Abstract
Obesity and type 2 diabetes mellitus (T2DM) are common, chronic metabolic disorders with associated significant long-term health problems at global epidemic levels. It is recognised that gut microbiota play a central role in maintaining host homeostasis and through technological advances in both animal and human models it is becoming clear that gut microbiota are heavily involved in key pathophysiological roles in the aetiology and progression of both conditions. This review will focus on current knowledge regarding microbiota interactions with short chain fatty acids, the host inflammatory response, signaling pathways, integrity of the intestinal barrier, the interaction of the gut-brain axis and the subsequent impact on the metabolic health of the host.
Keywords: Gut microbiota, Obesity, Type two diabetes mellitus
Background
Obesity is an increasing global challenge with a current worldwide estimation of 1.9 billion adults being classed as overweight (body mass index (BMI) > 25), 650 million of which are obese (BMI > 30) with the total currently outnumbering those with malnutrition [1, 2]. Key factors contributing to the current obesity epidemic include:—1) an increased availability and intake of energy-dense foods that are high in fat and sugars; 2) a significant decrease in physical activity; and 3) the evolving interactions of host-intestinal microbiota and environment. Obesity is a significant risk factor for prevalent non-communicable conditions such as cardiovascular disease, diabetes, musculoskeletal disorders and several types of malignancy. The risk for these disorders increases exponentially with increasing BMI underlying the need for early recognition and intervention.
The number of people being diagnosed globally with type 2 diabetes mellitus (T2DM) is projected to reach beyond 700 million within the next twenty-five years [3]. Importantly, the prevention of T2DM has been identified and declared a target priority by the World Health Organization (WHO) [4] and the United Nations (UN) [5]. Increasing weight, central body fat distribution and BMI play an integral role in the development of T2DM, a chronic metabolic disease characterised by hyperglycaemia and associated with insulin resistance and/or insufficient pancreatic insulin production [6]. Obesity alone accounts for 80–85% of the risk factors for T2DM [7]. Similar to obesity, the complications of T2DM include cardiovascular disease, kidney disease, limb amputations and blindness, which all subsequently can lead to disability and premature mortality with a global healthcare cost of greater than 1.3 trillion dollars [8].
Gut microbiota
Hippocrates, the ‘father of medicine’, claimed ‘all disease begins in the gut’ [9] indicating that the contribution to human health that the intestinal microbiota provides has been thought about for thousands of years. However, only recently, has the acknowledgement been stated that, as humans, we are in fact ‘supra-organisms’ composed of both human and microbial cells [10]. Humans carry two sets of genes, those encoded in our own genome and those encoded in our microbiota. Metchnikoff, in the early 1900s, first suggested the central importance of the intestine in host physiology and pathology [11]. Intestinal microbiota were viewed as essential modulators with the ability to influence human homeostasis and that disruption of this harmony by specific microbiota could result in a diseased state through accumulation of microbiota by-products.
The ‘human microbiome’ was first postulated by Joshua Lederberg, to signify the ecological community of commensal, symbiotic and pathogenic microorganisms that share our body space [12]. The genetic material of the intestinal microbes, collectively defined the ‘gut microbiome’, surpasses the magnitude of the human genome over one hundred times [13, 14]. Many bacterial species cannot be cultured, but with the recent technological advancement of modern molecular methods has led to the evolution of next-generation sequencing technology for the study of microbial deoxyribonucleic acid (DNA) from faecal samples. This has provided the ability to examine the entire genomic content of a community, by using direct sequencing of microbial RNA without the need for prior amplification [15–17].
An adult human is colonized by approximately one hundred trillion microbes, most of which are predominantly found in the gastrointestinal tract (GIT). Accommodating this enormous number of microorganisms in the GIT has often led to the intestinal microbiota being referred to as the ‘hidden organ’ [18]. The GIT hosts bacteria, archaea, viruses and fungi, with the largest population of microbiota being found to inhabit the colon. Gastric acid, bile and pancreatic secretions prevent the colonization of the stomach and proximal small intestine by most bacteria. Bacterial density begins to increase towards the distal small intestine (108 bacteria per gram content in the distal ileum), and increases rapidly throughout the colon rising to an estimated 1011 – 1012 bacteria per gram of colonic content, contributing roughly 60% of faecal mass [15, 19] estimated to weigh approximately 1.5 kg (kg) [20].
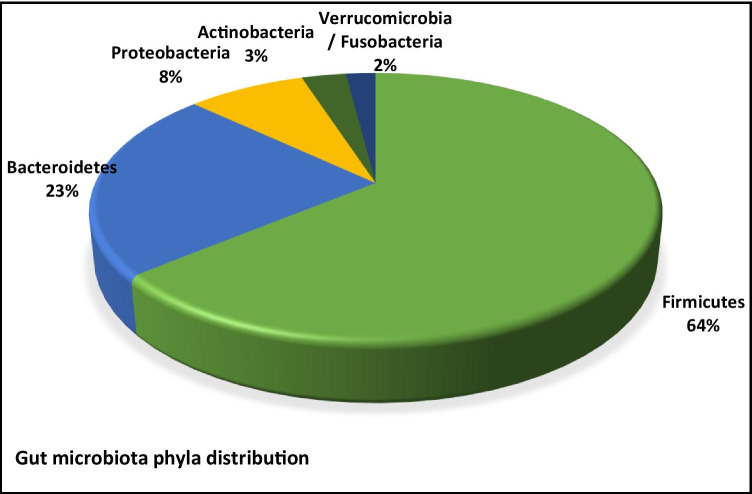
Over 90% of all phylotypes of colonic bacteria belong to just two known phyla; Gram-positive Firmicutes and Gram-negative Bacteroidetes. Bacteria are estimated to belong to over five hundred different species with 99% belonging to just thirty to forty genera from the four main phyla: Firmicutes 64% (e.g. Clostridium, Enterococcus, Lactobacillus, Ruminococcus); Bacteroidetes 23% (e.g. Bacteroides, Prevotella); Proteobacteria 8% (e.g. Helicobacter, Escherichia); and Actinobacteria 3% (e.g. Bifidobacterium) [21–23]. Other, much smaller phyla include Verrucomicrobia (Akkermansia) and Fusobacteria (Fusobacterium) (Fig. 1) [24]. It has been shown that the proximal GIT is enriched in bacteria belonging to the phyla Firmicutes and Proteobacteria, in particular the genus Lactobacilli, compared to the distal GIT which mainly comprises of bacteria belonging to the phyla Bacteroidetes and Firmicutes, with particular attention to the Akkermansia muciniphilia species [25].
Fig. 1.
Colonic microbiota belong to the main phyla in the following order: Firmicutes (64%), Bacteroidetes (23%), Proteobacteria (8%), Actinobacteria (3%), Verrucomicrobia and Fusobacteria (2%)
Intestine microbial composition is highly variable between individuals and is being continuously modified by both endogenous and exogenous factors [26]. The host genome has a central role in determining the composition of gut microbiota but many geographic and environmental factors such as diet, illness, lifestyle, hygiene and medication can contribute to changes in the population [27–29]. Reports have shown dietary modification can account for 57% of the variations in gut microbiota compared to host genetic variations that may account for as little as 12% [30]. There is now a considerable amount of data to suggest that disruption of gut microbiota may give rise to many inflammatory diseases such as obesity, inflammatory bowel disease, T2DM, arthritis and cancer [31].
As mentioned previously, the gut microbiota are becoming increasingly recognized as having a fundamental role in human physiology and health, [32] with resulting direct or indirect host effects [33]. Mechanisms include:- the fermentation of non-digestible substrates like dietary fibre and endogenous intestinal mucus to produce short chain fatty acids (SCFAs); the modulation of the immune and inflammatory response; regulation of neuronal signaling; regulation of integrity and mobility of the gut barrier; biosynthesis of vitamins, steroid hormones, and neurotransmitters; metabolism of branched chain amino acids (BCAAs), bile salts, and drugs; and the regulation of the hepatic production of triglycerides by suppressing lipoprotein lipase inhibitors [34–37].
Microbiome influence on obesity and T2DM
Diversity is key to a healthy gut allowing for microbe redundancy with multiple microbes capable of performing similar functions. Disruption in specific host microbial populations may be more important than overall phylogenetic ratios, resulting in changes in the production of SCFAs and metabolites that directly influence glucose and insulin regulation [38–40]. Suggested mechanisms for this development which will be discussed throughout this review and include immune dysregulation; altered energy regulation; altered gut hormone regulation; and pro-inflammatory mechanisms [41]. Body weight is not controlled by the calories that are ingested but rather by the calories that are absorbed [42]. When in calorie excess, adipose tissue cannot maintain its buffering capacity to store excess energy in the form of triglycerides, resulting in an overflow of lipids into the systemic circulation [43]. Increased lipid availability to non-adipose tissues such as the liver, skeletal muscle and pancreas contribute to ectopic fat storage and the development of insulin resistance. Secondly, adipose tissue generates inflammation triggering an increase in the production and secretion of pro-inflammatory cytokines and adipokines such as tumour necrosis factor.
(TNF), interleukins (IL-6) and monocyte chemoattractant proteins (MCP-1) which may also accelerate the development of peripheral insulin resistance and altered glucose homeostasis [43].
Gut microbiota metabolites: the importance of SCFAs
SCFAs are small organic monocarboxylic acids and are the major microbial metabolites produced during anaerobic carbohydrate fermentation in the intestine by acting as vital components in microbe-to-host signaling pathways [44]. Acetate, butyrate and propionate constitute greater than 95% of the total SCFA content [35, 45]. By-products of carbohydrate fermentation include the colonic gases (hydrogen (H2), carbon dioxide (CO2) and methane (CH4)) which can have inhibitory effects, limiting SCFA production [35, 45, 46]. The species Methanobrevibacter Smithii (a H2-using methanogen) prevents the accumulation of H2 by combining together H2 and CO2, producing CH4, allowing for the continued carbohydrate fermentation resulting in greater SCFA production and the availability of calories to the host [47].
