Abstract
Notch receptors participate in a conserved signaling pathway that controls the development of diverse tissues and cell types, including lymphoid cells. Signaling is normally initiated through one or more ligand-mediated proteolytic cleavages that permit nuclear translocation of the intracellular portion of the Notch receptor (ICN), which then binds and activates transcription factors of the Su(H)/CBF1 family. Several mammalian Notch receptors are oncogenic when constitutively active, including Notch1, a gene initially identified based on its involvement in a (7;9) chromosomal translocation found in sporadic T-cell lymphoblastic leukemias and lymphomas (T-ALL). To investigate which portions of ICN1 contribute to transformation, we performed a structure-transformation analysis using a robust murine bone marrow reconstitution assay. Both the ankyrin repeat and C-terminal transactivation domains were required for T-cell leukemogenesis, whereas the N-terminal RAM domain and a C-terminal domain that includes a PEST sequence were nonessential. Induction of T-ALL correlated with the transactivation activity of each Notch1 polypeptide when fused to the DNA-binding domain of GAL4, with the exception of polypeptides deleted of the ankyrin repeats, which lacked transforming activity while retaining strong transactivation activity. Transforming polypeptides also demonstrated moderate to strong activation of the Su(H)/CBF1-sensitive HES-1 promoter, while polypeptides with weak or absent activity on this promoter failed to cause leukemia. These experiments define a minimal transforming region for Notch1 in T-cell progenitors and suggest that leukemogenic signaling involves recruitment of transcriptional coactivators to ICN1 nuclear complexes.
Notch receptors are highly conserved type I transmembrane glycoproteins that regulate the morphogenesis of multicellular animals through a novel signaling pathway with pleiotropic effects of apoptosis, proliferation, and cellular differentiation (1, 11). An enlarging body of evidence suggests that normal signaling is triggered by several ligand-induced proteolytic cleavages that release the intracellular domain of Notch (ICN) from the cell membrane (6, 16, 18, 23, 25, 34, 48). This permits ICN to translocate to the nucleus, where it up-regulates the activity of downstream transcription factors of the Su(H)/CBF1 family (4, 7, 10, 18, 28, 50).
Pathophysiologic alterations in Notch signaling have been linked to several diseases, including certain forms of leukemia. The human Notch1 gene was originally identified by analysis of a recurrent (7;9) chromosomal translocation [t(7;9)] found in a subset of T-cell lymphoblastic leukemias and lymphomas (T-ALL) (9). The t(7;9) disrupts the Notch1 gene, fusing the portion encoding its intracellular domain (ICN1) to enhancer and promoter elements of the T-cell receptor β (TCRβ) gene. The resultant TCRβ-Notch1 fusion gene encodes a series of truncated t(7;9)-specific Notch1 polypeptides that localize to the nucleus and structurally resemble ICN1 (2). The leukemogenic potential of Notch1 was formally proven in our previous studies in which lethally irradiated mice were reconstituted with bone marrow (BM) cells transduced ex vivo by retroviruses encoding activated forms of Notch1. About 50% of animals reconstituted with BM cells expressing “gain-of-function” Notch alleles developed CD4+-CD8+ T-ALLs closely resembling their human counterparts by 10 to 40 weeks posttransplantation, whereas animals reconstituted with cells expressing full-length Notch1, which lacks intrinsic signaling activity, remained healthy (38). These findings are consistent with the hypothesis that leukemogenesis stems from a pathophysiologic increase in one or more Notch signals.
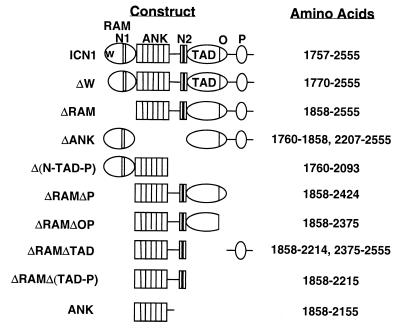
ICN1, like the intracellular portions of other Notch receptors, is composed of a series of distinct structural domains (see Fig. 1), a feature that lends itself to deletional analysis of function. The N-terminal portion of human ICN1 consists of an ∼110 amino acid RAM domain (amino acids 1757 to 1865), which contains a high-affinity binding site for Su(H)/CBF1 (51), and a functional nuclear localization signal sequence (3, 19). C terminal to the RAM domain lie six interated ankyrin repeats (ANK) (amino acids 1860 to 2155), a highly conserved domain necessary for all known Notch functions (1). Immediately C terminal to the ANK is a stretch of ∼100 amino acids that has been implicated in functional interaction with cytokine-signaling pathways (5), which is followed by a second functional nuclear localization sequence (3, 24). Amino acids 2155 to 2374 encompass a transcriptional activation domain (TAD) bounded at its C terminus by an OPA sequence (27), while C-terminal amino acids 2375 to 2555 include a PEST sequence. Although PEST sequences frequently target polypeptides for rapid degradation (47), the function of the PEST sequence in Notch1 is unknown.
FIG. 1.
Schematic representation of Notch1 polypeptides encoded by expression constructs. N1, nuclear localization signal sequence 1; N2, nuclear localization signal sequence 2; O, OPA sequence; P, PEST sequence.
Several different classes of activities that result in up-regulation of Su(H)/CBF1 appear to be potentially relevant to Notch1 induction of T-ALL. One involves displacement of transcriptional corepressors, such as CIR (14) and N-Cor/SMRT (21), from Su(H)/CBF1 upon binding of ICN1, thereby producing transcriptional activation through derepression (13). This activity has been investigated primarily using reporter genes containing artificial promoters consisting of iterated Su(H)/CBF1-binding sites and requires both the high-affinity RAM Su(H)/CBF1-binding domain and the ANK region (8, 35, 49). Once bound to transcription factors on DNA, several domains of ICN can also recruit transcriptional coactivators. The ANK of both GLP-1, a Caenorhabditis elegans Notch receptor, and human ICN1 (2) have the capacity to activate transcription in yeast when expressed as GAL4 fusion proteins, and mutations that abolish this activity inhibit function in vivo (24, 45). The C-terminal TAD of ICN1 has been shown to associate with the transcriptional coactivators PCAF and GCN5 (26). Genetic and structural analyses have also suggested the existence of another poorly understood ICN1 activity that is Su(H)/CBF1 independent (29, 30, 32, 33, 35, 36, 49). One proposed component of this pathway is Deltex, a modulator of Notch signaling that inhibits the activity of certain basic helix-loop-helix (bHLH) transcription factors (32, 36).
To begin to dissect the signals that contribute to leukemogenesis, we made modifications to our BM transplant (BMT) model that resulted in induction of Notch1-dependent T-ALL in 100% of animals by 15 weeks posttransplantation. This improved model has allowed us to perform a structure-transformation analysis of human ICN1 in vivo. We find that both the ANK and TAD are necessary for induction of T-ALL, while the RAM and PEST domains are dispensable. The minimal transforming region of ICN1 is a strong activator of transcription when fused to the DNA-binding domain of GAL4 and retains the capacity to activate transcription from the HES-1 promoter element in cultured cells. These findings indicate that Notch1 transformation of T-cell progenitors likely requires the recruitment of transcriptional coactivators to ICN1 nuclear complexes.
