hs3st3b1 in adult gland does not affect the overall GAG composition of HS, CS and HA. Although there was a reduction in N-sulfated and an increase in non-sulfated HS disaccharides in the Hs3st3a1 KO SMGs, no differences were observed in staining of 10E4 antibody and FGF10/FGFR2b complex binding to endogenous HS. Overall, the Hs3st3b1 KO SMGs were comparable to WT SMGs in their HS disaccharides, and 10E4 antibody and FGF10/FGFR2b complex binding to the basement membrane HS.
Hs3st3a1 and Hs3st3b1 KO drink significantly more water than the wildtype mice, but stimulated saliva flow is not affected
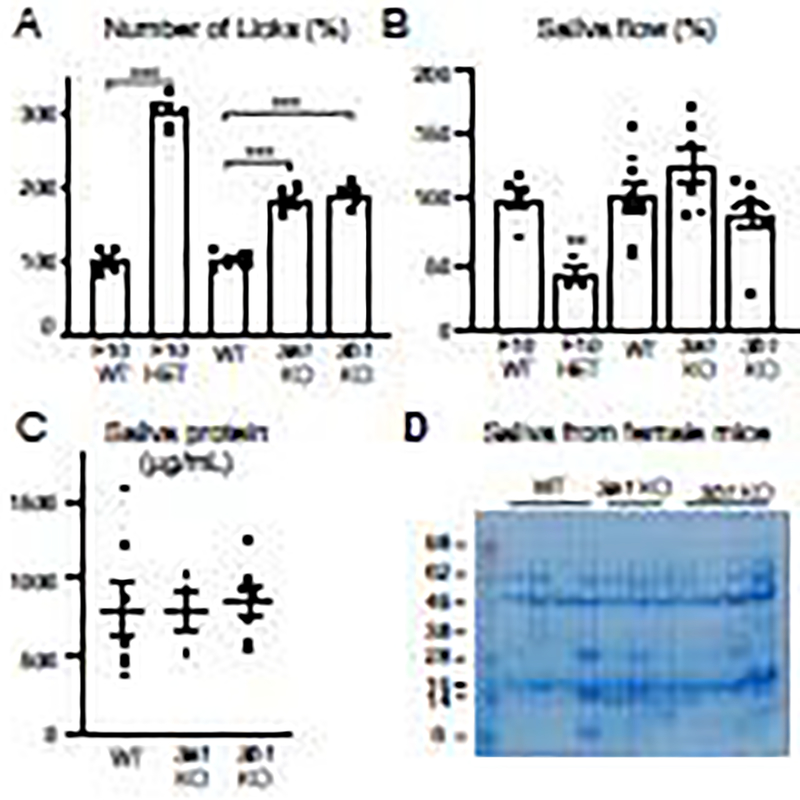
The changes we detected in both fetal and adult gene expression, 3-O-sulfated epitope staining and disaccharide analysis, and potential for alterations in myoepithelial function led us to analyze adult salivary gland function in the knockout mice. Salivary function involves both a basal level of secretion, primarily from the SMGs that maintain oral moisture, and stimulated secretion, that involves neuronal input and a large volume of fluid secretion primarily from the parotid glands. It is difficult to reliably measure the low basal saliva volume in mice, as mice must be handled and or anesthetized, which may both alter salivary secretion. Therefore, we assessed basal salivary function by monitoring the drinking behavior of the mice using a Lickometer test. As a positive control in this assay, we used Fgf10 HET mice, which have salivary gland hypoplasia, reduced saliva production, drink more water than the wild-type controls and are proposed as a model for xerostomia [25], although xerostomia in humans is a subjective perception of dry mouth. Adult mice were tested for 1 h using the Lickometer with free access to water and the number of licks were recorded. As expected, the Fgf10 HET mice showed significant increase in the number of water licks compared to their WT littermate controls (Fig. 8A). Surprisingly, both Hs3st3a1 KO and Hs3st3b1 KO also showed significant increase in their drinking behavior compared to the WT littermates (Fig. 8A). These data suggest that basal salivary gland function or possibly saliva composition may be affected in these knockout mice.
Figure 8. Hs3st3a1 and Hs3st3b1 KO mice drink significantly more water than the wildtype mice, but saliva flow and salivary proteins are not affected.
(A) Female adult mice were tested for 1 h using the Lickometer with free access to water and the number of licks are expressed as a percentage to their respective WT controls for each mouse strain. Data is presented as mean of four mice per groups that was tested 5 different times. Error bars: SD. One-way ANOVA compared to WT, * p < 0.05, ** p < 0.01, *** p < 0.001 and **** p < 0.0001. (B) Salivary flow rates in adult female mice were determined as described in Materials and Methods. Whole saliva was collected after pilocarpine stimulation. Saliva flow normalized to the WT and shown as %. Mean ± SEM. Dots represent saliva measurements of individual mice. Unpaired t-test for FGF10 HET, ** P < 0.01, as compared to FGF10 WT. One-way ANOVA for Hs3st3a1 KO and Hs3st3b1 KO compared to WT, not significant (C) Quantification of protein concentration in saliva from female mice assessed using BCA assay. Graph shows Mean ± SEM. Dots represent measurements of individual mice. No significant differences detected compared to WT littermates.
To determine whether there are changes in stimulated salivary function we measured whole saliva production after pilocarpine stimulation in anesthetize mice. This was expressed as a ratio of saliva volume to body weight, then normalized to that of the WT littermates for each strain. As expected, Fgf10 HET mice showed reduced saliva flow in both male and female compared to the wildtype littermates (Fig. 8B and Supplementary Fig. 3A). No significant changes in stimulated saliva production were measured in either Hs3st3a1 or Hs3st3b1 knockout in both male and female compared to the wildtype mice (Fig. 8B and Supplementary Fig. 3A), also suggesting the innervation of the gland is not affected in the KO mice. In addition, evaluation of total protein concentration in the stimulated saliva did not show any differences between the WT and KO mice (Fig. 8C and Supplementary Fig. 3B). Similarly, no obvious differences were observed in the protein profiles on Coomassie gels of the stimulated saliva from mice of the three genotypes, although there is individual variability among mice (Supplementary Fig. 3C and D).
Together these data show that loss of Hs3st3a1 and Hs3st3b1 enzymes results in increased drinking behavior in the mice, similar to xerostomia, a clinical condition when a patient’s perception of a dry mouth may result in taking frequent sips of water. The role of myoepithelial cells in regulating basal secretion from SMGs is not well-studied and it is assumed they play a mechanical role, wrapping around acinar cells and contracting, resulting in the expulsion of saliva into the oral cavity. The role of myoepithelial cell HS in this process is not known and remains to be investigated in the future.
