Abstract
Members of the marine Roseobacter group are key players in the global carbon and sulfur cycles. While over 300 species have been described, only 2% possess reduced genomes (mostly 3–3.5 Mbp) compared to an average roseobacter (>4 Mbp). These taxonomic minorities are phylogenetically diverse but form a Pelagic Roseobacter Cluster (PRC) at the genome content level. Here, we cultivated eight isolates constituting a novel Roseobacter lineage which we named ‘CHUG’. Metagenomic and metatranscriptomic read recruitment analyses showed that CHUG members are globally distributed and active in marine pelagic environments. CHUG members possess some of the smallest genomes (~2.6 Mb) among all known roseobacters, but they do not exhibit canonical features of typical bacterioplankton lineages theorized to have undergone genome streamlining processes, like higher coding density, fewer paralogues and rarer pseudogenes. While CHUG members form a genome content cluster with traditional PRC members, they show important differences. Unlike other PRC members, neither the relative abundances of CHUG members nor their relative gene expression levels are correlated with chlorophyll a concentration across the global samples. CHUG members cannot utilize most phytoplankton-derived metabolites or synthesize vitamin B12, a key metabolite mediating the roseobacter-phytoplankton interactions. This combination of features is evidence for the hypothesis that CHUG members may have evolved a free-living lifestyle decoupled from phytoplankton. This ecological transition was accompanied by the loss of signature genes involved in roseobacter-phytoplankton symbiosis, suggesting that relaxation of purifying selection owing to lifestyle shift is likely an important driver of genome reduction in CHUG.
Subject terms: Bacterial evolution, Marine microbiology
Introduction
The marine Roseobacter group is a subfamily-level lineage in Alphaproteobacteria and plays an important role in global carbon and sulfur cycling [1, 2]. It is highly abundant in the coastal environments, accounting for up to 20% of all bacterial cells [3–5]. Over 300 species and 100 genera have been described [6], the vast majority of which harbor large and variable genomes and grow readily on nutrient-rich solid media which are not representative of the niches found in the oligotrophic oceans. Early culture-independent 16S rRNA gene surveys showed that the oceanic roseobacters are represented by a few uncultivated lineages [1, 7]. Recently, novel cultivation techniques and single-cell genomics have made available (partial) genome sequences of several previously uncultivated lineages including NAC11-7 [8], DC5-80-3 (also called RCA) [9, 10], and CHAB-I-5 [11, 12]. Although these lineages are spottily distributed throughout the Roseobacter phylogeny, they together form a pelagic Roseobacter cluster (PRC). The PRC members consistently harbor smaller genomes and show more similar genome content compared to other roseobacters [11]. Learning their evolutionary histories is essential to understand how the genetic and metabolic diversity of the pelagic Roseobacter lineages has formed, which in turn helps appreciate their roles in oceanic carbon and sulfur cycles. However, most PRC members form orphan branches and lack closely related reference genomes, which hampers our further understanding of their evolutionary trajectories.
Here, we isolated eight closely related roseobacters from several ocean regions that consistently possess some of the smallest genomes (~2.6 Mb) among all known roseobacters. They together formed a novel Roseobacter lineage which we named ‘CHUG’ (Clade Hidden and Underappreciated Globally) that is abundant and active in global oceans. Unlike other PRC lineages, CHUG members are uncorrelated with chlorophyll a (Chl-a) concentration in their global distribution and they cannot de novo synthesize vitamin B12, which is often the metabolite roseobacters supply to phytoplankton during their symbiosis [2, 13–15]. In contrast to the model roseobacter Ruegeria pomeroyi DSS-3, which is known to interact with phytoplankton species [16, 17], CHUG members cannot use many metabolites commonly released by marine phytoplankton groups. Therefore, the reductive evolution of CHUG may also indicate a dissociation with phytoplankton, a feature so far unique to CHUG among pelagic roseobacters.
Materials and methods
Detailed methods are described in Supplementary Text 1. Briefly, samples were collected from surface water of the South China Sea, the East China Sea and the northern Gulf of Mexico. Eight CHUG isolates were retrieved following different dilution cultivation procedures and sequenced with Illumina platforms. The raw reads were quality trimmed with Trimmomatic v0.36 [18] and assembled with SPAdes v3.10.1 [19]. Only contigs with length >2000 bp and sequencing depth >5x were retained. Among these, the isolate HKCCA1288 was further sequenced with PacBio Sequel platform and assembled using Unicycler v0.4.6 [20] to obtain a complete and closed genome. Protein-coding genes were predicted with Prokka v1.12 [21].
The average nucleotide identity (ANI) between genomes was calculated using fastANI v1.3 [22]. The assembled genome size, gene number, coding density, and GC content of each genome were predicted using CheckM v1.0.7 [23], whereas the estimated genome size was adjusted as (assembled genome size)/(completeness + contamination) [24]. Pseudogenes were predicted following our recent study [25], and other genomic features were summarized using custom scripts (see Code availability). The phylogenetic ANOVA analyses were performed to compare the analyzed genomic traits while controlling for the evolutionary history of those traits using the ‘phylANOVA’ function of the ‘phytools’ R package [26].
To characterize the global occurrence and activity of CHUG, the TARA Ocean metagenomic and metatranscriptomic sequencing data with size fractions up to 3 µm (prokaryote-enriched) [27, 28] and additional metagenomic sequencing data with size fraction of 5–20 µm (nanoplankton-enriched) [29] were mapped to all 79 roseobacters studied here using bowtie v2.3.2 [30] and BLASTN v2.9.0+ [31]. Only reads sharing >95% similarity and >80% alignment to their best hit were kept for the calculation of relative abundance and activity, which is approximated by Reads Per Kilobase per Million mapped reads (RPKM). The relative abundances of CHUG and other PRC lineages across the global oceans were compared using the Wilcox test. The correlation analysis between their relative abundances and environmental factors was performed using the ‘rcorr’ function in the ‘Hmisc’ R package [32], and the significance level was adjusted using stringent Bonferroni correction for multiple comparisons. These analyses were performed for the CHUG lineage as an entirety instead of each individual genome because these isolates are very closely related and performed equally well in metagenomic read recruitment.
The Roseobacter phylogeny was constructed based on 120 bacterial marker genes [33], and sampling the reference Roseobacter genomes included in the phylogeny followed a previous study [34]. Marker genes were each aligned at the amino acid sequence level using MAFFT v7.222 [35] and trimmed using trimAl v1.4.rev15 [36]. Next, a maximum likelihood (ML) phylogenomic tree was built using IQ-TREE v1.6.2 [37] based on the concatenated alignments. To characterize the similarity between roseobacters at the genome content level, a binary matrix showing the presence and absence pattern of orthologous gene families predicted by OrthoFinder v2.2.1 [38] was used to construct the genome content dendrogram with IQ-TREE v1.6.2 [37]. The gene copy number of each orthologous family was further used to estimate the ancestral genome content for CHUG, its sister group and the outgroup using BadiRate v1.35 [39] on top of the ML phylogenomic tree. A potential role of random genetic drift in driving CHUG genome reduction was approximated by comparing the ratio of radical nonsynonymous nucleotide substitution rate (dR) to conservative nonsynonymous nucleotide substitution rate (dC) on the ancestral branch leading to the CHUG cluster with this ratio on the ancestral branch leading to its sister clade, following our previous protocol [40]. The phylogenetic signal of a specific gene (e.g., coxL, pdo, sox) or trait (e.g., light utilization) was predicted with the ‘phylosig’ function of the ‘phytools’ R package [26]. The association with a particular category (e.g., PRC or non-PRC) of a gene or a trait with or without a strong phylogenetic signal was performed using ‘binaryPGLMM’ analysis in the ‘ape’ R package [41] and χ2 test, respectively.
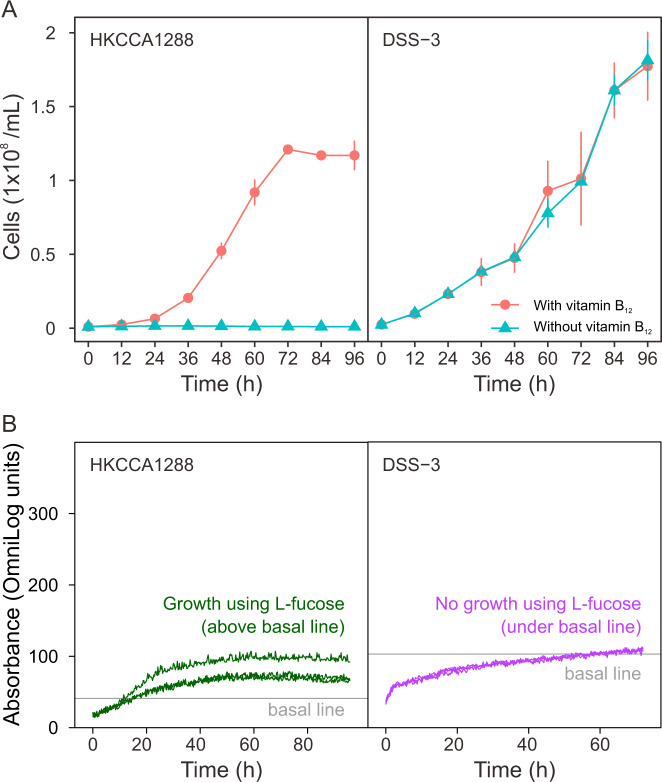
To validate the vitamin B12 auxotrophy in CHUG members, a growth assay was performed separately on HKCCA1288 as the experimental CHUG strain and on the model roseobacter strain Ruegeria pomeroyi DSS-3 [16] as the positive control. Strains were each cultivated in a defined medium for 96 h in the presence or absence of vitamin B12, and samples were collected every 12 h for cell counting using a flow cytometer (Guava EasyCyte Plus, MA, USA) equipped with a fluorescence detector. To test the differences in substrate (190 carbon sources) utilization, the two strains were assayed with the phenotype microarray (PM) technology from BiOLOG following our recent study [42]. All experiments were performed in triplicate.
