Abstract
Nucleus-encoded tRNAs are selectively imported into the mitochondrion of Leishmania, a kinetoplastid protozoan. An oligoribonucleotide constituting the D stem-loop import signal of tRNATyr(GUA) was efficiently transported into the mitochondrial matrix in organello as well as in vivo. Transfer through the inner membrane could be uncoupled from that through the outer membrane and was resistant to antibody against the outer membrane receptor TAB. A number of mutations in the import signal had differential effects on outer and inner membrane transfer. Some mutants which efficiently traversed the outer membrane were unable to enter the matrix. Conversely, restoration of the loop-closing GC pair in reverse resulted in reversion of transfer through the inner, but not the outer, membrane, and binding of the RNA to the inner membrane was restored. These experiments indicate the presence at the two membranes of receptors with distinct specificities which mediate stepwise transfer into the mitochondrial matrix. The combination of oligonucleotide mutagenesis and biochemical fractionation may provide a general tool for the identification of tRNA transport factors.
Mitochondria are genetic parasites presumed to have evolved from endosymbiotic bacteria. The recent sequencing of a large number of mitochondrial genomes has revealed an unexpected diversity in size and gene content (5). The loss of most of the original bacterial protein-coding genes, or their transfer to the nucleus, has been explained in terms of a “big bang” radiation of the different eukaryotic lineages from a single (monophyletic) endosymbiont (5). What is not so easily explained is the loss of a variable number of tRNA genes apparently randomly in different protists, higher plants, and at least one invertebrate lineage (6). At least 24 different tRNA species are required to read the universal genetic code. In kinetoplastid protozoa, including leishmanias and trypanosomes, a complete set of tRNAs are apparently imported in order to compensate for the total lack of mitochondrial tRNA genes (7, 8, 11, 16, 21–23). The mitochondria of tetrahymena import about two-thirds of their tRNAs (2), while in budding yeast only a single tRNALys species is imported (15). In higher plants, different species mitochondrially import different sets of tRNAs; for example, mitochondria from wheat but not maize import tRNAHis (9). Thus, there appears to be a species-specific selectivity, reflecting perhaps the presence of different mitochondrial receptors recognizing individual or groups of import signals on tRNAs.
To study the basis of the selectivity of tRNA import, we have developed an in organello system using leishmania mitochondria (1, 12, 14). Similar systems from trypanosomes have been recently reported (18, 25). Our experiments showed that tRNA import is selective, e.g., tRNATyr is efficiently imported whereas tRNAGln(CUG) is not (1), and that a conserved sequence motif in the D arm of tRNATyr is necessary and sufficient for import (13). The importance of the D loop of tRNAIle for import in vivo has also been demonstrated (10). Importable RNA interacts directly with the outer mitochondrial membrane (12), and a 15-kDa RNA binding protein, TAB, associated with the outer membrane (OM) was purified and shown to function as an import receptor (1). Using specific antibody, it was further shown that TAB interacts with tRNATyr but not with tRNAGln (13). Thus, the TAB-tRNA interaction accounts for the selectivity of import in this system.
More recent studies have indicated that, in addition to sequence discrimination at the OM, a second level of selection possibly occurs at the inner membrane (IM). Analysis of the distribution of imported tRNATyr and other transcripts in the different intramitochondrial compartments, i.e., OM, intermembrane space (IMS), IM, and matrix (MX), showed that import occurs in a stepwise fashion, with a distinct kinetic separation of the OM and IM transfer steps (17). While OM transfer requires ATP, IM transfer is driven by both the electrical and chemical components of the electromotive force generated at the IM of energized mitochondria (17). These results lead to the hypothesis that distinct receptors occur at the OM and IM which concertedly determine sequence selectivity for MX entry of tRNAs. However, nothing is known about the specificity of the interaction at the IM or the identity of any of the components of IM transport machinery.
Here we show that a short oligoribonucleotide containing the D arm of tRNATyr(GUA) rapidly and efficiently transits to the mitochondrial MX in vitro. Site-specific mutagenesis of the D arm was employed to detect similarities and differences in the sequence and/or structural requirements for OM and IM transfer.
MATERIALS AND METHODS
Cell culture and preparation of mitochondria.
Promastigotes of Leishmania tropica strain UR6 were cultured at 22°C on solid blood agar medium (3) supplemented with 150 μg of biopterin and 50 μg of adenine per ml. Mitochondria were purified from DNase I-treated lysates by Percoll gradient centrifugation and stored in a 50% glycerol storage buffer, as described previously (14). Before use, mitochondria were diluted with cold isotonic sucrose-Tris-EDTA (STE) buffer (14), washed by centrifugation, and resuspended in STE at a final protein concentration of 8 to 10 mg/ml.
Submitochondrial fractionation.
Separation of submitochondrial compartments was performed as previously described (17). Briefly, mitochondria were treated with 320 μM digitonin in STE buffer for 15 min on ice to selectively solubilize the OM (19). Mitoplasts were separated from the soluble fraction (OM plus IMS) by centrifugation. To further fractionate the mitoplasts, they were suspended in a solution of 0.6 M sucrose, 10 mM Tris-HCl (pH 7.5), and 1 mM EDTA and subjected to three freeze-thaw cycles. Soluble MX and particulate IM fractions were subsequently separated by centrifugation.
Preparation of import substrates.
32P-labeled tRNATyr(GUA) transcripts were synthesized by runoff transcription of the plasmid pSKB1 (1), which contains a cloned genomic copy of the corresponding Leishmania gene, using T7 RNA polymerase and [α-32P]UTP, as described previously (4). Wild-type and mutant D arm minihelix RNAs were synthesized by runoff transcription of the corresponding double-stranded oligonucleotides containing a T7 RNA polymerase promoter. To prepare the templates, the promoter primer GGAATTCTAATACGACTCACTATAGGGACTGTAGCTC, containing an EcoRI linker, a T7 RNA polymerase promoter, and nucleotides 5 to 13 of tRNATyr(GUA) (11) (see Fig. 1), was annealed to the following oligonucleotides, each containing sequences complementary to positions 5 to 27 of tRNATyr(GUA): wild type, ATGCTCTACCAATTGAGCTACAGTC; U17→A, ATGCTCTACCTATTGAGCTACAGTC; G18→C, ATGCTCTACGAATTGAGCTACAGTC; A21→C, ATGCTCGACCAATTGAGCTACAGTC; G22→C, ATGCTGTACCAATTGAGCTACAGTC; G22→C, C13→G, ATGCTGTACCAATTCAGCTACAGTC; A23→U, ATGCACTACCAATTGAGCTACAGTC; A23→U, U12→A, ATGCACTACCAATTGTGCTACAGTC; C11→A, U12→C, ATGCTCTACCAATTGGTCTACAGTC. The resulting partially double-stranded molecule was end-filled with Moloney murine leukemia virus reverse transcriptase (13) and purified by ethanol precipitation. The sequences of the 27-mer transcripts were verified by two-dimensional oligonucleotide fingerprinting (24).
