Summary
The interaction of plants with complex microbial communities is the result of co‐evolution over millions of years and contributed to plant transition and adaptation to land. The ability of plants to be an essential part of complex and highly dynamic ecosystems is dependent on their interaction with diverse microbial communities. Plant microbiota can support, and even enable, the diverse functions of plants and are crucial in sustaining plant fitness under often rapidly changing environments. The composition and diversity of microbiota differs between plant and soil compartments. It indicates that microbial communities in these compartments are not static but are adjusted by the environment as well as inter‐microbial and plant–microbe communication. Hormones take a crucial role in contributing to the assembly of plant microbiomes, and plants and microbes often employ the same hormones with completely different intentions. Here, the function of hormones as go‐betweens between plants and microbes to influence the shape of plant microbial communities is discussed. The versatility of plant and microbe‐derived hormones essentially contributes to the creation of habitats that are the origin of diversity and, thus, multifunctionality of plants, their microbiota and ultimately ecosystems.
Keywords: endosphere, microbiota, plant development, plant evolution, rhizosphere, symbiosis
Significance Statement
Plants and all their tissues are inhabited by, surrounded by and, thus, in interactions with complex microbial communities. Plant hormones drive the establishment of these communities, termed microbiota, which add a diversity of functions to plants that are crucial for plant fitness under changing environments since the time of land colonisation.
INTRODUCTION
Plants, like all multicellular organisms, do not live in a sterile environment. The in‐ and outsides of plants are populated by specific and often selectively assembled microbial communities, called microbiota (Bulgarelli et al., 2013; Goodrich et al., 2016; Orozco‐Mosqueda et al., 2018). These microbial communities can be vast in the range of species/taxa (diversity) and number of individuals present (abundance). The sheer quantity of microbial species found to be associated with plant tissues (e.g. leaves, roots) together with the genetic and functional diversity of those microbial communities has given rise to the term microbiome (the collective genomes of an organism’s microbiota) (Handelsman et al., 2007). The soil biota represents the origin of plant‐associated microbiomes. Microbiome compositions differ between plant organs and are therefore defined, for example, as phyllosphere or rhizosphere microbiomes for communities attached to the outside of leaves and roots, respectively, or endosphere microbiomes for communities found inside plant tissues (Knief et al., 2012; Bulgarelli et al., 2013; Turner et al., 2013a; Berg et al., 2014; Bai et al., 2015; Cregger et al., 2018). The best‐characterised members of plant microbiomes are bacteria and fungi. They can live in neutral, beneficial or pathogenic interaction within or outside of the plant (Raaijmakers et al., 2009; Turner et al., 2013a). Unsurprisingly, in addition to environmental niche effects and edaphic factors, plants have evolved mechanisms to shape such communities, benefit from and even exploit microbiomes as a huge genetic resource to expand their ability to cope with changing environmental conditions (Bulgarelli et al., 2012; Lundberg et al., 2012; Philippot et al., 2013; Reinhold‐Hurek et al., 2015; Liu et al., 2019; Brown et al., 2020). The ability of plant roots to modify microbial communities is strongest in the endosphere but can reach well beyond the rhizosphere. Under leaf pathogen attacks, for instance, plant roots excrete metabolites to change the composition of the soil biota as a strategy to recruit beneficial microbes that activate effective defence against the leaf invaders (Lakshmanan et al., 2012; Chaparro et al., 2013; Berendsen et al., 2018; Stringlis et al., 2018). Consequently, as for humans, the plant microbiome has been referred to as the extended or secondary genome of plants, as it encodes huge numbers of genes and may thus provide additional genetic and functional diversity to the host (Grice and Segre, 2012; Berendsen et al., 2012; Mendes et al., 2013; Turner et al., 2013a). The versatile effects of the plant microbiota can be roughly divided into two fractions (Figure 1) (Mendes et al., 2013). Firstly, microbial communities can improve the environmental adaptability and fitness of plants by contributing to plant protection against abiotic stress (e.g. drought) or pathogens (incl. herbivores) as well as by fostering nutrient and water supply. The latter also involves pedogenetic effects of microbes. Secondly, microbes support plant and root system architecture. In addition to stimulating lateral root growth and root hair formation, which further improves water and nutrient accessibility, microbes can promote growth and plant regeneration. Together, this indicates the ability of microbes to steer plant intrinsic and extrinsic processes that support the sessile nature of plants and help them to grow and reproduce under changing environments (Figure 1). The questions arise how and to what extent can plants shape microbiomes to their own benefit. Within the colonised zones, plants and their microbial communities strongly influence each other within the given environmental conditions (Fitzpatrick et al., 2020). Various mechanisms describe how plants communicate with and, thus, change their living environment. Chemical cues contribute strongly to antagonistic (e.g. through the production of antimicrobial metabolites) or mutually supportive (e.g. through the production of probiotics) microbe–microbe interactions (Bulgarelli et al., 2015; Hacquard et al., 2015; Pieterse et al., 2016; Hassani et al., 2018). For instance, volatiles have been very versatile for plants and even plant communities to protect them against insects (Sharifi et al., 2018; Hammerbacher et al., 2019). Primary and secondary metabolites produced and exuded by the plant can selectively attract or repel members of microbial communities (Leach et al., 2017; Nobori et al., 2018). Vice versa, microbial metabolites can alter plant development and responses to environmental cues, often contributing to increased plant health and fitness (Strehmel et al., 2014; Venturi and Keel, 2016; Sasse et al., 2018; Fitzpatrick et al., 2020; O’Banion et al., 2020). In this whole process of plant–microbiota interactions, plant hormones take a central role. Hormones are chemical messengers that are involved in cellular and physiological processes in an endocrine (function distant to biosynthesis site) or paracrine (function in cells adjacent to biosynthesis site) manner. In addition to edaphic factors, hormones are part of host factors (e.g. nutrients, signalling processes) and microbial adaptation processes to their hosts. In this way, hormones can contribute to the microbial diversity in the endosphere and different root compartments by regulating plant defence and development, or in the rhizosphere by direct or indirect activities of excreted hormones on microbes (Figure 2). This review presents our current knowledge of hormone effects on the composition and diversity of plant microbiomes. Taking three perspectives, we will describe to what extent (i) hormone signalling inside plants, (ii) hormones excreted by plants and (iii) microbe‐derived hormones affect microbial communities (Figure 2). Taking different perspectives also allows us to understand that plants and microbes often employ the same hormones for completely different purposes. We introduce the function of hormones as common chemical language, whose purposeful utilisation by plants and microbes characterises them as versatile go‐betweens to establish species‐rich and functionally diverse biocenoses.
Figure 1.
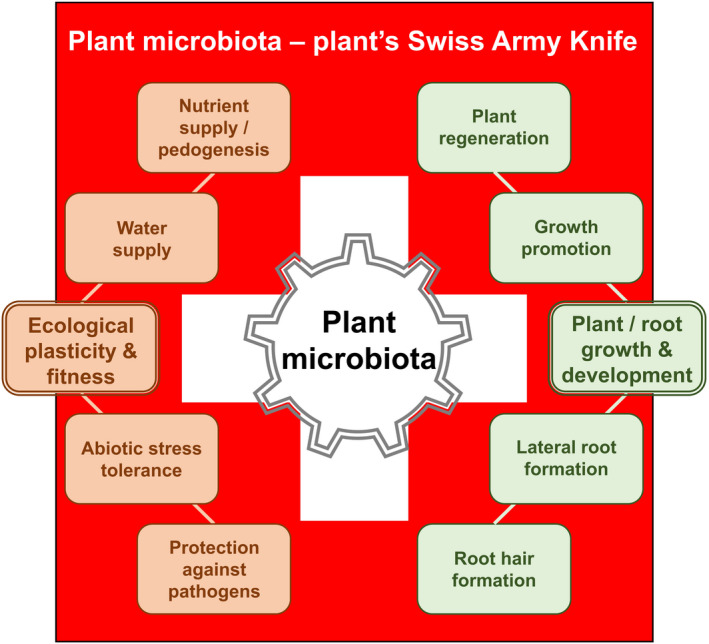
Plant microbiota are a plant’s Swiss Army Knife. To access the benefits of the microbiota, plants need to have some control over their assembly, and hormones play a crucial role therein. The benefits can be divided into two branches: (i) enhanced ecological plasticity and fitness based on improved nutrient and water supply as well as protection against biotic and abiotic stress and (ii) optimised plant/root growth and development due to microbial support in plant regeneration, growth and the formation of lateral roots and root hairs.
Figure 2.
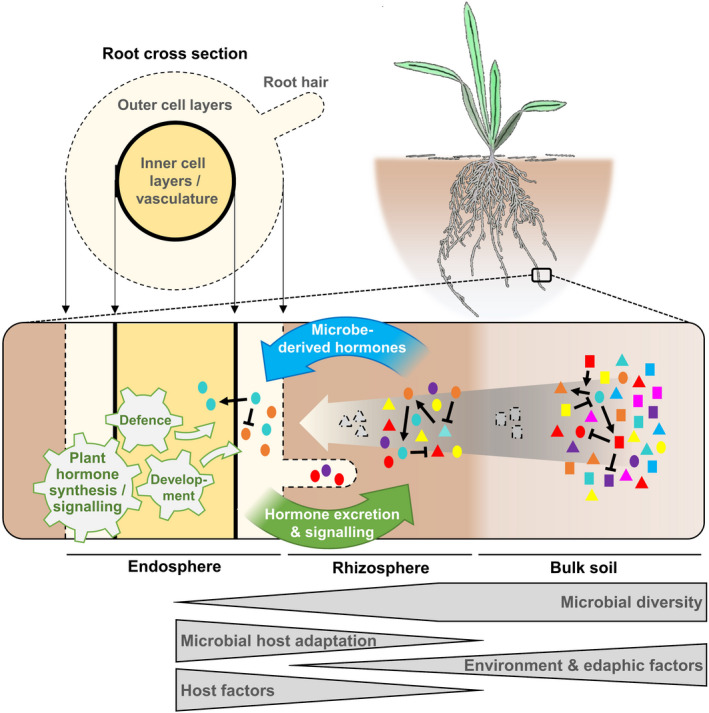
Hormone function in the assembly of microbiota. The ability of soil microbes to colonise the rhizosphere or endosphere is dependent on their degree of specialisation (indicated by grey arrow in the inner scheme). While the abiotic environment and edaphic factors (e.g. soil properties) affect the composition of microbial communities in bulk soil, microbe‐derived and plant‐excreted hormones contribute to their assembly in the plant rhizosphere. Hormone‐dependent developmental and defence‐related processes inside the plant shape the endosphere community. Microbe–microbe interactions contribute to the assembly of microbiomes across all habitats (indicated by ↑ and ↓). While, in general, microbial diversity is found to be lower in rhizosphere versus bulk soil, metatranscriptome analyses revealed similar diversities in both compartments but differences in microbial composition (Turner et al., 2013b). Due to further evolution‐driven microbial host adaptation processes and host factors, microbial diversity is lower in the endosphere and only highly host‐adapted microbes are able to colonise outer and/or inner root layers (incl. vasculature).
THE EFFECTS OF HORMONE SIGNALLING INSIDE PLANTS ON PLANT MICROBIOMES
To comprehend hormone function in shaping microbiomes requires looking back to the origin and evolution of plant–microbe interactions. Plants started to colonise land at the mid‐late Ordovician period (470–443 million years ago [mya]). Terrestrial colonisation involved contending with a range of stresses and nutrient acquisition literally without a root system. Complex interactions between hormone signalling pathways have been posited as key for dealing with and fine‐tuning responses to combined biotic and abiotic stresses whilst also limiting growth trade‐offs (Yasuda et al., 2008; Mosher et al., 2010; Vos et al., 2015). For more details we refer to excellent reviews (Pieterse et al., 2009; Berens et al., 2017). Regulatory networks of hormones – specifically auxin, abscisic acid (ABA), cytokinins (CKs), gibberellic acid (GA), jasmonic acid (JA), salicylic acid (SA) and strigolactones (SL) – are conserved across embryophyte lineages, whose common ancestors were some of the earliest colonisers of land (Wang et al., 2015). GA perception and signalling, for instance, diversified as part of early land plant evolution and their adaptation to the environment (Yasumura et al., 2007). In this process of plant transition and adaptation to land, beneficial symbionts played some important role. Fossil records revealed the presence of plant symbioses since the Silu‐Devonian period (443–419 mya) (Remy et al., 1994; Martin et al., 2017). The presence of hormone signalling in early plants and the functional involvement of hormone networks in the outcome of plant–microbe interactions readily positions hormones as tools to be co‐opted for the regulation of plant symbioses and microbiome assembly. In addition to protection against environmental cues, an eminent task of beneficial symbionts was to serve plants in nutrient and water supply, especially under the fluctuating Silu‐Devonian climate (Selosse and Tacon, 1998).
The importance of symbionts for ecological plasticity of plants is indicated by the ability of plants to shape the microbiome to some extent (Bulgarelli et al., 2012; Lundberg et al., 2012; Yu and Hochholdinger, 2018). This suggests the existence and evolution of heritable plant traits determining the assembly of plant microbiota. Consistent with this, host phylogenetic studies have revealed an evolution‐based trajectory that can partially explain microbiota assembly (Escudero‐Martinez and Bulgarelli, 2019). Environmental stress might substantiate underlying traits as a kind of biased microbial enrichment under stress conditions. Although plant domestication can affect microbiome assembly (reviewed in Pérez‐Jaramillo et al., 2016), stress‐driven effects appeared to be independent of host phylogeny and domestication (Naylor et al., 2017; Santos‐Medellín et al., 2017; Fitzpatrick et al., 2018). This suggests a hierarchy in the mechanisms plants use to modify microbiomes and indicates the importance of stress responses (e.g. against pathogens) in microbiome assembly. Common to all those plant processes is the tight dependency on hormones.
Hormone‐dependent plant defence processes shape endosphere microbiomes
The very effective and highly plastic way plants regulate a diversity of processes under an often rapidly changing environment is intimately linked to the function of plant hormones. Almost all hormones have been shown to participate in the plant immune system and thereby help to stop pathogen infections and to balance the interaction with beneficial symbionts (Jacobs et al., 2011; Pieterse et al., 2012; Pozo et al., 2015). Pathogenic microbes often use disturbances of plant hormone homeostasis to manipulate host defence responses to promote pathogenicity and virulence but also to induce cell growth and division for nutrition (Chanclud and Morel, 2016; Kunkel and Harper, 2018; Han and Kahmann, 2019). While mostly considered in bilateral plant–microbe interactions, underlying processes have fundamental impacts on the assembly and diversity of plant microbiomes. Per se, plant immune receptors do not distinguish between pathogenic and beneficial microbes but perceive them as potential intruders by conserved molecular patterns such as bacterial flagellin or fungal chitin (Jones and Dangl, 2006; Antolín‐Llovera et al., 2014; Teixeira et al., 2019; Zhou and Zhang, 2020). It is therefore not surprising that plant immunity, as a central process in the control of plant–microbe interactions, has a direct impact on the composition of microbiomes. Bilateral plant–pathogen or plant–beneficial symbiont systems have provided deep insights into the function of plant hormones as determinants of such interactions (reviewed in Robert‐Seilaniantz et al., 2011; Pieterse et al., 2012; Bürger and Chory, 2019). Reductionist plant–microbe approaches helped to define hormone functions in local and systemic plant immune responses at the site of pathogen attacks. SA, JA and ethylene (ET) are best studied for their function in plant–microbe interactions (Van Wees et al., 2008; Pieterse et al., 2009). Canonically, JA and ET signalling are involved in response to necrotrophic pathogens and specifically JA signalling in response to herbivory and wounding. SA signalling, conversely, is involved in defence responses to biotrophs (Li et al., 2019). In addition, those hormones were found to be instrumental in mediating systemic protection of whole plants in response to local interactions with microbes (Pieterse et al., 2009). SA participates in systemic acquired resistance (SAR), a defence strategy where a local pathogen attack at leaves results in plant‐wide protection against subsequent pathogen infection attempts (Kachroo et al., 2020). In contrast, JA and ET function in induced systemic resistance (ISR). ISR is triggered by (beneficial) rhizobacteria, which upon root interaction activate a systemic signalling process to protect the whole plant (Pieterse et al., 2014). Taken together, this designates hormones as part of the plant’s tool kit to keep colonisation by pathogenic and beneficial microbes under control. In this setting, plant hormones emerge as important chemical signals that, in addition to governing internal processes, are instrumental in the multidirectional communication between plants and their associated microbial communities as the most (functionally) diverse entity of their living environment (Berendsen et al., 2012; Lemanceau et al., 2017; Fitzpatrick et al., 2020). Information on their impact on microbiomes (particularly the endosphere), however, is only just emerging (Lebeis et al., 2015; Carvalhais et al., 2017). SA is involved in the assembly of epiphytic and endophytic root microbial communities (Kniskern et al., 2007; Doornbos et al., 2011; Lebeis et al., 2015). Higher alpha diversity values indicate a greater diversity and variety of species in any given microbial community. This characteristic has been shown to improve several ecosystem functions important for plant health, that is, nutrient cycling (Wagg et al., 2019). Alterations in alpha diversity, particularly in the endosphere and rhizosphere, could indicate an impairment in plants’ ability to recruit and maintain a typical microbial community, possibly through an inability to signal to and recognise beneficial partners in the soil or an inability to restrict colonisation by undesirable microbes. Arabidopsis treated with exogenous SA or mutants constitutively producing SA show reduced endosphere alpha diversity as well as a reduction in actinobacteria (Kniskern et al., 2007; Lebeis et al., 2015). In fact, the taxonomic profile at‐large of bacterial endosphere communities appears to be strongly driven by SA accumulation/insensitivity, as revealed by several immune signalling mutant genotypes (Lebeis et al., 2015). Similarly, endosphere communities of tomato (Solanum lycopersicum) plants constitutively degrading the ET precursor 1‐aminocyclopropane‐1‐carboxylic acid (ACC) showed a reduced alpha diversity compared to wild‐type plants (French et al., 2019).
Those hormone activities are not restricted to SA, JA and ET, which are historically considered as hormones that substantiate the immune response to stop pathogens. It is now obvious that hormones that were formerly considered to act only in plant development have deep‐rooted functions in shaping plant–microbe interactions and, most likely, the composition and diversity of microbiomes (Robert‐Seilaniantz et al., 2011; Bürger and Chory, 2019).
Plants adjust root development to facilitate interactions with the microbiome
Plant microbiota differ between plant tissues such as flowers, leaves and roots (Ottesen et al., 2013). In addition, plant development affects assemblage of the rhizosphere microbiome (Chaparro et al., 2014). Among the plant traits that are crucial for the establishment of root‐associated microbial communities are root morphology and architecture. Root exudation differs between the different root zones, thereby attracting different microbes (Haichar et al., 2008; Dennis et al., 2010). Microbes, in turn, have specialised to colonise specific root regions and zones (Saleem et al., 2016). The evolution of roots therefore likely proceeded in line with the co‐evolution of plant–symbiont interactions (Selosse and Tacon, 1998; Strullu‐Derrien et al., 2018). True roots of higher plants are effective tissues for nutrient acquisition that have developed root apical meristems and root caps to penetrate soil in an efficient way. Roots have evolved gradually and several times independently in lycophytes and euphyllophytes (Hetherington and Dolan, 2018; Fujinami et al., 2020). At the time of transition to land, early vascular plants (e.g. Zosterophyllum, Cooksonia, Rhynia) had rhizoid‐based systems or very rudimentary root axes with limited functionalities and abilities to penetrate the top soil surface (few mm to cm) (Kenrick and Strullu‐Derrien, 2014; Xue et al., 2016). The association with microbes and complex microbial communities was therefore essential for plant survival in terrestrial ecosystems (Martin et al., 2017; Strullu‐Derrien et al., 2018). However, there is currently no fossil‐based evidence for a direct influence of beneficial symbionts on root evolution. The question arises to what extent are alterations in root system architecture launched by plants, under unfavourable conditions, with the aim of generating a habitat for microbes. Such a strategy would enable plants to access their secondary genomes for stress protection and nutrient or water acquisition. An answer might lie in the function and utilisation of hormones.
As sessile organisms, plants rely on hormones to orchestrate the different developmental stages, and to add plasticity to plant development under changing environments. Abiotic stress responses, such as drought stress or nutrient deficiencies, require adjustments in root system architecture to penetrate the soil matrix systematically in order to access minerals and water. The primary challenge for plants might be (i) to attract beneficial microbes and (ii) to provide the necessary accommodation infrastructure (habitat). Plant hormones take a central role in the regulation of root system architecture (excellently reviewed in Vanstraelen and Benková, 2012; Petricka et al., 2012) (Figure 3). In brief, auxin and CK are among the main regulators of primary root growth through their involvement in cell division and differentiation at the root meristem, respectively. Brassinosteroids (BRs), GA and SL act synergistically to auxin, while JA, ET and ABA support CK‐mediated cell differentiation. Subsequent root cell elongation involves ABA, auxin, CK, ET, JA and SL as inhibiting hormones and GA as an activating hormone. Lateral root development, in turn, is stimulated by auxin, while ABA, CK and ET act antagonistically (Lavenus et al., 2013). Auxin, together with ET, is also important for root hair initiation and elongation, which are processes inhibited by BR and CK (Vissenberg et al., 2020). The underlying hormone signalling networks are responsive to different environmental stimuli to adjust root system architecture. Nitrogen (N) starvation, for instance, induces primary root growth, whereas enhanced lateral root density and elongation as well as root hair formation help plants to overcome both, low phosphorus (P) and N levels (Li et al., 2016; Jia and von Wirén, 2020). Developing horizontal root systems might be especially helpful under stress to recruit beneficial microbes from microbiota‐enriched top‐soil layers. For instance, SLs, which are known to support root colonisation by arbuscular mycorrhizal fungi (AMF), participate in enhanced lateral root formation under P starvation (Lavenus et al., 2013). SL activities are likely to represent the initial step to recruit and accommodate AMF, which enhance P supply via their hyphal network upon root colonisation (Parniske, 2008; Li et al., 2016). By this means, root symbionts sustain plant fitness by improving nutrient use efficiencies. This further indicates that hormones link the regulation of root development and the establishment of beneficial symbioses.
