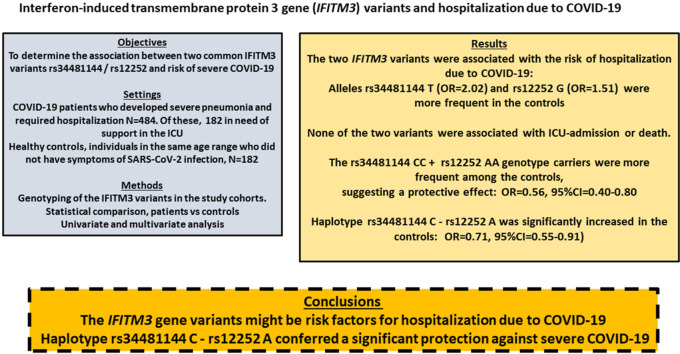
Abstract
The interferon induced transmembrane-protein 3 (IFITM3) plays an important role in the defence against viral infection. IFITM3 gene variants have been linked to differences in expression and associated with the risk of severe influenza by some authors. More recently, these variants have been associated with the risk of COVID-19 after SARS-CoV-2 infection. We determined the effect of two common IFITM3 polymorphisms (rs34481144 C/T and rs12252 A/G) on the risk of hospitalization due to COVID-19 by comparing 484 patients (152 required support in thr intensive care unit, ICU) and 182 age and sex matched controls (no disease symptoms). We found significantly higher frequencies of rs34481144 T and rs12252 G carriers among the patients (OR = 2.02 and OR = 1.51, respectively). None of the two variants were associated with ICU-admission or death. We found a significantly higher frequency of rs34481144 CC + rs12252 AA genotype carriers among the controls, suggesting a protective effect (p = 0.001, OR = 0.56, 95%CI = 0.40–0.80). Moreover, haplotype rs34481144 C - rs12252 A was significantly increased in the controls (p = 0.008, OR = 0.71, 95%CI = 0.55–0.91).
Our results showed a significant effect of the IFITM3 variants in the risk for hospitalization after SARS-CoV-2 infection.
Keywords: Interferon induced transmembrane proteins, Ifitm3, Gene variant, COVID-19, Genetic association
Graphical abstract
1. Introduction
The human Interferon-induced transmembrane proteins (IFITMs) are a family of small proteins with five members, three of them (IFITM1-3) with a role in the innate immunity against microorganism infection (Smith et al., 2014; Liao et al., 2019; Zhao et al., 2019; Ren et al., 2020). IFITMs localize at the plasmatic and cell-vesicle membranes (Bailey et al., 2014), and block the fusion of enveloped-viruses with the cell membranes (Weidner et al., 2010; Perreira et al., 2013; Compton et al., 2014; Desai et al., 2014; Smith et al., 2019; Suddala et al., 2019). These proteins contain an amphipathic helix domain that would alter the cell lipid membrane disfavouring virus fusion (Li et al., 2013; Chesarino et al., 2017; Suddala et al., 2019).
IFITMs play a role in the response to coronavirus (CoV) mainly as inhibitors of infection (Huang et al., 2011; Wrensch et al., 2014; Zheng et al., 2020), although they could also enhance CoV infections (Zhao et al., 2014, 2018). In the case of IFITM3 the capacity to inhibit or enhance SARS-CoV or MERS-CoV infection seems to depend on the localization in the plasma membrane (increased infection) or in the endosomal membrane (reduced infection) (Zhao et al., 2018). Interestingly, polymorphisms in the IFITM3 genes that determine differences in theplasma membrane localization would affect the susceptibilty to viral infection (Everitt et al., 2012; Zhao et al., 2018).
Due to their role in infection, variants in the IFITM genes are candidates to modify the risk for viral infection and disease severity (Zhao et al., 2019). Two IFITM3 single nucleotide polymorphisms (SNPs) have been well characterised, rs12252 and rs34481144. SNP rs12252 A/G has been associated with the risk of severe influenza by some authors but not others, and the functional mechanism remains controversial (Chen et al., 2018; Prabhu et al., 2018; Qin et al., 2018; Martins et al., 2020). The G allele was described as a risk factor for the severity of disease after avian and H1N1/09 infection (Zhang et al., 2013). The GG homozygotes showed a lower rate of seroconversion after trivalent vaccination with inactivated viruses (Lei et al., 2020). These authors reported that healthy adult volunteers with the rs12252 C/C genotype showed lower specific antibodies against H1N1, H3N2 and B viruses compared to rs12252 A-carriers. Moreover, IFITM3-deletion mice (Ifitm3−/−) showed lower total IgG against these viruses compared to wild type littermates. The authors concluded that the genetic mechanisms linked to IFITM3 reduction might attenuate the antibody response against influenza, increasing the risk of adverse events in infected (Lei et al., 2020).
