Abstract
Interferon A (IFN-A) genes are differentially expressed after virus induction. The differential expression of individual IFN-A genes is modulated by substitutions in the proximal positive virus responsive element A (VRE-A) of their promoters and by the presence or absence of a distal negative regulatory element (DNRE). The functional feature of the DNRE is to specifically act by repression of VRE-A activity. With the use of the yeast one-hybrid system, we describe here the identification of a specific DNRE-binding protein, the pituitary homeobox 1 (Ptx1 or Pitx1). Ptx1 is detectable in different cell types that differentially express IFN-A genes, and the endogenous Ptx1 protein binds specifically to the DNRE. Upon virus induction, Ptx1 negatively regulates the transcription of DNRE-containing IFN-A promoters, and the C-terminal region, as well as the homeodomain of the Ptx1 protein, is required for this repression. After virus induction, the expression of the Ptx1 antisense RNA leads to a significant increase of endogenous IFN-A gene transcription and is able to modify the pattern of differential expression of individual IFN-A genes. These studies suggest that Ptx1 contributes to the differential transcriptional strength of the promoters of different IFN-A genes and that these genes may provide new targets for transcriptional regulation by a homeodomain transcription factor.
The multifunctional secreted interferon (IFN) proteins mediate antiviral defense, immune and cell growth regulation. After virus induction, type I IFN (IFN-A and IFN-B) genes are expressed in a large variety of human and murine cells. The IFN-B gene and the individual subtypes of IFN-A genes are transcribed at various levels depending on cell type or virus inducers, reflecting differences in the transcriptional activity of the corresponding gene promoter in a particular cell type (10, 18, 19). Virus-responsive element B (VRE-B) of the human IFN-B promoter mediates both induction and repression. The positive control of VRE-B depends on many activators, including NF-κB, ATF-2 (c-jun), IRF-3, IRF-7, and HMG I(Y). In association with the transcriptional coactivator CBP (p300), these factors play an essential role in the assembly of a higher-order transcription enhancer complex named the enhanceosome (12, 22, 36, 40, 42). The negative control of VRE-B is also the result of different repressors. IRF-2 is able to antagonize activators of the IRF family by competing for their binding (9), NRF represses the activation due to NF-κB (23), and PRDI-BF1 is a postinduction repressor of the gene (26).
The IFN-B and IFN-A genes are transcriptionally activated and repressed through either common or specific mechanisms. IRF-binding sites are conserved in both the VRE-A and VRE-B regions, but binding sites for the other factors regulating IFN-B gene expression were not found within VRE-A. Furthermore, even if IFN-A genes are structurally related, differences in the expression of the individual subtypes of the multigenic IFN-A gene family after virus induction are observed. Most attention has been paid to comparisons of some of the different murine IFN-A promoters (5, 7, 25). The murine IFN-A11 gene is poorly expressed upon Newcastle disease virus (NDV) induction, whereas the IFN-A4 gene is strongly inducible. The lack of transcriptional activity of the IFN-A11 promoter could be related in part to substitutions in IRF-binding sites which are present in VRE-A4 of the IFN-A4 promoter. Recent data suggest that IRF-3 and IRF-7 are also involved in the transcription of the murine IFN-A genes (3, 11, 20, 28, 42).
Whereas a large number of repressors binding to the IFN-B promoter elements have been identified, repression of the IFN-A genes is not well characterized. In addition to substitutions in the proximal VRE-A, the repression of the IFN-A11 gene after virus induction is due to the presence of a distal negative regulatory element (DNRE) of 20 bp, which is delimited upstream of VRE-A (17, 27). This element exerts an inhibitory effect on proximal VRE-A promoters after virus induction, whatever its orientation or position, and is therefore considered a silencer. On the other hand, the DNRE on its own has no effect on VRE-B promoter after virus induction or any constitutive repressive effect on heterologous promoters. Therefore, the functional feature of the particular silencer DNRE is that its silencing activity is strictly dependent on the presence of a functional VRE-A and that it does not function as a general negative regulator. Similar DNREs are present in some IFN-A promoters, and the presence or the absence of DNRE may contribute to the differential expression of the IFN-A genes after virus induction. Furthermore, a DNRE (4DNRE) is also present in the highly inducible IFN-A4 promoter but a central antisilencer region located between the silencer and the VRE-A4 overrides the silencer activity. Two DNRE-binding factors have been observed before virus induction of murine L929 cells and human HeLa S3 cells and are still maintained even following induction. One of these factors corresponds to the HMG I(Y) protein but does not modulate the binding to DNRE of a second, uncharacterized factor related to the silencer activity (17).
In this study we have used the yeast one-hybrid system to clone a cDNA encoding the human homologue of the murine bicoid-related pituitary homeobox 1 (Ptx1 or Pitx1) that specifically recognizes the DNRE. The DNRE and the Ptx1-binding element are able to repress to the same extent the virus-induced transcriptional level of VRE-A promoters. We have shown that the Ptx1 gene is constitutively transcribed in cell types that differentially express IFN-A genes after virus induction and that the Ptx1 protein specifically binds the DNRE. Upon virus induction, overexpression of Ptx1 negatively regulates the IFN-A promoters containing the DNRE, and the C-terminal region, as well as the homeodomain of the protein, is required for the trans repression. The central antisilencer region in the highly inducible IFN-A4 promoter overrides the repressive activity of Ptx1. Ptx1 antisense RNA experiments showed that endogenous IFN-A expression is quantitatively increased and the pattern of differential gene expression is qualitatively influenced. These data suggest that Ptx1 may exert a modulation on the differential transcriptional strength of the promoters of different IFN-A genes.
MATERIALS AND METHODS
Cloning with the use of the yeast one-hybrid system.