Discussion
Here, we knocked out two of the seven isoforms of HS 3-O-sulfotransferases to compare their function during both SMG development and during adult homeostasis. Both enzymes are expressed in the basal duct and myoepithelial cells throughout development, with Hs3st3b1 having higher expression than Hs3st3a1. Our analyses reveal that FGF10-dependent epithelial morphogenesis is reduced in ex vivo culture in both KO’s and that branching morphogenesis of the intact gland is reduced in the Hs3st3b1−/− SMGs along with reduced staining for 3-O-sulfated HS in the basement membrane. Analysis of HS content highlighted that the composition of HS disaccharides in Hs3st3a1−/− SMG HS is altered, although both enzymes have similar sulfotransferase domains and are thought to act on the same types of disaccharides, there may be functional redundancy occurring with loss of Hs3st3b1 that maintains HS sulfation. We analyzed the transcription of HS biosynthetic enzymes during development, which suggests some transcriptional compensation, mainly in the Hs3st3b1−/− HS. We analyzed the function of adult salivary glands measuring drinking behavior, which suggests they have xerostomia. We propose that reduced 3-O-sulfation of the myoepithelial basement membrane reduces SMG basal secretory function. We begin to unravel the complexity 3-O-sulfated HS and its biological functions.
It is not surprising that loss of a single 3-O-sulfotransferase would be compensated for by other sulfotransferase enzyme family members. We showed that siRNA-mediated knockdown of four Hs3st isoforms (Hs3st1, Hs3st3a1, Hs3s3b1 and Hs3st6) was needed in ex vivo SMG epithelia to reduce FGF10-dependent morphogenesis, and that individual knockdown of either Hs3st3a1 or Hs3st3b1 was without effect [10]. In terms of 3-O-sulfated HS biology and the need to generate highly sulfated epitopes on HS to regulate function we know that the enzyme isoforms are classified into two subgroups due to the function of the 3-O-sulfated epitope [4]; HS modified by Hs3st1 and Hs3st5 have anticoagulant activity whereas HS modified by Hs3st2, Hs3st3 (a1 and b1), Hs3st5 and Hs3st6 are receptors for herpes simplex virus [7, 26, 27]. In addition, we are not able to simply cross the Hs3st3a1−/− and Hs3st3b1−/− mice to generate a double KO, due to the unlikelihood of interallelic crossover between the two mutant alleles that arose as a gene duplication about 500kb apart on chromosome 11. Interestingly, a double knockout of Hs3st3a1 and Hs3st3b1 generated by Crisper is viable, and fertile and has a partially penetrant kidney phenotype [28], however no analysis of SMG growth or HS was reported. Knockout mice with multiple Hs3st isoforms deleted may present with more developmental phenotypes, although, compensation from other Hs3sts as shown by the scRNAseq DotPlots of the five SMG stages (Fig. 1) may confound analysis. Thus, we plan to generate multiple isoforms KO’s.
Similarly, the KO of Hs3st1, which was thought to be the primary enzyme responsible for the assembly of the 3-O-sulfated AT-binding sequence, had mild phenotypes and coagulation was not affected. Instead, they experience intrauterine growth retardation and spontaneous eye degeneration, which is dependent on the mouse background [29]. It is also reported that Hs3st1 KO mice exhibit a pro-inflammatory phenotype being more susceptible to LPS- and TNF-induced death [30]. Mice deficient in Hs3st2 are viable and do not display any abnormalities [31]. The mild or lack of phenotypic defects associated with deletion of individual Hs3st enzymes may suggest that other Hs3sts can functionally compensate. Although, we did find some transcriptional compensation in the expression of other sulfotransferases, but not Hs3sts, when Hs3st3a1 was deleted in the adult (Fig. 6 and Supplementary Fig. 1). However, the regulation of HS sulfation may not only be due to increased gene transcription but may involve enzymatic compensation of another enzyme if multiple isoforms are expressed in one cell type. In addition, it is not clear if changes in overall sulfation, i.e., charge of HS selectively alters sulfotransferase expression. A future goal is to systematically generate mice with multiple 3-O-sulfotransferase knockouts to investigate specific functions in specific cell types.
Although adult Hs3st3a1 or the Hs3st3b1 mice displayed fairly normal SMG phenotypes in vivo, when isolated fetal SMG epithelia were cultured ex vivo with FGF10, laminin-111 ECM and minimal media, we measured a reduction in FGF10-dependant growth. Since multiple growth factors can enhance epithelial proliferation in vivo, it was necessary to culture the epithelia in minimal conditions to exacerbate the epithelial phenotype. It is possible that these knockout mice may present with observable phenotypes if subjected to stress or damage, such as gland irradiation, ductal ligation or partial gland extirpation. An example of this occurs in the FGF1 KO mouse, which displays no significant phenotype [32], but develops an aggressive diabetic phenotype coupled with an aberrant adipose expansion when challenged with a high-fat diet [33].
An increase in the number of water licks was recorded with the KO mice suggest may have dry mouth or xerostomia. This could be due to a reduction in unstimulated salivary flow, which is mainly due to SMG secretions, whereas stimulated saliva flow is due to parotid secretion. Basal salivary flow is produced mainly by SMG acinar cells with mechanical help from myoepithelial cells, both of which are innervated by parasympathetic fibers from the superior salivatory nucleus. Whereas stimulated saliva is produced by parotid glands, innervated by parasympathetic fibers from the inferior salivatory nucleus [34]. A clinical example where a lower basal saliva flow occurs was reported in patients with burning mouth syndrome [35], although, their stimulated saliva flow was not affected. Other causes of an increase in water intake could be diabetes mellitus or Huntington’s disease. Increased thirst and drinking have been reported in Huntington’s disease patients and the R6/2 mouse model of Huntington’s disease [36]. Although this may be a result of increases in blood glucose levels due to diabetes. It is possible that the Hs3st3a1 and Hs3st3b1 may have a subtle pancreas or kidney phenotype, which was not grossly apparent. Both Hs3st3a1 and Hs3st3b1 are enriched in progenitor cells within the ureteric branch tips of the arborizing ureteric epithelium of the fetal kidney [22]. Similarly, when kidneys from E14 Hs3st3a1 and Hs3st3b1 KO mice were stained with beta galactosidase, LacZ staining was detected in the ureteric branch tips of kidneys (Fig. 2F–G). The embryonic kidneys in the Hs3st3a1:Hs3st3b1 double knockout form variable double ureters, however this phenotype was observed with a low penetrance [28].
We conclude that generation of knockout mice where multiple Hs3st isoforms are deleted may present with abnormal developmental phenotypes which will enable better understanding of how 3-O-sulfated HS controls HS composition and regulates progenitor cell proliferation. Further analysis of other organ systems and crossing these mice with other sulfotransferase knockout mice will be important to explore the role of the 3-O-sulfate HS code in organ growth and function. Understanding how sulfotransferase enzymes regulate HS function, particularly as co-receptor complexes with growth factors, may provide tools to improve therapeutic approaches to organ regeneration.