Figure 3.
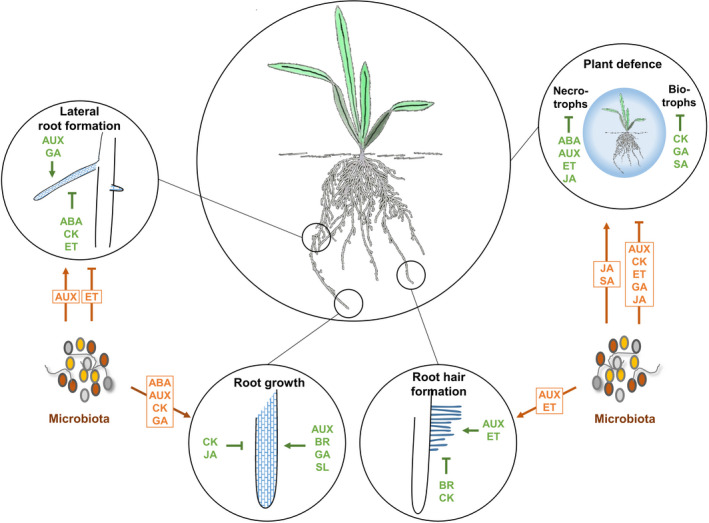
Simplified summary of the functional diversity of plant‐derived and microbial hormone activities on root system architecture and plant defence. Hormone signalling inside plants (in green) activates (↑) or suppresses (↓) lateral root formation, root growth and root hair formation, as well as plant defence against biotrophic and necrotrophic pathogens. Bacterial and fungal microbes are able to produce hormones or modify hormone signalling in plants (in orange), thus altering different aspects of root development and manipulating plant defence. Please note that indicated hormone activities change and can even have opposite effects depending on hormone concentration, hormone homeostasis, plant species, plant developmental stage, environmental stimuli, etc. ABA, abscisic acid; AUX, auxin; BR, brassinosteroid; CK, cytokinin; GA, gibberellic acid; ET, ethylene; JA, jasmonic acid; SA, salicylic acid; SL, strigolactone.
In terms of plant–microbiome co‐evolution, changes in root system architecture might have been part of plant developmental programmes to generate habitats for beneficial symbionts, a strategy especially important under environmental stress. Various microbes can either produce or otherwise change the levels of phytohormones in the rhizosphere or within a plant and, in doing so, impinge on plant development and stress responses (Hacquard et al., 2015; Chanclud and Morel, 2016; Ludwig‐Müller, 2020). Indole acetic acid (IAA), CKs, GAs, ABA and ET have been isolated from microbial culture media (Dodd et al., 2010; Spaepen, 2015). Especially in the rhizosphere, hormone‐producing microbes are often non‐pathogenic and even beneficial to plants. Among the well‐known phytohormone‐producing microbes are plant growth‐promoting bacteria (PGPBs) and plant growth‐promoting fungi (PGPFs) (Glick, 2012; Zamioudis and Pieterse, 2012; Bakker et al., 2018). Developmental changes caused by microbe‐derived hormones can include alterations in root and shoot growth, as well as in root system architecture and potentially the modification of flowering time (Dodd et al., 2010; Spaepen, 2015; Lu et al., 2018). Finally, microbes produce hormones that can otherwise benefit a plant, for example, by protecting it against pathogens (hormones as antibiotics or as defence inducers) or by conferring resistance to abiotic stresses (Tsukanova et al., 2017; Liu et al., 2017a; Rosier et al., 2018; Kudoyarova et al., 2019). In line with that, root symbionts such as PGPBs affect root cell division and differentiation thereby changing determinants of root system architecture: meristem‐driven indeterminate primary root growth, lateral root and root hair formation (Verbon and Liberman, 2016). PGPB species such as Pseudomonas simiae increase lateral root and root hair formation in an auxin‐dependent and JA/ET‐independent manner (Zamioudis et al., 2013), while Pseudomonas putida produces auxin for elongation of primary roots (Patten and Glick, 2002). Bacillus megaterium promotes root system architecture via plant CK signalling independently of plant ET and auxin pathways (López‐Bucio et al., 2007; Ortíz‐Castro et al., 2008). PGPFs such as Trichoderma spp. produce ET and/or auxin to increase root hair formation and primary and lateral root growth, respectively (Contreras‐Cornejo et al., 2015, 2009). In addition, the ectomycorrhizal fungus Laccaria bicolor releases auxin to trigger lateral root development in poplar (Populus tremula × Populus alba) and Arabidopsis (Felten et al., 2009). Those studies exemplify that microbes produce hormones and/or activate plant hormone signalling to alter root system architecture presumably to facilitate their accommodation. This further indicates that changes in root system architecture can originate from sophisticated communication between plants and microbes. It is therefore not surprising that hormones, which were initially thought to exclusively regulate developmental processes, affect interactions with microbes (Robert‐Seilaniantz et al., 2011; Pieterse et al., 2012; Bürger and Chory, 2019). This might be explained in part by their effects on root development and the provision of habitats for beneficial symbionts.
The fact that microbes can often produce more than one phytohormone, frequently in conjunction with other traits that confer physiological effects to plants or other microbes, makes it difficult to disentangle direct effects by individual hormones. This is probably exemplified by the extreme plant growth‐promoting properties of the bacterium Pantoea phytobeneficialis MSR2, which derive from the combined abilities to fix nitrogen, solubilise phosphate, degrade the ET precursor ACC, metabolise JA and produce the plant hormones auxin and CK (Nascimento et al., 2020). Altogether, this suit of studies clearly indicates the importance of hormone‐mediated defence signalling and plant developmental processes in shaping the composition of microbiomes.
PLANT HORMONE EXCRETION AND SIGNALLING CAN SHAPE THE RHIZOSPHERE MICROBIOME
Some functional analogies have been noted between the rhizosphere and the mammalian gut; both constitute the location of nutrient absorption, and contribute to plant/animal health and development. These functions are strongly supported by the organisms’ associated microbiomes (Ramírez‐puebla et al., 2013; Hacquard et al., 2015). The community composition and identity of microbes in the rhizosphere enhance the plant’s functional capabilities (especially in terms of biotic and abiotic stress resistance) vastly, and it is therefore prudent that plants tightly control the microbial flora therein (Figures 1 and 2) (Turner et al., 2013a; Pieterse et al., 2016; Bakker et al., 2018). The ‘cry‐for‐help’ hypothesis, for example, suggests that plant exposure to pathogens triggers modification of root exudates to signal to the microbial community and promote the accumulation of beneficial microbes in the rhizosphere (Bakker et al., 2018; Rolfe et al., 2019). This is exemplified by the occurrence of disease suppressive soils after heavy disease outbreaks in the field, which are enriched in microbes with plant‐protective properties. These microbes are able to confer pathogen resistance to subsequent crop generations (Raaijmakers and Mazzola, 2016; Bakker et al., 2018; Berendsen et al., 2018). In addition, disease‐resistant genotypes often accumulate beneficial microbes in their rhizosphere (Kwak et al., 2018; Mendes et al., 2018), further suggesting a functional link between plant immunity and microbiome composition. Plant hormones are a key component in the perception of (pathogenic) microbes and subsequent plant immune signalling (for more details of hormone function in plant immunity see Pieterse et al., 2012). However, there is an emerging role of plant hormones in shaping root microbiome composition either directly or indirectly, in order to support plant growth under biotic and abiotic stress conditions (Haney and Ausubel, 2015; Carvalhais et al., 2017). The capacity for microbes to subvert plant hormone signalling and immune responses is complicating the picture. Effectors secreted by some microbes are capable of targeting specific plant proteins involved in mounting hormone‐dependent immune responses. Readers interested in this subject are referred to (Nobori et al., 2018; Han and Kahmann, 2019). Specific plant hormones can be released into the rhizosphere and may have a direct impact on plant‐interacting microbes and the root associated microbiome at large (Xu et al., 2018b; Carvalhais et al., 2019; Nasir et al., 2019; Liu et al., 2020). Evidence exists for the presence of most plant‐derived hormones in root exudates (Torrey, 1976; Reddy et al., 1989; Faure et al., 2009). On the other hand, there are potential indirect influences hormones have on shaping root exudates used to communicate with the microbial community (Schreiner et al., 2011; Carvalhais et al., 2015). The direct versus indirect effect of plant hormones on shaping the microbiome is not always clear and should be taken into careful consideration when designing and interpreting studies. The quantitative aspect of hormones and their interaction with microbes is also important, particularly when considering hormones released into the rhizosphere, and often not well represented in microbiome research. Quantification of plant hormones is challenging, with hormones present at pg g−1 or ng g−1 concentrations in fresh plant tissue (Pan et al., 2010). Methods for more accurate quantification are however improving (Fu et al., 2011; Wang et al., 2020).
Direct and indirect effects of plant hormones on microbial communities
Strigolactones – more than facilitators of specific plant symbioses
SLs are a group of compounds relatively recently recognised as plant hormones with functions in shoot branching, root system architecture, parasitic weed germination and plant–microbe communication (Yoneyama et al., 2008; Al‐Babili and Bouwmeester, 2015; Clear and Hom, 2019; Aliche et al., 2020). They also display complex cross‐talk with other hormones and are therefore involved in many aspects of plant growth and development (Cheng et al., 2013; Omoarelojie et al., 2019). SLs have a role in classical plant symbioses with AMF and nodulating rhizobia (Steinkellner et al., 2007; Foo and Davies, 2011; Clear and Hom, 2019). Although an SL receptor has not been characterised in AMF, so far, treatment with SL induces a variety of fungal responses, including morphological and transcriptional changes, and the stimulation of the release of secreted proteins, which support plant colonisation (Lanfranco et al., 2018). SLs are long known to induce hyphal branching of AMF, a process initiated before colonisation of plant roots (Akiyama et al., 2005, 2010; Akiyama and Hayashi, 2006; Yoneyama et al., 2008). The hyphal branching phenotype is generally not observed in other soil‐borne fungi (Steinkellner et al., 2007). Higher concentrations of SLs (>10 μm) have, however, been shown to reduce hyphal branching with some phytopathogenic fungi (Dor et al., 2011), reduce the growth rate of a beneficial fungus (Mucor sp.) and also be required for a Mucor sp. to promote plant growth (Rozpadek et al., 2018).
Soybean (Glycine max) and Lotus japonicus mutants deficient in SL synthesis display reduced nodule numbers and this phenotype can be rescued by exogenous SL application (10 μm) (Foo and Davies, 2011; Rehman et al., 2018). Additionally, rhizobial infection has been shown to alter the expression of SL biosynthesis genes (Rehman et al., 2018). Further, SLs have been implicated in increased swarming and motility of rhizobia (Tambalo et al., 2014; Peláez‐Vico et al., 2016). In light of the importance of SLs in plant–microbe symbiosis formation, several studies have used community profiling, by amplicon sequencing, to address the impact of SLs on the wider bacterial and fungal microbiomes (Carvalhais et al., 2019; Nasir et al., 2019; Liu et al., 2020). Studies suggest that SL signalling is involved in shaping fungal and bacterial rhizosphere communities. Several specific fungal, but not bacterial taxa were differentially abundant in the Arabidopsis SL biosynthesis mutant more axillary growth 4 (max4). The abundance of biocontrol Penicillum and Epicoccum sp. and the Plectosphaerella cucumerina pathogen was reduced, while that of species from two other pathogen groups (Hypocreales and Ramularia) was increased in the mutant rhizosphere (Carvalhais et al., 2019). In rhizospheres of soybean plants overexpressing SL biosynthesis and signalling genes, representatives of Fusarium solani, Rhizobiaceae, predatory Bdellovibrio and Shinella were more abundant (Liu et al., 2020). Conversely, SL biosynthesis‐ and signalling‐deficient rice (Oryza sativa) mutants showed a reduction in several beneficial groups of bacteria (plus Bdellovibrio) in the mutant rhizospheres but also a decrease in the pathogenic fungus Olpidium brassicae (Nasir et al., 2019). Taken together, SL clearly affects the shape of both fungal and bacterial rhizosphere communities but without an obvious positive or negative influence.
Abscisic acid and auxin – carbon source for microbes and stimulator of microbial signalling
ABA is canonically involved in abiotic stress tolerance and seed dormancy, the antagonistic relationship with GA breaking dormancy in favourable conditions (reviewed in Verma et al., 2016). Auxins, such as IAA, are implicated in plant growth and development but more recently their involvement in stress tolerance has become apparent (Shani et al., 2017). ABA also supports plant–AMF interactions (Herrera‐Medina et al., 2007; Martín‐Rodríguez et al., 2011; reviewed by Stec et al., 2016) and shows a positive correlation with the abundance of SL. ABA affects nodulation (Suzuki et al., 2004), likely by inhibition of CK biosynthesis (Ding et al., 2008), which is vital for nodule organogenesis. ABA and IAA can be used as the sole carbon source by some rhizobacteria, for example, a Rhodococcus sp. and a Novoshingobium sp. can use ABA (Belimov et al., 2014), while P. putida strain 1290 can feed on IAA (Leveau and Lindow, 2005). In these studies the concentrations of ABA and IAA are far greater than those previously reported to be present in plant tissues (Belimov et al., 2014; Rehman et al., 2018); however, no attempt was made to establish a lower limit. Secretion of ABA or IAA into the rhizosphere could be a mechanism for plants to select microbes capable of using these as a carbon source. Exogenous ABA application has been shown to induce vast gene expression changes in the endophyte Aspergillus nidulans (Xu et al., 2018a), and IAA induced production of invasive filaments in Saccharomyces cerevisiae (Prusty et al., 2004), providing evidence for the perception of ABA by microbes and subsequent changes in growth processes. Most interestingly, IAA application increased the antimicrobial activity of several strains of the actinobacterial Streptomycetaceae family isolated from Arabidopsis roots towards Escherichia coli and Bacillus subtilis (van der Meij et al., 2018). Actinobacteria are one of the most dominant phyla in the plant root microbiome and are known for their antimicrobial compound‐producing capabilities.
Jasmonic acid, salicylic acid and ethylene – defence hormones with wider effects on microbial communities
SA is a well‐studied defence hormone and perceived by SA immune signal receptors NPR1, NPR3 and NPR4. The latter two act in conjunction with NPR1 to regulate different SA‐mediated immune responses (Dong, 2004; Ding et al., 2018; Li et al., 2019). Furthermore, SA and NRP1 are known to interact with JA signalling pathways to adjust immune signalling in response to pathogen attack (Li et al., 2004; Spoel et al., 2007; Leon‐Reyes et al., 2010). Mutants deficient in NPR1 function have reduced endosphere alpha diversity, and can restrict colonisation by endophytes (Hein et al., 2008; Chen et al., 2020a). These mutants maintain their ability to synthesise SA and in fact may have elevated levels of SA under some conditions (Rayapuram and Baldwin, 2007). The SA‐associated alpha diversity reduction that was observed in root endospheres extended into the rhizosphere (Hein et al., 2008; Doornbos et al., 2011) while constitutive degradation of SA reduced alpha diversity in the endosphere but not rhizosphere of tomato or Arabidopsis (Doornbos et al., 2011; French et al., 2019). Moreover, direct SA application affected microbes in bulk soil indicating SA plant signalling‐independent effects on microbial communities (Lebeis et al., 2015). Collectively, these results suggest that SA may act via canonical signalling pathways, via interaction with other hormones such as JA or directly on community members to promote or inhibit their growth.
Exogenous JA application, used to activate JA signalling, has been shown to increase Arabidopsis rhizosphere alpha diversity along with an enrichment of several potentially beneficial microbial taxa (Carvalhais et al., 2013), whilst Arabidopsis mutants deficient in JA conversion to its bioactive form (JA‐isoleucine [JA‐Ile]) show a reduced diversity (Doornbos et al., 2011). Results are not always consistent across plant species and tissues. A contrasting role of JA in epiphytic Arabidopsis leaf communities and wheat (Triticum aestivum) root endosphere community composition has been reported (Kniskern et al., 2007; Liu et al., 2017b). Again, however, actinobacteria were implicated as the major component of community changes. JA‐mediated microbiome assembly is likely realised, indirectly, via root exudate composition changes. Mutants deficient in JA signalling display reductions, in their root exudate profiles, of several positive bacterial chemotaxis compounds and compounds mediating inter‐bacterial interactions (Carvalhais et al., 2015).
Such changes in actinobacteria abundance were also observed to be affected by ET, which often acts synergistically with JA in defence signalling. However, ET activity appears not to be restricted to the root endosphere but can also alter additional plant–microbiome processes (Ravanbakhsh et al., 2018). When grown in an intercropping system, cyanide released by cassava (Manihot esculenta) roots was perceived by neighbouring peanut (Arachis hypogaea) roots to trigger ET excretion. This exogenous ET resulted in an increase in rhizosphere alpha diversity, especially again the abundance of actinobacteria, whilst the abundance of acidobacteria was reduced (Chen et al., 2020b). In addition to reassembling the rhizosphere communities, those actinobacteria improved nutrient supply and seed production in peanuts.
Taken together, classical defence hormones have functions in microbiome assembly that go beyond the endosphere and intrinsic plant defence signalling. It highlights how microbiome studies enable us to assign previously unknown functions to defence hormones such as direct effects or plant‐independent effects on microbe fitness.
Cytokinin, gibberellic acid and brassinosteroids – known developmental regulators that steer plant–microbe interactions
Plant hormones such as CK, GA and BR are underrepresented in microbiome research. Many studies, however, consider the influence these hormones have on specific beneficial and pathogenic plant–microbe interactions (Nakashita et al., 2003; Choi et al., 2010; Jiang et al., 2013; Reusche et al., 2013; Yu et al., 2018), with potentially broader implications in shaping plant microbiomes.
CKs are known to be key for the formation of nodules in legumes. Exogenous CK induces the formation of pseudonodules in non‐rhizobia‐infected nodulating L. japonicus (Heckmann et al., 2011) and several other nodulating legumes, but not in non‐nodulating legumes or non‐legumes (Gauthier‐Coles et al., 2019). This effect can be blocked by ET (Heckmann et al., 2011). The enhancement of pathogen resistance promoted by CK has been shown in several plant–pathogen systems, which seems to be synergistic with, and dependent on, SA‐responsive pathways (Choi et al., 2010; Jiang et al., 2013; Reusche et al., 2013). Contrastingly, CK appears to support the progression of fungal biotrophic pathogens and stimulate beneficial AMF interactions in pea (Pisum sativum) (Walters and McRoberts, 2006; Chanclud et al., 2016; Morrison et al., 2017; Goh et al., 2019).
GA and BR are involved in nodulation and AMF formation (excellently reviewed by McGuiness et al., 2019). While GA has a rather suppressive effect on arbuscule formation, BRs seem to support it (Foo et al., 2016, 2013). The impact of these two hormones on nodulating rhizobia is far less clear – with promotion or inhibition being dependent on concentration and/or species – but it has been speculated that cross‐talk with ET is, in part, responsible (McGuiness et al., 2019). Outside of classical mycorrhizal and rhizobial symbioses, BRs have been shown to promote disease resistance of plants to a broad range of plant pathogens (Nakashita et al., 2003). Earlier studies considered the influence of spray treatment of several species of flowering plants with varying concentrations of GA or IAA on fungal load, assessed by weighing culturable fungi isolated from soils (Sullia, 1968; Gupta, 1971). The response of soil fungi was extremely plant species‐ and hormone concentration‐dependent. Often an intermediate hormone concentration provided an increase in fungal load and higher concentrations reduced the load back to basal levels. In these two studies hormones were applied as foliar sprays, indicating that some signal must be systemically propagated from shoots to roots to impact soil fungi and promote their growth, possibly via root exudates. In conclusion, given the importance of these hormones in interactions with both beneficial and pathogenic microbes, future studies using NGS techniques (e.g. metatranscriptomics with microbiome profiling and plant RNA sequencing) should help to assess their impact on the wider microbiome.
Hormone‐dependent changes in root exudates
There is a huge array of research considering the effect of specific root exudates on root‐associated microbiomes (Chaparro et al., 2013; Stringlis et al., 2018; Huang et al., 2019; Voges et al., 2019). Readers more widely interested in this topic are referred to some excellent reviews (Dennis et al., 2010; Doornbos et al., 2012; van Dam and Bouwmeester, 2016; Sasse et al., 2018; Guerrieri et al., 2019; Preece and Peñuelas, 2020), while we focus here only on hormone effects on exudation. Root exudates can impact microbial communities and selectively modify them by changing the spatial organisation, gene expression or abundance of particular taxa. In addition to serving as carbon sources, exudates can have antimicrobial activity, or they can act as signalling molecules. These are likely indirectly affected by plant hormone signalling, and examples exist of the impact hormones have on root exudates (Schreiner et al., 2011; Carvalhais et al., 2015).