The IFITM3 rs12252 G was also associated with the progression of HIV-1 infection toward reduced CD4+ T-cells count (Zhang et al., 2015). This SNP was linked to an alternative splicing of the transcript that could results in truncation and altered localization of the IFITM3, but this effect was not confirmed by some authors (Everitt et al., 2012; Allen et al., 2017; Randolph et al., 2017). SNP rs34481144 C/T is in the 5′ UTR of the gene and the T allele has been associated with an increased risk of severe influenza (Allen et al., 2017). The urderlying mechanism seems to be an impaired binding of transcription factors to the promoter with the T allele, that would results in a reduced mRNA expression compared to the C allele. Furthermore, the change C > T would disrupt a methylation site and T-carriers had reduced CD8 T-cells in their airways during natural influenza infection. IFITM3 would promotes the accumulation of airway CD8 T-cells at mucosal sites, and a reduction of the expression might result in lower CD8 levels in the airways and increased risk for severe influenza (Allen et al., 2017).
In reference to SARS-CoV-2 the effect of IFITMs on infection remains unclear but might depend on the celular localization, as reported for IFITM3 in mouse and humans. A recent study with IFITM3 mutants identified amino acids with pro- and anti-viral effects in terms of distinct cellular locations and mechanisms of action (Shi et al., 2021). The expression of IFITMs in human lung cells would facilitate the SARS-CoV-2 infection, in a mechanism that could involves the interaction between the spike (S) viral protein and IFITMs (Prelli Bozzo et al., 2021). Moreover, the entry and replication of SARS-CoV-2 into several cell types was inhibited by IFITM-derived peptides and targeting antibodies. In agreement with this role in infection, variants in the IFITM3 gene have been associated with the clinical outcomes after SARS-CoV-2 infection (COVID-19). Most of the studies analysed the rs12252 SNP and concluded an association between the G allele and disease severity or a lack of association, but were based on small sizes that impairs the statistical power (Zhang et al., 2020; Schönfelder et al., 2021; Gómez et al., 2021; Alghamdi et al., 2021).
Our aim was to determine the assocation between IFITM3 rs34481144 and rs12252 SNPs and haplotypes and severe COVID-19. For this purpose we compared patients with pneumonia that required hospitalization and population controls without symptoms of SARS-CoV-2 infection. We also compared patients who required or not admission in the Intensive Care Unit (ICU), and the effect of these gene variants in the risk of death due to COVID-19.
2. Methods
We studied 484 COVID-19 patients hospitalysed in the period March–December 2020 due severe pneumonia and positive for SARS-CoV-2 in a PCR-test. Of these, 152 needed critical care support (ICU) with high-flow oxygen, positive-pressure ventilation or vasoactive drugs. The controls were 182 individuals aged 55 years or older without disease symptoms. These controls were negative for SARS-CoV-2 infection with a serological test. Patients and controls were of European ancestry from the region of Asturias, Northern Spain (total population approx 1 million), and gave their informed consent to participate in the study approved by the Ethical Committee of Medical Research of Asturias.
We isolated the DNA from blood-leukocytes from all the patients and controls. The IFITM3 rs34481144 C/T and rs12252 A/G SNPs were genotyped by real-time PCR with Taqman probes (Fisher Scientific, assays id. C_26288451_10 and C_175677529_10). The method was validated by sequencing DNA samples representing the three genotypes of the two SNPs. These nucleotide changes are defined according to either the genomic coding (plus) or the cDNA strands. In this work the two SNPs were named according to the standard Human Genome Variation Society guidelines, rs34481144 C/T (c.-23 G > A) and rs12252 A/G (c.42T > C, p.Ser14 = ).
To confirm the adjustment of the observed genotype frequencies to the Hardy-Weinberg equilibrium we used an online programme (https://wpcalc.com/en/equilibrium-hardy-weinberg).The statistical analysis was performed with the r-program (www.r-project.org). The linear regression was used to compare the frequencies between the groups, and multiple logistic regression (linear generalised model, LGM) to determine the association between COVID-19 severity and sex, age, hypertension, and the rs34481144 and rs12252 variants. For all the comparisons we considered a p < 0.05 as statistically significant, and Odds ratios (OR) and 95% confidence intervals (CI) were also calculated. Ther statistical power was calculated online (https://clincalc.com/stats/samplesize.aspx).
3. Results and discussion
In Table 1 we summarised the main values in COVID-19 patients and healthy controls. Hospitalization with ICU admission was associated with male sex (p = 0.01), advanced age (p = 0.0002), and hypertension (p = 0.0006), but not with the IFITM3 SNPs (Table 1). Compared to healthy age and sex matched controls, COVID-19 hospitalization was increased among carriers of rs34481144 T-genotypes (p = 0.04; OR = 1.43; 95%CI = 1.02–2.03) and rs12252 G-genotypes (p = 0.04; OR = 2.02; 95%CI = 1.01–4.08). The frequency of deaths was significantly higher in the ICU patients (24% vs 2%). The risk of death among the ICU patients was associated with advanced age (p = 0.04; OR = 1.04, 95%CI = 1.01–1.08), but not with male sex or the IFITM3 SNPs (Table 2 ). There was a higher frequency of hypertensives among the death cases, without statistical significance (p = 0.25). Because there were only 7 deaths among the no-ICU patients we did not compare the studied variables between deceased and not in the no-ICU group.
