Abstract
African swine fever virus (ASFV) belongs to the family of Asfarviridae to the genus Asfivirus. ASF virus causes hemorrhage illness with a high mortality rate and hence, commercial loss in the swine community. The ASFV has been categorized by variation in codon usage that is caused by high mutation rates and natural selection. The evolution is caused mainly due to the mutation pressure and regulating the protein gene expression. Based on publicly accessible nucleotide sequences of the ASFV and its host (pig & tick), codon usage bias analysis was performed since an approved effective vaccination is not available to date, it is very important to analyze the codon usage bias of the p30, p54, and p72 proteins of ASFV to produce an effective and efficient vaccine to control the disease. Even though the codon usage bias analyses have been evaluated earlier, the evaluation of the codon usage pattern specific to p30, p54, and p72 of ASFV is inadequate. In all the protein-coding sequences, nucleotide base and codons terminating with base T were most frequent and the mean effective number of codons (Nc) was high, indicating the presence of codon usage bias. The GC contents and dinucleotide frequencies also indicated the codon usage bias of the ASFV pig and tick. The Nc plot, parity plot, neutrality plot analysis, revealed natural selection, as well as mutation pressure, were the major constraints in altering the codon bias of ASF virus. codon usage bias analysis was performed with no substantial differences in codon usage of the ASFV in pig and tick.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13337-021-00719-x.
Keywords: African swine fever virus, p30, p54, p72, Codon usage, Pig, Tick
Introduction
African swine fever is a transboundary transmittable disease found infecting animals, causing high-level mortal rates in the swine industry and it is presently reflected as a severe constraint to the swine industry and food safety worldwide [20]. African swine fever virus (ASFV) which causes African swine fever is the only identified arboviral DNA virus. It is found that ASFV is an endemic disease in various regions affecting globally. ASFV results in very high illness and high death rates in swine and also has severe consequences affecting the production of domestic swine globally [20, 22]. This endemic disease is found infecting wild and domestic pigs that in turn affects the losses in trade, production, and eradication plans. ASFV disease is also a World Organization for Animal Health (OIE) listed viral disease. As of now, there is no approved vaccine in use for ASF [2, 22].
African swine fever virus is a large and enclosed virus with a linear genome of 189 kb in length enclosing 180 and more genes that belongs to the family of Asfarviridae and causes hemorrhages in domestic pigs and boars [2, 18]. ASFV is a hereditarily double-stranded complex DNA virus that holds a chain of genes utilizes for viral infection, evades the host’s immune, and alters the processing of cells [29]. The virus is found infecting the phagocytes and replication is found in the cytoplasmic site. The p30, p54, and p72 are important components of viral particles that play roles in attachment, entrance, and reproduction [9]. p30 is an ASFV encoded by CP204L gene 30-kDa phosphoprotein that is produced, membrane-localized, and released into the culture medium shortly after infection. Phosphorylation, glycosylation, and membrane attachment sites are predicted by sequence analysis of the p30 open reading frame [1]. The p30 is one of the most immunogenic structural proteins in the ASFV virion, and its production during the early stages of infection makes it a suitable target for ASFV infection diagnostic assays [21].
p54 is one of the most essential ASFV proteins, p54 encoded by the gene E183L is a very important ASFV antigenic structural protein with a relative molecular weight of 25 kDa [9]. According to several studies, and it plays a vital role in virus morphology and viral infection. Anti-p54 antibodies were discovered to prevent ASFV from attaching to susceptible cells, implying that it plays a function in virus invasion [7]. Similarly, the p54 gene is required for viral survival and envelop precursor recruitment to assembly sites [23, 24].
P72 is the major structural antigenic protein of the ASF virus with a relative molecular weight of 73.2 kDa, which is a very decisive protein encoded by the gene B646L [1]. The p72 protein is extremely antigenic and immunogenic, which serves as the foremost component of viral icosahedrons and a very important in developing the capsid for the virus which expresses virulence in the later stages of infection [17]. Beyond that, it has been demonstrated that the p72 protein is a virulence determinant factor and contributes to the adsorption of the virus to the host cell [13, 14].
The viruses have been categorized by variations in codon usage that is caused by high mutation rates and natural selection. The evolution is caused mainly due to the mutation pressure and regulating the protein gene expression [31]. The amino acids can be coded by more than one codon except for methionine and tryptophan, due to the redundancy in the genetic code that is termed as synonymous codon usage. Conversely, the usage of the codons that code for an amino acidare not in an orderly manner and these codon's usage is most frequent, this is termed as codon usage bias [16]. The factors that affect the codon usage bias are the length of the gene, natural selection, mutation pressure, and the structure of the virus. The hosts and virus can also influence the usage of codons that might be an impact for the virus existence from the factors such as immunity, aptness, host resistance, and evolution. The usage of synonymous triplet codons is non-random and the deviance from the equal usage of synonymous codons is mainly due to the mutation pressure and natural selection [34].
Since, to date there are no approved vaccines for the ASFV, analyzing the patterns of codon usage in the virus might provide information that emphasizes understanding the gene regulation, gene expression, molecular evolution, and in the field of vaccinology to design drugs that require high-level protein expression to induce resistance to the virus. Taking into view the consequences of the three structural viral proteins p30, p54, and p72 the high virulence of ASFV, it is imperative to know about the pattern of codon usage in the p30, p54, and p72 protein and its molecular evolution. An extensive analysis of the codon usage pattern and the factors that influence the evolution of ASFV have been evaluated in this study.
Materials and methods
Data elucidation and alignment
The ASFV coding sequences of the CP204L, E183L and, B646L genes that encode for p30, p54 and, p72 proteins respectively, were fetched from the GenBank database, NCBI (https://www.ncbi.nlm.nih.gov/nucleotide/). The ASFV coding sequences of Sus Scrofa (pig) and Ornithodorus spp (tick) for all three proteins were downloaded in FASTA format and were used for further analysis. The multiple alignment and editing of sequences were performed using the MEGA-X software [12].
Overall nucleotide content and composition analysis of codons
To study the pattern of codon usage the following descriptive analysis was carried out:
1. The overall frequency of nucleotide bases Adenine, Cytosine, Guanine, and Thymine was obtained from the Seqinr library of R software.
2. The frequency of nucleotide bases at third codon sites A3, C3, G3, and T3 was obtained from the MEGA-X software.
3. The composition of G + C contents GC, GC content at first codon site GC1, GC content at second codon site GC2, and GC content at third codon site GC3 and the average of GC contents at first and second codon sites GC12 were calculated with Seqinr library of R software [26].
