Abstract
Salinity stress is one of the most serious environmental stresses which limit plant growth, development and productivity. In this study, we screened 25 bacterial isolates based on the biochemical activity of ACC deaminase. Two potent PGPR namely Bacillus marisflavi (CHR JH 203) and Bacillus cereus (BST YS1_42) having the highest ACC deaminase (ACCD) activity were selected for further analyses such as polymerase chain reaction (PCR), salt tolerance assay, expression analysis, antioxidant assay, etc. The structural gene for ACCD activity was further confirmed by PCR showing the amplicon size ~ 800 bp. The acdS positive isolates exhibited optimum growth at 3% w/v (NaCl), indicating its ability to survive and thrive in induced saline soil. Inoculation of acdS+ strain on pea plants was found to be efficient and ameliorated the induced NaCl-stress by enhancing the various parameters like plant-biomass, carbohydrates, reducing sugars, protein, chlorophylls, phenol, flavonoids content and increasing antioxidants enzymes levels in plants. Moreover, the expression of ROS scavenging genes (PsSOD, PsCAT, PsPOX, PsNOS, PsAPX, PsChla/bBP), defense genes and cell rescue genes (PsPRP, PsMAPK, PsFDH) were analyzed. Inoculated plants exhibited a higher gene expression level and salt tolerance under 1%NaCl concentration. Thus, our results indicate that CHR JH 203 and BST YS1_42 strain showed the highest plant growth-promoting attributes could be used as bio-inoculants for crops under saline stress in the field towards sustainable crop development.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-03047-5.
Keywords: Enzymatic antioxidants, Gene expression, PGPR and acdS gene, Pisum sativum, Salinity stress
Introduction
Establishing a sustainable agricultural system is one of the major challenges currently faced the world over due to increasing food demand. Several abiotic stresses (ASs) such as salinity, drought, cold, metal, and flooding negatively impact on growth, development and production of crop plants worldwide (Gontia-Mishra et al. 2016; Sapre et al. 2019). Salinity stress (SS) is one of the major ASs which causes a wide range of physiological, molecular and biological changes in plants (Manchanda and Garg 2008; Numan et al. 2018).Worldwide, due to SS, more than 20% of irrigated land is negatively affected with an average yield loss of greater than 50% for major crops (Pitman and Läuchli 2002; Tuteja 2007; Mustafa et al. 2019). Chaves et al. (2009) stated that SS itself affected 6% of total and 30% of the irrigated land area worldwide, and this resulted in a huge economic loss (Flowers et al. 2010). Salinity stress causes a decline in the photosynthetic pigments (e.g. Carotenoids and Chlorophylls), protein synthesis, respiration, as well as changes in their morphological and anatomical features, lipid metabolism, energy transformation, ionic imbalance which ultimately resulting in yield loss of plant (Parida and Das 2005; Tuteja 2007).The secondary effects of SS include oxidative stress; thereby causes damage to the cellular components like membrane proteins, lipids, nucleic acids and metabolic dysfunction (Hossein and Rezvani 2006).
Ethylene is a gaseous hormone involved in the regulation of plant growth and development, also a principal modulator between plant response to diverse environmental stresses (ESs) and normal growth (Abeles 1992). Among several ASs, ethylene is one of the key positive mediators for salinity tolerance (ST) in Arabidopsis thaliana, grapevines, maize, and tomato (Munns and Tester 2008; Siddikee et al. 2011; Freitas et al. 2018). Ethylene production is quickly induced under SS (Morgan and Drew 1997; Yang et al. 2009). This higher ethylene level reduced seed germination as well as root growth (Belimov et al. 2001; Saravanakumar and Samiyappan 2007) conferring inhibitory effects on plants. Various bacterial isolates possess dissimilar levels of enzymatic activity under different ESs. However, 1-aminocyclopropane-1-carboxylate deaminase (ACCD) containing bacteria can hydrolyze 1-aminocyclopropane-1-carboxylate (ACC), the precursor of ethylene, thereby sinking the excess ethylene content and protects the plant from its inhibitory effect (Habib et al. 2016; Glick and Glick 2020). When plants were inoculated with ACCD-producing bacterial isolates, this resulted from an increase in biomass, photosynthetic pigment content, a number of flowers and buds compared to non-inoculated control plants (Ali et al. 2014a) thereby considered as plant growth-promoting bacteria (PGPR). These beneficial rhizobacteria were termed PGPR (Kloepper 1981). PGPR are best defined as beneficial rhizospheric bacteria that aggressively colonize the rhizosphere and promote the growth of the plants. ACC deaminase-producing isolate namely Achromobacter piechaudii strain ARV8 was found to be well organized in fostering the growth of tomato crop under elevated SS conditions (Shailendra Singh 2015). Here are several published pieces of literature suggesting that plants inoculated with PGPR containing ACC deaminase exhibit greater resistance to a variety of stresses including salinity, flood, drought, and against numerous pathogens (Ravanbakhsh et al. 2017; Saikia et al. 2018; Ghosh et al. 2018).
The acdS gene containing bacterium Variovorax paradoxus strain 5C-2 showed enhanced growth and photosynthesis in pea plants under 130 mM NaCl treatment (Wang et al. 2016). Moreover, this ACC deaminase-producing (containing acdS gene) bacterial isolates can confer high amounts of antioxidants such as SOD and CAT in plants which play a significant protective role against the deleterious effect of ROS, generated under SS conditions (Habib et al. 2016). The effect of acdS+ strain on plant growth under abiotic stress has been also reported by various researchers (Cheng et al. 2007; Singh et al. 2015; Li et al. 2019; Gupta et al. 2021b). The ACCD encoding the acdS gene is highly conserved among various micro-organisms and therefore, can be used to evaluate the diversity and phylogeny among closely relate acdS+ isolates (Bouffaud et al. 2018). Although horizontal gene transfer (HGT) of acdS+ isolates was suspected due to in-congruence between 16S rRNA and acdS gene-based on bacterial phylogenies (Nascimento et al. 2014).To elucidate the exact mechanism of action, these bacteria employ in protecting plants, it is essential to understand the alteration in the biochemical profile as well as the expression of the stress-related genes in plants. Pea (Pisum sativum) plants are very sensitive towards salinity stress especially in the early stage of development thereby affecting their productivity and production (Barnawal et al. 2014; Gupta et al. 2021a).
In our study, the acdS+ isolate possessed an acdS gene also have the ability to produce the ACCD enzyme throughout Bacillus isolates. Henceforth, the present study aims to reveal the beneficiary effect of ACCD producing bacteria on Pea plants under salt stress by studying the physiological as well as molecular aspects of the expression of some key protective genes in Pea.
Materials and methods
Bacterial strains and culture conditions
Rhizospheric micro-organisms were isolated from soil of leguminous crops collected from different regions of North India such as Uttar Pradesh (Lucknow-26.85° N,80.95° E, Barabanki-26.99° N, 81.25° E, Bareilly-28.37° N, 79.43° E, Allahabad-25.44° N, 81.85° E, Moradabad-28.84° N, 78.77° E, Meerut-28.98° N, 77.71° E, Shahjahanpur-27.88° N, 79.91° E, Badaun-28.03° N, 79.12° E, Sitapur-27.58° N, 80.67° E, Basti-26.82° N, 82.76° E, Gorakhpur-26.73° N, 83.35° E, Varanasi-25.32° N, 82.97° E, Kushinagar-26.74° N, 83.89° E); Bihar (Patna-25.59° N, 85.14° E, Gaya-24.80° N, 84.99° E); and Jharkhand (Ranchi-23.34° N, 85.31° E, Hazaribag-23.99° N, 85.37° E, Chatra-24.21° N, 84.87° E)and cultured on nutrient-enriched Rhizobium minimal media (RMM) containing ACC as a sole supply of nitrogen and its consumption analysis was performed (Penrose and Glick 2003). ACCD positive bacterial strains were selected, lyophilized in 10% glycerol stock and preserved in the culture collections of the IIRC-3, Plant–Microbe Interaction and Molecular Immunology Laboratory, Department of Biosciences, Integral University, Lucknow, Uttar Pradesh, India. These bacterial strains have been earlier screened using 16S rRNA gene sequencing and sequences were submitted to NCBI database (NCBI accession no. given in Supplementary Table S2).
ACC deaminase (ACCD) assay and PCR amplification of acdS gene
The ACCD activity of selected bacterial isolates was evaluated based on the capability of the particular isolate to utilize ACC as a sole source of nitrogen (Glick et al. 1995). Selected strains of bacteria were cultured in 15 ml of nutrient broth (NB) medium at 28 ± 2 °C for 24 h. Centrifuge the bacterial cultures at 11,000 RPM for 5 min followed by pellet washing twice in 1 ml of normal saline solution. After that, spot inoculation was done on Burks’ media supplemented with 3 mM ACC (Sigma-Aldrich, United States) as the sole nitrogen source. ACC-free media without and with 0.2% Ammonium Sulfate ((NH4)2SO4 were used as a negative and positive control, respectively. The cultures were further incubated for 7 days at 28 ± 2 °C and the growth of these bacterial strains on ACC-supplemented media was compared with the positive and negative controls (Ali et al. 2014b).Quantitative estimation of ACCD activity was carried out by measuring the production of α-ketobutyrate and ammonia as a by-product generated by the break-down of ACC through the ACCD enzyme (Penrose and Glick 2003).
PCR amplification of the acdS gene was carried out in positive and negative bacterial isolates using gene-specific primers (GSP) to produce ∼ 800 base pairs (bp) of amplicon according to the preceding report (Jha et al. 2012). To amplify acdS gene primers were synthesized from conserved regions of a sequence of acdS+ isolates using Primer Express software v2.0 (Applied Biosystems, United States) along with two universal primers of acdS7 for subsequent analysis (Primers sequences used for PCR are mentioned in (Supplementary Table S1).
