Abstract
Nicotinamide adenine dinucleotide (NAD+) is a key player in many metabolic pathways as an activated carrier of electrons. In addition to being the cofactor for redox re-actions, NAD+ also serves as the substrate for various enzymatic transformations such as adenylation and ADP-ribosylation. Maintaining cellular NAD+ homeostasis has been suggested as an effective anti-aging strategy. Given the importance of NAD+ in regulating a broad spectrum of cellular events, small molecules targeting NAD+ metabolism have been pursued as therapeutic interventions for the treatment of mitochondrial disorders and age-related diseases. In this article, small molecule regulators of NAD+ biosynthetic enzymes will be reviewed. The focus will be given to the discovery and development of these molecules, the mechanism of action as well as their therapeutic potentials.
Keywords: Nicotinamide adenine dinucleotide (NAD+), metabolic pathways, electrons, redox reactions, enzymatic transformations, therapeutic potentials
1. INTRODUCTION
Nicotinamide adenine dinucleotide (NAD+), best known for its role in redox biology, serves as the substrate for a group of NAD+-utilizing enzymes such as sirtuins (NAD+-dependent protein deacylases) [1], poly(ADP-ribose) polymerases (PARPs) [2, 3], and ADP-ribosyl cyclases [4]. These enzymes catalyze the cleavage of the N-glycosidic bond to remove the nicotinamide (NAM) moiety and subsequently transfer the ADP-ribosyl group to other small molecules or protein targets. NAD+-utilizing enzyme-mediated cellular processes play important roles in maintaining genomic integrity [5], regulating gene transcription [6], promoting DNA repair, as well as controlling cell proliferation and differentiation [7, 8]. Additionally, NAD+ and its reduced form, NADH, have been suggested as inhibitors of mitochondrial permeability transition pore (mPTP), the opening of which leads to the disruption of mitochondrial membrane homeostasis and triggers cell death through apoptosis or necrosis [9–11]. Both matrix [12, 13] and external NAD(H) [9] suppressed the opening of mPTP at millimolar concentrations to maintain the integrity of the mitochondrial membrane.
Given the importance of NAD+ in a myriad of cellular events, it is not surprising that the intracellular NAD+ content needs to be tightly and precisely regulated. In mammalian cells, NAD+ level is maintained through the coordinated actions of complementary de novo [14], salvage [15], and nicotinamide riboside (NR) [16] biosynthetic pathways. The de novo pathway (Fig. 1), also known as the kynurenine pathway, starts with tryptophan which can be oxidized to N-formylkynurenine by either tryptophan 2,3-dioxygenase (TDO) [17] or indoleamine 2,3-dioxygenase (IDO) [18]. Further hydrolysis of N-formylkynurenine by arylformamidase leads to the formation of L-kynurenine. Three consecutive reactions catalyzed by kynurenine 3-monooxygenase (KMO), kynureninase (KYU), and 3-hydroxyanthranilate 3,4-dioxygenase (HAO) are required to convert kynurenine to 2-amino-3-carboxymuconate semialdehyde, which undergoes spontaneous cyclization to quinolinic acid [19]. Subsequently, quinolinic acid is coupled with 5-phosphoribosyl-1-pyrophosphate (PRPP) by the action of quinolinate phosphoribosyl transferase (QPRTase) to produce nicotinic acid mononucleotide (NaMN) [20]. The adenylation of NaMN catalyzed by nicotinamide/nicotinic acid mononucleotide adenylyltransferase (NMNAT) forms nicotinic acid adenine dinucleotide (NaAD). Completion of NAD+ synthesis is accomplished by amidation of NaAD to form NAD+ catalyzed by NAD+ synthetase.
Fig. (1).
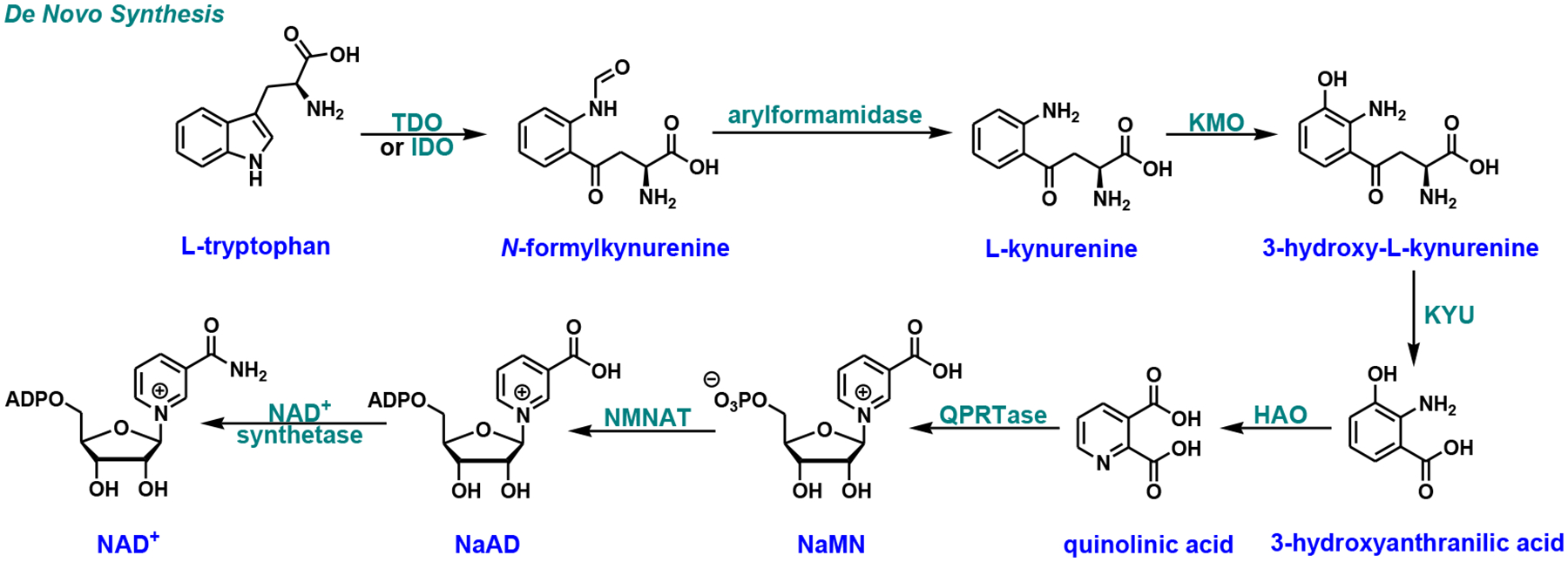
De novo NAD+ biosynthetic pathway in mammals. TDO, tryptophan 2,3-dioxygenase; IDO, indoleamine 2,3-dioxygenase; KMO, kynurenine 3-monooxygenase; KYU, kynureninase; HAO, 3-hydroxyanthranilate 3,4-dioxygenase; QPRTase, quinolinate phosphoribosyl transferase; NMNAT, nicotinamide/nicotinic acid mononucleotide adenylyltransferase; NaMN, nicotinic acid mononucleotide; NaAD, nicotinic acid adenine dinucleotide.
The salvage pathway recycles NAD+ degradation products to reconstitute NAD+ (Fig. 2). NAM is coupled with PRPP by nicotinamide phosphoribosyl transferase (NAMPT), the rate-limiting enzyme of the pathway, to form nicotinamide mononucleotide (NMN). Ultimately, NMN can be adenylated to NAD+ by NMNAT. Similarly, nicotinic acid (NA) is avidly recycled by nicotinic acid phosphoribosyl transferase (NaPRTase) to NaMN through the Preiss-Handler pathway [21]. NaMN thus formed can then enter the de novo pathway to be further transformed.
Fig. (2).
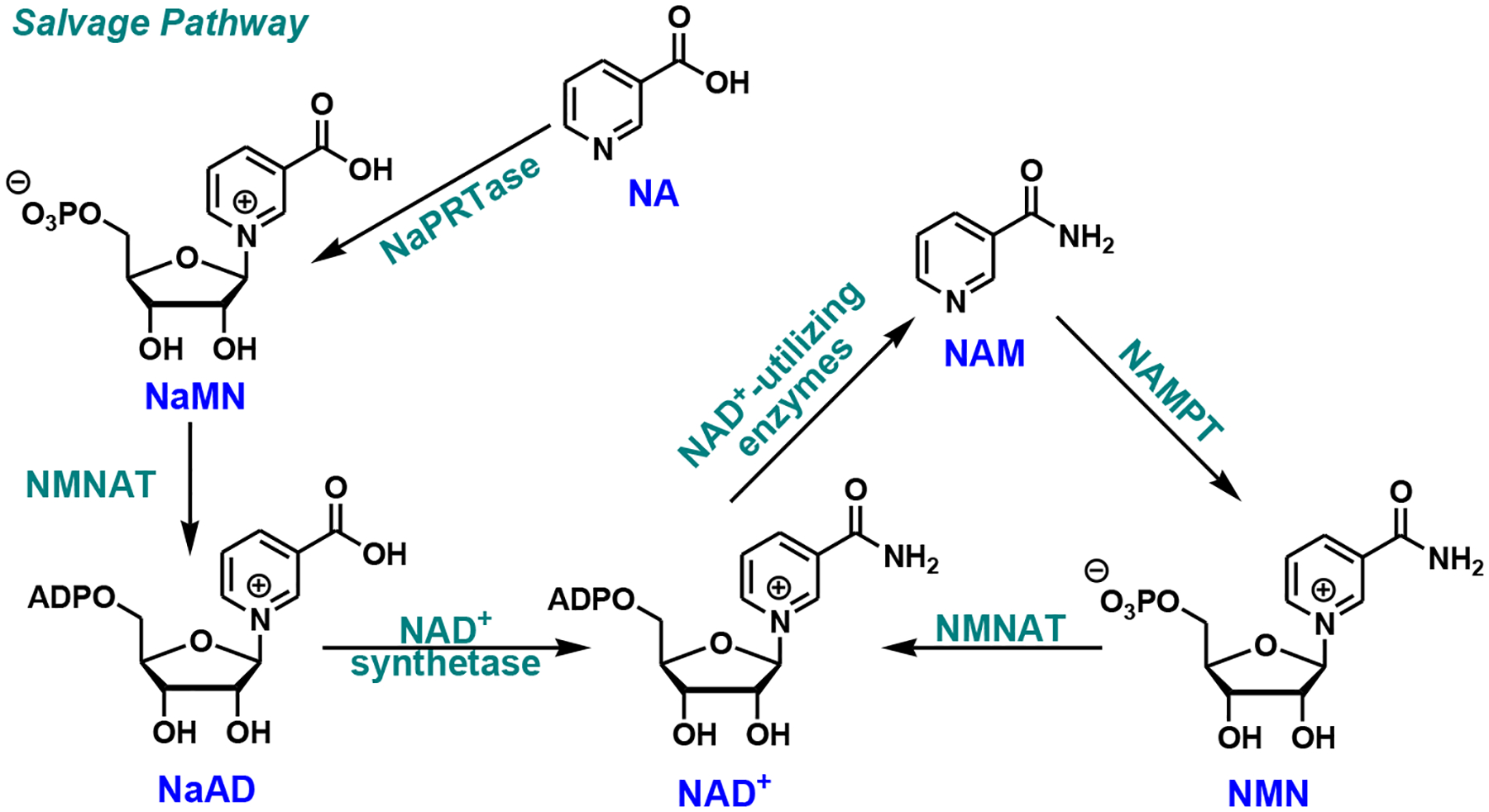
Mammalian salvage pathway. NAMPT, nicotinamide phosphoribosyl transferase; NaPRTase, nicotinic acid phosphoribosyl transferase; NAM, nicotinamide; NA, nicotinic acid.
The NR pathway was discovered more than a decade ago (Fig. 3) [16, 22]. NR, a natural metabolite and an ingredient in milk, is phosphorylated by nicotinamide riboside kinases (NRK1/NRK2) to NMN and eventually to NAD+. Recent studies suggest that NR is one of the most potent agents to stimulate NAD+ production in cells [23–25]. NR is water-soluble and cell-permeable with no apparent toxicity [26]. All of these features render NR an ideal candidate for the NAD+ boosting campaign.
Fig. (3).
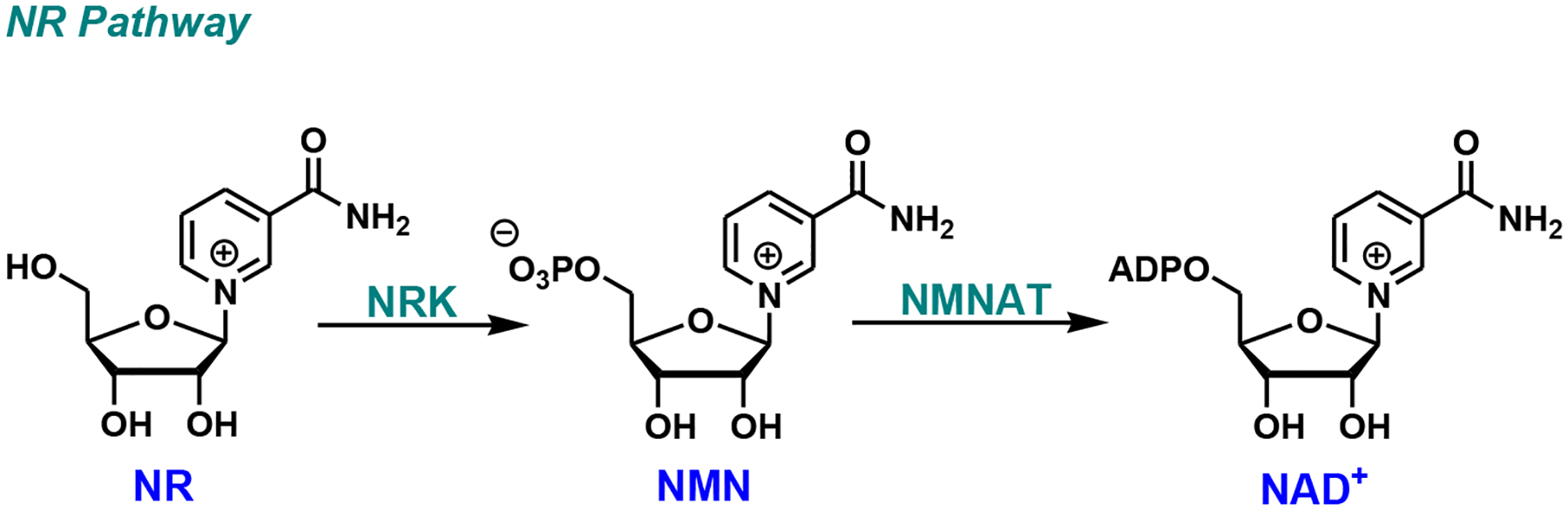
NR pathway. NRK, nicotinamide riboside kinase; NR, nicotinamide riboside.
There is a renewed interest in understanding NAD+ metabolism because increasing cellular NAD+ contents has been suggested as a potential anti-aging strategy [27–29]. During the normal aging process, the NAD+ level decreases as a result of oxidative stress, reduced mitochondrial biogenesis, and imbalanced synthesis and consumption of NAD+ [30–32]. This decline has been associated with age-related diseases such as metabolic disorders, neurodegenerative diseases, and cancer. Exercise, calorie restriction (CR), and supplementation with NAD+ precursors are known to augment intracellular NAD+ concentrations [33–37], which may slow down or even reverse the aging process and delay the progression of age-associated disorders. From herein, we will provide an overview of the discovery and development of small molecule modulators targeting NAD+ biosynthetic enzymes. The design and optimization of some representative cases and their potential therapeutic applications will also be discussed.
2. MOLECULES TARGETING NAD+ BIOSYNTHETIC PATHWAYS
Maintaining NAD+ homeostasis is essential for cell survival and growth. Pharmacological interventions of the above-mentioned NAD+ biosynthetic pathways have been intensely pursued as therapeutic treatments for mitochondrial dysfunction and age-related disorders. It is important to point out that although NR has attracted great interest as a potent NAD+ boosting agent, there have not been many advances in identifying small molecule regulators of NRKs. Our discussion will be focusing on de novo and salvage pathway enzymes, particularly those with known regulators.
2.1. Compounds targeting de novo NAD+ biosynthesis
2.1.1. IDO Inhibitors
The heme enzyme IDO catalyzes the oxidation of tryptophan by molecular oxygen to form N-formylkynurenine [38, 39]. IDO is distinct from TDO in its tissue expression pattern, substrate specificity, and inducibility. It is highly expressed in non-hepatic tissues [40] and has a broader specificity as it triggers the oxidative cleavage of indole moiety in tryptophan, 5-hydroxytryptophan, and serotonin [41]. IDO can be induced by inflammatory stimuli such as interleukin-1 (IL-1) [42], tumor necrosis factor (TNF) [43], and bacterial lipopolysaccharide (LPS) [44]. It has been implicated in mediating tumor immune escape and suggested as an oncology target [45, 46].