Materials and Methods
Evaluation of scRNAseq gene expression
SMGs from ICR and C3H control mice strains were previously analyzed by scRNAseq analysis and the ready to use rds objects corresponding to the salivary gland development atlas repository (GSE150327) were downloaded from the associated publication (https://sgmap.nidcr.nih.gov) [20]. R was used for scripting and the SEURAT package [37, 38] used for visualization of gene expression. The downloaded SEURAT objects contained integrated datasets from embryonic and postnatal submandibular glands. Individual time points were extracted using the SplitObject function from SEURAT and DotPlots were generated with the DotPlot function of the same package. A minimum Sample scripts for visualization of gene expression and exploration of this dataset were obtained from the associated GitHub repository (https://github.com/chiblyaa/Salivary-Gland-Development).
Generation of the Hs3st3a1 and Hs3st3b1 single knockout mice
Both Hs3st3a1 (VG19780) and Hs3st3b1 (VG13569) knockout mice strain were created from ES cell clones obtained from the trans-NIH Knock-Out Mouse Project (KOMP Repository) (www.komp.org) and generated by Regeneron Pharmaceuticals, Inc. Methods used to create the Velocigene targeted alleles have previously been described in [39]. Homologous recombination of this vector in ES cells produced a knockout allele with a PGK-neomycin cassette and a LacZ reporter gene inserted between the second and the Hs3st3b1 exons or a deleted sequence (553bp) directly downstream of the ATG site in Hs3st3a1. The targeting vector was introduced by electroporation in 129/Ola embryonic stem cells (ES14Tg2a). Embryonic stem cells containing the recombined allele were injected into C57BL/6 blastocysts and re-implanted into CD1 pseudo pregnant females. Germ-line chimeric males bearing the recombined allele was mated with C57Bl/6 female mice to obtain heterozygote pups.
Maintenance of mice
Fgf10 heterozygous mice were provided by Dr. M Lewandowski at National Cancer Institute, NIH, Frederick, USA. All mice were maintained and treated according to guidelines approved by the National Institute of Dental and Craniofacial Research and National Institutes of Health Animal Care and Use Committee.
Genotyping
DNA from mice tail snips was extracted at weaning (21 days) using Genotyping buffer containing 0.1M Tris PH7.5, 0.05M EDTA, 0.2 % (v/v) SDS, 0.2M NaCl and proteinase K solution incubated overnight at 55 °C and DNA precipitated with isopropanol. Dried DNA pellet was then resuspended in DEPC water. Amplification of the LacZ gene was performed using the LacInF primer (GGTAAACTGGCTCGGATTAGGG) and LacInR primer (TGACTGTAGCGGCTGATGTTG). To amplify the Hs3st3b1 gene, Hs3st3b1 Fwd (TCACAGCTCCGAATGAGACATC) and Hs3st3b1 Rev (CCCAGCGCCTACTGTCTTATC) primers were used. For Hs3st3a1 amplification, Hs3st3a1 Fwd (ACTGTCGCGCAGCATCTTCAG) and Hs3st3a1 Rev (GCCAGGCAGTAGAAGACGTAG) primers were used. Amplification of both genes was performed using cycling conditions recommended by the KOMP consortium. PCR reaction: 12.5mL 2× JumpStart REDTaq ReadyMix reaction mix (Sigma), 0.125mM of each primer with 100 ng of extracted DNA to a total of 25 mL with DEPC H20. PCR cycling parameters used were the same for the detection of the Hs3st3a1 and Hs3st3b1 mutants: Initial denaturing 94 °C for 5 min; Denaturing: 94 °C for 15 sec, annealing: 55 °C for 30 sec, extension: 72 °C for 40 sec. Repeat thirty cycles. Final extension: 72 °C for 5 min. PCR products were fractionated on a 2.0 % (w/v)TBE agarose gel with ethidium bromide. For the genotyping of Hs3st3b1, a 95 bp DNA band identifies wildtype mice, a 210 bp band identifies knockout mice and presence of both the 95 bp and 210 bp bands identify hemizygous mice.
For the genotyping of Hs3st3a1, an 81 bp DNA band identifies wildtype mice, a 210 bp band identifies knockout mice and presence of both the 81 bp and 210 bp bands identify hemizygous mice. For the genotyping of Hs3st3b1, a 95 bp DNA band identifies wildtype mice, a 210 bp band identifies knockout mice and presence of both the 95 bp and 210 bp bands identify hemizygous mice.
Genotyping of the FGF10 mice was performed using the primers (Neo Fwd- TATCGCCTTCTTGACGAGTTCTTCTGA and Fgf10 Rev- TATCGCCTTCTTGAC GAGTTCTTCTGA). PCR conditions that included an initial denaturing at 94 °C for 5 min; denaturing at 94 °C for 15 sec, annealing at 65 °C for 30 sec (decreased 1 °C every cycle), extension at 72 °C for 40 sec. Repeated for ten cycles. Denaturing at 94 °C for 15 sec, annealing at 55 °C for 30 sec, extension at 72 °C for 40 sec. Repeated thirty cycles. Final extension at 72 °C for 5 min. A single band was detected in DNA from FGF10 heterozygous and homozygous mice/pups. Homozygous FGF10 pups died at birth.
Ex Vivo SMG Organ Culture
Fetal SMGs were cultured on polycarbonate membranes (13 mm, 0.1-μm pore size) (Whatman) or mesenchyme-free SMG epithelia were obtained as previously described [40, 41]. The culture medium used for the isolated epithelium was DMEM/F12 (ThermoFisher Scientific) supplemented with streptomycin (100 μg/mL), vitamin C (150 μg/mL), transferrin (50 μg/mL) and FGF10 (400 ng/ml: R&D Systems). Glands were photographed at 1, 24 and 48 hr, and the morphogenic index (AU × 103) was measured as described previously [42]. Each experiment was repeated at least three times. Explants were either lysed for RNA or fixed for immunostaining. The endbud number was counted at the beginning of the experiment (T1) and at the end of the assay at 48-hours (T48) and expressed as a ratio (T48/T1) and normalized to the wildtype control. All experiments were repeated at least three times.
Quantitative PCR (qPCR)
Real time PCR was perform ed as previously described [42]. RNA was isolated from embryonic and adult SMGS using the RNAqueous-Micro and RNAqueous-4PCR kits (both from Thermo Fish er Scientific), respectively. cDNA was synthesized from DNAse-treated RNA using the iScript Reverse Transciption Supermix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) according to the manufacturer’s instructions. For qPCR, 1ng of embryonic cDNA or 10 ng of adult cDNA per well was used for amplification with 0.4 μM of primer mix and iQ SYBR® Green Super mix with fluorescein (Bio-Rad) using the CFX qPCR system (Bio-Rad). The qPCR primers were designed using Beacon Designer software (Premier Biosoft, San Francisco, CA) and the sequences have been listed in the Key Resource Table associated with this manuscript. Primers were validated to efficienciently amplify and generate a single amplicon. Gene expression levels were normalized to housekeeping gene Rsp 29 and wild type cDNA. Aleast three biological replicates for each group were processed in duplicates.