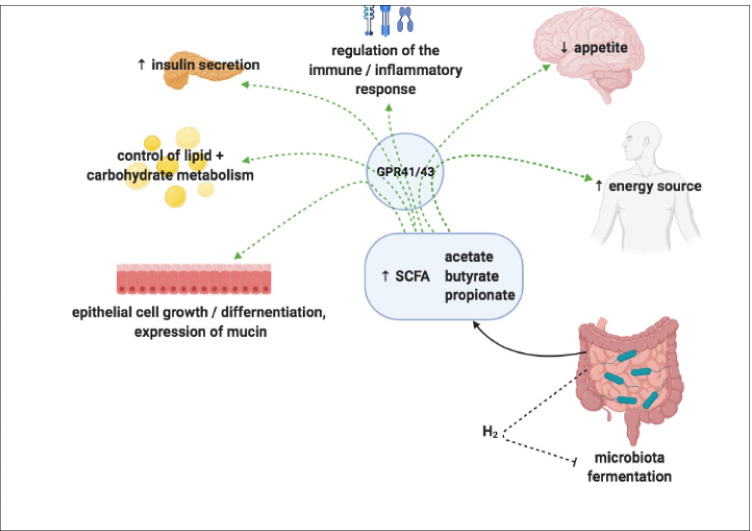
The roles of SCFAs in the host include constituting an important energy source providing as much as 10% of the daily energy requirement [48, 49]; facilitate hepatic control of lipids and carbohydrates; aid the transportation and metabolism of epithelial cells; positively influence epithelial cell growth and differentiation [50, 51]; promote the expression of mucin to strengthen the intestinal barrier [52]; have anti-inflammatory properties reducing the secretion of pro-inflammatory cytokines and chemokines [53]; and serve as immune stimulators to condition tissue and immune cells to better eliminate pathogens [54] (Fig. 2).
Fig. 2.
Overview of SCFAs in host metabolism. SCFAs provide an important energy source; facilitate hepatic control of lipids and carbohydrates; aid the transportation and metabolism of epithelial cells; influence epithelial cell growth and differentiation; promote the expression of mucin to strengthen the intestinal barrier; stimulate host inflammatory pathways; and regulate the immune system for the elimination of pathogens
SCFAs are predominantly produced from gut microbiota such as the genera Prevotella, Ruminococus, Coprococcus, and Roseburia, and the species Akkermansia muciniphilia and Eubacterium rectale [55]. During host fasting periods, intestinal microbes have adapted such that Akkermansia muciniphilia can degrade intestinal mucus, increasing the local availability of N-Acetylglucosamine, N-acetylgalactosamine, fructose and galactose which can serve as essential substrates for continued microbial fermentation [45, 56].
SCFAs in metabolic pathways
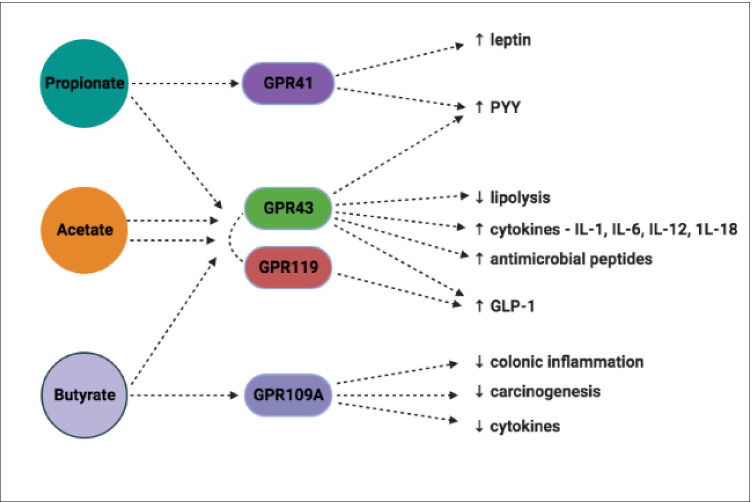
SCFAs perform a central role in metabolic pathways acting as signaling molecules by linking with selected G-protein-coupled receptors (GPRs): GPR41, GPR43 (also termed FFAR3 and FFAR2) GPR119 and GPR109A which are abundant in adipocytes, intestinal immune cells, gut epithelial cells and pancreatic β-cells [57–61]. Propionate primarily activates GPR41, butyrate activates GPR109A however GPR43 and GPR119 can be activated by acetate, butyrate and propionate at similar rates [61, 62] (Fig. 3).
Fig. 3.
Main SCFAs and their roles in host signaling pathways. SCFAs perform a central role in metabolic pathways linking with selected GPRs. GPRs are abundant in adipocytes, intestinal immune cells, gut epithelial cells and pancreatic β-cells. Propionate primarily activates GPR41 and butyrate activates GPR109A. GPR43 and GPR119 can be activated by acetate, butyrate and propionate at similar rates
Activation of GPR41 and GPR43 induces the secretion of peptide tyrosine-tyrosine (PYY), a short peptide that is released into the ileum and colon to reduce appetite by directly affecting the central nervous system. GPR41 coupling can stimulate adipocytes to express leptin [63, 64], resulting in the inhibition of the release of neuropeptide Y (NPY), a hypothalamic neurotransmitter that stimulates eating. This was first demonstrated using GPR41 deficient mice that display significantly lower leptin levels than corresponding wild-type mice [65].
GPR43 activation leads to positive homeostatic effects on several metabolic pathways:- the release of glucagon-like peptide 1 (GLP-1) from entero-endocrine L-cells which stimulate glucose-medicated insulin release from pancreatic β-cells, suppress pancreatic α-cell glucagon secretion, protect β-cells from apoptosis, promote β-cell proliferation and increase GIT transit time [66–68]; acetate activation directly reduces lipolysis in adipocytes leading to decreased plasma-free fatty acid levels [69]; promotion of the production of the antimicrobial peptides RegIIIγ and β-defensin in intestinal epithelial cells; and the generation of immunity-related cytokines such as IL-1, IL-6, IL-12 and IL-18 [70, 71].
GPR43 is mainly expressed in immune tissue and white adipose tissue as demonstrated in wild-type mice. Activation of GPR43 in white adipocytes reduces insulin-induced protein kinase B (AKT)-activation consequently suppressing fat accumulation. GPR43-deficient mice are phenotypically obese in contrast to mice who overexpress GPR43, specifically in adipose tissue. This cohort maintain their lean status even when subjected to high calorific feeding. Germ-free (GF) and antibiotic treated mice both had normal phenotypes suggesting the importance of gut microbiota as deliverers of GPR activating SCFAs [72].
GPR119 agonists reduce circulating blood glucose levels by promoting the intestinal secretion of GLP-1, improving pancreatic β-cell function and insulin secretion [60]. GPR109A stimulation suppresses colonic inflammation and carcinogenesis, promoting anti-inflammatory aspects of colonic macrophages and dendritic cells inducing the differentiation of regulatory and IL-10 producing T-cells. GPR109A expression decreases in the absence of gut microbiota [73, 74].
Acetate
The most abundant SCFA in the GIT is acetate which is readily absorbed, transported to the liver and acts as a substrate in cholesterol synthesis. Very little metabolism occurs within the colon. Acetate behaves as a substrate and signaling molecule in the processes of cholesterol synthesis, lipogenesis, host appetite and glucose homeostasis. Methanobrevibacter smithii can further exacerbate lipogenesis to the detriment of host adiposity, as it is heavily involved in the bacterial fermentation of fructans producing large amounts of acetate [75].
On entering the systemic circulation, the presence of acetyl-CoA synthetase in adipose tissue allows for the immediate usage as a substrate in lipogenesis [76]. Acetate may have the ability to influence host appetite by manipulating the expression profiles of regulatory neuropeptides situated in the hypothalamus through activation of the Krebs cycle [43, 77]. Using a rodent model, Perry et al. demonstrated that increased levels of acetate led to an elevated production of ghrelin, the stimulation of parasympathetic activity which thus increased food intake and the promotion of glucose-stimulated insulin secretion [78].
Butyrate
Butyrate has many important properties in host homeostasis:—acts as the main substrate in colonocyte metabolism [76, 79]; regulates cell proliferation and differentiation [62, 79]; induces apoptosis of colonic cancer cells [59, 62, 80, 81]; activates intestinal gluconeogenesis (80); induces the inflammation cascade; provides protection against oxidative stress [81]; and maintains the permeability of the gut barrier [59, 62, 82].
Butyrate is oxidised in the mitochondria of colonocytes and contributes as a substrate in the Krebs cycle for adenosine triphosphate (ATP) production. Catalysing enzymes in this process are down-regulated in GF-mice, leaving significantly decreased levels of ATP in GF-colonocytes. This highlights the potential stimulating role for butyrate-producing microbiota [83–85].
Butyrate can affect DNA methylation, proliferation and differentiation in colonic epithelial cells by inhibiting histone deacetylase and suppressing nuclear factor kappa B (NF-kB) activation [86]. NF-kB is a transcription factor in control of gene expression encoding pro-inflammatory cytokines, chemokines, inducible inflammatory enzymes, adhesion molecules, growth factors and some acute phase proteins and immune receptors [59, 87].
Butyrate exerts anti-inflammatory effects through the inhibition of interferon-y production, signaling pathways and the up-regulation of peroxisome proliferator-activated receptor gamma (PPARγ). PPARγ is a ligand-activated transcription factor that is highly expressed in colonic epithelial cells and is thought to activate the anti-inflammatory cascade [59].
Propionate
Propionate influences aspects of glucose homeostasis, inhibits hepatic cholesterol synthesis [62, 88], exerts anti-inflammatory effects by the promotion of regulatory T-cell differentiation and IL-10 production [53] and has the ability to reduce host appetite. It has been demonstrated to have competing and opposite effects on gluconeogenesis [89] acting as both a substrate and as an inhibitor [88]. The inhibiting effect may be related to its metabolic intermediaries, methymalonyl CoA and succinyl CoA, which are specific inhibitors of pyruvate carboxylase. Propionate may influence hepatic glucose metabolism indirectly by lowering plasma fatty acid concentration [90], which is closely related to the rate of gluconeogenesis [91].
Propionate stimulates the intestinal release of the satiety hormone PYY and GLP-1 coupling with GPRs [43]. De Vadder et al., demonstrated propionate supplementation can result in the reduction of weight, abdominal adipose tissue, hepatic fat and assist with insulin sensitivity maintenance [80]. Alhabeeb et al., also displayed the beneficial effects of propionate supplementation in healthy volunteers exhibiting increased satiety and reduced appetite, as measured by visual analogue scales [92].
Other gut microbiota fermentation products
BCAAs, succinate, ammonia, amines, phenol and indole are other important gut microbiota metabolites which also behave as nutrients, messenger molecules and have the ability to shape host pathophysiology. They are mainly derived from protein metabolism, fermentation of aromatic amino acids and dietary fibre [93]. The most abundant amino acid fermenting bacteria belong to the genera Clostridium, Bacteroides, Lactobacillus, Streptococcus, Propionibacterium and Fusobacterium [94, 95].
