Abstract
The non-structural proteins (nsPs) of the chikungunya virus (CHIKV) form the virus’s replication complex. They are known to participate in several functions that allow efficient replication of the virus in diverse host systems. One such function is evading the host defense system such as RNA interference (RNAi). Two nsPs of CHIKV, namely, nsP2 and nsP3, were found to suppress the host/vector RNAi machinery and exhibit RNAi suppressor activity. The present study was undertaken to identify interacting partners of CHIKV-nsP3 in Aedes aegypti. We performed pull-down assays with the mass spectrometry approach and showed the interaction of CHIKV-nsP3 with several Aedes proteins. Further co-immunoprecipitation assays revealed that CHIKV-nsP3 interacts with RM62F, a DEAD-box containing RNA known to play roles in multiple gene regulatory processes such as alternative splicing, RNA release, and also is a component of Ago2-RISC complex.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13337-021-00734-y.
Keywords: Chikungunya virus, Aedes aegypti, nsP3, RNA interference, Arboviruses
Introduction
Chikungunya virus (CHIKV), a positive-sense single-stranded RNA virus, is a member of the alphavirus family [38] and is responsible for causing outbreaks to human populations globally [31]. The CHIKV genome is ~11.8 kb which contains 3′ polyadenylation and 5′ capping. It consists of two open reading frames encoding three structural proteins (capsid, envelope glycoproteins E1 and E2), two small cleavage peptides (E3 and 6 K/TF), and four non-structural proteins (nsP1, nsP2, nsP3, and nsP4). Structural proteins are involved in facilitating the entry and fusion of viral particles to the membrane of hosts [38], and the nsPs are engaged in the various steps of virus replication [38]. These proteins exhibit multiple functions and enzymatic activities, e.g., methyltransferase (nsP1), protease and helicase (nsP2), ADP-ribosylhydrolase (nsP3), and RNA-dependent RNA polymerase (nsP4). They have been shown to interact actively with many host proteins to execute their functions ranging from viral RNA capping, proteolysis, replication complex formation, and RNA replication [6] to evade the host immune system [3, 28, 41].
Viral proteins interact with host proteins to help in viral replication and restricting host immune pathways. Among the immune pathways, RNA interference (RNAi) serves as the primary antiviral defense mechanism in insects and is a phenomenon of post-transcriptional gene silencing in a sequence-specific manner [2, 13]. Upon viral infection, Dicer (DCR), a host RNase III endonuclease, recognizes viral dsRNA and processes the dsRNA into siRNA or viRNA [11]. These viRNAs/siRNAs interact with a highly specialized complex called the RNA interference silencing complex (RISC), which ultimately results in the viral genome's degradation [30, 39]. Genome-wide screening of RNAi factors in insects revealed that Argonaute, Dicer, Drosha/Pasha, Aubergine, R2D2, and Loquacious form a core RNAi component while several other proteins, i.e., DEAD-box family of RNA helicases, chromatin factors, and proteins associated with cell cycle, ABC transporter family, metabolic factors and translational machinery components comprise auxiliary components of RNAi complex [16]. These proteins act in a complementary manner with each other to mount an antiviral response against the invading virus. To counteract this response, viral proteins act as viral suppressors of RNAi (VSRs) to create conditions favorable for viral replication by targeting components of the immune pathways [30, 32].
Previous research from our laboratory has identified CHIKV-nsP2 and nsP3 to possess VSR activity [24]. The present study is undertaken to identify potential interacting partners of CHIKV-nsP3 that could be part of the Aedes aegypti (Ae. aegypti) RNAi machinery. Our preliminary interaction study revealed an RNAi factor, RM62F, as one of the interacting partners of nsP3.
Materials and methods
Cell lines and viruses
Aag2 cell line was a kind gift from Dr. Alain Kohl (University of Glasgow, Scotland, UK). Cells were propagated in Leibovitz’s L-15 medium (Cat no. AL011S, HiMedia, India) supplemented with 10% fetal bovine serum (FBS), glutamine, tryptose phosphate broth, and penicillin/streptomycin at 28 °C and 5% CO2. The Vero cell line (ATCC-CCL-81) and C6/36 cells were maintained in Dulbecco’s Modified Eagle Media (DMEM) (Cat no: AL007A, HiMedia, India) supplemented with 10% FBS, penicillin/streptomycin at 37 °C and 5% CO2. CHIKV (Accession no. JF950631.1) was isolated and purified from a patient serum collected during an outbreak in 2010 [34] and propagated in both C6/36 and Vero cell lines in alternate cycles. Virus purified from Vero cells was used for Aag2 infection.
Virus infection
Aag2 cells were grown in 6-well plates in complete L-15 media. Cells were infected with CHIKV strain at multiplicity of infection (MOI) 0.1, 1, and 10. The virus was diluted in sera and antibiotic-free L-15 media, and cells were incubated at 28 °C. After 2 h post-infection (hpi), cells were washed with phosphate-buffered saline (PBS) (pH 7.4) and grown in complete L-15 media. The cells and media were collected at different time points, and cell pellets were washed once with PBS and either resuspended in TRIzol for RNA isolation or lysis buffer for western blot.
