Abstract
Objective:
Breast cancer is the most common cancer in American women, except for skin cancers. In this meta-analysis, the associations of polymorphisms within paraoxonase 1 (PON1), leptin (LEP) and leptin receptor (LEPR) genes with susceptibility to breast cancer were comprehensively evaluated.
Methods:
A universal search in PubMed, Scopus, CNKI, SID, Web of Knowledge and Google Scholar was performed to identify relevant studies up to 01 May, 2021. The strength of the associations was estimated by Odds ratios (ORs) with 95% confidence intervals (95% CIs).
Results:
A total of 39 case-control studies including 7 studies with 2005 cases and 2748 controls were on PON1 rs662, 6 studies with 2,031 cases and 1,973 controls on PON1 rs854560, 12 studies with 3,444 cases and 3,583 controls on LEP rs7799039, and 14 studies with 5,330 cases and 6,188 controls on LEPR rs1137101 were selected. Pooled data showed that PON1 rs662 and rs854560 polymorphisms were associated with risk of breast cancer in overall population, but not LEP rs7799039 and LEPR rs1137101.
Conclusions:
Our pooled data revealed that the PON1 rs662 and rs854560 polymorphisms were significantly associated with an increased risk of breast cancer in the overall population. However, LEP rs7799039 and LEPR rs1137101 polymorphisms were not associated.
Key Words: Breast cancer, paraoxonase 1, leptin, leptin receptor, polymorphism
Introduction
Global facts and figures about the cancer revealed that breast cancer still key public health concern and leading cause of deaths among women globally (Jafari-Nedooshan et al., 2017; Moghimi et al., 2018). Heightened awareness of breast cancer risk in the past decades has led to an increase in the detection methods which can be used to detect the breast cancer in the early stages (Dinegde and Xuying, 2017). In the more affluent countries, mammography screening has been in place for a few decades and has successfully reduced mortality (Motamedi et al., 2012; Najminejad et al., 2020; Esmaeili et al., 2021). However, in developing countries, screening and paid little attention to fight with breast cancer is one of the lowest priorities in health policy makers (da Costa Vieira et al., 2017). Breast cancer is most likely triggered and/or promoted by multiple risk factors. The two strongest risk factors for breast cancer are gender and age (Feng et al., 2018). The etiological make-up of a heterogeneous and complex disease such as breast cancer is diverse and includes genetics and environmental factors. Breast cancer susceptibility gene 1 (BRCA1) and breast cancer susceptibility gene 2 (BRCA2) are the two major genes associated with hereditary breast and ovarian cancer (Forat-Yazdi et al., 2015; Neamatzadeh et al., 2015). However, there are more than 30 instances of SNPs identified as breast cancer susceptibility loci in the genome by GWAS (Kaklamani et al., 2011). Paraoxonase 1 (PON1), leptin (LEP) and leptin receptor (LEPR) genes are good example of a GWAS-identified locus that has been implicated in development of breast cancer (Gallicchio et al., 2007; Liu and Liu, 2011).
PON1, also called serum aromatic esterase 1, is the main means of protection of the nervous system against the neurotoxicity of organophosphates in serum (Richard et al., 2013; Mackness and Sozmen, 2020). Moreover, PON1 hydrolysis numerous exogenous and endogenous esters, such as arylesters, homocysteine thiolactone (HTL), other lactones, and cyclic carbonates (Costa et al, 2011; Seow et al., 2016). The human PON1 (MIM#602720) gene is a member of a multigene family consisting of three members including PON2 and PON3, which share ≈60% sequence identity with PON1 (Gallicchio et al., 2007; Liu and Liu, 2011). However, PON1 remains the most popular member of this family. The PON1 gene is located on chromosome 7q21.22, consisting 9 exons and spans 33.2 kb (Li et al., 1997). Of the PON1 polymorphisms, PON1 rs662 and rs854560 are most widely studied for their association with susceptibility to different cancers (Seow et al., 2016). Moreover, human LEP gene plays a critical role in energy expenditure as well as the progression of carcinogenesis (Tang et al., 2019). It is also reported that LEP may affect angiogenesis, inflammation, thrombosis, and tumor growth, invasion, and metastasis (Tang et al., 2019). It is revealed that the LEP signal may be transmitted through several signaling pathways such as JAK/STAT, MAPK, PI3K, Wnt/β-catenin, and ERK (Kavitha et al., 2013). The human LEP (MIM#164160) is located on chromosome 7q31.3, consists of three exons and spans approximately 16.4 kb (Funcke et al, 2014). It is highly polymorphic and the LEP rs7799039 G>A SNP is the most widely studied for its role in development of different human diseases (Tang et al., 2019).
Over the past decade, several molecular epidemiological studies have been performed to identify the association of PON1 rs662, rs854560, LEP rs7799039G>A, and LEPR rs1137101 polymorphisms with susceptibility to breast cancer, but the findings have been conflicting. Thus, we performed a systematic review and updated meta-analysis to obtain a more precise assessment of the association between PON1, LEP and LEPR polymorphisms and the risk of breast cancer.
Materials and Methods
Search strategy
This meta-analysis was reported based on the Preferred Reporting Items for Meta-analyses (PRISMA) guideline. In this meta-analysis, we carried out electronic literature retrieval in Medicine’s PubMed, Scopus, EMBASE, Web of Knowledge, Cochrane Library, Google Scholar, Scientific Information Database (SID), WanFang, VIP, Chinese Biomedical Database (CBD), Scientific Electronic Library Online (SciELO) and China National Knowledge Infrastructure (CNKI) database up to 01 May, 2021. The following keywords and terms were used to search: (‘’breast cancer’’ OR “breast tumor” OR “breast neoplasm” OR “breast malignant tumor” OR “breast carcinoma’’) AND (‘’ Paraoxonase 1’’ OR ‘’Serum Paraoxonase/Arylesterase’’ OR ‘’Serum Aryldialkylphosphatase’’ OR ‘’Aromatic Esterase’’ OR ‘’Arylesterase’’ OR ‘’A-Esterase’’ OR ‘’Esterase’’ OR ‘’PON1) AND (‘’Leptin’’ OR ‘’Obesity Factor’’ OR ‘’Obese Protein’’ OR ‘’LEP’’) AND (‘’Leptin Receptor’’ OR ‘’LEPR’’ OR ‘’OBR’’ ‘’OB Receptor’’ OR ‘’HuB219’’ OR ‘’CD295’’) AND (‘’Q192R’’ OR ‘’rs662’’ OR ‘’L55M’’ OR ‘’rs854560’’ OR ‘’LEP G2548A’’OR ‘’rs7799039’’ OR ‘’LEPR Q223R’’ OR ‘’rs1137101’’ OR ‘’LEPR Lys109Arg’’ OR ‘’rs1137100’’ OR ‘’rs1137101’’ OR ‘’c.668A>G’’ OR ‘’p.Gln223Arg’’ OR ‘’Arg223Gln’’ OR ‘’R223Q’’ OR ‘’Q223R’’ OR ‘’rs7799039’’ OR ‘’2548G/A’’) AND (‘’Gene’’ OR ‘’Genotype’’ OR ‘’Allele’’ OR ‘’Polymorphism’’ OR ‘’ Single nucleotide polymorphisms’’ OR ‘’SNP’’ OR ‘’Variation’’ OR ‘’Mutation’’). No restrictions were placed on the language, year of publication, ethnicity, and sample size. The references in included studies and reviewers were carefully checked for other potential data. When a publication involved some subgroups, it was treated separately.
Selection and Exclusion Criteria
The major selection criteria were as follows: 1) studies with case-control or cohort design; 2) studies that assessed the association of genetic variants within PON1, LEP and LEPR gene with risk of breast cancer; and (2) presented sufficient data to calculate the pooled-estimating. Accordingly, the major exclusion criteria were: 1) Studies did not evaluate the association of LEP, LEPR and PON1 polymorphisms and risk of breast cancer; 2) studies focusing on animals or in vitro; 3) Studies that did not provide usable or sufficient data for pooling; 4) case only studies or no controls; 5) linkage studies and family based studies (twins and sibling); 6) case reports, abstracts, comments, conference abstracts, editorials, reviews, meta-analysis; and 7) duplicated studies or data. When duplicated studies were published by the same author obtained from the same patient sample, only the one with the largest sample size was included in this meta-analysis.
