Abstract
SARs (scaffold attachment regions) are candidate DNA elements for partitioning eukaryotic genomes into independent chromatin loops by attaching DNA to proteins of a nuclear scaffold or matrix. The interaction of SARs with the nuclear scaffold is evolutionarily conserved and appears to be due to specific DNA binding proteins that recognize SARs by a mechanism not yet understood. We describe a novel, evolutionarily conserved protein domain that specifically binds to SARs but is not related to SAR binding motifs of other proteins. This domain was first identified in human scaffold attachment factor A (SAF-A) and was thus designated SAF-Box. The SAF-Box is present in many different proteins ranging from yeast to human in origin and appears to be structurally related to a homeodomain. We show here that SAF-Boxes from four different origins, as well as a synthetic SAF-Box peptide, bind to natural and artificial SARs with high specificity. Specific SAR binding of the novel domain is achieved by an unusual mass binding mode, is sensitive to distamycin but not to chromomycin, and displays a clear preference for long DNA fragments. This is the first characterization of a specific SAR binding domain that is conserved throughout evolution and has DNA binding properties that closely resemble that of the unfractionated nuclear scaffold.
In the eukaryotic nucleus, chromosomes occupy individual, nonoverlapping territories and reactions of DNA and RNA metabolism are confined to discrete structures in the nuclear interior (for review, see reference 40). Despite many efforts to elucidate the molecular basis for nuclear architecture, a clear conception of higher-order structures in the nucleus has not yet emerged. One much debated possibility is that structure and function of the nucleus are determined by a proteinaceous, skeletonlike entity called the nuclear scaffold or nuclear matrix and its interaction with architectural DNA elements in the genome (18). Attachment of chromatin to the nuclear scaffold seems to occur via specialized AT-rich DNA elements that have been found in all eukaryotic organisms investigated and have been proposed to partition the genome into distinct, topologically independent loops of variable size (30). Termed SARs (scaffold attachment regions) or MARs (matrix attachment regions) (8, 17), these DNA elements are bound by nuclear scaffolds in an evolutionarily conserved manner (9), presumably because of one or more conserved binding proteins present in these scaffolds. The recognition of SARs by their cognate binding proteins is not yet understood in molecular terms but apparently does not depend on a precise recognition sequence because a consensus sequence common to all SARs could not be identified. Instead, SARs may be recognized by structural features and/or short sequence motifs clustered in SAR but not non-SAR DNA. In fact, most characterized SARs contain homopolymeric runs of A or T (A-tracts) that result in a characteristically narrow minor groove of DNA (reviewed in reference 4). The importance of these A-tracts for the interaction of SARs with the nuclear scaffold has been demonstrated by experiments with distamycin. This minor-groove-binding peptide antibiotic selectively binds to (dA · dT)n sequences, thereby suppressing or dissociating interactions of SARs with the nuclear scaffold (24). In addition to the presence of A-tracts and other AT-rich sequence motifs, such as unwinding elements (3) or the recently described matrix attachment region recognition signature (MRS) (50), SARs need to have a certain length to exhibit a specific interaction. Natural SARs are usually between 600 and 3,000 bp long, suggesting a requirement for cooperative interactions between the SAR and cognate binding proteins in the nuclear scaffold.
In addition to their presumed role in nuclear architecture, SARs have also been implicated in the regulation of gene expression, as they are frequently observed close to enhancers (8, 17), stimulate gene expression of heterologous reporter genes when integrated into the genome (48), and can regulate chromatin accessibility (22). SARs delimit individual units of gene expression in some cases (17, 41) but may also be located in the introns of large genes, where they appear stably bound to the nuclear scaffold yet dynamic enough not to impair transcription (23, 45). Intronic SARs do not differ from gene-flanking SARs with respect to their nucleotide composition or their effect on reporter gene expression. It is therefore likely that both types of SARs perform the same function in vivo, the anchorage of chromatin loops to the nuclear scaffold, and thereby, presumably, affect the expression of adjacent genes.
Several SAR binding proteins have been identified and characterized in the last years. These proteins include ubiquitous, abundant proteins like topoisomerase II (1), histone H1 (20), lamin B1 (34), HMG I/Y (52), and nucleolin (12) but also proteins that are expressed primarily in certain cell types, like SATB1 (11) or p114 (51). We have isolated and characterized the human nuclear proteins SAF-A (scaffold attachment factor A), also known as hnRNP-U because of its association with hnRNP particles (14, 15, 25, 44), and SAF-B (43). Interestingly, even though many of these proteins have been thoroughly characterized, it has not been possible to gain an understanding of the mechanism of binding specificity in molecular terms, and it has also been difficult to decide which of the proteins are required for nuclear architecture in vivo. We have therefore focused our interest on one of the major SAR binding proteins in human cells, SAF-A, to investigate the mode of recognition and binding to SARs in molecular detail.
In this report, we demonstrate that SAF-A binds to SAR DNA through a novel, evolutionarily conserved protein domain. This domain, which we call SAF-Box, is found in proteins ranging from yeast to human in origin and recognizes SAR DNA through a multitude of weak interactions that collectively result in high-specificity binding. Binding properties of the isolated domain closely resemble those of the unfractionated nuclear scaffold, and its presence in all eukaryotes may be one reason for the evolutionary conservation of SAR-scaffold interactions.
MATERIALS AND METHODS
Cell culture, transfection, and proliferation assay.
COS7 and MCF-7 cells were cultured in Dulbecco's modified Eagle medium containing 10% fetal calf serum and 0.6 μg of insulin/ml (MCF-7 only) at 37°C in a humidified atmosphere and were passaged every 3 days by fivefold dilution into fresh medium. Absolute cell numbers were determined with an automatic cell counter (Coulter) according to the manufacturer's protocol. For the analysis of protein localization in vivo, cells cultured on coverslips were transfected with expression vectors encoding enhanced-green-fluorescent-protein (EGFP) fusion proteins of wild-type SAF-A or of the deletion mutant ΔN45, using SuperFect reagent (Qiagen) as recommended by the manufacturer. Cells were observed by fluorescent microscopy 30 h posttransfection.
For proliferation assays, cells were transfected with increasing amounts of the expression vectors by electroporation (3 to 12 μg of DNA in 800 μl of complete medium containing 5 × 105 cells; 4-mm cuvette, 400 V, 960 μF). Cells were placed in 10 ml of prewarmed medium immediately after the pulse, and 2.5 ml of the suspension was plated on six-well dishes. With MCF-7 cells, this method yields >60% of transfected cells and less than 5% cell death. Transfection efficiency and cell death were indistinguishable for the different types and amounts of DNA used in our studies. At 30 h posttransfection, the medium was removed by aspiration and replaced by medium containing 0.5 μCi of [3H]thymidine/ml. After 12 h, cells were washed twice with phosphate-buffered saline, lysed in 0.5 ml of phosphate-buffered saline–0.1% sodium dodecyl sulfate (SDS), and diluted to 5 ml with water. The lysate was vortexed vigorously, and DNA was precipitated by the addition of trichloroacetic acid (TCA) to a final concentration of 20% for 30 min on ice. Precipitated DNA was quantified by scintillation counting after filtration through Millipore GF/C glass fiber filters and extensive washing with 10% TCA and methanol.
Cloning, expression, and purification of recombinant protein fragments.