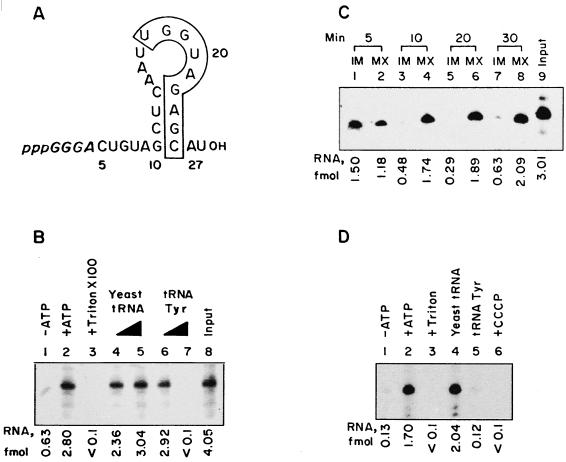
FIG. 1.
Import of D arm minihelix in vitro. (A) Sequence of the wild-type minihelix. Numbers correspond to positions on the intact tRNATyr molecule (11). The secondary structure shown was derived by energy minimization using the FOLDRNA program (26). Bases in italics are derived from the T7 RNA polymerase initiation sequence on the template. The boxed region contains the conserved nonanucleotide motif found in importable RNAs (13). (B) Total uptake of the wild-type minihelix by intact mitochondria. Reaction mixtures were incubated without (lane 1) or with (lanes 2 through 7) 4 mM ATP. In lane 3, Triton X-100 (0.5%) was added after the import incubation. In lanes 4 through 7, unlabeled yeast tRNA (lanes 4 and 5) or low-specific-activity tRNATyr transcript (lanes 6 and 7) was added as the competitor at concentrations of 0.1 (lanes 4 and 6) or 1 (lanes 5 and 7) pmol. After RNase treatment, the total internalized RNA was analyzed. (C) Intramitochondrial distribution of D arm minihelix. Intact mitochondria were incubated with wild-type RNA and ATP at 37°C for the time intervals shown. After each incubation, the mitochondria were RNase treated and subfractionated, and the contents of RNA in the IM and MX fractions were analyzed. (D) Import of D arm minihelix into mitoplasts. Mitoplasts (100 μg of protein) were incubated with the wild-type minihelix in the absence (lane 1) or presence (lanes 2 through 6) of 4 mM ATP. In lane 3, Triton X-100 (0.5%) was added after the incubation. Reactions 4 and 5 contained 1 pmol of yeast tRNA or tRNATyr, respectively, as the competitor. In reaction 6, mitoplasts were preincubated with 50 μM CCCP before the import incubation. RNase-resistant RNA was recovered for analysis. The region of the major minihelix band in each lane was excised and counted; after background subtraction, femtomole values were computed from the specific activity.
In organello import assays.
Unless otherwise stated, mitochondria or mitoplasts (100 μg of protein) were incubated in 20-μl reaction mixtures containing 10 mM Tris-HCl (pH 8), 10 mM magnesium acetate, 2 mM dithiothreitol, and 4 mM ATP with 100 fmol of 32P-labeled substrate for 15 min at 37°C (17). Then RNase A (2.5 μg/ml) and RNase T1 (50 U/ml) were added, and incubation continued for an additional 15 min at 37°C. The vesicles were washed in cold STE buffer by centrifugation. To measure total uptake, RNase-treated mitochondria or mitoplasts were lysed in guanidium isothiocyanate, and 32P-labeled RNA was recovered by isopropanol precipitation, as described previously (13). Alternatively, intramitochondrial RNAs were assayed by subjecting postimport RNase-treated mitochondria to digitonin treatment, followed by freeze-thaw lysis to separate the submitochondrial compartments (as described above) and recovery of 32P-labeled RNA from each fraction. Antibody inhibition experiments were carried out with mitochondria or mitoplasts successively incubated with 4 mg of bovine serum albumin/ml in STE for 1 h at 0°C and then with 100 μg of normal or anti-TAB immunoglobulin G (IgG) per ml (1) for 30 min at 0°C and finally washed with STE. To study the effect of uncouplers, mitoplasts were preincubated with 50 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) for 10 min on ice before the import reaction. 32P-labeled RNA obtained in each case was analyzed by urea–10% polyacrylamide gel electrophoresis (PAGE) followed by autoradiography. Quantitation was performed by liquid scintillation counting of the dried gel band and/or scanning in a Bio-Rad model GS 710 densitometer.
In vivo matrix localization assay.
32P-labeled minihelix RNA (1 pmol) was added to 5 × 108 promastigotes in 0.4 ml of HEPES-buffered saline (21 mM HEPES, 137 mM NaCl, 5 mM KCl, 0.7 mM NaH2PO4; pH 7.4) and the cells were electroporated at 450 V and 500 μF in a Bio-Rad gene pulser. Electroporated cells were incubated on ice for 5 min, centrifuged, resuspended in 1 ml of medium M199 containing 10% fetal bovine serum, and incubated at 22°C for 10 min. The cells were returned to ice and 0.2-ml aliquots were added to 0.8 ml of phosphate-buffered saline. After pelleting by centrifugation, the cells were suspended in 1 ml of hypotonic lysis buffer (14), lysed by two syringe passages, and returned to isotonicity by sucrose addition, as described previously (14). The particulate fraction containing mitochondrial vesicles was suspended in 0.1 ml of STE containing 10 mM MgCl2 and incubated with DNase I (50 U/ml), RNase A (2.5 to 5 μg/ml), and RNase T1 (50 to 100 U/ml) for 15 min at 37°C. The mitochondria were then washed with STE, lysed in guanidinium isothiocyanate, and processed for RNA isolation (13). Quantitations were performed by densitometry.
Binding assay.
