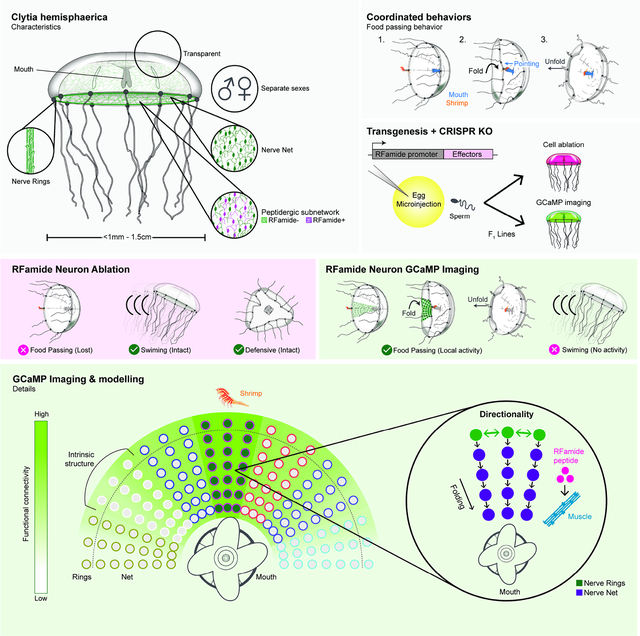
Summary
Jellyfish are radially symmetric organisms without a brain that arose more than 500 million years ago. They achieve organismal behaviors through coordinated interactions between autonomously functioning body parts. Jellyfish neurons have been studied electrophysiologically, but not at the systems level. We introduce Clytia hemisphaerica as a transparent, genetically tractable jellyfish model for systems neuroscience. We generate stable F1 transgenic lines for cell type-specific conditional ablation and whole-organism GCaMP imaging. Using these tools and computational analyses, we find that a diffuse network of RFamide-expressing neurons is functionally subdivided into a series of anatomically cryptic, spatially localized subassemblies whose selective activation controls directional food transfer from the tentacles to the mouth, revealing an unanticipated degree of structured neural organization in this species. Clytia affords a platform for systems-level studies of neural function, behavior, and evolution within a clade of marine organisms with growing ecological and economic importance.
Graphical Abstract
In Brief
A jellyfish species is developed as a genetically tractable neuroscience model, in which the application of GCaMP imaging and cell type-specific ablation has revealed spatially restricted neuronal subnetworks controlling feeding behaviors.
Introduction
Jellyfish offer insights into the structure, function, and evolution of nervous systems: they are apparent “living fossils,” whose last common ancestor with bilaterians emerged just after the appearance of neurons (Figure 1A; Arendt et al., 2016; Cartwright et al., 2007). Jellyfish use neurons homologous to our own (Arendt et al., 2016; Bosch et al., 2017), but lack centralization, i.e., “brains”. How such organisms are able to feed themselves, navigate, escape from predators, and even sleep (Mackie, 2004; Nath et al., 2017; Lewis and Long, 2005; Meech, 2019) in the absence of a central brain poses an important problem in the field of evolutionary neurobiology, with implications for autonomous systems engineering (Nawroth et al., 2012). Jellyfish are also attracting growing interest as critical components of ocean ecosystems, in part due to jellyfish blooms and their negative economic impact (Condon et al., 2013; Graham et al., 2014; Hays et al., 2018).
Figure 1: Introduction to Clytia.
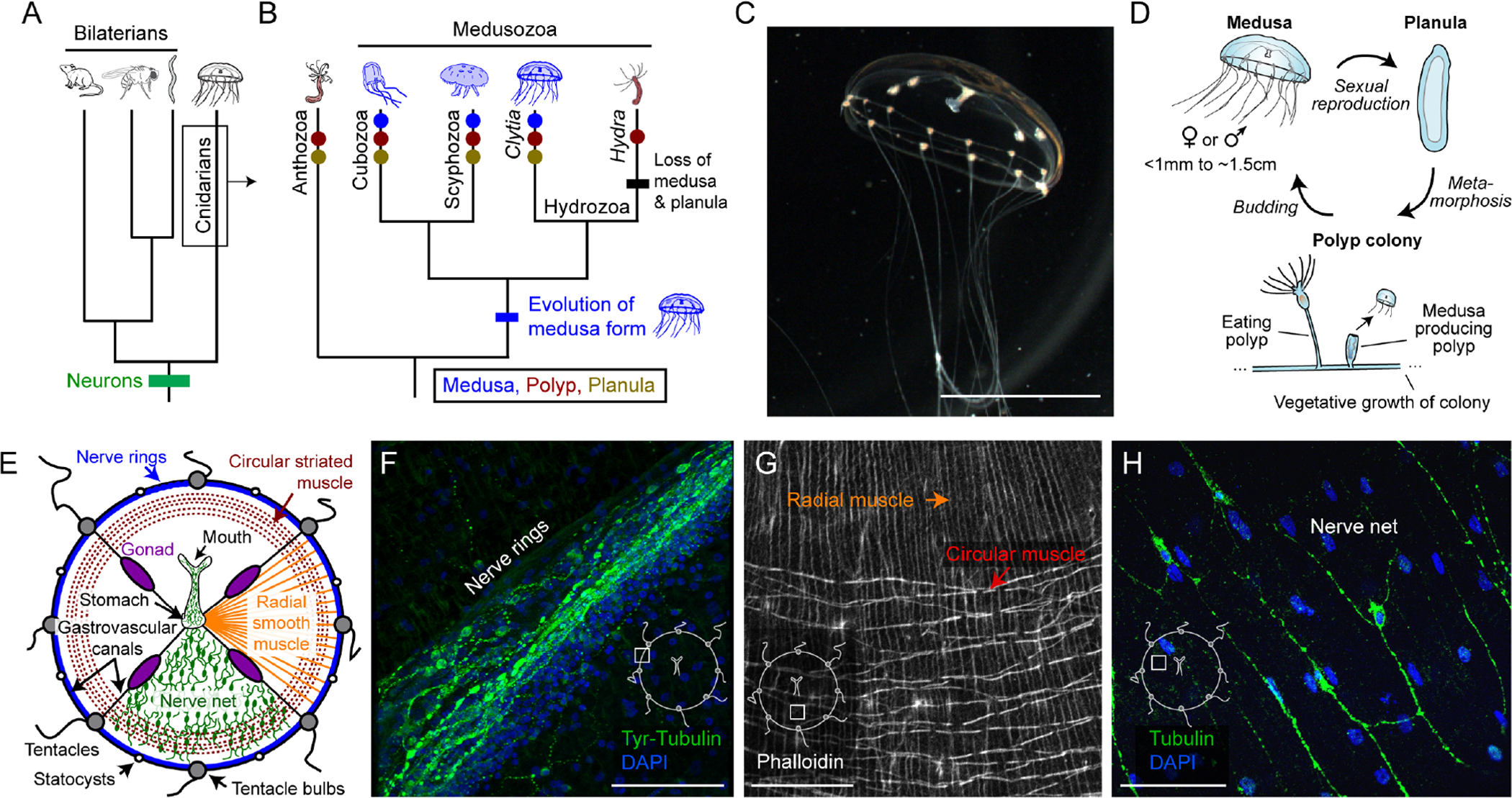
(A) Relationship between cnidarians, bilaterian lineages, and the appearance of neurons.
(B) Specific cnidarian lineages and transitions in body form. Text: taxonomic class; italics: genus.
(C) Clytia medusa. Scale: ~0.5cm.
(D) Clytia life cycle: medusa (jellyfish), planula (larva), and polyp stages.
(E) Medusa anatomy. Nerve net and radial muscle are shown in only one quadrant for clarity.
(F) Nerve rings. Green: anti-Tyrosinated Tubulin. Blue: DAPI. Scale: 50μm. Inset here and in G-H indicates anatomical location.
(G) Radial vs. circular muscle visualized with phalloidin. Scale: 50μm.
(H) Nerve net. Green: anti-aTubulin. Blue: DAPI. Scale: 50μm.
Despite the importance of jellyfish to evolution, ecology, and economics, remarkably little is known about the neural control of their behavior. Jellyfish neurons have been studied extensively using single-unit electrophysiological recordings (Meech, 2019; Satterlie, 2002), but systems-level analysis has been absent due to the lack of a genetically tractable model. Attractive features of jellyfish for systems neuroscience include their small size, relative planarity, and transparency, facilitating optical approaches (Katsuki and Greenspan, 2013; Meech, 2019; Bosch et al., 2017). Existing cnidarian genetic neuroscience models, such as Hydra (Dupre and Yuste, 2017; Tzouanas et al., 2021; Badhiwala et al., 2021), lack a jellyfish life cycle stage (Künzel et al., 2010; Renfer et al., 2010; Wittlieb et al., 2006).
Here we introduce the hydrozoan jellyfish Clytia hemisphaerica, originally established to study early development and evolution (Houliston et al., 2010), as a genetic model for systems neuroscience. The Clytia genome has been sequenced (Leclère et al., 2019) and an atlas of its cell types generated using single-cell RNA sequencing (Chari et al., 2021). CRISPR/Cas9-mediated gene knockout has been reported (Momose et al., 2018) but transgenesis has not yet been established.
In this inaugural study, we describe the generation of stable Clytia F1 transgenic lines for population neural imaging and neuronal cell type-specific ablation. Using these tools, we have investigated the neural control of feeding, in which captured food is vectorially transferred from the margin of the umbrella to the central mouth. We find that directional infolding of the umbrella is controlled by anatomically cryptic neural subassemblies that tile the umbrella, uncovering a surprising degree of structural organization within a superficially diffuse neural net. This work introduces Clytia as a genetically tractable model for systems neuroscience, affording a platform for understanding the neural control of its decentralized behavior and internal states, and for comparative studies across phylogeny.
Results
Jellyfish (medusae) are a free swimming life stage within the phylum Cnidaria and subphylum Medusozoa (Figure 1A–D; Leclère et al., 2019). Clytia medusae are small (~1mm–1.5 cm), optically transparent, and have approximately 10,000 neurons in a 1cm adult (Figure 1C; Chari et al., 2021). Their anatomy exhibits the hallmarks of the hydrozoan medusa body plan (Figure 1E): nerve rings (Figure 1F), circular and radial muscle (Figure 1G), and a subumbrellar nerve net (Figure 1H). Importantly, the tri-phasic sexual reproductive cycle (Figure 1D) can be recapitulated in the laboratory (Methods; Houliston et al., 2010; Lechable et al., 2020). Following controlled fertilization, zygotes develop into a planula (larval) stage. Planulae attach to a substrate (microscope slide) where they form clonal polyp colonies that release free-swimming medusae. The entire life cycle takes ~6–8 weeks.
Identification and genetic targeting of a neuronal subpopulation marked by RFamide
Neurons immunoreactive for an RFamide (RFa) peptide have previously been identified in Clytia (Mackie et al., 1985), and have been suggested to play a role in medusa defensive and feeding behaviors (Mackie, 2003; Satterlie, 2008, 2002; Weber, 1989). We confirmed RFa immunoreactivity in the nerve net, mouth, nerve rings, and tentacles (Figure 2A–C and Figure S1A–G). RFa+ neurons comprised a large subpopulation (~80%) of nerve net neurons identified using tyrosinated tubulin, a generic neuronal marker (Gröger and Schmid, 2000; Figure 2A; Figure S1A–B). They were radially oriented, with varicosities apposed to radial muscle fibers (Figure 2B; Figure S1C–D). In contrast, RFa-negative neurons in the nerve net were generally smaller and lacked a clear radial orientation (Figure 2A, arrow; Figure S1A–B). RFa+ neurons in other body parts were less abundant than in the nerve net (~10% of total neurons in Clytia by scRNA-seq; Chari et al., 2021).
Figure 2: RFamide neurons are necessary for margin folding.
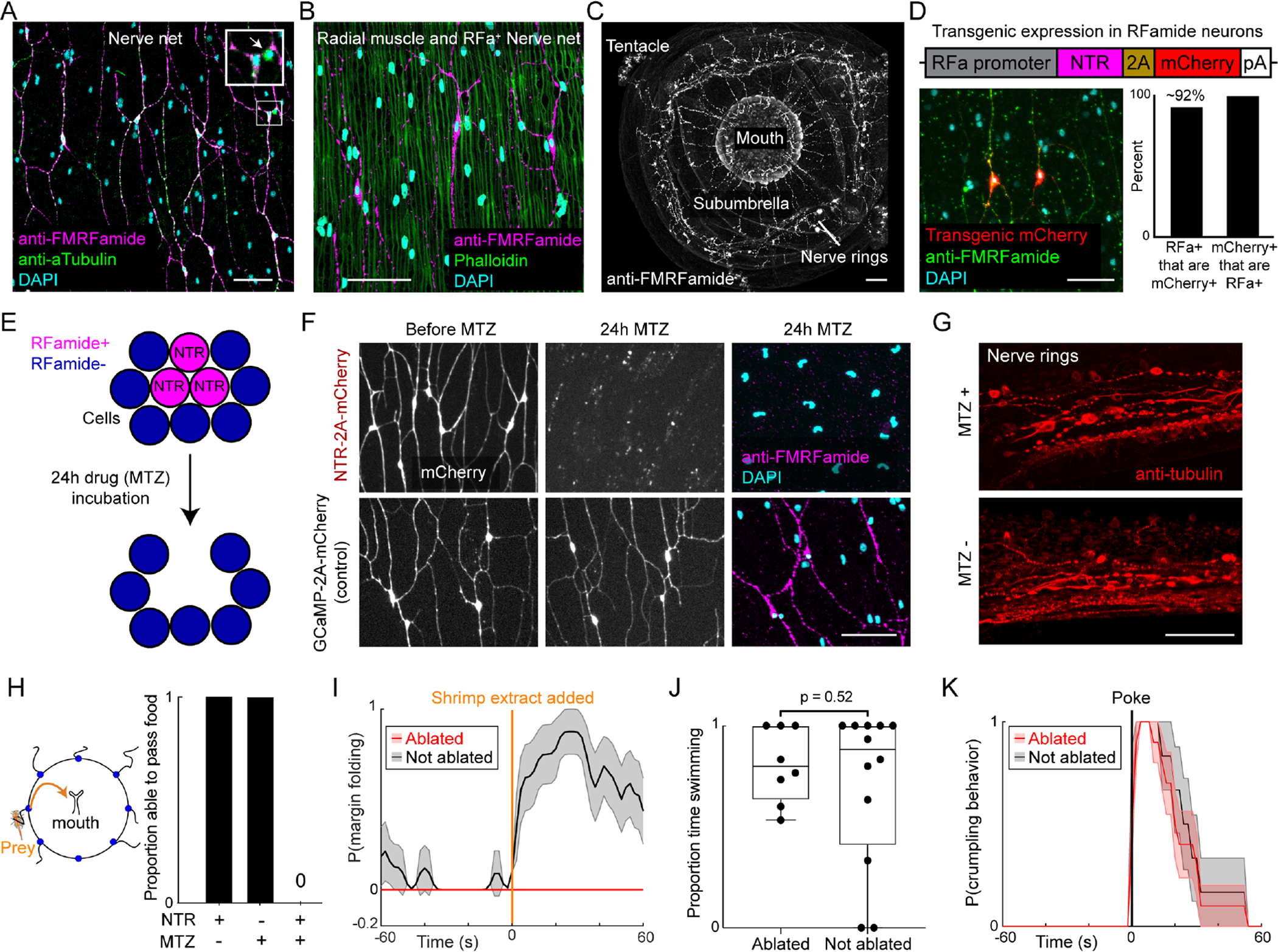
(A) FMRFamide-immunoreactive subnetwork (magenta, “RFa”). Inset: magnification of white box. Arrow: RFa-negative neuron.
(B) RFa+ neurons (magenta) are oriented along the radial muscle (green).
(C) Whole jellyfish showing distribution of RFa+ neurons.
(D) Transgenic cassette (top) showing genomic fragment (“promoter”) driving Nitroreductase (NTR) and mCherry. Image and quantification of specificity shown below. n ≥ 3 locations from each of 4 animals.
(E) Conditional genetic ablation of RFa+ neurons. MTZ, metronidazole. Adapted from Curado et al., 2008.
(F) Effect of MTZ incubation on RFa+ neurons in NTR-expressing (top) and control non-NTR-expressing (bottom; see Fig. 4) transgenic animals.
(G) Intact neurons in the nerve rings of NTR transgenic animals following incubation in MTZ, demonstrating specificity of ablation.
(H) Proportion of animals that passed food to the mouth within 5min of prey capture. Experimental (NTR transgenic + MTZ), n=10 animals. Controls, n=6 each.
(I) Probability of margin folding in experimental (NTR + MTZ, red line) and control (NTR without MTZ, grey line) animals, aligned to addition of shrimp extract. n=8 each, mean ± SEM.
(J) RFa+ neurons are not necessary for swimming. Proportion time swimming in NTR transgenics with (MTZ+, n=12) or without (MTZ−, n=8) MTZ, 30-seconds/each.
(K) Stimulus-triggered average of crumpling behavior following a mechanical stimulus. NTR transgenics with (red line, n=10) or without (grey line, n=6) MTZ; mean ± SEM. Scale: 50μm. See also Figure S1 and Figure S2.
To gain genetic access to RFa+ neurons, we established transgenesis in Clytia (Methods). Plasmid and Tol2 transposase were co-injected into Clytia eggs (Figure S1H; Koga et al., 1996; Ni et al., 2016) and polyp colonies screened for expression of an mCherry reporter. Strongly expressing colonies were cultured until they produced F0 medusae, which were backcrossed to parental strains. ~100% of these backcrosses yielded germline transmission. Stable F1 progeny were maintained and expanded vegetatively as clonal polyp colonies which released non-mosaic transgenic medusae that could be collected daily (Figure 1D; Figure S1H).
We first ablated the RFa+ neurons to determine the effect on the organism’s behavior. To this end, we cloned 6.6kb of 5’ flanking DNA from the Clytia RFamide precursor gene and inserted it upstream of a bi-cistronic construct encoding nitroreductase (NTR; Curado et al., 2008; Tabor et al., 2014) and mCherry, separated by a 2A peptide (Figure 2D; Daniels et al., 2014). Addition of the drug Metronidazole (MTZ) causes autonomous ablation of cells expressing NTR, which converts MTZ to its toxic form (Figure 2E; White and Mumm, 2013).
The RFamide 5’ genomic fragment successfully drove strong and specific mCherry expression in the umbrellar RFa+ network (~92% of RFa+ neurons targeted, 100% of targeted neurons were RFa+, Figure 2D). A 24-hr incubation in MTZ efficiently eliminated RFa+ neurons in the TGRFa∷NTR-2A-mCherry line (Figure 2D, F). Other neuronal populations were intact, demonstrating that ablation of RFa+ neurons was specific (Figure 2G). No ablation was observed in controls with either MTZ or the NTR transgene omitted (Figure 2F).