Table 1.
Main values in the COVID-19 patients and healthy sex and age matched controls (individuals without COVID-19 symptoms).
| COVID19 N = 484 |
CONTROLS N = 182 |
ICU N = 152 |
No-ICU N = 332 |
|
|---|---|---|---|---|
| Male | 276 (57%) | 93 (51%) | 113 (74%) | 198 (59%) |
| Mean age, years | 66 ± 16 | 67 ± 11 | 67 ± 16 | 64 ± 16 |
| Interquartil range, years | 55–75 | 70–76 | 60–76 | 52–75 |
| Hypertension | 224 (46%) | 49 (27%) | 88 (58%) | 136 (41%) |
| Death | 45 (9%) | – | 38 (24%) | 7 (2%) |
| rs34481144 C/T | ||||
| CC | 181 (37%) | 84 (46%) | 61 (40%) | 120 (36%) |
| CT | 235 (49%) | 80 (44%) | 69 (46%) | 166 (50%) |
| TT | 68 (14%) | 18 (10%) | 22 (14%) | 46 (14%) |
| TT + CT vs CC | p = 0.04; OR = 1.43; 95%CI = 1.02–2.03 | |||
| T-frequency | 371 (0.38) | 116 (0.32) | 113 (0.37) | 258 (0.39) |
| T vs C | p = 0.03; OR = 1.33; 95%CI = 1.03–1.72 | |||
| rs12252 A/G | ||||
| AA | 433 (89%) | 172 (92%) | 133 (88%) | 300 (90%) |
| AG | 47 (10%) | 10 (8%) | 17 (11%) | 30 (9%) |
| GG | 4 (1%) | 0 | 2 (1%) | 2 (1%) |
| AG + GG vs AA | p = 0.04; OR = 2.02; 95%CI = 1.01–4.08 | |||
| G-frequency | 55 (0.06) | 14 (0.03) | 14 (0.07) | 34 (0.05) |
| p = 0.18; OR = 1.51; 95%CI = 0.83–2.74 | ||||
Table 2.
Main values in the ICU patients according to death.
| ICU N = 152 |
||
|---|---|---|
|
Death N = 38 |
No-death N = 114 |
|
| Age range | 33–84 | 32–84 |
| Mean age | 68.79 ± 11.84 | 64.54 ± 10.58 |
| Male | 28 (74%) | 85 (75%) |
| Hypertension | 25 (66%) | 63 (55%) |
| rs34481144 C/T | ||
| CC, 61 | 18 (47%) | 43 (38%) |
| CT, 69 | 14 (37%) | 55 (48%) |
| TT, 22 | 6 (16%) | 16 (14%) |
| T-frequency | 26 (0.34) | 87 (0.38) |
| rs12252 A/G | ||
| AA, 133 | 32 (86%) | 101 (88%) |
| AG, 17 | 5 (14%) | 12 (11%) |
| GG, 2 | 1 (0 | 1 (1%) |
| G-frequency | 0.07 | 0.07 |
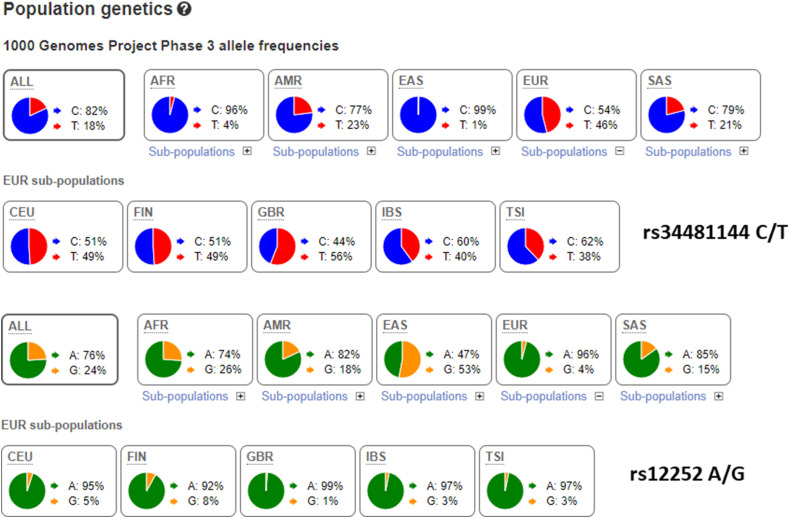
According to our results the rs34481144 T was associated with an increased risk of severe COVID-19 in need of hospitalization. The reported frequency among Spanish (accesssed at LD-Link, https://ldlink.nci.nih.gov/?tab=home) was T = 0.40, similar to the frequency among our patients but higher than in the healthy controls (Fig. 1 ). This supported the finding that carriers of the rs34481144 T were at increased risk of COVID-19, with the CC as a protective. Similarly, the rs12252 G showed a higher frequency among the patients (0.06) compared to the Spanish (0.03) and the healthy controls (0.03). For this variant the AA genotype might be protective.