Bacterial fermentation of dietary fibre produces large amounts of succinate, reported as an unsuspected bacterial metabolite with the ability to improve glycaemic control through the activation of intestinal gluconeogenesis [96]. Increased BCAA plasma levels correlate with specific bacterial species such as Prevotella copri and Bacteroides vulgatus and have been shown to be characteristic of individuals with insulin resistance [97]. Increases in a small number of essential amino acids including the BCAAs (leucine, valine, and isoleucine), and the aromatic amino acids (phenylalanine and tyrosine), have been reported to be associated with a five-fold increased risk of developing T2DM [98].
Indolepropionic acid, generated from bacterial aromatic amino acid catabolism, is strongly correlated with host dietary fibre intake and appears to reduce the risk of developing T2DM. It exerts potent anti-oxidative activity and has radical scavenging properties in vitro, suggesting it has the ability to provide protection for the pancreatic β-cell and possibly from amyloid accumulation [99]. Indolepropionic acid may also aid the modulation of incretin secretion from entero-endocrine L-cells by inhibiting voltage-gated potassium channels. This affects the action potential properties of L-cells resulting in enhanced calcium entry, triggering GLP-1 secretion. However, when stimulated over a longer period of time, it leads to the inhibition of mitochondrial metabolism creating a reduction in intracellular ATP concentration. This induces the opening of ATP-sensitive potassium channels, hyperpolarising the plasma membrane and slowing GLP-1 secretion [100, 101].
Lastly, imidazole propionate, produced from the degradation of histidine by gut microbiota impairs the ability of cells to correctly respond to insulin. Imidazole propionate inhibits the intracellular insulin receptor signalling cascade by activating the p38γ–p62– mammalian target of rapamycin complex 1 (mTORC1) pathway which inhibits the formation of the insulin receptor substrate protein and mTORC1 complex. mTORC1 is an integral part of the intracellular cascade and regulates various metabolic pathways, including the insulin receptor cascade [102].
Bile acids and gut microbiota and the beneficial effects on the host
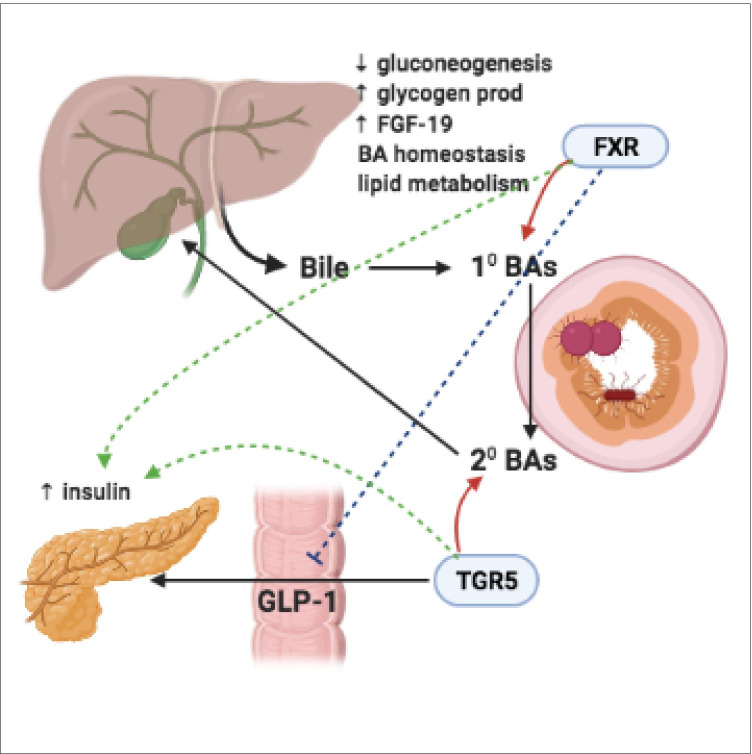
Bile acids (BAs) are steroid carboxylic acids produced in perivenous hepatocytes primarily from the hydroxylation of cholesterol which is controlled by cytochrome P450 enzyme cholesterol 7α hydroxylase (CYP7A1). Before being secreted for storage in the gallbladder, primary BAs are conjugated to glycine further enhancing their hydrophilicity. Entero-hepatic circulation enables 95% of BAs to be reabsorbed from the distal ileum, allowing time for the interaction of gut microbiota and primary BAs to produce secondary BAs [103]. BAs have multiple functions including the facilitation of the digestion and absorption of dietary fats and lipid-soluble vitamins in the small intestine; maintenance of the intestinal barrier; and controlling metabolic pathways by acting as signaling molecules for the regulation of triglyceride, cholesterol, glucose and energy homeostasis [104].
Gut microbiota play a key role in BA synthesis, modification and signaling by converting host-derived primary BAs into secondary BAs, and by deconjugation through the enzymatic activity of bile salt hydrolases [105]. The predominant microbiota that contribute in BA pathways are from the genera Lactobacillus, Bifidobacteria, Enterobacter, Bacteroides and Clostridium. Primary BAs bind to the nuclear hormone farnesoid X receptor (FXR) whereas secondary BAs bind to G protein-coupled BA receptor 1 (TGR5) [103, 106]. BAs, acting through FXR signaling can decrease gluconeogenesis and promote glycogen production in the liver. FXR stimulation results in the secretion of gut-derived hormones, such as fibroblast growth factor 19 (FGF-19) which in turn regulates BA synthesis as well as lipid and glucose metabolism [103, 107, 108]. FGF-19 induces the synthesis of glycogen and inhibits glucose production [109]. TGR5 receptor activation results in GLP-1 secretion from intestinal L-cells, whereas FXR signalling inhibits GLP-1. Both BA-TGR5 and BA-FXR signalling stimulates insulin production from β-cells in the pancreas. Glucose-stimulated insulin release is additionally promoted by BA-TGR5 signalling in α-cells, which promotes the conversion of pro-glucagon to GLP-1 and GLP-1 release (Fig. 4).
Fig. 4.
BA synthesis and signalling pathway. BAs are produced in perivenous hepatocytes primarily from the hydroxylation of cholesterol. Before being secreted for storage in the gallbladder, primary BAs are conjugated to glycine further enhancing their hydrophilicity. Entero-hepatic circulation enables 95% of BAs to be reabsorbed from the distal ileum, allowing time for the interaction of gut microbiota and primary BAs. Gut microbiota convert host-derived primary BAs into secondary BAs. Primary BAs bind to the FXR receptor, secondary BAs bind to the TGR5 receptor. FXR signaling can decrease gluconeogenesis, promote glycogen production and enhance the secretion of gut-derived hormones such as FGF-19. FGF-19 has the ability to regulate BA synthesis, lipid and glucose metabolism. TGR5 receptor activation results in GLP-1 secretion from intestinal L-cells. FXR signaling inhibits GLP-1. BA-TGR5 and BA-FXR receptor signaling stimulates insulin production from pancreatic β-cells
Gut microbiota can control BA synthesis by metabolising naturally occurring FXR antagonist tauro-β-muricholic acid resulting in the development of obesity, steatosis and impaired tolerance to glucose and insulin [110, 111]. Increased BA synthesis contributes to greater energy expenditure by the stimulation of brown adipose tissue and skeletal muscle via TGR5, and by increasing thyroid hormone production by activating type 2 deiodinase [106, 112]. In the hypothalamus, BA-TGR5 signalling mediates satiety. BA-TGR5 activation in immune cells results in the inhibition of the nucleotide-binding domain-like receptor protein 3 (NLRP3)-inflammasome (a multimeric protein complex) and attenuated inflammation [106].
Disruption of the intestinal mucosal barrier by microbiota
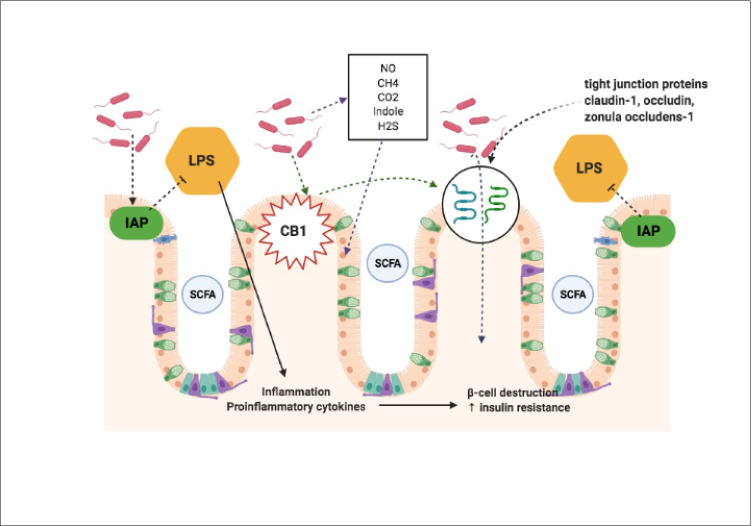
The mucosal lining of the GIT acts as a natural barrier preventing undesirable interactions between the colonic epithelium, viruses, toxins and pathogenic bacteria [113]. Disruption of the GIT wall integrity allows for the translocation of toxins into the systemic circulation leading to metabolic endotoxaemia and results in low-grade inflammation, autoimmunity and oxidative stress, which have the potential for β-cell destruction and insulin resistance [38, 114]. Gut microbiota produce numerous organic compounds like nitrous oxide (NO), CH4, CO2, indole and hydrogen sulphide which possess pro and anti-inflammatory properties with the capability to alter GIT permeability [115].
GIT barrier function is maintained via several mechanisms:—appropriate localisation and distribution of tight junction proteins (claudin-1, zonula occludens-1 and occludin); the presence of a thick mucus layer covering the epithelial cells; up-regulation of the secretion of mucus from goblet cells by butyrate; the presence of mucin-associated bacteria [116, 117]; a normal endocannabinoid system tone; and lipopolysaccharide (LPS) detoxification by intestinal alkaline phosphatase (Fig. 5 and 6). The presence of SCFAs enhance gut barrier integrity [114, 118, 119]. Gut microbiota have the ability to disrupt intestinal tight junction proteins and alter alkaline phosphatase activity resulting in increased gut permeability. Gut microbes selectively act to modulate colonic expression of endocannabinoid receptor type-1 (CB1), which strongly influences gut permeability through effects on zonula occludens-1 and occludin [120, 121].
Fig. 5.
