Abstract
IMP dehydrogenase (IMPDH) is the rate-limiting enzyme in the de novo synthesis of guanine nucleotides. It is a target of therapeutically useful drugs and is implicated in the regulation of cell growth rate. In the yeast Saccharomyces cerevisiae, mutations in components of the RNA polymerase II (Pol II) transcription elongation machinery confer increased sensitivity to a drug that inhibits IMPDH, 6-azauracil (6AU), by a mechanism that is poorly understood. This phenotype is thought to reflect the need for an optimally functioning transcription machinery under conditions of lowered intracellular GTP levels. Here we show that in response to the application of IMPDH inhibitors such as 6AU, wild-type yeast strains induce transcription of PUR5, one of four genes encoding IMPDH-related enzymes. Yeast elongation mutants sensitive to 6AU, such as those with a disrupted gene encoding elongation factor SII or those containing amino acid substitutions in Pol II subunits, are defective in PUR5 induction. The inability to fully induce PUR5 correlates with mutations that effect transcription elongation since 6AU-sensitive strains deleted for genes not related to transcription elongation are competent to induce PUR5. DNA encompassing the PUR5 promoter and 5′ untranslated region supports 6AU induction of a luciferase reporter gene in wild-type cells. Thus, yeast sense and respond to nucleotide depletion via a mechanism of transcriptional induction that restores nucleotides to levels required for normal growth. An optimally functioning elongation machinery is critical for this response.
Nucleotides are essential for DNA and RNA synthesis and therefore for cellular growth. Ribonucleotide pool size is carefully regulated in accordance with cellular growth rate in prokaryotic and eukaryotic cells (10, 13, 16, 18). IMP dehydrogenase (IMPDH) is a rate-limiting enzyme in the de novo synthesis of guanine nucleotides. Changes in IMPDH activity are found in some transformed cells and human tumors, and modulation of ribonucleotide levels can trigger p53-mediated cell cycle arrest (27, 29, 51). IMPDH is a medically important target of antimetabolite drugs aimed at reducing cellular proliferation and is the drug target for antimicrobial, immunosuppressive, and antitumor compounds. Such inhibitors include mycophenolate, 6-azauracil (6AU), tiazofurin, and ribavirin (15, 58). In cultured mammalian cells, IMPDH expression parallels guanine nucleotide levels (7, 22). Addition of guanosine to the growth medium results in a 5- to 10-fold reduction of IMPDH mRNA. In contrast, inhibitors of IMPDH that depress the intracellular guanine nucleotide pools induce a fourfold increase in mRNA levels in mammalian cells (17). Changes in IMPDH levels are regulated during growth and differentiation, and tumor cells can become resistant to the growth-inhibiting effect of IMPDH-directed drugs by mutating and/or overproducing the enzyme (43).
One process in which nucleoside triphosphate (NTP) levels play a critical role is transcription elongation by RNA polymerase II (Pol II). In vitro, elongation is strongly influenced by both NTP levels and regulatory elongation factors (48). When NTP substrate concentrations are low, Pol II's elongation rate is slowed and its propensity to become arrested increases. SII is an elongation factor that releases Pol II from the arrested state and enables it to resume elongation (53). Deletion of the gene encoding SII in Saccharomyces cerevisiae, DST1 (also known as PPR2), results in increased sensitivity to the growth inhibitor 6AU (31). This drug reduces intracellular GTP and UTP levels by inhibiting the biosynthetic enzymes IMPDH and orotidylate decarboxylase, respectively (12). One interpretation of the drug-sensitive phenotype is that the elongation stress put upon Pol II by limited intracellular NTP levels causes frequent arrest of Pol II which can be overcome, at least to some extent, by SII (3, 12, 26). In the absence of SII, elongation becomes limiting for growth in the presence of the drug (3). This view is supported by findings that mutations in genes encoding Pol II subunits (RP021 [also known as RPB1], RPB2, and RPB6) that also confer increased sensitivity to 6AU (6AUs) manifest a biochemical elongation defect (24, 37, 57). Mutations in genes encoding elongation factors or proteins implicated in elongation (ELP1, ELP3, SPT4, SPT5, SPT6, and SPT16) also confer increased 6AUs to yeast or otherwise modify 6AUs phenotypes (21, 33, 35, 55). There are, however, 6AUs mutations in genes that lack apparent roles in elongation, including PPR1, SNO1, and SNZ1 (30, 36). PPR1 encodes a DNA-binding transcriptional regulator of the pyrimidine biosynthetic pathway, whereas the latter two genes encode members of a family of stationary-phase proteins that may be involved in pyridoxine biosynthesis (11, 34, 36).
In previous work, we showed that combining a 6AUs allele of rpb2 with a disruption of DST1 resulted in synergistic 6AU sensitivity (26). This allele, rpb2-10, is a point mutation that reduces the average elongation rate of Pol II in vitro (37). The mutant polymerase is also prone to arrest in vitro at well-characterized arrest sites from which it can be rescued by SII (37). Hence, this strain has an elongation-compromised Pol II and lacks an elongation factor that assists arrested Pol II. Drug treatment resulted in a large reduction of total poly(A)+ RNA levels and specific transcripts (26). This suggested that SII is involved in transcription of many if not all genes in yeast and offered direct evidence that 6AU inhibits mRNA synthesis. This drug-induced shutoff of transcription elongation is similar in magnitude and kinetics to the loss of poly(A)+ RNA seen in yeast defective in transcription initiation due to mutation of RP021 and other genes encoding the general initiation machinery (8, 19, 39, 46, 47, 52). Interestingly, wild-type cells also showed a strong reduction in mRNA synthesis after 6AU treatment; however, this change was transient, implying that wild-type but not mutant cells can compensate for depressed intracellular NTP pools.
To understand the biological consequence of perturbing the elongation machinery, we have analyzed the impact of 6AU upon gene expression in wild-type and mutant yeast strains. Here we report the novel finding that 6AU treatment provokes a transcriptional induction of PUR5, a gene encoding an IMPDH homologue in S. cerevisiae. The time course of induction was consistent with an upregulation of IMPDH enzyme and activity and restoration of normal NTP levels, suggesting that yeast cells compensate for depressed intracellular NTP pools, at least in part, via the transcriptional induction of specific genes. Five strains carrying 6AUs mutations in elongation-related genes were defective in this response. 6AUs strains not known to be elongation defective retained the ability to induce PUR5 transcription. PUR5 sequences upstream of the open reading frame (ORF) conferred 6AU dependence upon a heterologous reporter gene. This suggests that yeast can respond to the intracellular loss of GTP by induction of a rate-limiting enzyme. PUR5 transcription may be particularly sensitive to lowered GTP levels and an optimally functioning elongation machinery.
MATERIALS AND METHODS
Strains and plasmids.