Fig. 1.
Reported frequencies of the IFITM3 variants in populations worldwide. Data accessed at the ensemble web (www.ensembl.org).
Afr: Africans, Amr: Americans, EAS: East-Asians, EUR: Europeans, SAS: South-Asians; CEU: Utah residents of European ancestry, FIN: Finns, GBR: Great Britain Caucasians, IBS: Spanish, TSI: Toscany.
Our study was based on a limited size, and we thus calculated the required size to reach a statistical power of 80 at a p = 0.05. For a case-control study comparing patients with healthy controls the post-hoc power was 57% for the rs34481144 T-carriers. For a power of 80 at least 500 healthy subjects should be genotyped.
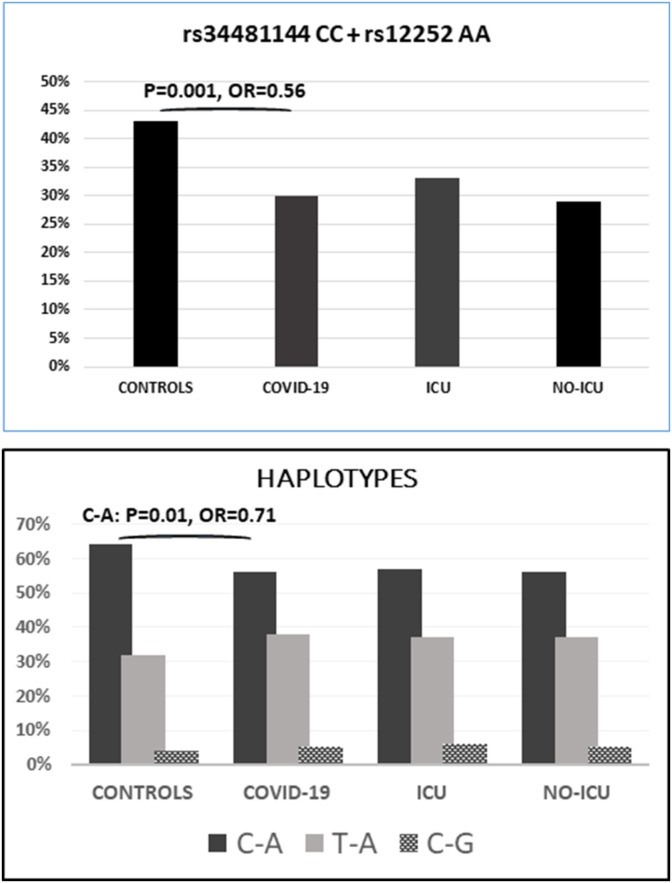
We determined the asociation between the rs34481144 - rs12252 haplotypes and the risk of hospitalization due to COVID-19 (suppl. Table 1). Haplotype C-A was decreased in the patients compared to controls (57% vs 64%), with a significant protective effect: p = 0.04, OR = 0.72, 95%CI = 0.53–0.98 (Fig. 2 ).The haplotype distribution in our population was consistent with the reported in the human genome database (https://ldlink.nci.nih.gov), with the rs34481144 C/C genotype (protective) always inherited (in complete linkage disequilibrium) with rs12252 G/G (risk genotype). The rs12252 A was inherited with either rs34481144 C or T, indicating that the risk alleles were inherited on opposite haplotypes. This finding was in agreement with the reported effect among severe influenza patients, where the risk allele of rs34481144 was inherited with the protective allele of rs12252 suggesting that these IFITM3 variants provided non-overlapping risks (Allen et al., 2017). Of note, the CA haplotype was more common among the patients and also among the Spanish unselected controls (LD-link), supporting a resistence to develop severe COVID-19 among carriers of this haplotype (suppl. Table).
Fig. 2.
Above, frequencies of the rs3448114 CC + rs12252 AA genotypes. This combination was significantly more frequent in the controls compared to COVID-19 hospitalized patients: p = 0.001, OR = 0.56 (95%CI = 0.40–0.80).
Below, frequencies of the three rs3448114 + rs12252 haplotypes. Haplotype C-A was significantly more frequent in the controls (protective): p = 0.008, OR = 0.71, 95%CI = 0.55–0.91.
The raw numbers are presented in the supplementary file.
Individuals double homozygotes CC-AA were more frequent in the controls (43% vs 30%), with p = 0.001 (OR = 0.56; 95%CI = 0.40–0.80). This genotype combination was non-significantly more common in the ICU than in no-ICU patients (33% vs 29%; p = 0.31) (Fig. 2).