KEY RESOURCE TABLE.
| REAGENT/RESOURCE | SOURCE | IDENTIFIER | Dilution |
|---|---|---|---|
| Antibodies | |||
| HS4C3V single-chain HS Ab | Toin H. van Kuppevelt (University of Nijmegen, Netherlands) | N/A | 1:10 |
| Anti-VSV glycoprotein-Cy3 | Sigma | C7706 | 1:100 |
| Anti-heparan sulfate proteoglycan (perlecan) | Millipore Sigma | mAb1948 | 1:200 |
| Anti-E-cadherin (2AE10) | Cell Signaling | mAb#3195 | 1:100 |
| Anti-Smooth muscle actin | Millipore Sigma | A2547 | 1:200 |
| Anti-keratin 5 | Covance | PRB-160P | 1:2000 |
| Anti-Mucin10 (Prol1) | Everest Biotech | EB10617 | 1:200 |
| Anti-10E4 | Amsbio | 370255-1 | 1:100 |
| Anti-Collagen TIV | Millipore | AB769 | 1:100 |
| All dye-conjugated secondary antibodies | Jackson ImmunoResearch Laboratories | 1:200 | |
| Alexa Fluor® 488 AffiniPure Donkey Anti-Mouse IgM, μ chain specific antibody | Jackson ImmunoResearch Laboratories | 715-545-020 | 1:200 |
| Alexa Fluor® 488 AffiniPure F(ab’)2 Fragment Donkey Anti-Human IgG, Fcγ fragment specific antibody | Jackson ImmunoResearch Laboratories | 709-546-098 | 1:200 |
| Chemicals, recombinant proteins, and enzymes | |||
| DAPI (Dihydrochloride) | Millipore Sigma | 268298 | 1:10000 |
| Recombinant mouse FGFR2 beta (IIIb) Fc chimera protein | R & D | 708-MF-050 | 50 nM |
| Recombinant human FGF10 protein | R &D | 345-FG-025 | 50 nM |
| Heparinase III (heparitinase 1) Flavobacterium heparinum | Amsbio | AMS.HEP-ENZ III-S | 0.020 IU/mL |
| X-GAL | Millipore Sigma | 10745740001 | 20 mg/mL |
| Pronase® Protease, Streptomyces griseus | Millipore Sigma | 53702 | |
| proteinase K solution | Bioline | BIO-37084 | 0.5 mg/mL |
| Xylene substitute | Millipore Sigma | A5597 | |
| Mouse on Mouse blocking reagent | Vector Laboratories | MKB-2213-1 | 1:12.5 |
Whole mount immunofluorescence staining
Whole mount immunofluorescence was performed with cultured embryonic SMGs as previously described [10]. Briefly, SMGs were fixed with 4% (v/ v) PFA in PBS and permeabilized with 0.1 % (v/v) triton-X-100. SMGs were blocked for 1 hour at room temperature with 10% (v/v) heat-in activated donkey serum (Jackson Immuno Research Laboratories, Westgrove, PA, USA), 1% (v/v) BSA (Sigma, St Louis, MO, USA) and Mouse on Mouse blocking reagent (Vector Laboratories, Burlingame, CA, USA). Then incubated overnight at 4 °C with the appropriate primary antibody (anti-perlecan (Millipore Sigma, St Louis, MO, USA), anti-E-cadherin (Cell Signaling Technology, Danvers, MA, USA) and HS4C3V single-phage HS antibody) then washed with PBS containing 0.1% (v/v) Tween −20 (PBST) and incubated with the respective dye-conjugated secondary antibodies (all from Jackson Immuno Research Laboratories, Westgrove, PA, USA) an d DAPI stain (Millipore Sigma, MA, USA) for 1 hour at room temperature. Staining for anti-heparan sulfate single phage HS4C3 antibody (1:10) was performed as above and detected using monoclonal anti-VSV glycoprotein-Cy3 labeled antibody (P5D4) (Sigma, St. Louis, MO, USA). To evaluate specificity of HS4C3V antibody binding, SMGs were pretreated with 0.020IU/mL heparinase III (AMSBIO, Cambridge, MA, USA) in 0.01M Hepes (p H 7.4) with 0.15M NaCl and 5mM CaCl2 for 2hours at 37°C to degrade the HS. Following washes with PBST, SMGs were mounted with Fluro-Gel mounting medium (Electron Microscopy Sciences, Hatfield, PA, USA). All slides were imaged using the Zeiss 880 confocal microscope.
Fluorescence intensity of each antibody channel was quantified using ImageJ software (NIH) in the region of interest (ROI) that included the epithelial region within the basement membrane by measuring the integrated density. This was normalized to the fluorescence intensity of nuclei in the ROI, and then the data was normalized to control. An average of five images with five 1μm Z-stacks were taken per SMG and a minimum of three glands were used for each group.
Immunofluorescence staining of adult SMG
Immuno fluorescence staining on adult SMGs was performed using paraffin-embedded tissue sections as previously described [43]. Briefly, the adult glands were fixed with 4%(v/v) paraform aldehyde overnight at RT and were processed for paraffin-sections. Tissue sections were de-paraffinized three times for 5 minutes each in xylene substitute (Millipore Sigma, St Louis, MO, USA), followed by re-hydration in graded ethanol solutions (100%, 95%, 70%, 50% and 30% (v/v)with dH2O) for 5 minutes each, then subjected to heat-mediated antigen retrieval using R-universal epitope recovery buffer (Electron Microscopy Sciences, Hatfield, PA, USA) for 10 minutes at high-pressure. Following blocking with 10% (v/v) heat-in activated donkey serum (Jackson Immuno Research Laboratories, Westgrove, PA, USA), 1% (v/v) BSA (Sigma, St Louis, MO, USA) and Mouse on Mouse blocking reagent (Vector Laboratories, Burlingame, CA, USA), the following primary antibodies specific for SMA (Millipore Sigma, St Louis, MO, USA. #A2547), Keratin 5 (Covance, CA, USA), and Mucin 10/Prol1 (Everest Biotech, Oxford shire, UK) were used, and sections were then incubated with the respective dye-conjugated secondary antibodies (all from Jackson Immuno Research Laboratories, Westgrove, PA, USA) and DAPI stain (Millipore Sigma, MA, USA). A complete list of antibodies used and their respective dilutions is shown in the Key Resource Table associated with this manuscript.
