Abstract
Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) transcripts involves the mutually exclusive usage of exons IIIb and IIIc to produce two different receptor isoforms. Appropriate splicing of exon IIIb in rat prostate cancer DT3 cells requires a previously described cis element (ISAR, for “intronic splicing activator and repressor”) which represses the splicing of exon IIIc and activates the splicing of exon IIIb. This element is nonfunctional in rat prostate AT3 cells, which repress exon IIIb inclusion and splice to exon IIIc. We have now identified an intronic element upstream of exon IIIb that causes repression of exon IIIb splicing. Deletion of this element abrogates the requirement for ISAR in order for exon IIIb to be spliced in DT3 cells and causes inappropriate inclusion of exon IIIb in AT3 cells. This element consists of two intronic splicing silencer (ISS) sequences, ISS1 and ISS2. The ISS1 sequence is pyrimidine rich, and in vitro cross-linking studies demonstrate binding of polypyrimidine tract binding protein (PTB) to this element. Competition studies demonstrate that mutations within ISS1 that abolish PTB binding in vitro alleviate splicing repression in vivo. Cotransfection of a PTB-1 expression vector with a minigene containing exon IIIb and the intronic splicing silencer element demonstrate PTB-mediated repression of exon IIIb splicing. Furthermore, all described PTB isoforms were equally capable of mediating this effect. Our results support a model of splicing regulation in which exon IIIc splicing does not represent a default splicing pathway but rather one in which active repression of exon IIIb splicing occurs in both cells and in which DT3 cells are able to overcome this repression in order to splice exon IIIb.
Alternative splicing represents a commonly used pathway through which different gene products can be produced from a single gene. In many cases of alternative splicing, the splicing pattern is tightly regulated such that distinct cell types differentially splice a given pre-mRNA to produce different protein isoforms. cis-acting elements which, when located in an exon, can act either to activate or repress splicing have been identified and characterized (13, 19, 33, 36, 51, 55, 56, 60–62, 66). In addition, intron sequences which either activate or block splicing of adjacent exons have been described (reference 6 and references therein; 9, 11, 45, 53). In more than one case, it has been demonstrated that both positive and negative regulatory cis elements are present within a single alternatively spliced transcript (6, 8, 11, 45). Pre-mRNA splicing is known to take place in the spliceosome, a large multicomponent enzymatic machine which consists of the U1, U2, U4/6, and U5 small nuclear RNAs (snRNAs) along with associated small nuclear ribonucleoproteins (snRNPs) and non-snRNP proteins (3, 59). The mechanisms which operate to direct this spliceosomal apparatus to yield alternatively spliced RNAs have been poorly defined in mammalian systems to date (59). Well-described examples of cell-specific factors in Drosophila which can act positively or negatively to alter the splicing of specific exons have been proposed to be models for alternative splicing in mammals (reviewed in reference 40). Nonetheless, such purely cell-specific factors have not been identified in mammals, and ongoing debate centers around the question whether analogous cell-specific alternative splicing factors will be found to modulate the processing of mammalian gene transcripts. It has been proposed that mammals have adapted mechanisms which rely on relative differences in the levels of multiple factors, which regulate pre-mRNA splicing in a combinatorial manner (28, 45).
A number of nonspliceosomal proteins that are not tissue restricted are capable of altering the splicing of a number of different pre-mRNA substrates. Several SR protein family members bind exonic enhancer sequences to increase the inclusion of the corresponding exon (33, 35, 36, 51, 54, 55). In addition, SR proteins have differential effects on splice site selection. ASF/SF2, for example, promotes the use of a proximal 5′ splice site upstream of a defined 3′ splice site, an effect which can be counteracted by heterogeneous nuclear RNP A1 (hnRNPA1) (4, 17, 20, 39). Two other hnRNPs, hnRNP F and hnRNP H, are components of a complex that forms on a neural cell-specific intronic enhancer element, resulting in the increased splicing of the N1 exon of c-src (11, 43). KH-type splicing regulatory protein (KSRP) is a component of this complex, although its expression, like that of hnRNP F and hnRNP H, is not neural cell specific (44). In contrast to its role in activating the splicing of the N1 exon, hnRNP H binds to an exonic splicing silencer in β-tropomyosin and has been proposed to cause the exclusion of exon 7 in nonmuscle cells (9).
Polypyrimidine tract binding protein (PTB) was originally purified based on its ability to bind to an adenovirus polypyrimidine tract and was subsequently also described as hnRNP-I (2, 18, 21, 22, 47). A role for PTB in alternative splicing was first proposed by Mulligan et al. studying the alternative splicing of β-tropomyosin transcripts (46). They demonstrated that mutations within cis elements upstream of the skeletal muscle-specific exon 7 of the β-tropomyosin pre-mRNA resulted in its inclusion in HeLa cells in vivo, and these mutations were demonstrated to disrupt the binding of PTB in vitro. The propensity of PTB to bind to stretches of pyrimidines has led to the hypothesis that it may compete with the splicing factor U2AF65 for the polypyrimidine tract upstream of a regulated exon, thus causing skipping of that exon by blocking the recognition of the branch point from the splicing machinery (52). This would appear to be the case in certain regulated pre-mRNAs such as β-tropomyosin and γ-aminobutyric acid type A (GABAA) γ2 receptor (1, 27, 46, 65). It has been demonstrated, however, for α-tropomyosin and recently for c-src, that PTB binding sites also reside outside of the branch point-associated polypyrimidine tract (bp/py) (8, 12, 24, 25, 48). In both of these cases, PTB binding sites are located on both sides of the regulated exon, giving rise to a potentially more complex mechanism of exon repression. This has been elegantly demonstrated in the repression of the N1 exon in nonneural cell types, where PTB binding to an upstream conserved CUCU motif is dependent on its ability to bind to a downstream CUCU located within an intronic enhancer element termed the downstream control sequence (12).
PTB, like many other factors implicated in alternative splicing, is not expressed in a strictly tissue-specific fashion. We have previously identified a switch in the ratio of the major PTB isoforms in DT3 and AT3 cell lines but have not found any functional difference between the isoforms (57). In cases of neural cell-specific splicing, the overall level of expression of PTB is lower than in nonneural cell types (1, 8, 65). This could potentially allow for a shift to splicing inclusion by counteractive enhancing factors; however, recently it has been shown that a different form of PTB enriched in the neural tissues, termed brain PTB, can inhibit the ability of Nova-1, a neural cell-specific alternative splicing factor, to increase the inclusion of GlyRα2 exon E3A (50). Furthermore, it has been suggested that the CUG binding protein may counteract PTB in neural cells (65). In vitro studies on the silencer element upstream of exon 7 of the β-tropomyosin pre-mRNA suggest that the interaction of PTB with other cellular cofactors may be RNA dependent (26). It is thus possible that the levels of proteins that interact with PTB may be expressed only in certain tissues.
Alternative splicing of rat fibroblast growth factor receptor 2 (FGF-R2) transcripts, in which a mutually exclusive splicing choice produces two forms of the C-terminal half of the third extracellular immunoglobulin-like domain, results in two different receptors with distinct ligand binding profiles (Fig. 1A) (7, 30–32, 41). The mutually exclusive exons, IIIb and IIIc, are 148 and 145 nucleotides, respectively, and the choice of either exon is highly specific for a given cell type. We have utilized cell lines derived from the rat Dunning prostate cancer model to study the splicing of FGF-R2 transcripts (6). The isoform of FGF-R2 that results when exon IIIb is used, FGF-R2 (IIIb), is the exclusive gene product observed in normal prostatic epithelia and in a well-differentiated, androgen-dependent DT3 (or DT-E) prostate tumor or cell line. A poorly differentiated and androgen-independent AT3 tumor exhibits a change in alternative splicing that results in loss of FGF-R2 (IIIb) and in expression of FGF-R2 (IIIc). The markedly different growth factor specificities of the resulting receptors alter signal transduction pathways, which have been proposed to be involved in the progression of prostate cancer (5, 16, 37, 63, 64).
FIG. 1.
Schematic representation of the alternative splice variants of FGF-R2. (A) Diagram showing the relative positions of exons IIIb and IIIc in the context of a protein domain map of FGF-R2. Abbreviations denote the immunoglobulin-like domains (Ig 1 to Ig 3), transmembrane domain (TM), and both intracellular tyrosine kinase domains (TK). (B) The AT3 and DT3 cells splice to the mutually exclusive exons IIIc and IIIb, respectively. In DT3 cells, the previously described cis element, ISAR, functions both to activate IIIb inclusion and to repress IIIc inclusion. In AT3 cells, there has been no detected function for ISAR, and so IIIc is included regardless of the presence of ISAR.
