Abstract
The outbreak of a new coronavirus (SARS-CoV-2) was first identified in Wuhan, People's Republic of China, in 2019, which has led to a severe, life-threatening form of pneumonia (COVID-19). Research scientists all around the world have been trying to find small molecule drugs to treat COVID-19. In the present study, a conserved macrodomain, ADP Ribose phosphatase (ADRP), of a critical non-structural protein (Nsp3) in all coronaviruses was probed using large-scale Molecular Dynamics (MD) simulations to identify novel inhibitors. In our virtual screening workflow, the recently-solved X-ray complex structure, 6W6Y, with a substrate-mimics was used to screen 17 million ZINC15 compounds using drug property filters and Glide docking scores. The top twenty output compounds each underwent 200 ns MD simulations (i.e. 20 × 200 ns) to validate their individual stability as potential inhibitors. Eight out of the twenty compounds showed stable binding modes in the MD simulations, as well as favorable drug properties from our predctions. Therefore, our computational data suggest that the resulting top eight out of twenty compounds could potentially be novel inhibitors to ADRP of SARS-CoV-2.
Keywords: SARS-CoV-2, COVID-19, Nsp3, Macrodomain, Docking, MD simulation
1. Introduction
Since the first cases in Wuhan, People's Republic of China, at the end of year 2019, the spread of Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2), which causes the disease COVID-19, has escalated worldwide. It has infected approximately 212 M people and killed approximately 4.43 M, leading the World Health Organization (WHO) to classify it as a pandemic. SARS-CoV-2, belonging to a group of beta-coronavirus, resembles the other two members: SARS-CoV and Middle East Respiratory Syndrome (MERS-CoV). SARS-CoV and MERS-CoV were also responsible for earlier outbreaks of deadly respiratory diseases, but there is no single drug to treat them. Currently, only one drug, Remdesivir, has been approved by the US food and drug administration (FDA) to treat SARS-CoV2, but its efficacy is not very high. The demand for therapeutics keeps increasing as the delta variant leads to an increase in breakthrough infections among vaccinated people.
The genome of SARS-CoV-2 encodes an approximately 7096-residue-long polyprotein, which consists of many structural and non-structural proteins (NSPs) [1]. Like other coronaviruses, SARS-CoV-2 uses positive-sense RNA genome to code NSPs and structural proteins, such as the spike glycol-protein, envelope, and membrane [2]. The 16 NSPS found in SARS-CoV-2 form a large, membrane-bound replicase complex, in which Nsp3 is the largest component, with a residue range between 1 and 1922 [2]. Nsp3, which exists in all CoVs, is a multidomain (macrodomain) protein, ADP ribose phosphatase domain (ADRP, also known as macrodomain), which contains the N-terminal ubiquitin-like domain (Ubl), SARS-unique domain, Papain-like proteinase (PLpro), nucleic acid binding (NAB) domain, also containing transmembrane domain and Y-domain (Fig. 1 A). Macrodomain (Mac1) prevents host immune response to viral infection by removing ADP-Ribosylation from modified host protein [3]. Host poly-ADP ribose polymerase (PARP) enzymes catalyze the transfer of the ADP-ribose phosphate group to their target proteins, primarily attached to the Mac1 to perform its function (assisting in DNA damage repair, cellular stress, and proper immune response) which allows the host to recognize and attack viruses like SAS-COV2 [2,4]. The binding of SARS-CoV-2 to Mac1 is crucial, as it initiates virulence and RNA replication [5]. Type I interferons (IFN–I) impel the intrinsic induction by blocking the phosphorylation dimerization and resulting in nuclear translocation of the host IFN regulatory factor 3 (IRF3), which is a transcription regulator important for innate immunity [2,6]. The role of Mac1 is crucial, as their inhibition helps to reduce the viral load, facilitate recovery and interferes with the host's immune response to SARS-CoV-2 (which makes it an attractive protein target) [2,6]. Although SARS-CoV-2 has higher transmission efficiency from human to human than previous viruses (for instance, SARs and MERs), the Mac1 protein exists in both SARs and MERs, which makes it a common drug target for both viruses [7,8].
Fig. 1.
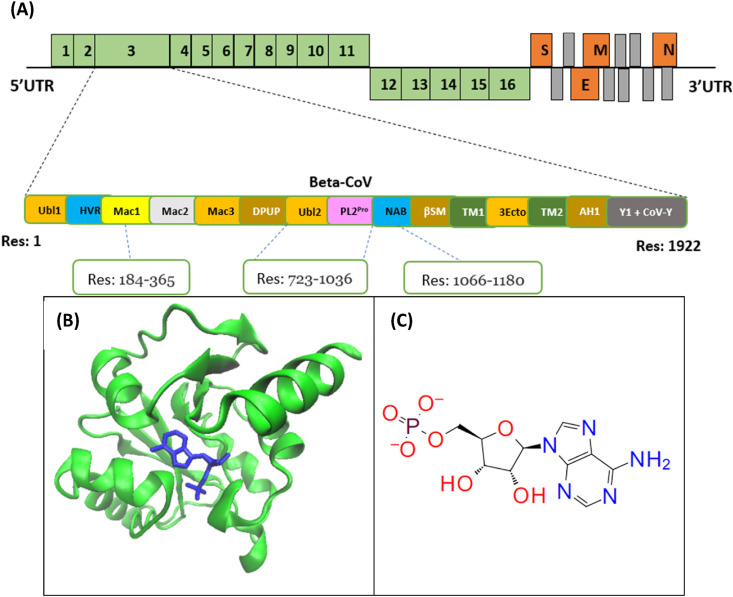
(A) Genome organization of Non-structural proteins (green) and structural proteins (orange) of a beta-coronavirus; NSP3 and its domains: Ubl1 (Res: 1–112), HVR (Res: 113–183),Mac1(Res: 184–365), Mac2 (Res: 389–524), Mac3 (Res: 525–652), DPUP (Res: 653–720), Ubl2- PL2Pro(Res: 723–1036), NAB (Res: 1066–1180), βSM (G2M) (Res: 1203–1318), TM1 (Res:1391–1413), 3Ecto (Res:1414–1495), TM2 (Res: 1496–1518), AH1 (Res: 1523–1545), Y1 + CoV–Y (Res: 1546–1922).(B) ADP Ribose phosphatase of NSP3 in complex with AMP (PDB ID: 6W6Y), Residue range of 207–379. (C) Chemical structure of AMP displayed. AMP is used instead of ADP because it is more stable and has the least amount of stored energy.
Although there have been studies (Table 1 ) which focused on identifying novel drugs targeting viral macrodomains using the virtual screening and pharmacophore approach, there have been no such studies conducted which utilized elaborate virtual screening methods. In a 2020 study conducted by Babar et al., a total of 64,043 drugs were screened, in which potential inhibitors were chosen based on their docking score and high binding affinity for key active site residues [9]. In a 2020 study conducted by Debnath, virtual screening was performed on the 113K MolPort database, from which six candidates were selected based on their XP glide score range, whose binding affinities were validated using free energy calculations and MD simulations [10]. However, Debnath did not analyze the stability of the top potential inhibitors using the MD simulation [9,10]. Using a similar approach, the present study uses a much larger database and longer MD simulations for more compounds.
Table 1.
