Abstract
HLA-G is a promiscuous immune checkpoint molecule. The HLA-G gene presents substantial nucleotide variability in its regulatory regions. However, it encodes a limited number of proteins compared to classical HLA class I genes. We characterized the HLA-G genetic variability in 4640 individuals from 88 different population samples across the globe by using a state-of-the-art method to characterize polymorphisms and haplotypes from high-coverage next-generation sequencing data. We also provide insights regarding the HLA-G genetic diversity and a resource for future studies evaluating HLA-G polymorphisms in different populations and association studies. Despite the great haplotype variability, we demonstrated that: (1) most of the HLA-G polymorphisms are in introns and regulatory sequences, and these are the sites with evidence of balancing selection, (2) linkage disequilibrium is high throughout the gene, extending up to HLA-A, (3) there are few proteins frequently observed in worldwide populations, with lack of variation in residues associated with major HLA-G biological properties (dimer formation, interaction with leukocyte receptors). These observations corroborate the role of HLA-G as an immune checkpoint molecule rather than as an antigen-presenting molecule. Understanding HLA-G variability across populations is relevant for disease association and functional studies.
Subject terms: Bioinformatics, Genotype, Haplotypes, Immunogenetics, Population genetics, Immunogenetics
Introduction
The human leukocyte antigen G (HLA-G) belongs to the family of non-classical HLA class I molecules, first identified at the maternal–fetal interface, especially on the placenta’s cytotrophoblast cells, where immunotolerance contributes to the fetus maintenance. The HLA-G molecule is considered a promiscuous immune checkpoint molecule, inhibiting antigen-presenting cells, T, B, and NK lymphocytes, through the interaction with several leukocyte receptors, including ILT2, ILT4, CD160, KIR2DL4, and CD8 receptors1. ILT2 and ILT4 also interact with some classical class I HLA molecules but have more affinity for HLA-G2, and KIR2DL4 is a specific receptor for HLA-G3,4. HLA-G ligation with these receptors modulates T cytotoxic and NK cell activity, inducing the expression of inhibitory receptors containing Tyrosine motifs5.
HLA-G deserves special attention in pregnancy because HLA-G plays a role in immunosuppression and homeostasis maintenance during pregnancy6. Therefore, the non-expression of classical HLA class I molecules by the trophoblastic cells make them potential targets for the lysis mediated by NK cells and the interaction of the HLA-G molecule (trophoblast) with uterine NK inhibitor receptors hampers the trophoblast cell lysis7. HLA-G also activates the production of cytokines that promote remodeling of the vascularization at the maternal–fetal region, of major importance for oxygen supply to the fetus8–10. HLA-G deregulation is also important in pathological situations such as tumors and autoimmune diseases. HLA-G polymorphisms and expression levels have been associated with susceptibility to infections, tumors, and autoimmune diseases11–16.
At least seven HLA-G isoforms have been described by alternative splicing of the primary transcript, encoding four membrane-bound (G1-G4) and three soluble (G5-G7) isoforms, and their presence in tissue cells may be benefic or pernicious, depending on the underlying condition15,17. However, rare isoforms have been detected in tumor samples18.
The HLA-G coding region presents similarities to the classical HLA class I genes. While most HLA genes are extremely polymorphic, HLA-G is highly conserved in its coding nucleotide sequence, presenting a much lower number of different haplotypes, defined mainly by polymorphisms that span around putative regulatory regions and introns. In the IPD-IMGT/HLA database19, release 3.45.1 from July 2021, HLA-G exhibited 88 alleles (different sequences), encoding 26 allotypes, but few are frequent, as demonstrated later in this study. The HLA-G coding region is relatively conserved, exhibiting few functional polymorphisms (e.g., non-synonymous variants); however, the HLA-G regulatory regions are more variable, displaying target sites for many transcriptional and post-transcriptional regulatory elements20. Thus, some polymorphic sites at these segments have been associated with different HLA-G expression profiles.
While the HLA-G gene regulatory region variability may impact the magnitude of gene/protein expression, the coding region variability may impact the HLA-G biological properties. Differential transcriptional and post-transcriptional HLA-G activities have been associated with nucleotide variability at the promoter and 3’ untranslated regions21–24. Additional sites potentially involved in HLA-G expression have also been reported for the HLA-G distal-promoter (located between − 2635 and − 1406 positions) and the proximal-promoter (located between − 1406 and − 1)25. Several regulatory elements can induce HLA-G expression, including progesterone responsive elements (PRE), heat shock element (HRE), interferon-sensitive response element (ISRE), while others may repress protein expression (RREB1—Ras-responsive element-binding protein 1; GLI3—Glioma Associated Oncogene Homologue 3)17. Besides the distal and proximal promoters, a 121-bp region 12 Kb upstream the HLA-G gene (Enhancer L) is reported to be a target for transcription factors (CEBP and GATA) associated with the placentation process26.
A previous initiative addressing HLA-G diversity in worldwide samples was reported27; however, this survey used a different sequencing technology and distinct genotyping and haplotyping methods; i.e., it was based on low-coverage data from the 1000 Genomes consortium phase I, which included a much lower number of samples27. Other studies addressed specific samples from Brazil, Cyprus, and Benin, and characterized the full gene sequence and the proximal promoter but disregarded the distal promoter region28–30. Many studies addressed HLA-G polymorphisms using Sanger sequencing in association studies, usually evaluating only a segment of the gene or specific exons (e.g.31–35, and many others).
This study explored the HLA-G genetic diversity and the evolutionary aspects in worldwide population samples by using a state-of-the-art method to characterize polymorphisms and haplotypes from high-coverage next-generation sequencing (NGS) data. We also provide the most comprehensive survey of the HLA-G genetic diversity across worldwide populations and resources for future analyses evaluating HLA-G polymorphisms in terms of disease association and functional studies.
Samples and methods used to characterize HLA-G genetic diversity
We evaluated 4738 individuals from 88 different population samples from four datasets, all of them sequenced in high-coverage (> 30×) and processed through the same computational pipeline. The first dataset was the new high-coverage sequencing data of the 1000 Genomes consortium36, the second consisted of 831 samples from the International Genome Sample Resource (IGSR) consortium37, the third encompassed 1323 elderly Brazilians from the Saúde, Bem-estar e Envelhecimento (SABE) cohort38, and the fourth comprised the high coverage sequencing of 80 individuals from Southeast Asia and Oceania, including the Agta hunter-gatherer people from the Northern Luzon island, Philippines. The population samples and their sample sizes are in Table S1. In all cases, we started from a genome-wide BAM file with reads aligned to the reference hg38 genome (version hg38DH) by using BWA MEM (https://sourceforge.net/projects/bio-bwa/).
The method used to extract the HLA-G data is described in detail in a file (the supplementary methods) with a description of all methods and variables we used. This method also follows the same strategy used to characterize HLA class I genes in Brazilian samples38. Using a single pipeline to extract the data from all samples is essential to avoid biases in the genotyping and haplotyping procedures. In brief, we used the hla-mapper version 4.0 to optimize read alignment in the HLA complex, minimizing alignment errors and cross-alignments that are commonly observed among HLA genes39. We have called genotypes using GATK HaplotypeCaller in the GVCF mode (https://gatk.broadinstitute.org) with a further refinement step using vcfx (www.castelli-lab.net/apps/vcfx). This method allows the detection of any previously unknown HLA-G variant. The haplotype inference used both read-aware phasing (GATK ReadBackedPhasing) and probabilistic models with the workflow phasex/shapeit4 (www.castelli-lab.net/apps/phasex). Please refer to the file (supplementary methods) for details regarding the mapping, genotyping, and haplotyping procedures. Applying the bioinformatic pipeline, 4640 samples (97.9%) passed the quality control and the haplotyping procedure, as presented in Table S1.
To support the quality of the data presented here, we evaluated departures of the Hardy–Weinberg expectations (HWE) at the SNP level in population samples with at least 10 individuals (63 populations out of 88). Of those, 55 (87.3%) present no HWE deviation, and 8 populations present just two deviations. Thus, most SNPs fit HWE.
Resources used to evaluate the HLA diversity
The method described above allowed us to retrieve SNP and haplotype data from approximately 7870 nucleotides surrounding the HLA-G gene, from 4328 bases upstream of the first translated ATG to 100 nucleotides downstream HLA-G. This large region includes most of the regulatory elements, all exons and all introns. We extracted coding (exons + introns), exonic (the CDS), promoter, and 3’UTR sequences. We also translated CDS sequences into proteins (the allotypes).
In the following sections, we will discuss the HLA-G polymorphisms across the world, providing insights regarding the HLA-G genetic diversity, and present a resource for future studies evaluating HLA-G polymorphisms and disease association studies in different populations. We also provide several useful tools to evaluate HLA-G diversity (Table 1) that can be downloaded from the website www.castelli-lab.net/HLA-G or are included as supplementary material.
Table 1.
Resources available for download or listed as supplementary material regarding the HLA-G gene and its polymorphisms.
| Resource | Description | Availability |
|---|---|---|
| VCF listing all variants | A VCF file containing all variants, the reference and alternative alleles, and global counts, to be used as support for variant refinement when genotyping HLA-G in NGS procedures or variant annotation | For downloada |
| Phased VCF | A VCF file with the phased genotypes for each sample | Upon request |
| FASTA file with full sequences | A copy of every full-length sequence observed in this survey (approximately 12 Kb each), and their global counts | For downloada |
| FASTA file with gene sequences | The full sequence of each HLA-G allele (exons + introns) observed in this survey, with their global counts. Each sequence is identified with its official name according to the IPD-IMGT/HLA database, or as a novel sequence | For downloada |
| FASTA file with CDS sequences | We extracted all different exonic sequences (CDS). This file contains a copy of each different sequence, with their global counts. Each sequence is identified with its official name according to the IPD-IMGT/HLA database, or as a novel sequence | For downloada |
| FASTA file with protein sequences | We translated all exonic sequences into proteins (allotypes). This file contains a copy of each different sequence, with their global counts. Each sequence is identified with its official name according to the IPD-IMGT/HLA database, or as a novel sequence | For downloada |
| FASTA file with 3’UTR sequences | The sequence of each 3’UTR haplotype we have detected, with their names and global counts | For downloada |
| SNP frequencies | This table provides the frequency for the reference and alternative alleles in the global population, in XLSX format | Supplementary material (Table S2) |
| Genomic alleles (4-digit resolution) frequencies | This table provides the frequency for each genomic allele (4-digit resolution) in each population we have studied, in XLSX format. There are 3 sheets, one for biogeographic regions, one for countries, and one for specific population samples | Supplementary material (Table S3) |
| Allotypes frequencies (2-digit resolution) | This table provides the frequency for each allotype (2-digit resolution, full-length protein) in each population we have studied, in XLSX format. There are 3 sheets, one for biogeographic regions, one for countries, and one for specific population samples | Supplementary material (Table S4) |
| 3’UTR frequencies | This table provides the frequency for 3’UTR haplotype in each population we have studied, in XLSX format. There are 4 sheets, one for biogeographic regions, one for countries, one for specific population samples, and one describing the combination between genomic alleles and 3’UTR haplotypes in the global sample | Supplementary material (Table S5) |
aAvailable for download in www.castelli-lab.net/HLA-G.
