Abstract
Lithium is the gold standard therapy for Bipolar Disorder (BD) but its effectiveness differs widely between individuals. The molecular mechanisms underlying treatment response heterogeneity are not well understood, and personalized treatment in BD remains elusive. Genetic analyses of the lithium treatment response phenotype may generate novel molecular insights into lithium’s therapeutic mechanisms and lead to testable hypotheses to improve BD management and outcomes. We used fixed effect meta-analysis techniques to develop meta-analytic polygenic risk scores (MET-PRS) from combinations of highly correlated psychiatric traits, namely schizophrenia (SCZ), major depression (MD) and bipolar disorder (BD). We compared the effects of cross-disorder MET-PRS and single genetic trait PRS on lithium response. For the PRS analyses, we included clinical data on lithium treatment response and genetic information for n = 2283 BD cases from the International Consortium on Lithium Genetics (ConLi+Gen; www.ConLiGen.org). Higher SCZ and MD PRSs were associated with poorer lithium treatment response whereas BD-PRS had no association with treatment outcome. The combined MET2-PRS comprising of SCZ and MD variants (MET2-PRS) and a model using SCZ and MD-PRS sequentially improved response prediction, compared to single-disorder PRS or to a combined score using all three traits (MET3-PRS). Patients in the highest decile for MET2-PRS loading had 2.5 times higher odds of being classified as poor responders than patients with the lowest decile MET2-PRS scores. An exploratory functional pathway analysis of top MET2-PRS variants was conducted. Findings may inform the development of future testing strategies for personalized lithium prescribing in BD.
Subject terms: Bipolar disorder, Pharmacogenomics
Introduction
Bipolar affective disorder (BD) is a severe and often chronic psychiatric illness, characterized by recurrent dysregulation of mood with alternating episodes of mania and depression. BD affects an estimated 48.8 million people globally and is associated with an early disease onset accounting for 9.9 million years of life lived with disability worldwide [1]. All-cause mortality and risk of suicide [2] are substantially increased in people with the disorder. Both genetic and environmental factors have been identified that contribute to the pathogenesis of BD [3] but the underlying molecular biology remains poorly understood, largely due to substantial genetic and clinical heterogeneity.
Lithium occupies a status as the ‘gold standard’ treatment amongst the mood-stabilizing medicines used for acute and maintenance therapy in BD [4–6]. It possesses strong anti-manic properties [7, 8] and is protective against further episodes of both manic and depressive polarity6, making it more effective in preventing re-hospitalizations than other BD medicines [9]. Furthermore, lithium has proven anti-suicidal properties [10]. Lithium is recommended as a first-line option for anti-manic and maintenance treatment by several clinical practice guidelines [11–14]. However, lithium therapy comes with several caveats. First, the therapeutic response to lithium in BD is highly heterogeneous. In acute mania, about 65% of patients respond at least partially to lithium monotherapy while 35% are refractory [15, 16]. In maintenance treatment, an excellent long-term response is reported only for ~30% of patients, whereas 30% have intermediate outcomes and 30% respond poorly [17]. Second, lithium is toxic at excessive doses and plasma levels need to be carefully monitored for the duration of treatment [18]. Third, lithium has long-term adverse effects including suppression of thyroid function and is associated with an elevated risk of renal failure [5].
To facilitate more personalized prescribing of lithium or alternative BD medicines, there is a need to define clinical and biological markers that could stratify BD patients into treatment response groups, ensuring that lithium is recommended at the earliest opportunity to patients most likely to respond, and is avoided in those who will likely derive no clinical benefits. Certain clinical factors (e.g. absence of physical co-morbidity [19, 20]; manic index episode [19]; family history of favourable lithium treatment [21] are associated with better treatment response, but none of these variables are sufficient to guide lithium prescribing on an individual basis. Among biological markers, both individual genetic variants (i.e. single nucleotide polymorphisms, SNPs [17] and the cumulative burden of many genetic variants (i.e. polygenic risk scores, PRS) have been identified that differentiate lithium response groups. We recently reported that BD patients with high genetic loading for schizophrenia, as calculated by PRS analysis, are less likely to have favourable lithium treatment outcomes than BD patients with low schizophrenia PRS [22]. Similarly, patients with high loadings for major depression (MDD) PRS were less likely to respond well to lithium than those with low MDD-PRS loading [23].
From these findings, several new research questions arise. First, does a combination of highly correlated genetic traits improve the predictive accuracy of the PRS approach? Previous studies have demonstrated that SCZ, BD, and MDD show substantial genetic overlap [24–26] and share biological pathways involved in disease pathophysiology [24, 25, 27]. The generation of multi-trait genomic risk scores from these genetically related disorders (SCZ, MDD, and BD) has been shown to improve diagnostic prediction accuracy [28], and therefore may be an avenue towards better translation of genetic findings into practice [29]. Second, while high SCZ and MDD-PRS were independently shown to predict poorer overall treatment response [22], other genomic signatures could predict better response, or different forms of response (e.g. level of agitation, depression, or cognition); hence, various combinations of PRS might help stratify or refine the response prediction. Third, related traits that are enriched with variants that predict the same direction of response (i.e. high load = poor response) might share certain key genetic variants, which influence response, enabling their molecular dissection.
In the current study, we investigated the contribution of variants from three genome-wide association studies (GWAS) in SCZ, MDD, and BD to lithium response in BD patients, and examined whether PRSs that are derived from cross-disorder meta-analysis of individual disorder GWAS improve genetic prediction. Further, we explored the relative individual contribution of disease-specific PRS to the combined PRS and investigated whether these were characterized by overlapping or divergent sets of genetic variants. Finally, to derive insights into molecular pathways that might influence lithium pharmacodynamics in BD, we conducted a pathway analysis of the top SNPs from the combined PRS analysis.