Specific root exudates can selectively modify the microbiome by exploiting the substrate preferences of different bacterial species (Badri et al., 2013; Zhalnina et al., 2018). Badri et al. (2013) showed that certain compounds from Arabidopsis root exudates promote the growth of groups of operational taxonomic units (OTUs), which are clusters of sequences, identified from amplicon sequencing data, and considered as representative of the same species. Some compounds are able to differentially promote and inhibit different OTUs. Phenolic compounds were found to be of particular importance, as they are able to modify the growth of more OTUs than other compounds. Caffeic acid and phenolic compounds have also been implicated as exudates that can aid in disease resistance (Ling et al., 2013; Gu et al., 2016). Caffeic acid and chlorogenic acid in approximately the 10–100 μm range inhibited the growth of a fungal pathogen (Ling et al., 2013). Moreover, SA was a major component of root exudates and posited as involved in the repression of fungal growth (Wu et al., 2009; Ling et al., 2013).
JA or SA application has been shown to induce higher abundance of glucosinolates in exudates of Brassica rapa (Schreiner et al., 2011), a secondary metabolite with antimicrobial activity against Brassica napus to Plasmodiophora brassicae (Xu et al., 2018b). Although not specifically in root exudates, lower aliphatic glucosinolate levels (and biosynthesis gene expression levels thereof) have also been detected in Arabidopsis IAA perception mutants, after drought stress induction (Salehin et al., 2019). This implicates a function of auxins, not only JA and SA, in the production of glucosinolates and illustrates again the complexity and abundance of cross‐talk between hormone signalling pathways and their regulation of metabolite production. Arabidopsis mutants deficient in JA signalling showed changes in their root exudate profiles, which correlated with shifts in microbiome composition. In addition to a lower abundance of several compounds known to act as chemotactic signals, kaempferol with its known antimicrobial activity was less abundant in mutant lines. In addition, the abundance of several specific compounds including sugars and amino acids was found to correlate significantly with the abundance of specific OTUs assigned to genera such as Bacillus and Pseudomonas, or to the Clostridiales order (Carvalhais et al., 2015).
Benzoxazinoids (BXs) are a group of compounds present in many cereal crops (such as maize [Zea mays], rye [Secale cereale] and wheat) and known for their role in plant defence (Hu et al., 2018). JA and ET application has been implicated in the production of BX in maize (Dafoe et al., 2011). Several studies have shown the impact of BXs in fungal and bacterial microbial communities in the maize rhizosphere. The overall effect on pathogen abundance is however ambiguous. While some studies report decreases in pathogenic microbes (Kudjordjie et al., 2019; Cotton et al., 2019), others report increases (Cadot et al., 2020). The BX‐deficient phenotype (herbivory susceptibility, reduced JA and reduced SA) has been shown to be mediated by the BX breakdown product 6‐methoxy‐benzoxazolin‐2‐one (MBOA) and is dependent on microbiome changes (Hu et al., 2018). These changes could be caused directly by MBOA in exudates or by signalling to condition soils with a broader range of metabolites (Cotton et al., 2019).
Taken together, belonging to the class of low‐molecular weight compounds, plant hormones and secondary metabolites are highly mobile and can affect processes by their direct biological activity (Erb and Kliebenstein, 2020). Importantly, hormones can mediate their function through metabolite synthesis and excretion and we are just beginning to understand the impact of this mechanism on microbiome assembly and plant fitness (Leach et al., 2017; Nobori et al., 2018; Huang et al., 2019).
MICROBES PRODUCE PHYTOHORMONES AND MANIPULATE HORMONE SIGNALLING TO INTERACT WITH PLANTS AND OTHER MICROBES
Plant‐associated microbes produce a broad range of hormones and hormone‐like substances, or possess enzyme activities, which alter hormone levels in the plant endosphere, phyllosphere and rhizosphere (Dodd et al., 2010; Spaepen, 2015). Some of these microbe‐derived hormones have obvious effects on plant physiology or support host colonisation, for example, by interfering with plant defence responses (Figure 3). Other microbial hormones can serve as a nutrient source or have antimicrobial activities, and may thus influence neighbouring microbial communities directly. However, these direct effects on microbes may be locally restricted and confined to specific niches, making them hard to trace. In addition, such effects might be masked by plant‐derived hormones. Hence, available information on broader impacts of microbe‐derived hormones on soil‐resident or plant‐associated microbiomes is currently rather limited.
Auxin – growth hormone widely produced by bacterial and fungal microbes
Auxin and CK affect plant growth, which can either be beneficial, if they support general plant growth especially under unfavourable conditions, or detrimental, if they are being exploited for feeding a pathogen (Boivin et al., 2016). Several pathogenic and non‐pathogenic bacterial and fungal microorganisms are capable of auxin (mostly IAA) biosynthesis. The existence of biosynthesis pathways for auxin has been proposed based on the detection of IAA and metabolic intermediates in culture media and the presence of IAA biosynthesis genes in bacterial and fungal genomes. In most of these pathways, the aromatic amino acid tryptophan serves as precursor (Spaepen et al., 2007; Spaepen and Vanderleyden, 2011; Patten et al., 2013; Duca et al., 2014; Chanclud and Morel, 2016; Kunkel and Harper, 2018; Han and Kahmann, 2019; Meents et al., 2019; Morffy and Strader, 2020). Microbe‐derived auxin may serve several functions especially in interaction with plants. Some evidence suggests that in microbes, auxin has a physiological role and serves as signalling molecule (Spaepen et al., 2007; Patten et al., 2013). The IAA precursor tryptophan, IAA itself and other auxins have been shown to induce bacterial IAA biosynthesis genes. In a similar way, plant‐derived metabolites such as flavonoids can induce IAA biosynthesis, for example, in rhizobia (Spaepen et al., 2007; Spaepen and Vanderleyden, 2011; Patten et al., 2013). Microbial IAA production and secretion may aid in regulation of pH homeostasis. In bacteria, IAA may serve as signalling molecule in biofilm formation, population growth and behaviour especially under unfavourable conditions such as nutrient limitation, temperature changes or acidic pH (Spaepen et al., 2007; Spaepen and Vanderleyden, 2011; Patten et al., 2013; Duca et al., 2014; Kunkel and Harper, 2018). Antimicrobial activity has been attributed to the weak acid IAA, and some bacteria may perceive IAA as a signal to induce their antibiotic production, possibly in order to increase the ability to compete with other microbes for limited resources (Duca et al., 2014). Some bacteria can actively degrade IAA and may use it as a carbon and nitrogen source. IAA degradation may also serve as a means to protect bacteria from the antimicrobial activity of IAA, and has been observed in conjunction with chemotaxis towards IAA in P. putida (Scott et al., 2013; Duca et al., 2014). Among competing microorganisms in a complex chemical environment such as the rhizosphere, this trait may thus help to open up ecological niches. On the other hand, the growth‐promoting properties, for example, of the PGPB Burkholderia phytofirmans depend upon its abilities to degrade auxin, especially in an environment with high IAA levels (Zúñiga et al., 2013). High levels of IAA produced by a bacterial community can affect Arabidopsis root growth. Interestingly, strains of auxin‐degrading bacteria can interfere with this negative chemical signalling and restore root growth (Leveau and Lindow, 2005; Finkel et al., 2020).
Auxin can contribute to pathogenesis and virulence of gall‐forming and other plant pathogenic bacteria and fungi (Spaepen et al., 2007; Kunkel and Harper, 2018; Han and Kahmann, 2019). The tumour‐forming soil bacterium Agrobacterium tumefaciens can change plant hormone biosynthesis and activate cell proliferation directly by integrating genes for auxin (and CK) biosynthesis into the host genome, resulting in the formation of the typical crown galls (Gohlke and Deeken, 2014). Other gall‐forming plant pathogens, such as Pseudomonas savastanoi pv. savastanoi, which causes knot disease in olive (Olea europaea), are able to produce and secrete these hormones for tumour formation on their own (Dodueva et al., 2020). Fungi which cause tumours or other plant organ deformations (e.g. Ustilago maydis, Taphrina spp.) possess auxin biosynthesis genes, but the function of these genes in infection and disease symptom development is not entirely clear (Tsai et al., 2014; Ludwig‐Müller, 2015; Chanclud and Morel, 2016; Han and Kahmann, 2019; Dodueva et al., 2020). The hemibiotrophic rice blast fungus Magnaporthe oryzae and some Colletotrichum spp., as well as the biotrophic rust fungus Puccinia graminis f. sp. tritici, produce IAA during their biotrophic growth phase (Ludwig‐Müller, 2015; Chanclud and Morel, 2016). In addition, local accumulation of IAA in plants either through microbial secretion or after microbe‐induced elevation of plant IAA levels supports plant colonisation, most likely through the suppression of (SA‐mediated) host defence responses. Through the induction of expansins, local accumulation of auxin can also contribute to cell wall loosening and thus provides a microbial means to create host entry sites or induce cellular hypertrophy to feed the invader (Spaepen et al., 2007; Kazan and Manners, 2009; Patten et al., 2013; Duca et al., 2014; Kunkel and Harper, 2018; McClerklin et al., 2018).
As shown for PGPBs, microbial auxin contributes to changes in plant physiology such as enhanced root growth and root hair formation and altered root system architecture (Figure 3). The resulting surface area extension may lead to the provision of more root exudates, which could serve as growth substrate for microbial communities in the rhizosphere (Barea et al., 2005; Spaepen et al., 2007; Dodd et al., 2010; Vacheron et al., 2013; Cassán et al., 2014; Duca et al., 2014; Spaepen, 2015; Kudoyarova et al., 2019). The capability of PGPBs to maintain plant growth even under nutrient deficiency or other abiotic stress conditions has been attributed to the microbes’ ability to alter root development through the production of auxin (Marulanda et al., 2009; Belimov et al., 2015; Rolli et al., 2015; Zhou et al., 2016; Kudoyarova et al., 2019). Genetically modified strains of Azospirillum brasilense, in which IAA biosynthesis was enhanced, increased shoot biomass in wheat seedlings. Interestingly, the IAA overproducing strains had a significant impact on rhizosphere microbiota, whereby the effects on rhizobacteria and fungi were different, depending on the promoter (constitutive versus root exudate‐responsive) by which bacterial IAA biosynthesis was driven. Whether the observed effects of bacterial IAA on the microbial communities was direct or indirect via plant‐mediated effects needs to be determined (Baudoin et al., 2010).
Changes in Arabidopsis root system architecture also accompany colonisation by the beneficial, plant growth‐promoting root endophyte Serendipita indica (formerly Piriformospora indica) (Sirrenberg et al., 2007). Serendipita indica can produce IAA in culture media at levels that may be sufficient to impact on primary and especially lateral root formation (Sirrenberg et al., 2007; Meents et al., 2019). In interaction with barley (Hordeum vulgare), S. indica‐derived IAA was shown to be required for root colonisation during biotrophic growth, but not for plant growth promotion (Hilbert et al., 2012). It has been shown recently that other fungus‐derived metabolites can induce local auxin responses and may thus be responsible for initiation of lateral root formation (Inaji et al., 2020). Other beneficial plant symbionts such as many Rhizobium spp. can secrete IAA, and it has been assumed that they use it together with altering plant auxin transport to locally change auxin homeostasis during root nodule formation in legumes (Spaepen and Vanderleyden, 2011; Boivin et al., 2016; Mathesius, 2020). In a similar way, ectomycorrhizal fungi can produce auxin, which seems to support fungal colonisation, and can alter host root morphology (Boivin et al., 2016; Chanclud and Morel, 2016). In contrast, although IAA also promotes fungal invasion and AM formation especially at early stages, AMF seem to be unable to synthesise auxin (Das and Gutjahr, 2020; Ludwig‐Müller, 2020).
Cytokinins – used by microbes as antimicrobial agents and for host colonisation
Similar to auxin, CKs regulate cell division and differentiation, and have a major effect on plant growth processes (Figure 3) (Werner and Schmülling, 2009; Cammarata et al., 2019). It is thus not surprising that microbe‐associated IAA biosynthesis is often accompanied by the ability to produce CKs (Morris, 1986; Costacurta and Vanderleyden, 1995; Jameson, 2000; Boivin et al., 2016; Kudoyarova et al., 2019). Adenine molecules serve as backbones in microbial CK biosynthesis. Like auxin, CKs produced directly or indirectly by bacterial pathogens such as A. tumefaciens or P. savastanoi contribute to gall or tumour formation (Costacurta and Vanderleyden, 1995; Jameson, 2000; Kazan and Lyons, 2014; Hinsch et al., 2015; Spaepen, 2015; Sørensen et al., 2018; Spallek et al., 2018). The presence of CK biosynthesis genes and the production of CKs or CK‐like molecules have also been shown for fungal pathogens, for example, for Claviceps purpurea, U. maydis, Leptosphaeria maculans, M. oryzae, and Fusarium pseudograminearum (Bruce et al., 2011; Hinsch et al., 2015; Chanclud et al., 2016; Trdá et al., 2017; Sørensen et al., 2018). In addition, bacterial and fungal genomes harbour histidine kinases with similarities to plant ET or CK receptors (Hérivaux et al., 2017; Kabbara et al., 2018). The histidine kinase PcrK of the plant pathogenic bacterium X. campestris pv. campestris can sense a plant CK, and, upon perception, supports bacterial growth under oxidative stress. PcrK orthologs are conserved in other plant‐associated bacterial genera, for example, Pseudomonas or Dickeya (Wang et al., 2017) and it has been proposed that PcrKs act as a bacterial virulence factor by providing a means to cope with plant defence‐related oxidative stress. Consistent with this, analysis of microbial mutants revealed that fungal‐derived CKs often have virulence functions and likely suppress host defence responses (Spallek et al., 2018; Han and Kahmann, 2019). CKs are often found in (hemi)biotrophic fungi and have been linked to the formation of so called ‘green islands’, that is, areas around fungal infection sites, where senescence processes are thought to be inhibited and nutrient fluxes redirected in order to support the biotrophic growth phase of the intruder (Walters and McRoberts, 2006; Walters et al., 2008; Spallek et al., 2018). However, CK production is not restricted to pathogenic fungi, and may support some general physiological processes, for example, during hyphal growth, nutrient uptake, growth under unfavourable conditions and sexual reproduction (Chanclud and Morel, 2016). CKs accumulate in AMF‐colonised plants, but it is unclear if they derive from the fungus or the host plant, and how much they contribute to the infection process (Boivin et al., 2016; Chanclud and Morel, 2016; Bedini et al., 2018; Das and Gutjahr, 2020). CK production has been assigned to some ectomycorrhizal fungi, but whether there is a functional role in establishing the symbiosis is not known (Morrison et al., 2015; Boivin et al., 2016). Rhizobia can produce CKs alongside IAA, and, through the release of Nod factors, induce endogenous CK accumulation and nodule formation. However, bacteria‐produced CKs alone seem not to be sufficient for nodule formation (Frugier et al., 2008; Kisiala et al., 2013; Boivin et al., 2016; Miri et al., 2016; Foo, 2020).
The contribution of CK biosynthesis of PGPBs to alter plant development is less well documented, and probably often masked by the effects of concomitantly produced IAA or GAs (Ortíz‐Castro et al., 2009; Sgroy et al., 2009; Dodd et al., 2010; Vacheron et al., 2013; Spaepen, 2015; Kudoyarova et al., 2019). At least at high levels, exogenous application of CKs may even inhibit root growth, potentially through the induction of ET production, and in some cases plant growth inhibition has been connected to bacterial CK biosynthesis (Dodd et al., 2010). Bacterial CK can act as biocontrol agent against pathogens: When applied to plant leaves, Pseudomonas fluorescens G20‐18 can activate plant resistance to pathogenic Pseudomonas syringae, and this is dependent on the bacterium’s ability to produce CK and the plant’s ability to perceive it (Großkinsky et al., 2016; Akhtar et al., 2020). Microbial production of CKs may also improve abiotic stress resistance, for example, against drought (Xu et al., 2012; Liu et al., 2013; Kudoyarova et al., 2019). Moreover, it has been reported that microbial‐produced CK can increase the release of root exudates, for example, amino acids, into the rhizosphere, and may thus have a broader impact on the rhizosphere microbiome (Kudoyarova et al., 2014).
Ethylene – a balancing act between microbial production and degradation
The inhibitory effects of high levels of (microbe‐produced) auxin or CK on root growth are often linked to simultaneously elevated levels of the gaseous hormone ET (Dodd et al., 2010; Glick, 2014; Gamalero and Glick, 2015). ET can inhibit the cell cycle and negatively affect cell division and meristem size in leaves and roots. ET inhibits root elongation and lateral root formation, but can increase the number of root hairs (Figure 3) (Dodd et al., 2010; Street et al., 2015; Van de Poel et al., 2015). Plants produce ET in response to a variety of stresses including pathogen attack, high salinity, flooding, heat, drought, nutrient deficiency and heavy metal toxicity, and can impair leaf and root growth as well as yield (Glick, 2012, 2014; Dubois et al., 2018). In plants, ET is synthesised from the amino acid methionine through sequential conversion into S‐adenosyl methionine and then into ACC through ACC synthase. ET production can be adjusted through the regulation of ACC synthase abundance/activity or the availability of the ET precursor ACC (Dubois et al., 2018; Nascimento et al., 2018). ET can have positive or negative effects on fungal spore germination and hyphal growth, and can restrict root colonisation by specific endophytes or symbionts (Tudzynski and Sharon, 2002; Zhu et al., 2017; Liu et al., 2017a). Exogenous application of ET restricts plant colonisation by mycorrhizal fungi and limits arbuscule formation. At various stages, ET can also interfere with the formation and function of nodules (Guinel, 2015; Bedini et al., 2018; Das and Gutjahr, 2020; Foo, 2020; Ludwig‐Müller, 2020; Mathesius, 2020). Therefore, rhizobia have developed strategies to manipulate the plant ET biosynthesis pathway likely in order to reduce ET levels within the root. Several rhizobial species contain ACC deaminase enzymes, which can catabolise exuded ACC into ammonia and α‐ketobutyrate. In this way, rhizobia may be able to reduce the local amount of the ET precursor ACC, thus limiting its negative effect on nodule formation and/or function (Ma et al., 2002; Ma et al., 2003; Gamalero and Glick, 2015). In fact, overexpression of ACC deaminase in Mesorhizobium loti led to an increase in the number of nodules formed in L. japonicus (Conforte et al., 2010). In a similar way, co‐infection of Bradyrhizobium japonicum with ACC deaminase‐expressing rhizobacteria improved nodulation in mung bean (Vigna radiata) (Shaharoona et al., 2006). ACC deaminase‐producing bacteria also supported colonisation and arbuscule formation by the AMF Gigaspora rosea in cucumber (Cucumis sativus) (Gamalero et al., 2008). Especially under stress conditions, for example, high salinity, plants produce high amounts of ACC and ET. If not converted into ET, ACC can be excreted from roots into the rhizosphere (Glick, 2014; Nascimento et al., 2018; Liu et al., 2019). High ACC deaminase activity is a frequent feature of free‐living or endophytic PGPBs and fungi (Ma et al., 2003; Glick, 2005; Dodd et al., 2010; Orozco‐Mosqueda et al., 2020). ACC deaminase‐producing bacteria are more abundant in the rhizosphere of plants grown under stress conditions (Nascimento et al., 2018). PGPBs may use their ACC deaminase activity to convert ACC and utilise it as a growth substrate outside the plant. It has been proposed that by using up ACC outside plant roots, PGPBs may create a sink that can protect plants from growth‐inhibiting levels of stress‐related ET and alleviate detrimental effects of unfavourable environmental conditions (Glick, 2005; Glick, 2014; Gamalero and Glick, 2015; Nascimento et al., 2018; Kudoyarova et al., 2019; Orozco‐Mosqueda et al., 2020). In PGPBs, the ability to produce IAA is often found together with ACC deaminase activity. Reducing growth‐inhibiting levels of IAA‐induced ET may help to increase the plant growth‐promoting potential of bacterial IAA in the presence and absence of plant stress (Glick, 2014; Belimov et al., 2015; Kudoyarova et al., 2019; Orozco‐Mosqueda et al., 2020). Whether ACC consumption by microbes affects rhizosphere microbiomes directly is not known.
Some pathogenic and non‐pathogenic bacterial and fungal microorganisms can produce ET, often using methionine as a precursor. Botrytis cinerea, Fusarium oxysporum f. sp. tulipae, Alternaria alternata, Ralstonia solanacearum and P. syringae pathovars are among the ET‐producing plant pathogens (Arshad and Frankenberger, 1990; Nagahama et al., 1992, 1994; Weingart and Völksch, 1997; Weingart et al., 2001; Tudzynski and Sharon, 2002; Valls et al., 2006; Kazan and Lyons, 2014; Zhu et al., 2017). In some Colletotrichum spp., which cause post‐harvesting diseases on fruits, (fruit‐derived) ET supports spore germination, hyphal growth and appressorium formation, thus supporting fungal virulence (Flaishman and Kolattukudy, 1994). The brown spot pathogen Cochliobolus miyabeanus produces ET to promote infection of rice plants, most likely through the suppression of cellular defence responses (Van Bockhaven et al., 2015). Some pathovars of P. syringae can produce ET in culture and during infection in planta. However, disruption of the bacterial gene encoding the ET‐forming enzyme reduced pathogen virulence in one pathovar, but not in another, suggesting that pathogen‐derived ET is not a general virulence factor in pathogenic plant–P. syringae interactions (Weingart and Völksch, 1997; Weingart et al., 2001). Despite the negative effect ET has on nodule formation, some rhizobia, for example, B. japonicum, can produce ET in culture media supplemented with methionine (Boiero et al., 2007). ET production has been shown for some ectomycorrhizal fungi, and the hormone seems to support symbiosis, potentially through the interference with host immunity or by inducing plant auxin production and lateral root formation (Splivallo et al., 2009; Boivin et al., 2016; Chanclud and Morel, 2016). Organic compounds such as carbohydrates, amino acids (especially methionine) and organic acids present in root exudates can stimulate microbial ET production. Consequently, the rhizosphere can be rich in ET‐producing microbes (Arshad and Frankenberger, 1990, 1991). ET biosynthesis has been shown for a broad range of rhizobacteria including Azospirillum, Azotobacter and Bacillus spp. (Nagahama et al., 1992; Cassán et al., 2014). Due to its potentially suppressive effect on other soil microbes, microbe‐produced ET may influence soil microbial populations directly (Smith, 1973; Smith and Cook, 1974) and rhizosphere ET levels can reach concentrations that can affect plant development (Arshad and Frankenberger, 1991). Although generally considered as root growth inhibitory, microbial ET production may enhance root hair formation/elongation (Figure 3) (Ribaudo et al., 2006; Galland et al., 2012; Vacheron et al., 2013).