Variation in the dinucleotide frequency that affects the codon usage
The relative abundance frequency of dinucleotides is also a factor that affects the codon usage bias. The 16 dinucleotides may affect both the natural selection and mutation pressure [3, 5, 11, 27, 30]. The dinucleotide abundance frequency is the ratio- observed to the expected frequencies. The frequencies (Pab) greater than 1.23 and lesser than 0.78 were indicated as overrepresented and underrepresented frequencies, respectively. Furthermore, dinucleotide abundant frequencies with values: Pab ≥ 1.50, 1.30 ≤ Pab < 1.50, 1.23 ≤ Pab < 1.30, 1.20 ≤ Pab < 1.23 were regarded as extremely overrepresented, very overrepresented, significantly overrepresented and marginally overrepresented respectively whereas dinucleotide abundant frequencies with values Pab ≤ 0.50, 0.50 ≤ Pab < 0.70, 0.70 ≤ Pab < 0.78, 0.78 ≤ Pab < 0.81 indicated very underrepresented, significantly underrepresented, and marginally underrepresented respectively [11].
The measure of usage of preferred synonymous codons encoding an amino acid with relative synonymous codon usage (RSCU)
The codon usage pattern is promptly reflected by the relative synonymous codon usage (RSCU), it has been a standardized method used to estimate the bias amongst the genes and within the genes that varies with their sizes and composition of codons that encodes a respective amino acid. The observed frequency observations to the expected frequency observation of a specific amino acid on every codon are termed as RSCU. Commonly, the stronger bias in the codon usage is specified with the RSCU estimates. The RSCU estimates with 1, lesser than 1, greater than 1 are being reflected as no bias, negative bias, and positive bias respectively. Also, the RSCU estimates lesser than 0.6 and greater than 1.6 are categorized as underestimated and overestimated respectively. The RSCU estimates were obtained with the Seqinr library of R software. [3, 5, 11, 27, 30, 31, 34]
Estimating the measure of codon usage bias and degree of absolute synonymous codon bias with the effective number of codons (Nc)
The effective number of codons (Nc) is estimated for each coding sequence that is the most efficient evaluator of the entire bias of the synonymous codons. The Nc estimates generally vary between 20 and 60. If the Nc value is closer to 20, then it indicates that amino acid is encoded by only one synonymous codon, if the Nc estimate is nearly 60 then it specifies that particular amino acid is encoded by all synonymous codons equally. There is no bias in codon usage if Nc estimates are high i.e. closer to 60 conversely, there is a high risk of bias if Nc estimates are low i.e. closer to 20 [32]. The Nc values were obtained with the library coRdon in R software [6].
Furthermore, to decide the major aspects influencing the bias in the codon usage an Nc plot was generated with the ggplot2 library of R software. The Nc plot is obtained when Nc estimated values are plotted against GC3 values. The Nc values would lie on or surrounding the standard Nc curve when the codon bias is influenced by GC3 estimated values. Conversely, the mutation pressure or natural selection pressure accords the change in codon usage when the obtained values lie far-off beneath the curve [3, 5, 11, 27, 30].
To measure the compositional bases bias in purines and pyrimidines usage with Chargaff’s second parity rule (PR2)
Chargaff’s second parity rule defines that the percentage of base composition adenine, cytosine, guanine, and thymine should be equal (%A = %T = %G = %C) and specifically adenine to be equal to thymine (A = T), guanine to be equal to cytosine (G = T). The ratio between the base compositions i.e. A = T and G = C should be 1:1, then it is concluded that there is no deviance between selection pressure and mutation pressure. The PR2 bias is plotted between the A-T bias and G-C bias at the third codon site. The relation between the nucleotide bases pyrimidine (T and C) at the third codon position and purine (A and G) at the third codon position, the plot was generated by calculating G3/(G3 + C3) which was used as ordinate and A3/(A3 + T3) which was taken as abscissa in the PR2 plot and both coordinates meet at 0.5 where (A = T and G = C) [30]. The PR2 bias plot was generated with library ggplot2 in R software with
The PR2 estimates degree of deviation indicates that the bias might be due to natural selection, mutation pressure, or both. If the PR2 estimates are found evenly plotted, then the bias is entirely due to mutation pressure [3, 5, 10, 11, 15, 27, 28, 30].
To measure the role of evolution by natural selection and mutational pressure with the neutrality plot analysis:
One of the factors that strongly shows the variance in the composition is GC3, though other parameters might also be responsible for the variance in the GC3 contents such as mutation and translational selection on the usage of synonymous codons. The neutrality plot analysis is a statistical method implemented to understand how the two variables correlate with each other. The neutrality plot was tested with GC12 as ordinate against GC3 as abscissa to obtain the correlation between them. It is described that if the value of slope was zero then bias is due to selective restraint conversely if the slope is equal to one then it indicates bias due to neutrality restraint [3, 5, 10, 11, 15, 27, 30]. The neutrality plot was generated with R software.
Results
Data elucidation and Alignment of ASFV
The ASFV coding sequences of proteins- p30, p54 and, p72 encoded by CP204L, E183L and, B646L genes respectively were fetched from the GenBank database. The number of p30, p54 and, p72 ASFV coding sequences of pig were 113, 102, 59 and, tick were 7, 8, 8 respectively. The sequences with homogeneity greater than 99% were excluded from the study. All the protein-coding sequences of ASFV were aligned with the MUSCLE algorithm and sequences were edited utilizing the MEGA-X software.
Overall nucleotide content and composition analysis of ASFV codons
To insight into how the nucleotide bases influence the codon usage pattern of ASFV proteins- p30, p54 and, p72 in pig and tick were considered for the codon usage bias analysis. The nucleotide bases composition of ASFV for all three genes have been calculated which are as follows:
The percentage mean of Sus scrofa (pig):
(1) A (Adenine), C (Cytosine), G (Guanine), and T (Thymine) of the three proteins was estimated as p30- 35.35, 17.05, 20.25, 2.36. p54- 29.32, 25.63, 20.98, 24.07 and p72-28.48, 25.23, 19.81, 26.47 respectively.
(2) Nucleotide bases at third codon position A3, C3, G3 and, T3 were estimated to be p30 -21.4, 25.58, 78.72, 34.28, p54- 31.74, 13.91,21.61, 32.71 and p72- 21.40, 25.58, 18.72, 34.28 respectively.
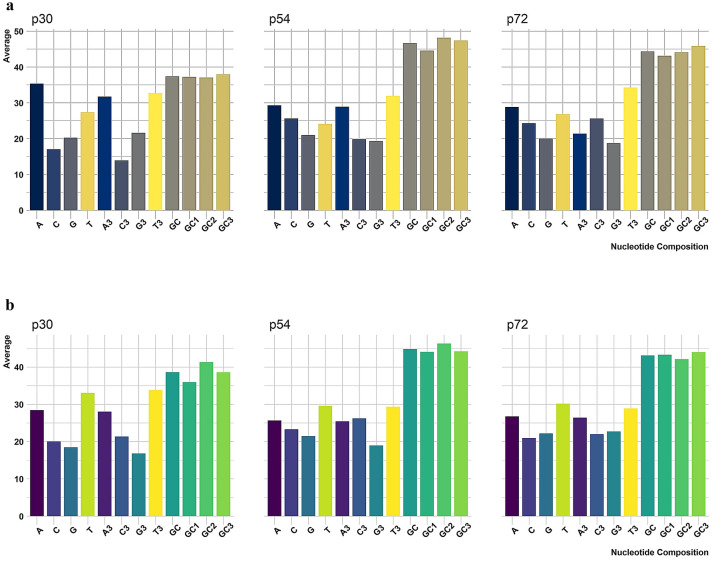
(3) The GC, GC1, GC2, GC3 contents of the entire coding sequences were found to be p30- 37.40, 37.25, 37.04, 37.92, p54- 46.67, 44.59, 48.16, 47.38 and p72 -44.35, 43.11, 44.10, 45.86 and 43.60 respectively. (Table.1) (Fig. 1a).