MATERIALS AND METHODS
cDNA expression constructs.
cDNAs encoding full-sized ICN1 and forms of ICN bearing various deletions (ΔW, ΔRAM, and ΔANK) have been previously described (3). Additional deletions were made in these cDNAs by digestion at unique restriction sites and ligation of compatible linker oligonucleotides. The amino acid sequences of polypeptides encoded by the resultant cDNAs are shown in Fig. 1. To permit the expression of GAL4 fusion proteins, in-frame restriction sites were introduced into the 5′ ends of cDNAs encoding various Notch1 polypeptides. These modified cDNAs were then ligated to the vector pM (Clontech), which permits expression of polypeptides that are fused at their amino termini to the DNA-binding domain of GAL4.
Production and characterization of retrovirus.
cDNAs ligated into the retroviral vector plasmid MigRI were transfected into the packaging cell line Bosc23 (41). Supernatants harvested 48 and 72 h after transfection were pooled and stored at −80°C until use. Retroviral titers were normalized by transduction of NIH 3T3 cells, which were assayed 48 h posttransduction for green fluorescent protein (GFP) positivity by flow cytometry, as previously described (43).
Harvest, transduction, and transplantation of murine BM cells.
All experiments were conducted in accordance with NIH guidelines for the care and use of animals and with an approved animal protocol from the University of Pennsylvania Animal Care and Use Committee. Transduction of BM cells from female BALB/c mice (Taconic Farms) with GFP-normalized retroviral supernatants and transplantation of these cells into lethally irradiated (900 rads) 4- to 8-week-old female syngeneic recipients was performed as previously described (40). Spinoculations were performed in medium containing interleukin-3 (6 ng/ml; R & D Systems), interleukin-6 (10 ng/ml; R & D Systems), SCF (100 ng/ml; R & D Systems), and 5% WEHI-conditioned supernatant as previously described (40). On day 3 following BM harvest, 3 × 105 cells were injected into syngeneic 4- to 8-week-old female mice that had been lethally irradiated (900 rads total in a split dose). Experiments with ΔW-, ΔRAMΔP-, ΔANK-, and ΔRAMΔOP-transduced BM cells were performed at least twice with two independently prepared aliquots of retroviral supernatants. In each cohort of animals reconstituted with BM cells transduced with these deleted forms of ICN1, positive and negative control transplants were also performed with BM cells transduced with leukemogenic forms of ICN1 (either ICN1 or ΔW) and empty MigRI, respectively. None of 20 MigRI control mice, derived from at least four independent experiments, have developed hematopoietic malignancy at >1 year posttransplantation.
Analysis of transplanted animals.
Mice were followed after transplantation by periodic peripheral blood sampling from retro-orbital sinuses and daily physical inspection. Peripheral blood (PB) was assessed beginning 2 weeks posttransplantation and every 2 weeks thereafter for the presence of GFP+ immature T cells by flow cytometric analysis using antibodies to CD4 and CD8 (see below). Tissues were harvested from diseased and unaffected animals after asphyxiation with CO2. Portions of BM, spleen, thymus, and tumor masses were used to prepare single-cell suspensions. For analyses on a Becton Dickinson FACSCalibur equipped with CellQuest software, suspensions of PB, BM, spleen, lymph node, or thymus cells were stained with the following antibodies (PharMingen): biotinylated CD11b/Mac-1 (M1/70), CD8α (53-6.7), Thy1.2 (53-2.1), or CD43 (Ly-48) and phycoerythrin-conjugated Gr-1 (RB6-8C5), CD4 (RM4-5), or CD45R (RA3-6B2). Biotinylated antibodies were revealed with streptavidin-CyChrome. Dead cells were identified by staining with TOPRO-III (Molecular Probes) and excluded from the analysis.
Expression of Notch1 polypeptides was determined by Western blot analysis of whole-cell detergent lysates with anti-Notch1 rabbit sera (3). DNA prepared from diseased and normal tissues by sodium dodecyl sulfate extraction and proteinase K digestion was analyzed on Southern blots hybridized to probes for the internal ribosomal entry site or murine TCRβ, as described previously (39, 40, 43).
Transient transfection assays.
cDNA inserts cloned into the plasmid pcDNA3 were transfected with Lipofectamine (Gibco BRL) into human 293A cells or murine NIH 3T3 cells. To assay activation of endogenous Su(H)/CBF1, cells were cotransfected with either HES-AB or HES-ΔAB plasmids containing firefly luciferase gene reporters (18) and the plasmid pRL-TK (Promega), which drives expression of a control sea pansy luciferase gene from the thymidine kinase reporter. The transactivation activity of GAL4 fusion polypeptides was assayed by cotransfection with various pM plasmids, GAL4X5 luciferase (a plasmid containing five Gal4 binding sites adjacent to a cDNA encoding firefly luciferase), and the pRL-TK sea pansy luciferase control plasmid. Normalized luciferase activities were determined in triplicate 48 h posttransfection in whole-cell lysates with the Promega Dual Luciferase Kit and a Turner Systems luminometer configured for dual assays.
RESULTS
ICN1 induces T-ALL in 100% BMT recipients.
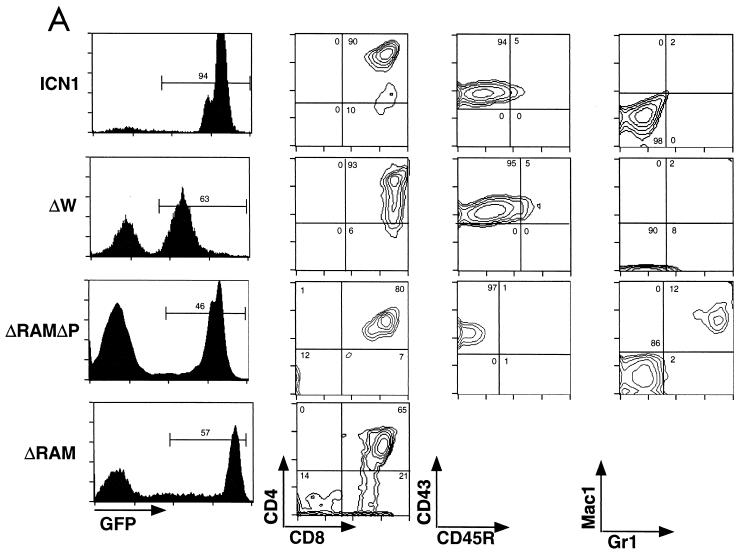
While our previous model established that ICN1 is an oncogene, T-ALL appeared in only 30 to 50% of BM-reconstituted mice, and cells transduced with the pGD retroviral vector were not readily identifiable, making it difficult to distinguish technical failures from true-negative results. To overcome these limitations, we have substituted a different retroviral vector, MigRI, which drives the expression of a single bicistronic transcript encoding proteins of interest and GFP, and altered the conditions of transduction of hematopoietic progenitors ex vivo (43). To assess the effects of these modifications, we initially assayed the ability of ICN1 to induce T-ALL. Six of six lethally irradiated recipient BALB/c mice receiving BM cells transduced with Mig ICN1 developed an abnormal CD4+-CD8+ double-positive (DP) GFP+ population of immature T cells in the PB as early as 36 days post-BMT. From 6 to 15 weeks following transplantation, the white blood cells (WBC) in each of the ICN1 mice increased, as did the number of GFP+ cells, with all animals succumbing to illness or becoming moribund by 15 weeks post-BMT. Gross and microscopic examination of tissues from diseased mice showed marked splenomegaly, hepatomegaly, and generalized lymphadenopathy due to extensive infiltration of organs by cells morphologically consistent with lymphoblasts, which also effaced and replaced the BM and variably involved the thymus, intestines, and kidneys (data not shown). Flow cytometry confirmed heavy infiltration of organs by DP cells expressing levels of GFP 2 to 3 logs higher than those of control cells (see Fig. 3A). None of six control mice receiving MigRI-transduced BM cells developed circulating DP cells or a hematopoietic malignancy in over 1 year of observation (data not shown). This robust model, in which 100% of mice receiving ICN1-transduced BM cells develop T-ALL with latencies of less than 16 weeks, permitted us to conduct an in vivo structure-transformation analysis of ICN1.