Mitochondria or mitoplasts (50 μg of protein) were incubated with 10 fmol of 32P-labeled RNA in 10 μl of binding buffer (10 mM Tris-HCl [pH 8], 10 mM magnesium acetate, 1 mM dithiothreitol, 0.1 M KCl) for 30 min on ice. The vesicles were then washed in 1 ml of cold STE, and bound RNA was recovered and analyzed as above. To obtain the dissociation constant (Kd) of the wild-type D arm-receptor complex, mitoplasts were titrated with increasing concentrations (t) of 32P-labeled RNA, and the bound (b) and free (f = t − b) RNA concentrations were determined. Scatchard plots of b/f versus b yielded Kd = −(1/slope) and the total receptor concentration, R0, as the intercept on the x axis. For each mutant, Kd was then derived from the equation Kd = [(t − b) (R0 − b)]/b.
RESULTS
Stepwise import of the D arm minihelix.
It was shown earlier (13, 17) that the tRNATyr(GUA) molecule can be replaced by progressively smaller derivatives containing the D arm region, with concomitant increases in the efficiencies of transfer across the outer and inner mitochondrial membranes. Such derivatives apparently use the same import pathway as the intact molecule, as demonstrated by competition and antibody inhibition experiments. This observation raises the attractive possibility of using small synthetic oligonucleotides to probe the transport systems on mitochondrial membranes. Accordingly, a 27-nucleotide RNA hairpin (minihelix) (Fig. 1A) containing the wild-type D arm sequence of the tRNATyr(GUA) was synthesized by T7 RNA polymerase-mediated transcription of the appropriate oligonucleotide template. The energy-minimized secondary structure of this molecule (Fig. 1A) contains the 4-bp stem and 8-base loop present in intact tRNA. Neither the 5′ terminal extension, containing the GGGA initiation sequence for T7 RNA polymerase, nor the 3′ tail forms any detectable base pair with the remainder of the molecule, and this is true for the wild type as well as for all the mutant sequences used in this study (see below).
Uptake of the minihelix by isolated leishmania mitochondria was dependent on ATP; entry into the membrane-bound organelle was evident from the susceptibility of the internalized RNA to RNase in the presence of detergent (Fig. 1B). Moreover, import of the minihelix was specifically competed out by the parental molecule tRNATyr(GUA), indicating the utilization of a common import pathway (Fig. 1B).
The distribution of the minihelix in various intramitochondrial compartments following entry through the OM was then studied by biochemical fractionation (17). Briefly, mitochondria were incubated with 32P-labeled RNA, excess RNA was digested with RNase, and then the washed vesicles were treated with digitonin to selectively permeabilize the OM (19). After centrifugal separation of the soluble fraction (OM plus IMS) from the insoluble mitoplasts, the latter was subjected to freeze-thaw cycles to liberate the soluble MX contents with the remaining insoluble fraction representing the IM. At shorter incubation times (5 min), more than 50% of the RNA was found to be associated with the IM, with the remainder being in the MX (Fig. 1C). By 10 min, most of the RNA was in the MX fraction (Fig. 1C). This kinetic pattern indicates that OM transfer is faster than IM transfer and is similar to what was previously observed with the parental molecule (17), except that the rate of IM transfer of the minihelix is noticeably higher, probably due to the absence of extra sequences which hinder import.
In the above system using intact mitochondria, IM transfer is dependent on OM transfer, making it difficult to independently assess the requirements of the former step. Therefore, mitoplasts obtained by digitonin permeabilization of intact mitochondria were incubated with the 32P-labeled D arm minihelix, treated with RNase, and analyzed for their RNA content. Transfer of the minihelix into the MX was dependent on ATP and inhibited by protonophore uncouplers such as CCCP (Fig. 1D), demonstrating the requirement of a proton motive force across the IM (17). Under otherwise identical conditions, the amount of wild-type oligonucleotide entering the MX was the same (2 to 2.5 fmol/100 μg of mitochondrial protein), irrespective of whether intact mitochondria or mitoplasts were used, indicating that the essential components of the IM machinery are not lost during mitoplast isolation.
To further assess the validity of the oligonucleotide import system, the transfer of the D arm minihelix across the OM and IM was directly compared with that of intact tRNATyr under a variety of conditions. ATP, temperature, high concentrations of monovalent cations, dissipation of IM electromotive force by protonophores (CCCP and nigericin), and disruption of membrane potential by K+ in the presence of valinomycin all had similar or identical quantitative effects on the transfer of either substrate (Table 1). Thus, except for the kinetics of transfer (see above), there is little or no biochemical distinction between full-length tRNA and the isolated D arm.
TABLE 1.
Comparison of import characteristics of tRNATyr and D arm minihelix
| Exptl variablea | Parameterb | Membrane | Value with substrate
|
|
|---|---|---|---|---|
| tRNATyr | Wild-type minihelix | |||
| ATP | Km | OM | 1.52 mM | 2.15 mM |
| KCl | IC50 | IM | 0.74 M | 0.80 M |
| CCCP | IC50 | IM | 3.0 μM | 3.0 μM |
| Nigericin | IC50 | IM | 2.6 μM | 3.5 μM |
| KCl + 5 μM valinomycin | IC50 | IM | 12.5 mM | 13.5 mM |
| Temp | Δυ/Δt | OM | 0.0066 | 0.0053 |
Import reactions were carried out with either mitochondria or mitoplasts (100 μg of protein) in the presence of 100 fmol of 32P-labeled RNA substrate for 15 min at 25°C (except for the temperature-dependence experiment, in which temperature was varied). The amount of RNA transferred was quantified by RNase protection, PAGE, autoradiography, and densitometric band scanning. Various concentrations of the indicated reagents or various temperatures were used.
Km, the concentration of ATP for half-maximal (50%) rate of import (determined from titration curves); IC50, the concentration of reagent for 50% inhibition of transfer; Δυ/Δt, increase in rate of transfer (Δυ) per degree Celsius rise in temperature (Δt) before saturation (between 25 and 37°C in both cases).
Effect of point mutations in the D arm on transfer through OM and IM in organello.
