Abstract
Neuroinflammation and neurodegeneration may represent two polarities of brain pathology. Brain myeloid cells, particularly microglia, play key roles in these conditions. Here, we employed single-cell mass cytometry (CyToF) to compare myeloid cell populations in the experimental autoimmune encephalomyelitis (EAE) model of neuroinflammation, the R6/2 model of Huntington’s disease (HD), and the superoxide dismutase 1 (mSOD1) model of amyotrophic lateral sclerosis (ALS). We identified three myeloid cell populations exclusive to the central nervous system (CNS) and present in each disease model. Blood-derived monocytes comprised five populations and migrated to the brain in EAE, but not in HD and ALS models. Single-cell analysis resolved differences in signaling and cytokine production within similar myeloid populations in EAE compared to HD and ALS models. Moreover, these analyses highlighted α5 integrin on myeloid cells as a potential therapeutic target for neuroinflammation. Together, these findings illustrate how neuropathology may differ between inflammatory and degenerative brain disease.
Introduction
The term “neuroinflammation” has been broadly applied to various neuropathological conditions1. A wide spectrum of neurological disorders ranging from those immunologically-driven such as acute disseminated encephalomyelitis and multiple sclerosis (MS)2, to degenerative diseases such as Alzheimer’s disease (AD)3 and Parkinson’s disease (PD)4, to genetic disorders such as Huntington’s disease (HD)5 and SOD1-driven amyotrophic lateral sclerosis (ALS)6, are often collectively called neuroinflammatory7. One rationale behind applying the “neuroinflammatory” label to these diverse neurological conditions resides in the empirical observation of microgliosis in these conditions8. From detection of inflammatory mediators in AD and PD autopsy brain sections to genomic and transcriptomic studies of brain specimens4,9 it has been suggested that neurodegeneration might be promoted in part by microglia responding to “inflammation” in the brain10,11.
Here we used Cytometry by Time-of-Flight (CyTOF) to enable a high dimensional analysis of cell surface markers, signaling molecules and cytokines on brain myeloid cells at the single-cell level12. We characterized the myeloid cell phenotypes in commonly used mouse models of neuroinflammation or neurodegeneration – Experimental Autoimmune Encephalomyelitis (EAE), a model of MS2, R6/2 mice, a model of HD expressing human mutant HTT exon 113, and mice overexpressing mutant superoxide dismutase1 (mSOD1), a model of ALS14.
Results
Phenotypic heterogeneity within the CNS-resident myeloid population.
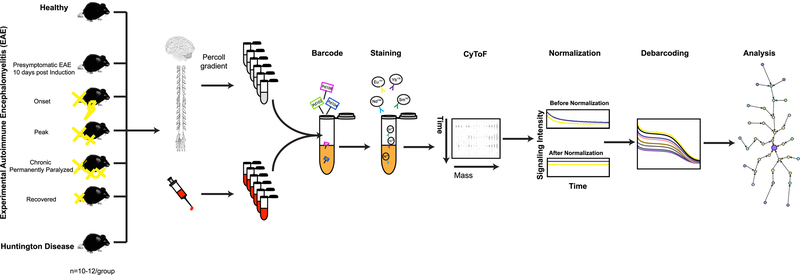
We used CyTOF to analyze the cellular phenotype, signaling properties, and cytokine production in single-cells of both central nervous system tissues (brain and spinal cord) and peripheral blood. We first compared different clinical stages of EAE, with R6/2 transgenic mice, a model of HD13, at a late stage of the disease when the R6/2 mice displayed tremor, irregular gait, abnormal movements and decreased survival15 (Fig. 1). The CyTOF panel was assembled based on a high-throughput screen of 255 antibodies to integral membrane proteins (Supplementary Table 1), proteins that regulate myeloid cell functions16, transcription factors and signaling molecules relevant in neuroinflammation (Supplementary Table 2a–c). Single-cell suspensions of CNS tissue and blood were prepared as described previously17 (Fig. 1).
Figure 1: Schematic representation of the experimental strategy.
Immune response profiles were analyzed in Healthy, five different clinical stages of EAE, and end-stage HD. Single-cell suspensions from CNS and whole blood of each condition were prepared as described in Methods. Individual samples were simultaneously processed by using the barcoding strategy (Methods). Barcoded samples were pooled, stained with a panel of 39 antibodies (Supplementary Table 2a–c and Methods), and analyzed by mass cytometry. Raw mass cytometry data were normalized for signal variation over time and debarcoded and analyzed using the X-shift algorithm, a nonparametric clustering method that automatically identifies cell populations by searching for local maxima of cell event density in the multidimensional marker space. The result is displayed as a MST layout. In each experiment, tissues from ten mice were pooled in order to provide enough cell number (Methods). Each experiment was performed seven to ten times independently.
In order to explore the phenotypic diversity of immune cell populations in the CNS and blood, we applied a K-nearest-neighbor density based clustering algorithm called “X-shift”18. The algorithm allows for the unsupervised clustering analysis of data from single cells18. To visualize the phenotypic continuum of cell populations, output is organized into a Minimum Spanning Tree (MST)18, creating a 2-dimensional layout. The size of nodes and the color coding is explained in the methods section.
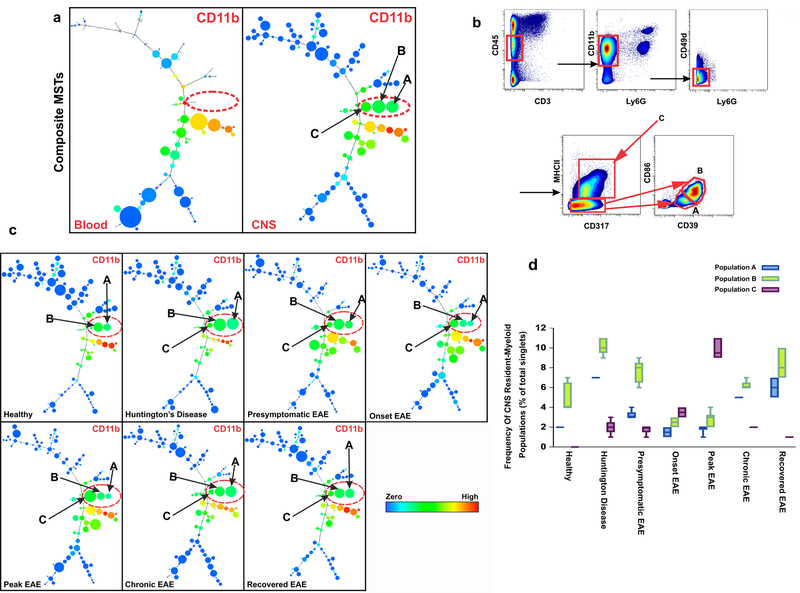
Unsupervised clustering of 1,800,183 single cells from the CNS and blood of 75 samples, created a detailed MST map of distinct cell populations (Supplementary Fig. 1). This analysis revealed three distinct CD11b+ myeloid populations in the CNS that were absent in peripheral blood (Fig. 2a). These populations, identified as CNS-resident myeloid populations, are designated A, B, and C (Fig. 2a, Supplementary Fig 1).
Figure 2: Data-driven, unsupervised clustering defines three distinct CNS-resident myeloid populations.
a) Composite CNS MST of X-shift clusters constructed by combining CNS samples from all the conditions and their biological replicates (n=37) in comparison to composite MST from peripheral blood samples (n=37) reveals three myeloid (CD11b+) populations that are unique to CNS (Populations A, B and C). All samples were barcoded and analyzed together on CyTOF. The color code shows the expression level of CD11b. b) Manual gating based on markers and threshold defined by the X-shift/DMT algorithm confirmed the existence of populations A, B and C. Live cells are identified by the lack of cleaved poly-(ADP)-ribose polymerase (c-PARP) binding (Supplementary Fig. 13b, Methods). This panel represents data from Peak EAE, where 7 independent experiments confirmed a similar gate. c) Visualization of clusters under different clinical conditions demonstrates that populations A, B and C are present in both EAE and HD models. The color code shows the expression level of CD11b (n=5). d) Frequency of populations A, B and C based on manual gating confirms that populations A, B and C are present in both EAE and HD models. Center line is average; boxes extend to 25th and 75th percentile; whiskers extend to 5th and 95th percentiles. (n= 5 independent experiments for healthy, end-stage HD, presymptomatic EAE, Chronic EAE; n=6 independent experiments for Onset EAE, Peak EAE).
In order to verify that populations A, B and C could be identified by manual gating, we applied a feature of the X-shift called a Divisive Marker Tree (DMT) algorithm18. This feature automatically constructs an optimal marker-based classification of clusters18. By setting the gates according to computationally defined thresholds we verified that populations A, B, and C were distinguishable by cell surface marker expression of CD45, CD11b, CD317 (BST2/PDCA-1), major histocompatibility complex class II (MHC-II), CD39, and CD86 (Fig. 2b). Population A represents CD317+, MHC-II−, CD39low, CD86−; population B is CD317+, MHC-II−, CD39hi, CD86+; and population C is CD317+, MHC-II+, CD39hi, CD86+ (Fig. 2b, Supplementary Fig. 2). MHC-II and CD86, among others, have previously been identified as myeloid activation markers19 therefore suggesting that populations B and C represent activated microglia populations.
Populations A, B, and C also expressed several other cell surface markers including the recently identified microglia markers 4D4 and FCRLS20, and low to medium levels of CD88 (complement component 5a receptor 1 (C5aR)), MHC class I (MHC-I, H-2), and TAM receptor tyrosine kinases Mer (MerTK) (Supplementary Fig. 3a).