FIG. 3.
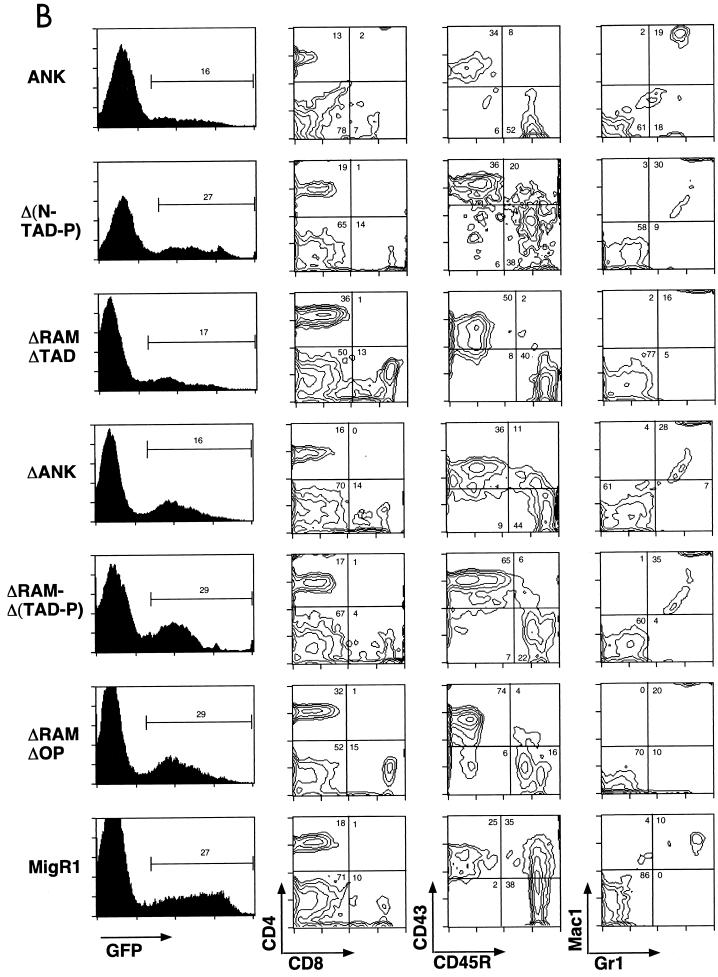
Immunophenotypes of splenic WBC from mice receiving transduced BM cells. (A) Presence of GFP+ DP T cells in mice reconstituted with BM transduced by leukemogenic Notch1 retroviruses. CD43, CD45R, Mac1, and Gr1 staining were not performed on cells obtained from ICNΔRAM animals. (B) Lack of GFP+ DP T cells in mice reconstituted with BM transduced by nonleukemogenic Notch1 retroviruses. GFP expression is shown on the left in both panels; gates used to identify the GFP+ population (given as percentages) are indicated. The antibodies used for staining are shown at the bottom adjacent to the relevant axis; numbers correspond to relative percentages within the GFP+ gate. The results shown are representative of individual analyses of each mouse included in the Table 1 summary. Similar results were also found in analyses of cells isolated from the PB, BM, and lymph nodes from each mouse.
Requirement for C-terminal Notch1 sequences.
Because certain Notch gain-of-function phenotypes in invertebrates and mammalian cells are produced by polypeptides consisting of only the ANK domain (46, 49), we first investigated the possibility that the Notch1 ANK would be sufficient to induce T-ALL. In the same cohort of animals, we also scored Notch1 polypeptides deleted of residues lying N terminal (ΔRAM) or C terminal of the ANK [Δ(N-TAD-P)] to assess the role of residues flanking the ANK (for structure of polypeptides, see Fig. 1). The activities of these polypeptides were compared to those of full-sized ICN1 and ΔW (3), a form of ICN1 lacking the first 13 N-terminal amino acids of the RAM domain.
All mice reconstituted with ICN1-, ΔW-, and ΔRAM-transduced BM cells developed an abnormal DP T-cell population in the PB between 23 and 107 days posttransplantation that expanded with time (Table 1 and Fig. 2). In this experiment and others, there was a trend toward slightly longer latencies in animals reconstituted with ΔW- and ΔRAM-transduced marrow as compared to the ICN1 cohort (Fig. 2 and data not shown). In contrast, DP T cells never appeared in the PB (or any organs outside of the thymus) of ANK and ΔN-TAD-P animals, despite the presence of 10 to 40% GFP+ circulating WBC (Fig. 3B). Disease progression, defined by cachexia and inactivity, were evident in all ICN1, ΔW, and ΔRAM animals, whereas all ANK and Δ(N-TAD-P) animals remained healthy until necropsy (up to 14 months post-BMT). Gross and microscopic examination of disease-free ANK and Δ(N-TAD-P) mice showed no evidence of hematologic malignancy (data not shown).
TABLE 1.
Summary of results of BMT experimentsa
| Notch1 construct | Mice
|
Onset (days)
|
||
|---|---|---|---|---|
| No. transplanted | No. developing leukemia | Mean | Range | |
| ICN | 6 | 6 | 47 | 23–55 |
| ΔW | 16 | 16 | 77 | 26–107 |
| ΔRAM | 8 | 8 | 67 | 50–75 |
| ΔRAMΔP | 15 | 15 | 89 | 75–172 |
| Δ(N-TAD-P) | 8 | 0 | 423 | NA |
| ΔANK | 12 | 0 | 242 | NA |
| ΔRAMΔOP | 16 | 0 | 375 | NA |
| ΔRAMΔTAD | 5 | 0 | 419 | NA |
| ANK | 8 | 0 | 242 | NA |
| ΔRAMΔ(TAD-P) | 5 | 0 | 390 | NA |
Mean onset indicates the time period that mice were followed posttransplantation without the development of leukemia. The development of leukemia was defined by the detection of CD4+-CD8+ T cells in peripheral blood. NA, not applicable.