Most of the IDO inhibitors are substrate analogs. For example, 1-methyltryptophan, β-(3-benzofuranyl) alanine, and β–(3-benzothienyl)alanine (Fig. 4), discovered in the early 1990s, were competitive inhibitors of IDO [47]. Using purified rabbit IDO, the Ki values were determined to be 6.6 ± 0.6 μM for 1-methyltryptophan, 70 ± 4 μM for β-(3-benzofuranyl) alanine, and 25 ± 2 μM for β-(3-benzothienyl)alanine, respectively [47]. The D-isomer of 1-methyltryptophan, indoximod (D-1MT/NLG-8189), was one of the first IDO inhibitors entering clinical trials as a combination therapy for breast cancer [48], prostate cancer [49], and melanoma [50]. Although the mechanism of action (MOA) of indoximod remains elusive, it has been suggested that it acts as a tryptophan mimic to restore tryptophan signaling to the mTOR pathway after tryptophan depletion by IDO [51].
Fig. (4).
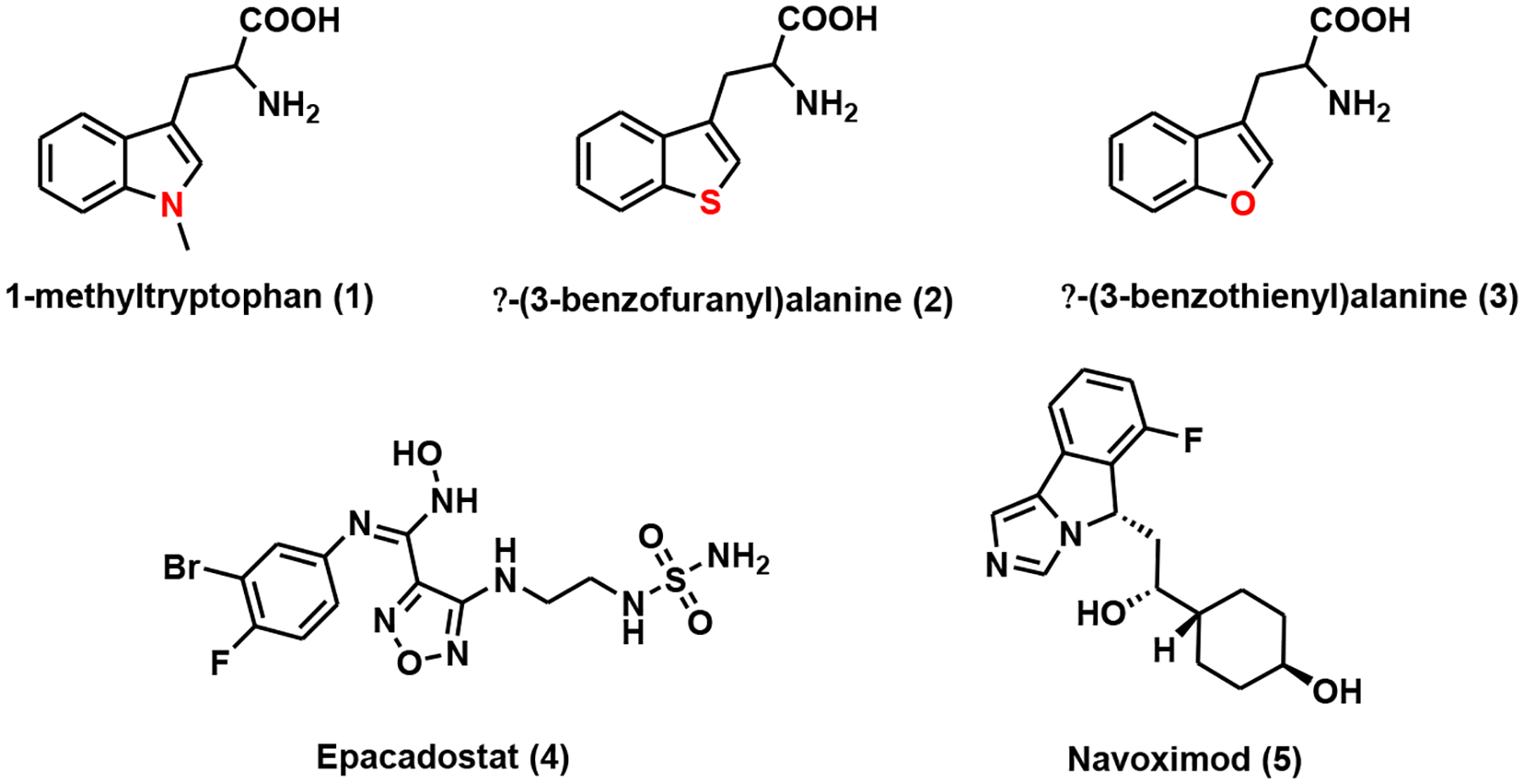
IDO inhibitors.
Other pharmacophores have also been explored for the discovery of IDO inhibitors. Epacadostat (or INCB24360 (Fig. 4), with a hydroxyamidine scaffold, is a potent and selective IDO1 inhibitor with an IC50 of 10 nM as determined by a cell-based assay [52]. Despite lacking classical drug-like properties, epacadostat demonstrated excellent cell permeability and pharmacokinetic (PK) profile in animal models [52]. This compound was used in combination with anti-PD1 monoclonal antibody (mAb) pembrolizumab in phase III clinical trial for the treatment of melanoma [53]. Navoximod (or NLG919, Fig. 4) was inspired by a series of structure-based drug development campaigns stemming from 4-phenylimidazole [54, 55]. Navoximod inherited phenylimidazole ability to bind to heme iron with improved active site occupancy and hydrogen bond interactions. Using cell-based assays, the EC50 of navoximod was determined to be 75 nM [56]. It was entered into clinical trials either as a monotherapy or combination therapy for solid tumors due to its favorable bioavailability, PK, and toxicity profiles [56–58].
2.1.2. KMO Inhibitors
KMO serves as a key branching point enzyme of the de novo NAD+ biosynthetic pathway. The inhibition of this enzyme funnels L-kynurenine to kynurenic acid, a metabolite with anticonvulsant and neuroprotective properties due to its antagonism against the ionotropic receptors such as N-methyl-D-aspartate (NMDA) receptor and nicotinic acetylcholine receptor (nAChR) [39, 59–61]. KMO is a FAD-dependent monooxygenase that converts L-kynurenine to 3-hydroxy-L-kynurenine, a free radical generator known to increase oxidative stress and induce neuronal cell death [62, 63]. Kmonull mice showed markedly reduced serum 3-hydroxy-L-kynurenine levels and improved phenotypes in response to extrapancreatic tissue insults, suggesting that pharmacological inhibition of KMO could serve as a novel therapeutic treatment for acute pancreatitis (AP) [64].
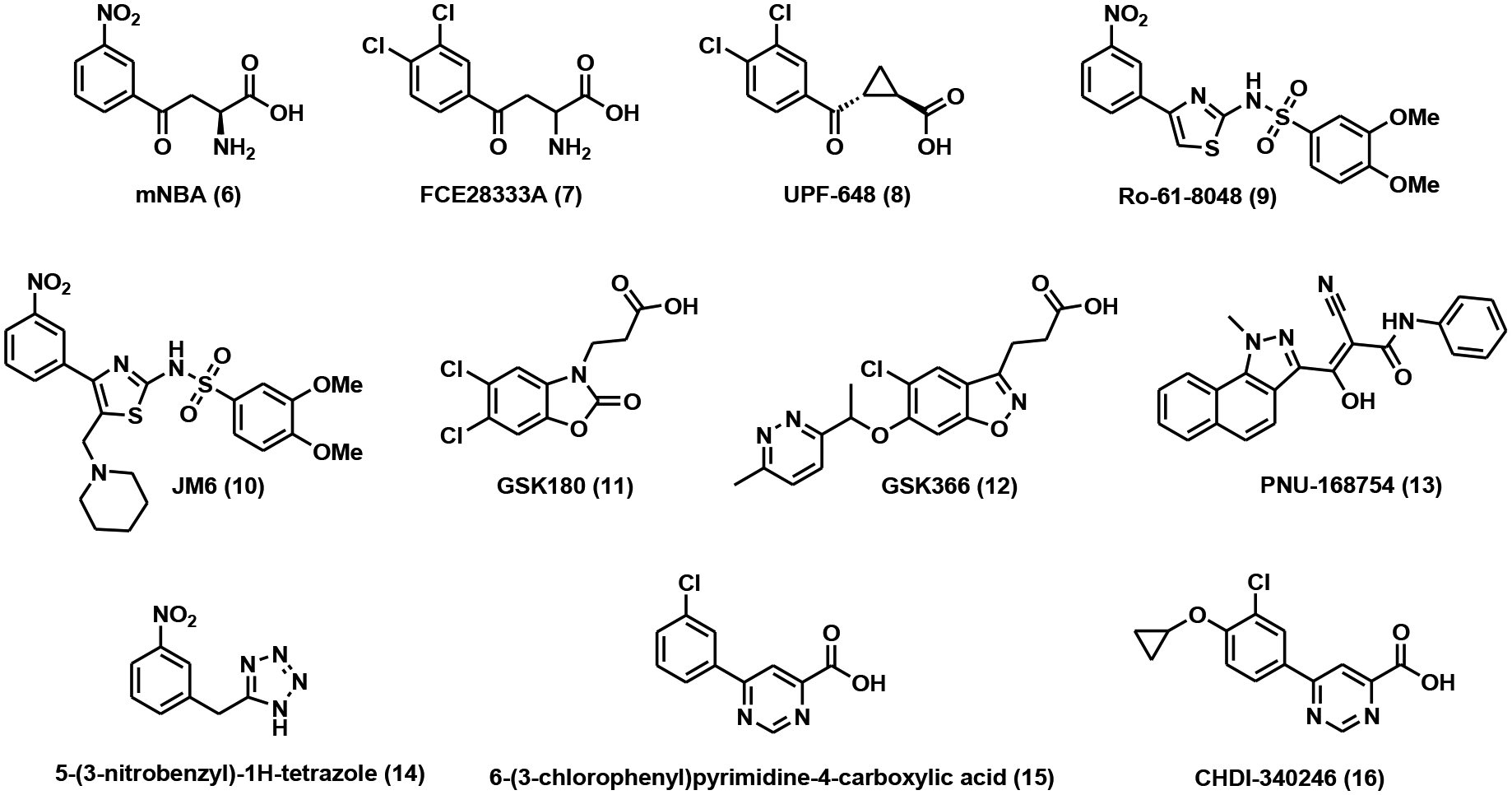
KMO inhibitors (Table 1) can be classified into several major categories, including substrate analogs (4-phenyl-4-oxobutanoic acids, sulfonamides, oxazolidinones), 3-oxo-propanitriles, tetrazoles, and arylpyrimidines.
Table 1.
KMO inhibitors.
| Compound | Name | IC50 (nM) | Assay | References |
|---|---|---|---|---|
| 6 | mNBA | 900 ± 100 | competitive inhibition assay (rat tissues) | [65] |
| 7 | FCE28333A | 200 ± 20 | inhibition assay (rat brain tissue) | [66] |
| 8 | UPF-648 | 40a | recombinant human KMO assay | [67] |
| 9 | Ro-61–8048 | 37 ± 3 | radioenzymatic assay (rat kidney mitochondria) | [69] |
| 10 | JM6 | ~4000 | enzyme coupled assay (mouse brain homogenate) | [59] |
| 11 | GSK180 | 6 | recombinant human KMO assay | [64] |
| 12 | GSK366 | 2.3 | Rapidfire MS assay (recombinant human KMO) | [70] |
| 13 | PNU-168754 | 40 | radiometric assay (rat liver mitochondrial extract) | [71] |
| 14 | 5-(3-nitrobenzyl)-1H-tetrazole | 6300 | Rapidfire MS assay (recombinant human KMO) | [72] |
| 15 | 6-(3-chlorophenyl)pyrimidine-4-carboxylic acid | 0.5 | human KMO assay | [73] |
| 16 | CHDI-340246 | 0.5 | human KMO assay | [73] |
IC50 for the racemic mixture.
2.1.2.1. Substrate Analogs
2.1.2.1.1. 4-Phenyl-4-oxobutanoic Acids
Substrate analogs were the first class of KMO inhibitors to be explored, even before the crystal structure of KMO was solved. m-Nitrobenzoyl-L-alanine (mNBA, Fig. 5), a close mimic of the endogenous substrate kynurenine, is a competitive inhibitor of KMO with an IC50 of 0.9 μM [65]. It potently increased the levels of kynurenine in the brain and blood of rats when administered at a dosage of 400 mg/kg intraperitoneally [65]. A series of structure-activity relationship (SAR) campaigns inspired by mNBA led to the identification of FCE28833A (Fig. 5), a dichloronated derivative of mNBA. It demonstrated improved potency with an IC50 of 0.2 μM [66]. Oral treatment of FCE28333A at 400 mg/kg resulted in a 14-fold increase of kynurenine in the rat brain [66]. A nanomolar KMO inhibitor, UPF-648 (IC50 = 40 nM) [67], was developed by inserting a cyclopropyl group between the carbonyl and carboxylic acid moieties in FCE28333A (Fig. 5). Significant elevation of kynurenine in the gerbil brain and plasma was observed after UPF-648 treatment [67]. The co-crystal structure of UPF-648 in complex with S. cerevisiae KMO was later determined [68]. The binding of UPF-648 to the active site of KMO induced a structural rearrangement to preclude substrate binding [68]. This study provided a blueprint for the future structure-based inhibitor design for the highly related human KMO.
Fig. (5).
KMO inhibitors.
2.1.2.1.2. Sulfonamides
Removal of the amino group in mNBA, which was known to not contribute significantly to the ligand binding, and replacing the carboxylic acid moiety with a sulfonamide isostere led to the discovery of a group of nanomolar KMO inhibitors [69]. Ro-61–8048 (Fig. 5), with an IC50 of 37 nM in vitro, increased brain kynurenine levels with an EC50 of 5.5 μmol/kg [69]. JM6 is an orally bioavailable prodrug of Ro-61–8048 (Fig. 5). The additional N-methylenepiperidine group can be cleaved off under acidic conditions to release Ro-61–8048. Chronic administration of JM6 not only raised the neuroprotective kynurenic acid levels in the brain but also ameliorated neurodegeneration in Alzheimer’s (AD) and Huntington’s disease (HD) mouse models [59]. Both Ro-61–8048 and JM6 showed poor blood-brain barrier (BBB) permeability, suggesting that the peripheral inhibition of KMO could serve as a therapeutic approach for neurodegenerations [59].
2.1.2.1.3. Oxazolidinones and Benzisoxazoles
GSK180 (Fig. 5), with an oxazolidinone core, is a competitive KMO inhibitor with an IC50 of 6 nM as determined by a recombinant KMO biochemical assay [64]. It preserves the carboxylic acid and hydrogen bond acceptor required for KMO inhibition. Moreover, GSK180 also demonstrates favorable physiochemical properties for intravenous administration [64]. Treatment with GSK180 raised the levels of kynurenine in mice and provided protective effects against multiple organ dysfunction syndrome (MODS) in a rat model of AP [64].
GSK366 is a benzisoxazole analog of GSK180 (Fig. 5). It is one of the most potent human KMO inhibitors with an IC50 of 2.3 nM [70]. The addition of the methylpyridazine group to the benzisoxazole core structure was thought to trap FAD in a “tilting” conformation, which further improved binding affinity and residence time with minimum peroxide formation [70]. Due to these favorable properties, GSK633 has been suggested as a suitable clinical candidate for the treatment of AP and MODS.
2.1.2.2. 3-Oxo-propanitriles
A group of tricyclic 3-oxo-propanitriles has been disclosed in a patent. They act as KMO inhibitors, among which PNU-168754 (Fig. 5) showed an IC50 of 40 nM [71]. Unfortunately, no additional information is available for these compounds.
2.1.2.3. Tetrazoles
Tetrazole analogs were uncovered using a high throughput mass spectrometry (MS) assay as human KMO inhibitors [72]. One representative compound, 5-(3-nitrobenzyl)-1H-tetrazole (Fig. 5), was reported to have an IC50 of 6.3 μM [72].
2.1.2.4. Arylpyrimidines
Arylpyrimidine carboxylic acids were considered as rigid cyclic analogs of kynurenine. They were designed to retain the favorable structural features of known KMO inhibitors. The structure of one of the lead compounds in this series, 6-(3-chlorophenyl)pyrimidine-4-carboxylic acid, is shown in Fig. (5). This remarkably simple compound demonstrated sub-nanomolar potency against human KMO (IC50 = 0.5 nM) [73]. Further elaboration of the phenyl ring with a paracyclopropoxy group led to the formation of CHDI-340246 (Fig. 5) as a potent KMO inhibitor both in vitro and in vivo [73]. This inhibitor showed limited ability to cross the BBB. Oral administration of CHDI-340246 at 10 mg/kg dosage caused pronounced elevation of plasma kynurenine levels and reduction of 3-hydroxy-L-kynurenine with excellent ADME and PK profiles in Sprague-Dawley rats [73]. In an HD mouse model, treatment of CHDI-340246 restored spiny projection neurons (SPNs) membrane excitability without significant improvement in behavioral phenotypes or disease progression [74].