Comparison between this study and previous studies.
| Authors | Protein PDB ID | Ligand Database | Method | Final Lead Compounds |
|---|---|---|---|---|
| Debnath et al. | 6W02 | MolPort (113K) | E-Pharmacophore based VS | 6 |
| Babar et al. | 6YWL | SwissSimularity1 NANPDB (6482) TCM (57K) |
Drug Simularity Search IF Docking for top 80 ligands MD Simulation(100ns) for top 6 ligands |
6 |
| Present study | 6W6Y | Zinc153 (17 M) | Glide XP Docking MD simulation (200ns) for top 20 ligands |
8 |
In the details, the structure of ADP-ribose phosphatase in the complex of AMP (PDB ID: 6W6Y, Fig. 1B–C) was used in the structure-based high throughput screening of zinc15 library with 17 million compounds. AMP as a substrate, which is used as a monomer in RNA, also plays an important role in intracellular signaling and cellular metabolic processes [9]. Next, we have utilized a long MD simulation (200 ns) to examine the top 20 hits from the virtual screening. Followed by advanced MD simulation and MM-GBSA binding energy calculation methods was used to obtain better estimate of the binding affinity. Eight compounds out of the 20 hits showed significantly improved binding free energy score and good drug properties. The present study adds important knowledge to the ongoing efforts of finding the potential drug target compounds and novel inhibitors to ADP-Ribose phosphatase of SARS-CoV-2.
2. Methods
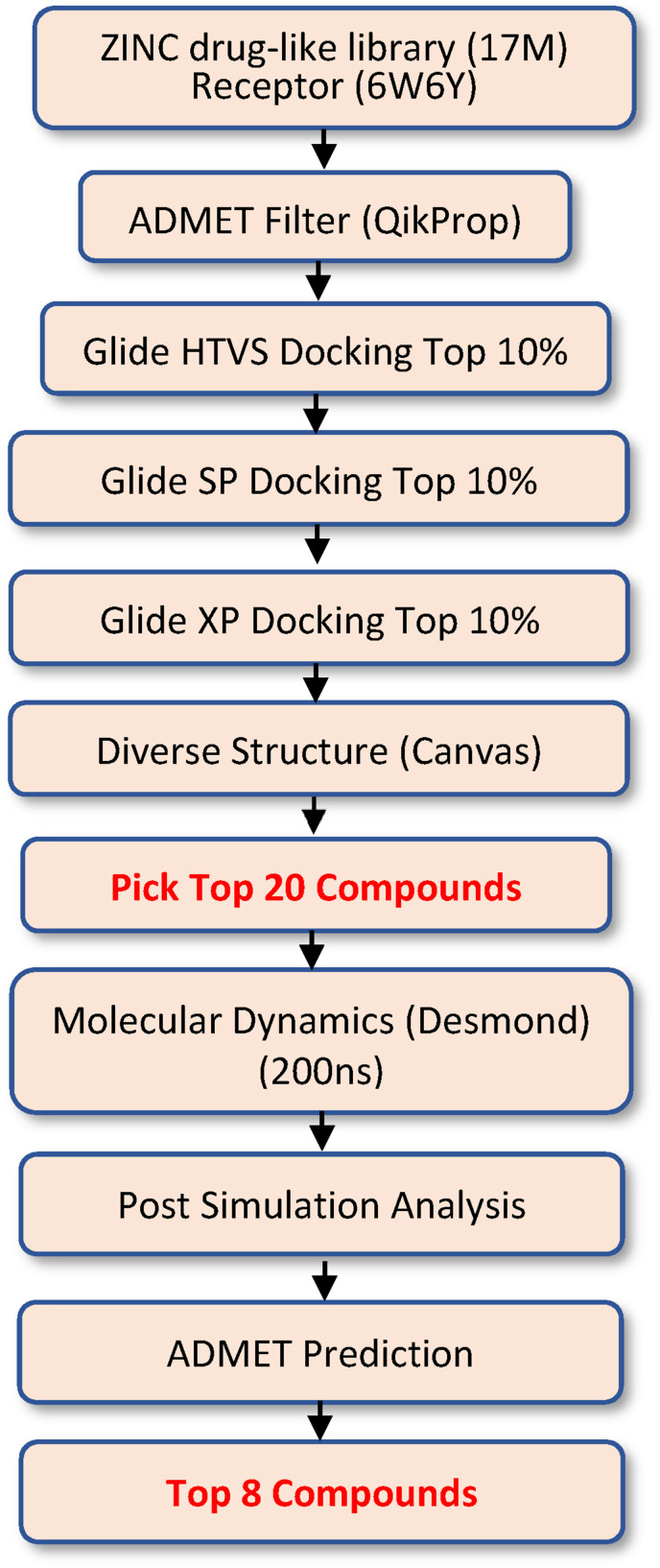
A virtual screening workflow (VSW) in Fig. 2 was developed to identify lead inhibitors to the ADP-Ribose phosphatase of SARS-CoV-2 from ZINC 15 drug-like library with 17 million entries. This VSW consists of ten essential steps including drug property prediction, molecular docking, and molecular dynamics simulation. The first step of the VSW was the input of the prepared protein structure and ligand library. Then, the compounds were filtered by drug property in setup 2 and docking with multiple Glide docking score functions with increasing accuracy (Glide HTVS, SP and XP) in step 2–5. Ligand similarity analysis was performed to identify different molecular scaffolds in step 6. In step 7, the ligands that have either a worse Glide XP score than the reference compound (the crystal ligand AMP) or more than one red flag in drug property (# of star, from QikProp) were removed, the top 20 compounds were manually selected from the remaining compounds by maximizing the number of molecular scaffolds (i.e., different ligand cluster IDs). In steps 8 and 9, the 200ns MD simulation was carried out, followed by post simulation analyses including MMGBSA binding free energy calculation, simulation interaction diagram analysis, and protein conformation clustering analysis. In step 9, the prediction of ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) was used to check the human oral bioavailability of potential drug candidates. Finally, the compounds that had better MMGBSA binding free energy than the reference compounds were selected and presented in the main text. The details of these ten steps are presented in the following six modules.
Fig. 2.
Virtual screening workflow to identify lead competitive inhibitors to the ADP-Ribose phosphatase of SARS-CoV-2 from ZINC 15 drug-like library.
2.1. Preparation of protein and ligand library
The crystal structure (6W6Y) of ADP ribose phosphatase of Nsp3 from SARS-CoV-2 was prepared using Maestro's Protein Preparation Wizard [11]. The protein was preprocessed to assign correct bond orders, add hydrogen atoms, create disulfide bonds, and to delete water beyond 5 Å (Å) from hetero groups. The charge state of the titratable residues was optimized using PROPKA at a pH of 7. A restrained minimization was done to relax the protein using an OPLS3 force field [12]. Epik, a tool based on accurate methodologies from Hammer and Taft, was used to generate the proper ionization state of each ligand [11]. The lowest tautomeric state for each ligand structure was selected and minimized to relax the ligands to a best fit structure. Lastly, a geometry optimization was performed using quantum mechanics methods in Jaguar.
2.2. Filtering and docking
The prepared merged protein-ligand complex was put through Schrodinger's Virtual Screening Interface, where it was prefiltered through Lipinski's Rule and filtered with ADMET risk parameter assessments through QikProp. The parameters and cut-off values employed when screening using QikProp were described in Table S1 [13]. The active binding site of ADP ribose phosphatase was defined using the center of the ligand, from which the grid file was generated using a van der Waals scaling factor of 1 and a partial charge cutoff of 0.25. The prepared compounds were docked into the generated grid of the protein receptor using an OPLS3 force field and their docking scores were calculated using both SP and XP scoring functions [12]. The default settings were used as the parameters for the scoring function: Active Epik state penalties to the docking score, dock sampling was flexible with sample nitrogen-inversions, same ring conformations, and bias sampling of torsions for amides, which only penalized non-planar conformations [14,15]. The results of docking concluded with 20 top compounds with high docking scores, indicating that they all had high affinity for the receptor.