Table 1.
Composition of Nucleotides in p30, p54, and p72 of the ASFV (Pig and Tick) sequences
| Components | ASFV (Pig) | ASFV (Tick) | ||||
|---|---|---|---|---|---|---|
| p30 | p54 | p72 | p30 | p54 | p72 | |
| A | 35.35 | 29.32 | 28.84 | 28.44 | 25.66 | 26.75 |
| C | 17.05 | 25.63 | 24.31 | 20.03 | 23.31 | 20.94 |
| G | 20.25 | 20.98 | 20.00 | 18.46 | 21.45 | 22.16 |
| T | 27.36 | 24.07 | 26.82 | 33.07 | 29.58 | 30.16 |
| T3 | 31.74 | 28.93 | 21.4 | 33.82 | 29.36 | 28.88 |
| C3 | 13.91 | 19.85 | 25.58 | 21.35 | 26.24 | 21.99 |
| A3 | 21.61 | 19.31 | 18.72 | 28.03 | 25.45 | 26.43 |
| G3 | 32.71 | 31.89 | 34.28 | 16.80 | 18.95 | 22.70 |
| GC | 37.40 | 46.67 | 44.35 | 38.60 | 44.86 | 43.13 |
| GC1 | 37.25 | 44.59 | 43.11 | 35.94 | 44.07 | 43.32 |
| GC2 | 37.04 | 48.16 | 44.10 | 41.30 | 46.32 | 42.09 |
| GC3 | 37.92 | 47.38 | 45.86 | 38.58 | 44.20 | 43.97 |
| Nc | 53.44 | 53.63 | 53.88 | 50.84 | 48.96 | 53.86 |
A, C, G, T represent base nucleotide. A3, C3, G3, T3 represent base nucleotides at the third codon position
GC (overall G + C contents), GC1 (G + C contents at 1st codon position), GC2 (G + C contents at 2nd codon position),
GC3 (G + C contents at 3rd codon position), Nc- the effective number of codons
Fig.1.
a&b Overall Nucleotide composition frequencies (%) in p30, p54, and p72 of the ASFV (Pig and Tick) sequences
The percentage mean of Ornithodorus spp (tick):
(1) A (Adenine), C (Cytosine), G (Guanine), and T (Thymine) of the three proteins was estimated as p30- 28.43, 20.03, 18.46, 33.07. p54- 25.64, 23.30, 2.44, 29.57 and p72- 26.74, 20.93, 22.15, 30.15 respectively.
(2) Nucleotide bases at third codon position A3, C3, G3 and, T3 were estimated to be p30 – 28.02, 21.34, 16.80, 33.82, p54- 25.44, 26.24, 18.95, 29.35 and p72- 26.42, 21.95, 22.70, 28.88 respectively.
(3) The GC, GC1, GC2, GC3 contents of the entire coding sequences were found to be p30- 38.59, 35.94, 41.30, 38.58, p54- 44.85, 44.06, 46.31, 44.19 and p72 – 43.12, 43.31, 42.08, and 43.97 respectively. (Table.1) (Fig. 1b).
Variation in the dinucleotides frequency that affects the codon usage of ASFV
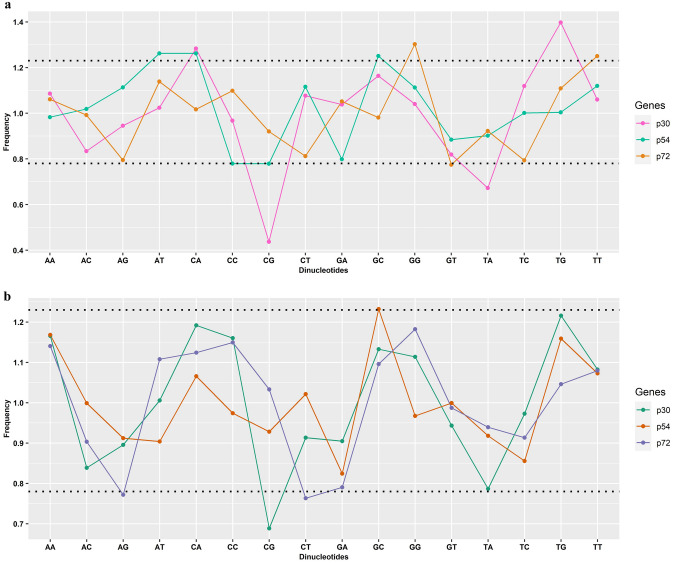
The variations in the dinucleotides frequencies affect the codon usage bias of the many organisms. It has been suggested that dinucleotides bias can affect overall codon usage bias in several organisms. To evaluate the differences in the frequencies of the 16 dinucleotides from the coding sequences of ASFV p30, p54 and, p72 protein with the odds ratio bias the relative abundance frequencies were calculated and obtained. The dinucleotides frequencies were not as expected frequencies, they showed variations in the frequencies of dinucleotides. It was also found that 80% of frequencies among the 16 dinucleotides were found among normal range between 0.78 and 1.25 (underrepresented to overrepresented).
The abundant dinucleotides analysis of ASFV (pig).
p30- indicated that CA and TG were overrepresented whereas, CG and TA were underrepresented dinucleotides respectively. p54- indicated that CA, AT and GC were overrepresented whereas, CG and CC were underrepresented dinucleotides respectively. p72- indicated that GG and TT were overrepresented whereas, GT was underrepresented dinucleotides respectively. (Fig. 2a).
Fig.2.
a&b The overall dinucleotide abundance frequencies in the p30, p54, and p72 of the ASFV (Pig and Tick) sequences and the dotted line represents overrepresentation (> 1.23) and underrepresentation (< 0.78)
The abundant dinucleotides analysis of ASFV (tick).
p30- no overrepresented dinucleotides whereas, CG was underrepresented dinucleotides respectively. p54- indicated that GC was overrepresented whereas, no underrepresented dinucleotides respectively. p72- no overrepresented dinucleotides whereas, CT and AG were underrepresented dinucleotides respectively. (Fig. 2b).