Data extraction
Two authors independently extracted the data from each eligible study and if the extracted data was different, they would review the publication again and reached consensus. If they could not get a consistent assessment, third author would be invited to resolve the dispute and a final decision was made. The following data were extracted from each study: first author name, year of publication, country of origin, ethnicity (Asian, Caucasians, Africans and Mixed populations), numbers of cases and controls, source of control, genotype and allele frequencies, genotyping method, minor allele frequency (MAFs) and Hardy-Weinberg equilibrium (HWE) in controls.
Statistical Analysis
All of the statistical calculations were performed using Comprehensive Meta-Analysis (CMA) software version 2.0 (Biostat, USA). Two-sided P-values < 0.05 were considered statistically significant. The strength of association between genetic variants at PON1, LEP and LEPR genes and risk of breast cancer was estimated by Odds ratios (ORs) with 95% confidence intervals (95% CIs). The significance of the pooled effect size was determined by Z-test, in which P<0.05 was considered statistically significant. The associations was evaluated under all five genetic models, i.e., allele (B vs. A), homozygote (BB vs. AA), heterozygote (BA vs. AA), dominant (BB+BA vs. AA), and the recessive (BB vs. BA+AA), in which ‘’B’’ presents mutant and ‘’A’’ wild allele (Jafari-Nedooshan et al., 2019; Jafari et al., 2020). Between-study heterogeneity was estimated using a Cochran-based Q statistical test, with P-values less than 0.1 indicated the absence of indicated heterogeneity among studies. Moreover, a quantitative measure of between-study heterogeneity was tested using the I2 statistic (range of 0 to 100%), in which the heterogeneity was considered low, moderate, and high based on I2 values of 25%, 50%, and 75%, respectively. Thus, there was no heterogeneity (P > 0.1 or I2 < 50%) the fixed-effect model (Mantel-Haenszel method) was applied. There was heterogeneity (P <0.1 and I2 > 50%) the random-effect (DerSimonian-Laird method) model was used for analysis. Stratified analysis was carried out on the basis of ethnicity and source of controls. The Hardy-Weinberg equilibrium (HWE) for controls in each study was evaluated using the χ2 test and P >0.05 was considered to be consistent with HWE (Bahrami Dastgheib et al., 2020; Bahrami Shajari et al., 2020). To explore the influence of an individual study on the pooled data, sensitivity analysis was also used to confirm the stability of the results under all genetic models. Begg’s funnel plot test was used to assess possible publication bias, with P <0.05 being considered to present statistical significance.
Results
Selected Studies Characteristics
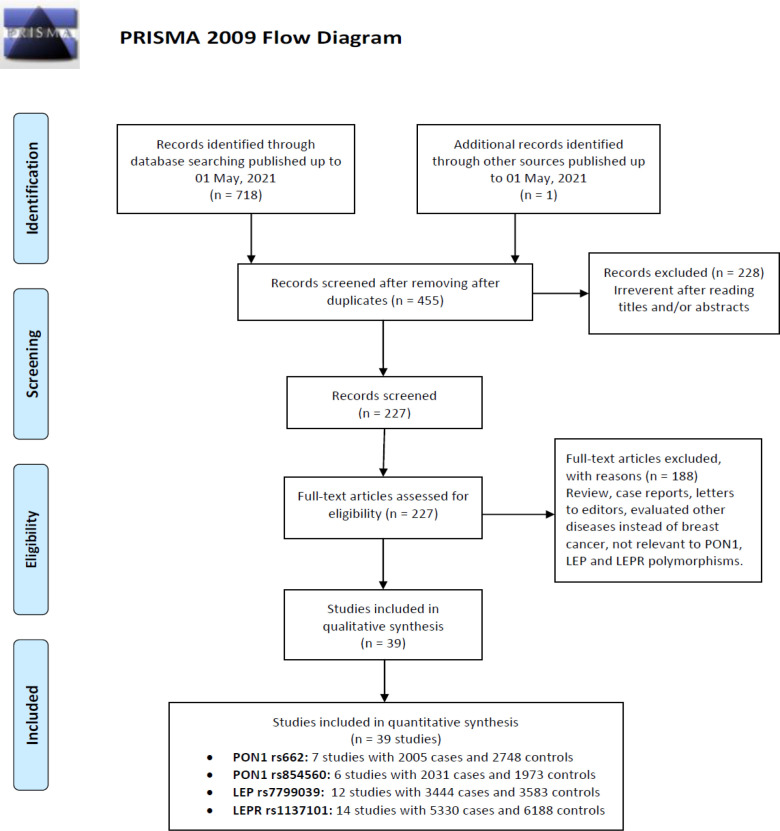
The selection process of eligible studies is presented in Figure 1. Initially, 719 studies were obtained through publication search in electronic databases and other sources. Irrelevant articles were excluded by evaluating the titles and abstracts. Therefore, 76 publications were deleted for obvious irrelevance. Finally, 39 case-control studies including 7 studies with 2005 cases and 2,748 controls were on PON1 rs662, 6 studies with 2,031 cases and 1,973 controls on PON1 rs854560, 12 studies with 3,444 cases and 3,583 controls on LEP rs7799039, and 14 studies with 5,330 cases and 6,188 controls on LEPR rs1137101 were selected. Pooled data showed that PON1 rs662 and rs854560 polymorphisms were associated with risk of breast cancer in overall population, but not LEP rs7799039 and LEPR rs1137101. Table 1 describes principal characteristics of included studies. The studies have been carried out in USA, Brazil, Italy, Malaysia, Egypt, turkey, China, Iran, Mexico, Sri Lanka, India, Tunisia, Nigeria, and Korea. Among these studies, eight studies were conducted among Asians, two studies among Caucasians and two studies Africans. Seven different genotyping methods were used: PCR, PCR-RFLP, TaqMan, SNPstream, and TOFMS. The genotype, allele and minor allele frequency (MAF) in each study for PON1 rs662, rs854560, LEP rs7799039 and LEPR rs1137101 are shown in Table 1. Moreover, the distribution of genotypes in the controls was in agreement with Hardy-Weinberg equilibrium (HWE) for all selected studies, except for one study on IL-8 -251T>A polymorphism (Table 1).
Figure 1.
Flowchart of Literature Search and Selection Process
Table 1.