2.3. Ligand similarity clustering
The ligand similarity clustering was done on the Canvas program. First, digital fingerprints of 3D ligand structures were generated using 3-point pharmacophore [16]. Next, hierarchical clustering with default parameters was performed to group similar compounds into different clusters using their fingerprints and a cluster ID was assigned to each compound [17,18].
2.4. MD simulation
The twenty prepared receptor-ligand complexes were used to construct MD simulation systems. The complexes were solvated in an orthorhombic water box with a buffer distance of 10 Å using a predefined SPC water model [19]. A 0.15 M NaCl salt concentration was added to neutralize the system. The systems were built with an OPLS3 force field using Desmond System Builder in Maestro on a Linux operating system [12]. The relaxation/minimization and production runs were set up using the Desmond module following our previous procedure [20].
2.5. Post simulation analysis
2.5.1. Simulation interaction diagram (SID) analysis
The Desmond SID tool in Maestro was used to calculate the Root-Mean-Square Deviation (RMSD), the Root-Mean-Square Fluctuation (RMSF), the Secondary Structural Elements (SSE), and the residue-ligand interactions and contacts throughout the course of the simulation. The protein Cα and ligand RMSD plots obtained from the SID analysis were analyzed to ensure the convergence of each of the MD simulations. A relatively flat plot would imply that a steady state was reached.
2.5.2. Trajectory clustering analysis
The Desmond clustering tool was utilized to organize the complex structures from the trajectories [21]. The parameters included using the backbone RMSD matrix as the structural similarity metric and using a 2.5 Å merging distance cutoff and average linkage for the hierarchical clustering. The centroid structure, the structure with the most neighbors in the structural family, was chosen to represent each structural family. Of the centroids, the most abundant structures of the populated structural families were extracted and analyzed further. Clustering is used to identify the most abundant conformations and reduces complexity.
2.5.3. Binding energy calculations and decompositions
The ligand-binding affinities on the frames obtained in the last 50ns of each MD simulation were calculated using the surface-area-based Generalized Born model [22,23] with an implicit membrane solvation model (VSGB 2.0) [24]. as explained in our previous works [20].
2.6. ADMET prediction
Prediction of ADMET properties for the eight best ZINC compounds were performed on the SwissADME web server (http://www.swissadme.ch/). This server is developed by the Swiss Institute of Bioinformatics, used to provide physiochemical descriptors, ADMET parameters, pharmacokinetic properties, and drug-like small molecule inhibitors to support drug discovery [25]. The SMILE codes for each compound were inserted into the webserver to receive their ADMET properties.
3. Results
MM-GBSA results reveal the top eight compounds according to their ligand binding energy. A total of 17 M zinc compounds were screened from which compounds with docking scores higher than −9.7 kcal/mol were considered, discarding the others. The top twenty compounds were picked based on their SMILE code, numbers of STARs, cluster IDs, and centroid (Fig. S3). The various properties including the glide XP docking, MM-GBSA, and MD simulation helped in picking the top eight compounds out of twenty total compounds. The MM-GBSA method was used to estimate the binding free energy of the twenty compounds. The binding interaction between ligand-protein receptors is specified by the free energy binding. Using the crystal ligand as the control, the top picked compounds possess significantly higher binding energy against the Mac1. The results of other energy terms such as Van Der Waals energy (VDW), electrostatic energy (ELE), hydrophobic, and ligand-receptor RMSD strongly indicate that these top-picked compounds could be targeted against SARS-CoV-2. The hydrophobic value (−46.7 ± 3.6 kcal/mol) was favorable for the binding of ZINC000096223736, followed by other compounds. ZINC000096223736 had superior binding energy than the other compounds which is visible in (Table 2 ). The protein-ligand contact pattern shows the protein binding pocket, and the main residues responsible for the interaction. Protein-ligand contacts of the top eight compounds during MD simulation are shown in (Table 3 ), the most abundant residues display the highest number of interactions throughout the results section.
Table 2.
Various properties of the top 20 compounds and the crystal reference ligand from the Glide XP docking and the MD simulations.
| # | ZINC ID | Docking Score (kcal/mol) | VDW (kcal/mol) | ELE (kcal/mol) | Hydrophobic (kcal/mol) | MM-GBSAa (kcal/mol) | Receptor RMSDa (Å) | Ligand RMSDa (Å) |
|---|---|---|---|---|---|---|---|---|
| Ref. | PDB ID: 6W6Y | −9.752 | −31.2 ± 4.1 | 2.9 ± 3.6 | −11.2 ± 2.1 | −39.5 ± 6.5 | 1.1 ± 0.3 | 2.5 ± 0.5 |
| 1 | ZINC082673 | −11.7 | −43.6 ± 1.9 | −4.4 ± 2.7 | −34.2 ± 1.9 | −82.1 ± 3.8 | 1.7 ± 0.1 | 1.7 ± 0.3 |
| 2 | ZINC036569382 | −11.6 | −40 ± 4.2 | 6.1 ± 3.4 | −21.2 ± 2.0 | −55.1 ± 6.2 | 2.5 ± 0.1 | 3.4 ± 0.2 |
| 3 | ZINC003830180 | −11.1 | −30.0 ± 3.7 | 4.8 ± 2.2 | −8.9 ± 1.6 | −34.2 ± 4.2 | 2.4 ± 0.2 | 3.2 ± 0.5 |
| 4 | ZINC006112607 | −11 | −36.4 ± 4.0 | −0.3 ± 4.8 | −26.4 ± 3.4 | −63.2 ± 9.5 | 2.2 ± 0.2 | 5.7 ± 0.3 |
| 5 | ZINC014116837 | −10.9 | −48.2 ± 2.7 | −4.2 ± 2.7 | −36.1 ± 2.2 | −88.4 ± 3.6 | 2.4 ± 0.1 | 2.3 ± 0.3 |
| 6 | ZINC121003678 | −10.8 | −43.5 ± 3.9 | 1.7 ± 2.1 | −42.8 ± 3.6 | −84.6 ± 7.8 | 1.4 ± 0.3 | 0.8 ± 0.2 |
| 7 | ZINC217844024 | −10.8 | −42.9 ± 2.8 | −7.9 ± 3.7 | −31.5 ± 1.9 | −82.3 ± 4.9 | 1.7 ± 0.2 | 0.8 ± 0.2 |
| 8 | ZINC096223736 | −10.5 | −50.2 ± 4.0 | −5.0 ± 3.4 | −46.7 ± 3.6 | −101.9 ± 6.6 | 1.2 ± 02 | 1.1 ± 0.3 |
| 9 | ZINC097036564 | −10.4 | −39.3 ± 3.2 | 9.2 ± 2.5 | −28.1 ± 1.9 | −58.2 ± 4.4 | 1.1 ± 0.2 | 1.0 ± 0.2 |
| 10 | ZINC237938532 | −10.4 | −19.4 ± 8.2 | −4.9 ± 3.6 | −14.5 ± 7.1 | −29.0 ± 13.7 | 1.8 ± 0.2 | 23.7 ± 3.7 |
| 11 | ZINC096232566 | −10.4 | −41.2 ± 3.5 | −10.7 ± 3.6 | −33.5 ± 2.5 | −85.4 ± 5.5 | 1.5 ± 0.2 | 0.7 ± 0.2 |
| 12 | ZINC003869813 | −10.4 | −24.1 ± 6.5 | 1.7 ± 6.0 | −7.7 ± 3.3 | −30.1 ± 7.3 | 1.4 ± 0.2 | 9.8 + 5.1 |
| 13 | ZINC097036607 | −10.4 | −39.6 ± 4.1 | 9.3 ± 4.8 | −29.8 ± 2.4 | −60.1 ± 5.3 | 1.0 ± 0.2 | 2.3 ± 0.3 |
| 14 | ZINC097036605 | −10.3 | −29.2 ± 7.0 | 3.6 ± 4.1 | −16.6 ± 9.6 | −39.2 ± 15.1 | 2.3 ± 0.1 | 22.9 ± 3.0 |
| 15 | ZINC079784201 | −10.3 | −46.8 ± 3.1 | −5.3 ± 2.9 | −39.6 ± 4.2 | −91.7 ± 6.9 | 1.5 ± 0.1 | 1.0 ± 0.2 |
| 16 | ZINC426746041 | −10.3 | −45.4 ± 3.6 | −5.1 ± 3.1 | −41.3 ± 2.6 | −91.8 ± 5.3 | 1.2 ± 0.1 | 1.4 ± 0.3 |
| 17 | ZINC113844870 | −10.2 | −26.4 ± 6.9 | 2.1 ± 4.1 | −8.0 ± 3.6 | −32.4 ± 11.1 | 1.5 ± 0.2 | 11.4 ± 2.6 |
| 18 | ZINC000001793 | −10.2 | −30.2 ± 2.1 | −4.6 ± 2.8 | −17.4 ± 1.9 | −52.2 ± 4.0 | 1.3 ± 0.1 | 0.9 ± 0.2 |
| 19 | ZINC009504042 | −10.2 | −37.7 ± 4.3 | −2.2 ± 3.9 | −29.7 ± 3.4 | −69.7 ± 7.4 | 2.4 ± 1.6 | 7.9 ± 0.6 |
| 20 | ZINC005615258 | −9.9 | −41.0 ± 4.0 | −8.9 ± 8.0 | −18.9 ± 2.9 | −68.9 ± 10.9 | 1.7 ± 0.1 | 1.7 ± 0.3 |
2Top 8 compounds are represented in bold font.