PCR reaction mixture was prepared in a volume of 100 µl reaction mixture containing 60 ng genomic DNA as a template, 400 ng each of forward and reverse primers of universal primers, 2.5 mM of each dNTPs (Genei, Banglore, India), 10 μl of 10× Taq DNA polymerase Assay buffer added with 1.5 mM MgCl2 and 0.5 μl of 3U/µl Taq DNA polymerase enzyme (Genei, Banglore, India). The reaction conditions of PCR encompassed an initial denaturation step at 95 °C for 5 min, 35 amplification cycles of denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s and extension at 72 °C for 45 s, followed by a final extension at 72 °C for 5 min. The PCR reaction was carried out in a thermal cycler (T100, BioRad, USA). The amplified products were further analyzed by gel electrophoresis and visualized using a gel documentation system (Biorad, USA). The amplicon of PCR product of acdS gene was purified using a PCR purification kit (Wizard® SV Gel and PCR clean-Up System) and sequenced (Applied Biosystems, Bangalore, India). The identification of acdS gene has been confirmed using the BLAST algorithm available at https://blast.ncbi.nlm.nih.gov/Blast.cgi and the sequence was submitted to the NCBI database. An analysis of phylogenetic relationships was carried out using MEGA 7.0 (Kumar et al. 2016b) and sequence alignment was carried out using Clustal Omega online tool.
Salt sensitivity assay of PGPR
Salt tolerance (ST) assay of bacterial isolates was performed by growing on NB medium amended with 0%, 1%, 2%, 3% and 5% of NaCl (w/v) at 28 ± 2 °C for 20 h with 150 RPM in incubator shaker (Remi CIS-24 Plus, India). The quantification of bacterial growth was analyzed by measuring optical density (O.D.) at 600 nm using UV–Vis spectrophotometer (Eppendorf, Germany). All the bacterial cultures were inoculated in triplicate.
Effect of acdS positive PGPR on plant growth promotion under salinity stress condition
Bacterial inoculums were prepared by centrifuging overnight-grown bacteria at 11,000 rpm for 10 min to harvest them. For bacterial inoculum preparation, overnight (O/N) grown bacterial cells were harvested by centrifugation at 11,000 rpm for 10 min followed by pellet washing twice with 50 mM phosphate saline buffer followed by pellet washing twice with 50 mM phosphate saline buffer. Pellets were then re-suspended in double-distilled water (ddw) at a ratio of 1:1to maintain the uniform cell density of 1 × 108colony forming unit (CFU) ml−1.
For evaluating the effect of bacterial isolates on plant growth promotion, 150 seeds of Pisum sativum were surface sterilized using 0.1% of HgCl2 for 3 min followed by washing with sterilized water 3 times. Seed bacterization was carried out as described by (Nautiyal 1997) using suspension culture already prepared from selected PGPR strains namely CHR JH 203 and BST YS1_42. Thereafter, the seeds were air-dried for 20 min under sterile conditions and then sown in sterile thermocoal pots of 6.95 cm × 6.3 cm × 5.2 cm size filled with garden soil collected from Integral University, Lucknow, India (latitude/longitude 26.9585° N, 80.9992° E) to perform greenhouse experiments.
Greenhouse experiment
Seeds inoculated with PGPR strains CHRJH 203 and BST YS1_42 along with uninoculated control were grown for up to 30 days in greenhouse conditions. Five pots of each control and PGPR inoculated were taken for the salt stress treatment. The SS was artificially induced by adding 1% NaCl to the soil w/v followed by irrigating pot with 1% NaCl containing water on an interval of 15 days. Control seedlings were irrigated with normal tap water alternate days. The plants were grown for up to 30 days after sowing (DAS) thereafter harvested to determine the root length, shoot length, root, shoot fresh and dry biomass parameters. The harvested plants were further studied for any changes in biochemical parameters.
Measurement of total sugar, reducing sugar and protein contents
Total 100 mg of shoot sample from each treated and non-treated seedlings were homogenized in 1 ml of a 50 mM ice-chilled phosphate buffer (pH 8.0) using pre-chilled mortar and pestle. The homogenate was then centrifuged at 15,000×g for 12 min at 4 °C. Enzyme assays were conducted using the supernatant. The total sugars content was estimated using the method described by Dubois et al. (1951) and the reducing sugars content was estimated using Somogy's method as modified by (Nelson 1944). Glucose was used as a standard. The protein concentration was determined in line with (Lowry et al. 1994) using bovine serum albumin (BSA) as standard.
Measurement of photosynthetic pigments (chlorophyll a, chlorophyll b, total chlorophyll and carotenoids), proline, phenol and flavonoid contents
According to Arnon (1949), chlorophyll content was estimated in the leaves. Proline content was measured using ninhydrin (Bates et al. 1973). The standard curve was prepared using l-proline (Sigma-Aldrich, USA) for comparison. The total phenolic content in plant samples was determined by the Folin–Ciocalteu method given by (McDonald et al. 2001) using gallic acid as a standard. The quantitative estimation of flavonoid contents was accomplished spectrophotometrically by means of aluminium chloride method based on the formation of complex flavonoid-aluminium (Chang et al. 2002) using quercetin as a standard.
Estimation of antioxidant enzymatic activities
Estimation of antioxidants enzymatic activities of leaf samples were performed using established standardized protocols. A method described by (Giannopolitis and Ries 1977) for measuring the activity of Superoxide dismutase (SOD). Catalase (CAT) activity was assessed by monitoring the disappearance of H2O2 at 240 nm after adding enzyme extract to the reaction mixture (Aebi 1984). Peroxidase (POX) assay was determined by (Chance and Maehly 1955). The activity of Ascorbate peroxidase (APX) was estimated at 290 nm using the method described by (Nakano and Asada 1981). As ascorbate is oxidized, the absorbance decreases at 290 nm, indicating the activity of APX.
RNA extraction and quantification
Pea shoot samples were harvested in liquid nitrogen and used for RNA extraction using TRIZOL reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The quality and quantity of RNA samples were analyzed by agarose gel electrophoresis and Nanodrop One Spectrophotometer (Thermo Fisher Scientific USA). Good quality total RNA samples having 260/280 ratio between 1.8 and 1.9, A260/A230 ratio between 2.0 and 2.4 were considered for cDNA synthesis. RNA was extracted from each sample in their biological triplicates.
Preparation of cDNA and quantitative real-time PCR (qRT-PCR) analysis
The cDNA was synthesized from 1 µg of total RNA. The reaction was prepared by adding oligo (dT) 18 primer (100 µM), 5× reaction buffer with MgCl2 (250 Mm Tris–HCl (pH 8.3), 250 Mm KCl; 20 mM MgCl2), RiboLock RNase Inhibitor (20 U/µL), 10 mM dNTP mix, Revert Aid M-MuLV RT (200 U/µL) and nuclease-free water was added to an RNase-free microfuge tube to make the final volume of 20 µL. The reaction mixture was incubated for 60 min at 42 °C followed by incubation for 5 min at 70 °C. After that, cDNA was stored at − 20 °C for further use.
For expressional studies, the sequences of primers were retrieved from NCBI Genbank, and then the primers were designed using Primer 3 Plus software (Roy 2019) and listed in Supplementary Table S2.
Gene expressional analysis
Quantitative real-time PCR (qRT-PCR) analysis was carried out to measure mRNA transcripts accumulation of antioxidant genes such PsSOD, PsPOX, PsAPX, PsNOS, PsCAT and PsChla/bBP) defense and cell rescue gene (PsPRP, PsMAPK and, PsFDH) mediating the salt tolerance in pea. Nuclease-free water was used for the negative control, asa no-template control reaction. 15 ng of cDNA sample was used as a template for the relative gene expression analysis. The reaction conditions were set for cDNA amplification at 94 °C for 3 min, 35 cycles of 94 °C for 30 s, Tm ± 5 °C for 30 s, 72 °C for 45 s and 72 °C for 15 min. Actin gene of Pisum sativum was used as the internal control for normalization with primer sequences forward as 5′-CCTTTCAGAGGGAACAACCA-3′ and reverse as 5′-GTGCACAATTGATGGACCAG-3′ (PsActin Gene bank Accession number: U81049). Fold-change expression of transcripts accumulation in pea samples was calculated using the standard 2-∆∆CT method according to Livak and Schmittgen (2001). Patterns of transcripts expression obtained from qPCR analysis were plotted in Microsoft Excel 2010.
Statistical analysis
A minimum of three biological replicates were used for the experiments and each data point shown in the results was the mean of these replicates. A standard error of the mean is represented by the error bars (± SEM). Statistical evaluation of all the means was performed using two-way ANOVA analysis and statistically meaningful data were compared using "Multiple Comparison Test" performed using GraphPad Software (GraphPad In-Stat version 5.00, San Diego, CA, USA). Different letters in graphs indicate significant differences between treatments (P < 0.05) while the same values indicate the non-significant between the samples.
Results
ACCD activity of selected isolates
ACC deaminase activity is responsible for plant growth promotion by means of direct way. Therefore, the ACCD activity of acdS+ isolates was quantified by determining the α-ketobutyrate production through deamination reaction using ACC as a nitrogen source in DF (Dworkin2 and Foster 1958) minimal salt broth media at 540 nm. All the twenty-five isolates showed variation in their ACCD activity that ranges between 0.016 ± 0.001 and 0.395 ± 0.003 mmol mg−1 h−1 ACC concentration (Table 1). Among them, the highest ACCD activity 0.395 ± 0.003 mg−1 h−1 was found in Bacillus marisflavi CHR JH 203 strain followed by 0.368 ± 0.001 mg−1 h−1 in Bacillus cereus sp. strain BST YS1_42 and 0.360 ± 0.009 mg−1 h−1in Bacillus subtilis strain BLY 71 (Table 1). ACC deaminase activity was performed in each ACC-utilizing bacterial isolates which were measured by the 2,4-dinitrophenylhydrazine assay after induction in the minimal medium with ACC as a source of Nitrogen.
Table 1.
