Abstract
Cytochrome bd-type oxygen reductases (cytbd) belong to one of three enzyme superfamilies that catalyze oxygen reduction to water. They are widely distributed in Bacteria and Archaea, but the full extent of their biochemical diversity is unknown. Here we used phylogenomics to identify three families and several subfamilies within the cytbd superfamily. The core architecture shared by all members of the superfamily consists of four transmembrane helices that bind two active site hemes, which are responsible for oxygen reduction. While previously characterized cytochrome bd-type oxygen reductases use quinol as an electron donor to reduce oxygen, sequence analysis shows that only one of the identified families has a conserved quinol binding site. The other families are missing this feature, suggesting that they use an alternative electron donor. Multiple gene duplication events were identified within the superfamily, resulting in significant evolutionary and structural diversity. The CydAA’ cytbd, found exclusively in Archaea, is formed by the co-association of two superfamily paralogs. We heterologously expressed CydAA’ from Caldivirga maquilingensis and demonstrated that it performs oxygen reduction with quinol as an electron donor. Strikingly, CydAA’ is the first isoform of cytbd containing only b-type hemes shown to be active when isolated from membranes, demonstrating that oxygen reductase activity in this superfamily is not dependent on heme d.
Subject terms: Molecular evolution, Metabolism
Introduction
The predominance of oxygen in our atmosphere determines the bioenergetic importance of oxygen as an electron acceptor and the prevalence of aerobic respiratory chains. There are only three enzyme superfamilies capable of acting as terminal respiratory oxygen reductases—heme-copper oxygen reductases, alternative oxidases and cytochrome bd-type oxygen reductases (cytbd) [1]. While enzymes from this superfamily have been characterized from a number of Bacteria, their role in archaeal respiration has not yet been determined. Archaeal aerobic respiratory chains share some similarities with bacterial respiratory chains, however they often differ in their composition of respiratory enzymes and are adapted to use different co-factors. Complexes that are typically involved in bacterial respiration such as succinate-quinone oxidoreductases [2], cytochrome bc1 complexes [3], and heme-copper oxygen reductases have been previously characterized in Archaea [4, 5], while NADH:quinone oxidoreductases (complex I) and alternative complex III are either rare or absent from this domain [6]. The presence of cytbd has been noted in archaeal genomes [7], metagenomes and metaproteomes [8–11] but, no functional member of the cytbd superfamily in Archaea has ever been demonstrated.
Cytochrome bd-type oxygen reductase is a respiratory enzyme that converts oxygen to water using three hemes, unlike the heme-copper oxygen reductases that have two hemes and a copper in the active sites [1]. Purified cytbd accepts electrons from quinols using a low-spin heme b558 and transfers these electrons to a di-heme active site containing two high-spin hemes. In some of the characterized cytochrome bd enzymes, these active site hemes were shown to be heme b595 and heme d, but some other isoforms were shown to contain only hemes b. Those cytochrome bd family members that contain only hemes b are usually referred to as cyanide insensitive oxidases (CIO) or cytochrome bb’–type oxygen reductase, and have been identified in Pseudomonas aeruginosa, Bacillus subtilis and others [12–15]. It is unclear whether the presence of only hemes b is physiologically relevant but it has been suggested that these enzymes are less sensitive to inhibition by cyanide [13]. There is no sequence signature that distinguishes those enzymes in the superfamily that only contain heme b, and no CIO has ever been isolated and characterized.
The canonical cytochrome bd oxygen reductases contain a minimum of two subunits, CydA and CydB, but often have additional “auxiliary” subunits [16–18] such as CydX, a single-transmembrane subunit that is associated with cytochrome bd-I from E. coli that has been implicated in the stability of the enzyme [19]. Cytochrome bd-type oxygen reductases (cytbd) have a high affinity for oxygen [20] and the previously characterized cytbds have been associated with roles in oxygen detoxification, respiratory protection of nitrogenases and as part of sulfide oxidizing respiratory chains [21–25]. Cytochrome bd catalytic turnover generates a proton motive force by translocation of protons using a conserved proton channel from the cytoplasm to the site of oxygen reduction located near the periplasmic side (electrically positive) of the membrane [26]. Yet, cytochrome bd is not as energetically efficient as the heme-copper oxygen reductases that pump protons in addition to translocating “chemical” protons from the cytoplasm to the periplasmic active site [27, 28]. Expression of cytochrome bd has often been associated with microoxic conditions where a high-affinity oxygen reductase would be required [29].
In this work, we used phylogenomics to determine the diversity and distribution of this high-affinity oxygen reductase in Archaea and Bacteria. We determined that there are three distinct families of cytbd—one of which contains the quinol binding characteristics present in the structures of cytbd from Escherichia coli and Geobacillus thermodenitrificans [30–32] and two which do not—and discussed their evolutionary relationships. The distribution of these families even within Archaea involve significant variation and include the two distinct isoforms CydAB and CydAA’, the latter of which appears to have been created by gene duplication. We evaluate the relative distribution of the CydAA’ and CydAB within the domain Archaea and consider the likely role of CydAA’ variants within their ecological context. In addition, we show that the CydAA’ from Caldivirga maquilingensis is a highly active oxygen reductase with unique biochemical and structural characteristics. This combined phylogenomic and experimental approach has significantly expanded our knowledge of the evolutionary and biochemical diversity within the superfamily, which has important implications for the role of the cytbd superfamily in novel respiratory pathways.
Results and discussion
Diversity of cytochrome bd-type oxygen reductases
The molecular structures of cytbd from Escherichia coli and Geobacillus thermodenitrificans have been determined and showed that cytbd typically has two conserved subunits CydA and CydB, along with a third subunit, CydX or CydS which is a single-transmembrane subunit. The cydX or cydS genes are not well conserved or always found along with cydA and cydB in the genome [30–33]. Of the two main subunits, CydA is better conserved in all known cytbd while CydB is very divergent and is hypothesized to have evolved at faster rates than CydA [34]. The CydA subunit is composed of nine transmembrane helices and contains almost all the conserved amino acids known to be important for catalyzing oxygen reduction and proton translocation, including the ligands for three hemes and the residues forming a proton channel [35, 36]. The first four helices of CydA contain all of the amino acids that form the active site. These include the proton channel and ligands to bind the active site heme b595 and heme d (these ligands have only been verified in the isoforms containing hemes b and d, and not the ones containing only hemes b). While H19 is a ligand to heme d (in E. coli cytbd), R9 appears to help stabilize the heme b595 [31]. The other five helices (V–IX) form the quinol binding site in the biochemically characterized bd-type oxygen reductases and include the ligands to heme b558, the point of entry for electrons from quinols [30–32]. With this structural framework in mind, we performed a sequence analysis of cytochrome bd-type oxygen.
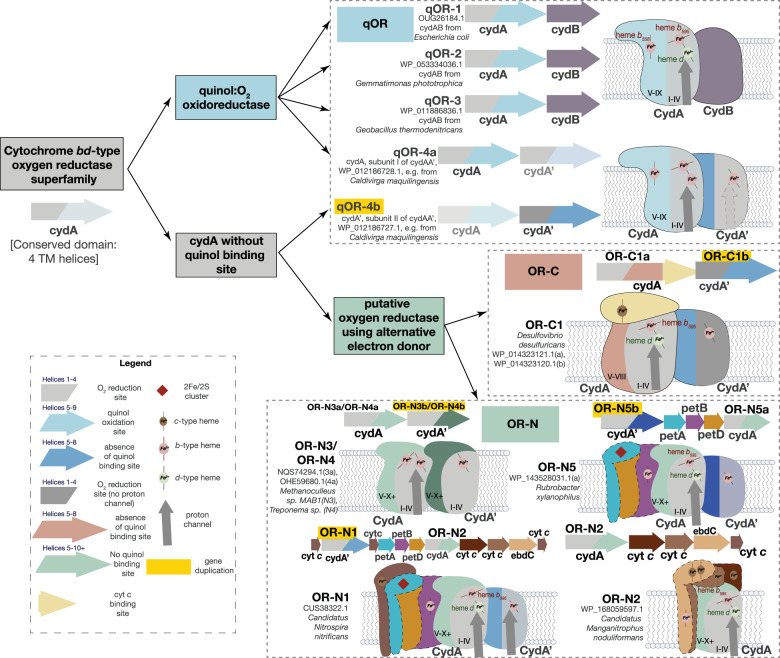
An analysis of 24,706 genomes available in the Genome Taxonomy Database (release89) [37, 38], revealed the presence of 17938 CydA homologs. Of these, 13,018 genomes contained at least one CydA homolog, suggesting that this enzyme family is widely distributed and important (Supplementary Table1). Phylogenomic analysis of CydA homologs revealed 15 clades of CydA that could be distinguished on the basis of unique sequence characteristics. (Supplementary Fig. S1, Supplementary Tables 1–3). Four of these clades contain the features that are considered part of the quinol binding site—for e.g., conserved residues Lys252 and Glu257 (E. coli CydA numbering) while the remaining do not. We inferred that the former four CydA clades were quinol:O2 oxidoreductases and we named them qOR1, qOR2, qOR3 and qOR4a. While CydA of the families qOR1, qOR2 and qOR3 associate with CydB to form CydAB, CydAA’ is formed by the co-association of two distinct CydA clades, qOR4a and qOR4b. qOR4b does not possess quinol binding site characteristics and is likely the result of a gene duplication event. Sequence analysis of CydA sequences from all 15 CydA clades demonstrated that 2 of the remaining clades are missing quinol binding sites and instead contain a number of heme c binding motifs (CxxCH) (Supplementary fig. S1, Supplementary multiple sequence alignments MSA1, MSA2). We named these enzymes OR-C1a and OR-C1b because of the presence of heme c binding motifs. The ‘a’ and ‘b’ attachment to the names signifies that they likely co-associate to form one enzyme OR-C1, just like qOR4a-CydA and qOR4b-CydA’. Eight of the remaining CydA clades were related and named OR-N1, OR-N2, OR-N3a, OR-N3b, OR-N4a, OR-N4b, OR-N5a and OR-N5b. OR-N is named for Nitrospirota because of predominance of these enzymes in that phylum (Supplementary Figs. S1, S2 Supplementary multiple sequence alignments MSA1, MSA2, MSA3). Their close relationship is also supported by the likely structure of the proteins of which they are a part and their genomic context (Fig. 1). We have attempted to develop a nomenclature for the cytochrome bd-type oxygen reductase family that can be easily expanded upon. We designate three large families of cytbd—qOR, OR-C and OR-N—based on their phylogenetic placement, presence/absence of biochemical signatures such as quinol and heme c binding site features, genomic operon context and taxonomic origin. We have designated subfamilies numerically starting from 1 and attached an ‘a’ or ‘b’ subscript if it is likely that two CydA subfamilies co-associate to form one enzyme. Most of the ‘a’-type subfamilies include the proton channel residues E99 and E107 or one of the two (E. coli numbering) while ‘b’-type subfamilies mostly do not. It appears that ‘a’ and ‘b’-type subfamilies are also the result of multiple independent gene duplication events within this superfamily. We will discuss the unusual number of gene duplication events within the cytbd superfamily and the OR-C and OR-N families later in the text and in Supplementary Material but begin with the quinol oxidizing qOR family. Sequences from this family contain all the amino acids that were previously identified as forming the heme ligands, proton channel, oxygen reduction site and quinol binding site [30–32, 36].