Gut microbiota interactions with the intestinal mucosal barrier. The mucosal lining of the GIT acts as a natural barrier preventing undesirable interactions. Disruption of the GIT wall integrity allows for the translocation of toxins into the systemic circulation leading to metabolic endotoxaemia resulting in low-grade inflammation, autoimmunity and oxidative stress. Gut microbiota produce numerous organic compounds like NO, CH4, CO2, indole and hydrogen sulphide with the capability to disrupt GIT permeability. GIT barrier function is maintained using:—appropriate localisation and distribution of tight junction proteins; the presence of a thick mucus layer covering the epithelial cells; up-regulation of the secretion of mucus from goblet cells; the presence of mucin-associated bacteria; a normal endocannabinoid system tone; and LPS detoxification by intestinal alkaline phosphatase
Fig. 6.
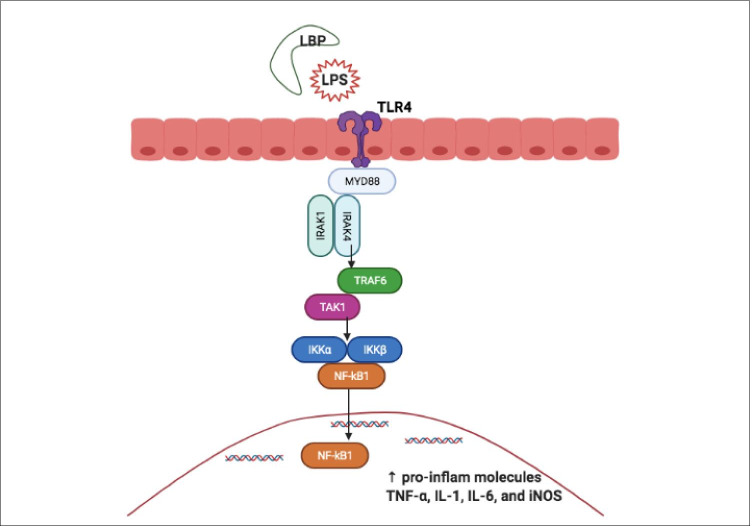
LPS triggering the inflammatory pathway. LPS is recognised and bound by LBP. Bacterial fragments and LPS are recognised by TLRs. LPS binds and activates TLR4, triggering dimerisation, and the recruitment of downstream adaptor molecules such as MyD88/MAL to mount an inflammatory response. Activated MyD88/MAL recruits IRAK), TRAF6, TAK1, JNK and IKK complexes. The IKK complex converges at NF-kB, which is maintained in the inactive state by IKKB. This, in turn, is degraded by proteasomes, resulting in the translocation of NF-kB into the nucleus, activating the release of pro-inflammatory molecules TNF-α, IL-1β, IL-6, and iNOS
Obesity and T2DM are both conditions characterised by GIT barrier disruption leading to a significant increase in permeability, which correlates, with greater levels of LPS in the blood [122]. Brun et al., analysed cross-sectional samples of intestine obtained from obese mice that clearly demonstrated a decrease in the tight junction protein occludin and an irregular distribution of zonula occludens-1 [123]. Several species have been shown to benefit gut barrier function. Faecalibacterium prausnitzii and Roseburia intestinalis are important butyrate-producing microbes and are believed to protect against bacterial translocation either directly or through their ability to produce butyrate [124]. Increased levels of the genera Bifidobacterium have also been associated with reduced gut leakiness, allowing less LPS to translocate into the serum [122]. GIT motility and gut microbiota are closely interrelated and can significantly affect one another. Transplanting human gut microbes into GF-mice significantly shortened GIT transit time when subjected to a polysaccharide-rich diet [125]. SCFAs modulate colonic motility by stimulating the secretion of serotonin from gut enterochromaffin cells, in part through activation of the vagus nerve via the serotonin (5-HT3) receptor [126].
Gut microbiota influence on the inflammatory response
Obesity and T2DM are characterised by chronic low-grade inflammation with abnormal expression and production of multiple inflammatory mediators such as increased levels of TNF, C-reactive protein (CRP), plasminogen activator inhibitor-1 and interleukins (IL-1, IL-6) [127–129]. The concept of inflammation in metabolic conditions was first proposed by Hotamisligil et al. who demonstrated that adipocytes can express the cytokine TNF-α and that this expression is particularly increased within obese animals. Neutralisation of TNF-α in these animals led to a decrease in insulin resistance. This is believed to be the first experiment to potentially expose a relationship between the expression and plasma concentration of a pro-inflammatory cytokine and insulin resistance [130].
Gut microbiota, acting through LPS activity can influence inflammation and insulin resistance. LPS is an essential component of the cell walls of Gram-negative bacteria such as the phylum Bacteroidetes [131–133]. The lipid A portion of LPS contains the relevant endotoxin activity and has differing levels of pro-inflammatory activity owing to the variability in the detailed lipid A structure. LPS from members of the families Enterobacteriaceae and Desulfovibrionaceae (phylum: Proteobacteria), exhibit an endotoxin activity that is 1,000-fold that of LPS from the family Bacteroideaceae (phylum: Bacteroidetes of which members of this phylum are the most numerous LPS producers in the gut) [134].
Dietary fat is transported from the intestine after being incorporated as triglycerides into chylomicrons which have a high affinity for LPS. Thus, the formation of chylomicrons aids the movement of LPS from intestinal cells into the circulation [131].
‘Metabolic endotoxemia’ is a condition characterised by a two to three-fold increase in circulating LPS levels [135]. After entering the circulation, LPS is recognised and bound by lipopolysaccharide binding protein (LBP), an acute-phase protein synthesised in the liver [136]. Bacterial fragments and LPS are recognised by toll-like receptors (TLRs) that are a family of key pattern recognition receptors that aid cells in the recognition of ligands such as endotoxin [132, 137, 138]. LPS binds and activates TLR4, triggering dimerisation, and the recruitment of downstream adaptor molecules such as myeloid differentiation primary response 88 adaptor-like (MyD88/MAL) to mount an inflammatory response [26] (Fig. 6).
Activated MyD88/MAL recruits the interleukin-1 receptor-associated kinase (IRAK), tumour necrosis factor receptor-associated factor 6 (TRAF6), transforming growth factor B-activated kinase 1 (TAK1), jun N-terminal kinase (JNK) and inhibitor of nuclear factor-kB kinase (IKK) complexes. The IKK complex converges at NF-kB, which is maintained in the inactive state by IkB. This, in turn, is degraded by proteasomes, resulting in the translocation of NF-kB into the nucleus, activating the release of pro-inflammatory molecules TNF-α, IL-1, IL-6, and inducible nitric oxide synthase (iNOS). The activation of serum kinases (JNK and IKK) can induce insulin receptor substrate (IRS-1) serine phosphorylation, resulting in insulin resistance [27].
TNF-α expression upregulates the transcription of suppressor of cytokine signaling 3 (SOCS-3) which binds to tyrosine 960 of the insulin receptor, preventing IRS-1 binding to the insulin receptor. IRS-1 is subsequently degraded leading to the disruption of the insulin signaling pathway and glucose transport (via GLUT-4) [136, 139]. The importance of the TLR-4 pathways in worsening metabolic disease was confirmed by inducing a deletion of TLR-4 which subsequently prevented high fat diet (HFD) induced insulin resistance [140, 141].
LPS can also activate a local immune response via high-affinity binding to the NLRP3 inflammasome and NLRs expressed at high levels on the surface of macrophages and dendritic cells [142]. They are believed to play a role in the development of leptin resistance [143], resulting in hyperphagia and weight gain further increasing fat intake, raising LPS and ongoing inflammation [144].
HFD feeding significantly alters the gut microbial composition by reducing the numbers of the genera Bifidobacterium, which have many physiologically positive effects [145]. Mice who consume a HFD supplemented with oligo-fructose have restored quantities of Bifidobacterium with associated decreased endotoxemia suggesting Bifidobacterium may improve intestinal permeability and lower circulating levels of endotoxin. The increase in Bifidobacterium correlates with improved glucose tolerance, glucose-induced insulin secretion, lower body weight and decreased production of inflammatory mediators [133, 146].
Obese rodents have two to three times greater levels of plasma LPS than non-obese counterparts and display low-grade systemic inflammation. Injection of the species Escherichia coli LPS subcutaneously into wild-type mice fed on normal chow led to the development of inflammation, obesity, fasted glycaemia and insulinaemia. Importantly, in cluster of differentiation 14 (CD14)-knockout mice, in whom LPS cannot be recognised, there was a delay or even a complete lack of development of most features of metabolic disease induced by a HFD or LPS infusion [135].
Antibiotic induced disruption of the microbiota
Antibiotics can strongly influence the composition of the gut microbiota for up to two years after administration [147]. Disruption caused by antibiotic treatment can induce a stress response which facilitates the transfer of drug resistant genes to virulent species leading to drug resistance [148]. A recently performed meta-analysis concluded an increased risk of childhood obesity in those children exposed to more than one antibiotic treatment within their first six months of life [149]. A population-wide case–control study in Denmark demonstrated a positive relationship between antibiotic exposure and the development of T2DM years later and a relationship between a T2DM diagnosis and the number of antibiotic prescriptions. Antibiotics may predispose patients to T2DM, however the authors suggest caution because T2DM patients are possibly more vulnerable to developing infections in the years prior to diagnosis [150].
Vrieze et al., analysed the effects of antibiotic treatment on gut microbiota and its effect on metabolic parameters in patients diagnosed with obesity and insulin resistance. Vancomycin significantly reduced microbial diversity, with particular decreases in the abundance of Firmicutes, mainly butyrate-producers, with corresponding increases in the phylum Proteobacteria, specifically the genera Lactobacillus. These microbial changes were accompanied by an overall decrease in peripheral insulin sensitivity. No effect was observed with amoxicillin [151].