ACC deaminase activity assay of ACC-utilizing bacterial isolates
| S. no | Bacterial isolates | Genus affiliation | 16 s rRNA gene sequence Accession No | ACC deaminase activity (mmol mg−1 h−1) |
|---|---|---|---|---|
| 1 | ALD 33 | Pseudomonas pseudoalcaligenes | KT429582.1 | 0.016 ± 0.001 |
| 2 | BBK 2 | Pseudomonas monteilii | KT429580.1 | 0.049 ± 0.002 |
| 3 | BBK 30 | Pseudomonas mendocina | KT429581.1 | 0.018 ± 0.003 |
| 4 | BLY 48 | Bacillus cereus | KT429584.1 | 0.038 ± 0.002 |
| 5 | BLY 71 | Bacillus subtilis | KT429585.1 | 0.360 ± 0.009 |
| 6 | BME ES1_65 | Bacillus cereus strain | KT429590.1 | 0.264 ± 0.002 |
| 7 | BSB KS2_34 | Bacillus subtilis | KT429598.1 | 0.041 ± 0.001 |
| 8 | BSB KS4_1 | Bacillus cereus | KT429599.1 | 0.338 ± 0.002 |
| 9 | BST YS1_42 | Bacillus cereus | KT429601.1 | 0.368 ± 0.001 |
| 10 | CHR JH 203 | Bacillus marisflavi | KT429593.1 | 0.395 ± 0.003 |
| 11 | CHR JH_206 | Bacillus anthracis | KT751334.1 | 0.262 ± 0.009 |
| 12 | GAY P130 | Pseudomonas moraviensis | KT751336.1 | 0.078 ± 0.001 |
| 13 | GKP KS2_14 | Bacillus megaterium | KT429596.1 | 0.318 ± 0.002 |
| 14 | GKP KS2_7 | Pseudomonas aeruginosa | KT429595.1 | 0.344 ± 0.003 |
| 15 | HTG JH204 | Bacillus amyloliquefaciens | KT751335.1 | 0.102 ± 0.002 |
| 16 | IU-UP-BST-YS1 43 | Bacillus cereus | KT751337.1 | 0.041 ± 0.002 |
| 17 | LKO 36 | Bacillus brenneri | KT751326.1 | 0.011 ± 0.001 |
| 18 | LKO 45 | Bacillus cereus | KT429583.1 | 0.024 ± 0.002 |
| 19 | MBD 133 | Bacillus subtilis | KT429586.1 | 0.341 ± 0.001 |
| 20 | MUT 223 | Bacillus safensis | KT751329.1 | 0.301 ± 0.003 |
| 21 | PTN Br7 | Bacillus thuringiensis | KT429588.1 | 0.217 ± 0.002 |
| 22 | RCH JH_1 | Bacillus altitudinis | KT429592.1 | 0.341 ± 0.002 |
| 23 | HTGJH5 | Bacillus cereus | KT429594.1 | 0.295 ± 0.004 |
| 24 | SPN ES4_60 | Pseudomonas sp.strain | KT429591.1 | 0.023 ± 0.003 |
| 25 | VI28_13_NF_E02 | Bacillus stratosphericus | KF717365.1 | 0.044 ± 0.002 |
Gene amplification and sequencing analysis
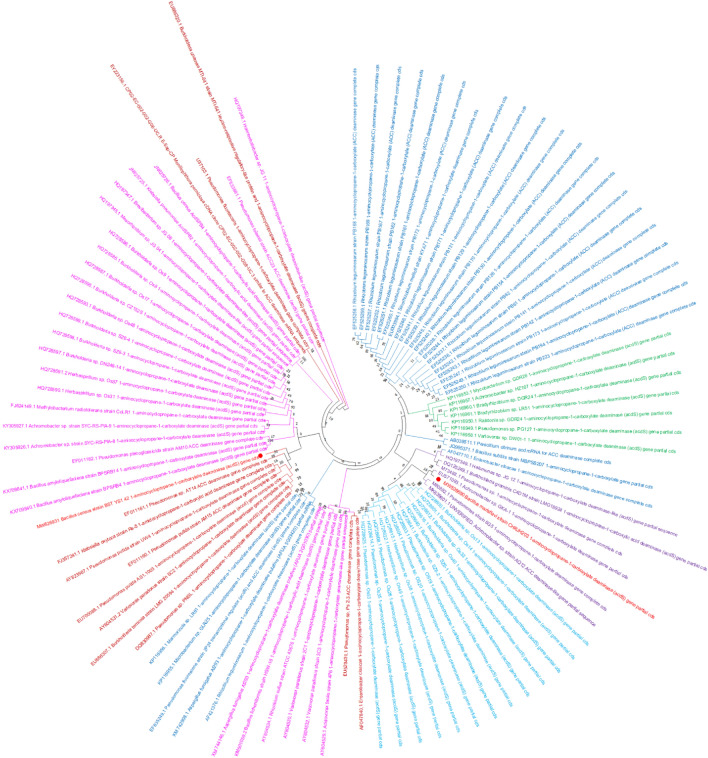
All the selected 25 isolates were able to degrade ACC and thereby utilizing nitrogen in the selective media in which two rhizospheric strains CHRJH 203 and BST YS1_42 showed the highest ACCD activity (Table 1). Further confirmation of the strains was conducted via PCR using bacterial gDNA as a template for amplification of the acdS gene. For this, we developed the primer sets for acdS gene amplification. All four primer sets were further discarded as they were unable to amplify the acdS gene from pure bacterial genomic DNA sample, as they give non-specific amplification (Supplementary Table S1). PCR results produced the acdS amplicon of about 800 bp in both CHR JH 203 and BST YS1_42 strains using the universal set of primer of acdS7 (Fig. 1b), and it was also confirmed by Sanger sequencing of amplified product. Sequencing results were BLASTed with genomic DNA sequences which resulted that both the isolates belonged to the Bacilli class and are closely related to Acidovorax, Bacillus, Achromobacter, Pseudomonas, Variovorax, Klebsiella, and Burkholderia species (Fig. 2, Table 2).
Fig. 1.
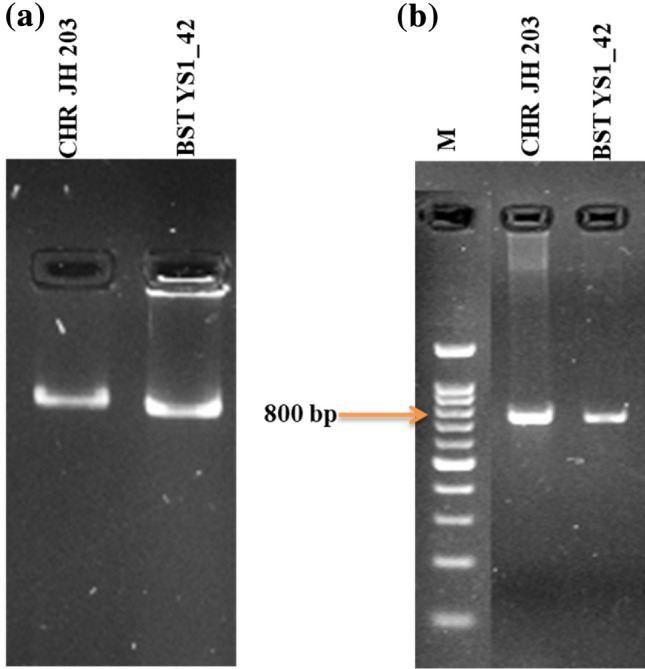
PCR amplification of acdS gene in CHRJH 203—Bacillus marisflavi and BST YS1_42—Bacillus cereus strains from gDNA using a universal set of acdS7primers. a genomic DNA of two bacterial isolates CHRJH 203—Bacillus marisflavi and BST YS1_42- Bacillus cereus strains; b PCR amplicon of acdS gene sizes ~ 800 bp. M: 1 kb ladder (G-biosciences, USA)
Fig. 2.
Phylogenetic tree analysis based on acdS gene sequences of Bacillus marisflavi CHRJH203and Bacillus cereus BST YS1_42 strains was constructed using neighbor-joining method between 98 acdS+ isolates by MEGA 7 program. The sequence of Bacillus marisflavi CHRJH203showed 80% nucleotide sequence similarity to Achromobacter sp. GK-1strain (Genbank accession number EU571093.1) while Bacillus cereus BST YS1_42 showed 86% nucleotide sequence similarity with Variovorax paradoxus strain 5C2, Pseudomonas sp. PNSL, Pseudomonas putida strain AS1.1003, Pseudomonas putida strain AM15, Pseudomonas putida strain UW4, Klebsiella oxytoca strain Rs-5 and Burkholderia terricola strain LMG 20594 (Genbank accession number AY604531.2, DQ830987.1, EU700088.1, EF011160.1, AY823987.1, FJ357241.1, and EU886307.1). The bootstrap values (≥50%) are concluded from 1000 replicates and shown at branch nodes
Table 2.
Universal acdS primers used to amplify acdS gene
| Primer pairs | Amplicon length (bp) | Troubleshoot on tested acdS+ isolates |
|---|---|---|
| acdS1* (F + R) | 856 bp | Not specific |
| acdS2* (F + R) | 808 bp | Non-specific |
| acdS3* (F + R) | 856 bp | Negative |
| acdS4* (F + R) | 1000 bp | Not universal (4 tested acdS+ isolate never amplified) |
| acdS5 (F + R) | 650 bp | Non-specific amplification (with 5 isolates) |
| acdS6 (F + R) | 1017 bp | Non-specific amplification (tested with 10 acdS + isolates) |
| acdS7 (F + R) | 800 bp | Positive (with four acdS+ isolates) |
The acdS gene was PCR amplified using acdS7 (F+R) set of primers which was used for further study—highlighted in bold
*Tested bacterial strains were Bacillus subtilis MBD 133, Pseudomonas pseudoalcaligenes ALD 33, Bacillus subtilis BLY 71, Bacillus cereus strain BME ES1_65, Bacillus cereus BST YS1_42, Bacillus marisflavi CHR JH 203, Pseudomonas moraviensis GAY P130, Pseudomonas aeruginosa GKP KS2_7, Bacillus cereus HTG JH5, and Pseudomonas sp. strainSPN ES4_60
Salt tolerant assay and determination of maximum tolerance level
Based on the ACCD activity and salt-tolerant capability, two isolates were further selected to evaluate the salt-tolerant assay. Bacillus cereus BST YS1_42 and Bacillus marisflavi CHR JH 203 strains were grew in the Nutrient Broth (NB) media supplemented with 0%, 1%, 2%, 3%, and 5% NaCl (w/v). Results show that these isolates were found decreasing their CFU counts by increasing the salt concentration beyond 3% NaCl (Supplementary Figure S1). Thus, both strains have the ability to grow up to 3% NaCl and showed salt-tolerant ability. Therefore these isolates have the maximum salt-tolerant ability of 3% NaCl (w/v).