For immunostaining with 10E4 antibody, paraffin-embedded samples were treated with Pronase Protease, Streptomyces griseus enzyme (Millipore Sigma, MA, USA) as previously described [44]. Briefly, rehydrated sections were treated for 10 minutes at 37 °C with 0.5 mg/mL Pronase to expose the HS epitopes, washed with PBS and blocked with 2% (v/v) BSA/PBS for 30 minutes. The sections were then incubated overnight at 4°C with 10E4 antibody and anti-collagen IV, then washed three times with PBS for 10 minutes each, and then incubated with Alexa Fluor® 488 Affin iPure Donkey Anti-Mouse IgM, μ chain specific and Alexa Fluor® 594 Affin iPure F(ab’)2 Fragment Donkey Anti-goat IgG (both from Jackson Immuno Research Laboratories, Inc) for 1h at room temperature to detect the 10E4 antibody and anti-collagen IV antibody, respectively. All slides were imaged using the Zeiss 880 confocal microscope. Similar settings for laser power and detector gain were maintained with i each experiment.
Fluorescence intensity of each antibody channel was quantified using the ImageJ software (NIH) by measuring the integrated density value of the region of interest (ROI) from at least five Z-stacks consisting of five 1μm slices from each group. The ratio of each fluorescent channel to the nuclei (DAPI) fluorescence was calculated and then the averaged ratio was normalized to that of the control. A minimum of three SMGs were imaged per group.
Ligand and carbohydrate engagement (LACE) assay of embryonic and adult SMGs
A modification of the LACE assay [45] was performed using recombinant mouse FGFR2 beta (IIIb) Fc chimera protein (referred to as rFGFR2b-Fc) and recombinant human FGF10 (all from R&D Systems Inc., MN) as previously described [46]. Embryonic SMGs were fixed, permeabilized and blocked as described earlier under “ Whole mount immunofluorescence staining”. Fetal SMGs were incubated overnight at 4 °C with 50 nM soluble recombinant rFGFR2b-Fc, 50 nM recombinant FGF10, anti-E-cadherin (Cell signaling) and anti-perlecan (Millipore Sigma). Whereas for adult glands, paraffin section were de-paraffinized and rehydrated as described earlier. Heat-activated antigen retrieval using a homemade Tris-EDTA pH 9 buffer prepared with Tris Base (1.21g), EDTA (0.37g) in 100 mL of distilled water was performed. The samples were blocked for 1 hr at RT with 10 % (v/v) heat-inactivated donkey serum, 1 % BSA (v/v), and 1.8 % (v/v) MOM IgG blocking reagent, incubated overnight at 4 °C with 50 nM soluble recombinant rFGFR2b-Fc, 50 nM recombinant FGF10, anti-E-cadherin (Cell signaling) and anti-collagen Type IV (Millipore). After washing Cy dye-conjugated secondary antibodies were added for 1hr. The rFGFR2b-Fc was detected with Alexa Fluor® 488 AffiniPure F(ab’)2 Fragment Donkey Anti-Human IgG, Fcγ fragment specific secondary antibody (Jackson ImmunoResearch Laboratories, Inc). Immunofluorescence was analyzed with a Zeiss LSM880 microscope.
Lickometer testing
The licking behavior in female mice was monitored for 1 h using a Lickometer (Habitest system, Coulbourn Instruments, Whitehall, PA), which is a computer-operated system as previously described [47]. Prior to the experiment, the mice underwent a training session for 20 minutes and then the number of licks of 4 mice were recorded for 1 h. Controls and knockout mice were tested at the same time and then retested 5 times on separate days.
Saliva collection
Mice were weighed and anesthetized with ketamine (60 mg/kg) and xylazine (8 mg/kg) intramuscularly. This was followed by a subcutaneous injection of pilocarpine at 0.25 mg/kg body weight to stimulate saliva secretion. Whole saliva was collected gravimetrically with a 75-mm hematocrit tube (Drummond) into 1.5 mL pre-weight Eppendorf tubes for 20 min. The amount of saliva collected was calculated as micrograms per gram of body weight and then normalized as percentage to the wildtype littermates.
X-GAL staining
The Hs3st3a1 or Hs3st3b1 genes were disrupted by targeted insertion of the lacZ gene. Consequently, expression of LacZ is driven by the Hs3st3a1 or Hs3st3b1 promoter, permitting detection of the endogenous their expression pattern. Beta galactosidase staining was performed as previously described [48]. Briefly, embryos or tissues were fixed in 0.2 % (v/v) glutaraldehyde in 2 % (v/v) PFA/PBS for 45 minutes at room temperature and rinsed with PBS. Then incubated with Tissue Rinse Solution A (Millipore) for 5 minutes at room temperature followed by a 30 minute wash with Tissue Rinse Solution B (Millipore). The samples were then incubated with X-GAL (20 mg/mL) diluted in fresh staining buffer (0.1 M phosphate buffer (pH7.5), 2 mM MgCl2, 0.02 % (v/v) NP-40, 20 mM Tris-HCl (pH7.5), 8 mM potassium hexacyanoferrate (II) trihydrate, 13 mM potassium hexacyanoferrate (III)) at 37°C, then washed in PBS and refixed in 4 % (v/v) PFA for 10 minutes.
GAG composition, HS and CS disaccharide analysis
GAGs from female mice SMGs were prepared and analyzed for HS and CS disaccharides as previously described in Sarnaik et al., 2019 and Yan et al.,2021 [49, 50]. Briefly, freeze-dried SMGs were proteolyzed at 55 °C with 10 mg/mL actinase E (Kaken Biochemicals, Japan), lyophilized and denatured using 8 M urea containing 2 % (v/v) CHAPS. The extract was passed over the Vivapure Q Mini H spin column, the column washed twice with 8 M urea/2 % (v/v) CHAPS buffer followed by three washes with 0.2M NaCl. GAG components were eluted with 16 % (v/v) NaCl then concentrated using 3-kDa molecular weight cut off spin column, washed with distilled water and lyophilized. GAGs recovered from 1 mg of freeze-dried tissue was used for digestion with either recombinant heparin lyases I, II and III (10 mU each) or chondroitin lyase ABC (10 mU) and disaccharide analysis.
The unsaturated standards for HS (ΔUA-GlcNAc; ΔUA-GlcNS; ΔUA-GlcNAc6S; ΔUA2S-GlcNAc; ΔUA2S-GlcNS; ΔUA-GlcNS6S; ΔUA2S-GlcNAc6S; ΔUA2S-GlcNS6S), CS (ΔUA-GalNAc; ΔUA-GalNAc4S; ΔUA-GalNAc6S; ΔUA2S-GalNAc; ΔUA2S-Gal-NAc4S; ΔUA2S-GalNAc6S; ΔUA-GalNAc4S6S; ΔUA2S-GalNAc4S6S) and HA (ΔUA-GlcNAc), where ΔUA is 4-deoxy-α-L-threo-hex-4-enopyranosyluronic acid (all purchased from Iduron, UK) as well as the dried disaccharides were labeled with 2-Aminoacridone (AMAC) (Sigma-Aldrich, St. Louis, MO, USA) and liquid chromatography- mass spectrometry analysis was performed as described in Sarnaik et al., 2019.
