Abstract
Peroxisomes are independent organelles found in virtually all eukaryotic cells. Genetic studies have identified more than 20 PEX genes that are required for peroxisome biogenesis. The role of most PEX gene products, peroxins, remains to be determined, but a variety of studies have established that Pex5p binds the type 1 peroxisomal targeting signal and is the import receptor for most newly synthesized peroxisomal matrix proteins. The steady-state abundance of Pex5p is unaffected in most pex mutants of the yeast Pichia pastoris but is severely reduced in pex4 and pex22 mutants and moderately reduced in pex1 and pex6 mutants. We used these subphenotypes to determine the epistatic relationships among several groups of pex mutants. Our results demonstrate that Pex4p acts after the peroxisome membrane synthesis factor Pex3p, the Pex5p docking factors Pex13p and Pex14p, the matrix protein import factors Pex8p, Pex10p, and Pex12p, and two other peroxins, Pex2p and Pex17p. Pex22p and the interacting AAA ATPases Pex1p and Pex6p were also found to act after Pex10p. Furthermore, Pex1p and Pex6p were found to act upstream of Pex4p and Pex22p. These results suggest that Pex1p, Pex4p, Pex6p, and Pex22p act late in peroxisomal matrix protein import, after matrix protein translocation. This hypothesis is supported by the phenotypes of the corresponding mutant strains. As has been shown previously for P. pastoris pex1, pex6, and pex22 mutant cells, we show here that pex4Δ mutant cells contain peroxisomal membrane protein-containing peroxisomes that import residual amounts of peroxisomal matrix proteins.
To form a functional peroxisome, peroxisome membranes must be generated and the subsequent import of both membrane and matrix proteins must occur. Various studies have established that both peroxisomal membrane proteins (PMPs) and peroxisomal matrix proteins are made in the cytosol and delivered posttranslationally to the peroxisome (26). However, the similarities between PMP import and peroxisomal matrix protein import appear to end there. Peroxisomal matrix proteins are targeted to the peroxisome lumen via the presence of either a PTS1 or a PTS2 targeting signal (40). The PTS1 signal, which consists of a -SKL sequence (or a conserved variant thereof) at the extreme carboxy terminus of the protein (16), is present on the vast majority of peroxisomal matrix proteins. The PTS2 signal consists of the sequence R/K-L/V/I-X5-H/Q-L/A near the amino terminus (41) and has so far been identified in only a handful of proteins. Both types of signals are recognized by specific receptors: Pex5p for the PTS1 signal (6, 29) and Pex7p for the PTS2 signal (27, 31). After binding to the receptor, matrix proteins are taken to the peroxisome surface and inserted into the organelle lumen through an as yet unknown mechanism. For Pex5p, it has been proposed that the receptor is then recycled back to the cytosol and can undergo further rounds of import (7). In contrast, PMPs lack PTS1 and PTS2 motifs (40), are imported independently of Pex5p and Pex7p (2), and require a distinct PMP-binding protein, Pex19p, for their import (19, 28, 34).
In addition to Pex5p, Pex7p, and Pex19p, a variety of other proteins required for peroxisome biogenesis have been identified (19). These include peroxins necessary for the biogenesis of peroxisome membranes (Pex3p), docking factors for the PTS receptors (Pex13p and Pex14p), proteins that act downstream of receptor docking but are required for translocation of proteins across the peroxisome membrane (Pex8p, Pex10p, and Pex12p), and other proteins whose functions are less clear (Pex1p, Pex2p, Pex4p, Pex6p, Pex17p, and Pex22p).
Despite the identification of so many components of peroxisome biogenesis, there is still little direct evidence as to the order in which these peroxins act. In the course of investigating the effects that different peroxins have on Pex5p, the PTS1 receptor, we observed that the loss of Pex1p or Pex6p in either yeast or human cells leads to an accelerated turnover of Pex5p and a significant drop in steady-state levels of Pex5p (7, 49). Koller et al. (25) have recently reported a similar phenotype for Pichia pastoris pex4 and pex22 mutants. Here we show that the phenotype of reduced Pex5p abundance in P. pastoris is unique to selected pex mutants and can be used to determine the epistatic relationships among the different peroxins. The results of our epistasis analysis suggest that Pex4p, Pex22p, Pex1p, and Pex6p act in the terminal steps of peroxisomal matrix protein import. This hypothesis is consistent with the reported phenotypes of P. pastoris pex1, pex6, and pex22 mutants, all of which contain peroxisomes and import detectable levels of peroxisomal matrix proteins. Similarly, we show here that the pex4 mutant also contains peroxisomes that are capable of peroxisomal matrix protein import.
MATERIALS AND METHODS
Yeast media.
The various media used were based on those described earlier for P. pastoris (17). Specifically, YPD (1% yeast extract, 2% peptone, 2% dextrose) was used as rich medium. YPM (1% yeast extract, 2% peptone, 0.5% methanol) was used as rich methanol medium. YPOLT (1% yeast extract, 2% peptone, 0.18% oleic acid, 0.02% Tween 40) was used as rich oleic acid medium. SD + histidine (0.17% yeast nitrogen base without amino acids or ammonium sulfate, 0.5% ammonium sulfate, 0.1% l-histidine, 2% dextrose) was used as minimal-dextrose medium. SM + histidine + arginine (0.17% yeast nitrogen base without amino acids or ammonium sulfate, 0.5% ammonium sulfate, 0.1% l-histidine, 0.1% l-arginine, 0.5% methanol) was used as minimal-methanol medium. Sporulation medium (1% potassium chloride, 0.5% sodium acetate, 1% dextrose) was used to induce mating and sporulation. All cultures and plates were grown at 30°C.
Yeast strains.
The yeast strains used in this study are presented in Table 1. The pex his4Δ strains were created by mating each pex mutant with SGY55 (4), sporulating the resultant diploids, and screening the haploid progeny for the pex his phenotype. The double-mutant strains created for this study were made as follows. The single-mutant strains in question were grown to saturation in YPD, and 0.5 ml of each was mixed and allowed to grow overnight on a YPD plate. On the following day, cells were scraped off the YPD plate, resuspended in 1 ml of sterile water, and plated on solid sporulation medium to induce allow mating. After 24 h, the sporulation plates were replica plated to minimal-methanol plates to select for diploids. Three days later, cells were transferred to fresh minimal-methanol plates and grown for another 3 days. The resulting diploid cells were then transferred to a 3-ml liquid YPD culture and grown overnight. The yeast cells from this culture were harvested, resuspended in 0.5 ml of sterile water, and seeded at high density onto solid sporulation medium. The cells were incubated for 5 days to allow the formation of a significant number of asci. The asci were then scraped from the plates, pelleted, and resuspended in 2 ml of sterile water. To separate asci, 650 μl of the resuspended pellet was incubated with 350 μl of 100% ethanol at room temperature for exactly 30 min. Cells were then plated at 10-fold serial dilutions to obtain single haploid spores. Spores were colony purified and assayed for growth on minimal-methanol medium. Spores that could not grow on methanol were then assayed by complementation analysis to distinguish the double-mutant progeny from the single-mutant progeny.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| SGY22 | arg4-1 pex3-1 his4Δ::ARG4 | 23 |
| SGY23 | arg4-1 pex6-1 his4Δ::ARG4 | 23 |
| SGY24 | arg4-1 pex13Δ::ARG4 his4Δ::ARG4 | 15 |
| SGY26 | arg4-1 pex8-3 his4Δ::ARG4 | 23 |
| SGY27 | arg4-1 pex2-2 his4Δ::ARG4 | 23 |
| SGY29 | arg4-1 leu2::ARG4 pex5Δ::LEU2 his4Δ::ARG4 | Laboratory stock |
| SGY55a | arg4-1 his4Δ::ARG4 | 4 |
| SGY132 | arg4-1 leu2::ARG4 pex10Δ::LEU2 his4Δ::ARG4 | 23a |
| SGY311 | arg4-1 pex4Δ::ARG4 his4Δ::ARG4 | Laboratory stock |
| SGY443 | arg4-1 pex12Δ::ARG4 his4Δ::ARG4 | 23a |
| SGY501 | arg4-1 pex1Δ::ARG4 his4Δ::ARG4 | Laboratory stock |
| STK11 | arg4 pex22Δ::Zeocin his4 | 25 |
| CYPP01 | arg4-1 pex4Δ::ARG4 pex10Δ::LEU2 his4Δ::ARG4 | This study |
| CYPP03 | arg4-1 pex1Δ::ARG4 pex10Δ::LEU2 his4Δ::ARG4 | This study |
| CYPP04 | arg4-1 pex6-1 pex10Δ::LEU2 his4Δ::ARG4 | This study |
| CYPP05 | arg4-1 pex1Δ::ARG4 pex4Δ::ARG4 his4-1 | This study |
| CYPP06 | arg4-1 pex4Δ::ARG4 pex8-3 his4Δ::ARG4 | This study |
| CYPP07 | arg4-1 pex4Δ::ARG4 pex6-1 his4Δ::ARG4 | This study |
| CYPP08 | arg4-1 pex2-2 pex4Δ::ARG4 his4Δ::ARG4 | This study |
| CYPP09 | arg4-1 pex4Δ::ARG4 pex12Δ::ARG4 his4Δ::ARG4 | This study |
| CYPP10 | arg4-1 pex4Δ::ARG4 pex13Δ::ARG4 his4Δ::ARG4 | This study |
| CYPP11 | arg4-1 pex4Δ::ARG4 pex14Δ::ARG4 his4Δ::ARG4 | This study |
| CYPP12 | arg4-1 pex4Δ::ARG4 pex17Δ::ARG4 his4Δ::ARG4 | This study |
| CYPP14 | arg4-1 pex3-1 pex4Δ::ARG4 his4Δ::ARG4 | This study |
| CYPP15 | arg4-1 pex4Δ::ARG4 pex22Δ::Zeocin his4 | This study |
| CYPP16 | arg4-1 pex10Δ::LEU2 pex22Δ::Zeocin his4 | This study |
| CYPP18 | arg4-1 pex1Δ::ARG4 pex22Δ::Zeocin his4 | This study |
| CYPP19 | arg4-1 pex6-1 pex22Δ::Zeocin his4 | This study |
| CYPP20 | arg4-1 pex14Δ::ARG4 his4Δ::ARG4 | Laboratory stock |
| CYPP21 | arg4-1 pex17Δ::ARG4 his4Δ::ARG4 | Laboratory stock |
The SGY55 strain was used in the wild type in the experiments described here.