The Saccharomyces cerivisiae strains used in this study were all derived from strain YM954 (MATa ade2 his3 leu2 lys2 trp1 ura3 gal4 gal80). The double-stranded motifs containing three oriented copies of the wild-type DNRE element from the IFN-A11 promoter, i.e., (5′-cggATTTAAGTCTAATTTAAAGTcggATTTAAGTCTAATTTAAAGTcggATTTAAGTCTAATTTAAGTcgg-3′), and mutated-DNRE motifs DM1 (5′-cggATTTAAGTAGGATTTAAAGTcggATTTAAGTAGGATTTAAAGTcggATTTAAGTAGGATTTAAGTcgg-3′), DM2 (5′-cggATTTACAGCTACTGTAAAGTcggATTTACAGCTACTGTAAAGTcggATTTACAGCTACTGTAAGTcgg-3′), or DM3 (5′-cggATTTACAGCTAATTTAAAGTcggATTTACAGCTAATTTAAAGTcggATTTACAGCTAATTTAAGTcgg-3′) were made by PCR using single oligonucleotides from Eurogentec as templates and different oligonucleotide primers containing a XhoI site (unrelated nucleotides are written in lowercase, and nucleotide mutations are underlined). The resulting double-stranded oligonucleotides were cloned into the pYi2267OHIS plasmid containing the URA3 marker and bearing the CYC1-HIS3 gene fusion (4), 200 bp upstream of the CYC1 TATA box owing to the unique XhoI site. All constructions were checked by nucleotide sequencing on the double-stranded DNA template. The resulting plasmids were linearized with Stu1 (within URA3) and used to transform the YM954 strain. Stable prototroph transformants were selected, and correct integration events were checked by Southern blot analysis.
About 109 cells of strain YMDNRE (URA3::DNRE-HIS3::URA3) were transformed with 5 μg of the HeLa S3 cDNA-Gal4 fusion library (cloned into the pGAD plasmid [Clontech]) as described previously (8). Cells capable of growing in the absence of both histidine (reporter gene) and leucine (selection marker of the library vector) and in the presence of 20 mM aminotriazole were directly selected. From a screen of about 5 × 106 transformants, 46 colonies resistant to aminotriazole appeared over the course of 8 days. The corresponding plasmids were recovered and used to retransform the YMDNRE strain as well as to transform strains harboring, integrated in the genome, a HIS3 gene placed downstream of either the DM1, DM2, or DM3 mutant of the DNRE motif. This allowed us to isolate one plasmid expressing a Gal4 fusion protein which activated the transcription of the HIS3 gene placed downstream of the DNRE, the DM1 mutant, or the DM3 mutant but not the DM2 mutant. This plasmid was used for further analysis.
Plasmid constructions.
Deletions and mutations in the native promoters of IFN-A11 and IFN-A4 were made by the PCR method using plasmids already described or previous PCR products as templates (17). All constructions were checked by nucleotide sequencing on double-stranded DNA template. Different oligonucleotide primers from Eurogentec were used (data not shown). The oligonucleotide containing three copies of the DNRE element was subcloned into the PstI site of -119A4wt. The human PTX1 and the murine Ptx1 cDNAs containing their complete open reading frame or cDNAs containing deleted or mutated forms of Ptx1 subcloned in expression plasmids were used. The PTX1 and Ptx1 cDNAs were subcloned into the pRc-CMV2 vector (Invitrogen) which contains a T7 promoter sequence to allow the transcription and translation of the cDNA inserts.
RT-PCR.
Fresh peripheral blood leukocytes (PBL) from healthy donors were obtained by Ficoll separation. Monocytes (more than 95% monocytes) were separated from lymphocytes (more than 75% of lymphocytes) by an adherence procedure. The human cell lines HeLa S3, HL60, U937, and KG1 or the murine cell lines L929, L929 Ptx1 knockdown cell lines, and AtT-20 were used. To study Ptx1, RT-PCR primers were designed to detect both murine and human PTX1 or Ptx1 mRNA. Total RNA (5 μg) was subjected to reverse transcriptase PCR (RT-PCR) by standard methods using, successively, a 24-mer sense oligonucleotide (5′-AAGAAGAAGAAACAGCGGCGGCAA-3′) and a 24-mer antisense oligonucleotide (5′-GTACACGTCCTCGTAGGGCTGCAC-3′) located in the second and third exons of the murine Ptx1 gene, respectively (the nucleotide substitution between murine Ptx1 and human PTX1 is underlined). These primers were also designed to exclude amplification of other members of the Ptx family. For the IFN-A study, consensus conserved primers annealing with all IFN-A subtypes were used as previously described (20), i.e., a 23-mer sense oligonucleotide (5′-ATGGCTAGGCTCTGTGCTTTCCT-3′) and a 24-mer antisense oligonucleotide (5′-AGGGCTCTCCAGACTTCTGCTCTG-3′). For the IFN-B study, specific primers for IFN-B mRNA were used, i.e., a 19-mer sense oligonucleotide (5′-TTCCTGCTGTGCTTCTCCA-3′) and a 20-mer antisense oligonucleotide (5′-GTTTTGGAAGTTTCTGGTAA-3′). The level of IFN mRNA was quantified by using serial dilution RT-PCR as previously described (20), and PCRs were quantified by PhosphorImager analysis. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) primers were used as positive controls, i.e., a 33-mer sense oligonucleotide (5′-TGAAGGTCGGAGTCAACGGATTTGGTCGTATTG-3′) and a 29-mer antisense oligonucleotide (5′-ATGTGGGCCATGAGGTCCACCACCCTGTT-3′). After the reverse transcription step using the antisense oligonucleotide, 30 cycles of amplification were performed. The amplification products of Ptx1 and PTX1 (272 bp) were analyzed by Southern blot hybridization using a murine 26-mer internal antisense oligonucleotide (5′-CCCTTGCACAGGTCCAACTGCTGGTT-3′) probe (the nucleotide substitution between murine Ptx1 and human PTX1 is underlined). The amplification products of Ptx1, PTX1, and IFN-A genes were also analyzed by DNA sequencing of the purified RT-PCR fragment cloned into plasmid vectors.
In vitro transcription and translation.