Impact of the gut microbiota on the gut-brain axis
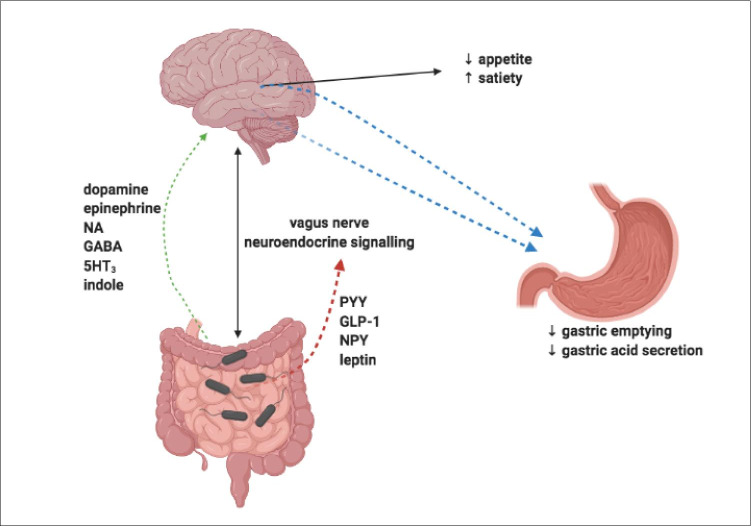
The gut-brain axis is a bi-directional signalling pathway regulating metabolism through balancing food intake and energy expenditure [152] whilst also influencing behaviour and brain function [153]. The gut signals to the brain via the central nervous system or acting through microbiota-derived metabolites, which have a role in controlling appetite directly or indirectly. Metabolites such as GLP-1, PYY, leptin and ghrelin modify gut hormone secretion, which can impact hypothalamic neuroendocrine pathways [78, 154, 155]. In particular, GLP-1 and PYY have receptors expressed in the brain involved in the regulation of host energy balance [155].
PYY modulates appetite and satiety by the suppression of NPY and activating proopiomelanocortin (POMC) neurons in the arcuate nucleus (ARC) or by delaying gastric emptying [156, 157]. GLP-1 also regulates appetite via effects on POMC and NPY neurons in the ARC and contributes to the inhibition of gastric emptying and gastric acid secretion [158, 159] (Fig. 7).
Fig. 7.
Gut microbiota effects on the gut-brain axis. The gut-brain axis is a bi-directional signalling pathway influencing food intake, energy expenditure, behaviour and brain function. The gut signals to the brain via a combination of the central nervous system and microbiota-derived metabolites. PYY modulates appetite and satiety by the suppression of NPY and activating POMC neurons in the ARC or by delaying gastric emptying. GLP-1 also regulates appetite via effects on POMC and NPY neurons in the ARC and contributes to the inhibition of gastric emptying and gastric acid secretion. Dopamine, epinephrine, norepinephrine, gamma-aminobutyric acid, serotonin and indole are derived by gut microbiota and may affect a persons’ dietary preference
There is growing evidence that dopamine, epinephrine, norepinephrine, gamma-aminobutyric acid, serotonin, indole metabolites and other neurotransmitters derived by gut microbiota could affect a persons’ dietary preference [153].
Future Direction
The discovery of a link between gut microbiota and global metabolic disorders such as obesity and T2DM is undeniably exciting with the tantalizing possibility of targeting therapeutics for the production of personalised treatments. Well-designed, tightly controlled prospective human and animal studies are still required to establish causality between microbiota and metabolic disease. The vast majority of current gut microbiota human studies represent simple associations only; from which it is extremely difficult to interpret any firm causal conclusions. However, by using next-generation technology such as metagenomics and metabolomics to target microbial combinations with similar functions, we should develop a better understanding of the relationship between gut microbiota and metabolic disease.
Interventional studies surrounding microbiota transfer (faecal microbiota transplant, FMT) for metabolic disorders remain in their infancy, by using mainly GF-rodent models. These models provide crucial mechanistic information, however, it remains difficult to directly translate this data to humans with their inherent heterogeneity. We have already seen that gut microbiota can be disrupted by both host and environmental factors, so further investigations are required to fully appreciate this relationship.
Furthermore, by fully understanding the interplay between gut microbiota and metabolic disease, and by using methods such as microbiota transfer, pre/probiotic supplementation and dietary manipulation, we can design well-defined prospective studies to develop personalised treatments.
Conclusion
The increasing worldwide epidemic of obesity and T2DM is a cause of significant morbidity and cost. Both conditions are complex metabolic disorders with varying aetiology and early scientific evidence suggests that gut microbiota play a pivotal role in their development. The advancement of microbial analysis techniques and the combined usage of rodent and human models has allowed the ongoing investigation of the gut microbiota, as part of the patho-aetiology of T2DM and obesity, to become more achievable.
Gut microbiota acting both directly and indirectly through the degradation products of intestinal fermentation have the ability to manipulate host homeostasis, metabolic and inflammatory pathways. This influence can be both beneficial and detrimental to the host. This review has provided valuable insight into the many complex interactions that the gut microbiota initiate. The review appreciates the importance of the SCFAs, more specifically, acetate, butyrate and propionate, as well as bile acids, in initiating the inflammatory cascade, the disruption caused to microbiota associated with antibiotic treatments, their ability to control the permeability of the mucosal barrier and subsequent manipulation of the gut-brain axis to alter host behaviour.
Further detailed scientific work is necessary towards establishing the gut microbiota’s role in its ability to control the disease processes for the future purpose of treating both obesity and T2DM.
Acknowledgements
Figures have been produced using Biorender.com.
Abbreviations
- AKT
Protein kinase B
- ARC
Arcuate nucleus
- ATP
Adenosine triphosphate
- BA
Bile acid
- BCAA
Branched chain amino acid
- BMI
Body mass index
- CB1
Cannabinoid receptor type-1
- CD14
Cluster of differentiation 14
- CH4
Methane
- CO2
Carbon dioxide
- CoA
Coenzyme A
- CRP
C-reactive protein
- CYP7A1
Cytochrome P450 enzyme cholesterol 7α-hydroxylase
- DNA
Deoxyribonucleic acid
- FFAR
Free fatty acid receptor
- FGF-19
Fibroblast growth factor 19
- FXR
Farnesoid X receptor
- GF
Germ free
- GIT
Gastrointestinal tract
- GLUT-4
Glucose transporter type-4
- GLP-1
Glucagon-like peptide 1
- GPR
G protein coupled receptor
- H2
Hydrogen
- HFD
High fat diet
- 5-HT3
Serotonin
- IKK
Inhibitor of nuclear factor-kB kinase
- IL
Interleukin
- iNOS
Inducible nitric oxide synthase
- IRAK
Interleukin-1 receptor-associated kinase
- IRS-1
Insulin receptor substrate 1
- JNK
Jun N-terminal Kinase
- Kg
Kilograms
- LBP
Lipopolysaccharide binding protein
- LPS
Lipopolysaccharide
- MCP-1
Monocyte chemoattractant protein-1
- mTORC1
Mammalian target of rapamycin complex 1
- MyD88
Myeloid differentiation primary response 88
- MyD88 MAL
Myeloid differentiation primary response 88 adaptor-like
- NF-kB
Nuclear factor kappa B
- NLRP3
Nucleotide-binding domain-like receptor protein 3
- NLRs
Nucleotide-binding oligomerization domain-like receptors
- NO
Nitrous oxide
- NPY
Neuropeptide Y
- POMC
Proopiomelanocortin
- PPARy
Peroxisome proliferator-activated receptor gamma
- PYY
Peptide tyrosine-tyrosine
- RNA
Ribonucleic acid
- SCFA
Short chain fatty acid
- SOCS-3
Suppressor of cytokine signalling 3
- TAK1
Transforming growth factor B-activated kinase 1
- TGR5
G protein-coupled bile acid receptor 1
- TLRs
Toll-like receptors
- T2DM
Type two diabetes mellitus
- TNF
Tumour necrosis factor
- TRAF6
Tumour necrosis factor receptor-associated factor 6
- UN
United Nations
- WHO
World Health Organization
Author’s contributions
AC performed the relevant background / literature searches for the major construction of this review. DH and JS were major contributors in the editing of this paper. All authors read and approved the final manuscript.
Data availability
Data sharing not applicable to this article as no datasets were generated or analysed during the completion of this review.
Code availability
Not applicable.
Declarations
Conflicts of interest / Competing interests
The authors declare that they have no financial or non-financial competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors consent to this publication.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
A. L. Cunningham, Email: cunninghamal@doctors.org.uk
J. W. Stephens, Email: J.W.Stephens@swansea.ac.uk
D. A. Harris, Email: Dean.A.Harris@wales.nhs.uk
References
- 1.Obesity and overweight. World Health Organisation; 2018–02–09. Available from: http://www.who.int/mediacentre/factsheets/fs311/en. Accessed 01 March 2021.
- 2.Power ML, Schulkin J. The evolution of obesity. Baltimore, MD: The Johns Hopkins University Press; 2009
- 3.International Diabetes Federation. IDF Diabetes Atlas, Ninth Edition 2019 (IDF, 2019). https://www.diabetesatlas.org/en. Accessed 01 March 2021
- 4.World Health Organisation. Global Report on Diabetes (WHO, 2016). https://www.who.int/publications/i/item/9789241565257. Accessed 01 March 2021.
- 5.United Nations General Assembly. Political declaration of the high-level meeting of the general assembly on the prevention and control of non-communicable diseases (UN, 2012). https://www.who.int/nmh/events/un_ncd_summit2011/political_declaration_en.pdf. Accessed 01 March 2021.
- 6.American Diabetes Association. Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010;33 Suppl 1(Suppl 1):S62-S69. 10.2337/dc10-S062 [DOI] [PMC free article] [PubMed]
- 7.Ma R, Chan J. Metabolic complications of obesity. In: Williams G, Fruhbeck G, eds. Obesity: Science to Practice. John Wiley & Sons Ltd, 2009: 235–270
- 8.Bommer C, Sagalova V, Heesemann E, et al. Global Economic Burden of Diabetes in Adults: Projections From 2015 to 2030. Diabetes Care. 2018;41(5):963–970. 10.2337/dc17-1962 [DOI] [PubMed]
- 9.Aydin Ö, Nieuwdorp M, Gerdes V. The gut microbiome as a target for the treatment of type 2 diabetes. Curr Diab Rep. 2018;18(8):55. 10.1007/s11892-018-1020-6 [DOI] [PMC free article] [PubMed]
- 10.Lederberg J. Infectious history. Science. 2000;288(5464):287–293. 10.1126/science.288.5464.287 [DOI] [PubMed]
- 11.Bested AC, Logan AC, Selhub EM. Intestinal microbiota, probiotics and mental health: from Metchnikoff to modern advances: Part I - autointoxication revisited. Gut Pathog. 2013;5(1):5. 10.1186/1757-4749-5-5 [DOI] [PMC free article] [PubMed]
- 12.Lederberg J, McCray A. "Ome SweetOmics--A genealogical treasury of words." The scientist. 2001;15.7: 8–8.