Results and discussion
The CHUG diversity
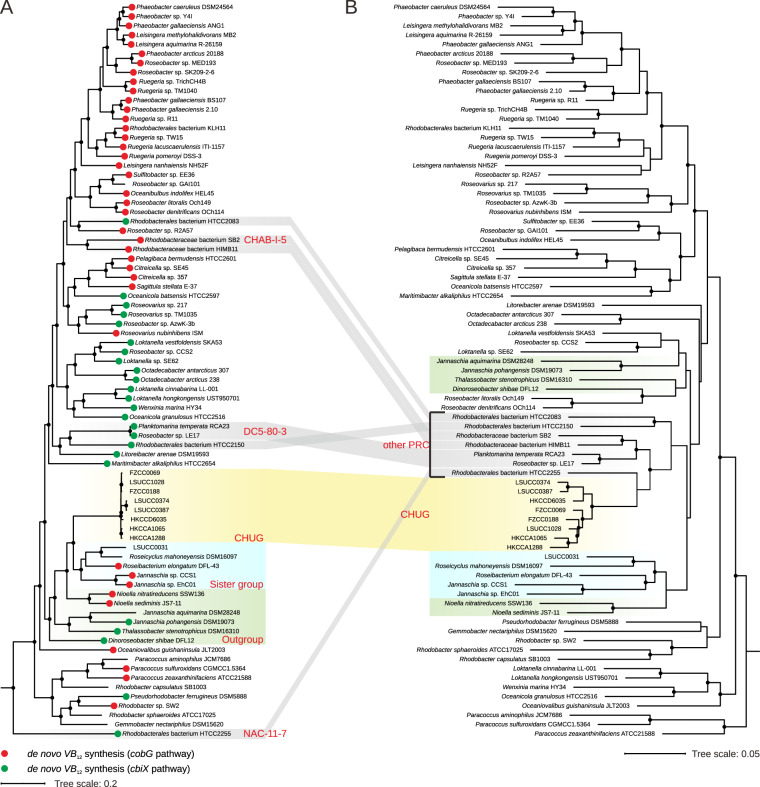
The eight CHUG genomes share ≥99.7% 16S rRNA gene identity and ≥93% ANI. The CHUG cluster further exhibit ≥98.2% 16S rRNA gene identity when sequences of a few uncultivated members are included (Fig. S1), which is comparable to other pelagic Roseobacter lineages, such as 98% [10] for DC5-80-3 and 96% [43] or 98% [7] for CHAB-I-5. CHUG genomes are relatively distantly related to their sister group (Fig. 1A), showing ≤96.5% 16S rRNA gene identity and ≤71% ANI. Unlike their sister group and the outgroup members isolated from various habitats (e.g., saline lake, algal culture, and coastal sediment; Table S1) other than the pelagic environment, CHUG members were collected exclusively from seawater. There are some important differences regarding the source ecosystems for the eight CHUG isolates, though. While five isolates were collected from regular coastal seawater, two (HKCCA1065 and HKCCA1288) and one (HKCCD6035) were isolated from the ambient seawater of the brown alga Sargassum hemiphyllum and of the coral Platygyra acuta, respectively (Table S1).
Fig. 1. Phylogenomic tree and gene content dendrogram of roseobacters.
A Maximum likelihood phylogenomic tree showing the position of CHUG in the Roseobacter group. The phylogeny was inferred using IQ-TREE v1.6.2 [37] based on a concatenation of 45,904 amino acid sites over 120 conserved bacterial proteins [33]. Solid circles in the phylogeny indicate nodes with bootstrap values >95%. The potential of aerobic (key gene cobG, red) and anaerobic (key gene cbiX, green) cobinamide synthesis (the first stage of Vitamin B12 synthesis) is labeled at the tips. B Dendrogram of the same Roseobacter genomes based on the presence/absence pattern of orthologous gene families.
Although not monophyletic in the phylogenomic tree (Fig. 1A), CHUG and seven other genomes from taxa previously sampled from pelagic environments form a coherent group called the ‘Pelagic Roseobacter Cluster’ (PRC; Fig. 1B) [11]. One previously identified PRC member, Roseobacter sp. R2A57 (4.13 Mb), was not affiliated with PRC in the present study. To facilitate our analysis, we divided the 79 roseobacters used here into five groups: CHUG, its sister group, the outgroup of CHUG and its sister group, other PRC members and other reference roseobacters, with eight, five, six, seven and 53 genomes, respectively.
Genomic features
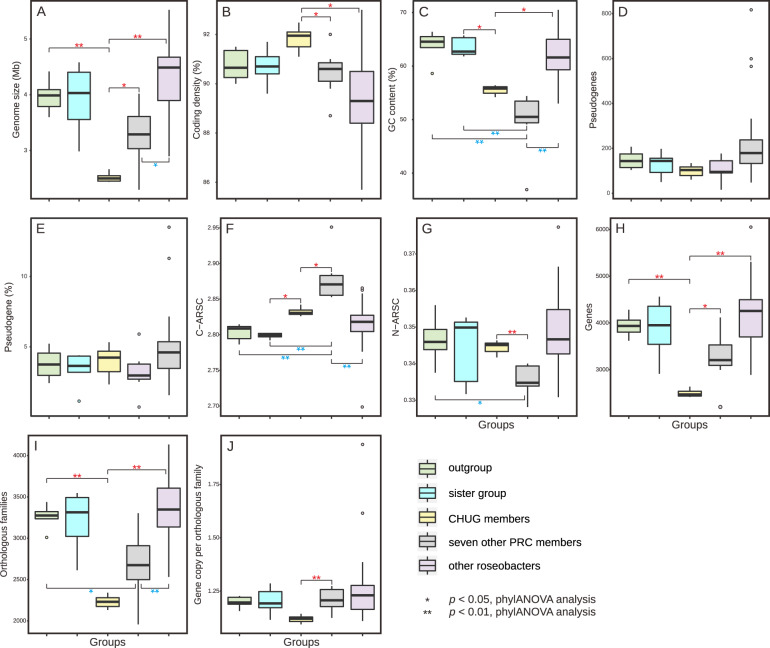
Among the eight CHUG genomes, one (HKCCA1288) is closed with 2.66 Mb and the remaining draft genomes are nearly complete (≥98.5%) according to CheckM predictions (Table S1). Among other roseobacter genomes under comparison, at least 17 genomes are closed and the remaining are nearly complete (≥96.5%) (Table S1). Based on the assembled genome sizes, CHUG members possess much smaller genomes (2.52 ± 0.07 Mb, Fig. 2A) than an average roseobacter (4.16 ± 0.68 Mb). Further, their genome sizes are comparable to those of the NAC11-7 cluster represented by the strain HTCC2255 (estimated complete size to be 2.34 Mb), which is a phylogenetically basal roseobacter with the smallest genome among all known roseobacters [44]. As in HTCC2255, no plasmids were found in CHUG. However, the coding density of CHUG (91.7 ± 0.5%, Fig. 2B) show no significant difference from its sister group and the outgroup based on the phylogenetic ANOVA analysis (p > 0.05, ‘phylANOVA’; the same test used below unless stated otherwise). CHUG genomes have a lower genomic GC content (55.4 ± 0.8%, Fig. 2C) compared to their sister group (63.5 ± 1.6%, p < 0.05), although no significant difference was identified compared to the outgroup. In terms of pseudogenes, the number (99 ± 24, Fig. 2D) and ratio (3.9 ± 0.9%, Fig. 2E) in CHUG members are not significantly different from those of the sister group and outgroup. The seven other PRC members also have smaller genomes (3.26 ± 0.51 Mb, Fig. 2A) and a reduced GC content (49.6 ± 5.5%, Fig. 2C) compared to the 53 other reference roseobacters (genome size: 4.32 ± 0.64 Mb, GC content: 61.9 ± 4.1%; p < 0.01), but no significant differences were identified between the two groups in the coding density (Fig. 2B), the number (Fig. 2D) and ratio of pseudogenes (Fig. 2E).