The D arm of tRNATyr(GUA) contains the motif UGGUAGAGC (Fig. 1A), which is conserved in the corresponding region of tRNAs imported in leishmania and trypanosome mitochondria as well as in a synthetic transcript imported through the same pathway (13). To determine the role of the primary sequence and secondary structure in the import signal, point mutations were introduced within this region. Mutant RNAs synthesized by in vitro transcription of the corresponding oligonucleotide templates were assayed for intramitochondrial distribution after uptake in the presence of ATP. In this assay, total uptake (i.e., the amount of RNA in the IM-plus-MX fractions, whereas the OM-plus-IMS fractions contained negligible amounts of RNA [data not shown]), is a measure of OM transfer, while the fraction of the total internalized RNA present in the MX represents IM transfer. For those mutants which are deficient in OM transfer, it is difficult to accurately assess IM transfer using intact mitochondria, since the amount internalized is very low. Therefore, these mutants were directly assayed for their import into mitoplasts.
The observed effects on OM and IM transfer (Fig. 2 and Table 2) may be summarized as follows. (i) Single mutations in the loop (G18→C, U17→A, A21→C) resulted in a decline of OM transfer by four- to eightfold compared to the wild-type sequence. The effect of these mutations on IM transfer was somewhat less, being reduced to 30 to 40% of the wild-type level. (ii) The mutation G18→C at the loop-closing pair of the D arm also reduced OM and IM transfer by about 10-fold. (iii) However, if the loop-closing pair was restored by a second mutation, C13→G, OM transfer remained low (13% of wild type), but at least half of the RNA internalized into intact mitochondria was found in the MX, indicating efficient IM transfer. This was confirmed with mitoplasts; the G22→C, C13→G mutant was transferred across the IM with about 80% of the efficiency of the wild-type sequence. (iv) The mutation A23→U reduced both OM and IM transfer to 20 to 30% of that of the wild type. When the AU pair was restored by a second mutation, U12→A, OM transfer increased from 29 to 64% (compared to wild type), but IM transfer was not restored. (v) A double mutation in the stem, C11→U, U12→C, which destabilizes the stem without altering the primary sequence of the conserved motif (Fig. 1), resulted in a significant amount of OM transfer (59% of wild type) but had a more severe effect on IM transfer (reduced to 30% of wild type). These results indicate that certain mutations in the import signal result in different efficiencies of transfer across the IM and OM.
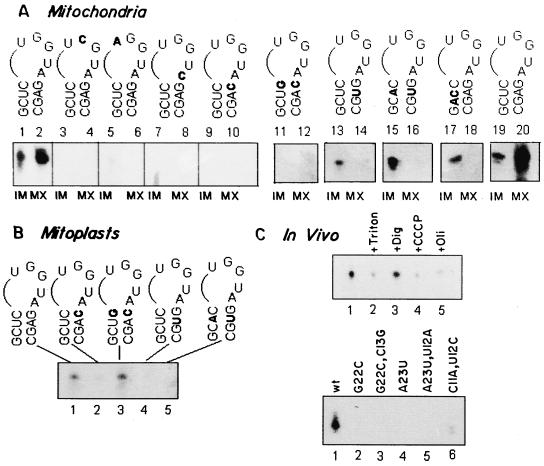
FIG. 2.
Effect of mutations on intramitochondrial location of D arm minihelix. (A) 32P-labeled wild-type or mutant minihelix (100 fmol) was incubated with mitochondria (100 μg of protein). After 15 min at 37°C, RNase was added, and the washed mitochondria were fractionated into IM and MX compartments. Lanes 1 through 10 and 11 through 20 show the results of two different experiments. The RNAs used were as follows. Lanes 1 and 2, 19 and 20, wild type; lanes 3 and 4, G18→C; lanes 5 and 6, U17→A; lanes 7 and 8, A21→C; lanes 9 and 10, G22→C; lanes 11 and 12, G22→C, C13→G; lanes 13 and 14, A23→U; lanes 15 and 16, A23→U, U12→A; and lanes 17 and 18, C11→A, U12→C. (B) 32P-labeled wild-type (lane 1), G22→C (lane 2), G22→C, C13→G (lane 3), A23→U (lane 4), and A23→U, U12→A (lane 5) RNAs (100 fmol of each) were incubated with mitoplasts (100 μg of protein) in the presence of ATP for 15 min at 37°C, and the RNase-resistant RNA was analyzed. Band quantitation was performed as in Fig. 1; the smear at the bottom of lane 7 was disregarded. (C) Matrix targeting in vivo. (Upper panel) Promastigotes were transfected with 32P-labeled wild-type minihelix (1 pmol) in the absence (lanes 1 to 3) or presence of 10 μM CCCP (lane 4) or of 50 μM oligomycin (lane 5). Aliquots of the transfected cells were lysed, and mitochondrial fractions were treated with RNase and DNase in the absence (lanes 1, 4, and 5) or presence of 1% Triton X-100 (lane 2) or 320 μM digitonin (lane 3). (Lower panel) Promastigotes were transfected with the wild type (lane 1) or the G22→C (lane 2), G22→C, C13→G (lane 3), A23→U (lane 4), A23+→U, U12→A (lane 5), and C11→A, U12→C (lane 6) mutants, and the RNase-resistant RNA associated with the mitochondrial fraction was analyzed. Quantitation was performed by densitometry.
TABLE 2.
Effect of mutations on outer and inner membrane transfer
| Mutation(s) | OM transfer (% of w.t.)a | Intramitochondrial distribution (fraction of total)b
|
IM transfer (% of w.t.)c | |
|---|---|---|---|---|
| IM | MX | |||
| None (w.t.) | 100 | 0.30 | 0.70 | 100 |
| U17→A | 12.2 | 0.67 | 0.33 | 28.8 |
| G18→C | 23.9 | 0.73 | 0.27 | 30.3 |
| A21→C | 11.9 | 0.80 | 0.20 | 38.5 |
| G22→C | 9.0 | 0.82 | 0.18 | 12.7 |
| G22→C, C13→G | 12.8 | 0.47 | 0.53 | 88.3 |
| A23→U | 29.3 | 0.84 | 0.16 | 19.2 |
| A23→U, U12→A | 64.3 | 0.82 | 0.18 | 10.5 |
| C11→A, U12→C | 58.7 | 0.81 | 0.19 | 29.7 |
Measured by total uptake into intact mitochondria and/or the sum of the distribution in intramitochondrial compartments. Quantitations were performed by gel band counting (Fig. 2). The amount of wild-type (w.t.) sequence imported (2.0 to 2.8 fmol) was taken as 100%.
Ratio of the amount of RNA in a particular compartment to the total amount internalized (i.e., IM + MX).
Measured by uptake into mitoplasts; quantitation performed as described for OM transfer.
Targeting of the D arm minihelix to the mitochondrial MX in vivo.