Nomenclature, SNPs, nucleotide diversity, and linkage disequilibrium across HLA-G
Due to the lack of a standardized nomenclature for the regulatory regions, individual regulatory region haplotypes have been previously designated as “distal/proximal promoter regions”, and “3’UTR haplotypes”, as proposed by our group29,40,41. Because the HLA-G region evaluated in this study includes an extensae region (from − 4328 upstream the first translated ATG to 100 nucleotides downstream the transcription end site), and there is no designated nomenclature to nominate the sequences we have characterized (for instance, when the sequence includes the complete promoter, all introns, and the complete 3’UTR sequence), we used the term GEMBIO_HLA-G_H1 (complete haplotype number 1, for HLA-G, available in the GeMBio laboratory Database) to designate this large gene sequence, which is available for download (Table 1). These names can be converted to whatever future nomenclature will be available and standardized. Whenever this complete nomenclature is used in this text, the previous names of the regulatory (Distal-G, PROMO-G, and 3’UTR) and coding region (as defined by the IPD-IMGT/HLA database) haplotypes are also mentioned.
We detected 442 different variants encompassing approximately 7870 nucleotides surrounding the HLA-G locus. The majority are bi-allelic single nucleotide exchanges, but there are some indels, multi-allelic variants and combinations of them. The list of each variant, with its reference and alternative alleles, and their global frequencies, is available in Table S2. The VCF containing the phased genotypes of every individual is available upon request.
These 442 variants are arranged into 514 different haplotypes or full-length sequences (Table 1) and some of these haplotypes are quite common. The two most frequent ones are: (1) GEMBIO_HLA-G_H1 presents a frequency of 21.6% and is associated with known haplotypes, such as the distal promoter Distal-010101a, the proximal promoter PROMO-G010101a, the coding allele G*01:01:01:01, and the 3’UTR haplotype known as UTR-01, and (2) GEMBIO_HLA-G_H2 presents a frequency of 10.8% and carries the distal promoter Distal-010102a, the proximal promoter PROMO-G010102a, the coding allele G*01:01:02:01, and UTR-0229,30. We observed 20 full-length haplotypes presenting global frequencies higher than 1%, exhibiting a summed frequency of 76.85% (from GEMBIO_HLA-G_1 to GEMBIO_HLA-G_20). Thus, despite the presence of many variants, there are few full-length frequent haplotypes, reflecting the high Linkage Disequilibrium (LD) along the HLA-G locus. In Fig. 1, we present the LD pattern along the HLA-G locus for bi-allelic variants with a minor allele frequency (MAF) > 0.1% and fitting HWE (106 variants).
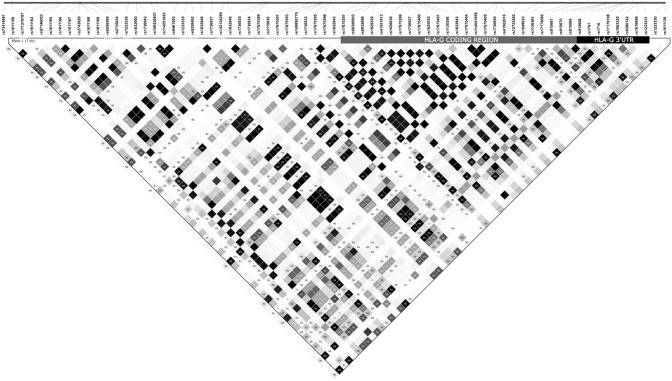
Figure 1.
Linkage disequilibrium between pairs of 106 bi-allelic SNPs at the HLA-G gene region, starting from 4 Kb upstream the gene up to 100 nucleotides downstream, all presenting a global minimum allele frequency of 1%. The image was generated in the Haploview and further edited by using Inkscape. Areas in black indicate strong LD (r2 > 0.8), shades of gray indicate moderate LD, and white indicates low LD.
The entire HLA-G gene is included within a single segregation block. Although Fig. 1 illustrates the global LD pattern, we observed similar patterns when considering only samples from Europe, Africa, Asia, and American. Most of the pairwise comparisons present D’ = 1 (Fig. 1 and S1), and many comparisons present r2 > 0.8. We demonstrate that this high LD extends at least 4 kb upstream HLA-G to 100 nucleotides downstream. Moreover, there are many variants in complete LD (r2 = 1.00). Of the 106 variants plotted in Fig. 1, 43 (40.6%) present another variant in complete LD. For instance, rs17875397 (position − 56) is in absolute LD with nine other variants, including rs17875400 (+ 507) and rs17875405 (+ 1534). There are also many variants in almost complete LD (measured by r2 > 0.95). For instance, considering the threshold r2 > 0.95, 55 selected SNPs can tag all 106 frequent variants along HLA-G. Because of that, LD among HLA-G variants must be considered when performing any association study to disregard hitchhiking effects. This high LD explains the presence of a few haplotypes that are frequent globally when analyzing almost 8 Kb surrounding HLA-G. This high LD was previously proposed evaluating other population samples29,40, and it may extend at least more 20 Kb downstream the gene, where an Alu insertion accompanies the G*01:01:01:01 allele42, or even extend up the HLA-A locus43.
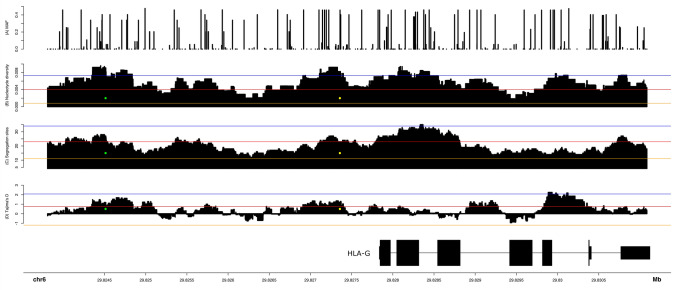
Figure 2 illustrates the frequency of each HLA-G variant (panel A), nucleotide diversity (panel B), SNP density (Panel C), and Tajima’s D (panel D) across the HLA-G region, in windows of 500 nucleotides and step size of 1, starting from 4 kb upstream HLA-G (the promoter region) to 200 nucleotides downstream it, considering all 1000Genomes samples pooled together. To evaluate the significance of the parameters estimated from HLA-G, we built a null distribution considering the patterns observed in chromosome 6. For that purpose, we computed these statistics in 10,000 random windows of 500 nucleotides from chromosome 6. The values above the blue and red horizontal lines are higher than 99.9% and 99% of the ones observed in the null distribution for chromosome 6, respectively. The orange line represents the average observed for each statistic. Similar patterns can be observed in all biogeographic regions. This analysis has revealed interesting insights. Nucleotide diversity across HLA-G is usually higher than 0.4% (the red line), which is much higher than the average observed from chromosome 6 (0.082%). The regions with the highest nucleotide diversity (around 1%), which are among the 0.1% highest observed in chromosome 6 (blue line), coincides with the promoter region around positions − 3614 (green dot, rs1611163) and − 725 (yellow dot, rs1233334). Another very polymorphic region coincides with intron 2. Likewise, the strongest positive Tajima’s D signals in the promoter region, which is compatible with balancing selection, coincide with these two markers. The region surrounding marker − 3614 (rs1611163) presents many frequent variants, including rs1611164, rs1611165, rs1611166, rs1611167, rs2743926, rs1611168, rs11169, in a short segment of 336 bp. Likewise, the region surrounding maker − 725 presents many frequent variants, including rs2249863 (− 716), rs2735022 (− 689), rs35674592 (− 666), rs17875391 (− 646), and rs1632944 (− 633), all previously described elsewhere40. The position − 725 is a tri-allelic variant, which has been described as influencing HLA-G expression levels possibly by epigenetic mechanisms44. Moreover, except for rs17875391, all these polymorphisms are listed as eQTLs for HLA-G expression by the GTEX portal (https://gtexportal.org). It is not clear whether these variants directly influence HLA-G expression or if they are in LD with other regulatory elements, such as enhancer L, located 12 Kb upstream HLA-G26. However, the strongest signal of balancing selection coincides with intron 5 (Fig. 2, panel D), with the Tajima’s D values higher than 99.9% of the ones observed for chromosome 6.
Figure 2.
Frequency of each variant (panel A), nucleotide diversity (panel B), number of segregation sites (panel C), and Tajima’s D (panel D) across HLA-G, considering all samples from the 1000Genomes project pooled together, starting from approximately 4 kb upstream the gene (the promoter region) to 100 nucleotides downstream HLA-G. Panels B, C, and D were computed in sliding windows of 500 nucleotides and a step size of 1. The HLA-G exon/intron structure is indicated in the bottom panel, with fine lines indicating introns and thick lines indicating exons. To evaluate the significance of the parameters estimated from HLA-G, we built a null distribution considering the patterns observed in chromosome 6, computing these statistics in 10,000 random windows of 500 nucleotides from chromosome 6. The values above the blue and red horizontal lines are higher than 99.9% and 99% of the ones observed for chromosome 6, respectively. The orange line represents the average observed in chromosome 6 for each statistic. The green and the yellow dots represent polymorphisms -3614 (rs1611163) and − 725 (rs1233334), respectively.