Plaque assay
CHIKV was quantified with plaque assay using a previously described protocol [35]. Briefly, Vero cells were seeded in 96-well plate in DMEM with 10% FBS. Supernatant media collected of MOI 0.1, 1, and MOI 10 at different time points from CHIKV infected Aag2 cells was added at different dilutions and incubated for 1 h. After the incubation, viruses containing media were removed from the wells and covered with 1% carboxymethylcellulose (CMC) in sera-free media. Cells were incubated for 48 h at 37 °C. Post washing with PBS, cells were fixed with 4% formaldehyde and stained with 0.25% crystal violet in 30% methanol. Virus titer in infected cells was calculated by plaque-forming units (pfu) = (No. of plaques)/(Dilution × volume of the virus).
Quantitative real-time PCR
Expression profiles of RNAi factors and CHIKV genes were performed from infected cells from different time points. The sequences of RNAi factors were retrieved from ImmunoDB, and the CHIKV-nsP3 sequence was taken from CHIKV/IND/DEL/01 strain (Accession no. JF950631.1) [34]. Primers for qPCR were made using the Prime Quest tool of Integrated DNA Technologies (IDT) with amplicon length ranging from 90–110 nucleotides.
| Primer names | Forward primer | Reverse primer |
|---|---|---|
| Ago-2 | TACCCGGCTCCAACCTATTA | CTGCATTCTCTCGTACTCCTTG |
| Dicer-2 | TTGCTACCGTTGGGAGTTATG | GTGACAGTCGAAGGGTTGAATA |
| RM62F | GACCACAGGTTCGGATTT | CAACGACGCAAGTTGGTAATG |
| RPS17 | GTGAGCGCAGAGACAACTAC | TCCAGCTGCTTCAACATCTC |
| TSN | CACTGTGGTGGAAGTGTTCA | GCCTCACGTGGAGGTTTAAT |
| VIG | GCCGAAAGACGCCAGATTA | CGTTTGTTTCCACTCTGTGATT |
| R2D2 | GACGAATTGCTCTGGACGAA | CGCTGTTGTCTCGTTTCATTTC- |
| FMR1 | AGAGCTGAACAGTGTTGATAGG | CTGGACATTTCCGTTGGATTTG |
| CHIKV-E1 | TACCCATTTATGTGGGGC | GCCTTTGTACACCACGATT |
| CHIKV-nsP3 | AAGGCGCACTGTACTCATATC | TAGGCAGACTTGCTCATTGG |
Total RNA was extracted from cells using Trizol and quantified with Nanodrop. We used 300 ng RNA per reaction. The qPCR was carried out using the Quantitect SYBR green one-step real-time PCR kit (Cat. no. 204243 Qiagen). The qPCR was done with the protocol as: cDNA synthesis 50 °C for 30 min, initial denaturation at 95 °C for 2 min, followed by 40 cycles as denaturation at 95 °C for 15 s, annealing at 56 °C for 30 s and extension at 72 °C for 30 s, data acquisition and followed by melt curve analysis. Expression values were normalized to RPS17 housekeeping control mRNA levels. Relative log-fold expression levels were calculated using 2ˆ(–ΔΔ CT) method [33] and represented as mean ± SD. Statistical analysis of experimental data was conducted using GraphPad Prism (version 6). Two-tailed student’s t-test and Fisher’s Least Significant Difference (LSD) test were performed to check the significance of the data and p values < 0.001 were considered significant.
Generation of plasmid constructs
RM62F sequence was retrieved from ImmunoDB database and primers were designed from N-terminal region of RM62F (10 to 576 amino acids of total 911 amino acids). Total Aedes RNA was isolated from Ae. aegypti mosquitoes with Trizol (Invitrogen). To clone full-length CHIKV-nsP3 protein, the viral RNA was isolated using High Pure Viral RNA Kit (Cat. no. 11858882001, Roche). Both RM62F and CHIKV-nsP3 were amplified using gene-specific primers such as CHIKV-nsP3: (Forward: 5′-GTAATGGGATGTGCACCGTCGTACCGG-3′), (Reverse: 5′-TTACTCGTCGTCCGTGTCTG-3′) and RM62F: (Forward: 5′-ATAGGTACCATGGATCCGGGAGGTTTCCGCCCAC-3′ and Reverse: 5′-TATGGATCCCCGTACGAAGAGCAACGGCC-3′) with the PrimeScript One-Step RT PCR kit (Cat. no. RR055A, Takara, Japan) as per manufacturer instructions and cloned in pET28a respectively and RM62F-pET29a vector which have S-Tag at N-terminal and His-Tag in C-terminal for protein expression.