From the preceding in organello analysis, it is evident that mutations in the conserved D arm motif result in deficiency of transport into the mitochondrial MX, due to a defect in either OM or IM transfer. To examine whether this is also true in vivo, 32P-labeled minihelices were introduced into leishmania promastigotes by electroporation. Transfected cells were lysed by syringe passage in hypotonic medium, the mitochondrial fraction was recovered and treated with DNase and RNase, and the nuclease-resistant RNA was analyzed. Hypotonic treatment results in leakage of the contents of the IMS; this assay therefore scores exclusively for MX-localized RNA and does not distinguish between OM and IM transfer.
Approximately 20 to 40 molecules of the wild-type minihelix per transfected cell were recovered from the mitochondrial fraction under optimal conditions (Fig. 2C). Entrapment of the RNA within a membrane vesicle was evident from an enhanced RNase sensitivity in the presence of Triton X-100, but RNase resistance was unaffected by treatment with digitonin at a concentration which selectively permeabilizes the OM (17), indicating MX localization. Transfection in the presence of the mitochondrial inhibitors CCCP and oligomycin resulted in reduction of MX transfer in vivo (Fig. 2C), consistent with the sensitivity of in organello IM transfer to these agents (17) (Fig. 1). About 20% of the RNA remained RNase resistant in the presence of Triton X-100 or the inhibitors; this could be due to sequestration into an unknown subcellular component present in the crude mitochondrial fractions.
The effect of mutations on targeting in vivo was analyzed. All the mutants examined were deficient in MX localization (Fig. 2C). The C11→A, U12→C mutant was targeted at about 15% of the wild-type level. These data confirm that the D arm signal is necessary and sufficient for transport into the mitochondrial MX in vivo as well as in vitro.
Sequence-specific binding of D arm variants to OM and IM.
To determine whether the above effects on OM and IM transfer reflect altered binding to membrane-bound receptors, 32P-labeled minihelices were allowed to interact with intact mitochondria or mitoplasts under conditions favoring specific binding but discouraging translocation (i.e., in the presence of 0.1 M KCl at 0°C, in the absence of ATP) (12). The vesicles were then washed, and bound RNA was analyzed by gel electrophoresis. Control experiments (data not shown) demonstrated that the bound RNA was completely sensitive to RNase, i.e., it was associated with the membrane surface.
Both the G22→C and G22→C, C13→G mutants were bound inefficiently to the OM, but the second mutation restored IM binding to about 60% of that of the wild type (Fig. 3A). In contrast, restoration of the second base pair in the stem (i.e., A23→U, U12→A) increased binding to the OM but not to the IM (Fig. 3A). The wild-type sequence bound to the OM and IM with apparent Kd values of 0.93 and 3.86 nM, respectively (Table 3). The mutation G22→C increased KdOM nearly 8-fold, while KdIM was increased 12-fold. But whereas KdOM remained high for the double mutant G22→C, C13→G, KdIM was restored to near-wild-type levels (Table 3). The converse effect of base pair restoration at the next position (A23) was also evident: reformation of the AU pair reduced KdOM by a factor of 2 but increased KdIM by a factor of 3.5 (Table 3). Finally, the double mutation C11→A, U12→C had a much more severe effect on KdIM (8-fold) than on KdOM (2-fold). Thus, the binding efficiencies parallel the transfer efficiencies and indicate the presence of distinct receptors on the two membranes which recognize different structural features of the D arm.
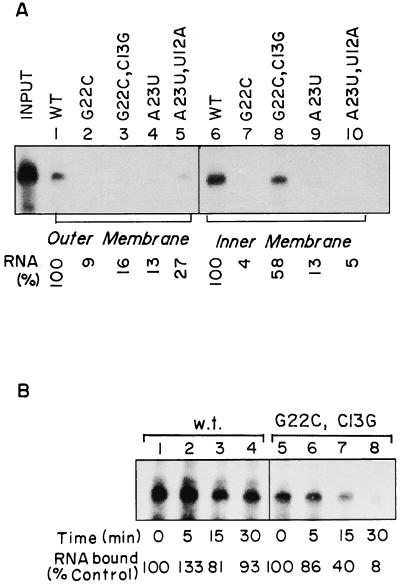
FIG. 3.
Binding of minihelices to the OM and IM. (A) The following 32P-labeled RNAs were incubated with mitochondria (lanes 1 to 5) or mitoplasts (lanes 6 to 10): lanes 1 and 6, wild type; lanes 2 and 7, G22→C; lanes 3 and 8, G22→C, C13→G; lanes 4 and 9, A23→U; and lanes 5 and 10, A23→U, U12→A. After washing with STE, the membrane-bound RNA was recovered and analyzed as in Fig. 1. The amounts of wild-type RNA (taken as 100%) bound to mitochondria (lane 1) and mitoplasts (lane 6) were 0.64 and 1.12 fmol, respectively. (B) Stability of inner membrane complexes of wild-type (lanes 1 to 4) and G22→C, C13→G mutant (lanes 5 to 8) minihelices. Binding reactions with mitoplasts were performed with the incubation intervals shown. Control reactions without incubation (lanes 1 and 5) yielded 1.80 and 1.01 fmol, respectively. Quantitations were performed by densitometric scanning of the major band in each lane.
TABLE 3.
Dissociation constants for complexes between D arm mutants and mitochondrial membranes
| Mutation | Stability of secondary structure (−ΔG [kcal mol−1])a |
Kd (nM)b
|
|
|---|---|---|---|
| OM | IM | ||
| Wild type | 9.9 | 0.9 | 3.9 |
| G22→C | 2.2 | 7.3 | 46.3 |
| G22→C, C13→G | 9.8 | 7.0 | 5.1 |
| A23→U | 1.0 | 6.9 | 22.8 |
| A23→U, U12→A | 9.2 | 3.2 | 77.9 |
| C11→A, U12→C | 0.7 | 1.7 | 32.2 |
Free energy of formation, obtained by using the FOLDRNA program (28).
Obtained by Scatchard analysis.
Although the net yields of the complexes formed with the wild-type sequence and the G22→C, C13→G mutant were comparable, there was a notable difference in their rates of dissociation (Fig. 3B). Thus, while the wild-type complex remained stable for at least 30 min at 0°C, more than 90% of the mutant complex was dissociated within this time period.
Competition between D arm variants at OM and IM.