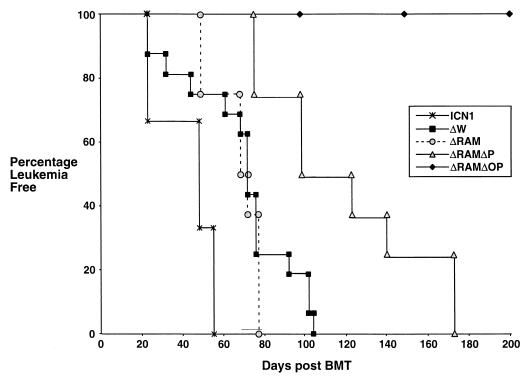
FIG. 2.
Onset of leukemia in mice reconstituted with retrovirally transduced BM cells. Time of leukemia onset is defined by the time post-BMT that CD4+-CD8+ DP GFP+ T cells were identified in the peripheral circulation. Mice typically survived for approximately 1 to 2 months after the onset of leukemia, during which time the fraction and number of DP GFP+ cells continued to increase (data not shown). None of the mice reconstituted with the nontransforming viral constructs or the empty MigRI virus developed circulating DP GFP+ cells or any evidence of neoplasia. A representative survival curve for a cohort of mice reconstituted with a nontransforming virus (ΔRAMΔOP) is shown. Although this figure shows leukemia-free survival to 200 days, these mice remained leukemia free for the duration of this study (see Table 1).
Examination of tissues from diseases ΔW and ΔRAM animals showed findings similar to those observed in ICN1 animals, with the most consistent findings being splenomegaly, hepatomegaly, lymphadenopathy, and BM replacement by lymphoblasts (not shown). Flow cytometric analyses confirmed that these tumor cells had a GFP+ DP phenotype (Fig. 3A).
Analysis of C-terminally deleted forms of ICN1.
To further define the C-terminal sequences that are necessary for leukemogenesis and identify a minimal transforming region, a series of additional deletions were made in ΔRAM (Fig. 1). The resultant cDNAs encoded polypeptides lacking all sequences C terminal of the bipartite nuclear localization sequence [ΔRAMΔ(TAD-P)], the PEST sequence (ΔRAMΔP), the entire TAD (ΔRAMΔTAD), or the OPA and PEST sequences (ΔRAMΔOP). To demonstrate a role for the ANK region in leukemogenesis, an ICN1 cDNA encoding a polypeptide deleted of the ANK (ΔANK) was also made.
All 15 mice reconstituted with ΔRAMΔP-transduced BM cells developed extrathymic GFP+ DP T cells followed by T-ALL (Fig. 3A). The latency of T-ALL induction with ΔRAMΔP was consistently longer than that seen with other leukemogenic forms of ICN (Fig. 2), indicating that deletion of C-terminal amino acids 2375 to 2555 attenuated transforming activity. In contrast, constructs encoding polypeptides with all or a portion of the C-terminal TAD deleted [ΔRAMΔ(TAD-P), ΔRAMΔTAD, and ΔRAMΔOP] uniformly failed to cause disease at up to 14 months after reconstitution (Table 1). ΔANK also failed to cause disease, indicating that C-terminal sequences are insufficient for leukemogenesis in the absence of the ANK domain. Extrathymic GFP+ DP T cells were never detected in mice receiving nontransforming constructs, even though these constructs were successfully transduced into cells, as judged by the presence of a persistent population of circulating GFP+ cells (Fig. 3B).
Detection of transforming and nontransforming polypeptides in tissue extracts.
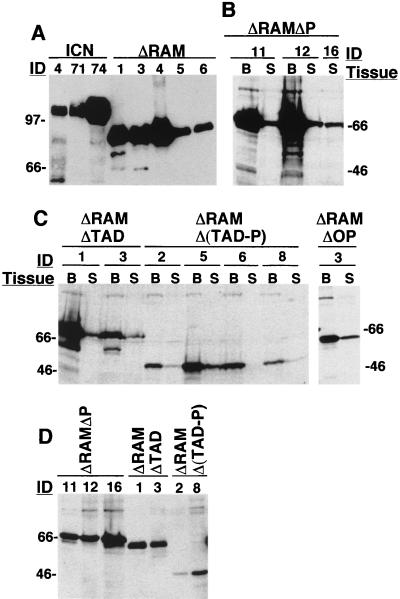
Although we previously noted that GFP and Notch1 polypeptide levels correlated in MigRI-transduced cells (43), it remained possible that certain deletions destabilized Notch1 polypeptides in vivo. To exclude this possibility, tissue extracts were analyzed on Western blots (Fig. 4). ICN1 and ΔRAM lymph node tissues contained abundant Notch1 polypeptides of the appropriate size (Fig. 4A). The presence of other Notch polypeptides were assessed and compared in extracts prepared from BM cells, splenocytes, and thymocytes; representative results are shown (Fig. 4B to D). Extracts prepared from BM and spleen showed that polypeptides of the expected size for transforming ΔRAMΔP (Fig. 4B) and nontransforming ΔRAMΔTAD, ΔRAMΔOP, and ΔRAMΔ(TAD-P) (Fig. 4C) Notch1 were readily detected. In each animal, the levels of Notch1 polypeptides were higher in BM than in spleen, a finding compatible with our prior observation that transduced cells are first detected in the BM, then in the spleen, and then in other peripheral sites (43). We also observed that nontransforming ΔRAMΔTAD and ΔRAMΔ (TAD-P) polypeptides were detected in thymocyte extracts (Fig. 4D), indicating that T-cell progenitors were successfully transduced even in animals that remained well. In some extracts, the level of nontransforming polypeptides (e.g., ΔRAMΔTAD in thymocytes (Fig. 4D) was approximately equal to that of transforming polypeptides such as ΔRAMΔP, despite apparent selection for high-level expression in clones that grew out as leukemias. These findings suggest that the failure of particular polypeptides to cause T-ALL is unlikely to stem from inadequate expression.
FIG. 4.
Western blot analyses of tissue extracts. (A) Extracts of nodal masses in ICN1 and ΔRAM animals. (B and C) Extracts of BM (B), splenic cells (S), or (D) thymic cells from animals expressing transforming ΔRAMΔP or nontransforming ΔRAMΔTAD, ΔRAMΔOP, and ΔRAMΔ(TAD-P) polypeptides. Portions of lymph node tumors, spleens, and thymuses were snap frozen in liquid nitrogen, pulverized with a pestle, and then lysed in sodium dodecyl sulfate-polyacrylamide gel electrophoresis loading buffer (∼100 μl/mg of tissue) at 95°C for 10 min. BM cells were flushed from femurs with normal saline, pelleted, and immediately lysed in loading buffer. Blots shown in panels A and B were stained with αTC rabbit serum (12) raised against the TAD of human Notch1, while blots shown in panels C and D were stained with αT6 rabbit serum (2) raised against the human Notch1 ANK and revealed by a chemiluminescent method.