The proteins were translated with the TNT coupled transcription and translation kit from Promega as specified by the manufacturer. Plasmids pRc-CMV-PTX1 and pRc-CMV-Ptx1 were used for translation of human PTX1 and murine Ptx1 proteins, respectively.
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed as described previously (17). In vitro-translated reticulocyte lysates were preincubated with 1 μg of poly(dG-dC)-poly(dG-dC) in the presence of 100 ng of sonicated salmon sperm DNA for 10 min on ice. The mixture was then added to the binding buffer containing 10 fmol of 32P-end-labeled probe (50,000 cpm, 0.1 ng) either with or without competitor oligonucleotides in a final volume of 20 μl, and the incubation was carried out for a further 30 min at room temperature. Complexes were resolved by electrophoresis on prerun 5% native polyacrylamide gels. After being dried, the gels were autoradiographed overnight. The following chemically synthesized double-stranded oligonucleotides were used as probes: DNRE, 5′-cagATTTAAGTCTAATTTAAAGTcgt-3′; 4DNRE, 5′-cagATTTAAGTGTAATTTAAAGAcgt-3′; DM1, 5′-cagATTTAAGTAGGATTTAAAGTcgt-3′; DM2, 5′-cagATTTACAGCTACTGTAAAGTcgt-3′, and DM3, 5′-cagATTTACAGCTAATTTAAAGTcgt-3′ (unrelated nucleotides are written in lowercase and nucleotide mutations are underlined); CE3, 5′-ACCAGGATGCTAAGCCTCTGTC-3′; CE3M, 5′-ACCAGGATGCGTCGCCTCTGTC-3′; and CE3M2, 5′-ACCAGGATGCTAATAATCTGTC-3′ (nucleotide mutations are underlined); and for the related Drosophila bicoid target site (Db), 5′-ACCAGGATGCTAATCCTCTGTC-3′.
Ptx1 EMSA using nuclear extracts were performed with 1 μg of poly(dI-dC)-poly(dI-dC) as described previously (13). YY1 EMSA was performed as described previously (31). Competitor and supershift experiments were performed as described previously (13).
DNA transfection, viral induction, and CAT and luciferase assays.
L929 and HeLa S3 cells were transfected as previously described (17) and by the standard calcium phosphate precipitation method. NDV induction was carried out 48 h later. The mock-induced cells were set up as above except that no NDV was added. Cells were harvested 24 h postinduction, and cytoplasmic extracts were prepared. The chloramphenicol acetyltransferase (CAT) assay was carried out as previously described (17). Luciferase activities were measured in cell lysates by using commercial reagents (Promega). Transfection efficiency was determined by the β-galactosidase activity assay with a chemiluminescence kit (Tropix). In each experiment, a given construction was transfected in duplicate and two different clones of each construction were tested.
Stably transfected cell lines.
To construct the corresponding stably transfected cell lines, the control pRc-CMV2 vector (Invitrogen) and the antisense Ptx1 vector (containing the full-length Ptx1 cDNA in reverse orientation) were used. The pRc-CMV2 vector contains the neomycin (G418) resistance gene. L929 cells (5 × 105 cells/100-mm dish), seeded in minimum essential medium supplemented with antibiotics, l-glutamine, nonessential amino acids, and 10% fetal calf serum, were transfected by the calcium phosphate precipitation method with 20 μg of plasmid. At 4 h after transfection, the cells were glycerol shocked with 10% glycerol for 1 min and washed three times with phosphate-buffered saline. The transfected cells were then selected for 3 weeks in a medium containing G418 (600 μg/ml; GIBCO). Clones were isolated, propagated, and tested for Ptx1 DNA-binding activity. Three Ptx1 antisense clones were pooled. Ptx1 antisense clones were also transfected as previously described.
RESULTS
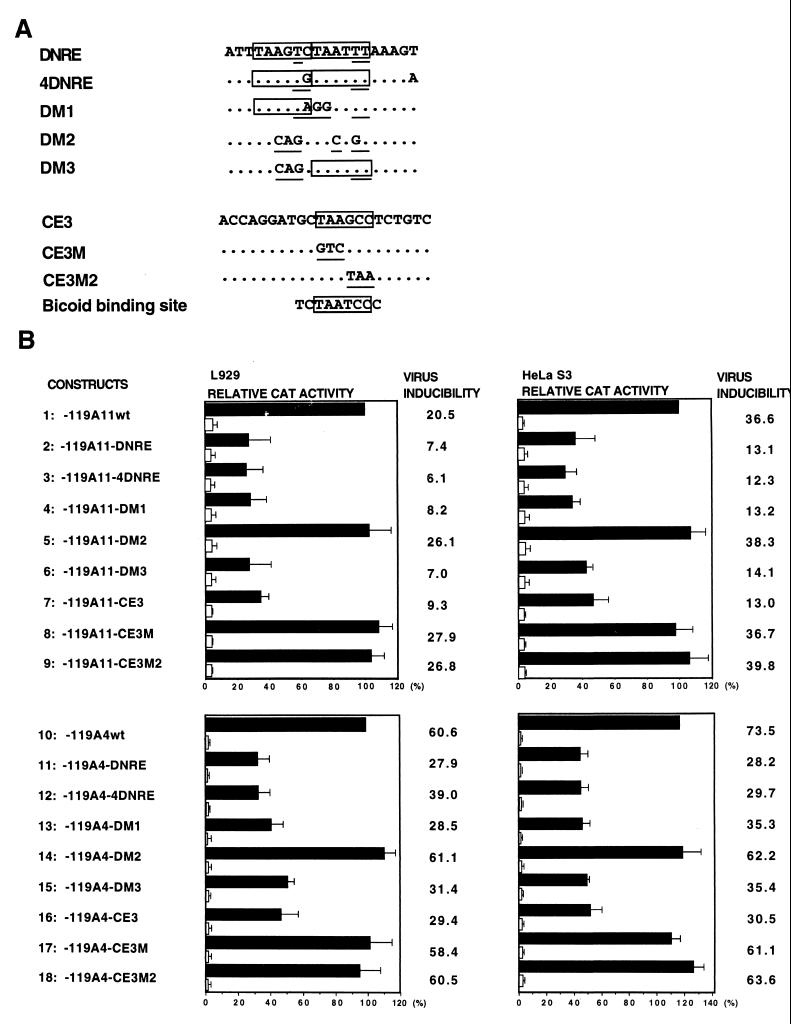
We have previously identified a silencer element (DNRE) within the murine IFN-A11 promoter which is responsible for the distal repression of IFN-A11 gene after NDV induction in both murine L929 and human HeLa S3 cell lines (17, 27). The similarity of the results obtained with the two cell lines suggests that the factor(s) involved in this repression may be present in both murine and human cell lines. Furthermore, the isolated DNRE of the IFN-A11 promoter or similar elements of DNRE found in other IFN-A promoters are able to reduce the inducibility of these different IFN-A promoters. These results suggest that DNRE may play a general role in the differential transcriptional strength of the IFN-A gene promoters. To confirm the functional role of DNRE, we previously introduced a series of mutations within a DNA-protein interaction site of the IFN-A11 promoter established by DNase I footprinting. For example, while DM1 and DM3 mutations were shown to maintain the negative effect of DNRE after virus induction, the DM2 mutant caused the loss of repression of the promoter (17) (see also Fig. 2).