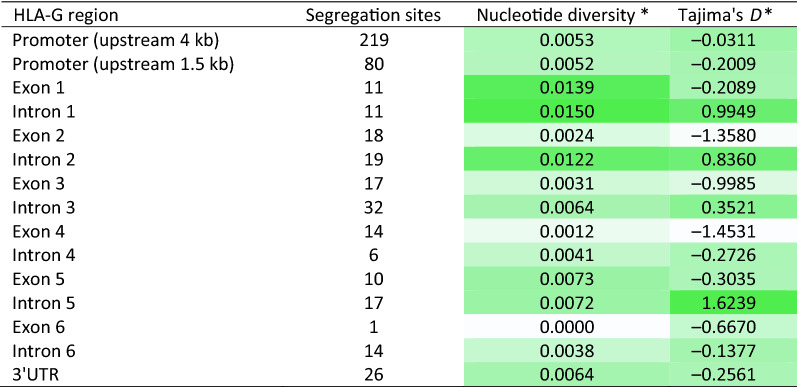
When we consider the transcribed HLA-G segment, illustrated in the bottom panel at Fig. 2, and the positions and frequencies of variants across HLA-G (panel A), we noticed that most frequent variants coincide with intronic regions and that exon 4 is quite conserved. Table 2 presents the number of segregation sites, nucleotide diversity, and Tajima’s D in each HLA-G exon and intron. Introns 1 and 2 present two of the highest nucleotide diversity indexes across HLA-G (Table 2, Fig. 2) and the highest number of segregation sites in the sliding window analysis (Fig. 2). Some of the intronic SNPs are also considered eQTLs for HLA-G expression, according to GTEX data (https://gtexportal.org). It is not clear whether these intronic variants directly influence HLA-G expression. However, as addressed throughout the following sections, these intronic variants (as haplotypes) are in almost complete LD with some promoter and 3’UTR haplotypes.
Table 2.
The number of segregation sites, nucleotide diversity, and Tajima’s D across different HLA-G regions in 4640 samples from 88 different populations.
*The indexes are scaled in shades of green, from white (the lowest value) to dark green (the highest value).
On the other hand, the nucleotide diversity drops to low levels in exons, except for exon 1, which is very short (Table 2) and presents two frequent synonymous variable sites. Additionally, the highest Tajima’ D coincides with introns, mostly intron 1, intron 2, and intron 5, all presenting positive Tajima’s D, which is compatible with balancing selection. In contrast, all exons present negative values, which is compatible with purifying selection (Table 2). Exon segments are conserved, and most of the exonic variants are synonymous mutations. Exon 2 presents only two frequent variants, one non-synonymous associated with the allotype G*01:03, and the other a synonymous mutation. Exon 3 presents four common variants, two synonymous mutations, one non-synonymous associated with allotype G*01:04, and one frameshift associated with allotype G*01:05. Exon 4 presents only three frequent variants, and one configures a non-synonymous mutation associated with allotype G*01:06. Other rare variants in exons are usually synonymous mutations.
We also calculated the ratio of substitution rates at non-synonymous and synonymous sites in exons (Table 3). Interestingly, none of the exons, when isolated, presented evidence of purifying selection. However, when pooled together, there is evidence of purifying selection at exons 2, 3, and 4 (Table 3). This phenomenon was observed in a previous study that evaluated a reduced number of sequences45. Taken together, the nucleotide diversity and Tajima’s D patterns (Fig. 2 and Table 2), and the ratio of substitutions (Table 3), made it clear that HLA-G is indeed conserved in exons, although not necessarily conserved in introns and regulatory sequences. Nevertheless, nucleotide diversity for introns and the regulatory sequences are much higher than the human average (0.075%)46.
Table 3.
Synonymous and nonsynonymous nucleotide substitution test of neutrality, positive, and purifying selection for analysis over 77 HLA-G sequences defined by exon mutations, in 4640 samples from 88 different populations.
| dN/dS neutrality test | Number of sequences | Number of codons | HA = neutrality (dN ≠ dS)* | HA = positive selection (dN > dS) | HA = purifying selection (dN < dS)* |
|---|---|---|---|---|---|
| Exon 1 | 11 | 24 | − 0.8576, P = 0.3928 | − 0.9059, P = 1.0000 | 0.8804, P = 0.1902 |
| Exon 2 | 19 | 90 | − 1.1546, P = 0.2505 | − 1.1160, P = 1.0000 | 1.1258, P = 0.1312 |
| Exon 3 | 20 | 92 | − 0.6806, P = 0.4973 | − 0.6930, P = 1.0000 | 0.6790, P = 0.2491 |
| Exon 4 | 17 | 92 | − 1.0288, P = 0.3056 | − 1.007, P = 1.0000 | 0.9609, P = 0.1692 |
| Exon 5 | 10 | 39 | − 1.5091, P = 0.1338 | − 1.3480, P = 1.0000 | 1.4232, P = 0.0786 |
| All exons | 77 | 339 | − 2.5808, P = 0.0110 | − 2.5069, P = 1.0000 | 2.6134, P = 0.0050 |
| Exons 2 and 3 | 39 | 182 | − 1.3036, P = 0.1948 | − 1.2308, P = 1.0000 | 1.2635, P = 0.1044 |
| Exons 2, 3, and 4 | 55 | 274 | − 1.7453, P = 0.0834 | − 1.7482, P = 1.0000 | 1.6929, P = 0.0465 |
*Significant P-values are marked in boldface.
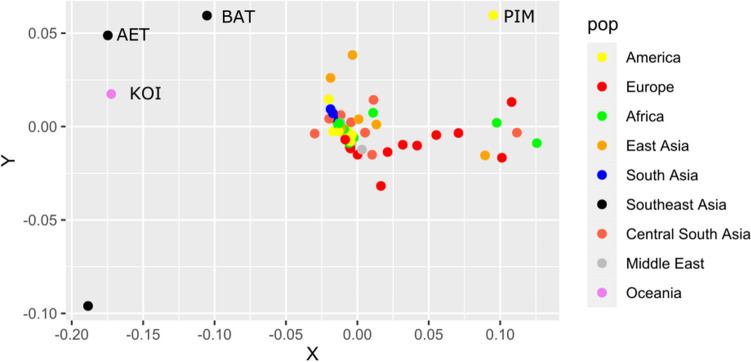
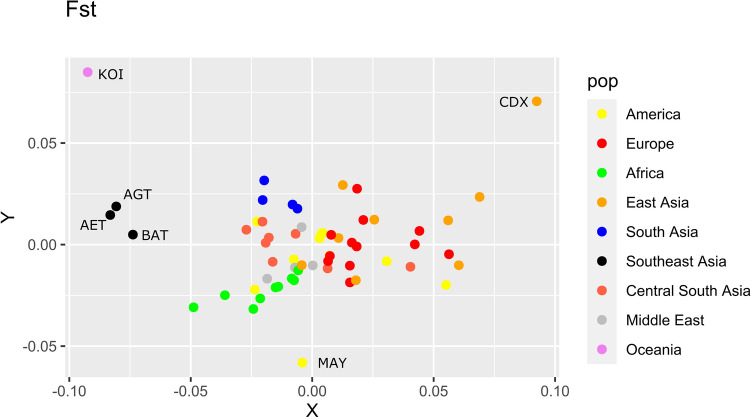
Most of the HLA-G common SNPs are shared worldwide. Because of that, there is a low population differentiation (Fig. 3), except for very isolated populations that may have experienced strong bottleneck effects, such as the Aeta, Batak, and Pima.
Figure 3.
Multidimensional scaling (MDS) illustrating the distance (measured by FST estimated from HLA-G SNPs) among population samples with at least 10 individuals. The name of each population is available at Table S1. We have indicated the names of some outliers.
The HLA-G coding region diversity
We considered the diversity observed between position −300 and + 838, which is the region tracked by the IPD-IMGT/HLA database19, and included, in this survey, 191 variation sites. All coding sequences we have observed, together with their names and global counts, are available for download (Table 1). We detected 190 different HLA-G alleles; 36 of them were already reported (fully or partially) in the IPD-IMGT/HLA database19, with a summed frequency of 95.54%. The 154 remaining sequences configure possible new HLA-G alleles. Because these new sequences have not been cloned and submitted to the IPD-IMGT/HLA, we named all new sequences as GEMBIO_HLA-G_G followed by a number that identifies this sequence in the GeMBio laboratory. Interestingly, at least 41 of these new alleles occurred more than once, and some are very frequent, such as GEMBIO_HLA-G_G144, with 51 copies detected in Africa, America, and the Middle East. We also confirmed the existence of many rare HLA-G alleles that have been submitted to the IPD-IMGT/HLA database, such as G*01:01:01:14Q (4 copies), G*01:09 (1 copy), G*01:11 (11 copies), G*01:14 (6 copies), G*01:21 N (2 copies), G*01:26 (2 copies), and others (Table S3).
In terms of global diversity, there are 14 HLA-G alleles with global frequencies higher than 1%: G*01:01:01:01, G*01:01:01:04, G*01:01:01:05, G*01:01:01:06, G*01:01:01:08, G*01:01:02:01, G*01:01:03:03, and G*01:01:22:01 (all of them encoding the G*01:01 allotype), G*01:03:01:02 (encoding the G*01:03 allotype), G*01:04:01:01, G*01:04:01:02, and G*01:04:04 (encoding the G*01:04 allotype), G*01:05 N (the null allele), and G*01:06:01:01 (encoding the G*01:06 allotype). These alleles represent 91% of all HLA-G sequences detected worldwide, and their frequencies differ in each biogeographic region (Fig. 4), and in each population sample (Table S3). For instance, the frequency of G*01:01:01:01 is high in East Asia, low in Africa, and rare in Oceania, while G*01:01:01:04 is quite rare in Asia and frequent in Africa (Fig. 4). The differentiation among populations, evaluated by FST, became clearer when evaluated using the haplotypes/genomic alleles (Fig. 5) compared to SNPs (Fig. 3), with African, European, South Asian, and Southeast Asian population samples clustering together. However, most of the population samples amalgamate in the center of the plot, because the common HLA-G alleles are present worldwide (Fig. 5).
Figure 4.
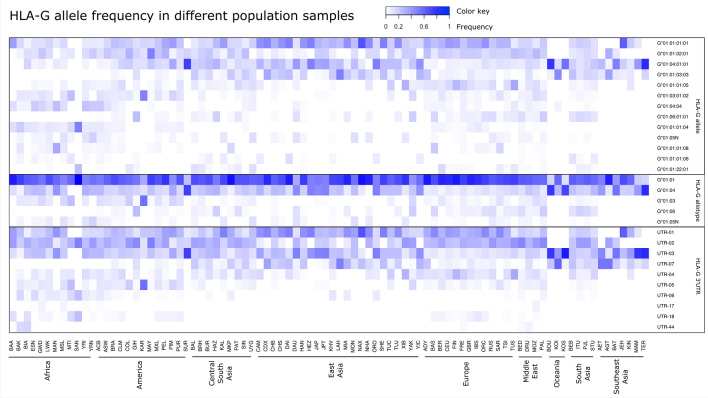
Frequencies of the most common HLA-G alleles, allotypes, and 3’UTR haplotypes in different population samples across the world. Tables S3, S4, and S5 present all frequency values.