2.1.3. KYU Inhibitors
KYU is a PLP-dependent enzyme that facilitates the hydrolytic cleavage of the Cβ-Cγ bond in either kynurenine or 3-hydroxykynurenine to generate anthranilic acid or 3-hydroxyanthranilic acid, respectively [75]. Human KYU demonstrated a 256-fold substrate preference toward 3-hydroxykynurenine [76]. When KMO is inhibited, KYU provides an alternative pathway to form quinolinic acid.
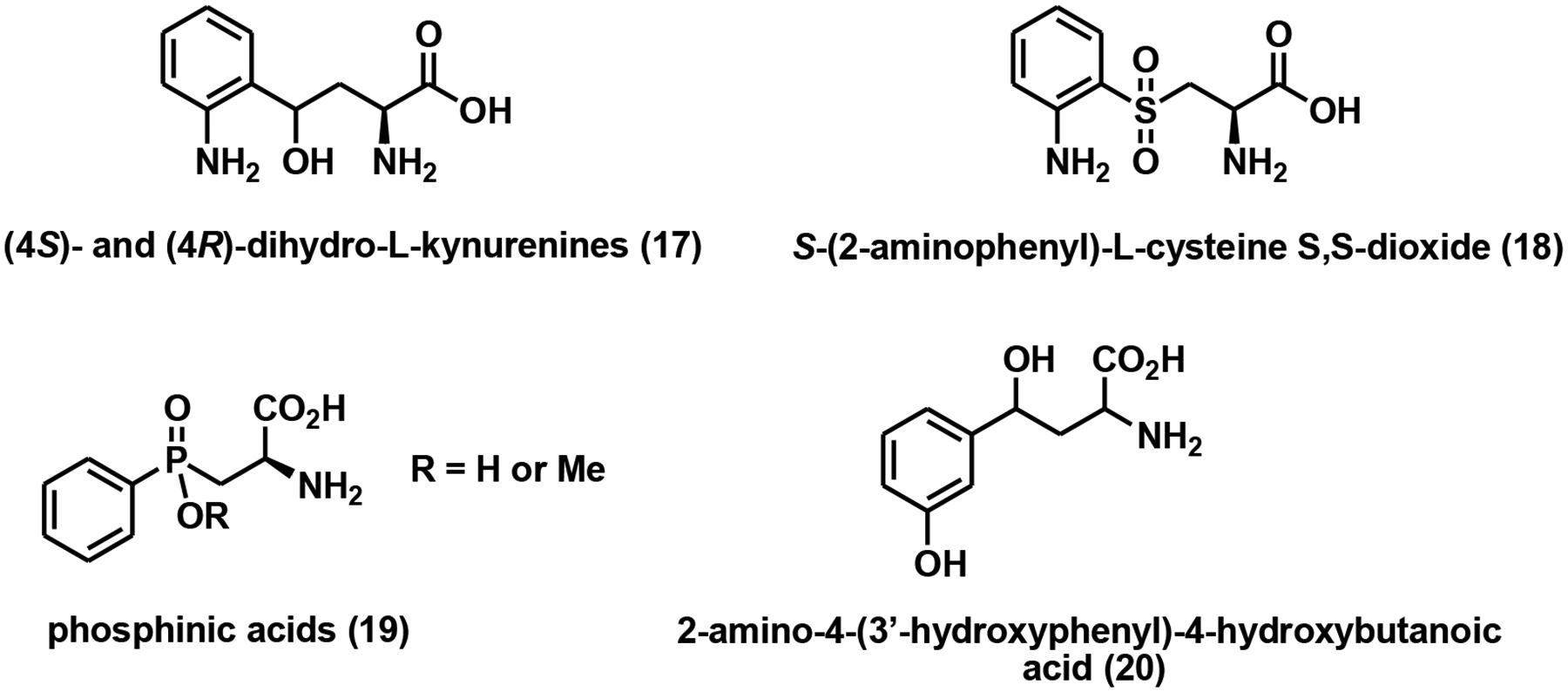
So far, only a handful of potent and specific KYU inhibitors have been discovered, most of which are transition state (TS) mimics of this enzyme. (4S)- and (4R)-dihydro-L-kynurenines (Fig. 6) are competitive inhibitors of KYU with the Ki values of 0.3 and 1.4 μM, respectively [77]. Based on this work, a group of S-aryl-L-cysteine S,S-dioxides were also developed as KYU inhibitors. The most potent compound in this group, S-(2-aminophenyl)-L-cysteine S,S-dioxide (Fig. 6), has a Ki of 70 nM against P. fluorescens KYU [78]. Chemically stable TS mimics are powerful inhibitors by trapping catalysis energy as binding energy. A couple of phosphinic acid analogs (Fig. 6) were thus designed and synthesized with the Ki values in the micromolar range [79]. A nanomolar human KYU inhibitor, 2-amino-4-(3’-hydroxyphenyl)-4-hydroxybutanoic acid (Fig. 6), was identified in 2002. It inhibited rat, human, and bacterial KYU with Ki values of 130 nM, 100 nM, and 10 μM, respectively [80].
Fig. (6).
KYU inhibitors.
2.1.4. QPRTase Inhibitors
In mammals, QPRTase is primarily localized in the liver and kidney, and to a lesser extent, the brain and spleen [81]. As the rate-limiting enzyme of the de novo pathway, QPRTase converts quinolinic acid to NAD+ even after the salvage pathway is inhibited [82]. QPRTase has been suggested as a therapeutic target for the treatment of gliomas [83]. Overexpression of QPRTase was thought to confer poor prognosis and increase resistance to oxidative stress through stimulating NAD+ de novo production [83].
The only known QPRTase inhibitor is phthalic acid. This compound structurally mimics the native substrate and serves as a competitive inhibitor of human QPRTase with a Ki of 2.8 μM as determined by a continuous UV assay [84]. Treatment of phthalic acid in primary astrocytes and neurons decreased intracellular NAD+ levels in a concentration-dependent manner, and in turn, reduced NAD+-dependent SIRT1 activity, both of which were thought to compromise cell viability [85].
2.1.5. NMNAT Modulators
NMNAT is a common enzyme involved in all three NAD+ biosynthetic pathways. In mammals, there are three NMNAT isoforms with distinct subcellular localizations: NMNAT1 is mainly localized in the nucleus, NMNAT2 resides primarily in the Golgi complex, and NMNAT3 is a mitochondrial protein [86]. The three isoforms also demonstrated differential substrate selectivity, with NMNAT3 having the highest substrate tolerance [86]. The NMNATs were thought to play key roles in regulating compartment-specific “NAD pools” [86]. Overexpression of NMNATs extended lifespans with improved health profiles in model organisms [87–89]. More strikingly, NMNATs also exhibited neuronal protective effects, presumably through stimulating NAD+-dependent sirtuin activity [90–92]. Recently, NMNAT2 has been suggested as an anti-cancer therapeutic target. Overexpression of NMNAT2 in colorectal cancer (CRC) patients showed positive correlations with tumor invasiveness and p53 expression [93]. At the transcriptional level, NMNAT2 expression can be induced by p53 in response to DNA damage [94]. At the protein level, NMNAT2 is deacetylated and activated by SIRT3 [95]. The SIRT3-NMNAT2 axis has been proposed as the key regulator of cell growth and proliferation in non-small cell lung cancer (NSCLC) cells [95].
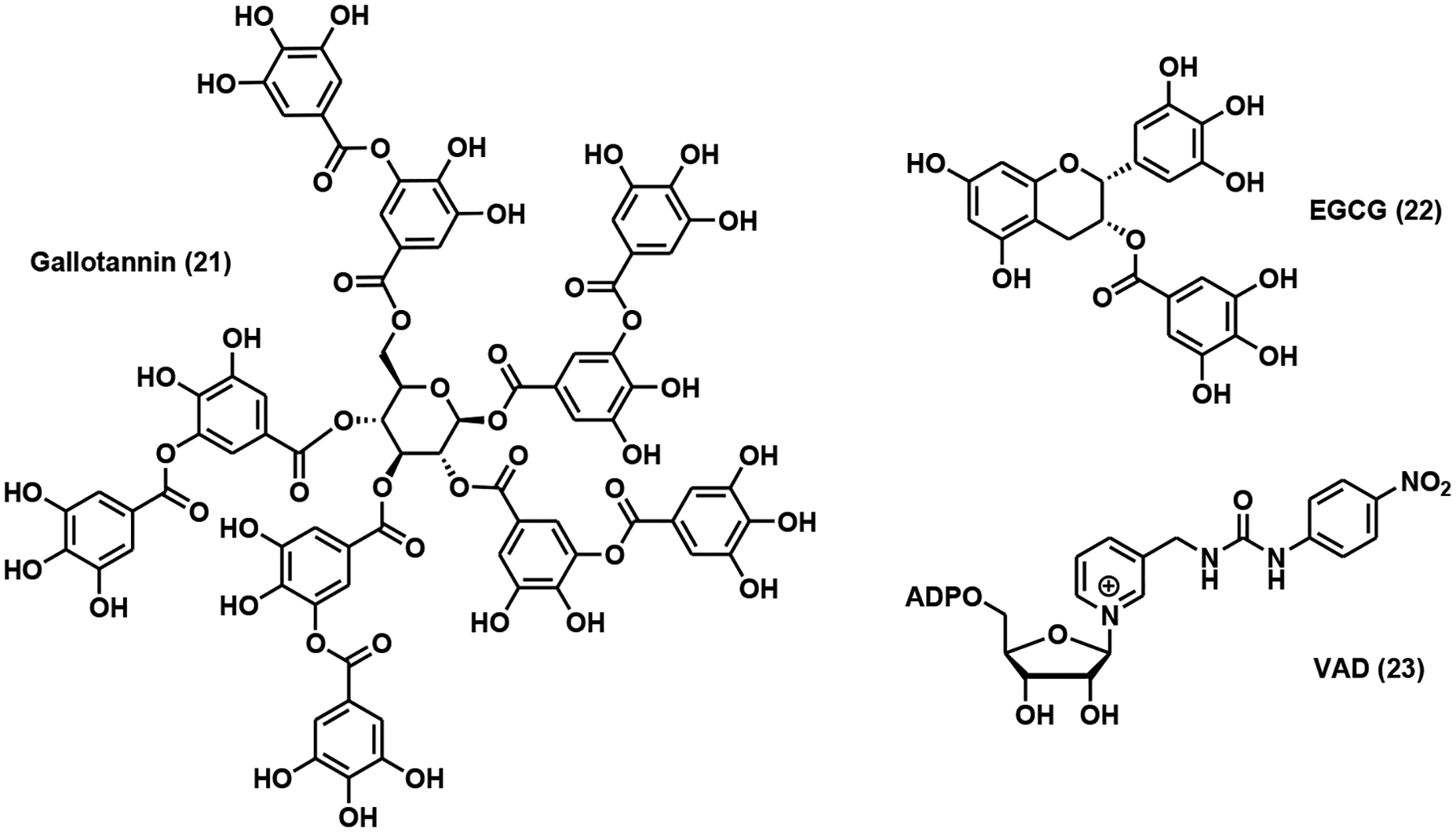
Natural products were first explored for the identification of NMNAT modulators. Gallotannin (Fig. 7), a known PARP inhibitor, inhibited all three NMNAT isoforms with IC50 values of 10, 55, and 2 μM [86]. Epigallocatechin gallate (EGCG, Fig. 7), at 50 μM concentration, doubled NMNAT2 activity and also activated NMNAT3 to a lesser extent [86]. Vacor adenine dinucleotide (VAD, Fig. 7), a metabolite of Vacor and an NAD+ analog, inhibited NMNAT2 and NMNAT3 with IC50 values of 20 and 463 μM, respectively [82]. Unfortunately, VAD also inhibited other NAD+-dependent dehydrogenases with micromolar potency [82]. Treatment of VAD in NMNAT2-expressing cancer cells caused significant NAD+ depletion, reduced glycolysis, and energy metabolism, and ultimately necrosis [82].
Fig. (7).
NMNAT modulators.
2.2. Compounds Targeting NAD+ Salvage Pathway
2.2.1. NAMPT Modulators
NAMPT is the rate-limiting enzyme of the salvage pathway that recycles NAM directly to NMN. Mammalian NAMPT is expressed in intracellular and extracellular forms [96]. Unlike the intracellular NAMPT (iNAMPT), which has been confirmed as an NAD+ biosynthetic enzyme, the extracellular form of NAMPT is a multifaceted protein. It serves as a cytokine named pre-B cell colony enhancing factor (PBEF) [97], and as an insulin-like adipocytokine termed visfatin [98] in addition to its role in extracellular NAD+ biosynthesis (eNAMPT). eNAMPT demonstrates robust NAD+ biosynthetic activity, even higher than that of iNAMPT, and acts coordinately with iNAMPT to regulate pancreatic insulin secretion [99]. Overexpression of NAMPT has been closely correlated with poor prognosis in astrocytoma/glioblastoma [100], increased tumor-igenesis and metastases in melanoma [101], and increased aggressiveness in malignant lymphoma [102]. All of these suggested NAMPT as an anti-cancer therapeutic target.
Co-crystal structures of NAMPT in complex with known inhibitors, GMX1778 and FK866, suggested that an ideal NAMPT inhibitor should be a heterocyclic NAM analog conjugated to a tail group through a hydrogen bond-forming spacer and a narrow hydrophobic linker (Fig. 8) [103, 104]. FK866 (Fig. 8), initially designed as a substrate mimic, represents the first noncompetitive yet highly specific NAMPT inhibitor with nanomolar potency as determined by cell viability assays [105, 106]. It induced apoptosis in cancer cells via blockade of the NAD+ salvage pathway [105, 106]. FK866 is one of the few NAMPT inhibitors that advanced into clinical trials for the treatment of T-cell lymphoma [107], advanced melanoma (ClinicalTrials.org identifier: NCT00432107), and B-cell chronic lymphocytic leukemia (ClinicalTrials.org identifier: NCT00435084). GMX1778 (Fig. 8), structurally distinct from FK866, is a competitive NAMPT inhibitor [108, 109]. It exhibited low nanomolar cytotoxicity, which can be rescued by exogenously added NA or NAM [108, 109]. To improve the water solubility of GMX1778, a polyethylene glycol (PEG) linker was tethered to the parent compound through an esteraselabile carbonate, leading to the formation of GMX1777 (Fig. 8) [110]. Intravenous administration of GMX1777 at 75 mg/kg led to tumor regression in several models [110]. FK866 and GMX1778/GMX1777 laid the foundation for further medicinal chemistry campaigns of NAMPT inhibitors. The following discussion will be grouped into three different sections: 1) FK866 and GMX1778 inspired inhibitors; 2) Inhibitors with novel pharmacophores; and 3) Dual NAMPT inhibitors. All of these NAMPT inhibitors are summarized in Table 2.
Fig. (8).