A measure of usage of preferred synonymous codons encoding an amino acid with relative synonymous codon usage (RSCU) of ASFV
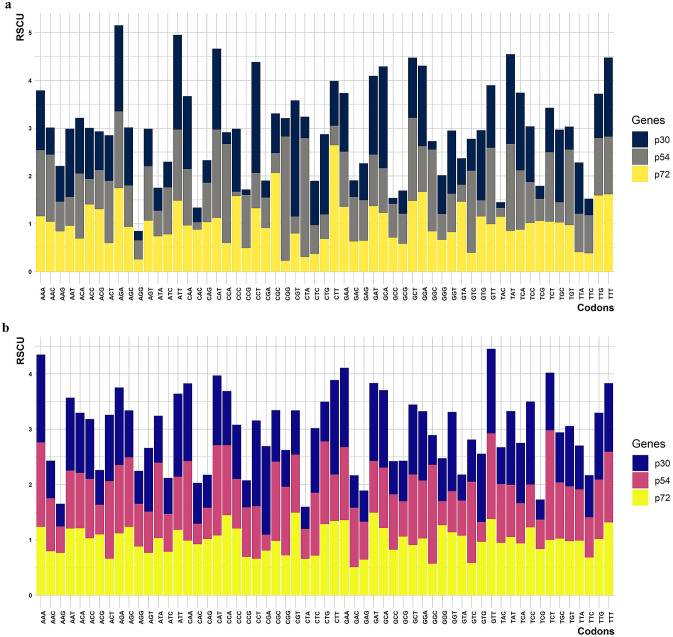
The RSCU values of the coding sequences of ASFV were assessed to estimate the synonymous codons that encode a particular amino acid. Among the 64 codons only 59 codons were used to estimate the RSCU values, the other 5 codons were excluded because ATG and TGG codes only for a single amino acid, and TAG, TAA, TGA stop codons that don’t code for any amino acid.
The estimated RSCU values of ASFV (pig):
p30- Among the 59 codons, 27 codons were positively biased and 32 codons were negatively biased. 11 codons were overrepresented and 20 codons were underrepresented.
p54- 26 codons were positively biased and 33 codons were negatively biased. 9 codons were overrepresented and 17 codons were underrepresented.
p72- 26 codons were positively biased, 32 codons were negatively biased and, 1 codon had no bias. 5 codons were overrepresented and 9 codons were underrepresented. (Table.2) (Fig. 3a).
Table.2.
Relative synonymous codons usage of each amino acid in p30, p54, and p72 of the ASFV (Pig and Tick) sequences
| Codon | ASFV (Pig) | ASFV (Tick) | ||||
|---|---|---|---|---|---|---|
| p30 | p54 | p72 | p30 | p54 | p72 | |
| GCA | 2.13 | 0.933 | 1.229 | 1.4 | 1.091 | 1.214 |
| GCC | 0.124 | 0.703 | 0.711 | 0.6 | 1 | 0.821 |
| GCG | 0.488 | 0.626 | 0.581 | 0.733 | 0.636 | 1.06 |
| GCT | 1.258 | 1.737 | 1.479 | 1.267 | 1.273 | 0.906 |
| TGC | 1.52 | 0.421 | 1.028 | 0.906 | 1.013 | 1.023 |
| TGT | 0.48 | 1.579 | 0.972 | 1.094 | 0.987 | 0.977 |
| GAC | 0.352 | 0.926 | 0.63 | 0.588 | 1.071 | 0.508 |
| GAT | 1.648 | 1.074 | 1.37 | 1.412 | 0.929 | 1.492 |
| GAA | 1.227 | 1.155 | 1.353 | 1.436 | 1.316 | 1.357 |
| GAG | 0.773 | 0.845 | 0.647 | 0.564 | 0.684 | 0.643 |
| TTC | 0.345 | 0.796 | 0.383 | 0.76 | 0.725 | 0.683 |
| TTT | 1.655 | 1.204 | 1.617 | 1.24 | 1.275 | 1.317 |
| GGA | 1.686 | 0.952 | 1.668 | 1.254 | 1.046 | 1.025 |
| GGC | 0.179 | 1.708 | 0.841 | 0.537 | 1.785 | 0.57 |
| GGG | 0.813 | 0.539 | 0.664 | 0.776 | 0.431 | 1.266 |
| GGT | 1.322 | 0.801 | 0.827 | 1.433 | 0.738 | 1.139 |
| CAC | 0.307 | 0.155 | 0.876 | 0.739 | 0.37 | 0.921 |
| CAT | 1.693 | 1.845 | 1.124 | 1.261 | 1.63 | 1.079 |
| ATA | 0.48 | 0.534 | 0.738 | 0.849 | 1.361 | 1.032 |
| ATC | 0.537 | 0.981 | 0.778 | 0.651 | 0.681 | 0.786 |
| ATT | 1.983 | 1.485 | 1.484 | 1.5 | 0.958 | 1.182 |
| AAA | 1.252 | 1.381 | 1.157 | 1.588 | 1.525 | 1.235 |
| AAG | 0.748 | 0.619 | 0.843 | 0.412 | 0.475 | 0.765 |
| CTA | 0.452 | 2.485 | 0.304 | 0.404 | 0.537 | 0.657 |
| CTC | 0.927 | 0.601 | 0.369 | 1.169 | 1.134 | 0.715 |
| CTG | 1.682 | 0.507 | 0.685 | 0.719 | 1.493 | 1.285 |
| CTT | 0.938 | 0.407 | 2.642 | 1.708 | 0.836 | 1.343 |
| TTA | 1.073 | 0.799 | 0.409 | 0.793 | 0.925 | 0.986 |
| TTG | 0.927 | 1.201 | 1.591 | 1.207 | 1.075 | 1.014 |
| AAC | 0.569 | 1.401 | 1.043 | 0.68 | 0.955 | 0.795 |
| AAT | 1.431 | 0.599 | 0.957 | 1.32 | 1.045 | 1.205 |
| CCA | 0.247 | 2.067 | 0.599 | 0.98 | 1.263 | 1.446 |
| CCC | 1.317 | 0.089 | 1.58 | 0.98 | 0.895 | 1.205 |
| CCG | 0.115 | 1.108 | 0.494 | 0.49 | 0.895 | 0.691 |
| CCT | 2.321 | 0.735 | 1.327 | 1.551 | 0.947 | 0.659 |
| CAA | 1.527 | 1.178 | 0.965 | 1.404 | 1.436 | 0.986 |
| CAG | 0.473 | 0.822 | 1.035 | 0.596 | 0.564 | 1.014 |
| AGA | 1.802 | 1.603 | 1.746 | 1.404 | 1.231 | 1.119 |
| AGG | 0.198 | 0.397 | 0.254 | 0.596 | 0.769 | 0.881 |
| CGA | 0.356 | 0.635 | 0.912 | 1.6 | 0.286 | 0.807 |
| CGC | 0.83 | 0.413 | 2.064 | 0.933 | 1.429 | 0.982 |
| CGG | 0.385 | 2.598 | 0.226 | 0.667 | 1.238 | 0.719 |
| CGT | 2.43 | 0.354 | 0.797 | 0.8 | 1.048 | 1.491 |
| AGC | 1.215 | 0.865 | 0.934 | 0.85 | 1.255 | 1.235 |
| AGT | 0.785 | 1.135 | 1.066 | 1.15 | 0.745 | 0.765 |
| TCA | 1.628 | 1.239 | 0.876 | 1.091 | 0.723 | 0.937 |
| TCC | 1.166 | 0.853 | 1.018 | 1.5 | 0.771 | 1.228 |
| TCG | 0.273 | 0.459 | 1.061 | 0.364 | 0.53 | 0.835 |
| TCT | 0.932 | 1.45 | 1.044 | 1.045 | 1.976 | 1 |
| ACA | 1.165 | 1.358 | 0.692 | 1.086 | 1 | 1.21 |
| ACC | 1.068 | 0.531 | 1.404 | 1.086 | 1.067 | 1.031 |
| ACG | 0.812 | 0.813 | 1.307 | 0.629 | 0.533 | 1.1 |
| ACT | 0.955 | 1.298 | 0.597 | 1.2 | 1.4 | 0.66 |
| GTA | 0.555 | 0.353 | 1.462 | 0.471 | 0.634 | 1.075 |
| GTC | 0.668 | 1.714 | 0.394 | 0.765 | 1.465 | 0.583 |
| GTG | 1.468 | 0.337 | 1.152 | 1.235 | 0.356 | 0.965 |
| GTT | 1.309 | 1.596 | 0.993 | 1.529 | 1.545 | 1.377 |
| TAC | 0.121 | 0.182 | 1.149 | 0.667 | 1.059 | 0.948 |
| TAT | 1.879 | 1.818 | 0.851 | 1.333 | 0.941 | 1.052 |
Bold values indicate the overrepresented codons
The Average of all the 59 synonymous codons in p30, p54, and p72 of the ASFV (Pig and Tick)
Fig.3.