Fig. 2. Genomic feature comparisons between CHUG, their sister group, the outgroup, seven other PRC members, and other reference roseobacters.
The significance level in genomic features between CHUG and the other four groups is shown in red, while that between seven other PRC members and the remaining groups are shown in blue. Statistical tests were performed using phylANOVA analysis [26] for genome size (A), coding density (B), GC content (C), number of pseudogenes (D), ratio of pseudogenes (E), C-ARSC (F), N-ARSC (G), number of genes (H), number of orthologous families (I), and gene copy number per orthologous family (J), respectively. The markers *p < 0.05 and **p < 0.01, respectively. C-ARSC carbon atoms per amino-acid-residue side chain, N-ARSC nitrogen atoms per amino-acid-residue side chain.
CHUG genomes show increased use of carbon atoms per amino-acid-residue side chain (C-ARSC, 2.833 ± 0.005, Fig. 2F) compared to the sister group (2.799 ± 0.004, p < 0.05). However, no significant difference was found in CHUG members in the use of C-ARSC compared to the outgroup, nor that of nitrogen atoms per amino-acid-residue side chain (N-ARSC, 0.345 ± 0.002, Fig. 2G) compared to the sister group or the outgroup. Likewise, the seven other PRC genomes have significantly higher C-ARSC (2.879 ± 0.031, Fig. 2F) than the 53 other reference roseobacters (2.817 ± 0.026, p < 0.01), but no significant difference was found between their N-ARSC values (Fig. 2G).
Consistent with their genome size differences, CHUG genomes contain a significantly smaller number of coding genes (2486 ± 78, Fig. 2H) compared to the outgroup (3939 ± 214, p < 0.01) and the seven other PRC genomes (3253 ± 545 genes, p < 0.05). The CHUG genomes contain 2215 ± 70 orthologous gene families (Fig. 2I) with 1.12 ± 0.01 gene copy per family (Fig. 2J). By comparison, the outgroup genomes contain 3259 ± 130 orthologous gene families (p < 0.01) with 1.20 ± 0.04 (p > 0.05) gene copy per family, while the seven other PRC genomes possess 2678 ± 398 orthologous gene families (p > 0.05) with 1.21 ± 0.05 (p < 0.01) gene copy per family. No significant difference occurs between CHUG and the sister group in the number of genes, orthologous gene families and gene copies per family. Additionally, while the number of genes and gene copies per family of the seven other PRC genomes is not significantly different from those in the 53 other reference roseobacters (Fig. 2H, J), the seven other PRC genomes have fewer orthologous families compared to the 53 other reference roseobacters (3362 ± 362, p < 0.01, Fig. 2I).
Global distribution and ecological drivers
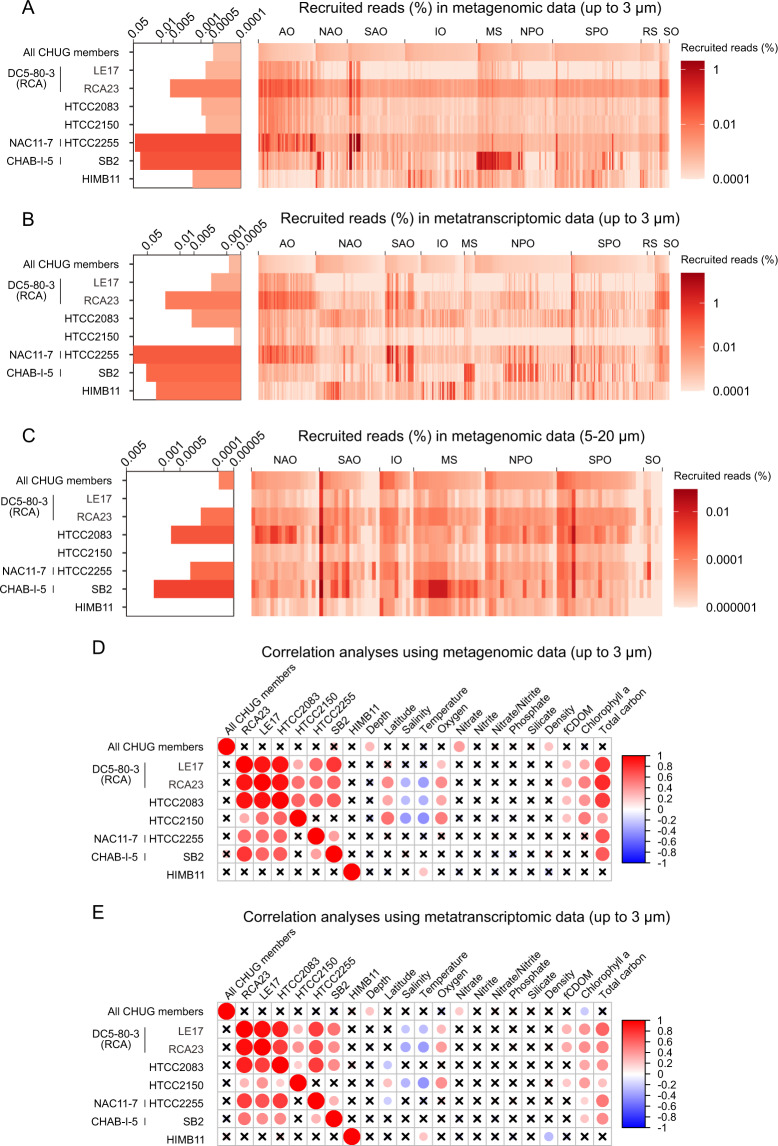
The eight CHUG members recruited 0.0005% and 0.0008% of all metagenomic (Fig. 3A) and metatranscriptomic (Fig. 3B) reads from the global TARA Ocean datasets with size fractions up to 3 µm (prokaryote-enriched) [27, 28], respectively. The CHUG members appear to be less abundant and less active than a few other PRC members such as the strain HTCC2255 (NAC11-7), the strain SB2 (CHAB-I-5) and Planktomarina temperata RCA23 (RCA or DC5-80-3) (Welch’s t-test, p < 0.01 for each). A similar pattern was also found using TARA Ocean metagenomic sequencing data with the size fraction of 5–20 µm (nanoplankton-enriched; Fig. 3C) [29]. From a gene-centric perspective, 58.6% ± 1.2% and 88.3% ± 12.7% genes from the eight CHUG genomes and seven other PRC members recruited TARA metatranscriptomic reads, respectively (see Supplementary Text 2.1 for more details).
Fig. 3. The global distribution of CHUG and its ecological correlation with environmental factors.
A–C The relative abundance of CHUG and other PRC members in the bacterial communities based on recruitment analysis using the metagenomic TARA Ocean sequencing samples with size fractions up to 3 µm (A), and metatranscriptomic sequencing samples with size fractions up to 3 µm (B), and metagenomic sequencing samples with size fraction of 5–20 µm (C). D, E Correlation analysis between the relative abundance of CHUG and other PRC members and environmental parameters measured in the TARA Ocean metagenomic (D) and metatranscriptomic (E) samples. The p value is adjusted using stringent Bonferroni correction. Nonsignificant correlations are indicated by crosses for p > 0.05 after adjusting. AO Arctic Ocean, NAO North Atlantic Ocean, SAO South Atlantic Ocean, IO Indian Ocean, MS Mediterranean Sea, NPO North Pacific Ocean, SPO South Pacific Ocean, RS Red Sea, SO Southern Ocean, fCDOM fluorescence, colored dissolved organic matter.
The relative abundances of typical PRC lineages are positively correlated with each other, with Chl-a concentration (a proxy for phytoplankton biomass [45]), and with total carbon in both metagenomic and metatranscriptomic samples (Fig. 3D, E; Bonferroni corrected p < 0.05). This is interesting but not entirely new. DC5-80-3 and NAC11-7, for example, were previously shown to be positively correlated with phytoplankton blooms [1, 4, 46–49]. In the case of CHAB-I-5, while such a correlation was not evident in a previous study with a more limited sampling effort [11], a few uncultivated members carry signature genes mediating organismal interactions (e.g., type VI secretion system and quorum sensing) [12], potentially enabling them to explore microenvironments including phytoplankton. On the other hand, the relative abundance and activity of the CHUG are not correlated with other PRC members, chlorophyll a (Chl-a) concentration, or the total carbon (Fig. 3D, E; Bonferroni corrected p > 0.05). The same result was also obtained for Rhodobacteraceae bacterium HIMB11 [50], another PRC member whose ecology and evolution has been studied. These results suggest that CHUG (and perhaps HIMB11) may take a free-living lifestyle decoupled from marine phytoplankton.