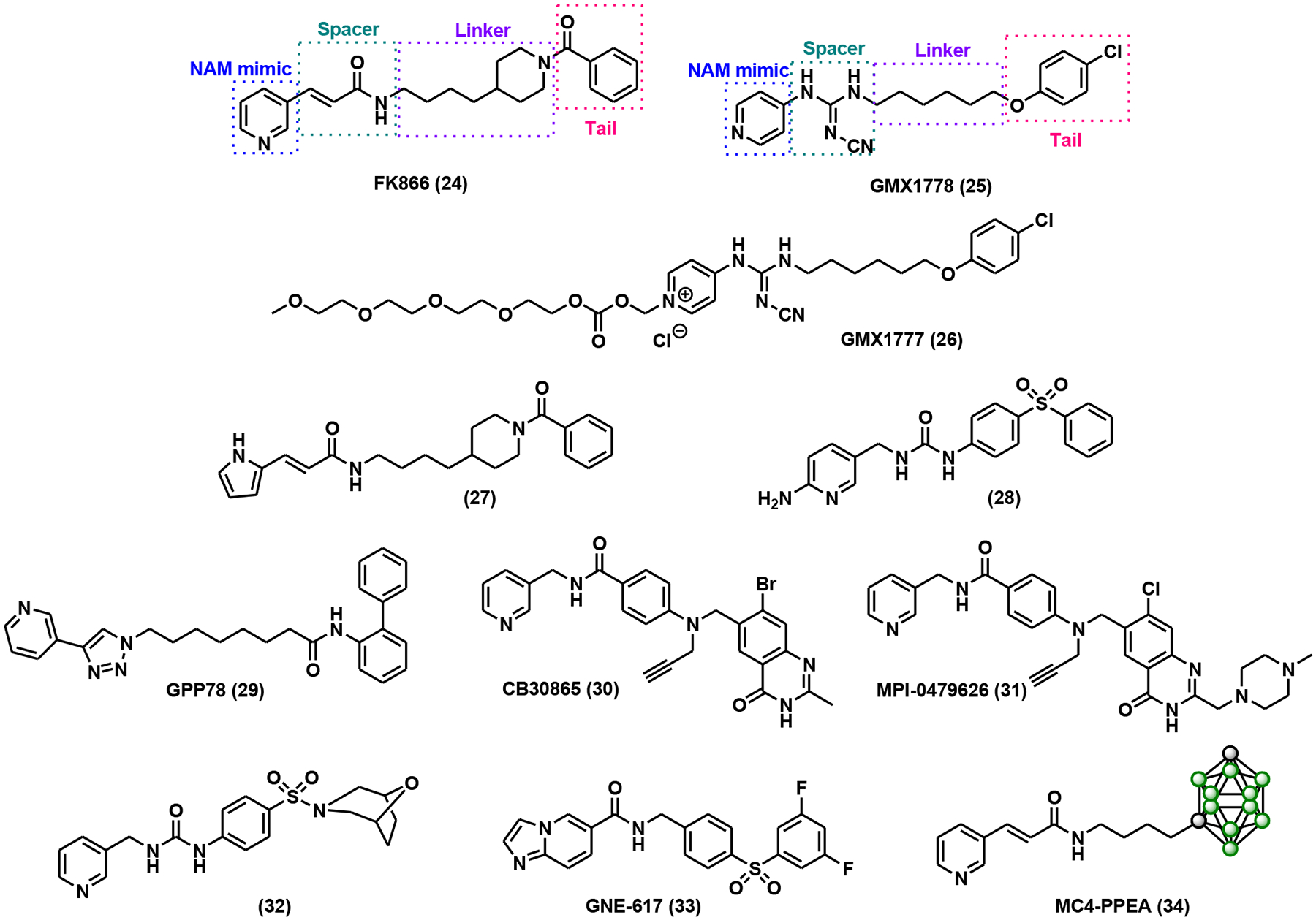
FK866 and GMX1778 inspired NAMPT inhibitors.
Table 2.
NAMPT inhibitors
| Compound | Name | IC50 (nM) | Assay | References |
|---|---|---|---|---|
| 24 | FK866 | 4.90 ± 0.55 | MTT assay (NIH-3T3 cells) | [146] |
| 1.60 ± 0.32 | human NAMPT assay | [147] | ||
| 25 | GMX1778 | < 25 | coupled enzyme NAMPT assay | [109] |
| Kd = 120 nM | recombinant human NAMPT | [109] | ||
| 27 | 1290±50 | SRB assay | [111] | |
| 20300 | HPLC-based NAMPT assay | [111] | ||
| 28 | 3 | NAMPT biochemical inhibition assay | [112] | |
| 70 | SRB assay (A2780 cells) | [112] | ||
| 29 | GPP78 | 3.8 ± 0.3 | MTT assay (SH-SY5Y cells) | [114] |
| 30 | CB30865 | 2.1 ± 0.44 | MTT assay (CH1 cells) | [148] |
| 31 | MPI-0479626 | 0.38 ± 0.08 | NAD depletion assay | [117] |
| 0.23 ± 0.12 | NAMPT inhibition assay | [117] | ||
| 32 | 7 | NAMPT biochemical inhibition assay | [113] | |
| 32 | SRB assay (A2780 cells) | [113] | ||
| 33 | GNE-617 | 5 | NAMPT biochemical inhibition assay | [118] |
| 2 | SRB assay (A2780 cells) | [118] | ||
| 34 | MC4-PPEA | 0.31 | MTT assay (DLD1 cells) | [119] |
| 35 | Ki = 80 nM | TR-FRET assay | [120] | |
| 36 | 4162 | 777 ± 84 | NAMPT biochemical inhibition assay | [115] |
| 37 | 6443 | 352 ± 45 | NAMPT biochemical inhibition assay | [115] |
| 38 | 3909 | 504 ± 124 | NAMPT biochemical inhibition assay | [115] |
| 39 | OT-82 | 41 | NAMPT biochemical inhibition assay | [121] |
| 40 | KPT-9274 | 120 | NAMPT biochemical inhibition assay | [124] |
| 570 | MTT assay (Caki-1 cells) | [124] | ||
| 41 | STF-31 | 19 | NAMPT biochemical inhibition assay | [149] |
| 120 | SRB assay (A2780 cells) | [149] | ||
| 42 | 15 | NAMPT biochemical inhibition assay | [127] | |
| 5200 ± 480 | MTT assay (A549 cells) | [127] |
2.2.1.1. FK866 and GMX1778 Inspired NAMPT inhibitors
2.2.1.1.1. NAM Mimic Modifications
The pyridine moiety is the common structural feature of FK866 and GMX1778. Attempts have been made to replace it with other nitrogen-containing heterocycles such as pyrimidine, pyrrole, and indole with very little success [111–113]. A pyrrole-based inhibitor (27, Fig. 8) exhibited comparable or mildly improved cytotoxicity in MCF7 and K562 cells [111]. 2-Aminopyridine analog (28, Fig. 8) demonstrated nanomolar IC50s in both recombinant NAMPT (3 nM) and A2780 cell proliferation (70 nM) assays with good PK profile and efficacy in mouse models [112]. Accumulating data suggested that the pyridine ring is still the preferred NAM mimic for a potent NAMPT inhibitor.
2.2.1.1.2. Spacer Modifications
“Click” chemistry-mediated library construction has led to the development of a group of NAMPT inhibitors with triazole-containing spacers [114]. One representative compound from this library, GPP78 (Fig. 8), significantly reduced intracellular NAD+ content with an IC50 of 3 nM and induced cytotoxicity with an IC50 of 3.8 nM in SH-SY5Y cells [114]. In the same study, linker length was also investigated. Compounds with seven carbon linkers were the most potent ones across the panels [114]. In addition to vinylogous amide (such as the one in FK866), cyanoguanidine (such as the one in GMX1778) and triazole, amide, urea, thiourea spacers can also be tolerated by NAMPT [112, 115, 116].
2.2.1.1.3. Tail Group Modifications
CB30865 (Fig. 8), a quinazolinone derivative, was uncovered as a potent NAMPT inhibitor in a chemical proteomics study [117]. The methylpiperazinyl analog of CB30865, MPI-0479626 (Fig. 8), showed sub-nanomolar potency in a recombinant NAMPT inhibition assay as well as an HCT116 cell cytotoxic assay [117]. MPI-0479626 treatment caused remarkable NAD+ depletion and subsequent PARP inhibition in response to oxidative damage [117].
An extensive SAR study resulted in compound 32 (Fig. 8) featuring a sulfonamide tail group [113]. In addition to its nanomolar IC50s in in vitro assays, this inhibitor also demonstrated excellent ADME profiles [113]. Also noteworthy in this study was that the classic long hydrophobic linker was condensed to a benzene ring without significant loss of binding affinity and potency [113].
Diphenylsulfone can also be used as the tail group in NAMPT inhibitors. For example, GNE-617 (Fig. 8) possessed robust NAMPT inhibitory activity and in vivo efficacy [118]. It was proposed that the intracellular metabolism of GNE-617 produced a phosphoribosylated GNE-617, the true tight-binding inhibitor of NAMPT [118]. This was further confirmed by structural biology analysis.
Interestingly, FK866 analogs with carborane tail groups were explored as NAMPT inhibitors, taking advantage of carborane’s hydrophobic nature and its potential to form hydrogen bond interactions with the protein target [119]. MC4-PPEA (Fig. 8), in particular, exhibited an IC50 of 410 pM in A549 cells, four times more potent than FK866 [119].
2.2.1.2. Inhibitors with Novel Pharmacophores
In a recent study, fragment-based NMR and FRET screenings were coupled with structure-based drug design for the discovery of NAMPT inhibitors [120]. From two libraries of roughly 15,200 fragments, 6 ligands were initially identified as strong NAMPT binders [120]. Subsequently, structural biology-guided inhibitor design led to the synthesis and characterization of compound 35 (Fig. 9) with a Ki of 80 nM as determined by FRET assay [120].
Fig. (9).
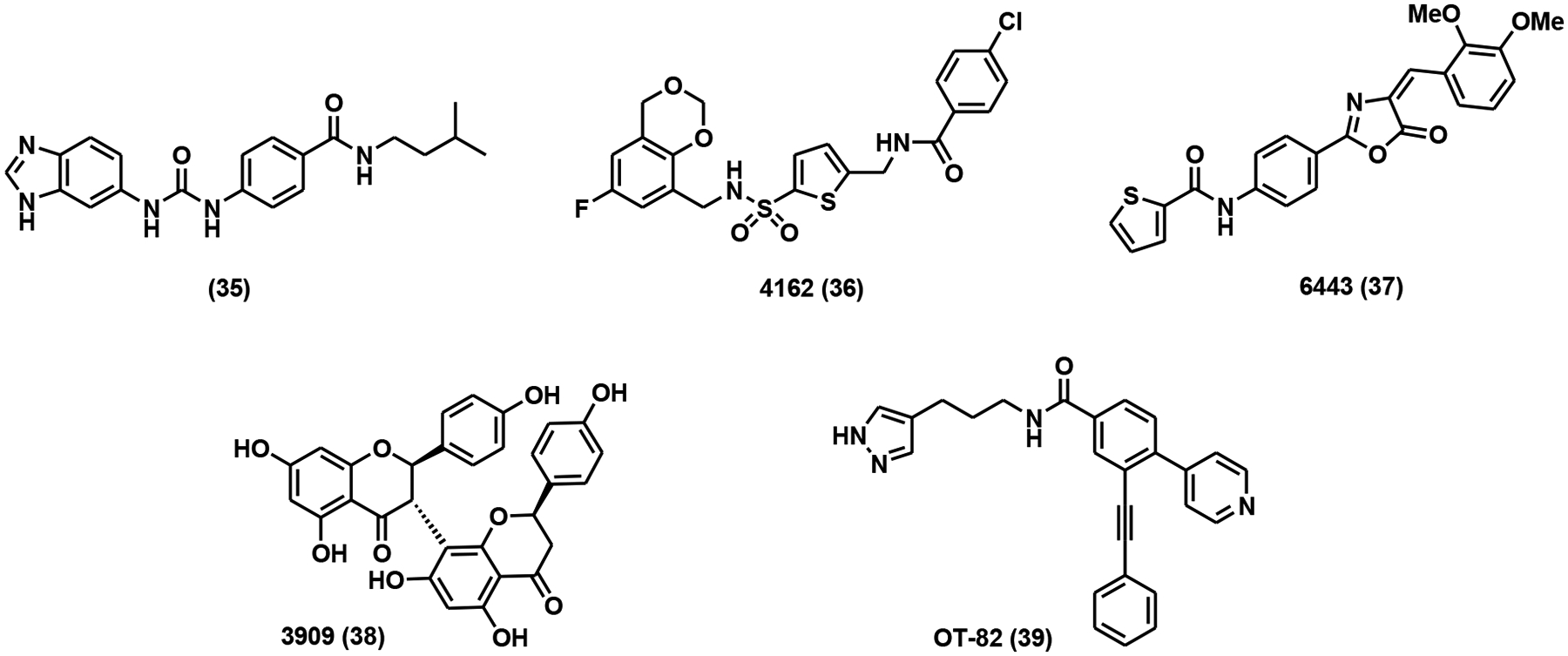
NAMPT inhibitors with novel pharmacophores.
High-throughput screenings (HTSs) have also yielded some potent NAMPT inhibitors with novel pharmacophores. An HTS using recombinant NAMPT uncovered several non-classical NAMPT inhibitors (compounds 36 to 38, Fig. 9), all of which had nanomolar potency [115]. These compounds clearly provided new templates for future inhibitor optimizations. OT-82 (Fig. 9), a lead compound discovered in a cell-based HTS, showed nanomolar potency in a panel of hematological malignancies and non-hematological malignancies derived cell lines [121]. No apparent toxicity of OT-82 at 55 mg/kg was observed in multiple mouse models. Currently, OT-82 has entered a Phase I clinical trial for the treatment of relapsed or refractory lymphoma (Clinical-Trials.gov identifier: NCT03921879) [121].
2.2.1.3. Dual NAMPT Inhibitors
Dual inhibitors are small molecule inhibitors targeting two protein targets simultaneously. The power of dual inhibitors has been increasingly appreciated because of their improved therapeutic efficacy and reduced risk of drug-drug interactions [122, 123]. KPT-9274 (Fig. 10) is a dual inhibitor of NAMPT and p21-activated kinase 4 (PAK4), both of which are essential for renal cell carcinoma (RCC) [124]. KPT-9274 not only reduced invasiveness and metastases of RCC cell lines but also inhibited tumor growth in a concentration-dependent manner in a human RCC xenograft model [124]. STF-31 (Fig. 10) serves as a dual inhibitor of both NAMPT and glucose transporter 1 (GLUT1) [125]. It inhibited glucose uptake and cell proliferation with distinct dosage windows in tumor cells [125]. Inhibitors targeting NAMPT and epigenetically important histone deacetylase (HDAC) have been developed using Cu(I)-catalyzed “click” reaction [126, 127]. A representative compound from these studies is shown in Fig. (10) (compound 42). It features a classical NAMPT inhibitor structure as well as the hydroxamate “warhead” targeting HDACs. It exhibited strong inhibitory effects on both targets with IC50 of 15 nM against NAMPT and 2 nM against HDAC1, respectively [127]. At 25 mg/kg intraperitoneal dosage, this compound potently inhibited tumor growth in an HCT116 xenograft mouse model, outperforming FK866 and SAHA (a known HDAC inhibitor) [127].
Fig. (10).
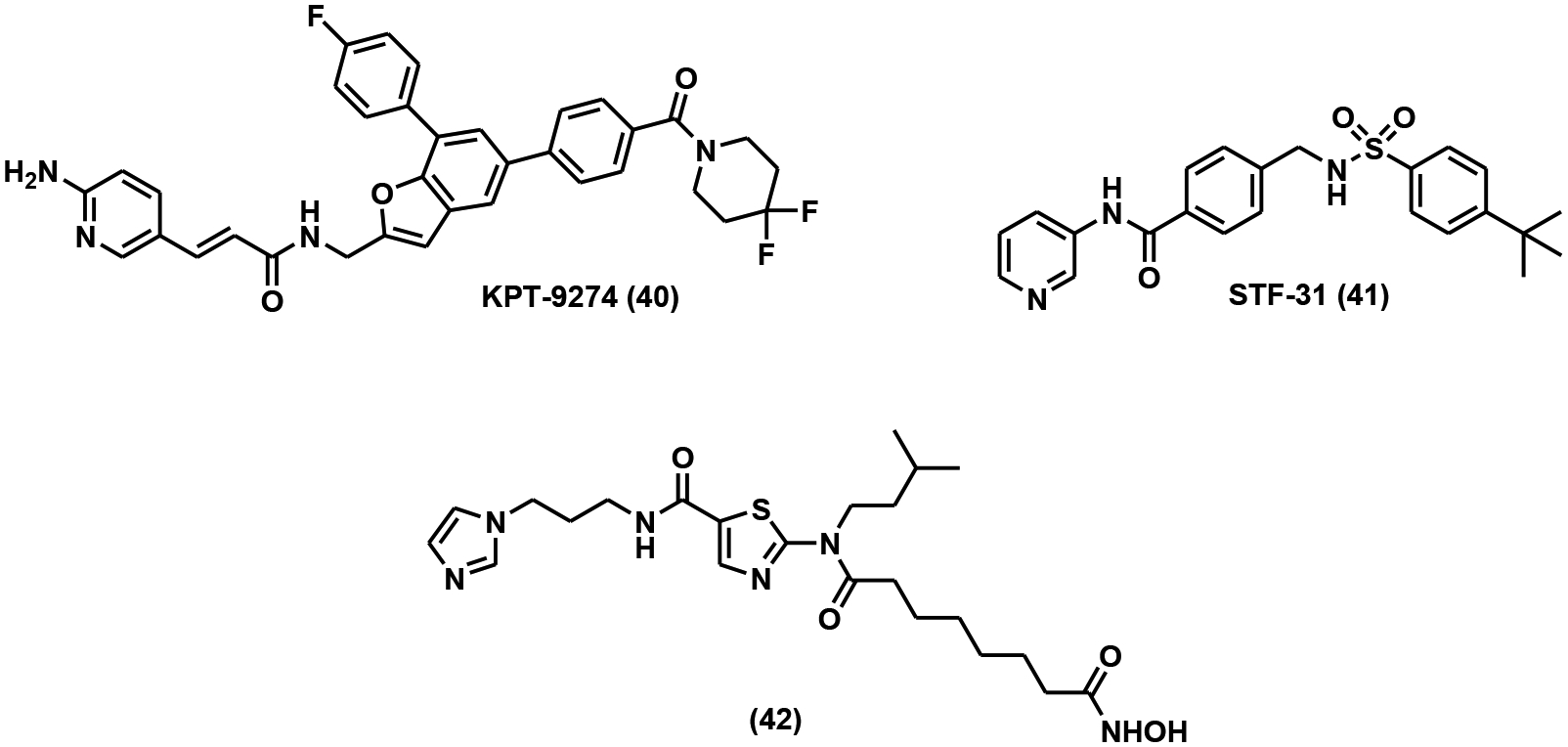
Dual NAMPT inhibitors.