Populations A, B, and C lacked expression of lymphoid lineage markers such as CD3 (T cells), CD45R/B220 (B cells), Ly6C (monocytes), and Ly6G (granulocytes). These three CNS-specific populations were also characterized by the differential expression of a number of markers. Populations B and C expressed different levels of CD80, TAM receptor Axl, T-cell immunoglobulin mucin protein 4 (TIM4), CD274 (PD-L1), CD195 (CCR5), CD194 (CCR4), and low levels of CD206 (mannose receptor) and TREM2, while population A lacked the expression of all these markers (Supplementary Fig. 3b). The expression level of these markers changed depending on disease conditions (Supplementary Fig. 4a–c).
We distinguished CNS-resident myeloid cells from infiltrating myeloid populations, with a genetic tool21. In addition to defining populations A, B, and C as CNS-resident myeloid cells based on their presence in only the CNS (not in peripheral blood) coupled with the expression of other markers including low CD45 (traditionally believed to mark microglia in the CNS), 4D4 and FCRLS (Supplementary Fig. 5), we further confirmed them as CNS-resident myeloid cells by analyzing the CNS of conditional Cx3cr1CreER Rosa26-YFP mice21. In this model, YFP is conditionally-expressed exclusively in long-lived myeloid cells of the CNS, such as microglia after tamoxifen withdrawal due to the irreversible recombination event mediated by the Cre recombinase21. Here, we were able to identify these populations in conditional Cx3cr1CreER Rosa26-YFP healthy mice and confirm that they express YFP (Supplementary Fig. 6a). In this paper, we avoid calling these cells microglia and refer to them as CNS–resident myeloid cells, which include microglia, meningeal macrophages, and perivascular macrophages.
Neuroinflammatory and neurodegenerative mouse models display congruent CNS myeloid cell populations
To investigate whether disease-specific cues modulate the presence and the frequency of the three CNS-resident myeloid cell populations, we analyzed the MSTs and confirmed the findings by manual gating in healthy mice, R6/2 mice modeling HD, as well as five different states of EAE: presymptomatic, onset, peak, chronic, and recovered. Quantification of each population (Fig. 2c) and representative nodes in the MST (Fig. 2d) in independent biological replicates of healthy samples compared to each disease state, demonstrated that population C was generated during disease states (both EAE and HD disease conditions) but was barely detectable in the healthy CNS (Fig. 2c, d; Supplementary Fig. 6b). In EAE, population C continued to expand from the presymptomatic stage (frequency of 1.8%) to the peak of disease (frequency 9.7%). Thereafter, the frequency of population C declined in chronic EAE animals with permanent paralysis and in recovered EAE mice (0.9% and 1.7% respectively) (Fig. 2d). Of note, of the three CNS-resident populations, only population C expresses CD11c. While very low expression of CD11c is detectable in the R6/2 HD model, CD11c expression levels are increased as EAE progresses to peak disease and then becomes downregulated in both the EAE chronic and recovered groups (Supplementary Fig. 7a). Given that population C expresses MHC-II and upregulates CD11c during EAE, a T cell driven inflammatory CNS disease, antigen presentation may be a key role for population C, a CNS-resident population22.
To capture the expansion rate of each population under each disease condition, we also analyzed the percentage of cells positive for the proliferation marker Ki-67. The percentage of Ki-67+ cells in all three CNS-resident myeloid populations is increased as EAE disease progresses from presymptomatic through the chronic stages, and is then decreased during recovery (Supplementary Fig. 7b). Interestingly, the percentage of Ki-67+ cells in these population in the HD model was significantly lower than in the different clinical stages of EAE. This suggests that populations A, B and C are all expanding at different rates, depending on each disease condition.
Signaling phenotypes distinguish CNS-resident myeloid cells in neuroinflammatory versus neurodegenerative models
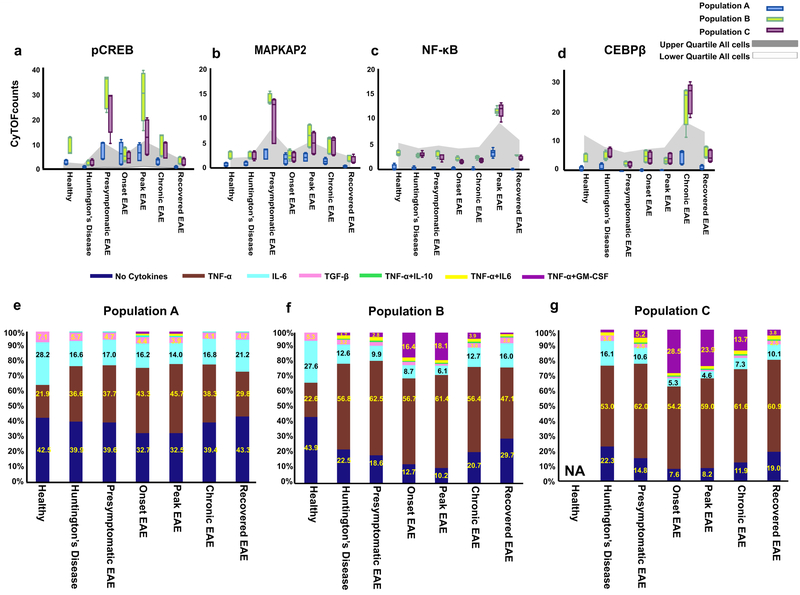
To understand the signaling differences in CNS-resident myeloid populations A, B, and C in the HD model versus the different stages of EAE progression, we next focused on the transcription factors and signaling markers such as pSTAT1, pSTAT3, pSTAT5, cAMP response element-binding protein (pCREB), pMAPKAPK2, NF-κB (p65), and CCAAT/enhancer-binding protein and alpha and beta (C/EBPα, C/EBPβ) proteins (Supplementary Table 2a).
First, this revealed substantial differences in the expression patterns of these signaling proteins across all three CNS-resident myeloid populations. Where populations B and C showed a high level of signaling, population A differed substantially from these two populations with very low expression levels of signaling proteins, potentially highlighting a different functional role for these CNS-resident myeloid populations (Fig. 3a–d). These results are consistent with the fact that population A does not express myeloid cell activation markers, such as CD86, MHC-II and CD11c, and could therefore represent a functionally less active population under these conditions (Fig. 2b, Supplementary Fig. 7a).
Figure 3: CyTOF analysis reveals the signaling and cytokine molecular signatures in the three CNS-resident myeloid populations under different clinical conditions.
a-d) Dynamic of key signaling molecules of immune activation pathways in the three CNS-resident myeloid populations. Box-and-whisker plots show median raw CyTOF signal intensity per population. The grey area represents the interquartile range of the given signaling molecule in all cells in a sample, averaged across replicates, and thus indicates the overall expression range for each marker. Center line is average; boxes extend to 25th and 75th percentile; whiskers extend to 5th and 95th percentiles. (n= 5 independent experiments for healthy, end-stage HD, presymptomatic EAE, Chronic EAE; n=6 independent experiments for Onset EAE, Peak EAE).
(e-g) Single-cell analysis of cytokine production by the three CNS-resident myeloid populations in response to different disease conditions. Analysis of cytokine co-expression in CNS-resident myeloid populations in healthy and diseased states demonstrating heterogeneous subsets in each population. Percentages of single-cells expressing zero, one or two cytokines are represented in a stacked bar graph (n=3 independent experiments).
Second, this analysis revealed the progression of key signaling pathways within the CNS in populations B and C during the development of EAE. In the presymptomatic stage of EAE, a significantly increased level of pCREB and pMAPKAPK2 expression represents the only signaling signature in populations B and C (Fig. 3a, b). At the peak of EAE, a second wave of increased expression of pCREB and pMAPKAPK2 in populations B and C emerged as a hallmark similar to what we observed in the presymptomatic stage and in agreement with previous studies23 (Fig. 3a, b). Interestingly, in chronic EAE, where animals never recovered from paralysis, upregulation of NF-κB(p65) in concert with C/EBPβ in populations B and C was identified, (Fig. 3c–d). These data indicate that there is sequential signaling in EAE.
Third, these inflammatory signaling signatures were noticeably absent in populations A, B, and C in the HD model compared to EAE. This emphasizes the considerable differences in signaling properties in the HD model compared to EAE in CNS-resident myeloid cell populations (Fig. 3a–d). Though similar CNS-resident myeloid cell populations were identified in both models, the nature of their signaling properties under these conditions was vastly different.
Multiple cytokine-producing myeloid cells in models of neuroinflammation versus neurodegeneration
We sought to determine the capacity for cytokine production of these myeloid cells, without imposition of any ex vivo stimulation24. To test cytokine production, the panel included tumor necrosis factor-α (TNF-α), interferon-γ (IFN-γ), IFN-α, interleukin-10 (IL-10), IL-6, IL-17A, granulocyte-macrophage colony-stimulating factor (GM-CSF), and transforming growth factor-β (TGF-β) (Supplementary Table 2b). CNS-resident myeloid populations were manually gated as defined above (Fig. 2b). We calculated the fraction of cells detected that secrete a given cytokine, defined by expression values exceeding the 90th percentile of a healthy sample for each cluster (Supplementary Fig. 8 a,b,).