Fig. 1. Families within the cytochrome bd-type oxygen reductase superfamily.
The cytochrome-bd type oxygen reductase superfamily is divided into three families—qOR, OR-C and OR-N, primarily defined by the presence of the quinol binding site in the first, the presence of heme c binding site in the second and the abundance of OR-N enzymes in Nitrospirota. The above schematic represents the various subfamilies within each family that are defined by the phylogenetic clustering shown in Supplementary Fig. S1. The operon context and putative complex arrangement of each CydA-containing enzyme is also shown with a reference protein accession number and source microorganism. The potential gene duplication events are highlighted in yellow. A legend is also provided to mark the related conserved domains in the same colors and redox co-factors such as hemes and iron-sulfur clusters.
Evolution of the quinol-oxidizing cytochrome bd-type oxygen reductases (qOR)
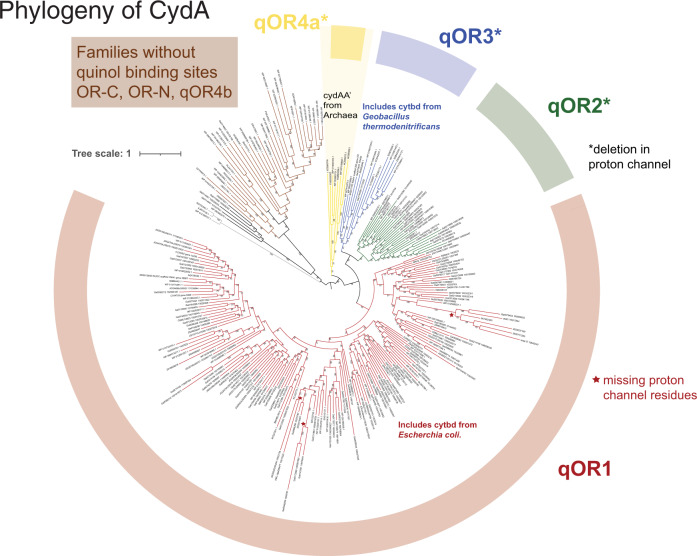
To explore the evolutionary relationship between the qOR1, qOR2, qOR3 and qOR4a subfamilies, which are true orthologs, we generated a maximum likelihood phylogenetic tree using RAxML and IQ-TREE with the OR-C and OR-N family sequences as outgroup (Fig. 2, Supplementary Fig. S3). This topology is also similar to that inferred by the phylogenetic inference software, MrBayes (Supplementary Fig. S4). Sequence features can be identified to distinguish these families and to validate the above identified monophyletic clades as meaningfully distinct; some of which are outlined below while the remaining features are mentioned in Supplementary Table 3. All CydA sequences from the qOR1 subfamily have seven amino acids between the two conserved glutamates (ex. Glu99 and Glu107 such as in Escherichia coli CydA [26, 39]) in the proton channel, while CydA sequences from qOR2, qOR3 and qOR4a typically have six amino acids between the two conserved glutamates (ex. Glu101 and Glu108 in qOR3-subfamily cytbd from Geobacillus thermodenitrificans [30]). This insertion/deletion has been hypothesized to lead to a reversal in the position of hemes from the qOR1-bd in Escherichia coli to the qOR3-bd in Geobacillus thermodenitrificans although further research is required to establish whether the reversal of heme positions is universal (further discussion on the insertion/deletion in the proton channel and the Q-loop is included in Supplementary Material). Sequence features that distinguish the qOR4a-subfamily cytbd are insertions between helices V and VI, as well as insertions in helix VIII (Supplementary alignment MSA1, Supplementary Table 3). Conserved tyrosines (Tyr115 and Tyr117 in Geobacillus thermodenitrificans CydA) are present in qOR2 and qOR3 families but not in the qOR4a-subfamily, which is consistent with the close evolutionary relationship between the qOR2 and qOR3-subfamilies observed in the tree topology. Other conserved sequence features that are unique to each family are listed in Supplementary alignments MSA1 and MSA3 and Supplementary Table 3.
Fig. 2. Phylogeny of quinol-oxidizing cytochrome bd-type oxygen reductases.
Protein sequences of cytbd subunit I, CydA were extracted from a taxonomically diverse set of genomes and metagenomes from IMG, filtered with UCLUST using a percentage identity cut-off of 0.6 and aligned using MUSCLE. The multiple sequence alignment, MSA4 was used to infer a phylogenetic tree using RAxML. The RAxML tree topology was similar to that inferred by IQ-tree. The CydA sequences that do not contain the quinol binding site, from the OR-C and OR-N subfamilies as well as qOR4b, were used as the outgroup. At least four monophyletic clades of typical CydA sequences that contain the O2- and quinol binding site could be defined—qOR1, qOR2, qOR3 and qOR4a. The long branch within the qOR1 clade comprises a number of cytbd that are highly similar to enzymes from this clade but are missing the proton channel. Subunit I of CydAA’ is from the qOR4a-subfamily while subunit II or CydA’ is from the qOR4b-subfamily.
Comparing the CydA phylogenetic tree and the distribution of cytbd across Archaea and Bacteria provides some insight into the relative age of these families. The qOR1 subfamily, which includes the Escherichia coli enzyme, appears to be the most widely distributed with enzymes in over 60 bacterial phyla, (Supplementary Table 4) but it is only sparsely distributed in Archaea. In fact, there are only a very few representatives in Euryarchaeota and Asgardarchaeota (Fig. 3). It is only widely distributed in Halobacterota, whose oxidative metabolism is expected to have evolved relatively late [40] (Fig. 3). This strongly suggests that the qOR1 subfamily is the oldest of the extant families and that cytbd likely originated in Bacteria. While CydA from the qOR2 subfamily is also fairly well-distributed and found in over 20 bacterial phyla, the qOR3-subfamily enzymes are almost exclusive to the Firmicutes and Firmicutes_I phyla with a few enzymes in Archaea. The qOR4a-subfamily enzymes appear to be specific to the Archaea (Supplementary Tables 2, 4). A close evolutionary relationship between the qOR2 and qOR3-subfamilies is suggested by CydA tree topology and identifiable sequence characteristics. However, other trees we inferred (~2 out of 30 trees) have modelled a closer relationship between the qOR2 and qOR1 subfamilies than between qOR3 and qOR1 (Supplementary Fig. S5). This alternative topology might suggest that qOR2 evolved from qOR1 before qOR3, but a majority of our tree topologies suggest that qOR2 and qOR3 are more closely related to each other than they are to qOR1. Therefore, we do not infer anything about the relative ancestry of qOR2 and qOR3. The qOR1 subfamily has seven amino acids between the conserved glutamates in the proton channel, while the qOR4a, qOR2 and qOR3 subfamilies consistently have six amino acids. Lastly, enzymes from the qOR4a, qOR2 and qOR3 subfamilies are almost completely absent from Proteobacteria. This suggests that the qOR2, qOR3 and qOR4a subfamilies diverged from the qOR1 family, before Proteobacteria diverged from other bacterial phyla. While our dataset and phylogenetic analysis is consistent with the above discussion, it must be noted that many lateral gene transfers have been observed within the cytochrome bd-type oxygen reductase [1] that complicate evolutionary analysis. In addition, while our dataset has a large number of species, it is ~3% of the number of bacterial OTUs in the Global Prokaryotic Census [41]. While the phyla estimated to have the greatest global diversity [41] are well represented in the GTDB, our analysis is still limited by the number of available genomes, especially for rare taxa and this represents an additional constraint on the analysis we have presented here.