Protein purification
Full-length CHIKV-nsP3 protein was purified using a previously published protocol [15]. Briefly, CHIKV-nsP3 and RM62F cloned plasmids were transformed into E. coli codon plus cells. CHIKV-nsP3 and RM62F expressing E. coli cells were grown separately at 37 °C and induced overnight with 1 mM IPTG at 18 °C. The cells were collected and lysed in lysis buffer (PBS (pH 7.4), 150 mM NaCl, 5% glycerol, 3 mM β-mercaptoethanol, with 1 mM PMSF and lysozyme). After sonication and centrifugation, the lysates were loaded onto an affinity column (Ni–NTA) and washed with lysis buffer containing 20 mM Imidazole. Proteins were eluted with lysis buffer having 300 mM imidazole. The elution fraction was confirmed with SDS-PAGE and Western blot for proteins. The CHIKV-nsP3 protein was further purified on the Q-sepharose matrix and was washed with PBS lysis buffer (PBS (pH 7.4), 150 mM NaCl, 5% glycerol, 3 mM β-mercaptoethanol), followed by elution with different pH conditions e.g., pH 7.0, pH 6.0, and pH 5.0. Eluted fractions containing purified CHIKV-nsP3 and RM62F protein were confirmed by western blot and concentrated. The proteins were used for further assays after buffer exchange in lysis buffer.
Antibody generation
Antibodies against proteins were raised in-house. Purified CHIKV-nsP3 and RM62F proteins were injected into rabbit and mice with Freund’s complete adjuvant. Booster doses of respective proteins with Freund’s incomplete adjuvant were given in regular intervals and sera were collected. Antibody specificity was checked against purified protein and later against cell lysate to confirm the reactivity.
Pull-down assay
To examine the interaction between CHIKV-nsP3 and A. aegypti host factors, pull-down assay was performed using purified nsp3 protein and Aag2 cell line. Briefly, 100 µl Ni–NTA beads were washed with 1X PBST and incubated with 100 µg purified nsP3 protein for 2 h at 4 °C. Aag2 lysate was prepared in IP lysis buffer, and 1 mg lysate was added to the column, followed by overnight binding at 4 °C. The beads were washed thrice with 20 mM Imidazole, and the bound proteins were eluted in 300 mM Imidazole and processed for mass spectrometry (MS).
Co-immunoprecipitation (Co-IP)
To confirm the interaction of CHIKV-nsp3 with Ae. aegypti RM62F, co-immunoprecipitation of Aag2 lysate was performed with purified nsp3 protein using Pierce co-immunoprecipitation kit (Thermo Scientific, USA) as per the manufacturer’s instructions. Briefly, 100ul Protein A beads were incubated with CHIKV-nsP3 specific antibody, and after wash, these were incubated with purified nsP3 protein (100ug) and allowed to bind overnight to antibody at 4 °C. Beads were washed with IP wash buffer and incubated with Aag2 lysate (1 mg) for 4 h. After washing, elution fractions were taken for Western blot analysis using nsP3 and RM62F specific antibodies.
Sample preparation and mass spectrometry
Pull-down elutes were processed with in-solution trypsin digestion. The elutes were reduced with 10 mM DTT (Dithiothreitol) for an hour at RT and alkylated with 40 mM IAA (iodoacetamide) for an hour. Proteins were digested using trypsin-Gold (Promega) at 1:50 proportion. Samples were incubated in water bath at 37 °C for 16 h. Trypsin digestion reaction was stopped by formic acid (0.1%).
LC–MS/MS analysis was performed Orbitrap Velos Pro MS coupled to Easy n-LC 1000 (Thermo Fisher Scientific). Tryptic digested peptide mixtures were first loaded onto a reverse-phase C-18 pre-column (Acclaim PepMap, 75 μm x 2 cm, 3 μm, 100 Å, Thermo Fisher Scientific) and then separated using an analytical column (Acclaim PepMap, 50 μm x 15 cm, 2 μm, 100 Å) using a gradient of 5% solvent B (0.1% formic acid in 95/5 acetonitrile/water) to 35% solvent B in 35 min. The eluted peptides were injected into the mass spectrometer and MS1 data were acquired using mass range from 350-2000 Da in Full scan mode at 60,000 resolution. The raw data were analyzed using Proteome Discoverer 1.4 with the SEQUEST algorithm against lab generated Ae. aegypti database. The identification of peptides was validated using Percolator at 5% FDR.
Western Blot assay
Protein sample concentration was determined by Bradford's reagent and plotted against bovine serum albumin (BSA) standards of known concentrations. An equal amount of protein samples was loaded in 10% SDS-PAGE gels and run at 100 V till the dye front reaches the other end. Proteins were then transferred onto the nitrocellulose membrane. The membrane was then blocked with PBS (pH 7.4) + 5% BSA for 1 h at RT (room temperature). The membrane was then incubated with primary antibodies (1:2000 dilution) in PBS + 2.5% BSA overnight at 4 °C. The membrane was then washed with PBS + 0.1% Tween-20 for 10 min and repeated thrice. It was then incubated with anti-mouse HRP secondary antibodies (1:5000 dilution) in PBST + 2.5% BSA for 1 h at RT. The membrane was then washed with PBS + 0.1% Tween-20 for 3 X 10 min. The membrane was then visualized in Chemidoc MP (Bio-Rad) after brief incubation with SuperSignal West Pico Chemiluminescent Substrate (Thermo scientific).