Statistical analysis
Data were log transformed before all statistical analysis and analyzed with an unpaired Student’s test (two-tailed) for comparing two groups or a One-way ANOVA with Dunnett’s multiple comparison test for more than two groups. Treatment groups were compared to the wildtype control using Prism 8 Software (GraphPad, La Jolla, CA, USA). Graphs show the mean ±SEM or ±SD for each group from three or more experiments.
Supplementary Material
| Primers | ||
|---|---|---|
| Gene | Forward primer | Reverse primer |
| Acta2 | GCATGGATGGCATCAATCAC | ACCTATCTGGTCACCTGTATGTA |
| Aqp5 | TCTACTTCTACTTGCTTTTCCCCTCCTC | CGATGGTCTTCTTCCGCTCCTCTC |
| Ccnd1 | CTTAATGTGATTACCGCTGTATTCC | CCTGACTGCTGTGATGCTATG |
| Cnn1 | CGCACAACTACTACAACTC | CCCAAACCGTAACCCTATA |
| Etv5 | AAGCCCTTCAAAGTGATAGCGGAGAC | GTGTCCACAAACTTCCTCTTTCTGTCAATC |
| Glce | CATTTCTTGAGAGGGAGTGAGCATTTGTTG | GCTTATGTATGTGACCGTGAAACCTGAAC |
| Gstt1 | GCGGCAGTTCACAACTCACAGTTCACAAT | TGGAGCTGAGCCTGAGAGCCATCAT |
| Hs3st1 | CCATCCGCCTGCTGCTTATCCTGAG | AGCCGACCGTCCCGCATTAGG |
| Hs3st3a1 | GGTGATGTCTCCTCCCTTCCCTGTC | CGTGCTCCTCGCTAAACCAATTTAATTCC |
| Hs3st3b1 | GCGGGCATTGCTGGAGTTCCTG | GGGTTCTGGGCATCAAGTCTCGGTAC |
| Hs3st6 | GCCATTCAACCGCAAGTTCTACCAG | GTCAGCCAGCAGACAGACATAAATTAAAGG |
| Hs2st1 | GCTCTGCTGTCACCTTCCTGCTG | GCCATCTTCCTTAGTCCTCACAACATCC |
| Hs6st1 | TGAGAGGAATTTGTTTAGATGCCCAGTTTAG | TGACAGAAGCAGCAGCAACCAAC |
| Hs6st2 | CGGCGGTGGTGGATGGCAAG | GGCTTTGTGGAGGATGGAGAGTTGG |
| Kit | CCTCAGCCTCAGCACATAGC | GAACACTCCAGAATCGTCAACTC |
| Krt5 | TCCTGTTGAACGCCGCTGAC | CGGAAGGACACACTGGACTGG |
| Krt14 | GCTGCTACATGCTGCTCAGGCTTAGG | CCAGGAAGGACAAGGGTCAAGTAAAGAGAGTG |
| Krt19 | GCCACCTACCTTGCTCGGATTG | GTCTCTGCCAGCGTGCCTTC |
| Lamb3 | TCAGTGCTATCCCAGACCAAACAAGACATC | CCGACCACATCATCTACCTGCCCTTC |
| Myc | GCAGTGAGCGGACGGTTGGAAGAG | AGCGGCGGCGAGGGTTGC |
| Myh11 | GTCAGGAGCCACAGTCACCAGCAA | GGCAGGCAGGAAAGGGAAGGGAAT |
| Mylk | CCATCCTGCGGTGTCTCA | AATGTCCTCCTTGCTGTAACTCA |
| Ndst1 | TGGCTGGTTTCTGTTTGGATTCTGTTTCTG | AATGGCTGGTGGACACTGGACTGG |
| Ndst2 | ACAGAAGACAGGCACCACGGCTATTC | TGGCATTGGAAGGAACAGGGAAGAAGTC |
| Postn | CTGGTATCAAGGTGCTATCTGCGGGAAG | ACATCGGAGTAGTGCTGAGTGGTAGTGG |
| Prol1 | ACC ACA CCA GCA ACA ACC ACA A | TGG CTG TAG AGG TGC TAG GCT TAG |
| Rps29 | GGAGTCACCCACGGAAGTTCGG | GGAAGCACTGGCGGCACATG |
| Thbs1 | CACCGCCAAACAACCTCTGACAT | AGTACCGAACAGCTCCTCCACATT |
| Hs3st3a1 KO | ACTGTCGCGCAGCATCTTCAG | GCCAGGCAGTAGAAGACGTAG |
| Hs3st3b1 KO | TCACAGCTCCGAATGAGACATC | CCCAGCGCCTACTGTCTTATC |
HIGHLIGHTS.
Genetic deletion of either Hs3st3a1 or Hs3st3b1 in mice reduces epithelial morphogenesis
Loss of 3-O-sulfotranserases reduces basement membrane sulfation and alters HS composition
Loss of specific 3-O-sulfotransferases alters specific heparan sulfate biosynthetic transcriptional programs, suggesting Hs3st3a1 regulates Hs3st3b1.
Deletion of Hs3st3a1 or Hs3st3b1, which are highly expressed in myoepithelial cells, impairs basal salivary hypofunction
Acknowledgements
We would like to thank the NIH Gene Transfer Core for generating the chimeras of the knockouts, the NIDCR Imaging Core for help with imaging analysis (ZIC DE000750-01) and the NIDCR Veterinary Resources Core for assistance with the murine procedures (ZIC DE000740-05). We would like to thank Dr. Marit H. Aure for her helpful discussion. The research was supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research at the National Institutes of Health.