RFamide neurons are required for feeding
The umbrella is involved in several behaviors, including swimming, feeding, and defensive crumpling (Hyman, 1940; Romanes, 1885). While swimming and crumpling utilize symmetrical umbrellar contractions, feeding employs asymmetric contractions that vectorially transfer captured food (brine shrimp, Artemia) from contracted tentacles at the umbrella margin to the elongated feeding organ (“mouth”), which extends from the center of the umbrella (Figure 1C, E and Supplemental Video 1). Swimming stops (Figure S2F), and the mouth bends (“points”) towards the infolding portion of the margin to receive the food (Figure 3A–B; “pointing”). Food transfer is robust: 96% of first transfer attempts occurred within one minute of prey capture, of which 88% were successful (Figure S2G–H). 96.3% (52/54) of caught prey were eventually eaten.
Figure 3: Margin folding is a coordinated, directionally targeted behavior.
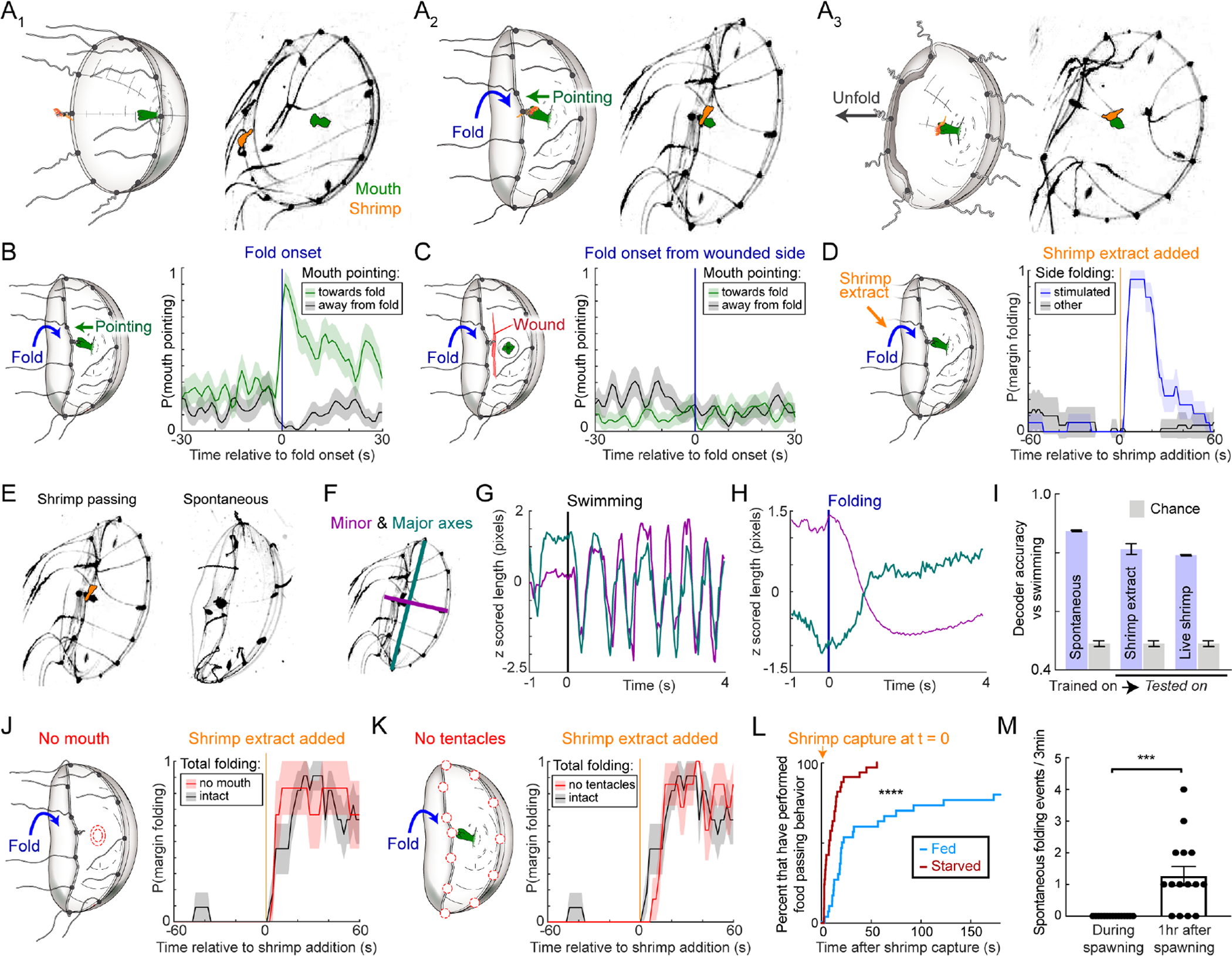
(A) Pseudo-colored video frames (right) and illustrations (left) showing margin folding to pass prey from a tentacle to the mouth. First, a prey is captured by a tentacle (A1). Next, the corresponding portion of the margin folds inwards, and the mouth points in the direction of folding (A2). Lastly, the prey is transferred to the mouth, and the margin relaxes to the open position (A3).
(B) The mouth points towards the infolding margin (green) but not in the opposite direction (gray). These data from the unwounded side of the animals shown in (C) also serve as an internal control for that experiment. n=52 folding events from 5 animals. In all such plots in this figure, mean ± SEM.
(C) Mouth pointing requires communication with the margin: pointing towards the folding margin was eliminated following a subumbrellar wound (green line; cf. B). NB, the mouth can point spontaneously in the direction of the wound (see baseline). n=52 folding events, 5 animals.
(D) Margin folding was triggered by local application of shrimp extract (blue line). Gray line: mean of unstimulated quadrants. n=18 trials, 2 animals.
(E) Video frames showing shrimp-evoked (left, repeated from A2) and spontaneous (right) folding.
(F) Video frame (repeated from E) with the major (longest) and minor (shortest) axes of the bell indicated.
(G) Symmetric bell contraction is characteristic of swimming. Major and minor axis lengths aligned to the onset of a swim bout.
(H) Asymmetric bell contraction is characteristic of margin folding. Major and minor axis lengths aligned to the onset of folding during food passing. Mean ± SEM; n=29 folding events, 3 animals.
(I) Decoder performance (folding vs. swimming) using axis lengths (F) as training features. The decoder was trained on spontaneous folding, yet could still identify induced folding. n=3 animals.
(J) Margin folding does not require the mouth. Extract-triggered margin folding with (grey line) or without (red line) a mouth; n=11 control animals, n=6 with no mouth.
(K) Margin folding does not require tentacle bulbs. Stimulus-triggered average, n=7 animals. Control data repeated from panel L.
(L) Fed animals took longer to pass food than 24h starved animals. Plot shows time between prey capture and food passing (n=30 fed, 33 starved animals; p<0.0001, Mantel-Cox Test).
(M) Spontaneous folding was significantly reduced during spawning (n=15 animals each; p = 0.0002, unpaired t-test).
See also Figure S2.
Remarkably, RFa+ neuron ablation completely eliminated asymmetric inward folding and hence the transfer of shrimp captured by the tentacles (Figure 2H). Ablation also prevented folding induced by chemosensory stimulation using shrimp extract (Figure 2I). Thus, RFa+ neurons are required for both food- and chemically-induced margin folding. In contrast, swimming and crumpling were unperturbed (Figure 2J–K; Figure S2A–C), suggesting that other neural cell types control these behaviors (King and Spencer, 1981).
Bath application of synthetic RFa peptide to transgenic medusae following RFa+ neuron ablation caused radial muscle contraction, confirming that the muscle was functionally intact (Figure S2D). Local infusion of RFa but not control peptide into the subumbrella caused local muscle contraction and margin infolding (Figure S2E). These data suggest that local release of peptide from RFa+ neurons activates the muscle, either directly or via an intermediate cell population, to determine the site of margin folding.
Margin folding requires coordination between autonomously functioning body parts and is influenced by internal state
These results prompted us to investigate margin folding behavior in more detail. First, we tested the necessity and sufficiency of different body parts using surgical manipulations. Mouth pointing towards the infolding margin was blocked if the umbrella was cut between the mouth and the margin, while margin folding was not (Figure 3C). Following excision of the mouth, the body swims, captures prey, and tries to pass prey to the hole where the mouth formerly was (Supplemental Video 3). The mouth-less umbrella also performed margin folding in response to shrimp extract (Figure 3J), which can trigger directional folding when locally applied (Figure 3D; S2J; Supplemental video 2). Shrimp extract could also trigger margin folding when tentacles and tentacle bulbs were removed (Figure 3K; S2K). Removal of other body parts revealed a similar theme of modular functional organization, confirming earlier studies (Romanes, 1885; Passano, 1973; Quiroga Artigas et al., 2018).
Clytia also performed margin folding in the absence of any added stimuli (13.7±15% of time observed, mean±SD), which was visually indistinguishable from evoked folding behavior (Figure 3E, Figure S2I and Supplemental Video 1). To quantify margin folding and compare spontaneous to prey-evoked behavior, we trained an automated classifier to discriminate margin folding from swimming with high accuracy, using the major and minor axes of the umbrella as features (Figure 3F–I). When trained on spontaneous folding alone, the same classifier could identify episodes of induced folding evoked by live shrimp or shrimp extract, with similar accuracy (Figure 3I).
To investigate whether margin folding behavior was a fixed-action stimulus-response reflex or was modulated by internal state (Anderson and Adolphs, 2014), we examined the effect of food deprivation. Animals starved for 24 hrs performed food passing significantly faster than ad libitum fed controls, following prey capture (Figure 3L). In addition, spontaneous folding behavior failed to occur when the animals were spawning (Figure 3M). Lastly, when multiple shrimp were captured simultaneously they were transferred to the mouth sequentially rather than coordinately, at a rate higher than chance (Figure S2L–M). Margin folding is therefore modulated by at least two internal states (metabolic and reproductive) and involves coordination across different sectors of the umbrella.
RFamide neurons are active during margin folding behavior in multiple contexts
To determine how RFa+ neuronal activity correlates with margin folding behavior, we generated a transgenic F1 line bi-cistronically expressing the calcium indicator GCaMP6s and mCherry (TGRFa∷GCaMP6s-2A-mCherry; Figure 4A; Chen et al., 2013). This transgenic line also had a high efficiency and specificity of targeting RFa+ neurons (~94% of RFa+ neurons targeted, 100% of targeted neurons were RFa+). Because GCaMP imaging was partially obscured by umbrellar expression of the endogenous GFP1 gene (Fourrage et al., 2014), we knocked out this gene using CRISPR/Cas9 (Momose et al., 2018) in the TGRFa∷GCaMP6s-2A-mCherry genetic background (Figure S3A).
Figure 4: Activity of RFamide neurons is spatio-temporally correlated with margin folding.
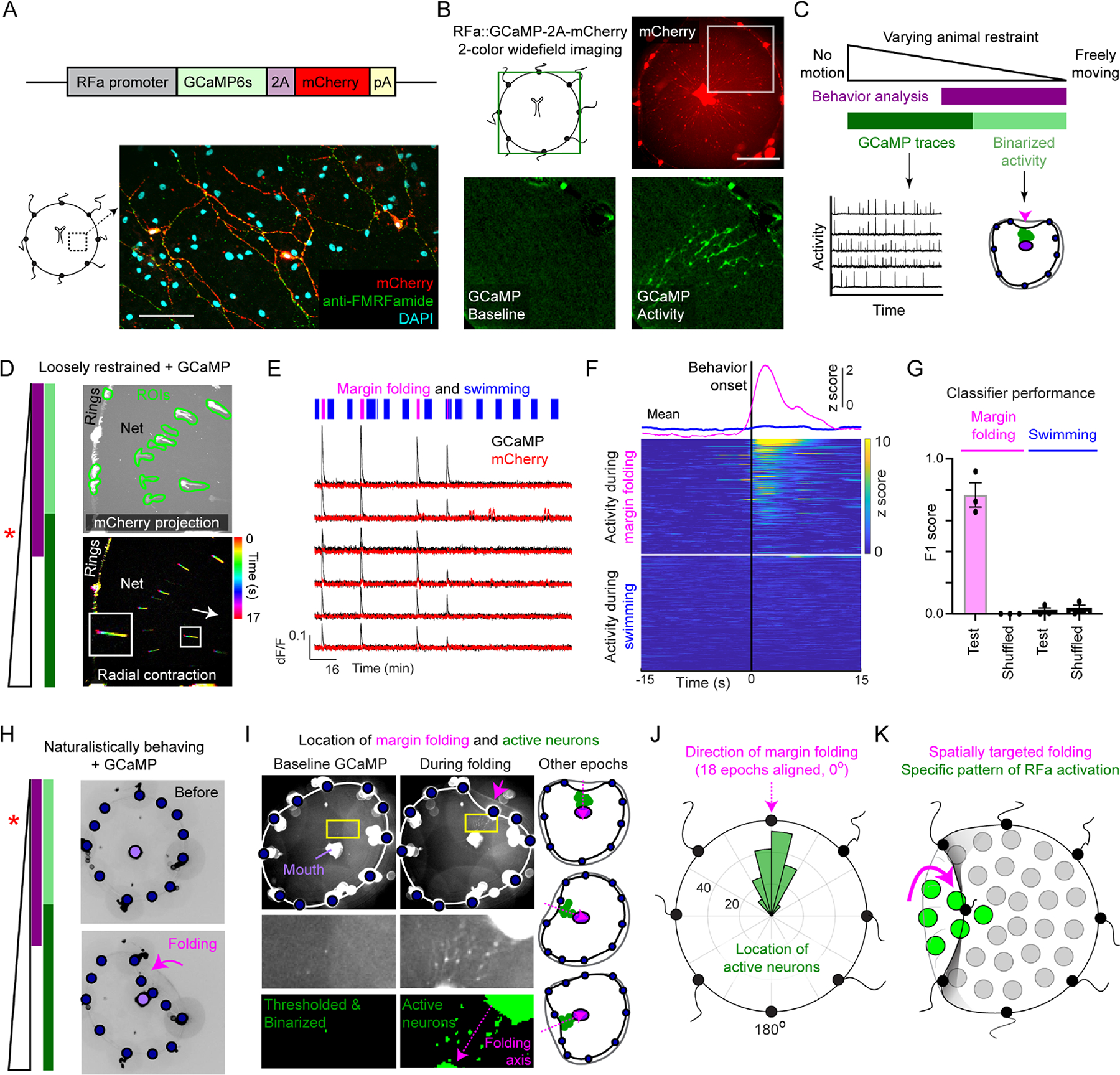
(A) Transgene (top) and expression (bottom) of GCaMP6s and mCherry in RFa+ neurons. Scale: 50μm.
(B) Video frames showing mCherry (upper right) and GCaMP signal (bottom; magnified view of inset box in top panel). Scale: ~1mm.
(C) Different neural and behavioral information could be extracted using varying restraint.
(D) Example in which behavior identity and GCaMP traces were extracted. Animal restraint diagrammed (left, asterisk, cf. panel C). Top: ROIs enclosing individual neurons superimposed on a maximum projection of the mCherry channel; white streaks show extent of motion. Bottom: video segment with frames colored temporally showing radial movement characteristic of folding. Arrow: direction of the mouth. FOV: ~3.5mm2.
(E-F) RFa+ neurons are active during folding, not swimming. (E) Behavior identity (top raster) and example traces extracted simultaneously from GCaMP and mCherry channels in loosely restrained animals. (F) Behavior-triggered average of GCaMP activity during folding behavior (magenta) vs. swimming (blue). Top traces, mean; heatmaps, individual units. n=222 (folding) and 371 (swimming) neuron-event pairs from 4 animals.
(G) Performance of Random Forest classifier trained on population neural activity predicting margin folding (magenta) vs. swimming (blue) behaviors (as well as quiescence). Dots represent individual animals.
(H) Animal restraint diagrammed (left, asterisk, cf. panel C). Example frames of GCaMP imaging in a naturalistically behaving animal before (top) and during (bottom) margin folding.
(I) RFa+ neurons were active at the location of folding. Example activity during baseline (left column) and folding (right column). Bottom panels: magnifications of yellow boxes (top). Annotated example frames (right) show active neurons (green dots) during folding events from different directions (magenta arrow). Blue dots: tentacle bulbs, purple dot: mouth. Shrimp extract diffusely present in water. FOV, ~3.5mm2.
(J) Polar histogram showing the location of active neurons (green bars) relative to the location of inward folding (magenta arrow). Folding from different directions aligned to 0° (n=175 neurons from 18 folding events).
(K) Schematic spatial location of activity during folding.
See also Figure S3
We performed wide-field, two-color, in vivo imaging in TGRFa∷GCaMP6s-2A-mCherry/+; GFP1−/− transgenic jellyfish, using either restrained or unrestrained preparations to balance GCaMP signal extraction and naturalistic behavior (Figure 4B–C; Figure S3B–D). In the unrestrained preparation, animals could behave freely in the small imaging chamber, allowing identification and localization of active neurons within the subumbrellar nerve net. However extraction of GCaMP traces from individual cells was not possible due to movement (“Naturalistically behaving”, Figure 4H–J; Supplemental video 3). Preparations in which the jellyfish were relatively motionless (“restrained”) using agarose embedding allowed extraction of high-quality GCaMP traces from single neurons (Figure 5A–C; Figure S4B; Supplemental video 5). Due to variation in the agarose-embedded preparations (see Methods), in some animals there was both sufficient restraint to extract single-cell traces and sufficient freedom of movement to identify apparent attempts at swimming or folding (“Loosely restrained”, Figure 4D–G).
Figure 5: Functional parcellation of the umbrellar RFamide network.