2.2.1.4. NAMPT Activators
The first purported NAMPT activator was discovered through an in vivo screen for the identification of chemicals that could elevate hippocampal neurogenesis in mice [128]. The initial screening of a compound library of 1,000 chemicals produced eight compounds with neurogenic properties, out of which one had a promising pharmacological profile. The lead compound, P7C3 (Fig. 11), was found to reduce the death of newborn neurons rather than stimulating neuron birth. Using shotgun mass spectrometry, NAMPT was determined to be the cellular target of P7C3 [129]. The same study further examined the effects of P7C3 on the NAD+ levels in living cells. NAD+ deficiency caused by doxorubicin was alleviated with P7C3 treatment, presumably through NAMPT activation to stimulate NAD+ biosynthesis [129]. The direct activation of NAMPT by P7C3 has been controversial due to its inability to shift the melting temperature of NAMPT [130].
Fig. (11).
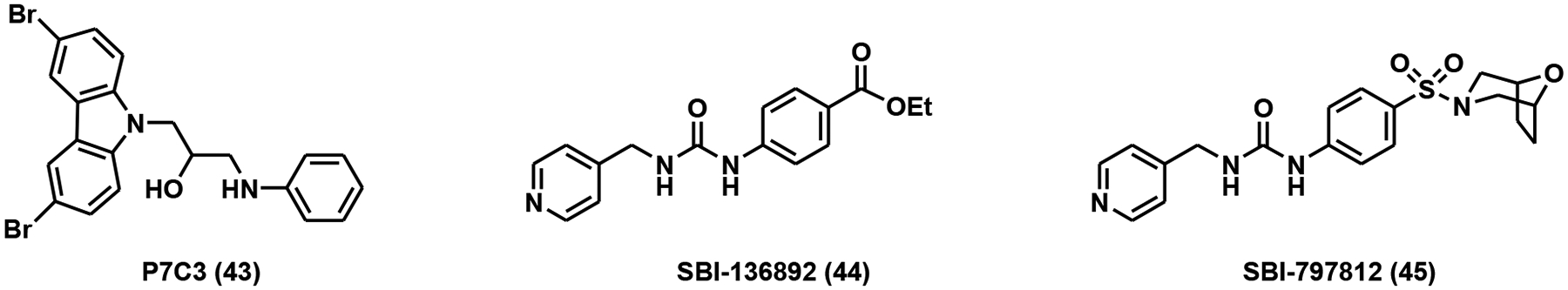
NAMPT activators.
A second NAMPT activator was identified through HTS using a protein thermal shift assay. The lead compound, SBI-136892 (Fig. 11), was able to increase the melting temperature of NAMPT in a dose-dependent manner [130]. Intriguingly, the structure of SBI-136892 was similar to a known NAMPT inhibitor, GNI-50. Based on the lead compound, an improved analog was generated. The analog, SBI-797812 (Fig. 11), had an EC50 of 0.37 ± 0.06 μM and could increase the formation of NMN by 2.1-fold compared to the control [130]. It was observed that NAMPT activation by SBI-797812 required ATP and could be nullified through the addition of NAMPT inhibitors. SBI-797812 was shown to increase the affinity of NAMPT for ATP, stabilize the pHis247 intermediate, reduce the levels of product pyrophosphate, and diminish the NAD+ feedback loop [130]. Treatment of mice with SBI-797812 led to a 1.3-fold increase of NAD+ levels in the liver, despite mouse NAMPT having an 8-fold lower affinity for the drug than human NAMPT [130].
2.2.2. NaPRTase Inhibitors
NaPRTase catalyzes the phosphoribosylation of NA to form NaMN, merging the salvage pathway with the de novo NAD+ synthesis (Fig. 2). NaPRTase is ubiquitously expressed in normal tissues such as heart, kidney and pancreas [131], but is significantly downregulated in certain tumors, including neuroblastoma, glioblastoma [109], and lymphomas [102]. On the other hand, overexpression of NaPRTase was observed in ovarian cancer and CRC [132, 133].
Although human NaPRTase shares a high degree of structural similarity with human NAMPT [134], it cannot be inhibited by FK866 [135]. NaPRTase lacks the “tunnel” structure in NAMPT to accommodate FK866 [134]. It is not surprising that most classical NAMPT inhibitors failed to demonstrate any appreciable inhibitory effects on NaPRTase. It was reported that NA analogs such as 2-pyrazinoic acid, 2-hydroxynicotinic acid and 2-fluoronicotinic acid inhibited NaPRTase activity in human platelet lysate with apparent Ki values in the micromolar range [136]. 2-Hydroxynicotinic acid (Fig. 12), in particular, sensitized ovarian cancer cells to FK866 treatment both in vitro and in vivo through NaPRTase inhibition [132]. A group of non-steroidal anti-inflammatory drugs (NSAIDs) was suggested as competitive NaPRTase inhibitors [137]. The most potent one in the group, flufenamic acid (Fig. 12), showed a Ki of 46 μM [137].
Fig. (12).
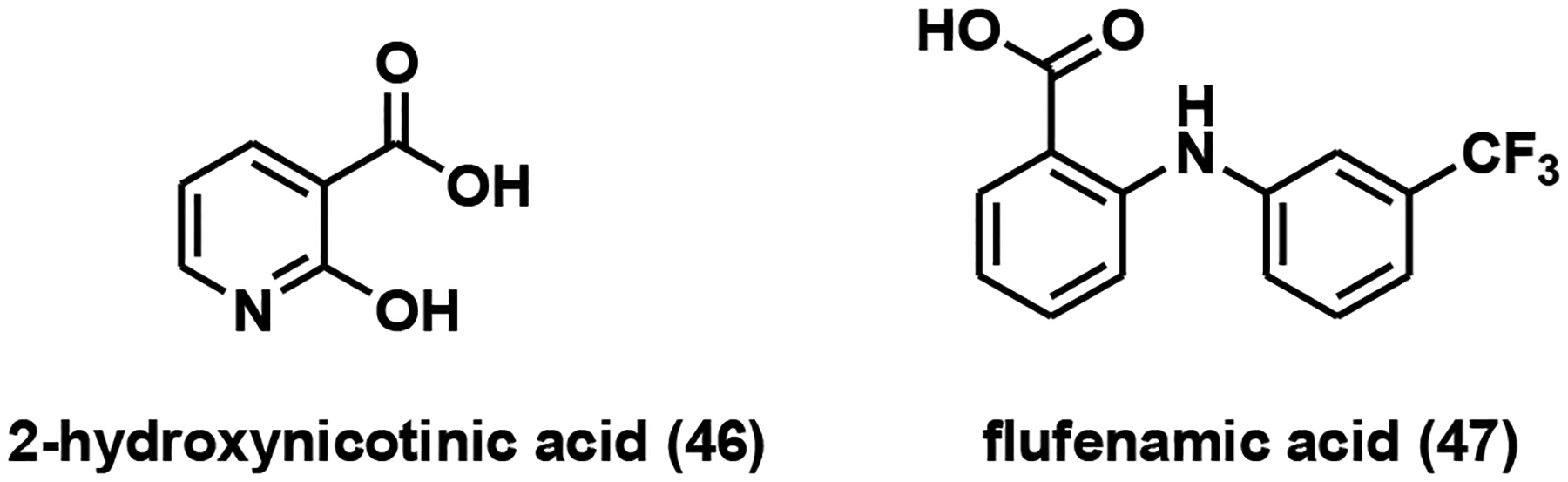
NaPRTase inhibitors.
CONCLUSION AND PERSPECTIVES
NAD+ is an essential metabolite that couples cellular energy status to signaling. Intracellular NAD+ levels have been linked to organismal health and are maintained by a complex network of biosynthetic pathways [15]. There have been many excellent review articles on the physiological relevance of NAD+ metabolism in normal biology as well as disease pathogenesis [138–141]. In this article, our discussion focuses on the small molecule regulators targeting NAD+ biosynthetic enzymes, especially the enzymes involved in the de novo and salvage pathways.
De novo pathway generates NAD+ from an amino acid precursor, tryptophan. IDO catalyzes the first and rate-limiting step in this pathway. It has been suggested as a therapeutic target for cancer immunotherapy [142]. Earlier inhibitors targeting IDO are mainly tryptophan mimics with micromolar potency [47]. Recently, several IDO inhibitors with novel pharmacophores and nanomolar potency have entered clinical trials [53, 56–58].
KMO serves as the other important control point of the de novo pathway. Inhibition of KMO leads to the reduction of the free radical generator 3-hydroxy-L-kynurenine. As such, KMO has been implicated as a viable target for the treatment of neurodegenerative diseases such as Alzheimer’s Disease and Huntington’s Disease [59]. Given the physiological relevance of KMO, intense efforts have been made to the development of several series of KMO inhibitors, ranging from substrate analogs to compounds with novel chemical entities. The discovery of KMO inhibitors has also been facilitated by the advance in structural biology [68], which guided the design of some of the most potent inhibitors known so far. The limitation of the currently available KMO inhibitors is their inability to pass through the blood-brain barrier (BBB). New drug delivery technologies, diverse formulations as well as structural modifications have been proposed as effective strategies to improve the BBB permeability of KMO inhibitors.
In humans, the salvage pathway is the predominant NAD+ biosynthetic pathway in which NAMPT acts as the rate-limiting enzyme [143]. Consequently, NAMPT has been intensively studied for the regulation of intracellular NAD+ levels. FK866, GMX1778, and GMX1777, three nanomolar NAMPT inhibitors, have advanced into clinical trials as cancer therapeutics [107, 110]. However, all of them demonstrated dose-limiting toxicities such as thrombocytopenia. Despite their limitations, they provided the blueprint for the development of new generations of NAMPT inhibitors. Most recently, co-administration of NA, the substrate of NAPRTase, has emerged as an alternative approach to mitigate NAMPT inhibitor toxicity in normal cells [109, 110].
Elevating intracellular NAD+ concentrations has provided therapeutic benefits in various pre-clinical studies [27–29, 144, 145]. Although small molecule activators targeting NAD+ biosynthetic enzymes hold great therapeutic potentials, the reports of these compounds have been rather sporadic. Unlike inhibitors, there are no general and systematic strategies to identify activators. Furthermore, the discovery of the NR pathway is relatively new, and our understanding of NRKs is still in its infancy. Detailed interrogation of the biological functions and cellular regulations of these NAD+ biosynthetic enzymes will open new avenues for the development of selective and potent small-molecule regulators.
ACKNOWLEDGEMENTS
The authors acknowledge the support received in part by CHE-1846785 from NSF (to Y.C.), 2020 VCU CCTR Endowment Fund (sub-award of UL1TR002649 from National Center for Advancing Translational Sciences to VCU) (to Y.C.), and Start-up funds from VCU (to Y.C.).
FUNDING
This work has been supported in part by CHE-1846785 from NSF (to Yana Cen), 2020 VCU CCTR Endowment Fund (sub-award of UL1TR002649 from National Center for Advancing Translational Sciences from VCU) (to Yana Cen), and start-up funds from VCU (to Yana Cen).
LIST OF ABBREVIATIONS
- AD
Alzheimer’s Disease
- BBB
Blood-brain Barrier
- CR
Calorie Restriction
- CRC
Colorectal Cancer
- GLUT1
Glucose Transporter 1
- HAO
3-hydroxyanthranilate 3,4-dioxygenase
- HD
Huntington’s Disease
- HDAC
Histone Deacetylase
- HTSs
High-throughput Screenings
- IDO
Indoleamine 2,3-dioxygenase
- KMO
Kynurenine 3-monooxygenase
- KYU
Kynureninase
- LPS
Lipopolysaccharide
- mAb
Monoclonal Antibody
- MOA
Mechanism of Action
- mPTP
Mitochondrial Permeability Transition Pore
- MS
Mass Spectrometry
- NA
Nicotinic Acid
- NaAD
Nicotinic Acid Adenine Dinucleotide
- nAChR
Nicotinic Acetylcholine Receptor
- NAD+
Nicotinamide Adenine Dinucleotide
- NAM
Nicotinamide
- NaMN
Nicotinic Acid Mononucleotide
- NAMPT
Nicotinamide Phosphoribosyl Transferase
- NaPRTase
Nicotinic Acid Phosphoribosyl Transferase
- NMDA
N-methyl-D-aspartate
- NMN
Nicotinamide Mononucleotide
- NMNAT
Nicotinamide/Nicotinic Acid Mononucleotide Adenylyltransferase
- NR
Nicotinamide Riboside
- NRK1/NRK2
Nicotinamide Riboside Kinases
- NSAIDs
Non-steroidal Anti-inflammatory Drugs
- NSCLC
Non-small Cell Lung Cancers
- PARPs
Poly(ADP-ribose) Polymerases
- PBEF
Pre-B Cell Colony Enhancing Factor
- PK
Pharmacokinetic
- PRPP
5-phosphoribosyl-1-pyrophosphate
- QPRTase
Quinolinate Phosphoribosyl Transferase
- RCC
Renal Cell Carcinoma
- TDO
Tryptophan 2,3-dioxygenase
- TNF
Tumor Necrosis Factor
- TS
Transition State
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
Publisher's Disclaimer: DISCLAIMER:
Publisher's Disclaimer: The above article has been published, as is, ahead-of-print, to provide early visibility but is not the final version. Major publication processes like copyediting, proofing, typesetting and further review are still to be done and may lead to changes in the final published version, if it is eventually published. All legal disclaimers that apply to the final published article also apply to this ahead-of-print version.