Figure 5.
Multidimensional scaling (MDS) illustrating the distance (measured by FST estimated from HLA-G alleles—coding haplotypes) in populations samples with at least 10 individuals. The name of each population is available at Table S1. We have indicated the names of some outliers.
There are 74 alleles that have occurred at least twice. For instance, the new genomic allele named GEMBIO_HLA-G_G128 presents a frequency of 30% among the Aeta and around 10% in Agta and the Bouganville Island samples (Table S3). The hunter-gatherers in the Philippines (Agta) have been linked to Bouganville and Highland Papua New Guinea and Australian hunter-gatherers, possibly associated with a previous out of Africa47. The sequence GEMBIO_HLA-G_G31 occurred in many countries, achieving a frequency of 16.7% in the Central African Republic (Table S3). Both these new alleles encode the allotype G*01:01, which is the most frequent worldwide (Fig. 4). As explained earlier, these names represent sequences stored in the GeMBio laboratory Database (www.castelli-lab.net/HLA-G). Thus, HLA-G genetic diversity is higher than we currently acknowledge, although only a few different allotypes can be found worldwide. This observation reinforces the conservation in terms of protein diversity, but not necessarily in terms of DNA diversity. In fact, as demonstrated in the previous section, HLA-G nucleotide diversity is much higher than the one observed across chromosome 6, particularly in intronic regions. In addition, there is evidence of purifying selection acting on HLA-G exons, reinforcing the evidence of protein conservation in worldwide populations.
We have characterized the pair of full sequences of every individual using the cutting-edge sequencing technology, and we have described here, for the first time, the complete sequences (DNA and protein) of some alleles that are only partially reported in the IPD-IMGT/HLA Database. Additionally, we have provided the sequences of many new HLA-G alleles. Sequence characterization was simplified because many individuals presented the same full-length sequence for alleles G*01:01:14, G*01:01:15, G*01:01:19, G*01:04:05, G*01:11, and G*01:14, which are only partially characterized in the IPD-IMGT/HLA Database. Likewise, some new sequences occurred in more than ten individuals. This worldwide data clearly reveals that the IPD-IMGT/HLA is outdated in terms of stored data for HLA-G. It should be emphasized that, as new NGS initiatives grow, invaluable high-quality and large-scaled HLA data will be shortly produced. Thus, it is of utmost importance that the IPD-IMGT/HLA considers these data to update the official database.
The HLA-G allotype/protein diversity
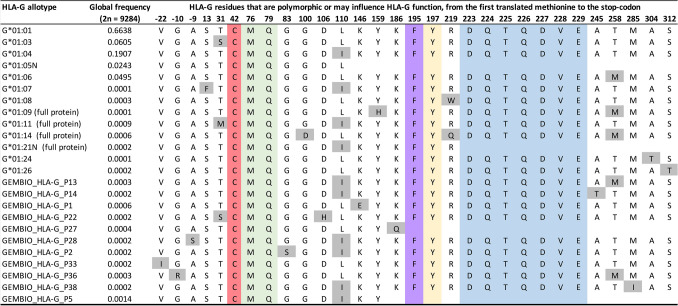
We have detected 53 different full-length proteins (allotypes), 13 of them already reported by the IPD-IMGT/HLA database (Table S4). As already observed in many studies (www.allelefrequencies.net), only five allotypes are globally frequent (G*01:01, G*01:03, G*01:04, G*01:05 N, and G*01:06), with a summed frequency of 98.9%.
Some allotypes are particularly frequent in specific population samples (Table S4, Fig. 4). G*01:01 is the most frequent in all regions, except Oceania. G*01:06, for instance, is the product of a nucleotide exchange + 1799 C > T at codon 258 in exon 4, exchanging Threonine/Methionine (rs12722482, Table S1), and is frequent in the Middle East and South Asia, with intermediate frequencies in Europe and America, and almost absent in East Asia and Africa. Likewise, this allele was frequent in Cyprus30, absent in other Amerindian tribes from Brazil45 and Benin28, and very rare in other Amerindians from South America48. G*01:06 was previously associated with a risk for allograft rejection49 and pregnancy complications31,50,51, albeit the mechanisms underlying these associations are unknown. G*01:03, which results from an A > T exchange at position + 292 at exon 2, in codon 31, exchanging a Threonine for Serine (rs41551813, Table S1), is common among Africans, Americans (including Brazil), and in the Middle East, but rare among other biogeographic regions. Conversely, it is the most frequent allele among Karitiana (Table S4) and quite frequent in other Amerindian tribes from the Brazilian Amazon45. The frequency of G*01:04, which is a consequence of the nucleotide exchange + 755 C > A at exon 3, exchanging Leucine/Isoleucine (rs12722477, Table S1), is particularly high in Asia and Oceania, especially among Japanese. It is also high in Africa, Agta, and many Amerindians from South America28,45,48 and less frequent in Europe. G*01:05 N, a truncated HLA-G protein caused by a deletion at exon 3 leading to a premature stop codon further in intron 4 (rs41557518, Table S1), is rare in most regions but frequent in Africa. European populations present the highest frequency of allotype G*01:01, and usually the lowest frequency of other allotypes. Because of that, Europeans (especially Finns) present the lowest observed heterozygosity for allotypes across the world (data not shown).
Some HLA-G allotypes only occurred in specific populations. For example, G*01:11 occurred among Caribbeans, African Americans, Puerto Ricans, and Brazilians, and G*01:14 among Caribbeans, Sierra Leoneans, and African Americans. Moreover, we detected at least 40 new allotypes. These new allotypes are usually rare and restricted to single populations. However, some have occurred more than once, such as the new allotype here named GEMBIO_HLA-G_P5 (P for protein sequence), which occurred in many samples from South East Asia and Oceania (Table S4), and it is a truncated protein sequence. The sequence of each allotype is available as a resource for download (Table 1).
In Fig. 6, we highlight all the important amino acid residues that may influence HLA-G function, and the polymorphic residues for allotypes that occurred at least twice in our dataset. Among these critical residues for HLA-G function, we may cite the Cysteine at the α1 domain (position 42 in the mature protein), which is responsible for dimer formation (C42-C42) among HLA-G membrane-bound and soluble isoforms, and dimers interact with more affinity with HLA-G receptors4. The Methionine and Glutamine (76 and 79 at the mature protein, in green in Fig. 6) residues, also at the α1 domain, interact with KIR2DL452. The residues 195 and 197, at the α3 domain, interact with ILT-2 and ILT-4 leukocyte receptors (purple and yellow in Fig. 7, respectively)53. The motif DQTQDVE, also at the α3 domain, interacts with the TCD8 receptor (in blue in Fig. 6)54. As can be observed, there is no variability at residues associated with HLA-G major biological properties in a worldwide sample. Note that truncated proteins lacking the α3 domain (G*01:05 N, G*01:21 N, and some new allotypes) present limited interaction with HLA-G receptors. However, the global frequency of these truncated allotypes is low (2.6%), mostly represented by G*01:05 N.
Figure 6.
HLA-G residues that are polymorphic or may influence HLA-G function, considering known allotypes detected in 4642 samples from across the world and new allotypes that have occurred at least twice. Polymorphic residues are marked in shades of gray. Important residues for the HLA-G function are marked in other colors. For allotypes with premature stop-codons, there is no amino acid indication after the frameshift mutation. In red, the Cysteine responsible for dimer formation. In green, the Methionine and Glutamine that interact with KIR2DL4. In purple and yellow, the important residues for ILT-2 and ILT-4 interaction. In blue, the motif DQTQDVE, which interacts with the TCD8 receptor.
Figure 7.
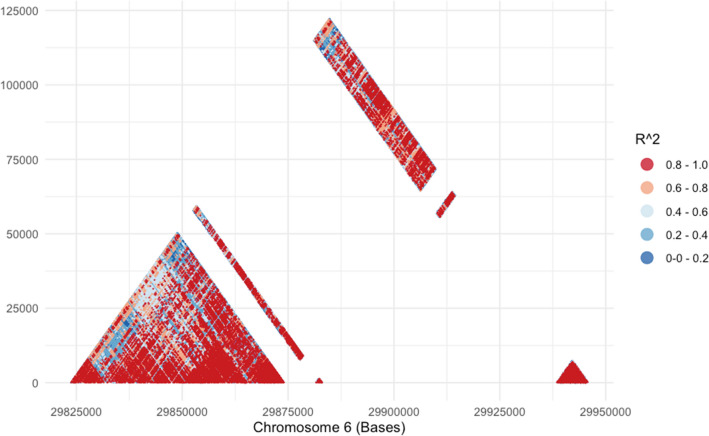
Linkage disequilibrium encompassing the HLA-G and HLA-A region, considering 5347 individuals from worldwide population samples and SNPs with minimum allele frequency higher than 1%. We have removed variants that coincides with known structural variants, producing three continuous segments: chr6:29,823,675–29,874,064, starting from the HLA-G promoter up to 43 Kb downstream HLA-G, chr6:29,881,527–29,883,079, a 1.5 kb region between HLA-A and HLA-G, and chr6:29,938,412–29,945,862, starting 4 kb upstream HLA-A to the end of the HLA-A 3’UTR.
The HLA-G 3’ untranslated region
The 3’ untranslated region (3’UTR) of most of the mRNA transcripts is the primary target for microRNAs (miRNA), which are small non-coding RNA molecules that post-transcriptionally modulate gene expression levels. MiRNAs modulate gene expression by two different mechanisms, mRNA degradation and translation blockage55. Variants in both miRNA coding gene and miRNA target sequence may influence miRNA/mRNA binding strength and thus influence gene expression levels56.
The HLA-G 3’UTR segment was fully characterized in 2010 by Castelli and colleagues, analyzing a large admixed Brazilian sample using Sanger sequencing41. There were eight frequent haplotypes in that survey, named UTR-01 to UTR-08, in order of frequency. Each of these 3’UTR haplotypes was in LD with specific HLA-G coding region alleles. The existence of these frequent haplotypes and their associated alleles were further confirmed by many other studies, using Sanger sequencing and NGS in different populations24,27,28,30,40,57–67. There is a high LD between haplotypes from the HLA-G 3’UTR and the coding region (Fig. 1, Table 4), with each 3’UTR haplotype following specific HLA-G coding alleles, such as G*01:01:01:01-UTR-01, G*01:01:01:04-UTR-06, G*01:01:01:05-UTR-04, G*01:03-UTR-05, G*01:04-UTR-03, and G*01:01:03-UTR-0741.
Table 4.