FIG. 2.
Repression of the virus-induced transcriptional level of VRE-A11 and VRE-A4 by the Ptx1 element CE3. (A) Homologies between the core consensus recognition DNA sequence for the binding of Ptx or bicoid proteins and CE3 in the DNRE sequences. The nucleotide substitutions are underlined, and the homologies are boxed. (B) Effect of CE3 in the IFN-A11- and IFN-A4-proximal promoters. Plasmid constructs with the CAT gene under the control of the indicated promoter fragments were tested by measuring the CAT activity. L929 and HeLa S3 cells were transfected and mock induced (open bars) or NDV induced (solid bars) as described in Materials and Methods. Since the data are pooled from several experiments, they are presented in arbitrary units of CAT activity. CAT activity values represent CAT/β-galactosidase activity ratios relative to the induced activity -119A11wt or -119A4wt, which was arbitrarily set at 100%. CAT activities for each plasmid are the means and standard errors (SE) for at least five separate transfections with at least two separate plasmids. Error bars indicate SEs. Virus inducibility is the ratio of the NDV-induced activity over the mock-induced activity.
Cloning of the PTX1 gene by the yeast one-hybrid system.
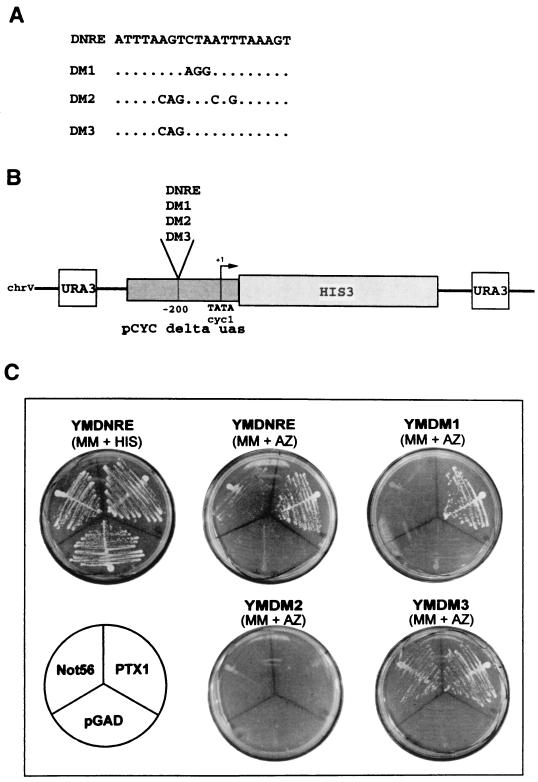
To identify the gene(s) encoding the factor(s) that could recognize DNRE, the multimerized DNRE element (three copies) was used in a yeast one-hybrid system (Fig. 1A and B), similar to that previously described (4). For the cloning of DNA-binding proteins, a cDNA-Gal4 fusion library from HeLa S3 RNA was used. The first screening yielded different families of positive cDNA-Gal4 fusion clones classified by sequence analysis. Different mutated DNRE were used in the second screening. The mutated plasmids were used to transform strains harboring, integrated in the genome, a HIS3 gene placed downstream of either the DM1, DM2, or DM3 mutant (three copies [Fig. 1A]) of the DNRE motif (YMDM1, YMDM2, and YMDM3). As shown in Fig. 1C, these transformations allowed us to isolate one plasmid expressing a Gal4 fusion protein which activated the transcription of the HIS3 gene placed downstream of the DNRE or the DM1 or DM3 mutants but not the DM2 mutant. The sequence of this 1-kb cDNA indicated a partial open reading frame of 678 bp. Sequence comparison with databases indicated that it is a human protein that contains a homeodomain (HD) which is identical to PTX1 (30). Another plasmid was isolated by this screening. This plasmid expressed the neighbour of tid (Not) 56 protein (GenBank accession no. Y09022), whose function remains unknown. This Gal4 fusion protein activated the transcription of the HIS3 gene placed downstream of the DNRE or the DM3 mutant but not the DM1 or DM2 mutants and was not further investigated.
FIG. 1.
Identification of the PTX1 gene by the one-hybrid system. (A) The DNRE and the different mutated DNRE sequences used after multimerization in the one-hybrid system. (B) Structure of the His3 reporter gene constructs used for the one-hybrid experiments. (C) Aminotriazole resistance of transformed YMDNRE, YMDM1, YMDM2, and YMDM3 cells. YMDNRE, YMDM1, YMDM2, and YMDM3 cells were transformed either by plasmids expressing the PTX1-Gal4 or Not56-Gal4 fusion proteins or by the parental vector pGAD. Transformants were streaked on histidine (HIS)-containing medium or a medium lacking histidine (MM) but containing 20 mM aminotriazole (AZ).