REFERENCES
- [1].Michan S; Sinclair D Sirtuins in mammals: insights into their biological function. Biochem. J, 2007, 404(1), 1–13. 10.1042/BJ20070140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].D’Amours D; Desnoyers S; D’Silva I; Poirier GG Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J, 1999, 342(Pt 2), 249–268. 10.1042/bj3420249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Schreiber V; Dantzer F; Ame JC; de Murcia G Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol, 2006, 7(7), 517–528. 10.1038/nrm1963 [DOI] [PubMed] [Google Scholar]
- [4].Lee HC Physiological functions of cyclic ADP-ribose and NAADP as calcium messengers. Annu. Rev. Pharmacol. Toxicol, 2001, 41, 317–345. 10.1146/annurev.pharmtox41.1.317 [DOI] [PubMed] [Google Scholar]
- [5].Lombard DB; Chua KF; Mostoslavsky R; Franco S; Gostissa M; Alt FW DNA repair, genome stability, and aging. Cell, 2005, 120(4), 497–512. 10.1016/j.cell.2005.01.028 [DOI] [PubMed] [Google Scholar]
- [6].Blander G; Guarente L The Sir2 family of protein deacetylases. Annu. Rev. Biochem, 2004, 73, 417–435. 10.1146/annurev.biochem.73.011303.073651 [DOI] [PubMed] [Google Scholar]
- [7].Hassa PO; Haenni SS; Elser M; Hottiger MO Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol. Mol. Biol. Rev, 2006, 70(3), 789–829. 10.1128/MMBR.00040-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Herceg Z; Wang ZQ Functions of poly(ADP-ribose) polymerase (PARP) in DNA repair, genomic integrity and cell death. Mutat. Res, 2001, 477(1–2), 97–110. 10.1016/S0027-5107(01)00111-7 [DOI] [PubMed] [Google Scholar]
- [9].Kharechkina ES; Nikiforova AB; Teplova VV; Odinokova IV; Krestinina OV; Baburina YL; Kruglova SA; Kruglov AG Regulation of permeability transition pore opening in mitochondria by external NAD(H). Biochim. Biophys. Acta, Gen. Subj, 2019, 1863(5), 771–783. 10.1016/j.bbagen.2019.01.003 [DOI] [PubMed] [Google Scholar]
- [10].Rasola A; Bernardi P The mitochondrial permeability transition pore and its involvement in cell death and in disease pathogenesis. Apoptosis, 2007, 12(5), 815–833. 10.1007/s10495-007-0723-y [DOI] [PubMed] [Google Scholar]
- [11].Kwong JQ; Molkentin JD Physiological and pathological roles of the mitochondrial permeability transition pore in the heart. Cell Metab, 2015, 21(2), 206–214. 10.1016/j.cmet.2014.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hunter DR; Haworth RA The Ca2+-induced membrane transition in mitochondria. I. The protective mechanisms. Arch. Biochem. Biophys, 1979, 195(2), 453–459. 10.1016/0003-9861(79)90371-0 [DOI] [PubMed] [Google Scholar]
- [13].Haworth RA; Hunter DR Allosteric inhibition of the Ca2+-activated hydrophilic channel of the mitochondrial inner membrane by nucleotides. J. Membr. Biol, 1980, 54(3), 231–236. 10.1007/BF01870239 [DOI] [PubMed] [Google Scholar]
- [14].Kurnasov O; Goral V; Colabroy K; Gerdes S; Anantha S; Osterman A; Begley TP NAD biosynthesis: identification of the tryptophan to quinolinate pathway in bacteria. Chem. Biol, 2003, 10(12), 1195–1204. 10.1016/j.chembiol.2003.11.011 [DOI] [PubMed] [Google Scholar]
- [15].Belenky P; Bogan KL; Brenner C NAD+ metabolism in health and disease. Trends Biochem. Sci, 2007, 32(1), 12–19. 10.1016/j.tibs.2006.11.006 [DOI] [PubMed] [Google Scholar]
- [16].Tempel W; Rabeh WM; Bogan KL; Belenky P; Wojcik M; Seidle HF; Nedyalkova L; Yang T; Sauve AA; Park HW; Brenner C Nicotinamide riboside kinase structures reveal new pathways to NAD+. PLoS Biol, 2007, 5(10), e263. 10.1371/journal.pbio.0050263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Schutz G; Feigelson P Purification and properties of rat liver tryptophan oxygenase. J. Biol. Chem, 1972, 247(17), 5327–5332. 10.1016/S0021-9258(20)81108-9 [DOI] [PubMed] [Google Scholar]
- [18].Shimizu T; Nomiyama S; Hirata F; Hayaishi O Indoleamine 2,3-dioxygenase. Purification and some properties. J. Biol. Chem, 1978, 253(13), 4700–4706. 10.1016/S0021-9258(17)30447-7 [DOI] [PubMed] [Google Scholar]
- [19].Magni G; Amici A; Emanuelli M; Raffaelli N; Ruggieri S Enzymology of NAD+ synthesis. Adv. Enzymol. Relat. Areas Mol. Biol, 1999, 73, 135–182, xi. [xi.]. [DOI] [PubMed] [Google Scholar]
- [20].Nishizuka Y; Hayaishi O Enzymic synthesis of niacin nucleotides from 3-hydroxyanthranilic acid in mammalian liver. J. Biol. Chem, 1963, 238, 483–485. 10.1016/S0021-9258(19)84026-7 [DOI] [PubMed] [Google Scholar]
- [21].Preiss J; Handler P Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J. Biol. Chem, 1958, 233(2), 488–492. 10.1016/S0021-9258(18)64789-1 [DOI] [PubMed] [Google Scholar]
- [22].Belenky P; Racette FG; Bogan KL; McClure JM; Smith JS; Brenner C Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell, 2007, 129(3), 473–484. 10.1016/j.cell.2007.03.024 [DOI] [PubMed] [Google Scholar]
- [23].Fletcher RS; Ratajczak J; Doig CL; Oakey LA; Callingham R; Da Silva Xavier G; Garten A; Elhassan YS; Redpath P; Migaud ME; Philp A; Brenner C; Canto C; Lavery GG Nicotinamide riboside kinases display redundancy in mediating nicotinamide mononucleotide and nicotinamide riboside metabolism in skeletal muscle cells. Mol. Metab, 2017, 6(8), 819–832. 10.1016/j.molmet.2017.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Tran A; Yokose R; Cen Y Chemo-enzymatic synthesis of isotopically labeled nicotinamide riboside. Org. Biomol. Chem, 2018, 16(19), 3662–3671. 10.1039/C8OB00552D [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ratajczak J; Joffraud M; Trammell SA; Ras R; Canela N; Boutant M; Kulkarni SS; Rodrigues M; Redpath P; Migaud ME; Auwerx J; Yanes O; Brenner C; Cantó C NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat. Commun, 2016, 7, 13103. 10.1038/ncomms13103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Trammell SA; Schmidt MS; Weidemann BJ; Redpath P; Jaksch F; Dellinger RW; Li Z; Abel ED; Migaud ME; Brenner C Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat. Commun, 2016, 7, 12948. 10.1038/ncomms12948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ryu D; Zhang H; Ropelle ER; Sorrentino V; Mázala DA; Mouchiroud L; Marshall PL; Campbell MD; Ali AS; Knowels GM; Bellemin S; Iyer SR; Wang X; Gariani K; Sauve AA; Cantó C; Conley KE; Walter L; Lovering RM; Chin ER; Jasmin BJ; Marcinek DJ; Menzies KJ; Auwerx J NAD+ repletion improves muscle function in muscular dystrophy and counters global PARylation. Sci. Transl. Med, 2016, 8(361), 361ra139. 10.1126/scitranslmed.aaf5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Vaur P; Brugg B; Mericskay M; Li Z; Schmidt MS; Vivien D; Orset C; Jacotot E; Brenner C; Duplus E Nicotinamide riboside, a form of vitamin B3, protects against excitotoxicity-induced axonal degeneration. FASEB J, 2017, 31(12), 5440–5452. 10.1096/fj.201700221RR [DOI] [PubMed] [Google Scholar]
- [29].Wu LE; Sinclair DA Restoring stem cells - all you need is NAD(.). Cell Res, 2016, 26(9), 971–972. 10.1038/cr.2016.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Camacho-Pereira J; Tarragó MG; Chini CCS; Nin V; Escande C; Warner GM; Puranik AS; Schoon RA; Reid JM; Galina A; Chini EN CD38 Dictates Age-Related NAD Decline and Mitochondrial Dysfunction through an SIRT3-Dependent Mechanism. Cell Metab, 2016, 23(6), 1127–1139. 10.1016/j.cmet.2016.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Covarrubias AJ; Kale A; Perrone R; Lopez-Dominguez JA; Pisco AO; Kasler HG; Schmidt MS; Heckenbach I; Kwok R; Wiley CD; Wong HS; Gibbs E; Iyer SS; Basisty N; Wu Q; Kim IJ; Silva E; Vitangcol K; Shin KO; Lee YM; Riley R; Ben-Sahra I; Ott M; Schilling B; Scheibye-Knudsen M; Ishihara K; Quake SR; Newman J; Brenner C; Campisi J; Verdin E Senescent cells promote tissue NAD+ decline during ageing via the activation of CD38+ macrophages. Nat Metab, 2020, 2(11), 1265–1283. 10.1038/s42255-020-00305-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Massudi H; Grant R; Braidy N; Guest J; Farnsworth B; Guillemin GJ Age-associated changes in oxidative stress and NAD+ metabolism in human tissue. PLoS One, 2012, 7(7), e42357. 10.1371/journal.pone.0042357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cantó C; Auwerx J Caloric restriction, SIRT1 and longevity. Trends Endocrinol. Metab, 2009, 20(7), 325–331. 10.1016/j.tem.2009.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cantó C; Jiang LQ; Deshmukh AS; Mataki C; Coste A; Lagouge M; Zierath JR; Auwerx J Interdependence of AMPK and SIRT1 for metabolic adaptation to fasting and exercise in skeletal muscle. Cell Metab, 2010, 11(3), 213–219. 10.1016/j.cmet.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cantó C; Auwerx J Targeting sirtuin 1 to improve metabolism: all you need is NAD(+)? Pharmacol. Rev, 2012, 64(1), 166–187. 10.1124/pr.110.003905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cantó C; Houtkooper RH; Pirinen E; Youn DY; Oosterveer MH; Cen Y; Fernandez-Marcos PJ; Yamamoto H; Andreux PA; Cettour-Rose P; Gade-mann K; Rinsch C; Schoonjans K; Sauve AA; Auwerx J The NAD(+) precursor nicotinamide riboside enhances oxidative metabolism and protects against high-fat diet-induced obesity. Cell Metab, 2012, 15(6), 838–847. 10.1016/j.cmet.2012.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yoshino J; Mills KF; Yoon MJ; Imai S Nicotinamide mononucleotide, a key NAD(+) intermediate, treats the pathophysiology of diet- and age-induced diabetes in mice. Cell Metab, 2011, 14(4), 528–536. 10.1016/j.cmet.2011.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Castro-Portuguez R; Sutphin GL Kynurenine pathway, NAD+ synthesis, and mitochondrial function: Targeting tryptophan metabolism to promote longevity and health-span. Exp. Gerontol, 2020, 132, 110841. 10.1016/j.exger.2020.110841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Phillips RS; Iradukunda EC; Hughes T; Bowen JP Modulation of Enzyme Activity in the Kynurenine Pathway by Kynurenine Monooxygenase Inhibition. Front. Mol. Biosci, 2019, 6(3), 3. 10.3389/fmolb.2019.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yamazaki F; Kuroiwa T; Takikawa O; Kido R Human indolylamine 2,3-dioxygenase. Its tissue distribution, and characterization of the placental enzyme. Biochem. J, 1985, 230(3), 635–638. 10.1042/bj2300635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yuasa HJ; Sugiura M; Harumoto T A single amino acid residue regulates the substrate affinity and specificity of indoleamine 2,3-dioxygenase. Arch. Biochem. Biophys, 2018, 640, 1–9. 10.1016/j.abb.2017.12.019 [DOI] [PubMed] [Google Scholar]
- [42].Hu B; Hissong BD; Carlin JM Interleukin-1 enhances indoleamine 2,3-dioxygenase activity by increasing specific mRNA expression in human mononuclear phagocytes. J. Interferon Cytokine Res, 1995, 15(7), 617–624. 10.1089/jir.1995.15.617 [DOI] [PubMed] [Google Scholar]
- [43].Babcock TA; Carlin JM Transcriptional activation of indoleamine dioxygenase by interleukin 1 and tumor necrosis factor alpha in interferon-treated epithelial cells. Cytokine, 2000, 12(6), 588–594. 10.1006/cyto.1999.0661 [DOI] [PubMed] [Google Scholar]
- [44].Yoshida R; Hayaishi O Induction of pulmonary indoleamine 2,3-dioxygenase by intraperitoneal injection of bacterial lipopolysaccharide. Proc. Natl. Acad. Sci. USA, 1978, 75(8), 3998–4000. 10.1073/pnas.75.8.3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Muller AJ; DuHadaway JB; Donover PS; Sutanto-Ward E; Prendergast GC Inhibition of indoleamine 2,3-dioxygenase, an immunoregulatory target of the cancer suppression gene Bin1, potentiates cancer chemotherapy. Nat. Med, 2005, 11(3), 312–319. 10.1038/nm1196 [DOI] [PubMed] [Google Scholar]
- [46].Muller AJ; Malachowski WP; Prendergast GC Indoleamine 2,3-dioxygenase in cancer: targeting pathological immune tolerance with small-molecule inhibitors. Expert Opin. Ther. Targets, 2005, 9(4), 831–849. 10.1517/14728222.9.4.831 [DOI] [PubMed] [Google Scholar]
- [47].Cady SG; Sono M 1-Methyl-DL-tryptophan, beta-(3-benzofuranyl)-DL-alanine (the oxygen analog of tryptophan), and beta-[3-benzo(b)thienyl]-DL-alanine (the sulfur analog of tryptophan) are competitive inhibitors for indoleamine 2,3-dioxygenase. Arch. Biochem. Biophys, 1991, 291(2), 326–333. 10.1016/0003-9861(91)90142-6 [DOI] [PubMed] [Google Scholar]
- [48].Jackson E; Dees EC; Kauh JS; Harvey RD; Neuger A; Lush R; Antonia SJ; Minton SE; Ismail-Khan R; Han HS; Vahanian NN; Ramsey WJ; Link CJ; Streicher H; Sullivan D; Soliman HH A phase I study of indoximod in combination with docetaxel in metastatic solid tumors. J. Clin. Onco, 2013, 31(15_suppl), 3026–3026. 10.1200/jco.2013.31.15_suppl.3026 [DOI] [Google Scholar]
- [49].Jha GG; Gupta S; Tagawa ST; Koopmeiners JS; Vivek S; Dudek AZ; Cooley SA; Blazar BR; Miller JS A phase II randomized, double-blind study of sipuleucel-T followed by IDO pathway inhibitor, indoximod, or placebo in the treatment of patients with metastatic castration resistant prostate cancer (mCRPC). J. Clin. Onco, 2017, 35(15_suppl), 3066–3066. [Google Scholar]
- [50].Zakharia Y; Rixe O; Ward JH; Drabick JJ; Shaheen MF; Milhem MM; Munn D; Kennedy EP; Vahanian NN; Link CJ; McWilliams RR Phase 2 trial of the IDO pathway inhibitor indoximod plus checkpoint inhibition for the treatment of patients with advanced melanoma. J. Clin. Onco, 2018, 36(15_suppl), 9512–9512. 10.1200/JCO.2018.36.15_suppl.9512 [DOI] [Google Scholar]
- [51].Prendergast GC; Metz R A perspective on new immune adjuvant principles: Reprogramming inflammatory states to permit clearance of cancer cells and other age-associated cellular pathologies. OncoImmunology, 2012, 1(6), 924–929. 10.4161/onci.21358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yue EW; Sparks R; Polam P; Modi D; Douty B; Wayland B; Glass B; Takvorian A; Glenn J; Zhu W; Bower M; Liu X; Leffet L; Wang Q; Bowman KJ; Hansbury MJ; Wei M; Li Y; Wynn R; Burn TC; Koblish HK; Fridman JS; Emm T; Scherle PA; Metcalf B; Combs AP INCB24360 (Epacadostat), a highly potent and selective Indoleamine-2,3-dioxygenase 1 (IDO1) Inhibitor for Immuno-oncology. ACS Med. Chem. Lett, 2017, 8(5), 486–491. 10.1021/acsmedchemlett.6b00391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Long GV; Dummer R; Hamid O; Gajewski TF; Caglevic C; Dalle S; Arance A; Carlino MS; Grob JJ; Kim TM; Demidov L; Robert C; Larkin J; Anderson JR; Maleski J; Jones M; Diede SJ; Mitchell TC Epacadostat plus pembrolizumab versus placebo plus pembrolizumab in patients with unresectable or metastatic melanoma (ECHO-301/KEYNOTE-252): a phase 3, randomised, double-blind study. Lancet Oncol, 2019, 20(8), 1083–1097. 10.1016/S1470-2045(19)30274-8 [DOI] [PubMed] [Google Scholar]