Construction of expression clones for partial proteins ZZ-N45, ZZ-N247, and ZZ-N247ΔN45 from human SAF-A were described in reference 19. Expression clones encoding the SAF-Boxes from proteins C43E11.1 (from Caenorhabditis elegans), mlo1+ (from Schizosaccharomyces pombe), and T19P19.70 (from Arabidopsis thaliana) were constructed by cloning PCR-amplified fragments into the pEZZ18 vector (Pharmacia), using primer-incorporated restriction sites. All PCRs were performed with Pfu polymerase (Stratagene), a 5′ primer with an EcoRI site, and a 3′ primer with a stop codon and a HindIII site. For the C. elegans protein, the template was Cosmid C43E11 (kindly supplied by the C. elegans Sequencing Consortium, Cambridge, United Kingdom) and primers were CATGAATTCCGCGGACGAGGATATTTTA and ACTGAAGCTTTCATAATTTGGCAAGTACCTCCTT. For the S. pombe protein, the template was a cloned cDNA fragment (cDNA118, kindly supplied by Jean-Paul Javerzat, Bordeaux, France) and primers were CATGAATTCCATGTCAGATTACAAGAGTCTT and ACTGAAGCTTTCAAGTATTTTCATCGTTACTCTC. For the A. thaliana protein, the template was BAC T19P19 (kindly supplied by the Arabidopsis Biological Resource Center, Columbus, Ohio) and primers were CATGAATTCCTCGTCATCGCCTTTTCCA and ACTGAAGCTTTCACTCAGCACGAAGTGCTTCATC. All expression constructs were verified by sequencing and encoded proteins of 45, 49, 45, and 50 amino acids (aa) from SAF-A, C43E11.1, mlo1+, and T19P19.70, respectively, plus an amino-terminal ZZ-tag of 14 kDa.
Purification of recombinant proteins with a ZZ-tag was performed by chromatography on immunoglobulin G (IgG)-Sepharose and Mono-Q (both from Pharmacia), as described previously (19). Only protein fractions with >90% purity were used for the binding assays.
Peptide synthesis and purification.
The SAF-Box peptide was synthesized on an ABIMED EPS 221 semiautomated peptide synthesizer using a Novasyn TGR resin (Novabiochem), 9-fluoroenylmethoxy carbonyl chemistry with PyBOP activation, double coupling strategy, and capping of unreacted Nα groups with acetic anhydride (36). After synthesis, the peptide was cleaved from the resin in 90% (vol/vol) trifluoroacetic acid (TFA), 5% (vol/vol) water, and 5% (vol/vol) triethylsilane for 3.25 h at room temperature, precipitated with 5 volumes of cold t-butyl-methylether for 15 h at −20°C, and redissolved in buffer A (100 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA). For purification, the peptide was coupled to Thiopropyl-Sepharose 6B (Pharmacia) via disulfide bond formation with the N-terminal cysteine for 1.5 h in buffer A. As this cysteine was coupled last, it was present only in full-length peptides and allowed for the removal of shorter synthesis by-products by washing the column with 50 volumes of buffer A. The peptide was eluted with 10 mM dithiothreitol, diluted 20-fold in buffer B (200 mM acetate buffer [CH3COOH-CH3COONa] [pH 4.0]), and applied to a fast protein liquid chromatography Mono-S column (Pharmacia; volume, 1 ml). The peptide was eluted in a linear gradient from 0 to 1,000 mM NaCl in buffer B. Fractions containing the peptide (approximately at 550 mM NaCl) were pooled, applied to a high-performance liquid chromatography (HPLC) C18 column (YMC-pack ODS-AQ; YMC), and eluted with a linear gradient from water–0.1% (vol/vol) TFA to acetonitrile–0.07% TFA. The crude synthesis product as well as fractions of all purification steps was analyzed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry as described (5). Only fractions containing peptide of >90% purity were used for DNA binding assays.
DNA binding assays.
For pull-down DNA binding assays, recombinant protein or synthetic peptide was coupled to IgG-Sepharose or Thiopropyl-Sepharose, respectively, at room temperature for 1 h in 100 mM Tris-HCl (pH 7.5)–100 mM NaCl–1 mM EDTA and washed three times in the same buffer. Beads were either used for binding assays immediately or stored in coupling buffer at 4°C for several days without noticeable changes in binding activity. A standard DNA binding assay was done with 10 μl of settled beads and 30 ng of radioactively end-labeled DNA in 200 μl of binding buffer (10 mM Tris-HCl [pH 8.0], 80 mM NaCl, 2 mM EDTA) and was incubated on a rocking platform for 1 h at room temperature. Sheared Escherichia coli DNA (average fragment size, between 500 and 1,000 bp) was used as an unlabeled, unspecific competitor where indicated. Unbound DNA was removed by washing six times with binding buffer, and DNA binding was quantified by scintillation counting. For gel analysis, bound DNA was eluted from the drained beads in 50 μl of binding buffer with 3% SDS, immediately followed by the addition of 380 μl of Tris-EDTA (TE) (10 mM Tris [pH 8.0], 1 mM EDTA), purified by phenol-chloroform extraction, and precipitated with ethanol. DNA was redissolved in TE, separated on 1% standard agarose or 3% Resophor Agarose (Eurobio) gels, and visualized by autoradiography after gel drying. DNAs used for binding assays were EcoRI-BamHI-digested pMII (human SAR MII cloned into pUC18 [45]) and XbaI-HindIII-digested pGN1.5 (petunia SAR GN1.5 cloned into pGEM3 [13]. For the experiment shown in Fig. 8, two unphosphorylated complementary 45-mer oligonucleotides, AATTCAGAAAATAATAAAATAAAACTAGCTATTTTATATTTTTTC and GTCTTTTATTATTTTATTTTGATCGATAAAATATAAAAAAGTTAA (containing EcoRI-compatible overhangs), were annealed by boiling in 10 μl of TE for 5 min and cooling to 70°C over approximately 30 min. The mixture (2 μg of annealed oligonucleotides) was allowed to cool to room temperature before phosphorylation of 5′ ends with 10 U of T4 polynucleotide kinase for 30 min at 37°C in ligase buffer and ligation by 1 U of T4 ligase for 15 h at room temperature. Ligated oligonucleotides were radioactively labeled by a fill-in reaction with Klenow polymerase and [32P]dATP, mixed with an equal amount of MII-pUC18 (as internal specificity control) and increasing amounts of E. coli competitor DNA, and tested for binding to the SAF-Box in a standard assay. For control experiments, substrates were prepared in an identical manner using unrelated oligonucleotides.
FIG. 8.
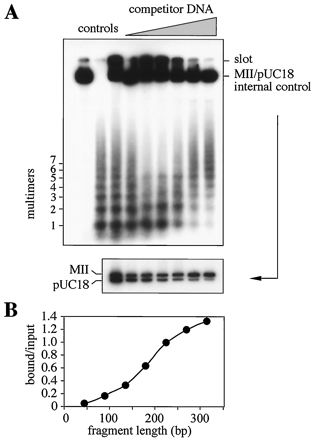
The SAR binding peptide prefers to bind to long DNA fragments. (A) Synthetic oligonucleotides containing the MRS sequence (50) were multimerized by ligation, radioactively end labeled, and used as the substrate in pull-down DNA binding assays with the immobilized synthetic peptide. Labeled MII-pUC18 mixture was added as internal specificity control. DNA bound in assays with increasing amounts of competitor DNA was recovered from the beads, split into two equal parts, and analyzed by gel electrophoresis through 3% Resophor agarose (upper panel) to visualize the multimers or 1% agarose to resolve the internal control (lower panel). Samples were normalized on the basis of scintillation counting, and identical radioactivity was applied to each lane. Controls: MII-pUC18 mixture alone, multimers alone, and the input mixture. (B) The gel shown in the upper part of panel A was scanned to quantify bound DNA by densitometry. Lane 3 (input) and lane 9 (bound in the presence of a 500-fold excess of competitor DNA) (shown in panel A, from the left) were used to calculate the bound-to-input ratio separately for each multimer; ratios exceeding 1 result from the application of the same radioactivity to each lane, demonstrating an overrepresentation of higher multimers. Note the clear preference for fragments that are >200 bp.
Other methods.
SDS-polyacrylamide gel electrophoresis of recombinant proteins was performed as described by Laemmli (29), and electrophoresis of peptides was performed according to the method of Schägger and von Jagow (46). Protein gels were stained with Coomassie brilliant blue (47). Protein concentrations were determined using the Bio-Rad protein assay and verified by comparison with samples of known protein content on SDS-polyacrylamide gels.