Plasmid construction and yeast transformations.
The plasmids expressing the wild-type pex4 gene and the pex4-C133A mutant have been described previously (5). All bacterial manipulations were carried out with the Escherichia coli DH10B host (18). Purified plasmid DNA was introduced into the yeast strains by electroporation (5).
Electron and immunoelectron microscopy.
For transmission electron microscopy, strains were grown in YPD to an optical density at 600 nm (OD600) of 1.0, diluted to an OD600 of 0.5, and incubated in YPM for 18 h to induce peroxisomal enzymes. Cells were fixed and processed as described previously (17). Samples were sectioned at 70 to 80 kV with a 35° angle diatome diamond knife and placed on Formvar-coated copper grids (Ted Pella, Inc.). The sections were poststained with 2% uranyl acetate and 0.3% lead citrate and observed on a Zeiss 10B transmission electron microscope.
For immunoelectron microscopy, log-phase cells were induced in minimal-methanol medium for 18 h and fixed in suspension for 15 min by adding an equal volume of freshly prepared 8% formaldehyde contained in 1× phosphate-buffered saline (PBS), pH 7.4. The cells were pelleted and resuspended in 4% formaldehyde contained in 1× PBS, pH 7.4, and fixed for an additional 18 to 24 h at 4°C. The cells were then washed briefly in PBS and resuspended in 1% low-temperature-gelling agarose. After cooling, the agarose blocks were trimmed into 1-mm3 pieces, cryoprotected by infiltration with a mixture of 2.3 M sucrose–20% polyvinylpyrrolidone (10K; pH 7.4) for 2 h, mounted onto cryo-pins, and rapidly frozen in liquid nitrogen. Ultrathin cryosections were cut on a Leica UCT ultramicrotome equipped with an FC-S cryo-attachment and collected onto Formvar-carbon-coated nickel grids. The grids were washed with several drops of 1× PBS containing 2.5% fetal calf serum–10 mM glycine, pH 7.4.; blocked in 10% fetal calf serum for 30 min; and incubated overnight in affinity-purified anti-pex10 polyclonal antibody (diluted 1:50). After washing, the grids were incubated for 2 h in 5-nm gold donkey anti-rabbit conjugates (available from Jackson Immunoresearch Labs). The grids were then washed with several drops of PBS, followed by several drops of double-distilled H2O, and subsequently embedded in an aqueous solution containing 3.2% polyvinyl alcohol (10K), 0.2% methylcellulose (400 centiposes), and 0.1% uranyl acetate. The grids were observed and photographed on a Philips 410 transmission electron microscope at 80 kV.
Differential centrifugation, subcellular fractionation, and enzyme assays.
All fractionation experiments were performed as previously described (5). For differential centrifugation, yeast cells were grown in YPD to mid-logarithmic phase, diluted to an OD600 of 0.5, and shifted to rich oleic acid medium or minimal-methanol medium to induce proliferation of peroxisomal enzymes. After an 18- to 20-h incubation period, a postnuclear supernatant (PNS) of each strain was prepared. Organelles were then isolated by centrifugation of the PNS at 25,000 × g. Equal proportions of the organelle pellet and cytosolic supernatant were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed for the presence of the protein of interest by standard Western blot techniques. For subcellular fractionation, the 25,000 × g organelle pellet containing the mitochondria and peroxisomes (generated as described above) was fractionated over a 32 to 60% linear sucrose gradient as described previously (5). Fractions of 1 ml were collected from the bottom of the gradient and analyzed for catalase activity and succinate dehydrogenase activity.
RNA extraction and Northern blot analysis.
The pex4Δ and pex10Δ strains were grown to an OD600 of 0.6 in YPD, pelleted, and shifted to minimal-methanol medium. The methanol-induced cultures were incubated overnight at 30°C with agitation. On the following day, approximately 250 OD600 units of cells were collected by centrifugation and total RNA was extracted as previously described (13). Poly(A)+ RNA was extracted from the total RNA by magnetic bead separation (Dynabeads; Dynal Inc.) in accordance with the manufacturer's directions.
For Northern blot analysis, 0.6 μg of poly(A)+ RNA was loaded per lane on a denaturing agarose gel (each sample was loaded twice to check for consistency). Following separation by electrophoresis, RNA was transferred to GeneScreen Plus filters (NEN Life Science Products). Probe hybridization and autoradiography were performed using standard protocols (35). Briefly, the filter was hybridized to a 32P-labeled probe specific for the PEX5 gene (GenBank accession no. U59222), washed, and exposed to X-ray film (Kodak X-Omat AR). The filter was then stripped (boiled for 15 min in 0.5% SDS) and hybridized to a 32P-labeled probe specific for the P. pastoris actin gene (ACT1; GenBank accession no. AF216956). The probe for the PEX5 gene was generated by PCR amplification of a plasmid containing the PEX5 cDNA (forward primer, 5′-GTCCATGTTGAACAGTAAAACCC-3′; reverse primer, 5′-TCCTCCGGTTTGTTGTAATTAGC-3′). The resulting 809-bp fragment was labeled as described below. The ACT1 probe consisted of a 1,090-bp segment generated by colony PCR using whole P. pastoris cells as a template (forward primer, 5′-CAATGGTTCCGGTATGTGTAAGG-3′; reverse primer, 5′-ACACTTGAGGTGCACAATGGATG-3′). The PEX5 PCR product was purified using the QiaQuick PCR Purification System (Qiagen), and the ACT1 PCR product was purified using the QiaQuick Gel Extraction System (Qiagen). Purified PCR products were radiolabeled using the Prime-It II kit (Stratagene) in accordance with the manufacturer's directions. The amount of signal produced in each lane was quantitated by analyzing the density of each band (from a scan of the original film) using MacBAS V2.5 software.
Antibodies, protein extracts, and Western blotting.
We have previously described the polyclonal antibodies recognizing P. pastoris Pex4p (5), Pex5p (15), and Pex10p (23). Anti-thiolase antibodies were raised against recombinant Saccharomyces cerevisiae thiolase and affinity purified. Secondary horseradish peroxidase-conjugated goat anti-rabbit antibodies were obtained from Sigma and Jackson ImmunoResearch.
For the production of whole-cell lysates, yeast strains were grown in YPD to mid-logarithmic phase, pelleted, and resuspended in an equal volume of minimal-methanol medium. Cultures were incubated with agitation at 30°C for 14 h to allow the induction of peroxisomal proteins. Approximately five OD600 units of methanol-induced cells were pelleted and resuspended in 1 ml of ice-cold sterile water. To lyse the cells, 150 μl of an ice-cold 2 M NaOH–1.2 M β-mercaptoethanol solution (freshly made) was added to the resuspended cells. The cells were vortexed briefly to mix and incubated on ice for 10 min. To precipitate cellular proteins, 150 μl of ice-cold 50% trichloroacetic acid was added to the lysate. Following a 15-min incubation on ice, the proteins were pelleted by centrifugation at 12,000 × g for 5 min. The supernatant was discarded, and the pellet was patiently resuspended in 100 μl of 5% SDS in TBS (25 mM Tris base [pH 8.0], 137 mM NaCl, 3 mM KCl). The protein concentration in each lysate was quantitated using the Micro BCA Protein Assay Reagent Kit (Pierce) in accordance with the manufacturer's directions.
To detect Pex5p levels, 30 μg of the total protein was separated by SDS–10% PAGE. Western blotting and chemiluminescent detection were performed as detailed by Crane et al. (5). Equal loading was confirmed on all blots by staining the protein remaining on the gel after transfer with Coomassie blue. The amount of Pex5p detected relative to the wild type was quantitated for each sample by analyzing the density of each band (from a scan of the original film) using MacBAS V2.5 software.
Detection of initial synthesis of Pex5p.