Yeast strains used in these studies are listed in Table 1. Where indicated, 6AU (75 μg/ml) and mycophenolic acid (15 μg/ml) were included in the medium. Strains DY700 and DY706 were generated from Z96 (R. Young, Massachusetts Institute of Technology) and ABG-G11 (D. Kaback, UMDNJ-New Jersey Medical School), respectively, by transformation with pPur5P800luc. Strains DY731 and DY732 were generated from ABG-G11 and ABG-G12, respectively, by transformation with pRS316 (42). Strains DY741, DY742, DY743, DY746, DY760, and DY761 were generated from BY4741, BY4742, BY515, BY11569 (all from Research Genetics, Huntsville, Ala.), FY120 (21), and FY1638 (21), respectively, by transformation with pRS316. DY2050 was generated from DY173 by transformation with pC1016. DY173 was generated from DY100 by a two-step allele replacement of RP021 with rpo21-18 after subcloning the mutant allele from the plasmid pYF1504 (J. Friesen, University of Toronto) into pRS306 (42). DY100 was generated from Z96 by disrupting DST1 using the hisG recombination cassette as described elsewhere (1, 26). DY190 was generated from DY100 by transformation with pC1016.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype |
|---|---|
| ABGG10a | MATa ade1Δ2 LEU2Δ4 HIS3 his5 leu2 |
| DY103b | MATα ura3-52 leu2-3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (LEU2 RPB2)][pRS316 (URA3)] |
| DY105b | MATα ura3-52 leu2-3,112 his3Δ200 rpb2Δ297::HIS3 [pRP2-10L (LEU2 rpb2-10)][pRS316 (URA3)] |
| DY106b | MATα ura3-52 leu2-3, 112 his3Δ200 rpb2Δ297::HIS3 dst1::hisG [pRP214 (RPB2 LEU2)][pRS316 (URA3)] |
| DY108b | MATα ura3-52 leu2-3,112 his3Δ200 rpb2Δ297::HIS3 dst1::hisG [pRP2-10L (LEU2 rpb2-10)][pRS316 (URA3)] |
| DY190 | MATα ura3-52 leu2-3,112 his3Δ200 rpb2Δ297::HIS3 dst1::hisG [pRP214 (RPB2 LEU2)][pC1016 (DST1 URA3)] |
| DY700 | MATα ura3-52 leu2-3,112 his3Δ200 rpb2Δ297::HIS3 [pRP214 (LEU2 RPB2)][pPur5P800luc (URA3)] |
| DY706 | MATα Δ4HIS3 trp1 ura3-1 his3-11,15 leu2-3,112 [pPur5P800luc (URA3)] |
| DY731 | MATα Δ4HIS3 trp1 ura3-1 his3-11,15 leu2-3,112 [pRS316 (URA3)] |
| DY732 | MATα Δ3LEU2 ade1 ura3-1 his3-11,15 leu2-3,112 [pRS316 (URA3)] |
| DY741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 [pRS316 (URA3)] |
| DY742 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 [pRS316 (URA3)] |
| DY743 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Δyml056c::kanMX4 [pRS316 (URA3)] |
| DY746 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Δppr1::kanMX4 [pRS316 (URA3)] |
| DY760 | MATa his4-912δ lys2-128δ leu2 Δ1 ura3-52 [pRS316 (URA3)] |
| DY761 | MATa his4-912δ lys2-128δ leu2Δ1 ura3-52 rpb1-221 [pRS316 (URA3)] |
| DY2050 | MATα ura3-52 leu2-3,112 his3Δ200 rpb2Δ297::HIS3 rpo21-18 dstl::hisG [pRP214 (RPB2 LEU2)] [pC1016 (URA DST1)] |
| MW926cd | MATα snz1Δ2 leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 |
| MW980cd | MATa snz1Δ3 sno1Δ3 snz2Δ3 sno2Δ3 snz3Δ3 sno3Δ3 leu2-3,112 trp1-1 ura3-1 ade2-1 his3-11,15 can1-100 |
| MW1072c | MATa ade2-1 his3-11 trp1-1 |
| Z196e | MATa ura3-52 leu2-3,112 his3Δ200 his4-912 lys2-128 rpb1Δ187::HIS3 [pRP112(CEN URA3 RPB1)] |
| Z460e | MATa ura3-52 leu2-3,112 his3Δ200 his4-912 lys2-128 rpb1Δ187::HIS3 [pRP1-1L(CEN URA3 rpb1-1)] |
pC1016 was constructed by inserting a PCR product representing the DST1 locus prepared with the primers 5′-GGCACTGGACTCTAAATCTC-3′ and 5′-AAAGATTTTACGTGAGACAGAC-3′ into the SmaI site of pRS316. The reporter plasmid pPur5P800luc was derived from pGAL-Luc (6) by excision of the GAL promoter with BamHI and HindIII digestion and insertion of a BamHI and HindIII-digested PCR product generated using the primers 5′-CTGATCAGGATCAGGATCCGGCCATTGCTTTTGCTACTT-3′ and 5′-GGGGTACCAAGCTTGTTAACAACAAACACAGTCCA-3′, which amplify the upstream region of PUR5 from genomic DNA.
RNA analysis.
Total RNA was isolated from thawed cell pellets by the hot phenol extraction method and quantitated by measuring absorbance at 260 nm (4). For Northern analysis, 15 μg of total RNA was resolved on a 1% formaldehyde-agarose gel and blotted onto Zeta-Probe GT nylon membrane (Bio-Rad, Hercules, Calif.). Filters were rinsed with 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), baked at 75 to 80°C for 1 h, and cross-linked in a Stratalinker 1800 (Stratagene, La Jolla, Calif.) for 1 min. Filters were prehybridized for a minimum of 3 h at 42°C in 5× SSC–5× Denhardt's solution (4)–50% (vol/vol) formamide–1% (wt/vol) sodium dodecyl sulfate (SDS)–100 μg of salmon sperm DNA per ml and hybridized under the same conditions with ≈108 cpm of 32P-labeled DNA probe for 15 to 18 h. Filters were washed at 22°C twice in 0.2× SSC–0.1% SDS for 5 min each time and twice in 0.2× SSC–0.1% SDS for 5 min each time, followed by two 0.2× SSC–0.1% SDS washes at 42°C for 15 min each, exposed to Kodak X-Omat film, and quantitated with a Fuji BAS1000 imaging system. DNA probes were prepared by PCR using a wild-type yeast genomic DNA template (S288C; Research Genetics) and the corresponding ORF primer pairs obtained from Research Genetics except for primers complementary to YHR216W, which were 5′-GTGGTATGTTGGCCGGTACTACCG-3′ and 5′-TCAGTTATGTAAACGCTTTTCGTA-3′. Probes were labeled to a specific activity of ≈107 to 108 cpm/μg with Klenow DNA polymerase (Promega Life Sciences, Madison, Wis.), random hexamer primers (Gibco BRL, Rockville, Md.), and [α-32P]dATP (Amersham Pharmacia Biotech, Piscataway, N.J.).
Luciferase assay.
Cells from a saturated culture were diluted to an optical density at 600 nm (OD600) of 0.05 to 0.1 and grown to an OD600 of 0.5 at 30°C with aeration. The culture was treated with 6AU, and triplicate samples of 0.5 OD unit of cells were collected at the indicated times and frozen. Cell pellets were stored at −80°C, thawed at 4°C for assay, and lysed by vortexing (six times, 20 s each) with 40 to 50 μl of acid-washed glass beads in 55 μl of luciferase lysis buffer (Promega luciferase assay system; product no. E1501). Samples were spun for 15 s at 15,000 × g in a microcentrifuge. Forty microliters of the soluble cell extract was moved to 100 μl of assay substrate at 22°C, and luminescence was scored in a Optocomp I Luminometer (GEM Biomedical, Pineville, N.C.).
RESULTS
6AU induces IMPDH expression.