Calculated from the snapshots from the last 10 ns simulation.
Table 3.
Protein-Ligand contacts during MD simulations for the top eight compounds and the reference ligand.
| REF COMP | ZINC096223736 | ZINC426746041 | ZINC079784201 | ZINC014116837 | ZINC096232566 | ZINC121003678 | ZINC217844024 | ZINC082673 |
|---|---|---|---|---|---|---|---|---|
| A21 | A21 | A21 | A21 | |||||
| D22 | D22 | D22 | D22 | D22 | D22 | |||
| I23 | I23 | I23 | I23 | I23 | I23 | I23 | I23 | I23 |
| V24 | V24 | V24 | ||||||
| A38 | A38 | A38 | A38 | A38 | A38 | A38 | A38 | |
| G48 | G48 | G48 | G48 | |||||
| V49 | V49 | V49 | V49 | V49 | V49 | V49 | V49 | V49 |
| A52 | A52 | A52 | A52 | A52 | A52 | A52 | ||
| K55 | ||||||||
| A124 | ||||||||
| P125 | P125 | P125 | P125 | P125 | P125 | P125 | ||
| L126 | L126 | L126 | L126 | L126 | L126 | L126 | L126 | L126 |
| A129 | A129 | A129 | A129 | A129 | A129 | A129 | A129 | A129 |
| GI130 | G130 | G130 | ||||||
| I131 | I131 | I131 | I131 | I131 | I131 | I131 | I131 | I131 |
| F132 | F132 | F132 | F132 | F132 | F132 | F132 | ||
| P136 | P136 | |||||||
| A154 | A154 | A154 | A154 | A154 | A154 | A154 | A154 | A154 |
| V155 | V155 | V155 | V155 | V155 | V155 | V155 | ||
| F156 | F156 | F156 | F156 | F156 | F156 | F156 | F156 | F156 |
| D157 | D157 | D157 | D157 | D157 | D157 | |||
| L160 | L160 | L160 | L160 | L160 | L160 | L160 | L160 | |
| L164 |
*Reference Compound is the Crystal Ligand in the PDB ID: 6W6Y.
3.1. ADMET properties show good human oral bioavailability
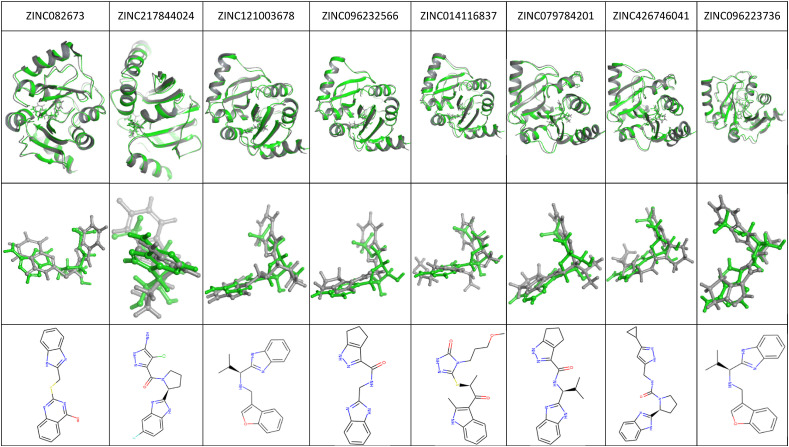
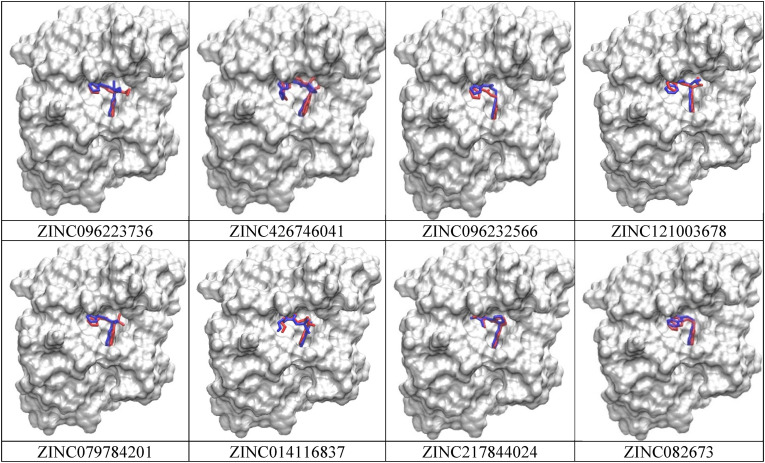
The predicted ADMET properties for the eight compounds show that there is high intestinal absorption with only one of them showing chances to distribute into the brain. However, some of the compounds do inhibit the cytochrome P450 enzymes (CYPs) including CYP1A2, CYP2C19, CYP2C9, CYP2D6, and CYP3A4 inhibitors, which indicated that these compounds could be metabolized. CYP3A4 possesses the highest activity in the small intestine and liver and metabolizes 50% of the medicines [26]. The CYP inhibitors also indicate an increase in the plasma concentration of the drug. BBB permeability protects the exposure of molecules that are toxic to the neurons in the brain. All eight compounds fulfill the conditions of drug-likeness properties without violation of Lipinski rule of five including MW < 500, calculated octanol-water partition coefficient (LogP) ≤ 5, some hydrogen bonding acceptors ≤10, as well as several hydrogen bonding donors ≤5. The PAINS (pan-assay interference compounds) alert system also gave off zero alerts to all eight compounds, which indicates a low chance of false positives from occurring (Table 4 ). The docked complex of the top eight ligands compared with the crystal complex structure is performed to view the closest interaction-based measures and pose predictions (Fig. 3 ). The top eight ligands show a good binding pose with the crystal structure, further validating the top eight hits.