Characteristics of the Case-Control Studies Included in the Meta-Analyses
| First Author | Country (Ethnicity) |
SOC | Genotyping Method |
Cases/ controls |
Cases | Controls | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Allele | Genotype | Allele | |||||||||||
| PON1 rs662 | AA | AG | GG | A | G | AA | AG | GG | A | G | ||||
| Stevens 2006 | USA (Caucasian) | PB | PCR-RFLP | 483/483 | 259 | 182 | 42 | 700 | 266 | 238 | 198 | 47 | 674 | 292 |
| Gallicchio 2007 | Brazil (Mixed) | PB | PCR-RFLP | 58/904 | 38 | 15 | 5 | 91 | 25 | 469 | 353 | 82 | 1291 | 517 |
| Antognelli 2009 | Italy (Caucasian) | PB | PCR-RFLP | 547/544 | 484 | 50 | 13 | 1018 | 76 | 340 | 152 | 52 | 832 | 256 |
| Naidu 2010 | Malaya (Asian) | HB | PCR-RFLP | 387/252 | 200 | 158 | 29 | 558 | 216 | 115 | 115 | 22 | 345 | 159 |
| Hussein 2011 | Egypt (African) | PB | PCR-RFLP | 100/100 | 51 | 41 | 8 | 143 | 57 | 46 | 42 | 12 | 134 | 66 |
| Kaya 2016 | Turkey (Caucasian) | HB | TaqMan | 32/35 | 10 | 11 | 11 | 31 | 33 | 5 | 13 | 17 | 23 | 47 |
| Wu 2017 | China (Asian) | HB | TaqMan | 365/378 | 155 | 156 | 54 | 466 | 264 | 167 | 156 | 55 | 490 | 266 |
| Agachan 2019 | Turkey (Caucasian) | PB | PCR-RFLP | 33/52 | 17 | 4 | 12 | 38 | 28 | 6 | 29 | 17 | 41 | 63 |
| PON1 rs854560 | TT | TA | AA | T | A | TT | TA | AA | T | A | ||||
| Stevens 2006 | USA (Caucasian) | PB | PCR-RFLP | 483/493 | 176 | 230 | 77 | 582 | 384 | 202 | 233 | 58 | 637 | 349 |
| Antognelli 2009 | Italy (Caucasian) | PB | PCR-RFLP | 547/607 | 107 | 115 | 325 | 329 | 765 | 188 | 188 | 231 | 564 | 650 |
| Naidu 2010 | Malaya (Asian) | HB | PCR-RFLP | 387/269 | 159 | 178 | 50 | 496 | 278 | 126 | 126 | 17 | 378 | 160 |
| Hussein 2011 | Egypt (African) | PB | PCR-RFLP | 100/76 | 19 | 21 | 60 | 59 | 141 | 35 | 35 | 6 | 105 | 47 |
| Wu 2017 | China (Asian) | HB | TaqMan | 483/483 | 284 | 72 | 9 | 640 | 90 | 346 | 30 | 2 | 722 | 34 |
| Farmohammadi 2019 | Iran (Asian) | HB | PCR-RFLP | 150/150 | 47 | 65 | 38 | 159 | 141 | 66 | 59 | 25 | 191 | 109 |
| LEP rs7799039 | GG | GA | AA | G | A | GG | GA | AA | G | A | ||||
| Snoussi 2006 | Tunisia (Caucasian) | HB | PCR-RFLP | 308/222 | 37 | 152 | 119 | 226 | 390 | 11 | 99 | 112 | 37 | 152 |
| Vairaktaris 2008 | Greece (Caucasian) | HB | PCR | 150/152 | 32 | 78 | 40 | 142 | 158 | 112 | 99 | 11 | 32 | 78 |
| Teras 2009 | USA (Caucasian) | PB | SNPstream | 1077/1086 | 445 | 445 | 187 | 1335 | 819 | 442 | 442 | 202 | 445 | 445 |
| Cleveland 2010 | USA (Caucasian) | PB | PCR | 1059/1101 | 226 | 492 | 341 | 944 | 1174 | 180 | 561 | 360 | 226 | 492 |
| Morris 2013 | Mexico (Mixed) | HB | PCR | 130/189 | 22 | 71 | 37 | 115 | 145 | 46 | 95 | 48 | 22 | 71 |
| Rostami 2015 | Iran (Asian) | HB | PCR-RFLP | 203/171 | 115 | 64 | 24 | 294 | 112 | 63 | 77 | 31 | 115 | 64 |
| Mahmoudi 2015 | Iran (Asian) | PB | PCR-RFLP | 45/41 | 27 | 11 | 7 | 65 | 25 | 17 | 19 | 5 | 27 | 11 |
| Karakus 2015 | Turkey (Caucasian) | PB | PCR | 199/185 | 49 | 105 | 45 | 203 | 195 | 47 | 98 | 40 | 49 | 105 |
| Mohammadzadeh 2015 | Iran (Asian) | HB | PCR-RFLP | 100/100 | 36 | 55 | 9 | 127 | 73 | 52 | 45 | 3 | 36 | 55 |
| Rodrigo 2017 | Sri Lanka (Asian) | PB | PCR | 80/80 | 32 | 43 | 5 | 107 | 53 | 53 | 24 | 3 | 32 | 43 |
| Liu 2018 | China (Asian) | HB | TOFMS | 434/442 | - | 182 | 252 | - | 686 | - | 206 | 236 | - | 182 |
| Geriki 2019 | India (Asian) | HB | PCR-RFLP | 93/186 | 15 | 45 | 33 | 75 | 111 | 54 | 75 | 57 | 15 | 45 |
| LEPR rs1137101 | AA | AG | GG | A | G | AA | AG | GG | A | G | ||||
| Snoussi 2006 | Tunisia (African) | NS | PCR-RFLP | 308/222 | 98 | 145 | 65 | 341 | 275 | 102 | 90 | 30 | 294 | 150 |
| Woo 2006 | Korea (Asian) | HB | PCR | 45/45 | 0 | 12 | 33 | 12 | 78 | 0 | 8 | 37 | 8 | 82 |
| Gallicchio 2007 | USA (Caucasian) | PB | TaqMan | 53/872 | 14 | 24 | 15 | 52 | 54 | 278 | 443 | 151 | 999 | 745 |
| Han 2008 | China (Asian) | HB | PCR | 240/500 | 33 | 41 | 166 | 107 | 373 | 12 | 78 | 410 | 102 | 898 |
| Okobia 2008 | Nigeria (African) | HB | PCR-RFLP | 209/209 | 46 | 107 | 56 | 199 | 219 | 56 | 107 | 46 | 219 | 199 |
| Teras 2009 | USA (Caucasian) | PB | SNP stream | 648/659 | 128 | 332 | 181 | 588 | 694 | 125 | 314 | 211 | 564 | 736 |
| Cleveland 2010 | USA (Caucasian) | PB | PCR | 1059/1098 | 173 | 521 | 355 | 867 | 1231 | 187 | 551 | 360 | 925 | 1271 |
| Nyante 2011 | USA (Caucasian) | PB | PCR | 1972/1775 | 494 | 952 | 526 | 1940 | 2004 | 416 | 847 | 485 | 1679 | 1817 |
| Kim 2012 | Korea (Asian) | HB | PCR | 390/447 | 8 | 88 | 294 | 104 | 676 | 6 | 91 | 350 | 103 | 791 |
| Mohammadzadeh 2014 | Iran (Asian) | HB | PCR-RFLP | 100/100 | 25 | 56 | 19 | 106 | 94 | 54 | 40 | 6 | 148 | 52 |
| Mahmoudi 2015 | Iran (Asian) | PB | PCR-RFLP | 45/41 | 19 | 25 | 1 | 63 | 27 | 17 | 18 | 6 | 52 | 30 |
| Wang 2015 | China (Asian) | PB | PCR-RFLP | 150/128 | 20 | 25 | 105 | 65 | 235 | 3 | 19 | 106 | 25 | 231 |
| Rodrigo 2017 | Sri Lanka (Asian) | PB | PCR-RFLP | 80/80 | 65 | 9 | 6 | 139 | 21 | 60 | 6 | 14 | 126 | 34 |
| El-Hussiny 2017 | Egypt (African) | NS | PCR-RFLP | 48/79 | 24 | 15 | 9 | 63 | 33 | 22 | 24 | 2 | 68 | 28 |
SOC, Source Of Controls; HB, Hospital Based; PB, Population Based; RFLP, Restriction Fragment Length Polymorphism; MAF, Minor Allele Frequency; HWE, Hardy-Weinberg Equilibrium
Quantitative Data Synthesis
PON1 rs662
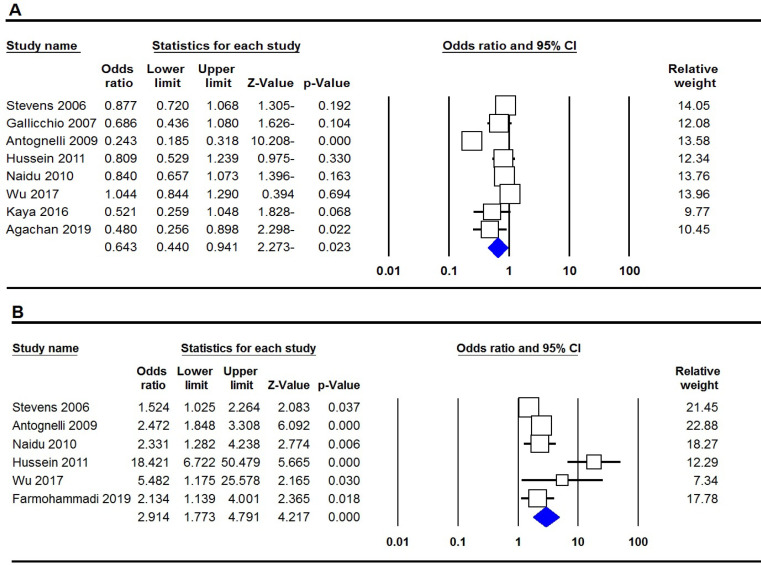
Table 2 listed the main results of the meta-analysis of PON1 rs662 polymorphism and breast cancer risk. When all the eligible studies were pooled into the meta-analysis, a significant association was found between PON1 rs662 and breast cancer under all three genetic models, i.e., allele (G vs. A: OR= 0.719, 95% CI: 0.648-0.798; p≤0.001, Figure 1A), homozygote (GG vs. AA: OR= 0.542, 95% CI: 0.332-0.885; p=0.014) and dominant (GG+GA vs. AA: OR= 0.720, 95% CI: 0.330-0.864; p=0.011). When subgroup analysis by ethnicity performed the results showed that the PON1 rs662 polymorphism was associated with breast cancer risk among Caucasian women under two genetic models, i.e., homozygote (GG vs. AA: OR= 0.341, 95% CI: 0.134-0.866; p=0.024) and dominant (GG+GA vs. AA: OR= 0.317, 95% CI: 0.119-0.839; p=0.021), but not among Asians. Moreover, subgroup analysis by source of controls showed that the variant was associated with breast cancer in PB group of studies.