CHUG genome reduction and vitamin B12 auxotrophy
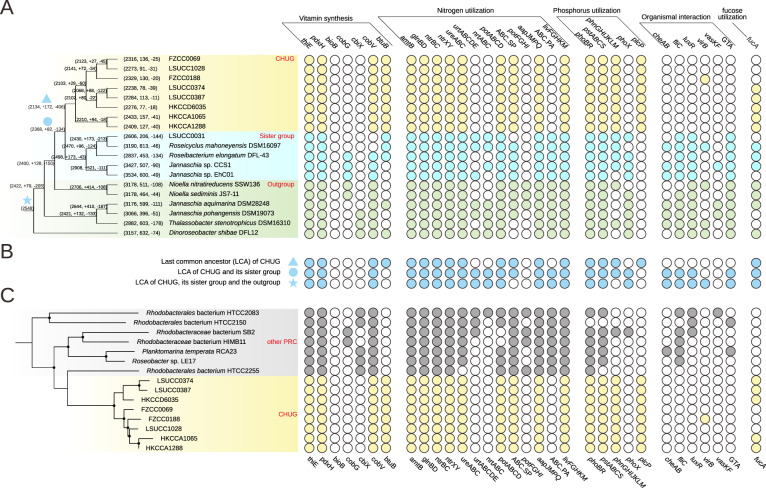
The last common ancestor (LCA) of the CHUG cluster was estimated to have 2320 genes, 2134 orthologous gene families (1.09 gene copy per family), and a genome size of 2.35 Mb. There were 172 families (185 genes) gained and 406 families (425 genes) lost on the ancestral branch leading to the LCA of CHUG, while 28 and 52 families (30 and 79 genes) underwent copy number increase and decrease, respectively. Compared to its sister group and the outgroup, CHUG members lost 412 Kb (9.8%) on the ancestral branch leading to its LCA (filled triangle in Fig. 4A).
Fig. 4. The phyletic pattern of select genes.
The solid and open circles in the right panel represent the presence/absence of the genes, respectively. A The phyletic pattern in the CHUG, its sister group and its outgroup. The phylogenomic tree shown in the left panel is pruned from the full phylogenomic tree shown in Fig. 1A, and branch length is ignored for better visualization. The ancestral genome reconstruction was performed with BadiRate v1.35 [39]. Each ancestral and leaf node is associated with three numbers, representing the total number of orthologous gene families at this node, and the number of orthologous gene families gained and lost on the branch leading to this node. The last common ancestor (LCA) of CHUG, the LCA shared by CHUG and its sister group, and the LCA shared by CHUG, its sister group and the outgroup are marked with a filled triangle, a filled circle, and a filled star, respectively. B The estimated phyletic pattern of the above-mentioned three LCAs. C The gene presence and absence pattern in the CHUG and other seven PRC genomes. The dendrogram in the left panel is pruned from that shown in Fig. 1B. thiE thiamine-phosphate pyrophosphorylase, pdxH pyridoxamine 5′-phosphate oxidase, bioB biotin synthase, cobG precorrin-3B synthase, cbiX sirohydrochlorin cobaltochelatase, cobV adenosylcobinamide-GDP ribazoletransferase, btuB vitamin B12 transporter, amtB ammonium transport system, glnBD nitrogen regulatory protein P-II, ntrBC nitrogen regulation two-component system, ntrXY nitrogen regulation two-component system, ureABC urease, urtABCDE urea transport system, nrtABC nitrate/nitrite transport system, phoBR two-component phosphate regulatory system, pstABCS phosphate transport system (high affinity), phnGHIJKLM carbon-phosphorus (C-P) lyase, phoX alkaline phosphatase, plcP phospholipase C, cheAB chemotaxis family protein, fliC flagellin, luxR quorum-sensing system regulator, virB type IV secretion system protein, vasKF type VI secretion system protein, GTA gene transfer agent, fucA l-fuculose-phosphate aldolase.
Since the CHUG genomes experienced net DNA and gene losses, we explored whether metabolic auxotrophy (i.e., inability to synthesize a compound required for the growth) arose as a result of these losses. The extant CHUG genomes harbor the complete pathways for the synthesis of all 20 amino acids, many of which, such as the synthesis of lysine (dapD) and methionine (ahcY), were transcribed in the wild (Fig. S2). Specifically, genes encoding both vitamin B12-dependent (metH) and -independent methionine synthase (metE) were expressed, and the expression level of the former was approximately twice the latter one (Fig. S2). CHUG members further encode the key genes for thiamine (vitamin B1) synthesis (thiamine-phosphate pyrophosphorylase, thiE) and pyridoxine (vitamin B6) synthesis (pyridoxamine 5′-phosphate oxidase, pdxH). Nevertheless, the key gene for biotin (vitamin B7) synthesis (biotin synthase, bioB) was not found in CHUG nor in the sister group and the outgroup, suggesting that the biotin auxotrophy in CHUG was not part of their net gene losses.
Intriguingly, CHUG is auxotrophic for cobalamin (vitamin B12) biosynthesis, which can be synthesized by most roseobacters [2]. This was validated using a growth assay, in which the CHUG strain HKCCA1288 did not grow in the defined medium lacking vitamin B12 but grew well when vitamin B12 was supplemented (Fig. 5A). As a comparison, the model roseobacter Ruegeria pomeroyi DSS-3 grew equally well in the presence or absence of vitamin B12 (Fig. 5A). Mapping of the vitamin B12 de novo synthesis pathway to the phylogeny (Fig. 1A) indicates that the loss of this capability was most likely associated with the genome reduction leading to the LCA of the CHUG cluster. On the other hand, no genome content changes were inferred related to vitamin B12 synthesis by the ancestral genome reconstruction (Fig. 4B). This controversy can be ascribed to the facts that the de novo synthesis of cobinamide, the key precursor of vitamin B12, has two non-homologous pathways (i.e., aerobic and anaerobic synthesis of cobinamide via key genes cobG and cbiX, respectively), and that distinct pathways are maintained in the CHUG sister lineages (Fig. 1A). The ancestral genome reconstruction further inferred that the loss of vitamin B12 de novo synthesis capability is compensated with the coincidental gain of a putative transporter (btuB) for vitamin B12 and its related compounds such as cobinamide [51] (Fig. 4B), which is absent from all other PRC members capable of de novo vitamin B12 synthesis (Fig. 4C). Taken together, the loss of de novo synthesis capability, the gain of a putative transporter, and the increased expression of the vitamin B12-dependent (metH) methionine synthase gene indicates that CHUG may have to acquire vitamin B12 or its precursor from the environment.
Fig. 5. Growth assay of CHUG strain HKCCA1288 and the model roseobacter Ruegeria pomeroyi DSS-3.
A Growth assay performed on defined marine ammonium mineral salts (MAMS) medium with and without supplementing vitamin B12 are plotted in red and blue, respectively. Three triplicates were performed for each condition and error bars denote standard deviation. B Growth assay under phenotype microarray (PM) plates with l-fucose as the sole carbon source. The horizontal gray line represents the basal line as defined in the negative control (well A01 on PM1, Fig. S3A, C) without any carbon source. Curves above and below the basal line indicate the strain can and cannot use l-fucose for growth, respectively. Three replicates were performed for each condition.
Utilization of organic compounds including metabolites released by marine phytoplankton and other marine organisms
Of the 190 organic compounds assayed through the phenotype microarrays, 43 are experimentally verified metabolites secreted by select species of the dominant eukaryotic marine phytoplankton groups including diatom [52], dinoflagellate [53], and coccolithophore [54, 55]. The CHUG isolate HKCCA1288 is limited in using these phytoplankton-related substrates for growth compared to R. pomeroyi DSS-3. Specifically, while both can use five of these substrates and neither can use eight of them (Table S2 and Fig. S3), the remaining 30 compounds were exclusively used by R. pomeroyi DSS-3.
Most of the remaining 147 substrates covered by the phenotype microarrays cannot differentiate the two strains. Specifically, 16 of these compounds can support both strains and 90 supported neither (Table S2 and Fig. S3). Among the compounds differentially utilized by the two strains, 32 and 9 were exclusively used by R. pomeroyi DSS-3 and HKCCA1288, respectively (Table S2 and Fig. S3). The latter includes l-fucose, which can support HKCCA1288 as a sole carbon source (Fig. 5B). l-fucose is the degradation product of fucoidan made by marine brown algae [56, 57]. Consistently, the gene encoding l-fuculose-phosphate aldolase (fucA) responsible for l-fucose degradation was found in HKCCA1288 and four other CHUG genomes (Fig. 4) but not in R. pomeroyi DSS-3 (Table S2). Notably, the early-branching CHUG lineages represented by HKCCA1288 and HKCCA1065 were collected from the ambient seawater of the brown algae Sargassum hemiphyllum. These lines of evidence are consistent with the hypothesis that the ambient seawater of marine brown algae is likely an important microenvironment supporting CHUG members. Another compound of potential interest is d-fucose, since it also specifically supported HKCCA1288 and it is a component of the glycosphingolipid [58] and glycoside [59] in some sponges, tentatively suggesting sponge ambient seawater as another microenvironment for some CHUG members.