From the above experiments, two mutants were identified which are transferred through the OM, but not IM, with good efficiency: the A23→U, U12→A mutant and the C11→A, U12→C mutant. Conversely, the G22→C, C13→G mutant is transferred efficiently through the IM but not the OM. Only the wild-type sequence is transferred through both the membranes efficiently. To examine whether the different structures use the same or distinct receptors at either membrane, competition assays were performed in which one labeled oligonucleotide was challenged with an excess of another oligonucleotide (either unlabeled or labeled to low specific activity). OM transfer of both the A23→U, U12→A and C11→A, U12→C mutants was efficiently competed out by the wild-type sequence, although in the latter case competition was somewhat less effective than in the self-self situation (Fig. 4A). At the IM, the wild-type structure and the G22→C, C13→G mutant also competed with each other, and again, cross competition was quantitatively less than self-competition (Fig. 4B). Competition for transfer was paralleled by competition for binding at either membrane (data not shown). These results are consistent with the binding of the different structures to the same or similar receptors at either membrane, albeit with different affinities (Table 3).
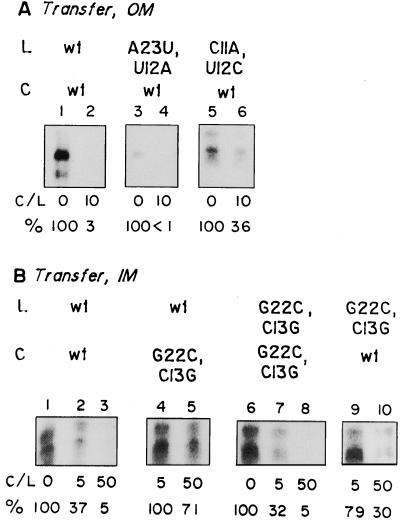
FIG. 4.
Cross competition between wild-type and mutant minihelices for transfer across OM and IM. Mitochondria (A) or mitoplasts (B) were incubated with high-specific-activity 32P-labeled RNA (L) in the absence or presence of low-specific-activity competitor (C; C/L, ratio of competitor to substrate), and total uptake was assayed with RNase protection. Panel A, lanes 1 to 2, 3 to 4, and 5 to 6, contained high-specific-activity wild-type, A23→U, U12→A, and C11→A, A12→U RNA (100 fmol), respectively. Wild-type competitor (1 pmol) was included in reactions 2, 4, and 6. In panel B, high-specific-activity wild-type (lanes 1 to 5) or G12→C, C13→G (lanes 6 to 10) RNA (100 fmol) was incubated without competitor (lanes 1 and 6), with 0.5 pmol (lanes 2 and 9) or 5 pmol (lanes 3 and 10) of wild-type competitor, or with 0.5 pmol (lanes 4 and 7) or 5 pmol (lanes 5 and 8) of G22→C, C13→G competitor. Quantitations were performed by densitometric scanning of the major band in each lane.
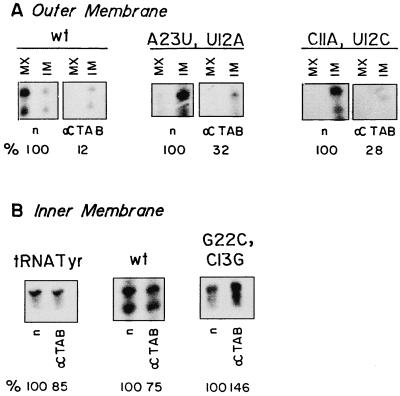
Dependence of OM and IM transfer on TAB.
TAB is an OM-associated RNA binding protein required for the uptake of tRNATyr into leishmania mitochondria (1). To examine the role of TAB in the uptake and intramitochondrial distribution of D arm minihelices, antibody inhibition experiments were performed (Fig. 5A). Anti-TAB antibody, but not normal IgG (Fig. 5A) or antibody against total leishmania antigen (data not shown), inhibited OM transfer of the wild-type sequence as well as that of the A23→U, U12→A, and C11→A, U12→C mutants. Thus, the mutants, like the wild type, interact with TAB at the OM.
FIG. 5.
Effect of anti-TAB antibody on transfer of RNA across the OM and IM. (A) Intact mitochondria (100 μg of protein) preincubated with normal (n) or anti-TAB (α) IgG were incubated with 100 fmol of wild-type D arm minihelix (left), A23→U, U12→A mutant (center), or C11→A, U12→C mutant (right) in the presence of ATP. The amounts of total internalized RNA (IM + MX) are expressed as percentages of the control values obtained in the presence of normal IgG. (B) Mitoplasts were incubated with normal (n) or anti-TAB (α) IgG and then with 100 fmol of tRNATyr(GUA) transcript (left), wild-type D arm minihelix (center), or G22→C, C13→G mutant (right) for 15 min at 37°C and treated with RNase, and the internalized RNA was recovered. IM transfer is expressed as the percentage of the control value obtained with normal IgG. Quantitations were performed as for Fig. 4. In panel B, middle, both bands are of nearly equal intensity and yield similar values for extent of inhibition.
In marked contrast, transfer of tRNATyr as well as transfer of the wild-type D arm minihelix and the G22→C, C13→G mutant across the IM of isolated mitoplasts was resistant to anti-TAB antibody (Fig. 5B). This observation indicates that IM receptors that are distinct from TAB mediate transport into the matrix.
DISCUSSION
In this report, a simple method for probing the structural basis of the selectivity of mitochondrial tRNA import is described. The use of synthetic oligonucleotides as import substrates is based on our observation that the D arm of tRNATyr is necessary and sufficient for import in organello (13) as well as in vivo (Fig. 2), and this approach also greatly facilitates the generation of mutations for the study of structure-function relationships. Furthermore, fractionation of the submitochondrial compartments allows an assessment of the effect of an individual mutation on the precise location of the corresponding structure within the mitochondrion. By all the criteria examined, i.e., competition by intact tRNA (Fig. 1), effect of anti-TAB antibody on import into mitochondria or mitoplasts (Fig. 5), dependence on ATP and temperature (Table 1), and inhibition of transfer by inhibitors and uncouplers (Fig. 1 and Table 1), the D arm minihelix appears to be using the same pathway for import as the parental molecule, although the rate and extent of import of the minihelix are noticeably higher (Fig. 1), presumably due to its smaller size and/or the lack of inhibitory sequences.