Together, the combined potential of soil microbes to elevate or reduce ET levels in and around plants may provide a greater phenotypic plasticity, especially in response to various stress factors (Ravanbakhsh et al., 2018). Given the potential antimicrobial activity of ET and the usability of its precursor ACC as nutrient source, microbial activities that change local concentrations of ET or ACC may impact on co‐habitation in microbial niches in the rhizosphere or have broader implications in rhizobiome community composition.
Gibberellic acid – stimulant bridging microbial development and host colonisation
GAs regulate primary root elongation, increase the number of lateral roots (Dodd et al., 2010; Vanstraelen and Benková, 2012) and are thereby involved in the creation of microbial habitats (Figure 3). GAs were first isolated from the phytopathogenic fungus Fusarium fujikuroi (syn. Gibberella fujikuroi). Only a few bioactive GAs (e.g. GA1, GA3, GA4, GA7) affect plant growth and development, with major impacts on cell division and elongation, stem and root elongation, seed germination and flower and seed development (Yamaguchi, 2008). The typical symptoms of the ‘bakanae’ or ‘foolish seedlings’ disease of rice caused predominantly by F. fujikuroi are an excessive elongation and yellowing of diseased leaves, which can be directly attributed to the hormonal function of bioactive GAs produced by the fungus (Wulff et al., 2010; Jeon et al., 2013; Suga et al., 2019). Interestingly, although GA biosynthesis gene clusters are present in related plant‐colonising Fusarium spp., GA biosynthesis is not a general feature, but is limited to F. fujikuroi (Bömke and Tudzynski, 2009; Wiemann et al., 2013). The fact that a F. fujikuroi mutant lacking GA biosynthesis was strongly confined in its ability to invade rice cells points to a role of GAs as virulence effectors in F. fujikuroi (Wiemann et al., 2013). Whether the virulence of a F. fujikuroi strain is dependent on the amount of GAs it produces is still not entirely clear. The biosynthesis of bioactive GAs has been detected in a number of other plant pathogenic and non‐pathogenic fungi (e.g. Phaeosphaeria sp., Aspergillus niger, Neurospora crassa, Penicillium spp.) and in bacteria (e.g. some strains of Acetobacter, Azospirillum, Bacillus, Bradyrhizobium and Rhizobium spp.) (MacMillan, 2001; Bottini et al., 2004; Bömke and Tudzynski, 2009; Dodd et al., 2010; Khan et al., 2015; Spaepen, 2015; Tsukanova et al., 2017; Salazar‐Cerezo et al., 2018). Strikingly, the biosynthetic gene cluster for the GA precursor ent‐kaurene was identified to be significantly enriched in genomes of plant‐associated bacteria, suggesting that GA may support bacterial colonisation of plant environments (Levy et al., 2018a). GAs can support spore germination and hyphal growth in different fungal species, for example, F. fujikuroi, Rhizophagus irregulare and Penicillium spp., but do not seem to have a broader physiological function in fungi (Nakamura et al., 1988; Rademacher, 1994; Mercy et al., 2017). It is also unclear why some AMF produce GAs, as exogenous GA application inhibits AM formation, and GAs are generally considered to have a negative impact on this symbiosis (Barea and Azcón‐Aguilar, 1982; Bedini et al., 2018; Das and Gutjahr, 2020; Foo, 2020). As in other fungi, GAs may support hyphal growth of AMF. This is possibly supported by the observation that, although it inhibited arbuscule formation, treatment of L. japonicus with GA3 promoted hyphal growth and branching inside the root (Takeda et al., 2015). In addition to F. fujikuroi, several endophytic fungi can produce GAs in culture media. Some of these fungi transfer beneficial traits to their host plants, including growth promotion and increased stress tolerance (Khan et al., 2015). Whether these beneficial effects of endophytic fungi can be associated with their ability to produce GAs remains to be determined.
Rhizobia such as B. japonicum, Sinorhizobium fredii and Rhizobium phaseoli possess GA biosynthesis gene clusters, and can produce GAs (Atzorn et al., 1988; Boiero et al., 2007; Bano et al., 2010; Méndez et al., 2014; Nett et al., 2017). During nodulation, GAs inhibit initial colonisation events, but, likely due to their function in cell division and elongation, support nodule organogenesis (Foo, 2020). Although expression of rhizobial GA biosynthesis genes and the presence of presumably bacteria‐derived GAs was demonstrated in symbiotic bacteroids and nodules, GA operon knockouts in B. japonicum did not affect nodule formation and symbiosis, and therefore the physiological function of GA biosynthesis in rhizobia–plant interactions remains unclear (Tully and Keister, 1993; Rademacher, 1994; Nett et al., 2017). Interestingly, the bacterial leaf pathogen Xanthomonas oryzae pv. oryzicola possesses a GA biosynthesis operon, which is homologous to the one in rhizobia (Lu et al., 2015; Nett et al., 2017). The insertional disruption of genes within this operon led to reduced virulence of the bacterial pathogen on rice plants, and this was accompanied by an increased expression of marker genes for JA‐mediated defence. It has been suggested that X. oryzae pv. oryzicola likely produces a GA that can antagonise JA‐mediated defence, thus supporting bacterial virulence on its host plant.
PGPBs can produce bioactive forms of GA, or release them enzymatically from inactive conjugated forms (Bottini et al., 2004; Bömke and Tudzynski, 2009; Dodd et al., 2010; Glick, 2012; Spaepen, 2015). Low levels of available N stimulate GA production by bacteria in culture, while impaired gas exchange and osmotic stress reduce it (Bottini et al., 2004). Often an increase in root and/or shoot growth and germination can be observed after infection with GA‐producing PGPBs (Cassán et al., 2014; Shahzad et al., 2016). Inoculation with a GA‐producing Bacillus amyloliquefaciens increased growth of rice plants and suppressed endogenous levels of JA and ABA (Shahzad et al., 2016). Plants treated with GA‐producing bacteria can also be more tolerant to abiotic stress factors. Pepper (Capsicum annuum) plants inoculated with a GA‐producing strain of Serratia nematodiphila accumulated more GA and ABA under cold treatment (Kang et al., 2015). However, unless bacterial hormone biosynthesis mutants become available, it will be hard to evaluate which direct effects the microbial‐derived GAs have on plant growth.
Abscisic acid – driver of the microbe–plant fitness alliance
The plant hormone ABA regulates developmental processes, is involved in biotic and abiotic stress responses, adjusts (root) growth and controls transpiration under stress conditions such as drought, high salinity or heavy metal toxicity (Ton et al., 2009; Cutler et al., 2010; Pieterse et al., 2012; Vishwakarma et al., 2017). Many plant colonising and saprophytic fungi can produce ABA. Botrytis cinerea, M. oryzae, U. maydis, Verticillium dahliae and Alternaria brassicicola are among the plant pathogenic fungi capable of ABA biosynthesis (Kettner and Dörffling, 1995; Siewers et al., 2006; Hartung, 2010; Bruce et al., 2011; Hauser et al., 2011; Spence and Bais, 2015; Spence et al., 2015; Shi et al., 2017; Han and Kahmann, 2019; Meents et al., 2019). ABA supports spore germination and fungal growth, and fungus‐derived ABA was required for appressorium formation and supported virulence of M. oryzae on rice plants (Spence and Bais, 2015; Spence et al., 2015; Chanclud and Morel, 2016). Exogenous ABA application at relatively low levels supports plant colonisation by AMF and arbuscule formation, while higher concentrations have negative effects on colonisation (Bedini et al., 2018; Das and Gutjahr, 2020; Foo, 2020). ABA treatment reduced the germination rate of spores of the AMF Rhizoglomus irregulare, but increased hyphal branching (Mercy et al., 2017). AMF not only increase endogenous levels of plant ABA during colonisation, they can also synthesise ABA, but it is unclear how much fungus‐derived ABA contributes to AM symbiosis (Esch et al., 1994; Chanclud and Morel, 2016; Bedini et al., 2018; Ludwig‐Müller, 2020). ABA production was also shown for some ectomycorrhizal fungi and some rhizobia, for example, B. japonicum (Boiero et al., 2007; Bano et al., 2010; Morrison et al., 2015). However, ABA is generally known to inhibit nodule formation, apparently by interfering with Nod factor signalling (Foo, 2020; Mathesius, 2020). Several rhizobacteria species can produce and secrete ABA in culture media, and some of these have been shown to increase ABA levels in plants (Forchetti et al., 2007; Sgroy et al., 2009; Dodd et al., 2010; Cohen et al., 2015; Tsukanova et al., 2017; Rosier et al., 2018). The physiological function of ABA in bacteria is unclear. Endophytic bacteria of sunflower (Helianthus annuus) produce more ABA (and JA) under osmotic stress. Azospirillum strains isolated from arid/semi‐arid or water‐stressed areas contained more ABA than those isolated from well‐watered areas, and wheat plants performed better under water stress when they were inoculated with the high‐ABA level Azospirillum strains (Forchetti et al., 2007; Ilyas and Bano, 2010). An ABA‐producing Azospirillum strain also increased Arabidopsis root length and ABA levels, even in an ABA biosynthesis mutant. Wild‐type plants inoculated with the ABA producer were also more tolerant to drought stress (Cohen et al., 2015). Conversely, some rhizobacteria possess the ability to degrade ABA (Hasegawa et al., 1984; Belimov et al., 2014). Two of these strains were shown to use ABA as a sole carbon source when grown in media, change root morphology and lower ABA levels in roots and/or shoots when applied to plants. However, the lack of bacterial mutants defective in ABA biosynthesis or metabolism makes it difficult to determine whether the changes in plant ABA levels and root morphology, as well as increased plant stress tolerance, are direct effects of bacterial ABA production or degradation (Dodd et al., 2010; Kudoyarova et al., 2019).
Salicylic acid and jasmonic acid – targets for microbial manipulation of host communication
SA and JA are major regulators of mutually inhibiting defence signalling pathways against (hemi)biotrophic and necrotrophic pathogens, respectively (Glazebrook, 2005; Pieterse et al., 2012). SA and JA can also have direct negative effects on fungal or bacterial growth (Miersch et al., 1999; Hao et al., 2019). Many plant pathogens can interfere with either SA (e.g. U. maydis, Phytophthora sojae, V. dahliae) or JA (e.g. M. oryzae) accumulation or defence signalling in order to support virulence (Patkar et al., 2015; Lanver et al., 2017). Depending on its own lifestyle, a pathogen may make use of the negative cross‐talk between SA and JA signalling and activate one of the pathways in order to suppress the other. Several pathogen effectors have been identified, which interfere with SA or JA defence signalling components (Kazan and Lyons, 2014; Gimenez‐Ibanez et al., 2016; Han and Kahmann, 2019). Some pathogenic fungi or oomycetes can already prevent the formation of or degrade plant‐derived SA: P. sojae and V. dahliae secrete isochorismatases, which hydrolyse the SA precursor isochorismate, in order to suppress SA‐mediated defence responses (Liu et al., 2014). Ustilago maydis secretes both a chorismate mutase, which converts the SA precursor chorismate, and an SA hydroxylase, which converts SA into catechol (Djamei et al., 2011; Rabe et al., 2013). However, while the former enzyme is associated with lower SA levels inside the host plant and better colonisation, the latter can help the fungus to use SA as carbon source, albeit without any apparent virulence function. SA hydroxylases have been identified in other (phytopathogenic) fungi and soil bacteria (Rabe et al., 2013; Hao et al., 2019). Some pathogenic microbes can also produce SA, JA, JA conjugates or molecular mimics. SA has, for example, been detected in mycelia of the biotrophic fungal pathogen V. dahliae and the necrotroph A. brassicicola (Gimenez‐Ibanez et al., 2016; Meents et al., 2019). Gibberella fujikuroi, M. oryzae, some species of the genus Lasiodiplodia and certain ff. spp. of F. oxysporum are among the JA‐producing pathogenic fungi (Miersch et al., 1992; Cole et al., 2014; Gimenez‐Ibanez et al., 2016). At least the causal agent of the witch’s broom disease of cocoa (Theobroma cacao), Moniliophthora perniciosa, can even produce both, SA and JA (plus IAA and ABA), and both hormones supported fungal growth in vitro (Kilaru et al., 2007). However, it is not always clear how or to what extent the microbial production of SA and/or JA contributes to pathogen virulence (Thatcher et al., 2009; Cole et al., 2014; Chanclud and Morel, 2016). Gibberella fujikuroi and some ff. spp. of F. oxysporum can produce the bioactive JA conjugate JA‐Ile, which can bind to the COI1–JAZ co‐receptor to activate plant JA signalling, and may therefore alter JA signalling directly, to promote virulence (Cole et al., 2014; Kazan and Lyons, 2014). In a similar way, some strains of the bacterial plant pathogen P. syringae produce coronatine, which mimics JA‐Ile, and can therefore bind to the COI1–JAZ co‐receptor as well. Coronatine supports pathogen infection by activating JA signalling, thus re‐opening stomata for bacterial entry, suppressing SA‐mediated defence responses and promoting disease symptom development (Brooks et al., 2005; Melotto et al., 2006; Gimenez‐Ibanez et al., 2016; Kunkel and Harper, 2018). The grapevine (Vitis vinifera) pathogen Lasiodiplodia mediterranea produces an inactive JA ester, lasiojasmonate A, during the late stages of infection, and apparently lets the plant convert it into bioactive JA‐Ile in order to activate cell death‐related JA responses. This may support the fungus’ necrotrophic growth phase (Chini et al., 2018). In contrast, M. oryzae secretes a monooxygenase during plant colonisation, probably in order to convert fungus‐ and plant‐derived JA, thus avoiding activation of JA‐mediated defence responses (Patkar et al., 2015).
SA production has been detected in the beneficial root endophytic fungi S. indica and Mortierella hyalina, although it is not known whether this is required for plant colonisation or the beneficial effects on the host plants (Meents et al., 2019). The ectomycorrhizal fungus Pisolithus tinctorius can synthesise and metabolise JA, but it is not clear to what extent this affects mycorrhisation (Miersch et al., 1999).
PGPBs and plant growth‐promoting fungi can activate ISR against a broad spectrum of pathogens via SA‐ or JA (and ET)‐dependent signalling pathways and several (ISR‐inducing) PGPBs produce SA or JA (Forchetti et al., 2007; Van der Ent et al., 2009; Dodd et al., 2010; Bakker et al., 2014). Although in a few cases ISR activation has been attributed to some PGPBs’ ability to synthesise SA, bacterial SA biosynthesis does not seem to be a general requirement for it (De Meyer and Höfte, 1997; Maurhofer et al., 1998; De Meyer et al., 1999; Ran et al., 2005; Dodd et al., 2010; Bakker et al., 2014; Tsukanova et al., 2017; Rosier et al., 2018). Bacteria‐produced SA has also been discussed as a precursor for siderophores and its biosynthesis may support bacterial growth under iron‐limiting conditions (Bakker et al., 2014). Some antibacterial and antifungal activity has been attributed to SA and, furthermore, SA was shown to negatively affect biofilm formation and quorum sensing. These effects could impact the composition of soil microbial communities (Van Duy et al., 2007; Bakker et al., 2014; Lebeis et al., 2015). Conversely, SA degradation has been observed in several strains of the genera Arthrobacter, Bacillus and Pseudomonas (Dodd et al., 2010; Tsukanova et al., 2017). JA production has not been studied very intensively; however, JA (and ABA) production was detected in endophytic bacteria isolated from plants grown under drought stress, and it has been suggested that this may help plants to cope with stress conditions such as drought or salinity (Forchetti et al., 2007).
In summary, the ability to perceive, produce and/or degrade hormones is commonly found in a broad range of microbes. Depending on microbial lifestyles and niche preferences, the usage of hormone‐based communication serves at least two aims. In addition to enhancing their competitiveness in the rhizosphere by inhibiting growth or fitness of competing microbes, microbes employ hormones to prepare the plant endosphere for colonisation by adjusting root development and manipulating plant defence.
CONCLUSION AND OUTLOOK
Plants and microbes have co‐evolved diverse strategies to communicate and thus interact with each other (Leach et al., 2017). From a plant perspective, it is a balancing act. Plants invest significant resources such as nutrients to create an attractive rhizosphere habitat for microbes. The merit to annex beneficial microbes and their beneficial activities comes with the risk of wasting costly resources to unhelpful or even detrimental microbes. It is therefore crucial for plants to have selective mechanisms in place to have some control over the access to the resource‐rich rhizosphere and to balance interactions. Hormones apparently take an essential role here and despite our limited knowledge seem to be as important for microbiome assembly as they are for the outcome of bilateral plant–microbe interactions.
For plants, hormones represent evolution‐driven, effective mediators to adapt to a diversity of environmental stimuli and to coordinate complex developmental processes (e.g. root system architecture). Considering the versatile roles of hormones, it is not surprising that microbes have adapted to and even hijacked hormone signalling. As a kind of common chemical language, hormones have evolved into go‐betweens of microbes and plants to implement conditions for the establishment of niches at the rhizosphere and to facilitate targeted host adaptation for endosphere colonisation. Astonishingly, microbes do not only synthesise hormones but perceive hormones to activate signalling processes to outcompete competitors and build alliances, suggesting the adoption of hormones in steering microbe–microbe interactions in a plant‐independent manner.
While the importance of hormones in the outcome of plant–microbe interactions is well known, we are just starting to understand the go‐between role of plant‐ as well as microbe‐derived hormones in root microbiome assembly. Our efforts to employ microbial communities, for instance for more sustainable and biodiversified crop production systems, require a better understanding of hormonal activities and their prevalence in plant–microbiome interactions. In this respect, it is essential to detect the origin of hormone synthesis and signalling (e.g. plant, tissue, cell type, microbe, species, rhizosphere, endosphere, etc.) and to determine their effect on plants and the assembly of microbial communities under different or changing environments. Recent advances in omics‐based technologies such as metatranscriptomics, metaproteomics or metabolomics (Levy et al., 2018a, 2018b,2018a, 2018b) combined with amplicon sequencing can help us in cataloguing hormonal processes in plant/root holobionts. Metabolomic profiling of root exudates will facilitate the identification of those compounds impacted by hormone signalling with a potential to shape the microbiome whilst also giving greater insight into the functional molar ranges of exuded plant‐derived and microbe‐derived hormones. These techniques are already being employed with some success (Dafoe et al., 2011; Badri et al., 2013; Chaparro et al., 2013; Zhalnina et al., 2018; Kudjordjie et al., 2019). These technologies do not replace amplicon sequencing but provide an additional, richer layer of information describing the broad functional nature of communities, not limited to only taxonomic classifications. To further assign hormone activities to individual members and to gain mechanistic insights requires functional approaches such as cell‐based assays with plants also lacking defined hormone synthesis and signalling components (Yoo et al., 2007; Lehmann et al., 2020). In addition to reductionist approaches with single microbes those functional analyses can be applied to characterise and assemble synthetic communities. Finally, we need to advance techniques to culture microbes and share knowledge and microbial strains (isolates) through existing resources and infrastructures. Culture collections curated by non‐profit biological resource centres can host valuable information (e.g. origin [soil type, plant association, etc.], activities, links to omics data, etc.) to support efforts to generate synthetic microbial communities and to validate their functionality in different ecosystems (e.g. as part of detoxification, renaturation, crop cultivation, etc.). Understanding the communication patterns of plants and microbes is of outstanding importance in this process to access the full genetic and ecological potential of microbiomes.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGEMENTS
This work was funded by a research grant (BB/S012877/2 to RE and PS) from the Biotechnological and Biological Research Council (BBSRC) of the United Kingdom. LR was funded by the Biotechnology and Biological Sciences Research Council through MIBTP. Open Access funding enabled and organized by ProjektDEAL.