RESULTS
Identification of the SAF-Box.
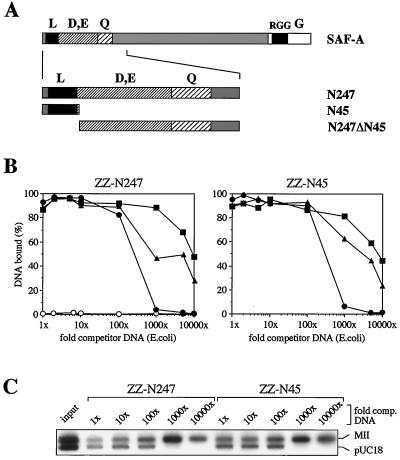
SAF-A is an abundant component of the nuclear scaffold that binds to SAR DNA with great specificity (14, 15, 44). In a recent report, we have shown that the DNA binding activity of SAF-A is destroyed upon proteolytic cleavage during apoptosis and could map the DNA binding domain of SAF-A to the amino-terminal 247 residues (19, 26). In a Southwestern blotting procedure, recombinant partial proteins shorter than 247 aa did not bind to DNA and two regions in these 247 aa of the protein appeared to be necessary for DNA binding. The two regions, aa 1 to 45 and aa 158 to 247, are separated by a stretch of acidic residues. This acidic stretch is not involved in DNA binding, because it can be deleted from a recombinant protein—resulting in a direct fusion of the two necessary regions—without affecting DNA binding properties in Southwestern blots (not shown). During these experiments we felt that the protein denaturation inherent in the Southwestern blotting approach might be a severe limitation for defining protein-DNA interactions in detail. To eliminate this experimental problem, we employed a pull-down assay using native, purified partial proteins. Recombinant amino-terminal proteins, expressed as secreted fusion proteins with a ZZ-tag (19, 33), were immobilized on IgG-Sepharose and tested for binding to labeled DNA. When investigated with this more sensitive method, the DNA binding domain of SAF-A was mapped to the amino-terminal 45 residues of SAF-A (Fig. 1), containing the spaced leucine motif described in our previous publication (19). Surprisingly, the second “necessary” region identified in those original experiments was found to be dispensable for DNA binding itself. This region seems to be involved in refolding of the DNA binding domain after denaturation, because it is necessary in Southwestern blots but not in pull-down assays with native proteins. On the other hand, the protein fragment with aa 1 to 45 retained DNA binding characteristics indistinguishable from those of the shortest DNA binding fragment known at that time, N247, and its removal from N247 abrogated DNA binding completely (Fig. 1B). Thus, the amino-terminal 45 residues of SAF-A are necessary and sufficient for specific binding to SAR DNA.
FIG. 1.
A SAR binding domain maps to the extreme amino terminus of human SAF-A. (A) Schematic representation of complete SAF-A and the protein fragments used for DNA binding assays. The RNA binding RGG-Box (25) and regions rich in leucine (L), acidic residues (D, E), glutamine (Q), and glycine (G) are indicated. (B) Pull-down DNA binding assays with recombinant constructs of human SAF-A. Proteins ZZ-N247 and ZZ-N45 were overexpressed, purified, immobilized on IgG-Sepharose, and incubated with the human SAR element MII (filled squares), non-SAR pUC18 (filled circles), or an equimolar mixture of both DNAs (filled triangles) in the presence of increasing amounts of E. coli competitor DNA. Bound DNA was quantified by scintillation counting and expressed as percentage of input. Note that the amount of immobilized protein (1 μg) was chosen to be saturating up to a 100-fold excess of competitor DNA. A control experiment with ZZ-N247 lacking the amino-terminal 45 residues (ZZ-N247ΔN45) is shown in the left panel (open circles). (C) A pull-down DNA binding assay was performed with an equimolar mixture of a SAR (MII) and non-SAR DNA (pUC18) and increasing amounts of unspecific competitor DNA. Bound DNA was eluted from the beads, and aliquots of identical radioactivity were analyzed on agarose gels. Note the high specificity for SAR DNA under stringent conditions.
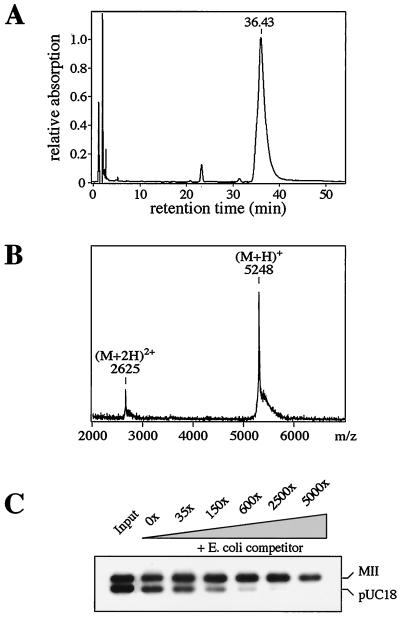
In complementary experiments, we have synthesized a peptide containing the amino-terminal 45 residues of human SAF-A and an additional amino-terminal cysteine. This single amino acid tag was used for chromatographic purification over Thiopropyl-Sepharose and for immobilization on the same material for use in pull-down DNA binding assays. The purified peptide eluted as a symmetric peak in reverse-phase HPLC (Fig. 2A) and had the expected molecular mass, as determined by MALDI-TOF mass spectrometry (Fig. 2B). In DNA binding assays, SAR-specific binding of the peptide was indistinguishable from that of the recombinant ZZ-N45 fusion protein (Fig. 2C, compare to Fig. 1C). This indicates that the synthetic peptide spontaneously folds into an active conformation and, by this definition, represents an independent protein domain.
FIG. 2.
A synthetic SAR binding peptide. (A) A 46-residue peptide with the SAF-Box from human SAF-A was synthesized and purified by chromatography. The last purification step, reverse-phase HPLC on a C18 column, shows the peptide elutes as a symmetric peak in a water-acetonitrile gradient. (B) MALDI-TOF mass spectrometry demonstrates the integrity of the peptide. The calculated molecular weight is 5,245. (C) The purified peptide (100 ng) was immobilized on Sepharose beads and tested for DNA binding to the MII-pUC18 mixture in the presence of increasing amounts of competitor DNA.
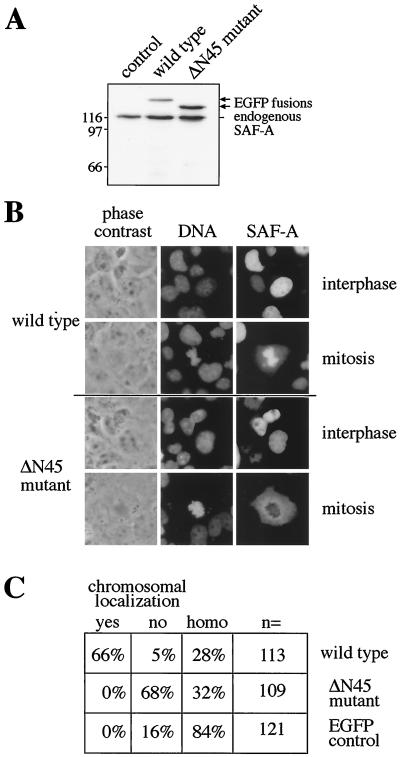
To investigate the in vivo relevance of the novel DNA binding domain, we constructed expression vectors for human SAF-A with and without the first 45 aa, amino-terminally fused to EGFP for direct visualization in live cells. Transient transfection experiments demonstrate that both the wild-type and the ΔN45 deletion mutant are expressed at levels slightly lower than or comparable with those for endogenous SAF-A (Fig. 3A) and localize to the nucleus of interphase cells (Fig. 3B). In mitotic cells, however, the localization of both proteins differs markedly. While wild-type SAF-A localizes to mitotic chromosomes in two-thirds of cells, the deletion mutant does not associate with chromosomes at all (Fig. 3B and C). Rather, the region of condensed chromosomes appears negative for the deletion mutant in over two-thirds of cells. For both proteins, approximately one-third of cells display homogenous cellular staining reminiscent of the EGFP control cells, suggesting that the SAF-A constructs are neither specifically concentrated in nor kept out of the region of condensed chromosomes in these cells. We do not presently know the reason for this different behavior of the same protein in different cells but found it independent of expression levels when individual cells were compared for signal strength and protein localization. However, the clear effect of the deletion of the amino-terminal domain strongly suggests that chromosomal localization is due to the DNA binding of SAF-A.