Footnotes
Competing Interest Statement. The authors have no competing interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Mooij HL, Cabrales P, Bernelot Moens SJ, Xu D, Udayappan SD, Tsai AG, van der Sande MA, de Groot E, Intaglietta M, Kastelein JJ, Dallinga-Thie GM, Esko JD, Stroes ES, Nieuwdorp M, Loss of function in heparan sulfate elongation genes EXT1 and EXT 2 results in improved nitric oxide bioavailability and endothelial function, J Am Heart Assoc 3(6) (2014) e001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Thacker BE, Xu D, Lawrence R, Esko JD, Heparan sulfate 3-O-sulfation: a rare modification in search of a function, Matrix Biol 35 (2014) 60–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Colliec-Jouault S, Shworak NW, Liu J, de Agostini AI, Rosenberg RD, Characterization of a cell mutant specifically defective in the synthesis of anticoagulantly active heparan sulfate, J Biol Chem 269(40) (1994) 24953–8. [PubMed] [Google Scholar]
- [4].Shworak NW, Liu J, Petros LM, Zhang L, Kobayashi M, Copeland NG, Jenkins NA, Rosenberg RD, Multiple isoforms of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. Isolation, characterization, and expression of human cdnas and identification of distinct genomic loci, J Biol Chem 274(8) (1999) 5170–84. [DOI] [PubMed] [Google Scholar]
- [5].Liu J, Shriver Z, Blaiklock P, Yoshida K, Sasisekharan R, Rosenberg RD, Heparan sulfate D-glucosaminyl 3-O-sulfotransferase-3A sulfates N-unsubstituted glucosamine residues, J Biol Chem 274(53) (1999) 38155–62. [DOI] [PubMed] [Google Scholar]
- [6].Liu J, Shworak NW, Sinay P, Schwartz JJ, Zhang L, Fritze LM, Rosenberg RD, Expression of heparan sulfate D-glucosaminyl 3-O-sulfotransferase isoforms reveals novel substrate specificities, J Biol Chem 274(8) (1999) 5185–92. [DOI] [PubMed] [Google Scholar]
- [7].Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG, A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry, Cell 99(1) (1999) 13–22. [DOI] [PubMed] [Google Scholar]
- [8].Deligny A, Denys A, Marcant A, Melchior A, Mazurier J, van Kuppevelt TH, Allain F, Synthesis of heparan sulfate with cyclophilin B-binding properties is determined by cell type-specific expression of sulfotransferases, J Biol Chem 285(3) (2010) 1701–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McKeehan WL, Wu X, Kan M, Requirement for anticoagulant heparan sulfate in the fibroblast growth factor receptor complex, J Biol Chem 274(31) (1999) 21511–4. [DOI] [PubMed] [Google Scholar]
- [10].Patel VN, Lombaert IM, Cowherd SN, Shworak NW, Xu Y, Liu J, Hoffman MP, Hs3st3-modified heparan sulfate controls KIT+ progenitor expansion by regulating 3-O-sulfotransferases, Dev Cell 29(6) (2014) 662–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pempe EH, Xu Y, Gopalakrishnan S, Liu J, Harris EN, Probing structural selectivity of synthetic heparin binding to Stabilin protein receptors, J Biol Chem 287(25) (2012) 20774–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Thacker BE, Seamen E, Lawrence R, Parker MW, Xu Y, Liu J, Vander Kooi CW, Esko JD, Expanding the 3-O-Sulfate Proteome--Enhanced Binding of Neuropilin-1 to 3-O-Sulfated Heparan Sulfate Modulates Its Activity, ACS Chem Biol 11(4) (2016) 971–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zhao J, Zhu Y, Song X, Xiao Y, Su G, Liu X, Wang Z, Xu Y, Liu J, Eliezer D, Ramlall TF, Lippens G, Gibson J, Zhang F, Linhardt RJ, Wang L, Wang C, 3-O-Sulfation of Heparan Sulfate Enhances Tau Interaction and Cellular Uptake, Angew Chem Int Ed Engl 59(5) (2020) 1818–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Neugebauer JM, Cadwallader AB, Amack JD, Bisgrove BW, Yost HJ, Differential roles for 3-OSTs in the regulation of cilia length and motility, Development 140(18) (2013) 3892–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Samson SC, Ferrer T, Jou CJ, Sachse FB, Shankaran SS, Shaw RM, Chi NC, Tristani-Firouzi M, Yost HJ, 3-OST-7 regulates BMP-dependent cardiac contraction, PLoS Biol 11(12) (2013) e1001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tecle E, Diaz-Balzac CA, Bulow HE, Distinct 3-O-sulfated heparan sulfate modification patterns are required for kal-1-dependent neurite branching in a context-dependent manner in Caenorhabditis elegans, G3 (Bethesda) 3(3) (2013) 541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Guo Y, Li Z, Lin X, Hs3st-A and Hs3st-B regulate intestinal homeostasis in Drosophila adult midgut, Cell Signal 26(11) (2014) 2317–25. [DOI] [PubMed] [Google Scholar]
- [18].Ferreras L, Moles A, Situmorang GR, El Masri R, Wilson IL, Cooke K, Thompson E, Kusche-Gullberg M, Vives RR, Sheerin NS, Ali S, Heparan sulfate in chronic kidney diseases: Exploring the role of 3-O-sulfation, Biochim Biophys Acta Gen Subj 1863(5) (2019) 839–848. [DOI] [PubMed] [Google Scholar]
- [19].Maiza A, Sidahmed-Adrar N, Michel PP, Carpentier G, Habert D, Dalle C, Redouane W, Hamza M, van Kuppevelt TH, Ouidja MO, Courty J, Chantepie S, Papy-Garcia D, Stettler O, 3-O-sulfated heparan sulfate interactors target synaptic adhesion molecules from neonatal mouse brain and inhibit neural activity and synaptogenesis in vitro, Sci Rep 10(1) (2020) 19114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hauser BR, Aure MH, Kelly MC, Genomics, C. Computational Biology, Hoffman MP, Chibly AM, Generation of a Single-Cell RNAseq Atlas of Murine Salivary Gland Development, iScience 23(12) (2020) 101838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ninche N, Kwak M, Ghazizadeh S, Diverse epithelial cell populations contribute to the regeneration of secretory units in injured salivary glands, Development 147(19) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rutledge EA, Benazet JD, McMahon AP, Cellular heterogeneity in the ureteric progenitor niche and distinct profiles of branching morphogenesis in organ development, Development 144(17) (2017) 3177–3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ten Dam GB, Kurup S, van de Westerlo EM, Versteeg EM, Lindahl U, Spillmann D, van Kuppevelt TH, 3-O-sulfated oligosaccharide structures are recognized by anti-heparan sulfate antibody HS4C3, J Biol Chem 281(8) (2006) 4654–62. [DOI] [PubMed] [Google Scholar]
- [24].Mani K, Cheng F, Sandgren S, Van Den Born J, Havsmark B, Ding K, Fransson LA, The heparan sulfate-specific epitope 10E4 is NO-sensitive and partly inaccessible in glypican-1, Glycobiology 14(7) (2004) 599–607. [DOI] [PubMed] [Google Scholar]
- [25].May AJ, Chatzeli L, Proctor GB, Tucker AS, Salivary Gland Dysplasia in Fgf10 Heterozygous Mice: A New Mouse Model of Xerostomia, Curr Mol Med 15(7) (2015) 674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu J, Shworak NW, Fritze LM, Edelberg JM, Rosenberg RD, Purification of heparan sulfate D-glucosaminyl 3-O-sulfotransferase, J Biol Chem 271(43) (1996) 27072–82. [DOI] [PubMed] [Google Scholar]