Table 2.
Meta-Analysis Results of Association between PON1 rs662 Polymorphism and Breast Cancer Risk
| Polymorphism | Genetic Model | Type of Model | Heterogeneity | Odds Ratio | Publication Bias | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | Ztest | POR | PBeggs | PEggers | |||
| Overall | G vs. A | Random | 91.39 | ≤0.001 | 0.719 | 0.648-0.798 | -6.234 | ≤0.001 | 0.063 | 0.467 |
| GG vs. AA | Random | 73.8 | ≤0.001 | 0.542 | 0.332-0.885 | -2.446 | 0.014 | 0.035 | 0.221 | |
| GA vs. AA | Fixed | 14.09 | 0.32 | 1.011 | 0.800-1.278 | 0.092 | 0.926 | 0.901 | 0.374 | |
| GG+GA vs. AA | Random | 90.55 | ≤0.001 | 0.534 | 0.330-0.864 | -2.554 | 0.011 | 0.107 | 0.428 | |
| GG vs. GA+AA | Random | 62.05 | 0.01 | 0.72 | 0.492-1.053 | -1.696 | 0.09 | 0.173 | 0.576 | |
| Ethnicity | ||||||||||
| Caucasian | G vs. A | Random | 94.69 | ≤0.001 | 0.48 | 0.220-1.047 | -1.846 | 0.065 | 1 | 0.694 |
| GG vs. AA | Random | 82.08 | 0.001 | 0.341 | 0.134-0.866 | -2.262 | 0.024 | 1 | 0.479 | |
| GA vs. AA | Random | 57.75 | 0.069 | 0.894 | 0.481-1.661 | -0.355 | 0.723 | 0.734 | 0.486 | |
| GG+GA vs. AA | Random | 93.93 | ≤0.001 | 0.317 | 0.119-0.839 | -2.312 | 0.021 | 1 | 0.578 | |
| GG vs. GA+AA | Random | 78.94 | 0.003 | 0.594 | 0.278-1.269 | -1.345 | 0.179 | 0.734 | 0.872 | |
| Asian | G vs. A | Fixed | 41.99 | 0.189 | 0.951 | 0.810-1.116 | -0.617 | 0.537 | NA | NA |
| GG vs. AA | Fixed | 0 | 0.378 | 0.943 | 0.663-1.341 | -0.326 | 0.744 | NA | NA | |
| GA vs. AA | Fixed | 0 | 0.952 | 1.027 | 0.721-1.462 | 0.146 | 0.884 | NA | NA | |
| GG+GA vs. AA | Fixed | 50.42 | 0.159 | 0.931 | 0.751-1.153 | -0.658 | 0.511 | NA | NA | |
| GG vs. GA+AA | Fixed | 0 | 0.607 | 0.959 | 0.688-1.337 | -0.247 | 0.805 | NA | NA | |
| Source of Controls | ||||||||||
| HB | G vs. A | Fixed | 54.8 | 0.109 | 0.923 | 0.789-1.079 | -1.01 | 0.313 | 0.296 | 0.332 |
| GG vs. AA | Fixed | 36.58 | 0.207 | 0.878 | 0.625-1.233 | -0.75 | 0.453 | 0.296 | 0.105 | |
| GA vs. AA | Fixed | 0 | 0.918 | 1.05 | 0.750-1.470 | 0.284 | 0.777 | 0.296 | 0.163 | |
| GG+GA vs. AA | Fixed | 52.79 | 0.12 | 0.904 | 0.732-1.117 | -0.933 | 0.351 | 0.296 | 0.429 | |
| GG vs. GA+AA | Fixed | 0 | 0.514 | 0.907 | 0.662-1.243 | -0.609 | 0.543 | 0.296 | 0.007 | |
| PB | G vs. A | Random | 93.28 | ≤0.001 | 0.563 | 0.311-1.020 | -1.895 | 0.058 | 0.462 | 0.793 |
| GG vs. AA | Random | 77.27 | 0.001 | 0.445 | 0.215-0.920 | -2.186 | 0.029 | 0.462 | 0.645 | |
| GA vs. AA | Fixed | 49.26 | 0.096 | 0.976 | 0.704-1.353 | -0.147 | 0.883 | 0.22 | 0.354 | |
| GG+GA vs. AA | Random | 92.42 | ≤0.001 | 0.429 | 0.209-0.878 | -2.316 | 0.021 | 0.462 | 0.657 | |
| GG vs. GA+AA | Random | 73.1 | 0.005 | 658 | 0.351-1.232 | -1.307 | 0.191 | 0.806 | 0.96 | |
PON1 rs854560
Table 2 listed the main results of the meta-analysis of PON1 rs854560 polymorphism and breast cancer risk. Pooled data showed that the PON1 rs854560 polymorphism was significantly associated with risk of breast cancer under all four genetic models, i.e., allele (A vs. T: OR=2.107, 95% CI: 1.401-3.167; p≤0.001), homozygote (AA vs. TT: OR= 3.214, 95% CI: 1.757-5.879; p≤0.001, Figure 2B), heterozygote (AT vs. TT: OR= 0.379, 95% CI: 0.208-0.691; p=0.002), dominant (AA+AT vs. TT: OR= 1.868, 95% CI: 1.293-2.700; p=0.001) and recessive (AA vs. AT+TT: OR= 3.067, 95% CI: 1.687-5.575; p≤0.001). Subgroup analysis by ethnicity revealed that PON1 rs854560 polymorphism was a significantly associated with breast cancer among Asian and Caucasian women.
Figure 2.
Forest Plot for Association of the PON1 Polymorphisms with Breast Cancer Risk in Overall Population. A, rs662 (allele model); B, rs854560 (homozygote model)
LEPR rs1137101
Table 2 listed the main results of the meta-analysis of LEPR rs1137101 polymorphism and breast cancer risk. When all the eligible studies were pooled into the meta-analysis, no significant association was found between LEPR rs1137101 and breast cancer under all five genetic models in overall population. Subgroup analysis by ethnicity revealed that the variant was a significantly associated with breast cancer among African women under all four genetic models, i.e., allele (A vs. G: OR= 0.772, 95% CI: 1.161-1.654; p≤0.001), homozygote (AA vs. GG: OR= 0.772, 95% CI: 1.339-2.786; p≤0.001), heterozygote (AG vs. GG: OR= 0.772, 95% CI: 1.010-1.772; p=0.043), and dominant (AA+AG vs. GG: OR= 0.772, 95% CI: 1.268-2.137; p≤0.001), but not among Caucasians and Asians.
LEP rs7799039G>A
Table 2 listed the main results of the meta-analysis of LEP rs7799039G>A polymorphism and breast cancer risk. Pooled data showed that this polymorphism was not associated with risk of breast cancer under all four genetic models in overall population. Moreover, subgroup analysis by ethnicity and source of controls revealed that LEP rs7799039G>A polymorphism was not significantly associated with breast cancer.
Test of Heterogeneity and sensitivity analyses
As shown in Tables 2 and 4, there was a significant heterogeneity existed under most genetic models for PON1 rs662, rs854560, LEP rs7799039 and LEPR rs1137101 polymorphisms. Thus, stratified analyses by ethnicity and source of controls carried out to find the potential source of heterogeneity. Results showed that ethnicity and source of controls have overall effect on the heterogeneity for these polymorphisms. We carried out the sensitivity analyses to assess the robustness of the results by removing each study in turn and all the results were not essentially altered, suggesting that the results of the present meta-analysis were statistically stable.
Table 4.