CHUG members take an evolutionary path decoupled from phytoplankton
We have provided three independent lines of evidence for the hypothesis that CHUG takes a free-living lifestyle decoupled from phytoplankton. First, unlike other PRC lineages, the global distribution of CHUG is not correlated with marine phytoplankton biomass (approximated by Chl-a concentration). Moreover, when the TARA Ocean metagenomic sequencing reads at the nanoplankton-enriched size fraction (5–20 µm) were recruited, CHUG members exhibited a lower relative abundance than other PRC representatives by approximately one order of magnitude (Fig. 3C). Second, unlike all other PRC members and most other reference roseobacters, all CHUG members lack the genes for de novo vitamin B12 synthesis. The auxotrophy for vitamin B12 was also validated for HKCCA1288 – for which we generated a complete genome sequence – in a growth assay (Fig. 5A). The marine eukaryotic algae are predominantly vitamin B12 auxotrophs [60], whereas most roseobacters have the potential to synthesize vitamin B12 [2]. This complementarity is one of the major mechanisms that facilitate mutualistic interactions between roseobacters and phytoplankton [2, 13–15]. One more evidence is from the high-throughput growth assays, which demonstrated limited capacity of using phytoplankton-derived metabolites as carbon sources by CHUG compared to that by R. pomeroyi DSS-3 known to interact with marine phytoplankton groups (Table S2).
These observations are unusual because members of the Roseobacter group are known as the dominant bacterial lineages associated with marine phytoplankton groups [17] and their evolutionary history was likely correlated with phytoplankton diversification [2, 61]. They usually benefit from the fixed carbon or other excretions released by phytoplankton and, in return, produce secondary metabolites (e.g., vitamins, indole-3-acetic acid) to promote phytoplankton growth [15, 62, 63]. These interactions likely occur in microzones immediately surrounding phytoplankton cells, which may create gene flow barriers and facilitate population differentiation of associated roseobacters [42, 64]. Therefore, the ecology and evolution of the Roseobacter group in the pelagic ocean are generally shaped by marine phytoplankton, making the possible separation from this ecological pattern in the CHUG lineage unique.
Other important metabolic potential of CHUG
Nitrogen (N) is a primarily limiting nutrient in surface oceans [65]. Genes encoding the nitrogen regulatory protein P-II (glnBD) were highly expressed in the wild CHUG populations (Fig. S2). Genes encoding the high-affinity ammonium transporter (amtB) and the two-component regulatory system (ntrBC and ntrXY) were found in CHUG genomes. Genes encoding urease (ureABC) were also identified in CHUG members, though the urea transport system (urtABCDE) was not found. It is possible that urea is assimilated via passive diffusion across the cell membrane in CHUG as shown in other bacteria [66], or that urea is taken up by another promiscuous transporter. The genes encoding the transporter for nitrate/nitrite assimilation (nrtABC) are also missing in CHUG genomes. CHUG members retain the genes for the spermidine/putrescine transporter (potABCD and ABC.SP) (Table S3), and the latter was among the most highly expressed genes in the oceanic CHUG populations (Fig. S2). However, CHUG members do not carry genes for other polyamine transport systems, such as potFGHI for putrescine acquisition. CHUG members also retain and highly expressed aapJMPQ for the general l-amino acid transporter (Fig. S2), but lost genes encoding the polar amino acid transport system ABC.PA, which is prevalent in all other roseobacters studied here. CHUG members further have a reduced number of genes (only one copy) encoding the branched-chain amino acid transport system (livFGHKM) compared to its sister group (at least three copies), the outgroup (at least three copies) and other PRC members (at least two copies; Table S3). Overall, fewer genes involved in the acquisition of amino acids were found in CHUG (Table S3), but they may remain efficient due to the high expression level of the retained genes.
Phosphorus (P) is often a co-limiting nutrient in surface oceans [65]. To deal with P limitation, CHUG members may be assisted by the essential regulatory and metabolic pathways known to be induced by P limitation including the two-component regulatory system (phoBR), the high-affinity phosphate transporter (pstABCS) and the C-P lyase (phnGHIJKLM) for phosphonate utilization. However, they lost the phoX encoding an alkaline phosphatase for phosphodiester utilization [67] during the genome reduction process (Fig. 4A, B). A notable evolutionary innovation upon the emergence of the CHUG cluster was a gain of the gene encoding phospholipase C (plcP) (Fig. 4A, B), which is missing from all the traditional PRC members (Fig. 4C). The plcP is the key gene of the pathway for phospholipid substitution with non-phospholipids in response to P starvation, and is prevalent in marine bacterioplankton [68].
CHUG members have also lost genes for chemotaxis (cheAB) and flagellar assembly (fliC). These genes were essential to mediate roseobacter-phytoplankton interactions [69], but may become dispensable when switching to a planktonic lifestyle [70]. Consistent with this, the quorum-sensing (QS) system (luxR), type IV secretion system (virB), and type VI secretion system (vasKF) involved in organismal interactions were rarely found in CHUG genomes (Fig. 4A). CHUG members also lost the gene cluster encoding gene transfer agent (GTA), which resembles small double-stranded DNA (dsDNA) bacteriophages that increase horizontal gene transfer and metabolic flexibility at high population density [71].
In terms of the strategies for energy conservation, CHUG members maintain a complete (Fig. S4) and actively expressed (Fig. S2) photosynthesis gene cluster enabling aerobic anoxygenic photosynthesis (e.g. puf, bch, and crt). Some of them further carry gene clusters for the oxidation of carbon monoxide (cox) and sulfide/thiosulfate (sox) as energy sources, but lack genes involved in dissimilatory nitrate/nitrite reduction. For important substrates commonly used by roseobacters, CHUG members are limited in their catabolic pathways. For the utilization of aromatic compounds degradation, for example, CHUG members possess protocatechuate ring cleavage pathway (pcaGH) but lost genes for the ring cleavage of phenylacetate (paaABCDE) and homogenisate (hmgA). CHUG carry both demethylation (dmdA) and cleavage pathways (dddD and dddL) to utilize dimethylsulfoniopropionate (DMSP), but lack genes to degrade other important methylated compounds including trimethylamine N-oxide (tmd), trimethylamine (tmm), and taurine (tauABC and xsc). More details are provided in Supplementary Text 2.1 and 2.2.
Potential evolutionary forces driving genome reduction of CHUG
The most abundant marine bacterioplankton, such as the Pelagibacterales (also called the SAR11 clade) in Alphaproteobacteria and the Prochlorococcus in Cyanobacteria, are often equipped with very small genomes [72]. The evolutionary mechanisms driving their genome reduction have been discussed extensively. Among these, selection for metabolic efficiency under extreme nutrient limitation (i.e., ‘genome streamlining’) has been theorized as the dominant force driving their genome reduction [72, 73]. Although CHUG members possess smaller genomes and lower GC content compared to the sister group and the outgroup (Fig. 2A, C), they do not show other features of genome streamlining (Fig. 2B, E, J), such as higher coding density, fewer paralogues, or rarers pseudogenes [72, 74]. Likewise, these genomic features are also missing in other abundant PRC members (Fig. 2B, E, J) such as NAC11-7, DC5-80-3, and CHAB-I-5, though they have smaller genomes compared to other reference roseobacters (Fig. 2A). Therefore, the genome reduction process of CHUG and other PRC members does not meet the canonical definition of genome streamlining.
Other important evidence against the genome streamlining explanation for CHUG genome reduction is from the genomic proxies for nutrient acquisition and saving strategies used by marine bacterioplankton. Among the selective factors that may drive bacterioplankton genome reduction in the pelagic ocean, N limitation is considered as the dominant factor [44, 72, 75, 76]. Although the relative abundance of gene transcripts (but not the genes) in the wild CHUG populations is positively correlated with the nitrate concentration (Fig. 3E; Bonferroni corrected p < 0.05), which provides marginal evidence for a role of N limitation, other key evidence is missing. For example, we did not observe a reduced use of N in the amino acid sequences (approximated by N-ARSC) in CHUG compared to the sister group and the outgroup. Similar observation is used as evidence against the hypothesis that N limitation is a strong driver of genome streamlining in other marine bacterioplankton lineages [77, 78]. A second potential ecological factor driving genome streamlining is P limitation [79], though this theory has been debated [80]. Genome reduction likely leads to a sizable decrease in cellular P requirement and thus may confer a competitive advantage in the P-limited marine environments [81]. Although a few important genes for P acquisition (pst for high-affinity phosphate transporter and phn for C-P lyase) are retained and a gene encoding phospholipase C (plcP) responsible for cell membrane phospholipid substitution for non-phosphorus lipids [68] was even acquired, the key P scavenging gene encoding PhoX alkaline phosphatase was lost during the CHUG genome reduction (Fig. 4). Therefore, available evidence for either N or P limitation as a driver of CHUG genome reduction is self-contradictory.
Because evidence for genome streamlining is weak, we examined neutral evolutionary forces as potential explanations for CHUG genome reduction. In fact, neutral mechanisms have recently been considered to play important roles in driving genome reduction of marine bacterioplankton lineages [40, 82, 83]. Most of the prior studies focused on Prochlorococcus (see references cited in the following paragraphs). While some extended their discussions to Pelagibacterales [40, 84], knowledge on the evolutionary mechanisms driving genome reduction of most other marine bacterioplankton lineages is rather limited.