Among the eight cytokines evaluated, TNF-α was the most prominently produced in the three identified CNS-resident myeloid populations in both neuroinflammatory and neurodegenerative conditions compared to healthy cells (Supplementary Fig. 8 a,b). Most notably, in populations B and C during early stages of EAE disease (presymptomatic, onset, peak), and in the case of population C, also during chronic EAE, the majority of cells (up to 80%) produced TNF-α whereas the percentage of TNF-α expressing cells ranged from 30%–50% in the neurodegenerative R6/2 model of HD. In addition to TNF-α, a modest percentage of cells in these three populations expressed GM-CSF, IL-6, IL-10, and TGF-β (Supplementary Fig. 8 a,b).
To analyze the multifunctional nature of each population at a single-cell level25, we applied the X-shift clustering algorithm18. Each population was clustered based on expression patterns of cytokines only, and the frequency of cells that produce each cytokine alone or in any combination at the single-cell level in each disease condition was assessed. Seven distinct subsets of cytokine-producing cells were delineated in populations A, B, and C at the single-cell level based on producing either a) single cytokines-TNF-α, or IL-6, or TGF-β, or b) a combination of TNF-α with IL-6, GM-CSF, IL-10 or c) a lack of cytokine production (Fig. 3e–g).
Quantifying the fraction of each of these seven subsets within each population under different disease conditions, we found that in a healthy state, most of the cells within each population produced no cytokines at all or just a single cytokine (Fig. 3e–g). The frequency of single-positive TNF-α–producing cells increased significantly in comparison to the healthy state in both neuroinflammatory and neurodegenerative models whereas the frequency of single-positive IL-6 or TGF-β -producing cells decreased (Fig. 3e–g).
We identified three subsets where more than one cytokine was produced within a single cell: dual TNF-α and GM-CSF producing cells, dual TNF-α and IL-10-producing cells, and dual TNF-α and IL-6-producing cells (Fig. 3e–g). Most noticeably, the frequency of the subset that coexpressed both GM-CSF and TNF-α in populations B and C significantly increased during neuroinflammatory conditions especially at the onset and peak of EAE disease, making this the second most abundant subset among cytokine-producing cells (up to 18% and 29%, respectively) (Fig. 3e–g).
Conversely, in the neurodegenerative HD model, the frequency of this GM-CSF and TNF-α subset was very low in all three CNS-resident myeloid populations (0% to 2%). With respect to other multifunctional subsets, both neuroinflammatory and neurodegenerative models also displayed the emergence of a low frequency of TNF-α+ IL-6+ and TNF-α+ IL-10+ multifunctional cells (2–3%). By comparing the cytokine profile in these neuroinflammatory and neurodegenerative models, we can identify the GM-CSF, TNF-α dual producing subset as a signature of neuroinflammation (Fig. 3e–g).
Moreover, in contrast to populations B and C, a significant fraction of cells in population A produced no cytokines in healthy and disease conditions, and the cytokine producing subsets were dominated by single cytokine producing cells even during disease conditions. Multi-functional subsets producing more than one cytokine comprised a very small percentage of cells (only 1%) (Fig. 3e–g). This result is consistent with the analysis above, where population A shows a lack of myeloid cell activation markers and has a lower expression level of signaling molecules compared to the other two populations (Fig. 3e–g).
To capture the cellular and cytokine profile of myeloid cells in the earlier stages of the model of HD prior to development of severe symptoms (week 13–14), we next analyzed R6/2 mouse CNS tissues at 4 weeks, 7 weeks and 10 weeks. The frequency of CNS-resident myeloid populations (Fig. 2b), showed that all three populations A, B, and C exist pre-disease and during the progression of HD (Supplementary Fig. 9a). Over the course of the disease, the frequency of populations A and B increased, whereas population C remained relatively low and unchanged (Supplementary Fig. 9a).
To gain increased resolution into the cytokine profile of each population at a single-cell level, we applied the unsupervised X-shift analysis (Fig 3e–g,). This revealed six distinct subpopulations of cytokine-producing cells at the single cell level based on the production of IL-10, TNF-α, IL-6, TGF-β, GM-CSF and the lack of cytokine production (Supplementary Fig. 9b). Of note, these subpopulations have a “single-cytokine only” producing phenotype and there is a notable lack of cells that secrete multiple cytokines (Supplementary Fig. 9b).
Remarkably, during early stages of HD-like pathology, there is the presence of a distinct subpopulation that only secretes IL-10. In all three CNS-resident myeloid populations, the fraction of IL-10 only secreting cells ranged from 30 to 60%, depending on the population (Supplementary Fig. 9b). This IL-10 expressing subpopulation was not detected at a late stage in the HD model (13–14 weeks) (Fig 3e–g). IL-10 is most often considered an anti-inflammatory cytokine26.
We then investigated the frequency and cytokine profile of populations A, B and C in a second mouse model of a progressive neurodegenerative disorder, ALS, using transgenic mSOD1 mice27. Employing the same CyTOF panel and analysis strategy as above, the cellular and cytokine profiles were analyzed at two time points: onset (95 days of age) when decline in motor performance has been reported, and a late stage (140 days) when mice were completely paralyzed14. All three populations A, B and C defined by the biaxial dot plot as above were observed at both onset and late stages of the mSOD1 ALS model (Supplementary Fig. 10).
The frequency of populations A and B increased as the symptoms progressed to disease late-stage of the ALS mouse model, which is consistent with previous reports of the increase in the number of microglia in mSOD1 mice14. The frequency of population C remained the same during both disease time points (Supplementary Fig. 10a). Analysis of the cytokine profiles of the three CNS-resident myeloid populations at the single cell level in the ALS model at onset and late-stage revealed eight distinct subsets based on cells with no cytokine production, cells with individual secretion of either TNF-α, IL-6, IL-10, TGF-β, or GM-CSF alone, and those cells expressing a combination of TNF-α and IL-6, TNF-α and IL-10 (Supplementary Fig. 10b). The frequency of subsets expressing either no cytokine, GM-CSF alone or TGF-β alone decreased as the disease progressed to ALS late-stage, whereas the frequency of TNF-α producing cells sharply increased (Supplementary Fig. 10b). Similar to early stages in the HD model, we observed the presence of IL-10 expressing cells across all three CNS-resident myeloid populations at the onset in the ALS model, albeit at a much lower frequency (2–10%). However, in contrast to the HD model, these IL-10 expressing cells increased (populations A and C) or remained constant (population B) in the ALS model (Supplementary Fig. 10b). Interestingly, similar to the end-stage of HD, there is a very small subset of cells producing multiple cytokines in end-stage ALS (0.5–2%) (Supplementary Fig. 10b). Thus, in both HD and ALS models, the majority of cells are not multifunctional cytokine producers.
Collectively, these data highlight a fundamental property of three identified CNS-resident myeloid cell populations, by demonstrating that each population, although defined as relatively homogeneous by common cell surface markers, in fact, contains heterogeneous functional subsets based on their cytokine secretion profile.
Importantly, we have identified two distinct cytokine-secreting subsets that represent the signature of neuroinflammatory conditions in contrast to neurodegenerative models. Although both neuroinflammatory and neurodegenerative models displayed the development of double positive TNF-α and GM-CSF producing cells, the high frequency of this subset correlated best with the height of neuroinflammatory conditions in EAE—peak and onset—in populations B and C, whereas the frequency of cells in these same populations was extremely low or absent in both models of HD and ALS. Conversely, both neurodegenerative conditions show the presence of a remarkable subset of cells expressing IL-10, often considered an anti-inflammatory cytokine26, across all three CNS-resident myeloid populations prior to late stage disease symptoms.
Blood-derived monocyte subsets exhibit different kinetics of migration to the CNS in inflammatory versus HD and ALS models
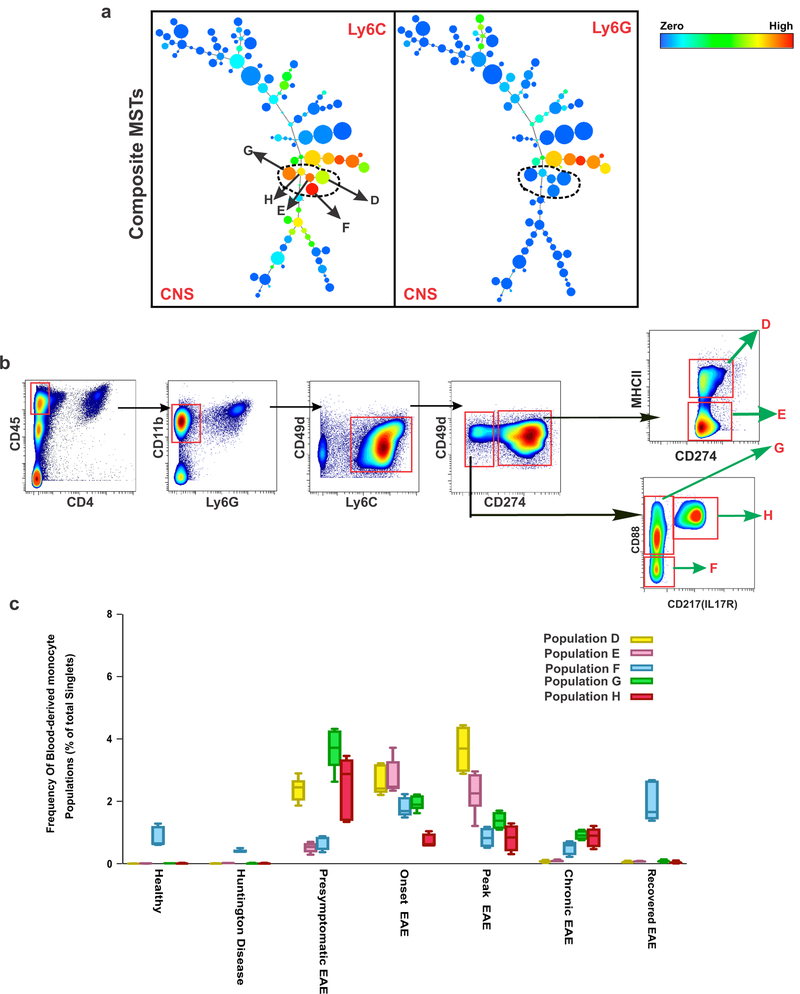
The inflammatory response in the CNS is due in part to the entry of peripherally-derived myeloid cells28,29. We next characterized the properties of these infiltrating cells in both EAE and HD mice. Peripheral monocytes were distinguished from other myeloid cells (CD11b+ cells) based on expression of their key surface marker Ly6C and lack of Ly6G expression. A composite MST from all samples combined revealed five discrete Ly6C+Ly6G− cell clusters in CNS samples (Fig. 4a). The X-shift algorithm separated the Ly6C compartment into five separate clusters (D, E, F, G, and H), and the DMT visualization revealed that the main markers driving the separation are CD274 (PD-L1), CD88 (C5aR1), CD217 (IL-17R), and MHC-II (Fig. 4b). Population D is characterized as Ly6C+, CD274+, MHC-II+ monocytes, population E as Ly6C+, CD274+, MHC-IIneg monocytes, population F as Ly6C+, CD274neg,, CD88neg, CD217neg monocytes, population G as Ly6C+, CD274neg, CD88+, CD217neg monocytes and population H as Ly6C+, CD274neg, CD88+, CD217+ monocytes.