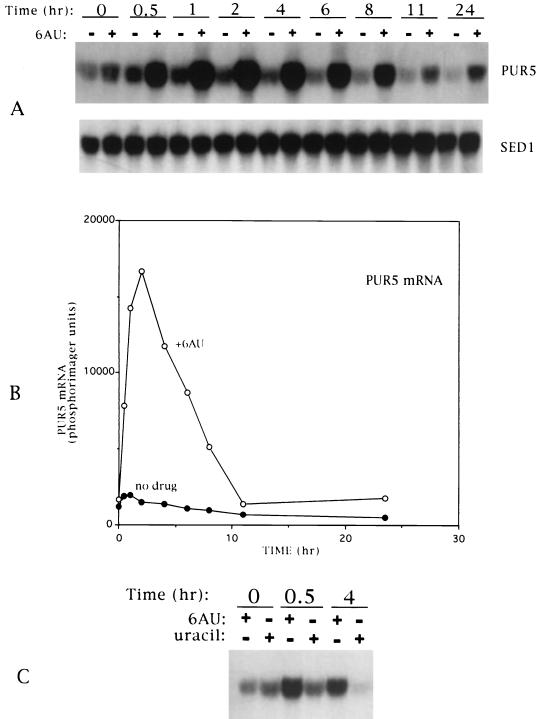
We have previously shown that both wild-type and 6AUs yeast strains show a reduction in mRNA levels when challenged with 6AU (26). For wild-type cells, this was a transient reduction with a maximal depression at 30 min. Cells were able to regenerate normal levels of poly(A)+ RNA by 4 h posttreatment (26). To test the idea that recovery is due to upregulation of the amount of the drug's target, IMPDH, we compared the levels of PUR5 mRNA in 6AU-treated and untreated wild-type yeast cells. Equivalent total RNA was extracted from an equal amount of cells and subjected to Northern blotting. An ≈10-fold induction of PUR5 mRNA was observed 2 h after a 6AU challenge (Fig. 1A and B). This level declined to base line by 10 h posttreatment. As a control for RNA loading, we probed for SED1 mRNA. SED1 encodes an abundant cell wall protein whose expression continues throughout the growth of a culture and into stationary phase (41). Its level is relatively resistant to change during the otherwise general 6AU-induced transcriptional shutoff seen in elongation mutants (26). As expected, SED1 mRNA was detected in all samples, at levels that remained relatively unchanged by 6AU (Fig. 1A). Hence, PUR5 transcript levels dramatically increase in the presence of 6AU during a period in which levels of GTP and total mRNA synthesis are low (Fig. 1 and reference 26). The response was dependent on the inhibitory activity of 6AU and not simply high extracellular pyrimidine levels, since the addition of uracil did not result in induction (Fig. 1C). No 6AU induction was observed for DST1 mRNA, which encodes SII, or for GUA1, which encodes GMP synthase, the next enzyme in the GTP biosynthetic pathway (data not shown).
FIG. 1.
(A) Time course of IMPDH mRNA induction by 6AU. Yeast strain DY103 was diluted to an OD600 of 0.1 in SC-Ura and grown to an OD600 of ≈0.5 at 30°C. The culture was split, and 6AU (75 μg/ml) was added to half; at the indicated times, RNA was harvested and subjected to Northern blotting. (B) Quantitation of Northern blot in panel A. (C) Underivatized uracil does not induces PUR5 mRNA synthesis. Cells were treated with 6AU or uracil (each at 75 μg/ml); RNA was harvested and subjected to Northern blotting.
IMPDH induction arises from one of four related genes.
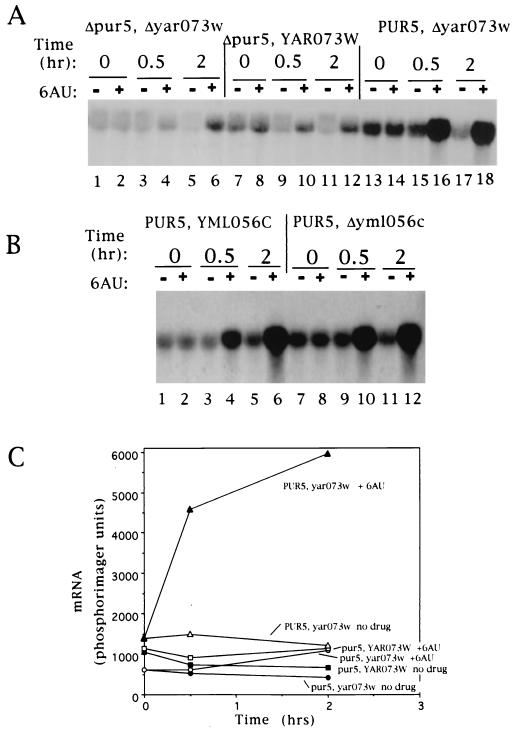
An inspection of the yeast genome revealed that S. cerevisiae has four ORFs that encode a set of homologous IMPDH-like proteins. Two of these genes, located on chromosomes I (YAR073W) and VIII (PUR5; YHR216W) are related (96% amino acid identity) by an apparent ancient chromosomal duplication (5, 56). The copy on chromosome I is near the telomere and thought to be transcriptionally silent. The copy on chromosome VIII (PUR5), on the other hand, has previously been shown to be transcriptionally active (5). Another pair, on chromosomes XII (YLR432W) and XIII (YML056C) (89% amino acid identity) are also thought to be related by duplication, but their transcriptional activity has not been examined directly. None of the four genes are essential (5, 54). To learn which of the genes contributed to the observed induction, we examined mutant strains with deletions in these genes. The probe used cross-hybridizes to mRNA from all four genes. Strains with a deletion of a region of chromosome VIII including PUR5 destroyed most of the response (Fig. 2A, lanes 7 to 12). Induction was readily observed in cells deleted for a region of chromosome I containing YAR073W (Fig. 2A, lanes 13 to 18) or in a strain with a deletion of YML065C on chromosome XIII (Fig. 2B, compare lanes 1 to 6 with lanes 7 to 12). Trace amounts of mRNA remained detectable in a double deletant lacking PUR5 and YAR073W (Fig. 2A, lanes 1 to 6), suggesting that the chromosome XII and/or XIII homologues contribute to a small extent to the steady-state levels of mRNA. A slight induction of this residual expression is also apparent after 2 h of 6AU treatment in the double deletant (Fig. 2A, lanes 5 and 6). Since the majority of the IMPDH mRNA induced by 6AU was derived from transcription of PUR5, we focused on the regulation of this member of the IMPDH family.
FIG. 2.
IMPDH mRNA induction is derived largely from the PUR5 gene. Strains lacking YAR073W and PUR5 (ABGG10; A, lanes 1 to 6), PUR5 (DY731; A, lanes 7 to 12), YAR073W (DY732; A, lanes 13 to 18), or YML056C (DY743; B, lanes 7 to 12) were treated with 6AU (75 μg/ml) for the indicated times; RNA was isolated and subjected to Northern blotting. A control strain (DY741; B, lanes 1 to 6) otherwise isogenic to DY743 was also analyzed. Filters were quantitated by phosphorimaging and plotted (C).