a&b Overall frequencies of Relative synonymous codon usage (RSCU) in p30, p54, and p72 of the ASFV (Pig and Tick) sequences
The estimated RSCU values of ASFV (tick):
p30- Among the 59 codons, 29 codons were positively biased and 30 codons were negatively biased. 1 codon was overrepresented and 10 codons were underrepresented.
p54- 27 codons were positively biased, 30 codons were negatively biased and 2 codons had no bias. 2 codons were overrepresented and 7 codons were underrepresented.
p72- 30 codons were positively biased, 28 codons were negatively biased and, 1 codon had no bias. None of the codons were overrepresented and 2 codons were underrepresented.(Table.2) (Fig. 3b)
Estimating the measure of codon usage bias and degree of absolute synonymous codon bias with effective number of codons (Nc) of ASFV.
The degree of codon usage bias of ASFV was quantified with the effective number of codons (Nc).
The Nc values for ASFV (pig).
p30- The Nc was ranging from 52.02 to 57.64 and had an average value of 53.46 P54- The Nc was ranging from 51.60 to 55.15 and had an average value of 53.63 p72- The Nc was ranging from 53.25 to 54.84 and had an average value of 53.88.
The Nc values for ASFV (tick).
p30- The Nc was ranging from 46.28 to 56.24 and had an average value of 50.83 P54- The Nc was ranging from 47.24 to 53.99 and had an average value of 48.96 p72- The Nc was ranging from 52.43 to 56.40 and had an average value of 53.86. (Table.1).
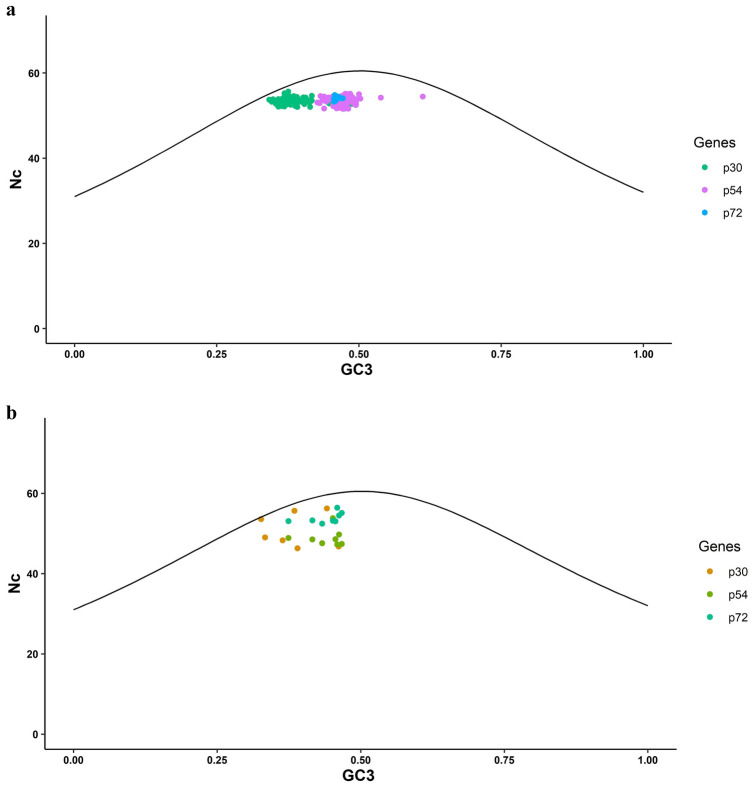
Further, to understand the synonymous codon usage pattern the Nc plot was generated, wherein the Nc values at ordinate were plotted against GC3 at abscissa along with the standard curve to obtain the role of mutational pressure in shaping the codon usage bias of ASFV. The Nc plot revealed that the Nc scores were found lying under the standard Nc curve, signifying that the codon usage bias is mainly affected by natural selection over mutation pressure (Fig. 4a, b).
Fig.4.