Methods
Discovery GWAS summary datasets
The GWAS summary statistics for SCZ, MDD, and BD were each obtained from the Psychiatric Genetics Consortium (PGC; http://www.med.unc.edu/pgc/). The SCZ dataset included 36,989 patients with SCZ and 113,075 healthy controls, including a subset of individuals with East Asian ancestry [26]. The MDD dataset was produced from a meta-analysis of seven cohorts (deCODE, Generation Scotland, GERA, iPSYCH, UK Biobank, PGC29 and 23andMe) containing 135,458 MD cases and 344,901 healthy controls [30]. The BD dataset was obtained from Stahl et al., 2019 [31], and the discovery sample was produced from a meta-analysis of 19,112 BD cases and 31,356 healthy controls after excluding 1242 individuals (1240 cases, 2 controls) who constituted the ConLi+Gen cohorts in the original data. Cross-disorder meta-analysis of the above summary statistics was performed using METACARPA [32], which accounts for unknown sample overlaps across the studies. If the summary statistics of multiple GWAS involve overlapping samples then the beta values across the GWAS are expected to be correlated (these correlations are functions of overlapping sample sizes) and failure to account these correlations would inflate the fixed effect inverse variance weighted met-analysis beta values [33]. When the exact number of samples overlapping across the studies are unknown, the correlations among betas can be empirically estimated. In a p-value based meta-analysis, the correlations between summary statistics were estimated using tetrachoric coefficients of truncated GWAS z-statistics instead of the Pearson correlations [34]. METACARPA uses this correlation matrix of beta values to account for the covariance between the GWAS beta values and implements computed meta-analysis statistics using Lin and Sullivan’s [33] method. The meta-summary statistics of MD, SCZ, and BD (MET3) and MD and SCZ (MET2) and Wald p-values produced METACARPA were employed for PRS calculation.
Target study sample
For the PRS analysis, clinical data on lithium treatment response and genetic information were obtained from the International Consortium on Lithium Genetics (ConLi+Gen; www.ConLiGen.org). A sample of n = 2283 BD patients with complete phenotype information was included in the analysis [23].
Target outcome
Lithium treatment response was assessed using the validated “Retrospective Criteria of Long-Term Treatment Response in Research Subjects with Bipolar Disorder” scale (the Alda scale) [35–37]. The instrument quantifies symptom improvement in BD by lithium over the course of treatment. Global improvement is rated with an “A” score (range 0–10), which is then weighted against five criteria (Alda B score) that assess the quality of evidence for the response score [17], to arrive at a total Alda score. For dichotomized assessment of treatment response, patients with a total score of 7 or higher were categorized as “good responders”, and the remainder were categorized as poor responders [17, 37]. For continuous assessment of treatment response, total Alda scores were used [38].
Genotyping, quality control, and imputation
Genotyping and imputation in the ConLi+Gen cohorts have been described in detail elsewhere [17]. Briefly, genotyping across multiple cohorts was performed using several commercial SNP arrays (Affymetrix 6·0, Human610/660 W, HumanOmniExpress, HumanOmni1-Quad, HumanOmni2.5). The 1000 Genomes reference panel was used to impute additional genotypes as implemented in SHAPEIT2 [39] and minimac2 [40]. Quality control filters for the SNPs, including minor allele frequency (MAF) < 0.01, imputation quality score R-square < 0.6 and Hardy-Weinberg test of equilibrium for genotype frequencies (HWE) p-value ≤ 1e-6, were applied to the imputed data of each cohort. From the imputed dosage score, genotype calls for the filtered SNPs were derived and common sets of SNPs across the cohorts were merged using PLINK [41]. Merged genotype data from all the cohorts underwent additional post-merge quality control, and were used for creating PRSs.
Polygenic Risk Score (PRS) analysis
The posterior effect sizes of GWAS and meta-analysis summary statistics were first obtained using the software PRS-CS [32]. PRS-CS uses the Bayesian regression framework to obtain the posterior effect sizes of the GWAS summary statistics with continuous shrinkage priors on the effect sizes and utilizing the linkage disequilibrium (LD) among the SNPs [42]. This approach results in a pruned set of posterior effect sizes for the summary statistics without the need to threshold the p-values for PRS calculation. For the current analysis, the precomputed LD pattern of the 1000 Genomes European reference panel and the default priors for the effect sizes were used. The PRS scores were generated using PLINK based on the posterior effect sizes instead of the actual effect sizes of the GWAS and meta-analysis.
Statistical analysis
To assess the association of PRSs of individual traits (SCZ, BD, and MD), the cross-disorder meta-summary PRSs of SCZ, MD and BD (MET3), and SCZ and MD (MET2) with lithium treatment response, a binary logistic regression model was applied for the binary outcome (lithium response versus non-response). For association with Alda total, a tobit analysis model (censored regression) was used to account for the floor effect at zero. We used the following covariates for analysis: age, gender, 4 ethnicity principal components (PCs), site, and SNP chip type. The proportion of phenotypic variance explained by PRSs (partial R2) was calculated using the Nagelkerke method, as implemented in R package rsq [43]. R (version 4.0.0) [44] was used for data manipulations and statistical analyses. We also examined the joint effect of MDD-PRS and SCZ-PRS in a multivariate regression model with all the relevant covariates for comparison with MET2-PRS. The incremental R2 for the two PRS scores was computed as the difference in R2 of the models fit with and without the two PRS scores.
For all PRSs, we divided the study sample into deciles, ranging from the lowest polygenic load (1st decile, reference group) to the highest polygenic load (10th decile). Then, we compared lithium response rates in BD patients in the higher polygenic load deciles (2nd–10th deciles) with patients in the lowest polygenic load decile (1st decile).
We performed a power analysis for each PRS using the R package avengeme [45] with the input parameters heritability on liability, the number of independent SNPs, disease prevalence, and proportion of null markers. Heritability and prevalence for schizophrenia and bipolar disorder were obtained from Table 1 in Wray et al., 2010 [46]; the number of independent SNPs as produced by the PRS-CS package and the proportion of null markers were set as 90%.
Table 1.
Characteristics of the patient sample.
| Characteristic | Lithium responders (Alda Total Score ≥ 7) | Lithium non-responders (Alda Total Score ≤ 6) |
|---|---|---|
| N (%) | 637 (27.9) | 1646 (72.1) |
| Female N (%) | 352 (55.3) | 950 (57.7) |
| Age mean (SD) | 50.23 (14.71) | 45.92 (13.47) |
| Alda A mean (SD) | 9.23 (0.79) | 5.07 (2.7) |
| Alda B mean (SD) | 1.14 (0.94) | 3.04 (1.61) |
SD standard deviation.