Studies of rat and human FGF-R2 splicing have revealed the presence of several cis elements, which can alter the splicing of mutually exclusive alternative exons IIIb and IIIc. We have previously characterized ISAR (for “intronic splicing activator and repressor”), a cis element located in the intron between exons IIIb and IIIc of the rat gene. This element dramatically increases the splicing efficiency of the upstream IIIb exon in DT3 cells and represses the splicing of the downstream IIIc exon (6) (Fig. 1B). ISAR was not shown to have an effect on the relative inclusion of either exon in AT3 cells (Fig. 1B). A highly homologous sequence from human FGF-R2, IAS-3, was likewise shown to increase exon IIIb splicing, together with two other intronic elements, IAS-1 and IAS-2 (13, 23). A putative secondary structure involving interactions between IAS-2 and IAS-3 has been proposed to be involved in mediating the effects of these cis elements (13). However, no specific proteins that bind to these elements have been identified which appear to mediate effects on alternative splicing of these exons. An exonic splicing silencer element has also been identified in exon IIIb, and it has been suggested that hnRNP A1 may play a role in mediating the repression of exon IIIb splicing (14).
In the present study, we characterize an element that causes profound repression of FGF-R2 exon IIIb inclusion. Deletion or substitution of an intronic region upstream of exon IIIb results in nearly complete inclusion of exon IIIb in both DT3 and AT3 cells. In AT3 cells, deletion of this element results in a spliced RNA that contains both exons IIIb and IIIc. In DT3 cells, deletion of this element allows efficient inclusion of exon IIIb even in the absence of ISAR. Further analysis of this silencer demonstrates two separate regions, ISS1 and ISS2, that together exert most of the repressive effects on IIIb splicing. One of these subelements, ISS1, contains a pyrimidine-rich sequence that binds to PTB; deletion, replacement, or mutations of ISS1 cause the loss of PTB binding. The same sequence disruptions cause the loss of splicing repression in vivo. In addition, we have used cotransfection of an FGF-R2 minigene, which contains this silencer element, together with a PTB expression vector to demonstrate increasing repression of exon IIIb with progressive increases in PTB protein expression. In addition, this PTB-mediated repression was observed with all described PTB splice variants. This study suggests that FGF-R2 exon IIIb may be constitutively repressed by PTB in both AT3 and DT3 cells but that this repression is overcome in DT3 cells by factors that interact with ISAR and possibly other cis elements.
MATERIALS AND METHODS
Plasmid construction.
The plasmid DNA constructs used in this study were all made using standard cloning techniques such as those described previously (6). The pI-11 and pI-11-FS splicing constructs were obtained as described previously (6). Plasmids pI-11-FS:+ISS+ISAR and pI-11-FS:+ISS−ISAR were previously described as pI-11-FS ΔBclI/NdeI and pI-11-FS ΔBclI/NsiI, respectively (6). Plasmids pI-11-IIIb:+ISS+ISAR and pI-11-IIIb:+ISS−ISAR were heretofore referred to as pI-11-IIIb-plus and pI-11-IIIb-minus, respectively (6). Plasmids pI-11-IIIb:−ISS+ISAR and pI-11-IIIb:−ISS−ISAR were obtained by PCR amplification with primers Int3BFS and Int2R2 from templates pI-11-FS:+ISS+ISAR and pI-11-FS:+ISS−ISAR, respectively. The sequence of Int 3BFS is 5′-ccggactagttccttcctggttggccgtta-3′, and primer Int2R2 was described in a past publication (6). These PCR products were digested with SpeI and XhoI and cloned into the XbaI and XhoI sites in the intron of pI-11. Plasmids pI-11-FS:−ISS+ISAR and pI-11-FS:−ISS−ISAR were obtained in a similar manner except that the PCR primers used were Int3BFS and Int3CR (the latter primer also as previously described). Plasmids pI-11-IIIb(−ISAR):Δ1, pI-11-IIIb(−ISAR):Δ2, pI-11-IIIb(−ISAR):Δ3, and pI-11-IIIb(−ISAR):Δ4 were obtained following amplification with forward primers 3BF-One, 3BF-Two, 3BF-Three, and 3BF-Four and reverse primer Int 2R2 using template pI-11-FS:+ISS−ISAR. These products were also digested with SpeI and XhoI and cloned into the XbaI and XhoI sites of pI-11. The sequences of these forward primers are as follows: 3BF-One, 5′-ccggactagtctcattgtgatctcctccct-3′; 3BF-Two, 5′-ccggactagtcagctctttaggtgcaattca-3′; 3BF-Three, 5′-ccggactagtgattgttttcttgtggtgtg-3′; and 3BF-Four, 5′-ccggactagttggtgggaccataggcagca-3′. Full-length PTB1 isoform cDNA was subcloned from pGEX2TK-PTB1 into the mammalian expression vector pcDNA3-HisA (Invitrogen) utilizing the EcoRI restriction sites. The vector pcDNA3-HisC-LacZ was provided as a control by Invitrogen. Plasmids pDP-RC-ΔEX-ISS1, pDP-RC-ΔEX-ISS1mt, and pDP-RC-ΔEX-Globin were used for the synthesis of hot probes ISS1, ISS1mt, and Globin, respectively. These constructs were created by cloning annealed oligonucleotides F-WT and R-WT (for ISS1), F-MT and R-MT (for ISS1mt), and F-BG and R-BG (for Globin) into the EcoRI and SpeI sites of pDP-RC-ΔEX. pDP-RC-ΔEX was created using pDP19 (Ambion) but replacing the entire polylinker between the EcoRI and HindIII sites of this vector with the sequence 5′-tagaactagtgcggccgcatatcatcgatgctcgag-3′. Oligonucleotide sequences were as follows: F-WT, 5′-aattctcattgtgatctcc tccctcccacagctctttaggtgtaa-3′; R-WT, 5′-ctagttacacctaaagagct gtgggagggaggagatcacaatgag-3′; F-MT, 5′-aattctcattgtgatggagaaa ggaccacagctctttaggtgtaa-3′; R-MT, 5′-ctagttacacctaaagag ctgtggtcctttctccatcacaatgaG-3′; F-BG, 5′-aattggagaccaatagaaa ctgggcatgtggagacataggtgtaa-3′; R-BG: 5′-ctagttacacctatgtctcc acatgcccagtttctattggtctcc-3′.
PCR amplification and RT-PCR assay of transfected minigenes.
PCR from DNA templates for plasmid construction was done using standard reaction conditions as described previously (6). RNA for reverse transcription-PCR (RT-PCR) assays was isolated using the method of Chomczynski and Sacchi (10). RT-PCR using T7 and SP6 primers to analyze pooled stable transfections was performed as described previously (6). When RT-PCR was used to assay results from transient cotransfections, the RNAs were first treated with RQ1 DNase (Promega), as specified by the manufacturer, to eliminate background from residual plasmid DNA templates. Also, in this case, T7 was used with primer PIP11R, which corresponds to the sequence at the 3′ end of the second exon of pI-11: 5′-ccggactagtaagcttaggctcttggcgtt-3′. In all amplification reactions, a water control and a mock RT control were included, which resulted in no PCR product in all experiments. PCR products were either loaded directly onto 5% nondenaturing polyacrylamide gels or, when necessary, added to restriction endonuclease digestions with either AvaI or HincII (New England Biolabs). We always observed complete digestion when using this method. Aliquots representing equal amounts of each PCR mixture with undigested and digested PCR products were loaded onto 5% polyacrylamide gels. The gels were electrophoresed at 100 V for 3 to 4 h, dried, and exposed to Amersham Hyperfilm-MP or Molecular Dynamics phosphorimager screens. Analysis was performed with a Molecular Dynamics PhosphorImager. Quantifications were performed as follows. For DT3 and AT3 cells (see Fig. 2B and C), we quantified the percentages of the spliced products that contained both exons IIIb and IIIc (U-IIIb-IIIc-D), those that contained either exon IIIb or exon IIIc (U-IIIb/IIIc-D), and those that skipped exons IIIb and IIIc and spliced the adenovirus exons together (U-D) by adding the values for the bands representing these products from the undigested RT-PCR lanes (corrected for molar equivalents) and representing each as a percentage of the total (where U and D are the 5′ and 3′ exons, respectively, of pI-11). In addition, data were quantified to assess the percentage of the single-inclusion product that contained IIIb. This was done by using the quantification of the band at 380 bp, which remained following HincII digestion (U-IIIb-D), as the numerator. The denominator consisted of U-IIIb-D and the 377-bp band that remained following AvaI digestion (U-IIIc-D). Quantification of experiments using minigenes with only one internal exon (exon IIIb) (see Fig. 3B and C) was determined as the quantification of the 380-bp band (U-IIIb-D) divided by the sum of the same band and the exon IIIb-skipped 232-bp band. Quantifications for Fig. 6C were done by dividing the amount of IIIb inclusion (as calculated for Fig. 3) at 500 ng of cotransfected HisG-PTB by the amount of IIIb inclusion resulting from cotransfection with empty vector. This normalized percentage of IIIb inclusion was then subtracted from 100 to obtain a normalized percentage of IIIb skipping. Error bars represent the normalized error of triplicate cotransfections. Quantifications in Fig. 6D were done by averaging a set of triplicate transient cotransfections and setting the amount of IIIb inclusion in the vector-alone lane equal to 100; all other values are normalized to that value. In all cases, products of different sizes were corrected for molar equivalents.