FIG. 3.
The SAF-Box targets SAF-A to mitotic chromosomes in transient transfection experiments. (A) COS7 cells were transfected with expression vectors encoding fusion proteins of wild-type SAF-A or a SAF-Box deletion mutant with EGFP. Cells were analyzed 24 h posttransfection by SDS-polyacrylamide gel electrophoresis and immunoblotting of total cell extracts with SAF-A-specific antibodies. Control cells were not transfected. (B) COS7 cells cultivated on coverslips were transfected as above and analyzed microscopically. Typical images of interphase cells and mitotic cells are shown for both protein constructs. (C) Mitotic cells transfected with wild-type SAF-A–EGFP, ΔN45 mutant-EGFP, or EGFP alone were scored for localization of green fluorescence on chromosomes (yes or no) or homogeneous cellular staining (homo). In all cases, more than 100 mitotic cells were scored (n=).
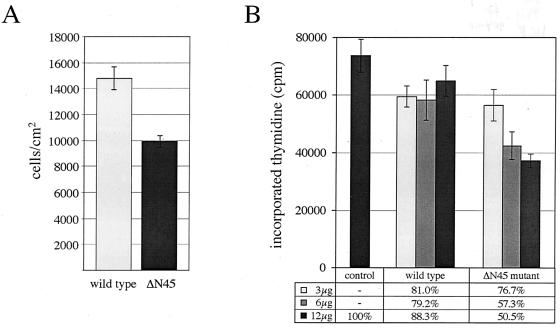
It is assumed that SARs are important regulatory elements for organizing higher-order chromatin domains. In accordance with such a role, one would predict that SAR binding proteins would have critical functions in DNA replication, gene transcription, or more global processes, such as proliferation or differentiation. In fact, transient transfection of cells with the SAF-A deletion mutant ΔN45 fused to EGFP results in a significant decrease in the proliferation of these cells. This is demonstrated by lower absolute cell numbers (Fig. 4A) and a marked, concentration-dependent decrease in the replicative incorporation of radioactive thymidine into genomic DNA (Fig. 4B). A comparable effect is observed neither in cells transfected with a construct encoding a wild-type SAF-A–EGFP fusion protein nor in control cells that had received the empty pEGFP-N1 vector only. Obviously, the mutant protein lacking the DNA binding domain exerts a dominant negative effect on the natural function of SAF-A and confirms the importance of the novel domain in vivo.
FIG. 4.
Expression of a mutant SAF-A lacking the SAF-Box exerts a dominant negative effect on proliferation. (A) MCF-7 cells were transfected with 12 μg of expression vectors for wild-type SAF-A–EGFP or the ΔN45 mutant-EGFP, and absolute cell numbers were determined 48 h posttransfection by using a Coulter cell counter. (B) MCF-7 cells were transfected with 12 μg of the empty pEGFP-N1 vector (control) or 3, 6, or 12 μg of the expression vectors described in panel A. After 30 h, cells were labeled by exchanging the medium with fresh medium containing 0.5 μCi of [3H]thymidine/ml. After 12 h, cells were harvested and the amount of radioactive thymidine incorporated into genomic DNA was determined by precipitation with TCA, filtration over GF/C fiberglass filters, and scintillation counting. All assays were done in triplicate; error bars indicate the standard deviations. The results are presented as absolute values in counts per minute (cpm), and relative values are normalized to the vector control. In all assays, transfection efficiency and cell death were approximately 60 and 4%, respectively.
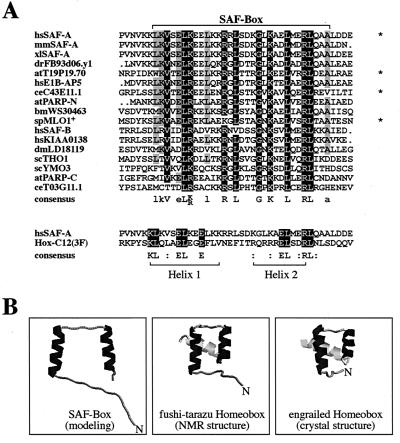
Database comparisons revealed that a protein motif homologous to 31 aa in this domain is present in many proteins from eukaryotic but not prokaryotic organisms. Interestingly, this motif is found in both SAF proteins identified in this laboratory, SAF-A and SAF-B, and is the only homologous sequence for these two proteins. We have therefore designated this region the SAF-Box (Fig. 5A). The SAF-Box shares significant homology with helix 1 and helix 2 of a homeodomain, e.g., from Hox-C12(3F), and all residues of the SAF-Box are compatible with the homeodomain consensus sequence derived by Bürglin (6). Hence, the SAF-Box appears to be structurally related to the corresponding region of a homeodomain known to fold into a hooklike structure composed of two α-helices separated by a turn (Fig. 5B). Interestingly, helix 3, which is common to all homeodomains and confers specific DNA binding, is neither present in the SAF-Box nor needed for its binding to SAR-DNA. Experiments are under way in this laboratory to elucidate the actual structure of the SAF-Box and how it interacts with DNA.
FIG. 5.
The SAF-Box is a conserved protein domain. (A) Alignment of 17 SAF-Boxes from proteins originating in human (hs), mouse (mm), Xenopus laevis (xl), zebra fish (dr), A. thaliana (at), C. elegans (ce), Bombyx mori (bm), S. pombe (sp), Drosophila melanogaster (dm), and S. cerevisiae (sc). The two SAF-Boxes from the A. thaliana PARP are indicated as PARP-N and PARP-C for the amino-terminal or carboxy-terminal box, respectively. Homologies to the homeodomain of Hox-C12(3F) are shown below. Asterisks denote proteins used for further studies. (B) Comparison of the putative structure of the SAF-Box as derived from secondary structure predictions and computer-assisted modeling with known structures of fushi tarazu (42) and engrailed (7, 27) homeoboxes. Helix 3 of a homeobox (light gray) is not present in the SAF-Box. NMR, nuclear magnetic resonance.
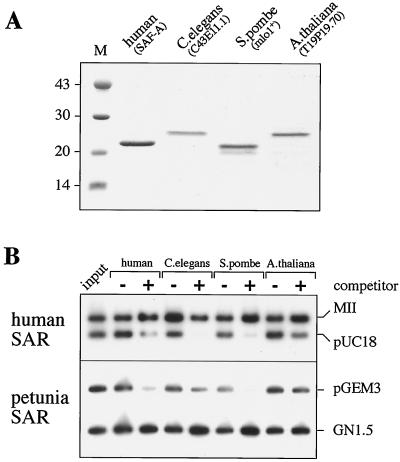
To investigate whether the conservation of the SAF-Box motif is accompanied by a conservation of specific SAR binding activity, we cloned, expressed, and purified recombinant SAF-Boxes from four different proteins originating in humans, C. elegans, A. thaliana, and S. pombe (Fig. 6A). The purified proteins were immobilized on IgG-Sepharose as described above and tested for binding to DNA using two different equimolar SAR–non-SAR DNA mixtures. Indeed, all tested proteins displayed specific binding to SARs from human and petunia but not to non-SAR vector controls (Fig. 6B). Thus, specific SAR binding is a conserved feature of the SAF-Box.