The clinical course of the SARS-CoV-2 infection is heterogeneous, ranging from individuals who remained asymtopmatic to severe COVID-19 with bilateral pneumonia and hospitalization. The most severe cases require attention in the ICU and are at a high risk of death. The variable manifestation depends on several factors, being advanced age, male gender, and hypertension and other preexisting cardiovascular complications well recognised risk factors for disease severity and mortality. Besides, a genetic predisposition has been reported and several loci have been associated with the risk of severe COVID-19 (Fricke-Galindo, 2021). In particular, the chromosome 3 chemokine-receptor gene cluster has been replicated in several studies, and there is also a strong evidence for a protective role among individuals with the O blood group and an increased risk for the A-group (Ellinghaus et al., 2021; Pairo-Castineira et al., 2021). Several authors have studied the association between COVID-19 and gene variants previously reported as risk factors for other viral diseases, such as influenza or HIV (Zhang et al., 2015). Many of these candidate variants are in the genes that encode proteins of the innate immunity, such as the IFITM3.
Everitt et found a statistically significant higher number of seasonal or pandemic influenza H1N1/09 hospitalized subjects with the minor IFITM3 rs12252-G allele (Everitt et al., 2012). The study was based on only 53 patients, but the authors provided evidence that the GG genotype reduced the influenza virus restriction in vitro. Zhang et al. also found a significant increased frequency of the GG genotype among patients with severe compared to mild influenza-disease, but the study was based on only 32 and 51 cases, respectively (Zhang et al., 2013). Mills et al. compared rs12252 genotypes in 34 patients with H1N1 influenza and severe pneumoniaand >5000 individuals with community-acquired mild lower respiratory tract infection and matched controls, and found an association between GG homozygotes and susceptibility to mild influenza but did not confirm the association with severe H1N1 infection (Mills et al., 2014).Gaio et al. compared 312 influenza patients with 624 matched non-hospitalized controls and found that GA/GG carriers had a higher risk of being hospitalized than AA patients (OR = 2.54, 95%CI = 1.54–4.19) (Gaio et al., 2016). Lopez Rodriguez et al. did not find a significant association between rs12252 and influenza virus infection (n = 148) or hospitalization due to influenza pneumonia (n = 60). The study compared these patients with 246 healthy Spanish individuals (López-Rodríguez et al., 2016). Interestingly, there were no GG homozygotes in the control group, and the rare G allele was more common in the patients with the highest frequency among the ICU-patients (n = 34). In a multicenter study with 275 cases of avian (H7N9) and pandemic (H1N1pdm09) influenza Lee et al. found an over-representation of rs12252 GG homozygotes among the patints compared to population controls (54.5% vs 33.2%; p = 0.02) (Lee et al., 2017).
Randolph et al. did not find a significant association between rs12252 and influenza infection in children (n = 358), and this variant was not associated with critical illness severity. These authors did not find alternative IFITM3 transcripts in the rs12252 genotypes, and this SNP was not associated with IFITM3 expression levels (Randolphet al., 2017). Interestingly, in this study the rs34481144 CC genotype was significantly more frequent in the controls, suggesting a protective effect against influenza infection. In addition to the above referenced studies the association between rs12252 GG and influenza has been confirmed by some authors (Pan et al., 2017) but not others (Carter et al., 2018).
Few studies have addressed other IFITM3 SNPs in influenza. The most relevant study reported a significant asociation between the rs34481144 T allele and disease severity, that was linked to reduced IFITM3 expression compared to the C-allele (Allen et al., 2017). These authors also found that influenza-infected carriers of the A (risk) allele had reduced CD8 T-cells in their airways, that was in agreement with a IFITM3 promotion of airway CD8 T-cell accumulation. IFITM3 could thus have an important role in promoting the persistence of immune cells at mucosal sites, and IFITM3 functional variants could thus contribute to the risk of viral infection and disease severity. Interestingly, the T risk allele was absent in East Asian populations, and its effect on influenza and other viral diseases cannot thus evaluated in these population groups.
In reference to SARS-CoV-2 infection and COVID-19 severity, to the best of our knowledge only the nest studies have addressed the association with IFITM3 SNPs. Zhang et al. compared 56 mild and 24 severe COVID-19 patients and found a higher frequency of the GG homozygotes among the severe cases (Zhanget al., 2020). In a study involving 880 Saudi patients, Alghamdi et al. found that rs12252-G was associated with hospital admission (OR = 1.65; 95% CI = 1.01–2.70) and mortality (OR = 2.2; 95% CI = 1.16–4.20) (Alghamdi et al., 2021). Our group also found a significantly higher frequency of the rs12252-G among COVID-19 patients who required hospitalization (n = 288) during the first pandemic wave (march–may 2020) in Spain, compared to age and sex matched population controsl (n = 440) (Gomez et al., 2021). These SNP did not differ between patients who required ICU or not. Schönfelder et al. did not find association between rs12252 or rs34481144 and SARS-CoV-2 infection risk or severity of COVID-19 in a German cohort (239 patients and 253 controls) (Schönfelder et al., 2021). However, these authors found the rs12252 GG genotype in two SARS-CoV-2-positive patients and none of the controls, and the study was thus underpowered to evaluate the association with genotype. Thay also found a non-significant higher frequency of rs34481144 T-allele in the patients who require hospitalization (severe cases, n = 75). Unfortunately these authors did not analyse the association with the rs12252 - rs34481144 haplotypes.