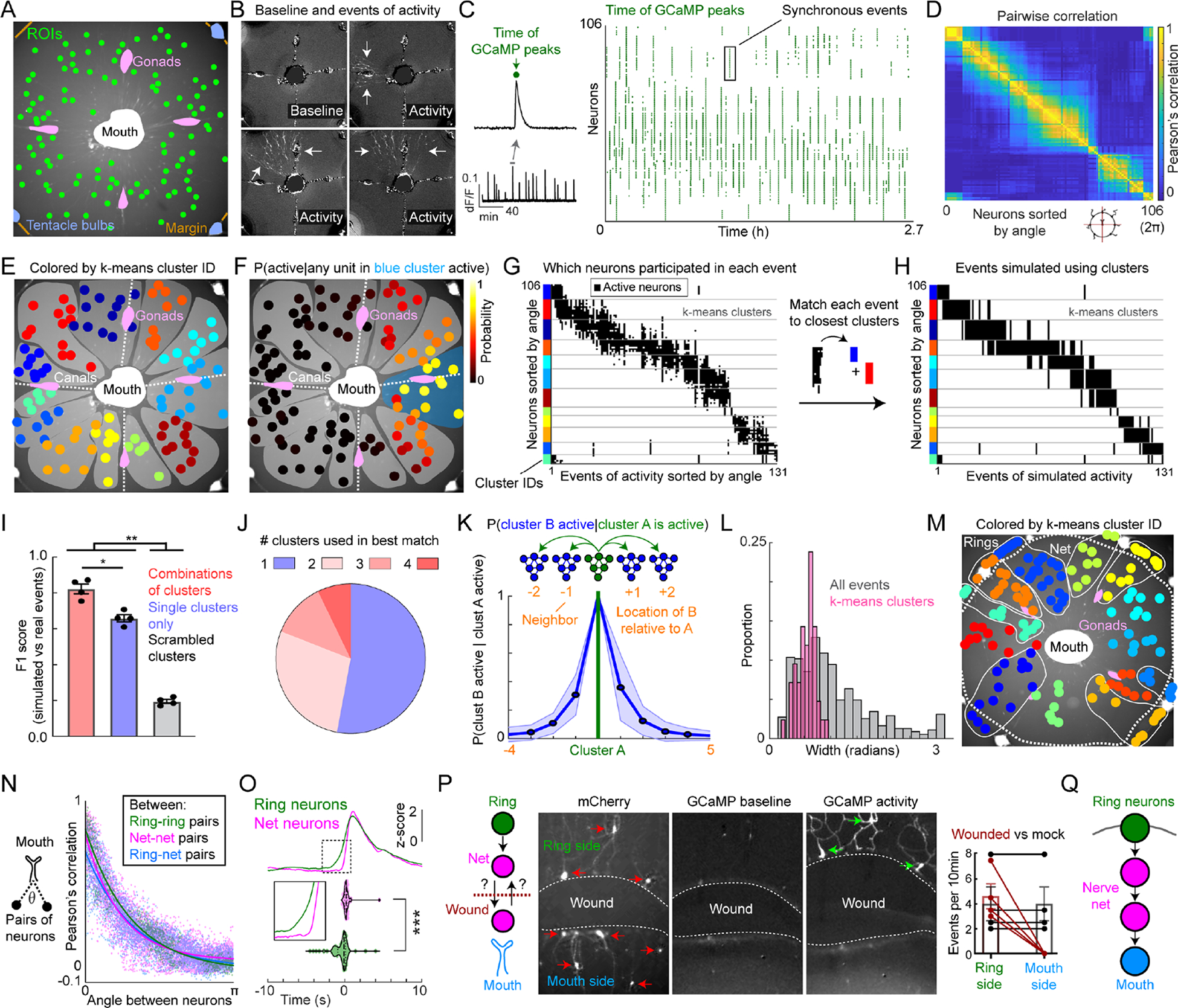
(A) Example field of view (FOV) for imaging spontaneous GCaMP activity in highly restrained jellyfish. Center of ROIs (green) superimposed on mCherry image. FOV, ~3.5mm2.
(B) Example GCaMP frames showing synchronous firing of neuronal ensembles. Baseline fluorescence (upper left) and representative activity (remaining panels).
(C) Example GCaMP trace (bottom left) and a magnified peak (top left); green dot indicates peak time here (top left) and for each neuron across the recording (raster, right). Neurons sorted by angular anatomical position relative to the mouth.
(D) Pairwise Pearson’s correlation of neuronal activity (i.e. time of GCaMP peaks).
(E) k-means clustering of neurons based on their activity; neurons colored by cluster identity.
(F-G) Spontaneous events of neural activity spread beyond single clusters in a stochastic manner. (F) Neurons colored by the probability that they were active given that any neuron in the blue-shaded cluster was active. Remaining clusters shaded in gray, cf. panel E. (G) Raster of events of activity, showing which neurons were active during each event. Neurons sorted by k-means cluster, with clusters sorted by angle relative to the mouth. Events sorted by median angular location of active neurons. Y-axis colors: cluster ID, cf. E.
(H) Simulation of the events in G, generated by matching each event to its closest combination of k-means clusters using the Hamming Distance.
(I) F1 scores comparing observed events to those simulated using single, scrambled, or combinations of clusters (n=4 jellyfish, ANOVA with Tukey’s post-hoc tests, mean±SEM).
(J) Number of clusters used in best matching simulated events. Maximum number of clusters that could be co-active was 4. n=599 events from 4 animals.
(K) Neighboring clusters were more likely to be co-active than distant ones. Conditional probability of a cluster “B” activating given that a cluster “A” activated. The comparison to self is centered (P = 1). Activation: >60% neurons in a cluster active within 2s. Blue line: mean ± standard deviation; n=3 animals.
(L) Angular width of clusters compared to widths of events. n=4 animals.
(M) Ring neurons cluster with their corresponding net domain (white outlines). FOV with neurons colored by cluster identity. FOV, ~3.5mm2.
(N) Pairwise activity correlation vs. angle between neurons in the net or ring relative to the mouth for the animal in panel M (Tau: Ring-ring: 0.97, Net-net: 0.75, Ring-Net: 0.99).
(O) Nerve ring activity precedes nerve net activity. Top traces, mean ring activity (green line) aligned to the onset of net activity (magenta line, t=0). Inset: magnification of boxed region. Violin plots (below) show onset times relative to t=0 for individual neurons to cross a threshold z-score (1.5). Onset time difference is significant (p < 0.0001, n=198 ring-event pairs, 135 net-event pairs, 4 animals).
(P) Wounding the subumbrella (left diagram) prevents centripetal spread of net activity. Images show mCherry (left) and GCaMP (right). Green arrows: active neurons. Quantification (far right): GCaMP events per 10min on either the ring or mouth side of wounded (red) or control (black) animals (n=4 each).
(Q) Activity flows unidirectionally from the rings through the net towards the mouth.
See also Figures S4 and S5.
Movements interpreted as attempted “swimming” comprised high frequency, circumferential contractions, while those interpreted as attempted “folding” comprised low frequency, radial movements (Figure 4D; Figure S3E; Figure 3G–H). To quantify these behaviors in such loosely restrained preparations, we extracted optic flow vectors across frames from the imaging videos. Classifiers trained on these features could distinguish behaviors in held-out test frames with high accuracy (swimming, accuracy = 0.85±0.04; folding, accuracy = 0.81±0.03, mean±SEM; Figure S3E). We used the mCherry channel to segment neurons and exclude that GCaMP signals reflected motion artifacts (Figure 4E; Figure S3H–I; Figure S4K).
In loosely restrained preparations in which both attempted margin folding and swimming movements occurred, behavioral epochs and neural activity could be aligned to examine their temporal relationship (Figure 4E–G). Patches of RFa+ neurons in both the nerve rings and net were strongly activated at the onset of folding, but not swimming (Figure 4E–F). Neural activity in the patches was relatively synchronous (Figure 4E). Using population neural activity, we trained a 3-way classifier to predict quiescence, swimming, or folding and found that folding episodes could be predicted with high accuracy, while swimming could not be distinguished from quiescence (Figure 4G). Principal component analysis (PCA) confirmed that the largest source of variance in neural activity occurred during folding (Figure S3F). These data indicate that RFa+ neuronal activity is temporally and specifically associated with margin folding behavior. The NTR ablation data (Figure 2H, I) indicate, moreover, that this activity is likely a cause and not a consequence of this behavior.
To examine the pattern of RFa+ neuronal activation more broadly across the nerve net under conditions where complete margin folding could be performed, we imaged GCaMP in the unrestrained preparation (Figure 4H) in three contexts: (1) spontaneously, i.e., no stimuli given (Figure S2I; Figure 3E), (2) with shrimp extract uniformly present, or (3) with intact shrimp presented to the tentacles. We manually extracted body shape and identified the location of active neurons during individual folding events. In both the spontaneous and evoked contexts, RFa+ neurons in the nerve net were activated in radially oriented, wedge-shaped patches located at the epicenter of margin infolding (Figure 4I–J; Figure S3H–K; Supplemental Video 4). Thus, RFa+ neuronal activity is temporally correlated with both evoked and spontaneous margin folding, and is spatially localized at the site of each folding event (Figure 4K).
The observation of local ensembles of synchronously active RFa+ neurons raised the question of whether these neurons activate each other, as well as the radial muscle, via release of RFamide peptide. To address this question, we perfused RFamide during GCaMP imaging. RFamide addition did not detectably activate RFa+ neurons (Figure S3G). This suggests that the peptide may not be used for excitatory, inter-RFa+ neurotransmission, but does not exclude the possibility that these neurons communicate via an unidentified co-transmitter or peptide.
Functional subdivisions within the umbrellar RFamide network
To further examine the pattern of ensemble activity in the RFa+ system (Figure 4J–K), we performed whole umbrella imaging in animals that were well restrained in agarose (Figure 5A; Figure S4A–B). These preparations revealed spatially localized ensembles of RFa+ neurons that exhibited repeated events of spontaneous, synchronous activity (Figure 5B–C; Figure S4B–C). These ensembles appeared as radially oriented, wedge-shaped populations that stretched between the margin and the mouth (Figure 5B; Supplemental Video 5), similar to the pattern observed in naturalistically behaving animals (Figure 4). The radial organization of ensembles was obvious along the diagonal of a neuronal activity correlation matrix in which neurons were sorted by their relative angular position (Figure 5D). However, the correlation pattern was not a series of sharply defined blocks, but rather patches of variable size with diffuse borders, indicating variability in ensemble membership across individual events.
Closer inspection of the correlation matrix revealed structure on multiple spatial scales, with very high correlations between neighboring neurons, as well as weaker, distance-dependent correlations (Figure 5D; Figure S4D–E). To identify core groups of highly correlated neurons active during repeated events of spontaneous activity, we used k-means clustering. This revealed striking spatial groupings that roughly tiled the animal (Figure 5E). However, ensemble membership between individual events was flexible, and many events were not restricted to single k-means clusters (Figure 5F). This variability suggests that active ensembles are not defined exclusively by deterministic connectivity rules (see below).
We next asked whether inter-episodic variability in ensemble membership reflected the flexible recruitment of cells from neighboring clusters. We first plotted the activity of individual neurons across multiple spontaneous events, grouping neurons by k-means cluster membership and sorting clusters by their relative radial positions (Figure 5G). We then simplified the results by graphing which clusters were active during each event, not which neurons were active (compare Figure 5G to 5H). Next, we asked how much of the observed activity could be explained by ensembles comprising single vs. multiple clusters.
Restricting ensembles to members of single k-means clusters explained much, but not all, of the observed activity over time (F1 score: ~0.66, Figure 5I, violet bar). However, allowing ensembles to flexibly incorporate multiple clusters better explained the observed activity (F1 score: ~0.82, Figure 5H–I). Spontaneous events restricted to single clusters were only slightly more frequent than those incorporating an additional cluster(s) (Figure 5J). Neighboring clusters were most often co-active (Figure 5K), although not all members of such clusters participated in each event (Figure 5F–G). Thus, inter-episodic variation in ensemble membership (Figure 5L) is best explained by a flexible incorporation of neurons from adjacent clusters into ensembles initiated by a “core” k-means cluster.
Because k-means forces neurons into single clusters, as an alternative way to capture flexibility in inter-episodic ensemble membership, we used Non-negative Matrix Factorization (NMF) followed by Independent Components Analysis (ICA) (Lopes-dos-Santos et al., 2013; See et al., 2018) to cluster active cells. In this method, individual neurons can be members of more than one cluster. Among neurons assigned to NMF-ICA clusters, the majority participated in only a single cluster (~89%), with fewer participating in 2 (~10%) or 3 clusters (<1%; n=489 neurons from 4 animals; Figure S4F–G). Thus, whether cluster membership is defined rigidly (k-means) or more flexibly (NMF/ICA), clusters emerge as principle units of spontaneous RFa+ network activity, with episodic variability in ensemble membership reflecting fluctuations in nearest-neighbor recruitment (Figure 5F).
A close examination of umbrellar anatomy did not any reveal identifiable structural correlates on the scale of these subassemblies (Figure S4H–J), and no obvious structure was evident in the mCherry channel (Figure S4K). Therefore, the spontaneously active ensembles within the RFa+ nerve net appear to be anatomically cryptic, at least at the level of light microscopy.
Ring neurons act upstream of subjacent net neurons
Spontaneous activity in the nerve rings encircling the umbrella also showed spatial clustering. Ring clusters were highly correlated with subjacent nerve net clusters (Figure 5M–N; Figure S4L–N). These observations suggested a possible flow of information between the RFa+ neurons in the nerve rings and net. We therefore examined the directionality of this flow. Nerve net activation always had a nerve ring correlate, but ring activity did not always coincide with a net event (Figure S5B). Correlated ring activity was initiated prior to net activity during spontaneous events (Figure 5O; Figure S5C). Furthermore, neurons in excised fragments of the ring responded to shrimp extract, but neurons in net fragments only responded if the margin was attached (Figure S5D), suggesting a dependence of net neurons on ring neurons for evoked activity. Consistent with this, excising the margin (containing the rings, Figure 1E) eliminated evoked folding behavior in the remaining umbrellar tissue (Figure S5A). Histology suggested that processes from ring and net neurons were intermingled at the margin (Figure S5E). Attempts to map process origin by focal dye labeling were unsuccessful.
We further examined information flow by generating transverse incisions in the subumbrella and monitoring whether spontaneous nerve net activity could be observed on the ring (peripheral) side of the wound, the mouth (central) side, or both (Figure 5P). We observed spontaneous activity peripheral but not central to the incisions (Figure 5P, Supplemental Video 6). Thus, information flows from the rings into the nerve net, where it continues centripetally towards the mouth (Figure 5Q).
The foregoing data suggested that radial umbrellar fragments containing tentacles, the mouth and RFa+ neurons in both the nerve rings and the net might perform food-passing behavior autonomously. Indeed, such isolated wedge-shaped sectors were able to pass shrimp from the margin to the mouth (Figure S5F; Supplemental Video 7).
Spontaneous and stimulus-evoked RFa+ ensemble activity exhibits similar patterns
The foregoing analysis raised the question of whether stimulus-evoked ensembles can emerge at arbitrary locations in the RFa+ subnetwork, or whether they are spatially constrained by the clusters revealed in our analysis of spontaneous events. To distinguish these alternatives, we quantitatively compared spontaneous to evoked ensemble activity in the same animal (Figure 6A–C; S5G–H), using the subspace alignment metric (Elsayed et al., 2016; Yoo and Hayden, 2020). This metric tests the variance in evoked activity epochs explained by the principal components of spontaneous epochs.
Figure 6: Ensemble activity arises from underlying structure.
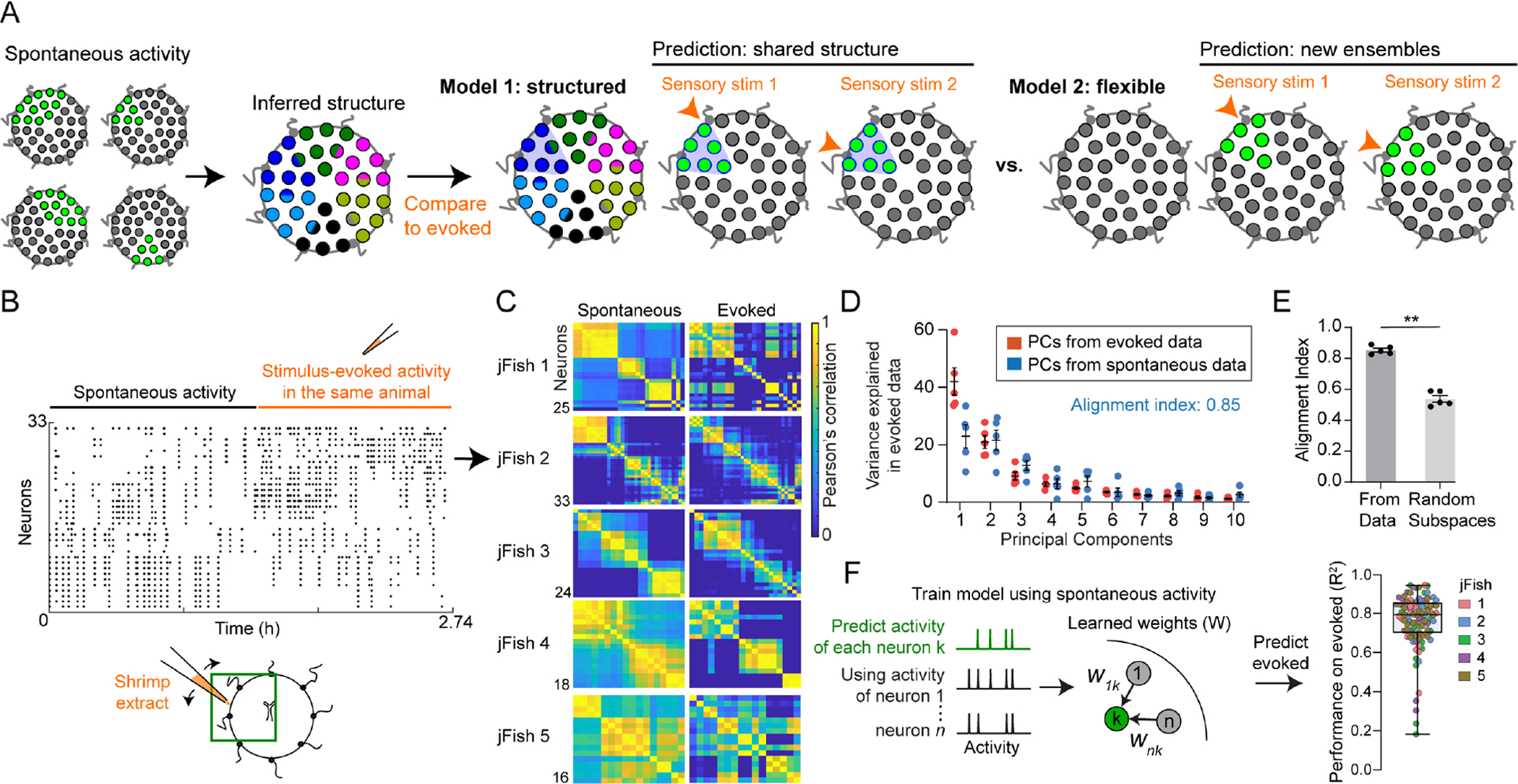
(A) Model 1 (“structured”): evoked activity uses spatial pre-patterning shared with spontaneous activity. Model 2 (“flexible”): assemblies generated de novo at each stimulus location.
(B) Example of spontaneous followed by evoked activity data. Dots indicate time of a GCaMP peak. Diagram: experiment and approximate FOV.
(C) Pairwise correlation during spontaneous (left panels) and corresponding evoked (right panels) epochs. Panel B corresponds to jellyfish 2.
(D) Variance explained in evoked data using its own principal components (PCs, orange) or those from spontaneous activity (blue). The first 5 PCs from spontaneous explain 85% of variance in evoked. Dots here and in E represent the 5 animals in C.