Functional analyses
To explore the biological context and potential mechanistic underpinnings of the SNPs discovered from the cross-disorder meta-analyses, we performed pathway analyses using Qiagen’s Ingenuity Pathway Analysis (IPA®, QIAGEN Redwood City, CA, USA, www.qiagen.com/ingenuity). To prepare the input genes for IPA®, we followed a two-step approach: first, all SNPs from the MET2-PRS that showed associations with both SCZ and MDD at meta-analysis threshold p < 5 × 10−8 were selected; second, SNPs from step 1 were mapped to their hosting genes using ANNOVAR software [47]. For intergenic SNPs, ANNOVAR provides distance to the nearest upstream and downstream gene. The SNPs were mapped to the closest genes using this distance (range: 0-796688 BP). This final list of mapped genes was entered into IPA® (content version: 52912811, 2020).
IPA compares the proportion of input genes mapping to a biological pathway to the reference gene list from the ingenuity databases. Molecule relationships previously experimentally observed in human, mouse, rat, and uncategorized species were included. The significance of the over-represented canonical pathways and functional networks is determined using the right-tailed Fisher’s exact test and later adjusted for multiple testing using the Benjamini-Hochberg (BH) method. Significant results are determined at BH adjusted p-value < 0.01.
Results
Sample
Details of the ConLi+Gen sample of patients with BD have been published previously [17]. For the current analyses, genetic and clinical data from n = 2283 patients were used (57% females). 27.9% of patients were classified as excellent lithium responders (Alda Total Score ≥ 7), whereas 72.1% were classified as non-responders (Alda Total Score ≤ 6) (Table 1).
Associations of individual and combined SCZ, MDD, and BD-PRS with lithium treatment response
The dichotomous lithium treatment response outcome (Alda total ≥ 7, indicating excellent vs non-responders) was significantly associated with SCZ-PRS (p = 0.0005) and MDD-PRS (p = 0.009), but not with BD-PRS (p = 0.24) (Fig. 1, Supplementary Table 1). Power analyses for each PRS indicated that the sample sizes required for the training dataset for 80% power were similar for schizophrenia (n = 5537) and bipolar disorder (n = 5741), suggesting that the observed lack of association between BD-PRS and lithium response may not be due to lack of power.
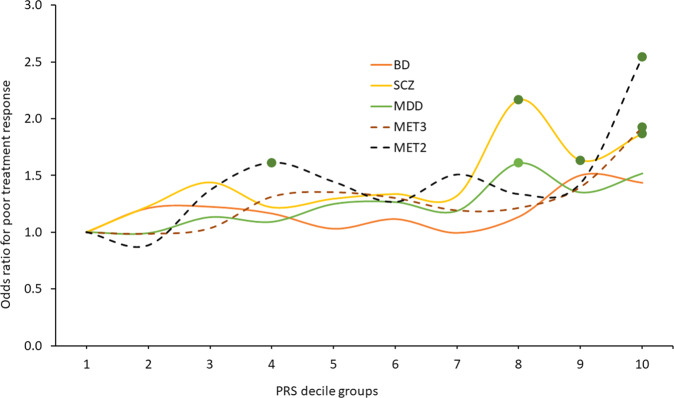
Fig. 1. The associations of polygenic risk scores (PRS) for bipolar disorder (BD), schizophrenia (SCZ), major depression (MDD), meta-SCZ/MDD/BD (MET3), and meta-SCZ/MDD (MET2) with dichotomous lithium treatment response (Alda total ≥ 7).
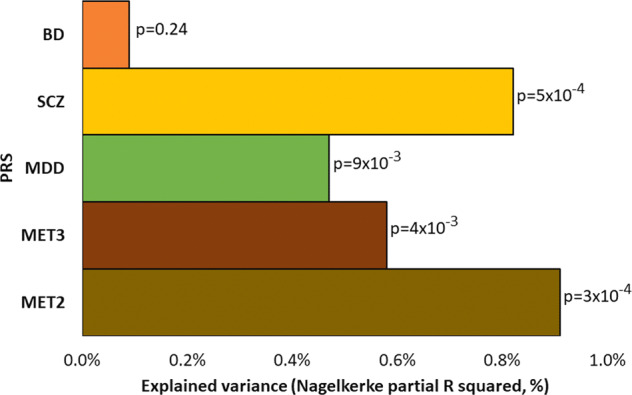
The x-axis refers to the percentage of explained variance in treatment response to lithium accounted for by the PRS (Nagelkerke partial R2). The y-axis plots the PRS for BD, SCZ, MDD, MET3, and MET2. Each bar is labelled with the p-values for the association between the PRS and lithium treatment response.
Significant associations were also shown for MET3-PRS (comprised of SCZ, MDD, and BD; p = 0.004) and MET2-PRS (comprised of SCZ and MDD; p = 0.0003). Each of these associations remained significant after adjustment for multiple testing (p-threshold = 0.05/5 = 0.01). The proportion of phenotype variance explained by MET2-PRS (partial R2 = 0.91%) was higher than for SCZ-PRS (partial R2 = 0.82%), MDD-PRS (partial R2 = 0.47%), and MET3-PRS (partial R2 = 0.58%) (Fig. 1, Supplementary Table 1).
We also examined the joint effect of MDD-PRS and SCZ-PRS in a multivariate regression model with all the relevant covariates for comparison with MET2-PRS. The incremental R2 for the two PRS scores was found to be 0.92%, similar to MET2-PRS R2. The joint p-value for the two scores was p = 0.0002, again similar to the MET2-PRS p-value (supplementary Table 1).
The continuous lithium response outcome Alda total, using a tobit regression model, was significantly associated with SCZ-PRS (p = 0.0095), MD-PRS (p = 0.0029), MET3-PRS (0.0044), and MET2-PRS (p = 0.0003). BD-PRS was not associated with Alda total (Supplementary Table 2). PRS beta values for all associations were negative, indicating a consistent direction of effect of PRS for SCZ, MDD, MET3, and MET2 on lithium response as measured by Alda total.