- [54].Kumar S; Jaller D; Patel B; LaLonde JM; DuHadaway JB; Malachowski WP; Prendergast GC; Muller AJ Structure based development of phenylimidazole-derived inhibitors of indoleamine 2,3-dioxygenase. J. Med. Chem, 2008, 51(16), 4968–4977. 10.1021/jm800512z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Röhrig UF; Majjigapu SR; Grosdidier A; Bron S; Stroobant V; Pilotte L; Colau D; Vogel P; Van den Eynde BJ; Zoete V; Michielin O Rational design of 4-aryl-1,2,3-triazoles for indoleamine 2,3-dioxygenase 1 inhibition. J. Med. Chem, 2012, 55(11), 5270–5290. 10.1021/jm300260v [DOI] [PubMed] [Google Scholar]
- [56].Mautino MR; Jaipuri FA; Waldo J; Kumar S; Adams J; Van Allen C; Marcinowicz-Flick A; Munn D; Vahanian N; Link CJ Abstract 491: NLG919, a novel indoleamine-2,3-dioxygenase (IDO)-pathway inhibitor drug candidate for cancer therapy. Cancer Res, 2013, 73(8)(Suppl.), 491–491. [Google Scholar]
- [57].Jung KH; LoRusso P; Burris H; Gordon M; Bang YJ; Hellmann MD; Cervantes A; Ochoa de Olza M; Marabelle A; Hodi FS; Ahn MJ; Emens LA; Barlesi F; Hamid O; Calvo E; McDermott D; Soliman H; Rhee I; Lin R; Pourmohamad T; Suchomel J; Tsuhako A; Morrissey K; Mahrus S; Morley R; Pirzkall A; Davis SL; Phase I Phase I Study of the Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Navoximod (GDC-0919) Administered with PD-L1 Inhibitor (Atezolizumab) in Advanced Solid Tumors. Clin. Cancer Res, 2019, 25(11), 3220–3228. 10.1158/1078-0432.CCR-18-2740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yamamoto N; Fujiwara Y; Kondo S; Iwasa S; Yonemori K; Shimomura A; Kitano S; Shimizu T; Koyama T; Ebata T; Sato N; Nakai K; Inatani M; Tamura K Phase I study of IDO1 inhibitor navoximod (GDC-0919) as monotherapy and in combination with atezolizumab in Japanese patients with advanced solid tumors. Ann. Oncol, 2018, 29(Suppl. 8), VIII138–VIII139. 10.1093/annonc/mdy279.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zwilling D; Huang SY; Sathyasaikumar KV; Notarangelo FM; Guidetti P; Wu HQ; Lee J; Truong J; Andrews-Zwilling Y; Hsieh EW; Louie JY; Wu T; Scearce-Levie K; Patrick C; Adame A; Giorgini F; Moussaoui S; Laue G; Rassoulpour A; Flik G; Huang Y; Muchowski JM; Masliah E; Schwarcz R; Muchowski PJ Kynurenine 3-monooxygenase inhibition in blood ameliorates neurodegeneration. Cell, 2011, 145(6), 863–874. 10.1016/j.cell.2011.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Perkins MN; Stone TW An iontophoretic investigation of the actions of convulsant kynurenines and their interaction with the endogenous excitant quinolinic acid. Brain Res, 1982, 247(1), 184–187. 10.1016/0006-8993(82)91048-4 [DOI] [PubMed] [Google Scholar]
- [61].Hilmas C; Pereira EF; Alkondon M; Rassoulpour A; Schwarcz R; Albuquerque EX The brain metabolite kynurenic acid inhibits alpha7 nicotinic receptor activity and increases non-alpha7 nicotinic receptor expression: physiopathological implications. J. Neurosci, 2001, 21(19), 7463–7473. 10.1523/JNEUROSCI.21-19-07463.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Okuda S; Nishiyama N; Saito H; Katsuki H Hydrogen peroxide-mediated neuronal cell death induced by an endogenous neurotoxin, 3-hydroxykynurenine. Proc. Natl. Acad. Sci. USA, 1996, 93(22), 12553–12558. 10.1073/pnas.93.22.12553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Okuda S; Nishiyama N; Saito H; Katsuki H 3-Hydroxykynurenine, an endogenous oxidative stress generator, causes neuronal cell death with apoptotic features and region selectivity. J. Neurochem, 1998, 70(1), 299–307. 10.1046/j.1471-4159.1998.70010299.x [DOI] [PubMed] [Google Scholar]
- [64].Mole DJ; Webster SP; Uings I; Zheng X; Binnie M; Wilson K; Hutchinson JP; Mirguet O; Walker A; Beaufils B; Ancellin N; Trottet L; Bénéton V; Mowat CG; Wilkinson M; Rowland P; Haslam C; McBride A; Homer NZ; Baily JE; Sharp MG; Garden OJ; Hughes J; Howie SE; Holmes DS; Liddle J; Iredale JP Kynurenine-3-monooxygenase inhibition prevents multiple organ failure in rodent models of acute pancreatitis. Nat. Med, 2016, 22(2), 202–209. 10.1038/nm.4020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Pellicciari R; Natalini B; Costantino G; Mahmoud MR; Mattoli L; Sadeghpour BM; Moroni F; Chiarugi A; Carpenedo R Modulation of the kynurenine pathway in search for new neuroprotective agents. Synthesis and preliminary evaluation of (m-nitrobenzoyl)alanine, a potent inhibitor of kynurenine-3-hydroxylase. J. Med. Chem, 1994, 37(5), 647–655. 10.1021/jm00031a015 [DOI] [PubMed] [Google Scholar]
- [66].Speciale C; Wu HQ; Cini M; Marconi M; Varasi M; Schwarcz R (R,S)-3,4-dichlorobenzoylalanine (FCE 28833A) causes a large and persistent increase in brain kynurenic acid levels in rats. Eur. J. Pharmacol, 1996, 315(3), 263–267. 10.1016/S0014-2999(96)00613-9 [DOI] [PubMed] [Google Scholar]
- [67].Pellicciari R; Amori L; Costantino G; Giordani A; Macchiarulo A; Mattoli L; Pevarello P; Speciale C; Varasi M Modulation of the kynurine pathway of tryptophan metabolism in search for neuroprotective agents. Focus on kynurenine-3-hydroxylase. Adv. Exp. Med. Biol, 2003, 527, 621–628. 10.1007/978-1-4615-0135-0_71 [DOI] [PubMed] [Google Scholar]
- [68].Amaral M; Levy C; Heyes DJ; Lafite P; Outeiro TF; Giorgini F; Leys D; Scrutton NS Structural basis of kynurenine 3-monooxygenase inhibition. Nature, 2013, 496(7445), 382–385. 10.1038/nature12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Röver S; Cesura AM; Huguenin P; Kettler R; Szente A Synthesis and biochemical evaluation of N-(4-phenylthiazol-2-yl)benzenesulfonamides as high-affinity inhibitors of kynurenine 3-hydroxylase. J. Med. Chem, 1997, 40(26), 4378–4385. 10.1021/jm970467t [DOI] [PubMed] [Google Scholar]
- [70].Hutchinson JP; Rowland P; Taylor MRD; Christodoulou EM; Haslam C; Hobbs CI; Holmes DS; Homes P; Liddle J; Mole DJ; Uings I; Walker AL; Webster SP; Mowat CG; Chung CW Structural and mechanistic basis of differentiated inhibitors of the acute pancreatitis target kynurenine-3-monooxygenase. Nat. Commun, 2017, 8, 15827. 10.1038/ncomms15827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Pevarello P; Varasi M; Amici R; Toma S; Speciale C Tricyclic 3-oxo-propanenitrile Compounds. WO 1999/016753 A2, 1999. [Google Scholar]
- [72].Lowe DM; Gee M; Haslam C; Leavens B; Christodoulou E; Hissey P; Hardwicke P; Argyrou A; Webster SP; Mole DJ; Wilson K; Binnie M; Yard BA; Dean T; Liddle J; Uings I; Hutchinson JP Lead discovery for human kynurenine 3-monooxygenase by high-throughput RapidFire mass spectrometry. J. Biomol. Screen, 2014, 19(4), 508–515. 10.1177/1087057113518069 [DOI] [PubMed] [Google Scholar]
- [73].Toledo-Sherman LM; Prime ME; Mrzljak L; Beconi MG; Beresford A; Brookfield FA; Brown CJ; Cardaun I; Courtney SM; Dijkman U; Hamelin-Flegg E; Johnson PD; Kempf V; Lyons K; Matthews K; Mitchell WL; O’Connell C; Pena P; Powell K; Rassoulpour A; Reed L; Reindl W; Selvaratnam S; Friley WW; Weddell DA; Went NE; Wheelan P; Winkler C; Winkler D; Wityak J; Yarnold CJ; Yates D; Munoz-Sanjuan I; Dominguez C Development of a series of aryl pyrimidine kynurenine monooxygenase inhibitors as potential therapeutic agents for the treatment of Huntington’s disease. J. Med. Chem, 2015, 58(3), 1159–1183. 10.1021/jm501350y [DOI] [PubMed] [Google Scholar]
- [74].Beaumont V; Mrzljak L; Dijkman U; Freije R; Heins M; Rassoulpour A; Tombaugh G; Gelman S; Bradaia A; Steidl E; Gleyzes M; Heikkinen T; Lehtimäki K; Puoliväli J; Kontkanen O; Javier RM; Neagoe I; Deisemann H; Winkler D; Ebneth A; Khetarpal V; Toledo-Sherman L; Dominguez C; Park LC; Munoz-Sanjuan I The novel KMO inhibitor CHDI-340246 leads to a restoration of electrophysiological alterations in mouse models of Huntington’s disease. Exp. Neurol, 2016, 282, 99–118. 10.1016/j.expneurol.2016.05.005 [DOI] [PubMed] [Google Scholar]
- [75].Phillips RS Structure and mechanism of kynureninase. Arch. Biochem. Biophys, 2014, 544, 69–74. 10.1016/j.abb.2013.10.020 [DOI] [PubMed] [Google Scholar]
- [76].Lima S; Kumar S; Gawandi V; Momany C; Phillips RS Crystal structure of the Homo sapiens kynureninase-3-hydroxyhippuric acid inhibitor complex: insights into the molecular basis of kynureninase substrate specificity. J. Med. Chem, 2009, 52(2), 389–396. 10.1021/jm8010806 [DOI] [PubMed] [Google Scholar]
- [77].Phillips RS; Dua RK Stereochemistry and mechanism of aldol reactions catalyzed by kynureninase. J. Am. Chem. Soc, 1991, 113(19), 7385–7388. 10.1021/ja00019a039 [DOI] [Google Scholar]
- [78].Dua RK; Taylor EW; Phillips RS S-Aryl-L-cysteine S,S-dioxides: design, synthesis, and evaluation of a new class of inhibitors of kynureninase. J. Am. Chem. Soc, 1993, 115(4), 1264–1270. 10.1021/ja00057a007 [DOI] [Google Scholar]
- [79].Ross FC; Botting NP; Leeson PD Synthesis of phosphinic acid transition state analogues for the reaction catalysed by kynureninase. Bioorg. Med. Chem. Lett, 1996, 6(22), 2643–2646. 10.1016/S0960-894X(96)00483-0 [DOI] [Google Scholar]
- [80].Walsh HA; Leslie PL; O’Shea KC; Botting NP 2-Amino-4-[3′-hydroxyphenyl]-4-hydroxybutanoic acid; a potent inhibitor of rat and recombinant human kynureninase. Bioorg. Med. Chem. Lett, 2002, 12(3), 361–363. 10.1016/S0960-894X(01)00758-2 [DOI] [PubMed] [Google Scholar]
- [81].Iwai K; Taguchi H Distribution of quinolinate phosphoribosyltransferase in animals, plants and microorganisms. J. Nutr. Sci. Vitaminol. (Tokyo), 1973, 19(6), 491–499. 10.3177/jnsv.19.491 [DOI] [PubMed] [Google Scholar]
- [82].Buonvicino D; Mazzola F; Zamporlini F; Resta F; Ranieri G; Camaioni E; Muzzi M; Zecchi R; Pieraccini G; Dolle C; Calamante M; Bartolucci G; Ziegler M; Stecca B; Raffaelli N; Chiarugi A Identification of the nicotinamide salvage pathway as a new toxification route for antimetabolites. Cell Chem Biol, 2018, 25(4), 471–482 e7. 10.1016/j.chembiol.2018.01.012 [DOI] [PubMed] [Google Scholar]
- [83].Sahm F; Oezen I; Opitz CA; Radlwimmer B; von Deimling A; Ahrendt T; Adams S; Bode HB; Guillemin GJ; Wick W; Platten M The endogenous tryptophan metabolite and NAD+ precursor quinolinic acid confers resistance of gliomas to oxidative stress. Cancer Res, 2013, 73(11), 3225–3234. 10.1158/0008-5472.CAN-12-3831 [DOI] [PubMed] [Google Scholar]
- [84].Malik SS; Patterson DN; Ncube Z; Toth EA The crystal structure of human quinolinic acid phosphoribosyltransferase in complex with its inhibitor phthalic acid. Proteins, 2014, 82(3), 405–414. 10.1002/prot.24406 [DOI] [PubMed] [Google Scholar]
- [85].Braidy N; Guillemin GJ; Grant R Effects of Kynurenine Pathway Inhibition on NAD Metabolism and Cell Viability in Human Primary Astrocytes and Neurons. Int. J. Tryptophan Res, 2011, 4, 29–37. 10.4137/IJTR.S7052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Berger F; Lau C; Dahlmann M; Ziegler M Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem, 2005, 280(43), 36334–36341. 10.1074/jbc.M508660200 [DOI] [PubMed] [Google Scholar]
- [87].Liu X; Liu M; Tang C; Xiang Z; Li Q; Ruan X; Xiong K; Zheng L Overexpression of Nmnat improves the adaption of health span in aging Drosophila. Exp. Gerontol, 2018, 108, 276–283. 10.1016/j.exger.2018.04.026 [DOI] [PubMed] [Google Scholar]
- [88].Gulshan M; Yaku K; Okabe K; Mahmood A; Sasaki T; Yamamoto M; Hikosaka K; Usui I; Kitamura T; Tobe K; Nakagawa T Overexpression of Nmnat3 efficiently increases NAD and NGD levels and ameliorates age-associated insulin resistance. Aging Cell, 2018, 17(4), e12798. 10.1111/acel.12798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Anderson RM; Bitterman KJ; Wood JG; Medvedik O; Cohen H; Lin SS; Manchester JK; Gordon JI; Sinclair DA Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J. Biol. Chem, 2002, 277(21), 18881–18890. 10.1074/jbc.M111773200 [DOI] [PubMed] [Google Scholar]
- [90].Ali YO; Li-Kroeger D; Bellen HJ; Zhai RG; Lu HC NMNATs, evolutionarily conserved neuronal maintenance factors. Trends Neurosci, 2013, 36(11), 632–640. 10.1016/j.tins.2013.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Yahata N; Yuasa S; Araki T Nicotinamide mononucleotide adenylyltransferase expression in mitochondrial matrix delays Wallerian degeneration. J. Neurosci, 2009, 29(19), 6276–6284. 10.1523/JNEUROSCI.4304-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tang BL Why is NMNAT Protective against Neuronal Cell Death and Axon Degeneration, but Inhibitory of Axon Regeneration? Cells, 2019, 8(3), E267. 10.3390/cells8030267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Cui C; Qi J; Deng Q; Chen R; Zhai D; Yu J Nicotinamide Mononucleotide Adenylyl Transferase 2: A promising diagnostic and therapeutic target for colorectal cancer. BioMed Res. Int, 2016, 2016, 1804137. 10.1155/2016/1804137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Pan LZ; Ahn DG; Sharif T; Clements D; Gujar SA; Lee PW The NAD+ synthesizing enzyme nicotinamide mononucleotide adenylyltransferase 2 (NMNAT-2) is a p53 downstream target. Cell Cycle, 2014, 13(6), 1041–1048. 10.4161/cc.28128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Li H; Feng Z; Wu W; Li J; Zhang J; Xia T SIRT3 regulates cell proliferation and apoptosis related to energy metabolism in non-small cell lung cancer cells through deacetylation of NMNAT2. Int. J. Oncol, 2013, 43(5), 1420–1430. 10.3892/ijo.2013.2103 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [96].Imai S Nicotinamide phosphoribosyltransferase (Nampt): a link between NAD biology, metabolism, and diseases. Curr. Pharm. Des, 2009, 15(1), 20–28. 10.2174/138161209787185814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Samal B; Sun Y; Stearns G; Xie C; Suggs S; McNiece I Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol. Cell. Biol, 1994, 14(2), 1431–1437. 10.1128/MCB.14.2.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Fukuhara A; Matsuda M; Nishizawa M; Segawa K; Tanaka M; Kishimoto K; Matsuki Y; Murakami M; Ichisaka T; Murakami H; Watanabe E; Takagi T; Akiyoshi M; Ohtsubo T; Kihara S; Yamashita S; Makishima M; Funahashi T; Yamanaka S; Hiramatsu R; Matsuzawa Y; Shimomura I Visfatin: a protein secreted by visceral fat that mimics the effects of insulin. Science, 2005, 307(5708), 426–430. 10.1126/science.1097243 [DOI] [PubMed] [Google Scholar]
- [99].Revollo JR; Körner A; Mills KF; Satoh A; Wang T; Garten A; Dasgupta B; Sasaki Y; Wolberger C; Townsend RR; Milbrandt J; Kiess W; Imai S Nampt/PBEF/Visfatin regulates insulin secretion in beta cells as a systemic NAD biosynthetic enzyme. Cell Metab, 2007, 6(5), 363–375. 10.1016/j.cmet.2007.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Reddy PS; Umesh S; Thota B; Tandon A; Pandey P; Hegde AS; Balasubramaniam A; Chandramouli BA; Santosh V; Rao MR; Kondaiah P; Somasundaram K PBEF1/NAmPRTase/Visfatin: a potential malignant astrocytoma/glioblastoma serum marker with prognostic value. Cancer Biol. Ther, 2008, 7(5), 663–668. 10.4161/cbt.7.5.5663 [DOI] [PubMed] [Google Scholar]
- [101].Maldi E; Travelli C; Caldarelli A; Agazzone N; Cintura S; Galli U; Scatolini M; Ostano P; Miglino B; Chiorino G; Boldorini R; Genazzani AA Nicotinamide phosphoribosyltransferase (NAMPT) is over-expressed in melanoma lesions. Pigment Cell Melanoma Res, 2013, 26(1), 144–146. 10.1111/pcmr.12037 [DOI] [PubMed] [Google Scholar]