References
- Akhtar, S.S. , Mekureyaw, M.F. , Pandey, C. and Roitsch, T. (2020) Role of cytokinins for interactions of plants with microbial pathogens and pest insects. Front. Plant Sci. 10, 1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama, K. and Hayashi, H. (2006) Strigolactones: chemical signals for fungal symbionts and parasitic weeds in plant roots. Ann. Bot. 97, 925–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama, K. , Matsuzaki, K. and Hayashi, H. (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature, 435, 824–827. [DOI] [PubMed] [Google Scholar]
- Akiyama, K. , Ogasawara, S. , Ito, S. and Hayashi, H. (2010) Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 51, 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Babili, S. and Bouwmeester, H.J. (2015) Strigolactones, a novel carotenoid‐derived plant hormone. Annu. Rev. Plant Biol. 66, 161–186. [DOI] [PubMed] [Google Scholar]
- Aliche, E.B. , Screpanti, C. , De Mesmaeker, A. , Munnik, T. and Bouwmeester, H.J. (2020) Science and application of strigolactones. New Phytol. 227, 1001–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antolín‐Llovera, M. , Petutsching, E.K. , Ried, M.K. , Lipka, V. , Nürnberger, T. , Robatzek, S. and Parniske, M. (2014) Knowing your friends and foes ‐ plant receptor‐like kinases as initiators of symbiosis or defence. New Phytol. 204, 791–802. [DOI] [PubMed] [Google Scholar]
- Arshad, M. and Frankenberger, W.T. (1990) Production and stability of ethylene in soil. Biol. Fertil. Soils, 10, 29–34. [Google Scholar]
- Arshad, M. and Frankenberger, W.T. (1991) Microbial production of plant hormones. Plant Soil, 133, 1–8. [Google Scholar]
- Atzorn, R. , Crozier, A. , Wheeler, C.T. and Sandberg, G. (1988) Production of gibberellins and indole‐3‐acetic acid by Rhizobium phaseoli in relation to nodulation of Phaseolus vulgaris roots. Planta, 175, 532–538. [DOI] [PubMed] [Google Scholar]
- Badri, D.V. , Chaparro, J.M. , Zhang, R. , Shen, Q. and Vivanco, J.M. (2013) Application of natural blends of phytochemicals derived from the root exudates of Arabidopsis to the soil reveal that phenolic‐related compounds predominantly modulate the soil microbiome. J. Biol. Chem. 288, 4502–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Y. , Müller, D.B. , Srinivas, G. et al . (2015) Functional overlap of the Arabidopsis leaf and root microbiota. Nature, 528, 364–369. [DOI] [PubMed] [Google Scholar]
- Bakker, P.A.H.M. , Pieterse, C.M.J. , de Jonge, R. and Berendsen, R.L. (2018) The soil‐borne legacy. Cell, 172, 1178–1180. [DOI] [PubMed] [Google Scholar]
- Bakker, P.A.H.M. , Ran, L.X. and Mercado‐Blanco, J. (2014) Rhizobacterial salicylate production provokes headaches!. Plant Soil, 382, 1–16. [Google Scholar]
- Bano, A. , Batool, R. and Dazzo, F. (2010) Adaptation of chickpea to desiccation stress is enhanced by symbiotic rhizobia. Symbiosis, 50, 129–133. [Google Scholar]
- Barea, J.M. and Azcón‐Aguilar, C. (1982) Production of plant growth‐regulating substances by the vesicular‐arbuscular mycorrhizal fungus Glomus mosseae . Appl. Environ. Microbiol. 43, 810–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barea, J.M. , Pozo, M.J. , Azcón, R. and Azcón‐Aguilar, C. (2005) Microbial co‐operation in the rhizosphere. J. Exp. Bot. 56, 1761–1778. [DOI] [PubMed] [Google Scholar]
- Baudoin, E. , Lerner, A. , Mirza, M.S. et al . (2010) Effects of Azospirillum brasilense with genetically modified auxin biosynthesis gene ipdC upon the diversity of the indigenous microbiota of the wheat rhizosphere. Res. Microbiol. 161, 219–226. [DOI] [PubMed] [Google Scholar]
- Bedini, A. , Mercy, L. , Schneider, C. , Franken, P. and Lucic‐Mercy, E. (2018) Unraveling the initial plant hormone signaling, metabolic mechanisms and plant defense triggering the endomycorrhizal symbiosis behavior. Front. Plant Sci. 9, 1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belimov, A.A. , Dodd, I.C. , Safronova, V.I. , Dumova, V.A. , Shaposhnikov, A.I. , Ladatko, A.G. and Davies, W.J. (2014) Abscisic acid metabolizing rhizobacteria decrease ABA concentrations in planta and alter plant growth. Plant Physiol. Biochem. 74, 84–91. [DOI] [PubMed] [Google Scholar]
- Belimov, A.A. , Dodd, I.C. , Safronova, V.I. , Shaposhnikov, A.I. , Azarova, T.S. , Makarova, N.M. , Davies, W.J. and Tikhonovich, I.A. (2015) Rhizobacteria that produce auxins and contain 1‐amino‐cyclopropane‐1‐carboxylic acid deaminase decrease amino acid concentrations in the rhizosphere and improve growth and yield of well‐watered and water‐limited potato (Solanum tuberosum). Ann. Appl. Biol. 167, 11–25. [Google Scholar]
- Berendsen, R.L. , Pieterse, C.M.J. and Bakker, P.A.H.M. (2012) The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. [DOI] [PubMed] [Google Scholar]
- Berendsen, R.L. , Vismans, G. , Yu, K. , Song, Y. , de Jonge, R. , Burgman, W.P. , Burmølle, M. , Herschend, J. , Bakker, P.A.H.M. and Pieterse, C.M.J. (2018) Disease‐induced assemblage of a plant‐beneficial bacterial consortium. ISME J. 12, 1496–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berens, M.L. , Berry, H.M. , Mine, A. , Argueso, C.T. and Tsuda, K. (2017) Evolution of hormone signaling networks in plant defense. Annu. Rev. Phytopathol. 55, 401–425. [DOI] [PubMed] [Google Scholar]
- Berg, G. , Grube, M. , Schloter, M. and Smalla, K. (2014) Unraveling the plant microbiome: looking back and future perspectives. Front. Microbiol. 5, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiero, L. , Perrig, D. , Masciarelli, O. , Penna, C. , Cassán, F. and Luna, V. (2007) Phytohormone production by three strains of Bradyrhizobium japonicum and possible physiological and technological implications. Appl. Microbiol. Biotechnol. 74, 874–880. [DOI] [PubMed] [Google Scholar]
- Boivin, S. , Fonouni‐Farde, C. and Frugier, F. (2016) How auxin and cytokinin phytohormones modulate root microbe interactions. Front. Plant Sci. 7, 1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bömke, C. and Tudzynski, B. (2009) Diversity, regulation, and evolution of the gibberellin biosynthetic pathway in fungi compared to plants and bacteria. Phytochemistry, 70, 1876–1893. [DOI] [PubMed] [Google Scholar]
- Bottini, R. , Cassán, F. and Piccoli, P. (2004) Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl. Microbiol. Biotechnol. 65, 497–503. [DOI] [PubMed] [Google Scholar]
- Brooks, D.M. , Bender, C.L. and Kunkel, B.N. (2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid‐dependent defences in Arabidopsis thaliana . Mol. Plant Pathol. 6, 629–639. [DOI] [PubMed] [Google Scholar]
- Brown, S.P. , Grillo, M.A. , Podowski, J.C. and Heath, K.D. (2020) Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula . Microbiome, 8, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce, S.A. , Saville, B.J. and Emery, R.J.N. (2011) Ustilago maydis prroduces cytokinins and abscisic acid for potential regulation of tumor formation in maize. J. Plant Growth Regul. 30, 51–63. [Google Scholar]
- Bulgarelli, D. , Garrido‐Oter, R. , Münch, P.C. , Weiman, A. , Dröge, J. , Pan, Y. , McHardy, A.C. and Schulze‐Lefert, P. (2015) Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe, 17, 392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli, D. , Rott, M. , Schlaeppi, K. et al . (2012) Revealing structure and assembly cues for Arabidopsis root‐inhabiting bacterial microbiota. Nature, 488, 91–95. [DOI] [PubMed] [Google Scholar]
- Bulgarelli, D. , Schlaeppi, K. , Spaepen, S. , van Themaat, E.V.L. and Schulze‐Lefert, P. (2013) Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838. [DOI] [PubMed] [Google Scholar]
- Bürger, M. and Chory, J. (2019) Stressed out about hormones: how plants orchestrate immunity. Cell Host Microbe, 26, 163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadot, S. , Guan, H. , Bigalke, M. , Walser, J.C. , Jander, G. , Erb, M. , van der Heijden, M. and Schlaeppi, K. (2020) Specific and conserved patterns of microbiota‐structuring by maize benzoxazinoids in the field. bioRxiv. 10.1101/2020.05.03.075135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammarata, J. , Roeder, A.H. and Scanlon, M.J. (2019) Cytokinin and CLE signaling are highly intertwined developmental regulators across tissues and species. Curr. Opin. Plant Biol. 51, 96–104. [DOI] [PubMed] [Google Scholar]
- Carvalhais, L.C. , Dennis, P.G. , Badri, D.V. , Kidd, B.N. , Vivanco, J.M. and Schenk, P.M. (2015) Linking jasmonic acid signaling, root exudates, and rhizosphere microbiomes. Mol. Plant Microbe Interact. 28, 1049–1058. [DOI] [PubMed] [Google Scholar]
- Carvalhais, L.C. , Dennis, P.G. , Badri, D.V. , Tyson, G.W. , Vivanco, J.M. and Schenk, P.M. (2013) Activation of the jasmonic acid plant defence pathway alters the composition of rhizosphere bacterial communities. PLoS One, 8, e56457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalhais, L.C. , Rincon‐Florez, V.A. , Brewer, P.B. , Beveridge, C.A. , Dennis, P.G. and Schenk, P.M. (2019) The ability of plants to produce strigolactones affects rhizosphere community composition of fungi but not bacteria. Rhizosphere, 9, 18–26. [Google Scholar]
- Carvalhais, L.C. , Schenk, P.M. and Dennis, P.G. (2017) Jasmonic acid signalling and the plant holobiont. Curr. Opin. Microbiol. 37, 42–47. [DOI] [PubMed] [Google Scholar]
- Cassán, F. , Vanderleyden, J. and Spaepen, S. (2014) Physiological and agronomical aspects of phytohormone production by model plant‐growth‐promoting rhizobacteria (PGPR) belonging to the genus Azospirillum . J. Plant Growth Regul. 33, 440–459. [Google Scholar]
- Chanclud, E. and Morel, J.B. (2016) Plant hormones: a fungal point of view. Mol. Plant Pathol. 17, 1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanclud, E. , Kisiala, A. , Emery, N.R.J. , Chalvon, V. , Ducasse, A. , Romiti‐Michel, C. , Gravot, A. , Kroj, T. and Morel, J.B. (2016) Cytokinin production by the rice blast fungus is a pivotal requirement for full virulence. PLoS Pathog. 12, e1005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro, J.M. , Badri, D.V. and Vivanco, J.M. (2014) Rhizosphere microbiome assemblage is affected by plant development. ISME J. 8, 790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro, J.M. , Badri, D.V. , Bakker, M.G. , Sugiyama, A. , Manter, D.K. and Vivanco, J.M. (2013) Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One, 8, e55731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X. , Marszałkowska, M. and Reinhold‐Hurek, B. (2020a) Jasmonic acid, not salicyclic acid restricts endophytic root colonization of rice. Front. Plant Sci. 10, 1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Bonkowski, M. , Shen, Y. , Griffiths, B.S. , Jiang, Y. , Wang, X. and Sun, B. (2020b) Root ethylene mediates rhizosphere microbial community reconstruction when chemically detecting cyanide produced by neighbouring plants. Microbiome, 8, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X. , Ruyter‐Spira, C. and Bouwmeester, H. (2013) The interaction between strigolactones and other plant hormones in the regulation of plant development. Front. Plant Sci. 4, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini, A. , Cimmino, A. , Masi, M. , Reveglia, P. , Nocera, P. , Solano, R. and Evidente, A. (2018) The fungal phytotoxin lasiojasmonate A activates the plant jasmonic acid pathway. J. Exp. Bot. 69, 3095–3102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, J. , Huh, S.U. , Kojima, M. , Sakakibara, H. , Paek, K.H. and Hwang, I. (2010) The cytokinin‐activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1‐dependent salicylic acid signaling in Arabidopsis . Dev. Cell, 19, 284–295. [DOI] [PubMed] [Google Scholar]
- Clear, M.R. and Hom, E.F.Y. (2019) The evolution of symbiotic plant–microbe signalling. Annu. Plant Rev Online, 2, 1–52. [Google Scholar]
- Cohen, A.C. , Bottini, R. , Pontin, M. , Berli, F.J. , Moreno, D. , Boccanlandro, H. , Travaglia, C.N. and Piccoli, P.N. (2015) Azospirillum brasilense ameliorates the response of Arabidopsis thaliana to drought mainly via enhancement of ABA levels. Physiol. Plant, 153, 79–90. [DOI] [PubMed] [Google Scholar]
- Cole, S.J. , Yoon, A.J. , Faull, K.F. and Diener, A.C. (2014) Host perception of jasmonates promotes infection by Fusarium oxysporum formae speciales that produce isoleucine‐ and leucine‐conjugated jasmonates. Mol. Plant Pathol. 15, 589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforte, V.P. , Echeverria, M. , Sánchez, C. , Ugalde, R.A. , Menéndez, A.B. and Lepek, V.C. (2010) Engineered ACC deaminase‐expressing free‐living cells of Mesorhizobium loti show increased nodulation efficiency and competitiveness on Lotus spp. J. Gen. Appl. Microbiol. 56, 331–338. [DOI] [PubMed] [Google Scholar]
- Contreras‐Cornejo, H.A. , López‐Bucio, J.S. , Méndez‐Bravo, A. , Macías‐Rodríguez, L. , Ramos‐Vega, M. , Guevara‐García, Á.A. and López‐Bucio, J. (2015) Mitogen‐activated protein kinase 6 and ethylene and auxin signaling pathways are involved in Arabidopsis root‐system architecture alterations by Trichoderma atroviride . Mol. Plant Microbe Interact. 28, 701–710. [DOI] [PubMed] [Google Scholar]
- Contreras‐Cornejo, H.A. , Macías‐Rodríguez, L. , Cortés‐Penagos, C. and López‐Bucio, J. (2009) Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin‐dependent mechanism in Arabidopsis. Plant Physiol. 149, 1579–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costacurta, A. and Vanderleyden, J. (1995) Synthesis of phytohormones by plant‐associated bacteria. Crit. Rev. Microbiol. 21, 1–18. [DOI] [PubMed] [Google Scholar]
- Cotton, T.E.A. , Pétriacq, P. , Cameron, D.D. , Meselmani, M.A. , Schwarzenbacher, R. , Rolfe, S.A. and Ton, J. (2019) Metabolic regulation of the maize rhizobiome by benzoxazinoids. ISME J. 13, 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregger, M.A. , Veach, A.M. , Yang, Z.K. , Crouch, M.J. , Vilgalys, R. , Tuskan, G.A. and Schadt, C.W. (2018) The Populus holobiont: dissecting the effects of plant niches and genotype on the microbiome. Microbiome, 6, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S.R. , Rodriguez, P.L. , Finkelstein, R.R. and Abrams, S.R. (2010) Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. [DOI] [PubMed] [Google Scholar]
- Dafoe, N.J. , Huffaker, A. , Vaughan, M.M. , Duehl, A.J. , Teal, P.E. and Schmelz, E.A. (2011) Rapidly induced chemical defenses in maize stems and their effects on short‐term growth of Ostrinia nubilalis . J. Chem. Ecol. 37, 984–991. [DOI] [PubMed] [Google Scholar]
- Das, D. and Gutjahr, C. (2020) Role of phytohormones in arbuscular mycorrhiza development. In The model legume Medicago truncatula ( de Bruijn, F. , ed). Hoboken, New Jersey: John Wiley & Sons, Inc., pp. 485–500. [Google Scholar]
- De Meyer, G. and Höfte, M. (1997) Salicylic acid produced by the rhizobacterium Pseudomonas aeruginosa 7NSK2 induces resistance to leaf infection by Botrytis cinerea on bean. Phytopathology, 87, 588–593. [DOI] [PubMed] [Google Scholar]
- De Meyer, G. , Audenaert, K. and Höfte, M. (1999) Pseudomonas aeruginosa 7NSK2‐induced systemic resistance in tobacco depends on in planta salicylic acid accumulation but is not associated with PR1a expression. Eur. J. Plant Pathol. 105, 513–517. [Google Scholar]
- Dennis, P.G. , Miller, A.J. and Hirsch, P.R. (2010) Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 72, 313–327. [DOI] [PubMed] [Google Scholar]
- Ding, Y. , Kalo, P. , Yendrek, C. , Sun, J. , Liang, Y. , Marsh, J.F. , Harris, J.M. and Oldroyd, G.E.D. (2008) Abscisic acid coordinates nod factor and cytokinin signaling during the regulation of nodulation in Medicago truncatula . Plant Cell, 20, 2681–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y. , Sun, T. , Ao, K. , Peng, Y. , Zhang, Y. , Li, X. and Zhang, Y. (2018) Opposite roles of salicylic acid receptors NPR1 and NPR3/NPR4 in transcriptional regulation of plant immunity. Cell, 173, 1454–1467. [DOI] [PubMed] [Google Scholar]
- Djamei, A. , Schipper, K. , Rabe, F. et al . (2011) Metabolic priming by a secreted fungal effector. Nature, 478, 395–398. [DOI] [PubMed] [Google Scholar]
- Dodd, I.C. , Zinovkina, N.Y. , Safronova, V.I. and Belimov, A.A. (2010) Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 157, 361–379. [Google Scholar]
- Dodueva, I.E. , Lebedeva, M.A. , Kuznetsova, K.A. , Gancheva, M.S. , Paponova, S.S. and Lutova, L.L. (2020) Plant tumors: a hundred years of study. Planta, 251, 82. [DOI] [PubMed] [Google Scholar]
- Dong, X. (2004) NPR1, all things considered. Curr. Opin. Plant Biol. 7, 547–552. [DOI] [PubMed] [Google Scholar]
- Doornbos, R.F. , Geraats, B.P.J. , Kuramae, E.E. , Van Loon, L.C. and Bakker, P.A.H.M. (2011) Effects of jasmonic acid, ethylene, and salicylic acid signaling on the rhizosphere bacterial community of Arabidopsis thaliana . Mol. Plant Microbe Interact. 24, 395–407. [DOI] [PubMed] [Google Scholar]
- Doornbos, R.F. , Van Loon, L.C. and Bakker, P.A.H.M. (2012) Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 32, 227–243. [Google Scholar]
- Dor, E. , Joel, D.M. , Kapulnik, Y. , Koltai, H. and Hershenhorn, J. (2011) The synthetic strigolactone GR24 influences the growth pattern of phytopathogenic fungi. Planta, 234, 419–427. [DOI] [PubMed] [Google Scholar]
- Dubois, M. , Van den Broeck, L. and Inzé, D. (2018) The pivotal role of ethylene in plant growth. Trends Plant Sci. 23, 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duca, D. , Lorv, J. , Patten, C.L. , Rose, D. and Glick, B.R. (2014) Indole‐3‐acetic acid in plant‐microbe interactions. Antonie Van Leeuwenhoek, 106, 85–125. [DOI] [PubMed] [Google Scholar]
- Erb, M. and Kliebenstein, D.J. (2020) Plant secondary metabolites as defenses, regulators, and primary metabolites: the blurred functional trichotomy. Plant Physiol. 184, 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch, H. , Hundeshagen, B. , Schneider‐Poetsch, H. and Bothe, H. (1994) Demonstration of abscisic acid in spores and hyphae of the arbuscular‐mycorrhizal fungus Glomus and in the N2‐fixing cyanobacterium Anabaena variabilis . Plant Sci. 99, 9–16. [Google Scholar]
- Escudero‐Martinez, C. and Bulgarelli, D. (2019) Tracing the evolutionary routes of plant‐microbiota interactions. Curr. Opin. Microbiol. 49, 34–40. [DOI] [PubMed] [Google Scholar]
- Faure, D. , Vereecke, D. and Leveau, J.H.J. (2009) Molecular communication in the rhizosphere. Plant Soil, 321, 279–303. [Google Scholar]
- Felten, J. , Kohler, A. , Morin, E. , Bhalerao, R.P. , Palme, K. , Martin, F. , Ditengou, F.A. and Legué, V. (2009) The ectomycorrhizal fungus Laccaria bicolor stimulates lateral root formation in poplar and Arabidopsis through auxin transport and signaling. Plant Physiol. 151, 1991–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, O.M. , Salas‐González, I. , Castrillo, G. , Conway, J.M. , Law, T.F. , Teixeira, P.J.P.L. , Wilson, E.D. , Fitzpatrick, C.R. , Jones, C.D. and Dangl, J.L. (2020) A single bacterial genus maintains root development in a complex microbiome. Nature, 587, 103–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, C.R. , Copeland, J. , Wang, P.W. , Guttman, D.S. , Kotanen, P.M. and Johnson, M.T.J. (2018) Assembly and ecological function of the root microbiome across angiosperm plant species. Proc. Natl Acad. Sci. USA, 115, E1157–E1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick, C.R. , Salas‐González, I. , Conway, J.M. , Finkel, O.M. , Gilbert, S. , Russ, D. , Teixeira, P.J.P.L. and Dangl, J.L. (2020) The plant microbiome: from ecology to reductionism and beyond. Annu. Rev. Microbiol. 74, 81–100. [DOI] [PubMed] [Google Scholar]
- Flaishman, M.A. and Kolattukudy, P.E. (1994) Timing of fungal invasion using host’s ripening hormone as a signal. Proc. Natl Acad. Sci. USA, 91, 6579–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo, E. (2020) Plant hormones play common and divergent roles in nodulation and arbuscular mycorrhizal symbioses. In The model legume Medicago truncatula ( de Bruijn, F. , ed). Hoboken, New Jersey: John Wiley & Sons, Inc., pp. 753–765. [Google Scholar]