Results
CHIKV regulates host RNAi machinery
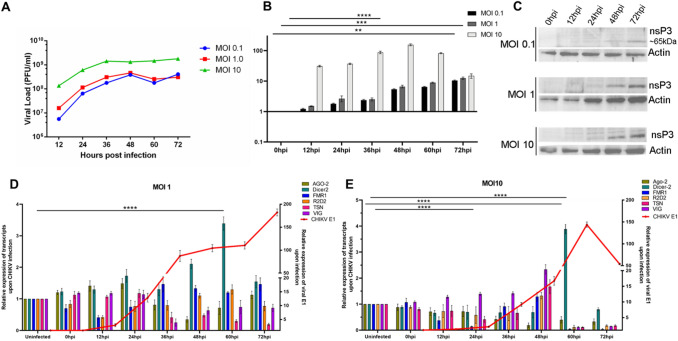
As a first step to understanding the relevance of CHIKV infection on the RNAi machinery, we sought to evaluate the expression of some important RNAi factors at different multiplicity of infections (MOIs) of CHIKV, namely, 0.1, 1 and 10 over several time points post-infection, namely, at 0hpi, 12hpi, 24hpi, 36hpi, 48hpi, 60hpi, and 72hpi using plaque assays. Analysis of viral kinetics revealed that the virus replicates vigorously till 48hpi at 0.1 and 1 MOI and thereafter growth kinetics reduced both at 0.1 and 1 MOI. In case of MOI 10, it replicates vigorously till 36hpi, and thereafter the viral growth reached a plateau (Fig. 1A).
Fig. 1.
Effects of CHIKV infection on RNAi machinery. A Viral l load (Y-axis) during CHIKV infection in Aag2 cells at MOI 0.1, 1, and 10 at different time points. B. Transcripts profiling of nsP3 at MOI 0.1, 1, and 10 at different time points and western blot analysis of nsP3 at MOI 0.1, 1, and 10 upon CHIKV infection. C. nsP3 protein expression profiling at MOI 0.1, 1, and 10 at different time points. D, E. Transcript profiling of Aedes RNAi factors such as Dicer-2, Ago-2, R2D2, TSN, VIG and CHIKV E1 at MOI 1 and 10 of CHIKV at different time points (**p value < 0.007, *** p value < 0.0008 and **** p value < 0.0001)
Next, we sought to profile CHIKV-nsP3 during these time points of infection at different MOIs to understand the expression of the viral transcript and the protein in relation to viral replication in Aag2 cells. Transcript expression profiling of nsP3 gene of CHIKV during infection in Aag2 cells showed that expression level of nsP3 begins to increase from 12hpi and peaked at 72hpi both in 0.1 and 1 MOIs and thereafter expression level decreases whereas in MOI 10 it peaked at 48hpi and then decreases. The expression of nsP3 protein during MOI 0.1, 1, and 10 increased steadily and peaked at 72hpi and was increased as MOI increased (Fig. 1B). It was observed that there were cytopathic effects on the cells post 60 h that might have contributed to the decrease in nsP3 transcript expression post this time point. Taken together, these results suggested that viral replication and viral protein expression were similar at lower MOIs of 0.1 and 1 and there was a more efficient expression of nsP3 at MOI 10 at earlier time points.
Further, we assessed the expression pattern of RNAi core and auxiliary factors such as Ago-2, Dicer-2, FMR1, R2D2, TSN, and VIG during CHIKV infection at MOI 1 and MOI 10. The expression level of these factors unveiled that in case of MOI 1. Dicer 2 was significantly upregulated from 24hpi till 60hpi while TSN, Ago-2 were significantly downregulated at 48hpi. At MOI 10, Dicer 2 was found to be significantly upregulated at 60hpi while Ago-2 was downregulated from 48hpi till 72hpi. TSN, VIG, and R2D2 are significantly upregulated at 48hpi and thereafter showed a sharp decrease in their expression.
Host interacting partners of CHIKV-nsP3
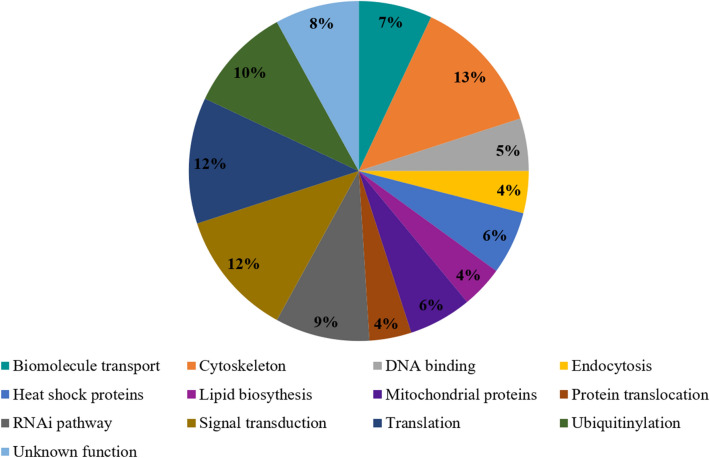
For a detailed understanding of the nsP3-evoked RNAi response, we next analyzed the interaction of nsP3 with the Ae. aegypti proteins and performed pull-down assay using purified nsP3 protein and Aag2 cell lysate. The IDs of nsP3 specific interacting proteins were fetched from ImmunoDB database. The host proteins that interacted with nsP3 were analyzed (Supplementary information 1), and pathway analysis of these host proteins revealed that 40% of the interacting host proteins were involved in cellular translation, ubiquitinylation, RNAi, and cell signaling pathways. Other pathways included heat shock proteins, translocation, DNA binding, endocytosis, biomolecule transport, lipid biosynthesis, protein translocation, mitochondrial proteins, and some unknown functions (Fig. 2).