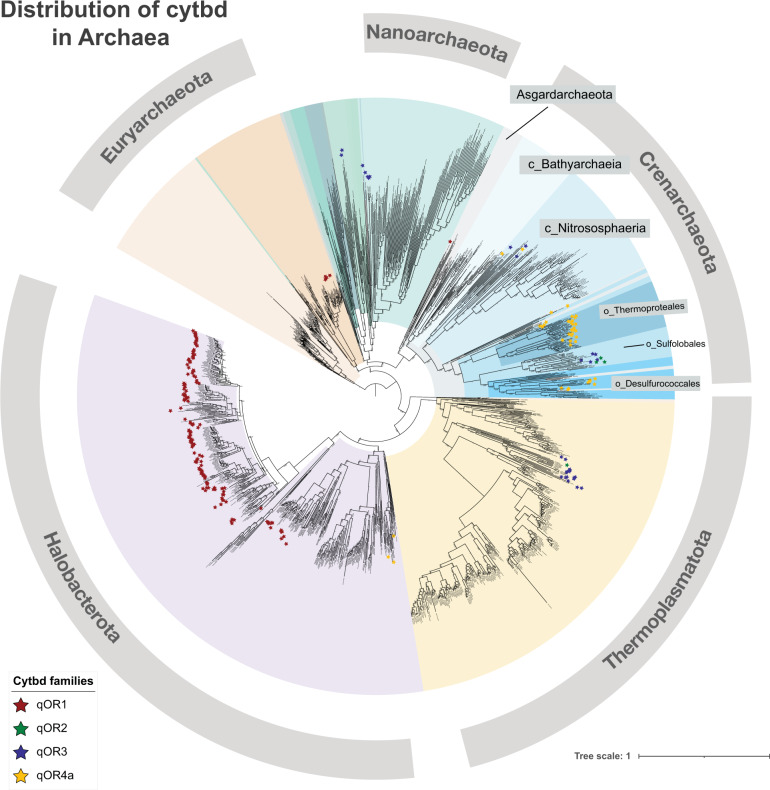
Fig. 3. Distribution of cytochrome bd-type oxygen reductases in Archaea.
Concatenated gene alignments were made from the archaeal genomes in GTDB using Anvi’o. A phylogenetic tree was made from the concatenated gene alignments using FastTree. All CydA sequences were extracted from GTDB genomes using BLAST with an e-value of 1e−1. The sequences were then filtered to remove CydA sequences without characteristics of the quinol binding site and then classified using a Hidden Markov Model (HMM)-based classifier trained to identify the families—qOR1, qOR2, qOR3 and qOR4a. CydA sequences from each family were then mapped back to each species, and visualized along with the species tree on the iTOL server. Most phyla of the domain Archaea were distinguished by color and a few classes of the phylum Crenarchaeota were labelled to emphasize the presence of CydAA’. It is clear that CydAA’ is almost exclusive to the order Thermoproteales and Desulfurococcales.
As mentioned above, most CydA subfamilies are widely distributed within Bacteria and Archaea, but the qOR4a-subfamily is unique in having sequences that belong only to Archaea. In addition, the qOR4a-subfamily is unique in having a completely different subunit II (CydA’), while the qOR1-, qOR2- and qOR3- subfamily members appear to have CydB homologs as their subunit II. CydB is either not homologous to CydA’ or is evolutionarily distant. The unique ancestry of the qOR4a-subfamily enzymes that are specific to Archaea raises a question about its distribution within that domain.
Distribution of cytochrome-bd type oxygen reductases in Archaea
To investigate the distribution of cytbd within Archaea and to contextualize the evolution of CydA within archaeal evolution, we mapped the presence of qOR4a, qOR1, qOR2 and qOR3 subfamilies of CydA onto a phylogenetic tree of all Archaea, using a concatenated gene alignment made from the archaeal genomes in GTDB [37] using Anvi’o [42], (Fig. 3). It is clear from this representation that most of the qOR4a-subfamily or CydAA’ belong to the class Thermoprotei within the phylum Crenarchaeota with a few CydAA’ in Nitrosphaeria, Thermoplasmatota and Archaeoglobi. Within the Thermoprotei, almost all members of the order Thermoproteales and family Acidilobaceae contain CydAA’ (Supplementary Table 5).
To place CydAA’ into an ecological context we looked at their environmental distribution (Supplementary Table 6). Microbes containing CydAA’ are largely found in thermal environments, including anoxic sediment from solfataric fields, hot springs and deep-sea vents, suggesting that CydAA’ might only be utilized by thermophiles, such as organisms from the genera Vulcanisaeta, Caldivirga, Thermofilum, Pyrobaculum and Thermocladium [43–47] (Supplementary Table 6). Within Yellowstone National Park (YNP), a number of these genera are found in hypoxic, sulfur/iron-rich ecosystems, although Pyrobaculum and Thermofilum have also been found in more oxygenated environments. It has been suggested that members of the Thermoproteales that are found in aerobic environments and have a heme-copper oxygen reductase are more likely to be using aerobic respiration as their primary energetic pathway [43]. This is consistent with what we observe in Pyrobaculum. Of the 8 Pyrobaculum genomes in the GTDB database, P. aerophilum, P. oguniense and P. caldifontis have a heme-copper oxygen reductase and are capable of respiring oxygen [48, 49], but are missing CydAA’. The three genomes that have CydAA’, but are missing a heme-copper oxygen reductase (P. neutrophilum, P. islandicum and Pyrobaculum sp001189275) are not capable of aerobic respiration and appear to be strict anaerobes [48]. This is suggestive of an adaptation based on oxygen availability in the environment, i.e., those Pyrobaculum which have heme-copper oxygen reductases respire oxygen and conserve energy using the more energetically efficient oxygen reductase [28], while those that have CydAA’ live in environments with less oxygen and use this high-affinity oxygen reductase for oxygen scavenging [20].
Expression of the cydAA’ genes have been demonstrated in the hot springs and sulfur-rich/iron-rich environments within YNP, using RT-PCR [10] and metatranscriptomics (Table 1 [50]). While the hot springs were typically hypoxic and sulfur-rich, the iron oxide mats had higher oxygen concentration at the surface and had <0.3 μM concentration of O2 within 1 mm. Nitrosphaeria, Acidilobaceae, Thermoproteales and Thermoplasmatota expressed cydAA’ in these environments, however it is not clear whether these microorganisms were exposed to high O2 concentrations. In fact, the Acidilobaceae are expected to be found in the middle and bottom layers of this mat where O2 concentrations are lower [10, 51]. All of the above observations are consistent with the presence of cydAA’ in microaerobic and hypoxic environments. The obvious question that needed to be addressed is whether CydAA’ actually functions as an oxygen reductase. This was accomplished by biochemically characterizing the CydAA’ from C. maquilingensis.
Table 1.
cydAA’ is expressed in many environments. ProteinGene expression is estimated based on read counts in metatranscriptomes.