Table 4.
The predicted drug properties for top 8 best compounds and the crystal ligand by the SwissSimilarity server.
| Compound | GI absorption | BBB permeant | CYP1A2 | CYP2C19 | CYP2C9 | CYP2D6 | CYP3A4 | Lipinksi rule | PAINS | Brenk |
|---|---|---|---|---|---|---|---|---|---|---|
| Crystal Ligand (PDB ID:6W6Y) | Low | No | No | No | No | No | No | Yes; 1 violation: NorO>10 | 0 alert | 1 alert: phosphor |
| ZINC000096223736 | High | No | Yes | Yes | Yes | Yes | Yes | 0 violation | 0 alert | 0 alert |
| ZINC000426746041 | High | No | Yes | Yes | No | yes | Yes | 0 violation | 0 alert | 0 alert |
| ZINC000079784201 | High | No | Yes | Yes | No | Yes | Yes | 0 violation | 0 alert | 0 alert |
| ZINC000014116837 | High | No | No | Yes | Yes | No | Yes | 0 violation | 0 alert | 0 alert |
| ZINC000096232566 | High | No | Yes | No | No | Yes | Yes | 0 violation | 0 alert | 0 alert |
| ZINC000121003678 | High | Yes | Yes | Yes | Yes | Yes | Yes | 0 violation | 0 alert | 0 alert |
| ZINC000217844024 | High | No | Yes | No | No | No | No | 0 violation | 0 alert | 0 alert |
| ZINC082673 | High | No | Yes | No | Yes | Yes | Yes | 0 violation | 0 alert | 0 alert |
Fig. 3.
Comparison between the docked complex (gray) of the top eight ligands and the crystal complex structure (green). The last column represents the ligand chemical structure.
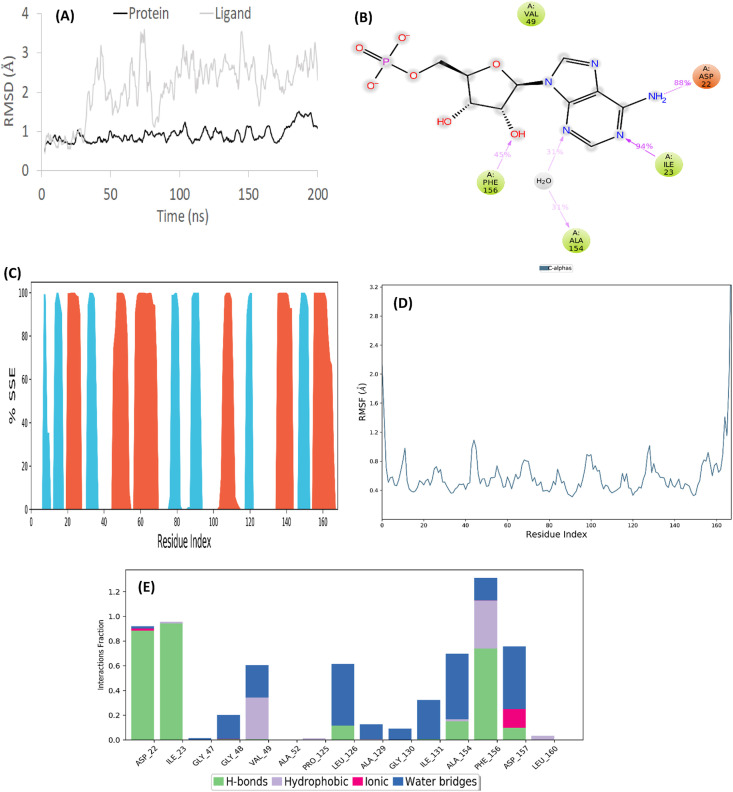
Crystal complex structure was stable in the MD simulation. The simulation interaction diagram of the crystal structure shows the RMSD plot obtained from the MD simulation. Monitoring the RMSD of the protein gives insight into the structural conformation obtained throughout the simulation. The RMSD plot has equilibrated and fluctuated towards the end of the simulation, it is acceptable for the crystal structure is a globular protein (Fig. 4 A). A ligand atom interaction with protein residues from the MD trajectory provides insight on four different hydrophobic interactions such as ASP32, PHE156, ALA154, and ILE23, which are the four residues interacting with the ligand atom (Fig. 4B). The protein SSE plot shows the recurring arrangements of close amino acids through distribution by residue position throughout the protein structure. The residue indicated 32.40% of the helix and 19.68% of the strand, which made the total percentages of SSE of the residue position to be 52.08%, with the rest of the area being random coil (Fig. 4C). The peaks in the RMSF plot analyses the portion of the protein that fluctuates the most during the simulation. The tails (N- and C-terminal) fluctuates more than any other parts of the protein (Fig. 4D). The interaction fraction plot indicates the ligand-protein interaction throughout the simulation. The four residues with the 2D ligand-protein interaction in Fig. 4B, show a greater amount of hydrogen bonds in the plot (Fig. 4E) during the simulation.
Fig. 4.
Detailed representation and various properties of simulation interaction diagrams after MD simulation of the crystal structure (PDB ID: 6W6Y). (A) RMSD plot from MD simulation of 200 ns. (B) 2D ligand-protein interaction diagram from the MD trajectory. The residues displayed interacted with ligand for at least 30% of the simulation time. (C) Protein Secondary Structure elements (SSE). Red represents alpha helices, blue represents beta strands, and the white places represent random coil. (D) RMSF graph of protein of the crystal structure docked complexes. (E) Protein-Ligand contacts during MD simulations. Interaction fraction greater than 1 is because of multiple contacts on one residue.