To detect Pex5p synthesis, the pex4Δ and pex10Δ strains were grown to log phase in YPD and subsequently shifted to minimal-methanol medium for 14 to 18 h. Cells were harvested and resuspended in 0.5 ml of minimal-methanol medium per 2.5 OD600 units of cells. The resuspension was incubated at 30°C with agitation for 10 min, and the cells were then labeled by the addition of 75 μCi of [35S]methionine per 0.5 ml (NEN EasyTag [35S]methionine). After 2 to 5 min (2 min for the experiment shown), 20 μl of chase solution (0.5 M methionine, 0.5 M cysteine) was added per 0.5 ml. A 0.5-ml aliquot was then immediately transferred to 0.5 ml of ice-cold azide stop solution (0.04 M methionine, 0.04 M cysteine, 0.13% sodium azide). Cells were then pelleted and resuspended in 1 ml of ice-cold water. The cells were lysed, and total cellular protein was precipitated as outlined above. Protein pellets were resuspended in 30 μl of SDS-PAGE loading buffer (NaOH was added as needed to neutralize the sample) and boiled for 5 min to solubilize Pex5p. One milliliter of ice-cold dilution buffer (1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 50 mM Tris [pH 7.5]) plus protease inhibitors (Boehringer Mannheim Complete Tablets) was added, and the samples were allowed to sit on ice for 40 min to assist solubility. At this point, any nonsoluble debris were removed and a saturating amount of polyclonal anti-Pex5p antibody was added (diluted into 0.5 ml of dilution buffer plus protease inhibitors per sample, distributed to each sample from a single mixture). After incubation overnight at 4°C, a saturating amount of protein A agarose beads (Santa Cruz Biotechnology) was added and samples were incubated for an additional 90 min at 4°C with end-over-end rotation. The beads were then pelleted and washed five times in ice-cold wash buffer (0.1% Triton X-100, 0.02% SDS, 150 mM NaCl, 5 mM EDTA, 50 mM Tris [pH 7.5]). The beads were resuspended in 15 μl of SDS-PAGE buffer and boiled for 5 min, and the entirety of the sample was separated by SDS-PAGE. The gel was then soaked in 0.5 M sodium salicylate for 30 min, dried, and exposed to film.
RESULTS
Reduced abundance of the PTS1 receptor in cells lacking Pex4p activity.
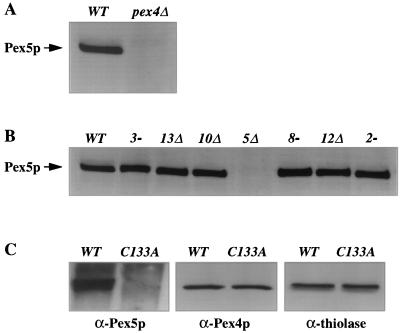
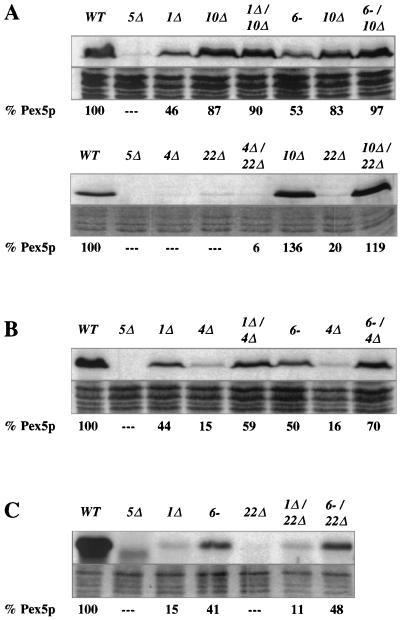
We have previously demonstrated that P. pastoris Pex4p is highly similar to known ubiquitin-conjugating enzymes, forms an adduct with ubiquitin in vivo, and requires its active-site cysteine (C133) for activity (5). Its likely function is therefore to ubiquitinate one or more target polypeptides. Given that the usual consequence of ubiquitination of target proteins is their degradation, we screened for peroxins that were stabilized by loss of Pex4p. We have yet to identify such a protein, but we did observe that pex4Δ cells contain extremely low levels of Pex5p, the PTS1 receptor (Fig. 1A). This phenotype has been previously observed by our lab for human and yeast cells lacking pex1 (7, 33) or pex6 (49) and has also been reported by Koller et al. for both the P. pastoris pex4 and pex22 mutants (25). Reduced Pex5p abundance is not a common phenotype of pex mutants (7), and the P. pastoris pex2-2, pex3-1, pex8-3, pex10Δ, pex12Δ, and pex13Δ mutants all contain normal levels of Pex5p (Fig. 1B). Normal Pex5p abundance was also observed in the pex14Δ and pex17Δ strains (data not shown). Although the pex1Δ and pex6-1 strains have reduced Pex5p levels, the phenotype is not as pronounced as in the pex4Δ or pex22Δ mutant. The average levels of Pex5p, compared to that of the wild type, were 35% for pex1Δ, 48% for pex6-1, 10% for pex4Δ, and 7% for pex22Δ. However, it was not unusual for the levels of Pex5p to vary by 20% of the wild-type level from one trial to the next.
FIG. 1.
Loss of Pex4p results in reduced steady-state levels of Pex5p. (A and B) Equal amounts of protein were extracted from methanol-induced cells, separated by SDS-PAGE, and blotted with anti-Pex5p antibodies. (A) Levels of Pex5p in wild-type (WT) and pex4Δ cells. (B) Normal Pex5p levels in pex3, pex13, pex10, pex8, pex12, and pex2 strains. (C) Total cellular proteins extracted from pex4Δ strains carrying a wild-type PEX4 expression plasmid and a plasmid that expresses an active-site mutant of PEX4, C133A. Levels of Pex5p (left), Pex4p (middle), and the peroxisomal matrix enzyme thiolase (right) present in these samples were determined by immunoblotting with specific antibodies.
The reduction of Pex5p levels in pex4Δ cells could be caused either by the absence of the Pex4p polypeptide or the absence of its enzymatic activity. To distinguish between these possibilities, we examined the steady-state levels of Pex5p in pex4Δ cells carrying plasmids that express either wild-type PEX4 or a mutant form of pex4 in which the active-site cysteine is replaced with alanine (PEX4-C133A [5]; the PEX4-C133A product is properly localized to the peroxisome [data not shown]). Total cellular protein was isolated from the two strains, equal amounts of the two protein samples were separated by SDS-PAGE, and the levels of Pex4p, Pex5p, and thiolase were determined by Western blot analysis. Although Pex4p and thiolase were present at similar levels in both strains, Pex5p levels were significantly reduced in the strain expressing PEX4-C133A, indicating that Pex4p enzyme activity is required for normal Pex5p abundance (Fig. 1C).
Pex5p is synthesized normally in pex4Δ cells.
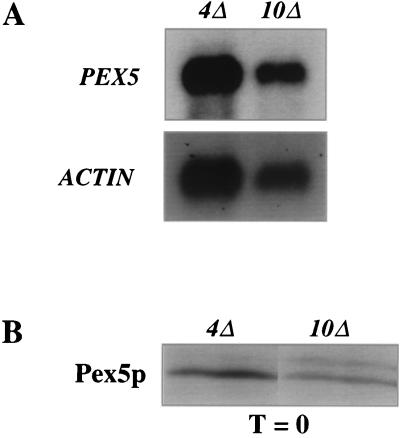
To determine if the reduced level of Pex5p in the pex4Δ strain was due to reduced levels of PEX5 transcript, we performed Northern blot analysis on pex4Δ and pex10Δ cells. Poly(A)+ RNAs from pex4Δ and pex10Δ cells were separated by denaturing agarose gel electrophoresis, transferred to membranes, and sequentially hybridized to radiolabeled probes for the P. pastoris PEX5 and ACT1 (actin) genes (Fig. 2A). Quantitation of the PEX5 and ACT1 mRNA levels revealed that the PEX5:ACT1 ratio in the pex4Δ strain was 1.3 times that in the pex10Δ strain. Similar results were obtained in two additional independent trials (data not shown). Thus, the reduction of Pex5p levels in the pex4Δ strain cannot be due to decreased levels of PEX5 mRNA synthesis.
FIG. 2.
PEX5 mRNA and Pex5p are synthesized normally in pex4Δ cells. (A) Northern blot analysis of poly(A)+ RNAs from pex4Δ and pex10Δ cells. The two RNA samples were separated by denaturing gel electrophoresis, transferred to membranes, and sequentially hybridized to radiolabeled probes specific for the PEX5 (top) and ACT1 (bottom) genes. Quantification revealed that the PEX5:ACT1 hybridization signal was 1.3-fold higher in the pex4Δ strain than in the pex10Δ strain. Similar results were observed in two additional independent trials. (B) Synthesis of equivalent levels of Pex5p during pulse-labeling of pex4Δ and pex10Δ cells. Each strain was incubated in the presence of [35S]methionine for 2 min and lysed in alkali, and the Pex5p present in each sample was immunoprecipitated using excess anti-Pex5p antibodies. The immunoprecipitates were separated by SDS-PAGE, and the amount of Pex5p was determined by fluorographic exposure to X-ray film. Similar results were observed in nine additional trials of this experiment.