To further evaluate the impact of the individual and combined PRS on lithium treatment response, we divided the study population into deciles based on their polygenic loading. Samples were grouped based on the deciles for PRS for BD, SCZ, MDD, MET3, and MET2 (n = 228 in each decile group). Decile group 1 (reference group) contained individuals with the lowest genetic loading for the respective PRS, whereas decile group 10 represented those with the highest loading.
Individuals with higher polygenic loading for any of the tested PRS except BD-PRS were at increased risk of experiencing a poor therapeutic response to lithium (Fig. 2). Compared to the low PRS (1st decile) reference groups, odds ratios (ORs) for poor response were significantly different (p < 0.05) for SCZ-PRS (8th decile: OR = 2.16, p = 0.001; 9th decile: OR 1.63, p = 0.04; 10th decile: OR = 1.87, p = 0.04), MDD-PRS (8th decile: OR = 1.61, p = 0.04), MET3-PRS (10th decile: OR = 1.92, p = 0.03), and MET2-PRS (4th decile: OR = 1.61, p = 0.03; 10th decile: OR = 2.54, p = 0.002). (Fig. 2, Supplementary Table 3).
Fig. 2. Odds ratios (ORs) for unfavourable treatment response to lithium (Alda score ≤ 6) in patients with BD.
ORs are derived by comparing patients with higher polygenic loads (PRS deciles 2–10) for BD (orange line), SCZ (yellow line), MDD (green line), meta-(SCZ/MDD/BD) (MET3, maroon dashed line), and meta-(SCZ/MDD) (MET2, brown dotted line) with patients with the lowest respective polygenic loads (PRS 1st decile). Dots indicate p < 0.05. Analyses were adjusted for gender, age, 4 PCs, site, and SNP chip type.
Pearson correlations between disease-specific and meta-PRS were calculated. As shown in Supplementary Table 4, the correlation between SCZ-PRS and MET2-PRS was r = 0.83, whereas MDD-PRS and MET2-PRS had a correlation of r = 0.6.
Exploratory functional analysis of top SCZ-MDD-PRS (MET2) variants
Eight thousands six hundred nineteen SNPs from the MDD and SCZ meta-analysis in METACARPA (MET2) at p ≤ 5 × 10−8 and in consistent direction in both GWAS studies were selected to explore the biological context of overlapping risk alleles (Supplementary Table 5). Of note, a PRS calculated from these selected 8619 SNPs was not by itself associated with lithium response (data not shown). By mapping SNPs to hosting- and nearby genes (distance to nearest gene is shown in Supplementary Table 5), we derived a list of 270 genes (Supplementary Table 6). 106 (41%) of these genes intersected with SCZ genes (GWAS p-value ≤ 5 × 10−8); 73 (29%) intersected with MDD genes (GWAS p-value ≤ 5 × 10−8); 37 (14%) intersected with both SCZ and MDD genes (Supplementary Table 6). Of the 270 MET2 genes, IPA® could unequivocally identify 256 genes in its database for enrichment and pathway analysis (Supplementary Table 7).
IPA® examined six categories of enrichment in which genes from the MET2 list were over-represented, including: top canonical pathways; upstream regulators; diseases & disorders; molecular & cellular functions; physiological system functions; and networks (Supplementary Table 8). The majority of these associations were driven by genes involved in histone biology (e.g. H4C1, H4C2, H4C3, H4C4, H4C8, H4C11, H4C12, H4C13; H3C2, H3C3, H3C8, H3C10, H3C11, H3C12; H1-1, H1-2, H1-4, H1-5, H1-6). Over-representation of MET2 genes was found for IPA® disease-related pathways in endocrine system disorders, gastrointestinal disease, immunological disease, metabolic disease, and organismal injury and abnormalities (Supplementary Table 8). Associations with the first three disease categories (endocrine, gastrointestinal, and immunological) was most strongly driven by MET2 genes over-represented in IPA®-defined genesets relating to insulin-dependent diabetes mellitus, diabetes mellitus, and glucose metabolism disorder (Supplementary Table 9).
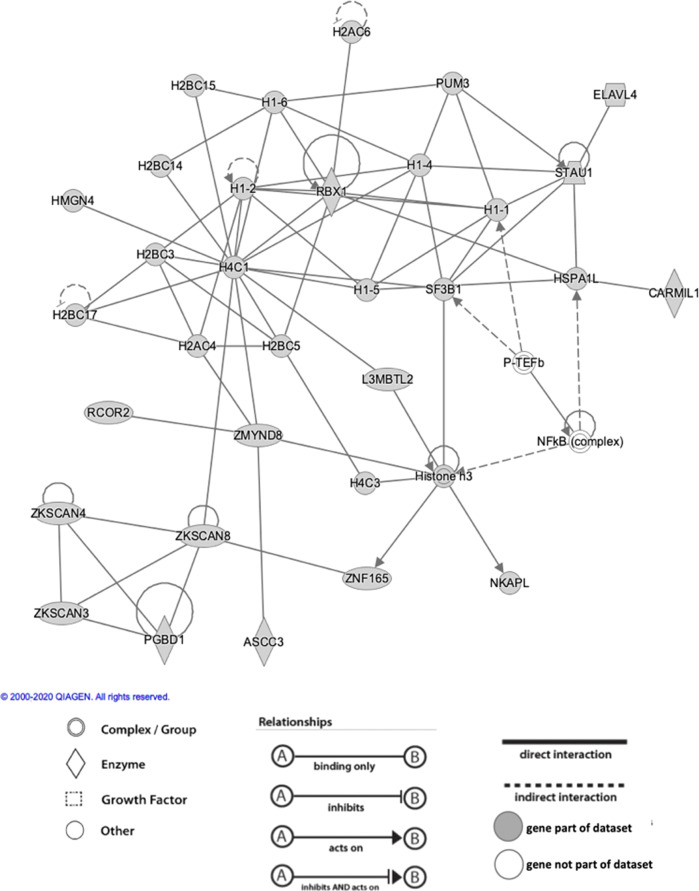
IPA® also calculated networks of the functionally most connected MET2 genes. Network 1 (IPA® score 67) was annotated to endocrine system disorders, gastrointestinal disease, and immunological disease, and identified H4 clustered histone 1 (H4C1), histone h3, and E3 ubiquitin-protein ligase (RBX1) as nodes with the highest number of network connections (Fig. 3, Supplementary Table 8). Network 2 (IPA® score 39) was annotated with cell cycle, cellular assembly and organization, and DNA replication, recombination, and repair. Adenosine triphosphate (ATP), estrogen receptor, and EP300 (encoding p300, a histone acetyltransferase) were identified as nodes with most network interactions (Supplementary Fig. 1). Networks 3, 4, and 5 (each with IPA® score 32), respectively, identified tumor necrosis factor (TNF), extracellular-signal-regulated kinases (ERK1/2), and heat shock protein family D member 1 (HSPD1) as most highly connected nodes (Supplementary Figs. 2–4).