- 13.Gill SR, Pop M, Deboy RT, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312(5778):1355–1359. 10.1126/science.1124234 [DOI] [PMC free article] [PubMed]
- 14.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed]
- 15.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7(7):688–693. 10.1038/sj.embor.7400731 [DOI] [PMC free article] [PubMed]
- 16.D'Argenio V. Human microbiome acquisition and bioinformatic challenges in metagenomic studies. Int J Mol Sci. 2018;19(2):383. 10.3390/ijms19020383 [DOI] [PMC free article] [PubMed]
- 17.Jovel J, Patterson J, Wang W, et al. Characterization of the gut microbiome using 16S or shotgun metagenomics. Front Microbiol. 2016;7:459. 10.3389/fmicb.2016.00459 [DOI] [PMC free article] [PubMed]
- 18.Harsch IA, Konturek PC. The role of gut microbiota in obesity and type 2 and type 1 diabetes mellitus: new insights into "Old" Diseases. Med Sci (Basel). 2018;6(2):32. 10.3390/medsci6020032 [DOI] [PMC free article] [PubMed]
- 19.Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22:283–307. 10.1146/annurev.nutr.22.011602.092259 [DOI] [PubMed]
- 20.Nicholson JK, Holmes E, Wilson ID. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3(5):431–438. 10.1038/nrmicro1152 [DOI] [PubMed]
- 21.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–810. 10.1038/nature06244 [DOI] [PMC free article] [PubMed]
- 22.Delzenne NM, Cani PD. Gut microbiota and the pathogenesis of insulin resistance. Curr Diab Rep. 2011;11(3):154–159. 10.1007/s11892-011-0191-1 [DOI] [PubMed]
- 23.Kootte RS, Vrieze A, Holleman F, et al. The therapeutic potential of manipulating gut microbiota in obesity and type 2 diabetes mellitus. Diabetes Obes Metab. 2012;14(2):112–120. 10.1111/j.1463-1326.2011.01483.x [DOI] [PubMed]
- 24.Blaut M. Ecology and physiology of the intestinal tract. Curr Top Microbiol Immunol. 2013;358:247–272. 10.1007/82_2011_192 [DOI] [PubMed]
- 25.Scheithauer TP, Dallinga-Thie GM, de Vos WM, Nieuwdorp M, van Raalte DH. Causality of small and large intestinal microbiota in weight regulation and insulin resistance. Mol Metab. 2016;5(9):759–770. 10.1016/j.molmet.2016.06.002 [DOI] [PMC free article] [PubMed]
- 26.:Meijnikman AS, Gerdes VE, Nieuwdorp M, Herrema H. Evaluating causality of gut microbiota in obesity and diabetes in humans. Endocr Rev. 2018;39(2):133–153. 10.1210/er.2017-00192 [DOI] [PubMed]
- 27.Saad MJ, Santos A, Prada PO. Linking gut microbiota and inflammation to obesity and insulin resistance. Physiology (Bethesda). 2016;31(4):283–293. 10.1152/physiol.00041.2015 [DOI] [PubMed]
- 28.Falony G, Joossens M, Vieira-Silva S, et al. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–564. 10.1126/science.aad3503 [DOI] [PubMed]
- 29.Murphy EF, Cotter PD, Healy S, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59(12):1635–1642. 10.1136/gut.2010.215665 [DOI] [PubMed]
- 30.Brown K, DeCoffe D, Molcan E, Gibson DL. Diet-induced dysbiosis of the intestinal microbiota and the effects on immunity and disease. Nutrients. 2012;4(8):1095–1119. 10.3390/nu4081095 [DOI] [PMC free article] [PubMed]
- 31.Guarner F, Malagelada JR. Gut flora in health and disease. Lancet. 2003;361(9356):512–519. 10.1016/S0140-6736(03)12489-0 [DOI] [PubMed]
- 32.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed]
- 33.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474(7351):327–336. 10.1038/nature10213 [DOI] [PMC free article] [PubMed]
- 34.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375(24):2369–2379. 10.1056/NEJMra1600266 [DOI] [PubMed]
- 35.Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol. 2006;40(3):235–243. 10.1097/00004836-200603000-00015 [DOI] [PubMed]
- 36.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science. 2005;307(5717):1915–1920. 10.1126/science.1104816 [DOI] [PubMed]
- 37.Koliwad SK, Kuo T, Shipp LE, et al. Angiopoietin-like 4 (ANGPTL4, fasting-induced adipose factor) is a direct glucocorticoid receptor target and participates in glucocorticoid-regulated triglyceride metabolism. J Biol Chem. 2009;284(38):25593–25601. 10.1074/jbc.M109.025452 [DOI] [PMC free article] [PubMed]
- 38.Sohail MU, Althani A, Anwar H, Rizzi R, Marei HE. Role of the gastrointestinal tract microbiome in the pathophysiology of diabetes mellitus. J Diabetes Res. 2017;2017:9631435. 10.1155/2017/9631435 [DOI] [PMC free article] [PubMed]
- 39.Singer-Englar T, Barlow G, Mathur R. Obesity, diabetes, and the gut microbiome: an updated review. Expert Rev Gastroenterol Hepatol. 2019;13(1):3–15. 10.1080/17474124.2019.1543023 [DOI] [PubMed]
- 40.Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16(6):341–352. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed]
- 41.Baothman OA, Zamzami MA, Taher I, Abubaker J, Abu-Farha M. The role of gut microbiota in the development of obesity and diabetes. Lipids Health Dis. 2016;15:108. 10.1186/s12944-016-0278-4 [DOI] [PMC free article] [PubMed]
- 42.Komaroff AL. The microbiome and risk for obesity and diabetes. JAMA. 2017;317(4):355–356. 10.1001/jama.2016.20099 [DOI] [PubMed]
- 43.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11(10):577–591. 10.1038/nrendo.2015.128 [DOI] [PubMed]
- 44.Roy CC, Kien CL, Bouthillier L, Levy E. Short-chain fatty acids: ready for prime time?. Nutr Clin Pract. 2006;21(4):351–366. 10.1177/0115426506021004351 [DOI] [PubMed]
- 45.Khan MT, Nieuwdorp M, Bäckhed F. Microbial modulation of insulin sensitivity. Cell Metab. 2014;20(5):753–760. 10.1016/j.cmet.2014.07.006 [DOI] [PubMed]
- 46.Krajmalnik-Brown R, Ilhan ZE, Kang DW, DiBaise JK. Effects of gut microbes on nutrient absorption and energy regulation. Nutr Clin Pract. 2012;27(2):201–214. 10.1177/0884533611436116 [DOI] [PMC free article] [PubMed]
- 47.Bauchop T, Mountfort DO. Cellulose fermentation by a rumen anaerobic fungus in both the absence and the presence of rumen methanogens. Appl Environ Microbiol. 1981;42(6):1103–1110. 10.1128/AEM.42.6.1103-1110.1981 [DOI] [PMC free article] [PubMed]
- 48.Rosenbaum M, Knight R, Leibel RL. The gut microbiota in human energy homeostasis and obesity. Trends Endocrinol Metab. 2015;26(9):493–501. 10.1016/j.tem.2015.07.002 [DOI] [PMC free article] [PubMed]
- 49.Bergman EN. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol Rev. 1990;70(2):567–590. 10.1152/physrev.1990.70.2.567 [DOI] [PubMed]
- 50.Shanahan F. The host-microbe interface within the gut. Best Pract Res Clin Gastroenterol. 2002;16(6):915–931. 10.1053/bega.2002.0342 [DOI] [PubMed]
- 51.Macfarlane GT, Macfarlane S. Bacteria, colonic fermentation, and gastrointestinal health. J AOAC Int. 2012;95(1):50–60. 10.5740/jaoacint.sge_macfarlane [DOI] [PubMed]
- 52.Willemsen LE, Koetsier MA, van Deventer SJ, van Tol EA. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E(1) and E(2) production by intestinal myofibroblasts. Gut. 2003;52(10):1442–1447. 10.1136/gut.52.10.1442 [DOI] [PMC free article] [PubMed]
- 53.Feuerer M, Herrero L, Cipolletta D, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15(8):930–939. 10.1038/nm.2002 [DOI] [PMC free article] [PubMed]
- 54.Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016;20(2):202–214. 10.1016/j.chom.2016.07.001 [DOI] [PMC free article] [PubMed]
- 55.Ohira H, Tsutsui W, Fujioka Y. Are short chain fatty acids in gut microbiota defensive players for inflammation and atherosclerosis?. J Atheroscler Thromb. 2017;24(7):660–672. 10.5551/jat.RV17006 [DOI] [PMC free article] [PubMed]
- 56.Belzer C, de Vos WM. Microbes inside--from diversity to function: the case of Akkermansia. ISME J. 2012;6(8):1449–1458. 10.1038/ismej.2012.6 [DOI] [PMC free article] [PubMed]
- 57.Le Poul E, Loison C, Struyf S, et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem. 2003;278(28):25481–25489. 10.1074/jbc.M301403200 [DOI] [PubMed]
- 58.Husted AS, Trauelsen M, Rudenko O, Hjorth SA, Schwartz TW. GPCR-Mediated Signaling of Metabolites. Cell Metab. 2017;25(4):777–796. 10.1016/j.cmet.2017.03.008 [DOI] [PubMed]
- 59.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27(2):104–119. 10.1111/j.1365-2036.2007.03562.x [DOI] [PubMed]
- 60.Mandøe MJ, Hansen KB, Hartmann B, Rehfeld JF, Holst JJ, Hansen HS. The 2-monoacylglycerol moiety of dietary fat appears to be responsible for the fat-induced release of GLP-1 in humans. Am J Clin Nutr. 2015;102(3):548–555. 10.3945/ajcn.115.106799 [DOI] [PubMed]
- 61.Ahmed K, Tunaru S, Offermanns S. GPR109A, GPR109B and GPR81, a family of hydroxy-carboxylic acid receptors. Trends Pharmacol Sci. 2009;30(11):557–562. 10.1016/j.tips.2009.09.001 [DOI] [PubMed]
- 62.Brown AJ, Goldsworthy SM, Barnes AA, et al. The orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem. 2003;278(13):11312–11319. 10.1074/jbc.M211609200 [DOI] [PubMed]
- 63.Karaki S, Tazoe H, Hayashi H, et al. Expression of the short-chain fatty acid receptor, GPR43, in the human colon. J Mol Histol. 2008;39(2):135–142. 10.1007/s10735-007-9145-y [DOI] [PubMed]
- 64.Xiong Y, Miyamoto N, Shibata K, et al. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc Natl Acad Sci U S A. 2004;101(4):1045–1050. 10.1073/pnas.2637002100 [DOI] [PMC free article] [PubMed]