Figure 4: Kinetics of Peripheral Monocytes in CNS under Inflammatory versus Degenerative conditions.
a) Composite MSTs of CNS samples (n=37) reveals five distinct Ly6C+Ly6G- myeloid populations (peripheral monocytes) in CNS. The color code shows the expression levels of Ly6C and Ly6G. b) Manual gating based on markers and threshold defined by the X-shift/DMT algorithm confirmed each population. This panel represents data from Peak EAE, where 7 independent experiments confirmed a similar gate. c) Frequency analysis based on manual gating demonstrates that there is a minimum accumulation of peripheral monocytes in healthy and neurodegenerative conditions. In EAE disease, different peripheral monocyte populations accumulated depending on the disease state. Center line is average; boxes extend to 25th and 75th percentile; whiskers extend to 5th and 95th percentiles. (n= 5 independent experiments for healthy, end-stage HD, presymptomatic EAE, Chronic EAE; n=6 independent experiments for Onset EAE, Peak EAE).
We analyzed the frequency of each of these five peripheral monocyte populations in the healthy state and in different stages in the disease models (Fig. 4c). In agreement with previous studies8, we observed no contribution of peripheral monocytes (an average of less than 0.4%) in the CNS in the HD model. In accordance with earlier reports28–30, there is a very low frequency of monocytes (0.8% to 1.2% respectively) and only population F was detected in the CNS of both healthy and recovered EAE mice. In contrast, the inflammatory stages of EAE—presymptomatic, onset, and peak—evoked the presence of all five identified peripheral monocyte subsets (Fig. 4c). In chronic EAE we observed a low frequency (0.5% to 0.9%) of monocyte populations F, G, and H (Fig. 4c).
Furthermore, the expression pattern of other cell surface markers demonstrated that CD80, CD86, CD38, CD39, MerTK, Axl, CD206, and TREM2 were upregulated in populations D and E. Populations F and G expressed low levels of these markers, and population H expressed medium levels of these cell surface markers (Supplementary Fig. 11).
With expression of costimulatory molecules (CD80, CD86), populations D and E most likely represent activated antigen presenting cells (APCs). Population E (MHC-II−) could be an independent population or may simply be a population that has transitioned from Population D. Both populations express CD274 (PD-L1) following activation, as a natural immunological brake, that results in the inactivation of newly activated T cells31.
Population F, (CD49d+CD88−CD217−MHC-II−) is upregulated during onset and recovered EAE. Populations G and H, present predominantly during presymptomatic EAE and further distinguished by CD49d+CD274−CD88+MHC-II− expression, are likely activated monocytes/macrophages generated during the innate and adaptive inflammatory response following EAE immunization. CD88 (C5aR1), the receptor for the complement peptide C5a, is expressed in EAE on activated monocytes/macrophages infiltrating into the CNS32. Population G (CD217-) may arise predominantly from activation of Th1 cells that is sustained during EAE onset; whereas population H (IL-17R+) is generated in response to the activation of Th17 cells following EAE immunization.
Discordance in expression of signaling molecules and cytokines in infiltrating versus resident myeloid cells
We next determined if the five peripheral monocyte populations had distinctive signaling states in response to the same disease model conditions compared to the three CNS-resident myeloid cell populations. Indeed several signaling proteins were differentially expressed under the same disease conditions (Fig. 5a).
Figure 5: Single-cell analysis of signaling molecules and cytokine production in different peripheral monocyte populations in response to different disease conditions.
a) Heat map representing the comparison of median raw CyTOF signal intensity for each signaling molecule between CNS-resident myeloid populations and peripheral monocyte populations in presymptomatic, onset and peak EAE when all five peripheral monocyte populations are present. The color representing the signaling molecule expression ranges from blue (undetectable) to white (intermediate) to red (maximum). Mass cytometry data are from five or six independent experiments (n= 5 independent experiments for healthy, end-stage HD, presymptomatic EAE, Chronic EAE; n=6 independent experiments for Onset EAE, Peak EAE). ND = not distinguishable. b) Single-cell analysis of cytokine production by different peripheral monocyte populations in response to different disease conditions. X-shift analysis of the co-expression of cytokines in peripheral monocyte populations suggests that each population contains heterogeneous subsets depending on each disease conditions. Percentages of single-cells expressing zero, one, two, three or four cytokines are represented in a stacked bar graph (n=3 independent experiments).
Expression of pSTAT3 was higher in several peripheral monocyte populations at the onset (populations D and E) and peak (populations D, E, and H) of EAE compared to all three CNS-resident myeloid cell populations (Fig. 5a). An increase in the transcription factor pSTAT3 is recognized as an important mediator of inflammation in MS patients33.
In contrast, pCREB expression was markedly higher in CNS-resident myeloid cells—particularly populations B and C—in relation to the peripheral monocyte populations (Fig. 5a); thus supporting a fundamental difference between infiltrating monocytes when compared to CNS-resident myeloid cells. The proliferation of CNS-resident myeloid cells but not peripheral monocytes, and the upregulation of proliferation-related genes such as fos during the course of EAE in CNS-resident myeloid cells, has recently been reported34. CREB is the main transcriptional regulator of the fos gene35. The present results demonstrating pCREB expression are concordant with patterns of microglial proliferation and fos expression, and suggest that CREB pathways promote proliferation of CNS-resident myeloid cells during EAE. NF-κB and C/EBPβ expression were also increased in CNS-resident myeloid cell populations but not in peripheral monocyte populations during EAE disease (Fig. 5a).
On the basis of these results, we hypothesized that different signaling properties of CNS-resident myeloid cells and peripheral monocytes should be reflected in distinct cytokine expression profiles during EAE pathology. Therefore, we next assessed the cytokine production capacity of each of the five peripheral monocyte populations. Peripheral monocyte and CNS-resident myeloid cell populations had similar cytokine expression profiles, predominantly producing TNF-α followed by IL-6, GM-CSF, IL-10, and TGF-β (Supplementary Fig. 12).
We next analyzed the profile of multiple cytokines produced by single-cell populations using the X-shift clustering algorithm. A comparative analysis of the five peripheral monocyte populations with the three CNS-resident myeloid cell populations revealed that in addition to seven distinct populations of cytokine-producing cells that were identified in CNS-resident myeloid cell populations (Fig. 3e–g), some of the peripheral monocyte populations have three additional multiple-cytokine-producing subsets in EAE (Fig. 5b). These three new multifunctional subsets consisted of triple cytokine producing cells, TNF-α+GM-CSF+IL-6+ and TNF-α+IL-6+ IL-10+, and quadruple cytokine producing cells, TNF-α+GM-CSF+IL-6+IL-10+ (Fig. 5b), whereas multifunctional subsets in CNS-resident myeloid populations were only double positive (Fig. 3e–g). These three additional subsets were only identified at the onset and peak of EAE and had a significantly higher frequency at the peak of the disease compared to the onset (Fig. 5b).
Differential expression of cell surface phenotype on infiltrating versus resident myeloid cells reveals therapeutic targets
Microglia and peripheral-derived myeloid cells have distinct developmental origins36, renewal mechanisms29, and as shown here, different signaling and cytokine profile under the same disease condition. Next, we explored these different cell types in reference to phenotypic surface proteins.
When comparing the cell surface markers in the CNS-resident myeloid cell populations (A, B, C) with the peripheral monocyte populations (D, E, F, G, H), we observed expression of adhesion molecules CD49d (α4 integrin) and CD49e (α5 integrin) was only on blood-derived myeloid populations and not on CNS-resident myeloid cell populations (Fig. 6a). CD49a-c were not present in any population in our pilot screen. While CD49d was also expressed by lymphocytes and dendritic cells, CD49e was only expressed by Ly6C+ subpopulations (Fig. 6a). CD49e binds fibronectin37, and is found in multiple sclerosis lesions, particularly around blood vessels38. The expression of CD49e on monocytes suggests that a CD49e–fibronectin interaction is critical in migration of these cells into the CNS parenchyma.