Fig. 3. IPA® top network of MET2 genes associated with SCZ and MDD at p < 5 × 10−8.
Annotated network functions include endocrine system disorders, gastrointestinal disease, immunological disease. H4 clustered histone 1 (H4C1), histone H3, and E3 ubiquitin-protein ligase (RBX1) are identified as nodes with most network interactions.
Discussion
To our knowledge, this is the first study analysing both individual and shared contributions of polygenic risk scores (PRS) for SCZ, MDD, and BD to the therapeutic response to lithium in individuals with bipolar disorder. Placing continuous shrinkage (CS) priors on SNP effect sizes for calculating PRS [42], we found that high genetic loadings for SCZ and MDD but not BD reduce patients’ likelihood of optimal clinical lithium response. We also found that a combined PRS derived from GWAS meta-analysis of MDD and SCZ (MET2-PRS) improves genetic response prediction, compared with individual disorder PRS and with a combined PRS comprising SCZ, MDD, and BD (MET3). The 228 patients with the highest MET2 loading in our cohort (top 10%) had 2.5 times greater odds of experiencing a poor clinical response to lithium, compared to the 228 patients with the lowest loading (bottom 10%). Exploratory bioinformatic pathway analysis (IPA®) of the genes containing MET2 variants most strongly associated with both MDD and SCZ (p < 5 × 10−8) implicated histone biology and genesets relating to metabolic disorders such as diabetes mellitus.
Our finding of negative associations between high loadings of SCZ-PRS and MDD-PRS with poor lithium response confirms our previous studies investigating these psychiatric traits in the ConLi+Gen cohort [22, 23]. Results are consistent with the clinical observation of poorer lithium response in BD patients who have a family history of SCZ, as opposed to a family history of BD [36], and support the idea that better lithium responsiveness is associated with a ‘core’ bipolar phenotype in the form of manic depression [5]. The specificity of SCZ- and MDD-PRS but not BD-PRS in predicting response is noteworthy, especially on the background of an 80% overlap in our study between SCZ-PRS and BD-PRS contrasting a 13% overlap between SCZ-PRS and MDD-PRS. Findings suggest that the genetic variants which relate to lithium responsiveness are operating independently to the risk of BD. How such trait-specificity is explained needs to be explored in future research. For example, there could be shared and separate sets of SCZ and MDD-risk alleles influencing lithium treatment response that are not contained within the BD-PRS. Alternatively, certain variants within the BD-PRS could be protective against the negative impact of shared SCZ and MDD-risk alleles.
The superior performance of MET2 in predicting lithium response, as compared to individual disorders, is consistent with previous work showing that multi-trait genomic risk scores can improve prediction accuracy [28]. It is notable that MET3, showing a 96% overlap with MET2, was not superior to PRS derived for individual diseases. Therefore, for genetic prediction of clinical outcomes, our study encourages a workflow that assesses single-trait associations first, and in a second step considers combined PRS using significantly associated traits only, irrespective of a priori single PRS correlations.
Our exploratory IPA® pathway analysis of top variants associated with both SCZ and MDD within the MET2-PRS proposes several overlapping biological mechanisms in these disorders. Some of these shared SCZ-MDD pathways may, in turn, play a role in lithium response. For example, the high prevalence of histone genes within our SCZ-MDD gene set may support findings from in vitro, animal and human studies demonstrating an influence of lithium on gene expression through epigenetic mechanisms including histone modification (for review, see ref. [48]) and the recent notion of epigenetic signatures as a potential molecular predictor of lithium response in BD [49]. Studies have shown that lithium treatment increases acetylation and methylation of histone H3 in rat hippocampus [50], and that it suppresses histone deacetylases (HDACs) [51]. Similarly, the high prevalence of genes within MET2 linked to diabetes mellitus and glucose metabolism echoes clinical-epidemiological studies showing that BD patients with comorbid diabetes mellitus or insulin resistance have poorer responses to lithium [20].
Our study has a number of limitations. First, the variance in lithium response that can be explained by the PRS method remains small (partial R2 = 0.91%), and it is unlikely that PRSs as stand-alone biological markers can be translated into clinical practice. However, genetic scores might be used in concert with other biological parameters and with clinical patient characteristics to form predictive algorithms of lithium treatment response. Machine-learning approaches to generate such multivariate algorithms have recently shown promise [52]. Second, our study used a lithium response measure (the Alda scale) focusing on long-term protection from manic and depressive relapses. Other forms of response to lithium (e.g. reduction of agitation in acute mania, reduction in overall suicidality) are less well captured by the Alda scale but are clinically important. Third, SZ-PRS and MDD-PRS are more statistically powerful than BD-PRS as they have been calculated on larger populations. This might partly explain the low contribution of BD-PRS in discriminating responders and non-responders, even though our power analysis indicated that the patient numbers required for each PRS are similar and were sufficient in our study. Forth, our functional analysis of top MET2 variants is only exploratory, as a direct association of these variants to lithium response (in addition to their association with SCZ and MDD) cannot be demonstrated through the methods employed in this study. Further, we chose a heuristic p-value cut-off for inclusion of MET2 variants into our bioinformatic pathways analysis, which employed the traditional threshold for genome-wide significance (p < 5 × 10−8). More generous cut-offs, resulting in larger gene lists for input into IPA® analysis may have identified different or additional functional links to MET2 genes.