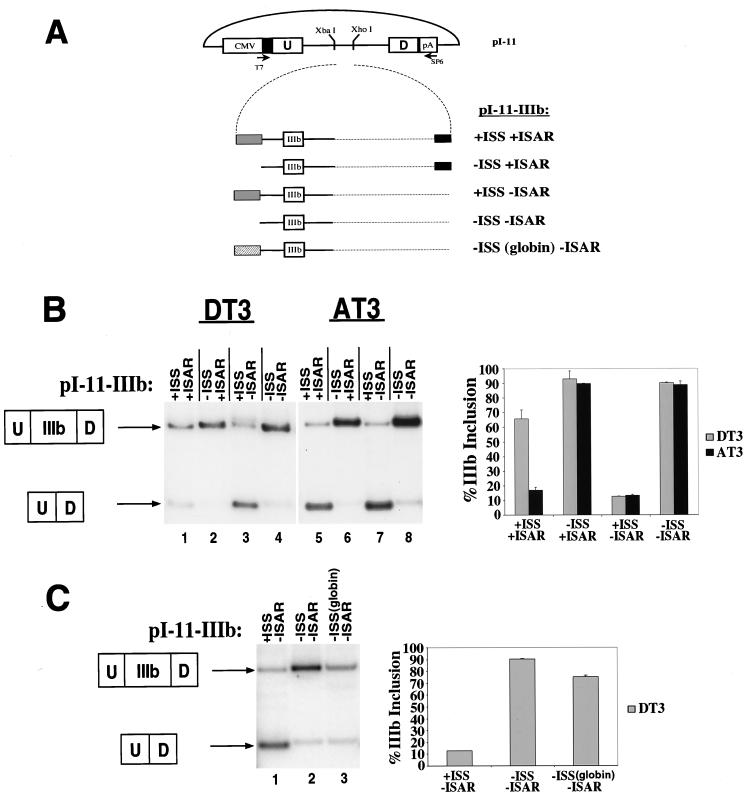
FIG. 2.
Identification of an ISS upstream of exon IIIb. (A) pI-11 (top) is a previously described adenovirus two-exon splicing construct adapted for eukaryotic expression. FGF-R2 genomic sequences were cloned into the XbaI and XhoI sites located upstream of the branch point sequence. CMV indicates the cytomegalovirus promoter, and pA indicates the bovine growth hormone polyadenylation sequence. The locations of the T7, SP6, and PIP11R primers are indicated. The FGF-R2 genomic sequences of pI-11-FS-derived minigenes are indicated at the bottom, together with the locations of the primers and restriction sites used to generate the four constructs shown, and are described in Results. Solid lines indicate intron sequences, open boxes indicate exons, and light dotted lines indicate intron sequences deleted during construction of the minigenes. The gray shaded box indicates the location of the ISS element, and the black box indicates ISAR. (B) Deletion of an intronic silencer element, ISS, increases exon IIIb inclusion in DT3 cells. Results from stable transfections and RT-PCR assays are indicated in the top panel. The uncut lanes (U) were used to quantify the amount of each type of splicing event and are tabulated in the center panel. The error is the standard deviation of the average of three separate stable transfections. The bottom panel is a quantification of the percentage of single-inclusion products that include exon IIIb. This is quantified by comparison of the IIIb-specific AvaI digestion (A) with the IIIc-specific HincII digestion (H). Deletion of ISS dramatically increases exon IIIb splicing. In DT3 cells, when ISS is located upstream of exon IIIb, efficient exon IIIb splicing requires ISAR. However, deletion of ISS results in an increase in exon IIIb inclusion irrespective of the presence of ISAR. (C) Deletion of ISS results in predominant splicing of both exon IIIb and exon IIIc in AT3 cells. In AT3 cells, no role for ISAR is seen. When ISS is present, the predominant product is nearly exclusively splicing to exon IIIc, as quantified in the bottom panel, although a significant amount of double-inclusion products is seen. When ISS is deleted, nearly all the products consist of both exons IIIb and IIIc, reflecting derepression of exon IIIb. Although a minimal amount of single-inclusion product is still seen, this product now also consists of a fivefold-greater amount of exon IIIb-containing product.
FIG. 3.
Deletion of ISS increases exon IIIb inclusion in both DT3 and AT3 cell lines when tested in the absence of exon IIIc. (A) Outline of the constructs used that contain exon IIIb. Designations are the same as in Fig. 2A, except that here a hatched box is added to indicate substitution of the β-globin intron sequence. (B) The boxed figures at the left indicate the two expected products in which either IIIb is included or exon IIIb is skipped and the upstream (U) and downstream (D) adenovirus exons are spliced. Quantifications graphed in the right panel were performed as described in Materials and Methods. (C) Substitution of β-globin intron sequence has the same effect as deletion of ISS.
FIG. 6.
Overexpression of PTB in DT3 cells leads to a reduction in exon IIIb inclusion. (A) Transient transfection of a HisG-tagged CMV-driven full-length PTB1 expression plasmid results in high levels of PTB expression in DT3 cells. Western analysis was performed following transfection with empty vector or 0.5, 1.0, or 2.0 μg of PTB. The gels were probed for nuclear protein CA150, as a loading control, HisG, or PTB as indicated. The formation of the PTB triplet seen at higher levels of overexpression could represent a degradation product, an internal translation start site, or an effect on endogenous PTB alternative splicing. (B) Transient transfection of progressively increasing amounts of HisG-PTB (in nanograms) results in progressively decreased amounts of IIIb inclusion. (C) Comparison of PTB-mediated repression using 500 ng of HisG-PTB in the presence (+ISS) or absence (ΔISS) of ISS. The results are normalized to represent the percentage of IIIb skipping relative to that in cells transfected with empty vector. (D) Quantified averaged experiment of a set of triplicate transient cotransfections of the three PTB splice variants with a minigene that contains ISS. Exon IIIb inclusion has been normalized by making the vector-alone level of inclusion equal to 100 and then representing all other values as a percentage of that amount. Averaged values of triplicate experiments are shown, and the standard deviation was less than 10% for all data points. These splice variants were HA tagged and lacked the first 56 amino acids; however, full-length splice variants yielded identical curves (not shown). (E) Transfection of the same expression plasmid expressing HisG-tagged LacZ reveals high levels of expression of an unrelated protein by immunoblotting. (F) Cotransfection of LacZ does not alter the level of exon IIIb inclusion when cotransfected with pI-11-IIIb:+ISS+ISAR.
Cell culture and transfection.
Stable transfections in AT3 and DT3 cells were performed as described previously (6). Transient cotransfections were performed with 50 ng of pI-11-IIIb:+/−ISS+ISAR as a test construct and the indicated amounts of expression plasmids for PTB and LacZ. The amount of plasmid DNA used in each transfection was kept constant by using empty plasmid PCDNA3.1-HisA to equalize the amount of expression plasmid in each experiment. The test minigene construct and expression construct were mixed and added to 100 μl of OptiMEM (Gibco). To this was added 5 μl of Lipofectamine (Gibco) premixed with 100 μl of OptiMEM. The samples were incubated at room temperature for 15 min, after which an additional 800 μl of OptiMEM was added, and the transfection mixture was placed over cells in six-well plates. The transfection mixtures were left on the cells for 2 h, after which the transfection mixture was replaced with Dulbecco modified Eagle medium with 10% fetal bovine serum. At 24 h from the start of transfections, total RNA was harvested for RT-PCR analysis.
UV cross-linking, immunoprecipitation, and competition assays.