a&b The Nc plot representing the relationship of Nc against GC3. Each point represents the p30, p54, and p72 sequences of ASFV (Pig and Tick)
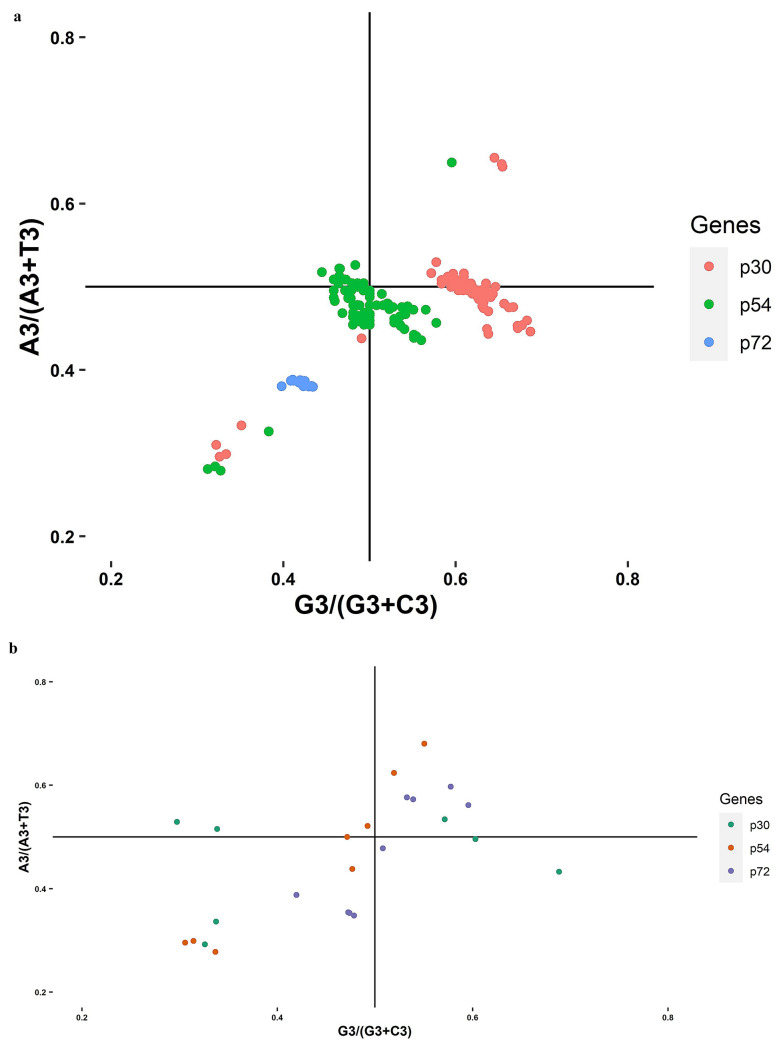
To measure the compositional bases bias in purines and pyrimidines usage with Chargaff’s second parity rule (PR2) of ASFV
The PR2 bias plot insists that the selection and mutation pressure have no bias if all the values lie on the center of the plot i.e. 0.5 where, both the ordinate and abscissa meet. The PR2 bias plot represents the relationship between pyrimidines (C & T) and purine (A & G bias at third codon position.p72p72.
PR2 analysis of ASFV (pig):
p30- The average value of AT and GC bias was 0.49 and 0.61, respectively. P54- The average value of AT and GC bias was 0.47 and 0.49, respectively. p72- The average value of AT and GC bias was 0.38 and 0.42, respectively. Here, in all three genes, pyrimidines are preferred over purines.
PR2 analysis of ASFV (tick):
p30- The average value of AT and GC bias was 0.44 and 0.45, respectively. P54- The average value of AT and GC bias was 0.45 and 0.43, respectively. p72- The average value of AT and GC bias was 0.48 and 0.51, respectively. Here, in p30 and p72 pyrimidines are preferred over purines whereas in p54 purines are preferred over pyrimidines.
Also, the plot shows that pressures are the effect in shaping the ASFV pig and tick codon use pattern because all the points lie far from the origin and are moderately biased. (Fig. 5a and Fig. 5b).
Fig.5.
a&b Parity rule 2 plots AT-bias against GC-bias. Each point represents the p30, p54, and p72 sequences of ASFV (Pig and Tick)
To measure the role of evolution by natural selection and mutational pressure with the neutrality plot analysis of ASFV
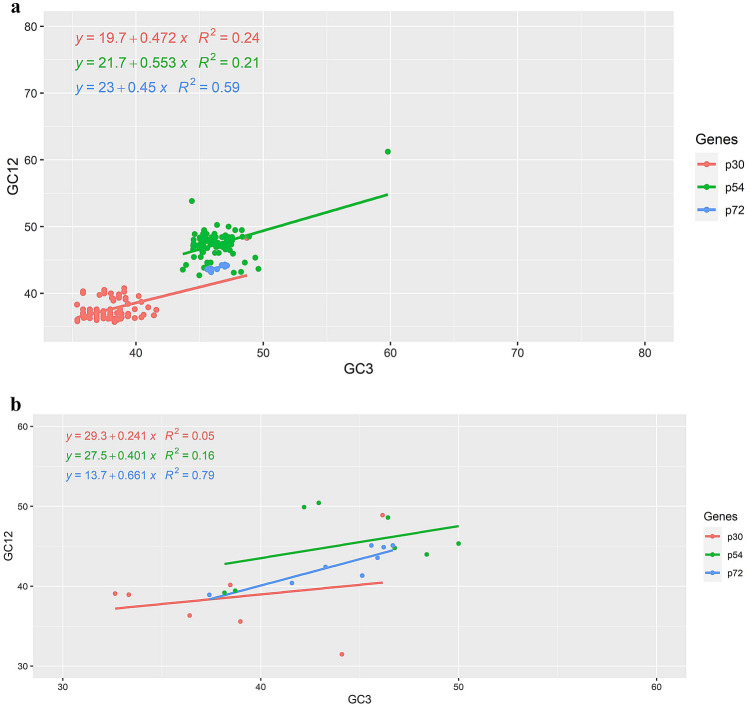
The neutrality plot analysis was represented to evaluate whether the codon usage bias of ASFV is influenced by the GC contents at the third codon position. The neutrality plot was obtained with GC3 against GC12, which resulted in a significant positive correlation. The neutrality plot highlights natural selection's dominance over mutational forces once again. The neutrality plot analysis of ASFV (pig).
p30-The comparative neutrality was 47.2% indicating mutation pressure and a 52.8% comparative constraint indicating natural selection. The obtained results indicated that natural selection is assertive over mutation pressure and has an effect on the codon usage of ASFV.
p54-The comparative neutrality was 55% indicating mutation pressure and a 44.7% comparative constraint indicating natural selection. The obtained results indicated that mutation pressure is assertive over the natural selection and has an effect on the codon usage of ASFV.
p72-The comparative neutrality was 45% indicating mutation pressure and a 55% comparative constraint indicating natural selection. The obtained results indicated that natural selection is assertive over mutation pressure and has an effect on the codon usage of ASFV (Fig. 6a).
Fig.6.
a&b Neutrality plot showing the relationship between %GC12 and %GC3 with the slope line indicating natural selection. The points represent the p30, p54, and p72 sequences of ASFV (Pig and Tick)
The neutrality plot analysis of ASFV (pig).
p30-The comparative neutrality was 24% indicating mutation pressure and a 75.9% comparative constraint indicating natural selection. The obtained results indicated that mutation pressure is assertive over natural selection and has an effect on the codon usage of ASFV.
p54-The comparative neutrality was 40.1% indicating mutation pressure and a 59.9% comparative constraint indicating natural selection. The obtained results indicated that mutation pressure is assertive over natural selection and has an effect on the codon usage of ASFV.
p72-The comparative neutrality was 66.1% indicating mutation pressure and a 33.9% comparative constraint indicating natural selection. The obtained results indicated that natural selection is assertive over mutation pressure and has an effect on the codon usage of ASFV (Fig. 6b).
In both the species only p72 had higher correlation compared to the other two genes and all the three genes were biased indicating both natural selection and mutational pressure had major role n the codon usage pattern of ASFV.