Conclusions
The application of multi-trait genomic risk scores revealed that a meta-PRS generated from a meta-GWAS of SCZ- and MDD-risk alleles (MET2-PRS), but not including BD-risk alleles, improves genetic prediction accuracy of the lithium response phenotype in BD compared to single-disorder PRS. High MET2-PRS loading is associated with poorer lithium response. Multi-trait PRS may inform the development of future biological testing strategies for personalized lithium prescribing in BD and may assist in the search for central therapeutic molecular mechanisms of this important drug.
Supplementary information
Author contributions
KOS, AT, ATA, JF, FS, MR, and BTB designed the study. KOS, AT, ATA, and BB conducted the analyses and drafted the manuscript. MA, NA, KA, RA, BA, J-MA, LB, AKB, FB, AB, SB, JMB, AB, CM-C, MC, PC, H-CC, CC, SC, SRC, CC, PMC, ND, AD, FD MDZ, JRD, BE, PF, AJF, LF, MAF, JMF, SG, JSG, FSG, MG-S, PG, RH, JH, UH, SH, PH, LH, Y-HH, SJ, EJ, J-PK, LK, P-HK, TK, JK, SK-S, EF-R, BK, IK, GL, ML, CL, ML, SGL, MM, MM, LM, MJM, SM, FC, MM, FMM, PM, CMN, MMN, TN, CO’D, NO, UO, SP, AP, CP, JBP, JBP, AR, ER, GAR, JKR, MS, PRS, BWS, GS, TS, PDS, KS, CS, CMS, AS, TS, PS, FT-A, AT, GT, JV, EV, SHW, GR, PPZ, MA, MB, FJM, PBM, and TGS contributed data, reviewed the manuscript, and made critical contributions to the revisions.
Funding
The study was supported by the German Federal Ministry of Education and Research (BMBF) through ERA-NET NEURON grants “SynSchiz—Linking synaptic dysfunction to disease mechanisms in schizophrenia—a multilevel investigation” (01EW1810 to MR) and “Impact of Early life MetaBolic and psychosocial strEss on susceptibility to mental Disorders; from converging epigenetic signatures to novel targets for therapeutic intervention” (01EW1904 to MR); and by the German Research Foundation (DFG grants FOR2107; RI908/11-2 to MR; WI 3439/3-2 to SHW). ATA is supported by 2019–2021 National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Grant from the Brain & Behaviour Research Foundation (BBRF) and National Health and Medical Research Council (NHMRC) Emerging Leadership Investigator Grant 2021 – 2008000. Open Access funding enabled and organized by Projekt DEAL.
Competing interests
ATA is supported by 2019–2021 National Alliance for Research on Schizophrenia and Depression (NARSAD) Young Investigator Grant from the Brain & Behaviour Research Foundation (BBRF). EV has received grants and served as consultant, advisor or CME speaker for the following entities: AB-Biotics, Allergan, Angelini, AstraZeneca, Bristol-Myers Squibb, Dainippon Sumitomo Pharma, Farmindustria, Ferrer, Forest Research Institute, Gedeon Richter, Glaxo-Smith-Kline, Janssen, Lundbeck, Otsuka, Pfizer, Roche, Sanofi-Aventis, Servier, Shire, Sunovion, Takeda, the Brain and Behaviour Foundation, the Spanish Ministry of Science and Innovation (CIBERSAM), and the Stanley Medical Research Institute. MB has received grants from the Deutsche Forschungsgemeinschaft (DFG), and Bundesministeriums für Bildung und Forschung (BMBF), and served as consultant, advisor or CME speaker for the following entities: Allergan, Aristo, Janssen, Lilly, Lundbeck, neuraxpharm, Otsuka, Sandoz, Servier, and Sunovion outside the submitted work. SK-S has received grants and served as consultant, advisor or speaker for the following entities: Medice Arzneimittel Pütter GmbH and Shire. BB has received grants and served as consultant, advisor or CME speaker for the following entities: AstraZeneca, Bristol-Myers Squibb, Janssen, Lundbeck, Otsuka, Servier, the National Health and Medical Research Council, the Fay Fuller Foundation, the James and Diana Ramsay Foundation. TK received honoraria for lectures, manuscripts, and/or consultancy, from Kyowa Hakko Kirin Co., Ltd., Eli Lilly Japan K.K., Otsuka Pharmaceutical Co., Ltd., GlaxoSmithKline K.K., Taisho Toyama Pharmaceutical Co., Ltd., Dainippon Sumitomo Pharma Co., Ltd., Meiji Seika Pharma Co., Ltd., Pfizer Japan Inc., Mochida Pharmaceutical Co., Ltd., Shionogi & Co., Ltd., Janssen Pharmaceutical K.K., Janssen Asia Pacific, Yoshitomiyakuhin, Astellas Pharma Inc., Wako Pure Chemical Industries, Ltd., Wiley Publishing Japan, Nippon Boehringer Ingelheim Co. Ltd., Kanae Foundation for the Promotion of Medical Science, MSD K.K., Kyowa Pharmaceutical Industry Co., Ltd. and Takeda Pharmaceutical Co., Ltd. TK also received a research grant from Takeda Pharmaceutical Co., Ltd. PF has received grants and served as consultant, advisor or CME speaker for the following entities Abbott, GlaxoSmithKline, Janssen, Essex, Lundbeck, Otsuka, Gedeon Richter, Servier and Takeda as well as the German Ministry of Science and the German Ministry of Health. ER has received grants and served as consultant, advisor or CME speaker for the following entities: Janssen and Institut Allergosan. ML declares that, over the past 36 months, he has received lecture honoraria from Lundbeck and served as a scientific consultant for EPID Research Oy; no other equity ownership, profit-sharing agreements, royalties, or patent. KA has received consulting honoraria from Taisho Toyama Pharmaceutical Co., Ltd. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Klaus Oliver Schubert, Anbupalam Thalamuthu, Azmeraw T. Amare.
A list of members and their affiliations appears in the Supplementary Information.
Change history
7/11/2022
A Correction to this paper has been published: 10.1038/s41398-022-02037-2
Supplementary information
The online version contains supplementary material available at 10.1038/s41398-021-01702-2.