Fig. 2.
Major pathways of Aedes aegypti proteins interacting with CHIKV-nsP3 protein. Representation of major pathways of host proteins that interact with CHIKV- nsP3 protein. These includes cytoskeleton, singal transduction, and translation
Further analysis with emphasis on the RNAi pathway revealed three proteins, namely 60S ribosomal protein L23 (RPL23) (AAEL013097), DEAD Box helicase RM62F (AAEL008738), and a hypothetical protein (AAEL012416), to be interacting with CHIKV-nsP3 (Supplementary information 1). Of these three RNAi pathway proteins, the DEAD Box helicase was of considerable interest owing to the role DEAD/DExH box helicases such as DDX1, DHX9, DDX5, and DDX18 play in viral replication and are known to interact with viral non-structural proteins [20, 25, 29, 43]. Thus, RM62F was taken further in the present study to evaluate its interaction with CHIKV-nsP3.
CHIKV-nsP3 interacts with RM62F
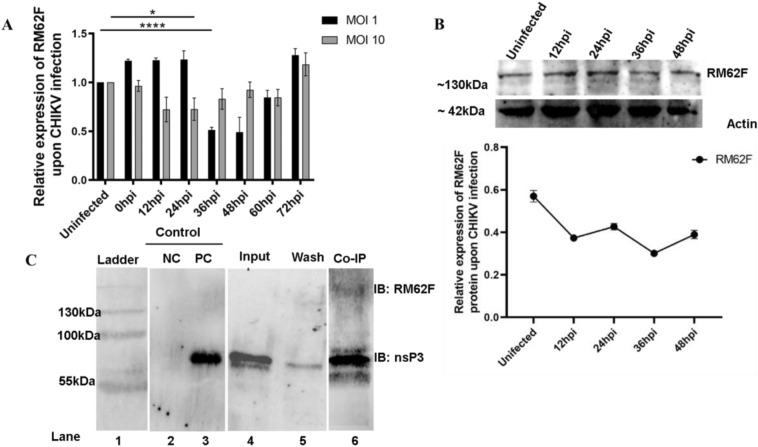
To understand the interaction of RM62F with CHIKV-nsP3, we first sought to evaluate the expression of RM62F transcript during CHIKV infection in Aedes cells at MOI 1 and 10 over 72 hpi at 12 h intervals (Fig. 3A). At the transcript level, at MOI 1, RM62F was overexpressed at the initial time points of infection till 24 hpi and then reduced at 36 hpi and then increased till 72 hpi. In the case of MOI 10, the expression reduced till 36 hpi and then increased till 72 hpi. In the case of protein expression, it was found that the protein level was reduced till 36 hpi as the infection proceeds and resumed slightly at 48 hpi (Fig. 3B). The profiling results at the transcript level and at protein revealed that RM62F was getting temporally affected during CHIKV infection in Aedes cells.
Fig. 3.
Interaction of Aedes aegypti host factor RM62F with CHIKV-nsP3 protein. A. Expression profiling of RM62F transcript upon CHIKV post-infection in Aag2 cells. 0 hpi samples along with different infection time points like 12 h, 24 h, 36 h, 48 h, 60, and 72 hpi at MOI 10 were checked with the antibody against RM62F, and RPS17 as an internal control. Relative comparison of RM62F expression levels at different infection time points. B Expression profiling of RM62F protein upon CHIKV post-infection in Aag2 cells. Control (Uninfected) samples along with different infection time points like 12 h, 24 h, 36 h, and 48 hpi at MOI 10 were checked with the antibody against RM62F, and actin (as a loading control). C. Co-IP of Aag2 lysate with recombinant nsP3 using anti-CHIKV nsP3 antibody. Immunoblotting was done with anti-CHIKV-nsP3 antibody and anti RM62F antibody. Lane1: pre-stained Protein ladder, lane 2: negative control (NC) represent elution of uninfected Aag2 lysate incubated with anti nsP3 antibody immobilized beads, and then probing with nsP3 specific antibody lane 3: positive control (PC) represent elution of recombinant nsP3 with anti-nsP3 antibody immobilized beads and probing with nsP3 specific antibody, lane 4: Pure CHIKV-nsP3 protein, probed with nsP3 specific antibody, lane 5: wash fraction after CHIKV-nsP3 incubation and lane 6: Co-IP of Aag2 lysate with recombinant nsP3 which is bound to anti-nsP3 antibody immobilized beads and blotted wit nsP3 and RM62F specific antibodies
As the next step, we sought to validate the pulldown assay by performing a co-IP assay using antibodies against CHIKV-nsP3 in uninfected Aag2 cells. Purified protein of CHIKV-nsP3 incubated with nsP3 antibodies immobilized onto beads served as postitive control while Aag2 lysate incubated with beads only served as negative control in the immunoprecipitation assay. Post incubation of the lysate with CHIKV-nsP3 protein, the sample assessed for the protein confirmed specificity of CHIKV-nsP3 as shown in lane 4 of Fig. 3c. Co-IP assay of nsP3 using the CHIKV-nsP3 specific antibodies in Aag2 lysate and immunoblotting using CHIKV-nsP3 and RM62F specific antibodies revealed direct interaction between these two proteins (Fig. 3C).