Homeotic genes encode transcriptional factors involved in the positive and negative regulation of target genes during development. These genes contain a highly conserved sequence of 180 bp, the homeobox, which encodes a 60-amino-acid polypeptide, the HD, which represents the DNA-binding domain of these factors. The partial cDNA cloned by the yeast one-hybrid system contains the complete HD of the protein between nucleotides 264 and 444. The amino acid sequence of the human transcription factor showed 96% homology to that of murine Ptx1 (13). Ptx1 was first described as a transcriptional activator binding the CE3 element of the pituitary pro-opiomelanocortin (POMC) gene promoter (13). It activates the transcription of other pituitary genes (37). Studies with Ptx1-deficient mice indicate that hindlimb patterning and mandible and pituitary development require this gene (16, 35). Three members of the Ptx family have been described: the Ptx1 (13) and the Ptx2 (29) genes, which are homologous and have an overlapping pattern of expression, and the Ptx3 gene (33). To date, HD transcription factors have not been shown to be involved in the regulation of the expression of the IFN genes.
The Ptx1-binding site of POMC, as well as DNRE, can repress virus-induced transcription of VRE-A promoters.
The pituitary Ptx1-binding sites are related to the bicoid-binding site, a positive regulator of the hunchback gene in Drosophila, and Ptx1 is a member of a subgroup (which also includes Otx and goosecoid) of paired-related factors which have a DNA-binding specificity similar to that of bicoid (32). Ptx1 binds to the core consensus recognition DNA sequence for bicoid-related proteins TAATCC (41) and to the Ptx-binding site of the POMC promoter, CE3, containing the TAAGCC sequence (13). After the cloning of PTX1, two sequences partially homologous to the core consensus recognition DNA sequence for the binding of Ptx and bicoid proteins were identified in the DNRE and the 4DNRE (Fig. 2A). Furthermore, the DM2 mutant, but neither DM1 nor DM3, altered the two sequences which in DNRE are homologous to the core consensus recognition DNA sequence for the binding of Ptx and bicoid proteins. The obvious functional experiments were then to test whether a Ptx-binding element like CE3 and DNRE can repress to the same extent the VRE of the IFN-A11 and IFN-A4 promoters after virus induction. DNRE, 4DNRE, DM1, DM2, DM3, CE3, and two mutated CE3 sequences, CE3M or CE3M2, affecting the core consensus recognition DNA sequence for Ptx were inserted directly upstream of the VRE of both IFN-A11 and IFN-A4 gene promoters. In the experiments in Fig. 2B, DNRE, 4DNRE, DM1, DM3, and CE3 conferred a similar repression of the virus-induced transcriptional level and similar inducibility to both promoters in L929 and HeLa S3 cells (solid bars). The IFN genes are not expressed before virus induction. Although CE3 leads to the activation of the POMC gene (13), it has no activating effect on the transcriptional activity of the proximal −119/+19 promoter of both the IFN-A11 and IFN-A4 genes in the absence of virus induction (Fig. 2B, lanes 7 and 16, open bars). After virus induction, the transcriptional activity of the promoters was released by deletions (lanes 1 and 10, solid bars) or by mutations that affected nucleotides present in the core consensus recognition DNA sequence for the binding of Ptx proteins of DNRE, e.g., DM2, and of CE3, e.g., CE3M or CE3M2 (lanes 5, 8, 9, 14, 17, and 18, solid bars). The results suggest that DNRE and CE3 are able to repress to the same extent the virus-induced transcriptional level of the two proximal IFN-A11 and IFN-A4 promoters in both murine and human cell lines. The negative effect of the DNRE element could be due to the binding of Ptx or related proteins.
In vitro-translated Ptx1 binds specifically to the DNRE elements.
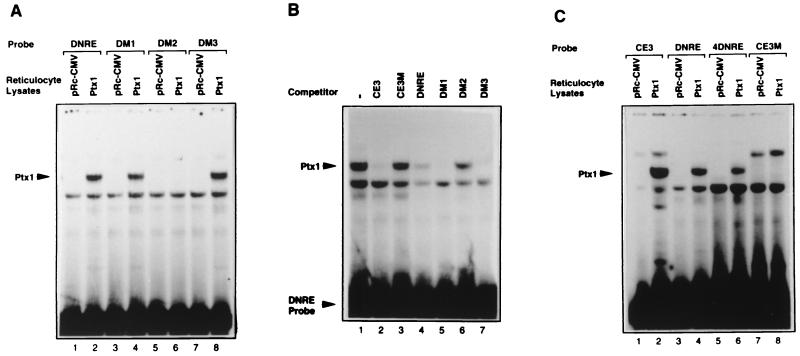
The results of the yeast one-hybrid system indicated that the partial fusion PTX1 protein could bind in vivo to DNRE and to the DM1 and DM3 mutants but not to the DM2 mutant. EMSA was used to test the DNA-binding properties of the intact protein in vitro. PTX1 and Ptx1 were transcribed and translated in rabbit reticulocyte lysates, and expression was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (data not shown). The lysates were used for EMSA. As shown in Fig. 3A, specific binding to DNRE appeared when Ptx1 was used in EMSA. Ptx1 protein binds to the wild-type DNRE and to the DM1 and DM3 mutants used as probes but not to the DM2 mutant probe. With DNRE as a probe (Fig. 3B), Ptx1 binding was competed by CE3, DNRE, DM1, and DM3 used as cold competitors, but competitors such as CE3M and DM2 had a poor effect. Human PTX1 protein gave similar results (data not shown). In conclusion, Ptx1 binds specifically to DNRE both in vivo and in vitro and its binding properties are consistent with the activities of the DNRE mutants. The same specific binding of Ptx1 was detected using the 4DNRE of the IFN-A4 promoter (Fig. 3C and data not shown), suggesting that different DNREs may be the bicoid-binding site for this factor. Essentially identical results were obtained for the CE3 sequence as a probe but with a higher affinity than for the DNRE. CE3M had no effect. These results, showing that the Ptx1 protein binds specifically to the DNRE and CE3, are in agreement with the ability of these elements to repress the virus-induced transcriptional level of IFN-A promoters. Then the pattern of PTX1 or Ptx1 gene expression was assessed in cell types that express IFN-A genes.