References
- 1.Ferrari AJ, Stockings E, Khoo JP, Erskine HE, Degenhardt L, Vos T, et al. The prevalence and burden of bipolar disorder: findings from the Global Burden of Disease Study 2013. Bipolar Disord. 2016;18:440–50. doi: 10.1111/bdi.12423. [DOI] [PubMed] [Google Scholar]
- 2.Chesney E, Goodwin GM, Fazel S. Risks of all-cause and suicide mortality in mental disorders: a meta-review. World Psychiatry. 2014;13:153–60. doi: 10.1002/wps.20128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grande I, Berk M, Birmaher B, Vieta E. Bipolar disorder. Lancet. 2016;387:1561–72. doi: 10.1016/S0140-6736(15)00241-X. [DOI] [PubMed] [Google Scholar]
- 4.Miura T, Noma H, Furukawa TA, Mitsuyasu H, Tanaka S, Stockton S, et al. Comparative efficacy and tolerability of pharmacological treatments in the maintenance treatment of bipolar disorder: a systematic review and network meta-analysis. Lancet Psychiatry. 2014;1:351–9. doi: 10.1016/S2215-0366(14)70314-1. [DOI] [PubMed] [Google Scholar]
- 5.Malhi GS, Tanious M, Das P, Berk M. The science and practice of lithium therapy. Aust NZ J Psychiatry. 2012;46:192–211. doi: 10.1177/0004867412437346. [DOI] [PubMed] [Google Scholar]
- 6.Malhi GS, Adams D, Berk M. Is lithium in a class of its own? A brief profile of its clinical use. Aust NZ J Psychiatry. 2009;43:1096–104. doi: 10.3109/00048670903279937. [DOI] [PubMed] [Google Scholar]
- 7.Yildiz A, Vieta E, Leucht S, Baldessarini RJ. Efficacy of antimanic treatments: meta-analysis of randomized, controlled trials. Neuropsychopharmacology. 2011;36:375–89. doi: 10.1038/npp.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cipriani A, Barbui C, Salanti G, Rendell J, Brown R, Stockton S, et al. Comparative efficacy and acceptability of antimanic drugs in acute mania: a multiple-treatments meta-analysis. Lancet. 2011;378:1306–15. doi: 10.1016/S0140-6736(11)60873-8. [DOI] [PubMed] [Google Scholar]
- 9.Joas E, Karanti A, Song J, Goodwin GM, Lichtenstein P, Landén M. Pharmacological treatment and risk of psychiatric hospital admission in bipolar disorder. Br J Psychiatry. 2017;210:197–202. doi: 10.1192/bjp.bp.116.187989. [DOI] [PubMed] [Google Scholar]
- 10.Tondo L, Hennen J, Baldessarini RJ. Lower suicide risk with long-term lithium treatment in major affective illness: a meta-analysis. Acta Psychiatr Scand. 2001;104:163–72. doi: 10.1034/j.1600-0447.2001.00464.x. [DOI] [PubMed] [Google Scholar]
- 11.NICE. Bipolar Disorder: The Management of Bipolar Disorder in Adults, Children and Adolescents, in Primary and Secondary Care. Leicester (UK): NICE; 2006. [PubMed]
- 12.Yatham LN, Kennedy SH, Parikh SV, Schaffer A, Beaulieu S, Alda M, et al. Canadian Network for Mood and Anxiety Treatments (CANMAT) and International Society for Bipolar Disorders (ISBD) collaborative update of CANMAT guidelines for the management of patients with bipolar disorder: update 2013. Bipolar Disord. 2013;15:1–44. doi: 10.1111/bdi.12025. [DOI] [PubMed] [Google Scholar]
- 13.Malhi GS, Bassett D, Boyce P, Bryant R, Fitzgerald PB, Fritz K, et al. Royal Australian and New Zealand College of Psychiatrists clinical practice guidelines for mood disorders. Aust NZ J Psychiatry. 2015;49:1087–206. doi: 10.1177/0004867415617657. [DOI] [PubMed] [Google Scholar]
- 14.Goodwin GM, Haddad PM, Ferrier IN, Aronson JK, Barnes T, Cipriani A, et al. Evidence-based guidelines for treating bipolar disorder: revised third edition recommendations from the British Association for Psychopharmacology. J Psychopharmacol. 2016;30:495–553. doi: 10.1177/0269881116636545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller F, Tanenbaum JH, Griffin A, Ritvo E. Prediction of treatment response in bipolar, manic disorder. J Affect Disord. 1991;21:75–77. doi: 10.1016/0165-0327(91)90052-T. [DOI] [PubMed] [Google Scholar]
- 16.Machado-Vieira R, Luckenbaugh DA, Soeiro-de-Souza MG, Marca G, Henter ID, Busnello JV, et al. Early improvement with lithium in classic mania and its association with later response. J Affect Disord. 2013;144:160–4. doi: 10.1016/j.jad.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou L, Heilbronner U, Degenhardt F, Adli M, Akiyama K, Akula N, et al. Genetic variants associated with response to lithium treatment in bipolar disorder: a genome-wide association study. Lancet. 2016;387:1085–93. doi: 10.1016/S0140-6736(16)00143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schubert KO, Wisdom A. Should the Australian Therapeutic Goods Administration recommend rapid dosing of lithium carbonate in acute mania? Aust NZ J Psychiatry. 2017. https://pubmed.ncbi.nlm.nih.gov/29216733/. [DOI] [PubMed]
- 19.Kessing LV, Hellmund G, Andersen PK. Predictors of excellent response to lithium: results from a nationwide register-based study. Int Clin Psychopharmacol. 2011;26:323–8. doi: 10.1097/YIC.0b013e32834a5cd0. [DOI] [PubMed] [Google Scholar]
- 20.Calkin CV, Ruzickova M, Uher R, Hajek T, Slaney CM, Garnham JS, et al. Insulin resistance and outcome in bipolar disorder. Br J Psychiatry. 2015;206:52–57. doi: 10.1192/bjp.bp.114.152850. [DOI] [PubMed] [Google Scholar]
- 21.Grof P, Duffy A, Cavazzoni P, Grof E, Garnham J, MacDougall M, et al. Is response to prophylactic lithium a familial trait? J Clin Psychiatry. 2002;63:942–7. doi: 10.4088/JCP.v63n1013. [DOI] [PubMed] [Google Scholar]
- 22.International Consortium on Lithium Genetics. Amare AT, Schubert KO, Hou L, Clark SR, Papiol S, et al. Association of polygenic score for schizophrenia and HLA antigen and inflammation genes with response to lithium in bipolar affective disorder: a genome-wide association study. JAMA Psychiatry. 2018;75:65–74. doi: 10.1001/jamapsychiatry.2017.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amare AT, Schubert KO, Hou L, Clark SR, Papiol S, Cearns M, et al. Association of polygenic score for major depression with response to lithium in patients with bipolar disorder. Mol Psychiatry. 2020. https://pubmed.ncbi.nlm.nih.gov/32203155/. [DOI] [PubMed]