Clonality of Notch-induced leukemias.
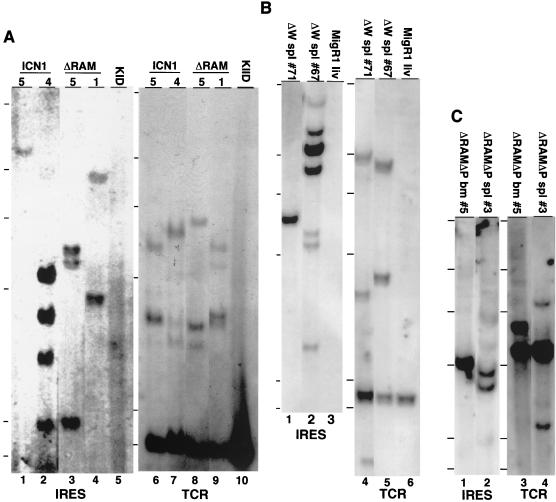
Southern blot analyses of DNA extracted from tissues infiltrated by Notch1-induced leukemias showed the presence of one to four MigRI proviruses; representative results are shown in Fig. 5A to C. Similarly, TCRβ gene rearrangement studies typically showed one to four non-germ line bands (Fig. 5A to C). In contrast, Southern blot analysis of DNAs obtained from the tissues of mice expressing nonleukemic forms of Notch1 showed a polyclonal pattern of proviral integration (not shown). This combination of findings, together with the 100% penetrance of the leukemogenic phenotype, suggests that Notch1-induced leukemias result from the selective outgrowth of one to a few transduced cells, possibly due to the stochastic acquisition of additional genetic aberrations.
FIG. 5.
Analysis of proviral integration and TCRβ chain rearrangement in leukemias from mice receiving transduced BM cells. (A) Southern blots of EcoRI-digested genomic DNA isolated from ICN1 spleens (mouse 4 and 5), ΔRAM spleens (mouse 1 and 5), and a kidney (KID) obtained from a control BALB/c animal. (B) Southern blots of EcoRI-digested genomic DNA isolated from ΔW spleens (spl) (mouse 67 and 71) and a liver (liv) from a MigRI control animal. (C) Southern blots of either EcoRI-digested (IRES) or HindIII-digested TCRβ (TCR) genomic DNA obtained from ΔRAMΔP BM (mouse 5) or spleen (mouse 3). To determine the number of proviral integration sites, DNAs were digested with EcoRI, which cleaves once within the provirus, and analyzed on Southern blots hybridized with a 592-bp encephalomyelitis virus IRES probe. A second set of digestions with XbaI, which cleaves once in each retroviral long terminal repeat, confirmed the presence of intact proviral DNA in all samples (data not shown). To determine the configuration of TCRβ genes, DNAs digested with either EcoRI or HindIII were analyzed on blots incubated with a TCR-Jβ2-specific DNA probe (10) that hybridizes to a 2.2-kb EcoRI fragment or a 5-kb HindIII fragment in germ line DNA. Two to five micrograms of DNA was loaded in each lane (A to C) except the BALB/c kidney lanes (A), which contained 15 μg. The hybridization probes are listed below the blots. The HindIII-digested lambda size markers are indicated adjacent to each blot. Sizes (in kilobases) are 23.1, 9.4, 6.6, 4.4, 2.3, and 2.0, reading from top to bottom.
Correlation of transforming and transcriptional activation activities.
To correlate transformation with known Notch1 activities, each polypeptide was scored in tissue culture cells for its ability to activate the HES-1 promoter and to function as a transcriptional activator when fused to the DNA-binding domain of GAL4. We focused most closely on analysis of C-terminal deletions spanning the ANK domain or TAD, since these regions appeared critical for transforming activity.
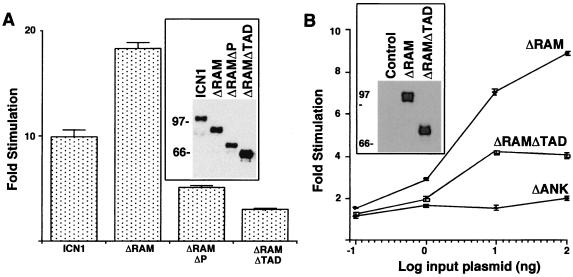
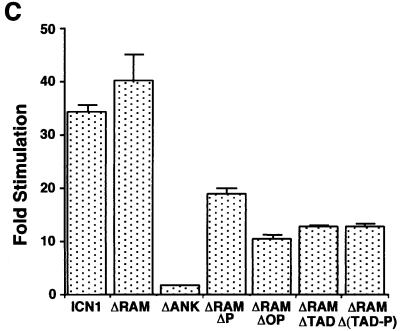
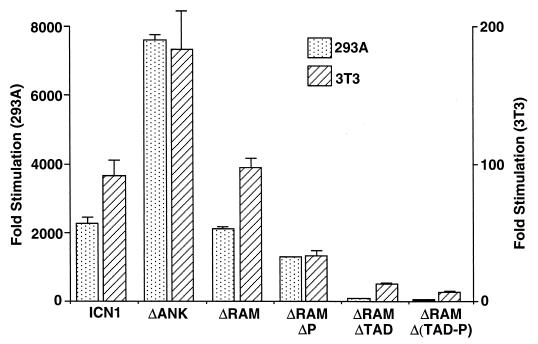
Both ICN1 and ΔRAM polypeptides were strong activators of the Su(H)/CBF1-sensitive HES1 promoter in 293A and NIH 3T3 cells, while the transforming ΔRAMΔP polypeptide produced intermediate levels of activation (Fig. 6A and C). In contrast, the nontransforming ΔRAMΔTAD, ΔRAMΔOP, and ΔRAMΔ(TAD-P) polypeptides were weaker activators in both cell types, while the nontransforming ΔANK polypeptide was devoid of activity (Fig. 6C). None of the constructs had any effect on a mutated HES-1 luciferase reporter, HESΔAB, lacking Su(H)/CBF1-binding sites (data not shown), consistent with the observed effects being mediated primarily through Su(H)/CBF1.
FIG. 6.
Transcriptional activation of the HES-1 promoter by Notch1 polypeptides. (A and C) Activation in acutely expressing 293A cells and NIH 3T3 cells, respectively. (B) Dose response in 293A cells. In the experiments shown in panels A and C, six-well dishes were transfected with 250 ng of pcDNA3 plasmids containing the indicated Notch1 cDNAs, 500 ng of HES-AB firefly luciferase plasmid, and 10 ng of pRL-TK Renilla luciferase plasmid. In the experiments shown in panel B, the amounts of pcDNA3 plasmids were varied. Cell lysates were prepared ∼48 h posttransfection. Firefly luciferase activities were normalized to the corresponding Renilla luciferase activities. Transcriptional activities were expressed relative to the normalized activities observed in extracts prepared from cells transfected with empty pcDNA3 plasmid. The results shown represent the means of three experiments.