FIG. 6.
SAR binding is a conserved activity of the SAF-Box. (A) Proteins containing the SAF-Boxes from four different proteins originating in humans, C. elegans, S. pombe, and A. thaliana, each with an amino-terminal ZZ-tag, were bacterially overexpressed, purified, and analyzed by SDS-polyacrylamide gel electrophoresis. (B) DNA binding assays. Identical amounts of the four proteins were immobilized on IgG-Sepharose and incubated with two different SAR–non-SAR mixtures in the absence (−) or presence (+) of a 500-fold excess of competitor DNA. Note that all proteins specifically bind to the SARs MII (human) and GN1.5 (petunia) in the presence of competitor DNA but not to plasmid controls (pUC18, pGEM3), although slight differences in specificity are apparent.
The SAF-Box binds to SAR DNA through mass binding.
Earlier investigations on the properties of native purified SAF-A from human cells had revealed that DNA binding of this protein was intimately linked to a self-assembly into long filamentous or globular complexes (14, 15). Under no experimental conditions could DNA binding be observed in the absence of self-assembly or vice versa, suggesting that protein-protein interactions played a central role in the binding of SAF-A to DNA. It was therefore not possible to characterize the DNA binding activity independent of protein self-assembly. Identification of the SAF-Box enabled us now to investigate the binding mode of this domain to DNA in more detail and thereby gain insight into the mechanism that may govern SAR binding to the nuclear scaffold. As a first step, we performed pull-down DNA binding assays with increasing amounts of recombinant ZZ-N247 and ZZ-N45 coupled to IgG-Sepharose. DNA binding occurred with a sigmoidal binding curve identical for both proteins when expressed in molar terms, indicating a cooperative binding mode (Fig. 7A). When similar experiments were performed with a mixture of SAR and non-SAR DNA and bound DNA was analyzed by agarose gel electrophoresis, SAR DNA was clearly preferred to non-SAR DNA at low protein concentrations (Fig. 7B). Saturating amounts of the SAF-Box bound to non-SAR DNA as well as to SAR DNA, reflecting a general DNA binding activity previously described for full-length SAF-A (14). Very similar binding properties were obtained with the recombinant SAF-Box from the S. pombe mlo1+ protein (Fig. 7B, lower panel) and the synthetic SAF-Box peptide (not shown). From these experiments, we determined a stoichiometry of approximately 300 SAF-Boxes necessary to bind to one molecule of the human MII SAR (3,000 bp) or roughly one SAF-Box per 10 bp of DNA. This is certainly an overestimate, because not all coupled protein molecules may be available for DNA binding due to steric constraints on the surface of beads, but clearly showed that many SAF-Boxes must cooperate to bind to a single molecule of DNA. This conclusion was further supported by our finding that DNA binding of the isolated SAF-Box was almost undetectable when tested in solution, e.g., in filter binding or gel mobility shift assays (data not shown), suggesting that the interaction of a single SAF-Box with DNA is unstable or of low affinity. The strong and specific binding of SARs in pull-down assays therefore appears to result from protein immobilization that brings individual protein molecules into close proximity. Thus, SAR binding of the SAF-Box may be due to the mass binding mechanism of Zuckerkandl and Villet (53), characterized by the binding of a large number of individual protein molecules to sequence motifs periodically recurring on one DNA chain. Although each interaction has relatively low affinity and specificity, many interactions are collectively turned into high-affinity, high-specificity binding through cooperative effects. With intact, full-length SAF-A, these cooperative effects seem to be induced by protein multimerization (see above), whereas the isolated SAF-Box needs immobilization on a surface to mimic the close proximity of individual domains in natural protein complexes.
FIG. 7.
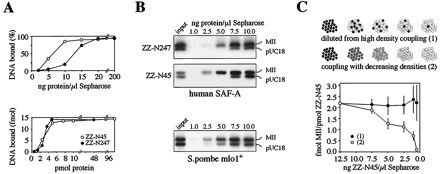
The DNA binding mode of the SAF-Box. (A) Increasing amounts of ZZ-N247 and ZZ-N45 from human SAF-A were immobilized on Sepharose beads and tested for binding to the isolated human MII SAR element. Note that both proteins have identical binding curves when expressed in molar terms (lower panel). (B) An experiment similar to that shown in panel A was performed with an equimolar mixture of a SAR (MII) and non-SAR (pUC18) DNA and the SAF-Box proteins ZZ-N247 and ZZ-N45 from human SAF-A and the SAF-Box from S. pombe mlo1+. Bound DNA was eluted from the beads and analyzed by agarose gel electrophoresis. (C) The human SAF-Box protein ZZ-N45 was immobilized on Sepharose beads under two different sets of conditions that result in identical absolute amounts of protein but different surface densities (upper panel). DNA binding assays with the isolated MII SAR demonstrate that the stoichiometry of bound DNA to protein is dependent on the density of coupled protein but not on the absolute amount of protein. Filled circles, constant density; open circles, decreasing density (lower panel). Note the reverse orientation of the x axis.
The mass binding mode of DNA-protein interaction has the testable consequence that it should depend on the distance between individual protein molecules, because these must be close enough to make contacts with the same DNA fragment. We have therefore compared DNA binding to beads that were coated with SAF-Box protein under two different sets of conditions (Fig. 7C). First, SAF-Box protein ZZ-N45 was immobilized at high density and diluted stepwise with empty beads to yield decreasing absolute amounts of protein but identical surface density. Second, protein was coupled to an increasing volume of beads, resulting in amounts identical to those of the first assay but with different surface densities. We found that the ratio of DNA molecules bound per molecule of protein was unaffected in the first assay with identical surface density but was strongly affected in the second assay. Thus, DNA binding by the SAF-Box occurs through mass binding.
We used the synthetic SAR binding peptide to further characterize the mode of DNA binding and compare it with known features of the interaction between SARs and the unfractionated nuclear scaffold. In the first experiment, we investigated whether DNA binding by the SAF-Box peptide was affected by DNA fragment length and might thus reproduce the known length effect of SAR-scaffold interactions (see above). To this end, we used artificial SARs of variable length created by ligation of oligonucleotides containing the recently described MRS (50). This sequence is the core element of the well-studied Drosophila histone cluster SAR originally identified by Laemmli and coworkers (38) and specifically binds to nuclear scaffold preparations (50). Multimeric constructs of the 45-bp MRS oligonucleotide were labeled, mixed with the pUC18-MII substrate as internal specificity control, and tested for binding to the immobilized SAF-Box peptide. Bound DNA was eluted and analyzed on a high-resolution agarose gel, revealing a shift in the population of bound multimers when increasing amounts of competitor DNA were added to the reaction (Fig. 8A). Thus, DNA binding by the SAF-Box is clearly length dependent under stringent conditions (where the internal control confirms specific binding), with a sigmoidal curve saturating at DNA fragment lengths greater than 300 bp (Fig. 8B). Control experiments using oligonucleotides without the MRS sequence did not show significant binding of either the monomer or multimers at stringent conditions (not shown).