| Genome_ID | Genome_Name | Locus_Tag | Reads | Best BLAST Hit NCBI | Organism | IMG Study ID | References for IMG study iD |
|---|---|---|---|---|---|---|---|
| 3300003719 | Ferric microbial mat communities from Yellowstone National Park, Wyoming, USA - One Hundred Spring Plain (OSP_B) (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0040881_1063211 | 246 | ESQ23817.1 | Acidilobus sp. OSP8 | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| Ga0040881_1110491, Ga0099831_11874701 |
106 42 |
AMD31390.1 | Acidilobus sp. 7A | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0040881_1111231 | 27 | WP_081246098.1 | Thermoproteus sp. CP80 | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0040881_1134581 | 25 | WP_066793619.1 | Caldivirga sp. MU80 | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0040881_1142191, Ga0040881_1205091 |
2174 12 |
NAY81522.1 | Thaumarchaeota archaeon | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0040881_1202971 | 5 | KUO89797.1 | Caldivirga sp. CIS_19 | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0040881_1228021, Ga0040881_1244931 |
8 5 |
KUO80886.1 | Vulcanisaeta sp. JCHS 4 | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| 3300003723 | Hypersaline microbial mat communities from Yellowstone National Park, Wyoming, USA - Beowulf (BE_B) (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0040873_1002501 | 4784 | AMD31390.1 | Acidilobus sp. 7A | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| Ga0040873_1003761 | 1921 | EQB65596.1 | Thermoplasmatales archaeon I-plasma | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0040873_1011521 | 2174 | KUO93148.1 | Thermocladium sp. ECH_B | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0040873_1013971 | 707 | PMP91269.1 | Caldisphaera sp. | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0040873_1028371 | 244 | NAY81522.1 | Thaumarchaeota archaeon | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| 3300003709 | Hypersaline microbial mat communities from Yellowstone National Park, Wyoming, USA - Grendel (GRN_C) (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0040875_1014081 | 163 | AMD31390.1 | Acidilobus sp. 7A | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| Ga0040875_1034841 | 11 | KUO93148.1 | Thermocladium sp. ECH_B | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| 3300007166 | Iron oxide microbial mat communities from Yellowstone National Park, Wyoming, USA - BED_top_diel_T=1 metaT (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0099835_1035811 | 192 | NAY81522.1 | Thaumarchaeota archaeon | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| Ga0099835_1427011, Ga0099835_1431661 |
4 3 |
KUO93148.1 | Thermocladium sp. ECH_B | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0099835_1456681 | 2 | PMP93815.1 | Nitrosphaera sp. | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0099835_1510461 | 17 | KUO89797.1 | Caldivirga sp. CIS_19 | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| 3300007164 | Iron oxide microbial mat communities from Yellowstone National Park, Wyoming, USA - BED_top_diel_T=3 metaT (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0099836_1305913 | 60 | NAY81522.1 | Thaumarchaeota archaeon | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| Ga0099836_1428691 | 2 | WP_066793619.1 | Caldivirga sp. MU80 | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0099836_1428931, Ga0099836_1472421 |
3 3 |
KUO93148.1 | Thermocladium sp. ECH_B | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| 3300007168 | Iron oxide microbial mat communities from Yellowstone National Park, Wyoming, USA - BED_top_diel_T=7 metaT (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0099838_1606901 | 3 | KUO93148.1 | Thermocladium sp. ECH_B | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| Ga0099838_1754862 | 40 | NAY81522.1 | Thaumarchaeota archaeon | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| 3300007161 | Iron oxide microbial mat communities from Yellowstone National Park, Wyoming, USA - BED_top_diel_T=8 metaT (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0099839_1320821 | 2 | NAZ28310.1 | Caldivirga sp. | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| Ga0099839_1444841, Ga0099839_1444842, Ga0099839_1513781 |
19 4 |
NAY81522.1 | Thaumarchaeota archaeon | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| 3300007574 | Iron oxide microbial mat communities from Yellowstone National Park, Wyoming, USA - ECH_B_top_diel_T=1 metaT (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0099840_1194611, Ga0099840_1228901 | 6 | KUO93148.1 | Thermocladium sp. ECH_B | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| Ga0099840_1208591, Ga0099840_1293341 Ga0099840_1307751, |
29 27 7 |
NAY81522.1 | Thaumarchaeota archaeon | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0099840_1212061 | 16 | AMD31390.1 | Acidilobus sp. 7A | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| 3300007486 | Iron oxide microbial mat communities from Yellowstone National Park, Wyoming, USA - ECH_B_top_diel_T=5 metaT (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0099841_1138611 | 5 | WP_117355195.1 | Acidilobus sp. 7A | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| Ga0099841_1140711, Ga0099841_1141101 |
23 12 |
NAY81522.1 | Thaumarchaeota archaeon | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0099841_1256151 | 7 | NAZ28310.1 | Caldivirga sp. | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| Ga0099841_1293981 | 15 | KUO93148.1 | Thermocladium sp. ECH_B | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| 3300007575 | Iron oxide microbial mat communities from Yellowstone National Park, Wyoming, USA - ECH_C_top_diel_T=7 metaT (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0099844_1215141, Ga0099844_1308701 |
10 10 |
NAY81522.1 | Thaumarchaeota archaeon | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| 3300021851 | Metatranscriptome of extremophilic microbial mat communities from Yellowstone National Park, Wyoming, USA - CONBC_RNA (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0187838_10229991 | 154 | WP_014289867.1 | Pyrobaculum ferrireducens | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| Ga0187838_11515921 | 155 | KUK06933.1 | Archaeoglobus fulgidus | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) | ||
| 3300037625 | Metatranscriptome of soil microbial communities from Old Woman Creek estuary, Ohio, United States - Aug_M1_C1_D5_B (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0401895_061319_2_361 | – | HGD34321.1 | Candidatus Korarchaetoa archaeon | Study ID: Gs0135942 | Data used with permission from PI |
| 3300003730 | Thermal spring microbial communities from Beowulf Spring, Yellowstone National Park, Wyoming, USA - Beowulf (BE_D) (Metagenome Metatranscriptome) (*) (MER-FS) (assembled) | Ga0040879_1097101 | 63 | NAY81522.1 | Thaumarchaeota archaeon | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
| Ga0040879_1273451 | 7 | NAZ28310.1 | Caldivirga sp. | Study ID: Gs0067861 | D’haeseleer, P. et al. Stand. Genome Sci. (2017) |
Expression of cydAA’ in the environment estimated using publicly available metatranscriptomes. cydA homologs from the metatranscriptomic data available on the IMG website were extracted using a BLASTP cutoff of 1e-5. The short fragments found in metatranscriptomic data were matched with the full corresponding protein sequence based on the best hit in the NCBI database.
Partial purification and spectroscopic characterization of the CydAA’ from C. maquilingensis
The cydAA’ operon from Caldivirga maquilingensis consists of two genes—cydA and cydA’. There are no additional subunits encoded within the operon corresponding to cydX/cydY or cydS, which are associated, respectively, with E. coli cydAB and G. thermodenitrificans cydAB. Homologues of these subunits are not apparent in the C. maquilingensis genome. We cloned the operon into the pET22b vector and expressed it in an Escherichia coli strain in which both bd-I and bd-II were deleted (CBO - C43, ΔcydA ΔappB) [17]. The enzyme, cytochrome bb’ oxygen reductase from Caldivirga was engineered to have numerous different tags—6xHistidine, FLAG, GST and GFP. None of these tags were successful, either because of a poor yield of protein or because of the inability of the affinity-tagged proteins to bind to columns with their corresponding epitopes. A GFP-tagged protein was used to verify the expression of CydAA’ from Caldivirga maquilingensis in E. coli. The presence of the protein could be observed by following the fluorescence of the protein under UV light. Since subunit II was tagged with GFP, it confirms the presence of subunit II in the preparation (Supplementary Fig. S6). In addition, the purified protein was verified by mass spectrometry with many peptides recovered from subunit I. (Supplementary Fig. S7). Gel electrophoresis of a partially purified CydAA’ shows two bands of the sizes expected for CydA and CydA’ (Supplementary Fig. S8).
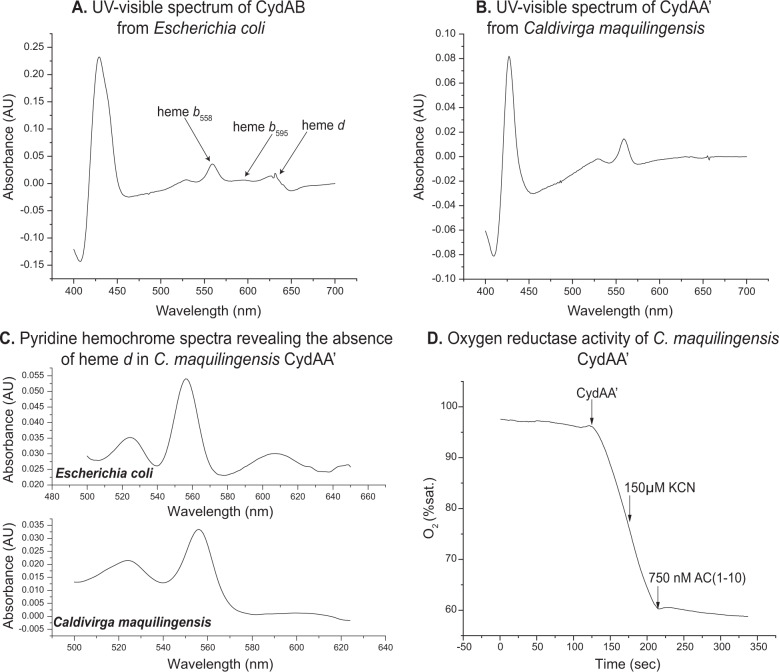
A UV-visible spectrum of CydAA’ in the reduced-minus-oxidized state reveals the absence of the heme d absorbance peak. The presence of heme b595 is also not apparent in the spectrum since the maxima at 595 nm and the Soret peak at 440 nm are also missing. This could indicate that heme b595 is low spin in this preparation. Hemes were extracted from CydAA’ of Caldivirga maquilingensis as described previously [52]. Only b-type hemes are present in the enzyme (Fig. 4). This was verified by analyzing the hemes in the protein by LC-MS (data not shown).
Fig. 4. Biochemical characteristics of CydAA’ from Caldivirga maquilingensis.
A. and B. UV-visible spectra of cytochrome bd-type oxygen reductase purified from Escherichia coli and Caldivirga maquilingensis, respectively. C. Pyridine hemochrome spectra of CydAA’ from Caldivirga maquilingensis revealing the absence of heme d in the partially purified enzyme. D. Oxygen reductase activity of CydAA’ from C. maquilingensis shows that it is highly active and cyanide insensitive. It is sensitive to Aurachin C1-10, a quinol binding site inhibitor that also inhibits E. coli cytochrome bd.
CydAA’ from Caldivirga maquilingensis has oxygen reduction activity
The oxygen reduction activity of CydAA’ was tested using a Clark electrode, with reduced coenzyme Q1 (reduced using DTT) as the electron donor. (Table 2, Fig. 4) At 37 °C the specific activity is ~330 e-/s (/heme b). While this is not as high as the activity of E. coli bd at the same temperature (over 1000 e-/s), the enzymatic activity is substantial, particularly considering the fact that the source of the enzyme is a thermophilic organism whose growth is optimum at 65 °C. The oxygen reductase activity of CydAA’ was insensitive to the presence of 250 µM KCN, a concentration of cyanide that would completely inhibit the activity of heme-copper oxygen reductases [1]. Since CydAA’ was expressed in the bd-deletion mutant, E. coli strain CBO, the only other potential oxygen reductase in this preparation is bo3 ubiquinol oxygen reductase [53] so the lack of cyanide sensitivity confirms that our purification protocol has separated the two enzymes. The enzyme is also susceptible to Aurachin AC1-10, a known inhibitor of cytochrome bd at concentrations as low as 250 nM [54]. We did not test for other possible functions for CydAA’ such as catalase activity [55] or peroxidase activity [56].