Site-specific mutagenesis of the D arm minihelix revealed a number of distinct phenotypes (Fig. 2 and Table 1). In the first group of mutants (U17→A, G18→C; A21→C, G22→C; and A23→U), both OM and IM transfer were reduced relative to the wild type. In the second group (A23→U, U12→A and C11→A, U12→C), OM transfer was reasonably efficient (about 60% of the wild type), but IM transfer was considerably reduced, with the RNA accumulating at the IM (Fig. 2). The third type of mutation, i.e., G22→C, C13→G, had the opposite property: efficient IM but defective OM transfer (Fig. 2). These results imply that the RNA sequence and/or structural specificities at the two membranes are nonidentical.
The differences are particularly pronounced when bases in the stem of the D arm (Fig. 1A) are altered. Each of the single mutations (G22→C and A23→U), or the double mutation on the same side of the helix (C11→A, U12→C), results in a drastic reduction of secondary structure stability (Table 3); the FOLDRNA program (26) predicts relatively unstable energy-minimized structures with the canonical 4-bp stem replaced by loops, bulges, and non-Watson-Crick pairs (data not shown). Restoration of the loop-closing pair by the second C13→G mutation, or of the neighboring pair by the U12→A mutation, results in restoration of the wild-type conformation. It would seem from this limited structure-function study that the loop-closing pair (G22:C13) is critical at both membranes, but for different reasons: while the G base is specifically recognized at the OM, base pairing is more important at the IM, since restoration of the pair in reverse results in reappearance of IM transfer (Fig. 2) and an increase in binding affinity for the IM receptor (Table 3). Conversely, the A23 residue critically interacts with the IM receptor(s) but may be replaced by U at the OM. This would explain the lack of reversion for IM transfer in the A23→U, U12→A mutant (Fig. 2); in fact, the binding affinity of this mutant for the IM is further lowered (Table 3). The importance of base pairing for IM transfer is further illustrated by the effects of the double mutation of C11 and U12 in the stem, which results in considerable weakening of the secondary structure (Table 3), while leaving the critical A23 and other residues in the conserved region intact. This mutation, however, only marginally affects OM transfer (Fig. 2) or OM binding (Table 3), suggesting recognition of primary sequence, rather than secondary structure, by the OM receptor TAB. In fact, synthetic transcripts containing the nonanucleotide conserved motif but lacking significant secondary structure are transferred efficiently through the OM by the TAB-dependent pathway (13, 14). Regardless of the precise nature of the interactions, it is evident from these experiments that distinct receptors exist for OM and IM transfer. This conclusion was reinforced by the resistance of IM transfer to antibody against TAB (Fig. 5).
Almost nothing is currently known about the nature of tRNA import factors. A major problem in systems such as leishmania, ciliates, and higher plants is the nonavailability of mutants with defective mitochondrial function. The use of oligonucleotide probes, coupled with submitochondrial fractionation, constitutes an alternative and general approach for the identification of the components of the membrane transport machinery. Previous experiments using intact tRNA molecules indicated the occurrence of nonproductive binding to mitochondrial membranes, with only about 10% of RNA bound to the OM being internalized (12); moreover, tRNAGln binds efficiently to the OM in a TAB-independent manner but is poorly imported (13). This occurrence of nonproductive binding makes it difficult to decide whether a particular tRNA binding membrane protein is an import factor or is unrelated to import. In contrast, binding of D arm oligonucleotides to the IM shows a strict correlation with importability. The most striking example of this correlation is the loss of binding as well as import in the G22→C mutant, and their simultaneous restoration by the second mutation, C13→G (Fig. 2 and 3 and Table 3). This should allow the identification of putative IM receptors by photochemical cross-linking and other methods.
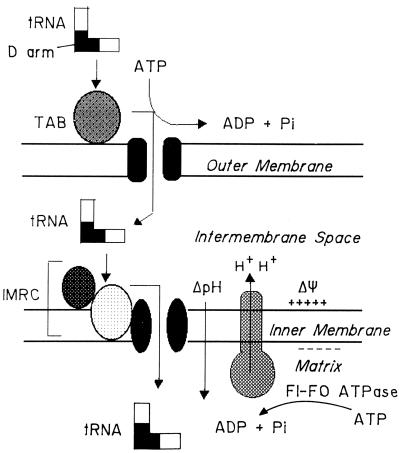
The observed differential effects of the same mutation on OM and IM transfer support and refine the stepwise transport model (17) of mitochondrial tRNA import (Fig. 6). In this model, a tRNA species containing the conserved D arm import signal is proposed to bind to TAB at the OM and to be transferred in an ATP-dependent manner into the IMS, where it interacts with a different receptor or receptor complex on the IM. Subsequent translocation into the MX is driven by both the electrical (ΔΨ) and chemical (ΔpH) components of the proton motive force across the IM. In the in vitro system, the proton motive force is generated by F1F0 ATPase-catalyzed ATP hydrolysis, with concomitant translocation of protons across the IM (Fig. 6).
FIG. 6.
Stepwise transfer of tRNA through the two mitochondrial membranes. For details see the text. TAB, outer membrane receptor; IMRC, inner membrane receptor complex components (two components are shown); ΔΨ, membrane potential; ΔpH, proton gradient; F1-F0 ATPase, oligomycin-sensitive proton pump.
Mitochondrial translation is dependent on the presence of an adequately balanced pool of different tRNA species in the MX. Independent evolution of OM and IM receptor specificities may be a means of ensuring this balance. Since the MX concentration of an individual species is dependent on the efficiencies of OM and IM transfer, a low efficiency of OM transfer would be compensated for by a high efficiency of IM transfer, and vice versa. There is also evidence that other domains of the tRNA molecule besides the D arm contain import signals. In tetrahymena, the anticodon of tRNAGln(UUG) functions as an import signal (20). Moreover, a tRNAIle derivative containing a nonfunctional D arm sequence from tRNAGln is nonetheless imported into leishmania mitochondria in vivo (10), indicating the presence of a signal elsewhere in the molecule, possibly in the anticodon arm, in addition to the one in the D arm. The presence of distinct OM and IM transport machineries would allow the sequential use of different signals on the same molecule for MX entry.
ACKNOWLEDGMENTS
We are indebted to Subhagata Ghosh for technical assistance, Chanchal Dasgupta for nucleotides, and Tapas Ghosh of the Bioinformatics Centre, Bose Institute, for running the FOLDRNA program.
This work was supported by a grant from the Department of Science and Technology, Government of India. S.N.B. and S.M. were supported by fellowships from the Council of Scientific and Industrial Research.