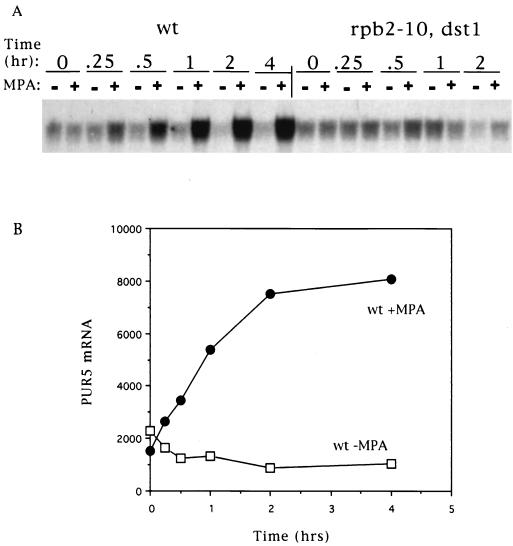
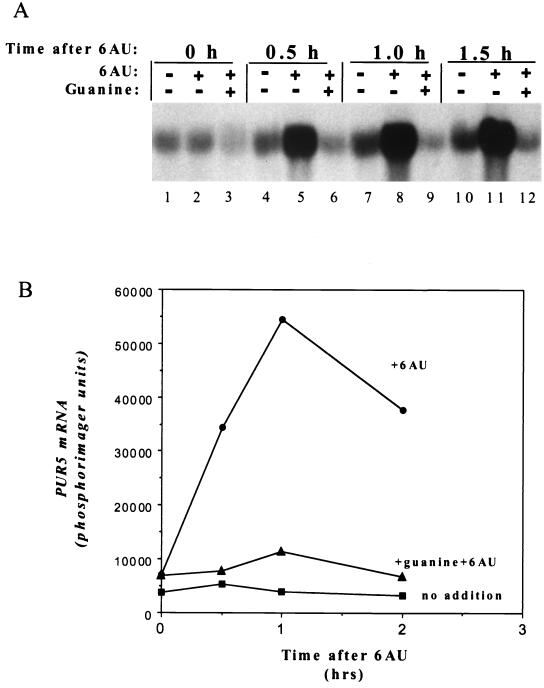
Growth of yeast strains deleted for DST1 is also sensitive to mycophenolic acid, a well-characterized IMPDH inhibitor that is structurally distinct from 6AU (12). We examined whether PUR5 transcription in wild-type cells could be activated by exposure to this drug. A strong induction was observed with mycophenolic acid (Fig. 3, wt). Yeast can import free guanine from the medium and convert it to GTP via salvage synthesis. Consequently, addition of guanine to the growth medium obviates the need for IMPDH activity and ameliorates the 6AUs phenotype of DST1 mutants (3, 12). When cells were pretreated with guanine for 30 min before the addition of 6AU, induction was no longer observed (Fig. 4). Taken together, these data strongly suggest that yeast can sense intracellular GTP depletion and compensate by inducing transcription of PUR5.
FIG. 3.
Induction of PUR5 transcription by mycophenolic acid. Yeast strains DY103 (wt [wild type]) and DY108 (rpb2-10, dst1) were treated with mycophenolic acid (MPA; 15 μg/ml); RNA was isolated and subjected to Northern blotting. Results were quantitated by phosphorimaging and plotted (B).
FIG. 4.
Guanine suppresses PUR5 induction by 6AU. Cultures of DY103 were grown for 30 min in medium containing (lanes 3, 6, 9, and 12) or lacking (lanes 1, 2, 4, 5, 7, 8, 10, and 11) 1 mM guanine; 6AU (75 μg/ml) was added; samples were withdrawn at the indicated times and analyzed by Northern blotting. A control culture lacking guanine and 6AU was also tested (lanes 1, 4, 7, and 10). Filters were phosphorimaged; the data are plotted in panel B.
6AUs mutants in the elongation machinery are defective in PUR5 induction.
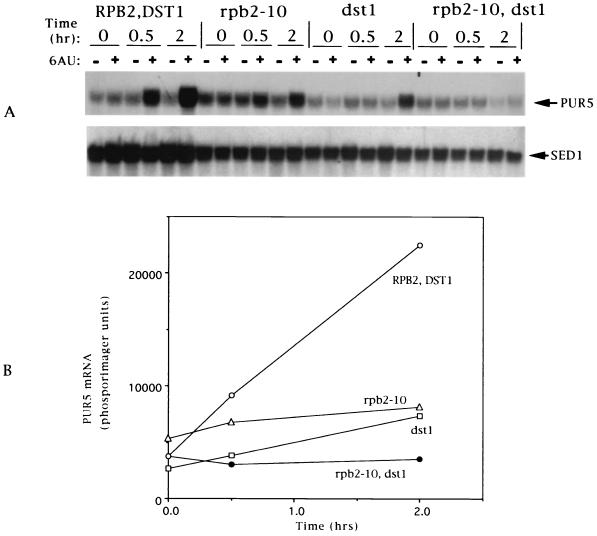
The foregoing results suggest that the previously observed loss of poly(A)+ mRNA seen in a yeast strain bearing two 6AUs mutations, the rpb2-10 point mutation and a disruption of DST1, could be due to the strain's inability to transcribe PUR5 and therefore to regenerate nucleotide levels. Indeed, strains with either the rpb2-10 mutation or a disruption of DST1 were compromised in the ability to induce PUR5, although a limited induction capacity remained (Fig. 5). This residual activity was obliterated when both mutations were introduced into a single strain (Fig. 5), consistent with prior genetic data demonstrating supersensitivity to 6AU of the double mutant relative to the single mutants (26). Induction of PUR5 by mycophenolic acid was also impaired in both the rpb2-10 and the dst1 single mutants (data not shown), as well as in the rpb2-10 dst1 double mutant, which was severely compromised in its response to mycophenolic acid (Fig. 3, rpb2-10, dst1).
FIG. 5.
6AUs elongation mutants fail to induce PUR5 transcription. Strains DY103 (RPB2,DST1), DY105 (rpb2-10), DY106 (dst1), and DY108 (rpb2-10, dst1) were treated with 6AU (75 μg/ml) for the indicated times. RNA was prepared, subjected to Northern blotting (A), and quantitated by phosphorimaging (B). Only the data for drug-treated cells are plotted presented in panel B.
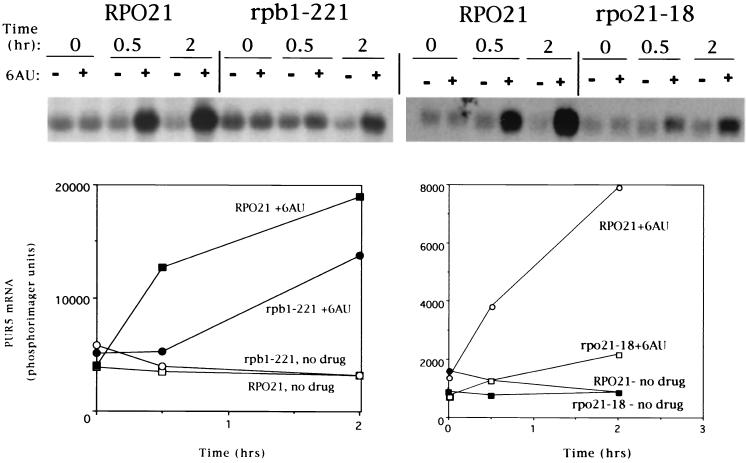
To examine further the correlation between 6AUs mutations in the elongation machinery and the inability to induce transcription of PUR5, we studied the response in two additional strains with 6AUs mutations in RP021. Both exhibited a reduced ability to induce PUR5 transcription in response to a 6AU challenge (Fig. 6). These two mutations, rpo21-18 and rpb1-221, are known to be involved in transcription elongation (3, 21, 57). The rpo21-18 mutation is a linker insertion that results in the addition of five amino acids to the largest subunit of Pol II (3). The resulting enzyme binds SII poorly and confers the 6AUs phenotype (57). The rpb1-221 mutation was identified as 6AUs allele of RP021 that suppresses a cold-sensitive mutation in elongation factor gene SPT5 (21). It is synthetically temperature sensitive when combined with a deletion of DST1. Induction of PUR5 was hindered (although still observable) when cells were grown at 30°C (Fig. 6). These data extend the correlation between 6AUs mutations in the elongation machinery and the inability to induce PUR5 transcription.