Table 2.
Oxygen reduction activity of E. coli CydAB and C. maquilingensis CydAA’ in the presence of 350 µM coenzyme Q1 and 5 mM DTT.
| Protein | Oxygen reduction activity (e-/s) |
|---|---|
| Caldivirga maquilingensis cytbb’ (CydAA’) | 333 20 |
| Escherichia coli cytbd (CydAB) | 1065 73 |
We previously noted that cydAA’ is typically found in organisms that perform sulfur-based chemistry such as sulfur reduction and sulfate reduction (Supplementary Table 6) and use DMSO-reductase like enzymes that contain molybdopterin as a cofactor [10]. Combining the above observation with the demonstrated oxygen reductase activity of CydAA’, it is likely that the role of CydAA’ is to detoxify oxygen to protect oxygen-sensitive enzymes involved in sulfur metabolism. This is similar to its expected role in Desulfovibrio [24] and in the protection of nitrogenases during the process of nitrogen fixation [22]. While its primary role may be that of oxygen detoxification, the presence of conserved residues corresponding to the proton channel in qOR4a-cydA implies that CydAA’ is likely to contribute to the generation of proton motive force, by proton translocation. The extent of its contribution would be limited by the prevailing oxygen concentration.
It is striking that oxygen reduction is conserved in the qOR4a-subfamily despite the replacement of CydB with CydA’ and therefore, it is worth considering the similarities and differences between E. coli CydAB and C. maquilingensis CydAA’.
Structural differences between CydAB and CydAA’ inferred from homology models
To aid in the understanding of differences between CydAA’ and other CydAB, we used multiple sequence alignments (Supplementary Figs. S9, S10) and structural models of CydA and CydA’ from Caldivirga maquilingensis (Supplementary Fig. S11, pdb files are available in supplementary material). The most drastic difference between the E. coli and C. maquilingensis enzymes is the absence of CydB. CydB in E. coli was shown to contain the oxygen diffusion channel [31, 32] and an additional proton channel leading to heme d, bound to subunit I [30–32]. In C. maquilingensis the second subunit is CydA’ is 26% similar to CydA. Only the first two helices are well conserved between these subunits in C. maquilingensis whereas other qOR4a-type CydA and qOR4b-type CydA’, such as in A. fulgidus are similar in the first 4 helices. To substitute for the proton channel that exists in CydB, conserved residues in CydA’ such as T71, T74 and H126 might form a different proton channel. CydA’ probably hosts an oxygen diffusion channel to substitute for the loss of the one in CydB but it is not possible to tell from the sequence alignment or structural model where in the subunit this might be. Interestingly, CydA’ retains H19, which has been implicated as a ligand to heme d and heme b595 in E. coli and G. thermodenitrificans cytbd respectively. This suggests that an additional heme might bind to the CydA’ subunit but we cannot verify or refute this from our protein preparation. A number of mutations are observed around the binuclear-active site in subunit I, substituting the polar residues in qOR3-CydA with non-polar residues in qOR4a-CydA (Supplementary Fig. S11) that might affect the midpoint potential of the heme or the proton-coupled electron transfer mechanisms.
Characteristics of the OR-C and OR-N families of the cytochrome bd superfamily
As mentioned earlier, a phylogenomic analysis of CydA homologs revealed two new families, OR-C and OR-N that share the first four helices containing the oxygen reduction site. The CydA subunit of OR-C bd-type oxygen reductases typically has eight transmembrane helices and an extended C-terminal periplasmic portion that binds hemes c (Supplementary Table 7), strongly suggesting that a cytochrome c could be an electron donor to this family. Adjacent to the OR-C1a-type cydA in the genome is OR-C1b, which also has 8 transmembrane helices. The OR-N3a/b, -N4a/b and -N5a/b family CydA typically have 10 helices while the OR-N2 and -N1 have 14 transmembrane helices. OR-N enzymes have been previously noted in Nitrospira [57] and Chloroflexi (N5a/b) [58]. They were recently shown to be expressed in manganese oxidizing autotrophic microorganism, Candidatus Manganitrophus noduliformans (N2/N1) from the phylum Nitrospira [59] and is implicated in oxygen reduction. Greater details on the OR-C and OR-N families, including distribution, alignments and conserved amino acids are found in Supplementary Material. The OR-C and OR-N families are widely distributed in Bacteria. OR-C is present only in Bacteria, while OR-N has very few representatives in Archaea (Supplementary Table 2, Supplementary Table 4).
A phylogenetic tree of all CydA clades suggest that the OR-C and OR-N families are more closely related to qOR4b than the other qOR subfamilies (Fig. 2, Supplementary Fig. S1). There are also conserved sequence features that suggest that the OR-C and OR-N families are more closely related to the qOR3 and qOR4a families than the qOR1 family including the deletion in the proton channel between the conserved glutamates (E101 and E108 like in G. thermodenitrificans cytbd), and the presence of nearby conserved tyrosines (Y123 and Y125 in G. thermodenitrificans cytbd). (Supplementary Table 3). This suggests that the OR-C and OR-N families diverged from either of these two families and evolved after the qOR reductases. The evolutionary analysis within this superfamily is complicated by the high number of independent gene duplication events. It appears that qOR4b, OR-N5b, OR-N3b, OR-N4b and OR-C1b subfamilies were the result of gene duplication events (Supplementary Fig. S1). In fact, OR-3a and OR-3b CydA share 50% amino acid sequence similarity. In addition, the presence of OR-N1-type and OR-N2-type cydA in the same operon in some Bacteria and the extent of similarity between them (up to 40%) suggest that they were part of yet another gene duplication. The importance of gene duplication in protein evolution and functional diversification is well known [60]. The nature this process has taken in the cytbd is interesting—a majority of the CydA paralogs have maintained the architecture associated with oxygen reduction and all of them have maintained the H19 ligand to the active site heme d (as per the E. coli structure). In addition, all the above-mentioned duplication events appear to have resulted in a complex of multiple CydA-like proteins with the possible exception of OR-N1 and OR-N2. OR-N2 is often found in operons without another CydA-like protein (Fig. 1). H19 and heme d are found near the interface of subunit I and subunit II in the CydAB structures. The complete conservation of these features with a change in their interacting partner is suggestive of the process of duplication and interface evolution recently investigated in hemoglobin [61]. Future work on the biochemical and structural characterization of the various cytbd families will help us develop insight into the driving forces behind the evolution of this superfamily. Presently, it is clear that the defining characteristic of the cytbd superfamily is the di-heme oxygen reduction site found in the first four helices of CydA homologs. Our analysis suggests that the bd protein scaffold was diversified multiple times to perform O2 chemistry in unique environments, possibly to function with different electron donors.
Conclusions
The cytbd superfamily is one of two oxygen reductase superfamilies that are widely distributed in Bacteria and Archaea. In the current work we have demonstrated the large diversity of this superfamily using phylogenomics. In addition, we biochemically characterized the CydAA’ from C. maquilingensis showing that CydAA’ is a robust oxygen reductase. The isolated CydAA’ contained only b hemes and no heme d. Hence, C. maquilingensis CydAA’ is a bb’-type oxygen reductase and is the first enzyme from the qOR4a-subfamily to be purified and demonstrated to be a functional oxygen reductase. Finally, we demonstrate that significant diversification of CydA has occurred with the conserved oxygen reduction site being adapted to multiple functions within various ecological niches.
Materials and methods
Phylogenomic analysis of cytochrome bd sequences
To investigate the diversity of the cytochrome bd oxygen reductase superfamily, we performed a large-scale analysis of CydA protein sequences. We first queried the NCBI and IMG databases with BLASTP using an e-value of 1e-3 to generate a database of CydA sequences that had at least some of the conserved amino acids identified in CydA sequences previously [36]. We then used the database of 13738 sequences, whose diversity is reflected in MSA1, and filtered it with a sequence cut-off of 50% to generate the tree in Fig. 2. The parameters used for the various tree-generating algorithms are detailed below. From this tree, we extracted seven CydA sequences from various families, listed below, to probe the full extent of the cytochrome bd family diversity within the GTDB database.