The relationship between HLA-G alleles and 3’UTR haplotypes, for combinations that have occurred at least twice in 4640 individuals across the globe.
| HLA-G allele | 3'UTR haplotype | Global frequencya | Internal frequencyb |
|---|---|---|---|
| G*01:01:01:01 | UTR-06 | 0.0027 | 0.0116 |
| G*01:01:01:01 | UTR-60 | 0.0002 | 0.0009 |
| G*01:01:01:01 | UTR-01 | 0.2277 | 0.9846 |
| G*01:01:01:04 | UTR-20 | 0.0036 | 0.1000 |
| G*01:01:01:04 | UTR-18 | 0.0198 | 0.5576 |
| G*01:01:01:04 | UTR-06 | 0.0121 | 0.3394 |
| G*01:01:01:05 | UTR-04 | 0.0704 | 0.9985 |
| G*01:01:01:06 | UTR-27 | 0.0002 | 0.0161 |
| G*01:01:01:06 | UTR-04 | 0.0131 | 0.9839 |
| G*01:01:01:08 | UTR-01 | 0.0204 | 1.0000 |
| G*01:01:01:09 | UTR-01 | 0.0033 | 1.0000 |
| G*01:01:01:13 | UTR-06 | 0.0066 | 1.0000 |
| G*01:01:01:14Q | UTR-01 | 0.0004 | 1.0000 |
| G*01:01:02:01 | UTR-02 | 0.1470 | 0.9956 |
| G*01:01:02:01 | UTR-10 | 0.0002 | 0.0015 |
| G*01:01:02:02 | UTR-02 | 0.0045 | 1.0000 |
| G*01:01:02:04 | UTR-02 | 0.0015 | 1.0000 |
| G*01:01:03:03 | UTR-07 | 0.0744 | 0.9957 |
| G*01:01:03:03 | UTR-31 | 0.0002 | 0.0029 |
| G*01:01:03:04 | UTR-07 | 0.0002 | 1.0000 |
| G*01:01:12 | UTR-02 | 0.0022 | 1.0000 |
| G*01:01:14 | UTR-02 | 0.0025 | 1.0000 |
| G*01:01:15 | UTR-06 | 0.0039 | 1.0000 |
| G*01:01:17 | UTR-02 | 0.0026 | 1.0000 |
| G*01:01:19 | UTR-02 | 0.0017 | 1.0000 |
| G*01:01:22:01 | UTR-02 | 0.0128 | 1.0000 |
| G*01:01:22:04 | UTR-02 | 0.0004 | 1.0000 |
| G*01:03:01:02 | UTR-56 | 0.0029 | 0.0510 |
| G*01:03:01:02 | UTR-17 | 0.0038 | 0.0662 |
| G*01:03:01:02 | UTR-48 | 0.0004 | 0.0076 |
| G*01:03:01:02 | UTR-05 | 0.0499 | 0.8752 |
| G*01:04:01:01 | UTR-03 | 0.1211 | 0.9799 |
| G*01:04:01:01 | UTR-02 | 0.0004 | 0.0035 |
| G*01:04:01:01 | UTR-13 | 0.0018 | 0.0148 |
| G*01:04:01:02 | UTR-53 | 0.0002 | 0.0227 |
| G*01:04:01:02 | UTR-03 | 0.0093 | 0.9773 |
| G*01:04:04 | UTR-23 | 0.0009 | 0.0171 |
| G*01:04:04 | UTR-03 | 0.0495 | 0.9829 |
| G*01:04:05 | UTR-03 | 0.0027 | 1.0000 |
| G*01:05 N | UTR-02 | 0.0243 | 1.0000 |
| G*01:06:01:01 | UTR-02 | 0.0484 | 1.0000 |
| G*01:06:01:02 | UTR-02 | 0.0009 | 1.0000 |
| G*01:08:02 | UTR-02 | 0.0003 | 1.0000 |
| G*01:11 | UTR-03 | 0.0009 | 1.0000 |
| G*01:14 | UTR-05 | 0.0006 | 1.0000 |
| G*01:21 N | UTR-03 | 0.0002 | 1.0000 |
| G*01:26 | UTR-02 | 0.0002 | 1.0000 |
aThe global frequency of this haplotype.
bConsidering all the 3’UTR haplotypes associated with a given genomic allele, this is the frequency in which this specific 3’UTR haplotype follows the given genomic allele. An internal frequency of 1.000 indicates that the given genomic allele always presents the same 3’UTR sequence.
Although two consecutive exons form the HLA-G 3’UTR, the sequence after the stop codon at exon 6 does not present any variant. Thus, for the 3’UTR analysis, we have considered only the last HLA-G exon. According to transcripts NM_001363567 and NM_002127, the last HLA-G exon extends up to position 6:29,831,122 (hg38), which corresponds to the HLA-G position + 3292, when we consider the presence of the 14 bp sequence15. Our data reinforce the presence of few nucleotide exchanges in the HLA-G 3’UTR, with 27 variants distributed across the last HLA-G exon. Of these, only nine are recognized as polymorphisms, with a global frequency superior to 1%. Supplementary Table S2 displays the global frequency of each of these variants.
The 27 variants form 35 different haplotypes, with different frequencies across the globe, as shown in Fig. 4 and Table S5. However, as initially described in 2010, only eight of these haplotypes are globally frequent (Fig. 4), with frequencies varying in different biogeographic regions. UTR-01 and UTR-02 are common in Europe, Africa, America, and parts of Asia, and less common in Southeast Asia and Oceania. UTR-01 and UTR-02 are the most divergent ones, but they are in LD with the HLA-G*01:01:01:01 and G*01:01:02:01 coding alleles (encoding the same G*01:01 allotype), which in turn are also the most divergent coding region alleles. Conversely, the frequencies of UTR-03 and -07 follow the opposite path in terms of worldwide distribution, with the first associated with allotype G*01:04 and the last with G*01:01. UTR-07 is absent in Africa (Fig. 4).
The HLA-G 3’UTR presents at least three variants that have been evaluated in terms of functional studies. Undoubtedly, the most studied is the presence or absence of a 14-bp fragment at the beginning of the last HLA-G exon, commonly referred to as the 14 bp INS/DEL (rs371194629) polymorphism. The ancestral allele is the presence of this sequence, which is detectable in most primates. The supplementary alignment provides its status in each 3’UTR sequence. The DEL allele is associated with the frequent UTR-01, UTR-03, UTR-04, UTR-06, and UTR-18 haplotypes (Fig. 4), among others. The ancestral allele (INS, presence) was associated with lower HLA-G production of the membrane-bound and soluble isoforms68–71, while other studies pointed the opposite72,73. Previous studies indicated that the 14-base sequence (or another variant in linkage with it) triggers alternative splicing removing from the mature HLA-G mRNA the first 92 bases of the last exon, which also includes the 14b sequence74. These alternative mRNA seems to be more stable75, but the fraction of mRNA that undergoes this alternative splicing varies in different cell lines and tissues. Moreover, the 14-bp polymorphism may influence the binding of a few miRNAs that target the 14b sequence directly, in addition to various miRNAs that target the 92b segment that may be spliced out when the 14b sequence is present76,77. However, with the exception of miR-133a78, none of these interactions have been proven functionally.
The + 3142 G > C variant (rs1063320) influences the binding of some miRNAs, with Guanine allele favoring the targeting of three miRNAs, miR-148a-3p, miR-148b-3p, and miR-15256,76. However, while studies confirm that these miRNAs down-regulate HLA-G expression79,80, apparently, they down-regulate HLA-G irrespectively of the nucleotide at position + 314280. Interestingly, miR-148a-3p and miR-148b also influence HLA-C expression, and both genes are co-expressed in the placenta during pregnancy81. The allele associated with lower miRNA binding, + 3142C, can be found worldwide and co-occurs with the frequent UTR-01, UTR-04, UTR-06, and UTR-18 haplotypes (Fig. 4, supplementary alignment).
The + 3187 A > G variant (rs9380142) coincides with an AU-rich motif in the HLA-G 3’UTR that may trigger HLA-G mRNA degradation. The presence of Guanine in this position reduces the size of this AU-rich motif, positively influencing mRNA stability and associated with higher HLA-G expression82. The only frequent 3’UTR carrying the high-expressing allele (Guanine) is UTR-01.
Many studies addressed the influence of the 3’UTR sequence on the HLA-G expression levels of both membrane-bound and soluble isoforms. Since HLA-G presents a high LD (Fig. 1), any study addressing HLA-G expression profile and polymorphism must consider how these polymorphisms interact with each other as haplotypes. For instance, while UTR-04 and UTR-06 present the high-expressing + 3142 > C allele, they also carry the low-expressing + 3187 > A allele. The 14 bp INS allele usually co-occurs with the low expressing + 3142 > G and + 3187 > A alleles, with rare exceptions. The most divergent haplotypes, UTR-01 and UTR-02, differ in all the variants functionally reported to influence HLA-G expression and in two other positions (supplementary alignment). The coding alleles associated with these haplotypes encode the same HLA-G*01:01 allotype. It seems that the frequent 3’UTR haplotypes detected in modern humans somehow present polymorphisms that tune HLA-G expression in a dependent manner, possibly resulting in either high or low expression levels depending on the stimuli and the physiological microenvironment. The influence of specific haplotypes on the HLA-G expression levels has already been demonstrated functionally24.
The relationship between the HLA-G 3’UTR haplotypes and HLA-G alleles was addressed in many studies24,27,28,30,40–42,57–67, and was confirmed and updated in this survey (Table 4). Some HLA-G alleles, such as G*01:01:01:08, G*01:01:22:01, G*01:05 N, and G*01:06:01:01, always carry the same 3’UTR haplotype, while others, such as G*01:01:01:01, G*01:01:01:05, and G*01:01:01:06, are mostly, but not exclusively, associated with a single 3’UTR haplotype. Conversely, the G*01:01:01:04 and G*01:03:01:02 alleles present more than one frequent 3’UTR haplotype. Because of the impact of the 3’UTR sequence on the HLA-G function, this region should be included in the IPD-IMGT/HLA database, and the names of alleles carrying more than one 3’UTR haplotype should be updated. Table 4 lists only the haplotypes with a global frequency higher than 0.5%, which achieved a summed frequency of 95.3%. Following the internal frequencies presented in Table 4, one may argue that it is possible to input the HLA-G allotypes by observing a few SNPs at the 3’UTR segment. Table S5 presents a complete list of all genomic HLA-G alleles and their 3’UTR sequences.