Two additional pieces of data indicated that the various deletions in ICN1 produced predominantly qualitative changes in function. First, Western blot analysis performed using protein loads normalized for transfection efficiency showed that transforming ICN1, ΔRAM, ΔRAMΔP, and nontransforming ΔRAMΔTAD polypeptides were all expressed at similar levels in 293A cells (Fig. 6A, inset). Secondly, a dose-response experiment in 293A cells showed that ΔRAM was more potent than ΔRAMΔTAD or ΔANK at all levels of plasmid input (Fig. 6B).
To assess transactivation activity per se, GAL4-Notch1 fusion polypeptides were assayed on a GAL4-responsive luciferase reporter gene (Fig. 7). All polypeptides retaining an intact TAD were intermediate to strong transcriptional activators. ICN1 and ICNΔRAM were equivalently strong, whereas ΔANK consistently produced the highest levels of transcriptional activation of any polypeptide analyzed. ΔRAMΔP produced intermediate levels of transcriptional activation, whereas polypeptides lacking any part of the TAD [ΔRAMΔ(TAD-P), ΔRAMΔOP, and ΔRAMΔTAD], all produced only weak transcriptional activation. In control Western blot analyses, each of these cDNAs resulted in the expression of roughly equivalent amounts of protein when normalized for transfection efficiency (data not shown), indicating that the observed variation in activity was due to qualitative rather than quantitative differences.
FIG. 7.
Transcriptional activation by GAL4-Notch1 fusion polypeptides. 293A or NIH 3T3 cells grown in six-well plates were transfected with 250 ng of pM plasmids encoding various GAL4-Notch1 fusion polypeptides, 500 ng of GAL4X5 firefly luciferase plasmid, and 10 ng of pRL-TK Renilla luciferase plasmid. Firefly luciferase activities were normalized to the corresponding Renilla luciferase activities. Transcriptional activities were expressed relative to the normalized activities observed in extracts prepared from cells transfected with empty pcDNA3 plasmid. The results shown represent the means of three experiments.
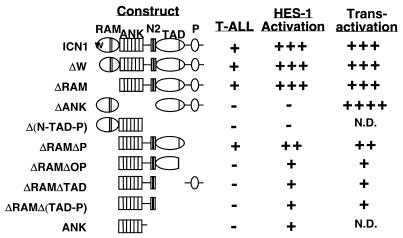
Together, these cell culture correlates revealed that transforming polypeptides were moderate to strong activators of the HES-1 promoter and moderate to strong transactivators when fused to the DNA-binding domain of GAL4. In contrast, nontransforming forms of Notch1 produced weaker or negligible activation of transcription from the HES-1 promoter and were weaker transactivators with the exception of ΔANK, which was a potent transactivator but devoid of activity on the HES-1 promoter. These correlates are summarized in Fig. 8.
FIG. 8.
Correlation of leukemogenesis with the transcriptional activities of various forms of Notch1. HES-1 activation refers to the capacity to activate transcription from a promoter element derived from the murine HES-1 gene; transactivation refers to the activity of Notch1-GAL4 DNA-binding domain fusion polypeptides on an artificial promoter containing five iterated GAL4 binding sites.
DISCUSSION
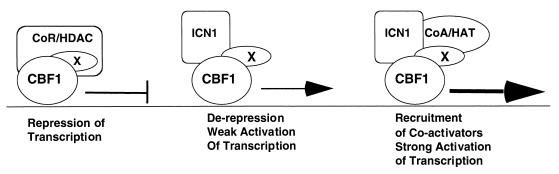
We have used a robust in vivo assay to define a minimal Notch1 transformation domain for T-cell progenitors consisting of the ANK region, a flanking sequence containing a functional nuclear localization signal sequence, and a TAD. This analysis serves as the basis for a working model of leukemogenic NOTCH1 signaling (Fig. 9), in which the ANK interact with downstream transcription factors, possibly displacing corepressors in doing so, and the TAD serves to recruit coactivator molecules.
FIG. 9.
Model for leukemogenic Notch1 signaling complex. CBF1, transcription factor Su(H)/CBF1; CoR/HDAC, corepressor-histone deacetylase complex; X, molecule capable of binding to Su(H)/CBF1 and ICN1, of which SKIP and LAG-3 are prototypes; CoA/HAT, coactivator-histone acetylase complex.
ANK domains generally serve as sites of protein-protein interaction, and multiple polypeptides genetically implicated in Notch signaling have been shown to bind this domain, including Su(H)/CBF1 (3, 10, 45), Deltex (31), SPT6 (also known as EMB5) (15), and LAG-3 (47). Because polypeptides with deletions spanning the Notch1 ANK remain strong transactivators when fused to the DNA-binding domain of GAL4, yet have no transforming activity and do not transactivate the HES-1 promoter element, it seems likely that this domain is crucial for the association of ICN1 with downstream signaling molecules.
In addition to the ANK, T-cell transformation by Notch1 also requires a TAD lying between amino acids 2215 and 2374. This conclusion is based on the finding that four different constructs encoding polypeptides either entirely lacking the TAD [ANK, ΔRAMΔ(TAD-P), and ΔRAMΔTAD] or just the OPA sequence portion of the TAD (ΔRAMΔOP) uniformly failed to cause T-cell leukemia. This dependency on the TAD for transformation correlates with cell culture assays in which the TAD is required for transactivation by GAL4-Notch1 fusion polypeptides and for strong activation of the HES-1 promoter. Recently, the TAD of murine ICN1 was shown to be required for interaction with two transcriptional coactivator complexes, PCAF and GCN5 (26), providing a mechanism for its protranscriptional effects in cultured cells, as well as its protransforming effects in vivo.
Our model for leukemogenic Notch1 signaling reflects several important remaining uncertainties. One concerns how “RAM-less” forms of ICN1 activate Su(H)/CBF1-sensitive promoter elements. All of the transforming forms of Notch1 that we have identified retain the capacity to moderately to strongly activate transcription from a 254-bp DNA sequence derived from the murine HES-1 promoter (18), a known downstream target of Notch1 signals. The activation of this promoter by Notch1 requires two iterated Su(H)/CBF1-binding sites, implying that it is mediated through Su(H)/CBF1. Independent evidence that the RAM domain is not strictly required for signaling through Su(H)/CBF1 comes from invertebrates, in which RAM-less forms of Notch can still produce Su(H)/CBF1-dependent phenotypes (45, 46). The ANK region contains a weak binding site for Su(H)/CBF1 that might be the basis for direct physical interaction in vivo (3, 10, 22, 45). Alternatively, the ANK of ICN1 could interact indirectly through a bridging molecule such as SKIP (55) or LAG-3 (42) (factor X in Fig. 9), each of which bind to both the ANK domain of Notch and Su(H)/CBF1-type transcription factors. Of note, SKIP also binds to the corepressors SMRT and N-CoR, which are competitively displaced by binding of the ANK to SKIP (63).