In order to reconcile the protein phylogeny of cytochrome bd oxygen reductases with species taxonomy, we identified and mapped all cytbd to their respective species within the GTDB database release89 [37]. All CydA protein sequences were extracted from GTDB genomes using BLAST [62] by querying with seven CydA sequences (YP_001729719.1-qOR1, MBA3917267.1-qOR2, Ga0063455_1000025672-qOR3, WP_012186727.1-qOR4b, WP_012186728.1-qOR4a, WP_014321499.1-OR-C, TKB90292.1-OR-N). The resulting 67,521 sequences were filtered for sequences >300 amino acids and for unique accession ids to obtain 18,212 sequences. These were then aligned using muscle [63] using the optional maxiters cut-off of 2. The alignment was visualized on Jalview [64] and sequences were filtered by manually deleting the CydA sequences without characteristics of the quinol binding site or the proton channel, as well as some sequences which did not align well with the rest of the sequences. This filtration step was used to remove 369 sequences but also resulted in the loss of 95 CydA sequences from within qOR1 that appear to have lost the proton channel. We added these 95 sequences back to the database after the HMM-based classification step and labelled them as qOR1*. Of the remaining sequences 274 sequences, 200 were cytochromes c that were homologous to the C-terminal portion of the OR-C1a subfamily. The characterization of the remaining divergent 74 sequences is beyond the scope of this work. The filtered set of 17,843 CydA sequences were then classified using a Hidden Markov Model (HMM)-based classifier trained to identify the families—qOR1, qOR2, qOR3, qOR4a, OR-C and OR-N. The HMMs for those subfamilies and families are available on Github (https://github.com/ranjani-m/cytbd-superfamily). The presence or absence of cytbd in each species was tabulated and is available as Supplementary Table 2. The all Archaea species tree used to analyze the distribution of cytochrome bd oxygen reductases in Archaea was generated using Anvi’o [42]. A multiple sequence alignment was created by extracting all ribosomal proteins from archaeal genomes using the HMM source Archaea_76. This alignment was then used to generate a phylogenetic tree using FastTree as per Anvi’o’s default settings. This tree was annotated using the data available in Supplementary Table 4 on the iTOL server [65].
The protein phylogeny of cytbd sequences was inferred using sequences of cytbd subunit I, CydA. These were extracted from a taxonomically diverse set of genomes and metagenomes from IMG [66], filtered with UCLUST [67] using a percentage identity cut-off of 0.5 and aligned using MUSCLE. The multiple sequence alignment was used to infer a phylogenetic tree using RAxML [68] on the CIPRES Science Gateway [69] with the PROTGAMMA model of rate heterogeneity, DAYHOFF matrix specification and a bootstrap analysis with 100 iterations. IQ-trees [70] were inferred using a Gamma model of rate heterogeneity, DAYHOFF substitution matrix and 1000 ultrafast bootstrap replicates while MrBayes [71] trees were inferred using an equal rate of variation across sites and the Jones substitution matrix. MCMC analysis was run for the default 4 chains and 1,000,000 generations.
Preparation of construct for of cytochrome bd oxidase from Escherichia coli
The genes encoding the bb’ oxygen reductase (Gene Object ID: 641276193-4) from C. maquilengensis were PCR amplified using primers purchased from Integrated DNA Technology. The genes were cloned into pET22b (Invitrogen) using 5′ NdeI and 3′ XhoI cut sites. The vector was engineered to use EGFP, GST or FLAG tags alternatively. The tag was added to subunit II in case of EGFP and FLAG; a tag on both subunit I and II was attempted for the His-tag and GST tag. The expression vector, along with pRARE (Novagen) was then transformed into (CBO ΔcydBΔappC::kan) for protein expression.
Cell growth and protein purification
A single colony was inoculated into 5 ml of LB (yeast extract and tryptone were purchased from Acumedia and NaCl from Sigma-Aldrich) with 100 µg/ml Ampicillin and incubated with shaking at 37 °C. The following day, the 5 ml culture was inoculated in 300 ml LB with 100 µg/ml Ampicillin and grown overnight at 37 °C. On the third day, 10 ml of the secondary culture was inoculated into 24 2 L flasks containing 1 L LB with 100 µg/ml Ampicillin, each. The flasks were incubated at 37 °C while shaking at 200 rpm, until the OD600 of the culture reached 0.6. Gene expression was then induced using 0.5 mM IPTG, the temperature was lowered to 30 °C, and the culture was incubated for 8 h or overnight.
The fully-grown cultures were then pelleted by spinning down at 8000 rpm for 8 min, in 500 ml centrifuge bottles. The harvested cells were then resuspended in 100 mM Tris-HCl, 10 mM MgS04, pH 8 with DNaseI and a protease inhibitor cocktail from Sigma. The cells were then homogenized using a Bamix Homogenizer, and passed through a Microfluidizer cell at 100 psi, three times, to lyse the cells. The soluble fraction of the lysate was then separated from the insoluble by spinning down the lysate at 8000 rpm. Membranes were extracted from the soluble fraction by centrifuging the soluble fraction at 42,000 rpm for 4 h.
Membranes were resuspended in 20 mM Tris, 300 mM NaCl, pH 8 and then solubilized with 1% DDM or 1% SML. The solubilized membranes were spun down at 42,000 rpm for 45 min to remove unsolubilized membranes. The supernatant was stirred with Ni-NTA resin for 1 h and then loaded onto a column. The flow through was shown to contain the CydAA’ because of its poor affinity for the nickel column. The flow through was then diluted in buffer to contain 50 mM salt and then loaded onto a DEAE column equilibriated with 20 mM Tris, pH 8, 0.05% DDM. An elution gradient was run between 0 and 500 mM NaCl and CydAA’ was partially purified from the fraction with higher absorbance at A412nm, corresponding to the soret peak for heme b and used for assays and spectroscopy. This is similar to the first step for purification of cytbd from Escherichia coli [33]. If the elute from the Ni-NTA resin was subject to two steps of purification, first with the Q-sepharose column with the same gradient as above and then the DEAE-Sepharose, a more pure cytbd preparation was obtained but the yield was drastically reduced.
UV-visible spectroscopy
Spectra of the protein were obtained using an Agilent DW-2000 Spectrophotometer in the UV-visible region. The cuvette used has a pathlength of 1 cm. The oxidized spectrum was taken of the air-oxidized protein. The enzyme was reduced with dithionite to obtain a reduced spectrum.
Collection of pyridine hemochrome spectra and heme analysis
For the wildtype or mutants enzymes, 35 μl of the enzyme solution was mixed with an equal volume of 40% pyridine with 200 mM NaOH. The oxidized spectra was measured in the presence of ferricyanide and the reduced in the presence of dithionite. The values of heme b were calculated according to the matrix suggested in [52]. The concentration of heme d was estimated using the extinction coefficient ε(629–670nm) = 25 mM−1 cm−1.
Measurement of oxygen reductase activity
Oxygen reductase activity was measured using the Mitocell Miniature Respirometer MT200A (Harvard Apparatus). 5 mM DTT and 350 µM Q1 were used as electron donors to measure oxygen reduction by C. maquilingensis CydAA’ and E. coli cytochrome bd. 150–250 µM KCN was used to test the cyanide sensitivity of the enzymes.
Structural modelling of CydAA’ from Caldivirga maquilingensis
Sequences of subunit I from Geobacillus thermodenitrificans and Caldivirga maquilingensis were aligned using a larger alignment comprising many hundreds of bb’ sequences made with the software MUSCLE. This alignment was used as to create a model of subunit I from Caldivirga using the Geobacillus subunit I as a template on the Swiss Model server. A model of subunit II was also created using subunit I as a template. The model was then visualized using VMD 1.9.2beta1 [72].
Supplementary information
Supplementary Table 1. Total number of various CydA families and subfamilies in the GTDB.
Supplementary Table 2. Distribution of CydA subfamilies by genome in GTDB.
Supplementary Table 3. Conserved features residues identified in cytbd families without quinol oxidizing features.
Supplementary Table 4. Distribution of CydA subfamilies by phyla in GTDB.
Supplementary Table 5. Distribution of quinol oxidizing cytbd in archaeal genomes found in the GTDB
Supplementary Table 6. Growth conditions and temperature for organisms containing cydAA’.
Supplementary Table 7. Number of heme c binding sites in cytbd sequences from OR-C1a/OR-C1b subfamilies.
Supplementary multiple sequences alignment MSA1.
Supplementary multiple sequences alignment MSA2.
Supplementary multiple sequences alignment MSA3.
Supplementary multiple sequences alignment MSA4.
Supplementary multiple sequences alignment MSA5.
Homology model of qOR4a-CydA from Caldivirga maquilingensis
Acknowledgements
We would like to thank NIH for funding this research (Grant # R01AI148160, Principle Investigator: Dr Robert Gennis). We were inspired by the work of Phil Hugenholtz and his lab in reconciling protein phylogenies with organismal taxonomy and, we thank them for a very productive discussion session on the evolution of aerobic respiratory enzymes. We thank Hank Yu for his valuable comments on the initial paper and, Daan Speth for helping us by organizing the genome data from GTDB into an easily searchable file. We thank Bijoy Desai, Ambika Nadkarni and Rajashekar Iyer for useful discussions on the best naming scheme for the cytochrome bd superfamily. We would also like to thank Kelly Wrighton, Adrienne Narrowe and Reb Daly from the Wrighton lab at the Colorado State University for allowing us to use some of their metatranscriptomic data to demonstrate the expression of cydAA’ in the environment.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Ranjani Murali, Email: m.ranjani@gmail.com.
James Hemp, Email: james.hemp@metrodora.org.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-021-01019-4.