(E) Alignment indices, using the first 5 PCs from spontaneous to explain variance in evoked. Index is significantly higher than using PCs from random subspaces (p = 0.0079, Mann-Whitney Test).
(F) GLMs trained using only the spontaneous activity epoch; evoked neural activity could still be predicted with high accuracy (right, R2, dots: neurons from all jellyfish in C).
See also Figure S5.
We observed high subspace alignment between spontaneous and evoked epochs (85 ± 1.21%, mean ± SEM, Figure 6D), which was similar to the alignment between two sets of spontaneous epochs (83 ± 4%, mean+/−SEM) and was significantly different from the alignment between randomly generated subspaces (Figure 6E; p=0.0079). These high alignment indices suggest that evoked ensemble structure may be constrained by an intrinsic pre-pattern exhibited during spontaneous episodes.
As an independent method for comparing spontaneous to evoked ensembles, we trained generalized linear models (GLMs) to predict the activity of each imaged neuron using the weighted activity of all other imaged neurons (Efron et al., 2004; Mishchenko et al., 2011; Pillow et al., 2008). GLMs trained and tested on spontaneous activity recapitulated the activity of a given cell with high accuracy (83 ± 1.3% mean ± SEM; Figure 6F). More importantly, GLMs trained on spontaneous activity and tested on stimulus-evoked neural activity exhibited similarly high accuracy (80 ± 1.4% mean ± SEM, Figure 6F; Figure S5I). These results further argue that spontaneous and evoked RFa+ ensembles share common structural constraints.
Modeling supports a partially structured nerve net organization
To examine possible circuit implementations of observed ensemble activity, we undertook a modeling approach. We constructed a series of spiking neural network models, each of which incorporated different assumptions about underlying connectivity (Figure 7A–D). At one extreme, we assumed that connectivity strength between a given RFa+ neuron and its neighbors declined continuously as a function of their angular distance (“continuous”; Figure 7B). At the opposite extreme, we assumed all neurons were hard-wired into non-interacting subnetworks, with each neuron connected to all other members of its subnetwork with equal strength (“structured”; Figure 7C). Finally, we generated hybrid models using weighted combinations of “continuous” vs. “structured” connectivity (Figure 7D). For each type of model, we tuned both the strength, and angular extent of, network connectivity to best fit two observed statistics: the circumferential width of each spontaneous event, and the percent of neurons recruited per event (see Methods; Figure S6A–E).
Figure 7: Modeling supports a partially structured nerve net organization.
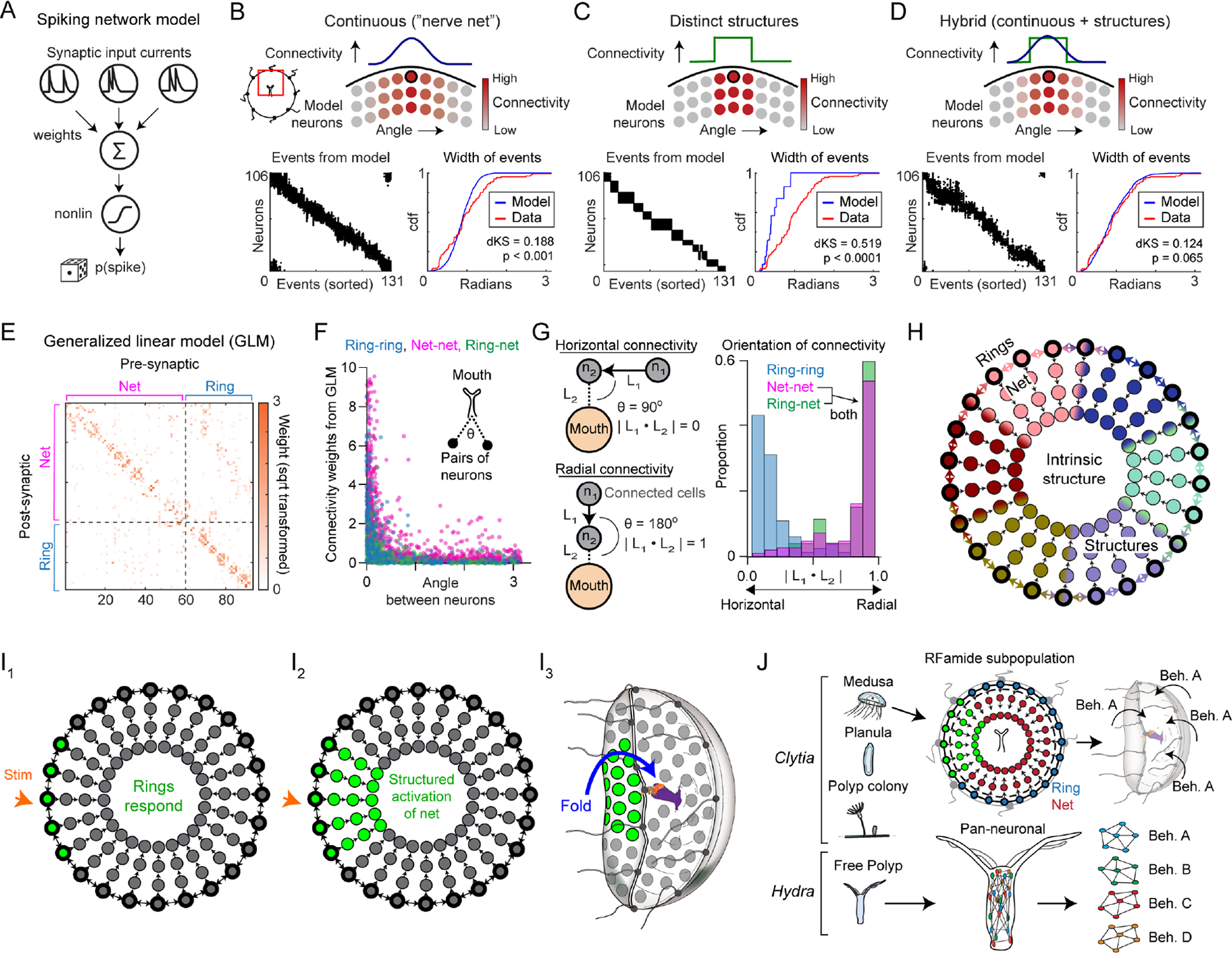
(A) A spiking neural network model was used to simulate ensemble events. The activity of each neuron depended on the weighted sum of its inputs, with a hyperbolic tangent response nonlinearity and an element of stochastic spike generation.
(B-D) Diagram of model connectivity (top), events generated by the model (bottom left), and the angular width of those events (bottom right). Hybrid models best fit the data (D). In the hybrid model shown here, 22% of inputs per neuron were from the structured connectivity but models with 85% structured connectivity gave similar results (Figure S6C).
(E) Example matrix of weights from GLMs trained on spontaneous activity.
(F) GLM weights were highest for pairs of neurons with a small angle between them relative to the mouth.
(G) The orientation of connectivity relative to the mouth (i.e. absolute value of the dot product) between pairs of neurons with strong connectivity (GLM weights >2). Net-net and ring-net: high radial connectivity; ring-ring: high circumferential connectivity (net-net vs ring-ring, p = 3.5e–70; ring-net vs ring-ring, p = 5.7e–15; net-net vs ring-net, p = 0.8, Kolmogorov–Smirnov tests).
(H) Model of the organization of the umbrellar RFa+ system, in which sparsely and radially connected nerve net neurons act downstream of horizontally connected nerve ring neurons, forming a combination of continuous and structured connectivity.
(I) Working model of RFa+ system function. Sensory stimuli activate nerve ring neurons (I1), which recruit the nerve net (I2), activating the radial muscle and causing inward margin folding.
(J) In Clytia medusae, an RFa+ subnetwork is active at different times and locations to drive directional behavior. Pan-neuronal imaging in Hydra revealed non-overlapping subnetworks for different behaviors (Dupre and Yuste, 2017). Hydra diagram adapted from Dupre and Yuste, 2017.
See also Figures S6 and S7.
The best fit to the data was achieved using hybrid models combining both continuous and structured connectivity (Figure 7D; Figure S6C–D). A model using only discontinuous structures had the lowest performance (Figure 7C); some degree of continuous connectivity was essential for the model to exhibit inter-episodic flexibility in event boundaries, as observed experimentally. The relative contribution of structured vs. continuous connectivity did not strongly effect performance, as long as structured connectivity was present (Figure S6C–D). In all best-fit models, continuous connectivity was sparse and local, with each neuron only forming synapses with a small number of its nearest neighbors (Figure S6C–E). These findings support a model in which flexible ensembles are generated by combining “core” networks with continuously graded, local connectivity.
The foregoing model assumes that local connectivity between net neurons, as well as input from ring neurons, accounts for the synchronous activity of ensemble members. To evaluate the relative contributions of ring→net vs net→net connectivity to this synchronicity, we first trained GLMs using recordings that included both ring and net neurons (Figure 5). These GLMs performed with high accuracy (76 ± 1.2% mean ± SEM; Figure S7A). Matrices of fitted GLM weights between RFa+ neurons, i.e. inferences of connection strengths (Mishchenko et al., 2011), revealed that the GLMs used only sparse, local weights (Figure 7E–F), which was not due to GLM regularization (Figure S7B). This pattern of sparse connectivity was similar to our best fit network models (Figure S6C1–D1).
To determine whether synchronous firing in the umbrellar net ensembles could be explained purely by coordinated ring→net input, we digitally “ablated” intra-umbrellar network connectivity from our GLMs, either after or before training but prior to testing (Figure S7C–F). These “ablated” GLMs performed significantly less well than the “intact” GLMs (Ablated, 54 ± 2%; Intact, 82 ± 0.6%; Figure S7G–J). These results strengthen the idea that sparse local net→net connectivity contributes to the synchronicity of ensemble activity.
Finally, we examined the relationship between GLM weights (connection strength), and the angle of connections relative to the mouth. Interestingly, both ring→net and net→net connectivity was primarily oriented radially, whereas ring→ring connectivity was primarily horizontally/circumferentially (Figure 7G; net-net vs ring-ring, p = 3.5e–70; ring-net vs ring-ring, p = 5.7e–15; net-net vs ring-net, p = 0.8). This suggests that the radial and circumferential extent of ensembles may reflect intrinsic synaptic biases within the net and ring RFa+ subnetworks, respectively.
Discussion
Here we introduce the jellyfish Clytia hemisphaerica as a model for systems neuroscience. Clytia combines optical accessibility and genetic tractability with a varied behavioral repertoire. We report germline transgenesis in this organism and a systems-level interrogation of neural activity. Clytia offers opportunities to study neural development, function, evolution, and behavior in an organism that provides a window into the first nervous systems on the planet.
A jellyfish model for neuroscience
Among cnidarian genetic models, Clytia is complementary in several respects. First and foremost, Clytia has a medusa stage, while the others (e.g Hydra) are polyps (Bosch et al., 2017). Understanding the particulars of jellyfish behavioral control is relevant to their growing ecological and economic importance. Second, the medusa stage offers a relatively rich behavioral repertoire in comparison to polyps (e.g. Costello et al., 2021). Finally, Clytia affords several advantages over Hydra as a genetically tractable model. It allows routine generation of F1 lines, whereas in Hydra only F0 transgenics have been used for neural imaging (Dupre and Yuste, 2017). Furthermore, Clytia husbandry permits inter-strain genetic crosses, which are currently difficult to achieve in Hydra (Klimovich et al., 2019). Finally, transgenesis is more easily scaled up in Clytia, where thousands of unfertilized eggs can be generated for injection daily. In contrast, the Hydra lifecycle limits the availability of embryos (Figure S1H; Klimovich et al., 2019).
Imaging results in Clytia provide an interesting contrast to those obtained in Hydra (Dupre and Yuste, 2017). The Hydra neural net is divided into several functionally distinct subnetworks, whose activity is correlated with different types of polyp movement (contraction, extension, bending) (Dupre and Yuste 2017). These subnetworks are not spatially separated, but overlapping. As in Hydra, the Clytia umbrellar neural net comprises multiple subnetworks, including the RFa+ subnetwork. In contrast to Hydra, however, this subnetwork is further parcellated into functional subassemblies (Figure 7H). These spatially distinct subassemblies therefore indicate a higher order of neural structural organization in Clytia than in Hydra (Figure 7J). Further comparisons between Clytia, Hydra, and other cnidarian model organisms should provide important insights into neural network evolution (Bosch et al., 2017).
Decentralized, modular neural control of organismal behavior
A striking feature of Clytia behavior is its extreme functional modularity: for example, an isolated mouth can ingest food, and margin folding can occur in a “mouth-less” umbrella. Nevertheless, in intact jellyfish the mouth points towards the infolding margin during food transfer, revealing coordination between these modules. Such coordination is also observed during food-passing in wedge-shaped strips of Clytia umbrella containing the mouth (Figure S5F). These data suggest that functionally autonomous behavioral modules (e.g., mouth, umbrella) are combined to form coordinated “super-modules,” copies of which are arrayed circumferentially around the umbrella. Such a mechanism accommodates the continuous growth that jellyfish exhibit.
If this hierarchical view is correct, coordinated behaviors in organisms lacking a central brain may have emerged by duplication and modification of smaller autonomous modules to form functionally interacting super-modules. How these interactions are achieved remains to be determined. Over time, modification of super-modules might give rise to new structures and organism-level behaviors. Such a nested modular organization could therefore be an important substrate for the evolution of complex behaviors.
Clytia have a remarkable ability to regenerate and recover behaviorally following injury (Kamran et al., 2017; Sinigaglia et al., 2020). They also continuously integrate new stem cell-derived neurons into their nervous system as they grow, without disrupting organismal behavior (Chari et al., 2021). The local network interactions and modular organization that we have described may facilitate such continuous growth and repair. Clytia affords a genetically tractable model to investigate the dynamics of such regeneration in real time, a process at the interface of neural development and systems-level function.
Limitations of the study
Our conclusions are largely based on analysis of spontaneous episodes of neural activity in immobilized animals. These episodes likely correspond to spontaneous activity with margin folding events observed in freely moving animals, since spontaneous activity without folding was not seen. We cannot exclude that such “spontaneous” events are triggered by microscopic exogenous stimuli, rather than endogenously generated. Furthermore, activity in immobilized animals may exhibit subtle differences from that in freely moving jellyfish. Our analyses relied on a finite number of spontaneous ensemble events (~100/animal); significantly more events could change the observed correlation structure. Lastly, imaging was performed in juvenile jellyfish: the functional organization described here may change as the animal grows.
STAR Methods
Resource Availability
Lead Contact
Requests for resources and reagents should be addressed to lead contact, David J. Anderson (wuwei@caltech.edu).
Materials availability
The plasmids and transgenic jellyfish generated in this study are available upon request. Plasmids will also be deposited to Addgene.
Data and code availability
Source data reported in this paper will be shared by the lead contact upon request.
Code used for analyses in this paper will be shared by the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Culture of the Clytia life cycle was carried out in accordance with published protocols (Lechable et al., 2020), with the exception of the culture tank design. Circulating systems used here had the same overall flow design as in Lechable et al., 2020, but used modifications of zebrafish tanks (Pentair) for animal housing. Polyp slides were held in glass slide racks (Fisher, cat#02-912-615) in small tanks with high water flow. Jellyfish were cultured in large-sized tanks with a curved plastic insert placed at the back of the tank with a nylon mesh outlet at the bottom. A slow drip from the circulating system into these tanks allowed for water turnover without risking sweeping the jellyfish through the outlet. A constant speed 5rpm DC motor (Uxcell) attached to the lid of a multi-well tissue culture plate was then used to create a constant circular current. A dimmer switch was used to tune the rotation speed of the motor, with reduced speeds as the jellyfish grew. Jellyfish used for transgenesis, and all polyps, were maintained in these circulating systems, while smaller jellyfish were maintained in beakers. In beakers, current was generated using stirring with a DC motor, as above. All artificial sea water for culture and experiments was made using Red Sea Salts (Bulk Reef Supply, cat# 207077) diluted into building deionized water to 36ppt.
Unless otherwise indicated, behavior experiments were performed using sexually mature animals of the Z4B strain of Clytia, which are female. Transgenesis was performed by crossing Z4B females to Z13 males. For generating experimental F1 lines, NTR and GCaMP lines were backcrossed to Z4B. A single F1 polyp colony was then chosen to maintain for each strain to control for genetic background. Clonal experimental animals were then collected from these polyp colonies. Experiments and culture were performed at room temperature.
Method Details
Histology
For antibody staining, Clytia were fixed for 2h at room temperature in 4% PFA in 0.2um filtered artificial sea water (Red Sea Salts, Bulk Reef Supply, cat# 207077, diluted into building deionized water to 36ppt). They were then washed 3x in PBS followed by blocking for 1h in PBS with 0.1% Triton (PBST) and 10% normal donkey serum (NDS). Animals were then incubated for 1–3 nights in primary antibody with 5% NDS in PBST at 4°. Primary antibodies used in this study were: anti-FMRF (Immunostar, cat# 20091), anti-Tyrosine Tubulin (Sigma cat# T9028–100UL), 647-conjugated anti-aTubulin, clone DM1a (Millipore Sigma cat# 05–829-AF647), and anti-aTubulin (YL1/2; Thermofisher cat# MA1–80017, Figure S5E). Following primary antibody incubation, one short (~5min) and then 3–4 long (~30min+) washes were performed in PBST, and then animals were transferred into secondary antibodies and/or Phalloidin-488 (Thermo Fisher, cat# A12379) for 2h at room temperature or overnight at 4°. Secondary antibodies: donkey anti-rabbit conjugated to Alexa Fluor 647 or 488 (Jackson ImmunoResearch cat# 711-605-152 and Thermofisher cat# A-21206), or donkey anti-mouse conjugated to Alexa Fluor 647 (Thermofisher cat# A-31571). Animals were stained with DAPI (BD cat#564907) and mounted onto glass slides for imaging. For staining shown in Figure 1F, animals were dehydrated stepwise into methanol and then rehydrated prior to the blocking step. Quantification of overlap related to images in Figure 2 was from at least 3 separate locations/each from at least 3 animals. In situ hybridization was carried out as described in (Chari et al., 2021), including the RFamide (pp5) probe, which was the same as the one used in that study. Imaging of histological specimens was carried out using an Olympus FV3000 confocal. Multicolor micrographs shown throughout are pseudocolored composites, with brightness and contrast adjusted individually for clarity and aesthetics using ImageJ (NIH; Schneider et al., 2012).
Cloning
To generate the Actin::mCherry plasmid used to establish transgenesis, codon-optimized mCherry cDNA was designed using COOL (Yu et al., 2017). The ACT2 promoter was cloned from upstream of a non-muscle actin gene (XLOC_011689) using primers: TTTGCTGCGTACAACAACAACGACC and TCGACTTATGTCCTGATAGTTCGGA. The 3’UTR used in all constructs was from a different actin gene (XLOC_021750) and was amplified using primers: CCAACAGATGTGGATCTCCAAACA and ACTGGAAGCCTGAGTTCCATCAAA. This was assembled into the pT2AL200R150G backbone (Urasaki et al., 2006; licensed under MTA - N° K2010–008).