- 24.Cross-Disorder Group of the Psychiatric Genomics Consortium. et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brainstorm Consortium, et al. Analysis of shared heritability in common disorders of the brain. Science. 2018;360. https://pubmed.ncbi.nlm.nih.gov/29930110/. [DOI] [PMC free article] [PubMed]
- 26.Schizophrenia Working Group of the Psychiatric Genomics C. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gandal MJ, Haney JR, Parikshak NN, Leppa V, Ramaswami G, Hartl C, et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science. 2018;359:693–7. doi: 10.1126/science.aad6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maier R, Moser G, Chen G-B, Ripke S, Cross-Disorder Working Group of the Psychiatric Genomics Consortium. Coryell W, et al. Joint analysis of psychiatric disorders increases accuracy of risk prediction for schizophrenia, bipolar disorder, and major depressive disorder. Am J Hum Genet. 2015;96:283–94. doi: 10.1016/j.ajhg.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Rheenen W, Peyrot WJ, Schork AJ, Lee SH, Wray NR. Genetic correlations of polygenic disease traits: from theory to practice. Nat Rev Genet. 2019;20:567–81. doi: 10.1038/s41576-019-0137-z. [DOI] [PubMed] [Google Scholar]
- 30.Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat Genet. 2018;50:668–81. doi: 10.1038/s41588-018-0090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. 2019;51:793–803. doi: 10.1038/s41588-019-0397-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Southam L, Gilly A, Süveges D, Farmaki A-E, Schwartzentruber J, Tachmazidou I, et al. Whole genome sequencing and imputation in isolated populations identify genetic associations with medically-relevant complex traits. Nat Commun. 2017;8:15606. doi: 10.1038/ncomms15606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin DY, Sullivan PF. Meta-analysis of genome-wide association studies with overlapping subjects. Am J Hum Genet. 2009;85:862–72. doi: 10.1016/j.ajhg.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Province MA, Borecki IB. A correlated meta-analysis strategy for data mining “OMIC” scans. Pac Symp Biocomput. 2013:236–46. https://pubmed.ncbi.nlm.nih.gov/23424128/. [PMC free article] [PubMed]
- 35.Duffy A, Alda M, Milin R, Grof P. A consecutive series of treated affected offspring of parents with bipolar disorder: is response associated with the clinical profile? Can J Psychiatry. 2007;52:369–76. doi: 10.1177/070674370705200606. [DOI] [PubMed] [Google Scholar]
- 36.Garnham J, Munro A, Slaney C, Macdougall M, Passmore M, Duffy A, et al. Prophylactic treatment response in bipolar disorder: results of a naturalistic observation study. J Affect Disord. 2007;104:185–90. doi: 10.1016/j.jad.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 37.Manchia M, Adli M, Akula N, Ardau R, Aubry J-M, Backlund L, et al. Assessment of response to lithium maintenance treatment in bipolar disorder: A Consortium on Lithium Genetics (ConLiGen) Report. PLoS ONE. 2013;8:e65636. doi: 10.1371/journal.pone.0065636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott J, Etain B, Manchia M, Brichant-Petitjean C, Geoffroy PA, Schulze T, et al. An examination of the quality and performance of the Alda scale for classifying lithium response phenotypes. Bipolar Disord. 2019. https://pubmed.ncbi.nlm.nih.gov/31466131/. [DOI] [PubMed]
- 39.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 40.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–4. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ge T, Chen CY, Ni Y, Feng YA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.rsq: R-Squared and Related Measures. R package version 2.0. https://CRAN.R-project.org/package=rsq, 2020.
- 44.Team RC. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2019.
- 45.Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genet. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wray NR, Yang J, Goddard ME, Visscher PM. The genetic interpretation of area under the ROC curve in genomic profiling. PLoS Genet. 2010;6:e1000864. doi: 10.1371/journal.pgen.1000864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gardea-Resendez M, Kucuker MU, Blacker CJ, Ho AM-C, Croarkin PE, Frye MA, et al. Dissecting the epigenetic changes induced by non-antipsychotic mood stabilizers on schizophrenia and affective disorders: a systematic review. Front Pharmacol. 2020;11:467. doi: 10.3389/fphar.2020.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marie-Claire C, Lejeune FX, Mundwiller E, Ulveling D, Moszer I, Bellivier F, et al. A DNA methylation signature discriminates between excellent and non-response to lithium in patients with bipolar disorder type 1. Sci Rep. 2020;10:12239. doi: 10.1038/s41598-020-69073-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee RS, Pirooznia M, Guintivano J, Ly M, Ewald ER, Tamashiro KL, et al. Search for common targets of lithium and valproic acid identifies novel epigenetic effects of lithium on the rat leptin receptor gene. Transl Psychiatry. 2015;5:e600. doi: 10.1038/tp.2015.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ookubo M, Kanai H, Aoki H, Yamada N. Antidepressants and mood stabilizers effects on histone deacetylase expression in C57BL/6 mice: brain region specific changes. J Psychiatr Res. 2013;47:1204–14. doi: 10.1016/j.jpsychires.2013.05.028. [DOI] [PubMed] [Google Scholar]
- 52.Nunes A, Ardau R, Berghöfer A, Bocchetta A, Chillotti C, Deiana V, et al. Prediction of lithium response using clinical data. Acta Psychiatr Scand. 2020;141:131–41. doi: 10.1111/acps.13122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.