- Foo, E. and Davies, N.W. (2011) Strigolactones promote nodulation in pea. Planta, 234, 1073–1081. [DOI] [PubMed] [Google Scholar]
- Foo, E. , McAdam, E.L. , Weller, J.L. and Reid, J.B. (2016) Interactions between ethylene, gibberellins, and brassinosteroids in the development of rhizobial and mycorrhizal symbioses of pea. J. Exp. Bot. 67, 2413–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foo, E. , Ross, J.J. , Jones, W.T. and Reid, J.B. (2013) Plant hormones in arbuscular mycorrhizal symbioses: an emerging role for gibberellins. Ann. Bot. 111, 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchetti, G. , Masciarelli, O. , Alemano, S. , Alvarez, D. and Abdala, G. (2007) Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 76, 1145–1152. [DOI] [PubMed] [Google Scholar]
- French, E. , Ghaste, M. , Widhalm, J. and Iyer‐Pascuzzi, A. (2019) Defense hormones modulate root microbiome diversity and composition in tomato. bioRxiv. 10.1101/656769. [DOI] [Google Scholar]
- Frugier, F. , Kosuta, S. , Murray, J.D. , Crespi, M. and Szczyglowski, K. (2008) Cytokinin: secret agent of symbiosis. Trends Plant Sci. 13, 115–120. [DOI] [PubMed] [Google Scholar]
- Fu, J.H. , Sun, X.H. , Wang, J.D. , Chu, J.F. and Yan, C.Y. (2011) Progress in quantitative analysis of plant hormones. Chin. Sci. Bull. 56, 355–366. [Google Scholar]
- Fujinami, R. , Yamada, T. and Imaichi, R. (2020) Root apical meristem diversity and the origin of roots: insights from extant lycophytes. J. Plant Res. 133, 291–296. [DOI] [PubMed] [Google Scholar]
- Galland, M. , Gamet, L. , Varoquaux, F. , Touraine, B. , Touraine, B. and Desbrosses, G. (2012) The ethylene pathway contributes to root hair elongation induced by the beneficial bacteria Phyllobacterium brassicacearum STM196. Plant Sci. 190, 74–81. [DOI] [PubMed] [Google Scholar]
- Gamalero, E. and Glick, B.R. (2015) Bacterial modulation of plant ethylene levels. Plant Physiol. 169, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalero, E. , Berta, G. , Massa, N. , Glick, B.R. and Lingua, G. (2008) Synergistic interactions between the ACC deaminase‐producing bacterium Pseudomonas putida UW4 and the AM fungus Gigaspora rosea positively affect cucumber plant growth. FEMS Microbiol. Ecol. 64, 459–467. [DOI] [PubMed] [Google Scholar]
- Gauthier‐Coles, C. , White, R.G. and Mathesius, U. (2019) Nodulating legumes are distinguished by a sensitivity to cytokinin in the root cortex leading to pseudonodule development. Front. Plant Sci. 9, 1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Chini, A. and Solano, R. (2016) How microbes twist jasmonate signaling around their little fingers. Plants, 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook, J. (2005) Contrasting mechanisms of defense against niotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. [DOI] [PubMed] [Google Scholar]
- Glick, B.R. (2005) Modulation of plant ethylene levels by the bacterial enzyme ACC deaminase. FEMS Microbiol. Lett. 251, 1–7. [DOI] [PubMed] [Google Scholar]
- Glick, B.R. (2012) Plant growth‐promoting bacteria: mechanisms and applications. Scientifica, 2012, 963401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick, B.R. (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. [DOI] [PubMed] [Google Scholar]
- Goh, D.M. , Cosme, M. , Kisiala, A.B. , Mulholland, S. , Said, Z.M.F. , Spíchal, L. , Emery, R.J.N. , Declerck, S. and Guinel, F.C. (2019) A stimulatory role for cytokinin in the arbuscular mycorrhizal symbiosis of pea. Front. Plant Sci. 10, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke, J. and Deeken, R. (2014) Plant responses to Agrobacterium tumefaciens and crown gall development. Front. Plant Sci. 5, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J.K. , Davenport, E.R. , Waters, J.L. , Clark, A.G. and Ley, R.E. (2016) Cross‐species comparisons of host genetic associations with the microbiome. Science, 352, 532–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice, E.A. and Segre, J.A. (2012) The human microbiome: our second genome. Annu. Rev. Genomics Hum. Genet. 13, 151–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Großkinsky, D.K. , Tafner, R. , Moreno, M.V. , Stenglein, S.A. , García de Salamone, I.E. , Nelson, L.M. , Novák, O. , Strnad, M. , van der Graaff, E. and Roitsch, T. (2016) Cytokinin production by Pseudomonas fluorescens G20–18 determines biocontrol activity against Pseudomonas syringae in Arabidopsis . Sci. Rep. 6, 23310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y. , Wei, Z. , Wang, X. , Friman, V.P. , Huang, J. , Wang, X. , Mei, X. , Xu, Y. , Shen, Q. and Jousset, A. (2016) Pathogen invasion indirectly changes the composition of soil microbiome via shifts in root exudation profile. Biol. Fertil. Soils, 52, 997–1005. [Google Scholar]
- Guerrieri, A. , Dong, L. and Bouwmeester, H.J. (2019) Role and exploitation of underground chemical signaling in plants. Pest Manag. Sci. 75, 2455–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinel, F.C. (2015) Ethylene, a hormone at the center‐stage of nodulation. Front. Plant Sci. 6, 1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, P.C. (1971) Foliar spray of gibberellic acid and its influence on the rhizosphere and rhizoplane mycoflora. Plant Soil, 34, 233–236. [Google Scholar]
- Hacquard, S. , Garrido‐Oter, R. , González, A. et al . (2015) Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe, 17, 603–616. [DOI] [PubMed] [Google Scholar]
- Haichar, F.E.Z. , Marol, C. , Berge, O. , Rangel‐Castro, J.I. , Prosser, J.I. , Balesdent, J. , Heulin, T. and Achouak, W. (2008) Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2, 1221–1230. [DOI] [PubMed] [Google Scholar]
- Hammerbacher, A. , Coutinho, T.A. and Gershenzon, J. (2019) Roles of plant volatiles in defence against microbial pathogens and microbial exploitation of volatiles. Plant Cell Environ. 42, 2827–2843. [DOI] [PubMed] [Google Scholar]
- Han, X. and Kahmann, R. (2019) Manipulation of phytohormone pathways by effectors of filamentous plant pathogens. Front. Plant Sci. 10, 822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handelsman, J. , Tiedje, J. , Alvarez‐Cohen, L. et al . (2007) The New Science of Metagenomics: Revealing the Secrets of our Microbial Planet. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- Haney, C.H. and Ausubel, F.M. (2015) Plant microbiome blueprints. Science, 349, 788–789. [DOI] [PubMed] [Google Scholar]
- Hao, G. , Naumann, T.A. , Vaughan, M.M. , McCormick, S. , Usgaard, T. , Kelly, A. and Ward, T.J. (2019) Characterization of a Fusarium graminearum salicylate hydroxylase. Front. Microbiol. 9, 3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung, W. (2010) The evolution of abscisic acid (ABA) and ABA function in lower plants, fungi and lichen. Funct. Plant Biol. 37, 806–812. [Google Scholar]
- Hasegawa, S. , Poling, S.M. , Maier, V.P. and Bennett, R.D. (1984) Metabolism of abscisic acid: bacterial conversion to dehydrovomifoliol and vomifoliol dehydrogenase activity. Phytochemistry, 23, 2769–2771. [Google Scholar]
- Hassani, M.A. , Durán, P. and Hacquard, S. (2018) Microbial interactions within the plant holobiont. Microbiome, 6, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, F. , Waadt, R. and Schroeder, J.I. (2011) Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckmann, A.B. , Sandal, N. , Bek, A.S. , Madsen, L.H. , Jurkiewicz, A. , Nielsen, M.W. , Tirichine, L. and Stougaard, J. (2011) Cytokinin induction of root nodule primordia in Lotus japonicus is regulated by a mechanism operating in the root cortex. Mol. Plant Microbe Interact. 24, 1385–1395. [DOI] [PubMed] [Google Scholar]
- Hein, J.W. , Wolfe, G.V. and Blee, K.A. (2008) Comparison of rhizosphere bacterial communities in Arabidopsis thaliana mutants for systemic acquired resistance. Microb. Ecol. 55, 333–343. [DOI] [PubMed] [Google Scholar]
- Hérivaux, A. , Dugé de Bernonville, T. , Roux, C. et al . (2017) The identification of phytohormone receptor homologs in early diverging fungi suggests a role for plant sensing in land colonization by fungi. MBio, 8, e01739–e1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera‐Medina, M.J. , Steinkellner, S. , Vierheilig, H. , Ocampo Bote, J.A. and García Garrido, J.M. (2007) Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytol. 175, 554–564. [DOI] [PubMed] [Google Scholar]
- Hetherington, A.J. and Dolan, L. (2018) Stepwise and independent origins of roots among land plants. Nature, 561, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert, M. , Voll, L.M. , Ding, Y. , Hofmann, J. , Sharma, M. and Zuccaro, A. (2012) Indole derivative production by the root endophyte Piriformospora indica is not required for growth promotion but for biotrophic colonization of barley roots. New Phytol. 196, 520–534. [DOI] [PubMed] [Google Scholar]
- Hinsch, J. , Vrabka, J. , Oeser, B. , Novák, O. , Galuszka, P. and Tudzynski, P. (2015) De novo biosynthesis of cytokinins in the biotrophic fungus Claviceps purpurea . Environ. Microbiol. 17, 2935–2951. [DOI] [PubMed] [Google Scholar]
- Hu, L. , Robert, C.A.M. , Cadot, S. et al . (2018) Root exudate metabolites drive plant‐soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9, 2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, A.C. , Jiang, T. , Liu, Y.X. , Bai, Y.C. , Reed, J. , Qu, B. , Goossens, A. , Nützmann, H.W. , Bai, Y. and Osbourn, A. (2019) A specialized metabolic network selectively modulates Arabidopsis root microbiota. Science, 364, eaau6389. [DOI] [PubMed] [Google Scholar]
- Ilyas, N. and Bano, A. (2010) Azospirillum strains isolated from roots and rhizosphere soil of wheat (Triticum aestivum L.) grown under different soil moisture conditions. Biol. Fertil. Soils, 46, 393–406. [Google Scholar]
- Inaji, A. , Okazawa, A. , Taguchi, T. , Nakamoto, M. , Katsuyama, N. , Yoshikawa, R. , Ohnishi, T. , Waller, F. and Ohta, D. (2020) Rhizotaxis modulation in Arabidopsis is induced by diffusible compounds produced during the cocultivation of arabidopsis and the endophytic fungus Serendipita indica . Plant Cell Physiol. 61, 838–850. [DOI] [PubMed] [Google Scholar]
- Jacobs, S. , Zechmann, B. , Molitor, A. , Trujillo, M. , Petutschnig, E. , Lipka, V. , Kogel, K.‐H. and Schafer, P. (2011) Broad‐spectrum suppression of innate immunity Is required for colonization of Arabidopsis roots by the fungus Piriformospora indica . Plant Physiol. 156, 726–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson, P.E. (2000) Cytokinins and auxins in plant‐pathogen interactions ‐ an overview. Plant Growth Reg. 32, 369–380. [Google Scholar]
- Jeon, Y.A. , Yu, S.H. , Lee, Y.Y. , Park, H.J. , Lee, S. , Sung, J.S. , Kim, Y.G. and Lee, H.S. (2013) Incidence, molecular characteristics and pathogenicity of Gibberella fujikuroi species complex associated with rice seeds from Asian countries. Mycobiology, 41, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Z. and von Wirén, N. (2020) Signaling pathways underlying nitrogen‐dependent changes in root system architecture: from model to crop species. J. Exp. Bot. 71, 4393–4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, C.J. , Shimono, M. , Sugano, S. , Kojima, M. , Liu, X. , Inoue, H. , Sakakibara, H. and Takatsuji, H. (2013) Cytokinins act synergistically with salicylic acid to activate defense gene expression in rice. Mol. Plant Microbe Interact. 26, 287–296. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kabbara, S. , Schmülling, T. and Papon, N. (2018) CHASEing cytokinin receptors in plants, bacteria, fungi, and beyond. Trends Plant Sci. 23, 179–181. [DOI] [PubMed] [Google Scholar]
- Kachroo, P. , Liu, H. and Kachroo, A. (2020) Salicylic acid: transport and long‐distance immune signaling. Curr. Opin. Virol. 42, 53–57. [DOI] [PubMed] [Google Scholar]
- Kang, S.M. , Khan, A.L. , Waqas, M. , You, Y.H. , Hamayun, M. , Joo, G.J. , Shahzad, R. , Choi, K.S. and Lee, I.J. (2015) Gibberellin‐producing Serratia nematodiphila PEJ1011 ameliorates low temperature stress in Capsicum annuum L. Eur. J. Soil Biol. 68, 85–93. [Google Scholar]
- Kazan, K. and Lyons, R. (2014) Intervention of phytohormone pathways by pathogen effectors. Plant Cell, 26, 2285–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan, K. and Manners, J.M. (2009) Linking development to defense: auxin in plant‐pathogen interactions. Trends Plant Sci. 14, 373–382. [DOI] [PubMed] [Google Scholar]
- Kenrick, P. and Strullu‐Derrien, C. (2014) The origin and early evolution of roots. Plant Physiol. 166, 570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettner, J. and Dörffling, K. (1995) Biosynthesis and metabolism of abscisic acid in tomato leaves infected with Botrytis cinerea . Planta, 196, 627–634. [Google Scholar]
- Khan, A.L. , Hussain, J. , Al‐Harrasi, A. , Al‐Rawahi, A. and Lee, I.J. (2015) Endophytic fungi: resource for gibberellins and crop abiotic stress resistance. Crit. Rev. Biotechnol. 35, 62–74. [DOI] [PubMed] [Google Scholar]
- Kilaru, A. , Bailey, B.A. and Hasenstein, K.H. (2007) Moniliophthora perniciosa produces hormones and alters endogenous auxin and salicylic acid in infected cocoa leaves. FEMS Microbiol. Lett. 274, 238–244. [DOI] [PubMed] [Google Scholar]
- Kisiala, A. , Laffont, C. , Emery, R.J.N. and Frugier, F. (2013) Bioactive cytokinins are selectively secreted by sinorhizobium meliloti nodulating and nonnodulating strains. Mol. Plant Microbe Interact. 26, 1225–1231. [DOI] [PubMed] [Google Scholar]
- Knief, C. , Delmotte, N. , Chaffron, S. , Stark, M. , Innerebner, G. , Wassmann, R. , von Mering, C. and Vorholt, J.A. (2012) Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 6, 1378–1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kniskern, J.M. , Traw, M.B. and Bergelson, J. (2007) Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana . Mol. Plant Microbe Interact. 20, 1512–1522. [DOI] [PubMed] [Google Scholar]
- Kudjordjie, E.N. , Sapkota, R. , Steffensen, S.K. , Fomsgaard, I.S. and Nicolaisen, M. (2019) Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome, 7, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoyarova, G. , Arkhipova, T. , Korshunova, T. , Bakaeva, M. , Loginov, O. and Dodd, I.C. (2019) Phytohormone mediation of interactions between plants and non‐symbiotic growth promoting bacteria under edaphic stresses. Front. Plant Sci. 10, 1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoyarova, G.R. , Melentiev, A.I. , Martynenko, E.V. , Timergalina, L.N. , Arkhipova, T.N. , Shendel, G.V. , Kuz’mina, L.Y. , Dodd, I.C. and Veselov, S.Y. (2014) Cytokinin producing bacteria stimulate amino acid deposition by wheat roots. Plant Physiol. Biochem. 83, 285–291. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N. and Harper, C.P. (2018) The roles of auxin during interactions between bacterial plant pathogens and their hosts. J. Exp. Bot. 69, 245–254. [DOI] [PubMed] [Google Scholar]
- Kwak, M.J. , Kong, H.G. , Choi, K. et al . (2018) Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1109. [DOI] [PubMed] [Google Scholar]
- Lakshmanan, V. , Kitto, S.L. , Caplan, J.L. , Hsueh, Y.H. , Kearns, D.B. , Wu, Y.S. and Bais, H.P. (2012) Microbe‐associated molecular patterns‐triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol. 160, 1642–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfranco, L. , Fiorilli, V. , Venice, F. and Bonfante, P. (2018) Strigolactones cross the kingdoms: plants, fungi, and bacteria in the arbuscular mycorrhizal symbiosis. J. Exp. Bot. 69, 2175–2188. [DOI] [PubMed] [Google Scholar]
- Lanver, D. , Tollot, M. , Schweizer, G. et al . (2017) Ustilago maydis effectors and their impact on virulence. Nat. Rev. Microbiol. 15, 409–421. [DOI] [PubMed] [Google Scholar]
- Lavenus, J. , Goh, T. , Roberts, I. , Guyomarc'h, S. , Lucas, M. , De Smet, I. , Fukaki, H. , Beeckman, T. , Bennett, M. and Laplaze, L. (2013) Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 18, 450–458. [DOI] [PubMed] [Google Scholar]
- Leach, J.E. , Triplett, L.R. , Argueso, C.T. and Trivedi, P. (2017) Communication in the Phytobiome. Cell, 169, 587–596. [DOI] [PubMed] [Google Scholar]
- Lebeis, S.L. , Paredes, S.H. , Lundberg, D.S. et al . (2015) Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science, 349, 860–864. [DOI] [PubMed] [Google Scholar]
- Lehmann, S. , Dominguez‐Ferreras, A. , Huang, W.J. , Denby, K. , Ntoukakis, V. and Schäfer, P. (2020) Novel markers for high‐throughput protoplast‐based analyses of phytohormone signaling. PLoS One, 15, e0234154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemanceau, P. , Barret, M. , Mazurier, S. , Mondy, S. , Pivato, B. , Fort, T. and Vacher, C. (2017) Plant communication with associated microbiota in the spermosphere, rhizosphere and phyllosphere. Adv. Bot. Res. 82, 101–133. [Google Scholar]
- Leon‐Reyes, A. , Van der Does, D. , De Lange, E.S. , Delker, C. , Wasternack, C. , Van Wees, S.C. , Ritsema, T. and Pieterse, C.M.J. (2010) Salicylate‐mediated suppression of jasmonate‐responsive gene expression in arabidopsis is targeted downstream of the jasmonate biosynthesis pathway. Planta, 232, 1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leveau, J.H.J. and Lindow, S.E. (2005) Utilization of the plant hormone indole‐3‐acetic acid for growth by Pseudomonas putida strain 1290. Appl. Environ. Microbiol. 71, 2365–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, A. , Conway, J.M. , Dangl, J.L. and Woyke, T. (2018a) Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe. 24, 475–485. [DOI] [PubMed] [Google Scholar]
- Levy, A. , Salas Gonzalez, I. , Mittelviefhaus, M. et al . (2018b) Genomic features of bacterial adaptation to plants. Nat. Genet. 50, 138–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Brader, G. and Palva, E.T. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate‐mediated and salicylate‐mediated signals in plant defense. Plant Cell, 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, N. , Han, X. , Feng, D. , Yuan, D. and Huang, L.J. (2019) Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? Int. J. Mol. Sci. 20, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Zeng, R. and Liao, H. (2016) Improving crop nutrient efficiency through root architecture modifications. J. Integr. Plant Biol. 58, 193–202. [DOI] [PubMed] [Google Scholar]
- Ling, N. , Zhang, W. , Wang, D. , Mao, J. , Huang, Q. , Guo, S. and Shen, Q. (2013) Root exudates from grafted‐root watermelon showed a certain contribution in inhibiting Fusarium oxysporum f. sp. niveum . PLoS One, 8, e63383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , Hewezi, T. , Lebeis, S.L. , Pantalone, V. , Grewal, P.S. and Staton, M.E. (2019) Soil indigenous microbiome and plant genotypes cooperatively modify soybean rhizosphere microbiome assembly. BMC Microbiol. 19, 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. , Rice, J.H. , Lopes, V. , Grewal, P. , Lebeis, S. , Hewezi, T. and Staton, M. (2020) Overexpression of strigolactone‐associated genes exerts fine‐tuning selection on soybean rhizosphere bacterial and fungal microbiome. Phytobiomes J. 4, 239–251. [Google Scholar]
- Liu, F. , Xing, S. , Ma, H. , Du, Z. and Ma, B. (2013) Cytokinin‐producing, plant growth‐promoting rhizobacteria that confer resistance to drought stress in Platycladus orientalis container seedlings. Appl. Microbiol. Biotechnol. 97, 9155–9164. [DOI] [PubMed] [Google Scholar]