UV cross-linking and immunoprecipitation were done essentially as previously described, unless otherwise noted (57). UV cross-linking and competition studies were done using 20 μg of DT3 nuclear extracts. Direct cross-linking experiments were performed using hot RNA probes generated with T7 polymerase and plasmids pDP-RC-ΔEX-ISS1, pDP-RC-ΔEX-ISS1mt, and pDP-RC-ΔEX-Globin after linearization with NotI. For these experiments, binding-reaction mixtures were supplemented with 0.01 mg of heparin (Sigma) per ml and 0.01 mg of tRNA (Sigma) per ml as nonspecific competitors, incubated at 30°C for 20 min, and cross-linked as described below. Cold competitor RNA was in vitro transcribed, gel purified, and incubated in nuclear extract for 8 min at 30°C under conditions previously described (57). Transcription of all other probes was done using templates generated from the PCR by using pI-11-IIIb as a template in all reactions. The ISS1 PCR template was generated by using the forward and reverse primers F-5′-ggatcctaatacgactcactatagggagactcattgt gatctcctccctcc-3′ and R-5′-acctacactgctgtg-3′, respectively. The ISS1mut was generated by using the same reverse primer but a different forward primer: F-5′-ggatcctaatacgactcactatagggagactcattgtgatggagaaaggaccacagctctttaggt-3′. It is important to note that the ISS1 and ISS1mt cold competitors contained an additional 2 nucleotides at the 3′ end of the transcript. In addition, the β-globin sequence used for cold competition was nearly twice as long as that used in direct cross-linking. The reason for this minor discrepancy is to have a nonspecific competitor that was the average size of all the different cold competitors used. Sec RNA was in vitro transcribed, and 15 fmol (100,000 cpm) was added to each preincubated reaction mixture and further incubated for 15 min at 30°C. The reaction mixtures were then put onto ice and UV cross-linked in a Stratagene Stratalinker using two pulses of 500 mJ each. RNase A was added to a final concentration of 0.1 mg/ml, and the reaction mixtures were incubated at 37°C for 30 min. An equivalent volume of 2× sodium dodecyl sulfate (SDS) loading buffer was added to each reaction mixture, and the products were further analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (12.5% polyacrylamide). Gels were run until the bromophenol blue dye front had reached the bottom of the gel, and then they were dried and exposed on Hyperfilm-MP. The nuclear extracts from AT3 and DT3 cells were made by following a previously described modification of the original protocol used for HeLa nuclear extracts (15, 42). However, because DT3 cells could not be grown in suspension, large-volume DT3 monolayers were grown in roller bottles, harvested with trypsin, and washed three times with phosphate-buffered saline (PBS) prior to subsequent preparative steps.
Western blot analysis.
All cotransfections done in tissue culture were performed in duplicate, with one sample used for Western analysis. Transfected cells were harvested and cell lysates were prepared as described previously (57). Protein lysates were quantified using the Bradford assay, and 25 μg of each lysate was analyzed by SDS-PAGE (12.5% polyacrylamide). For LacZ transfections, 15 μg of lysate was loaded onto a gradient SDS-PAGE minigel (Bio-Rad). The protein was electrophoresed at 100 V for 5 h and transferred overnight onto Immobilon-P polyvinylidene difluoride membranes. The membranes were blocked for 5 h at room temperature with a blocking buffer (PBS, 5% dry milk, 0.1% Tween 20), probed for 2 h at room temperature with respective antibodies diluted in blocking buffer (1:2,000 PTB antiserum, 1:5,000 HisG monoclonal antibody, and 1:2,000 anti-CA150 antibodies), washed three times for 10 min in wash buffer (PBS, 0.1% Tween 20), and incubated for 1 h at room temperature with the respective HPer-conjugated secondary antibodies (Amersham-Pharmacia). The blots were again washed three times for 10 min with wash buffer, and the signal was detected using Hyperfilm-MP.
RESULTS
Deletion of an intronic region upstream of exon IIIb results in derepression of splicing of this exon in DT3 and AT3 cells.
We used stable transfection of minigenes derived from rat FGF-R2 genomic sequences to characterize the cis elements required for regulated splicing in DT3 and AT3 cells as described previously (6). Briefly, genomic FGF-R2 sequences were placed within the intron of an adenovirus-derived splicing construct, pI-11 (Fig. 2A). Splicing of exons IIIb and/or IIIc was assayed by RT-PCR using T7 and SP6 primers (Fig. 2A). Because exon IIIb contains an AvaI site not present in exon IIIc and exon IIIc contains two HincII sites not present in exon IIIb, we digested the RT-PCR products to identify minigene transcripts which contain either exon IIIb or IIIc. When our original minigenes containing exons IIIb and IIIc were transfected into DT3 and AT3 cells, we observed a predominant product that consisted of either exon IIIb or exon IIIc spliced between the adenovirus upstream (U) exon and downstream (D) exon, respectively. In subsequent descriptions, this product will be designated the single-inclusion product. However, we also observed some products that contained both exon IIIb and exon IIIc (designated U-IIIb-IIIc-D, henceforth designated the double-inclusion product) as well as some products in which the adenovirus exons were directly ligated (U-D, henceforth designated the skipped product). A minigene, pI-11-FS:+ISS+ISAR, recapitulated the splicing pattern of the endogenous gene. When this minigene was transfected into DT3 cells, the single-inclusion product (Fig. 2A) consisted nearly exclusively of exon IIIb; transfection into AT3 cells resulted in the single-inclusion product consisting nearly entirely of exon IIIc (Fig. 2B and C, lanes 1 to 3). However, when ISAR was deleted, the resulting minigene, pI-11-FS:+ISS−ISAR, demonstrated a dramatic decrease in exon IIIb splicing in DT3 cells: the single-inclusion product now consisted mostly of exon IIIc, and an increase in the amount of the skipped product was also noted (Fig. 2B, compare lanes 1 to 3 with lanes 7 to 9 [quantified in the chart and graph]) (6). Thus, the function of ISAR is dual; it activates the splicing of the upstream IIIb exon and represses the splicing of the downstream exon IIIc. To adequately quantify the effect of these deletions on splicing in DT3 cells, we present the data in Fig. 2B in two ways. First, using bands from the undigested lanes, we quantify the percentage of double-inclusion products (U-IIIb-IIIc-D), single-inclusion products (U-IIIb/IIIc-D), and skipped products (U-D). We also graphically present the percentage of single-inclusion products that contain exon IIIb. In AT3 cells, ISAR had no observable effect; exon IIIb was present in less than 10% of the single-inclusion products and the levels of double-inclusion products and skipped products did not significantly change (Fig. 2C, compare lanes 1 to 3 with lanes 7 to 9 [quantified in the chart and graph]).
We created additional minigenes in which we deleted 121 nucleotides from the intron upstream of exon IIIb (Fig. 2A). As a result, the intron between the adenovirus exon and exon IIIb was shortened from 206 to 85 nucleotides, still including 40 nucleotides derived from the adenovirus intron. In DT3 cells, deletion of this region from a minigene which still contained ISAR (pI-11-FS:−ISS+ISAR) resulted in an increase in exon IIIb inclusion in the single-inclusion product from 77 to 91% (Fig. 2B, compare lanes 1 to 3 with lanes 4 to 6 [quantified in the graph]). Deletion of this region from a minigene without ISAR (pI-11-FS:−ISS−ISAR) also resulted in an increase in the single-inclusion IIIb product, from 39 to 75% (Fig. 2B, compares lanes 7 to 9 with lanes 10 to 12 [quantified in the graph]). It can also be seen that deleting ISAR in a minigene that lacks ISS results in an increase in double-inclusion products from 12.8 to 28.8%, consistent with the ability of ISAR to repress IIIc splicing. This effect of ISAR appears to be independent of the sequences upstream of exon IIIb. In AT3 cells, the predominant product contains exon IIIc and there are very low levels of exon IIIb-containing product regardless of the presence of ISAR (Fig. 2C, compare lanes 1 to 3 with lanes 7 to 9 [quantified in the graph]). However, when the intronic region upstream of exon IIIb was deleted, either with or without ISAR, the levels of double-inclusion products increased to 81.9 and 82.9%, respectively (Fig. 2C, lanes 4 and 10 [quantified in the chart]). Although the most obvious effect of this deletion in AT3 cells was a switch toward nearly exclusive splicing to yield the double-inclusion product, the small amount of single-inclusion product also showed a fivefold increase in exon IIIb inclusion, as demonstrated graphically (Fig. 2C). Thus, the net effect of this sequence appears to be to repress, or silence, the splicing of exon IIIb in both DT3 and AT3 cells while having no apparent effect on IIIc splicing. Based on these findings, we refer to this element as an intronic splicing silencer (ISS).
Deletion or substitution of ISS in minigenes containing only exon IIIb results in high levels of IIIb inclusion.