Linkage disequilibrium between HLA-G and HLA-A
HLA-A is a classical class I gene that encodes a key molecule for antigen presentation. HLA-A is the second most variable histocompatibility gene, with more than 6000 alleles already described in the IPD-IMGT/HLA Database. As antigen presentation is the main function of HLA-A, it is common sense that natural selection has shaped the genetic variability in its coding region, increasing variability at the peptide binding site, thus allowing a great diversity of antigen presentation83,84.
HLA-A and HLA-G are only 100 Kb apart from each other. This proximity results in high LD and hitchhiking effects might even involve common regulatory regions. Linkage disequilibrium between HLA-G and HLA-A has been previously demonstrated using low HLA-A typing resolution43. Here, we demonstrate the LD level between these two genes by using forefront technology. First, we processed the region encompassing HLA-G and HLA-A using the same pipeline, as described in the supplementary methods. Then, we removed variants laying on regions with known structural variants and rare variants (MAF < 1%), resulting in 1873 frequent SNPs covering the HLA-G, HLA-A, and their intergenic region. With this data, we calculated the r2 between pairs of bi-allelic SNPs (Fig. 7). Because we removed SNPs in regions with structural variants, we have 3 blocks of continuous variants: (a) chr6:29,823,675–29,874,064, starting from the HLA-G promoter up to 43 Kb downstream HLA-G, (b) chr6:29,881,527–29,883,079, a 1.5 kb region between HLA-A and HLA-G, and (c) chr6:29,938,412–29,945,862, starting 4 kb upstream HLA-A to the end of the HLA-A 3’UTR. Figure 7 demonstrates that the high LD across HLA-G extends from its promoter region up to HLA-A, with most pairwise r2 higher than 0.6.
This analysis also revealed some new insights involving the relationship between HLA-G alleles and HLA-A allotypes. For instance, there is an absolute LD (or almost absolute) between G*01:01:01:05 and A*03 (particularly A*03:01), G*01:01:01:08 and A*30:02, G*01:01:01:13 and A*29:01, G*01:01:03:03 and A*11 (particularly A*11:01), G*01:01:02:01 and A*32:01, G*01:05 N and A*30:01, and G*01:06:01:01 and A*01:01. Conversely, the most frequent HLA-G alleles G*01:01:01:01, G*01:01:02:01, G*01:03:01:02, and G*01:04:01:01 are associated with multiple non-overlapping HLA-A allotypes. Therefore, it becomes clear that the LD between HLA-G and HLA-A is stronger than originally perceived.
Balancing selection has been well documented for both HLA-A83,85 and HLA-G86, but at different fashions. While the most polymorphic segment at HLA-A is the coding sequence, associated with the presence of frequent and divergent coding alleles in worldwide populations, the most polymorphic sites at HLA-G are the regulatory regions (i.e., promoter, introns, and the 3’UTR, Fig. 2). Many studies have detected signatures of balancing selection at the HLA-G promoter and 3’UTR40,66,86,87, while the coding segment is conserved, as we demonstrated here, being under purifying selection. However, it is not clear whether balancing selection is indeed operating at the HLA-G promoter (for instance, in the region encompassing polymorphism − 725), or whether these findings are due to a hitchhiking effect caused by the selective pressures acting the neighboring HLA-A gene.
Since HLA-A is a crucial molecule for triggering adaptive immune responses, HLA-A is usually a target for natural selection. Balancing selection is an important force that maintains advantageous genetic diversity in populations, including variations responsible for long-term adaptation to the environment88. For the coding region, it is well established that balancing selection deals with an excess of non-synonymous substitutions and an increase of functionally relevant variability.
A previous study from our group has proposed that the HLA-A locus is not influencing HLA-G, and therefore HLA-G is a direct target of selection87. However, the study above addressed this issue using the 1000Genomes phase 1 data, which was the only dataset available at the time. Since then, several studies have revealed that the 1000genomes phase I dataset at the HLA region is strongly affected by mapping bias. This mapping bias, together with the fact that those samples were sequenced in a low coverage fashion, and no specific tools for HLA were applied, might have led to a wrong conclusion that HLA-A does not influence HLA-G.
Here, we demonstrate a strong LD between HLA-G and HLA-A in a way that the most frequent HLA-G alleles are associated with specific HLA-A alleles. Because there is a strong LD between any HLA-G allele and specific promoter and 3’UTR sequences, balancing selection operating at the HLA-A locus may also shapes the HLA-G promoter frequencies, downplaying its direct role on HLA-G. The same rationale can be applied to the HLA-G intronic sequences. High heterozygosity in HLA-A, which is commonly observed in worldwide populations, would lead to high heterozygosity in the HLA-G coding region (at the DNA level) and, thus, to high heterozygosity in the HLA-G promoter and intronic regions30,40. We cannot disregard the possibility that part of the genetic signature of balancing selection on HLA-A is due to HLA-G since the signatures of Natural Selection observed in both genes are not independent, as previous stated.
Implication of the HLA-G variability in worldwide populations
After fully sequencing HLA-G and its promoter region in 4640 individuals from 88 different populations samples, we have concluded that this gene is highly conserved in the protein level, with no variation in the residues responsible for the biological function of the protein, i.e., dimer formation, alternative splicing, peptide coupling, and interaction with HLA-G receptors. Considering that HLA-G dimer interaction with HLA-G receptors is more stable than with monomers17,53, and considering that dimer formation primarily depends on the disulfide bridge formed between Cysteine residues (C42–C42)53, as seen in Fig. 6, all complete proteins detected in worldwide samples exhibit Cysteine at residue 42. Then, theoretically, all proteins and the most common HLA-G isoforms exhibiting the α1 domain may form dimers. Since the mechanisms of HLA-G alternative splicing have not been completely defined, the frequent variants in the HLA-G introns may influence splicing, yielding different isoforms with different properties. In this context, some of these isoforms may not present the α1 domain17, avoiding dimer formation. Most of the HLA-G coding region variability coincides with introns (Fig. 2 and Table 2), and it is possible that these variants influence HLA-G regulation. All proteins we have detected exhibit no variability at residue motifs described to interact with ILT-2 (Phe195), ILT-4 (Phe195, Tyr197), KIR2DL4 (Met76, Gln79) and CD8 (DQTQDVE, residues 223–229)2,54,89. Nevertheless, isoforms presenting an incomplete α1 domain cannot interact with KIR2DL4, particularly the truncated HLA-G proteins encoded by the HLA-G*01:05 N, *01:13 N, and 01:21 N alleles (Fig. 6), as well as with ILTs and CD8 receptors.
Because HLA-G is a promiscuous immune checkpoint molecule, the modulation of its function may be tuned according to specific therapeutical approaches. The use of monoclonal antibodies against HLA-G in situations in which its expression is undesirable (for instance, in cancers and chronic viral disorders) would have many side effects since HLA-G is constitutively expressed in thymus, pancreas, and hematopoietic stem cells17. On the other hand, the use of recombinant HLA-G would be a plausible alternative for allograft and autoimmune disease treatment. Since few coding allotypes (proteins) are most frequently observed in worldwide populations, the production of a small number of recombinant HLA-G proteins would be sufficient to individualize treatment, according to the patient HLA-G coding region typing. Besides targeting the molecule, it is also possible to modulate its expression taking advantage of the knowledge of the target sites for transcription and post-transcription factors that act on the HLA-G gene expression. Considering the canonical and non-canonical miRNA and transcription factor22,24,56,76,90–93 targeting HLA-G, a comprehensive survey of the HLA-G genetic diversity as provided in this study may help to unveil regulatory factors and strategies to differentially modulate HLA-G production whenever necessary.
Taken together, the results of this investigation indicate that: (1) most of the HLA-G polymorphic sites are located in the regulatory and intronic regions, (2) although some promoter segments present the highest nucleotide diversity and Tajima’s D, the strongest signals of balancing selection come from intronic segments, particularly intron 5 (3) a high LD is observed across HLA-G, from 4 Kb upstream the gene up to the HLA-A locus, (4) it is possible that balancing selection acting on HLA-A may be influencing the high heterozygosity observed in the regulatory and intronic sequences of HLA-G, and vice-versa, (5) the presence of few proteins frequently observed in worldwide populations corroborates the role of the HLA-G as an immune checkpoint molecule rather than as an antigen-presenting molecule; (6) the lack of variation in residues responsible for major HLA-G biological functions reinforces the immunomodulatory properties of the molecule, maintained throughout evolution; (7) most common variation sites within HLA-G regulatory and coding region have been maintained as haplotypes, as observed in worldwide populations, (8) motifs associated with HLA-G interaction with receptors may be engineered modified to impede or enhance receptor interaction according to the underlying disorder, (9) possible therapeutic use of HLA-G as an immune checkpoint agent may be accomplished with the production of a limited number of recombinant proteins.
Concluding remarks
This is the most comprehensive study of the genetic diversity involving HLA-G performed so far. This large dataset, composed of populations from different ancestries and biogeographical regions and using sequencing data produced with cutting-edge sequencing technology and bioinformatics pipelines, may provide a broad picture of worldwide patterns of HLA-G haplotype distribution and linkage disequilibrium. This landmark achievement on HLA-G genetic diversity that will hardly be overridden in the near future provides all the basis for in-depth association and functional studies, which is of utmost importance given the immunomodulatory properties of the molecule.
Supplementary Information
Acknowledgements
We acknowledge Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq/Brazil, #302060/2019-7), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES/Brazil, Finance code: 001), INCTC/FAPESP (Grant #465539/2014-9) for financial support, and FAPESP/Brazil (#Grant 2017/19223-0).
Author contributions
Conceptualization, E.C.C., B.S.A., C.T.M.J., and E.A.D.; methodology, E.C.C., N.S.B.S., M.R.S.P., A.S.S., M.S.N., M.O.S., J.W., and C.T.M.J.; software, E.C.C.; resources, A.E.P., M.D., D.S., G.A., J.B., A.B.M., Y.A.O.D., M.R.P.B., and M.Z.; writing—original draft preparation, E.C.C., B.S.A., and E.A.D.; writing—review and editing, Y.C.N.M., M.O.S., and M.S.N. All authors have read and agreed to the published version of the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Erick C. Castelli, Email: erick.castelli@unesp.br
Eduardo A. Donadi, Email: eadonadi@fmrp.usp.br
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-02106-4.