Study limitations. Our study has several limitations. First, it was based on a limited number of patients and controls, that reduced the statistical power. Second, we compared patients whith severe COVID-19 who required hospitalization and healthy individuals without symptoms of SARS-CoV-2 infection. The association between IFITM3 and mild disease was not evaluated in our study. It is thus possible that these variants are associated with an overall risk for infection rather than with disease severity. However, we have previously reported the rs12252 G frequency among elderly population controls (n = 440) recruited before the SARS-CoV-2 pandemic. The risk G allele had the same frequency (0.03) than in the SARS-CoV-2 asymptomatics, suggesting a non-significan effect on the risk of viral infection (Gómez et al., 2021). More studies to clarify theses issues are necessary.
Conclusions. Carriers of the rs34481144 T and rs12252 G were significantly more frequent in COVID-19 patients who required hospitalization, compared to age matched individuals without signs of SARS-CoV-2 infection. The rs34481144 C - rs12252 A haplotype, previously reported as protective against influenza, was also significantly more frequent in the healthy controls. Beyond its limitations our study supports a role for IFITM3 gene variants on susceptibility to develop COVID-19. Our study was in agreement with previous reports that concluded a significant association between IFITM3 variants and severe COVID-19. Studies focused to determine the functional mechanisms that link these SNPs with disease severity are of major interest.
Contributorship
All the authors contributed to this work by recruiting the patients and performing the genetic and statistical analysis. All the authors approved the submission of this ms.
Writing original draft and review: E.C., J.G., G.M.A.
Conceptualization: E.C., J.G., G.M.A.
Statistical analysis: E.C.
Supervision: J.G., G.M.A., E.C.
Genetica analysis: E.C, J.G., IDH, SM, MEAA, JAB, SRA
Other authors: patients data acquisition
Funding information
No external funding was received for this work.
Data accessibility
The full raw data are available as excell files upon request to the corresponding authors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crviro.2021.100016.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Alghamdi J., Alaamery M., Barhoumi T., Rashid M., Alajmi H., Aljasser N., et al. Interferon-induced transmembrane protein-3 genetic variant rs12252 is associated with COVID-19 mortality. Genomics. 2021 Jul;113(4):1733–1741. doi: 10.1016/j.ygeno.2021.04.002. 33838280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E.K., Randolph A.G., Bhangale T., Dogra P., Ohlson M., Oshansky C.M., et al. SNP-mediated disruption of CTCF binding at the IFITM3 promoter is associated with risk of severe influenza in humans. Nat. Med. 2017 Aug;23(8):975–983. doi: 10.1038/nm.4370. 28714988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey C.C., Zhong G., Huang I.C., Farzan M. IFITM-family proteins: the cell's first line of antiviral defense. Annu Rev Virol. 2014 Nov 1;1:261–283. doi: 10.1146/annurev-virology-031413-085537. 25599080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter T.C., Hebbring S.J., Liu J., Mosley J.D., Shaffer C.M., IvacicLC, et al. Pilot screening study of targeted genetic polymorphisms for association with seasonal influenza hospital admission. J. Med. Virol. 2018 Mar;90(3):436–446. doi: 10.1002/jmv.24975. Epub 2017 Nov 11, 29053189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Xiao M., Yang J., Chen Y.K., Bai T., Tang X.J., Shu Y.L. Association between rs12252 and influenza susceptibility and severity: an updated meta-analysis. Epidemiol. Infect. 2018 Nov 13;147:e39. doi: 10.1017/S0950268818002832. 30421689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarino N.M., Compton A.A., McMichael T.M., Kenney A.D., Zhang L., Soewarna V., et al. IFITM3 requires an amphipathic helix for antiviral activity. EMBO Rep. 2017 Oct;18(10):1740–1751. doi: 10.15252/embr.201744100. 28835547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton A.A., Bruel T., Porrot F., Mallet A., Sachse M., Euvrard M., et al. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe. 2014 Dec 10;16(6):736–747. doi: 10.1016/j.chom.2014.11.001. 25464829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai T.M., Marin M., Chin C.R., Savidis G., Brass A.L., Melikyan G.B. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014 Apr 3;10(4) doi: 10.1371/journal.ppat.1004048. 24699674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., et al. Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. 2020 Oct 15;383(16):1522–1534. doi: 10.1056/NEJMoa2020283. 