Meta-Analysis Results of Association between LEP rs7799039 Polymorphism and Breast Cancer Risk
| Polymorphism | Genetic Model | Type of Model | Heterogeneity | Odds Ratio | Publication Bias | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | Ztest | POR | PBeggs | PEggers | |||
| Overall | A vs. G | Random | 82.4 | ≤0.001 | 0.98 | 0.761-1.263 | -0.154 | 0.878 | 0.265 | 0.51 |
| AA vs. GG | Random | 71.3 | 0.001 | 0.905 | 0.572-1.432 | -0.428 | 0.669 | 0.386 | 0.405 | |
| AG vs. GG | Fixed | 12.18 | 0.335 | 0.993 | 0.857-1.150 | -0.095 | 0.924 | 0.901 | 0.57 | |
| AA+AG vs. GG | Random | 84.71 | ≤0.001 | 0.931 | 0.600-1.446 | -0.319 | 0.75 | 0.386 | 0.48 | |
| AA vs. AG+GG | Random | 46.09 | 0.072 | 0.931 | 0.723-1.197 | -0.56 | 0.576 | 0.71 | 0.506 | |
| Ethnicity | ||||||||||
| Asian | A vs. G | Random | 89.62 | ≤0.001 | 1.086 | 0.538-2.191 | 0.23 | 0.818 | 0.734 | 0.458 |
| AA vs. GG | Random | 76.03 | 0.006 | 1.303 | 0.404-4.203 | 0.443 | 0.658 | 0.308 | 0.092 | |
| AG vs. GG | Fixed | 0 | 0.424 | 0.832 | 0.506-1.368 | -0.724 | 0.469 | 1 | 0.294 | |
| AA+AG vs. GG | Random | 90.89 | ≤0.001 | 1.051 | 0.393-2.810 | 0.099 | 0.921 | 0.734 | 0.609 | |
| AA vs. AG+GG | Fixed | 52.94 | 0.095 | 0.914 | 0.576-1.450 | -0.382 | 0.702 | 0.308 | 0.068 | |
| Caucasian | A vs. G | Random | 69.07 | 0.039 | 0.847 | 0.675-1.063 | -1.431 | 0.153 | 1 | 0.815 |
| AA vs. GG | Random | 71.32 | 0.031 | 0.672 | 0.390-1.157 | -1.434 | 0.151 | 1 | 0.759 | |
| AG vs. GG | Fixed | 56.67 | 0.099 | 1.014 | 0.863-1.190 | 0.164 | 0.869 | 1 | 0.646 | |
| AA+AG vs. GG | Fixed | 64.42 | 0.06 | 0.708 | 0.466-1.076 | -1.616 | 0.106 | 1 | 0.837 | |
| AA vs. AG+GG | Fixed | 64.81 | 0.058 | 0.903 | 0.776-1.052 | -1.31 | 0.19 | 1 | 0.747 | |
| Source of Controls | ||||||||||
| HB | A vs. G | Random | 90.52 | ≤0.001 | 1.037 | 0.542-1.986 | 0.11 | 0.913 | 0.296 | 0.497 |
| AA vs. GG | Random | 85.4 | 0.001 | 1.286 | 0.377-4.386 | 0.402 | 0.688 | 0.296 | 0.501 | |
| AG vs. GG | Fixed | 0 | 0.443 | 0.94 | 0.638-1.385 | -0.313 | 0.754 | 1 | 0.259 | |
| AA+AG vs. GG | Random | 90.74 | ≤0.001 | 1.09 | 0.412-2.886 | 0.173 | 0.863 | 0.846 | 0.066 | |
| AA vs. AG+GG | Random | 67.46 | 0.046 | 1.089 | 0.530-2.240 | 0.232 | 0.816 | 1 | 0.582 | |
| PB | A vs. G | Random | 74.44 | 0.008 | 1.067 | 0.771-1.477 | 0.391 | 0.696 | 0.734 | 0.477 |
| AA vs. GG | Fixed | 21.62 | 0.281 | 0.82 | 0.658-1.023 | -1.761 | 0.078 | 0.308 | 0.175 | |
| AG vs. GG | Fixed | 0 | 0.711 | 0.919 | 0.770-1.096 | -0.945 | 0.345 | 0.734 | 0.53 | |
| AA+AG vs. GG | Random | 84.25 | ≤0.001 | 1.019 | 0.552-1.880 | 0.059 | 0.953 | 0.734 | 0.525 | |
| AA vs. AG+GG | Fixed | 0 | 0.842 | 1 | 0.847-1.180 | -0.005 | 0.996 | 0.089 | 0.025 | |
Publication bias
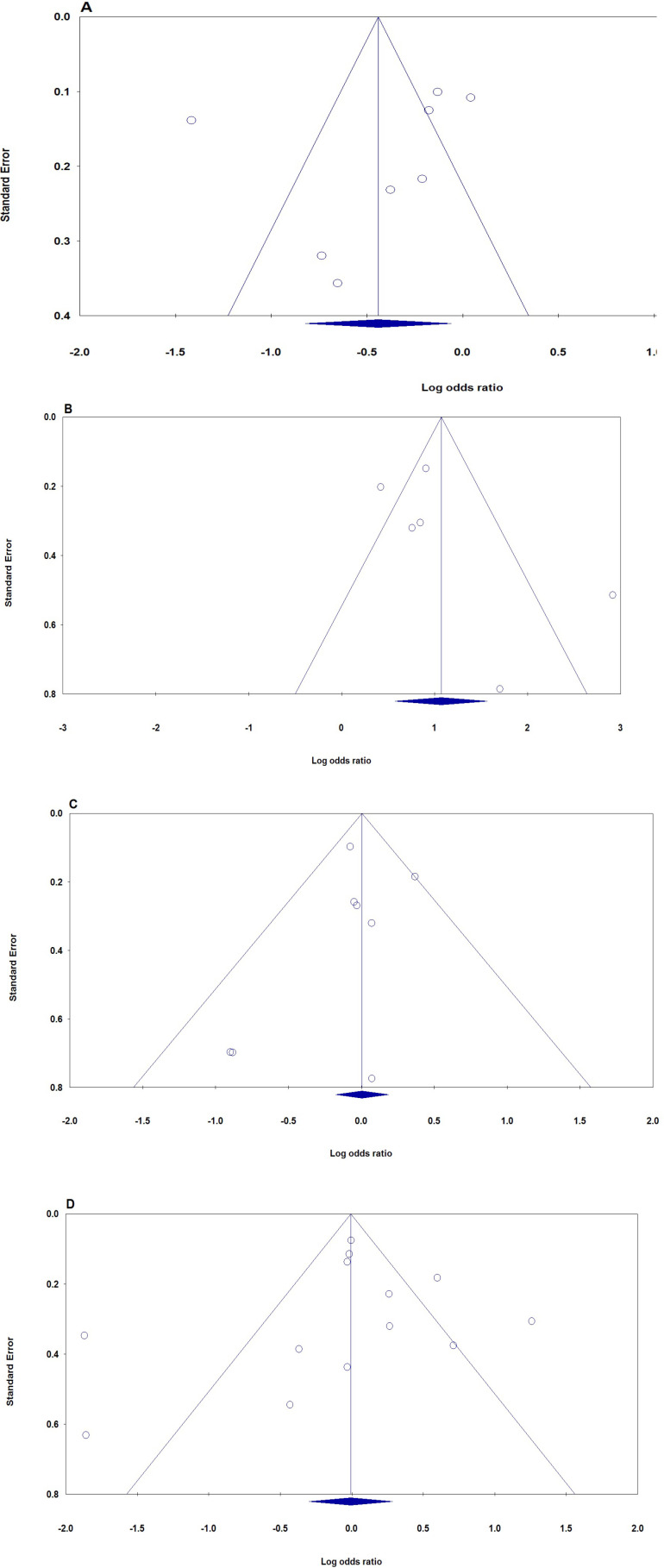
The publication bias of the studies was evaluated using the funnel plot and Egger’s test. Publication bias was not seen in the funnel plot (Figure 3). No statistically significant difference was discovered in the Egger’s test for PON1 rs662, rs854560, LEP rs7799039 and LEPR rs1137101 polymorphisms, indicating low publication bias in the current meta-analysis. Moreover, funnel plots’ shape of all comparison models did not reveal any obvious evidence of asymmetry and all P values of Egger’s tests were more than 0.05, providing statistical evidence of funnel plots’ symmetry.