References
- 1.Carosella ED, Moreau P, LeMaoult J, Rouas-Freiss N. HLA-G: From biology to clinical benefits. Trends Immunol. 2008;29:125–132. doi: 10.1016/j.it.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 2.Shiroishi M, et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8856–8861. doi: 10.1073/pnas.1431057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McIntire RH, Hunt JS. Antigen presenting cells and HLA-G: A review. Placenta. 2005;26:S104–S109. doi: 10.1016/j.placenta.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Boyson JE, et al. Disulfide bond-mediated dimerization of HLA-G on the cell surface. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16180–16185. doi: 10.1073/pnas.212643199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favier B, et al. Research on HLA-G: An update. Tissue Antigens. 2007;69:207–211. doi: 10.1111/j.1399-0039.2006.00757.x. [DOI] [PubMed] [Google Scholar]
- 6.Hviid TVF. HLA-G in human reproduction: Aspects of genetics, function and pregnancy complications. Hum. Reprod. Update. 2006;12:209–232. doi: 10.1093/humupd/dmi048. [DOI] [PubMed] [Google Scholar]
- 7.Ponte M, et al. Inhibitory receptors sensing HLA-G1 molecules in pregnancy: Decidua-associated natural killer cells express LIR-1 and CD94/NKG2A and acquire p49, an HLA-G1-specific receptor. Proc. Natl. Acad. Sci. U. S. A. 1999;96:5674–5679. doi: 10.1073/pnas.96.10.5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajagopalan S. HLA-G-mediated NK cell senescence promotes vascular remodeling: Implications for reproduction. Cell. Mol. Immunol. 2014;11:460–466. doi: 10.1038/cmi.2014.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajagopalan S, et al. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006;4:0070–0086. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parham P. NK Cells and Trophoblasts: Partners in Pregnancy. J. Exp. Med. 2004;200:951–955. doi: 10.1084/jem.20041783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amiot, L., Vu, N. & Samson, M. Immunomodulatory properties of HLA-G in infectious diseases. J. Immunol. Res.2014, (2014). [DOI] [PMC free article] [PubMed]
- 12.Rizzo R, Bortolotti D, Bolzani S, Fainardi E. HLA-G molecules in autoimmune diseases and infections. Front. Immunol. 2014;5:1–11. doi: 10.3389/fimmu.2014.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dias FC, Castelli EC, Collares CVA, Moreau P, Donadi EA. The role of HLA-G molecule and HLA-G gene polymorphisms in tumors, viral hepatitis, and parasitic diseases. Front. Immunol. 2015;6:2–11. doi: 10.3389/fimmu.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonon ASP, Garcia ISCTMA, Donadi EA. The role of HLA-G in parasitic diseases. HLA. 2017 doi: 10.1111/tan.13196. [DOI] [PubMed] [Google Scholar]
- 15.Donadi EA, et al. Implications of the polymorphism of HLA-G on its function, regulation, evolution and disease association. Cell. Mol. Life Sci. 2011;68:369–395. doi: 10.1007/s00018-010-0580-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contini P, Murdaca G, Puppo F, Negrini S. HLA-G Expressing immune cells in immune mediated diseases. Front. Immunol. 2020;11:1–9. doi: 10.3389/fimmu.2020.01613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carosella, E. D., Rouas-Freiss, N., Roux, D. T. Le, Moreau, P. & LeMaoult, J. HLA-G. An Immune Checkpoint Molecule. Advances in Immunology vol. 127 (Elsevier Inc., 2015). [DOI] [PubMed]
- 18.Tronik-Le Roux D, et al. Novel landscape of HLA-G isoforms expressed in clear cell renal cell carcinoma patients. Mol. Oncol. 2017;11:1561–1578. doi: 10.1002/1878-0261.12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson J, et al. The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res. 2015;43:D423–D431. doi: 10.1093/nar/gku1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castelli, E. C., Veiga-Castelli, L. C., Yaghi, L., Moreau, P. & Donadi, E. A. Transcriptional and posttranscriptional regulations of the HLA-G gene. J. Immunol. Res.2014, (2014). [DOI] [PMC free article] [PubMed]
- 21.Poras I, et al. Haplotypes of the HLA-G 3′ untranslated region respond to endogenous factors of HLA-G+ and HLA-G- cell lines differentially. PLoS ONE. 2017;12:1–18. doi: 10.1371/journal.pone.0169032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yaghi L, et al. Hypoxia inducible factor-1 mediates the expression of the immune checkpoint HLA-G in glioma cells through hypoxia response element located in exon 2. Oncotarget. 2016;7:63690–63707. doi: 10.18632/oncotarget.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garziera, M., Scarabel, L. & Toffoli, G. Hypoxic Modulation of HLA-G Expression through the Metabolic Sensor HIF-1 in Human Cancer Cells. J. Immunol. Res. 2017 (2017). [DOI] [PMC free article] [PubMed]
- 24.Verloes A, et al. The role of methylation, DNA polymorphisms and microRNAs on HLA-G expression in human embryonic stem cells. Stem Cell Res. 2017;19:118–127. doi: 10.1016/j.scr.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Alves CC, et al. Human leukocyte antigen-G 3’ untranslated region polymorphisms are associated with asthma severity. Mol. Immunol. 2018;101:500–506. doi: 10.1016/j.molimm.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 26.Ferreira LMR, et al. A distant trophoblast-specific enhancer controls HLA-G expression at the maternal-fetal interface. Proc. Natl. Acad. Sci. U. S. A. 2016;113:5364–5369. doi: 10.1073/pnas.1602886113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castelli, E. C. et al. Insights into HLA-G genetics provided by worldwide haplotype diversity. Front. Immunol.5, (2014). [DOI] [PMC free article] [PubMed]
- 28.Sonon P, et al. HLA-G, -E and -F regulatory and coding region variability and haplotypes in the Beninese Toffin population sample. Mol. Immunol. 2018;104:108–127. doi: 10.1016/j.molimm.2018.08.016. [DOI] [PubMed] [Google Scholar]
- 29.de Oliveira MLG, et al. Extended HLA-G genetic diversity and ancestry composition in a Brazilian admixed population sample: Implications for HLA-G transcriptional control and for case-control association studies. Hum. Immunol. 2018;79:790–799. doi: 10.1016/j.humimm.2018.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Castelli EC, et al. HLA-G variability and haplotypes detected by massively parallel sequencing procedures in the geographicaly distinct population samples of Brazil and Cyprus. Mol. Immunol. 2017;83:115–126. doi: 10.1016/j.molimm.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 31.Hviid TV, Hylenius S, Hoegh AM, Kruse C, Christiansen OB. HLA-G polymorphisms in couples with recurrent spontaneous abortions. Tissue Antigens. 2002;60:122–132. doi: 10.1034/j.1399-0039.2002.600202.x. [DOI] [PubMed] [Google Scholar]
- 32.Larsen MH, Hylenius S, Andersen AMN, Hviid TVF. The 3′-untranslated region of the HLA-G gene in relation to pre-eclampsia: Revisited. Tissue Antigens. 2010;75:253–261. doi: 10.1111/j.1399-0039.2009.01435.x. [DOI] [PubMed] [Google Scholar]
- 33.da Nardi FS, et al. Analysis of HLA-G polymorphisms in couples with implantation failure. Am. J. Reprod. Immunol. 2012;68:507–514. doi: 10.1111/aji.12001. [DOI] [PubMed] [Google Scholar]
- 34.Sipak O, et al. Molecular analysis of HLA-G in women with high-risk pregnancy and their partners with regard to possible complications. Int. J. Environ. Res. Public Health. 2019;16:1–13. doi: 10.3390/ijerph16060982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferguson R, et al. Human leukocyte antigen (HLA)-E and HLA-G polymorphisms in human papillomavirus infection susceptibility and persistence. Hum. Immunol. 2011;72:337–341. doi: 10.1016/j.humimm.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 36.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clarke L, et al. The international Genome sample resource (IGSR): A worldwide collection of genome variation incorporating the 1000 Genomes Project data. Nucleic Acids Res. 2017;45:D854–D859. doi: 10.1093/nar/gkw829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naslavsky, M. S. et al. Whole-genome sequencing of 1171 elderly admixed individuals from the largest Latin American metropolis (São Paulo, Brazil). Wagner CS Magalhães (20AD).