Plant growth analysis and greenhouse experiment
Growth assessment of Pisum sativum plants was carried out in pots under greenhouse conditions to evaluate the effect of two PGPR; CHRJH 203—Bacillus marisflavi; BST YS1_42 Bacillus cereus. The retarded growth was observed in the plants treated only with NaCl, compared to PGPR inoculated NaCl treated plants (Fig. 3, Table 3). The beneficial effects of these acdS+ PGPR isolates on Pisum sativum are investigated under saline and non-saline conditions through pot trials (greenhouse experiment) (Fig. 3). Measurement of plant biomass was done 25 days after sowing (DAS). The plants were harvested to determining the length and dry weights of the root and shoot (Table 3).
Fig. 3.
Effect of PGPR isolates on the growth of Pisum sativum plants under normal and 1% NaCl induced salinity stress on 25 days after sowing (DAS). a CT- control without PGPR inoculants and salt; CHR JH 203—Bacillus marisflavi; CHR JH 203+S-PGPR+NaCl; S—1% NaCl, b CT- control without inoculants and salt; BST YS1_42- Bacillus cereus bacterial treated; BST YS1_42+S-PGPR+NaCl; S—1% NaCl
Table 3.
Effect of ACCD-producing PGPR isolates on the pea growth under NaCl-induced salt stress conditions
| S. no | Treatments | Shoot FW (gm) | Root FW (gm) | Shoot DW (gm) | Root DW (gm) | Shoot length (cm) | Root length (cm) |
|---|---|---|---|---|---|---|---|
| 1 | CT | 0.77 ± 0.04b | 0.44 ± 0.04c | 0.071 ± 0.005c | 0.057 ± 0.003a | 9.98 ± 0.27b | 6.67 ± 0.33b |
| 2 | CHR JH 203 | 1.12 ± 0.1a | 0.73 ± 0.03a | 0.135 ± 0.010a | 0.067 ± 0.004ab | 13.17 ± 0.49a | 10.16 ± 0.38a |
| 3 | BST YS1_42 | 1.43 ± 0.05a | 0.62 ± 0.04ac | 0.118 ± 0.008ab | 0.061 ± 0.006abc | 10.70 ± 0.51bc | 9.17 ± 0.60a |
| 4 | S | 0.51 ± 0.02b | 0.31 ± 0.05c | 0.044 ± 0.005 cd | 0.041 ± 0.002d | 3.067 ± 0.29d | 4.40 ± 0.37b |
| 5 | CHR JH 203 + S | 0.65 ± 0.08b | 0.48 ± 0.04bc | 0.074 ± 0.005 cd | 0.069 ± 0.006abc | 7.43 ± 0.56e | 9.33 ± 0.50a |
| 6 | BST YS1_42 + S | 0.75 ± 0.07b | 0.65 ± 0.05ab | 0.090 ± 0.009bc | 0.052 ± 0.003 cd | 10.13 ± 0.46bc | 11.14 ± 0.46a |
Values are mean of five replicates ± standard error of the mean. CT Control; CHRJH 203- Bacillus marisflavi; BST YS1_42- Bacillus cereus; salt-stressed: S-1% NaCl;—CHRJH 203 + S and BST YS1_42 + S- PGPR + salt
Differences in letters between treatments indicate statistical significance (Two-way ANOVA, multiple comparison test P < 0.05)
*Data of root and shoot length was measured on 25 days of sowing (DAS)
Effect of PGPR on morphological parameters of Pisum sativum under NaCl induced salt stress condition
Exposure of NaCl stress on Pisum sativum led to a considerable decrease in plant growth and its biomass such as root length, shoot length, fresh weight, dry weight of root and shoot as compared to non-stressed control (Table 3). Results demonstrated that application of acdS+ PGPR strains B. marisflavi CHR JH 203 and B. cereus BST YS1_42 significantly (P < 0.05) increased root and shoot growth of Pisum sativum when compared to non-inoculated plants. The pots trial results indicated the beneficiary effect of these bacterial inoculants on plant growth promotion against control and under saline conditions. However, these acdS+ inoculated plants showed better results and tolerated the NaCl-induced stress significantly when compared to the control. In addition, the highest fresh weight and length (root and shoot) were observed in both Bacillus sp. (CHR JH 203 and BST YS1_42) under stress conditions (Table 3).
Effect on biochemical parameters
Carbohydrate, reducing sugar and protein
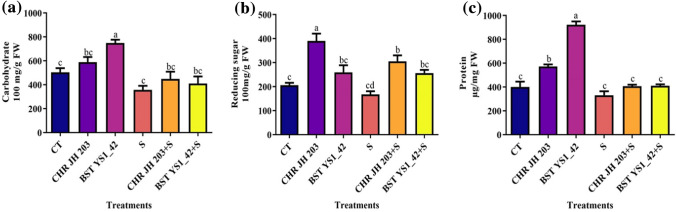
To examine the effect of PGPR inoculation on plant growth, the biochemical parameters such as carbohydrate, reducing sugar and protein contents were assayed. The analysis showed that NaCl-induced salinity stress is ameliorated when plants were treated with CHR JH 203 and BST YS1_42 strains, compared to control (normal soil) conditions thereby improved the plant growth (Fig. 4).
Fig. 4.
Effect of bacterial isolates on biochemical parameters under NaCl induced salinity stress condition in pea. a Total carbohydrate content (mg g−1 FW); b Total reducing sugar content (mg g−1 FW) and c Total protein content (µg-1 mg FW). Values are mean of five replicates ± standard error of the mean. Normal soil: CT- Control; CHRJH 203- Bacillus marisflavi; BST YS1_42- Bacillus cereus; Salt stressed: Salt stressed: S-1% NaCl; CHR JH 203+S and BST YS1_42+S-PGPR+NaCl. Differences in letters between treatments indicate statistical significance (Two-way ANOVA, multiple comparison test P < 0.05). The similar letter represents no significance while the different letter indicates the significance between them
Changes in photosynthetic pigments (chlorophyll a, chlorophyll b, total chlorophyll and carotenoids) under salt stress conditions
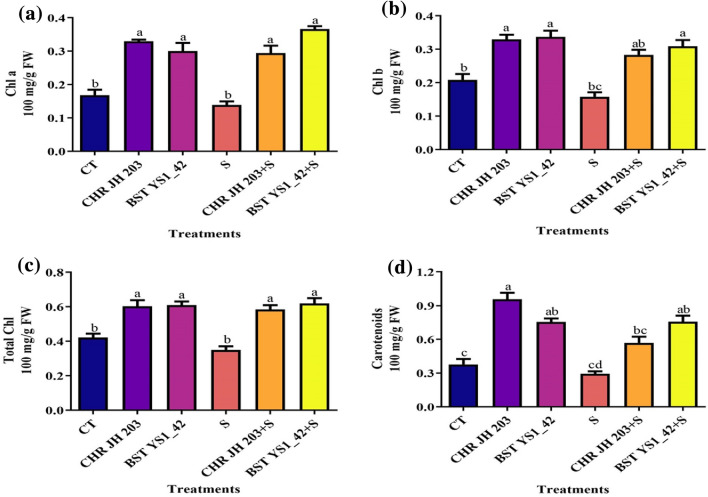
Bacillus cereus BST YS1_42 and Bacillus marisflavi CHR JH 203 inoculated plant showed significantly induced the biosynthesis of photosynthetic pigments such as chlorophyll a, chlorophyll b and total chlorophyll and carotenoids contents (Fig. 5). Under NaCl-induced stress conditions, Bacillus cereus BST YS1_42 strain inoculated pea plants significantly increases the chlorophylls and carotenoid content compared to non-inoculated plants. On the other hand, stress induced by 1% saline led to a decline in photosynthetic pigments compared to non-stressed control (Fig. 5).
Fig. 5.
Effect of ACCD-producing bacterial isolates on the physiology parameters of pea plants. a chlorophyll a, b chlorophyll b, c total chlorophyll contents, and d carotenoids content. Normal soil used as control: CT; CHR JH203 and BST YS1_42-acdS+ bacterial isolates; salt-stressed: S-1% NaCl; CHR JH203+S and BST YS1_42+S-PGPR+salt. Values are mean of five replicates ± standard error of the mean (SEM). Differences in letters between treatments indicate statistical significance (Two-way ANOVA, multiple comparison test P < 0.05). A similar letter represents no significant different letter indicates the significance between them
Modulations in flavonoids, phenol and proline contents
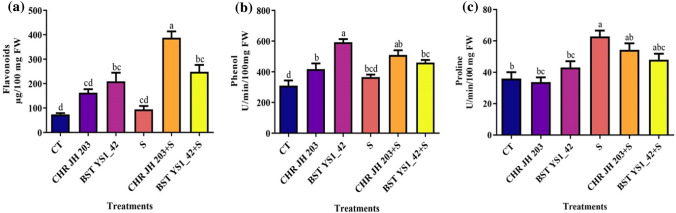
The non-enzymatic antioxidants such as flavonoids and phenolic content were observed to be higher in CHR JH 203 and BST YS1_42 inoculated plants. NaCl led to an increase in the flavonoids, phenol, and proline contents when compared to control. However acdS+ inoculants have further enhanced the flavonoids, and phenol content relative to non-inoculated control. But proline content did not increased on bacterial inoculation. Phenol and proline are other bio-markers of plants under salinity stress. The increases in phenolic and proline accumulation are extremely crucial for maintaining the osmotic-potential of plant tissues and thereby, protecting the plants from induced salinity stress. Inoculation ofacdS+ bacteria directly affected plant osmolytes (Fig. 6). The Proline content was observed to be higher when plants were inoculated with BST YS1_42 under normal soil conditions while in salt-stressed conditions there was a general reduction in proline content (Fig. 6). The results indicated that both the PGPR strains prevent plants from oxidative damage caused because of NaCl-induced salt stress.
Fig. 6.