References
- 1.Borisov VB, Gennis RB, Hemp J, Verkhovsky MI. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta BBA - Bioenerg. 2011;1807:1398–413. doi: 10.1016/j.bbabio.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moll R, Schäfer G. Purification and characterisation of an archaebacterial succinate dehydrogenase complex from the plasma membrane of the thermoacidophile Sulfolobus acidocaldarius. Eur J Biochem. 1991;201:593–600. doi: 10.1111/j.1432-1033.1991.tb16319.x. [DOI] [PubMed] [Google Scholar]
- 3.Kabashima Y, Sakamoto J. Purification and biochemical properties of a cytochrome bc complex from the aerobic hyperthermophilic archaeon Aeropyrum pernix. BMC Microbiol. 2011;11:52. doi: 10.1186/1471-2180-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anemüller S, Schäfer G. Cytochrome aa3 from Sulfolobus acidocaldarius. A single-subunit, quinol-oxidizing archaebacterial terminal oxidase. Eur J Biochem. 1990;191:297–305. doi: 10.1111/j.1432-1033.1990.tb19123.x. [DOI] [PubMed] [Google Scholar]
- 5.Bandeiras TM, Pereira MM, Teixeira M, Moenne-Loccoz P, Blackburn NJ. Structure and coordination of CuB in the Acidianus ambivalens aa3 quinol oxidase heme–copper center. JBIC J Biol Inorg Chem. 2005;10:625–35. doi: 10.1007/s00775-005-0012-6. [DOI] [PubMed] [Google Scholar]
- 6.Marreiros BC, et al. Exploring membrane respiratory chains. Biochim Biophys Acta BBA - Bioenerg. 2016;1857:1039–67. doi: 10.1016/j.bbabio.2016.03.028. [DOI] [PubMed] [Google Scholar]
- 7.Blank CE. Low Rates of Lateral Gene Transfer among Metabolic Genes Define the Evolving Biogeochemical Niches of Archaea through Deep Time. Archaea. 2012;2012:1–23. doi: 10.1155/2012/843539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inskeep WP, Jay ZJ, Herrgard MJ, Kozubal MA, Rusch DB, Tringe SG, et al. Phylogenetic and Functional Analysis of Metagenome Sequence from High-Temperature Archaeal Habitats Demonstrate Linkages between Metabolic Potential and Geochemistry. Front. Microbiol. 2013;4:1–21. doi: 10.3389/fmicb.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beam JP, Jay ZJ, Kozubal MA, Inskeep WP. Niche specialization of novel Thaumarchaeota to oxic and hypoxic acidic geothermal springs of Yellowstone National Park. ISME J. 2014;8:938–51. doi: 10.1038/ismej.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jay ZJ, Rusch DB, Tringe SC, Bailey C, Jennings RM, Inskeep WP. Predominant Acidilobus-Like Populations from Geothermal Environments in Yellowstone National Park Exhibit Similar Metabolic Potential in Different Hypoxic Microbial Communities. Appl Environ Microbiol. 2014;80:294–305. doi: 10.1128/AEM.02860-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jay ZJ, Beam JP, Dohnalkova A, Lohmayer R, Bodle B, Planer-Friedrich B, et al. Pyrobaculum yellowstonensis Strain WP30 Respires on Elemental Sulfur and/or Arsenate in Circumneutral Sulfidic Geothermal Sediments of Yellowstone National Park. Appl Environ Microbiol. 2015;81:5907–16. doi: 10.1128/AEM.01095-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azarkina N, Siletsky S, Borisov V, von Wachenfeldt C, Hederstedt L, Konstantinov AA. A Cytochrome bb′-type Quinol Oxidase in Bacillus subtilis Strain 168. J Biol Chem. 1999;274:32810–7. doi: 10.1074/jbc.274.46.32810. [DOI] [PubMed] [Google Scholar]
- 13.Cunningham L, Pitt M, Williams HD. The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol Microbiol. 1997;24:579–91. doi: 10.1046/j.1365-2958.1997.3561728.x. [DOI] [PubMed] [Google Scholar]
- 14.Sootsuwan K, Lertwattanasakul N, Thanonkeo P, Matsushita K, Yamada M. Analysis of the respiratory chain in Ethanologenic Zymomonas mobilis with a cyanide-resistant bd-type ubiquinol oxidase as the only terminal oxidase and its possible physiological roles. J Mol Microbiol Biotechnol. 2008;14:163–75. doi: 10.1159/000112598. [DOI] [PubMed] [Google Scholar]
- 15.Quesada A, Guijo MI, Merchán F, Blázquez B, Igeño MI, Blasco R. Essential role of cytochrome bd-related oxidase in cyanide resistance of Pseudomonas pseudoalcaligenes CECT5344. Appl Environ Microbiol. 2007;73:5118–24. doi: 10.1128/AEM.00503-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun YH, de Jong MF, den Hartigh AB, Roux CM, Rolán HG, Tsolis RM. The small protein CydX is required for function of cytochrome bd oxidase in Brucella abortus. Front Cell Infect Microbiol. 2012;2:1–15. doi: 10.3389/fcimb.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoeser J, Hong S, Gehmann G, Gennis RB, Friedrich T. Subunit CydX of Escherichia coli cytochrome bd ubiquinol oxidase is essential for assembly and stability of the di-heme active site. FEBS Lett. 2014;588:1537–41. doi: 10.1016/j.febslet.2014.03.036. [DOI] [PubMed] [Google Scholar]
- 18.Chen H, Luo Q, Yin J, Gao T, Gao H. Evidence for the requirement of CydX in function but not assembly of the cytochrome bd oxidase in Shewanella oneidensis. Biochim Biophys Acta BBA - Gen Subj. 2015;1850:318–28. doi: 10.1016/j.bbagen.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 19.VanOrsdel CE, Bhatt S, Allen RJ, Brenner EP, Hobson JJ, Jamil A, et al. The Escherichia coli CydX protein is a member of the CydAB cytochrome bd oxidase complex and is required for cytochrome bd oxidase activity. J Bacteriol. 2012;195:3640–50. doi: 10.1128/JB.00324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’mello R, Hill S, Poole RK. The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology. 1996;142:755–63. doi: 10.1099/00221287-142-4-755. [DOI] [PubMed] [Google Scholar]
- 21.Kaminski PA, Kitts CL, Zimmerman Z, Ludwig RA. Azorhizobium caulinodans uses both cytochrome bd (quinol) and cytochrome cbb3 (cytochrome c) terminal oxidases for symbiotic N2 fixation. J Bacteriol. 1996;178:5989–94. doi: 10.1128/jb.178.20.5989-5994.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juty NS, Moshiri F, Merrick M, Anthony C, Hill S. The Klebsiella pneumoniae cytochrome bd‘ terminal oxidase complex and its role in microaerobic nitrogen fixation. Microbiology. 1997;143:2673–83. doi: 10.1099/00221287-143-8-2673. [DOI] [PubMed] [Google Scholar]
- 23.Wakai S, Kikumoto M, Kanao T, Kamimura K. Involvement of sulfide:quinone oxidoreductase in sulfur oxidation of an acidophilic iron-oxidizing bacterium, Acidithiobacillus ferrooxidans NASF-1. Biosci Biotechnol Biochem. 2004;68:2519–28. doi: 10.1271/bbb.68.2519. [DOI] [PubMed] [Google Scholar]
- 24.Lamrabet O, Pieulle L, Aubert C, Mouhamer F, Stocker P, Dolla A, et al. Oxygen reduction in the strict anaerobe Desulfovibrio vulgaris Hildenborough: characterization of two membrane-bound oxygen reductases. Microbiology. 2011;157:2720–32. doi: 10.1099/mic.0.049171-0. [DOI] [PubMed] [Google Scholar]
- 25.Lemos RS, Gomes CM, Santana M, LeGall J, Xavier AV, Teixeira M. The ‘strict’ anaerobe Desulfovibrio gigas contains a membrane-bound oxygen-reducing respiratory chain. FEBS Lett. 2001;496:40–43. doi: 10.1016/s0014-5793(01)02399-7. [DOI] [PubMed] [Google Scholar]
- 26.Borisov VB, Belevich I, Bloch DA, Mogi T, Verkhovsky MI. Glutamate 107 in Subunit I of Cytochrome bd from Escherichia coli Is Part of a Transmembrane Intraprotein Pathway Conducting Protons from the Cytoplasm to the Heme b595/Heme d Active Site. Biochemistry. 2008;47:7907–14. doi: 10.1021/bi800435a. [DOI] [PubMed] [Google Scholar]
- 27.Bertsova YV, Bogachev AV, Skulachev VP. Generation of protonic potential by the bd-type quinol oxidase of Azotobacter vinelandii. FEBS Lett. 1997;414:369–72. doi: 10.1016/s0014-5793(97)01047-8. [DOI] [PubMed] [Google Scholar]
- 28.Borisov VB, Murali R, Verkhovskaya ML, Bloch DA, Han H, Gennis RB, et al. Aerobic respiratory chain of Escherichia coli is not allowed to work in fully uncoupled mode. Proc Natl Acad Sci. 2011;108:17320–4. doi: 10.1073/pnas.1108217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stolper DA, Revsbech NP, Canfield DE. Aerobic growth at nanomolar oxygen concentrations. Proc Natl Acad Sci. 2010;107:18755–60. doi: 10.1073/pnas.1013435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Safarian S, Rajendran C, Müller H, Preu J, Langer JD, Ovchinnikov S, et al. Structure of a bd oxidase indicates similar mechanisms for membrane integrated oxygen reductases. Science. 2016;352:583–6. doi: 10.1126/science.aaf2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Safarian S, Hahn A, Mills DJ, Radloff M, Elsinger ML, Nikolaev A, et al. Active site rearrangement and structural divergence in prokaryotic respiratory oxidases. Science. 2019;366:100–4. doi: 10.1126/science.aay0967. [DOI] [PubMed] [Google Scholar]
- 32.Theßeling A, Rasmussen T, Burschel S, Wohlwend D, Kägi J, Müller R, et al. Homologous bd oxidases share the same architecture but differ in mechanism. Nat Commun. 2019;10:5138. doi: 10.1038/s41467-019-13122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller MJ, Gennis RB. The purification and characterization of the cytochrome d terminal oxidase complex of the Escherichia coli aerobic respiratory chain. J Biol Chem. 1983;258:9159–65. [PubMed] [Google Scholar]
- 34.Voggu L, Schlag S, Biswas R, Rosenstein R, Rausch C, Götz F. Microevolution of Cytochrome bd oxidase in Staphylococci and its implication in resistance to respiratory toxins released by Pseudomonas. J Bacteriol. 2006;188:8079–86. doi: 10.1128/JB.00858-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Osborne J, Gennis R. Sequence analysis of cytochrome bd oxidase suggests a revised topology for subunit I. Biochim Biophys Acta BBA - Bioenerg. 1999;1410:32–50. doi: 10.1016/s0005-2728(98)00171-6. [DOI] [PubMed] [Google Scholar]
- 36.Mogi T, Akimoto S, Endou S, Watanabe-Nakayama T, Mizuochi-Asai E, Miyoshi H. Probing the Ubiquinol-Binding Site in Cytochrome bd by Site-Directed Mutagenesis. Biochemistry. 2006;45:7924–30. doi: 10.1021/bi060192w. [DOI] [PubMed] [Google Scholar]
- 37.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P, et al. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol. 2018;36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 38.Rinke C, Chuvochina M, Mussig AJ, Chaumeil P, Waite DW, Whitman WB et al. A rank-normalized archaeal taxonomy based on genome phylogeny resolves widespread incomplete and uneven classifications. bioRxiv. 2020. 2020.03.01.972265 10.1101/2020.03.01.972265.