- 65.Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008;105(43):16767–16772. 10.1073/pnas.0808567105 [DOI] [PMC free article] [PubMed]
- 66.Tazoe H, Otomo Y, Kaji I, Tanaka R, Karaki SI, Kuwahara A. Roles of short-chain fatty acids receptors, GPR41 and GPR43 on colonic functions. J Physiol Pharmacol. 2008;59 Suppl 2:251–262. [PubMed]
- 67.Tolhurst G, Heffron H, Lam YS, et al. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein-coupled receptor FFAR2. Diabetes. 2012;61(2):364–371. 10.2337/db11-1019 [DOI] [PMC free article] [PubMed]
- 68.Amato A, Cinci L, Rotondo A, et al. Peripheral motor action of glucagon-like peptide-1 through enteric neuronal receptors. Neurogastroenterol Motil. 2010;22(6):664-e203. 10.1111/j.1365-2982.2010.01476.x [DOI] [PubMed]
- 69.Ge H, Li X, Weiszmann J, et al. Activation of G protein-coupled receptor 43 in adipocytes leads to inhibition of lipolysis and suppression of plasma free fatty acids. Endocrinology. 2008;149(9):4519–4526. 10.1210/en.2008-0059 [DOI] [PubMed]
- 70.Zhao Y, Chen F, Wu W, et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018;11(3):752–762. 10.1038/mi.2017.118 [DOI] [PMC free article] [PubMed]
- 71.Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145(2). 10.1053/j.gastro.2013.04.056 [DOI] [PubMed]
- 72.Kimura I, Ozawa K, Inoue D, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. 10.1038/ncomms2852 [DOI] [PMC free article] [PubMed]
- 73.Cresci GA, Thangaraju M, Mellinger JD, Liu K, Ganapathy V. Colonic gene expression in conventional and germ-free mice with a focus on the butyrate receptor GPR109A and the butyrate transporter SLC5A8. J Gastrointest Surg. 2010;14(3):449–461. 10.1007/s11605-009-1045-x [DOI] [PubMed]
- 74.Singh N, Gurav A, Sivaprakasam S, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40(1):128–139. 10.1016/j.immuni.2013.12.007 [DOI] [PMC free article] [PubMed]
- 75.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc Natl Acad Sci U S A. 2006;103(26):10011–10016. 10.1073/pnas.0602187103 [DOI] [PMC free article] [PubMed]
- 76.Cook SI, Sellin JH. Review article: short chain fatty acids in health and disease. Aliment Pharmacol Ther. 1998;12(6):499–507. 10.1046/j.1365-2036.1998.00337.x [DOI] [PubMed]
- 77.Frost G, Sleeth ML, Sahuri-Arisoylu M, et al. The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun. 2014;5:3611. 10.1038/ncomms4611 [DOI] [PMC free article] [PubMed]
- 78.Perry RJ, Peng L, Barry NA, et al. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature. 2016;534(7606):213–217. 10.1038/nature18309 [DOI] [PMC free article] [PubMed]
- 79.Topping DL, Clifton PM. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol Rev. 2001;81(3):1031–1064. 10.1152/physrev.2001.81.3.1031 [DOI] [PubMed]
- 80.De Vadder F, Kovatcheva-Datchary P, Goncalves D, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014;156(1–2):84–96. 10.1016/j.cell.2013.12.016 [DOI] [PubMed]
- 81.Lazarova DL, Chiaro C, Wong T, et al. CBP activity mediates effects of the histone deacetylase inhibitor butyrate on WNT activity and apoptosis in colon cancer cells. J Cancer. 2013;4(6):481–490. 10.7150/jca.6583 [DOI] [PMC free article] [PubMed]
- 82.Kelly CJ, Zheng L, Campbell EL, et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17(5):662–671. 10.1016/j.chom.2015.03.005 [DOI] [PMC free article] [PubMed]
- 83.Maslowski KM, Vieira AT, Ng A, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461(7268):1282–1286. 10.1038/nature08530 [DOI] [PMC free article] [PubMed]
- 84.Martin FP, Wang Y, Sprenger N, et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol Syst Biol. 2008;4:157. 10.1038/msb4100190 [DOI] [PMC free article] [PubMed]
- 85.Donohoe DR, Garge N, Zhang X, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13(5):517–526. 10.1016/j.cmet.2011.02.018 [DOI] [PMC free article] [PubMed]
- 86.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133(7 Suppl):2485S-2493S. 10.1093/jn/133.7.2485S [DOI] [PubMed]
- 87.Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci U S A. 2014;111(6):2247–2252. 10.1073/pnas.1322269111 [DOI] [PMC free article] [PubMed]
- 88.Venter CS, Vorster HH, Cummings JH. Effects of dietary propionate on carbohydrate and lipid metabolism in healthy volunteers. Am J Gastroenterol. 1990;85(5):549–553. [PubMed]
- 89.Baird GD, Lomax MA, Symonds HW, Shaw SR. Net hepatic and splanchnic metabolism of lactate, pyruvate and propionate in dairy cows in vivo in relation to lactation and nutrient supply. Biochem J. 1980;186(1):47–57. 10.1042/bj1860047 [DOI] [PMC free article] [PubMed]
- 90.Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochim Biophys Acta. 2010;1801(11):1175–1183. 10.1016/j.bbalip.2010.07.007 [DOI] [PubMed]
- 91.Foley JE. Rationale and application of fatty acid oxidation inhibitors in treatment of diabetes mellitus. Diabetes Care. 1992;15(6):773–784. 10.2337/diacare.15.6.773 [DOI] [PubMed]
- 92.Alhabeeb H, Chambers ES, Frost G, Morrison DJ, Preston T. Inulin propionate ester increases satiety and decreases appetite but does not affect gastric emptying in healthy humans. Proceedings of the Nutrition Society. 2014;73(OCE1).
- 93.Mortensen PB, Clausen MR. Short-chain fatty acids in the human colon: relation to gastrointestinal health and disease. Scand J Gastroenterol Suppl. 1996;216:132–148. 10.3109/00365529609094568 [DOI] [PubMed]
- 94.Dai ZL, Wu G, Zhu WY. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front Biosci (Landmark Ed). 2011;16:1768–1786. 10.2741/3820 [DOI] [PubMed]
- 95.Macfarlane GT, Allison C, Gibson SA, Cummings JH. Contribution of the microflora to proteolysis in the human large intestine. J Appl Bacteriol. 1988;64(1):37–46. 10.1111/j.1365-2672.1988.tb02427.x [DOI] [PubMed]
- 96.De Vadder F, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Bäckhed F, Mithieux G. Microbiota-Produced Succinate Improves Glucose Homeostasis via Intestinal Gluconeogenesis. Cell Metab. 2016;24(1):151–157. 10.1016/j.cmet.2016.06.013 [DOI] [PubMed]
- 97.Pedersen HK, Gudmundsdottir V, Nielsen HB, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. 10.1038/nature18646 [DOI] [PubMed]
- 98.Wang TJ, Larson MG, Vasan RS, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–453. 10.1038/nm.2307 [DOI] [PMC free article] [PubMed]
- 99.de Mello VD, Paananen J, Lindström J, et al. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the Finnish Diabetes Prevention Study. Sci Rep. 2017;7:46337. 10.1038/srep46337 [DOI] [PMC free article] [PubMed]
- 100.Chimerel C, Emery E, Summers DK, Keyser U, Gribble FM, Reimann F. Bacterial metabolite indole modulates incretin secretion from intestinal enteroendocrine L cells. Cell Rep. 2014;9(4):1202–1208. 10.1016/j.celrep.2014.10.032 [DOI] [PMC free article] [PubMed]
- 101.Gribble FM, Reimann F. Function and mechanisms of enteroendocrine cells and gut hormones in metabolism. Nat Rev Endocrinol. 2019;15(4):226–237. 10.1038/s41574-019-0168-8 [DOI] [PubMed]
- 102.Koh A, Molinaro A, Ståhlman M, et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell. 2018;175(4):947–961.e17. 10.1016/j.cell.2018.09.055 [DOI] [PubMed]
- 103.Thomas C, Gioiello A, Noriega L, et al. TGR5-mediated bile acid sensing controls glucose homeostasis. Cell Metab. 2009;10(3):167–177. 10.1016/j.cmet.2009.08.001 [DOI] [PMC free article] [PubMed]
- 104.Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science. 2012;336(6086):1262–1267. 10.1126/science.1223813 [DOI] [PubMed]
- 105.Swann JR, Want EJ, Geier FM, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A. 2011;108 Suppl 1(Suppl 1):4523–4530. 10.1073/pnas.1006734107 [DOI] [PMC free article] [PubMed]
- 106.Shapiro H, Kolodziejczyk AA, Halstuch D, Elinav E. Bile acids in glucose metabolism in health and disease. J Exp Med. 2018;215(2):383–396. 10.1084/jem.20171965 [DOI] [PMC free article] [PubMed]
- 107.Kir S, Beddow SA, Samuel VT, et al. FGF19 as a postprandial, insulin-independent activator of hepatic protein and glycogen synthesis. Science. 2011;331(6024):1621–1624. 10.1126/science.1198363 [DOI] [PMC free article] [PubMed]
- 108.Cicione C, Degirolamo C, Moschetta A. Emerging role of fibroblast growth factors 15/19 and 21 as metabolic integrators in the liver. Hepatology. 2012;56(6):2404–2411. 10.1002/hep.25929 [DOI] [PubMed]
- 109.Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes Dev. 2012;26(4):312–324. 10.1101/gad.184788.111 [DOI] [PMC free article] [PubMed]
- 110.Lazar V, Ditu LM, Pircalabioru GG, et al. Gut microbiota, host organism, and diet trialogue in diabetes and obesity. Front Nutr. 2019;6:21. 10.3389/fnut.2019.00021 [DOI] [PMC free article] [PubMed]
- 111.Sayin SI, Wahlström A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–235. 10.1016/j.cmet.2013.01.003 [DOI] [PubMed]
- 112.Watanabe M, Houten SM, Mataki C, et al. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature. 2006;439(7075):484–489. 10.1038/nature04330 [DOI] [PubMed]
- 113.Hartmann P, Chen P, Wang HJ, et al. Deficiency of intestinal mucin-2 ameliorates experimental alcoholic liver disease in mice. Hepatology. 2013;58(1):108–119. 10.1002/hep.26321 [DOI] [PMC free article] [PubMed]