Figure 6: CyTOF analysis reveals a therapeutic target on Infiltrating myeloid Cells in inflammatory condition.
a) Cell Surface Phenotype analysis reveals high expression of CD49d (α4 integrin) and CD49e (α5 integrin) only on infiltrating myeloid cells compared to CNS-resident myeloid cells. CD49e is only expressed on myeloid cells whereas CD49d is also expressed on T cells and DCs. b) Average clinical score for EAE mice treated daily with an anti-CD49e antibody compared to an isotype control antibody. In this prophylactic regimen, treatment started at the time of EAE induction. Mice treated with anti-CD49e antibody exhibit a delay in disease onset and have significantly reduced overall disease severity. The experiment was concluded due to the high morbidity of control mice. Arrow indicates when the treatment was started. Each point represents the mean clinical disease score ± SEM. Each asterisk (*) represents a p-value < 0.05 by Mann-Whitney one-tailed test comparing between the two groups (n = 5 mice).
c) Average clinical score for EAE mice treated daily with anti-CD49e antibody compared to an isotype control starting when EAE disease is established. Mice treated with anti-CD49e antibody exhibit reduced overall disease severity. Arrow indicates when the treatment has started. Each point represents the mean ± SEM. Each asterisk (*) represents a p-value < 0.05 by Mann-Whitney one-tailed test comparing between the two groups (n = 10 mice). d and e) CyTOF analysis from CNS tissue of EAE mice treated with either isotype control antibody or anti-CD49e antibody (graph C experiment).
To investigate whether blocking the entry of monocytes into the CNS would affect the course of EAE disease, we treated EAE mice with either the MFR5 antibody specific to CD49e or its isotype antibody control. We used two treatment regimens to test the effect of anti-CD49e antibody treatment on EAE.
In the first regimen, we started the treatment on day one when we induced EAE in C57BL/6 mice and continued treatment every day. The onset of the disease in mice treated with anti-CD49e antibody in this prophylactic regimen was significantly delayed compared with the control group (Fig. 6b). Notably, antibody treatment reduced the severity of the disease and the animals never reached a severe paralytic stage (Fig. 6b). Fibronectin and α5β1 integrin have been reported to be upregulated in the endothelium at the height of vascular remodeling during pre-symptomatic EAE39. Therefore, starting anti-CD49e antibody treatment at the time of disease induction could function by both blocking monocyte trafficking into the CNS and inhibiting brain endothelial cell proliferation and angiogenic remodeling.
However, in the second treatment regimen, we started administration of anti-CD49e antibody once all mice exhibited clinical signs of EAE (day 15), and found that anti-CD49e antibody treatment also attenuates established EAE (Fig. 6c). Analyzing the cellular profile of mice treated with anti-CD49e antibody versus the isotype control group with CyTOF reveals a significant reduction in the size of each node representing each of the five monocyte populations following anti-CD49e antibody treatment (Fig. 6d,e). Taken together, treatment of EAE by blocking CD49e (α5-integrin) recruitment of peripheral myeloid cells to the CNS suggests that these infiltrating cells are essential for pathogenesis.
Discussion
For the last few decades, the concept of inflammation in response to brain pathologies has been under constant debate1,8,40,41. The results of our study challenge a view where any cellular and molecular activation of glial cells across various neuropathological conditions is simply labeled as “neuroinflammation”. In the current study, we characterized the myeloid cell compartment in two opposite polarities of CNS pathology using an animal model of neuroinflammatory disease, EAE, versus mouse models of neurodegenerative diseases such as HD and ALS with CyTOF coupled with an unbiased18 bioinformatics approach. We initially identified two CNS-resident myeloid populations (A and B) in healthy brains. Interestingly, in the EAE, HD, and ALS models, these two populations increased in total frequency in parallel with the expansion of a third disease-specific population, named C. These observations provide a basis for the contention that different CNS diseases involving microglia have similarities. However, delving further into the phenotype of these populations allowed us to demonstrate that the three CNS-resident myeloid populations have nuanced differences when comparing EAE, with the R6/2 model of HD and the mSOD1 model of ALS.
Single-cell analysis of several signaling markers revealed that in EAE, two of the CNS-resident myeloid populations (B and C) developed a closely coordinated series of signaling events with pCREB and pMAPKAPK2 as their signature. This occurred during the presymptomatic stage of disease, prior to clinical paralysis, and at the peak of disease when paralysis is manifest; whereas both NF-κB and C/EBPβ signaling pathways characterized the chronic disease state. By contrast, these two populations in the HD model did not exhibit any major expression of these same signaling pathways, contrary to previous reports42. The lack of similarity in signaling activity of CNS-resident myeloid cells between the HD model and chronic stage EAE is notable, considering that mice in both models developed permanent functional neurologic impairment. Chronic EAE, has been often described as the “neurodegenerative” phase of progressive MS in the literature43,44. Our results, showed that NF-κB and C/EBP signaling in CNS-resident myeloid cells in chronic EAE contrasts with the lack of any such signaling activity in an HD model.
The difference in the functional properties of CNS-resident myeloid cells was also reflected in their respective profiles of cytokine secretion in HD and ALS compared to EAE. While, from an analysis of the total population, these three populations in healthy and disease conditions demonstrated the ability to generate similar cytokines, albeit with different frequencies, analysis at the single-cell level confirmed that each population itself, in fact, contains different subsets based on their cytokine production profiles. The striking difference between EAE versus HD and ALS was the surge of cells that secrete multiple cytokines in EAE, such as TNF-α and GM-CSF. Such dual secretors constituted a substantial portion of the total cytokine producing cells in onset and peak of the EAE disease. In both HD and ALS models, the majority of cells are not multifunctional cytokine producers. Our data suggest that the analyses of cytokine levels as a marker of immune response should be interpreted in the context of whether single or multiple cytokines are produced within a cell population25.
Interestingly, we observed that a substantial fraction of cells within populations A, B and C that transitioned from pre-symptomatic to mild symptomatic HD (up to 10 weeks of age) express IL-10, which is often associated with anti-inflammatory properties26. IL-10 production was notably absent at the end-stage of the R6/2 HD model (13–14 weeks). Altered levels of IL-10 had been observed in the striatum of HD post-mortem brain samples45. This observation is interesting in the context of the protective (anti-inflammatory) and trophic support delivered directly by IL-10 to neurons46. These results emphasize the limitations of defining myeloid cells along single polarities like M1 M2, because as we see from these studies, cytokine profiles are highly nuanced47.
We next analyzed blood-derived myeloid cells. Myelomonocytic cells (Ly6C+Ly6G−) differentiate into five peripheral monocyte populations. Blood–derived myeloid cells do not contribute to myeloid population in the brain under neurodegenerative conditions such as ALS29. Similarly, we confirmed that the recruitment of myelomonocytic cells to the brain is absent in the HD model as well. By contrast, these five peripheral monocyte populations were present in all different clinical stages of EAE, but at varied frequencies.
We investigated the functional differences between CNS-resident myeloid cells versus recruited blood-derived myeloid cells in the pathogenesis of different CNS disease models. We showed that these two cell types have different signaling phenotypes under the same disease conditions. In addition, we were able to demonstrate that inflammatory signaling, such as pSTAT3 upregulation, occurs in peripheral monocytes during the active stages of EAE. Later in the chronic disease stage, the inflammatory signaling switches to CNS-resident myeloid cells that express NF-κB.
The implications of the shift in inflammatory signaling from peripheral myeloid cells to brain specific myeloid cells, with different signaling programs, may have broad implications for designing new drugs targeting the NF-kB pathway that lie beyond the blood brain barrier. The progressive stage of diseases like MS may be largely generated from cells within the CNS, and thus beyond the blood brain barrier.
An emerging theme from these data, in concert with our previous findings and those of others 28,34 is that the significant recruitment of monocytes is a transient event largely driven by classical inflammation. Once inflammation is significantly diminished or disappears, monocytes also largely vanish. The image of monocytes as the key cellular player that triggers the progression of EAE disease to the paralytic stage, a concept put forward by our own previous studies and others 28,48, now becomes even more complex and nuanced given the present discovery of the considerable heterogeneity of this monocyte population.
Blocking the entry of leukocytes to the brain has been explored as a therapeutic strategy 49. We have shown that blocking the homing of T lymphocytes and monocytes to the CNS with an antibody specific for CD49d (α4 integrin) suppressed EAE, and recent clinical studies confirmed reduced relapse rates in MS patients, leading to the approved therapeutic Natalizumab50. Unfortunately, this approved treatment for relapsing remitting MS that blocks T cells from entering the brain may result in the reactivation of the JC virus, leading to the sometimes fatal infection known as progressive multi-focal leukoencephalopathy (PML)49.
We also previously reported that by preventing the infiltration of blood-derived myeloid cells to the CNS, the activation of CNS-resident myeloid cells is required for the initiation of EAE and precedes the entry of blood-derived cells28. Furthermore, the progression of EAE is due to the entrance of blood-derived myeloid cells into the CNS28. Here we identified CD49e (α5 integrin) expression on only the peripheral monocyte populations and not on CNS-resident myeloid cell populations. Treatment with anti-CD49e antibody significantly reduced EAE disease severity and now provides a strong rationale for a novel therapeutic approach that specifically targets and inhibits monocyte trafficking into the CNS. Such a strategy might even have potentially fewer deleterious side effects than existing MS therapies.