Figure 3.
The Funnel Plots of Publication Bias for Association of the PON1, LEP and LEPR Polymorphism with Breast Cancer Risk in Overall Population. A: PON1 rs662 (allele mode); B: rs854560 (homozygote model), C: LEP rs7799039 (heterozygote model), and D: LEPR rs1137101 (dominant model)
Table 3.
Meta-Analysis Results of Association between PON1 rs854560 Polymorphism and Breast Cancer Risk
| Polymorphism | Genetic Model | Type of Model | Heterogeneity | Odds Ratio | Publication Bias | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | Ztest | POR | PBeggs | PEggers | |||
| Overall | A vs. T | Random | 92.43 | ≤0.001 | 2.107 | 1.401-3.167 | 3.582 | ≤0.001 | 0.22 | 0.21 |
| AA vs. TT | Random | 81.73 | ≤0.001 | 3.214 | 1.757-5.879 | 3.789 | ≤0.001 | 0.462 | 0.27 | |
| AT vs. TT | Random | 81.85 | ≤0.001 | 0.379 | 0.208-0.691 | -3.17 | 0.002 | 1 | 0.478 | |
| AA+AT vs. TT | Random | 81.83 | ≤0.001 | 1.868 | 1.293-2.700 | 3.326 | 0.001 | 0.22 | 0.12 | |
| AA vs. AT+TT | Random | 84.64 | ≤0.001 | 3.067 | 1.687-5.575 | 3.674 | ≤0.001 | 0.462 | 0.375 | |
| Ethnicity | ||||||||||
| Asian | A vs. T | Random | 82.54 | 0.003 | 1.785 | 1.150-2.772 | 2.581 | 0.01 | 0.296 | 0.248 |
| AA vs. TT | Fixed | 0 | 0.536 | 2.387 | 1.573-3.622 | 4.09 | ≤0.001 | 1 | 0.152 | |
| AG vs. TT | Random | 77.65 | 0.011 | 0.792 | 0.313-2.001 | -0.493 | 0.622 | 1 | 0.751 | |
| AA+AT vs. TT | Random | 93.29 | ≤0.001 | 1.212 | 0.469-3.132 | 0.397 | 0.691 | 1 | 0.802 | |
| AA vs. AT+TT | Fixed | 0 | 0.442 | 2.043 | 1.383-3.016 | 3.592 | ≤0.001 | 0.296 | 0.317 | |
| Caucasian | A vs. T | Random | 93.83 | ≤0.001 | 1.56 | 0.941-2.587 | 1.725 | 0.085 | NA | NA |
| AA vs. TT | Fixed | 73.12 | 0.054 | 2.086 | 1.650-2.638 | 6.143 | ≤0.001 | NA | NA | |
| AG vs. TT | Random | 79.06 | 0.029 | 0.559 | 0.331-0.946 | -2.169 | 0.03 | NA | NA | |
| AA+AT vs. TT | Random | 79.34 | 0.028 | 1.491 | 0.987-2.253 | 1.897 | 0.058 | NA | NA | |
| AA vs. AT+TT | Fixed | 81.38 | 0.02 | 1.878 | 1.134-3.109 | 2.45 | 0.014 | NA | NA | |
| Source of Controls | ||||||||||
| HB | A vs. T | Random | 82.54 | 0.003 | 1.785 | 1.150-2.772 | 2.581 | 0.01 | 0.296 | 0.248 |
| AA vs. TT | Random | 90.35 | ≤0.001 | 3.48 | 1.455-8.321 | 2.804 | 0.005 | 1 | 0.478 | |
| AG vs. TT | Random | 90.91 | ≤0.001 | 0.316 | 0.131-0.761 | -2.567 | 0.01 | 1 | 0.501 | |
| AA+AT vs. TT | Random | 93.29 | ≤0.001 | 1.212 | 0.469-3.132 | 0.397 | 0.691 | 1 | 0.802 | |
| AA vs. AT+TT | Random | 92.04 | ≤0.001 | 3.359 | 1.432-7.879 | 2.785 | 0.005 | 1 | 0.566 | |
| PB | A vs. T | Random | 95.14 | ≤0.001 | 2.254 | 1.228-4.140 | 2.622 | 0.009 | 1 | 0.482 |
| AA vs. TT | Random | 90.35 | ≤0.001 | 3.48 | 1.455-8.321 | 2.804 | 0.005 | 1 | 0.478 | |
| AG vs. TT | Random | 90.91 | ≤0.001 | 0.316 | 0.131-0.761 | -2.567 | 0.01 | 1 | 0.501 | |
| AA+AT vs. TT | Random | 82.19 | 0.004 | 1.84 | 1.137-2.976 | 2.483 | 0.013 | 0.296 | 0.413 | |
| AA vs. AT+TT | Fixed | 0 | 0.465 | 2.038 | 1.380-3.009 | 3.58 | ≤0.001 | 0.296 | 0.33 | |
Table 5.
Meta-Analysis Results of Association between LEPR rs1137101 Polymorphism and Breast Cancer Risk
| Polymorphism | Genetic Model | Type of Model | Heterogeneity | Odds Ratio | Publication Bias | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| I2 (%) | PH | OR | 95% CI | Ztest | POR | PBeggs | PEggers | |||
| Overall | A vs. G | Random | 86.48 | ≤0.001 | 0.943 | 0.780-1.139 | -0.614 | 0.539 | 0.661 | 0.781 |
| AA vs. GG | Random | 84.75 | ≤0.001 | 0.928 | 0.631-1.365 | -0.379 | 0.705 | 0.76 | 0.867 | |
| AG vs. GG | Random | 74.12 | ≤0.001 | 0.991 | 0.763-1.289 | -0.064 | 0.949 | 0.669 | 0.717 | |
| AA+AG vs. GG | Random | 83.33 | ≤0.001 | 0.994 | 0.742-1.331 | -0.041 | 0.967 | 0.427 | 0.761 | |
| AA vs. AG+GG | Random | 76.74 | ≤0.001 | 0.965 | 0.767-1.214 | -0.302 | 0.763 | 0.745 | 0.867 | |
| Ethnicity | ||||||||||
| Caucasian | A vs. G | Fixed | 41.76 | 0.161 | 0.977 | 0.916-1.043 | -0.688 | 0.491 | 0.734 | 0.349 |
| AA vs. GG | Fixed | 41.86 | 0.16 | 0.96 | 0.842-1.094 | -0.615 | 0.539 | 0.734 | 0.285 | |
| AG vs. GG | Fixed | 0 | 0.92 | 0.983 | 0.873-1.107 | -0.287 | 0.774 | 1 | 0.202 | |
| AA+AG vs. GG | Fixed | 0 | 0.859 | 0.996 | 0.892-1.112 | -0.071 | 0.944 | 1 | 0.273 | |
| AA vs. AG+GG | Fixed | 55.12 | 0.083 | 0.976 | 0.883-1.079 | -0.468 | 0.639 | 0.734 | 0.431 | |
| Asian | A vs. G | Random | 89.54 | ≤0.001 | 0.711 | 0.419-1.207 | -1.264 | 0.206 | 1 | 0.907 |
| AA vs. GG | Random | 87.83 | ≤0.001 | 0.442 | 0.124-1.570 | -1.262 | 0.207 | 1 | 0.762 | |
| AG vs. GG | Random | 86.29 | ≤0.001 | 0.749 | 0.273-2.054 | -0.562 | 0.574 | 0.452 | 0.394 | |
| AA+AG vs. GG | Random | 90.55 | ≤0.001 | 0.595 | 0.201-1.758 | -0.939 | 0.347 | 0.452 | 0.397 | |
| AA vs. AG+GG | Random | 72.07 | 0.001 | 0.664 | 0.413-1.066 | -1.697 | 0.09 | 0.763 | 0.888 | |
| African | A vs. G | Fixed | 3.035 | 0.357 | 1.386 | 1.161-1.654 | 3.612 | ≤0.001 | 1 | 0.813 |
| AA vs. GG | Fixed | 2.894 | 0.358 | 1.931 | 1.339-2.786 | 3.52 | ≤0.001 | 1 | 0.598 | |
| AG vs. GG | Fixed | 61.34 | 0.075 | 1.337 | 1.010-1.772 | 2.026 | 0.043 | 0.296 | 0.11 | |
| AA+AG vs. GG | Fixed | 0 | 0.423 | 1.647 | 1.268-2.137 | 3.747 | ≤0.001 | 1 | 0.916 | |
| AA vs. AG+GG | Fixed | 63.26 | 0.066 | 1.845 | 0.997-3.415 | 1.949 | 0.051 | 0.296 | 0.199 | |
| Source of Controls | ||||||||||
| HB | A vs. G | Random | 93.07 | ≤0.001 | 0.931 | 0.504-1.721 | -0.227 | 0.82 | 0.806 | 0.83 |
| AA vs. GG | Random | 93.33 | ≤0.001 | 0.964 | 0.209-4.445 | -0.047 | 0.963 | 1 | 0.815 | |
| AG vs. GG | Random | 90.34 | ≤0.001 | 0.866 | 0.285-2.629 | -0.254 | 0.799 | 0.734 | 0.597 | |
| AA+AG vs. GG | Random | 93.72 | ≤0.001 | 0.836 | 0.230-3.041 | -0.272 | 0.786 | 0.734 | 0.651 | |
| AA vs. AG+GG | Random | 81.48 | ≤0.001 | 0.963 | 0.567-1.635 | -0.141 | 0.888 | 0.806 | 0.43 | |
| PB | A vs. G | Random | 86.15 | ≤0.001 | 0.816 | 0.666-0.999 | -1.967 | 0.049 | 0.465 | 0.244 |
| AA vs. GG | Random | 72.44 | 0.001 | 0.893 | 0.758-1.051 | -1.361 | 0.173 | 0.367 | 0.271 | |
| AG vs. GG | Fixed | 8.403 | 0.364 | 0.979 | 0.871-1.100 | -0.359 | 0.719 | 0.763 | 0.753 | |