FIG. 3.
Specific binding of Ptx1 to DNRE probes. (A) Binding of Ptx1 to wild-type and point-mutated DNRE probes. Reticulocyte lysates containing the in vitro-transcribed and -translated protein from pRc-CMV (lanes 1, 3, 5, and 7) and pRc-CMV-Ptx1 (lanes 2, 4, 6, and 8) were used for EMSA in the presence of wild-type DNRE, DM1, DM2, or DM3 probes. (B) Specific binding of Ptx1 to the wild-type DNRE probe. Reticulocyte lysate containing Ptx1 was incubated with the wild-type DNRE probe and a 100-fold molar excess of unlabeled CE3, CE3M, DNRE, DM1, DM2, or DM3. (C) Binding of Ptx1 to the 4DNRE probe. Reticulocyte lysate containing Ptx1 (lanes 2, 4, 6, and 8) or not (lanes 1, 3, 5, and 7) was used for EMSA in the presence of the CE3, DNRE, 4DNRE, and CE3M probes.
PTX1 or Ptx1 genes are constitutively transcribed in cell types that are able to differentially express IFN-A genes.
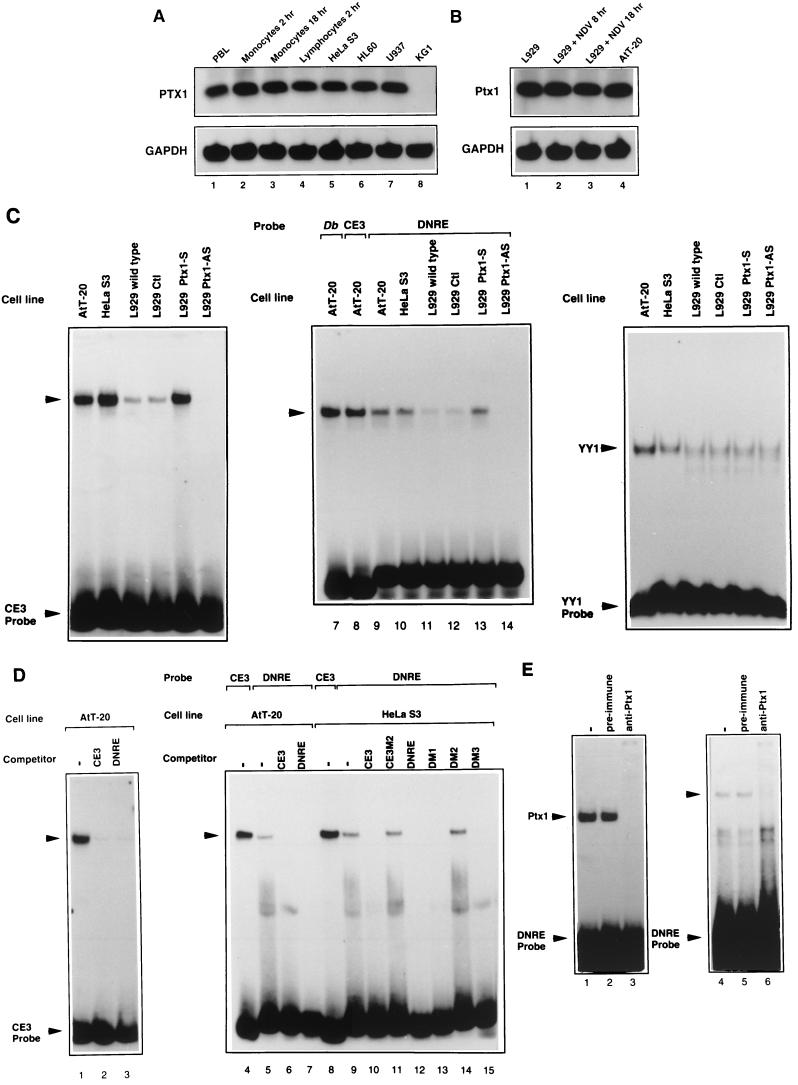
Ptx1 is expressed in adult anterior pituitary cells and during pituitary development (13, 15). However, the expression of the murine Ptx1 or the human PTX1 genes is not restricted to the pituitary cells; these genes are also expressed during embryogenesis and in adult tissues in derivatives of posterior lateral plate mesoderm (13, 14, 30). To date, transcription of murine Ptx1 and human PTX1 genes has not been described in cell types that are shown to differentially express IFN-A genes after virus induction, such as human PBL (10). In this study, the total RNAs of different cell types were isolated for detection of PTX1 or Ptx1 mRNA by RT-PCR. RT-PCR primers (exon 2 and exon 3 primers) were designed to detect both murine Ptx1 and human PTX1 mRNA. These primers were also designed to exclude amplification of other members of the Ptx family. RT-PCR products were cloned and analyzed by DNA sequencing. RT-PCR analysis of RNA from uninduced human PBL, monocytes, and lymphocytes and subsequent sequence analysis of RT-PCR products (data not shown) showed that PTX1 mRNAs are constitutively expressed in these cells (Fig. 4A, lanes 1 to 4). PTX1 transcripts were detected in the starting population of PBL in three experiments performed with cells from different donors (data not shown). PTX1 gene expression could also be detected by RT-PCR in different cell lines such as epithelial HeLa S3 cells, from which we have cloned PTX1 in this study, promyelocytic HL60 cells, and monoblastic U937 cells, but not in myeloblastic KG1 cells (lanes 5 to 8). No amplification was observed in the absence of the reverse transcription steps (data not shown). Our previous results suggest that the same type of murine IFN-A promoter regulation is observed in the murine fibroepithelial L929 and human HeLa S3 cell lines after virus induction. The murine corticotroph AtT-20 cells producing POMC and expressing the Ptx1 gene (13) were used as a positive control for RT-PCR analysis. RT-PCR of uninduced AtT-20 and L929 cells (Fig. 4B, lanes 1 and 4) and sequence analysis (data not shown) showed that Ptx1 mRNAs are constitutively present in both cell lines before virus induction. The presence of Ptx1 mRNA was still maintained in L929 cells after virus induction (lanes 2 and 3). No amplification was observed when the reverse transcription step was omitted (data not shown). In L929 and AtT-20 cell lines, a similar IFN activity was detected after virus induction (data not shown). Thus, PTX1 and Ptx1 genes are constitutively expressed in cell types that are able to differentially express IFN-A genes after virus induction.