Our data obtained with the HES-1 promoter differ from those observed in experiments using promoters consisting of iterated Su(H)/CBF1-binding DNA sites. In acute expression assays using such reporters, the RAM and ANK domains are both needed for transcriptional activation (8, 35, 49), which correlates with displacement of the transcriptional corepressors CIR and SMRT/N-CoR from Su(H)/CBF1 by ICN1 (14, 21). We have recently observed that our ΔRAM polypeptide, which is fully active on the HES-1 promoter in NIH 3T3 cells (Fig. 6) shows only limited activity on a promoter consisting of iterated Su(H)/CBF1-binding sites, confirming that this discrepancy applies to the leukemogenic form of ΔRAM used by us (J. C. Aster, unpublished data). Of potential importance in explaining these differences, the HES-1 promoter contains additional DNA sequences 5′ of the two Su(H)/CBF1-binding sites that bind unknown factors (17), and it is possible that such factors directly or indirectly provide additional contact points that permit recruitment of RAM-less forms of ICN1.
Although our results support a model in which Su(H)/CBF1-dependent signaling is required for transformation, we cannot rule out contributions from Su(H)/CBF1-independent pathways. Notch signaling through Su(H)/CBF1-independent pathways is supported by genetic data (33); however, the molecules and pathways mediating this signaling are poorly understood. One of the few molecules to be linked to Su(H)/CBF1-independent signaling is Deltex, which may inhibit the activity of certain bHLH transcription factors (32, 36). Initial experiments indicate that overexpression of Deltex is insufficient to cause T-cell transformation, as mice receiving human Deltex1-transduced bone marrow cells remain healthy (F. G. Karnell and W. S. Pear, unpublished). Additional work will be needed to determine whether Deltex-mediated signals are necessary for leukemogenesis.
Deletion of sequences C terminal of the TAD, including a PEST sequence, attenuated but did not prevent T-ALL induction in our murine BMT assay. Most prior genetic and biochemical analyses of Notch function have suggested that the C-terminal domain is a negative regulatory region, possibly due to its ability to interact with the Notch antagonist Numb (54). However, our work indicates that this domain also contributes modestly to transcriptional activation, possibly through interaction with currently unknown coactivators, as the ΔP deletion caused a significant reduction in transcriptional activation of the HES-1 promoter and transactivation by GAL4 fusion polypeptides in cell culture assays. Other groups have also observed that deletion of the corresponding region of murine Notch1 (amino acids 2398 to 2531) resulted in diminished transcriptional activation (8, 27), indicating that the requirement of this domain for full activity is a conserved feature of mammalian Notch1. Regardless of its basis, the attenuated phenotype that we observed with ΔRAMΔP serves to further highlight a theme emerging from our work: that transcriptional activation correlates with transforming activity.
The minimal ICN1-transforming domain we have identified in T-cell progenitors differs from the minimal transforming domain that two groups have identified independently in cultured baby rat kidney cells immortalized with E1A (8, 19). In both of these studies, a polypeptide consisting of the ANK domain and the conserved sequence extending to the C-terminal nuclear localization signal sequence [corresponding to our ΔRAMΔ(TAD-P) construct] was sufficient for transformation. It thus seems likely that the mechanism of T-cell progenitor transformation in vivo differs from that of cultured rat kidney cells in vitro. This would not be surprising, as structure-transformation differences have been observed with other oncoproteins, such as BCR-ABL and TEL-PDGFR, when transformation of cultured cells was compared to that of primary cells in vivo (37, 52).
The critical downstream events that contribute to ICN1-induced leukemogenesis are unknown but may be lineage specific, since transforming activity appears to be restricted among hematopoietic progenitors to T-cell precursors (43). One potentially important target is HES-1 (18, 20), a factor that, like Notch1 (43, 44), is important for commitment to T-cell fate (53). HES-1 belongs to the family of hairy enhancer-of-split factors that antagonize certain bHLH transcription factors that are required for lineage-specific differentiation. Although Notch1 promotes early T-cell differentiation, endogenous Notch1 is normally down-regulated in DP T cells and then reexpressed in single-positive thymocytes (12). The enforced expression of activated Notch1 in BM progenitors leads to the rapid appearance of an abnormal population of DP T cells (43), suggesting that down-regulation of Notch1 at this stage may be required for progression to later stages of T-cell maturation. Our development of a murine model that closely mimics its human disease counterpart provides an experimental framework to test such hypotheses in order to elucidate the mechanisms of Notch1-induced T-cell leukemia.
ACKNOWLEDGMENTS
We thank members of the Aster and Pear laboratories and members of the University of Pennsylvania John Morgan (IHGT) mouse and flow cytometry facilities. The flow cytometry studies were performed in the University of Pennsylvania Cancer Center Flow Cytometry and Cell Sorting Shared Resource (supported in part by the Lucille B. Markey Trust and the NIH).
This work was supported by NIH grant RO1CA82308 (J.C.A. and W.S.P.). This work was also supported in part by an award to W.S.P. from the University of Pennsylvania/Howard Hughes Program in Developmental Biology and a Scholar Award from the Leukemia and Lymphoma Society.