To simplify the study of the ISS element, we also studied the effect of deleting the ISS using FGF-R2 minigenes in which exon IIIc and flanking intron sequences are not included in the minigene (Fig. 3A). This minimal construct recapitulates the endogenous splicing pattern in both cell lines. When we transfected a minigene in which ISAR is present in the intron downstream of exon IIIb into DT3 cells (pI-11-IIIb:+ISS+ISAR [Fig. 3A]), exon IIIb was included in 68% of the spliced mRNAs, compared to products in which exon IIIb is skipped and the adenovirus exons are spliced (Fig. 3B, lane 1). When a minigene without ISAR was transfected into DT3 cells (pI-11-IIIb:+ISS−ISAR [Fig. 3A]), exon IIIb was almost completely skipped (Fig. 3B, lane 3). However, when the element upstream of exon IIIb, ISS, was deleted, exon IIIb was spliced nearly exclusively whether or not ISAR was present downstream (Fig. 3B, lanes 2 and 4). In AT3 cells, in which exon IIIb is nearly completely skipped regardless of the presence of ISAR (Fig. 3B, lanes 5 and 7), deletion of this element likewise dramatically increased exon IIIb inclusion (lanes 6 and 8).
A possible explanation for the results of deleting ISS is that the resultant shortening of the intron between the upstream exon and exon IIIb causes a non-sequence-specific effect on exon IIIb splicing by bringing the exons closer together. To investigate this possibility, we replaced the ISS sequence with an unrelated intron sequence derived from a β-globin intron 2 (Fig. 3C). Replacement of the ISS with this sequence resulted in 78% inclusion of exon IIIb compared to 89% inclusion when the ISS was deleted. Thus, deletion of the ISS resulted in an 8.5-fold increase in exon IIIb inclusion and replacement with the β-globin intronic sequence resulted in a 6.5-fold increase in exon IIIb inclusion (Fig. 3C [quantified in the graph]). While the replacement was slightly less effective than the deletion in increasing exon IIIb inclusion, these results suggest that the predominant effect of the deletion of ISS is due to the loss of specific sequence information.
These findings allowed us to refine our view of the requirement of ISAR for IIIb inclusion in DT3 cells. We previously suggested that exon IIIc inclusion represents a “default” splicing choice based on its stronger polypyrimidine tract than that of exon IIIb and that ISAR is required to overcome this weak polypyrimidine tract. These studies suggest that ISAR is required to derepress a non-cell-specific repression, which is exerted on exon IIIb in addition to the weakness of the neighboring splice sites. While a weaker polypyrimidine tract may play a role in regulated FGF-R2 splicing, it can be seen that it does not prevent highly efficient IIIb splicing when the intronic silencer element is deleted. In addition, the previous description of an exonic splicing silencer (ESS) within human exon IIIb suggests that this may also contribute to regulation, but again, this sequence alone clearly does not prevent efficient exon IIIb splicing when the upstream ISS element is deleted (14).
Sequential deletions of the ISS reveal two separate regions, ISS1 and ISS2, that are necessary for the observed splicing repression.
To further define the critical elements within the 121 nucleotides that encompass the ISS, we created minigenes with sequential deletions from the 5′ end of the ISS within a minigene that contained exon IIIb but had ISAR deleted (Fig. 4A). Because ISAR was not present downstream of exon IIIb, we were able to more easily assess the changes in IIIb silencing with progressive loss of sequence from this element. The resulting minigenes were transfected into DT3 cells, and the level of exon IIIb inclusion was determined. Whereas the Δ1 deletion had no appreciable effect on IIIb inclusion, the Δ2 deletion resulted in a twofold increase of the IIIb-containing product (Fig. 4B). Further deletions (including Δ3 and Δ4) resulted in no further significant increase in exon IIIb inclusion. We concluded that an element, which we named ISS1, has its 5′ border somewhere within the sequence deleted in Δ2 (Fig. 4). A second element was uncovered by deletion of the 3′-most end of ISS, which led to another near-twofold increase in IIIb inclusion (Fig. 4B, compare Δ4 with −ISS). Again we can assume that this second element, ISS2, has a 5′ border within this region of ISS. While the ISS2 sequence does not obviously resemble any previously described splicing silencers, it is conserved between rat and human FGF-R2 and has numerous UGG repeats. Analysis of the sequence comprising ISS1 was notable for the presence of a pyrimidine-rich region with some resemblance to previously described sequences that bind PTB (1, 48, 52). Since PTB has been implicated in the repression of splicing to adjacent exons in other mammalian systems, we sought to explore whether PTB might interact with ISS1 and, further, whether this interaction might be involved in repression of exon IIIb splicing.
FIG. 4.
Deletions of ISS sequence from the 5′ end uncover the existence of two silencer elements, ISS1 and ISS2. (A) A minigene that lacked ISAR was used as the backbone for a series of deletion mutants denoted Δ1 through Δ4. The relative positions of the two subelements are denoted as demonstrated in the lower panel. (B) Transfection of minigenes with progressive shortening of ISS reveals two regions with most of the splicing silencing effect. The percentage of IIIb inclusion is represented at the bottom. Sequences deleted in the Δ2 deletion mutant as well the sequences between Δ4 and −ISS displayed the most profound increase in exon IIIb inclusion.
PTB binds to sequences within ISS1 but not to other sequences comprising the silencer element upstream of exon IIIb.
To characterize specific PTB binding sequences within this region of the FGF-R2 pre-mRNA, we performed successive deletions within the RNA followed by cross-linking and immunoprecipitation using anti-PTB antibodies (data not shown). In particular, we found that PTB efficiently cross-linked to the ISS1 sequence in both DT3 and AT3 nuclear extracts (diagrammed in Fig. 5A and demonstrated in Fig. 5B, lanes 2 and 6). Immunoprecipitation of the cross-link using PTB antiserum confirmed its identity (data not shown). When the pyrimidine stretch was altered to contain a stretch of purines (ISS1mt), PTB cross-linking was greatly reduced (Fig. 5B, lanes 3 and 7). A size-matched nonspecific RNA derived from intron 2 of the human β-globin gene was also seen not to cross-link to PTB (lanes 4 and 8). All three probes cross-linked to a similar set of background bands that bound without specificity, unlike PTB, which depends on the presence of the pyrimidine stretch within ISS1. Although there is a canonical UCUU just downstream of the pyrimidine stretch, it is not sufficient for PTB cross-linking. The Sec RNA consists of an RNA sequence which has previously been shown to efficiently bind PTB and is used as a positive control for PTB cross-linking (lanes 1 and 5) (57).
FIG. 5.
ISS1 binds PTB and is required for repression of exon IIIb. (A) Diagram of the labeled RNAs used in these cross-linking studies. The pyrimidine stretch within ISS1 is highlighted in bold, while the purine mutations within that stretch in ISS1mt are underlined. The Globin sequence is an intronic sequence derived from the human β-globin gene. (B) Direct UV cross-linking of the ISS1, ISS1mt, and Globin RNAs in both DT3 and AT3 nuclear extracts. The Sec RNA is used as a positive control for PTB cross-linking. In all three cases, there are several bands common to all three RNAs; however, the PTB doublet cross-links specifically to the ISS1 and Sec RNAs, and not to the ISS mutant or substitution. The molecular weight markers (in thousands) are indicated at the left, and the cross-linked PTB bands are bracketed. (C) UV cross-linking and competition experiments demonstrated that PTB binding to the Sec RNA can be competed using self-competition or using a cold ISS1 competitor. However, the nonspecific globin competitor and ISS1mt cannot compete PTB cross-linking to the Sec RNA. For each panel, DT3 extracts were preincubated with the indicated molar fold excess of cold competitor RNA, after which the labeled Sec control probe was added prior to cross-linking. Molecular weights (in thousands) are indicated at the left, and PTB bands are bracketed. (D) Deletion, mutation, or replacement of ISS1 which abolishes PTB binding also results in the loss of exon IIIb splicing repression. These constructs were stably transfected in triplicate into DT3 cells, and the average percentage of exon IIIb inclusion is indicated.
Because direct UV cross-linking of protein to RNA does not always correlate precisely with binding, we used cross-linking and competition experiments to address the specificity of PTB binding to ISS1. We used the Sec RNA probe as the control PTB binding sequence in all competition experiments because it gave a cross-linking profile consisting primarily of the PTB doublet; however, in these experiments, lower heparin concentrations were used than in the direct cross-linking experiments, and thus other cross-linked proteins are more observable. We used self-competition with an unlabeled Sec RNA to define the levels of competition, which correlated with specific RNA binding (Fig. 5C). As a negative control, the nonspecific β-globin intron 2 RNA was likewise used to compete for binding to the radioactively labeled Sec probe (Fig. 5C). Thus, as expected, competition with unlabeled Sec RNA exhibited a dose-dependent reduction of PTB binding whereas the nonspecific β-globin RNA did not compete with binding of PTB to the labeled Sec probe. Of note, while PTB cross-linking was not decreased with the nonspecific competitor, we observed that the intensity of other, presumably less efficiently and specifically cross-linked proteins, was progressively decreased. As demonstrated, the ISS1 RNA sequence specifically competed PTB binding to Sec (Fig. 5C). When the nine consecutive pyrimidines within the ISS1 sequence were changed to purines (Fig. 5A) to yield the competitor sequence ISS1mt, competition for PTB binding was eliminated (Fig. 5C). Thus, these cross-linking competition studies verify that PTB binds in vitro to a region within the intronic splicing silencer that mediates splicing repression in vivo. These data additionally verify that the tract of polypyrimidines within ISS1 is indeed required for binding of PTB to this region of the FGF-R2 pre-mRNA.