FIG. 6.
PUR5 induction in 6AUs strains mutated in Pol II. Strains DY760 (RP021), DY761 (rpb1-221), DY190 (RP021), and DY2050 (rpo21-18) were grown in the presence or absence of 6AU (75 μg/ml), and RNA was prepared for Northern blotting at the indicated times. Results were quantitated by phosphorimaging and plotted.
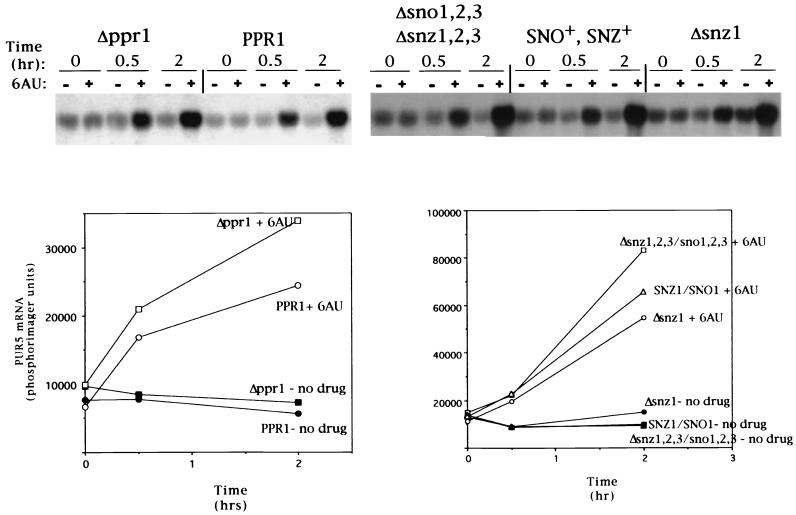
We next wished to determine whether all 6AUs mutants are defective in the PUR5 induction response. To address this question, we analyzed 6AUs strains lacking genes not known to be involved in transcription elongation, PPR1 and SNO1 (30, 36). A strain lacking PPR1 was obtained from the Saccharomyces Genome Deletion Project, and its 6AUs phenotype was confirmed (data not shown). PPR1 is a DNA-binding transcription factor that activates transcription from genes encoding pyrimidine biosynthetic enzymes such as URA3, which encodes orotidylate decarboxylase (12, 38). Loss of PPR1 function results in the underproduction of orotidylate decarboxylase (2), the need for which becomes more acute in the presence of 6AU (12, 30). PPR1 deletion strains grow poorly on uracil dropout medium in the absence of drug, consistent with the idea that cells lacking Ppr1p have a limited ability to trans activate the pyrimidine biosynthetic genes (2; data not shown). Like the wild-type strain, the PPR1 mutant was able to induce PUR5 transcription when challenged with 6AU (Fig. 7). Similarly, deletion of SNZ1 or all six members of the SNZ1-related genes (SNZ1,2,3 and SNO1,2,3) does not affect the ability of 6AU to induce PUR5 transcription (Fig. 7). The SNO/SNZ family of genes are coregulated in response to nutrient limitation and encode proteins with homology to glutamine amidotransferases and enzymes involved in vitamin B6 synthesis (11, 34, 36). Hence, not all 6AUs mutants are defective in PUR5 induction, indicating that drug sensitivity does not lead to failure to induce PUR5 but could be a consequence of a failure of its induction.
FIG. 7.
6AUs mutants in PPR1 and SNZ/SNO can induce PUR5. The experiment described for Fig. 6 was repeated with strains DY746 (Δppr1), DY742 (PPR1), MW980 (Δsno1,2,3, Δsnz1,2,3), MW1072 (SNO+, SNZ+), and MW926 (Δsnz1).
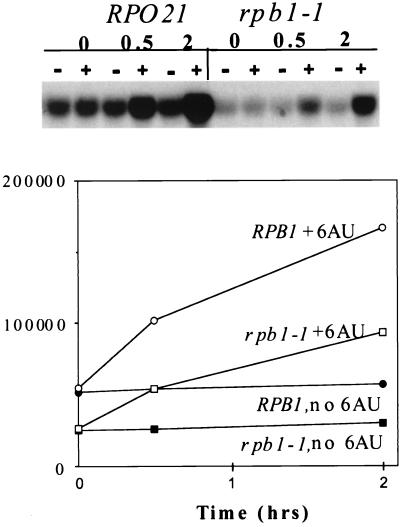
As an additional control, we examined the induction of PUR5 transcription in a strain bearing the well-characterized rpb1-1 mutation, which is not known to be 6AUs (32). This strain has a near-wild-type growth rate at 24°C. Although it possesses a reduced constitutive level of PUR5 mRNA, it is able to induce transcription of PUR5 following 6AU treatment (Fig. 8). Hence, not all Pol II mutations are equivalent in their impact on PUR5 induction.
FIG. 8.
PUR5 induction in the rpb1-1 strain. Strains Z196 (RP021) and Z460 (rpb1-1) were grown in the presence or absence of 6AU (75 μg/ml), and RNA was prepared for Northern blotting at the indicated times. Results were quantitated by phosphorimaging and plotted.
6AU inducibility is transferable.
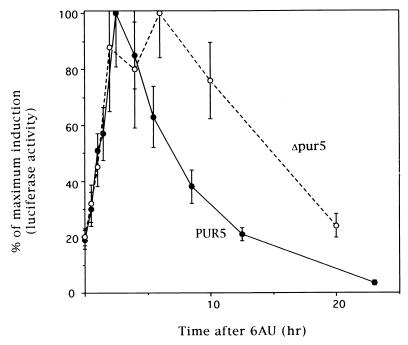
To test whether PUR5 sequences could confer the inductive response to a heterologous gene, we inserted 800 bp of PUR5 sequence into a selectable episomal plasmid upstream of the firefly luciferase coding sequence (6, 42). This DNA spans almost the entire region between the PUR5 ORF and the next upstream gene, PHO12. Although the transcription start site has not been mapped for PUR5, this region must include both the PUR5 promoter and the 5′ untranslated region. The plasmid was used to transform a strain which is wild type at DST1, RPB2, and RP021. Cells were challenged with 6AU, and induction was scored by assaying yeast extracts for luciferase activity. Luciferase activity was strongly induced by 6AU with kinetics comparable to that seen for PUR5 mRNA by Northern blotting (compare Fig. 9 and 1B). There was no response to 6AU when this strain harbored the same reporter construct lacking PUR5 sequences (data not shown).
FIG. 9.
PUR5 sequences confer 6AU inducibility on a luciferase reporter gene. Strains containing (PUR5) or lacking (Δpur5) PUR5 and harboring a reporter plasmid containing 800 bp of DNA of PUR5 sequence upstream of the firefly luciferase coding sequence were treated with 6AU (75 μg/ml) and frozen at the indicated times. Extracts were prepared and assayed for luciferase activity in a luminometer. Data for each strain were normalized to the maximal amount of relative light units obtained over the time course. Experiments were performed in triplicate, and the means ± standard deviations (error bars) were calculated and plotted.