Discussion
CHIKV replication is known to hijack the host machinery to regulate its own replication [37]. Owing to its ability to replicate in diverse organisms such as humans and the mosquito, the virus has evolved to manipulate both organisms to ensure its survival during adverse conditions. The nature of infection of the virus in both the host and the vector is hugely disparate; the virus multiplies exponentially in the human host cells that triggers a rapid launch of the host immune system, thereby resulting in faster virus clearance whereas the virus replication is much slower in the insect cells. The virus can survive in the insect cells throughout the life of the mosquito [9, 12]. The virus employs multiple strategies for its survival in both the host and the insect, depending on the host's kind of defense and the vector mounts on the virus. In the case of mosquitoes, RNAi is an important defense mechanism employed to counter viral infections [8, 22]. Several studies have shown arboviruses specifically activate the RNAi pathway in mosquitoes in addition to other innate immune systems such as the TOLL pathway, IMD pathway, and the JAK-STAT pathway [7, 10, 36]. Earlier reports have shown that in the case of arboviruses and CHIKV, RNAi was amongst the immune pathways to be activated upon virus infection [5, 26]. In the present study, transcript profiling of RNAi factors such as Ago-2, Dicer-2, FMR, TSN, and VIG upon the condition with different MOIs of the virus revealed that at lower MOIs, there was distinct regulation of RNAi factors. Some of the factors showing downregulation while others showed upregulation, whereas, in higher MOI, most of the RNAi factors were downregulated except for Dicer. Further in-depth studies to understand the possible role of Dicer-mediated cleavage of dsRNA during CHIKV infection are necessary as observed in other arboviruses.
CHIKV-nsP3 is a vital protein owing to its multiple functions in CHIKV replication in the host and vector [18]. In the present study, CHIKV-nsP3 was interacting with numerous proteins of various pathways that include translation, cell growth, signal transduction, and biomolecule transport. Such multiple interactions have been possible due to three domains in the protein, namely, macrodomain, Alphavirus unique domain (AUD), and hypervariable domain (HVD). Except for the macrodomain (which has ADP ribosyl hydrolase activity), other domains don’t have any enzymatic activity; however, all the domains are essential, and mutation of amino acids in these regions affects viral growth and viral replication [1, 14, 28].
Owing to our specific interest in evaluating those host proteins interacting with CHIKV-nsP3 that were involved in innate immune pathway, especially RNAi, we identified three proteins namely, a RPL23 (ribosomal protein large subunit 23), RM62F, a DEAD Box helicase and a putative protein. Ribosomal proteins are known to interact with nsP3 and are part of replication complex that is central for viral replication more in terms of RNA binding and processing [6]. The same holds good for the hypothetical protein that was a homolog of Drosophila Pumilio protein that is known to be a RNA binding protein involved in regulation mRNAs [40]. RM62F, on the other hand, is a DExH box helicase and DEAD/DExH helicases are known to be involved in RNA metabolisms such as transcription, translation, and small RNA generation [4, 17, 23, 25]. Further, studies have shown that RM62F is a member of the RNAi pathway in other insects such as Drosophila [44], Aedes, Anopheles [27] and its closest homologs are known to play a role in multiple processes such as alternative splicing, RNA transport, ribosome biogenesis, [42], micro RNA processing [29], RNAi mediated silencing [19] and antiviral RNAi [21, 44]. The existing knowledge regarding the role of this protein in host-virus interactions prompted us to choose RM62F for further evaluation with respect to its interaction with CHIKV-nsP3. The present work has served to establish that this protein interacts with CHIKV-nsP3; more comprehensive analyses may provide a functional relevance of this interaction.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Dr. Alain Kohl for providing the Aag2 cell line.
Funding
Ramesh kumar and Priyanshu srivastva were supported with Ph.D. fellowship by DBT and UGC respectively. This work was supported by the financial Grant from the Department of Biotechnology (BT/PR14725/AGR/36/672/2010). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Declarations
Conflicts of interest
The authors declare no conflict of interest.