These studies illustrate the power of mass cytometry for understanding previously undefined populations of CNS myeloid cells. The varieties of molecular programs in myeloid cells between inflammatory and degenerative conditions in these models of HD, ALS and MS, may allow us to further distinguish between neuroinflammation and neurodegeneration. Extrapolation of these studies to human material is one of the next steps. As we have shown here, unexpected therapeutic targets, like α5 integrin, have emerged by application of CyTOF for the analysis of neuropathology.
Methods
Mice.
C57BL/6J female mice were purchased from the Jackson Laboratory (Sacramento, CA) at 7 weeks. Animals were rested at Stanford University’s research animal facility for 2 weeks and were induced with EAE at 9 weeks of age.
R6/2 female mice were purchased from the Jackson Laboratory. The CNS tissues were harvested at the age of 4, 7 and 10 weeks old prior to development of severe symptoms, and at the age of 13– 14 weeks of age when they developed severe tremor, irregular gait, abnormal movements and seizures, and decreased survival15
Female mice overexpressing the G93A mutant human transgene for SOD1 were purchased from Jackson Laboratory. CNS tissues were harvested at the age of 95 days of age when a significant decline in motor function of mSOD mice compared to wt controls begins14 and 140 days disease end-stage being defined as the time when mice were completely paralyzed.
Animal experiments were approved by, and performed in compliance with, the National Institute of Health guidelines of the Institutional Animal Care and Use Committee at Stanford University. All animals were housed under a 12-hour light cycle. The maximum number of animals housed per cage was 5 mice. Animals were randomly selected and used in this study.
Induction of EAE in mice by immunization with MOG and adjuvant.
EAE was induced in female C57BL/6J mice (the Jackson Laboratory) at 9 weeks of age by subcutaneous immunization in the flank with an emulsion containing 200 μg myelin oligodendrocyte glycoprotein35–55 (MOG35–55; MEVGWYRSPFSRVVHLYR NGK) in saline and an equal volume of complete Freund’s adjuvant containing 4 μg/ml mycobacterium tuberculosis H37RA (Difco Laboratories Inc., Detroit, MI). All mice were administered 400 ng of pertussis toxin (List Biological Laboratories, Inc., Campbell, CA) intraperitoneally at 0 and 48 h post-immunization. The neurological impairment was scored as follows: presymptomatic; 10 days post EAE induction with no clinical disease; onset: loss of tail tone and hindlimb weakness, peak; complete hindlimb paralysis, recovered; recovery from hindlimb paralysis and sustaining the improvement, chronic; developed permanent functional impairment after 3–6 months and never recovered.
Although the inclusion of pertussis toxin (PTX) for EAE induction was originally thought to facilitate entry of immune cells into the CNS by increasing the permeability of the blood-brain barrier51, more recent work has shown that this is a byproduct of PTX. PTX has been reported to act as an adjuvant, acting on dendritic cells to drive the development of Th1 cells52,53, and suppressing the number and function of T regulatory cells54. Repeated PTX administration actually prevents EAE55.
Cx3cr1CreER Rosa26-YFP mice.
Cx3cr1CreER Rosa26-YFP mice were generated in Prof. Marco Prinz’ laboratory in Freiburg, Germany, by crossing Cx3cr1CreER (B6.129P2(C)-Cx3cr1tm2.1(cre/ERT2)Jung/J) with Rosa26-YFP (B6.129X1-Gt(ROSA)26Sortm1(EYFP)Cos/J), both available at Jackson Laboratory, and housed under SPF conditions in the local animal facility. Cx3cr1CreER mice were backcrossed for >9 generations onto the C57Bl/6 background21,56. Cx3cr1CreER mice were genotyped by PCR using the forward primer 5′- CCT CTA AGA CTC ACG TGG ACC TG −3′, the reverse primer 5′- GAC TTC CGA GTT GCG GAG CAC −3′ and a specific primer GCC GCC CAC GAC CGG CAA AC which amplify a 750 bp fragment wildtype or a 304 bp fragment from the transgenic Cx3cr1 locus. For induction of Cre recombinase, 5- to 7-week-old Cx3cr1CreER female mice were treated with 4 mg tamoxifen (TAM, Sigma) solved in 200 μl corn oil (Sigma) injected subcutaneously at two time points 48 h apart. The brain, spinal cord and blood of these mice were harvested at 15 weeks of age for CyToF analysis. Anti-GFP antibody (Supplementary Table. 2a) conjugated to a metal were used to detect the YFP expression. The lack of YFP expression in blood cell was confirmed by CyTOF using the same antibody panel. (Supplementary Fig. 13a).
Antibodies.
A high-throughput flow cytometric screen of 255 monoclonal antibodies against mouse cell surface markers was performed, using the LEGENDScreen™ Mouse PE Kit (Biolegend cat # 700005) (Supplementary Table 1). CNS tissues from healthy mice, EAE and HD mice were used for the initial screen. The CyTOF panel was assembled based on the high-throughput screen of 255 antibodies as well as proteins known to regulate myeloid cell functions (such as MerTK, Axl). transcription factors and signaling molecules known to be relevant in neuroinflammation.
A summary of antibodies used for each panel can be found in Supplementary Tables 2a–c, including their primary manufacturer, clone, corresponding metal conjugate, and final operating concentration. Antibodies were prepared in amounts varying from 100 to 500 μg at a time using the MaxPAR antibody conjugation kit (Fluidigm, Markham, ON, Canada) following the manufacturer’s protocol. After being labeled with their corresponding metal conjugate, the percent yield was determined by measuring their absorbance at 280nm using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE). Antibodies were diluted using Candor PBS Antibody Stabilization solution (Candor Bioscience GmbH, Wangen, Germany) to 0.3 mg/mL, and then stored at 4°C. Each antibody was titrated for optimal staining concentrations using primary murine samples and cell cultures.
Single-cell isolation.
Mice were deeply anesthetized and monitored. Upon the loss of nociceptive reflexes, animals were perfused transcardially with ice-cold PBS. Brains and spinal cords were removed and gently homogenized in cold HBSS (Life Technologies, 14175–095) on ice. Mononuclear cells were separated with a 30%/70% Percoll (GE Healthcare, Marlborough, MA) gradient centrifugation according to a previously reported protocol57.
Cell suspensions were washed in PBS with 2% FCS and 2 mM EDTA two times and were immediately fixed for 10 min at RT using a 1:1.4 proteomic stabilizer (in order to preserve the expression of signaling molecules) according to the manufacturer’s instructions (Smart Tube Inc., Palo Alto, CA) and frozen at −80 °C. Peripheral blood was collected via the retro-orbital prior to perfusion of the animal and transferred into sodium heparin-coated vacuum tubes 1:1 dilution in RMPI 1640 fixed immediately for 10 min at RT using 1:1.4 proteomic stabilizer according to the manufacturer’s instructions (Smart Tube Inc., Palo Alto, CA) and frozen at −80 °C. Sample preparation for cytokine analysis were performed as described previously24. Protein transporter inhibitor (ebioscience, 00–4980-93) was used to avoid the secretion of cytokines.
In each experiment, 10–12 mice were pooled in order to provide sufficient number of cells. Each experiment was repeated 7 to 10 times with separate immunization and cohort of mice.
Mass-Tag Cell Barcoding.
Samples from each condition were Mass-tag Cell Barcoded (MCB). In each sample a unique combination of six palladiumisotopes was used to encode 20 unique Mass-tag barcodes as previously described58,59. This technique allows all the samples to be pooled and stained within a single tube, eliminating tube-to-tube variability in antibody staining and minimizing the effect of variable instrument sensitivity. For each sample, 1.5 X 106 cells from each condition were barcoded. Methanol-permeabilized cells were washed once with Cell Staining Medium (CSM, PBS with 0.5% BSA, 0.02% NaN3) and then once with PBS. Different combinatorial mixtures of Palladium-containing MCB reagents in DMSO were then added to the individual samples at 1:100 DMSO with vortexing and then incubated at room temperature for 15 min, followed by three washes with CSM. The individual samples were then pooled for antibody staining and mass cytometry analysis. After data collection, each condition was deconvoluted using a mass cytometry debarcoding algorithm58.
Antibody Staining.
Barcoded cells were then resuspended in PBS with 0.5% BSA and 0.02% NaN3 and antibodies against CD16/32 were added at 20μg/ml for 10 min at RT on a shaker to block Fc receptors. Cells were stained with a cocktail of metal-conjugated surface marker antibodies (Supplementary Table 1), yielding 500 uL final reaction volumes and stained at room temperature for 30min at RT on a shaker. Following staining, cells were washed 2 times with PBS with 0.5% BSA and 0.02% NaN3. Next, cells were permeabilized with 4°C methanol for 10 min at 4°C. Cells were then washed twice in PBS with 0.5% BSA and 0.02% NaN3 to remove remaining methanol. Cells were then stained with intracellular antibodies (Supplementary Table 2b for signaling experiments and Supplementary Table 2c for cytokine experiments) in 500 μL for 30 min at RT on a shaker. Samples were then washed twice in PBS with 0.5% BSA and 0.02% NaN3. Cells were incubated overnight at 4 °C with 1 mL of 1:4000 191/193Ir DNA intercalator (DVS Sciences/Fluidigm, Markham, ON) diluted in PBS with 1.6% PFA overnight. The following day, cells were washed once with PBS with 0.5% BSA and 0.02% NaN3 and then two times with double-deionized water (ddH20).