Discussion
The analysis of usage in the codons of various DNA or RNA virus to their host species helps in understanding the genetic features and evolution of many organisms [34]. The present study revealed that there was moderate codon usage bias for the ASFV p30, p54, and p72 proteins. . Even though there are many analyses on the evolution and genetic assortment of ASFV that have already been studied in the previous research, further analysis is needed on codon usage bias of the ASFV specific to p30,p54, and p72 proteins. Eventually, the codon usage pattern of p30, p54, and p72 protein in ASFV hasn’t been analyzed yet. Also, recognizing these codon usage patterns helps in the phylogenetic study and by enhancing the codons that helps in target gene expression. The evolution of codon usage of various genes of a virus evolves at different periods [3]. Moreover, the codon usage pattern is notched with several factors such as the gene length, nucleotide composition, selection, mutation pressure, etc. [4, 25]. Though the codon usage analysis of numerous studies of different viruses was mainly determined by major indices such as RSCU, Nc, neutrality plot, parity rule plot, etc., In this study, we have focused on the three genes of ASFV (pig and tick) to observe variation in the codon usage pattern. The current study analyzed that the codon usage pattern of the ASFV p30, p54, and p72 proteins for two hosts: pig and tick. The obtained results revealed that in all the three genes in ASFV (pig) the frequency of nucleotide A was abundant compared to other nucleotide bases and the frequency of nucleotide at the third codon position, it was observed that both the ASFV had T3 was found abundant compared to others. The GC contents analysis revealed in p30 and p72, the GC3 was found to have a higher frequency and in p54, GC2 had a higher frequency. Similarly, in the ASFV (tick) it was observed that the frequency of nucleotide T was abundant. The frequency of nucleotide at the third codon position, it was observed that T3 was found abundant compared to others. The GC contents analysis revealed that the GC3 in p30 and p54, GC2 had higher frequency and in p72, GC3 was found to have a higher frequency. The dinucleotides frequencies analysis of ASFV in pig and tick also showed variations among the three genes and the relatively large abundance of dinucleotides in the ASFV gene has more metabolic building capacity, resulting in reduced transcription and replication rate. It was observed that all the nucleotide compositions are varying among the genes, indicating they are biased and shape the codon usage pattern of ASFV.
The RSCU analysis showed that A/T ending codons were seen as highly represented in both the ASFV pig and tick. Also, it was seen that the G/C ending codons were mostly underrepresented.
As it is seen that genes are mostly A/T biased, this suggests that selectional pressure resulting in low A/T frequencies is not actively implicated in the codon usage patterns of the ASFV pig and tick but these patterns are primarily regulated by compositional constraints mainly, which indicated that codon bias is a clear indication of dinucleotides bias in the ASFV genes.
The overall codon use bias has been calculated using the Nc values of three genes of the ASFV pig and tick. While observing the average Nc values of these three genes in pig and tick, the ASFV in the pig is lower biased compared to the ASFV in tick. Since the low codon usage bias could be beneficial for effective replication with more codon selection possibilities [8], this study suggests that ASFV’s gene replication and transcription may be facilitated by its low codon bias. Mutational pressure signifies if just GC3 content affects codon usage, this suggests; in such circumstances, the Nc values are somewhat over the projected Nc curve. In this study, it was observed that the Nc values were below the expected Nc curve in all three genes of ASFV pig and tick. The Nc values were distant from the curve, showing that selection pressure had the most important effect over mutational pressure in determining the codon usage pattern of ASFV genes.
After that, we used the neutrality plot to investigate the role of selection pressure. The major forces determining codon usage are thought to be mutational bias when the correlation between GC12 and GC3 is statistically significant and the slope of the regression line is near to 1. Slope approaching 0, on the other hand, imply that selection pressure is the most important factor influencing codon usage patterns. The neutrality plot showed that in the ASFV pig and tick, except for p54 in the ASFV pig and p72 in the ASFV pig, other proteins showed that mutational pressure influences the codon usage bias over the selection pressure.
In the parity analysis, a bias value greater than 0.5 implies that purine is preferred over pyrimidine. As a result, A will be chosen over T, and G will be preferred over C in this case. Except for p30 in ASFV pig, in all other gene sequences, pyrimidines were preferred over purine. The Nc plot, neutrality plot, and parity rule plot analyses showed that selection and mutational pressure were the major factors affecting the codon usage pattern of the ASFV pig and tick.
The data will aid in understanding the rising health risks to pigs as a result of living close to these animals, which may function as viral carriers and, despite being asymptomatic, could be a source of infection. The research will also form a basis for other animals to be evaluated and assessed for their ability to serve as virus hosts.
Conclusion
ASFV is an emerging virus and a major pig health problem in their production. Since there are no approved vaccines for ASFV yet, the development of an efficient vaccination, as well as the discovery of viable medications and the identification of prospective hosts, are all desperately required. These three genes' information could be valuable in understanding various prevention efforts to control the spread of diseases in the pig community. According to the genes studies, there are statistically significant differences in codon usage bias amongst the three genes in ASFV pig and tick. The mutational and selection factors, in addition to compositional considerations, were found to play a substantial role in shaping the ASFV codon usage pattern. Analyzing the codon usage pattern can help with protein and gene expression optimization analysis, it is also useful in the development of alternative viral vectored vaccine options. Similarly, codon usage information can also be used to inhibit viral protein synthesis during replication, in contrast to enhancing the protein expression. This study may also provide information on codon usage for other viruses, and investigations can be conducted for various applications. The codon use bias in ASFV is not high, and it was primarily influenced by natural selection and mutational pressure considering the three genes taken into this study. Consequently, these findings will benefit future ASFV surveillance and research activities, as well as provide useful information into the evolutionary analysis of the ASFV.
Supplementary Information
Below is the link to the electronic supplementary material.
Funding
Funding has been provided by ICAR-NIVEDI (Grant No. 4701317003).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Afonso CL, Alcaraz C, Brun A, Sussman MD, Onisk DV, Escribano JM, Rock DL. Characterization of p30, a highly antigenic membrane and secreted protein of African swine fever virus. Virology. 1992 doi: 10.1016/0042-6822(92)90718-5. [DOI] [PubMed] [Google Scholar]
- 2.Brown VR, Bevins SN. A review of African swine fever and the potential for introduction into the united states and the possibility of subsequent establishment in feral swine and native ticks. Front Vet Sci. 2018 doi: 10.3389/fvets.2018.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y, Li X, Chi X, Wang S, Ma Y, Chen J. Comprehensive analysis of the codon usage patterns in the envelope glycoprotein E2 gene of the classical swine fever virus. PLoS ONE. 2017 doi: 10.1371/journal.pone.0183646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clément Y, Sarah G, Holtz Y, Homa F, Pointet S, Contreras S, Nabholz B, Sabot F, Sauné L, Ardisson M, Bacilieri R, Besnard G, Berger A, Cardi C, De Bellis F, Fouet O, Jourda C, Khadari B, Lanaud C, Leroy T, Pot D, Sauvage C, Scarcelli N, Tregear J, Vigouroux Y, Yahiaoui N, Ruiz M, Santoni S, Labouisse JP, Pham JL, David J, Glémin S. Evolutionary forces affecting synonymous variations in plant genomes. PLoS Genet. 2017 doi: 10.1371/journal.pgen.1006799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comeron JM, Aguadé M. An evaluation of measures of synonymous codon usage bias. J Mol Evol. 1998 doi: 10.1007/pl00006384. [DOI] [PubMed] [Google Scholar]
- 6.Elek A, Kuzman M, Vlahoviček K. coRdon: codon usage analysis and prediction of gene expressivity. Bioconductor 3.8. 2019.