Effect of PGPR on non-enzymatic antioxidant activities in pea under salt stress. a Flavonoids content, b phenols content and c proline contents in Pea plants. CT Control without bacterial treatment; plants inoculated with CHR JH 203 and BST YS1_42—ACCD producing bacterial strains; S—1% NaCl concentration; CHR JH 203+Sand BST YS1_42+S—PGPR+NaCl. Values are the mean of five replicates ± standard error of mean. Differences in letters between treatments indicate statistical significance (Two-way ANOVA, multiple comparison test P < 0.05). A similar letter represents no significance between the samples
Antioxidants enzymatic activities assay
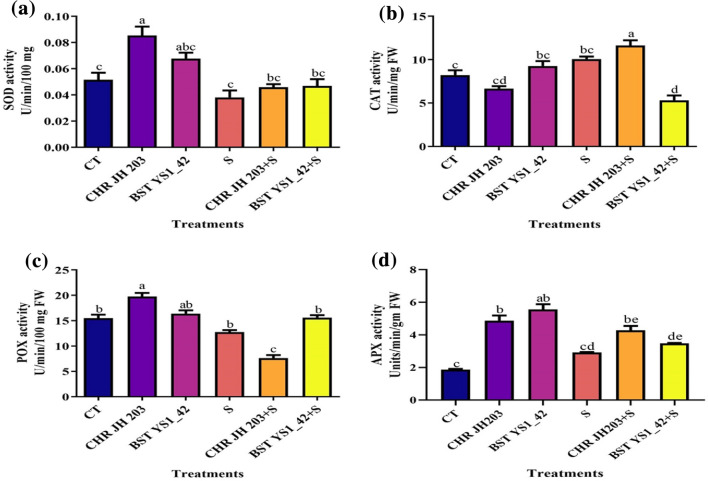
Assay of antioxidants enzymatic activities such as SOD, CAT, POX and APX was done on leaf samples. Superoxide dismutase activity was found to be increased in CHR JH 203 treated pea plants compared to control in normal soil conditions, whereas SOD was increased in both CHR JH 203 and BST YS1_42 treated plants under 1% saline condition compared to non-stressed plants (Fig. 7). Catalase activity was increased in BST YS1_42 inoculated plants compared to control under normal conditions, however, under saline stress conditions CAT activity was found enhanced which substantiates that CAT prevents the plant from oxidative stress. Inoculation of CHR JH 203 led to an increase in the CAT production under saline conditions thereby prevents the plant from stress conditions (Fig. 7). Peroxidase content was elevated in CHRJH 203 and BST YS1_42 inoculated pea plants in normal soil whereas under saline conditions, only BST YS1_42 increases the POX content. Ascorbate peroxidase content was found to be elevated in CHR JH 203 and BST YS1_42 treated pea plants under saline and non-saline conditions (Fig. 7).
Fig. 7.
Antioxidants enzymatic activities in pea under salinity stress conditions. a superoxide dismutase (SOD) activity; b catalase (CAT) activity, c peroxidases (POX) activity; and d Ascorbate peroxidase (APX) activity. Pea plants are grown in a greenhouse for four weeks in pots filled with garden soil and treated with NaCl-induced salt stress. CT Control without bacterial treatment; plants inoculated with CHRJH 203andBST YS1_42—bacterial strains; S—1% NaCl concentration; CHRJH 203+Sand BST YS1_42+S—PGPR+NaCl. Each value is a mean of five replicates ± SEM. Differences in letters between treatments indicate statistical significance (Two-way ANOVA, multiple comparison test P < 0.05). A similar letter represents no significance
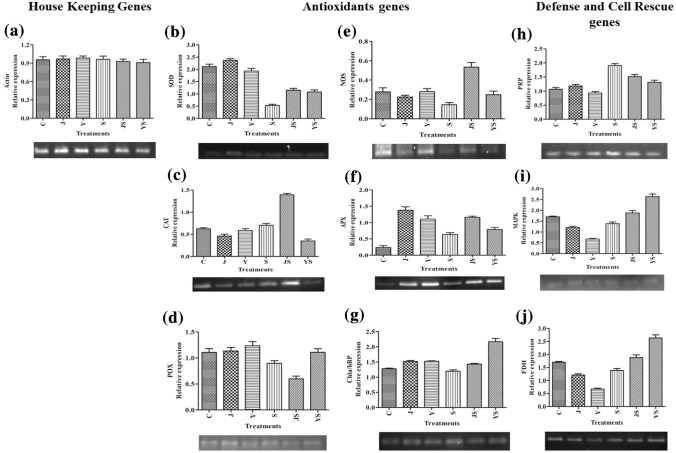
Expression levels of antioxidant genes conferring tolerance towards salinity
The expression analyses of defense-related genes were performed using cDNA synthesized from RNA extracted from salt-stressed pea seedlings. Gene expression patterns of PsSOD, PsCAT, PsPOX, PsNOS, PsAPX, PsChl a/b binding protein (Chla/bBP), PsPRP, PsMAPK and PsFDH genes were analyzed using semi-quantitative reverse transcriptase PCR (RT-PCR). The expression pattern of all the defense-related genes under salinity stress was found modulated compared to non-stressed control and PGPR inoculated plants. PGPR treated samples showed higher transcript accumulation of these antioxidants-related genes compared to inoculated and un-inoculated salt-treated plants. Under saline conditions, pea samples inoculated with Bacillus marisflavi CHR JH 203 and Bacillus cereus strain BST YS1_42 showed upregulation of relative gene expression level of PsSOD, PsAPX, PsNOS, PsMAPK, PsChla/bBP, and PsFDH compared to non-inoculated saline-treated control plants. The expression level of PsCAT gene was up-regulated by CHR JH 203 inoculated samples while BST YS1_42 strain increases the PsPOX expression under salt stress conditions. Other antioxidants and defense and cell rescue genes were also up-regulated under NaCl treated samples which substantiates that the pea plant itself tries to protect the cellular membrane from oxidative stress caused due to increased saline condition (Fig. 8). Bacillus cereus BST YS1_42 inoculated pea samples also showed up-regulation of the defense and cell rescue genes compared to samples grown under salt stress conditions. A significant reduction in proline accumulation in PGPR treated plants when compared to salt-treated plants showing the PGPR sensitivity towards salinity stress in the proline pathway. The acdS+ PGPR-inoculated plants beneath salinity stress recorded a 1.1–2.7 fold increase in the gene expression is observed compared to uninoculated non-stressed crops (Fig. 8).
Fig. 8.
Semi-quantitative reverse transcriptase PCR (RT-PCR) analysis of defense responsive genes of pea under salinity stress. The expressional analysis of pea a actin (PsActin), b superoxide dismutase (PsSOD), c catalase (PsCAT), d peroxidase (PsPOX), e nitric synthase (PsNOS), f ascorbate peroxidase (PsAPX), g chlorophyll a/b binding protein (PsChla/bBP), h proline-rich protein (PsPRP), i mitogen-activated protein kinases (PsMAPK), and j fiddlehead protein (PsFDH) on 15 days old pea seedlings were analyzed. The actin gene was taken as a control for normalization. RNA extracted from pea seedlings inoculated with CHR JH 203 and BST YS1_42 strains under both non-saline (CT) and saline conditions (1% NaCl) was used to synthesize cDNA for expression analysis. C Control; CHR JH 203 and BST YS1_42- acdS+ isolate; S-1% NaCl; CHR JH 203+Sand BST YS1_42+S- PGPR+NaCl. Bar graphs depicted the qRT-PCR whereas semi-quantitative RT-PCR results are shown in gel under the relevant graphs. Values are mean of three replicates ± standard error of mean; error bars represented SEM.
Discussion
Pea is one of the major agricultural crops grown in India. Salinity stress widely affects the growth and yield of pea plants (Osman and rady 2015). This study also witnessed a marked reduction in pea germination under induced salinity conditions. Several reports showed that salinity stress adversely affected plant growth parameters such as length, biomass, nutrient uptake, or photosynthetic pigments due to alteration in various physiological, biochemical and molecular processes (Ahanger and Agarwal 2017). Certain acdS+ PGPR exhibit an increase in germination rate, and improve the seedling emergence that can have the capacity to strengthen the plants against various external stress factors (Lugtenberg et al. 2002). For example Mukhtar et al. (2020) showed the effect of ACC deaminase producing PGPR in Solanum lycopersicum. Islam et al. (2015) also demonstrated that Bacillus cereus promotes growth in Vigna radiata and confers salt tolerance. Therefore, we have targeted those bacteria that are ACCD positive and evaluate their effect on plant growth under salinity stress. The PGPR application improved seed germination and seedlings stand over the control as salinity tends to decrease it. Similarly, it was reported previously that PGPR improves seed germination in sorghum and pearl millet (Niranjan Raj et al. 2003), wheat (Noumavo et al. 2013), and also provide tolerance to drought stress in chickpea (Kumar et al. 2016a). There have been claims that some PGPR increase seed emergence up to 100% when applied to plants (Noumavo et al. 2013).
In this study, the two highest ACCD producing PGPR isolates viz. CHR JH 203 and BST YS1_42 were found implicated in plant growth promotion in normal soil conditions and also found to be effective in ameliorating the adverse effects of NaCl-induced salinity stress, consequently improving the growth of garden Pea plants. The molecular identity of the acdS gene was further confirmed as it shares high sequence similarities with its orthologs from the other acdS+ bacterial isolates. To further validate the presence of the acdS gene encoding ACCD enzyme, the PCR was carried out using the universal set of primers selected from a previous study (Jha et al. 2012), and amplification with these sets of primers resulted in the desired amplified product of ~ 800 bp in the selected isolates i.e. CHR JH 203 and BST YS1_42. The amplified product was further sequenced and compared with the acdS+ gene sequences available in NCBI database (Fig. 2). The results have further validated the variation in the nucleotide sequences among different acdS+ isolates of different species thereby confirming the horizontal gene transfer (Hontzeas et al. 2005; Blaha et al. 2006; Nascimento et al. 2014). The obtained sequences also show similarity with pre-existing partial as well as complete acdS sequences. The acdS+ isolates sequence of Bacillus marisflavi CHRJH 203 and Bacillus cereus BST YS1_42 was further submitted in the Genbank (Accession No. MW539690 and MW626931). The acdS gene of CHRJH 203 isolate consists of 701 bp whereas the isolate BST YS1_42 contains 621 base pairs. The sequence analysis of CHRJH 203 was further revealed that a large open reading frame (ORF1) starting with ATG codon at position 3 and showed a partial sequence while the BST YS1_42 revealed a large ORF3 starting with ATG at codon position 33 and terminating with a TAA codon at position 653. The large ORF of CHRJH 203 encodes for a polypeptide of 231 amino acids while that of BST YS1_42 encodes a polypeptide of 207 amino acids with the calculated molecular weight of approximately 25.16 kDa (CHRJH 203) and 23.5 kDa (BST YS1_42). The CHRJH 203 and BST YS1_42 alignment of the deduced amino acid sequences showed a maximum identity (99.6% and 97.6%) with the Acidovorax wautersii and minimum (28%) identity with Pseudomonas aeruginosa strain PAO1. The alignment also confirmed that all of the amino acid residues that are known to be important were conserved, and the B. marisflavi strain CHR JH 203 and B. cereus strain BST YS1_42 deaminase active sites were somewhat identical. A Phylogenetic tree of acdS+ isolates were constructed based on the nucleotide sequences in the NCBI database as illustrated in (Fig. 2). Expectedly, the isolate CHR JH 203 and BST YS1_42 were in different clusters and thereby confirms the identity of the sequences. Moreover, the bacteria Pseudomonas sp., Klebsiella oxytoca, Variovorax paradoxus, Burkholderia sp., and Achromobacter sp. are in one cluster and which indicates that these acdS genes might have been inherited through horizontal and vertical gene transfer which supported the earlier findings as proposed by (Hontzeas et al. 2005; Blaha et al. 2006; Nascimento et al. 2014). In conclusion, the present study revealed a bacterium containing ACCD production led to elongation of the root and shoot length in the Pisum sativum plant. The production of α-ketobutyrate, deduced amino acid and nucleotide sequences identity with known acdS gene (retrieved from NCBI) further confirmed the enzymatic activity in B. marisflavi strain CHR JH 203 and B. cereus strain BST YS1_42. Alignment of the deduced amino acid sequences also revealed that the amino acids are known to be conserved and are somewhat identical with their closely related species.