32558485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., et al. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012 Mar 25;484(7395):519–523. doi: 10.1038/nature10921. 22446628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke-Galindo I., Falfán-Valencia R. Genetics insight for COVID-19 susceptibility and severity: a review. Front. Immunol. 2021 Apr 1;12:622176. doi: 10.3389/fimmu.2021.622176. 33868239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaio V., Nunes B., Pechirra P., Conde P., Guiomar R., Dias C.M., Barreto M. Hospitalization risk due to respiratory illness associated with genetic variation at IFITM3 in patients with influenza A(H1N1)pdm09 infection: a case-control study. PLoS One. 2016 Jun 28;11(6) doi: 10.1371/journal.pone.0158181. 27351739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez J., Albaiceta G.M., Cuesta-Llavona E., García-Clemente M., López-Larrea C., Amado-Rodríguez L., et al. The Interferon-induced transmembrane protein 3 gene (IFITM3) rs12252 C variant is associated with COVID-19. Cytokine. 2021 Jan;137:155354. doi: 10.1016/j.cyto.2020.155354. 33113474. [DOI] [PubMed] [Google Scholar]
- Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011 Jan 6;7(1) doi: 10.1371/journal.ppat.1001258. 21253575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Cao B., Ke C., Lu H., Hu Y., Tam C.H.T., et al. IFITM3, TLR3, and CD55 gene SNPs and cumulative genetic risks for severe outcomes in Chinese patients with H7N9/H1N1pdm09 influenza. J. Infect. Dis. 2017 Jul 1;216(1):97–104. doi: 10.1093/infdis/jix235. 28510725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei N., Li Y., Sun Q., Lu J., Zhou J., Li Z., et al. IFITM3 affects the level of antibody response after influenza vaccination. Emerg. Microb. Infect. 2020;9:976–987. doi: 10.1080/22221751.2020.1756696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Markosyan R.M., Zheng Y.M., Golfetto O., Bungart B., Li M., Ding S., et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013 Jan;9(1) doi: 10.1371/journal.ppat.1003124. 23358889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Goraya M.U., Yuan X., Zhang B., Chiu S.H., Chen J.L. Functional involvement of interferon-inducible transmembrane proteins in antiviral immunity. Front. Microbiol. 2019 May 16;10:1097. doi: 10.3389/fmicb.2019.01097. 31156602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Rodríguez M., Herrera-Ramos E., Solé-Violán J., Ruíz-Hernández J.J., Borderías L., Horcajada J.P., et al. IFITM3 and severe influenza virus infection. No evidence of genetic association. Eur. J. Clin. Microbiol. Infect. Dis. 2016 Nov;35(11):1811–1817. doi: 10.1007/s10096-016-2732-7. 27492307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins J.S.C., Oliveira M.L.A., Garcia C.C., Siqueira M.M., Matos A.R. Investigation of human IFITM3 polymorphisms rs34481144A and rs12252C and risk for influenza A(H1N1)pdm09 severity in a Brazilian cohort. Front. Cell Infect. Microbiol. 2020 Jul 10;10:352. doi: 10.3389/fcimb.2020.00352. 32754450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills T.C., Rautanen A., Elliott K.S., Parks T., Naranbhai V., Ieven M.M., et al. IFITM3 and susceptibility to respiratory viral infections in the community. J. Infect. Dis. 2014 Apr 1;209(7):1028–1031. doi: 10.1093/infdis/jit468. 23997235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., et al. Genetic mechanisms of critical illness in COVID-19. Nature. 2021 Mar;591(7848):92–98. doi: 10.1038/s41586-020-03065-y. 33307546. [DOI] [PubMed] [Google Scholar]
- Pan Y., Yang P., Dong T., Zhang Y., Shi W., Peng X., et al. IFITM3 Rs12252-C variant increases potential risk for severe influenza virus infection in Chinese population. Front. Cell Infect. Microbiol. 2017 Jun 30;7:294. doi: 10.3389/fcimb.2017.00294. 28713779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreira J.M., Chin C.R., Feeley E.M., Brass A.L. IFITMs restrict the replication of multiple pathogenic viruses. J. Mol. Biol. 2013 Dec 13;425(24):4937–4955. doi: 10.1016/j.jmb.2013.09.024. 24076421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu S.S., Chakraborty T.T., Kumar N., Banerjee I. Association between IFITM3 rs12252 polymorphism and influenza susceptibility and severity: a meta-analysis. Gene. 2018 Oct 20;674:70–79. doi: 10.1016/j.gene.2018.06.070. 29940276. [DOI] [PubMed] [Google Scholar]