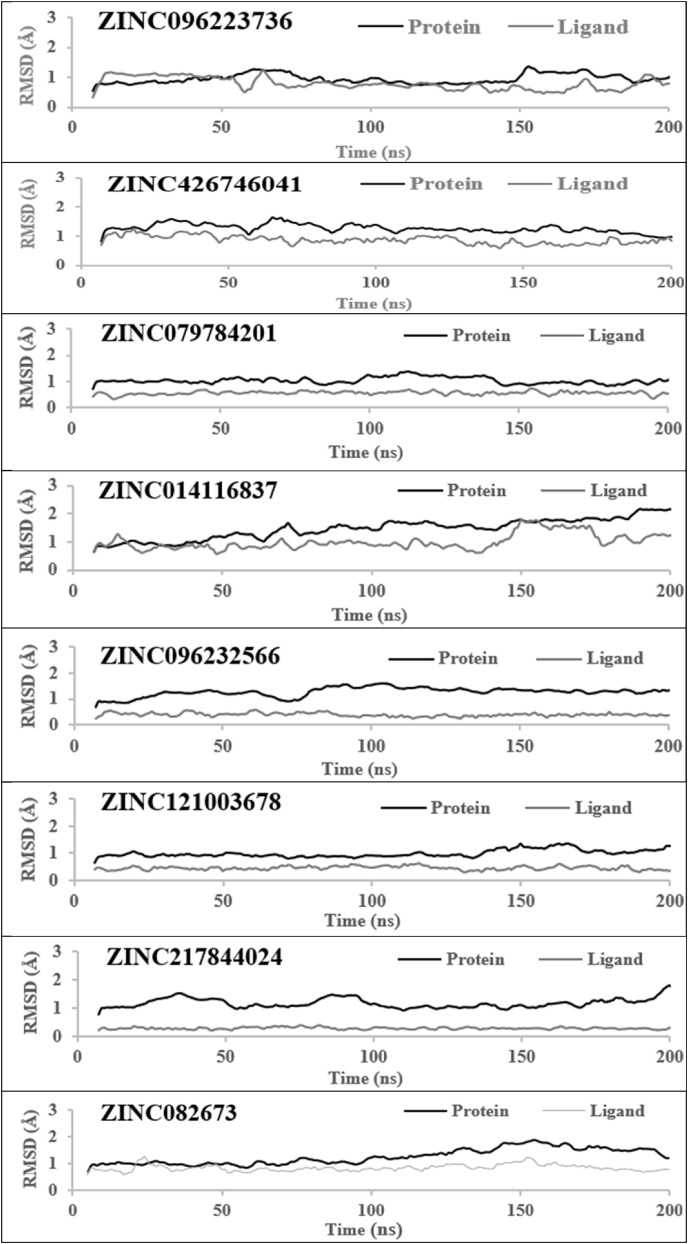
RMSD analysis of protein and ligand is stabilized. After the screening of all drugs, the best hits were subjected to MD simulation to better understand their behavior. RMSD is used to measure the crystallographic binding pose. Structural fluctuation patterns during 200ns MD simulation are described in terms of C RMSD of the top eight compounds. The structures are compared by the binding interaction and the energy between protein and ligand (Fig. 5 ). In this case, a low RMSD score is acceptable. Protein RMSD gives insight into its structural conformation throughout the simulation, whereas ligand RMSD indicates how stable the ligand is concerning the protein and its binding pocket. Most changes are shown in between 1 and 3 Å which is perfectly acceptable for a small protein like these. The ligand RMSD values stayed within the RMSD of the protein range, which indicates that the ligand has not diffused away from its initial binding site. The average RMSD value of the compound (ZINC014116837) was found to be relatively high. The protein-ligand convergence of the compound (ZINC096223736) was observed between 50 and 150ns. It was observed to be at 1 Å. The convergence was observed between 0 and 50ns in the compound (ZINC082673). All eight compounds showed minor fluctuations throughout the graphs. These results show that the lower the RMSD fluctuates, the better the model is in comparison to the target structure.
Fig. 5.
C RMSD of the top eight zinc compound ligands during 200 ns MD simulation.
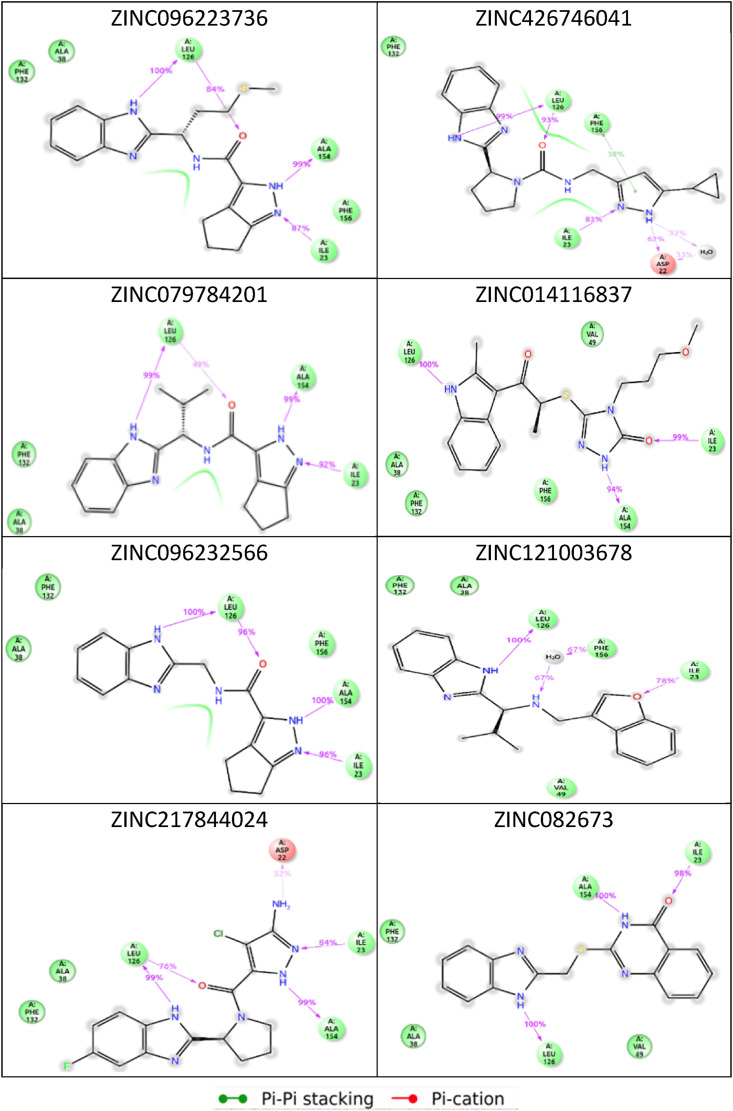
MD simulation shows improvement in the binding pose of the top eight ligands. The trajectories of the receptor-binding domain (RBD) of the top eight ligands were analyzed by comparing the ligand XP docking binding pose before and after MD simulation (Fig. 6 ). MD simulation is utilized to find the most dissimilar conformational changes. During the simulation, a ligand may significantly change from the originally bound conformation to optimize the overall interactions with the receptor. Rotatable bonds in the ligand may lead to high RMSD concerning initial bound conformation. The compound ZINC096223736 shows a stronger binding pose before and after the simulation followed by ZINC426746041 and ZINC079784201 also according to its MM-GBSA score shown in (Table 2). The 2D interaction of the ligand atom with protein residues from the MD trajectory of the top eight ligands with different active site amino acids is shown (Fig. 7 ). The results show that the selected hits give a good binding affinity towards the active site of the protein.
Fig. 6.
Comparison of ligand XP docking binding pose before (Red) and after (Blue) MD simulation for the top eight ligands.
Fig. 7.
2D ligand interaction diagrams from the MD trajectory for the top eight compounds. Residues displayed interacted with ligand for at least 30% of the simulation time.