The transient nature of the induction seen for endogenous PUR5 mRNA (Fig. 1) and reporter activity (Fig. 9) could result in part from successful negative feedback by GTP after the burst of PUR5 transcriptional activity that yields Pur5p and IMPDH activity. To test this idea, we placed the reporter plasmid in a strain lacking PUR5 (Fig. 2A). If feedback from IMPDH activity is involved in shutoff of the reporter, there should be a reduction in turnoff of the luciferase reporter. The luciferase induction kinetics in the PUR5 deletant were identical to that seen in cells with an intact PUR5 gene (Fig. 9). However, the strain lacking PUR5 showed a significant slowing in the time needed for luciferase activity to return to baseline, although the rate of its loss was comparable between strains (Fig. 9). The fact that reporter transcription was not indefinitely sustained in the PUR5 deletant might be explained by other additional mechanisms that repress the response (see Discussion). This strain may also have residual inductive capacity in the other three IMPDH homologues (Fig. 2A), which could provide a sufficient accumulation of IMPDH activity to regenerate GTP and repress reporter activity. This reporter will be useful in dissecting the sequence requirement of the 6AU response.
DISCUSSION
We have documented a transcriptional induction system in yeast in which depletion of intracellular nucleotide pools prompts the increased expression of a rate-limiting enzyme. The temporal pattern of expression suggests that a burst of de novo IMPDH synthesis and activity is responsible for the recovery of mRNA levels observed for wild-type cells (26). Remarkably, yeast mutants altered in the transcription elongation machinery, but not 6AUs mutations in genes not known to be involved in transcription elongation, failed to activate PUR5. This could explain the inability of a previously characterized elongation-defective strain to carry out mRNA synthesis (26). PUR5 would appear to be unusual in that it is highly transcribed when the intracellular GTP level is lowered to ≈10% of normal, a condition in which cellular mRNA synthesis is depressed (12, 26). Restricted substrate availability has profound effects on elongation in vitro and in vivo (48, 49). This appears to be exacerbated for mutant Pol II such as that bearing the rpb2-10 substitution (37). The ability of PPR1 and SNO/SNZ deletants to induce PUR5 is consistent with the idea that the elongation defect is causally involved in the inability to activate this gene during GTP depletion. It also suggests that defective induction of PUR5 is a primary problem in some elongation mutants and that this is one mechanism that leads to the 6AUs phenotype. Further analysis of mutant strains will be required to learn how strong this correlation is. The 6AUs phenotype of PPR1 mutants is likely due to defects in induction of the enzymes in pyrimidine nucleotide biosynthesis (12). Although the SNO/SNZ-encoded proteins resemble enzymes involved in nucleotide and pyridoxine metabolism (11, 34, 36), the molecular basis for the 6AUs phenotype in the SNO/SNZ mutants is less obvious.
Our findings regarding PUR5 induction are similar to previous results obtained in mammalian cells in which IMPDH mRNA levels increase after treatment with mycophenolic acid and fall when guanine is added to the medium (7, 17). mRNA accumulation in that case was through a posttranscriptional mechanism (17). In humans, two IMPDH genes have been identified (reviewed in reference 58). Stimulation of T cells requires de novo nucleotide synthesis and is accompanied by increased levels of both type I and type II mRNAs through a transcriptional induction (9, 59, 60). The mechanism involved is not known.
Transcriptional regulation of genes encoding enzymes of pyrimidine biosynthesis has been described in yeast (38). Induction involves direct activation of PPR1 by pyrimidine metabolic intermediates (14). Given this precedent, we must consider that the IMPDH induction that we observe may operate via a purine metabolite sensing mechanism that culminates with the activation of a PUR5 promoter-binding factor. Neither the cis-acting sequences nor transcriptional start sites have been characterized for any of the yeast IMPDH-like genes. The reporter plasmid analysis will serve as a first step in learning if induction operates through upstream promoter sequences, the identity of the regulatory factor(s) that may be involved, and why the other yeast IMPDH homologues are not responsive to the same extent as PUR5.
Sensitivity of yeast growth to 6AU has become widely used as a marker for defects in transcription elongation. This is due in part to the spectrum of genes in which most mutations are found, i.e., elements of the transcription elongation machinery. Mutations in DST1 and PPR1 were identified through a 6AUs screen (23, 30). 6AUs alleles of RP021 and RPB2 were identified following mutant hunts for conditional mutations and later linked to elongation through in vitro biochemistry (3, 26, 37, 57). Some 6AUs mutations, such as rpb1-221, were identified through genetic interactions, and they, or other alleles in those genes, were retrospectively shown to display the 6AUs phenotype (21). Still other genes, such as human SPT4, SPT5, and SPT16 and yeast ELP1 and ELP3, were shown to encode subunits of elongation factors for which mutant alleles were identified that were 6AUs or modified other 6AUs mutations (21, 33, 35, 50, 55). A comprehensive search for 6AUs mutants has not been completed, and it is possible that only a subset of genes that confer 6AU sensitivity has been identified. The existing set may be biased toward components of the elongation machinery, since those mutants are more likely to be tested for this phenotype. Some reports suggest that 6AU may inhibit enzymes involved in amino acid catabolism such as aminoisobutyrate-pyruvate aminotransferase, albeit at relatively high drug concentrations (25, 44, 45). Nevertheless, there is currently a good correlation between the inability to induce PUR5 and mutations that affect transcript elongation.
Both Northern and reporter analyses show a sharp peak in the inductive response; i.e., there is a signal terminating the response. Part of this may be transcriptional repression of PUR5 as the cells approach the diauxic shift (≈10 h [Fig. 1]). This is also suggested by the small but reproducible time-dependent reduction in the constitutive (uninduced) level of PUR5 mRNA seen in untreated cells (Fig. 1A, 1B, 2C, and 3B). PUR5 mRNA may also return to baseline due to a loss of the induction signal (low GTP) after adequate IMPDH and GTP levels have been achieved. This is inferred from the broadening of the peak seen when the endogenous PUR5 gene is deleted (Fig. 9, Δpur5). Additional mechanisms may also contribute to reduced transcription of PUR5 mRNA.
Is the transcription defect in 6AUs elongation mutants specific for PUR5? A common phenotype of yeast with a debilitated Pol II elongation machinery is a slowed response to gene induction signals (35, 40, 55). Even in the absence of 6AU, the rpb2-10 and dst1 mutants are defective in induction of GAL1 transcription in response to galactose (M. Wind and D. Reines, unpublished data). The inability of the 6AUs strains described here to induce PUR5 could arise from a pleiotropic gene induction problem at the level of elongation, and therefore transcriptional output from many or most genes is slowed. Alternatively, it could result from the unusual requirement for PUR5 to be transcribed during low GTP availability if, for example, Pol II has an unusually low Ks for GTP during transcription of PUR5. This property may be important during initiation, promoter clearance, or elongation on the PUR5 gene. In bacteria, regulation of a pyrimidine biosynthetic operon employs a mechanism in which the magnitude of the NTP pools is sensed by Pol II itself (20, 28). A version of this model can be postulated for PUR5 induction. Under GTP-poor conditions, promoter clearance or early elongation is possible on privileged genes such as PUR5, but the process would be particularly sensitive to elongationally crippled RNA polymerase and would require an optimally functioning enzyme and SII to support efficient elongation through the gene. If transcribed sequences determine the ability of Pol II to elongate through PUR5 at low GTP levels, such a requirement is not evident in the gross guanine content of the ORF part of the transcript, which is 23% guanine (about average for a yeast ORF [C. Ball {Saccharomyces Genome Database}, personal communication). Upstream regulatory sequences may play a sole or additional role in regulation of PUR5. A test of this hypothesis must await the identification of the transcription start site in order to assess the relative contributions from promoter elements and the 5′ untranslated region.