- Prelli Bozzo C., Nchioua R., Volcic M., Koepke L., Krüger J., Schütz D., et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 2021 Jul 28;12(1):4584. doi: 10.1038/s41467-021-24817-y. 34321474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L., Wang D., Li D., Zhao Y., Peng Y., Wellington D., et al. High level antibody response to pandemic influenza H1N1/09 virus is associated with interferon-induced transmembrane protein-3 rs12252-CC in young adults. Front. Cell Infect. Microbiol. 2018 May 15;8:134. doi: 10.3389/fcimb.2018.00134. 29868492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph A.G., Yip W.K., Allen E.K., Rosenberger C.M., Agan A.A., Ash S.A., et al. Evaluation of IFITM3 rs12252 association with severe pediatric influenza infection. J. Infect. Dis. 2017 Jul 1;216(1):14–21. doi: 10.1093/infdis/jix242. 28531322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L., Du S., Xu W., Li T., Wu S., Jin N., Li C. Current progress on host antiviral factor IFITMs. Front. Immunol. 2020 Nov 30;11:543444. doi: 10.3389/fimmu.2020.543444. 33329509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönfelder K., Breuckmann K., Elsner C., Dittmer U., Fistera D., Herbstreit F., et al. The influence of IFITM3 polymorphisms on susceptibility to SARS-CoV-2 infection and severity of COVID-19. Cytokine. 2021 Jun;142:155492. doi: 10.1016/j.cyto.2021.155492. 33711707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G., Kenney A.D., Kudryashova E., Zani A., Zhang L., Lai K.K., et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 2021 Feb 1;40(3) doi: 10.15252/embj.2020106501. 33270927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Weston S., Kellam P., Marsh M. IFITM proteins-cellular inhibitors of viral entry. Curr. Opin. Virol. 2014 Feb;4:71–77. doi: 10.1016/j.coviro.2013.11.004. 24480526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.E., Busse D.C., Binter S., Weston S., Diaz Soria C., Laksono B.M., et al. Interferon-induced transmembrane protein 1 restricts replication of viruses that enter cells via the plasma membrane. J. Virol. 2019 Mar 5;93(6) doi: 10.1128/JVI.02003-18. e02003-e02018, 30567988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suddala K.C., Lee C.C., Meraner P., Marin M., Markosyan R.M., Desai T.M., et al. Interferon-induced transmembrane protein 3 blocks fusion of sensitive but not resistant viruses by partitioning into virus-carrying endosomes. PLoS Pathog. 2019 Jan 14;15(1) doi: 10.1371/journal.ppat.1007532. 30640957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidner J.M., Jiang D., Pan X.B., Chang J., Block T.M., Guo J.T. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 2010 Dec;84(24):12646–12657. doi: 10.1128/JVI.01328-10. Epub 2010 Oct 13, 20943977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrensch F., Winkler M., Pöhlmann S. IFITM proteins inhibit entry driven by the MERS-coronavirus spike protein: evidence for cholesterol-independent mechanisms. Viruses. 2014 Sep 26;6(9):3683–3698. doi: 10.3390/v6093683. 25256397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Makvandi-Nejad S., Qin L., Zhao Y., Zhang T., Wang L., et al. Interferon-induced transmembrane protein-3 rs12252-C is associated with rapid progression of acute HIV-1 infection in Chinese MSM cohort. AIDS. 2015;29:889–894. doi: 10.1097/QAD.0000000000000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y.H., Zhao Y., Li N., Peng Y.C., Giannoulatou E., Jin R.H., et al. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 2013;4:1418. doi: 10.1038/ncomms2433. 23361009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Qin L., Zhao Y., Zhang P., Xu B., Li K., et al. Interferon-induced transmembrane protein 3 genetic variant rs12252-C associated with disease severity in coronavirus disease 2019. J. Infect. Dis. 2020 Jun 16;222(1):34–37. doi: 10.1093/infdis/jiaa224. 32348495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Guo F., Liu F., Cuconati A., Chang J., Block T.M., Guo J.T. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. U. S. A. 2014 May 6;111(18):6756–6761. doi: 10.1073/pnas.1320856111. 24753610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Sehgal M., Hou Z., Cheng J., Shu S., Wu S., et al. Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses. J. Virol. 2018 Feb 26;92(6) doi: 10.1128/JVI.01535-17. e01535-17, 29263263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Li J., Winkler C.A., An P., Guo J.T. IFITM genes, variants, and their roles in the control and pathogenesis of viral infections. Front. Microbiol. 2019 Jan 8;9:3228. doi: 10.3389/fmicb.2018.03228. 30687247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Zhao X., Zheng S., Chen D., Du P., Li X., et al. Bat SARS-Like WIV1 coronavirus uses the ACE2 of multiple animal species as receptor and evades IFITM3 restriction via TMPRSS2 activation of membrane fusion. Emerg. Microb. Infect. 2020 Dec;9(1):1567–1579. doi: 10.1080/22221751.2020.1787797. 32602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full raw data are available as excell files upon request to the corresponding authors.