- 39.Castelli EC, Paz MA, Souza AS, Ramalho J, Mendes-Junior CT. Hla-mapper: An application to optimize the mapping of HLA sequences produced by massively parallel sequencing procedures. Hum. Immunol. 2018;79:678–684. doi: 10.1016/j.humimm.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 40.Castelli EC, et al. A comprehensive study of polymorphic sites along the HLA-G gene: Implication for gene regulation and evolution. Mol. Biol. Evol. 2011;28:3069–3086. doi: 10.1093/molbev/msr138. [DOI] [PubMed] [Google Scholar]
- 41.Castelli EC, et al. The genetic structure of 3′untranslated region of the HLA-G gene: Polymorphisms and haplotypes. Genes Immun. 2010;11:134–141. doi: 10.1038/gene.2009.74. [DOI] [PubMed] [Google Scholar]
- 42.Santos KE, et al. Insights on the HLA-G evolutionary history provided by a nearby alu insertion. Mol. Biol. Evol. 2013;30:2423–2434. doi: 10.1093/molbev/mst142. [DOI] [PubMed] [Google Scholar]
- 43.Carlini F, et al. Association of HLA-A and non-classical HLA class i alleles. PLoS ONE. 2016;11:1–17. doi: 10.1371/journal.pone.0163570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ober C, Billstrand C, Kuldanek S, Tan Z. The miscarriage-associated HLA-G -725G allele influences transcription rates in JEG-3 cells. Hum. Reprod. 2006;21:1743–1748. doi: 10.1093/humrep/del036. [DOI] [PubMed] [Google Scholar]
- 45.Mendes-Junior CT, Castelli EC, Meyer D, Simões AL, Donadi EA. Genetic diversity of the HLA-G coding region in Amerindian populations from the Brazilian Amazon: A possible role of natural selection. Genes Immun. 2013;14:518–526. doi: 10.1038/gene.2013.47. [DOI] [PubMed] [Google Scholar]
- 46.Sachidanandam R, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 47.Rasmussen M, et al. An aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arnaiz-Villena A, et al. HLA-G in amerindians: Epidemiology and worldwide population comparison. Open Med. J. 2018 doi: 10.2174/1874220301805010001. [DOI] [Google Scholar]
- 49.Misra MK, et al. HLA-G gene expression influenced at allelic level in association with end stage renal disease and acute allograft rejection. Hum. Immunol. 2014;75:833–839. doi: 10.1016/j.humimm.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 50.Moreau P, et al. HLA-G gene polymorphism in human placentas: Possible association of G*0106 allele with preeclampsia and miscarriage. Biol. Reprod. 2008;79:459–467. doi: 10.1095/biolreprod.108.068874. [DOI] [PubMed] [Google Scholar]
- 51.Tan CY, et al. Paternal contribution of HLA-G*0106 significantly increases risk for pre-eclampsia in multigravid pregnancies. Mol. Hum. Reprod. 2008;14:317–324. doi: 10.1093/molehr/gan013. [DOI] [PubMed] [Google Scholar]
- 52.Gonen-Gross, T. et al. Inhibitory NK receptor recognition of HLA-G: Regulation by contact residues and by cell specific expression at the fetal-maternal interface. PLoS One5, (2010). [DOI] [PMC free article] [PubMed]
- 53.Shiroishi M, et al. Efficient leukocyte Ig-like receptor signaling and crystal structure of disulfide-linked HLA-G dimer. J. Biol. Chem. 2006;281:10439–10447. doi: 10.1074/jbc.M512305200. [DOI] [PubMed] [Google Scholar]
- 54.Gao GF, et al. Classical and nonclassical class I major histocompatibility complex molecules exhibit subtle conformational differences that affect binding to CD8αα. J. Biol. Chem. 2000;275:15232–15238. doi: 10.1074/jbc.275.20.15232. [DOI] [PubMed] [Google Scholar]
- 55.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 56.Tan Z, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am. J. Hum. Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Catamo E, et al. Comprehensive analysis of polymorphisms in the HLA-G 5′ upstream regulatory and 3′ untranslated regions in Brazilian patients with systemic lupus erythematosus. Tissue Antigens. 2015;85:458–465. doi: 10.1111/tan.12545. [DOI] [PubMed] [Google Scholar]
- 58.Garziera M, et al. Association of the HLA-G 3′UTR polymorphisms with colorectal cancer in Italy: A first insight. Int. J. Immunogenet. 2016;43:32–39. doi: 10.1111/iji.12243. [DOI] [PubMed] [Google Scholar]
- 59.Garcia A, et al. Association of HLA-G 3’UTR polymorphisms with response to malaria infection: A first insight. Infect. Genet. Evol. 2013;16:263–269. doi: 10.1016/j.meegid.2013.02.021. [DOI] [PubMed] [Google Scholar]
- 60.Garziera M, et al. HLA-G 3’UTR polymorphisms impact the prognosis of stage II-III CRC patients in fluoropyrimidine-based treatment. PLoS ONE. 2015;10:1–17. doi: 10.1371/journal.pone.0144000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sizzano F, et al. Genotypes and haplotypes in the 3′ untranslated region of the HLA-G gene and their association with clinical outcome of hematopoietic stem cell transplantation for beta-thalassemia. Tissue Antigens. 2012;79:326–332. doi: 10.1111/j.1399-0039.2012.01862.x. [DOI] [PubMed] [Google Scholar]
- 62.Carlini, F. et al. HLA-G UTR haplotype conservation in the Malian population: Association with soluble HLA-G. PLoS One8, (2013). [DOI] [PMC free article] [PubMed]
- 63.Catamo E, et al. HLA-G gene polymorphisms associated with susceptibility to rheumatoid arthritis disease and its severity in Brazilian patients. Tissue Antigens. 2014;84:308–315. doi: 10.1111/tan.12396. [DOI] [PubMed] [Google Scholar]
- 64.Di Cristofaro J, et al. HLA-G haplotype structure shows good conservation between different populations and good correlation with high, normal and low soluble HLA-G expression. Hum. Immunol. 2013;74:203–206. doi: 10.1016/j.humimm.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 65.Lucena-Silva N, et al. Haplotype frequencies based on eight polymorphic sites at the 3’ untranslated region of the HLA-G gene in individuals from two different geographical regions of Brazil. Tissue Antigens. 2012;79:272–278. doi: 10.1111/j.1399-0039.2012.01842.x. [DOI] [PubMed] [Google Scholar]
- 66.Sabbagh A, et al. Worldwide genetic variation at the 3′ untranslated region of the HLA-G gene: Balancing selection influencing genetic diversity. Genes Immun. 2014;15:95–106. doi: 10.1038/gene.2013.67. [DOI] [PubMed] [Google Scholar]
- 67.Dias FC, et al. The genetic diversity within the 1.4 kb HLA-G 5′ upstream regulatory region moderately impacts on cellular microenvironment responses. Sci. Rep. 2018;8:1–12. doi: 10.1038/s41598-018-24009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hviid TVF, et al. HLA-G and IL-10 in serum in relation to HLA-G genotype and polymorphisms. Immunogenetics. 2004;56:135–141. doi: 10.1007/s00251-004-0673-2. [DOI] [PubMed] [Google Scholar]
- 69.Hviid TVF, Rizzo R, Melchiorri L, Stignani M, Baricordi OR. Polymorphism in the 5′ Upstream Regulatory and 3′ Untranslated Regions of the HLA-G Gene in Relation to Soluble HLA-G and IL-10 Expression. Hum. Immunol. 2006;67:53–62. doi: 10.1016/j.humimm.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 70.Rebmann V, et al. Association of soluble HLA-G plasma levels with HLA-G alleles. Tissue Antigens. 2001;57:15–21. doi: 10.1034/j.1399-0039.2001.057001015.x. [DOI] [PubMed] [Google Scholar]
- 71.Rizzo R, et al. Role of HLA-G 14bp deletion/insertion and +3142C>G polymorphisms in the production of sHLA-G molecules in relapsing-remitting multiple sclerosis. Hum. Immunol. 2012;73:1140–1146. doi: 10.1016/j.humimm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 72.Svendsen SG, et al. The expression and functional activity of membrane-bound human leukocyte antigen-G1 are influenced by the 3’-untranslated region. Hum. Immunol. 2013;74:818–827. doi: 10.1016/j.humimm.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 73.Misra MK, et al. Association of HLA-G promoter and 14-bp insertion-deletion variants with acute allograft rejection and end-stage renal disease. Tissue Antigens. 2013;82:317–326. doi: 10.1111/tan.12210. [DOI] [PubMed] [Google Scholar]
- 74.Hiby SE, King A, Sharkey A, Loke YW. Molecular studies of trophoblast HLA-G: Polymorphism, isoforms, imprinting and expression in preimplantation embryo. Tissue Antigens. 1999;53:1–13. doi: 10.1034/j.1399-0039.1999.530101.x. [DOI] [PubMed] [Google Scholar]
- 75.Rousseau P, et al. The 14 bp Deletion-Insertion Polymorphism in the 3′ UT Region of the HLA-G Gene Influences HLA-G mRNA Stability. Hum. Immunol. 2003;64:1005–1010. doi: 10.1016/j.humimm.2003.08.347. [DOI] [PubMed] [Google Scholar]
- 76.Porto IOP, et al. MicroRNAs targeting the immunomodulatory HLA-G gene: A new survey searching for microRNAs with potential to regulate HLA-G. Mol. Immunol. 2015;65:230–241. doi: 10.1016/j.molimm.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 77.Altshuler DM, et al. An integrated map of genetic variation from 1092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang X, et al. Evidence that miR-133a causes recurrent spontaneous abortion by reducing HLA-G expression. Reprod. Biomed. Online. 2012;25:415–424. doi: 10.1016/j.rbmo.2012.06.022. [DOI] [PubMed] [Google Scholar]
- 79.Zhu, X. ming et al. Overexpression of miR-152 leads to reduced expression of human leukocyte antigen-G and increased natural killer cell mediated cytolysis in JEG-3 cells. Am. J. Obstet. Gynecol.202, 592.e1–592.e7 (2010). [DOI] [PubMed]
- 80.Manaster I, et al. MiRNA-mediated control of HLA-G expression and function. PLoS ONE. 2012;7:1–9. doi: 10.1371/journal.pone.0033395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kulkarni S, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–498. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yie SM, Li LH, Xiao R, Librach CL. A single base-pair mutation in the 3′-untranslated region of HLA-G mRNA is associated with pre-eclampsia. Mol. Hum. Reprod. 2008;14:649–653. doi: 10.1093/molehr/gan059. [DOI] [PubMed] [Google Scholar]
- 83.Hedrick PW, Thomson G. Evidence for balancing selection at HLA. Genetics. 1983;104:449–456. doi: 10.1093/genetics/104.3.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meyer D, Vitor VR, Bitarello BD, Débora DY, Nunes K. A genomic perspective on HLA evolution. Immunogenetics. 2018;70:5–27. doi: 10.1007/s00251-017-1017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brandt DYC, César J, Goudet J, Meyer D. The effect of balancing selection on population differentiation: A study with HLA genes. G3 Genes Genomes Genet. 2018;8:2805–2815. doi: 10.1534/g3.118.200367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tan Z, Shon AM, Ober C. Evidence of balancing selection at the HLA-G promoter region. Hum. Mol. Genet. 2005;14:3619–3628. doi: 10.1093/hmg/ddi389. [DOI] [PubMed] [Google Scholar]
- 87.Gineau L, et al. Balancing immunity and tolerance: Genetic footprint of natural selection in the transcriptional regulatory region of HLA-G. Genes Immun. 2015;16:57–70. doi: 10.1038/gene.2014.63. [DOI] [PubMed] [Google Scholar]
- 88.Andrés AM, et al. Targets of balancing selection in the human genome. Mol. Biol. Evol. 2009;26:2755–2764. doi: 10.1093/molbev/msp190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan WH, Fan LA. Residues Met76 and Gln79 in HLA-G α1 domain involved in KIR2DL4 recognition. Cell Res. 2005;15:176–182. doi: 10.1038/sj.cr.7290283. [DOI] [PubMed] [Google Scholar]
- 90.Gobin SJP, Van Den Elsen PJ. Transcriptional regulation of the MHC class Ib genes HLA-E, HLA-F and HLA-G. Hum. Immunol. 2000;61:1102–1107. doi: 10.1016/s0198-8859(00)00198-1. [DOI] [PubMed] [Google Scholar]
- 91.Flajollet S, Poras I, Carosella ED, Moreau P. RREB-1 Is a Transcriptional Repressor of HLA-G. J. Immunol. 2009;183:6948–6959. doi: 10.4049/jimmunol.0902053. [DOI] [PubMed] [Google Scholar]
- 92.Moreau P, Flajollet S, Carosella ED. Non-classical transcriptional regulation of HLA-G: An update. J. Cell. Mol. Med. 2009;13:2973–2989. doi: 10.1111/j.1582-4934.2009.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rousseau P, et al. In vivo, RFX5 binds differently to the human leucocyte antigen-E, -F, and -G gene promoters and participates in HLA class I protein expression in a cell type-dependent manner. Immunology. 2004;111:53–65. doi: 10.1111/j.1365-2567.2003.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.