- 7.Gómez-Puertas P, Rodríguez F, Oviedo JM, Brun A, Alonso C, Escribano JM. The African swine fever virus proteins p54 and p30 are involved in two distinct steps of virus attachment and both contribute to the antibody-mediated protective immune response. Virology. 1998 doi: 10.1006/viro.1998.9068. [DOI] [PubMed] [Google Scholar]
- 8.Jenkins GM, Holmes EC. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res. 2003 doi: 10.1016/s0168-1702(02)00309-x. [DOI] [PubMed] [Google Scholar]
- 9.Jia N, Ou Y, Pejsak Z, Zhang Y, Zhang J. Roles of African swine fever virus structural proteins in viral infection. J Vet Res. 2017 doi: 10.1515/jvetres-2017-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karlin S, Mrázek J, Campbell AM. Codon usages in different gene classes of the Escherichia coli genome. Mol Microbiol. 1998 doi: 10.1046/j.1365-2958.1998.01008.x. [DOI] [PubMed] [Google Scholar]
- 11.Khandia R, Singhal S, Kumar U, Ansari A, Tiwari R, Dhama K, Das J, Munjal A, Singh RK. Analysis of Nipah virus codon usage and adaptation to hosts. Front Microbiol. 2019 doi: 10.3389/fmicb.2019.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018 doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leblanc N, Cortey M, Fernandez Pinero J, Gallardo C, Masembe C, Okurut AR, Heath L, van Heerden J, Sánchez-Vizcaino JM, Ståhl K, Belák S. Development of a suspension microarray for the genotyping of African swine fever virus targeting the SNPs in the C-terminal end of the p72 gene region of the genome. Transbound Emerg Dis. 2013 doi: 10.1111/j.1865-1682.2012.01359.x. [DOI] [PubMed] [Google Scholar]
- 14.Liu Q, Ma B, Qian N, Zhang F, Tan X, Lei J, Xiang Y. Structure of the African swine fever virus major capsid protein p72. Cell Res. 2019 doi: 10.1038/s41422-019-0232-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malakar AK, Halder B, Paul P, Deka H, Chakraborty S. Genetic evolution and codon usage analysis of NKX-2 5 gene governing heart development in some mammals. Genomics. 2020 doi: 10.1016/j.ygeno.2019.07.023. [DOI] [PubMed] [Google Scholar]
- 16.Marín A, Bertranpetit J, Oliver JL, Medina JR. Variation in G + C-content and codon choice: differences among synonymous codon groups in vertebrate genes. Nucleic Acids Res. 1989 doi: 10.1093/nar/17.15.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazur-Panasiuk N, Woźniakowski G, Niemczuk K. The first complete genomic sequences of African swine fever virus isolated in Poland. Sci Rep. 2019 doi: 10.1038/s41598-018-36823-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazur-Panasiuk N, Żmudzki J, Woźniakowski G. African swine fever virus - persistence in different environmental conditions and the possibility of its indirect transmission. J Vet Res. 2019 doi: 10.2478/jvetres-2019-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan S, Mou C, Wu H, Chen Z. Phylogenetic and codon usage analysis of atypical porcine pestivirus (APPV) Virulence. 2020 doi: 10.1080/21505594.2020.1790282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Penrith ML, Vosloo W. Review of African swine fever: transmission, spread, and control. J S Afr Vet Assoc. 2009 doi: 10.4102/jsava.v80i2.172. [DOI] [PubMed] [Google Scholar]
- 21.Petrovan V, Yuan F, Li Y, Shang P, Murgia MV, Misra S, Rowland RRR, Fang Y. Development and characterization of monoclonal antibodies against p30 protein of African swine fever virus. Virus Res. 2019 doi: 10.1016/j.virusres.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Rendleman CM, Spinelli FJ. An economic assessment of the costs and benefits of African swine fever prevention. USA: Animal health insight; 1994. [Google Scholar]
- 23.Rodríguez JM, García-Escudero R, Salas ML, Andrés G. African swine fever virus structural protein p54 is essential for the recruitment of envelope precursors to assembly sites. J Virol. 2004 doi: 10.1128/jvi.78.8.4299-4313.2004[CrossRef][PubMed]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez F, Ley V, Gómez-Puertas P, García R, Rodriguez JF, Escribano JM. The structural protein p54 is essential for African swine fever virus viability. Virus Res. 1996 doi: 10.1016/0168-1702(95)01268-0[CrossRef]. [DOI] [PubMed] [Google Scholar]
- 25.Romero H, Zavala A, Musto H, Bernardi G. The influence of translational selection on codon usage in fishes from the family Cyprinidae. Gene. 2003 doi: 10.1016/s0378-1119(03)00701-7. [DOI] [PubMed] [Google Scholar]
- 26.R Development CORE TEAM. A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. R-project. org. 2010.
- 27.Sharp PM, Li WH. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986 doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 28.Sueoka N. Intrastrand parity rules of DNA base composition and usage biases of synonymous codons. J Mol Evol. 1995 doi: 10.1007/BF00163236. [DOI] [PubMed] [Google Scholar]
- 29.Tulman ER, Rock DL. Novel virulence and host range genes of African swine fever virus. Curr Opin Microbiol. 2001 doi: 10.1016/s1369-5274(00)00235-6. [DOI] [PubMed] [Google Scholar]
- 30.Wang L, Xing H, Yuan Y, Wang X, Saeed M, Tao J, Feng W, Zhang G, Song X, Sun X. Genome-wide analysis of codon usage bias in four sequenced cotton species. PLoS ONE. 2018 doi: 10.1371/journal.pone.0194372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong EH, Smith DK, Rabadan R, Peiris M, Poon LL. Codon usage bias and the evolution of influenza a viruses codon usage biases of influenza virus. BMC Evol Biol. 2010 doi: 10.1186/1471-2148-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wright F. The 'effective number of codons' used in a gene. Gene. 1990 doi: 10.1016/0378-1119(90)90491-9. [DOI] [PubMed] [Google Scholar]
- 33.Yu X, Liu J, Li H, Liu B, Zhao B, Ning Z. Comprehensive analysis of synonymous codon usage patterns and influencing factors of porcine epidemic diarrhea virus. Arch Virol. 2021 doi: 10.1007/s00705-020-04857-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou JH, Gao ZL, Sun DJ, Ding YZ, Zhang J, Stipkovits L, Szathmary S, Pejsak Z, Liu YS. A comparative analysis on the synonymous codon usage pattern in viral functional genes and their translational initiation region of ASFV. Virus Genes. 2013 doi: 10.1007/s11262-012-0847-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.