To determine whether PEX5 mRNA is translated in the pex4Δ strain, we compared the initial synthesis of Pex5p in pex4Δ and pex10Δ cells. Methanol-induced cells were labeled with [35S]methionine for 2 min, and total cellular protein was collected by alkaline lysis. Pex5p was immunoprecipitated from these lysates using excess anti-Pex5p antibodies, and the level of labeled Pex5p in each sample was determined by fluorography (Fig. 2B). The amounts of Pex5p synthesized in pex4Δ and pex10Δ cells were similar, with slightly higher levels of translation in the pex4Δ strain. This experiment was repeated 10 times, with similar results in all of the trials. These data demonstrate that the reduced level of Pex5p in pex4Δ cells occurs posttranslationally, presumably via accelerated degradation of Pex5p. Consistent with this, the reduced level of Pex5p in human pex1- and pex6-deficient cells is known to result from accelerated Pex5p degradation (7, 49). Unfortunately, we were unable to measure the half-life of Pex5p in pex4Δ and pex10Δ cells due to technical difficulties in chasing the [35S]methionine from the cells.
Epistasis analysis places Pex4p late in peroxisomal matrix protein import.
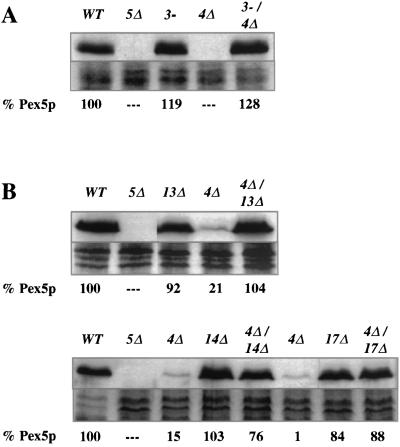
The variance in Pex5p stability displayed by different pex mutants allowed us to examine epistatic relationships among various pex mutants. An array of studies with both yeast (19, 21, 46, 48) and human (38) cells have established that PEX3 is required for synthesis of peroxisome membranes and that pex3 mutants are devoid of peroxisome-like structures. To test whether the reduced Pex5p levels seen in pex4Δ cells require the presence of peroxisome membranes and PEX3 function, we examined the levels of Pex5p in a pex3-1 pex4Δ double mutant. Total cellular protein was extracted from methanol-induced wild-type, pex5Δ, pex3-1, pex4Δ, and pex3-1 pex4Δ cells. The same amount of protein from each strain was then separated by SDS-PAGE and blotted with anti-Pex5p antibodies (Fig. 3A). In contrast to the low level of Pex5p detected in pex4Δ cells, the level of Pex5p observed in the pex3-1 pex4Δ double mutant was similar to those seen in the pex3-1 single-mutant and wild-type cells. These results demonstrate that Pex3p acts upstream of Pex4p. To visualize equivalency in loading, the protein remaining on the gel after transfer was stained with Coomassie blue (Fig. 3, 4, and 5).
FIG. 3.
The destabilization of Pex5p in the pex4Δ strain requires Pex3p, Pex13p, Pex14p, and Pex17p. For each panel, the strains were grown side by side and total cellular protein was extracted with alkali, separated by SDS-PAGE, and blotted with antibodies specific for Pex5p (top of each panel). Coomassie blue staining of each protein sample is also shown (bottom of each panel), and the quantity of Pex5p in each sample (as assessed by scanning densitometry of the resulting films) is shown below each lane. (A) Levels of Pex5p in wild-type (WT) cells; the pex5Δ, pex3-1, and pex4Δ mutants; and the pex3-1 pex4Δ double mutant. (B) Levels of Pex5p in wild-type cells; the pex5Δ, pex13Δ, and pex4Δ mutants; and the pex4Δ pex13Δ double mutant. The lower blot in panel B shows levels of Pex5p in wild-type cells; the pex5Δ, pex4Δ, and pex14Δ mutants; the pex4Δ pex14Δ double mutant; the pex4Δ and pex17Δ mutants; and the pex4Δ pex17Δ double mutant.
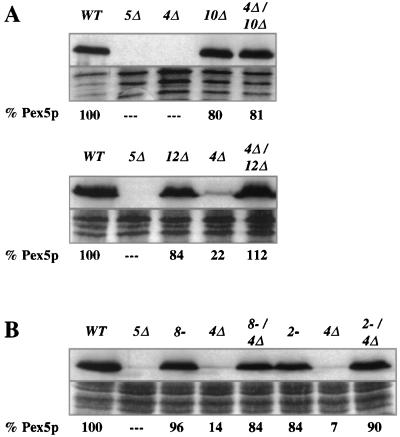
FIG. 4.
Destabilization of Pex5p in the pex4Δ strain requires Pex10p, Pex12p, Pex8p, and Pex2p. For each panel, the strains were grown side by side and total cellular protein was extracted with alkali, separated by SDS-PAGE, and blotted with antibodies specific for Pex5p (top of each panel). Coomassie blue staining of each protein sample is also shown (bottom of each panel), and the quantity of Pex5p in each sample (as assessed by scanning densitometry of the resulting films) is shown below each lane. (A) Levels of Pex5p in wild-type (WT) cells; the pex5Δ, pex4Δ, and pex10Δ mutants; and the pex4Δ pex10Δ double mutant. The lower blot in panel A shows levels of Pex5p in wild-type cells; the pex5Δ, pex12Δ, and pex4Δ mutants, and the pex4Δ pex12Δ double mutant. (B) Levels of Pex5p in wild-type cells; the pex5Δ, pex8-3, and pex4Δ mutants; the pex4Δ pex8-3 double mutant; the pex2-2 and pex4Δ mutants; and the pex2-2 pex4Δ double mutant.
FIG. 5.
Pex1p and Pex6p act downstream of Pex10p and upstream of Pex4p and Pex22p. For each panel, the strains were grown side by side and total cellular protein was extracted with alkali, separated by SDS-PAGE, and blotted with antibodies specific for Pex5p (top of each panel). Coomassie blue staining of each protein sample is also shown (bottom of each panel), and the quantity of Pex5p in each sample (as assessed by scanning densitometry of the resulting films) is shown below each lane. (A) Levels of Pex5p in wild-type (WT) cells; the pex5Δ, pex1Δ, and pex10Δ mutants; the pex1Δ pex10Δ double mutant; the pex6-1 and pex10Δ mutants; and the pex6-1 pex10Δ double mutant. The lower blot in panel A shows levels of Pex5p in wild-type cells; the pex5Δ, pex4Δ, and pex22Δ mutants; the pex4Δ pex22Δ double mutant; the pex10Δ and pex22Δ mutants; and the pex10Δ pex22Δ double mutant. (B) Levels of Pex5p in wild-type cells; the pex5Δ, pex1Δ, and pex4Δ mutants; the pex1Δ pex4Δ double mutant; the pex6-1 and pex4Δ mutants; and the pex6-1 pex4Δ double mutant. (C) Levels of Pex5p in wild-type cells; the pex5Δ, pex1Δ, pex6-1, and pex22Δ mutants; and the pex1Δ pex22Δ and pex6-1 pex22Δ double mutants.
One of the earliest steps in peroxisomal matrix protein import is the docking of PTS receptors to the peroxisome membrane. This process is mediated by Pex13p (8, 9, 14, 15) and Pex14p (1, 11) and may involve Pex17p (22), a peroxin that has also been implicated in PMP import (37). To assess the epistatic relationship of PEX4 to the PEX genes necessary for receptor docking, we examined Pex5p levels in the pex4Δ pex13Δ, pex4Δ pex14Δ, and pex4Δ pex17Δ double mutants, as well as in all relevant single mutants. In contrast to the pex4Δ mutant, the double mutants all contained high levels of Pex5p that were similar to those of the pex13Δ, pex14Δ, and pex17Δ single mutants (Fig. 3B). The fact that Pex13p, Pex14p, and Pex17p must act in order for the loss of Pex4p to cause destabilization of Pex5p indicates that these three peroxins act upstream of Pex4p.
The integral peroxisomal membrane proteins Pex8p, Pex10p, and Pex12p have been implicated in peroxisomal matrix protein import at a step downstream of receptor docking, most probably at matrix protein translocation (3, 32). Double mutants lacking pex4 and pex8, pex10, or pex12 were generated, and their Pex5p levels were examined. High Pex5p levels were detected in these double mutants, indicating that these matrix protein import factors act upstream of Pex4p (Fig. 4A and B). A specific role for PEX2 has yet to be elucidated, but the phenotype of pex2-deficient cells suggests that they also participate in matrix protein import downstream of receptor docking (7). The pex2-2 pex4Δ double mutant was generated and found to have normal Pex5p levels, indicating that PEX4 also acts after PEX2 (Fig. 4B).
Pex1p, Pex6p, and Pex22p also act late in peroxisomal matrix protein import.
The phenotype of Pex5p instability is also shared by the pex22Δ mutant and to lesser extents by the pex1Δ and pex6-1 mutants. While a previous study has established that Pex22p interacts physically with Pex4p (25), there is no evidence linking PEX4 or PEX22 function with PEX1 or PEX6 function. Therefore, we tested whether these mutants might also act late in peroxisomal matrix protein import. To do this, we generated double-mutant combinations of pex1Δ, pex6-1, and pex22Δ with pex10Δ, which blocks matrix protein import downstream of PTS receptor docking (3). Pex5p levels were examined in pex1Δ pex10Δ, pex6-1 pex10Δ, and pex10Δ pex22Δ double mutants, and in each case the levels of Pex5p were similar to that of the pex10Δ single mutant (Fig. 5A). The pex4Δ pex22Δ double mutant was also examined and found to have low Pex5p levels that were similar to those of both single mutants (Fig. 5A). The slight increase seen in the quantitation (6% for the double mutant versus undetectable for the single mutants) is not outside the sensitivity limit of the quantitation method.