Mutations or replacement in the upstream PTB binding region that abolish PTB binding result in a corresponding loss of repressor function on IIIb splicing in vivo.
The in vitro RNA binding studies demonstrated that PTB was capable of binding to the ISS1 region of the FGF-R2 pre-mRNA. To further investigate whether PTB binding is involved in mediating the observed exon IIIb repression, we tested the same sequence changes which caused a loss of PTB binding in vitro for the ability to disrupt exon IIIb repression in vivo. As shown in Fig. 5D, deletion, replacement, and mutation of ISS1 resulted in a loss of exon IIIb splicing repression in vivo. It should be pointed out that these constructs were all identical in size, ruling out the possibility that the effect of the deletion of ISS1 increased IIIb inclusion simply through shortening of this intron. Although ISS1mut did not enhance exon IIIb inclusion as well as deletion or replacement of the entire ISS1 sequence did, there was a clearly evident and reproducible decrease in splicing repression. The quantifications are representative of triplicate stable transfections. These data suggest that PTB binding to the ISS1 element is involved in decreased splicing efficiency of exon IIIb.
Overexpression of PTB in DT3 cells causes increased skipping of exon IIIb.
We anticipated that if exon IIIb inclusion in DT3 cells is dependent on the ability of ISAR and a factor(s) bound to it to overcome exon IIIb repression, which could be due to PTB, overexpression of the repressor might overwhelm the effect of ISAR. This would lead to decreased exon IIIb inclusion. To test this hypothesis, we cotransfected DT3 cells with increasing amounts of a plasmid expressing HisG-PTB1 and a constant amount of a minigene containing exon IIIb as well as ISS and ISAR (pI-11-IIIb:+ISS+ISAR). As seen in Fig. 6A, we were able to transiently overexpress HisG-PTB1. The same blot was probed for the HisG tag, which detects exogenous PTB, and for all PTB species by using a polyclonal PTB antiserum. When PTB was coexpressed with pI-11-IIIb:+ISS+ISAR, a decrease in the amount of IIIb inclusion product was observed as well as an equivalent increase in the amount of IIIb-skipped product (data not shown). Maximal repression of IIIb inclusion products occurred with 500 ng of HisG-PTB plasmid, and increasing the amounts of this expression plasmid above this level resulted in only modest increases in IIIb repression. A dose-dependent reduction in exon IIIb inclusion was observed from 125 to 500 ng of transfected DNA (Fig. 6B). This demonstrates that moderate PTB overexpression can overwhelm the activation pathway of IIIb.
To address whether this effect of PTB involved an interaction with ISS, we cotransfected 500 ng of HisG-PTB with minigenes that either contained the ISS (pI-11-IIIb:+ISS+ISAR) or had the ISS deleted (pI-11-IIIb:−ISS+ISAR). To compare the effects of PTB on the two different minigenes, normalized values of IIIb inclusion and exclusion were calculated. There was nearly a 3.5-fold increase in the ability of PTB to repress IIIb inclusion when the ISS was present versus when it was deleted (Fig. 6C). The fact that there was some degree of repression of IIIb even though the ISS was deleted was not completely unexpected (see Discussion).
Given that we have previously reported a change in the PTB isoform expression in DT3 and AT3 cells, we reasoned that perhaps the difference in splicing patterns could be attributed to the difference in the ability of the three PTB splice variants to repress exon IIIb inclusion. When the different PTB isoforms were overexpressed, very similar levels of exon IIIb repression were seen (Fig. 6D). This suggests that the isoforms do not differ in their repressor activity. It does not rule out, however, that the isoforms are differentially responsive to the derepression brought about by ISAR. We are currently investigating this possibility.
As a control, an equivalent amount of HisG-LacZ was transiently transfected into DT3 cells, resulting in a level of LacZ protein overexpression similar to that of PTB (Fig. 6E). There was no effect on IIIb inclusion at any level of LacZ overexpression (Fig. 6F). These experiments suggest that increasing the levels of PTB can counteract the activation effects of ISAR in DT3 cells and that this repression is dependent on the intronic splicing silencer.
DISCUSSION
In the present study, we have identified an intronic element upstream of exon IIIb in the FGF-R2 transcript which appears to exert repression, or silencing, of splicing to exon IIIb in both the DT3 and AT3 cell lines. While it is possible that cell-specific differences in the level of repression mediated by this sequence may contribute to splicing regulation, we were not able to appreciate such differences. Previous work of our laboratory as well as of others studying the human FGF-R2 gene has characterized several elements that affect the splicing of FGF-R2 exons IIIb and IIIc. An element within exon IIIb consisting of a critical UAGG sequence has been shown to inhibit the splicing of this exon and has thus been termed an exonic splicing silencer (ESS) (19). Counteracting this silencing of exon IIIb splicing are several intronic elements downstream of exon IIIb that are required to activate the splicing of exon IIIb in cells which include it (6, 13, 23). The role of one of these elements, ISAR, has further been shown to include repression of splicing to the downstream exon IIIc (6). In addition, there is evidence that other elements which are as yet not well defined are also required for regulation of FGF-R2 splicing (reference 6 and this study). The RNA element described here consists of two sequences, ISS1 and ISS2, which together exhibit potent splicing repressor activity. ISS1 contains a binding site for PTB and was shown in UV cross-linking and immunoprecipitation studies to bind PTB in both DT3 and AT3 cells. Competition studies showed that the binding is specific, since mutations which disrupt the polypyrimidine regions of ISS1 did not effectively compete PTB binding to the control RNA. We have furthermore shown that mutations that abolish PTB binding to this element in vitro also cause derepression of exon IIIb splicing in vivo. Thus, these results strongly suggest that interaction of PTB with a portion of this element is involved in the general repression of exon IIIb splicing.
Recently, it has been shown that depletion of PTB from HeLa nuclear extracts using either iterative oligonucleotide adsorption or immunodepletion can effectively derepress the splicing of the normally excluded SM exon of α-actinin in vitro as well as the N1 exon of c-src, respectively (12, 53). This effect was further shown to be reversed by the addition of recombinant PTB or purified PTB, respectively. It has also been recently reported that in three different neural-specific exons, a splicing switch could be induced in HeLa nuclear extracts by the addition of a cold RNA competitor containing cis elements that bind PTB (65). Conversely, the activation pathway of these exons in neural cell-derived extracts could be overwhelmed by the addition of recombinant GST-PTB1. In addition, it was shown that PTB purified from HeLa cells was capable of reversing a cold RNA competitor-induced derepression of the N1 exon in HeLa nuclear extracts (8). Collectively, these in vitro results suggest that PTB may play a prominent role in the regulation of a wide variety of alternatively spliced pre-mRNAs by repressing splicing in certain regions of the pre-mRNA transcript. Overexpression of PTB in vivo has been shown to promote exon inclusion of exon 4 of the CT/CGRP transcript, but these data are the first to demonstrate the repression of exon inclusion in vivo (34). In our in vivo experiments, repression was most robust when PTB was cotransfected with a minigene that contained the ISS, although some repression was still observed when this sequence was deleted. Analysis of sequences in the intron downstream of exon IIIb reveals a number of sequences, including seven UCUU motifs, which may also represent PTB binding sites (1, 48, 52). When we transfected a minigene that contained the entire ISS region and ISAR but lacked the downstream putative PTB binding sites into DT3 and AT3 cells, we noted that exon IIIb was included in at least 90% of spliced RNAs (data not shown). This high level of exon IIIb inclusion was seen both in the presence and absence of the ISS, although deletion of the ISS did result in a small increase in IIIb inclusion. These results suggest the presence of a sequence downstream of exon IIIb that can also act to silence splicing of exon IIIb. Based on these preliminary results, we suspect that the effect of PTB to decrease the efficiency of exon IIIb splicing may also involve the interaction of PTB at sequences in the downstream intron. Based on the results of other investigators, the existence of multiple PTB binding sites near an alternatively spliced exon is not unprecedented (12). Such is the case in splicing of exon 3 of α-tropomyosin and exon N1 of c-src, in which PTB binding to sequences both upstream and downstream of this alternative exon results in splicing repression (25, 48). Splicing repression by the Sxl protein in Drosophila has likewise been shown to involve interactions on both sides of a repressed exon (29). Further study is under way to characterize the elements downstream of exon IIIb, which are also likely to be involved in this repression.