- 114.Everard A, Cani PD. Diabetes, obesity and gut microbiota. Best Pract Res Clin Gastroenterol. 2013;27(1):73–83. 10.1016/j.bpg.2013.03.007 [DOI] [PubMed]
- 115.Tomasova L, Konopelski P, Ufnal M. Gut Bacteria and Hydrogen sulfide: the new old players in circulatory system homeostasis. Molecules. 2016;21(11):1558. 10.3390/molecules21111558 [DOI] [PMC free article] [PubMed]
- 116.Derrien M, Van Baarlen P, Hooiveld G, Norin E, Müller M, de Vos WM. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader akkermansia muciniphila. Front Microbiol. 2011;2:166. 10.3389/fmicb.2011.00166 [DOI] [PMC free article] [PubMed]
- 117.Everard A, Belzer C, Geurts L, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110(22):9066–9071. 10.1073/pnas.1219451110 [DOI] [PMC free article] [PubMed]
- 118.Wang HB, Wang PY, Wang X, Wan YL, Liu YC. Butyrate enhances intestinal epithelial barrier function via up-regulation of tight junction protein Claudin-1 transcription. Dig Dis Sci. 2012;57(12):3126–3135. 10.1007/s10620-012-2259-4 [DOI] [PubMed]
- 119.Burger-van Paassen N, Vincent A, Puiman PJ, et al. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J. 2009;420(2):211–219. 10.1042/BJ20082222 [DOI] [PubMed]
- 120.Muccioli GG, Naslain D, Bäckhed F, et al. The endocannabinoid system links gut microbiota to adipogenesis. Mol Syst Biol. 2010;6:392. 10.1038/msb.2010.46 [DOI] [PMC free article] [PubMed]
- 121.Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes. 2012;3(4):279–288. 10.4161/gmic.19625 [DOI] [PMC free article] [PubMed]
- 122.Cani PD, Possemiers S, Van de Wiele T, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58(8):1091–1103. 10.1136/gut.2008.165886 [DOI] [PMC free article] [PubMed]
- 123.Brun P, Castagliuolo I, Di Leo V, et al. Increased intestinal permeability in obese mice: new evidence in the pathogenesis of nonalcoholic steatohepatitis. Am J Physiol Gastrointest Liver Physiol. 2007;292(2):G518-G525. 10.1152/ajpgi.00024.2006 [DOI] [PubMed]
- 124.Hansen J, Gulati A, Sartor RB. The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Curr Opin Gastroenterol. 2010;26(6):564–571. 10.1097/MOG.0b013e32833f1195 [DOI] [PMC free article] [PubMed]
- 125.Kashyap PC, Marcobal A, Ursell LK, et al. Complex interactions among diet, gastrointestinal transit, and gut microbiota in humanized mice. Gastroenterology. 2013;144(5):967–977. 10.1053/j.gastro.2013.01.047 [DOI] [PMC free article] [PubMed]
- 126.Fukumoto S, Tatewaki M, Yamada T, et al. Short-chain fatty acids stimulate colonic transit via intraluminal 5-HT release in rats. Am J Physiol Regul Integr Comp Physiol. 2003;284(5):R1269-R1276. 10.1152/ajpregu.00442.2002 [DOI] [PubMed]
- 127.Dandona P, Aljada A, Bandyopadhyay A. Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol. 2004;25(1):4–7. 10.1016/j.it.2003.10.013 [DOI] [PubMed]
- 128.Reyes M, Quintanilla C, Burrows R, Blanco E, Cifuentes M, Gahagan S. Obesity is associated with acute inflammation in a sample of adolescents. Pediatr Diabetes. 2015;16(2):109–116. 10.1111/pedi.12129 [DOI] [PMC free article] [PubMed]
- 129.Delzenne NM, Cani PD, Everard A, Neyrinck AM, Bindels LB. Gut microorganisms as promising targets for the management of type 2 diabetes. Diabetologia. 2015;58(10):2206–2217. 10.1007/s00125-015-3712-7 [DOI] [PubMed]
- 130.Hotamisligil GS, Shargill NS, Spiegelman BM. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259(5091):87–91. 10.1126/science.7678183 [DOI] [PubMed]
- 131.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26(1):5–11. 10.1097/MOG.0b013e328333d751 [DOI] [PubMed]
- 132.Cani PD, Bibiloni R, Knauf C, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57(6):1470–1481. 10.2337/db07-1403 [DOI] [PubMed]
- 133.Cani PD, Neyrinck AM, Fava F, et al. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia. 2007;50(11):2374–2383. 10.1007/s00125-007-0791-0 [DOI] [PubMed]
- 134.Lindberg AA, Weintraub A, Zähringer U, Rietschel ET. Structure-activity relationships in lipopolysaccharides of Bacteroides fragilis. Rev Infect Dis. 1990;12 Suppl 2:S133-S141. 10.1093/clinids/12.supplement_2.s133 [DOI] [PubMed]
- 135.Cani PD, Amar J, Iglesias MA, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56(7):1761–1772. 10.2337/db06-1491 [DOI] [PubMed]
- 136.Zhao L. The gut microbiota and obesity: from correlation to causality. Nat Rev Microbiol. 2013;11(9):639–647. 10.1038/nrmicro3089 [DOI] [PubMed]
- 137.Chen X, Devaraj S. Gut microbiome in obesity, metabolic syndrome, and diabetes. Curr Diab Rep. 2018;18(12):129. 10.1007/s11892-018-1104-3 [DOI] [PubMed]
- 138.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. 10.1016/j.cell.2004.07.002 [DOI] [PubMed]
- 139.Rui L, Yuan M, Frantz D, Shoelson S, White MF. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J Biol Chem. 2002;277(44):42394–42398. 10.1074/jbc.C200444200 [DOI] [PubMed]
- 140.Tsukumo DM, Carvalho-Filho MA, Carvalheira JB, et al. Loss-of-function mutation in Toll-like receptor 4 prevents diet-induced obesity and insulin resistance. Diabetes. 2007;56(8):1986–1998. 10.2337/db06-1595 [DOI] [PubMed]
- 141.Shi H, Kokoeva MV, Inouye K, Tzameli I, Yin H, Flier JS. TLR4 links innate immunity and fatty acid-induced insulin resistance. J Clin Invest. 2006;116(11):3015–3025. 10.1172/JCI28898 [DOI] [PMC free article] [PubMed]
- 142.Li X, Watanabe K, Kimura I. Gut microbiota dysbiosis drives and implies novel therapeutic strategies for diabetes mellitus and related metabolic diseases. Front Immunol. 2017;8:1882. 10.3389/fimmu.2017.01882 [DOI] [PMC free article] [PubMed]
- 143.de Lartigue G, Barbier de la Serre C, Espero E, Lee J, Raybould HE. Diet-induced obesity leads to the development of leptin resistance in vagal afferent neurons. Am J Physiol Endocrinol Metab. 2011;301(1):E187-E195. 10.1152/ajpendo.00056.2011 [DOI] [PMC free article] [PubMed]
- 144.Raybould HE. Gut microbiota, epithelial function and derangements in obesity. J Physiol. 2012;590(3):441–446. 10.1113/jphysiol.2011.222133 [DOI] [PMC free article] [PubMed]
- 145.O'Mahony D, Murphy S, Boileau T, et al. Bifidobacterium animalis AHC7 protects against pathogen-induced NF-κB activation in vivo. BMC Immunol. 2010;11:63. 10.1186/1471-2172-11-63 [DOI] [PMC free article] [PubMed]
- 146.Amar J, Chabo C, Waget A, et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: molecular mechanisms and probiotic treatment. EMBO Mol Med. 2011;3(9):559–572. 10.1002/emmm.201100159 [DOI] [PMC free article] [PubMed]
- 147.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1(1):56–66. 10.1038/ismej.2007.3 [DOI] [PubMed]
- 148.Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest. 2014;124(10):4212–4218. 10.1172/JCI72333 [DOI] [PMC free article] [PubMed]
- 149.Rasmussen SH, Shrestha S, Bjerregaard LG, et al. Antibiotic exposure in early life and childhood overweight and obesity: A systematic review and meta-analysis. Diabetes Obes Metab. 2018;20(6):1508–1514. 10.1111/dom.13230 [DOI] [PubMed]
- 150.Mikkelsen KH, Knop FK, Frost M, Hallas J, Pottegård A. Use of antibiotics and risk of type 2 diabetes: a population-based case-control study. J Clin Endocrinol Metab. 2015;100(10):3633–3640. 10.1210/jc.2015-2696 [DOI] [PMC free article] [PubMed]
- 151.Vrieze A, Out C, Fuentes S, et al. Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J Hepatol. 2014;60(4):824–831. 10.1016/j.jhep.2013.11.034 [DOI] [PubMed]
- 152.de Clercq NC, Frissen MN, Groen AK, Nieuwdorp M. Gut microbiota and the gut-brain axis: new insights in the pathophysiology of metabolic syndrome. Psychosom Med. 2017;79(8):874–879. 10.1097/PSY.0000000000000495 [DOI] [PubMed]
- 153.Cryan JF, Dinan TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13(10):701–712. 10.1038/nrn3346 [DOI] [PubMed]
- 154.Torres-Fuentes C, Schellekens H, Dinan TG, Cryan JF. The microbiota-gut-brain axis in obesity. Lancet Gastroenterol Hepatol. 2017;2(10):747–756. 10.1016/S2468-1253(17)30147-4 [DOI] [PubMed]
- 155.Heiss CN, Olofsson LE. Gut microbiota-dependent modulation of energy metabolism. J Innate Immun. 2018;10(3):163–171. 10.1159/000481519 [DOI] [PMC free article] [PubMed]
- 156.De Silva A, Bloom SR. Gut hormones and appetite control: a focus on PYY and GLP-1 as therapeutic targets in obesity. Gut Liver. 2012;6(1):10–20. 10.5009/gnl.2012.6.1.10 [DOI] [PMC free article] [PubMed]
- 157.Savage AP, Adrian TE, Carolan G, Chatterjee VK, Bloom SR. Effects of peptide YY (PYY) on mouth to caecum intestinal transit time and on the rate of gastric emptying in healthy volunteers. Gut. 1987;28(2):166–170. 10.1136/gut.28.2.166 [DOI] [PMC free article] [PubMed]
- 158.Murphy KG, Bloom SR. Gut hormones and the regulation of energy homeostasis. Nature. 2006;444(7121):854–859. 10.1038/nature05484 [DOI] [PubMed]
- 159.Näslund E, Bogefors J, Skogar S, et al. GLP-1 slows solid gastric emptying and inhibits insulin, glucagon, and PYY release in humans. Am J Physiol. 1999;277(3):R910-R916. 10.1152/ajpregu.1999.277.3.R910 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analysed during the completion of this review.
Not applicable.