Salinity causes a reduction in plant growth as it tends to directly affect the biochemical profile of plants due to elevated ethylene levels. Ethylene is involved in various physiological responses (Noumavo et al. 2013) and is known as a stress hormone. It is synthesized at a rapid rate when the plant is under some stress (Stearns and Glick 2003) resulting in decreased seed germination and root development and eventually hindering plant growth (Saravanakumar and Samiyappan 2007). Salinity also led to increased levels of Reactive Oxygen Species (ROS) in plants which negatively influences cellular function and stimulates oxidative damage (Abdel Latef and Chaoxing, 2014; Merchante et al., 2013; Talaat, 2019). It can cause lipid peroxidation and chlorophyll degradation affecting photosynthesis, homeostasis and membrane permeability (Apel and Hirt 2004). The reduction in the photosynthetic rate leads to damage in chloroplast structure ultimately leading to cell death (Apel and Hirt 2004; Farooq et al. 2012; Tiwari et al. 2018). The decrease in chlorophyll content is the sign of a reduction in metabolic processes which inhibit plant growth and development (Farooq et al. 2012).Earlier studies have also shown that inoculation with acdS+ PGPR strains significantly promoted the growth of Pea seedlings under salinity-stressed conditions in pea (Burd et al. 2000; Wang et al. 2001). Plant Growth Promoting Bacteria (PGPB) synthesizing an enzyme i.e. ACC deaminase can cleave ACC, a precursor of ethylene, to α-ketobutyrate and ammonia, thereby reducing ethylene stress in plants (Sun et al. 2009). As per (Ali et al. 2014a) the total chlorophyll content was found to increase compared to ACC deaminase deficient mutants under salt-stressed rice (Bal et al. 2013) and cucumber (Kang et al. 2014). In this study, the decrease in chlorophyll due to salinity was almost ameliorated by PGPR as there was an increase in photosynthetic pigments such as chlorophyll a, chlorophyll b, total Chlorophyll and Carotenoids in CHR JH 203 and BST YS1_42 treated plants respectively. Henceforth from our study, we can conclude that Chla/bBP is the salt stress-regulated gene and could be used for augmenting abiotic stress tolerance in transgenic crops.
To conquer the oxidative damages the plants had developed antioxidants mechanisms as enzymes such as SOD, POX, CAT, NOS, MAPK, (PRP) Proline-rich protein, and molecules together with phenols, flavonoids, Fiddlehead protein and carotenoids (Gill and Tuteja 2010).The non-enzymatic antioxidants such as phenol and flavonoids content was observed to be higher in treated plants (CHR JH 203 and BST YS1_42) for both the cases but the Proline content was observed to be more when plants were exposed under salinity stress conditions, this gives a sign that plant itself try to prevent from oxidative stress caused by increased NaCl stress (Kaur and Asthir 2015).
Plants with high antioxidant enzyme activities have high levels of oxidative stress (Kumari et al. 2015). There was an increase in antioxidants enzyme activity in acdS+ treated plants as compared to control and 1% NaCl induced stressed condition which supported the above explanation. In this study, we took nine antioxidant genes to see their expressional analysis under salinity stress conditions. The acdS+ PGPR-inoculated plants under 1% saline stress recorded a 1.1–2.7 fold increase in the gene expression as compared to un-inoculated non-stressed plants. There was increased expression of genes responsible for antioxidant enzymes like SOD, POX, APX, NOS, MAPK, Chla/bBP and FDH protein. The gene for Fiddlehead protein which is similar to beta-ketoacyl-CoA synthases and chalcone synthases is involved in the biosynthesis of long-chain fatty acids in the cuticle and epidermal interactions. The Fiddlehead gene in the present study gave salinity tolerance to bacteria, but its transcript level in plants was low and hence it decreases in normal stress conditions. FDH-like genes may be directly responsible for cell-to-cell interactions that need to occur during pollen tube growth and carpel fusion (Efremova et al. 2004). In this study, the acdS+ isolates gave beneficiary effect and increase the FDH expression level under salinity stress environment. MAP kinases (MAPK) play a critical role in different types of signal transduction. MAPKs are involved in various pathways such as hormonal, developmental, biotic and abiotic stress signaling (Cristina et al. 2010). The stress signaling factor MAP kinases gene was up-regulated under saline stress is the sign that these MAPK pathways play an essential role in a different type of signal transduction which proves the strongest evidence for cross-talk during abiotic stress (Sinha et al. 2011). The above results give evidence that there are probably some genes that might be pre-committed in bacteria to overcome abiotic stress (Du et al. 2008). The CAT expression was found to be increased by B. marisflavi under saline stress, thereby showing the defensive role of acdS+ isolate under NaCl-induced stress. In our result PRP, shows a decrease in expressional activity in bacterial inoculated plants under salt stress, suggesting that these plants were under higher oxidative stress under saline conditions (Porcel and Ruiz-Lozano 2004; Kohler et al. 2008, 2009; Vardharajula et al. 2011) which tends to increase due to salinity.
Conclusion
In this study, seeds of Pisum sativum were coated with acdS+ bacterial isolates and were germinated for further NaCl-induced salinity stress. Plants inoculated with PGPR showed enhanced growth promotion and tolerance for salinity stress. Even though there are numerous studies on mitigating the adverse salinity effect, the present study has identified the two acdS+ strains of Bacillus genus that can be used to perform out further field research. Further biochemical and molecular evaluation of these ACCD-producing bacteria was conducted. PCR amplification of these ACCD containing isolates was done followed by sequencing of PCR product. The obtained sequence was used to construct the phylogenetic tree which revealed the close relationship of acdS+ strain among different genera. The NaCl induced maximum tolerance level of selected PGPR strains was performed followed by greenhouse experiments. Future studies should focus on field evaluation in different formulations. Concluding this study one can say that though strains CHR JH 203 (Bacillus marisflavi) and BST YS1_42 (Bacillus cereus) seem to be a more promising one, both of these PGPR can be developed into an effective bio-fertilizer which can promote plant growth of various crops even under saline stressed conditions. Therefore, both these isolates can also be used to make consortia and evaluate their effect under saline conditions. Hence the current study will provide an advantage towards sustainable crop development.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are grateful to Integral University for providing MCN number IU/R&D/2020-MCN001003 and DST-FIST for providing Financial Assistance to the Department of Bioscience, Integral University, Lucknow, Uttar Pradesh, India.
Author contributions
NP and SS conceived and coordinated the research. AG conducted the experiments and analyzed the data. AG and MK wrote the manuscript. AB, SR, JA, read and approved the manuscript.
Funding
This work was supported by Uttar Pradesh Council of Science and Technology, Lucknow [UPCST/D-514]; Department of Science and Technology, New Delhi [DST INSPIRE-IF160803].
Declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Contributor Information
Anmol Gupta, Email: anmolgupta632@gmail.com.
Ambreen Bano, Email: ambreenbano2408@gmail.com.
Smita Rai, Email: shiblyrai@gmail.com.
Manoj Kumar, Email: manojbiochem16@gmail.com.
Jasarat Ali, Email: jasaratt@gmail.com.
Swati Sharma, Email: ssharma@iul.ac.in, Email: sw_sh@rediffmail.com.
Neelam Pathak, Email: pathak.neelam007@gmail.com.