REFERENCES
- 1.Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Aster J, Pear W, Hasserjian R, Erba H, Davi F, Luo B, Scott M, Baltimore D, Sklar J. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harbor Symp Quant Biol. 1994;59:125–136. doi: 10.1101/sqb.1994.059.01.016. [DOI] [PubMed] [Google Scholar]
- 3.Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. doi: 10.1074/jbc.272.17.11336. [DOI] [PubMed] [Google Scholar]
- 4.Bailey A M, Posakony J W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 5.Bigas A, Martin D I, Milner L A. Notch1 and Notch2 inhibit myeloid differentiation in response to different cytokines. Mol Cell Biol. 1998;18:2324–2333. doi: 10.1128/mcb.18.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens J R, Cumano A, Roux P, Black R A, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Mol Cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 7.Christensen S, Kodoyianni V, Bosenberg M, Friedman L, Kimble J. lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H) Development. 1996;122:1373–1383. doi: 10.1242/dev.122.5.1373. [DOI] [PubMed] [Google Scholar]
- 8.Dumont E, Fuchs K P, Bommer G, Christoph B, Kremmer E, Kempkes B. Neoplastic transformation by Notch is independent of transcriptional activation by RBP-J signalling. Oncogene. 2000;19:556–561. doi: 10.1038/sj.onc.1203352. [DOI] [PubMed] [Google Scholar]
- 9.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 10.Fortini M E, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 11.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 12.Hasserjian R P, Aster J C, Davi F, Weinberg D S, Sklar J. Modulated expression of notch1 during thymocyte development. Blood. 1996;88:970–976. [PubMed] [Google Scholar]
- 13.Hsieh J J, Nofziger D E, Weinmaster G, Hayward S D. Epstein-Barr virus immortalization: Notch2 interacts with CBF1 and blocks differentiation. J Virol. 1997;71:1938–1945. doi: 10.1128/jvi.71.3.1938-1945.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh J J, Zhou S, Chen L, Young D B, Hayward S D. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbard E J, Dong Q, Greenwald I. Evidence for physical and functional association between EMB-5 and LIN-12 in Caenorhabditis elegans. Science. 1996;273:112–115. doi: 10.1126/science.273.5271.112. [DOI] [PubMed] [Google Scholar]
- 16.Huppert S S, Le A, Schroeter E H, Mumm J S, Saxena M T, Milner L A, Kopan R. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature. 2000;405:966–970. doi: 10.1038/35016111. [DOI] [PubMed] [Google Scholar]
- 17.Issack P S, Ziff E B. Genetic elements regulating HES-1 induction in Wnt-1-transformed PC12 cells. Cell Growth Differ. 1998;9:827–836. [PubMed] [Google Scholar]
- 18.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 19.Jeffries S, Capobianco A J. Neoplastic transformation by Notch requires nuclear localization. Mol Cell Biol. 2000;20:3928–3941. doi: 10.1128/mcb.20.11.3928-3941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jouve C, Palmeirim I, Henrique D, Beckers J, Gossler A, Ish-Horowicz D, Pourquie O. Notch signalling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development. 2000;127:1421–1429. doi: 10.1242/dev.127.7.1421. [DOI] [PubMed] [Google Scholar]
- 21.Kao H Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 23.Kidd S, Lieber T, Young M W. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kopan R, Nye J S, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 25.Kopan R, Schroeter E H, Weintraub H, Nye J S. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurooka H, Honjo T. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem. 2000;75:17211–17220. doi: 10.1074/jbc.M000909200. [DOI] [PubMed] [Google Scholar]
- 27.Kurooka H, Kuroda K, Honjo T. Roles of the ankyrin repeats and C-terminal region of the mouse notch1 intracellular region. Nucleic Acids Res. 1998;26:5448–5455. doi: 10.1093/nar/26.23.5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lecourtois M, Schweisguth F. Indirect evidence for Delta-dependent intracellular processing of notch in Drosophila embryos. Curr Biol. 1998;8:771–774. doi: 10.1016/s0960-9822(98)70300-8. [DOI] [PubMed] [Google Scholar]
- 29.Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 30.Ligoxygakis P, Yu S Y, Delidakis C, Baker N E. A subset of notch functions during Drosophila eye development require Su(H) and the E(spl) gene complex. Development. 1998;125:2893–2900. doi: 10.1242/dev.125.15.2893. [DOI] [PubMed] [Google Scholar]
- 31.Matsuno K, Diederich R J, Go M J, Blaumueller C M, Artavanis-Tsakonas S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development. 1995;121:2633–2644. doi: 10.1242/dev.121.8.2633. [DOI] [PubMed] [Google Scholar]
- 32.Matsuno K, Eastman D, Mitsiades T, Quinn A M, Carcanciu M L, Ordentlich P, Kadesch T, Artavanis-Tsakonas S. Human deltex is a conserved regulator of Notch signalling. Nat Genet. 1998;19:74–78. doi: 10.1038/ng0598-74. [DOI] [PubMed] [Google Scholar]
- 33.Matsuno K, Go M J, Sun X, Eastman D S, Artavanis-Tsakonas S. Suppressor of Hairless-independent events in Notch signaling imply novel pathway elements. Development. 1997;124:4265–4273. doi: 10.1242/dev.124.21.4265. [DOI] [PubMed] [Google Scholar]
- 34.Mumm J S, Schroeter E H, Saxena M T, Griesemer A, Tian X, Pan D J, Ray W J, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol Cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 35.Nofziger D, Miyamoto A, Lyons K M, Weinmaster G. Notch signaling imposes two distinct blocks in the differentiation of C2C12 myoblasts. Development. 1999;126:1689–1702. doi: 10.1242/dev.126.8.1689. [DOI] [PubMed] [Google Scholar]
- 36.Ordentlich P, Lin A, Shen C P, Blaumueller C, Matsuno K, Artavanis-Tsakonas S, Kadesch T. Notch inhibition of E47 supports the existence of a novel signaling pathway. Mol Cell Biol. 1998;18:2230–2239. doi: 10.1128/mcb.18.4.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pear W S. Signaling in leukemia: which messenger to kill? J Clin Investig. 2000;105:419–422. doi: 10.1172/JCI9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pear W S, Aster J C, Scott M L, Hasserjian R P, Soffer B, Sklar J, Baltimore D. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–2291. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pear W S, Cepko C. Transduction of genes using retrovirus vectors. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. 1, supplement 36. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 9.9.1–9.9.16. [Google Scholar]
- 40.Pear W S, Miller J P, Xu L, Pui J C, Soffer B, Quackenbush R C, Pendergast A M, Bronson R, Aster J C, Scott M L, Baltimore D. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 41.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petcherski A G, Kimble J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature. 2000;368:364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- 43.Pui J C, Allman D, Xu L, DeRocco S, Karnell F G, Bakkour S, Lee J Y, Kadesch T, Hardy R R, Aster J C, Pear W S. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 44.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald H R, Aguet M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–558. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 45.Roehl H, Bosenberg M, Blelloch R, Kimble J. Roles of the RAM and ANK domains in signaling by the C. elegans GLP-1 receptor. EMBO J. 1996;15:7002–7012. [PMC free article] [PubMed] [Google Scholar]
- 46.Roehl H, Kimble J. Control of cell fate by a GLP-1 peptide consisting primarily of ankyrin repeats. Nature. 1993;364:632–635. doi: 10.1038/364632a0. [DOI] [PubMed] [Google Scholar]
- 47.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 48.Schroeter E H, Kisslinger J A, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 49.Shawber C, Nofziger D, Hsieh J J, Lindsell C, Bogler O, Hayward D, Weinmaster G. Notch signaling inhibits muscle cell differentiation through a CBF1-independent pathway. Development. 1996;122:3765–3773. doi: 10.1242/dev.122.12.3765. [DOI] [PubMed] [Google Scholar]
- 50.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 51.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H) Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 52.Tomasson M H, Sternberg D W, Williams I R, Carroll M, Cain D, Aster J C, Ilaria R L, Jr, Etten R A, Gilliland D G. Fatal myeloproliferation, induced in mice by TEL/PDGFbetaR expression, depends on PDGFbetaR tyrosines 579/581. J Clin Investig. 2000;105:423–432. doi: 10.1172/JCI8902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomita K, Hattori M, Nakamura E, Nakanishi S, Minato N, Kageyama R. The bHLH gene Hes1 is essential for expansion of early T cell precursors. Genes Dev. 1999;13:1203–1210. doi: 10.1101/gad.13.9.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wakamatsu Y, Maynard T M, Jones S U, Weston J A. NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron. 1999;23:71–81. doi: 10.1016/s0896-6273(00)80754-0. [DOI] [PubMed] [Google Scholar]
- 55.Zhou S, Fujimuro M, Hsieh J J, Chen L, Miyamoto A, Weinmaster G, Hayward S D. SKIP, a CBF1-associated protein, interacts with the ankyrin repeat domain of NotchIC to facilitate NotchIC function. Mol Cell Biol. 2000;20:2400–2410. doi: 10.1128/mcb.20.7.2400-2410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]