| AA+AG vs. GG | Fixed | 40.64 | 0.12 | 0.974 | 0.875-1.086 | -0.471 | 0.638 | 0.133 | 0.23 | |
| AA vs. AG+GG | Random | 67.54 | 0.005 | 0.882 | 0.699-1.112 | -1.063 | 0.288 | 0.367 | 0.249 | |
Discussion
Genetics play an important role in development and progression breast cancer (Yazdi et al., 2015). There are more and more association studies searching susceptibility genes involved in breast cancer. To date, several variants within PON1 gene associated with susceptibility to breast cancer have been verified. rs662 and rs854560 polymorphism are the most characterized SNPs that are associated with development this disease. Our present work indicated that both rs662 and rs854560 polymorphisms at PON1 gene were associated with an increased risk of BC in the overall population. All previous meta-analysis have indicated that PON1 rs662 was associated with risk of breast cancer, but not rs854560. Two meta-analysis by Fang et al., (2012) and Saadat (2012) suggested that the PON1 rs662 is a risk factor for the development of breast cancer. Wu et al., (2017) evaluated the associations of PON1 rs662 and rs854560 polymorphisms with risk of breast cancer in 365 cases and 378 controls from the Guangxi region of southern China. Their results showed that PON1 rs854560 genetic polymorphisms may be associated with the risk of BC. However, they have found that rs662 polymorphism was not associated with breast cancer risk, or with any of the clinicopathological parameters. Pan et al., (2019) in meta-analysis reported that the PON1 rs662 is associated with decrease of breast cancer risk. Their results showed an increased risk in the Caucasian and Asian population as well as HB group and PB group. However, there was an association between rs854560 polymorphism and increased breast cancer risk. Liu et al., (2019) in a mate-analysis revealed that PON1 rs854560 polymorphism could be used to identify individual with elevated susceptibility to breast cancer. However, they have not found any positive association between PON1 rs662 polymorphism and breast cancer in polled analyses. In other meta-analysis, Zhang et al., (2015) found that PON1 rs662 polymorphism was associated with a decreased risk in breast cancer. Our meta-analysis supports the growing body of evidence that the PON1 rs662 and rs854560 polymorphisms is emerging as a RISK factor for breast cancer.
Our pooled data indicated that LEP rs7799039 variant was not associated with risk of breast cancer in overall population and ethnicity. Liu and Liu (2011) in a meta-analysis based on three studies with 2,003 cases and 1,967 controls revealed for LEP rs7799039G>A polymorphism and nine studies with 4,627 cases and 5,476 controls for LEPR rs1137101 revealed that these polymorphisms were not associated with breast cancer risk. However, Yan et al., (2016) in a meta-analysis suggests that the LEP rs7799039G>A plays an important role in breast cancer susceptibility, especially in Caucasian. Although previous meta-analyses have reported the association between rs7799039 and LEPR rs1137101 polymorphisms and susceptibility to breast cancer, the current meta-analysis was more in the number of studies included and larger in sample size, which comparatively reduced the influence of contingency on the pooled data. Therefore, our conclusions were more persuasive and accurate than previous meta-analysis.
The current meta-analysis has several limitations. Therefore, some conclusions of this study should be cautiously interpreted. First, only a small number of studies were found on PON1 polymorphisms. Further studies are still required to confirm the relationship of these polymorphisms with breast cancer in different populations, especially in African and mixed populations. Second, in this work there was a considerable heterogeneity in overall population studies. Differences of ethnicity, genotyping methods and source of controls may partially explain the significant heterogeneity. Moreover, various adjusted confounders, different study designs, and other undetected factors may also lead to the presence of heterogeneity. Finally, none of the included studies separately analyzed the relations of different confounders such as age, lifestyle, family history, hormone therapy, etc. in addition, breast cancer is a complex disease which is influenced by the environment, genetic factors, and genotype-environment interactions. Thus, these interactions in development of breast cancer should be considered.
In summary, this meta-analysis aimed to summarize association between the PON1 rs662, rs854560 LEP rs7799039 and LEPR rs1137101 polymorphisms and susceptibility to breast cancer. The pooled data revealed that rs662 and rs854560 polymorphisms were associated with risk of BC and could potentially serve as useful genetic markers for breast cancer. However, there was no association between LEP rs7799039 and LEPR rs1137101 polymorphisms and breast cancer risk. More studies among different ethnicities are required to be done to reinforce the results of the current study. Nevertheless, gene-gene or gene-environment interaction which is closely related to development of breast cancer should be considered in future studies.
Author Contribution Statement
Soheila Sayad, Meraj Farbod: conceptualization, investigation. Seyed Alireza Dastgheib, Mojgan Karimi-Zarchi: Software, original draft preparation. Seyedali Salari, Seyed Hossein Shaker: Investigation. Fatemeh Asadian: Investigation, writing. Fatemeh Asadian, Hossein Neamatzadeh: Methodology, software. Seyed Alireza Dastgheib: Formal analysis, investigation. Seyed Hossein Shaker: Project administration. Jalal Sadeghizadeh-Yazdi, Hossein Neamatzadeh: Writing, reviewing, editing
Ethics approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable for this manuscript.
Availability of data and material
The datasets generated during and/or analyzed during this study are available from the corresponding author on reasonable request.
Conflicts of interest/Competing interests
The authors declare that they have no conflict of interest.