ACKNOWLEDGMENTS
We acknowledge valuable suggestions and/or materials from C. Ball, A. Barton, A. Caplan, J. Friesen, G. Hartzog, D. Kaback, M. Werner-Washburne, R. Young, and F. Winston. We also thank S. Devine, R. Gourse, G. Hartzog, C. Moran, and G. Shadel for comments on the manuscript.
This work was supported by NIH grants GM46331 and CA77712.
ADDENDUM IN PROOF
The Saccharomyces Genome Database (http://genome-www.stanford.edu:80/Saccharomyces/) has recently updated the recommended gene names for the four IMP dehydrogenase-like genes (YAR073W, YHR216W [PUR5], YLR432W, and YML056C) described in this paper to IMD1, IMD2, IMD3, and IMD4, respectively.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio O M, Gottschling D E. Overcoming telomeric silencing: a trans-activator competes to establish gene expression in a cell cycle-dependent way. Genes Dev. 1994;8:1133–1146. doi: 10.1101/gad.8.10.1133. [DOI] [PubMed] [Google Scholar]
- 3.Archambault J, Lacroute F, Ruet A, Friesen J D. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol Cell Biol. 1992;12:4142–4152. doi: 10.1128/mcb.12.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates/Wiley-Interscience; 1988. [Google Scholar]
- 5.Barton A, Bussey H, Storms R, Kaback D. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: characterization of the 54 kb right terminal CDC14-FLO1-PHO11 region. Yeast. 1997;13:1251–1263. doi: 10.1002/(sici)1097-0061(199710)13:13<1251::aid-yea174>3.3.co;2-6. [DOI] [PubMed] [Google Scholar]
- 6.Brodsky J L, Lawrence J G, Caplan A J. Mutations in the cytosolic DnaJ homologue, YDJ1, delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry. 1998;37:18045–18055. doi: 10.1021/bi980900g. [DOI] [PubMed] [Google Scholar]
- 7.Collart F, Huberman E. Expression of IMP dehydrogenase in differentiating HL-60 cells. Blood. 1990;75:570–576. [PubMed] [Google Scholar]
- 8.Cormack B P, Struhl K. The TATA-binding protein is required for transcription by all three nuclear RNA polymerases in yeast cells. Cell. 1992;69:685–696. doi: 10.1016/0092-8674(92)90232-2. [DOI] [PubMed] [Google Scholar]
- 9.Dayton J S, Lindsten T, Thompson C B, Mitchell B S. Effects of human T lymphocyte activation on inosine monophosphate dehydrogenase expression. J Immunol. 1994;152:984–991. [PubMed] [Google Scholar]
- 10.Ditzelmuller G, Wohrer W, Kubicek C P, Rohr M. Nucleotide pools of growing, synchronized and stressed cultures of Saccharomyces cerevisiae. Arch Microbiol. 1983;135:63–67. doi: 10.1007/BF00419484. [DOI] [PubMed] [Google Scholar]
- 11.Ehrenshaft M, Bilski P, Li M Y, Chignell C F, Daub M E. A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci USA. 1999;96:9374–9378. doi: 10.1073/pnas.96.16.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Exinger G, Lacroute F. 6-Azauracil inhibition of GTP biosynthesis in Saccharomyces cerevisiae. Curr Genet. 1992;22:9–11. doi: 10.1007/BF00351735. [DOI] [PubMed] [Google Scholar]
- 13.Fairbanks L D, Bofill M, Ruckemann K, Simmonds H A. Importance of ribonucleotide availability to proliferating T-lymphocytes from healthy humans. Disproportionate expansion of pyrimidine pools and contrasting effects of de novo synthesis inhibitors. J Biol Chem. 1995;270:29682–29689. [PubMed] [Google Scholar]
- 14.Flynn P, Reece R. Activation of transcription by metabolic intermediates of the pyrimidine biosynthetic pathway. Mol Cell Biol. 1999;19:882–888. doi: 10.1128/mcb.19.1.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franchetti P, Grifantini M. Nucleoside and non-nucleoside IMP dehydrogenase inhibitors as antitumor and antiviral agents. Curr Med Chem. 1999;6:599–614. [PubMed] [Google Scholar]
- 16.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 17.Glesne D A, Collart F R, Huberman E. Regulation of IMP dehydrogenase gene expression by its end products, guanine nucleotides. Mol Cell Biol. 1991;11:5417–5425. doi: 10.1128/mcb.11.11.5417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grammatikos S I, Tobien K, Noe W, Werner R G. Monitoring of intracellular ribonucleotide pools is a powerful tool in the development and characterization of mammalian cell culture processes. Biotechnol Bioeng. 1999;64:357–367. doi: 10.1002/(sici)1097-0290(19990805)64:3<357::aid-bit12>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 19.Guzder S N, Qiu H, Sommers C H, Sung P, Prakash L, Prakash S. DNA repair gene RAD3 of S. cerevisiae is essential for transcription by RNA polymerase II. Nature. 1994;367:91–94. doi: 10.1038/367091a0. [DOI] [PubMed] [Google Scholar]
- 20.Han X, Turnbough C L., Jr Regulation of carAB expression in Escherichia coli occurs in part through UTP-sensitive reiterative transcription. J Bacteriol. 1998;180:705–713. doi: 10.1128/jb.180.3.705-713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartzog G A, Wada T, Handa H, Winston F. Evidence that SPT4, SPT5, and SPT6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 1998;12:357–369. doi: 10.1101/gad.12.3.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huberman E, Glesne D, Collart F. Regulation and role of inosine 5′ monophosphate dehydrogenase in cell replication, malignant transformation, and differentiation. In: Sahota A, Taylor M, editors. Purine and pyrimidine metabolism in man VIII. New York, N.Y: Plenum Press; 1995. pp. 741–746. [DOI] [PubMed] [Google Scholar]
- 23.Hubert J-C, Guyonvarch A, Kammerer B, Exinger F, Liljelund P, Lacroute F. Complete sequence of a eukaryotic regulatory gene. EMBO J. 1983;2:2071–2073. doi: 10.1002/j.1460-2075.1983.tb01702.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishiguro A, Nogi Y, Hisatake K, Muramatsu M, Ishihama A. The Rpb6 subunit of fission yeast RNA polymerase II is a contact target of the transcription elongation factor TFIIS. Mol Cell Biol. 2000;20:1263–1270. doi: 10.1128/mcb.20.4.1263-1270.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kontani Y, Kaneko M, Kikugawa M, Fujimoto S, Tamaki N. Identity of d-3-aminoisobutyrate-pyruvate aminotransferase with alanine-glyoxylate aminotransferase 2. Biochim Biophys Acta. 1993;1156:161–166. doi: 10.1016/0304-4165(93)90131-q. [DOI] [PubMed] [Google Scholar]