The fact that PEX10 is epistatic to PEX1, PEX4, PEX6, and PEX22 indicates that the products of these four genes act late in peroxisomal matrix protein import. However, it was apparent from our studies that the pex1Δ and pex6-1 mutants contain higher levels of Pex5p than the pex4Δ and pex22Δ mutants. To order these genes relative to one another, we examined Pex5p abundance in pex1Δ pex4Δ and pex4Δ pex6-1 double mutants. Pex5p levels in the pex1Δ pex4Δ and pex4Δ pex6-1 strains resembled those of the single pex1Δ and pex6-1 mutants, indicating that Pex1p and Pex6p act upstream of Pex4p (Fig. 5B). We also examined Pex5p levels in pex1Δ pex22Δ and pex6-1 pex22Δ double mutants, and the Pex5p levels again resembled those of the pex1Δ and pex6-1 single mutants (Fig. 5C). This indicates that Pex1p and Pex6p also act before Pex22p. As noted earlier, there is a noticeable variability in the relative amount of Pex5p present in a given strain. The range of variability is usually about 20% of the wild-type level, which likely explains why the double mutants often contained levels of Pex5p that were slightly higher or lower than that of either single mutant alone.
Peroxisomes of pex4Δ mutants contain residual levels of peroxisomal matrix proteins.
Several lines of evidence indicate that peroxisomal matrix protein import involves the cycling of PTS receptors between the cytoplasm and peroxisome, suggesting that the terminal step in matrix protein import is the release of PTS receptors back to the cytoplasm (42). Cells defective in this step would be expected to contain recognizable peroxisomes, import PMPs normally, and import residual levels of peroxisomal matrix proteins.
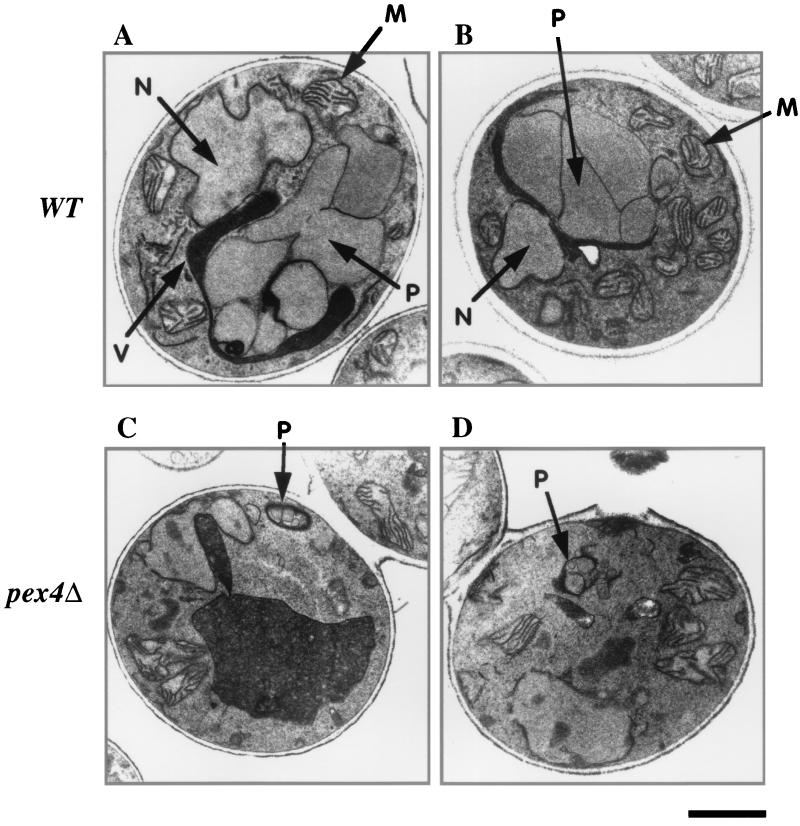
Previous studies of P. pastoris pex1-, pex6-, and pex22-deficient strains have indicated that these mutants have precisely this phenotype (20, 25, 39). As our prior analysis of the P. pastoris pex4Δ mutant did not address its phenotype at that level of detail (5), we proceeded to characterize the peroxisomes of pex4Δ cells. Previous studies have established that the yeast P. pastoris has numerous large peroxisomes when grown on energy sources that require peroxisomal metabolic pathways, particularly methanol or fatty acids (17, 23, 29). In particular, peroxisomes of methanol-grown cells can be easily detected in thin-section electron micrographs due to their large cuboidal shape, their electron-dense matrix, and the fact that they are typically attached to one another (Fig. 6A and B). Cells lacking Pex4p also contain peroxisomes with an electron-dense granular matrix and semicuboidal shape, though they are much smaller than peroxisomes of wild-type cells (Fig. 6C and D).
FIG. 6.
Transmission electron microscopy reveals the presence of small, matrix-containing peroxisomes in pex4Δ cells. Wild-type (WT) (A and B) and pex4Δ (C and D) cells were induced in methanol medium and processed for transmission electron microscopy. M, mitochondria; N, nucleus; P, peroxisome; V, vacuole. Bar, 1.0 μm.
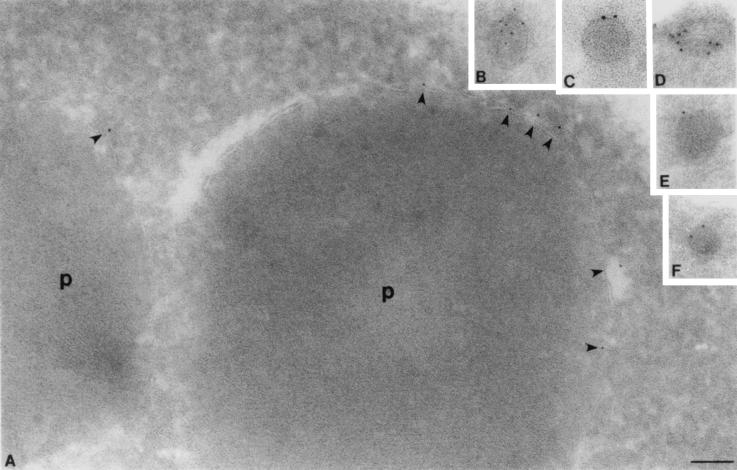
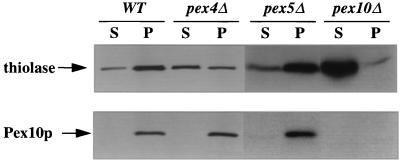
To confirm that these structures were indeed peroxisomes, we also performed immunoelectron microscopy on wild-type (Fig. 7A) and pex4Δ (Fig. 7B to F) cells. Cryosections of embedded cells were incubated with antibodies specific for Pex10p, a known integral PMP. The anti-Pex10p antibody was visualized by binding of a secondary gold particle conjugate. These experiments demonstrate that pex4Δ cells contain peroxisomes with a discernible electron-dense matrix but have only 1/10 of the diameter of normal peroxisomes.
FIG. 7.
Immunocryoelectron microscopy demonstrates that pex4Δ cells contain small, electron-dense peroxisomes. (A) Peroxisomes (p) of wild-type cells labeled with antibodies specific for Pex10p, shown here by immunogold labeling of the peroxisome membrane. Note the position of gold particles at the peroxisome membrane (shown by arrowheads) and the electron-dense nature of the protein-rich peroxisome matrix. (B to F) Peroxisomes of pex4Δ cells are also labeled with antibodies specific for Pex10p and contain an electron-dense, granular matrix. However, their radius is only 10% of the radius of wild-type peroxisomes, indicating that their volume may be only 1/1,000 of that of the wild-type peroxisome. Bar, 0.1 μm.
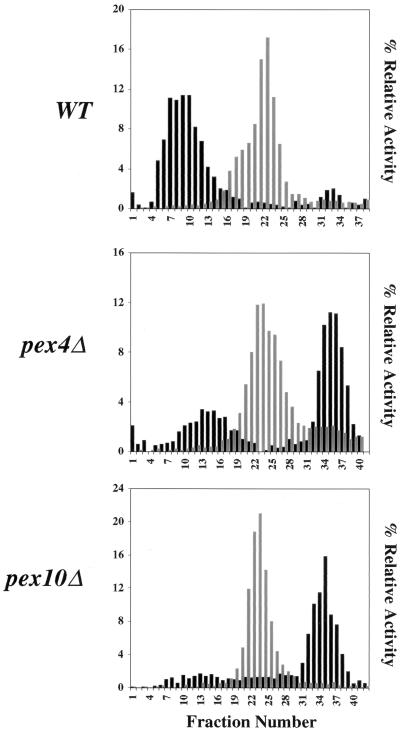
The morphology of peroxisomes in pex4Δ cells suggested that they were able to import low but significant levels of peroxisomal matrix proteins into the peroxisome lumen. We tested this hypothesis by examining pex4Δ cells for the presence of peroxisomes of normal density. Wild-type, pex4Δ, and pex10Δ cells were induced in methanol-containing medium and harvested, and a PNS was generated from each strain. Peroxisomes and other large organelles were collected by differential centrifugation and separated by sucrose density gradient centrifugation. The resulting fractions were assayed for the peroxisomal matrix protein catalase and the mitochondrial marker succinate dehydrogenase (Fig. 8). These experiments show that pex4Δ cells contain significant levels of catalase at approximately the same density as wild-type peroxisomes. Previous studies have shown that loss of Pex10p leads to a far more severe defect in peroxisomal matrix protein import (23), and this is reflected in the absence of a catalase peak at the normal density of peroxisomes. The low-density peak of catalase activity in each sample reflects enzyme that is nonspecifically trapped during collection of the organelle pellet, as well as enzyme that leaks from peroxisomes during manipulation of the organelle pellet.