References
- Bahrami R, Dastgheib SA, Niktabar SM, et al. Association of BMP4 rs17563 polymorphism with nonsyndromic cleft lip with or without cleft palate risk: Literature Review and Comprehensive Meta-Analysis. Fetal Pediatr Pathol. 2020;2020:1–15. doi: 10.1080/15513815.2019.1707916. [DOI] [PubMed] [Google Scholar]
- Bahrami R, Shajari A, Aflatoonian M, et al. Association of REarranged during Transfection (RET) c 73 + 9277T > C and c 135G > a polymorphisms with susceptibility to hirschsprung disease: A Systematic Review and Meta-Analysis. Fetal Pediatr Pathol. 2020;39:476–490. doi: 10.1080/15513815.2019.1672225. [DOI] [PubMed] [Google Scholar]
- Costa LG, Giordano G, Furlong CE. Pharmacological and dietary modulators of paraoxonase 1 (PON1) activity and expression: The hunt goes on. Biochemical Pharmacol. 2011;81:337–44. doi: 10.1016/j.bcp.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Costa Vieira RA, Biller G, Uemura G, Ruiz CA, Curado MP. Breast cancer screening in developing countries. Clinics. 2017;72:244–53. doi: 10.6061/clinics/2017(04)09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinegde NG, Xuying L. Awareness of breast cancer among female care givers in tertiary cancer hospital, China. Asian Pac J Cancer Prev. 2017;18:1977–83. doi: 10.22034/APJCP.2017.18.7.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeili R, Mohammadi S, Jafarbeik-Iravani N, et al. Expression of SCUBE2 and BCL2 predicts favorable response in ERα positive breast cancer. Arch Iran Med. 2021;24:209–17. doi: 10.34172/aim.2021.32. [DOI] [PubMed] [Google Scholar]
- Fang DH, Fan CH, Ji Q, et al. Differential effects of paraoxonase 1 (PON1) polymorphisms on cancer risk: Evidence from 25 published studies. Mol Biol Rep. 2012;39:6801–9. doi: 10.1007/s11033-012-1505-3. [DOI] [PubMed] [Google Scholar]
- Feng Y, Spezia M, Huang S, et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. 2018;5:77–106. doi: 10.1016/j.gendis.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forat-Yazdi M, Neamatzadeh H, Sheikhha MH, Zare-Shehneh M, Fattahi M. BRCA1 and BRCA2 common mutations in Iranian breast cancer patients: A meta analysis. Asian Pac J Cancer Prev. 2015:16. doi: 10.7314/apjcp.2015.16.3.1219. [DOI] [PubMed] [Google Scholar]
- Funcke J-B, von Schnurbein J, Lennerz B, et al. Monogenic forms of childhood obesity due to mutations in the leptin gene. Mol Cell Pediat. 2014:1. doi: 10.1186/s40348-014-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallicchio L, McSorley MA, Newschaffer CJ, et al. Body mass, polymorphisms in obesity-related genes, and the risk of developing breast cancer among women with benign breast disease. Cancer Detect Prev. 2007;31:95–101. doi: 10.1016/j.cdp.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Jafari-Nedooshan J, Dastgheib SA, Kargar S, et al. Association of IL-6 -174 G>C Polymorphism with Susceptibility to Colorectal Cancer and Gastric Cancer: a Systematic Review and Meta-Analysis. Acta Med (Hradec Kralove) 2019;62:137–146. doi: 10.14712/18059694.2020.2. [DOI] [PubMed] [Google Scholar]
- Jafari-Nedooshan J, Kargar S, Neamatzadeh H, et al. Lack of association of the fat mass and obesity associated (FTO) gene rs9939609 polymorphism with breast cancer Risk: a Systematic Review and Meta-Analysis Based on Case - Control Studies. Asian Pac J Cancer Prev. 2017;18:1031–7. doi: 10.22034/APJCP.2017.18.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari M, Jarahzadeh MH, Dastgheib SA, et al. Association of PAI-1 rs1799889 polymorphism with susceptibility to ischemic stroke: a Huge Meta-Analysis based on 44 Studies. Acta Med (Hradec Kralove) 2020;63:31–42. doi: 10.14712/18059694.2020.13. [DOI] [PubMed] [Google Scholar]
- Kaklamani V, Yi N, Sadim M, et al. The role of the fat mass and obesity associated gene (FTO) in breast cancer risk. BMC Med Genet. 2011;12:52. doi: 10.1186/1471-2350-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavitha K, Kowshik J, Kishore TKK, Baba AB, Nagini S. Astaxanthin inhibits NF-κB and Wnt/β-catenin signaling pathways via inactivation of Erk/MAPK and PI3K/Akt to induce intrinsic apoptosis in a hamster model of oral cancer. Biochimica et Biophysica Acta - General Subjects. 2013;1830:4433–44. doi: 10.1016/j.bbagen.2013.05.032. [DOI] [PubMed] [Google Scholar]
- Li WF, Matthews C, Disteche CM, Costa LG, Furlong CE. Paraoxonase (Pon1) gene in mice: Sequencing, chromosomal localization and developmental expression. Pharmacogenetics. 1997;7:137–44. doi: 10.1097/00008571-199704000-00007. [DOI] [PubMed] [Google Scholar]
- Liu C, Liu L. Polymorphisms in three obesity-related genes (LEP, LEPR, and PON1) and breast cancer risk: a meta-analysis. Int Soc Oncodevelopment Biol Med. 2011;32:1233–40. doi: 10.1007/s13277-011-0227-9. [DOI] [PubMed] [Google Scholar]
- Liu P, Wang Q, Cui Y, Wang J. A meta-analysis of the relationship between paraoxonase 1 polymorphisms and cancer. Free Radical Res. 2019;53:1045–50. doi: 10.1080/10715762.2019.1645956. [DOI] [PubMed] [Google Scholar]
- Mackness M, Sozmen EY. A critical review on human serum paraoxonase-1 in the literature: Truths and misconceptions. Turk J Biochem. 2020;46:1–6. [Google Scholar]
- Moghimi M, Kargar S, Jafari MA, et al. Angiotensin converting enzyme insertion/deletion polymorphism is associated with breast cancer risk: A Meta-Analysis. Asian Pac J Cancer Prev. 2018;19:3225–31. doi: 10.31557/APJCP.2018.19.11.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi S, Majidzadeh K, Mazaheri M, et al. Tamoxifen resistance and CYP2D6 copy numbers in breast cancer patients. Asian Pac J Cancer Prev. 2012;13:6101–4. doi: 10.7314/apjcp.2012.13.12.6101. [DOI] [PubMed] [Google Scholar]
- Najminejad H, Farhadihosseinabadi B, Dabaghian M, et al. Key regulatory miRNAs and their interplay with mechanosensing and mechanotransduction signaling pathways in breast cancer progression. Mol Cancer Res. 2020;18:1113–28. doi: 10.1158/1541-7786.MCR-19-1229. [DOI] [PubMed] [Google Scholar]
- Neamatzadeh H, Shiryazdi SM, Kalantar SM. BRCA1 and BRCA2 mutations in Iranian breast cancer patients: A systematic review. J Res Med Sci. 2015:20. [PMC free article] [PubMed] [Google Scholar]
- Pan X, Huang L, Li M, et al. The association between PON1 (Q192R and L55M) gene polymorphisms and risk of cancer: A Meta-Analysis Based on 43 Studies. Bio Med Res Int. 2019;2019:5897505. doi: 10.1155/2019/5897505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard SA, Frank EA, D’Souza CJM. Correlation between cholinesterase and paraoxonase 1 activities: Case series of pesticide poisoning subjects. BioImpacts. 2013;3:119–22. doi: 10.5681/bi.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadat M. Paraoxonase 1 genetic polymorphisms and susceptibility to breast cancer: A meta-analysis. Cancer Epidemiol. 2012;36:e101–3. doi: 10.1016/j.canep.2011.10.015. [DOI] [PubMed] [Google Scholar]
- Seow DCC, Gao Q, Yap P, et al. Profile of the paraoxonase 1 (PON1) gene 192Q/R polymorphism and clinical associations among older Singaporean Chinese with Alzheimer’s and Mixed Dementia. Dementia Geriatric Cognitive Disorders Extra. 2016;6:43–54. doi: 10.1159/000442382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kang M, Liu C, Qiu H. Leptin rs7799039 (G2548A) polymorphism is associated with cancer risk: a meta-analysis involving 25,799 subjects. OncoTargets Ther. 2019;12:2879–90. doi: 10.2147/OTT.S190093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Ma X, Gao X, Zhang S. Association between leptin (-2548G/A) genes polymorphism and breast cancer susceptibility: A meta-Analysis. Medicine (United States) 2016:95. doi: 10.1097/MD.0000000000002566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdi MF, Rafieian S, Gholi-Nataj M, et al. CYP2D6 Genotype and risk of recurrence in tamoxifen treated breast cancer patients. Asian Pac J Cancer Prev. 2015;16:6783–7. doi: 10.7314/apjcp.2015.16.15.6783. [DOI] [PubMed] [Google Scholar]
- Zhang M, Xiong H, Fang L, et al. Paraoxonase 1 (PON1) Q192R gene polymorphism and cancer risk: A meta-analysis based on 30 publications. Asian Pac J Cancer Prev. 2015;16:4457–63. doi: 10.7314/apjcp.2015.16.10.4457. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analyzed during this study are available from the corresponding author on reasonable request.