FIG. 4.
Human PTX1 and murine Ptx1 expression in cell types expressing IFN-A genes. Expression of PTX1 and Ptx1 genes in different cell types and cell lines was monitored by RT-PCR using primers designed to exclude amplification of other members of the Ptx family (see Materials and Methods). RT-PCR products were also cloned and analyzed by DNA sequencing (data not shown). (A) PTX1 gene expression. Total RNA was extracted from different human cell types as well as cell lines and was monitored by RT-PCR. As a control, expression of GAPDH mRNA is shown in the lower panel. (B) Ptx1 gene expression. Total RNA were extracted from NDV-induced L929 cells at 0, 8, and 18 h postinduction. AtT-20 cell line was used as a control for the expression of the Ptx1 gene. Expression of Ptx1 and GAPDH mRNA was monitored by RT-PCR. (C) Endogenous Ptx1-binding activity to DNRE in different cell lines. Nuclear extracts from different cell lines were used for EMSA in the presence of related Drosophila bicoid target site (Db), CE3 and DNRE probes (lanes 1 to 14). The AtT-20 cell line expressing the Ptx1 protein was used as a control. The binding activity was monitored using nuclear extracts from HeLa S3 cells, L929 wild-type cells, a L929 control clone (L929 Ctl), L929 cells transiently transfected with the Ptx1 sense expression vector (L929 Ptx1-S), and L929 clones stably transfected with the Ptx1 antisense RNA expression vector (L929 Ptx1-AS). The quality of nuclear extracts was tested by EMSA for YY1-binding activity (lanes 15 to 20). (D) Specific binding of Ptx1 to the wild-type DNRE probe. Nuclear extracts from AtT-20 and HeLa S3 cell lines were incubated with CE3 or DNRE probes and a 50-fold molar excess of unlabeled CE3, CE3M2, DNRE, DM1, DM2, and DM3. (E) The DNRE binding of recombinant Ptx1 (lanes 1 to 3) and nuclear protein of L929 cells (lanes 4 to 6) was supershifted by the addition of an antiserum against maltose-binding protein-Ptx1 (anti-Ptx1) but not by the addition of preimmune serum.
Endogenous Ptx1 protein binds specifically to the DNRE.
EMSA was used to test the DNA-binding properties of nuclear extracts from different cell lines. The AtT-20 cells expressing the Ptx1 protein were used as positive control under conditions previously described (13). As shown in Fig. 4C (lanes 1 to 14), a single band was observed with the related Drosophila bicoid target site (Db), CE3, and DNRE probes. This binding activity is present in AtT-20, HeLa S3, and, to a lesser extent, L929 cells. The binding was increased using extracts from L929 cells transiently transfected with Ptx1 expression vector (L929 Ptx1-S). We also generated Ptx1 knockdown cell lines by stably transfecting a Ptx1 antisense RNA expression vector in L929 cells (L929 Ptx1-AS). Three independent neomycin-resistant clones expressing Ptx1 antisense RNA were analyzed. A clone stably transfected with the same vector without the Ptx1 cDNA was chosen as a control (L929 Ctl). In the pool of three Ptx1 antisense clones, CE3- and DNRE-binding activities were almost undetectable, whereas another transcription factor such as YY1 was not significantly affected (lanes 15 to 20). The specificity of the DNA-binding activities of nuclear extracts from AtT-20 and HeLa S3 cell lines was also observed. Using CE3 as a probe (Fig. 4D, lanes 1 to 3), the binding activity present in AtT-20 cells was competed by CE3 and DNRE used as cold competitors. Using CE3 and DNRE as probes with nuclear extracts from AtT-20 and HeLa S3 cell lines (lanes 4 to 15), this binding was competed by CE3, DNRE, DM1, and DM3 used as cold competitors, but competitors such as inactive mutants CE3M2 and DM2 had no effect. The same specific binding was detected using the 4DNRE of the IFN-A4 promoter as probe (data not shown). The Ptx1 DNA-binding activity present in L929 nuclear extracts is supershifted in EMSA by addition of an antiserum against maltose-binding protein-Ptx1 (Fig. 4E). Taken together, these results suggest that the endogenous nuclear protein binding to CE3 and DNRE is immunologically related to or identical to the Ptx1 protein. This binding correlates with the ability of these elements to repress the virus-induced transcription of the IFN-A promoters.
Ptx1 inhibits the virus-induced transcriptional activity of the IFN-A promoters through DNRE.