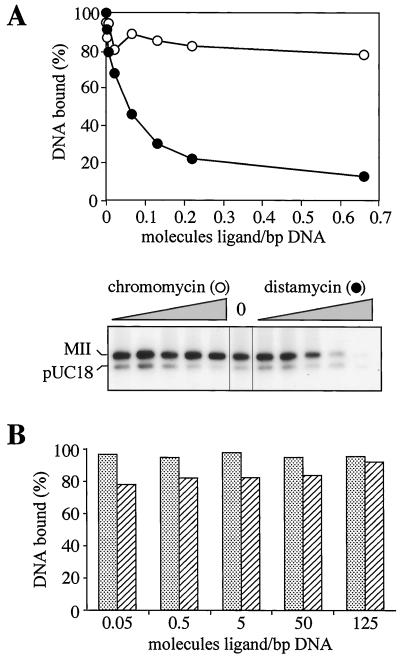
In a second set of experiments, we determined whether the interaction of SAR-DNA with the isolated SAF-Box was affected by DNA ligands, small molecules that bind to DNA at specific sites in vitro and in vivo. In line with experiments reported for SAR-scaffold interactions, we employed two minor-groove-binding peptide antibiotics, distamycin and chromomycin, that are well characterized with respect to their specific binding to A-tracts and G + C-rich sequence motifs, respectively (reference 24 and references therein). For the experiment shown in Fig. 9A, we performed pull-down DNA binding assays with an equimolar mixture of pUC18-MII and a 500-fold excess of unspecific E. coli competitor DNA. Under these conditions, specific SAR binding is observed in the absence of either drug (Fig. 9A). When distamycin and chromomycin were added in drug/DNA ratios known to result in highly specific binding of the drugs to DNA (24), distamycin effectively blocked SAR binding in a concentration-dependent manner, whereas chromomycin had no effect. Thus, the SAF-Box binds to SARs through interactions with A-tracts in the minor groove of DNA, like purified full-length SAF-A (reference 14 and unpublished observations) or unfractionated nuclear scaffolds (24). Interestingly, distamycin blocked binding of SAR DNA to the SAF-Box but was not able to disrupt preexistent interactions, even at very high drug/DNA ratios (Fig. 9B).
FIG. 9.
SAR binding of the synthetic peptide is sensitive to distamycin. (A) Pull-down DNA binding assays with the MII-pUC18 mixture and a 500-fold excess of unspecific competitor DNA in the presence of increasing amounts of distamycin or chromomycin. Bound DNA was quantified by scintillation counting (upper panel) and analyzed by agarose gel electrophoresis (lower panel, samples 3 to 8 from the upper panel). (B) Preexistent binding of SARs to the peptide are stable in the presence of distamycin. Pull-down DNA binding assays were performed as shown in panel A but in the absence of drugs. Distamycin (stippled bars) or chromomycin (hatched bars) was added after 1 h and was incubated with the DNA-peptide complexes for 30 min, before washing and quantification by scintillation counting.
DISCUSSION
In the present study we report the identification of the minimal SAR-DNA binding domain of human SAF-A and show that this domain is a novel, evolutionarily conserved domain also present in heterologous proteins. This SAF-Box appears to be structurally related to a homeodomain lacking the DNA recognition helix and binds to A-tracts in the minor groove of DNA with a characteristic mass binding mode.
Conservation of the SAF-Box.
The results of this study demonstrate that the minimal SAR binding domain of human SAF-A maps to the extreme amino terminus of the protein. Database comparisons and biochemical experiments have revealed that SAR-DNA binding is conferred by a highly conserved, novel protein domain that we call the SAF-Box (Fig. 5). We found that the SAF-Box is present in proteins from organisms as phylogenetically distant from each other as yeast, plants, and mammals. On the other hand, database searches for prokaryotic genomes did not reveal the existence of an equivalent sequence in either eubacteria or archaea. Thus, the SAF-Box appears to be common to all eukaryotes but absent from prokaryotes, compatible with its specific binding to SAR DNA (which has also been identified in eukaryotes only) and its inferred role in the architecture of the cell nucleus. Interestingly, the SAF-Box-containing proteins from evolutionarily distant eukaryotes are not orthologs, i.e., proteins of different species that can be traced back to a common ancestor. Rather, sequence homologies outside the SAF-Box are usually not detectable, suggesting that the box is an independent domain shared by several different proteins. The function of most of these proteins is not known, as they have been identified through genome sequencing projects rather than through biochemical or biological activities. Exceptions are the human proteins SAF-A, SAF-B, and E1B-AP5, which have been implicated in nuclear architecture and/or RNA metabolism (14, 16, 19, 43, 44), the S. pombe mlo1+ protein known to cause chromosome loss and lethality when overexpressed (21), and the enzyme poly(ADP-ribose) polymerase (PARP) from A. thaliana (31), which is involved in DNA repair mechanisms. Usually proteins contain only one SAF-Box, but the A. thaliana PARP has two SAF-Boxes arranged in tandem and at a location in the protein that is equivalent to that of the two zinc-finger DNA binding domains in a second isoform of A. thaliana PARP (M. Kazmaier et al., unpublished data) (GenBank accession no. AJ131705) and of PARPs from animals (49). Thus, the SAF-Box can replace a different, structurally unrelated DNA binding domain and may target this isoform of PARP to SAR DNA. In general, eukaryotic cells appear to have more than one SAF-Box-containing protein. In humans, five such proteins are known (two isoforms of SAF-A and SAF-B each and E1B-AP5); in A. thaliana, C. elegans, and Saccharomyces cerevisiae, there are two. It is likely that more SAF-Box-containing proteins will be found during genome sequencing projects in the future, and elucidating their function should be facilitated by the knowledge gathered with well-characterized proteins such as SAF-A.
Structure and DNA binding activity of the SAF-Box.
Sequence homology, secondary structure predictions, and computer-assisted modeling strongly suggest that the SAF-Box is structurally related to helix 1 and helix 2 of a homeodomain. Interestingly, the SAF-Box does not have helix 3, the DNA recognition helix common to all homeodomains, and is strikingly different from a homeodomain with respect to its DNA binding characteristics. While homeodomains bind to strictly defined nucleotide sequences in the major groove of DNA, DNA binding by the SAF-Box is not restricted to simple, easily detectable sequence motifs. Instead, binding occurs through a cooperative binding mode that recognizes SAR DNA through minor-groove interactions with multiple clustered A-tracts. In contrast to a homeodomain, binding of the SAF-Box to DNA is undetectable in solution but requires immobilization of the protein molecules on some surface, e.g., that of Sepharose beads. This suggests that DNA binding is governed by a mass binding mechanism requiring close proximity of individual DNA binding domains that collectively bind with high specificity to recurring sequence motifs on a contiguous DNA chain. This compares well with previously obtained data for purified full-length SAF-A, which showed an absolute requirement of protein self-assembly for DNA binding to occur (14, 15). The results presented here suggest that immobilization mimics this self-assembly by arranging the SAF-Boxes in a topologically constrained formation on a surface. In addition, this unusual binding mode is fully compatible with the presumed function of SAF-A and other SAF-Box-containing proteins in vivo, the binding of SAR DNA to the insoluble nuclear scaffold. Consequently, the recognition of SAR DNA is “fuzzy” in the sense that many different DNA sequences fulfil the criterion to be a SAR if they have a high number of binding sites (presumably A-tracts, the MRS, or unwinding elements) clustered on a DNA fragment of a certain length.
We were able to reproduce all characteristic DNA binding properties of the unfractionated nuclear scaffold in a simple three-component system of synthetic SAR binding peptide, synthetic oligonucleotides, and Sepharose beads. In this all-synthetic approach, the SAF-Box specifically interacts with the bipartite MRS sequence that was recently found associated with many—but not all—SARs (50), suggesting that the MRS might be one of the points of interaction between SAR DNA and the nuclear scaffold in vivo. This conclusion is supported by the elegant experiments of Laemmli and coworkers, who mapped the position of (anonymous) SAR binding proteins on the H1–H3 intergenic SAR of Drosophila using ExoIII (37). In this SAR, which is also the source of the MRS element used in our experiments, they found four strong stops for ExoIII that precisely map to the position of the 16-bp and 8-bp components of the two bipartite MRSs present in this SAR.
In our experiments, the SAF-Box displays a clear preference for DNA fragments longer than 200 bp and yields a length-dependence curve superimposable with that reported earlier for unfractionated scaffolds (2, 3). In addition, SAR binding of the SAF-Box peptide is effectively competed for by distamycin at drug/DNA ratios that result in highly specific binding to A-tracts in the minor groove and block SAR-scaffold interactions (24, 45). SAR binding is resistant to competition with prokaryotic DNA even at several-thousandfold excess but is highly conserved in eukaryotes because SAF-Boxes from humans, C. elegans, S. pombe, and A. thaliana all bind to SARs from sources as mutually distant as human and petunia.