Other Clytia transgenesis constructs were based on the miniTol2 backbone, a gift from Dr. Stephen Ekker (Addgene plasmid # 31829). To generate RFamide::NTR-2A-mCherry, the RFamide (XLOC_019434) fragment was amplified from Clytia genomic DNA using the following primers, ATCCCCATCCGCCATCTTTG, GTTGTGTTCTTTCTTGATTTGATGG, and inserted into the miniTol2 backbone using In-Fusion Cloning (Takara). This miniTol2-RFamide backbone was then used to insert different effectors, always using In-Fusion Cloning, following digestion with Spe1. To generate RFamide∷NTR-2A-mCherry and RFamide∷GCaMP6s-2A-mCherry: epNTR was amplified from the pCS2-epNTR plasmid, a gift from Dr. Harold Burgess (Addgene plasmid # 62213); both GCaMP6s and the 2A peptide used in this paper was derived from AAV-hSyn1-GCaMP6s-P2A-nls-dTomato, a gift from Jonathan Ting (Addgene plasmid # 51084).
Transgenesis
In order to establish and optimize transgenesis, we first used the actin promoter (see Cloning), which we found to drive strong, widespread expression in planula and in polyp tentacles. This enabled accurate estimates of efficiency during the critical early life stages following injection (Figure S1H). Using Actin-mCherry to test strategies, we established a protocol that now enables routine, efficient transgenesis, using microinjection of Tol2 transposase protein together with circular plasmid DNA into unfertilized eggs. Collection of eggs and sperm, and microinjection, was carried out as previously described (Momose et al., 2018). Briefly, Clytia medusa spawn ~2 hours after the onset of light. In order to collect eggs and sperm, animals were transferred into either dishes (for the females) or 6-well plates (for the males) within the first hour of light onset. After spawning, eggs were immediately collected and injected with a mixture of 6.25ng/ul Tol2 transposase protein and 10ng/ul plasmid DNA using a Femtojet (Eppendorf). Pulled glass capillaries were used for microinjection (WPI, TW100F-4). Rather than use the ‘inject’ function on the Femtojet, injections were carried out by puncturing eggs and allowing the backpressure in the capillary to fill to the desired volume (~1/4–1/3 egg diameter). Tol2 protein was produced by Creative Biomart using a plasmid generously provided by Dr. Stephen Ekker (Ni et al., 2016). Protein was then stored at −80 as 5ul aliquots and thawed directly prior to injection.
Immediately following injection – and within an hour of spawning (i.e. 3h after light onset) – eggs were fertilized and allowed to develop overnight into planula larvae. They were then transferred into a 12-well plate of artificial sea water containing penicillin/streptomycin, which prevents early metamorphosis of the planulae into polyps. On the second day following injection, we checked the expression of plasmids in the planulae under a fluorescent microscope to ensure that they were capable of driving sufficient expression. Importantly, at this stage expression is not dependent on integration. Planulae were then induced to metamorphose into primary polyps onto glass slides (Ted Pella, cat #260439) using a synthetic GLWamide neuropeptide (produced by Genscript; see Lechable et al., 2020 for details on culturing across the life cycle).
Primary polyps were hand-fed mashed shrimp until they began to grow into a colony. Mashed shrimp were generated by drawing brine shrimp into a 10ml syringe with a blunt-tipped needle attached and then expelling them while pressing the end of the needle against the bottom of a small dish or beaker. Once colonies had 3 or more polyps, they were screened for transgenic expression, with all but the most highly and broadly expressing polyp colonies removed. These colonies were then allowed to grow until they began releasing jellyfish. Jellyfish were then collected, raised to maturity, and backcrossed to parental strains in order to generate stable F1 colonies (see “Experimental Model…” section, above). F1s were seeded sparsely on slides, and were screened for several criteria: strong expression of the transgene, strong polyp colony growth and health, and the ability to release healthy jellyfish that were able to reach maturity. F1 colonies were maintained as single clonal colonies per slide, and once the best colony was identified, the rest were thrown away and that colony was expanded. This allows for the same genetic background to be used across experiments, as these colonies then release clonal experimental jellyfish as needed.
Behavioral analysis and NTR ablations
Acquisition.
Most behavioral analysis was carried out on videos acquired through an Olympus stereoscope (SZX2). Videos were manually annotated using the BENTO analysis suite (https://github.com/annkennedy/bento; Segalin et al., 2020). The exceptions are videos for tracking the major and minor axis (Figure 3F–I), and for NTR swimming controls (Figure S2A–C), both of which used a white LED tracing pad as a backlight rather than the stereoscope (Amazon ASIN# B01M26S3VY) and used automated rather than manual tracking (see below). All videos were acquired using Flea3 or Grasshopper USB3 cameras from FLIR, and the manufacturer’s acquisition software (FlyCapture).
Mouth pointing
Mouth pointing shown in Figure 3B–C was assayed by pinning animals to sylgard-coated plates (Dow Corning) using stainless steel minutien pins (Fine Science Tools). This prevented the margin from getting close to the mouth during a folding event, which would have the potential confound of causing directional mouth pointing by direct sensory stimulation of the mouth. The subumbrella was then wounded only on the right side of the animal to compare pointing in the intact (Figure 3B) versus the wounded (Figure 3C) direction. Having the internal control of the wounded direction ruled out the possibility that the mouth and margin were responding to a shared, directional sensory stimulus, or that the folding of the margin itself was a directional sensory stimulus for the mouth (e.g. by creating fluid flow). Videos were acquired at 15fps, and mouth pointing events 30s before and after a margin folding event were manually annotated using these videos. If the mouth was leaning in one direction at the start of the epoch, that lean was treated as a baseline for further pointing. All margin folding events were spontaneous (i.e. no stimuli were delivered) to avoid possible shared, directional sensory stimulation of the margin and mouth.
Sensory stimuli.
Shrimp extract used in all stimulation experiments was generated by homogenizing brine shrimp using a syringe with a blunt tipped needle followed by filtration through a 40μm cell strainer. Experiments used either this 40μm filtered extract, or extract that was passed through an additional 0.2μm filter. Mechanical stimuli in Figure S2J was delivered by gently touching the tentacles and margin using a glass Pasteur pipette. For experiments in which shrimp extract was used to trigger behavior in freely moving animals, animals were transferred into 6-well plates and 20ul of 40μm filtered shrimp extract was added to the well.
Directional folding and ablation experiments
Directional folding and ablation experiments shown in Figure 3: animals were pinned to sylgard plates, as described above for mouth pointing experiments. ~5ul of 0.2μm filtered shrimp extract was then pipetted directly onto either the top, bottom, left, or right portion of the margin. The timing of margin folding events from all quadrants, and the locations of sensory stimulation, were then manually annotated and compared. For physical ablation experiments, body parts were cut off using spring scissors (Fine Science Tools) or, to remove the mouth, by creating hole-punches using a blunt-tipped needle.
Automated behavior tracking
Automated behavior tracking of the major and minor axes, shown in Figure 3, was performed using custom Matlab (Mathworks) software. Briefly, following background subtraction, a convex hull was found around the dark pixels using the “regionprops” function, and the major and minor axes of the hull were extracted. To achieve high quality tracking, tentacles were trimmed and jellyfish were pinned to sylgard coated plates (Dow Corning) using a single stainless steel minutien pin (Fine Science Tools) through the mouth. This maintained their orientation relative to the camera. To behaviorally distinguish swimming from spontaneous margin-folding, we created binary classifiers using the length of the major and minor axes of jellyfish and the ratio between these measurements. Support vector machines (SVMs) with linear kernels were trained using these three features on equal samples of video frames where animals were either swimming or performing spontaneous margin-folding behavior, using cross-validation from data across animals. Classifier performance is reported as the average model accuracy across validation folds. Chance level performance was obtained by shuffling the identities of swimming and margin-folding behavioral frames before forming the same analysis. To assess the similarity between different forms of margin-folding, we tested the performance of SVMs trained to distinguish swimming vs. spontaneous margin-folding on margin-folding triggered by either shrimp extract or live shrimp (vs. swimming), using the same features of tracking data.
Comparison of fed to starved and spawning animals
Comparison of fed to starved and spawning animals shown in Figure 3. Jellyfish were split into two cohorts in the morning and one cohort was fed ad libitum during the day while the other was not (~24h starvation). Animals were then split into the wells of 6 well plates and passing times individually tested by manually placing shrimp in their tentacles. For comparison of animals during versus after spawning, jellyfish spawn ~2h after the onset of light. Mature medusa were recorded from ~1h50min-2h after light onset, and the number of spontaneous folding events was manually quantified. Egg release could be observed in each of the experimental animals.
Passing of multiple shrimp,
Passing of multiple shrimp, shown in Figure S2L: animals were fed many shrimp simultaneously such that most tentacles captured a shrimp nearly simultaneously. The time and direction of folding onset was then manually recorded for each, distinct shrimp passing event until most shrimp had been passed. A behavior-triggered average was then performed for each folding direction versus folding from any other region. These comparisons were then combined across folding directions.
NTR ablations:
animals were incubated overnight in 10mM Metronidazole (Fisher, cat# ICN15571005; Curado et al., 2008). The following morning, animals were washed several times in clean artificial sea water and behavioral testing was performed on the same day. For crumpling behavior, the subumbrella was gently poked with a glass pipette and crumpling duration was manually quantified. For control experiments shown in Figure S2, automated behavior tracking was performed as above, with the centroid of the convex hull used to calculate location, velocity, and turning angle.
The RFamide peptide
The RFamide peptide (QWLNGRF-amide) and scrambled-sequence control (FRGNLWQ-amide) used in Figure S2 and S3 were synthesized by Genscript and bath applied to a final concentration of 100um, or injected at 100um.
GCaMP imaging acquisition, processing, and data analysis
For highly restrained imaging experiments, ~4mm Clytia medusa were embedded in agarose, as follows. 3–4%, Type VII-A, low-melting point agarose (Sigma cat# A0701–25G) was first made in artificial sea water, with particular care to avoid evaporation. Agarose was then aliquoted and kept in a heat block set at 50-degrees until ready to use in screw-top tubes. Single tubes were then removed from the heat block and vortexed occasionally until reaching nearly room temperature. Medusa were then added into the tube, gently mixed, and rapidly transferred to a glass-bottomed dish (Ted Pella, 14036–20). They were then quickly spread out to make them as planar as possible before being briefly touched to a cold object to rapidly cool and harden the agarose. Agarose was then covered with a thin layer of mineral oil (Sigma M5310–1L) to avoid evaporation during imaging experiments. Following agarose embedding we observed a spectrum of animal restraint. This could be controlled, to a degree, by the extent of mixing of the jellyfish in the agarose prior to transferring to a glass bottom dish. The most well restrained animals were chosen for the analyses shown in Figure 5. Experiments in which both behavior and GCaMP traces could be analyzed, shown in Figure 4, were from animals in which agarose embedding was performed but significant freedom of movement was still visible. Having acquired videos, we then retrospectively chose animals that had the highest possible movement while still allowing extraction of traces by circling ROIs (see below). There was consistently a relatively small portion of the animal in the field of view in these experiments. For imaging experiments in naturalistically behaving jellyfish, animals were placed into a glass depression slide, and a coverslip was gently placed on top, using small amounts of Vaseline applied to each corner of the coverslip as a spacer. The coverslip was then gently pressed down to create a small chamber in which the jellyfish could still perform behaviors. Since the curvature of the jellyfish in the preparation prevented the entire animal from being in focus, the nerve net nearest to the mouth was focused on (rings and ring-adjacent net were out of focus and not measured). Chambers of this type were also used for peptide application during GCaMP imaging (Figure S3) and for application of shrimp extract to pieces of the ring or net (Figure S5D). In those experiments, flow-through was achieved using a Kimwipe (Kimberly-Clark Professional) applied to the edge of the coverslip.
Video acquisition and calcium trace extraction.
GCaMP imaging experiments always had synchronous acquisition of the red and green channels. This was achieved using an Olympus BX51WI microscope with two Photometrics Prime95B cameras connected using a W-View Gemini-2 Optical Splitter from Hamamatsu. Acquisition was controlled with Olympus Cellsens software. Images were acquired with downsampling during acquisition (to 600×600 pixels) and then further processed using ImageJ software, as follows (NIH; Schneider et al., 2012). First, images were re-sized to 400×400 pixels. An average Z-projection was then performed, and each frame in the video stack was divided by this projection to generate a normalized intensity over time for each pixel. If needed, a spatial filter was applied using ImageJ’s Bandpass Filter function to remove spatial light artifacts, e.g., from movement of the mouth, with custom parameters tuned to each video. Regions of Interest (ROIs) were then circled using the ImageJ ROI Manager, and the average pixel intensity within, and location of, these ROIs was then exported for further processing using Matlab (Mathworks). Signal from both the red and green channels were acquired simultaneously using two cameras in all experiments. We then could use the same ROIs as were used to extract GCaMP traces to extract fluorescence over time from the red channel to ensure that our downstream analyses are not the result of imaging or motion artifacts. Running the same analysis pipeline on traces extracted from the red channel did not result in correlated activity structure, shown in Supplemental Figure 4K. Red channel traces were not directly used for normalization of the GCaMP traces.
Spike inference and population activity metrics.
The raw traces, behavior-triggered averages, and behavior classifiers shown in Figure 4, and the relationship between ring and net neurons shown in Figure 5O, were computed using smoothed and z-scored GCaMP traces. For event-based analyses in Figures 5–6, and for computing correlation between neurons (where long-timescale GCaMP fluorescence changes affect results), we instead inferred peak times of neuronal events, and performed analyses on these inferred peaks to remove the effects of underlying noise or drift in the raw trace. Peaks were detected in one of two ways: either using Matlab’s “findpeaks” function or using spike inference from the CNMF_E software package (Zhou et al., 2018) with parameters manually adjusted for each trace, and inferred events manually validated. Following spike detection, the first spike in a bout of inferred spikes was used as the timing of the activity. For computing correlation between neurons, events were smoothed by sequential convolution with box filters of width 3-seconds and 1.5-seconds. A value of “1” was then added back in at the time of the inferred peak.
Detecting swimming events using optical flow.
Optical flow analysis shown in Figure S3 was performed using the Horn-Schunck method (Matlab function opticalFlowHS, default parameters) applied to cropped video frames of the nerve ring in the red channel. For Figure S3, we trained a classifier to distinguish video frames of manually annotated behaviors from all other frames in a trial, where input to the classifier was the mean orientation and magnitude of the optic flow vectors for each frame. We used a binary boosted decision tree classifier, trained using the LogitBoost algorithm with cross-validation. Classifier performance is reported as the average model accuracy across validation folds for each animal.
Predicting behavioral events from population activity.
Classifier analysis comparing GCaMP to behavior was performed in Matlab. Given a set of GCaMP traces, we used the first 1500 frames for training and held the remaining frames out for testing; this ranged from 15%–36% of the data in the training set, depending on the length of the recording. We trained a 3-way, Random Forest classifier with 60 bags, and used out-of-bag prediction to distinguish between epochs of margin folding, swimming, and quiescence.
Analysis of margin folding events.
For the loosely-restrained experiments in Figure 4, and imaging-with-stimulation experiments shown in Figure S5, neurons that responded to the stimulus and body shape were manually annotated using the ImageJ ROI Manager, and Adobe Illustrator, by comparing the fluorescence intensity of neurons on a frame before initiation of a folding event to a frame during the folding event (Figure 4I; Figure S3H). To generate a polar histogram of margin folding directions (Figure 4), we defined the folding axis as the line between the mouth and the center of the inward fold, and calculated the angular position of active neurons relative to this axis.
Reconstructing population events using k-means clusters.
In Figure 5, we asked what fraction of variance in spontaneous neural activity could be explained by activation of a small, fixed number of neural ensembles. First, we defined a population event as more than 2 neurons being active (using the single frame of peak detection, described above) within a 2-second window, and generated a matrix of neuronal participation in all observed ensembles for each animal. To identify a set of fixed neural ensembles, we performed k-means clustering on this matrix; we tested a range of values of k, and used the silhouette metric to pick the value of k that best captured the observed data. For each animal, we then generated a set of all possible combinations of up to 4 ensembles being coactive. For each observed event from that animal, we found the generated pattern of k-means cluster activation that minimized the Hamming distance between the observed activation and the cluster-based activation. This yielded a new matrix of events recreated using k-means clusters (shown in Figure 5H), which was compared to the observed events matrix to generate the F1 scores in Figure 5I.
NMF/ICA detection of cell ensembles.
While k-means clustering requires each cell to only be counted as a member of a single ensemble, we also wanted to visualize predicted ensemble membership if we allowed a cell to be part of more than one ensemble. Our method for detection of cell ensembles using NMF/ICA was based on a method described in (Lopes-dos-Santos et al., 2013) and used in (See et al., 2018). To determine the number of ensembles in each jellyfish, principal component analysis (PCA) was applied to data, processed as described above (i.e. extracted and smoothed peak times), to obtain the eigenvalue spectrum. Eigenvalues that exceed the upper bounds of the Marčenko-Pastur distribution (Marčenko and Pastur, 1967) were deemed significant and taken to represent the number of detected ensembles. We determined this bound using a statistical threshold based on surrogate data, obtained as follows. First, we shuffled the time bins of each neuron independently in order to destroy their temporal relations while maintaining the distribution of events. Second, we used the eigenvalues of correlation matrices obtain from shuffled data to construct a null distribution. We regarded as significant all eigenvalues of the original data matrix that were larger than the 99th percentile of the distribution of maximal eigenvalues computed from shuffled data (Lopes-dos-Santos et al., 2013).
Having identified the number of significant eigenvalues, we next used non-negative matrix factorization (NMF) to optimize the number of factors used for ensemble detection, so that the maximal number of overall neurons could be included in any ensemble. The resulting factors from NMF were then processed using the fast independent component analysis (fastICA) algorithm (Hyvärinen and Oja, 1997). The resulting independent components (ICs) represent the contribution of each neuron to each ensemble. We validated ensemble membership by applying the NMF/ICA method on shuffled data described above. This process was performed 100 times to obtain a normal distribution of IC weights for shuffled data. A neuron was considered a member of an ensemble if its IC weight was larger than 2 times the standard deviation of the IC weight distribution from shuffled data. With this cutoff, 79% of neurons were assigned to one or more ensembles.
Timing analysis of ring and net activation.