- [27].Xia G, Chen J, Tiwari V, Ju W, Li JP, Malmstrom A, Shukla D, Liu J, Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1, J Biol Chem 277(40) (2002) 37912–9. [DOI] [PubMed] [Google Scholar]
- [28].Rutledge EA, McMahon AP, Mutational analysis of genes with ureteric progenitor cell-specific expression in branching morphogenesis of the mouse kidney, Dev Dyn 249(6) (2020) 765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].HajMohammadi S, Enjyoji K, Princivalle M, Christi P, Lech M, Beeler D, Rayburn H, Schwartz JJ, Barzegar S, de Agostini AI, Post MJ, Rosenberg RD, Shworak NW, Normal levels of anticoagulant heparan sulfate are not essential for normal hemostasis, J Clin Invest 111(7) (2003) 989–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Smits NC, Kobayashi T, Srivastava PK, Skopelja S, Ivy JA, Elwood DJ, Stan RV, Tsongalis GJ, Sellke FW, Gross PL, Cole MD, DeVries JT, Kaplan AV, Robb JF, Williams SM, Shworak NW, HS3ST1 genotype regulates antithrombin’s inflammomodulatory tone and associates with atherosclerosis, Matrix Biol 63 (2017) 69–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hasegawa H, Wang F, Visualizing mechanosensory endings of TrkC-expressing neurons in HS3ST-2-hPLAP mice, J Comp Neurol 511(4) (2008) 543–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Miller DL, Ortega S, Bashayan O, Basch R, Basilico C, Compensation by fibroblast growth factor 1 (FGF1) does not account for the mild phenotypic defects observed in FGF2 null mice, Mol Cell Biol 20(6) (2000) 2260–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jonker JW, Suh JM, Atkins AR, Ahmadian M, Li P, Whyte J, He M, Juguilon H, Yin YQ, Phillips CT, Yu RT, Olefsky JM, Henry RR, Downes M, Evans RM, A PPARgamma-FGF1 axis is required for adaptive adipose remodelling and metabolic homeostasis, Nature 485(7398) (2012) 391–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ferreira JN, Hoffman MP, Interactions between developing nerves and salivary glands, Organogenesis 9(3) (2013) 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Spadari F, Venesia P, Azzi L, Veronesi G, Costantino D, Croveri F, Farronato D, Tagliabue A, Tettamanti L, Low basal salivary flow and burning mouth syndrome: new evidence in this enigmatic pathology, J Oral Pathol Med 44(3) (2015) 229–33. [DOI] [PubMed] [Google Scholar]
- [36].Wood NI, Goodman AO, van der Burg JM, Gazeau V, Brundin P, Bjorkqvist M, Petersen A, Tabrizi SJ, Barker RA, Morton AJ, Increased thirst and drinking in Huntington’s disease and the R6/2 mouse, Brain Res Bull 76(1–2) (2008) 70–9. [DOI] [PubMed] [Google Scholar]
- [37].Butler A, Hoffman P, Smibert P, Papalexi E, Satija R, Integrating single-cell transcriptomic data across different conditions, technologies, and species, Nat Biotechnol 36(5) (2018) 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R, Comprehensive Integration of Single-Cell Data, Cell 177(7) (2019) 1888–1902 e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, Chernomorsky R, Boucher M, Elsasser AL, Esau L, Zheng J, Griffiths JA, Wang X, Su H, Xue Y, Dominguez MG, Noguera I, Torres R, Macdonald LE, Stewart AF, DeChiara TM, Yancopoulos GD, High-throughput engineering of the mouse genome coupled with high-resolution expression analysis, Nat Biotechnol 21(6) (2003) 652–9. [DOI] [PubMed] [Google Scholar]
- [40].Hoffman MP, Kidder BL, Steinberg ZL, Lakhani S, Ho S, Kleinman HK, Larsen M, Gene expression profiles of mouse submandibular gland development: FGFR1 regulates branching morphogenesis in vitro through BMP- and FGF-dependent mechanisms, Development 129(24) (2002) 5767–78. [DOI] [PubMed] [Google Scholar]
- [41].Steinberg Z, Myers C, Heim VM, Lathrop CA, Rebustini IT, Stewart JS, Larsen M, Hoffman MP, FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis, Development 132(6) (2005) 1223–34. [DOI] [PubMed] [Google Scholar]
- [42].Patel VN, Likar KM, Zisman-Rozen S, Cowherd SN, Lassiter KS, Sher I, Yates EA, Turnbull JE, Ron D, Hoffman MP, Specific heparan sulfate structures modulate FGF10-mediated submandibular gland epithelial morphogenesis and differentiation, J Biol Chem 283(14) (2008) 9308–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lombaert IMA, Patel VN, Jones CE, Villier DC, Canada AE, Moore MR, Berenstein E, Zheng C, Goldsmith CM, Chorini JA, Martin D, Zourelias L, Trombetta MG, Edwards PC, Meyer K, Ando D, Passineau MJ, Hoffman MP, CERE-120 Prevents Irradiation-Induced Hypofunction and Restores Immune Homeostasis in Porcine Salivary Glands, Mol Ther Methods Clin Dev 18 (2020) 839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Simeonovic CJ, Popp SK, Starrs LM, Brown DJ, Ziolkowski AF, Ludwig B, Bornstein SR, Wilson JD, Pugliese A, Kay TWH, Thomas HE, Loudovaris T, Choong FJ, Freeman C, Parish CR, Loss of intra-islet heparan sulfate is a highly sensitive marker of type 1 diabetes progression in humans, PLoS One 13(2) (2018) e0191360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Allen BL, Rapraeger AC, Spatial and temporal expression of heparan sulfate in mouse development regulates FGF and FGF receptor assembly, J Cell Biol 163(3) (2003) 637–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Patel VN, Knox SM, Likar KM, Lathrop CA, Hossain R, Eftekhari S, Whitelock JM, Elkin M, Vlodavsky I, Hoffman MP, Heparanase cleavage of perlecan heparan sulfate modulates FGF10 activity during ex vivo submandibular gland branching morphogenesis, Development 134(23) (2007) 4177–86. [DOI] [PubMed] [Google Scholar]
- [47].Prochazkova M, Hall B, Hu M, Okine T, Reukauf J, Binukumar BK, Amin ND, Roque E, Pant HC, Kulkarni A, Peripheral and orofacial pain sensation is unaffected by the loss of p39, Mol Pain 13 (2017) 1744806917737205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Knosp WM, Knox SM, Lombaert IM, Haddox CL, Patel VN, Hoffman MP, Submandibular parasympathetic gangliogenesis requires sprouty-dependent Wnt signals from epithelial progenitors, Dev Cell 32(6) (2015) 667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yan L, Song Y, Xia K, He P, Zhang F, Chen S, Pouliot R, Weiss DJ, Tandon R, Bates JT, Ederer DR, Mitra D, Sharma P, Davis A, Linhardt RJ, Heparan sulfates from bat and human lung and their binding to the spike protein of SARS-CoV-2 virus, Carbohydr Polym 260 (2021) 117797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Sarnaik A, Abernathy MH, Han X, Ouyang Y, Xia K, Chen Y, Cress B, Zhang F, Lali A, Pandit R, Linhardt RJ, Tang YJ, Koffas MAG, Metabolic engineering of cyanobacteria for photoautotrophic production of heparosan, a pharmaceutical precursor of heparin., Algal Research 37 (2019) 57–63. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.