The qualitative and quantitative similarity of SAR binding by the isolated SAF-Box and the unfractionated nuclear scaffold strongly suggests that much of the DNA binding of the nuclear scaffold is due to SAF-Box-containing proteins like SAF-A. Unfortunately, direct approaches addressing this issue, e.g., an experiment to show whether a SAF-Box peptide blocks binding of SARs to a nuclear scaffold preparation, are not feasible due to the peculiar mass binding mode of the SAF-Box. However, SAF-A is one of the 10 most abundant proteins in conventional scaffold preparations and the only one with SAR binding activity (35). Other SAR binding proteins with or without a SAF-Box (SATB1, topoisomerase II, SAF-B, histone H1, lamins, HMG I/Y, or ARBP) contribute to SAR-scaffold interactions to various, possibly cell type-dependent, degrees. It is conceivable that some of these proteins fulfil more specialized roles in scaffold-related functions, such as transcription, splicing, DNA replication, or repair, rather than the highly conserved architectural attachment of chromatin. Such roles have already been demonstrated, e.g., for SATB1 and SAF-B, which are involved in the regulation of transcription or splicing, respectively (28, 32, 39). Interestingly, SATB1, a cell type-specific SAR binding protein predominantly expressed in thymocytes, has been reported earlier to also contain an atypical homeodomain that is involved in SAR binding (10). However, in contrast to the SAF-Box of SAF-A, the homeodomain of SATB1 binds to DNA poorly and with low specificity and is not sufficient for SAR binding. Rather, the homeodomain assists an independent SAR binding domain in recognizing the core unwinding element within the base-unpairing region of a SAR, leading to an increase in affinity of the SAR binding domain. It will be interesting to investigate in future experiments if the homeodomain of SATB1 itself has a SAR preference when tested in the pull-down approach described in this report.
ACKNOWLEDGMENTS
We thank Antje Pfeilstetter-Dietz and Nicola Arndt (Braunschweig, Germany), Jean-Paul Javerzat (Bordeaux, France), Alan Coulson (Cambridge, United Kingdom), and the Arabidopsis Biological Resource Center (Columbus, Ohio) for supplying valuable materials, Rolf Knippers for support and critically reading the manuscript, and Beate Schumacher for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft through grants Fa376-1/1 and Fa376-2/1 to F.O.F. and EMBO short-term fellowship ASTF 9180 to F.O.F.
REFERENCES
- 1.Adachi Y, Käs E, Laemmli U K. Preferential, cooperative binding of DNA topoisomerase II to scaffold-associated regions. EMBO J. 1989;8:3997–4006. doi: 10.1002/j.1460-2075.1989.tb08582.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benham C, Kohwi-Shigematsu T, Bode J. Stress-induced duplex DNA destabilization in scaffold/matrix attachment regions. J Mol Biol. 1997;274:181–196. doi: 10.1006/jmbi.1997.1385. [DOI] [PubMed] [Google Scholar]
- 3.Bode J, Kohwi Y, Dickinson L, Joh T, Klehr D, Mielke C, Kohwi-Shigematsu T. Biological significance of unwinding capability of nuclear matrix-associating DNAs. Science. 1992;255:195–197. doi: 10.1126/science.1553545. [DOI] [PubMed] [Google Scholar]
- 4.Boulikas T. Chromatin domains and prediction of MAR sequences. Int Rev Cytol. 1995;162A:279–388. doi: 10.1016/s0074-7696(08)61234-6. [DOI] [PubMed] [Google Scholar]
- 5.Bühler S, Michels J, Wendt S, Rück A, Brdiczka D, Welte W, Przybylski M. Mass spectrometric mapping of ion channel proteins (porins) and identification of their supramolecular membrane assembly. Proteins. 1998;1998(Suppl. 2):63–73. doi: 10.1002/(sici)1097-0134(1998)33:2+<63::aid-prot8>3.3.co;2-9. [DOI] [PubMed] [Google Scholar]
- 6.Bürglin T R. A comprehensive classification of homeobox genes. In: Duboule D, editor. Guidebook to the homeobox genes. Oxford, United Kingdom: Oxford University Press; 1994. pp. 25–71. [Google Scholar]
- 7.Clarke N D, Kissinger C R, Desjarlais J, Gilliland G L, Pabo C O. Structural studies of the engrailed homeodomain. Protein Sci. 1994;3:1779–1787. doi: 10.1002/pro.5560031018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cockerill P N, Garrard W T. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 9.Cockerill P N, Garrard W T. Chromosomal loop anchorage sites appear to be evolutionarily conserved. FEBS Lett. 1986;204:5–7. doi: 10.1016/0014-5793(86)81377-1. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson L A, Dickinson C D, Kohwi-Shigematsu T. An atypical homeodomain in SATB1 promotes specific recognition of the key structural element in a matrix attachment region. J Biol Chem. 1997;272:11463–11470. doi: 10.1074/jbc.272.17.11463. [DOI] [PubMed] [Google Scholar]
- 11.Dickinson L A, Joh T, Kohwi Y, Kohwi-Shigematsu T. A tissue-specific MAR/SAR DNA-binding protein with unusual binding site recognition. Cell. 1992;70:631–645. doi: 10.1016/0092-8674(92)90432-c. [DOI] [PubMed] [Google Scholar]
- 12.Dickinson L A, Kohwi-Shigematsu T. Nucleolin is a matrix attachment region DNA-binding protein that specifically recognizes a region with high base-unpairing potential. Mol Cell Biol. 1995;15:456–465. doi: 10.1128/mcb.15.1.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dietz A, Kay V, Schlake T, Landsmann J, Bode J. A plant scaffold attached region detected close to a T-DNA integration site is active in mammalian cells. Nucleic Acids Res. 1994;22:2744–2751. doi: 10.1093/nar/22.14.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fackelmayer F O, Dahm K, Renz A, Ramsperger U, Richter A. Nucleic-acid-binding properties of hnRNP-U/SAF-A, a nuclear-matrix protein which binds DNA and RNA in vivo and in vitro. Eur J Biochem. 1994;221:749–757. doi: 10.1111/j.1432-1033.1994.tb18788.x. [DOI] [PubMed] [Google Scholar]
- 15.Fackelmayer F O, Richter A. Purification of two isoforms of hnRNP-U and characterization of their nucleic acid binding activity. Biochemistry. 1994;33:10416–10422. doi: 10.1021/bi00200a024. [DOI] [PubMed] [Google Scholar]
- 16.Gabler S, Schütt H, Groitl P, Wolf H, Shenk T, Dobner T. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J Virol. 1998;72:7960–7971. doi: 10.1128/jvi.72.10.7960-7971.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gasser S M, Laemmli U K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986;46:521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- 18.Gasser S M, Laemmli U K. A glimpse at chromosomal order. Trends Genet. 1987;3:16–22. [Google Scholar]
- 19.Göhring F, Schwab B L, Nicotera P, Leist M, Fackelmayer F O. The novel SAR-binding domain of scaffold attachment factor A (SAF-A) is a target in apoptotic nuclear breakdown. EMBO J. 1997;16:7361–7371. doi: 10.1093/emboj/16.24.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izaurralde E, Käs E, Laemmli U K. Highly preferential nucleation of histone H1 assembly on scaffold-associated regions. J Mol Biol. 1989;210:573–585. doi: 10.1016/0022-2836(89)90133-2. [DOI] [PubMed] [Google Scholar]