For analysis of the timing of ring versus net activity, we first manually extracted groups of net neurons and their spatially corresponding, highly correlated ring neurons. We then identified events of nerve net activity for each group using the mean activity of net neurons by extracting the time when the mean crossed a threshold (z-score >1.5, followed by manual validation). We then examined the timing of individual ring and net neuron activity relative to this onset. To quantify the relative onset time within these “net-triggered averages”, any neuron in the ring or net within the same group that crossed a threshold of z-score > 1.5 within 5-seconds before, or 10 seconds after, the identified net onset was included, and that time to crossing was used for comparison.
Subumbrellar wound experiments.
For the wounding experiments in Figure 5P, wounds were generated in the subumbrella using either a scalpel or forceps, and the number of events on each side of the wound was quantified. Mock wounds were unwounded regions in similar locations; a line was drawn on the video after acquisition using ImageJ, and the number of events on either side of the line was quantified.
Comparing spontaneous to evoked events.
To examine whether the structure of neural activity was similar during spontaneous and evoked events, we used an approach that compares the neural subspace occupied during two conditions (Elsayed et al., 2016; Yoo and Hayden, 2020). We first performed PCA separately on the neurons by time matrix of smoothed events observed during each condition; we kept the top 5 principal components (PCs) in each condition, as we found that these captured over 80% of the variance of observed neural activity within a given condition. Next, we computed the similarity between spontaneous and evoked conditions, by projecting the matrix of raw evoked activity onto the top 5 PCs of the spontaneous condition, and computing the percent of variance captured by these 5 PCs relative to the total variance of the evoked epoch (and vice versa, projecting raw spontaneous activity onto the PCs of the evoked condition). Finally, we computed the alignment index (A) between conditions, defined as the ratio of the amount of variance in a condition explained by the top 5 PCs of the opposing condition, divided by the amount variance in a condition explained by its own top 5 PCs. For example, for the evoked condition, this can be written as:
Where Tr() is the matrix trace, DSpontaneous is the set of top 5 eigenvectors obtained by PCA in the spontaneous condition. Cevoked is the covariance matrix of the evoked condition, and σevoked(i) is the i-th singular value of Cevoked. Since the quantity in the denominator serves to normalize the alignment index, it ranges from 0 to 1, with 0 indicating perfect orthogonality between the subspaces and 1 indicating perfect alignment.
To obtain a statistical comparison, we generated random subspaces biased to the data covariance structure as described in (Elsayed et al., 2016). This was performed as a Monte-Carlo analysis applied to the covariance structure (C) from neural responses across all times (both spontaneous and evoked conditions). Random subspaces aligned to the structure of C are obtained as:
Where U and S are eigenvectors and eigenvalues of C respectively. V is a matrix with each element drawn independently from a normal distribution with mean zero and variance zero. Orth(Z) returns the orthonormal basis for a matrix Z. Thus, this procedure samples subspaces biased towards the space of neural activity, such that the sampled subspace will have the specified covariance matrix C. To calculate the alignment index of two sets of random subspaces ( and ) of dimension d = 5, we calculate:
We repeat the random sampling procedure 1000 times to obtain a distribution of random alignment matrices for each jellyfish.
Generalized linear model fitting and analysis.
To infer functional connectivity between neurons in the spontaneous epoch (Mishchenko et al., 2011; Pillow et al., 2008), we fit Gaussian-residual GLMs for each neuron k using data from spontaneous epochs as:
Where yi is the activity of target neuron k, X is the matrix of activity of all other neurons in a given jellyfish, β is a vector of coefficient weights (which can be interpreted as a proxy of the connectivity of all other neurons to neuron k), and φ is an error term. We regularized our model with an additional non-negativity constraint to impose sparseness and non-negativity on model weights β, implemented using a non-negative least angle regression of least absolute shrinkage and selection operator (LASSO, Efron et al., 2004) with the sparseness parameter determined using 10-fold cross validation. We found, however, that the sparseness that we found in the connectivity did not ultimately depend on the LASSO regularization (see below).
To assess whether data from the evoked epoch could be predicted using model weights found from the spontaneous epoch, we evaluated model performance of GLMs for each cell using held-out test data from the evoked epoch. Model performance in Fig 6F is measured using the Pearson’s correlation coefficient (PCC) between predicted activity and ground truth evoked epoch data. For the results shown in Figure 7, GLMs were additionally trained on the recordings shown in Figure 5 to examine the connectivity matrix obtained from model weights. These recordings were longer and had more of the jellyfish in the field of view as compared to the more complex sensory stimulation experiments. For this, we used the same GLM described above but assessed model performance on held-out test data from the same spontaneous epoch (rather than comparing to an evoked epoch), reporting model fit as the PCC between predicted activity and ground truth data. Model weights from GLMs of cells with a model fit R2 of at least 0.7 were used to construct the connectivity matrix in Figure 7E.
To examine the impact of LASSO regularization, which enforces sparsity, on sparsity of the connectivity matrix obtained, we retrained our models using elastic-net regularization. To predict the activity of a given neuron (y) from the activity of all other neurons simultaneously recorded (X), using weights (w), we created a GLM by maximizing the log-likelihood of y, X given W as shown below:
| (1.1) |
In the above equation (1.1), the highlighted components indicate the ridge and lasso terms in elastic net regularization. To assess the impact of the lasso term on sparsity generated in our model, we varied the term α in range [0 – 1] in 0.2 increments and quantified sparsity in our model as the percent of matrix elements with zero weights.
Simulation of spontaneous events.
To ask what forms of network architecture were consistent with the statistics of observed spontaneous population events, we simulated events in a model network of stochastic leaky integrate-and-fire neurons. We created a population of 500 model neurons, of which 250 were designated “ring” neurons and 250 “net” neurons; neurons were assigned evenly spaced angular positions around the model jellyfish. We assumed that a spontaneous event is initiated by a burst of spiking of a single ring neuron, which we randomly selected from among the model population. We then simulated propagation of this activity throughout the remainder of the model population, as follows. For each neuron, the membrane potential x evolves as:
where W is a matrix of synaptic weights between neurons (defined below) and pj is a synaptic current from neuron j that decays exponentially with time constant τ=0.2s. We implemented stochasticity in neuron spiking by setting pj(t) = 1 if U < xj(t), 0 otherwise, where U is sampled uniformly from 0 to 1.
We simulated population dynamics for 3 forms of the weight matrix W: one in which connection strength was proportional to angular distance between neurons (“continuous” or “nerve net” model), one in which blocks of neurons were all connected with equal strength (“distinct structures”), and a hybrid model that was the weighted sum of the two. In all models, we allowed connections between ring neurons, between net neurons, and from ring neurons to net neurons. In the continuous model, we defined , where D is the angular distance between neurons i and j, and A and σ are fit parameters determining connection strength and the width of connectivity respectively. In the “distinct structures” model, we defined Wij = A if neurons i and j were assigned to the same group using k-means clustering, and 0 otherwise; A is again a fit parameter determining connection strength. Finally, in the hybrid model, we defined , where Wijcontinuous is the weight matrix of the continuous model, Wijdistinct is the weight matrix of the “distinct structures” model, and g is a scalar between 0 and 1.
Parameter fitting in the model neural networks.
In the continuous version of the model, we fit weight matrix parameters A and σ, whereas in the distinct structures model we fit only parameter A; in the hybrid model, we fit A, σ, and g. In all cases, parameters A and σ were fit using a grid search, to minimize the sum of two Kolmogorov-Smirnov distances: the distance between the predicted and observed distribution of event angular extents, and the distance between distributions of number of active neurons per event (see next sections). For the hybrid model, we computed best-fit values of A and σ for g = {0.125, 0.25, 0.5, 0.75, 0.875}. All values of g were able to fit the distributions of event widths and fraction of activated cells well; g=0.125 and 0.75 are shown in Figure S6 to compare predicted correlation matrices.
Model evaluation metrics.
To evaluate model performance and fit model parameters, we computed two metrics on a sample of simulated model events; for each jellyfish, we generate an equal number of events as was observed experimentally, and averaged metric values across 5 repeated instantiations of the model. First, we calculated the distribution of event widths. Defining an event as a time when at least 2 (real or model) neurons were active, we computed the width of each event as the maximum angular distance between any two neurons active in that event. And second, we calculated the distribution of the fraction of cells activated, defined as the percentage of cells that participated in each event. To compare the distributions of observed and simulated events, we computed the Kolmogorov-Smirnov distance, defined as the maximum difference between the cumulative density function of the two distributions.
Quantification and Statistical Analysis
Statistics and quantification
Statistical analysis was performed using Prism (GraphPad) or MATLAB. The n number for each experiment is indicated in the figure legends, along with the statistical method used for each comparison and the p-value. The cutoff for significance was set as an α < 0.05. Quantification strategies for each experiment and modeling approach are separately described above (Method Details).
Supplementary Material
Related to Figure 2.
(A-B) The majority (>80%) of nerve net neurons (red) are FMRFamide immunoreactive (green, “RFa+ neurons”). White arrows indicate RFa+ nerve net neurons, and magenta arrows indicate RFa-negative nerve net neurons. Small panels to the right of panel A are magnifications of the inset white box showing the separate color channels.
(C-D) RFa+ neurons (magenta) are oriented along, and appose, the radial muscle (green). Circular, striated muscle is visible in the top half of the image in panel C, highlighting the radial orientation of the RFa+ neurons and their association with radial muscle.
(E) In situ hybridization (ISH) for the RFamide precursor gene showing its distribution across the organism.
(F) The distribution of RFa+ neurons across the organism (side view).
(G) The RFa+ neurons of the mouth, and their association with the nerve net.
(H) Diagrammatic overview of transgenesis procedures and genetics in Clytia. Clytia medusa are of separate sexes that spawn daily into the water 2 hours after the onset of light. Thousands of eggs can be collected and ‘in vitro’ fertilized for genetic crosses and/or injection. To generate transgenic Clytia, plasmid DNA containing Tol2 recognition sites was injected into eggs together with Tol2 protein. Larva were then induced to metamorphose into polyps, and polyp colonies were screened for the highest expression and lowest level of mosaicism. Jellyfish were then collected from the best colonies and backcrossed to parental strains to generate stable F1 lines, which were re-screened and maintained as clonal colonies. These could then be further used for genetic crosses. See Methods for further details.
(I) Comparison to transgenesis procedures in the cnidarian model Hydra (cf. panel H). Benefits of the Clytia lifecycle include the reliable, daily access to more oocytes, access to unfertilized eggs rather than fertilized embryos, and the ease of generation of inter-strain F1 lines. See Klimovich et al., 2019, for further details.
(A) RFa+ neuron ablation does not cause a loss of swimming behaviors. Location over time for example ablated (red; NTR+, MTZ+) and control (blue; NTR+, MTZ−) animals over 10 minutes.
(B) Velocity over time for NTR ablated (red) versus control (blue) jellyfish. Mean ± SEM. n=5 ablated and 6 control animals here and in panel C.
(C) The mean turning angle over a 10 minute duration (no significant difference, unpaired t-test).
(D) Diagrams and example images (left) showing radial muscle contraction after addition of synthetic RFamide peptide. Accompanying bar graph (right) shows the proportion of animals that display such radial contraction in response to the RFamide peptide (left bar, n=10 animals), a sequence-scrambled control (center bar, n=10 animals), and following genetic ablation of RFa+ neurons using the NTR system (right bar, n=7 animals). Peptide concentration: 100um.
(E) Microinjection of RFamide but not a scrambled control peptide triggers local inward folding of the margin (RFamide: n=9 trials from 4 animals; Scrambled: n=8 trials from 3 animals; unpaired t-tests).
(F) Swimming stops during margin folding. Mean ± SEM. n=218 passing events from 4 animals.
(G) Length of time between brine shrimp capture and attempted passing to the mouth. n=50 passing events from 17 animals.
(H) Success rate of passing prey from tentacles to the mouth on the first attempt. n=17 animals.
(I) Margin folding behavior can be observed with no visible prey present or stimuli provided. Micrograph (left) shows the mouth interacting directly with the folded-in margin (base and top of the feeding organ, “mouth”, indicated in green). Data on the right is pooled from 42 animals and shows the proportion of 2-minutes spent performing margin folding with no stimulus provided.
(J) Chemical but not mechanical stimuli triggered folding behavior. Proportion of time spent margin folding 2min after stimulus presentation – 2min before. Chemosensory stimulus: mashed, filtered brine shrimp. Mechanical stimulus: glass pipette applied to the tentacles and margin. Dots represent individual animals (n=4/each). Mean ± SEM.
(K) Summary diagram showing that excised body parts performed subsets of the complete food passing behavior autonomously.
(L) Diagram of the experiment (left) showing a jellyfish that has captured many shrimp simultaneously, and is passing one. The plot on the right shows the probability of initiating a passing event from a different location than the one folding at time zero. The probability of initiating passing from a different location is significantly lower during a passing event (unpaired t-test; n=9 animals, mean ± SEM).
(M) Example video frames showing rapid, sequential passing from different directions.
Related to Figure 4.
(A) Images of wildtype (top) and GFP1 knockout (bottom) animals. Other GFP genes are expressed in the tentacle bulbs (arrow) and mouth.
(B) Overview of imaging experiments using varying degrees of animal restraint. On one end of the spectrum, highly restrained, agarose-embedded animals allowed extraction of high quality GCaMP traces. On the other, minimally restrained animals behaving naturalistically in small chambers allowed for extraction of binarized GCaMP activity and body shape (pose) from individual frames. At the midpoint, we could discern the identity of the behavior being performed and extract GCaMP traces.
(C) Video frames of a freely behaving animal performing margin folding, adapted from Figure 3.
(D) Video frames of a naturalistically behaving animal performing margin folding during GCaMP imaging (compare to panels in C). This animal contributed to the data in Figure 4J. Adapted from Figure 4.
(E) Optic flow can be used to identify behaviors in partially restrained animals. (E1) Annotated behavior identity (top), the mean orientation of optic flow vectors for each frame (middle), and the spectrogram of that orientation (bottom). Swimming can be recognized by high frequency oscillations in the optic flow vectors, while low frequency, radial movements are characteristic of margin folding (compare behaviors in Figure 3G–H). (E2) Magnification of the region indicated with the grey box in panel E1. (E3) Performance of a classifier identifying behaviors using the optic flow signal (n=3 animals).
(F) The first two principal components over time of population neural activity, colored by the behavior being performed. Epochs of margin folding lead to trajectories primarily out along the first principal component, while bouts of swimming and quiescence remain intermingled at the origin.
(G) Bath application of RFamide peptide induced muscle contraction in patches of the subumbrella. This contraction was not accompanied by GCaMP activity (n=6 patches). Peptide concentration: 100uM.
(H-I) Examples of GCaMP activity in a naturalistically behaving animal. Fluorescence in the simultaneously acquired mCherry (left two columns) and GCaMP (right column) channels immediately before (H) or during (I) a margin folding event. GCaMP activation was visible in the green channel, but a similar change in fluorescence intensity was not observed in the red channel. Quantification (I, right) shows fluorescence from frames during/before the folding event for ROIs manually circled around individual neurons in this field of view (t-test). Line represents the median. FOV is ~3.5mm2.
(J-K) Polar histograms showing that the characteristic pattern of RFa+ neuron activation is also present during margin folding in other contexts, i.e. during spontaneous margin folding (J, 103 neurons, 8 events, 3 animals) or passing shrimp captured in the tentacles (K, 125 neurons, 9 events, 3 animals).
Related to Figure 5.
(A) Example field of view (FOV) for calcium imaging in a highly restrained jellyfish. Center of ROIs (green circles) shown superimposed on an image of the mCherry channel. FOV in panels A, G, K, and L is ~3.5mm2. Repeated from Figure 5A with example ROIs indicated and numbered in red, corresponding to traces in B.
(B) Example GCaMP traces corresponding to the five red ROIs designated in A. Zoomed in traces in B2 correspond to the region indicated with the green box in B1.
(C) 6065 single neuron activations form 551 single or ensemble events, pooled from recordings of spontaneous activity across the nerve net from 4 animals. Synchronicity (98.8%) is the number of individual neural activations (6065 total) where >1 neuron was active within a 3 second window (5990). There were 75 instances of a neuron firing alone.
(D) Pairwise correlation between neurons sorted according to hierarchical clustering (shown on the left). Neurons grouped together at different levels of the hierarchy are indicated with colored lines on the dendrogram and matching colored boxes on the correlation matrix.
(E) Pairwise correlation of neural activity versus the angle between those neurons relative to the mouth. Data pooled from, and colored by, 5 individual animals.
(F) Percent of neurons that are shared between functional groups using NMF/ICA. n=486 neurons from 4 animals.
(G) Cluster membership using NMF/ICA for the animal shown in Figure 5A. Neurons participating in multiple ensembles are indicated with multi-colored circles. Grey circles indicate neurons that were not assigned to an ensemble.
(H-J) Functional zones were not evident in the coarse anatomy of the nerve net. (H) Maximum projection of a confocal z-stack showing the distribution of FMRFamide-immunoreactive fibers across the animal. Scale: 500um. (I) Manual tracing of nerve net processes shown in panel H.
(J) Quantification of features of the anatomy compared to the width of k-means clusters of neural activity (n=3 animals).
(K) An analysis to control for imaging or motion artifacts in which we examined fluorescence over time extracted from the red channel using the same ROIs used for GCaMP analyses. Pairwise correlation between neuron’s fluorescence intensity over time (K2), the pairwise correlation vs angle (K3), and k-means clustering (K4) are shown. There is no similar structure in the red channel to what was observed in the green channel (compare to Figure 5).
(L) FOV and ROIs for the animal shown in panel M, and Figure 5M–N.
(M) Pairwise correlation between neurons for the animal shown in panel L. Whether those neurons are located in the ring or net is indicated above the correlation matrix.
(N) Pairwise correlation of neural activity versus the angle between neurons relative to the mouth. Each dot is colored based on whether it was a comparison between net neurons, between ring neurons, or between a ring neuron and a net neuron. Data is pooled from 5 animals, and is the same as in panel E, but colored by comparison type rather than individual animal.
(A) The margin is required for folding behavior. Stimulus-triggered average from 4 animals after removal of the margin (red line). Intact control data (grey line) is the same as shown in Figure 3J, repeated here for ease of comparison.
(B) Activity of nerve ring (top) and nerve net (bottom) neurons over time. Magenta boxes indicate nerve net events, and show that for each net event there was corresponding activity in the nerve rings. In contrast, white arrows point to example events in the nerve ring that do not have a nerve net correlate.
(C) Trial-by-trial activity corresponding to Figure 5O showing that ring neurons were active prior to net neurons. As in Figure 5O, top traces show mean of ring and net activity, aligned to the onset of net events (t=0, magenta line). The white line qualitatively highlights the earlier onset of ring neurons (compare the region between white and magenta lines in ring vs net).
(D) Nerve ring, but not net, neurons responded to food cues. The nerve net only responded if nerve ring was present. Regions containing the nerve ring (top left panels), nerve net (bottom left panels), or both (right panels) were surgically excised and shrimp extract was delivered during GCaMP imaging. Neurons in patches of the nerve net responded to KCL (but not shrimp extract) indicating that they were still capable of activation.