Mass cytometry Measurement.
Prior to analysis, the stained and intercalated cell pellet was resuspended in ddH2O containing polystyrene normalization beads containing lanthanum-139, praseodymium-141, terbium-159, thulium-169 and lutetium-175 as described previously59. Stained cells were analyzed on a CyTOF 2 (Fluidigm, Markham, ON) outfitted with a Super Sampler sample introduction system (Victorian Airship & Scientific Apparatus, Alamo, CA) at an event rate of 200 to 300 cells per second. All mass cytometry files were normalized together using the mass cytometry data normalization algorithm freely available for download from www.cytobank.org/nolanlab.
Analysis.
Samples are labeled with iridium nucleic acid (intercalator 191/193Ir DNA intercalator) (DVS Sciences/Fluidigm, Markham, ON). For each event, many features are recorded, including signal duration (called Event Length) and iridium intensity. Single events have lower iridium intensity (since they have less DNA) and lower Event Length values compared to aggregates. These characteristics enable gating of single cells (Supplementary Fig. 13b). Live cells are identified on the basis of lack of c-PARP (cleaved poly-(ADP)-ribose polymerase (PARP)) binding as described before60 (Supplementary Fig. 13b).
1). Clustering:
Visualization with Minimum Spanning Tree: Minimum Spanning Tree (MST)61, creates a 2-dimensional layout, Differences in cell frequency of each subpopulation across different conditions are visualized by varying the size of each node proportionally to the frequency of the respective cluster in a given condition. Differences in marker expression levels across populations are visualized by coloring the nodes according to condition-specific marker expression levels (blue= no expression; green, yellow and red= increasing levels of expression.
The raw CyTOF data was subject to arsinh(x/5) transformation18. We selected cells from each sample which were then pooled together for clustering, generating a dataset with a total of 1,800,183 cells for the signaling dataset and 1,967,893 cells for the cytokine dataset. These datasets were clustered with a novel density-based clustering method known as X-shift. X-shift was developed to compute large multidimensional datasets and automatically determine the optimal number of clusters. In short, X-shift uses the weighted K-nearest neighbor density estimation to find the local maxima of data-point (cell event) density in the multidimensional marker space. X-shift computes the density estimate for each data point and then searches for the local density maxima in a nearest-neighbor graph, which become cluster centroids. All the remaining data points are then connected to the centroids via density-ascending paths in the graph, thus forming clusters. Finally, the algorithm checks for the presence of density minima on a straight line segment between the neighboring centroids, merging closely aligned clusters as necessary. In summary, cells were assigned to different populations based on the/a local gradient of cell event density in the marker expression space. Two cell populations counted as separate if cell density in any point on a straight line between centers of populations was lower than the density in the population centers. In other words, the peaks of cell event density that represent two populations must be separated by a cleft. Furthermore, clusters separated by a Mahalonobis distance of less than 2.0 were merged together. The optimal nearest neighbor parameter, K, was chosen to be 70 in a data-driven manner, by finding the elbow-point of the plot of the number of clusters over K. All data processing was performed with the VorteX clustering environment (http://web.stanford.edu/~samusik/vortex).
2). Divisive Marker Tree (DMT) for gating:
In order to facilitate back-gating of X-shift clustered populations we organized the clusters into a Divisive Marker Tree (DMT)18. The DMT algorithm constructs a binary decision tree that starts with a root node encompassing all clusters; this set of clusters is then subject to iterative binary division. This process results in a hierarchical binary classification of cell types that resembles manual gating hierarchies. By tracing the sequence of marker divisions from the root, we were able to infer a concise marker-based signature for each cell population that differentiates it from other populations.
CD49e (α5 integrin) treatment.
Prophylactic administration of anti-CD49e antibody (Figs. 5b):
EAE mice (n = 5 per group) were treated daily with 200 ug of CD49e (α5 integrin) antibody (Clone= 5H10–27(MFR5)) or rat IgG2a, kappa isotype control (low endotoxin, azide-Free antibody, custom-made by Biolegend for this experiment). Treatment with antibody and isotype was initiated on day 0 after immunization via intraperitoneal injection.
EAE treatment with anti-CD49e antibody (Figs. 5c):
EAE mice (n = 10 per group) were treated daily with 200 ug of CD49e (α5 integrin) antibody (Clone= 5H10–27(MFR5)) or rat IgG2a, kappa isotype control (low endotoxin, azide-Free antibody, custom-made by Biolegend for this experiment). Treatment with antibody and isotype was initiated once immunized mice developed paralysis (representing clinical EAE) on day15.
EAE scores were assessed daily for clinical signs of EAE in a blinded fashion without knowing which mouse was receiving treatments. Mice were assessed daily and scored according to: 0, no clinical disease; 1, tail weakness; 2, hindlimb weakness; 3, complete hindlimb paralysis; 4, hindlimb paralysis and some forelimb weakness; 5, moribund or dead. The experiment was concluded due to high morbidity of control mice.
Statistics.
No statistical methods were used to pre-determine sample sizes, but our sample sizes are similar to those reported in previous publications62. Mice were randomized such that both treatment groups had the same average disease score. We did not exclude any animals for data point from the analysis. Data collection were randomized. Data distribution was assumed to be normal but this was not formally tested. Data collection were not performed blind to the conditions of the experiments. Analysis was performed blind to the conditions of the experiments.
Manny-Whitney one-tailed test was utilized for the CD49e treatment studies of EAE, comparing between isotype antibody control and CD49e antibody treatments. Cell population frequency data were completed with Prism GraphPad7 and shown as box-and-whisker charts where the center line is the mean, and boxes extend to 5th and 95th percentile; whiskers extend to 1.5x interquartile range.
Life Sciences Reporting Summary.
Further information on experimental design is available in the Life Sciences Reporting Summary.
Code availability.
All data processing performed with the VorteX clustering environment called X-shift algorithm can be found at http://web.stanford.edu/~samusik/vortex.
Data availability.
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements:
We thank Dr. Oleg Butovsky for generous gift of 4D4 and FCRLS antibody. We thank A. Trejo and A. Jager for mass cytometry quality control and maintenance; S. Douglas and V. Giangarra for technical assistance. This work was further supported by grants to GPN: U19 AI057229, 1U19AI100627, Department of Defense (CDMRP), Northrop-Grumman Corporation, R01CA184968, 1R33CA183654-01, R33CA183692, 1R01GM10983601, 1R21CA183660, 1R01NS08953304, OPP1113682, 5UH2AR067676, 1R01CA19665701, R01HL120724. GPN is supported by the Rachford & Carlotta A. Harris Endowed Chair. B.A was supported by a three-year postdoctoral fellowship from the Canadian Institute for Health Research (201102MFE-246400-166230).
Footnotes
Competing interests:
The authors declare no competing financial interest.