- 21.Javerzat J P, Cranston G, Allshire R C. Fission yeast genes which disrupt mitotic chromosome segregation when overexpressed. Nucleic Acids Res. 1996;24:4676–4683. doi: 10.1093/nar/24.23.4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenuwein T, Forrester W C, Fernandez-Herrero L A, Laible G, Dull M, Grosschedl R. Extension of chromatin accessibility by nuclear matrix attachment regions. Nature. 1997;385:269–272. doi: 10.1038/385269a0. [DOI] [PubMed] [Google Scholar]
- 23.Käs E, Chasin L A. Anchorage of the Chinese hamster dihydrofolate reductase gene to the nuclear scaffold occurs in an intragenic region. J Mol Biol. 1987;198:677–692. doi: 10.1016/0022-2836(87)90209-9. [DOI] [PubMed] [Google Scholar]
- 24.Käs E, Izaurralde E, Laemmli U K. Specific inhibition of DNA binding to nuclear scaffolds and histone H1 by distamycin. The role of oligo(dA) · oligo(dT) tracts. J Mol Biol. 1989;210:587–599. doi: 10.1016/0022-2836(89)90134-4. [DOI] [PubMed] [Google Scholar]
- 25.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kipp M, Schwab B L, Przybylski M, Nicotera P, Fackelmayer F O. Apoptotic cleavage of scaffold attachment factor A (SAF-A) by caspase-3 occurs at a noncanonical cleavage site. J Biol Chem. 2000;275:5031–5036. doi: 10.1074/jbc.275.7.5031. [DOI] [PubMed] [Google Scholar]
- 27.Kissinger C R, Liu B S, Martin-Blanco E, Kornberg T B, Pabo C O. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 28.Kohwi-Shigematsu T, Maass K, Bode J. A thymocyte factor SATB1 suppresses transcription of stably integrated matrix-attachment region-linked reporter genes. Biochemistry. 1997;36:12005–12010. doi: 10.1021/bi971444j. [DOI] [PubMed] [Google Scholar]
- 29.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 30.Laemmli U K, Käs E, Poljak L, Adachi Y. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr Opin Genet Dev. 1992;2:275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- 31.Lepiniec L, Babiychuk E, Kushnir S, Van Montagu M, Inze D. Characterization of an Arabidopsis thaliana cDNA homologue to animal poly(ADP-ribose) polymerase. FEBS Lett. 1995;364:103–108. doi: 10.1016/0014-5793(95)00335-7. [DOI] [PubMed] [Google Scholar]
- 32.Liu J, Bramblett D, Zhu Q, Lozano M, Kobayashi R, Ross S R, Dudley J P. The matrix attachment region-binding protein SATB1 participates in negative regulation of tissue-specific gene expression. Mol Cell Biol. 1997;17:5275–5287. doi: 10.1128/mcb.17.9.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Löwenadler B, Jansson B, Paleus S, Holmgren E, Nilsson B, Moks T, Palm G, Josephson S, Philipson L, Uhlen M. A gene fusion system for generating antibodies against short peptides. Gene. 1987;58:87–97. doi: 10.1016/0378-1119(87)90032-1. [DOI] [PubMed] [Google Scholar]
- 34.Luderus M E, de Graaf A, Mattia E, den Blaauwen J L, Grande M A, de Jong L, van Driel R. Binding of matrix attachment regions to lamin B1. Cell. 1992;70:949–959. doi: 10.1016/0092-8674(92)90245-8. [DOI] [PubMed] [Google Scholar]
- 35.Mattern K A, Humbel B M, Muijsers A O, De Jong L, Van Driel R. hnRNP proteins and B23 are the major proteins of the internal nuclear matrix of HeLa S3 cells. J Cell Biochem. 1996;62:275–289. doi: 10.1002/(sici)1097-4644(199608)62:2<275::aid-jcb15>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 36.Mayer-Fligge P, Volz J, Kruger U, Sturm E, Gernandt W, Schafer K P, Przybylski M. Synthesis and structural characterization of human-identical lung surfactant SP-C protein. J Pept Sci. 1998;4:355–363. doi: 10.1002/(sici)1099-1387(199808)4:5<355::aid-psc153>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 37.Mirkovitch J, Gasser S M, Laemmli U K. Relation of chromosome structure and gene expression. Philos Trans R Soc Lond Biol Sci. 1987;317:563–574. doi: 10.1098/rstb.1987.0081. [DOI] [PubMed] [Google Scholar]
- 38.Mirkovitch J, Mirault M E, Laemmli U K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 39.Nayler O, Strätling W, Bourquin J P, Stagljar I, Lindemann L, Jasper H, Hartmann A M, Fackelmayer F O, Ullrich A, Stamm S. SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements. Nucleic Acids Res. 1998;26:3542–3549. doi: 10.1093/nar/26.15.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nickerson J A, Blencowe B J, Penman S. The architectural organization of nuclear metabolism. Int Rev Cytol. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- 41.Phi-Van L, Strätling W H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988;7:655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qian Y Q, Furukubo-Tokunaga K, Resendez-Perez D, Müller M, Gehring W J, Wüthrich K. Nuclear magnetic resonance solution structure of the fushi tarazu homeodomain from Drosophila and comparison with the Antennapedia homeodomain. J Mol Biol. 1994;238:333–345. doi: 10.1006/jmbi.1994.1296. [DOI] [PubMed] [Google Scholar]
- 43.Renz A, Fackelmayer F O. Purification and molecular cloning of the scaffold attachment factor B (SAF-B), a novel human nuclear protein that specifically binds to S/MAR-DNA. Nucleic Acids Res. 1996;24:843–849. doi: 10.1093/nar/24.5.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Romig H, Fackelmayer F O, Renz A, Ramsperger U, Richter A. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 1992;11:3431–3440. doi: 10.1002/j.1460-2075.1992.tb05422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romig H, Ruff J, Fackelmayer F O, Patil M S, Richter A. Characterisation of two intronic nuclear-matrix-attachment regions in the human DNA topoisomerase I gene. Eur J Biochem. 1994;221:411–419. doi: 10.1111/j.1432-1033.1994.tb18753.x. [DOI] [PubMed] [Google Scholar]
- 46.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 47.Steck G, Leuthard P, Burk R R. Detection of basic proteins and low molecular weight peptides in polyacrylamide gels by formaldehyde fixation. Anal Biochem. 1980;107:21–24. doi: 10.1016/0003-2697(80)90486-8. [DOI] [PubMed] [Google Scholar]
- 48.Stief A, Winter D M, Strätling W H, Sippel A E. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature. 1989;341:343–345. doi: 10.1038/341343a0. [DOI] [PubMed] [Google Scholar]
- 49.Uchida K, Miwa M. Poly(ADP-ribose) polymerase: structural conservation among different classes of animals and its implications. Mol Cell Biochem. 1994;138:25–32. doi: 10.1007/BF00928439. [DOI] [PubMed] [Google Scholar]
- 50.van Drunen C M, Sewalt R G, Oosterling R W, Weisbeek P J, Smeekens S C, van Driel R. A bipartite sequence element associated with matrix/scaffold attachment regions. Nucleic Acids Res. 1999;27:2924–2930. doi: 10.1093/nar/27.14.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yanagisawa J, Ando J, Nakayama J, Kohwi Y, Kohwi-Shigematsu T. A matrix attachment region (MAR)-binding activity due to a p114 kilodalton protein is found only in human breast carcinomas and not in normal and benign breast disease tissues. Cancer Res. 1996;56:457–462. [PubMed] [Google Scholar]
- 52.Zhao K, Käs E, Gonzalez E, Laemmli U K. SAR-dependent mobilization of histone H1 by HMG-I/Y in vitro: HMG-I/Y is enriched in H1-depleted chromatin. EMBO J. 1993;12:3237–3247. doi: 10.1002/j.1460-2075.1993.tb05993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuckerkandl E, Villet R. Generation of high specificity of effect through low-specificity binding of proteins to DNA. FEBS Lett. 1988;231:291–298. doi: 10.1016/0014-5793(88)80836-6. [DOI] [PubMed] [Google Scholar]