In addition to the portion of ISS that binds PTB, the downstream region within this silencer, ISS2, appears to play a significant role in repressing splicing of exon IIIb. At present we have not identified any specific proteins which interact directly with this region. Analysis of the sequence would suggest that PTB does not bind to this portion of the repressor, and in fact the cross-linking competition studies have not demonstrated direct PTB binding to this region. Other hnRNP proteins are candidates for interacting with this region. Given the G-rich nature of this sequence, hnRNP H, which exhibits a preference for binding to G-rich RNA sequences, is one candidate. In fact, hnRNP H has previously been implicated in splicing repression of exon 7 of β-tropomyosin (9).
The mechanism by which PTB may function to repress splicing is at present poorly characterized. It has been suggested that PTB may prevent the binding of U2AF to the polypyrimidine tract of associated exons similar to the predicted mechanism that Sxl uses to repress the male-specific exon of the transformer pre-mRNA (52). However, this may be true only in some cases. Among the observations that question this general hypothesis are that the binding sites for Sxl and PTB are not always overlapping with the bp/py and have been shown to sometimes reside hundreds of nucleotides away (8, 29, 48). In the case of Sxl splicing autoregulation, the binding sites that are within the bp/py are less important than those located in the intron downstream of the male-specific exon (29). Furthermore, PTB binding downstream of N1 affects its ability to bind upstream, suggesting that PTB cooperativity plays a role in repression (12). There are no proteins that interact with PTB that have also been demonstrated to be functionally relevant for its role in alternative splicing. However, several proteins, including the FUSE-binding protein and the Sam68 tyrosine phosphoprotein, have been shown to exist in a complex with PTB assembled on a PTB binding sequence upstream of the repressed exon 7 of β-tropomyosin (26). Furthermore, like Sxl, PTB has been shown to interact with itself and to exist as a dimer in solution, raising the possibility that the dimerization of these proteins is a necessary event for interaction with other potential proteins and their repression of exon inclusion (49, 58). This is a logical possibility, given the existence of PTB or Sxl binding sites clustered on either side of certain regulated exons.
As a group, hnRNP proteins are known to rapidly associate with hnRNAs cotranscriptionally, and, in addition to packaging of RNA transcripts, these proteins may be involved in regulating which regions of the pre-mRNA are accessible to splicing factors and the spliceosome (38). However, formal evidence for such regulation by hnRNPs, as well as an understanding of the degree to which sequence-specific binding of RNAs by these proteins (other than PTB/hnRNP I) plays a role in maintaining splicing fidelity, is currently lacking. Certainly, these proteins could play a role not only in repressing splicing but also in activating splicing. hnRNP F and hnRNP H are required factors of an intronic splicing enhancer binding complex (11, 43, 44). In addition, the ability of hnRNPA1 to promote the use of a proximal 5′ splice site could be due to direct activation of a specific splicing pathway (4).
Study of mechanisms of alternative splicing in mammals has been hampered by the observation that, in examples studied to date, an increasing number of cis elements regulate the splicing of a given gene transcript. In many cases, positive and negative regulatory elements have been identified in the same gene transcript. Dissection of these elements and the factors that interact with them has not yet provided a definitive mechanism through which tissue-specific splicing is achieved. While several trans-acting factors have been identified that are necessary for splicing regulation, thus far no cell-type-specific factors have been identified which account for the observed differences in splicing of a given transcript. When more than one regulatory element is involved, the splicing pattern seen in a given study appears to represent an additive effect of the different elements. While some of these elements appear to exert their effects on splicing in a cell-specific manner, other elements have been observed that have non-cell-specific effects on splicing that appear to set a balance for cell-specific factors to shift. While a combinatorial model of splicing regulation might account for tissue specificity based on differences in the levels or activity of generally expressed factors interacting with several positive and negative elements, the existence of purely cell-specific factors which regulate alternative splicing is still a possibility.
In our previous study, we identified an element termed ISAR, which appears to function in a cell-specific manner in DT3 cells to activate the splicing of exon IIIb and repress the splicing of exon IIIc. We previously proposed that exon IIIc splicing may represent a “default” splicing pathway and that exon IIIb splicing was inefficient due to a weak polypyrimidine tract and possibly an exonic splicing silencer. Therefore, it was proposed that activation of IIIb splicing in DT3 cells probably depended on activation of splicing by intronic enhancer sequences downstream of exon IIIb. The present study, however, implies that this model is an oversimplification. While the strength of the exon IIIb polypyrimidine tract may be involved, there are clearly other cis elements that can independently repress the splicing of this exon. We suggest a model in which the regulated splicing of exons IIIb and IIIc in DT3 and AT3 cells represents a balance between positive and negative regulators of splicing (Fig. 7). Elucidation of the mechanism by which several regulatory cis elements are differentially recognized and processed during pre-mRNA splicing will require further characterization of the protein factors that interact with them.
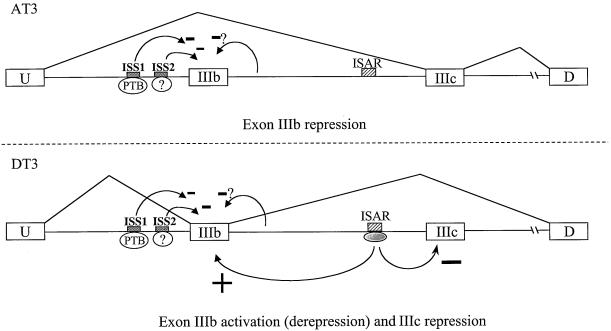
FIG. 7.
Model in which a combination of positive and negative effects leads to regulation of FGF-R2 alternative splicing. We propose that AT3 cells splice IIIc efficiently and skip exon IIIb as a result of the effect of ISS that is mediated in part by PTB. In DT3 cells, the same exon IIIb splicing silencing activity is present; however, it is counteracted by exon IIIb splicing activation, which includes factors that interact with ISAR. In addition, factors that interact with ISAR contribute to the repression of splicing of exon IIIc. Shaded boxes indicate ISS1 and ISS2 elements. Involvement of PTB with ISS1 is shown. Silencing effects, which involve ISS2 as well as less well characterized sequences downstream of exon IIIb are also proposed. The location and activity of ISAR and undefined protein(s) that bind to it are also shown.
ACKNOWLEDGMENTS
R. P. Carstens and E. J. Wagner have contributed equally to this manuscript and should therefore be considered co-first authors.
We thank members of the Garcia-Blanco laboratory for general advice and assistance. In particular, we thank Aaron Goldstrohm and George Pitoc for review and suggestions on the manuscript. We thank David Helfman and Chris Smith for providing antibodies and plasmids. We also thank Wallace L. McKeehan for providing the cell lines used in this study and for continued collaboration and support.
R.P.C. was supported by Public Health Service grant K08 CA72560-01 from the NCI. E.J.W. was supported by an NIH training grant per the CMB program, Duke University Medical Center. This work was supported by a grant from the American Cancer Society to M.A.G.-B. M.A.G.-B. was also supported by an Established Investigator Award from the American Heart Association.