Ethics approval
Scientific and ethical approval to carry out this study was obtained from the ICGEB-Institutional Animal Ethics Committee (ICGEB-IAEC). The IAEC approval number is ICGEB/IAEC/2017/01/VBD-3.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Abraham R, Hauer D, McPherson RL, Utt A, Kirby IT, Cohen MS, Merits A, Leung AKL, Griffin DE. ADP-ribosyl-binding and hydrolase activities of the alphavirus nsP3 macrodomain are critical for initiation of virus replication. Proc Natl Acad Sci U S A. 2018;115(44):E10457–E10466. doi: 10.1073/pnas.1812130115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agrawal N, Dasaradhi PV, Mohmmed A, Malhotra P, Bhatnagar RK, Mukherjee SK. RNA interference: biology, mechanism, and applications. Microbiol Mol Biol Rev. 2003;67(4):657–685. doi: 10.1128/mmbr.67.4.657-685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akhrymuk I, Kulemzin SV, Frolova EI. Evasion of the innate immune response: the Old World alphavirus nsP2 protein induces rapid degradation of Rpb1, a catalytic subunit of RNA polymerase II. J Virol. 2012;86(13):7180–7191. doi: 10.1128/JVI.00541-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ambrus AM, Frolov MV. The diverse roles of RNA helicases in RNAi. Cell Cycle. 2009;8(21):3500–3505. doi: 10.4161/cc.8.21.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair CD. Mosquito RNAi is the major innate immune pathway controlling arbovirus infection and transmission. Future Microbiol. 2011;6(3):265–277. doi: 10.2217/fmb.11.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bourai M, Lucas-Hourani M, Gad HH, Drosten C, Jacob Y, Tafforeau L, Cassonnet P, Jones LM, Judith D, Couderc T, Lecuit M, Andre P, Kummerer BM, Lotteau V, Despres P, Tangy F, Vidalain PO. Mapping of Chikungunya virus interactions with host proteins identified nsP2 as a highly connected viral component. J Virol. 2012;86(6):3121–3134. doi: 10.1128/JVI.06390-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carissimo G, Pondeville E, McFarlane M, Dietrich I, Mitri C, Bischoff E, Antoniewski C, Bourgouin C, Failloux AB, Kohl A, Vernick KD. Antiviral immunity of Anopheles gambiae is highly compartmentalized, with distinct roles for RNA interference and gut microbiota. Proc Natl Acad Sci USA. 2015;112(2):E176–185. doi: 10.1073/pnas.1412984112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng G, Liu Y, Wang P, Xiao X. Mosquito defense strategies against viral infection. Trends Parasitol. 2016;32(3):177–186. doi: 10.1016/j.pt.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coffey LL, Vignuzzi M. Host alternation of chikungunya virus increases fitness while restricting population diversity and adaptability to novel selective pressures. J Virol. 2011;85(2):1025–1035. doi: 10.1128/JVI.01918-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dietrich I, Jansen S, Fall G, Lorenzen S, Rudolf M, Huber K, Heitmann A, Schicht S, Ndiaye EH, Watson M, Castelli I, Brennan B, Elliott RM, Diallo M, Sall AA, Failloux AB, Schnettler E, Kohl A, Becker SC. RNA interference restricts rift valley fever virus in multiple insect systems. mSphere. 2017 doi: 10.1128/mSphere.00090-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130(3):413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrester NL, Coffey LL, Weaver SC. Arboviral bottlenecks and challenges to maintaining diversity and fitness during mosquito transmission. Viruses. 2014;6(10):3991–4004. doi: 10.3390/v6103991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gammon DB, Mello CC. RNA interference-mediated antiviral defense in insects. Curr Opin Insect Sci. 2015;8:111–120. doi: 10.1016/j.cois.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao Y, Goonawardane N, Ward J, Tuplin A, Harris M. Multiple roles of the non-structural protein 3 (nsP3) alphavirus unique domain (AUD) during Chikungunya virus genome replication and transcription. PLoS Pathog. 2019;15(1):e1007239. doi: 10.1371/journal.ppat.1007239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George A, Amrutha MS, Srivastava P, Sai VVR, Sunil S, Srinivasan R. Label-free detection of chikungunya non-structural protein 3 using electrochemical impedance spectroscopy. J Electrochem Soc. 2019;166(14):B1356–B1363. doi: 10.1149/2.1081914jes. [DOI] [Google Scholar]
- 16.Ghosh S, Kakumani PK, Kumar A, Malhotra P, Mukherjee SK, Bhatnagar RK. Genome wide screening of RNAi factors of Sf21 cells reveal several novel pathway associated proteins. BMC Genomics. 2014;15:775. doi: 10.1186/1471-2164-15-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goh PY, Tan YJ, Lim SP, Tan YH, Lim SG, Fuller-Pace F, Hong W. Cellular RNA helicase p68 relocalization and interaction with the hepatitis C virus (HCV) NS5B protein and the potential role of p68 in HCV RNA replication. J Virol. 2004;78(10):5288–5298. doi: 10.1128/jvi.78.10.5288-5298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotte B, Liu L, McInerney GM. The enigmatic alphavirus non-structural protein 3 (nsP3) revealing its secrets at last. Viruses. 2018 doi: 10.3390/v10030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishizuka A, Siomi MC, Siomi H. A drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16(19):2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin H, Zhou L, Ge X, Zhang H, Zhang R, Wang C, Wang L, Zhang Z, Yang H, Guo X. Cellular DEAD-box RNA helicase 18 (DDX18) promotes the PRRSV replication via interaction with virus nsp2 and nsp10. Virus Res. 2017;238:204–212. doi: 10.1016/j.virusres.2017.05.028. [DOI] [PubMed] [Google Scholar]