One potentially important neutral driver is random genetic drift due to a reduction of effective population size (Ne). A previous study showed that the major genome reduction event coincided with an accelerated rate of accumulating deleterious mutations in the early evolution of Prochlorococcus, providing important evidence that genetic drift is likely the primary mechanism of genome reduction in this lineage [40]. Specifically, the power of genetic drift (i.e., the inverse of Ne) of an ancestral lineage (e.g., the ancestral branch underlying the ancient genomic events) can be approximated by the ratio of the radical nonsynonymous nucleotide substitution rate (dR) to the conservative nonsynonymous nucleotide substitution rate (dC) [40]. Because a replacement by a physicochemically dissimilar amino acid (i.e., radical change) is likely to be more deleterious than a replacement by a similar amino acid (i.e., conservative change) [85, 86], the elevated dR/dC ratio is evidence for genetic drift acting to accumulate the deleterious type of mutations (i.e., the radical changes) in excess. In terms of the CHUG, the dR/dC ratio is not significantly elevated compared to its sister group (Fig. S5A) under two independent methods for biochemical classification of the 20 amino acids (Fig. S5B, C), suggesting that the deleterious type of mutations was not accumulated in excess at the ancestral branch leading to the LCA of the CHUG cluster (filled triangle in Fig. 4A). Since this ancestral branch corresponds to the time when major genome reduction occurred for CHUG, we can conclude that genetic drift is unlikely to be an important driver of CHUG genome reduction.
A second potentially important neutral driver of prokaryotic genome reduction is increased mutation rate, which has also been proposed to explain Prochlorococcus genome reduction [87]. Mathematical modeling predicts that not all auxiliary genes can be maintained by purifying selection when mutation rate is increased, and that an increase of 10 fold in mutation rate may lead to a 30% decrease in genome size [88]. More recently, this hypothesis was supported with empirical data from comparative genomics analyses [83], though whether increased mutation rate is a truly important driver of prokaryotic genome reduction is debated [89]. Given the potentially important role of increased mutation rate in driving prokaryotic genome reduction, determining the unbiased spontaneous mutation rate of the CHUG and the sister lineage using the mutation accumulation experiment followed by whole genome sequencing of the mutant lines becomes an urgent research need.
One more potentially important but rarely discussed neutral force leading to genome reduction is the loss of the genes that were important in the initial habitat but became dispensable after the bacteria switched to a new environment. This neutral loss mechanism, termed relaxation of purifying selection, may also have contributed to genome reduction in Prochlorococcus [90]. Importantly, the loss of dispensable genes under this mechanism is not related to the change of Ne but results instead from a change of habitat or lifestyle. In contrast to other PRC members and copiotrophic roseobacters such as R. pomeroyi DSS-3 sampled from the pelagic environments, CHUG members take a free-living lifestyle uncoupled from marine phytoplankton. This is supported by three lines of evidence: (i) unlike other PRC members, CHUG members do not exhibit a correlative pattern between their global distributions and Chl-a (Fig. 3D, E); (ii) vitamin B12, a fundamental metabolite many roseobacters produce and supply to phytoplankton, cannot be synthesized by CHUG; (iii) compared to R. pomeroyi DSS-3, CHUG members have a very limited capacity of using phytoplankton-derived metabolites as carbon sources. Since supplying vitamin B12 to phytoplankton in exchange for phytoplankton-derived carbon sources is an important mechanism underlying the symbiosis between roseobacters and phytoplankton [2, 13–15], the inability of the de novo synthesis of vitamin B12 and of using most phytoplankton-related metabolites indicates that the CHUG ancestor may have lost its ability to establish symbiosis with phytoplankton. As a consequence of this transition, other genes contributing to roseobacter-phytoplankton symbiosis (e.g., motility and chemotaxis), relying on population density (e.g., quorum sensing), and involved in interactions with other bacteria (e.g., gene transfer agent), may have become dispensable [70]. Indeed, the loss of these signature genes contributed to the genome reduction of CHUG (Fig. 4). We therefore propose that relaxation of purifying selection stemming from reduced interactions with marine phytoplankton may be one of the primary evolutionary forces leading to the major genome reduction of CHUG.
Concluding remarks
Although taking a planktonic lifestyle and having some of the smallest genomes among roseobacters, CHUG members lack some canonical features of typical marine bacterioplankton lineages undergoing genome streamlining. Because genome streamlining process is driven by natural selection under extreme nutrient limitation [72], the absence of many genomic features commonly found in streamlined bacterioplankton genomes indicates that the genome reduction of CHUG may have taken place in some pelagic microenvironments enriched in nutrients. While microalgal phycosphere has been thought as the most common microenvironments colonized by roseobacters inhabiting the pelagic ocean, it is unlikely to support CHUG. This is convincingly revealed by its auxotrophy for vitamin B12, its inability to use many phytoplankton-derived metabolites, and its decoupling from phytoplankton in its global distribution. Instead, the available evidence allows generating a testable hypothesis that the seawater surrounding marine brown algae and perhaps other marine macroorganisms is potentially important microenvironments that CHUG members may explore. The discovery of the CHUG cluster greatly expands the diversity of the marine Roseobacter group, and its unique evolutionary trajectory complements to our understanding of how the genomes of many marine bacterioplankton lineages become small.
Cultivar availability
One strain sampled from the East China Sea (FZCC0069) and two strains sampled from the South China Sea (HKCCA1065 and HKCCA1288) are available at the China General Microbiological Culture Collection Center (CGMCC) under the accession number CGMCC 1.19034, 1.19035, and 1.19036, respectively. Three strains sampled from the northern Gulf of Mexico (LSUCC1028, LSUCC0387, and LSUCC0374) are currently under deposition at the German Collection of Microorganisms and Cell Cultures (DSMZ).
Supplementary information
Acknowledgements
This research was funded by the National Science Foundation of China (41776129), the Hong Kong Research Grants Council General Research Fund (14163917), the Hong Kong Research Grants Council Area of Excellence Scheme (AoE/M-403/16), and the Direct Grant of CUHK (4053257 & 3132809). The research was also supported by a Louisiana Board of Regents grant (LEQSF(2014-17)-RD-A-06) and a Simons Early Career Investigator in Marine Microbial Ecology and Evolution Award to JCT.
Data availability
Genomic sequences of the eight CHUG genomes are available at the NCBI GenBank database under the accession number PRJNA574877.
Code availability
The custom scripts used in this study are available in the online repository (https://github.com/luolab-cuhk/CHUG-genome-reduction-project).
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01036-3.
References
- 1.Buchan A, González JM, Moran MA. Overview of the marine Roseobacter lineage. Appl Environ Microbiol. 2005;71:5665–77. doi: 10.1128/AEM.71.10.5665-5677.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo H, Moran MA. Evolutionary ecology of the marine Roseobacter clade. Microbiol Mol Biol Rev. 2014;78:573–87. doi: 10.1128/MMBR.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran MA, Belas R, Schell MA, González JM, Sun F, Sun S, et al. Ecological genomics of marine Roseobacters. Appl Environ Microbiol. 2007;73:4559–69. doi: 10.1128/AEM.02580-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giebel H-A, Kalhoefer D, Lemke A, Thole S, Gahl-Janssen R, Simon M, et al. Distribution of Roseobacter RCA and SAR11 lineages in the North Sea and characteristics of an abundant RCA isolate. ISME J. 2011;5:8–19. doi: 10.1038/ismej.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wemheuer B, Wemheuer F, Hollensteiner J, Meyer F-D, Voget S, Daniel R. The green impact: bacterioplankton response toward a phytoplankton spring bloom in the southern North Sea assessed by comparative metagenomic and metatranscriptomic approaches. Front Microbiol. 2015;6:805. doi: 10.3389/fmicb.2015.00805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pujalte MJ, Lucena T, Ruvira MA, Arahal DR, Macián MC. The family rhodobacteraceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. p. 439–512.