- Liu, H. , Carvalhais, L.C. , Crawford, M. , Singh, E. , Dennis, P.G. , Pieterse, C.M.J. and Schenk, P.M. (2017a) Inner plant values: diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 8, 2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Carvalhais, L.C. , Schenk, P.M. and Dennis, P.G. (2017b) Effects of jasmonic acid signalling on the wheat microbiome differ between body sites. Sci. Rep. 7, 41766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Song, T. , Zhang, X. et al . (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat. Commun. 5, 4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López‐Bucio, J. , Campos‐Cuevas, J.C. , Hernández‐Calderón, E. , Velásquez‐Becerra, C. , Farías‐Rodríguez, R. , Macías‐Rodríguez, L.I. and Valencia‐Cantero, E. (2007) Bacillus megaterium rhizobacteria promote growth and alter root‐system architecture through an auxin‐ and ethylene‐independent signaling mechanism in Arabidopsis thaliana . Mol. Plant Microbe Interact. 20, 207–217. [DOI] [PubMed] [Google Scholar]
- Lu, T. , Ke, M. , Lavoie, M. et al . (2018) Rhizosphere microorganisms can influence the timing of plant flowering. Microbiome, 6, 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X. , Hershey, D.M. , Wang, L. , Bogdanove, A.J. and Peters, R.J. (2015) An ent‐kaurene‐derived diterpenoid virulence factor from Xanthomonas oryzae pv. oryzicola . New Phytol. 206, 295–302. [DOI] [PubMed] [Google Scholar]
- Ludwig‐Müller, J. (2015) Bacteria and fungi controlling plant growth by manipulating auxin: balance between development and defense. J. Plant Physiol. 172, 4–12. [DOI] [PubMed] [Google Scholar]
- Ludwig‐Müller, J. (2020) Auxins and other phytohormones as signals in arbuscular mycorrhiza formation. In The model legume Medicago truncatula ( de Bruijn, F. , ed). Hoboken, New Jersey: John Wiley & Sons, Inc., pp. 766–776. [Google Scholar]
- Lundberg, D.S. , Lebeis, S.L. , Paredes, S.H. et al . (2012) Defining the core Arabidopsis thaliana root microbiome. Nature, 488, 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, W. , Penrose, D.M. and Glick, B.R. (2002) Strategies used by rhizobia to lower plant ethylene levels and increase nodulation. Can. J. Microbiol. 48, 947–954. [DOI] [PubMed] [Google Scholar]
- Ma, W. , Sebestianova, S.B. , Sebestian, J. , Burd, G.I. , Guinel, F.C. and Glick, B.R. (2003) Prevalence of 1‐aminocyclopropane‐1‐carboxylate deaminase in Rhizobium spp. Antonie Van Leeuwenhoek, 83, 285–291. [DOI] [PubMed] [Google Scholar]
- MacMillan, J. (2001) Occurrence of gibberellins in vascular plants, fungi, and bacteria. J. Plant Growth Regul. 20, 387–442. [DOI] [PubMed] [Google Scholar]
- Martin, F.M. , Uroz, S. and Barker, D.G. (2017) Ancestral alliances: plant mutualistic symbioses with fungi and bacteria. Science, 356, eaad4501. [DOI] [PubMed] [Google Scholar]
- Martín‐Rodríguez, J.Á. , León‐Morcillo, R. , Vierheilig, H. , Ocampo, J.A. , Ludwig‐Müller, J. and García‐Garrido, J.M. (2011) Ethylene‐dependent/ethylene‐independent ABA regulation of tomato plants colonized by arbuscular mycorrhiza fungi. New Phytol. 190, 193–205. [DOI] [PubMed] [Google Scholar]
- Marulanda, A. , Barea, J.M. and Azcón, R. (2009) Stimulation of plant growth and drought tolerance by native microorganisms (AM Fungi and bacteria) from dry environments: mechanisms related to bacterial effectiveness. J. Plant Growth Regul. 28, 115–124. [Google Scholar]
- Mathesius, U. (2020) Phytohormone regulation of Medicago truncatula‐rhizobia interactions. A review. In The model legume Medicago truncatula ( de Bruijn, F. , ed). Hoboken, New Jersey: John Wiley & Sons, Inc., pp. 743–752. [Google Scholar]
- Maurhofer, M. , Reimmann, C. , Schmidli‐Sacherer, P. , Heeb, S. , Haas, D. and Défago, G. (1998) Salicylic acid bitsynthetic genes expressed in Pseudomonas fluorescens strain P3 improve the induction of systemic resistance in tobacco against tobacco necrosis virus. Phytopathology, 88, 678–684. [DOI] [PubMed] [Google Scholar]
- McClerklin, S.A. , Lee, S.G. , Harper, C.P. , Nwumeh, R. , Jez, J.M. and Kunkel, B.N. (2018) Indole‐3‐acetaldehyde dehydrogenase‐dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog. 14, e1006811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuiness, P.N. , Reid, J.B. and Foo, E. (2019) The role of gibberellins and brassinosteroids in nodulation and arbuscular mycorrhizal associations. Front. Plant Sci. 10, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meents, A.K. , Furch, A.C.U. , Almeida‐Trapp, M. et al . (2019) Beneficial and pathogenic Arabidopsis root‐interacting fungi differently affect auxin levels and responsive genes during early infection. Front. Microbiol. 10, 380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto, M. , Underwood, W. , Koczan, J. , Nomura, K. and He, S.Y. (2006) Plant stomata function in innate immunity against bacterial invasion. Cell, 126, 969–980. [DOI] [PubMed] [Google Scholar]
- Mendes, L.W. , Raaijmakers, J.M. , Hollander, M.D. , Mendes, R. and Tsai, S.M. (2018) Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 12, 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes, R. , Garbeva, P. and Raaijmakers, J.M. (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. [DOI] [PubMed] [Google Scholar]
- Méndez, C. , Baginsky, C. , Hedden, P. , Gong, F. , Carú, M. and Rojas, M.C. (2014) Gibberellin oxidase activities in Bradyrhizobium japonicum bacteroids. Phytochemistry, 98, 101–109. [DOI] [PubMed] [Google Scholar]
- Mercy, L. , Lucic‐Mercy, E. , Nogales, A. , Poghosyan, A. , Schneider, C. and Arnholdt‐Schmitt, B. (2017) A functional approach towards understanding the role of the mitochondrial respiratory chain in an endomycorrhizal symbiosis. Front. Plant Sci. 8, 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miersch, O. , Brückner, B. , Schmidt, J. and Sembdner, G. (1992) Cyclopentane fatty acids from Gibberella fujikuroi . Phytochemistry, 31, 3835–3837. [Google Scholar]
- Miersch, O. , Regvar, M. and Wasternack, C. (1999) Metabolism of jasmonic acid in Pisolithus tinctorius cultures. Phyton Ann. Rei Bot. 39, 243–247. [Google Scholar]
- Miri, M. , Janakirama, P. , Held, M. , Ross, L. and Szczyglowski, K. (2016) Into the root: how cytokinin controls rhizobial infection. Trends Plant Sci. 21, 178–186. [DOI] [PubMed] [Google Scholar]
- Morffy, N. and Strader, L.C. (2020) Old town roads: routes of auxin biosynthesis across kingdoms. Curr. Opin. Plant Biol. 55, 21–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, R.O. (1986) Genes specifying auxin and cytokinin biosynthesis in phytopathogens. Annu. Rev. Plant Physiol. 37, 509–538. [Google Scholar]
- Morrison, E.N. , Emery, R.J.N. and Saville, B.J. (2017) Fungal derived cytokinins are necessary for normal Ustilago maydis infection of maize. Plant Pathol. 66, 726–742. [Google Scholar]
- Morrison, E.N. , Knowles, S. , Hayward, A. , Greg Thorn, R. , Saville, B.J. and Emery, R.J.N. (2015) Detection of phytohormones in temperate forest fungi predicts consistent abscisic acid production and a common pathway for cytokinin biosynthesis. Mycologia, 107, 245–257. [DOI] [PubMed] [Google Scholar]
- Mosher, S. , Moeder, W. , Nishimura, N. , Jikumaru, Y. , Joo, S.H. , Urquhart, W. , Klessig, D.F. , Kim, S.K. , Nambara, E. and Yoshioka, K. (2010) The lesion‐mimic mutant cpr22 shows alterations in abscisic acid signaling and abscisic acid insensitivity in a salicylic acid‐dependent manner. Plant Physiol. 152, 1901–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama, K. , Ogawa, T. , Fujii, T. and Fukuda, H. (1992) Classification of ethylene‐producing bacteria in terms of biosynthetic pathways to ethylene. J. Ferment. Bioeng. 73, 1–5. [Google Scholar]
- Nagahama, K. , Yoshino, K. , Matsuoka, M. , Sato, M. , Tanase, S. , Ogawa, T. and Fukuda, H. (1994) Ethylene production by strains of the plant‐pathogenic bacterium Pseudomonas syringae depends upon the presence of indigenous plasmids carrying homologous genes for the ethylene‐forming enzyme. Microbiology, 140, 2309–2313. [DOI] [PubMed] [Google Scholar]
- Nakamura, T. , Mukai, C. , Ozaki, Y. , Saotome, M. and Murayama, T. (1988) Effects of auxin and gibberellin on conidial germination and elongation of young hyphae in a cyclic 3′:5′ adenosine monophosphate‐dependent protein kinase mutant of Neurospora crassa . Plant Growth Regul. 7, 201–207. [Google Scholar]
- Nakashita, H. , Yasuda, M. , Nitta, T. , Asami, T. , Fujioka, S. , Arai, Y. , Sekimata, K. , Takatsuto, S. , Yamaguchi, I. and Yoshida, S. (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33, 887–898. [DOI] [PubMed] [Google Scholar]
- Nascimento, F.X. , Hernandez, A.G. , Glick, B.R. and Rossi, M.J. (2020) The extreme plant‐growth‐promoting properties of Pantoea phytobeneficialis MSR2 revealed by functional and genomic analysis. Environ. Microbiol. 22, 1341–1355. [DOI] [PubMed] [Google Scholar]
- Nascimento, F.X. , Rossi, M.J. and Glick, B.R. (2018) Ethylene and 1‐aminocyclopropane‐1‐carboxylate (ACC) in plant‐bacterial interactions. Front. Plant Sci. 9, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasir, F. , Shi, S. , Tian, L. , Chang, C. , Ma, L. , Li, X. , Gao, Y. and Tian, C. (2019) Strigolactones shape the rhizomicrobiome in rice (Oryza sativa). Plant Sci. 286, 118–133. [DOI] [PubMed] [Google Scholar]
- Naylor, D. , DeGraaf, S. , Purdom, E. and Coleman‐Derr, D. (2017) Drought and host selection influence bacterial community dynamics in the grass root microbiome. ISME J. 11, 2691–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett, R.S. , Montanares, M. , Marcassa, A. , Lu, X. , Nagel, R. , Charles, T.C. , Hedden, P. , Rojas, M.C. and Peters, R.J. (2017) Elucidation of gibberellin biosynthesis in bacteria reveals convergent evolution. Nat. Chem. Biol. 13, 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobori, T. , Mine, A. and Tsuda, K. (2018) Molecular networks in plant–pathogen holobiont. FEBS Lett. 592, 1937–1953. [DOI] [PubMed] [Google Scholar]
- O’Banion, B.S. , O’Neal, L. , Alexandre, G. and Lebeis, S.L. (2020) Bridging the gap between single‐strain and community‐level plant‐microbe chemical interactions. Mol. Plant Microbe Interact. 33, 124–134. [DOI] [PubMed] [Google Scholar]
- Omoarelojie, L.O. , Kulkarni, M.G. , Finnie, J.F. and Van Staden, J. (2019) Strigolactones and their crosstalk with other phytohormones. Ann. Bot. 124, 749–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco‐Mosqueda, M.D.C. , Glick, B.R. and Santoyo, G. (2020) ACC deaminase in plant growth‐promoting bacteria (PGPB): an efficient mechanism to counter salt stress in crops. Microbiol. Res. 235, 126439. [DOI] [PubMed] [Google Scholar]
- Orozco‐Mosqueda, M.D.C. , Rocha‐Granados, M.D.C. , Glick, B.R. and Santoyo, G. (2018) Microbiome engineering to improve biocontrol and plant growth‐promoting mechanisms. Microbiol. Res. 208, 25–31. [DOI] [PubMed] [Google Scholar]
- Ortíz‐Castro, R. , Contreras‐Cornejo, H.A. , Macías‐Rodríguez, L. and López‐Bucio, J. (2009) The role of microbial signals in plant growth and development. Plant Signal. Behav. 4, 701–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortíz‐Castro, R. , Valencia‐Cantero, E. and López‐Bucio, J. (2008) Plant growth promotion by Bacillus megaterium involves cytokinin signaling. Plant Signal. Behav. 3, 263–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen, A.R. , González Peña, A. , White, J.R. et al . (2013) Baseline survey of the anatomical microbial ecology of an important food plant: Solanum lycopersicum (tomato). BMC Microbiol. 13, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X. , Welti, R. and Wang, X. (2010) Quantitative analysis of major plant hormones in crude plant extracts by high‐performance liquid chromatography‐mass spectrometry. Nat. Protoc. 5, 986–992. [DOI] [PubMed] [Google Scholar]
- Parniske, M. (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775. [DOI] [PubMed] [Google Scholar]
- Patkar, R.N. , Benke, P.I. , Qu, Z. , Chen, Y.Y.C. , Yang, F. , Swarup, S. and Naqvi, N.I. (2015) A fungal monooxygenase‐derived jasmonate attenuates host innate immunity. Nat. Chem. Biol. 11, 733–740. [DOI] [PubMed] [Google Scholar]
- Patten, C.L. and Glick, B.R. (2002) Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl. Environ. Microbiol. 68, 3795–3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten, C.L. , Blakney, A.J.C. and Coulson, T.J.D. (2013) Activity, distribution and function of indole‐3‐acetic acid biosynthetic pathways in bacteria. Crit. Rev. Microbiol. 39, 395–415. [DOI] [PubMed] [Google Scholar]
- Peláez‐Vico, M.A. , Bernabéu‐Roda, L. , Kohlen, W. , Soto, M.J. and López‐Ráez, J.A. (2016) Strigolactones in the Rhizobium‐legume symbiosis: stimulatory effect on bacterial surface motility and down‐regulation of their levels in nodulated plants. Plant Sci. 245, 119–127. [DOI] [PubMed] [Google Scholar]
- Pérez‐Jaramillo, J.E. , Mendes, R. and Raaijmakers, J.M. (2016) Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 90, 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka, J.J. , Winter, C.M. and Benfey, P.N. (2012) Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63, 563–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot, L. , Raaijmakers, J.M. , Lemanceau, P. and van der Putten, W.H. (2013) Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , de Jonge, R. and Berendsen, R.L. (2016) The soil‐borne supremacy. Trends Plant Sci. 21, 171–173. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Leon‐Reyes, A. , Van der Ent, S. and Van Wees, S.C.M. (2009) Networking by small‐molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Van der Does, D. , Zamioudis, C. , Leon‐Reyes, A. and Van Wees, S.C.M. (2012) Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. [DOI] [PubMed] [Google Scholar]
- Pieterse, C.M.J. , Zamioudis, C. , Berendsen, R.L. , Weller, D.M. , Van Wees, S.C.M. and Bakker, P.A.H.M. (2014) Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. [DOI] [PubMed] [Google Scholar]
- Pozo, M.J. , López‐Ráez, J.A. , Azcón‐Aguilar, C. and García‐Garrido, J.M. (2015) Phytohormones as integrators of environmental signals in the regulation of mycorrhizal symbioses. New Phytol. 205, 1431–1436. [DOI] [PubMed] [Google Scholar]
- Preece, C. and Peñuelas, J. (2020) A return to the wild: root exudates and food security. Trends Plant Sci. 25, 14–21. [DOI] [PubMed] [Google Scholar]
- Prusty, R. , Grisafi, P. and Fink, G.R. (2004) The plant hormone indoleacetic acid induces invasive growth in Saccharomyces cerevisiae . Proc. Natl Acad. Sci. USA, 10, 4153–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers, J.M. and Mazzola, M. (2016) Soil immune responses. Science, 352, 1392–1393. [DOI] [PubMed] [Google Scholar]
- Raaijmakers, J.M. , Paulitz, T.C. , Steinberg, C. , Alabouvette, C. and Moënne‐Loccoz, Y. (2009) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil, 321, 341–361. [Google Scholar]
- Rabe, F. , Ajami‐Rashidi, Z. , Doehlemann, G. , Kahmann, R. and Djamei, A. (2013) Degradation of the plant defence hormone salicylic acid by the biotrophic fungus Ustilago maydis . Mol. Microbiol. 89, 179–188. [DOI] [PubMed] [Google Scholar]
- Rademacher, W. (1994) Gibberellin formation in microorganisms. Plant Growth Regul. 15, 303–314. [Google Scholar]
- Ramírez‐Puebla, S.T. , Servín‐Garcidueñas, L.E. , Jiménez‐Marín, B. , Bolaños, L.M. , Rosenblueth, M. , Martínez, J. , Rogel, M.A. , Ormeño‐Orrillo, E. and Martínez‐Romero, E. (2013) Gut and root microbiota commonalities. Appl. Environ. Microbiol. 79, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran, L.X. , van Loon, L.C. and Bakker, P.A.H.M. (2005) No role for bacterially produced salicylic acid in rhizobacterial induction of systemic resistance in Arabidopsis . Phytopathology, 95, 1349–1355. [DOI] [PubMed] [Google Scholar]
- Ravanbakhsh, M. , Sasidharan, R. , Voesenek, L.A.C.J. , Kowalchuk, G.A. and Jousset, A. (2018) Microbial modulation of plant ethylene signaling: ecological and evolutionary consequences. Microbiome, 6, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapuram, C. and Baldwin, I.T. (2007) Increased SA in NPR1‐silenced plants antagonizes JA and JA‐dependent direct and indirect defenses in herbivore‐attacked Nicotiana attenuata in nature. Plant J. 52, 700–715. [DOI] [PubMed] [Google Scholar]
- Reddy, M.N. , Pokojska, A. , Kampert, M. and Strezelczyk, E. (1989) Auxin‐, gibberellin‐like substances and cytokinins in the seed and root exudates of groundnut. Plant Soil, 113, 283–286. [Google Scholar]
- Rehman, N.U. , Ali, M. , Ahmad, M.Z. , Liang, G. and Zhao, J. (2018) Strigolactones promote rhizobia interaction and increase nodulation in soybean (Glycine max). Microb. Pathog. 114, 420–430. [DOI] [PubMed] [Google Scholar]
- Reinhold‐Hurek, B. , Bünger, W. , Burbano, C.S. , Sabale, M. and Hurek, T. (2015) Roots shaping their microbiome: global hotspots for microbial activity. Annu. Rev. Phytopathol. 53, 403–424. [DOI] [PubMed] [Google Scholar]
- Remy, W. , Taylor, T.N. , Hass, H. and Kerp, H. (1994) Four hundred‐million‐year‐old vesicular arbuscular mycorrhizae. Proc. Natl Acad. Sci. USA, 91, 11841–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusche, M. , Klásková, J. , Thole, K. , Truskina, J. , Novák, O. , Janz, D. , Strnad, M. , Spíchal, L. , Lipka, V. and Teichmann, T. (2013) Stabilization of cytokinin levels enhances Arabidopsis resistance against Verticillium longisporum . Mol. Plant Microbe Interact. 26, 850–860. [DOI] [PubMed] [Google Scholar]
- Ribaudo, C.M. , Krumpholz, E.M. , Cassán, F.D. , Bottini, R. , Cantore, M.L. and Curá, J.A. (2006) Azospirillum sp. promotes root hair development in tomato plants through a mechanism that involves ethylene. J. Plant Growth Regul. 24, 175–185. [Google Scholar]
- Robert‐Seilaniantz, A. , Grant, M. and Jones, J.D.G. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate‐salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. [DOI] [PubMed] [Google Scholar]
- Rolfe, S.A. , Griffiths, J. and Ton, J. (2019) Crying out for help with root exudates: adaptive mechanisms by which stressed plants assemble health‐promoting soil microbiomes. Curr. Opin. Microbiol. 49, 73–82. [DOI] [PubMed] [Google Scholar]
- Rolli, E. , Marasco, R. , Vigani, G. et al . (2015) Improved plant resistance to drought is promoted by the root‐associated microbiome as a water stress‐dependent trait. Environ. Microbiol. 17, 316–331. [DOI] [PubMed] [Google Scholar]
- Rosier, A. , Medeiros, F.H.V. and Bais, H.P. (2018) Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant‐microbe interactions. Plant Soil, 428, 35–55. [Google Scholar]
- Rozpadek, P. , Domka, A.M. , Nosek, M. , Wazny, R. , Jedrzejczyk, R.J. , Wiciarz, M. and Turnau, K. (2018) The role of strigolactone in the cross‐talk between Arabidopsis thaliana and the endophytic fungus Mucor sp. Front. Microbiol. 9, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar‐Cerezo, S. , Martínez‐Montiel, N. , García‐Sánchez, J. , Pérez‐y‐Terrón, R. and Martínez‐Contreras, R.D. (2018) Gibberellin biosynthesis and metabolism: a convergent route for plants, fungi and bacteria. Microbiol. Res. 208, 85–98. [DOI] [PubMed] [Google Scholar]
- Saleem, M. , Law, A.D. and Moe, L.A. (2016) Nicotiana roots recruit rare rhizosphere taxa as major root‐inhabiting microbes. Microb. Ecol. 71, 469–472. [DOI] [PubMed] [Google Scholar]
- Salehin, M. , Li, B. , Tang, M. , Katz, E. , Song, L. , Ecker, J.R. , Kliebenstein, D.J. and Estelle, M. (2019) Auxin‐sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 10, 4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos‐Medellín, C. , Edwards, J. , Liechty, Z. , Nguyen, B. and Sundaresan, V. (2017) Drought stress results in a compartment‐specific restructuring of the rice root‐associated microbiomes. MBio, 8, e00764–e00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasse, J. , Martinoia, E. and Northen, T. (2018) Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. [DOI] [PubMed] [Google Scholar]
- Schreiner, M. , Krumbein, A. , Knorr, D. and Smetanska, I. (2011) Enhanced glucosinolates in root exudates of Brassica rapa ssp. rapa mediated by salicylic acid and methyl jasmonate. J. Agric. Food Chem. 59, 1400–1405. [DOI] [PubMed] [Google Scholar]
- Scott, J.C. , Greenhut, I.V. and Leveau, J.H.J. (2013) Functional characterization of the bacterial iac genes for degradation of the plant hormone indole‐3‐acetic acid. J. Chem. Ecol. 39, 942–951. [DOI] [PubMed] [Google Scholar]