- 26.Lennon J C, III, Wind M, Saunders L, Hock M B, Reines D. Mutations in RNA polymerase II and elongation factor SII severely reduce mRNA levels in Saccharomyces cerevisiae. Mol Cell Biol. 1998;10:5771–5779. doi: 10.1128/mcb.18.10.5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linke S P, Clarkin K C, Di Leonardo A, Tsou A, Wahl G M. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 28.Liu C, Heath L S, Turnbough C L., Jr Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 1994;8:2904–2912. doi: 10.1101/gad.8.23.2904. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, Bohn S A, Sherley J L. Inosine-5′-monophosphate dehydrogenase is a rate-determining factor for p53-dependent growth regulation. Mol Biol Cell. 1998;9:15–28. doi: 10.1091/mbc.9.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loison G, Losson R, Lacroute F. Constitutive mutants for orotidine 5′ phosphate decarboxylase and dihydroorotic acid dehydrogenase in Saccharomyces cerevisiae. Curr Genet. 1980;2:39–44. doi: 10.1007/BF00445692. [DOI] [PubMed] [Google Scholar]
- 31.Nakanishi T, Nakuno A, Nomura K, Sekimizu K, Natori S. Purification, gene cloning, and gene disruption of the transcription factor SII in Saccharomyces cerevisiae. J Biol Chem. 1992;267:13200–13204. [PubMed] [Google Scholar]
- 32.Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orphanides G, Wu W H, Lane W S, Hampsey M, Reinberg D. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999;400:284–288. doi: 10.1038/22350. [DOI] [PubMed] [Google Scholar]
- 34.Osmani A H, May G S, Osmani S A. The extremely conserved pyroA gene of Aspergillus nidulans is required for pyridoxine synthesis and is required indirectly for resistance to photosensitizers. J Biol Chem. 1999;274:23565–23569. doi: 10.1074/jbc.274.33.23565. [DOI] [PubMed] [Google Scholar]
- 35.Otero G, Fellows J, Li Y, de Bizemont T, Dirac A M, Gustafsson C M, Erdjument-Bromage H, Tempst P, Svejstrup J Q. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell. 1999;3:109–118. doi: 10.1016/s1097-2765(00)80179-3. [DOI] [PubMed] [Google Scholar]
- 36.Padilla P A, Fuge E K, Crawford M E, Errett A, Werner-Washburne M. The highly conserved, coregulated SNO and SNZ gene families in Saccharomyces cerevisiae respond to nutrient limitation. J Bacteriol. 1998;180:5718–5726. doi: 10.1128/jb.180.21.5718-5726.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell W, Reines D. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J Biol Chem. 1996;271:6866–6873. doi: 10.1074/jbc.271.12.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy A, Exinger F, Losson R. cis- and trans-acting regulatory elements of the yeast URA3 promoter. Mol Cell Biol. 1990;10:5257–5270. doi: 10.1128/mcb.10.10.5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakurai H, Ohishi T, Fukasawa T. Promoter structure-dependent functioning of the general transcription factor IIE in Saccharomyces cerevisiae. J Biol Chem. 1997;272:15936–15942. doi: 10.1074/jbc.272.25.15936. [DOI] [PubMed] [Google Scholar]
- 40.Scafe C, Nonet M, Young R A. RNA polymerase II mutants defective in transcription of a subset of genes. Mol Cell Biol. 1990;10:1010–1016. doi: 10.1128/mcb.10.3.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimoi H, Kitagaki H, Ohmori H, Iimura Y, Ito K. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J Bacteriol. 1998;180:3381–3387. doi: 10.1128/jb.180.13.3381-3387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sikorski R, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Snyder F F, Lightfoot T, Hodges S D. Molecular characterization of IMP dehydrogenase in acquired resistance to mycophenolic acid. In: Sahota A, Taylor M, editors. Purine and pyrimidine metabolism in man VIII. New York, N.Y: Plenum Press; 1995. pp. 725–728. [DOI] [PubMed] [Google Scholar]
- 44.Tamaki N, Kubo K, Aoyama H, Funatsuka A. Inhibitory effect of 6-azauracil on purified rabbit liver 4-aminobutyrate aminotransferase. J Biochem. 1983;93:955–959. doi: 10.1093/oxfordjournals.jbchem.a134250. [DOI] [PubMed] [Google Scholar]
- 45.Tamaki N, Fujimoto S, Mizota C, Kaneko M, Kikugawa M. Inhibitory effect of 6-azauracil on beta-alanine metabolism in rat. J Nutr Sci Vitaminol. 1989;35:451–461. doi: 10.3177/jnsv.35.451. [DOI] [PubMed] [Google Scholar]
- 46.Thompson C M, Young R A. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tijerina P, Sayre M H. A debilitating mutation in transcription factor IIE with differential effects on gene expression in yeast. J Biol Chem. 1998;273:1107–1113. doi: 10.1074/jbc.273.2.1107. [DOI] [PubMed] [Google Scholar]
- 48.Uptain S, Kane C M, Chamberlin M J. Basic mechanisms of transcript elongation and its regulation. Annu Rev Biochem. 1997;66:117–172. doi: 10.1146/annurev.biochem.66.1.117. [DOI] [PubMed] [Google Scholar]
- 49.von Hippel P H. An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 50.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog G A, Winston F, Buratowski S, Handa H. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wahl G M, Linke S P, Paulson T G, Huang L C. Maintaining genetic stability through TP53 mediated checkpoint control. Cancer Surv. 1997;29:183–219. [PubMed] [Google Scholar]
- 52.Walker S S, Shen W C, Reese J C, Apone L M, Green M R. Yeast TAF(II)145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- 53.Wind M, Reines D. Transcription elongation factor SII. Bioessays. 2000;22:327–336. doi: 10.1002/(SICI)1521-1878(200004)22:4<327::AID-BIES3>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, Chu A M, Connelly C, Davis K, Dietrich F, Dow S W, El Bakkoury M, Foury F, Friend S H, Gentalen E, Giaever G, Hegemann J H, Jones T, Laub M, Liao H, Davis R W, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 55.Wittschieben B O, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis C D, Tempst P, Svejstrup J Q. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell. 1999;4:123–128. doi: 10.1016/s1097-2765(00)80194-x. [DOI] [PubMed] [Google Scholar]
- 56.Wolfe K H, Shields D C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 57.Wu J, Awrey D E, Edwards A M, Archambault J, Friesen J D. In vitro characterization of mutant yeast RNA polymerase II with reduced binding for elongation factor TFIIS. Proc Natl Acad Sci USA. 1996;93:1152–1157. doi: 10.1073/pnas.93.21.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zimmermann A G, Gu J J, Laliberte J, Mitchell B S. Inosine-5′-monophosphate dehydrogenase: regulation of expression and role in cellular proliferation and T lymphocyte activation. Prog Nucleic Acid Res Mol Biol. 1998;61:181–209. doi: 10.1016/s0079-6603(08)60827-2. [DOI] [PubMed] [Google Scholar]
- 59.Zimmermann A G, Spychala J, Mitchell B S. Characterization of the human inosine-5′-monophosphate dehydrogenase type II gene. J Biol Chem. 1995;270:6808–6814. doi: 10.1074/jbc.270.12.6808. [DOI] [PubMed] [Google Scholar]
- 60.Zimmermann A G, Wright K L, Ting J P, Mitchell B S. Regulation of inosine-5′-monophosphate dehydrogenase type II gene expression in human T cells. Role for a novel 5′-palindromic octamer sequence. J Biol Chem. 1997;272:22913–22923. doi: 10.1074/jbc.272.36.22913. [DOI] [PubMed] [Google Scholar]