- 39.Mogi T, Endou S, Akimoto S, Morimoto-Tadokoro M, Miyoshi H. Glutamates 99 and 107 in transmembrane helix III of subunit I of cytochrome bd are critical for binding of the heme b595-d binuclear center and enzyme activity. Biochemistry. 2006;45:15785–92. doi: 10.1021/bi0615792. [DOI] [PubMed] [Google Scholar]
- 40.Davín AA, Tannier E, Williams TA, Boussau B, Daubin V, Szöllósi J. Gene transfers can date the tree of life. Nat Ecol Evol. 2018;2:904–9. doi: 10.1038/s41559-018-0525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louca S, Mazel F, Doebeli M, Parfrey LW. A census-based estimate of Earth’s bacterial and archaeal diversity. PLOS Biol. 2019;17:e3000106. doi: 10.1371/journal.pbio.3000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, et al. Anvi’o: an advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jay ZJ, et al. The distribution, diversity and function of predominant Thermoproteales in high-temperature environments of Yellowstone National Park. Environ Microbiol. 2016;18:4755–69. doi: 10.1111/1462-2920.13366. [DOI] [PubMed] [Google Scholar]
- 44.Yim KJ, Cha I, Rhee J, Song H, Hyun D, Lee H, et al. Vulcanisaeta thermophila sp. nov., a hyperthermophilic and acidophilic crenarchaeon isolated from solfataric soil. Int J Syst Evol Microbiol. 2015;65:201–5. doi: 10.1099/ijs.0.065862-0. [DOI] [PubMed] [Google Scholar]
- 45.Itoh T, Suzuki K, Sanchez PC, Nakase T. Caldivirga maquilingensis gen. nov., sp. nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines. Int J Syst Evol Microbiol. 1999;49:1157–63. doi: 10.1099/00207713-49-3-1157. [DOI] [PubMed] [Google Scholar]
- 46.Toshchakov SV, Korzhenkov AA, Samarov NI, Mazunin IO, Mozhey OI, Shmyr IS, et al. Complete genome sequence of and proposal of Thermofilum uzonense sp. nov. a novel hyperthermophilic crenarchaeon and emended description of the genus Thermofilum. Stand Genom Sci. 2015;10:122. doi: 10.1186/s40793-015-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Itoh T, Suzuki K, Nakase T. Thermocladium modestius gen. nov., sp. nov., a new genus of rod-shaped, extremely thermophilic crenarchaeote. Int J Syst Bacteriol. 1998;48:879–87. doi: 10.1099/00207713-48-3-879. [DOI] [PubMed] [Google Scholar]
- 48.Cozen AE, Weirauch MT, Pollard KS, Bernick DL, Stuart JM, Lowe TM. Transcriptional Map of Respiratory Versatility in the Hyperthermophilic Crenarchaeon Pyrobaculum aerophilum. J Bacteriol. 2009;191:782–94. doi: 10.1128/JB.00965-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nunoura T, Sako Y, Wakagi T, Uchida A. Regulation of the aerobic respiratory chain in the facultatively aerobic and hyperthermophilic archaeon Pyrobaculum oguniense. Microbiology. 2003;149:673–88. doi: 10.1099/mic.0.26000-0. [DOI] [PubMed] [Google Scholar]
- 50.D’haeseleer P, Lee JZ, Prufert-Bebout L, Burow LC, Detweiler AM, Weber PK, et al. Metagenomic analysis of intertidal hypersaline microbial mats from Elkhorn Slough, California, grown with and without molybdate. Stand Genom Sci. 2017;12:67. doi: 10.1186/s40793-017-0279-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bernstein HC, Beam JP, Kozubal MA, Carlson RP, Inskeep WP. In situ analysis of oxygen consumption and diffusive transport in high-temperature acidic iron-oxide microbial mats. Environ Microbiol. 2013;15:2360–70. doi: 10.1111/1462-2920.12109. [DOI] [PubMed] [Google Scholar]
- 52.Berry EA, Trumpower BL. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal Biochem. 1987;161:1–15. doi: 10.1016/0003-2697(87)90643-9. [DOI] [PubMed] [Google Scholar]
- 53.Gennis RB. Some recent advances relating to prokaryotic cytochrome c reductases and cytochrome c oxidases. Biochim Biophys Acta BBA - Bioenerg. 1991;1058:21–24. doi: 10.1016/s0005-2728(05)80260-9. [DOI] [PubMed] [Google Scholar]
- 54.Meunier B, Madgwick SA, Reil E, Oettmeier W, Rich PR. New inhibitors of the quinol oxidation sites of bacterial cytochromes bo and bd. Biochemistry. 1995;34:1076–83. doi: 10.1021/bi00003a044. [DOI] [PubMed] [Google Scholar]
- 55.Borisov VB, Forte E, Davletshin A, Mastronicola D, Sarti P, Giuffre A. Cytochrome bd oxidase from Escherichia coli displays high catalase activity: an additional defense against oxidative stress. FEBS Lett. 2013;587:2214–8. doi: 10.1016/j.febslet.2013.05.047. [DOI] [PubMed] [Google Scholar]
- 56.Al-Attar S, Yu Y, Pinkse M, Hoeser J, Friedrich T, Bald D, et al. Cytochrome bd Displays Significant Quinol Peroxidase Activity. Sci Rep. 2016;6:27631. doi: 10.1038/srep27631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lücker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, et al. A Nitrospira metagenome illuminates the physiology and evolution of globally important nitrite-oxidizing bacteria. Proc Natl Acad Sci U S A. 2010;107:13479–84. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soo RM, Hemp J, Parks DH, Fischer WW, Hugenholtz P. On the origins of oxygenic photosynthesis and aerobic respiration in Cyanobacteria. Science. 2017;355:1436–40. doi: 10.1126/science.aal3794. [DOI] [PubMed] [Google Scholar]
- 59.Yu H, Leadbetter JR. Bacterial chemolithoautotrophy via manganese oxidation. Nature. 2020;583:453–8. doi: 10.1038/s41586-020-2468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohno S. Evolution by gene duplication. New York: Springer; 1970.
- 61.Pillai AS, Chandler SA, Liu Y, Signore AV, Cortez-Romero CR, Benesch JLP, et al. Origin of complexity in haemoglobin evolution. Nature. 2020;581:480–5. doi: 10.1038/s41586-020-2292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 63.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinforma. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]
- 66.Chen IA, Chu K, Palaniappan K, Pillay M, Ratner A, Huang J, et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019;47:D666–D677. doi: 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 68.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller MA, Pfeiffer W & Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE) 1–8 (2010). 10.1109/GCE.2010.5676129.
- 70.Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Haeseler A. v. et al. IQ-TREE 2: new Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol. 2020;37:1530–4. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 72.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Total number of various CydA families and subfamilies in the GTDB.
Supplementary Table 2. Distribution of CydA subfamilies by genome in GTDB.
Supplementary Table 3. Conserved features residues identified in cytbd families without quinol oxidizing features.
Supplementary Table 4. Distribution of CydA subfamilies by phyla in GTDB.
Supplementary Table 5. Distribution of quinol oxidizing cytbd in archaeal genomes found in the GTDB
Supplementary Table 6. Growth conditions and temperature for organisms containing cydAA’.
Supplementary Table 7. Number of heme c binding sites in cytbd sequences from OR-C1a/OR-C1b subfamilies.
Supplementary multiple sequences alignment MSA1.
Supplementary multiple sequences alignment MSA2.
Supplementary multiple sequences alignment MSA3.
Supplementary multiple sequences alignment MSA4.
Supplementary multiple sequences alignment MSA5.
Homology model of qOR4a-CydA from Caldivirga maquilingensis