FIG. 8.
The pex4Δ mutant contains peroxisomes of normal density. Wild-type (WT), pex4Δ, and pex10Δ cells were induced in methanol medium, a PNS was generated from each strain, and peroxisomes and mitochondria were collected by differential centrifugation. The resulting three organelle pellets were then separated by sucrose density gradient centrifugation. The same amount of each fraction was assayed for catalase (a PTS1-containing peroxisomal enzyme) activity (black bars), succinate dehydrogenase (mitochondrial enzyme) activity (grey bars), and density (data not shown). Enzyme activities are plotted as percentages of the total activity in the gradient. The densities of all of the gradient profile were similar, with the peak peroxisomal fraction of wild-type cells migrating at a density of 1.19 to 1.21 g/cm3 and the peak peroxisomal fraction of pex4Δ cells migrating at a density of 1.18 to 1.20 g/cm3.
To further address the ability of pex4Δ cells to import peroxisomal proteins, we assayed the import of a PTS2 protein and a PMP in wild-type, pex4Δ, pex5Δ, and pex10Δ cells. The PNS generated from each strain was separated into an organelle pellet and cytosolic supernatant by centrifugation at 25,000 × g, and the proportions of thiolase (PTS2 marker) and Pex10p (PMP marker) in the supernatant and pellet were quantitated. The pex4Δ cells import all of the detectable Pex10p, indicating that the strain has no defect in PMP import. The pex4Δ cells also imported 42% of the cellular thiolase, versus 75% for wild type, 69% for pex5Δ cells, and only 1% for pex10Δ cells (Fig. 9). The fact that pex4Δ cells import 1.5-fold less thiolase than pex5Δ cells indicates that the import defect of pex4Δ cells is not limited to the PTS1 pathway. This hypothesis is supported by the fact that overexpression of Pex5p is unable to suppress the growth defects of the pex4Δ strain (data not shown).
FIG. 9.
The pex4Δ mutant imports significant amounts of the PTS2 marker enzyme thiolase but less than wild-type (WT) or pex5Δ cells. Wild-type, pex4Δ, pex5Δ, and pex10Δ cells were induced to proliferate peroxisomal proteins, and a PNS was generated from each strain. This was then separated into an organelle pellet (p) and a cytosolic supernatant (s) by centrifugation at 25,000 × g. The same proportion of each fraction was separated by SDS-PAGE and blotted with antibodies specific for the PTS2-targeted enzyme thiolase (top) and the integral PMP Pex10p (bottom).
Subcellular distribution of Pex5p in the pex4Δ strain.
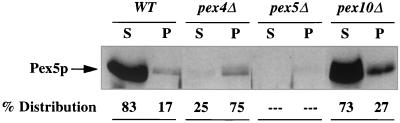
A previous study has established that P. pastoris Pex5p is a predominantly cytosolic, partly peroxisomal protein in wild-type cells (15). The hypothesis that Pex4p plays an important role in the recycling of Pex5p back to the cytoplasm predicts that loss of Pex4p should result in accumulation of Pex5p at the peroxisome. We examined the distribution of Pex5p in wild-type, pex4Δ, pex5Δ, and pex10Δ cells. Strains were induced in methanol medium, and postnuclear supernatants (PNS) were prepared and separated into an organelle pellet and a cytosolic supernatant. Equal proportions of these fractions were assayed Pex5p by Western blot analysis (Fig. 10). Virtually all of the Pex5p present in pex4Δ cells was associated with the organelle pellet, a stark contrast to the predominantly cytosolic distribution in wild-type and pex10Δ cells.
FIG. 10.
Most Pex5p is present in the organelle fraction of pex4Δ cells. Wild-type (WT), pex4Δ, pex5Δ, and pex10Δ cells were induced in methanol medium, and a PNS was generated from each strain. This was then separated into an organelle pellet (p) and a cytosolic supernatant (s) by centrifugation at 25,000 × g. The same proportion of each fraction was separated by SDS-PAGE and blotted with antibodies specific for Pex5p.
DISCUSSION
Genetic studies of peroxisome biogenesis have led to the identification of more than 20 different peroxins, each required for normal peroxisome biogenesis. Studies of these factors and of peroxisomal protein import mechanisms have shown that PMP import and peroxisomal matrix protein import occur through separate pathways, with each process controlled by a distinct set of genes. As all peroxisomal matrix proteins are translated in the cytosol, their proper localization is dependent on the action of the PTS receptors. Studies on Pex5p, the PTS1 receptor, indicate that it is a predominantly cytoplasmic, partly peroxisomal protein (6, 8, 15, 45) and may cycle between the cytoplasm and the peroxisome (7). Such results have inspired models of peroxisomal matrix protein import in which peroxins are required not only for the translocation of proteins through the peroxisome membrane but also to facilitate the movement of PTS receptors through their cycles. Such a view of peroxisomal matrix protein import suggests the existence of PTS receptor docking factors, matrix protein translocation factors, and PTS receptor recycling factors.
Within the context of this model, determining the order of action for different peroxisome biogenesis factors should be extremely useful for determining their functions. Obviously, biochemical approaches can be used to determine the order of peroxin action. Such studies have led to current models in which Pex14p acts in PTS receptor docking in peroxisomal matrix protein import (42) and Pex8p (32), Pex10p, and Pex12p (3) act downstream of receptor docking in peroxisomal matrix protein import. However, epistasis analysis is ideally suited for determining the order of gene action, provided that there are defining subphenotypes that can be used as markers of different steps in the process being studied. In this study, we used the subphenotype of reduced Pex5p abundance to examine the epistatic relationships among 12 different pex genes.
We have previously established that reduced Pex5p abundance is a reproducible phenotype for human pex1- and pex6-deficient cell lines but is not observed in human pex2-, pex7-, pex10-, pex12-, and pex16-deficient cells (7, 49). Furthermore, these studies revealed that the reduced abundance of Pex5p in pex1- and pex6-deficient cells was due to an increased rate of Pex5p degradation. Koller et al. recently reported that Pex5p abundance is reduced in P. pastoris pex4 and pex22 mutants (25). Here we show that Pex5p abundance is normal in P. pastoris pex2-2, pex3-1, pex8-3, pex10Δ, pex12Δ, pex13Δ, pex14Δ, and pex17Δ mutants and that the reduction in Pex5p abundance is more severe in pex4 and pex22 mutants than in pex1 and pex6 mutants. Furthermore, we demonstrated that pex4Δ cells have normal PEX5 mRNA abundance and are able to synthesize Pex5p. That pex4 and pex10 mutants have similar rates of Pex5p synthesis but different steady-state levels of Pex5p represents strong evidence that Pex5p is degraded at an accelerated rate in pex4Δ cells. These results are consistent with the increased rate of Pex5p degradation in human pex1- and pex6-deficient cells (7, 49).
Although technical difficulties in chasing labeled methionine from P. pastoris cells prevented us from measuring Pex5p stability in the pex4Δ and pex10Δ mutant strains, we were still able to use the variance in Pex5p abundance to order the pex mutants relative to one another by epistasis analysis. Our data show that Pex4p acts after the peroxisome membrane synthesis factor Pex3p, after the PTS receptor docking factors Pex13p and Pex14p, after the putative protein translocation factors Pex8p, Pex10p, and Pex12p, and after the other peroxins Pex2p and Pex17p. Furthermore, we found that Pex1p, Pex6p, and Pex22p also act downstream of Pex10p and that Pex1p and Pex6p act upstream of both Pex4p and Pex22p. This result suggests a model in which Pex1p, Pex4p, Pex6p, and Pex22p all act downstream of matrix protein translocation (Fig. 11).
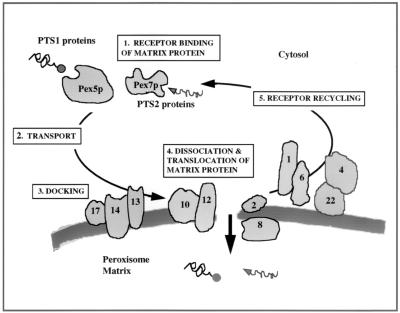
FIG. 11.
Model of peroxisomal matrix protein import. PTS-containing proteins are bound by their receptor after translation in the cytoplasm (step 1). The mechanism by which the receptor-bound matrix protein is transported to the peroxisome membrane remains unclear (step 2). The receptor-bound matrix protein then docks on the surface of the peroxisome membrane via interactions with the docking factors Pex13p, Pex14p, and possibly Pex17p (step 3). Following docking, the matrix protein must dissociate from the receptor and be translocated across the peroxisome membrane into the matrix space (step 4). It is not clear whether the receptor follows the cargo into the lumen or remains on the cytosolic surface of the peroxisome. However, in either scenario, the receptor is subsequently released from the translocation pore and recycled back to the cytoplasm to undergo further rounds of import (step 5).