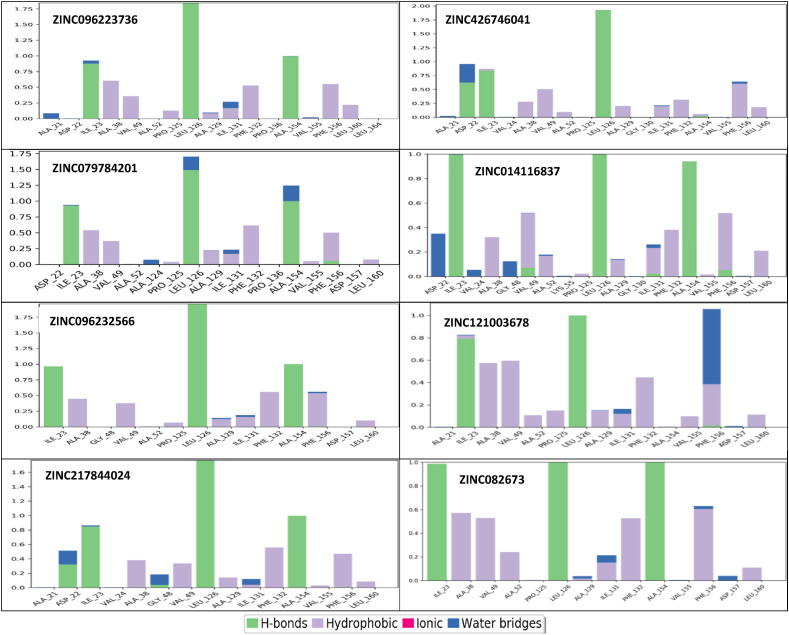
Protein-ligand interaction analysis reveals novel residue interactions in all eight compounds. Protein interactions with the ligand were monitored throughout the MD simulation. Protein-ligand interactions can be categorized into four different types as shown (Fig. 8 ). The highest amount of hydrogen bonding was observed on ZINC082673, ZINC014116837, and ZINC217844024. Leu126, ALA154, and ILE23 are the main residues containing the hydrogen bonds in all eight compounds. It can lead to a strong influence on drug specificity, metabolization, and absorption. Hydrophobic interaction generally involves a hydrophobic amino acid and an aromatic or aliphatic group on the ligand. ZINC082673, ZINC121003678, and ZINC014116837 are observed to have the highest number of hydrophobic contacts. No ionic or polar interactions were observed in the eight compound graphs. Water bridges were observed in every plot where there was a hydrogen-bonded protein-ligand interaction mediated by a water molecule. The last 50ns of each 200ns simulation show little deviation, indicating convergence (Fig. S5). All compounds showed higher hydrogen-bonding and hydrophobic interactions compared to the crystal structure, which leads to higher GI absorption of the compounds shown (Table 4).
Fig. 8.
Protein-Ligand contacts during MD simulations for the top eight compounds. Interaction fraction greater than 1 is possible because of multiple contacts on one residue.
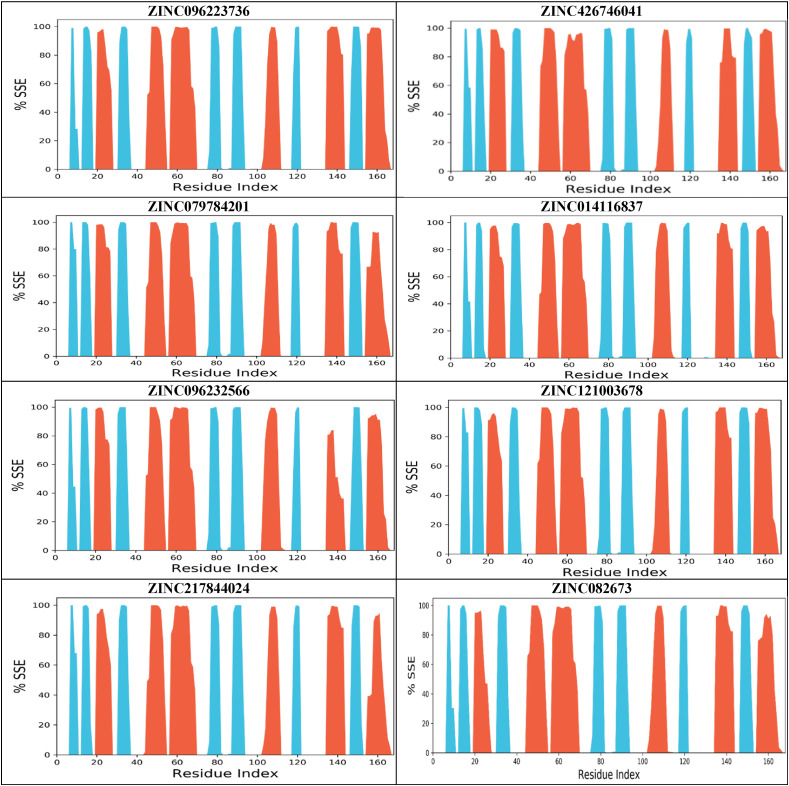
The secondary structure examination portrays minor differences. Protein SSE was monitored throughout the MD simulation. The plots summarize the SSE distribution by residue position throughout the protein structure. It is categorized into Alpha-helices, Beta-strands, and random coils (Fig. 9 ). Alpha-helices mainly have hydrophobic residues which are found in the core of the protein or are transmembrane proteins. On the other hand, beta-strands contain patterns of hydrophobic and polar amino acids. The random coil is a polymer conformation, where it is not one specific shape but instead a statistical distribution of all chains indicated by the white spaces in the plots. SSE is more rigid than the unstructured part of the protein, therefore they fluctuate less than the loop regions.
Fig. 9.
Protein Secondary Structure elements (SSE) for the top eight compounds during MD simulation. Red represents alpha helices, blue represents beta sheets, and the white places represent random coil.
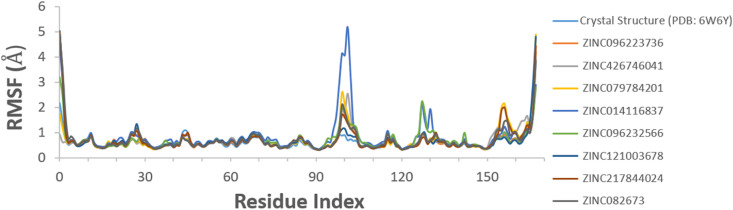
The RMSF shows fluctuation in localized regions of the protein and ligand. The flexibility of each compound was acquired by using RMSF. Protein C RMSF characterizes the local changes along the protein backbone during simulation (Fig. 10 ). It measures the deviation between the particle position and the reference position. The peaks indicate areas of protein that fluctuate the most during the simulation. The N- and C- terminal tails show more fluctuation than the other parts of the protein, due to the higher amount of proteins binding with the residues. ZINC014116837 showed the greatest fluctuation nearly at 5 Å around residue 100 positions. All the other compounds fluctuated around the same residue position nearly from 1 to 3 Å, which is also reflected in trajectory clustering analysis. Another small fluctuation of ZINC014116837 and ZINC096232566 was observed at residue position 130. Lastly, a fluctuation of ZINC217844024 and ZINC079784201 took place towards the end at the residue position 155. Using the crystal structure as a positive control, the complexes showed much higher residual fluctuation at different positions throughout the graph.
Fig. 10.
Protein C RMSF for the top eight compounds including the crystal structure during MD simulation.
4. Discussion
The cluster of pneumonia caused by SARS-CoV-2 created great challenges in public health all around the world. This pandemic has not only challenged people in terms of their health, but also in terms of the economy. To subsist with the virus, researchers and scientists are focusing on different aspects of the infections caused by it. The SARS-CoV-2 genome consists of 16 Nsps and 4 structural proteins. There are many studies that targeted domains like PLpro, MTase, NendoU, RdRp protein, and 3CLpro while others focused more on the host proteins of the SARS-CoV-2 genome. Out of the 16 Nsps, Nsp3 plays a major role in transcription and translation by seizing the host immune system. Nsp3 is the largest protein encoded by SARS-CoV-2, it binds to viral RNA as well as other viral proteins.
The Nsp3 is reported to interact with Nsp2 which leads to intervention at an earlier or later stage of viral replication. It has been stated that the macrodomain of Nsp3 enzyme activity plays an essential role in pathogenesis [27]. The 16 domains (∼Res: 1922) and regions of Nsp3 play an important role in transcription and translation, which interacts with the protein host by taking over the host immune system. There is an essential process required to bind ADRP to this domain. Targeting this domain with high receptor binding inhibitors could help to reduce the viral implications caused by the pandemic. Therefore, using different drug targeting approaches like computer-aided drug design or rational drug design could help find a drug to target this domain and redeem the host immune system.