- [102].Olesen UH; Hastrup N; Sehested M Expression patterns of nicotinamide phosphoribosyltransferase and nicotinic acid phosphoribosyltransferase in human malignant lymphomas. APMIS, 2011, 119(4–5), 296–303. 10.1111/j.1600-0463.2011.02733.x [DOI] [PubMed] [Google Scholar]
- [103].Galli U; Travelli C; Massarotti A; Fakhfouri G; Rahimian R; Tron GC; Genazzani AA Medicinal chemistry of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors. J. Med. Chem, 2013, 56(16), 6279–6296. 10.1021/jm4001049 [DOI] [PubMed] [Google Scholar]
- [104].Sampath D; Zabka TS; Misner DL; O’Brien T; Dragovich PS Inhibition of nicotinamide phosphoribosyltransferase (NAMPT) as a therapeutic strategy in cancer. Pharmacol. Ther, 2015, 151, 16–31. 10.1016/j.pharmthera.2015.02.004 [DOI] [PubMed] [Google Scholar]
- [105].Wosikowski K; Mattern K; Schemainda I; Hasmann M; Rattel B; Löser R WK175, a novel antitumor agent, decreases the intracellular nicotinamide adenine dinucleotide concentration and induces the apoptotic cascade in human leukemia cells. Cancer Res, 2002, 62(4), 1057–1062. [PubMed] [Google Scholar]
- [106].Hasmann M; Schemainda I FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res, 2003, 63(21), 7436–7442. [PubMed] [Google Scholar]
- [107].Goldinger SM; Gobbi Bischof S; Fink-Puches R; Klemke CD; Dréno B; Bagot M; Dummer R Efficacy and Safety of APO866 in Patients With Refractory or Relapsed Cutaneous T-Cell Lymphoma: A Phase 2 Clinical Trial. JAMA Dermatol, 2016, 152(7), 837–839. 10.1001/jamadermatol.2016.0401 [DOI] [PubMed] [Google Scholar]
- [108].Olesen UH; Christensen MK; Björkling F; Jäättelä M; Jensen PB; Sehested M; Nielsen SJ Anticancer agent CHS-828 inhibits cellular synthesis of NAD. Biochem. Biophys. Res. Commun, 2008, 367(4), 799–804. 10.1016/j.bbrc.2008.01.019 [DOI] [PubMed] [Google Scholar]
- [109].Watson M; Roulston A; Bélec L; Billot X; Marcellus R; Bédard D; Bernier C; Branchaud S; Chan H; Dairi K; Gilbert K; Goulet D; Gratton MO; Isakau H; Jang A; Khadir A; Koch E; Lavoie M; Lawless M; Nguyen M; Paquette D; Turcotte E; Berger A; Mitchell M; Shore GC; Beauparlant P The small molecule GMX1778 is a potent inhibitor of NAD+ biosynthesis: strategy for enhanced therapy in nicotinic acid phosphoribosyltransferase 1-deficient tumors. Mol. Cell. Biol, 2009, 29(21), 5872–5888. 10.1128/MCB.00112-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Beauparlant P; Bédard D; Bernier C; Chan H; Gilbert K; Goulet D; Gratton MO; Lavoie M; Roulston A; Turcotte E; Watson M Preclinical development of the nicotinamide phosphoribosyl transferase inhibitor prodrug GMX1777. Anticancer Drugs, 2009, 20(5), 346–354. 10.1097/CAD.0b013e3283287c20 [DOI] [PubMed] [Google Scholar]
- [111].You H; Youn HS; Im I; Bae MH; Lee SK; Ko H; Eom SH; Kim YC Design, synthesis and X-ray crystal-lographic study of NAmPRTase inhibitors as anti-cancer agents. Eur. J. Med. Chem, 2011, 46(4), 1153–1164. 10.1016/j.ejmech.2011.01.034 [DOI] [PubMed] [Google Scholar]
- [112].Gunzner-Toste J; Zhao G; Bauer P; Baumeister T; Buckmelter AJ; Caligiuri M; Clodfelter KH; Fu B; Han B; Ho YC; Kley N; Liang X; Liederer BM; Lin J; Mukadam S; O’Brien T; Oh A; Reynolds DJ; Sharma G; Skelton N; Smith CC; Sodhi J; Wang W; Wang Z; Xiao Y; Yuen PW; Zak M; Zhang L; Zheng X; Bair KW; Dragovich PS Discovery of potent and efficacious urea-containing nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with reduced CYP2C9 inhibition properties. Bioorg. Med. Chem. Lett, 2013, 23(12), 3531–3538. 10.1016/j.bmcl.2013.04.040 [DOI] [PubMed] [Google Scholar]
- [113].Zheng X; Bauer P; Baumeister T; Buckmelter AJ; Caligiuri M; Clodfelter KH; Han B; Ho YC; Kley N; Lin J; Reynolds DJ; Sharma G; Smith CC; Wang Z; Dragovich PS; Oh A; Wang W; Zak M; Gunzner-Toste J; Zhao G; Yuen PW; Bair KW Structure-based identification of ureas as novel nicotinamide phosphoribosyltransferase (Nampt) inhibitors. J. Med. Chem, 2013, 56(12), 4921–4937. 10.1021/jm400186h [DOI] [PubMed] [Google Scholar]
- [114].Colombano G; Travelli C; Galli U; Caldarelli A; Chini MG; Canonico PL; Sorba G; Bifulco G; Tron GC; Genazzani AA A novel potent nicotinamide phosphoribosyltransferase inhibitor synthesized via click chemistry. J. Med. Chem, 2010, 53(2), 616–623. 10.1021/jm9010669 [DOI] [PubMed] [Google Scholar]
- [115].Xu TY; Zhang SL; Dong GQ; Liu XZ; Wang X; Lv XQ; Qian QJ; Zhang RY; Sheng CQ; Miao CY Discovery and characterization of novel small-molecule inhibitors targeting nicotinamide phosphoribosyltransferase. Sci. Rep, 2015, 5, 10043. 10.1038/srep10043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Estoppey D; Hewett JW; Guy CT; Harrington E; Thomas JR; Schirle M; Cuttat R; Waldt A; Gerrits B; Yang Z; Schuierer S; Pan X; Xie K; Carbone W; Knehr J; Lindeman A; Russ C; Frias E; Hoffman GR; Varadarajan M; Ramadan N; Reece-Hoyes JS; Wang Q; Chen X; McAllister G; Roma G; Bouwmeester T; Hoepfner D Identification of a novel NAMPT inhibitor by CRISPR/Cas9 chemogenomic profiling in mammalian cells. Sci. Rep, 2017, 7, 42728. 10.1038/srep42728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Fleischer TC; Murphy BR; Flick JS; Terry-Lorenzo RT; Gao ZH; Davis T; McKinnon R; Ostanin K; Willardsen JA; Boniface JJ Chemical proteomics identifies Nampt as the target of CB30865, an orphan cytotoxic compound. Chem. Biol, 2010, 17(6), 659–664. 10.1016/j.chembiol.2010.05.008 [DOI] [PubMed] [Google Scholar]
- [118].Oh A; Ho YC; Zak M; Liu Y; Chen X; Yuen PW; Zheng X; Liu Y; Dragovich PS; Wang W Structural and biochemical analyses of the catalysis and potency impact of inhibitor phosphoribosylation by human nicotinamide phosphoribosyltransferase. ChemBioChem, 2014, 15(8), 1121–1130. 10.1002/cbic.201402023 [DOI] [PubMed] [Google Scholar]
- [119].Lee MW Jr; Sevryugina YV; Khan A; Ye SQ Carboranes increase the potency of small molecule inhibitors of nicotinamide phosphoribosyltranferase. J. Med. Chem, 2012, 55(16), 7290–7294. 10.1021/jm300740t [DOI] [PubMed] [Google Scholar]
- [120].Korepanova A; Longenecker KL; Pratt SD; Panchal SC; Clark RF; Lake M; Gopalakrishnan SM; Raich D; Sun C; Petros AM Fragment-based discovery of a potent NAMPT inhibitor. Bioorg. Med. Chem. Lett, 2018, 28(3), 437–440. 10.1016/j.bmcl.2017.12.023 [DOI] [PubMed] [Google Scholar]
- [121].Korotchkina L; Kazyulkin D; Komarov PG; Polinsky A; Andrianova EL; Joshi S; Gupta M; Vujcic S; Kononov E; Toshkov I; Tian Y; Krasnov P; Chernov MV; Veith J; Antoch MP; Middlemiss S; Somers K; Lock RB; Norris MD; Henderson MJ; Haber M; Chernova OB; Gudkov AV OT-82, a novel anticancer drug candidate that targets the strong dependence of hematological malignancies on NAD biosynthesis. Leukemia, 2020, 34(7), 1828–1839. 10.1038/s41375-019-0692-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Talevi A Multi-target pharmacology: possibilities and limitations of the “skeleton key approach” from a medicinal chemist perspective. Front. Pharmacol, 2015, 6, 205. 10.3389/fphar.2015.00205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Zimmermann GR; Lehár J; Keith CT Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov. Today, 2007, 12(1–2), 34–42. 10.1016/j.drudis.2006.11.008 [DOI] [PubMed] [Google Scholar]
- [124].Abu Aboud O; Chen CH; Senapedis W; Baloglu E; Argueta C; Weiss RH Dual and Specific Inhibition of NAMPT and PAK4 By KPT-9274 Decreases Kidney Cancer Growth. Mol. Cancer Ther, 2016, 15(9), 2119–2129. 10.1158/1535-7163.MCT-16-0197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Kraus D; Reckenbeil J; Veit N; Kuerpig S; Meisenheimer M; Beier I; Stark H; Winter J; Probstmeier R Targeting glucose transport and the NAD pathway in tumor cells with STF-31: a re-evaluation. Cell Oncol. (Dordr.), 2018, 41(5), 485–494. 10.1007/s13402-018-0385-5 [DOI] [PubMed] [Google Scholar]
- [126].Dong G; Chen W; Wang X; Yang X; Xu T; Wang P; Zhang W; Rao Y; Miao C; Sheng C Small Molecule Inhibitors Simultaneously Targeting Cancer Metabolism and Epigenetics: Discovery of Novel Nicotinamide Phosphoribosyltransferase (NAMPT) and Histone Deacetylase (HDAC) Dual Inhibitors. J. Med. Chem, 2017, 60(19), 7965–7983. 10.1021/acs.jmedchem.7b00467 [DOI] [PubMed] [Google Scholar]
- [127].Chen W; Dong G; Wu Y; Zhang W; Miao C; Sheng C Dual NAMPT/HDAC Inhibitors as a New Strategy for Multitargeting Antitumor Drug Discovery. ACS Med. Chem. Lett, 2017, 9(1), 34–38. 10.1021/acsmedchemlett.7b00414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Pieper AA; Xie S; Capota E; Estill SJ; Zhong J; Long JM; Becker GL; Huntington P; Goldman SE; Shen C-H; Capota M; Britt JK; Kotti T; Ure K; Brat DJ; Williams NS; MacMillan KS; Naidoo J; Melito L; Hsieh J; De Brabander J; Ready JM; McKnight SL Discovery of a proneurogenic, neuroprotective chemical. Cell, 2010, 142(1), 39–51. 10.1016/j.cell.2010.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Wang G; Han T; Nijhawan D; Theodoropoulos P; Naidoo J; Yadavalli S; Mirzaei H; Pieper AA; Ready JM; McKnight SL P7C3 neuroprotective chemicals function by activating the rate-limiting enzyme in NAD salvage. Cell, 2014, 158(6), 1324–1334. 10.1016/j.cell.2014.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Gardell SJ; Hopf M; Khan A; Dispagna M; Hampton Sessions E; Falter R; Kapoor N; Brooks J; Culver J; Petucci C; Ma C-T; Cohen SE; Tanaka J; Burgos ES; Hirschi JS; Smith SR; Sergienko E; Pinkerton AB; Boosting NAD Boosting NAD+ with a small molecule that activates NAMPT. Nat. Commun, 2019, 10(1), 3241. 10.1038/s41467-019-11078-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Duarte-Pereira S; Pereira-Castro I; Silva SS; Correia MG; Neto C; da Costa LT; Amorim A; Silva RM Extensive regulation of nicotinate phosphoribosyltransferase (NAPRT) expression in human tissues and tumors. Oncotarget, 2016, 7(2), 1973–1983. 10.18632/oncotarget.6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Piacente F; Caffa I; Ravera S; Sociali G; Passalacqua M; Vellone VG; Becherini P; Reverberi D; Monacelli F; Ballestrero A; Odetti P; Cagnetta A; Cea M; Nahimana A; Duchosal M; Bruzzone S; Nencioni A Nicotinic Acid Phosphoribosyltransferase Regulates Cancer Cell Metabolism, Susceptibility to NAMPT Inhibitors, and DNA Repair. Cancer Res., 2017, 77(14), 3857–3869. 10.1158/0008-5472.CAN-16-3079 [DOI] [PubMed] [Google Scholar]
- [133].Li XQ; Lei J; Mao LH; Wang QL; Xu F; Ran T; Zhou ZH; He S NAMPT and NAPRT, Key Enzymes in NAD Salvage Synthesis Pathway, Are of Negative Prognostic Value in Colorectal Cancer. Front. Oncol, 2019, 9, 736. 10.3389/fonc.2019.00736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Marletta AS; Massarotti A; Orsomando G; Magni G; Rizzi M; Garavaglia S Crystal structure of human nicotinic acid phosphoribosyltransferase. FEBS Open Bio, 2015, 5, 419–428. 10.1016/j.fob.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Khan JA; Tao X; Tong L Molecular basis for the inhibition of human NMPRTase, a novel target for anticancer agents. Nat. Struct. Mol. Biol, 2006, 13(7), 582–588. 10.1038/nsmb1105 [DOI] [PubMed] [Google Scholar]
- [136].Gaut ZN; Solomon HM Inhibition of nicotinate phosphoribosyltransferase in human platelet lysate by nicotinic acid analogs. Biochem. Pharmacol, 1971, 20(10), 2903–2906. 10.1016/0006-2952(71)90202-4 [DOI] [PubMed] [Google Scholar]
- [137].Gaut ZN; Solomon HM Inhibition of nicotinate phosphoribosyl transferase by nonsteroidal anti-inflammatory drugs: a possible mechanism of action. J. Pharm. Sci, 1971, 60(12), 1887–1888. 10.1002/jps.2600601230 [DOI] [PubMed] [Google Scholar]
- [138].Johnson S; Imai SI NAD + biosynthesis, aging, and disease. F1000 Res, 2018, 7, 132. 10.12688/f1000research.12120.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Yang Y; Sauve AA NAD(+) metabolism: Bioenergetics, signaling and manipulation for therapy. Biochim. Biophys. Acta, 2016, 1864(12), 1787–1800. 10.1016/j.bbapap.2016.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Yaku K; Okabe K; Nakagawa T NAD metabolism: Implications in aging and longevity. Ageing Res. Rev, 2018, 47, 1–17. 10.1016/j.arr.2018.05.006 [DOI] [PubMed] [Google Scholar]
- [141].Imai SI The NAD World 2.0: the importance of the inter-tissue communication mediated by NAMPT/NAD+/SIRT1 in mammalian aging and longevity control. NPJ Syst. Biol. Appl, 2016, 2, 16018. 10.1038/npjsba.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Qian S; Zhang M; Chen QL; He YY; Wang W; Wang ZY IDO as a drug target for cancer immunotherapy: recent developments in IDO inhibitors discovery. RSC Advances, 2016, 6(9), 7575–7581. 10.1039/C5RA25046C [DOI] [Google Scholar]
- [143].Houtkooper RH; Cantó C; Wanders RJ; Auwerx J The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr. Rev, 2010, 31(2), 194–223. 10.1210/er.2009-0026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Hong W; Mo F; Zhang Z; Huang M; Wei X Nicotinamide Mononucleotide: A Promising Molecule for Therapy of Diverse Diseases by Targeting NAD+ Metabolism. Front. Cell Dev. Biol, 2020, 8, 246. 10.3389/fcell.2020.00246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Bertoldo MJ; Listijono DR; Ho WJ; Riepsamen AH; Goss DM; Richani D; Jin XL; Mahbub S; Campbell JM; Habibalahi A; Loh WN; Youngson NA; Maniam J; Wong ASA; Selesniemi K; Bustamante S; Li C; Zhao Y; Marinova MB; Kim LJ; Lau L; Wu RM; Mikolaizak AS; Araki T; Le Couteur DG; Turner N; Morris MJ; Walters KA; Goldys E; O’Neill C; Gilchrist RB; Sinclair DA; Homer HA; Wu LE NAD(+) Repletion Rescues Female Fertility during Reproductive Aging. Cell Rep, 2020, 30(6), 1670–1681 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Billington RA; Travelli C; Ercolano E; Galli U; Roman CB; Grolla AA; Canonico PL; Condorelli F; Genazzani AA Characterization of NAD uptake in mammalian cells. J. Biol. Chem, 2008, 283(10), 6367–6374. 10.1074/jbc.M706204200 [DOI] [PubMed] [Google Scholar]
- [147].Zhang SL; Xu TY; Yang ZL; Han S; Zhao Q; Miao CY Crystal structure-based comparison of two NAMPT inhibitors. Acta Pharmacol. Sin, 2018, 39(2), 294–301. 10.1038/aps.2017.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Skelton LA; Ormerod MG; Titley JC; Jackman AL Cell cycle effects of CB30865, a lipophilic quinazoline-based analogue of the antifolate thymidylate synthase inhibitor ICI 198583 with an undefined mechanism of action. Cytometry, 1998, 33(1), 56–66. [DOI] [PubMed] [Google Scholar]
- [149].Dragovich PS; Zhao G; Baumeister T; Bravo B; Giannetti AM; Ho YC; Hua R; Li G; Liang X; Ma X; O’Brien T; Oh A; Skelton NJ; Wang C; Wang W; Wang Y; Xiao Y; Yuen PW; Zak M; Zhao Q; Zheng X Fragment-based design of 3-aminopyridine-derived amides as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT). Bioorg. Med. Chem. Lett, 2014, 24(3), 954–962. 10.1016/j.bmcl.2013.12.062 [DOI] [PubMed] [Google Scholar]