The placement of Pex4p very late in peroxisomal matrix protein import is consistent with the cellular phenotypes of pex4 mutants. Electron microscopy studies revealed that pex4 cells still have peroxisomes and that these peroxisomes resemble those of wild-type cells in all respects but their overall size. Peroxisomes of pex4Δ cells had the clustered, cuboidal, electron-dense appearance of normal methanol-induced P. pastoris peroxisomes, but their diameter was 10 times less. Biochemical studies revealed that peroxisomes of pex4Δ cells import wild-type levels of the PMP Pex10p and import reduced but significant levels of both PTS1- and PTS2-targeted peroxisomal matrix proteins. In contrast, pex10Δ cells displayed more severe peroxisomal matrix protein import defects, indicating that the peroxisomal matrix protein import defects of the pex4 mutant are unusually mild. The phenotypes of the pex4 mutant are not what we would expect for loss of a peroxin that plays an essential role in peroxisome membrane synthesis, PMP import, PTS receptor docking, or peroxisomal matrix protein translocation. However, they are consistent with the loss of a peroxin that participates in PTS receptor recycling, which is thought to be the final step in peroxisomal matrix protein import. This hypothesis for Pex4p function predicts that Pex5p should be trapped at or in the peroxisome in pex4Δ cells, and it is interesting that 75% of the Pex5p in pex4Δ cells is present in the organelle fraction (compared to only 17% in wild-type cells).
An analysis of Hansenula polymorpha Pex4p also concluded that Pex4p participates in the recycling of Pex5p to the cytoplasm (44). That study also found that Pex5p accumulates on or in peroxisomes in the absence of Pex4p. However, they also found that the peroxisomal protein import defect of pex4Δ cells could be partially suppressed by overexpressing PEX5 (44). Interestingly, H. polymorpha pex4 mutants import PTS2 proteins normally (44) whereas S. cerevisiae and P. pastoris pex4 mutants display a reproducible defect in PTS2 protein import (47). The mild PTS2 protein import defect and the severe reduction in Pex5p abundance in P. pastoris pex4Δ cells raise the possibility that the phenotypes of pex4Δ cells is simply due to reduced Pex5p abundance. However, the PTS2 protein import defect of pex4Δ cells is more severe than that of pex5Δ cells. Also, we tested whether PEX5 overexpression could rescue the growth defects of pex4Δ cells but failed to see any evidence of phenotypic rescue. Thus, it is unlikely that reduced Pex5p abundance can explain all of the phenotypes of pex4Δ cells.
Koller et al. (25) have recently reported that Pex22p physically interacts with Pex4p, that pex22 mutants show severely reduced Pex5p levels, and that Pex22p is required for Pex4p abundance. We previously established that Pex4p is peripherally associated with the outer surface of the peroxisome membrane (5), and Koller et al. (25) speculated that Pex22p may function as a docking site for Pex4p on the peroxisome membrane. Thus, it is was not surprising that our epistasis analysis placed Pex22p after Pex1p, Pex6p, and Pex10p and, by deduction, after all of the other peroxins we examined, except Pex4p. These multiple lines of evidence connect Pex22p and Pex4p at a terminal step in peroxisomal matrix protein import.
Our epistasis studies indicate that Pex1p and Pex6p also act late in peroxisomal matrix protein import, though upstream of Pex4p and Pex22p. The action of Pex1p and Pex6p at a common point in peroxisome biogenesis is altogether expected, given that genetic interactions have been described for PEX1 and PEX6 and that physical interactions have been reported for Pex1p and Pex6p (10, 12, 24). Furthermore, results that place Pex1p and Pex6p at a late step in peroxisomal matrix protein import are consistent with the phenotypes of pex1 and pex6 mutants in most species. In humans (33, 36, 49) and the yeasts P. pastoris (20, 39), H. polymorpha (24), and S. cerevisiae (19), cells lacking pex1 or pex6 contain numerous peroxisomes that are competent for PMP import. Studies of the yeasts P. pastoris (20, 39) and H. polymorpha (24) and human cells (33, 49) have also shown that peroxisomes of pex1- and pex6-deficient cells import residual levels of peroxisomal matrix proteins. Thus, the pex1 and pex6 mutants, like the pex4 and pex22 mutants, display phenotypes that we might expect from the loss of receptor recycling factors. The epistasis analysis presented here clearly places P. pastoris Pex1p and Pex6p downstream of Pex10p, a protein that acts downstream of receptor docking in peroxisomal matrix protein import, supporting the hypothesis that Pex1p and Pex6p act late in peroxisomal matrix protein import. However, studies of the yeast Yarrowia lipolytica have led Rachubinski and colleagues to propose a different model in which Pex1p and Pex6p participate in peroxisome membrane biogenesis (43). We have no model that can explain these differences in results or interpretation, and it may be that the roles of Pex1p and Pex6p are quite different in Y. lipolytica.
How the Pex1p-Pex6p step of peroxisome biogenesis relates to the subsequent step defined by Pex4p-Pex22p remains to be determined. However, the fact that reduced Pex5p abundance is observed in cells lacking any of these four factors indicates that there may be a functional connection between these two steps. Pex1p and Pex6p are a pair of interacting AAA ATPases, and it is well established that these proteins participate in the formation, organization, and/or dissociation of protein complexes (30). It is perhaps worthwhile to consider the possibilities that Pex1p and Pex6p can prepare or present substrates for ubiquitination by a Pex4p-Pex22p complex and that the ubiquitination event has a positive effect on Pex5p stability. However, the possibilities for how Pex4 and Pex22p protect Pex5p from degradation are numerous. For example, it could involve the destruction of a protein that degrades Pex5p or a modification of Pex5p that protects it from degradation by some general proteolysis machinery. Elucidation of the molecular mechanisms by which Pex1p, Pex6p, Pex4p, and Pex22p impact the stability of Pex5p is clearly a challenge but must be pursued if we are to understand the roles of these peroxins and the process of PTS receptor recycling.
ACKNOWLEDGMENTS
We thank Yifei Liu for affinity purifying the anti-Pex10p antibody.
C.S.C. was partially supported by the Predoctoral Program in Human Genetics at The Johns Hopkins University. This work was supported by NIH grant DK45787 to S.J.G.
REFERENCES
- 1.Albertini M, Rehling P, Erdmann R, Girzalsky W, Kiel J A K W, Veenhuis M, Kunau W-H. Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell. 1997;89:83–92. doi: 10.1016/s0092-8674(00)80185-3. [DOI] [PubMed] [Google Scholar]
- 2.Chang C C, South S, Warren D, Jones J, Moser A B, Moser H W, Gould S J. Metabolic control of peroxisome abundance. J Cell Sci. 1999;112:761–774. doi: 10.1242/jcs.112.10.1579. [DOI] [PubMed] [Google Scholar]
- 3.Chang C C, Warren D S, Sacksteder K A, Gould S J. PEX12 binds PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J Cell Biol. 1999;147:761–773. doi: 10.1083/jcb.147.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crane D I, Gould S J. The Pichia pastoris HIS4 gene: nucleotide sequence, creation of a non-reverting his4 deletion mutant, and development of HIS4-based replicating and integrating plasmids. Curr Genet. 1994;26:443–450. doi: 10.1007/BF00309932. [DOI] [PubMed] [Google Scholar]
- 5.Crane D I, Kalish J E, Gould S J. The Pichia pastoris PAS4 gene encodes a ubiquitin-conjugating enzyme required for peroxisome assembly. J Biol Chem. 1994;269:21835–21844. [PubMed] [Google Scholar]
- 6.Dodt G, Braverman N, Wong C, Moser A, Moser H W, Watkins P, Valle D, Gould S J. Mutations in the PTS1 receptor gene, PXR1, define complementation group 2 of the peroxisome biogenesis disorders. Nat Genet. 1995;9:115–124. doi: 10.1038/ng0295-115. [DOI] [PubMed] [Google Scholar]
- 7.Dodt G, Gould S J. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol. 1996;135:1763–1774. doi: 10.1083/jcb.135.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elgersma Y, Kwast L, Klein A, Voorn-Brouwer T, van den Berg M, Metzig B, America T, Tabak H F, Distel B. The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import of PTS1 containing proteins. J Cell Biol. 1996;135:97–109. doi: 10.1083/jcb.135.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erdmann R, Blobel G. Identification of Pex13p, a peroxisomal membrane receptor for the PTS1 recognition factor. J Cell Biol. 1996;135:111–121. doi: 10.1083/jcb.135.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faber K N, Heyman J A, Subramani S. Two AAA family proteins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol Cell Biol. 1998;18:936–943. doi: 10.1128/mcb.18.2.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fransen M, Terlecky S R, Subramani S. Identification of a human PTS1 receptor docking protein directly required for peroxisomal protein import. Proc Natl Acad Sci USA. 1998;95:8087–8092. doi: 10.1073/pnas.95.14.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geisbrecht B V, Collins C S, Reuber B E, Gould S J. Disruption of a PEX1-PEX6 interaction is the most common cause of the neurologic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease. Proc Natl Acad Sci USA. 1998;95:8630–8635. doi: 10.1073/pnas.95.15.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisbrecht B V, Zhu D, Schulz K, Nau K, Morrell J C, Geraghty M, Schulz H, Erdmann R, Gould S J. Molecular characterization of Saccharomyces cerevisiae D3,D2-enoyl-CoA isomerase. J Biol Chem. 1998;273:33184–33191. doi: 10.1074/jbc.273.50.33184. [DOI] [PubMed] [Google Scholar]