The emergency use of the FDA-approved drug Remdesivir is not as effective due to its associated problems, therefore its use is very limited. Therefore, there is still a need to either refine older drugs or search for new ones to treat SARS-CoV-2. In this regard, the use of structure-based high throughput virtual screening methods is the most useful way to find new drugs for COVID-19 based on their properties. In the present study, we used the virtual screening approach to discover the potential inhibitors to target ADP ribose phosphatase. Based on our findings using the bioinformatics tools, along with a drug similarity search, we analyzed compounds from Zinc15 and targeted novel inhibitors of ADP-Ribose phosphatase of SARS-CoV-2 using the virtual screening workflow which gave us a promising top twenty hits. The top twenty compounds were then further validated and shortlisted to eight of the inhibitors, which were further validated by the MM-GBSA score of binding free energy and MD simulation. The MM-GBSA score helped to specify the binding interaction between ligand-protein receptors.
The protein-ligand interaction further confirmed the top potential inhibitors. The large dataset and extended high throughput virtual screening method portray the best interactions between ligands of a molecular target to form a complex. The adverse effects and related articles of the selected compounds were checked through CAS Scifinder and PubChem, where the compounds showed no adverse effect. Moreover, the ZINC082673 compound is found to be useful in treatments of bacterial infection and the NadD inhibition leads to suppression of bacterial growth. NadD synthetase uses energy from ATP and is widely used as a drug target in various microorganisms. These results further validate the top hits to be the potential inhibitors.
Based on the results, the potential binding and inhibiting effects of the top eight compounds are indicated. The use of the structure-based high throughput virtual screening method is the most useful approach to find the potential molecules that could target the macrodomain. After examining 17 million Zinc15 compounds using structure-based high throughput virtual screening methods, the most potential hits were validated by MD simulation. This study has the potential to assist and repurpose the drug design. In vitro experiments can be carried out as this study can facilitate the global efforts in the speedy development of potential drug candidates against SARS-CoV-2.
5. Conclusion
This study may assist in further investigating and testing the novel inhibitors of SARS-CoV-2. We performed a thorough investigation of a total of 17 million Zinc15 compounds to examine using a structure-based high throughput virtual screening method which provides the most potential hits. The molecular dynamic simulation further validated the top hits. In this study, twenty potential compounds were selected from which eight potential inhibitors of SARS-CoV-2 showed commendable docking scores ranging from −9.9 to −11.7 kcal/mol. The computational pipeline gives us the best top eight hits, which exhibit good binding affinity towards the active site. Based on the XP glide docking, the binding affinity, and ADMET properties, the top eight Zinc compounds are the potential inhibitors of ADP-ribose phosphatase of Nsp3 of SARS-CoV-2. This paper could provide great knowledge about the potential inhibitors to target SARS-CoV-2.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
C.W acknowledges the support by the New Jersey Health Foundation (PC 76–21) and the National Science Foundation under Grants NSF ACI-1429467/RUI-1904797, and XSEDE MCB 170088. The Anton2 machine at the Pittsburgh Supercomputing Center (PSCA17017P) was generously made available by D. E. Shaw Research. MA acknowledges WCP and Penelitian Dasar Grants, Ministry of Education, Culture, Research, and Technology, Republic Indonesia.
Footnotes
Supplementary data to this article can be found online at doi:mmcdoino.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.Naqvi A.A., et al. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866(10) doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michalska K., et al. Crystal structures of SARS-CoV-2 ADP-ribose phosnhatase: from the apo form to ligand complexes. IUCrJ. 2020;7:814–824. doi: 10.1107/S2052252520009653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egloff M.P., et al. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 2006;80(17):8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmad Z. ATP synthase: a potential molecular drug target. Faseb. J. 2019;33:470–471. [Google Scholar]
- 5.Fehr A.R., et al. Viral macrodomains: unique mediators of viral replication and pathogenesis. Trends Microbiol. 2018;26(7):598–610. doi: 10.1016/j.tim.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yanai H., et al. Revisiting the role of IRF3 in inflammation and immunity by conditional and specifically targeted gene ablation in mice. 2018;115(20):5253–5258. doi: 10.1073/pnas.1803936115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., et al. A comparative overview of COVID-19, MERS and SARS: review article. Int. J. Surg. 2020;81:1–8. doi: 10.1016/j.ijsu.2020.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jian Lei Y.K., Rolf Hilgenfelda. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunn J.G.M. StatPearls Publishing; 2021. Physiology, Adenosine Triphosphate. [PubMed] [Google Scholar]
- 10.Debnath P., et al. In silico identification of potential inhibitors of ADP-ribose phosphatase of SARS-CoV-2 nsP3 by combining E-pharmacophore and receptor-based virtual screening of database. Chemistry. 2020;5(30):9388–9398. doi: 10.1002/slct.202001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sastry G.M., et al. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 12.Harder E., et al. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theor. Comput. 2016;12(1):281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 13.Schrödinger L. Schrödinger, LLC; New York, NY: 2015. QikProp, Version 4.4. [Google Scholar]
- 14.Friesner R.A., et al. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 15.Friesner R.A., et al. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006;49(21):6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 16.Wood D.J., et al. Pharmacophore fingerprint-based approach to binding site subpocket similarity and its application to bioisostere replacement. J. Chem. Inf. Model. 2012;52(8):2031–2043. doi: 10.1021/ci3000776. [DOI] [PubMed] [Google Scholar]
- 17.Zolfaghari F., et al. Hierarchical cluster analysis to identify the homogeneous desertification management units. PLoS One. 2019;14(12) doi: 10.1371/journal.pone.0226355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leal W., et al. How frequently do clusters occur in hierarchical clustering analysis? A graph theoretical approach to studying ties in proximity. J. Cheminf. 2016;8(1):4. doi: 10.1186/s13321-016-0114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. 2001;105(43):9954–9960. [Google Scholar]
- 20.Kumar V., Liu H., Wu C. Drug repurposing against SARS-CoV-2 receptor binding domain using ensemble-based virtual screening and molecular dynamics simulations. Comput. Biol. Med. 2021;135 doi: 10.1016/j.compbiomed.2021.104634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kevin J., Bowers E.C., Xu Huafeng, Dror Ron O., Eastwood Michael P., Gregersen Brent A., K J.L., Kolossvary Istvan, Moraes Mark A., Sacerdoti Federico D., Salmon John K., S Y., Shaw David E. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters; p. 84. [Google Scholar]
- 22.Ghosh A., Rapp C.S., Friesner R.A. Generalized born model based on a surface integral formulation. J. Phys. Chem. B. 1998;102(52):10983–10990. [Google Scholar]
- 23.Yu Z.Y.J., P M., Friesner R.A. What role do surfaces play in GB models? A new-generation of surface-generalized Born model based on a novel Gaussian surface for biomolecules. J. Comput. Chem. 2006;27:72–89. doi: 10.1002/jcc.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J.N., et al. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins-Structure Function and Bioinformatics. 2011;79(10):2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zoete V., et al. SwissSimilarity: a web tool for low to ultra high throughput ligand-based virtual screening. J. Chem. Inf. Model. 2016;56(8):1399–1404. doi: 10.1021/acs.jcim.6b00174. [DOI] [PubMed] [Google Scholar]
- 26.Bibi Z. Role of cytochrome P450 in drug interactions (Retraction of vol 5, 27, 2008) Nutr. Metabol. 2014;11:27. doi: 10.1186/1743-7075-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Frick D.N., et al. Molecular basis for ADP-ribose binding to the Mac1 domain of SARS-CoV-2 nsp3. Biochemistry. 2020;59(28):2608–2615. doi: 10.1021/acs.biochem.0c00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.