REFERENCES
- 1.Ashiya M, Grabowski P. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- 2.Bothwell A, Ballard D, Philbrick W, Lindwall G, Maher S, Bridgett M, Jamison S, Garcia-Blanco M A. Murine polypyrimidine tract binding protein. J Biol Chem. 1991;266:24657–24633. [PubMed] [Google Scholar]
- 3.Burge C, Tuschl T, Sharp P. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland R, editor. The RNA world. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- 4.Caceres J, Stamm S, Helfman D, Krainer A. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science. 1994;265:1706–1709. doi: 10.1126/science.8085156. [DOI] [PubMed] [Google Scholar]
- 5.Carstens R, Eaton J, Krigman H, Walther P, Garcia-Blanco M. Alternative splicing of fibroblast growth factor receptor 2 (FGF-R2) in human prostate cancer. Oncogene. 1997;15:3059–3065. doi: 10.1038/sj.onc.1201498. [DOI] [PubMed] [Google Scholar]
- 6.Carstens R, McKeehan W, Garcia-Blanco M. An intronic sequence element mediates both activation and repression of rat fibroblast growth factor receptor 2 pre-mRNA splicing. Mol Cell Biol. 1998;18:2205–2217. doi: 10.1128/mcb.18.4.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champion-Arnaud P, Ronsin C, Gilbert E, Gesnel M, Breathnach R. Multiple mRNAs code for proteins related to the BEK fibroblast growth factor. Oncogene. 1991;6:979–987. [PubMed] [Google Scholar]
- 8.Chan R, Black D. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen C, Kobayashi R, Helfman D. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat beta-tropomyosin gene. Genes Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Chou M, Rooke N, Turck C, Black D. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol Cell Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou M, Underwood J, Nikolic J, Luu M, Black D. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol Cell. 2000;5:949–957. doi: 10.1016/s1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- 13.DelGatto F, Plet A, Gesnel M, Fort C, Breathnach R. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol Cell Biol. 1997;17:5106–5116. doi: 10.1128/mcb.17.9.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DelGatto-Konczak F, Olive M, Gesnel M, Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol Cell Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dignam J, Lebovitz R, Roeder R. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng S, Wang F, Matsubara A, Kan M, McKeehan W. Fibroblast growth factor receptor 2 limits and receptor 1 accelerates tumorigenicity of prostate epithelial cells. Cancer Res. 1997;57:5369–5378. [PubMed] [Google Scholar]
- 17.Fu X, Mayeda A, Maniatis T, Krainer A. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci. 1992;23:11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Blanco M A, Jamison S, Sharp P. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes Dev. 1989;3:1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- 19.Gatto F D, Gesnel M, Breathnach R. The exon sequence TAGG can inhibit splicing. Nucleic Acids Res. 1996;24:2017–2021. doi: 10.1093/nar/24.11.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ge H, Manley J. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 21.Ghetti A, Pinol-Roma S, Michael W M, Morandi C, Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gil A, Sharp P, Jamison S, Garcia-Blanco M A. Characterization of cDNAs encoding the polypyrimidine tract-binding protein. Genes Dev. 1991;5:1224–1236. doi: 10.1101/gad.5.7.1224. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert E, Gatto F D, Champion-Arnaud P, Gesnal M, Breathnach R. Control of Bek and K-sam splice sites in alternative splicing of the fibroblast growth factor receptor 2 pre-mRNA. Mol Cell Biol. 1993;13:5461–5468. doi: 10.1128/mcb.13.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gooding C, Roberts G, Moreau G, Nadal-Ginard B, Smith C. Smooth muscle specific switching of α-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 1994;13:3861–3872. doi: 10.1002/j.1460-2075.1994.tb06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gooding C, Roberts G, Smith C. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated α-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- 26.Grossman J, Meyer M, Wang Y, Mulligan G, Kobayshi R, Helfman D. The use of antibodies to the polypyrimidine tract binding protein to analyze the protein components that assemble on alternatively spliced pre-mRNAs that use distant branch points. RNA. 1998;4:613–625. doi: 10.1017/s1355838298971448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo W, Mulligan G, Wormsley S, Helfman D. Alternative splicing of β-tropomyosin pre-mRNA: cis-acting elements and cellular factors that block the use of a skeletal muscle exon in nonmuscle cells. Genes Dev. 1991;5:2096–2107. doi: 10.1101/gad.5.11.2096. [DOI] [PubMed] [Google Scholar]
- 28.Hanamura A, Caceres J F, Mayeda A, Franza B R, Jr, Krainer A R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 29.Horabin J, Schedl P. Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol Cell Biol. 1993;13:7734–7746. doi: 10.1128/mcb.13.12.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaye M, Schlessinger J, Dionne C. Fibroblast growth factor receptor tyrosine kinases: molecular analysis and signal transduction. Biochim Biophys Acta. 1992;1135:185–199. doi: 10.1016/0167-4889(92)90136-y. [DOI] [PubMed] [Google Scholar]
- 31.Johnson D, Lu J, Chen H, Werner S, Williams L. The human fibroblast growth factor receptor genes: a common structural arrangement underlies the mechanisms for generating receptor forms that differ in their third immunoglobulin domain. Mol Cell Biol. 1991;11:4627–4634. doi: 10.1128/mcb.11.9.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson D, Williams L. Structural and functional diversity in the FGF receptor multigene family. Adv Cancer Res. 1993;60:1–41. doi: 10.1016/s0065-230x(08)60821-0. [DOI] [PubMed] [Google Scholar]
- 33.Lavigueur A, La B, Kornblihtt A, Chabot B. A splicing enhancer in the human fibronectin alternate EDI exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 34.Lou H, Helfman D, Gagel R, Berget S. Polypyrimidine tract-binding protein positively regulates inclusion of an alternative 3′-terminal exon. Mol Cell Biol. 1999;19:78–85. doi: 10.1128/mcb.19.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch K, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;16:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 36.Lynch K, Maniatis T. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 1995;3:284–293. doi: 10.1101/gad.9.3.284. [DOI] [PubMed] [Google Scholar]
- 37.Matsubara A, Kan M, Feng S, McKeehan W. Inhibition of growth of malignant rat prostate tumor cells by restoration of fibroblast growth factor receptor 2. Cancer Res. 1998;58:1509–1514. [PubMed] [Google Scholar]
- 38.Matunis E, Matunis M, Dreyfuss G. Association of individual hnRNP proteins and snRNPs with nascent transcripts. J Cell Biol. 1993;121:219–228. doi: 10.1083/jcb.121.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayeda A, Krainer A. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 40.McKeown M. Alternative mRNA splicing. Annu Rev Cell Biol. 1992;8:133–155. doi: 10.1146/annurev.cb.08.110192.001025. [DOI] [PubMed] [Google Scholar]
- 41.Miki T, Bottaro D, Fleming T, Smith C, Burgess W, Chan A, Aaronson S. Determination of ligand-binding specificity by alternative splicing: two distinct growth factor receptors encoded by a single gene. Proc Natl Acad Sci USA. 1992;89:246–250. doi: 10.1073/pnas.89.1.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller C, Jamison S, Garcia-Blanco M. HeLa nuclear extract: a modified protocol. New York, N.Y: Academic Press, Inc.; 1997. pp. 25–30. [Google Scholar]
- 43.Min H, Chan R, Black D. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 44.Min H, Turck C, Nikolic J, Black D. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- 45.Modafferi E, Black D. Combinatorial control of a neuron-specific exon. RNA. 1999;5:687–706. doi: 10.1017/s1355838299990155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mulligan G, Guo W, Helfman D. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of β-tropomyosin pre-mRNA. J Biol Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- 47.Patton J, Mayer S, Tempst P, Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- 48.Perez I, Lin C, McAfee J, Patton J. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- 49.Perez I, McAfee J G, Patton J G. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding. Biochemistry. 1997;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- 50.Polydorides A, Okano H, Yang Y, Stefani G, Darnell R. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc Natl Acad Sci USA. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramchatesingh J, Zahler A, Neugebauer K, Roth M, Cooper T. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Singh R, Valcarcel J, Green M. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268:1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 53.Southby J, Gooding C, Smith C. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutually exclusive exons. Mol Cell Biol. 1999;19:2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staknis D, Reed R. SR Proteins promote the first specific recognition of pre-mRNA and are present together with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun Q, Mayeda A, Hampson R, Krainer A, Rottman F. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 56.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner E, Carstens R, Garcia-Blanco M. A novel isoform ratio switch of the polypyrimidine tract binding protein. Electrophoresis. 1999;20:1082–1086. doi: 10.1002/(SICI)1522-2683(19990101)20:4/5<1082::AID-ELPS1082>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 58.Wang J, Bell L. The Sex-lethal amino terminus mediates cooperative interactions in RNA binding and is essential for splicing regulation. Genes Dev. 1994;8:2072–2085. doi: 10.1101/gad.8.17.2072. [DOI] [PubMed] [Google Scholar]
- 59.Wang J, Manley J. Regulation of pre-mRNA splicing in Metazoa. Curr Opin Genet Dev. 1997;7:205–211. doi: 10.1016/s0959-437x(97)80130-x. [DOI] [PubMed] [Google Scholar]
- 60.Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- 61.Wentz M, Moore B, Cloyd M, Berget S, Donehower L. A naturally arising mutation of a potential silencer of exon splicing in human immunodeficiency virus type I induces dominant aberrant splicing and arrests virus production. J Virol. 1997;71:8542–8551. doi: 10.1128/jvi.71.11.8542-8551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu R, Teng J, Cooper T. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yan G, Fukabori Y, McBride G, Nikolaropolous S, McKeehan W L. Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)-FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Mol Cell Biol. 1993;13:4513–4522. doi: 10.1128/mcb.13.8.4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yan G, Fukabori Y, Nikolaropoulos S, Wang F, McKeehan W L. Heparin-binding keratinocyte growth factor is a candidate stromal-to-epithelial-cell andromedin. Mol Endocrinol. 1992;12:2123–2128. doi: 10.1210/mend.6.12.1491693. [DOI] [PubMed] [Google Scholar]
- 65.Zhang L, Liu W, Grabowski P. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng Z, Huynen M, Baker C. A pyrimidine-rich exonic splicing suppressor binds multiple RNA splicing factors and inhibits spliceosome assembly. Proc Natl Acad Sci USA. 1998;95:14088–14093. doi: 10.1073/pnas.95.24.14088. [DOI] [PMC free article] [PubMed] [Google Scholar]