REFERENCES
- 1.Adhya S, Ghosh T, Das A, Bera S K, Mahapatra S. Role of an RNA binding protein in import of tRNA into Leishmania mitochondria. J Biol Chem. 1997;272:21396–21402. doi: 10.1074/jbc.272.34.21396. [DOI] [PubMed] [Google Scholar]
- 2.Chiu N, Chiu A, Suyama Y. Native and imported transfer RNA in mitochondria. J Mol Biol. 1975;94:37–50. doi: 10.1016/s0022-2836(75)80157-4. [DOI] [PubMed] [Google Scholar]
- 3.Das S, Adhya S. Organization and chromosomal localization of β-tubulin genes in Leishmania donovani. J Biosci. 1990;15:239–248. [Google Scholar]
- 4.Ghosh A, Ghosh T, Ghosh S, Das S, Adhya S. Interaction of small ribosomal and transfer RNAs with a protein from Leishmania donovani. Nucleic Acids Res. 1994;22:1663–1669. doi: 10.1093/nar/22.9.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gray M W, Burger G, Lang B F. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 6.Gray M W, Lang B F, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Brossard N, De Lage E, Littlejohn T G, Plante I, Rioux P, Saint-Louis D, Zhu Y, Burger G. Genome structure and gene content in protist mitochondrial DNAs. Nucleic Acids Res. 1998;26:865–878. doi: 10.1093/nar/26.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancock K, Hajduk S L. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J Biol Chem. 1990;265:19203–19215. [PubMed] [Google Scholar]
- 8.Hauser R, Schneider A. tRNAs are imported into the mitochondria of Trypanosoma brucei independent of their genomic context and of their genetic origin. EMBO J. 1995;14:4212–4220. doi: 10.1002/j.1460-2075.1995.tb00095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar R, Marechal-Drouard L, Akama K, Small I. Striking differences in mitochondrial tRNA import between different plant species. Mol Gen Genet. 1996;252:404–411. doi: 10.1007/BF02173005. [DOI] [PubMed] [Google Scholar]
- 10.Lima B D, Simpson L. Sequence dependent in vivo importation of tRNAs into the mitochondrion of Leishmania tarentolae. RNA. 1996;2:429–440. [PMC free article] [PubMed] [Google Scholar]
- 11.Lye L-F, Chen D-H T, Suyama Y. Selective import of nuclear-encoded tRNAs into mitochondria of the protozoan Leishmania tarentolae. Mol Biochem Parasitol. 1993;58:233–246. doi: 10.1016/0166-6851(93)90045-y. [DOI] [PubMed] [Google Scholar]
- 12.Mahapatra S, Adhya S. Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane-bound receptors. J Biol Chem. 1996;271:20432–20437. doi: 10.1074/jbc.271.34.20432. [DOI] [PubMed] [Google Scholar]
- 13.Mahapatra S, Ghosh S, Bera S K, Ghosh T, Das A, Adhya S. The D arm of tRNATyr is necessary and sufficient for import into Leishmania mitochondria in vitro. Nucleic Acids Res. 1998;26:2037–2041. doi: 10.1093/nar/26.9.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahapatra S, Ghosh T, Adhya S. Import of small RNAs into Leishmania mitochondria in vitro. Nucleic Acids Res. 1994;22:3381–3386. doi: 10.1093/nar/22.16.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin R P, Schneller J-M, Stahl A J, Dirheimer G. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry. 1979;18:4600–4605. doi: 10.1021/bi00588a021. [DOI] [PubMed] [Google Scholar]
- 16.Mottram J C, Bell S D, Barry D J. tRNAs of Trypanosoma brucei: unusual gene organization and mitochondrial importation. J Biol Chem. 1991;266:18313–18317. [PubMed] [Google Scholar]
- 17.Mukherjee S, Bhattacharyya S N, Adhya S. Stepwise transfer of tRNA through the double membrane of Leishmania mitochondria. J Biol Chem. 1999;274:31249–31255. doi: 10.1074/jbc.274.44.31249. [DOI] [PubMed] [Google Scholar]
- 18.Nabholz C E, Horn E K, Schneider A. tRNAs and proteins are imported into mitochondria of Trypanosoma brucei by two distinct mechanisms. Mol Biol Cell. 1999;10:2547–2557. doi: 10.1091/mbc.10.8.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ragan C I, Wilson M T, Darley-Usmar V M, Lowe P N. Sub-fractionation of mitochondria and isolation of the proteins of oxidative phosphorylation. In: Darley-Usmar V M, Rickwood D, Wilson M T, editors. Mitochondria, a practical approach. Oxford, England: IRL Press; 1987. pp. 79–112. [Google Scholar]
- 20.Rusconi C P, Cech T R. The anticodon is the signal sequence for mitochondrial import of glutamine tRNA in Tetrahymena. Genes Dev. 1996;10:2870–2880. doi: 10.1101/gad.10.22.2870. [DOI] [PubMed] [Google Scholar]
- 21.Schneider A, Martin J, Agabian N. A nuclear encoded tRNA of Trypanosoma brucei is imported into mitochondria. Mol Cell Biol. 1994;14:2317–2322. doi: 10.1128/mcb.14.4.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi X, Chen D H-T, Suyama Y. A nuclear tRNA gene cluster in the protozoan Leishmania tarentolae and differential distribution of nuclear-encoded tRNAs between the cytosol and mitochondria. Mol Biochem Parasitol. 1994;65:23–37. doi: 10.1016/0166-6851(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 23.Simpson A M, Suyama Y, Dewes H, Campbell D, Simpson L. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 1989;17:5427–5445. doi: 10.1093/nar/17.14.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volckaert G, Fiers W. Micro thin-layer techniques for rapid sequence analysis of 32P-labeled RNA: double digestion and pancreatic ribonuclease analyses. Anal Biochem. 1977;83:228–239. doi: 10.1016/0003-2697(77)90531-0. [DOI] [PubMed] [Google Scholar]
- 25.Yermovsky-Kammerer A E, Hajduk S L. In vitro import of a nuclearly encoded tRNA into the mitochondrion of Trypanosoma brucei. Mol Cell Biol. 1999;19:6253–6259. doi: 10.1128/mcb.19.9.6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuker M. Computer prediction of RNA structure. Methods Enzymol. 1989;180:262–268. doi: 10.1016/0076-6879(89)80106-5. [DOI] [PubMed] [Google Scholar]