References
- Abdel Latef AAH, Chaoxing H. Does Inoculation with Glomus mosseae improve salt tolerance in pepper plants? J Plant Growth Regul. 2014;33:644–653. doi: 10.1007/s00344-014-9414-4. [DOI] [Google Scholar]
- Abeles WF (1992) Roles and physiological effects of ethylene in plant physiology : dormancy, growth, and development. Ethyl Plant Biol
- Aebi H. [13] Catalase invitro. Methods Enzymol. 1984;105:121–126. doi: 10.1016/S0076-6879(84)05016-3. [DOI] [PubMed] [Google Scholar]
- Ahanger MA, Agarwal RM. Salinity stress induced alterations in antioxidant metabolism and nitrogen assimilation in wheat (Triticum aestivum L.) as influenced by potassium supplementation. Plant Physiol Biochem. 2017;115:449–460. doi: 10.1016/J.PLAPHY.2017.04.017. [DOI] [PubMed] [Google Scholar]
- Ahmad P. Salt-induced changes in photosynthetic activity and oxidative defense system of three cultivars of mustard (Brassica juncea L.) Afr J Biotechnol. 2012;11:2694–2703. doi: 10.5897/ajb11.3203. [DOI] [Google Scholar]
- Ali S, Charles TC, Glick BR. Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol Biochem. 2014;80:160–167. doi: 10.1016/j.plaphy.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Ali S, Charles TC, Glick BR. Plant physiology and biochemistry amelioration of high salinity stress damage by plant growth- promoting bacterial endophytes that contain ACC deaminase. Plant Physiol Biochem. 2014;80:160–167. doi: 10.1016/j.plaphy.2014.04.003. [DOI] [PubMed] [Google Scholar]
- Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- Arnon DI. Copper enzymes in isolated chloroplasts. polyphenoloxidase in Beta vulgaris. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal HB, Nayak L, Das S, Adhya TK. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant Soil. 2013 doi: 10.1007/s11104-012-1402-5. [DOI] [Google Scholar]
- Barnawal D, Bharti N, Maji D, et al. ACC deaminase-containing Arthrobacter protophormiae induces NaCl stress tolerance through reduced ACC oxidase activity and ethylene production resulting in improved nodulation and mycorrhization in Pisum sativum. J Plant Physiol. 2014;171:884–894. doi: 10.1016/j.jplph.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Bates LS, Waldren RP, Teare ID. Rapid determination of free proline for water-stress studies. Plant Soil. 1973;39:205–207. doi: 10.1007/BF00018060. [DOI] [Google Scholar]
- Belimov AA, Safronova VI, Sergeyeva TA, et al. Characterization of plant growth promoting rhizobacteria isolated from polluted soils and containing 1-aminocyclopropane-1-carboxylate deaminase. Can J Microbiol. 2001;47:642–652. doi: 10.1139/w01-062. [DOI] [PubMed] [Google Scholar]
- Blaha D, Prigent-Combaret C, Mirza MS, Moënne-Loccoz Y. Phylogeny of the 1-aminocyclopropane-1-carboxylic acid deaminase-encoding gene acdS in phytobeneficial and pathogenic Proteobacteria and relation with strain biogeography. FEMS Microbiol Ecol. 2006;56:455–470. doi: 10.1111/j.1574-6941.2006.00082.x. [DOI] [PubMed] [Google Scholar]
- Bouffaud ML, Renoud S, Dubost A, et al. 1-Aminocyclopropane-1-carboxylate deaminase producers associated to maize and other Poaceae species. Microbiome. 2018 doi: 10.1186/s40168-018-0503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd GI, Dixon DG, Glick BR. Plant growth-promoting bacteria that decrease heavy metal toxicity in plants. Can J Microbiol. 2000;46:237–245. doi: 10.1139/w99-143. [DOI] [PubMed] [Google Scholar]
- Chance B, Maehly AC. [136] Assay of catalases and peroxidases. {black small square} Methods Enzymol. 1955;2:764–775. doi: 10.1016/S0076-6879(55)02300-8. [DOI] [Google Scholar]
- Chang CC, Yang MH, Wen HM, Chern JC (2002) Estimation of total flavonoid content in propolis by two complementary colometric methods. J Food Drug Anal 10:178–182. 10.38212/2224-6614.2748
- Chaves MM, Flexas J, Pinheiro C. Photosynthesis under drought and salt stress: regulation mechanisms from whole plant to cell. Ann Bot. 2009;103:551–560. doi: 10.1093/aob/mcn125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Park E, Glick BR. 1-Aminocyclopropane-1-carboxylate deaminase from Pseudomonas putida UW4 facilitates the growth of canola in the presence of salt. Can J Microbiol. 2007;53:912–918. doi: 10.1139/W07-050. [DOI] [PubMed] [Google Scholar]
- Cristina M, Petersen M, Mundy J. Mitogen-activated protein kinase signaling in plants. Annu Rev Plant Biol. 2010;61:621–649. doi: 10.1146/annurev-arplant-042809-112252. [DOI] [PubMed] [Google Scholar]
- Du J, Huang YP, Xi J, et al. Functional gene-mining for salt-tolerance genes with the power of Arabidopsis. Plant J. 2008;56:653–664. doi: 10.1111/j.1365-313X.2008.03602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois M, Gilles K, Hamilton JK, et al. A colorimetric method for the determination of sugars. Nature. 1951;168:167. doi: 10.1038/168167a0. [DOI] [PubMed] [Google Scholar]
- Dworkin M, Foster JW. Experiments with some microorganisms which utilize ethane and hydrogen. J Bacteriol. 1958;75:592–603. doi: 10.1128/jb.75.5.592-603.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efremova N, Schreiber L, Bär S, et al. Functional conservation and maintenance of expression pattern of FIDDLEHEAD-like genes in Arabidopsis and Antirrhinum. Plant Mol Biol. 2004;56:821–837. doi: 10.1007/s11103-004-5576-y. [DOI] [PubMed] [Google Scholar]
- Farooq M, Hussain M, Wahid A, Siddique KHM. Plant responses to drought stress: from morphological to molecular features. Berlin, Heidelberg: Springer; 2012. Drought stress in plants: an overview; pp. 1–33. [Google Scholar]
- Flowers TJ, Galal HK, Bromham L. Evolution of halophytes: multiple origins of salt tolerance in land plants. Funct Plant Biol. 2010;37:604. doi: 10.1071/FP09269. [DOI] [Google Scholar]
- Freitas VS, Miranda RS, Freitas VS, et al. Ethylene triggers salt tolerance in maize genotypes by modulating polyamine catabolism enzymes associated with H2O2 production. Environ Exp Bot. 2018;145:75–86. doi: 10.1016/J.ENVEXPBOT.2017.10.022. [DOI] [Google Scholar]
- Ghosh PK, De TK, Maiti TK. Role of ACC deaminase as a stress ameliorating enzyme of plant growth-promoting rhizobacteria useful in stress agriculture: a review. Role Rhizospheric Microbes Soil Stress Manag Agric Sustain. 2018;1:57–106. doi: 10.1007/978-981-10-8402-7_3. [DOI] [Google Scholar]
- Giannopolitis CN, Ries SK. Superoxide dismutases: I. Occurrence in higher plants. Plant Physiol. 1977;59:309–314. doi: 10.1104/pp.59.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Glick BR, Glick BR. Beneficial plant-bacterial interactions. New York: Springer International Publishing; 2020. Modulating phytohormone levels; pp. 139–180. [Google Scholar]
- Glick BR, Karaturovic DM, Newell PC. A novel procedure for rapid isolation of plant growth promoting pseudomonads. Can J Microbiol. 1995;41:533–536. doi: 10.1139/m95-070. [DOI] [Google Scholar]
- Gontia-Mishra I, Sapre S, Sharma A, Tiwari S. Amelioration of drought tolerance in wheat by the interaction of plant growth-promoting rhizobacteria. Plant Biol. 2016;18:992–1000. doi: 10.1111/PLB.12505. [DOI] [PubMed] [Google Scholar]
- Gupta A, Bano A, Rai S et al (2021a) Plant growth promoting rhizobacteria (PGPR): a sustainable agriculture to rescue the vegetation from the effect of biotic stress: a review 10:2459–2465. 10.33263/LIANBS103.24592465
- Gupta A, Rai S, Bano A et al (2021b) Comparative evaluation of different salt-tolerant plant growth-promoting bacterial isolates in mitigating the induced adverse effect of salinity in Pisum sativum 11:13141–13154
- Habib SH, Kausar H, Saud HM. Plant growth-promoting rhizobacteria enhance salinity stress tolerance in okra through ros-scavenging enzymes. Biomed Res Int. 2016 doi: 10.1155/2016/6284547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hontzeas N, Richardson AO, Belimov A, et al. Evidence for horizontal transfer of 1-aminocyclopropane-1-carboxylate deaminase genes. Appl Environ Microbiol. 2005;71:7556–7558. doi: 10.1128/AEM.71.11.7556-7558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossein H, Rezvani MP (2006) Effect of water and salinity stress in seed germination on isabgol (PLANTAGO OVATA) 4:15–22
- Islam F, Yasmeen T, Arif MS, et al. (2015) Plant growth promoting bacteria confer salt tolerance in Vigna radiata by up-regulating antioxidant defense and biological soil fertility. Plant Growth Regul. 2015;801(80):23–36. doi: 10.1007/S10725-015-0142-Y. [DOI] [Google Scholar]
- Jha B, Gontia I, Hartmann A. The roots of the halophyte Salicornia brachiata are a source of new halotolerant diazotrophic bacteria with plant growth-promoting potential. Plant Soil. 2012;356:265–277. doi: 10.1007/s11104-011-0877-9. [DOI] [Google Scholar]
- Kang SM, Khan AL, Waqas M, et al. Plant growth-promoting rhizobacteria reduce adverse effects of salinity and osmotic stress by regulating phytohormones and antioxidants in Cucumis sativus. J Plant Interact. 2014;9:673–682. doi: 10.1080/17429145.2014.894587. [DOI] [Google Scholar]
- Kaur G, Asthir B. Proline: a key player in plant abiotic stress tolerance. Biol Plant. 2015;59:609–619. doi: 10.1007/s10535-015-0549-3. [DOI] [Google Scholar]
- Kloepper JW. Relationship of in vitro antibiosis of plant growth-promoting rhizobacteria to plant growth and the displacement of root Microflora. Phytopathology. 1981;71:1020. doi: 10.1094/PHYTO-71-1020. [DOI] [Google Scholar]
- Kohler J, Hernández JA, Caravaca F, Roldán A. Plant-growth-promoting rhizobacteria and arbuscular mycorrhizal fungi modify alleviation biochemical mechanisms in water-stressed plants. Funct Plant Biol. 2008;35:141. doi: 10.1071/FP07218. [DOI] [PubMed] [Google Scholar]
- Kohler J, Hernández JA, Caravaca F, Roldán A. Induction of antioxidant enzymes is involved in the greater effectiveness of a PGPR versus AM fungi with respect to increasing the tolerance of lettuce to severe salt stress. Environ Exp Bot. 2009;65:245–252. doi: 10.1016/j.envexpbot.2008.09.008. [DOI] [Google Scholar]