- 14.Girzalsky W, Rehling P, Stein K, Kipper J, Blank L, Kunau W H, Erdmann R. Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2-dependent protein import into peroxisomes. J Cell Biol. 1999;144:1151–1162. doi: 10.1083/jcb.144.6.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gould S J, Kalish J E, Morrell J C, Bjorkman J, Urquhart A J, Crane D I. PEX13p is an SH3 protein in the peroxisome membrane and a docking factor for the PTS1 receptor. J Cell Biol. 1996;135:85–95. doi: 10.1083/jcb.135.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gould S J, Keller G A, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould S J, McCollum D, Spong A P, Heyman J A, Subramani S. Development of the yeast Pichia pastoris as a model organism for a genetic and molecular analysis of peroxisome assembly. Yeast. 1992;8:613–628. doi: 10.1002/yea.320080805. [DOI] [PubMed] [Google Scholar]
- 18.Grant S G, Jessee J, Bloom F R, Hanahan D. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc Natl Acad Sci USA. 1990;87:4645–4649. doi: 10.1073/pnas.87.12.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hettema E H, Girzalsky W, van Den Berg M, Erdmann R, Distel B. Saccharomyces cerevisiae pex3p and pex19p are required for proper localization and stability of peroxisomal membrane proteins. EMBO J. 2000;19:223–233. doi: 10.1093/emboj/19.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heyman J A, Mononsov E, Subramani S. Role of the PAS1 gene of Pichia pastoris in peroxisome biogenesis. J Cell Biol. 1994;127:1259–1273. doi: 10.1083/jcb.127.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Höhfeld J, Veenhuis M, Kunau W H. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J Cell Biol. 1991;114:1167–1178. doi: 10.1083/jcb.114.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huhse B, Rehling P, Albertini M, Blank L, Meller K, Kunau W H. Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal protein translocation machinery. J Cell Biol. 1998;140:49–60. doi: 10.1083/jcb.140.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalish J E, Theda C, Morrell J C, Berg J M, Gould S J. Formation of the peroxisome lumen is abolished by loss of Pichia pastoris Pas7p, a zinc-binding integral membrane protein of the peroxisome. Mol Cell Biol. 1995;15:6406–6419. doi: 10.1128/mcb.15.11.6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Kalish J E, Keller G A, Morrell J C, Mihalik S J, Smith B, Cregg J M, Gould S J. Characterization of a novel component of the peroxisomal protein import apparatus using fluorescent peroxisomal proteins. EMBO J. 1996;15:3275–3285. [PMC free article] [PubMed] [Google Scholar]
- 24.Kiel J A, Hilbrands R E, van der Klei I J, Rasmussen S W, Salomons F A, van der Heide M, Faber K N, Cregg J M, Veenhuis M. Hansenula polymorpha Pex1p and Pex6p are peroxisome-associated AAA proteins that functionally and physically interact. Yeast. 1999;15:1059–1078. doi: 10.1002/(SICI)1097-0061(199908)15:11<1059::AID-YEA434>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 25.Koller A, Snyder W B, Faber K N, Wenzel T J, Rangell L, Keller G A, Subramani S. Pex22p of Pichia pastoris, essential for peroxisomal matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisomal membrane. J Cell Biol. 1999;146:99–112. doi: 10.1083/jcb.146.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazarow P B, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 27.Marzioch M, Erdmann R, Veenhuis M, Kunau W-H. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 1994;13:4908–4918. doi: 10.1002/j.1460-2075.1994.tb06818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuzono Y, Kinoshita N, Tamura S, Shimozawa N, Hamasaki M, Ghaedi K, Wanders R J, Suzuki Y, Kondo N, Fujiki Y. Human PEX19: cDNA cloning by functional complementation, mutation analysis in a patient with Zellweger syndrome, and potential role in peroxisomal membrane assembly. Proc Natl Acad Sci USA. 1999;96:2116–2121. doi: 10.1073/pnas.96.5.2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCollum D, Monosov E, Subramani S. The pas8 mutant of Pichia pastoris exhibits the peroxisomal protein import deficiencies of Zellweger syndrome cells. The PAS8 protein binds to the COOH-terminal tripeptide peroxisomal targeting signal and is a member of the TPR protein family. J Cell Biol. 1993;121:761–774. doi: 10.1083/jcb.121.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neuwald A F, Aravind L, Spouge J L, Koonin E U. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 31.Rehling P, Marzioch M, Wittke E, Veenhuis M, Kunau W H. The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7 gene. EMBO J. 1996;15:2901–2913. [PMC free article] [PubMed] [Google Scholar]
- 32.Rehling P, Skaletz-Rorowski A, Girzalsky W, Voorn-Brouwer T, Franse M M, Distel B, Veenhuis M, Kunau W H, Erdmann R. Pex8p, an intraperoxisomal peroxin of Saccharomyces cerevisiae required for protein transport into peroxisomes, binds the PTS1 receptor pex5p. J Biol Chem. 2000;275:3593–3602. doi: 10.1074/jbc.275.5.3593. [DOI] [PubMed] [Google Scholar]
- 33.Reuber B E, Germain Lee E, Collins C S, Morrell J C, Ameritunga R, Moser H W, Valle D, Gould S J. Mutations in PEX1 are the most common cause of the peroxisome biogenesis disorders. Nat Genet. 1997;17:445–448. doi: 10.1038/ng1297-445. [DOI] [PubMed] [Google Scholar]
- 34.Sacksteder K A, Jones J M, South S T, Li X, Liu Y, Gould S J. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol. 2000;148:931–944. doi: 10.1083/jcb.148.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Slawecki M, Dodt G, Steinberg S, Moser A B, Moser H W, Gould S J. Identification of three distinct peroxisomal protein import defects in patients with peroxisomal biogenesis disorders. J Cell Sci. 1995;108:1817–1829. doi: 10.1242/jcs.108.5.1817. [DOI] [PubMed] [Google Scholar]
- 37.Snyder W B, Koller A, Choy A J, Johnson M A, Cregg J M, Rangell L, Keller G A, Subramani S. Pex17p is required for import of both peroxisome membrane and lumenal proteins and interacts with Pex19p and the peroxisome targeting signal-receptor docking complex in Pichia pastoris. Mol Biol Cell. 1999;10:4005–4019. doi: 10.1091/mbc.10.12.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.South S T, Sacksteder K A, Li X, Liu Y, Gould S J. Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J Cell Biol. 2000;149:1345–1360. doi: 10.1083/jcb.149.7.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spong A P, Subramani S. Cloning and characterization of PAS5: a gene required for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J Cell Biol. 1993;123:535–548. doi: 10.1083/jcb.123.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramani S. Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- 41.Swinkels B W, Gould S J, Bodnar A G, Rachubinski R A, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10:3244–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabak H F, Braakman I, Distel B. Peroxisomes: simple function but complex in maintenance. Trends Cell Biol. 1999;9:447–453. doi: 10.1016/s0962-8924(99)01650-5. [DOI] [PubMed] [Google Scholar]
- 43.Titorenko V I, Chan H, Rachubinski R A. Fusion of small peroxisomal vesicles in vitro reconstructs an early step in the in vivo multistep peroxisome assembly pathway of Yarrowia lipolytica. J Cell Biol. 2000;148:29–44. doi: 10.1083/jcb.148.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Klei I J, Hibrands R E, Kiel J A, Rasmussen S W, Cregg J M, Veenhuis M. The ubiquitin-conjugating enzyme Pex4p of Hansenula polymorpha is required for efficient functioning of the PTS1 import machinery. EMBO J. 1998;17:3608–3618. doi: 10.1093/emboj/17.13.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Klei I J, Hibrands R E, Swaving G J, Waterham H R, Vrieling E G, Titorenko V I, Cregg J M, Harder W, Veenhuis M. The Hansenula polymorpha PER3 gene is essential for the import of PTS1 proteins into the peroxisome matrix. J Biol Chem. 1995;270:17229–17236. doi: 10.1074/jbc.270.29.17229. [DOI] [PubMed] [Google Scholar]
- 46.Waterham H R, Titorenko V I, Swaving G J, Harder W, Veenhuis M. Peroxisomes in the methyltrophic yeast Hansenula polymorpha do not necessarily derive from pre-existing organelles. EMBO J. 1993;12:4785–4794. doi: 10.1002/j.1460-2075.1993.tb06167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiebel F F, Kunau W-H. The Pas2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature. 1992;359:73–76. doi: 10.1038/359073a0. [DOI] [PubMed] [Google Scholar]
- 48.Wiemer E A C, Luers G H, Faber K N, Wenzel T, Veenhuis M, Subramani S. Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. J Biol Chem. 1996;271:18973–18980. doi: 10.1074/jbc.271.31.18973. [DOI] [PubMed] [Google Scholar]
- 49.Yahraus T, Braverman N, Dodt G, Kalish J E, Morrell J C, Moser H W, Valle D, Gould S J. The peroxisome biogenesis disorder group 4 gene, PXAAA1, encodes a cytoplasmic ATPase required for stability of the PTS1 receptor. EMBO J. 1996;15:2914–2923. [PMC free article] [PubMed] [Google Scholar]