- 7.Buchan A, Hadden M, Suzuki MT. Development and application of quantitative-PCR tools for subgroups of the Roseobacter clade. Appl Environ Microbiol. 2009;75:7542–7. doi: 10.1128/AEM.00814-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo H, Swan BK, Stepanauskas R, Hughes AL, Moran MA. Comparing effective population sizes of dominant marine alphaproteobacteria lineages. Environ Microbiol Rep. 2014;6:167–72. doi: 10.1111/1758-2229.12129. [DOI] [PubMed] [Google Scholar]
- 9.Giebel H-A, Kalhoefer D, Gahl-Janssen R, Choo Y-J, Lee K, Cho J-C, et al. Planktomarina temperata gen. nov., sp. nov., belonging to the globally distributed RCA cluster of the marine Roseobacter clade, isolated from the German Wadden Sea. Int J Syst Evol Microbiol. 2013;63:4207–17. doi: 10.1099/ijs.0.053249-0. [DOI] [PubMed] [Google Scholar]
- 10.Voget S, Wemheuer B, Brinkhoff T, Vollmers J, Dietrich S, Giebel H-A, et al. Adaptation of an abundant Roseobacter RCA organism to pelagic systems revealed by genomic and transcriptomic analyses. ISME J. 2015;9:371–84. doi: 10.1038/ismej.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billerbeck S, Wemheuer B, Voget S, Poehlein A, Giebel H-A, Brinkhoff T, et al. Biogeography and environmental genomics of the Roseobacter-affiliated pelagic CHAB-I-5 lineage. Nat Microbiol. 2016;1:16063. doi: 10.1038/nmicrobiol.2016.63. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Sun Y, Jiao N, Stepanauskas R, Luo H. Ecological genomics of the uncultivated marine Roseobacter lineage CHAB-I-5. Appl Environ Microbiol. 2016;82:2100–11. doi: 10.1128/AEM.03678-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner-Döbler I, Ballhausen B, Berger M, Brinkhoff T, Buchholz I, Bunk B, et al. The complete genome sequence of the algal symbiont Dinoroseobacter shibae: a hitchhiker’s guide to life in the sea. ISME J. 2010;4:61–77. doi: 10.1038/ismej.2009.94. [DOI] [PubMed] [Google Scholar]
- 14.Durham BP, Sharma S, Luo H, Smith CB, Amin SA, Bender SJ, et al. Cryptic carbon and sulfur cycling between surface ocean plankton. Proc Natl Acad Sci USA. 2015;112:453–7. doi: 10.1073/pnas.1413137112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper MB, Kazamia E, Helliwell KE, Kudahl UJ, Sayer A, Wheeler GL, et al. Cross-exchange of B-vitamins underpins a mutualistic interaction between Ostreococcus tauri and Dinoroseobacter shibae. ISME J. 2019;13:334–45. doi: 10.1038/s41396-018-0274-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran MA, Buchan A, González JM, Heidelberg JF, Whitman WB, Kiene RP, et al. Genome sequence of Silicibacter pomeroyi reveals adaptations to the marine environment. Nature. 2004;432:910–3. doi: 10.1038/nature03170. [DOI] [PubMed] [Google Scholar]
- 17.Seymour JR, Amin SA, Raina J-B, Stocker R. Zooming in on the phycosphere: the ecological interface for phytoplankton-bacteria relationships. Nat Microbiol. 2017;2:17065. doi: 10.1038/nmicrobiol.2017.65. [DOI] [PubMed] [Google Scholar]
- 18.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 22.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parks DH, Rinke C, Chuvochina M, Chaumeil P-A, Woodcroft BJ, Evans PN, et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat Microbiol. 2017;2:1533–42. doi: 10.1038/s41564-017-0012-7. [DOI] [PubMed] [Google Scholar]
- 25.Chu X, Li S, Wang S, Luo D, Luo H. Gene loss through pseudogenization contributes to the ecological diversification of a generalist Roseobacter lineage. ISME J. 2020;15:489–502. doi: 10.1038/s41396-020-00790-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Revell LJ. phytools: an R package for phylogenetic comparative biology (and other things) Methods Ecol Evol. 2012;3:217–23. [Google Scholar]
- 27.Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, et al. Ocean plankton. Structure and function of the global ocean microbiome. Science. 2015;348:1261359. doi: 10.1126/science.1261359. [DOI] [PubMed] [Google Scholar]
- 28.Salazar G, Paoli L, Alberti A, Huerta-Cepas J, Ruscheweyh H-J, Cuenca M, et al. Gene expression changes and community turnover differentially shape the global ocean metatranscriptome. Cell. 2019;179:1068–.e21. doi: 10.1016/j.cell.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vargas C, de, Audic S, Henry N, Decelle J, Mahé F, Logares R, et al. Eukaryotic plankton diversity in the sunlit ocean. Science. 2015;348:1261605. doi: 10.1126/science.1261605. [DOI] [PubMed] [Google Scholar]
- 30.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Harrell FE., Jr Package ‘Hmisc’. CRAN2018. 2019;2019:235–6. [Google Scholar]
- 33.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 34.Simon M, Scheuner C, Meier-Kolthoff JP, Brinkhoff T, Wagner-Döbler I, Ulbrich M, et al. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 2017;11:1483–99. doi: 10.1038/ismej.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen L-T, Schmidt HA, Haeseler A, von, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Librado P, Vieira FG, Rozas J. BadiRate: estimating family turnover rates by likelihood-based methods. Bioinformatics. 2012;28:279–81. doi: 10.1093/bioinformatics/btr623. [DOI] [PubMed] [Google Scholar]
- 40.Luo H, Huang Y, Stepanauskas R, Tang J. Excess of non-conservative amino acid changes in marine bacterioplankton lineages with reduced genomes. Nat Microbiol. 2017;2:17091. doi: 10.1038/nmicrobiol.2017.91. [DOI] [PubMed] [Google Scholar]
- 41.Paradis E, Schliep K. ape 5.0: an environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–8. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 42.Wang X, Zhang Y, Ren M, Xia T, Chu X, Liu C, et al. Cryptic speciation of a pelagic Roseobacter population varying at a few thousand nucleotide sites. ISME J. 2020;14:3106–19. doi: 10.1038/s41396-020-00743-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lekunberri I, Gasol JM, Acinas SG, Gómez-Consarnau L, Crespo BG, Casamayor EO, et al. The phylogenetic and ecological context of cultured and whole genome-sequenced planktonic bacteria from the coastal NW Mediterranean Sea. Syst Appl Microbiol. 2014;37:216–28. doi: 10.1016/j.syapm.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Luo H, Swan BK, Stepanauskas R, Hughes AL, Moran MA. Evolutionary analysis of a streamlined lineage of surface ocean Roseobacters. ISME J. 2014;8:1428–39. doi: 10.1038/ismej.2013.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roesler C, Uitz J, Claustre H, Boss E, Xing X, Organelli E, et al. Recommendations for obtaining unbiased chlorophyll estimates from in situ chlorophyll fluorometers: a global analysis of WET Labs ECO sensors. Limnol Oceanogr Methods. 2017;15:572–85. [Google Scholar]
- 46.Wagner-Döbler I, Biebl H. Environmental biology of the marine Roseobacter lineage. Annu Rev Microbiol. 2006;60:255–80. doi: 10.1146/annurev.micro.60.080805.142115. [DOI] [PubMed] [Google Scholar]
- 47.West NJ, Obernosterer I, Zemb O, Lebaron P. Major differences of bacterial diversity and activity inside and outside of a natural iron-fertilized phytoplankton bloom in the Southern Ocean. Environ Microbiol. 2008;10:738–56. doi: 10.1111/j.1462-2920.2007.01497.x. [DOI] [PubMed] [Google Scholar]
- 48.Rich VI, Pham VD, Eppley J, Shi Y, DeLong EF. Time-series analyses of Monterey Bay coastal microbial picoplankton using a ‘genome proxy’ microarray. Environ Microbiol. 2011;13:116–34. doi: 10.1111/j.1462-2920.2010.02314.x. [DOI] [PubMed] [Google Scholar]
- 49.Landa M, Blain S, Christaki U, Monchy S, Obernosterer I. Shifts in bacterial community composition associated with increased carbon cycling in a mosaic of phytoplankton blooms. ISME J. 2016;10:39–50. doi: 10.1038/ismej.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Durham BP, Grote J, Whittaker KA, Bender SJ, Luo H, Grim SL, et al. Draft genome sequence of marine alphaproteobacterial strain HIMB11, the first cultivated representative of a unique lineage within the Roseobacter clade possessing an unusually small genome. Stand Genomic Sci. 2014;9:632–45. doi: 10.4056/sigs.4998989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roth JR, Lawrence JG, Bobik TA. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–81. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 52.Ferrer-González FX, Widner B, Holderman NR, Glushka J, Edison AS, Kujawinski EB, et al. Resource partitioning of phytoplankton metabolites that support bacterial heterotrophy. ISME J. 2021;15:762–73. doi: 10.1038/s41396-020-00811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abreu AC, Molina-Miras A, Aguilera-Sáez LM, López-Rosales L, Del Cerón-García MC, Sánchez-Mirón A, et al. Production of amphidinols and other bioproducts of interest by the marine microalga amphidinium carterae unraveled by nuclear magnetic resonance metabolomics approach coupled to multivariate data analysis. J Agric Food Chem. 2019;67:9667–82. doi: 10.1021/acs.jafc.9b02821. [DOI] [PubMed] [Google Scholar]
- 54.Zhou C, Luo J, Ye Y, Yan X, Liu B, Wen X. The metabolite profiling of coastal coccolithophorid species Pleurochrysis carterae (Haptophyta) Chin J Ocean Limnol. 2016;34:749–56. [Google Scholar]
- 55.Bustamam MSA, Pantami HA, Azizan A, Shaari K, Min CC, Abas F, et al. Complementary analytical platforms of NMR spectroscopy and LCMS analysis in the metabolite profiling of isochrysis galbana. Mar Drugs. 2021;19:139. doi: 10.3390/md19030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rioux L-E, Turgeon SL, Beaulieu M. Effect of season on the composition of bioactive polysaccharides from the brown seaweed Saccharina longicruris. Phytochemistry. 2009;70:1069–75. doi: 10.1016/j.phytochem.2009.04.020. [DOI] [PubMed] [Google Scholar]
- 57.Ale MT, Mikkelsen JD, Meyer AS. Important determinants for fucoidan bioactivity: a critical review of structure-function relations and extraction methods for fucose-containing sulfated polysaccharides from brown seaweeds. Mar Drugs. 2011;9:2106–30. doi: 10.3390/md9102106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hada N, Nakashima T, Shrestha SP, Masui R, Narukawa Y, Tani K, et al. Synthesis and biological activities of glycosphingolipid analogues from marine sponge Aplysinella rhax. Bioorg Med Chem Lett. 2007;17:5912–5. doi: 10.1016/j.bmcl.2007.07.108. [DOI] [PubMed] [Google Scholar]
- 59.Kalinin VI, Ivanchina NV, Krasokhin VB, Makarieva TN, Stonik VA. Glycosides from marine sponges (Porifera, Demospongiae): structures, taxonomical distribution, biological activities and biological roles. Mar Drugs. 2012;10:1671–710. doi: 10.3390/md10081671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Helliwell KE. The roles of B vitamins in phytoplankton nutrition: new perspectives and prospects. New Phytol. 2017;216:62–8. doi: 10.1111/nph.14669. [DOI] [PubMed] [Google Scholar]
- 61.Luo H, Csuros M, Hughes AL, Moran MA. Evolution of divergent life history strategies in marine alphaproteobacteria. MBio. 2013;4:e00373–13. doi: 10.1128/mBio.00373-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Durham BP, Dearth SP, Sharma S, Amin SA, Smith CB, Campagna SR, et al. Recognition cascade and metabolite transfer in a marine bacteria-phytoplankton model system. Environ Microbiol. 2017;19:3500–13. doi: 10.1111/1462-2920.13834. [DOI] [PubMed] [Google Scholar]
- 63.Shibl AA, Isaac A, Ochsenkühn MA, Cárdenas A, Fei C, Behringer G, et al. Diatom modulation of select bacteria through use of two unique secondary metabolites. Proc Natl Acad Sci USA. 2020;117:27445–55. doi: 10.1073/pnas.2012088117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qu L, Feng X, Chen Y, Li L, Wang X, Hu Z et al. Metapopulation structure of diatom-associated marine bacteria. bioRxiv10.1101/2021.03.10.434754 (2021).