(E) Neuronal processes were visible between the nerve rings and nerve net. Whether these originated from the rings and/or net is unknown. White: anti-aTubulin. Scale: 20um.
(F) Diagrams and example images showing an experiment in which a strip of the animal was cut out, with a tentacle at one end and the mouth at the other. When a brine shrimp was fed to the tentacle, the strip was able to perform the complete food passing behavior.
(G) Local stimulation of the margin with shrimp extract activated a spatially localized ensemble, but did not activate distant neurons. Diagram of experiment (left), and results of individual trials (right), with the approximate location of the stimulus shown in orange and the responding neurons shown in green. Active neurons were annotated as in Figure 4I. We applied droplets of food cues to the margin of agarose-embedded Clytia using a Femtojet and pulled glass electrodes, while performing GCaMP imaging of the nerve net. n=195 neurons.
(H) Histogram and inset heatmap showing aligned summary data from the trials in panel G.
(I1) Left two panels show the correlation matrices from spontaneous versus evoked imaging epochs, from Figure 6C, together for comparison with the correlation matrix generated by GLMs predicting evoked activity after training on spontaneous activity, as described in Figure 6F. (I2) The weights between neurons that emerge from the GLMs after training on spontaneous activity epochs (left), and their performance on evoked data (right, dots: neurons).
Related to Figure 7.
(A) Pairwise correlation matrix from observed data, generated using the neurons-by-events matrix in Figure 5G. Compare to data generated by models in panels B-E.
(B-E) Output of models with different underlying connectivity structures. Hybrid models (C-D) best fit the data. (B1, C1, D1, E1) Diagram and connectivity matrix for each model. (B2, C2, D2, E2) Pairwise correlation between neurons generated by the models; compare to panel A. (B3, C3, D3, E3) The angular width of events generated by each model. (B4, C4, D4, E4) The fraction of cells activated during events generated by each model.
Related to Figure 7.
(A) GLM performance on held out data from recordings of spontaneous activity analyzed in Figure 5, which were used to generate the analyses shown in Figure 7E–G. n=339 neurons from 3 animals. Here and in E, F, H, I dots represent neurons.
(B) Lasso regularization was not the cause of inferred sparse connectivity. (B1) To create a GLM for estimating connectivity w to a target neuron y, given activity of all other neurons in matrix X, we maximized the likelihood shown in this formula, where lambda is a regularization parameter and alpha is the sparseness parameter. (B2) The sparsity of the connectivity, as defined by the percent of elements with zero weight in the matrix, for different values of alpha, showing that GLM regularization did not cause the sparse connectivity found here. Each dot is an individual jellyfish (n=4). (B3) Matrices of GLM weights at different values of alpha, showing their stability.
(C) Connectivity scheme in original model used in Fig 7E, which allows for net neurons to receive inputs from other net neurons as well as ring neurons.
(D) Net-net connections were ablated in GLMs to assess the importance of ring to net connectivity.
(E) Performance of GLMs for net neurons with all inputs (blue) vs GLMs where net-net connections were ablated (purple). p < 0.001, Mann Whitney test.
(F) Performance of GLMs for net neurons after ablation plotted against their Euclidean distance to the nearest ring neuron. Linear Regression, R2 = 0.24.
(G) A separate approach to test the importance of ring to net connectivity where new GLMs were trained for net neurons with only ring input.
(H) Performance of GLMs with ring and net inputs (blue) vs GLMs with only ring inputs (purple). p<0.001, Mann Whitney test.
(I) Performance of GLMs for net neurons trained with only ring input plotted against their Euclidean distance to the nearest ring neuron. Linear Regression, R2 = 0.4.
(J) Performance of GLMs for net neurons trained with only ring input shown on their anatomical position.
Supplemental Video 1. Spontaneous and prey-evoked margin folding behavior. Related to Figures 2 and 3.
Supplemental Video 2. Directionally appropriate folding triggered by local application of shrimp extract. Related to Figure 3.
Supplemental Video 3. Attempted food passing after excision of the mouth. Related to Figure 3.
Supplemental Video 4. Nerve net activation is visible at the location of inward folding in an unrestrained animal. Related to Figure 4.
Supplemental Video 5. GCaMP imaging of the nerve net in an agarose-restrained animal. Related to Figure 5.
Supplemental Video 6. Neural activity was only visible on the peripheral-side of an umbrellar wound. Related to Figure 5.
Supplemental Video 7. An excised strip of the umbrella can perform the complete food passing behavior. Related to Figure 5.
Highlights.
Transgenic jellyfish allow in toto imaging and ablation of specific neural subtypes (85)
Jellyfish feeding behavior coordinates autonomous body parts with no central brain (84)
A subnetwork of peptidergic neurons controls vectorial food transfer to the mouth (83)
Vectorial transfer activates latent subassemblies that tile the umbrellar network (83)
Acknowledgements
We thank the Marine Resources Center of the MBL (Woods Hole, MA) and J. Malamy for an introduction to Clytia, E. Houliston and R. Copley for Clytia resources and advice, M.D.C. Lamadrid for illustrations, S. Ekker for Tol2 reagents, J. Williams for software, and A. Collazo for imaging support, J. Parker, D. Prober, and Anderson lab members for feedback. This work was supported by the Caltech Center for Evolutionary Science, the Whitman Center of the MBL, the Life Sciences Research Foundation (to B.W.), NINDS (K99NS119749 to B.W.), and NIMH (K99MH117264 to A.K.). Content is the authors’ sole responsibility and not that of NIH. We thank the CRBM (FR 3761; EMBRC-France, managed by ANR-10-INBS-02) for support. T.M was supported by ANR (ANR-17-CE13-0016, i-MMEJ) and ASSEMBLE Plus JRA3 (EU Horizon 2020 No 730984). A.N. was supported by a Scholarship from the Agency of Science, Technology and Research, Singapore. D.J.A. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- Anderson DJ, Adolphs R, 2014. A Framework for Studying Emotions across Species. Cell 157, 187–200. 10.1016/j.cell.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendt D, Tosches MA, Marlow H, 2016. From nerve net to nerve ring, nerve cord and brain — evolution of the nervous system. Nat Rev Neurosci 17, 61–72. 10.1038/nrn.2015.15 [DOI] [PubMed] [Google Scholar]
- Badhiwala KN, Primack AS, Juliano CE, Robinson JT, 2021. Multiple neuronal networks coordinate Hydra mechanosensory behavior. eLife 10, e64108. 10.7554/eLife.64108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch TCG, Klimovich A, Domazet-Lošo T, Gründer S, Holstein TW, Jékely G, Miller DJ, Murillo-Rincon AP, Rentzsch F, Richards GS, Schröder K, Technau U, Yuste R, 2017. Back to the Basics: Cnidarians Start to Fire. Trends in Neurosciences 40, 92–105. 10.1016/j.tins.2016.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright P, Halgedahl SL, Hendricks JR, Jarrard RD, Marques AC, Collins AG, Lieberman BS, 2007. Exceptionally Preserved Jellyfishes from the Middle Cambrian. PLOS ONE 2, e1121. 10.1371/journal.pone.0001121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari T, Weissbourd B, Gehring J, Ferraioli A, Leclère L, Herl M, Gao F, Chevalier S, Copley RR, Houliston E, Anderson DJ, Pachter L, 2021. Whole Animal Multiplexed Single-Cell RNA-Seq Reveals Transcriptional Shifts Across Clytia Medusa Cell Types. In press, Science Advances. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, Looger LL, Svoboda K, Kim DS, 2013. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon RH, Duarte CM, Pitt KA, Robinson KL, Lucas CH, Sutherland KR, Mianzan HW, Bogeberg M, Purcell JE, Decker MB, Uye S.-i., Madin LP, Brodeur RD, Haddock SHD, Malej A, Parry GD, Eriksen E, Quinones J, Acha M, Harvey M, Arthur JM, Graham WM, 2013. Recurrent jellyfish blooms are a consequence of global oscillations. Proceedings of the National Academy of Sciences 110, 1000–1005. 10.1073/pnas.1210920110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JH, Colin SP, Dabiri JO, Gemmell BJ, Lucas KN, Sutherland KR, 2021. The Hydrodynamics of Jellyfish Swimming. Annu. Rev. Mar. Sci 13, 375–396. 10.1146/annurev-marine-031120-091442 [DOI] [PubMed] [Google Scholar]
- Curado S, Stainier DYR, Anderson RM, 2008. Nitroreductase-mediated cell/tissue ablation in zebrafish: a spatially and temporally controlled ablation method with applications in developmental and regeneration studies. Nat Protoc 3, 948–954. 10.1038/nprot.2008.58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels RW, Rossano AJ, Macleod GT, Ganetzky B, 2014. Expression of Multiple Transgenes from a Single Construct Using Viral 2A Peptides in Drosophila. PLOS ONE 9, e100637. 10.1371/journal.pone.0100637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre C, Yuste R, 2017. Non-overlapping Neural Networks in Hydra vulgaris. Current Biology 27, 1085–1097. 10.1016/j.cub.2017.02.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Hastie T, Johnstone I, Tibshirani R, 2004. Least angle regression. Ann. Statist 32, 407–499. 10.1214/009053604000000067 [DOI] [Google Scholar]
- Elsayed GF, Lara AH, Kaufman MT, Churchland MM, Cunningham JP, 2016. Reorganization between preparatory and movement population responses in motor cortex. Nature Communications 7, 13239. 10.1038/ncomms13239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourrage C, Swann K, Gonzalez Garcia JR, Campbell AK, Houliston E, 2014. An endogenous green fluorescent protein-photoprotein pair in Clytia hemisphaerica eggs shows co-targeting to mitochondria and efficient bioluminescence energy transfer. Open Biol 4, 130206. 10.1098/rsob.130206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham WM, Gelcich S, Robinson KL, Duarte CM, Brotz L, Purcell JE, Madin LP, Mianzan H, Sutherland KR, Uye S, Pitt KA, Lucas CH, Bøgeberg M, Brodeur RD, Condon RH, 2014. Linking human well-being and jellyfish: ecosystem services, impacts, and societal responses. Frontiers in Ecology and the Environment 12, 515–523. 10.1890/130298 [DOI] [Google Scholar]
- Gröger H, Schmid V, 2000. Nerve net differentiation in medusa development of Podocoryne carnea. Sci. Mar 64, 107–116. 10.3989/scimar.2000.64s1107 [DOI] [Google Scholar]
- Hays GC, Doyle TK, Houghton JDR, 2018. A Paradigm Shift in the Trophic Importance of Jellyfish? Trends in Ecology & Evolution 33, 874–884. 10.1016/j.tree.2018.09.001 [DOI] [PubMed] [Google Scholar]
- Houliston E, Momose T, Manuel M, 2010. Clytia hemisphaerica: a jellyfish cousin joins the laboratory. Trends in Genetics 26, 159–167. 10.1016/j.tig.2010.01.008 [DOI] [PubMed] [Google Scholar]
- Hyman LH, 1940. Observations and Experiments on the Physiology of Medusae. The Biological Bulletin 79, 282–296. 10.2307/1537823 [DOI] [Google Scholar]
- Hyvärinen A, Oja E, 1997. A Fast Fixed-Point Algorithm for Independent Component Analysis. Neural Computation 9, 1483–1492. 10.1162/neco.1997.9.7.1483 [DOI] [Google Scholar]
- Kakidani H, Ptashne M, 1988. GAL4 activates gene expression in mammalian cells. Cell 52, 161–167. 10.1016/0092-8674(88)90504-1 [DOI] [PubMed] [Google Scholar]
- Kamran Z, Zellner K, Kyriazes H, Kraus CM, Reynier J-B, Malamy JE, 2017. In vivo imaging of epithelial wound healing in the cnidarian Clytia hemisphaerica demonstrates early evolution of purse string and cell crawling closure mechanisms. BMC Dev Biol 17, 17. 10.1186/s12861-017-0160-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuki T, Greenspan RJ, 2013. Jellyfish nervous systems. Current Biology 23, R592–R594. 10.1016/j.cub.2013.03.057 [DOI] [PubMed] [Google Scholar]
- King MG, Spencer AN, 1981. The involvement of nerves in the epithelial control of crumpling behavior in a hydrozoan jellyfish. J. exp. Biol 94, 203–218. [Google Scholar]
- Klimovich A, Wittlieb J, Bosch TCG, 2019. Transgenesis in Hydra to characterize gene function and visualize cell behavior. Nat Protoc 14, 2069–2090. 10.1038/s41596-019-0173-3 [DOI] [PubMed] [Google Scholar]
- Koga A, Suzuki M, Inagaki H, Bessho Y, Hori H, 1996. Transposable element in fish. Nature 383, 30–30. 10.1038/383030a0 [DOI] [PubMed] [Google Scholar]
- Künzel T, Heiermann R, Frank U, Müller W, Tilmann W, Bause M, Nonn A, Helling M, Schwarz RS, Plickert G, 2010. Migration and differentiation potential of stem cells in the cnidarian Hydractinia analysed in eGFP-transgenic animals and chimeras. Developmental Biology 348, 120–129. 10.1016/j.ydbio.2010.08.017 [DOI] [PubMed] [Google Scholar]
- Lechable M, Jan A, Duchene A, Uveira J, Weissbourd B, Gissat L, Collet S, Gilletta L, Chevalier S, Leclère L, Peron S, Barreau C, Lasbleiz R, Houliston E, Momose T, 2020. An improved whole life cycle culture protocol for the hydrozoan genetic model Clytia hemisphaerica. Biology Open 9. 10.1242/bio.051268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclère L, Horin C, Chevalier S, Lapébie P, Dru P, Peron S, Jager M, Condamine T, Pottin K, Romano S, Steger J, Sinigaglia C, Barreau C, Quiroga Artigas G, Ruggiero A, Fourrage C, Kraus JEM, Poulain J, Aury J-M, Wincker P, Quéinnec E, Technau U, Manuel M, Momose T, Houliston E, Copley RR, 2019. The genome of the jellyfish Clytia hemisphaerica and the evolution of the cnidarian lifecycle. Nat Ecol Evol 3, 801–810. 10.1038/s41559-019-0833-2 [DOI] [PubMed] [Google Scholar]
- Lewis C, Long TAF, 2005. Courtship and reproduction in Carybdea sivickisi (Cnidaria: Cubozoa). Marine Biology 147, 477–483. 10.1007/s00227-005-1602-0 [DOI] [Google Scholar]
- Lopes-dos-Santos V, Ribeiro S, Tort ABL, 2013. Detecting cell assemblies in large neuronal populations. Journal of Neuroscience Methods, Latin American School on Computational Neuroscience (LASCON) 2012 220, 149–166. 10.1016/j.jneumeth.2013.04.010 [DOI] [PubMed] [Google Scholar]
- Mackie GO, 2004. Central Neural Circuitry in the Jellyfish Aglantha. Neurosignals 13, 5–19. 10.1159/000076155 [DOI] [PubMed] [Google Scholar]
- Mackie GO, 2003. Central circuitry in the jellyfish Aglantha digitale IV. Pathways coordinating feeding behaviour. Journal of Experimental Biology 206, 2487–2505. 10.1242/jeb.00450 [DOI] [PubMed] [Google Scholar]
- Mackie GO, Singla CL, Stell WK, 1985. Distribution of Nerve Elements Showing FMRFamide-like Immunoreactivity in Hydromedusae. Acta Zoologica 66, 199–210. 10.1111/j.1463-6395.1985.tb00840.x [DOI] [Google Scholar]
- Marčenko VA, Pastur LA, 1967. Distribution of Eigenvalues for Some Sets of Random Matrices. Math. USSR Sb 1, 457–483. 10.1070/SM1967v001n04ABEH001994 [DOI] [Google Scholar]
- Meech RW, 2019. Electrophysiology and Behavior of Cnidarian Nervous Systems, in: Oxford Research Encyclopedia of Neuroscience. Oxford University Press. 10.1093/acrefore/9780190264086.013.146 [DOI] [Google Scholar]
- Mishchenko Y, Vogelstein JT, Paninski L, 2011. A Bayesian approach for inferring neuronal connectivity from calcium fluorescent imaging data. Ann. Appl. Stat 5, 1229–1261. 10.1214/09-AOAS303 [DOI] [Google Scholar]
- Momose T, Cian AD, Shiba K, Inaba K, Giovannangeli C, Concordet J-P, 2018. High doses of CRISPR/Cas9 ribonucleoprotein efficiently induce gene knockout with low mosaicism in the hydrozoan Clytia hemisphaerica through microhomology-mediated deletion. Scientific Reports 8, 11734. 10.1038/s41598-018-30188-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath RD, Bedbrook CN, Abrams MJ, Basinger T, Bois JS, Prober DA, Sternberg PW, Gradinaru V, Goentoro L, 2017. The Jellyfish Cassiopea Exhibits a Sleep-like State. Current Biology 27, 2984–2990. e3. 10.1016/j.cub.2017.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawroth JC, Lee H, Feinberg AW, Ripplinger CM, McCain ML, Grosberg A, Dabiri JO, Parker KK, 2012. A tissue-engineered jellyfish with biomimetic propulsion. Nature Biotechnology 30, 792–797. 10.1038/nbt.2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni J, Wangensteen KJ, Nelsen D, Balciunas D, Skuster KJ, Urban MD, Ekker SC, 2016. Active recombinant Tol2 transposase for gene transfer and gene discovery applications. Mobile DNA 7, 6. 10.1186/s13100-016-0062-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passano LM, 1973. Behavioral Control Systems in Medusae: a Comparison Between Hydro- and Scyphomedusae. Publications of the Seto Marine Biological Laboratory 20, 615–645. [Google Scholar]
- Pillow JW, Shlens J, Paninski L, Sher A, Litke AM, Chichilnisky EJ, Simoncelli EP, 2008. Spatio-temporal correlations and visual signalling in a complete neuronal population. Nature 454, 995–999. 10.1038/nature07140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiroga Artigas G, Lapébie P, Leclère L, Takeda N, Deguchi R, Jékely G, Momose T, Houliston E, 2018. A gonad-expressed opsin mediates light-induced spawning in the jellyfish Clytia. eLife 7, e29555. 10.7554/eLife.29555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfer E, Amon-Hassenzahl A, Steinmetz PRH, Technau U, 2010. A muscle-specific transgenic reporter line of the sea anemone, Nematostella vectensis. Proceedings of the National Academy of Sciences 107, 104–108. 10.1073/pnas.0909148107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanes GJ, 1885. Jelly-fish, star-fish, and sea urchins: being a research on primitive nervous systems. D. Appleton, New York. [Google Scholar]
- Satterlie RA, 2008. Control of swimming in the hydrozoan jellyfish Aequorea victoria: subumbrellar organization and local inhibition. Journal of Experimental Biology 211, 3467–3477. 10.1242/jeb.018952 [DOI] [PubMed] [Google Scholar]
- Satterlie RA, 2002. Neuronal control of swimming in jellyfish: a comparative story. Canadian Journal of Zoology 80, 1654–1669. 10.1139/z02-132 [DOI] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW, 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- See JZ, Atencio CA, Sohal VS, Schreiner CE, 2018. Coordinated neuronal ensembles in primary auditory cortical columns. eLife 7, e35587. 10.7554/eLife.35587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalin C, Williams J, Karigo T, Hui M, Zelikowsky M, Sun JJ, Perona P, Anderson DJ, Kennedy A, 2020. The Mouse Action Recognition System (MARS): a software pipeline for automated analysis of social behaviors in mice. bioRxiv. 10.1101/2020.07.26.222299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinigaglia C, Peron S, Eichelbrenner J, Chevalier S, Steger J, Barreau C, Houliston E, Leclère L, 2020. Pattern regulation in a regenerating jellyfish. eLife 9, e54868. 10.7554/eLife.54868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor KM, Bergeron SA, Horstick EJ, Jordan DC, Aho V, Porkka-Heiskanen T, Haspel G, Burgess HA, 2014. Direct activation of the Mauthner cell by electric field pulses drives ultrarapid escape responses. Journal of Neurophysiology 112, 834–844. 10.1152/jn.00228.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouanas CN, Kim S, Badhiwala KN, Avants BW, Robinson JT, 2021. Hydra vulgaris shows stable responses to thermal stimulation despite large changes in the number of neurons. iScience 24, 102490. 10.1016/j.isci.2021.102490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urasaki A, Morvan G, Kawakami K, 2006. Functional Dissection of the Tol2 Transposable Element Identified the Minimal cis-Sequence and a Highly Repetitive Sequence in the Subterminal Region Essential for Transposition. Genetics 174, 639–649. 10.1534/genetics.106.060244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber C, 1989. Smooth muscle fibers of Podocoryne carnea (Hydrozoa) demonstrated by a specific monoclonal antibody and their association with neurons showing FMRFamide-like immunoreactivity. Cell Tissue Res. 255. 10.1007/BF00224109 [DOI] [Google Scholar]
- Webster N, Jin JR, Green S, Hollis M, Chambon P, 1988. The yeast UASG is a transcriptional enhancer in human hela cells in the presence of the GAL4 trans-activator. Cell 52, 169–178. 10.1016/0092-8674(88)90505-3 [DOI] [PubMed] [Google Scholar]
- White DT, Mumm JS, 2013. The Nitroreductase System of Inducible Targeted Ablation Facilitates Cell-specific Regenerative Studies in Zebrafish. Methods 62, 232–240. 10.1016/j.ymeth.2013.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittlieb J, Khalturin K, Lohmann JU, Anton-Erxleben F, Bosch TCG, 2006. Transgenic Hydra allow in vivo tracking of individual stem cells during morphogenesis. Proceedings of the National Academy of Sciences 103, 6208–6211. 10.1073/pnas.0510163103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SBM, Hayden BY, 2020. The Transition from Evaluation to Selection Involves Neural Subspace Reorganization in Core Reward Regions. Neuron 105, 712–724. e4. 10.1016/j.neuron.2019.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K, Ang KS, Lee D-Y, 2017. Synthetic Gene Design Using Codon Optimization On-Line (COOL), in: Hughes RA (Ed.), Synthetic DNA: Methods and Protocols, Methods in Molecular Biology. Springer, New York, NY, pp. 13–34. 10.1007/978-1-4939-6343-0_2 [DOI] [PubMed] [Google Scholar]
- Zhou P, Resendez SL, Rodriguez-Romaguera J, Jimenez JC, Neufeld SQ, Giovannucci A, Friedrich J, Pnevmatikakis EA, Stuber GD, Hen R, Kheirbek MA, Sabatini BL, Kass RE, Paninski L, 2018. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife 7, e28728. 10.7554/eLife.28728 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 2.