- 21.Kemp C, Imler JL. Antiviral immunity in drosophila. Curr Opin Immunol. 2009;21(1):3–9. doi: 10.1016/j.coi.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar A, Srivastava P, Sirisena P, Dubey SK, Kumar R, Shrinet J, Sunil S. Mosquito innate immunity. Insects. 2018 doi: 10.3390/insects9030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C, Ge LL, Li PP, Wang Y, Sun MX, Huang L, Ishag H, Di DD, Shen ZQ, Fan WX, Mao X. The DEAD-box RNA helicase DDX5 acts as a positive regulator of Japanese encephalitis virus replication by binding to viral 3' UTR. Antiviral Res. 2013;100(2):487–499. doi: 10.1016/j.antiviral.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mathur K, Anand A, Dubey SK, Sanan-Mishra N, Bhatnagar RK, Sunil S. Analysis of chikungunya virus proteins reveals that non-structural proteins nsP2 and nsP3 exhibit RNA interference (RNAi) suppressor activity. Sci Rep. 2016;6:38065. doi: 10.1038/srep38065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matkovic R, Bernard E, Fontanel S, Eldin P, Chazal N, Hassan Hersi D, Merits A, Peloponese JM, Jr, Briant L. The host DHX9 DExH-box helicase is recruited to Chikungunya virus replication complexes for optimal genomic RNA translation. J Virol. 2019 doi: 10.1128/JVI.01764-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McFarlane M, Arias-Goeta C, Martin E, O'Hara Z, Lulla A, Mousson L, Rainey SM, Misbah S, Schnettler E, Donald CL, Merits A, Kohl A, Failloux AB. Characterization of Aedes aegypti innate-immune pathways that limit Chikungunya virus replication. PLoS Negl Trop Dis. 2014;8(7):e2994. doi: 10.1371/journal.pntd.0002994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mead EA, Li M, Tu Z, Zhu J. Translational regulation of Anopheles gambiae mRNAs in the midgut during Plasmodium falciparum infection. BMC Genomics. 2012;13:366. doi: 10.1186/1471-2164-13-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meshram CD, Agback P, Shiliaev N, Urakova N, Mobley JA, Agback T, Frolova EI, Frolov I. Multiple host factors interact with the hypervariable domain of chikungunya virus nsP3 and determine viral replication in cell-specific mode. J Virol. 2018 doi: 10.1128/JVI.00838-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moy RH, Cole BS, Yasunaga A, Gold B, Shankarling G, Varble A, Molleston JM, tenOever BR, Lynch KW, Cherry S. Stem-loop recognition by DDX17 facilitates miRNA processing and antiviral defense. Cell. 2014;158(4):764–777. doi: 10.1016/j.cell.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Obbard DJ, Welch JJ, Kim KW, Jiggins FM. Quantifying adaptive evolution in the Drosophila immune system. PLoS Genet. 2009;5(10):e1000698. doi: 10.1371/journal.pgen.1000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pialoux G, Gauzere BA, Jaureguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis. 2007;7(5):319–327. doi: 10.1016/S1473-3099(07)70107-X. [DOI] [PubMed] [Google Scholar]
- 32.Samuel GH, Wiley MR, Badawi A, Adelman ZN, Myles KM. Yellow fever virus capsid protein is a potent suppressor of RNA silencing that binds double-stranded RNA. Proc Natl Acad Sci USA. 2016;113(48):13863–13868. doi: 10.1073/pnas.1600544113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3(6):1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 34.Shrinet J, Jain S, Sharma A, Singh SS, Mathur K, Rana V, Bhatnagar RK, Gupta B, Gaind R, Deb M, Sunil S. Genetic characterization of Chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol J. 2012;9:100. doi: 10.1186/1743-422X-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sirisena P, Kumar A, Sunil S. Evaluation of Aedes aegypti (Diptera: Culicidae) life table attributes upon Chikungunya virus replication reveals impact on egg-laying pathways. J Med Entomol. 2018;55(6):1580–1587. doi: 10.1093/jme/tjy097. [DOI] [PubMed] [Google Scholar]
- 36.Siu RW, Fragkoudis R, Simmonds P, Donald CL, Chase-Topping ME, Barry G, Attarzadeh-Yazdi G, Rodriguez-Andres J, Nash AA, Merits A, Fazakerley JK, Kohl A. Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs. J Virol. 2011;85(6):2907–2917. doi: 10.1128/JVI.02052-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Solignat M, Gay B, Higgs S, Briant L, Devaux C. Replication cycle of chikungunya: a re-emerging arbovirus. Virology. 2009;393(2):183–197. doi: 10.1016/j.virol.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strauss JH, Strauss EG. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58(3):491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat Chem Biol. 2007;3(1):36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 40.Weidmann CA, Goldstrohm AC. Drosophila Pumilio protein contains multiple autonomous repression domains that regulate mRNAs independently of Nanos and brain tumor. Mol Cell Biol. 2012;32(2):527–540. doi: 10.1128/MCB.06052-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong KZ, Chu JJH. The interplay of viral and host factors in chikungunya virus infection: targets for antiviral strategies. Viruses. 2018 doi: 10.3390/v10060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xing Z, Ma WK, Tran EJ. The DDX5/Dbp2 subfamily of DEAD-box RNA helicases. Wiley interdiscip Rev RNA. 2019;10(2):e1519. doi: 10.1002/wrna.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu L, Khadijah S, Fang S, Wang L, Tay FP, Liu DX. The cellular RNA helicase DDX1 interacts with coronavirus nonstructural protein 14 and enhances viral replication. J Virol. 2010;84(17):8571–8583. doi: 10.1128/JVI.00392-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster. Cell Microbiol. 2006;8(5):880–889. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.