References
- 1.Prinz M & Priller J The role of peripheral immune cells in the CNS in steady state and disease. Nat Neurosci 20, 136–144, doi: 10.1038/nn.4475 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Steinman L Immunology of relapse and remission in multiple sclerosis. Annu Rev Immunol 32, 257–281, doi: 10.1146/annurev-immunol-032713-120227 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Heppner FL, Ransohoff RM & Becher B Immune attack: the role of inflammation in Alzheimer disease. Nat Rev Neurosci 16, 358–372, doi: 10.1038/nrn3880 (2015). [DOI] [PubMed] [Google Scholar]
- 4.McGeer PL, Itagaki S, Boyes BE & McGeer EG Reactive microglia are positive for HLA-DR in the substantia nigra of Parkinson’s and Alzheimer’s disease brains. Neurology 38, 1285–1291 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Crotti A et al. Mutant Huntingtin promotes autonomous microglia activation via myeloid lineage-determining factors. Nat Neurosci 17, 513–521, doi: 10.1038/nn.3668 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao W, Beers DR & Appel SH Immune-mediated mechanisms in the pathoprogression of amyotrophic lateral sclerosis. J Neuroimmune Pharmacol 8, 888–899, doi: 10.1007/s11481-013-9489-x (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aguzzi A, Barres BA & Bennett ML Microglia: scapegoat, saboteur, or something else? Science 339, 156–161, doi: 10.1126/science.1227901 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graeber MB Neuroinflammation: no rose by any other name. Brain Pathol 24, 620–622, doi: 10.1111/bpa.12192 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers J et al. Inflammation and Alzheimer’s disease pathogenesis. Neurobiol Aging 17, 681–686 (1996). [DOI] [PubMed] [Google Scholar]
- 10.Sudduth TL, Schmitt FA, Nelson PT & Wilcock DM Neuroinflammatory phenotype in early Alzheimer’s disease. Neurobiol Aging 34, 1051–1059, doi: 10.1016/j.neurobiolaging.2012.09.012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang B et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell 153, 707–720, doi: 10.1016/j.cell.2013.03.030 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bendall SC et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696, doi: 10.1126/science.1198704 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mangiarini L et al. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87, 493–506 (1996). [DOI] [PubMed] [Google Scholar]
- 14.Solomon JN et al. Origin and distribution of bone marrow-derived cells in the central nervous system in a mouse model of amyotrophic lateral sclerosis. Glia 53, 744–753, doi: 10.1002/glia.20331 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Karpuj MV et al. Prolonged survival and decreased abnormal movements in transgenic model of Huntington disease, with administration of the transglutaminase inhibitor cystamine. Nat Med 8, 143–149, doi: 10.1038/nm0202-143 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Fourgeaud L et al. TAM receptors regulate multiple features of microglial physiology. Nature 532, 240–244, doi: 10.1038/nature17630 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cardona AE, Huang D, Sasse ME & Ransohoff RM Isolation of murine microglial cells for RNA analysis or flow cytometry. Nat Protoc 1, 1947–1951, doi: 10.1038/nprot.2006.327 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Samusik N, Good Z, Spitzer MH, Davis KL & Nolan GP Automated mapping of phenotype space with single-cell data. Nat Methods 13, 493–496, doi: 10.1038/nmeth.3863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brendecke SM & Prinz M Do not judge a cell by its cover--diversity of CNS resident, adjoining and infiltrating myeloid cells in inflammation. Semin Immunopathol 37, 591–605, doi: 10.1007/s00281-015-0520-6 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Butovsky O et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci 17, 131–143, doi: 10.1038/nn.3599 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldmann T et al. A new type of microglia gene targeting shows TAK1 to be pivotal in CNS autoimmune inflammation. Nat Neurosci 16, 1618–1626, doi: 10.1038/nn.3531 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Wlodarczyk A, Lobner M, Cedile O & Owens T Comparison of microglia and infiltrating CD11c(+) cells as antigen presenting cells for T cell proliferation and cytokine response. J Neuroinflammation 11, 57, doi: 10.1186/1742-2094-11-57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim H et al. Increased phosphorylation of cyclic AMP response element-binding protein in the spinal cord of Lewis rats with experimental autoimmune encephalomyelitis. Brain Res 1162, 113–120, doi: 10.1016/j.brainres.2007.05.072 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Liu F & Whitton JL Cutting edge: re-evaluating the in vivo cytokine responses of CD8+ T cells during primary and secondary viral infections. J Immunol 174, 5936–5940 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Darrah PA et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med 13, 843–850, doi: 10.1038/nm1592 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Steinman L A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat Med 13, 139–145, doi: 10.1038/nm1551 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Rosen DR Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 364, 362, doi: 10.1038/364362c0 (1993). [DOI] [PubMed] [Google Scholar]
- 28.Ajami B, Bennett JL, Krieger C, McNagny KM & Rossi FM Infiltrating monocytes trigger EAE progression, but do not contribute to the resident microglia pool. Nat Neurosci 14, 1142–1149, doi: 10.1038/nn.2887 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Ajami B, Bennett JL, Krieger C, Tetzlaff W & Rossi FM Local self-renewal can sustain CNS microglia maintenance and function throughout adult life. Nat Neurosci 10, 1538–1543, doi: 10.1038/nn2014 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Mildner A et al. Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat Neurosci 10, 1544–1553, doi: 10.1038/nn2015 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Salama AD et al. Critical role of the programmed death-1 (PD-1) pathway in regulation of experimental autoimmune encephalomyelitis. J Exp Med 198, 71–78, doi: 10.1084/jem.20022119 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reiman R, Gerard C, Campbell IL & Barnum SR Disruption of the C5a receptor gene fails to protect against experimental allergic encephalomyelitis. Eur J Immunol 32, 1157–1163, doi: (2002). [DOI] [PubMed] [Google Scholar]
- 33.Lu JQ, Power C, Blevins G, Giuliani F & Yong VW The regulation of reactive changes around multiple sclerosis lesions by phosphorylated signal transducer and activator of transcription. J Neuropathol Exp Neurol 72, 1135–1144, doi: 10.1097/NEN.0000000000000011 (2013). [DOI] [PubMed] [Google Scholar]
- 34.Yamasaki R et al. Differential roles of microglia and monocytes in the inflamed central nervous system. J Exp Med 211, 1533–1549, doi: 10.1084/jem.20132477 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Cesare D, Jacquot S, Hanauer A & Sassone-Corsi P Rsk-2 activity is necessary for epidermal growth factor-induced phosphorylation of CREB protein and transcription of c-fos gene. Proc Natl Acad Sci U S A 95, 12202–12207 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ginhoux F & Merad M [Microglia arise from extra-embryonic yolk sac primitive progenitors]. Med Sci (Paris) 27, 719–724, doi: 10.1051/medsci/2011278013 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Le Varlet B, Staquet MJ, Dezutter-Dambuyant C, Delorme P & Schmitt D In vitro adhesion of human epidermal Langerhans cells to laminin and fibronectin occurs through beta 1 integrin receptors. J Leukoc Biol 51, 415–420 (1992). [DOI] [PubMed] [Google Scholar]
- 38.Sobel RA & Mitchell ME Fibronectin in multiple sclerosis lesions. Am J Pathol 135, 161–168 (1989). [PMC free article] [PubMed] [Google Scholar]
- 39.Boroujerdi A, Welser-Alves JV & Milner R Extensive vascular remodeling in the spinal cord of pre-symptomatic experimental autoimmune encephalomyelitis mice; increased vessel expression of fibronectin and the alpha5beta1 integrin. Exp Neurol 250, 43–51, doi: 10.1016/j.expneurol.2013.09.009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estes ML & McAllister AK Alterations in immune cells and mediators in the brain: it’s not always neuroinflammation! Brain Pathol 24, 623–630, doi: 10.1111/bpa.12198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salter MW & Beggs S Sublime microglia: expanding roles for the guardians of the CNS. Cell 158, 15–24, doi: 10.1016/j.cell.2014.06.008 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Obrietan K & Hoyt KR CRE-mediated transcription is increased in Huntington’s disease transgenic mice. J Neurosci 24, 791–796, doi: 10.1523/JNEUROSCI.3493-03.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bannerman PG et al. Motor neuron pathology in experimental autoimmune encephalomyelitis: studies in THY1-YFP transgenic mice. Brain 128, 1877–1886, doi: 10.1093/brain/awh550 (2005). [DOI] [PubMed] [Google Scholar]
- 44.Steinman L Multiple sclerosis: a two-stage disease. Nat Immunol 2, 762–764, doi: 10.1038/ni0901-762 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Silvestroni A, Faull RL, Strand AD & Moller T Distinct neuroinflammatory profile in post-mortem human Huntington’s disease. Neuroreport 20, 1098–1103, doi: 10.1097/WNR.0b013e32832e34ee (2009). [DOI] [PubMed] [Google Scholar]
- 46.Zhou Z, Peng X, Insolera R, Fink DJ & Mata M Interleukin-10 provides direct trophic support to neurons. J Neurochem 110, 1617–1627, doi: 10.1111/j.1471-4159.2009.06263.x (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ransohoff RM A polarizing question: do M1 and M2 microglia exist? Nat Neurosci 19, 987–991, doi: 10.1038/nn.4338 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Mildner A et al. CCR2+Ly-6Chi monocytes are crucial for the effector phase of autoimmunity in the central nervous system. Brain 132, 2487–2500, doi: 10.1093/brain/awp144 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Steinman L The discovery of natalizumab, a potent therapeutic for multiple sclerosis. J Cell Biol 199, 413–416, doi: 10.1083/jcb.201207175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yednock TA et al. Prevention of experimental autoimmune encephalomyelitis by antibodies against alpha 4 beta 1 integrin. Nature 356, 63–66, doi: 10.1038/356063a0 (1992). [DOI] [PubMed] [Google Scholar]
Methods-only references
- 51.Linthicum DS, Munoz JJ & Blaskett A Acute experimental autoimmune encephalomyelitis in mice. I. Adjuvant action of Bordetella pertussis is due to vasoactive amine sensitization and increased vascular permeability of the central nervous system. Cell Immunol 73, 299–310 (1982). [DOI] [PubMed] [Google Scholar]
- 52.Hofstetter HH, Shive CL & Forsthuber TG Pertussis toxin modulates the immune response to neuroantigens injected in incomplete Freund’s adjuvant: induction of Th1 cells and experimental autoimmune encephalomyelitis in the presence of high frequencies of Th2 cells. J Immunol 169, 117–125 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Hou W et al. Pertussis toxin enhances Th1 responses by stimulation of dendritic cells. J Immunol 170, 1728–1736 (2003). [DOI] [PubMed] [Google Scholar]
- 54.Chen X et al. Pertussis toxin as an adjuvant suppresses the number and function of CD4+CD25+ T regulatory cells. Eur J Immunol 36, 671–680, doi: 10.1002/eji.200535353 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weber MS et al. Repetitive pertussis toxin promotes development of regulatory T cells and prevents central nervous system autoimmune disease. PLoS One 5, e16009, doi: 10.1371/journal.pone.0016009 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldmann T et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol 17, 797–805, doi: 10.1038/ni.3423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pino PA & Cardona AE Isolation of brain and spinal cord mononuclear cells using percoll gradients. J Vis Exp, doi: 10.3791/2348 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zunder ER et al. Palladium-based mass tag cell barcoding with a doublet-filtering scheme and single-cell deconvolution algorithm. Nat Protoc 10, 316–333, doi: 10.1038/nprot.2015.020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finck R et al. Normalization of mass cytometry data with bead standards. Cytometry A 83, 483–494, doi: 10.1002/cyto.a.22271 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fienberg HG, Simonds EF, Fantl WJ, Nolan GP & Bodenmiller B A platinum-based covalent viability reagent for single-cell mass cytometry. Cytometry A 81, 467–475, doi: 10.1002/cyto.a.22067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiu P et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 29, 886–891, doi: 10.1038/nbt.1991 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spitzer MH et al. IMMUNOLOGY. An interactive reference framework for modeling a dynamic immune system. Science 349, 1259425, doi: 10.1126/science.1259425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.