(A-B) The majority (>80%) of nerve net neurons (red) are FMRFamide immunoreactive (green, “RFa+ neurons”). White arrows indicate RFa+ nerve net neurons, and magenta arrows indicate RFa-negative nerve net neurons. Small panels to the right of panel A are magnifications of the inset white box showing the separate color channels.
(C-D) RFa+ neurons (magenta) are oriented along, and appose, the radial muscle (green). Circular, striated muscle is visible in the top half of the image in panel C, highlighting the radial orientation of the RFa+ neurons and their association with radial muscle.
(E) In situ hybridization (ISH) for the RFamide precursor gene showing its distribution across the organism.
(F) The distribution of RFa+ neurons across the organism (side view).
(G) The RFa+ neurons of the mouth, and their association with the nerve net.
(H) Diagrammatic overview of transgenesis procedures and genetics in Clytia. Clytia medusa are of separate sexes that spawn daily into the water 2 hours after the onset of light. Thousands of eggs can be collected and ‘in vitro’ fertilized for genetic crosses and/or injection. To generate transgenic Clytia, plasmid DNA containing Tol2 recognition sites was injected into eggs together with Tol2 protein. Larva were then induced to metamorphose into polyps, and polyp colonies were screened for the highest expression and lowest level of mosaicism. Jellyfish were then collected from the best colonies and backcrossed to parental strains to generate stable F1 lines, which were re-screened and maintained as clonal colonies. These could then be further used for genetic crosses. See Methods for further details.
(I) Comparison to transgenesis procedures in the cnidarian model Hydra (cf. panel H). Benefits of the Clytia lifecycle include the reliable, daily access to more oocytes, access to unfertilized eggs rather than fertilized embryos, and the ease of generation of inter-strain F1 lines. See Klimovich et al., 2019, for further details.
(A) RFa+ neuron ablation does not cause a loss of swimming behaviors. Location over time for example ablated (red; NTR+, MTZ+) and control (blue; NTR+, MTZ−) animals over 10 minutes.
(B) Velocity over time for NTR ablated (red) versus control (blue) jellyfish. Mean ± SEM. n=5 ablated and 6 control animals here and in panel C.
(C) The mean turning angle over a 10 minute duration (no significant difference, unpaired t-test).
(D) Diagrams and example images (left) showing radial muscle contraction after addition of synthetic RFamide peptide. Accompanying bar graph (right) shows the proportion of animals that display such radial contraction in response to the RFamide peptide (left bar, n=10 animals), a sequence-scrambled control (center bar, n=10 animals), and following genetic ablation of RFa+ neurons using the NTR system (right bar, n=7 animals). Peptide concentration: 100um.
(E) Microinjection of RFamide but not a scrambled control peptide triggers local inward folding of the margin (RFamide: n=9 trials from 4 animals; Scrambled: n=8 trials from 3 animals; unpaired t-tests).
(F) Swimming stops during margin folding. Mean ± SEM. n=218 passing events from 4 animals.
(G) Length of time between brine shrimp capture and attempted passing to the mouth. n=50 passing events from 17 animals.
(H) Success rate of passing prey from tentacles to the mouth on the first attempt. n=17 animals.
(I) Margin folding behavior can be observed with no visible prey present or stimuli provided. Micrograph (left) shows the mouth interacting directly with the folded-in margin (base and top of the feeding organ, “mouth”, indicated in green). Data on the right is pooled from 42 animals and shows the proportion of 2-minutes spent performing margin folding with no stimulus provided.
(J) Chemical but not mechanical stimuli triggered folding behavior. Proportion of time spent margin folding 2min after stimulus presentation – 2min before. Chemosensory stimulus: mashed, filtered brine shrimp. Mechanical stimulus: glass pipette applied to the tentacles and margin. Dots represent individual animals (n=4/each). Mean ± SEM.
(K) Summary diagram showing that excised body parts performed subsets of the complete food passing behavior autonomously.
(L) Diagram of the experiment (left) showing a jellyfish that has captured many shrimp simultaneously, and is passing one. The plot on the right shows the probability of initiating a passing event from a different location than the one folding at time zero. The probability of initiating passing from a different location is significantly lower during a passing event (unpaired t-test; n=9 animals, mean ± SEM).
(M) Example video frames showing rapid, sequential passing from different directions.
Related to Figure 4.
(A) Images of wildtype (top) and GFP1 knockout (bottom) animals. Other GFP genes are expressed in the tentacle bulbs (arrow) and mouth.
(B) Overview of imaging experiments using varying degrees of animal restraint. On one end of the spectrum, highly restrained, agarose-embedded animals allowed extraction of high quality GCaMP traces. On the other, minimally restrained animals behaving naturalistically in small chambers allowed for extraction of binarized GCaMP activity and body shape (pose) from individual frames. At the midpoint, we could discern the identity of the behavior being performed and extract GCaMP traces.
(C) Video frames of a freely behaving animal performing margin folding, adapted from Figure 3.
(D) Video frames of a naturalistically behaving animal performing margin folding during GCaMP imaging (compare to panels in C). This animal contributed to the data in Figure 4J. Adapted from Figure 4.
(E) Optic flow can be used to identify behaviors in partially restrained animals. (E1) Annotated behavior identity (top), the mean orientation of optic flow vectors for each frame (middle), and the spectrogram of that orientation (bottom). Swimming can be recognized by high frequency oscillations in the optic flow vectors, while low frequency, radial movements are characteristic of margin folding (compare behaviors in Figure 3G–H). (E2) Magnification of the region indicated with the grey box in panel E1. (E3) Performance of a classifier identifying behaviors using the optic flow signal (n=3 animals).
(F) The first two principal components over time of population neural activity, colored by the behavior being performed. Epochs of margin folding lead to trajectories primarily out along the first principal component, while bouts of swimming and quiescence remain intermingled at the origin.
(G) Bath application of RFamide peptide induced muscle contraction in patches of the subumbrella. This contraction was not accompanied by GCaMP activity (n=6 patches). Peptide concentration: 100uM.
(H-I) Examples of GCaMP activity in a naturalistically behaving animal. Fluorescence in the simultaneously acquired mCherry (left two columns) and GCaMP (right column) channels immediately before (H) or during (I) a margin folding event. GCaMP activation was visible in the green channel, but a similar change in fluorescence intensity was not observed in the red channel. Quantification (I, right) shows fluorescence from frames during/before the folding event for ROIs manually circled around individual neurons in this field of view (t-test). Line represents the median. FOV is ~3.5mm2.
(J-K) Polar histograms showing that the characteristic pattern of RFa+ neuron activation is also present during margin folding in other contexts, i.e. during spontaneous margin folding (J, 103 neurons, 8 events, 3 animals) or passing shrimp captured in the tentacles (K, 125 neurons, 9 events, 3 animals).
Related to Figure 5.
(A) Example field of view (FOV) for calcium imaging in a highly restrained jellyfish. Center of ROIs (green circles) shown superimposed on an image of the mCherry channel. FOV in panels A, G, K, and L is ~3.5mm2. Repeated from Figure 5A with example ROIs indicated and numbered in red, corresponding to traces in B.
(B) Example GCaMP traces corresponding to the five red ROIs designated in A. Zoomed in traces in B2 correspond to the region indicated with the green box in B1.
(C) 6065 single neuron activations form 551 single or ensemble events, pooled from recordings of spontaneous activity across the nerve net from 4 animals. Synchronicity (98.8%) is the number of individual neural activations (6065 total) where >1 neuron was active within a 3 second window (5990). There were 75 instances of a neuron firing alone.
(D) Pairwise correlation between neurons sorted according to hierarchical clustering (shown on the left). Neurons grouped together at different levels of the hierarchy are indicated with colored lines on the dendrogram and matching colored boxes on the correlation matrix.
(E) Pairwise correlation of neural activity versus the angle between those neurons relative to the mouth. Data pooled from, and colored by, 5 individual animals.
(F) Percent of neurons that are shared between functional groups using NMF/ICA. n=486 neurons from 4 animals.
(G) Cluster membership using NMF/ICA for the animal shown in Figure 5A. Neurons participating in multiple ensembles are indicated with multi-colored circles. Grey circles indicate neurons that were not assigned to an ensemble.
(H-J) Functional zones were not evident in the coarse anatomy of the nerve net. (H) Maximum projection of a confocal z-stack showing the distribution of FMRFamide-immunoreactive fibers across the animal. Scale: 500um. (I) Manual tracing of nerve net processes shown in panel H.
(J) Quantification of features of the anatomy compared to the width of k-means clusters of neural activity (n=3 animals).
(K) An analysis to control for imaging or motion artifacts in which we examined fluorescence over time extracted from the red channel using the same ROIs used for GCaMP analyses. Pairwise correlation between neuron’s fluorescence intensity over time (K2), the pairwise correlation vs angle (K3), and k-means clustering (K4) are shown. There is no similar structure in the red channel to what was observed in the green channel (compare to Figure 5).
(L) FOV and ROIs for the animal shown in panel M, and Figure 5M–N.
(M) Pairwise correlation between neurons for the animal shown in panel L. Whether those neurons are located in the ring or net is indicated above the correlation matrix.
(N) Pairwise correlation of neural activity versus the angle between neurons relative to the mouth. Each dot is colored based on whether it was a comparison between net neurons, between ring neurons, or between a ring neuron and a net neuron. Data is pooled from 5 animals, and is the same as in panel E, but colored by comparison type rather than individual animal.
(A) The margin is required for folding behavior. Stimulus-triggered average from 4 animals after removal of the margin (red line). Intact control data (grey line) is the same as shown in Figure 3J, repeated here for ease of comparison.
(B) Activity of nerve ring (top) and nerve net (bottom) neurons over time. Magenta boxes indicate nerve net events, and show that for each net event there was corresponding activity in the nerve rings. In contrast, white arrows point to example events in the nerve ring that do not have a nerve net correlate.
(C) Trial-by-trial activity corresponding to Figure 5O showing that ring neurons were active prior to net neurons. As in Figure 5O, top traces show mean of ring and net activity, aligned to the onset of net events (t=0, magenta line). The white line qualitatively highlights the earlier onset of ring neurons (compare the region between white and magenta lines in ring vs net).
(D) Nerve ring, but not net, neurons responded to food cues. The nerve net only responded if nerve ring was present. Regions containing the nerve ring (top left panels), nerve net (bottom left panels), or both (right panels) were surgically excised and shrimp extract was delivered during GCaMP imaging. Neurons in patches of the nerve net responded to KCL (but not shrimp extract) indicating that they were still capable of activation.
(E) Neuronal processes were visible between the nerve rings and nerve net. Whether these originated from the rings and/or net is unknown. White: anti-aTubulin. Scale: 20um.
(F) Diagrams and example images showing an experiment in which a strip of the animal was cut out, with a tentacle at one end and the mouth at the other. When a brine shrimp was fed to the tentacle, the strip was able to perform the complete food passing behavior.
(G) Local stimulation of the margin with shrimp extract activated a spatially localized ensemble, but did not activate distant neurons. Diagram of experiment (left), and results of individual trials (right), with the approximate location of the stimulus shown in orange and the responding neurons shown in green. Active neurons were annotated as in Figure 4I. We applied droplets of food cues to the margin of agarose-embedded Clytia using a Femtojet and pulled glass electrodes, while performing GCaMP imaging of the nerve net. n=195 neurons.
(H) Histogram and inset heatmap showing aligned summary data from the trials in panel G.
(I1) Left two panels show the correlation matrices from spontaneous versus evoked imaging epochs, from Figure 6C, together for comparison with the correlation matrix generated by GLMs predicting evoked activity after training on spontaneous activity, as described in Figure 6F. (I2) The weights between neurons that emerge from the GLMs after training on spontaneous activity epochs (left), and their performance on evoked data (right, dots: neurons).
Related to Figure 7.
(A) Pairwise correlation matrix from observed data, generated using the neurons-by-events matrix in Figure 5G. Compare to data generated by models in panels B-E.
(B-E) Output of models with different underlying connectivity structures. Hybrid models (C-D) best fit the data. (B1, C1, D1, E1) Diagram and connectivity matrix for each model. (B2, C2, D2, E2) Pairwise correlation between neurons generated by the models; compare to panel A. (B3, C3, D3, E3) The angular width of events generated by each model. (B4, C4, D4, E4) The fraction of cells activated during events generated by each model.
Related to Figure 7.
(A) GLM performance on held out data from recordings of spontaneous activity analyzed in Figure 5, which were used to generate the analyses shown in Figure 7E–G. n=339 neurons from 3 animals. Here and in E, F, H, I dots represent neurons.
(B) Lasso regularization was not the cause of inferred sparse connectivity. (B1) To create a GLM for estimating connectivity w to a target neuron y, given activity of all other neurons in matrix X, we maximized the likelihood shown in this formula, where lambda is a regularization parameter and alpha is the sparseness parameter. (B2) The sparsity of the connectivity, as defined by the percent of elements with zero weight in the matrix, for different values of alpha, showing that GLM regularization did not cause the sparse connectivity found here. Each dot is an individual jellyfish (n=4). (B3) Matrices of GLM weights at different values of alpha, showing their stability.
(C) Connectivity scheme in original model used in Fig 7E, which allows for net neurons to receive inputs from other net neurons as well as ring neurons.
(D) Net-net connections were ablated in GLMs to assess the importance of ring to net connectivity.
(E) Performance of GLMs for net neurons with all inputs (blue) vs GLMs where net-net connections were ablated (purple). p < 0.001, Mann Whitney test.
(F) Performance of GLMs for net neurons after ablation plotted against their Euclidean distance to the nearest ring neuron. Linear Regression, R2 = 0.24.
(G) A separate approach to test the importance of ring to net connectivity where new GLMs were trained for net neurons with only ring input.
(H) Performance of GLMs with ring and net inputs (blue) vs GLMs with only ring inputs (purple). p<0.001, Mann Whitney test.
(I) Performance of GLMs for net neurons trained with only ring input plotted against their Euclidean distance to the nearest ring neuron. Linear Regression, R2 = 0.4.
(J) Performance of GLMs for net neurons trained with only ring input shown on their anatomical position.
Supplemental Video 1. Spontaneous and prey-evoked margin folding behavior. Related to Figures 2 and 3.
Supplemental Video 2. Directionally appropriate folding triggered by local application of shrimp extract. Related to Figure 3.
Supplemental Video 3. Attempted food passing after excision of the mouth. Related to Figure 3.
Supplemental Video 4. Nerve net activation is visible at the location of inward folding in an unrestrained animal. Related to Figure 4.
Supplemental Video 5. GCaMP imaging of the nerve net in an agarose-restrained animal. Related to Figure 5.
Supplemental Video 6. Neural activity was only visible on the peripheral-side of an umbrellar wound. Related to Figure 5.
Supplemental Video 7. An excised strip of the umbrella can perform the complete food passing behavior. Related to Figure 5.
Data Availability Statement
Source data reported in this paper will be shared by the lead contact upon request.
Code used for analyses in this paper will be shared by the lead contact upon request.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