- Kumar M, Mishra S, Dixit V, et al. Synergistic effect of Pseudomonas putida and Bacillus amyloliquefaciens ameliorates drought stress in chickpea (Cicer arietinum L.) Plant Signal Behav. 2016 doi: 10.1080/15592324.2015.1071004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Vaishnav A, Jain S, et al. Bacterial-mediated induction of systemic tolerance to salinity with expression of stress alleviating enzymes in soybean (Glycine max L. Merrill) J Plant Growth Regul. 2015;34:558–573. doi: 10.1007/s00344-015-9490-0. [DOI] [Google Scholar]
- Li T, Zhang J, Shen C, et al. 1-Aminocyclopropane-1-carboxylate: a novel and strong chemoattractant for the plant beneficial Rhizobacterium Pseudomonas putida UW4. Mol Plant Microbe Interact. 2019;32:750–759. doi: 10.1094/MPMI-11-18-0317-R. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/METH.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lowry O, Schagger H, Cramer WA, Vonjagow G. Protein measurement with the folin phenol reagent. Anal Biochem. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Lugtenberg BJJ, Chin-A-Woeng TFC, Bloemberg GV. Microbe-plant interactions: principles and mechanisms. Antonie Van Leeuwenhoek Int J Gen Mol Microbiol. 2002;81:373–383. doi: 10.1023/A:1020596903142. [DOI] [PubMed] [Google Scholar]
- Manchanda G, Garg N. Salinity and its effects on the functional biology of legumes. Acta Physiol Plant. 2008;305(30):595–618. doi: 10.1007/S11738-008-0173-3. [DOI] [Google Scholar]
- McDonald S, Prenzler PD, Antolovich M, Robards K. Phenolic content and antioxidant activity of olive extracts. Food Chem. 2001;73:73–84. doi: 10.1016/S0308-8146(00)00288-0. [DOI] [Google Scholar]
- Merchante C, Alonso JM, Stepanova AN. Ethylene signaling: simple ligand, complex regulation. Curr Opin Plant Biol. 2013;16:554–560. doi: 10.1016/j.pbi.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Morgan PW, Drew MC. Ethylene and plant responses to stress. Physiol Plant. 1997;100:620–630. doi: 10.1111/J.1399-3054.1997.TB03068.X. [DOI] [Google Scholar]
- Mukhtar T, Rehman S, Smith D, et al. Mitigation of heat stress in Solanum lycopersicum L. by ACC-deaminase and exopolysaccharide producing Bacillus cereus: effects on biochemical profiling. Sustain. 2020;12:2159. doi: 10.3390/SU12062159. [DOI] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Mustafa G, Akhtar MS, Abdullah R. Global concern for salinity on various agro-ecosystems. Salt Stress Microbes Plant Interact Causes Solut. 2019;1:1–19. doi: 10.1007/978-981-13-8801-9_1. [DOI] [Google Scholar]
- Nakano Y, Asada K. Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in Spinach Chloroplasts. Plant Cell Physiol. 1981;22:867–880. doi: 10.1093/oxfordjournals.pcp.a076232. [DOI] [Google Scholar]
- Nascimento FX, Rossi MJ, Soares CRFS, et al. New insights into 1-Aminocyclopropane-1-Carboxylate (ACC) deaminase phylogeny, evolution and ecological significance. PLoS ONE. 2014 doi: 10.1371/journal.pone.0099168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nautiyal CS. Rhizosphere competence of Pseudomonas sp. NBRI9926 and Rhizobium sp. NBRI9513 involved in the suppression of chickpea (Cicer arietinum L.) pathogenic fungi. FEMS Microbiol Ecol. 1997;23:145–158. doi: 10.1111/J.1574-6941.1997.TB00398.X. [DOI] [Google Scholar]
- Nelson N. A photometric adaptation of the Somogyi method for the determination of glucose. J Biol Chem. 1944;153:375–380. doi: 10.1016/S0021-9258(18)71980-7. [DOI] [Google Scholar]
- Niranjan Raj S, Deepak SA, Basavaraju P, et al. Comparative performance of formulations of plant growth promoting rhizobacteria in growth promotion and suppression of downy mildew in pearl millet. Crop Prot. 2003;22:579–588. doi: 10.1016/S0261-2194(02)00222-3. [DOI] [Google Scholar]
- Noumavo PA, Kochoni E, Didagbé YO, et al. Effect of different plant growth promoting rhizobacteria on maize seed germination and seedling development. Am J Plant Sci. 2013;04:1013–1021. doi: 10.4236/ajps.2013.45125. [DOI] [Google Scholar]
- Numan M, Bashir S, Khan Y, et al. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: a review. Microbiol Res. 2018;209:21–32. doi: 10.1016/J.MICRES.2018.02.003. [DOI] [PubMed] [Google Scholar]
- Osman AS, Rady MM. Ameliorative effects of sulphur and humic acid on the growth, anti-oxidant levels, and yields of pea (Pisum sativum L.) plants grown in reclaimed saline soil. J Hortic Sci Biotechnol. 2015;87:626–632. doi: 10.1080/14620316.2012.11512922. [DOI] [Google Scholar]
- Parida AK, Das AB. Salt tolerance and salinity effects on plants: a review. Ecotoxicol Environ Saf. 2005;60:324–349. doi: 10.1016/j.ecoenv.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Penrose DM, Glick BR. Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plant. 2003;118:10–15. doi: 10.1034/j.1399-3054.2003.00086.x. [DOI] [PubMed] [Google Scholar]
- Pitman MG, Läuchli A. Global impact of salinity and agricultural ecosystems. Salin Environ Plants Mol. 2002 doi: 10.1007/0-306-48155-3_1. [DOI] [Google Scholar]
- Porcel R, Ruiz-Lozano JM. Arbuscular mycorrhizal influence on leaf water potential, solute accumulation, and oxidative stress in soybean plants subjected to drought stress. J Exp Bot. 2004;55:1743–1750. doi: 10.1093/jxb/erh188. [DOI] [PubMed] [Google Scholar]
- Pourbabaee AA, Bahmani E, Alikhani HA, Emami S. Promotion of wheat growth under salt stress by halotolerant bacteria containing ACC deaminase. J Agric Sci Technol. 2016;18:855–864. [Google Scholar]
- Ravanbakhsh M, Sasidharan R, Voesenek LACJ, et al. ACC deaminase-producing rhizosphere bacteria modulate plant responses to flooding. J Ecol. 2017;105:979–986. doi: 10.1111/1365-2745.12721. [DOI] [Google Scholar]
- Roy DG (2019) Scholarship @ Western A new algorithm for primer design
- Saikia J, Sarma RK, Dhandia R, et al. (2018) Alleviation of drought stress in pulse crops with ACC deaminase producing rhizobacteria isolated from acidic soil of Northeast India. Sci Rep. 2018;81(8):1–16. doi: 10.1038/s41598-018-21921-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapre S, Gontia-Mishra I, Tiwari S. ACC deaminase-producing bacteria: a key player in alleviating abiotic stresses in plants. Plant Growth Promot Rhizobacteria Agric Sustain. 2019 doi: 10.1007/978-981-13-7553-8_14. [DOI] [Google Scholar]
- Saravanakumar D, Samiyappan R. ACC deaminase from Pseudomonas fluorescens mediated saline resistance in groundnut (Arachis hypogea) plants. J Appl Microbiol. 2007;102:1283–1292. doi: 10.1111/j.1365-2672.2006.03179.x. [DOI] [PubMed] [Google Scholar]
- Shailendra Singh GG. Plant growth promoting rhizobacteria (PGPR): current and future prospects for development of sustainable agriculture. J Microb Biochem Technol. 2015;07:96–102. doi: 10.4172/1948-5948.1000188. [DOI] [Google Scholar]
- Siddikee MA, Chauhan PS. Sa T (2011) Regulation of ethylene biosynthesis under salt stress in red pepper (Capsicum annuum L.) by 1-Aminocyclopropane-1-Carboxylic Acid (ACC) Deaminase-producing Halotolerant Bacteria. J Plant Growth Regul. 2011;312(31):265–272. doi: 10.1007/S00344-011-9236-6. [DOI] [Google Scholar]
- Singh M, Srivastava PK, Verma PC, et al. Soil fungi for mycoremediation of arsenic pollution in agriculture soils. J Appl Microbiol. 2015;119:1278–1290. doi: 10.1111/jam.12948. [DOI] [PubMed] [Google Scholar]
- Sinha AK, Jaggi M, Raghuram B, Tuteja N. Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal Behav. 2011;6:196–203. doi: 10.4161/psb.6.2.14701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns JC, Glick BR. Transgenic plants with altered ethylene biosynthesis or perception. Biotechnol Adv. 2003;21:193–210. doi: 10.1016/S0734-9750(03)00024-7. [DOI] [PubMed] [Google Scholar]
- Sun Y, Cheng Z, Glick BR. The presence of a 1-aminocyclopropane-1-carboxylate (ACC) deaminase deletion mutation alters the physiology of the endophytic plant growth-promoting bacterium Burkholderia phytofirmans PsJN. FEMS Microbiol Lett. 2009;296:131–136. doi: 10.1111/j.1574-6968.2009.01625.x. [DOI] [PubMed] [Google Scholar]
- Talaat NB. Reactive oxygen, Nitrogen and sulfur species in plants. New York: Wiley; 2019. Role of reactive oxygen species signaling in plant growth and development; pp. 225–266. [Google Scholar]
- Tiwari G, Duraivadivel P, Sharma S, Hariprasad P. 1-Aminocyclopropane-1-carboxylic acid deaminase producing beneficial rhizobacteria ameliorate the biomass characters of Panicum maximum Jacq. by mitigating drought and salt stress. Sci Rep. 2018 doi: 10.1038/s41598-018-35565-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N. Methods in Enzymology. New York: Academic Press Inc; 2007. Mechanisms of high salinity tolerance in plants; pp. 419–438. [DOI] [PubMed] [Google Scholar]
- Vardharajula S, Zulfikar Ali S, Grover M, et al. Drought-tolerant plant growth promoting Bacillus spp.: effect on growth, osmolytes, and antioxidant status of maize under drought stress. J Plant Interact. 2011;6:1–14. doi: 10.1080/17429145.2010.535178. [DOI] [Google Scholar]
- Wang Y, Mopper S, Hasenstein KH. Effects of salinity on endogenous ABA, IAA, JA, and SA in Iris hexagona. J Chem Ecol. 2001;27:327–342. doi: 10.1023/A:1005632506230. [DOI] [PubMed] [Google Scholar]
- Wang Q, Dodd IC, Belimov AA, et al. Rhizosphere bacteria containing 1-aminocyclopropane-1-carboxylate deaminase increase growth and photosynthesis of pea plants under salt stress by limiting Na+ accumulation. Funct Plant Biol. 2016;43:161–172. doi: 10.1071/FP15200. [DOI] [PubMed] [Google Scholar]
- Yang J, Kloepper JW, Ryu CM. Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 2009;14:1–4. doi: 10.1016/j.tplants.2008.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.