- Selosse, M.A. and Tacon, F.L. (1998) The land flora: a phototroph‐fungus partnership? Trends Ecol. Evol. 13, 15–20. [DOI] [PubMed] [Google Scholar]
- Sgroy, V. , Cassán, F. , Masciarelli, O. , Del Papa, M.F. , Lagares, A. and Luna, V. (2009) Isolation and characterization of endophytic plant growth‐promoting (PGPB) or stress homeostasis‐regulating (PSHB) bacteria associated to the halophyte Prosopis strombulifera . Appl. Microbiol. Biotechnol. 85, 371–381. [DOI] [PubMed] [Google Scholar]
- Shaharoona, B. , Arshad, M. and Zahir, Z.A. (2006) Effect of plant growth promoting rhizobacteria containing ACC‐deaminase on maize (Zea mays L.) growth under axenic conditions and on nodulation in mung bean (Vigna radiata L.). Lett. Appl. Microbiol. 42, 155–159. [DOI] [PubMed] [Google Scholar]
- Shahzad, R. , Waqas, M. , Khan, A.L. , Asaf, S. , Khan, M.A. , Kang, S.M. , Yun, B.W. and Lee, I.J. (2016) Seed‐borne endophytic Bacillus amyloliquefaciens RWL‐1 produces gibberellins and regulates endogenous phytohormones of Oryza sativa . Plant Physiol. Biochem. 106, 236–243. [DOI] [PubMed] [Google Scholar]
- Shani, E. , Salehin, M. , Zhang, Y. et al . (2017) Plant stress tolerance requires auxin‐sensitive Aux/IAA transcriptional repressors. Curr. Biol. 27, 437–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharifi, R. , Lee, S.M. and Ryu, C.M. (2018) Microbe‐induced plant volatiles. New Phytol. 220, 684–691. [DOI] [PubMed] [Google Scholar]
- Shi, T.Q. , Peng, H. , Zeng, S.Y. , Ji, R.Y. , Shi, K. , Huang, H. and Ji, X.J. (2017) Microbial production of plant hormones: opportunities and challenges. Bioengineered, 8, 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewers, V. , Kokkelink, L. , Smedsgaard, J. and Tudzynski, P. (2006) Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea . Appl. Environ. Microbiol. 72, 4619–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirrenberg, A. , Göbel, C. , Grond, S. , Czempinski, N. , Ratzinger, A. , Karlovsky, P. , Santos, P. , Feussner, I. and Pawlowski, K. (2007) Piriformospora indica affects plant growth by auxin production. Physiol. Plant. 131, 581–589. [DOI] [PubMed] [Google Scholar]
- Smith, A.M. (1973) Ethylene as a cause of soil fungistasis. Nature, 246, 311–313. [DOI] [PubMed] [Google Scholar]
- Smith, A.M. and Cook, R.J. (1974) Implications of ethylene production by bacteria for biological balance of soil. Nature, 252, 703–705.4437621 [Google Scholar]
- Sørensen, J.L. , Benfield, A.H. , Wollenberg, R.D. et al. (2018) The cereal pathogen Fusarium pseudograminearum produces a new class of active cytokinins during infection. Mol. Plant Pathol. 19, 1140–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen, S. (2015) Plant hormones produced by microbes. In Principles of Plant‐Microbe Interactions ‐ Microbes for Sustainable Agriculture ( Lugtenberg, B. , ed). Cham: Springer, pp. 247–256. [Google Scholar]
- Spaepen, S. and Vanderleyden, J. (2011) Auxin and plant‐microbe interactions. Cold Spring Harb. Perspect. Biol. 3, a001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaepen, S. , Vanderleyden, J. and Remans, R. (2007) Indole‐3‐acetic acid in microbial and microorganism‐plant signaling. FEMS Microbiol. Rev. 31, 425–448. [DOI] [PubMed] [Google Scholar]
- Spallek, T. , Gan, P. , Kadota, Y. and Shirasu, K. (2018) Same tune, different song — cytokinins as virulence factors in plant–pathogen interactions? Curr. Opin. Plant Biol. 44, 82–87. [DOI] [PubMed] [Google Scholar]
- Spence, C. and Bais, H. (2015) Role of plant growth regulators as chemical signals in plant‐microbe interactions: a double edged sword. Curr. Opin. Plant Biol. 27, 52–58. [DOI] [PubMed] [Google Scholar]
- Spence, C.A. , Lakshmanan, V. , Donofrio, N. and Bais, H.P. (2015) Crucial roles of abscisic acid biogenesis in virulence of rice blast fungus Magnaporthe oryzae . Front. Plant Sci. 6, 1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splivallo, R. , Fischer, U. , Göbel, C. , Feussner, I. and Karlovsky, P. (2009) Truffles regulate plant root morphogenesis via the production of auxin and ethylene. Plant Physiol. 150, 2018–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel, S.H. , Johnson, J.S. and Dong, X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl Acad. Sci. USA, 104, 18842–18847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec, N. , Banasiak, J. and Jasiński, M. (2016) Abscisic acid ‐ an overlooked player in plant‐microbe symbioses formation? Acta Biochim. Pol. 63, 53–58. [DOI] [PubMed] [Google Scholar]
- Steinkellner, S. , Lendzemo, V. , Langer, I. , Schweiger, P. , Khaosaad, T. , Toussaint, J.P. and Vierheilig, H. (2007) Flavonoids and strigolactones in root exudates as signals in symbiotic and pathogenic plant‐fungus interactions. Molecules, 12, 1290–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Street, I.H. , Aman, S. , Zubo, Y. , Ramzan, A. , Wang, X. , Shakeel, S.N. , Kieber, J.J. and Schaller, G.E. (2015) Ethylene inhibits cell proliferation of the Arabidopsis root meristem. Plant Physiol. 169, 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strehmel, N. , Böttcher, C. , Schmidt, S. and Scheel, D. (2014) Profiling of secondary metabolites in root exudates of Arabidopsis thaliana . Phytochemistry, 108, 35–46. [DOI] [PubMed] [Google Scholar]
- Stringlis, I.A. , Yu, K. , Feussner, K. , de Jonge, R. , Van Bentum, S. , Van Verk, M.C. , Berendsen, R.L. , Bakker, P.A.H.M. , Feussner, I. and Pieterse, C.M.J. (2018) MYB72‐dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl Acad. Sci. USA, 115, E5213–E5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strullu‐Derrien, C. , Selosse, M.A. , Kenrick, P. and Martin, F.M. (2018) The origin and evolution of mycorrhizal symbioses: from palaeomycology to phylogenomics. New Phytol. 220, 1012–1030. [DOI] [PubMed] [Google Scholar]
- Suga, H. , Arai, M. , Fukasawa, E. , Motohashi, K. , Nakagawa, H. , Tateishi, H. , Fuji, S.I. , Shimizu, M. , Kageyama, K. and Hyakumachi, M. (2019) Genetic differentiation associated with fumonisin and gibberellin production in Japanese Fusarium fujikuroi . Appl. Environ. Microbiol. 85, e02414–e02418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullia, S.B. (1968) Effect of foliar spray of hormones on the rhizosphere mycoflora of leguminous weeds. Plant Soil, 29, 292–298. [Google Scholar]
- Suzuki, A. , Akune, M. , Kogiso, M. et al . (2004) Control of nodule number by the phytohormone abscisic acid in the roots of two leguminous species. Plant Cell Physiol. 45, 914–922. [DOI] [PubMed] [Google Scholar]
- Takeda, N. , Handa, Y. , Tsuzuki, S. , Kojima, M. , Sakakibara, H. and Kawaguchi, M. (2015) Gibberellins interfere with symbiosis signaling and gene expression and alter colonization by arbuscular mycorrhizal fungi in Lotus japonicus . Plant Physiol. 167, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tambalo, D.D. , Vanderlinde, E.M. , Robinson, S. , Halmillawewa, A. , Hynes, M.F. and Yost, C.K. (2014) Legume seed exudates and Physcomitrella patens extracts influence swarming behavior in Rhizobium leguminosarum . Can. J. Microbiol. 60, 15–24. [DOI] [PubMed] [Google Scholar]
- Teixeira, P.J.P. , Colaianni, N.R. , Fitzpatrick, C.R. and Dangl, J.L. (2019) Beyond pathogens: microbiota interactions with the plant immune system. Curr. Opin. Microbiol. 49, 7–17. [DOI] [PubMed] [Google Scholar]
- Thatcher, L.F. , Manners, J.M. and Kazan, K. (2009) Fusarium oxysporum hijacks COI1‐mediated jasmonate signaling to promote disease development in Arabidopsis. Plant J. 58, 927–939. [DOI] [PubMed] [Google Scholar]
- Ton, J. , Flors, V. and Mauch‐Mani, B. (2009) The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317. [DOI] [PubMed] [Google Scholar]
- Torrey, J.G. (1976) Root hormones and plant growth. Annu. Rev. Plant Physiol. 27, 435–459. [Google Scholar]
- Trdá, L. , Barešová, M. , Šašek, V. , Nováková, M. , Zahajská, L. , Dobrev, P.I. , Motyka, V. and Burketová, L. (2017) Cytokinin metabolism of pathogenic fungus Leptosphaeria maculans involves isopentenyltransferase, adenosine kinase and cytokinin oxidase/dehydrogenase. Front. Microbiol. 8, 1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, I.J. , Tanaka, E. , Masuya, H. , Tanaka, R. , Hirooka, Y. , Endoh, R. , Sahashi, N. and Kikuchi, T. (2014) Comparative genomics of Taphrina fungi causing varying degrees of tumorous deformity in plants. Genome Biol. Evol. 6, 861–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukanova, K.A. , Chebotar, V. , Meyer, J.J.M. and Bibikova, T.N. (2017) Effect of plant growth‐promoting Rhizobacteria on plant hormone homeostasis. S. Afr. J. Bot. 113, 91–102. [Google Scholar]
- Tudzynski, B. and Sharon, A. (2002) Biosynthesis, biological role and application of fungal phytohormones. In The Mycota (A Comprehensive Treatise On Fungi as Experimental Systems for Basic and Applied Research) ‐ Industrial Applications ( Osiewacz, H.D. , ed). Berlin, Heidelberg: Springer. [Google Scholar]
- Tully, R.E. and Keister, D.L. (1993) Cloning and mutagenesis of a cytochrome P‐450 locus from Bradyrhizobium japonicum that is expressed anaerobically and symbiotically. Appl. Environ. Microbiol. 59, 4136–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, T.R. , James, E.K. and Poole, P.S. (2013a) The plant microbiome. Genome Biol. 14, 209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, T.R. , Ramakrishnan, K. , Walshaw, J. , Heavens, D. , Alston, M. , Swarbreck, D. , Osbourn, A. , Grant, A. and Poole, P.S. (2013b) Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 7, 2248–2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacheron, J. , Desbrosses, G. , Bouffaud, M.L. , Touraine, B. , Moënne‐Loccoz, Y. , Muller, D. , Legendre, L. , Wisniewski‐Dyé, F. and Prigent‐Combaret, C. (2013) Plant growth‐promoting rhizobacteria and root system functioning. Front. Plant Sci. 4, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valls, M. , Genin, S. and Boucher, C. (2006) Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum . PLoS Pathog. 2, e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bockhaven, J. , Spíchal, L. , Novák, O. , Strnad, M. , Asano, T. , Kikuchi, S. , Höfte, M. and De Vleesschauwer, D. (2015) Silicon induces resistance to the brown spot fungus Cochliobolus miyabeanus by preventing the pathogen from hijacking the rice ethylene pathway. New Phytol. 206, 761–773. [DOI] [PubMed] [Google Scholar]
- van Dam, N.M. and Bouwmeester, H.J. (2016) Metabolomics in the rhizosphere: tapping into belowground chemical communication. Trends Plant Sci. 21, 256–265. [DOI] [PubMed] [Google Scholar]
- Van de Poel, B. , Smet, D. and Van Der Straeten, D. (2015) Ethylene and hormonal cross talk in vegetative growth and development. Plant Physiol. 169, 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ent, S. , Van Wees, S.C.M. and Pieterse, C.M.J. (2009) Jasmonate signaling in plant interactions with resistance‐inducing beneficial microbes. Phytochemistry, 70, 1581–1588. [DOI] [PubMed] [Google Scholar]
- van der Meij, A. , Willemse, J. , Schneijderberg, M.A. , Geurts, R. , Raaijmakers, J.M. and van Wezel, G.P. (2018) Inter‐ and intracellular colonization of Arabidopsis roots by endophytic actinobacteria and the impact of plant hormones on their antimicrobial activity. Antonie Van Leeuwenhoek, 111, 679–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duy, N. , Mäder, U. , Tran, N.P. , Cavin, J.F. , Tam, L.T. , Albrecht, D. , Hecker, M. and Antelmann, H. (2007) The proteome and transcriptome analysis of Bacillus subtilis in response to salicylic acid. Proteomics, 7, 698–710. [DOI] [PubMed] [Google Scholar]
- Van Wees, S.C. , Van der Ent, S. and Pieterse, C.M.J. (2008) Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448. [DOI] [PubMed] [Google Scholar]
- Vanstraelen, M. and Benková, E. (2012) Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 28, 463–487. [DOI] [PubMed] [Google Scholar]
- Venturi, V. and Keel, C. (2016) Signaling in the rhizosphere. Trends Plant Sci. 21, 187–198. [DOI] [PubMed] [Google Scholar]
- Verbon, E.H. and Liberman, L.M. (2016) Beneficial microbes affect endogenous mechanisms controlling root development. Trends Plant Sci. 21, 218–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma, V. , Ravindran, P. and Kumar, P.P. (2016) Plant hormone‐mediated regulation of stress responses. BMC Plant Biol. 16, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vishwakarma, K. , Upadhyay, N. , Kumar, N. et al . (2017) Abscisic acid signaling and abiotic stress tolerance in plants: a review on current knowledge and future prospects. Front. Plant Sci. 8, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissenberg, K. , Claeijs, N. , Balcerowicz, D. and Schoenaers, S. (2020) Hormonal regulation of root hair growth and responses to the environment in Arabidopsis. J. Exp. Bot. 71, 2412–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges, M.J.E.E.E. , Bai, Y. , Schulze‐Lefert, P. and Sattely, E.S. (2019) Plant‐derived coumarins shape the composition of an Arabidopsis synthetic root microbiome. Proc. Natl Acad. Sci. USA, 116, 12558–12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, I.A. , Moritz, L. , Pieterse, C.M.J. and Van Wees, S.C.M. (2015) Impact of hormonal crosstalk on plant resistance and fitness under multi‐attacker conditions. Front. Plant Sci. 6, 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagg, C. , Schlaeppi, K. , Banerjee, S. , Kuramae, E.E. and van der Heijden, M.G.A. (2019) Fungal‐bacterial diversity and microbiome complexity predict ecosystem functioning. Nat. Commun. 10, 4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, D.R. and McRoberts, N. (2006) Plants and biotrophs: a pivotal role for cytokinins? Trends Plant Sci. 11, 581–586. [DOI] [PubMed] [Google Scholar]
- Walters, D.R. , McRoberts, N. and Fitt, B.D.L. (2008) Are green islands red herrings? Significance of green islands in plant interactions with pathogens and pests. Biol. Rev. Camb. Philos Soc. 83, 79–102. [DOI] [PubMed] [Google Scholar]
- Wang, F.F. , Cheng, S.T. , Wu, Y. , Ren, B.Z. and Qian, W. (2017) A bacterial receptor PcrK senses the plant hormone cytokinin to promote adaptation to oxidative stress. Cell Rep. 21, 2940–2951. [DOI] [PubMed] [Google Scholar]
- Wang, C. , Liu, Y. , Li, S.S. and Han, G.Z. (2015) Insights into the origin and evolution of the plant hormone signaling machinery. Plant Physiol. 167, 872–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Zou, Y. , Kaw, H.Y. , Wang, G. , Sun, H. , Cai, L. , Li, C. , Meng, L.Y. and Li, D. (2020) Recent developments and emerging trends of mass spectrometric methods in plant hormone analysis: a review. Plant Methods, 16, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingart, H. and Völksch, B. (1997) Ethylene production by Pseudomonas syringae pathovars in vitro and in planta . Appl. Environ. Microbiol. 63, 156–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingart, H. , Ullrich, H. , Geider, K. and Völksch, B. (2001) The role of ethylene production in virulence of Pseudomonas syringae pvs. glycinea and phaseolicola . Phytopathology, 91, 511–518. [DOI] [PubMed] [Google Scholar]
- Werner, T. and Schmülling, T. (2009) Cytokinin action in plant development. Curr. Opin. Plant Biol. 12, 527–538. [DOI] [PubMed] [Google Scholar]
- Wiemann, P. , Sieber, C.M. , von Bargen, K.W. et al . (2013) Deciphering the cryptic genome: genome‐wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 9, e1003475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H. , Liu, D. , Ling, N. , Bao, W. , Ying, R. and Shen, Q. (2009) Influence of root exudates of watermelon on Fusarium oxysporum f. sp. niveum . Soil Sci. Soc. Am. J. 73, 1150–1156. [Google Scholar]
- Wulff, E.G. , Sørensen, J.L. , Lübeck, M. , Nielsen, K.F. , Thrane, U. and Torp, J. (2010) Fusarium spp. associated with rice Bakanae: ecology, genetic diversity, pathogenicity and toxigenicity. Environ. Microbiol. 12, 649–657. [DOI] [PubMed] [Google Scholar]
- Xu, G. , Yang, S. , Meng, L. and Wang, B.G. (2018a) The plant hormone abscisic acid regulates the growth and metabolism of endophytic fungus Aspergillus nidulans . Sci. Rep. 8, 6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Li, X.L. and Luo, L. (2012) Effects of engineered Sinorhizobium meliloti on cytokinin synthesis and tolerance of alfalfa to extreme drought stress. Appl. Environ. Microbiol. 78, 8056–8061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, L. , Yang, H. , Ren, L. , Chen, W. , Liu, L. , Liu, F. , Zeng, L. , Yan, R. , Chen, K. and Fang, X. (2018b) Jasmonic acid‐mediated aliphatic glucosinolate metabolism is involved in clubroot disease development in Brassica napus L. Front. Plant Sci. 9, 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue, J. , Deng, Z. , Huang, P. et al . (2016) Belowground rhizomes in paleosols: the hidden half of an Early Devonian vascular plant. Proc. Natl Acad. Sci. USA, 113, 9451–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, S. (2008) Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. [DOI] [PubMed] [Google Scholar]
- Yasuda, M. , Ishikawa, A. , Jikumaru, Y. et al . (2008) Antagonistic interaction between systemic acquired resistance and the abscisic acid‐mediated abiotic stress response in Arabidopsis . Plant Cell, 20, 1678–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasumura, Y. , Crumpton‐Taylor, M. , Fuentes, S. and Harberd, N.P. (2007) Step‐by‐step acquisition of the gibberellin‐DELLA growth‐regulatory mechanism during land‐plant evolution. Curr. Biol. 17, 1225–1230. [DOI] [PubMed] [Google Scholar]
- Yoneyama, K. , Xie, X. , Sekimoto, H. , Takeuchi, Y. , Ogasawara, S. , Akiyama, K. , Hayashi, H. and Yoneyama, K. (2008) Strigolactones, host recognition signals for root parasitic plants and arbuscular mycorrhizal fungi, from Fabaceae plants. New Phytol. 179, 484–494. [DOI] [PubMed] [Google Scholar]
- Yoo, S.‐D. , Cho, Y.‐H. and Sheen, J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu, M.H. , Zhao, Z.Z. and He, J.X. (2018) Brassinosteroid signaling in plant–microbe interactions. Int. J. Mol. Sci. 19, 4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, P. and Hochholdinger, F. (2018) The role of host genetic signatures on root–microbe interactions in the rhizosphere and endosphere. Front. Plant Sci. 9, 1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis, C. and Pieterse, C.M.J. (2012) Modulation of host immunity by beneficial microbes. Mol. Plant Microbe Interact. 25, 139–150. [DOI] [PubMed] [Google Scholar]
- Zamioudis, C. , Mastranesti, P. , Dhonukshe, P. , Blilou, I. and Pieterse, C.M.J. (2013) Unraveling root developmental programs initiated by beneficial Pseudomonas spp. bacteria. Plant Physiol. 162, 304–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhalnina, K. , Louie, K.B. , Hao, Z. et al . (2018) Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 3, 470–480. [DOI] [PubMed] [Google Scholar]
- Zhou, C. , Guo, J. , Zhu, L. , Xiao, X. , Xie, Y. , Zhu, J. , Ma, Z. and Wang, J. (2016) Paenibacillus polymyxa BFKC01 enhances plant iron absorption via improved root systems and activated iron acquisition mechanisms. Plant Physiol. Biochem. 105, 162–173. [DOI] [PubMed] [Google Scholar]
- Zhou, J.M. and Zhang, Y. (2020) Plant immunity: danger perception and signaling. Cell, 181, 978–989. [DOI] [PubMed] [Google Scholar]
- Zhu, P. , Xu, Z. , Cui, Z. , Zhang, Z. and Xu, L. (2017) Ethylene production by Alternaria alternata and its association with virulence on inoculated grape berries. Phytoparasitica, 45, 273–279. [Google Scholar]
- Zúñiga, A. , Poupin, M.J. , Donoso, R. , Ledger, T. , Guiliani, N. , Gutiérrez, R.A. and González, B. (2013) Quorum sensing and indole‐3‐acetic acid degradation play a role in colonization and plant growth promotion of Arabidopsis thaliana by Burkholderia phytofirmans PsJN. Mol. Plant‐Microbe Interact. 26, 546–553. [DOI] [PubMed] [Google Scholar]