- 65.Moore CM, Mills MM, Arrigo KR, Berman-Frank I, Bopp L, Boyd PW, et al. Processes and patterns of oceanic nutrient limitation. Nat Geosci. 2013;6:701–10. [Google Scholar]
- 66.Veaudor T, Cassier-Chauvat C, Chauvat F. Genomics of urea transport and catabolism in Cyanobacteria: biotechnological implications. Front Microbiol. 2019;10:2052. doi: 10.3389/fmicb.2019.02052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo H, Benner R, Long RA, Hu J. Subcellular localization of marine bacterial alkaline phosphatases. Proc Natl Acad Sci USA. 2009;106:21219–23. doi: 10.1073/pnas.0907586106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sebastián M, Smith AF, González JM, Fredricks HF, van Mooy B, Koblížek M, et al. Lipid remodelling is a widespread strategy in marine heterotrophic bacteria upon phosphorus deficiency. ISME J. 2016;10:968–78. doi: 10.1038/ismej.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Geng H, Belas R. Molecular mechanisms underlying Roseobacterphytoplankton symbioses. Curr Opin Biotechnol. 2010;21:332–8. doi: 10.1016/j.copbio.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 70.Luo H, Moran MA. How do divergent ecological strategies emerge among marine bacterioplankton lineages? Trends Microbiol. 2015;23:577–84. doi: 10.1016/j.tim.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 71.Biers EJ, Wang K, Pennington C, Belas R, Chen F, Moran MA. Occurrence and expression of gene transfer agent genes in marine bacterioplankton. Appl Environ Microbiol. 2008;74:2933–9. doi: 10.1128/AEM.02129-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giovannoni SJ, Cameron Thrash J, Temperton B. Implications of streamlining theory for microbial ecology. ISME J. 2014;8:1553–65. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D, et al. Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–5. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- 74.Swan BK, Tupper B, Sczyrba A, Lauro FM, Martinez-Garcia M, González JM, et al. Prevalent genome streamlining and latitudinal divergence of planktonic bacteria in the surface ocean. Proc Natl Acad Sci USA. 2013;110:11463–8. doi: 10.1073/pnas.1304246110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Luo H, Thompson LR, Stingl U, Hughes AL. Selection maintains low genomic GC content in marine SAR11 lineages. Mol Biol Evol. 2015;32:2738–48. doi: 10.1093/molbev/msv149. [DOI] [PubMed] [Google Scholar]
- 76.Mende DR, Bryant JA, Aylward FO, Eppley JM, Nielsen T, Karl DM, et al. Environmental drivers of a microbial genomic transition zone in the ocean’s interior. Nat Microbiol. 2017;2:1367–73. doi: 10.1038/s41564-017-0008-3. [DOI] [PubMed] [Google Scholar]
- 77.Grzymski JJ, Dussaq AM. The significance of nitrogen cost minimization in proteomes of marine microorganisms. ISME J. 2012;6:71–80. doi: 10.1038/ismej.2011.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee MD, Ahlgren NA, Kling JD, Walworth NG, Rocap G, Saito MA, et al. Marine Synechococcus isolates representing globally abundant genomic lineages demonstrate a unique evolutionary path of genome reduction without a decrease in GC content. Environ Microbiol. 2019;21:1677–86. doi: 10.1111/1462-2920.14552. [DOI] [PubMed] [Google Scholar]
- 79.Hessen DO, Jeyasingh PD, Neiman M, Weider LJ. Genome streamlining and the elemental costs of growth. Trends Ecol Evol. 2010;25:75–80. doi: 10.1016/j.tree.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 80.Vieira-Silva S, Touchon M, Rocha EPC. No evidence for elemental-based streamlining of prokaryotic genomes. Trends Ecol Evol. 2010;25:319–20. doi: 10.1016/j.tree.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 81.Thingstad T, Rassoulzadegan F. Conceptual models for the biogeochemical role of the photic zone microbial food web, with particular reference to the Mediterranean Sea. Prog Oceanogr. 1999;44:271–86. [Google Scholar]
- 82.Batut B, Knibbe C, Marais G, Daubin V. Reductive genome evolution at both ends of the bacterial population size spectrum. Nat Rev Microbiol. 2014;12:841–50. doi: 10.1038/nrmicro3331. [DOI] [PubMed] [Google Scholar]
- 83.Bourguignon T, Kinjo Y, Villa-Martín P, Coleman NV, Tang Q, Arab DA, et al. Increased mutation rate is linked to genome reduction in prokaryotes. Curr Biol. 2020;30:3848–.e4. doi: 10.1016/j.cub.2020.07.034. [DOI] [PubMed] [Google Scholar]
- 84.Viklund J, Ettema TJG, Andersson SGE. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol Biol Evol. 2012;29:599–615. doi: 10.1093/molbev/msr203. [DOI] [PubMed] [Google Scholar]
- 85.Zuckerkandl E, Pauling L, Bryson V, Vogel HJ. Evolving genes and proteins. Science American Association for the Advancement of Science; 1965. p. 68–71.
- 86.Dayhoff MO. Atlas of Protein Sequence And Structure. Silver Spring, MD, USA: National Biomedical Research Foundation; 1972. p. 89–100.
- 87.Dufresne A, Garczarek L, Partensky F. Accelerated evolution associated with genome reduction in a free-living prokaryote. Genome Biol. 2005;6:R14. doi: 10.1186/gb-2005-6-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Marais GAB, Calteau A, Tenaillon O. Mutation rate and genome reduction in endosymbiotic and free-living bacteria. Genetica. 2008;134:205–10. doi: 10.1007/s10709-007-9226-6. [DOI] [PubMed] [Google Scholar]
- 89.Gu J, Wang X, Ma X, Sun Y, Xiao X, Luo H. Unexpectedly high mutation rate of a deep-sea hyperthermophilic anaerobic archaeon. ISME J. 2021;15:1862–9. [DOI] [PMC free article] [PubMed]
- 90.Luo H, Friedman R, Tang J, Hughes AL. Genome reduction by deletion of paralogs in the marine cyanobacterium Prochlorococcus. Mol Biol Evol. 2011;28:2751–60. doi: 10.1093/molbev/msr081. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genomic sequences of the eight CHUG genomes are available at the NCBI GenBank database under the accession number PRJNA574877.
The custom scripts used in this study are available in the online repository (https://github.com/luolab-cuhk/CHUG-genome-reduction-project).