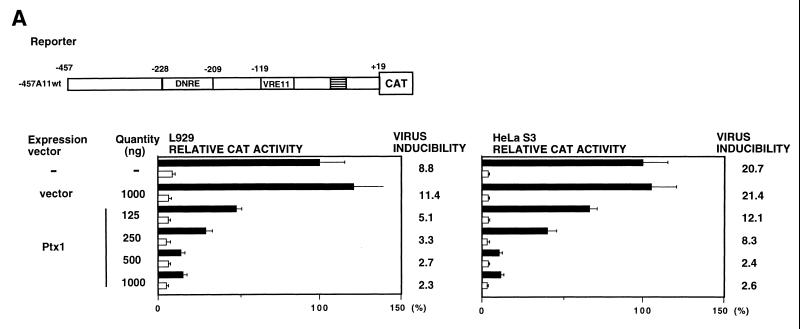
To ascertain the functional role of Ptx1 in the regulation of the IFN-A promoters, Ptx1-cDNAs were inserted into expression vectors and transfected along with different reporter constructs in HeLa S3 and L929 cells (Fig. 5). Whereas it has been previously demonstrated that Ptx1 overexpression led to activation of the POMC gene through the CE3 element (13), cotransfection of Ptx1-expression plasmids with the native promoter of IFN-A11 containing DNRE had no effect on the transcriptional activity of the promoter in the absence of virus induction (Fig. 5A, open bars). In contrast, after virus induction of both murine L929 and human HeLa S3 cell lines, Ptx1 overexpression led to a significant decrease (more than 80%) in the transcriptional activity of this promoter (solid bars). Titration of the Ptx1 plasmid suggested that the level of native IFN-A11 promoter repression after virus induction was maximal. Similar results were observed when using human PTX1 overexpression (data not shown).
FIG. 5.
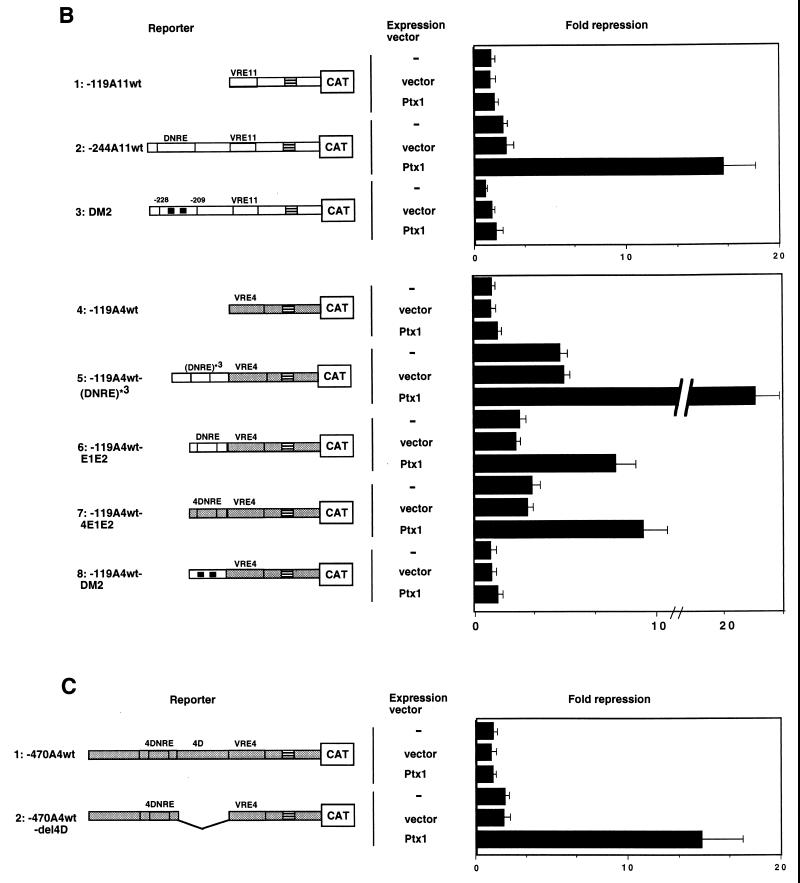
Repression of IFN-A transcription by Ptx1. (A) Repression of IFN-A11 promoter activity by Ptx1 after virus induction. L929 and HeLa S3 cells were cotransfected with the IFN-A11/CAT reporter construct and with empty expression vector or vector encoding Ptx1, as indicated. CAT expression in cotransfected cells was determined relative to the induced activity of -457A11wt alone, which was set at 100%. Assay conditions were as described in the legend to Fig. 2B. (B) Effect of Ptx1 on DNRE and deleted or mutated DNRE of the IFN-A11 and IFN-A4 promoters. HeLa S3 cells were cotransfected with various constructs and with empty expression vector or vector encoding Ptx1. The shaded and open boxes correspond to the IFN-A4 and IFN-A11 promoters, respectively. Assay conditions were as described in the legend to Fig. 2B. CAT activity after virus induction is reported as fold repression relative to -119A11wt for the IFN-A11 constructs or to -119A4wt for the IFN-A4 constructs, which were set at 1, respectively. (C) Effect of Ptx1 on the deleted antisilencer region 4D of the native IFN-A4 promoter. Cotransfections were performed as described in the legend to Fig. 5B. Assay conditions were as described in the legend to Fig. 2B. CAT activity after virus induction is reported as fold repression relative to -470A4wt, which was set at 1.
After virus induction, Ptx1 overexpression led to a significant (16-fold) repression of constructs containing the DNRE upstream of the IFN-A11 proximal promoter (Fig. 5B, lane 2) and Ptx1 repressed neither the promoter lacking the DNRE nor the promoter containing the mutated DNRE, DM2 (lanes 1 and 3). A repression was also observed with constructs containing three copies of the DNRE, the 4DNRE, or the DNRE (20-, 8-, and 10-fold repression, respectively) upstream of the proximal promoter of the IFN-A4 gene, which is strongly inducible (lanes 5 to 7). No effect was observed in the absence of the DNRE or the 4DNRE or in the presence of DM2 (lanes 4 and 8). Similar results were observed using human PTX1 overexpression (data not shown). In addition, viral Rous sarcoma virus, cytomegalovirus, and thymidine kinase promoters, as well as the elongation factor promoter, were poorly or not sensitive to Ptx1 overexpression (data not shown). Thus, these results suggest that after virus induction, the Ptx1 protein specifically represses the transcription of the IFN-A11 and IFN-A4 proximal promoters in the presence of DNRE. The fact that the intact IFN-A4 gene promoter remains highly inducible upon virus induction has been previously explained by the presence of a third element in the IFN-A4 promoter. This element is a central region located between the distal 4DNRE and the proximal VRE-A of the IFN-A4 promoter and is able to overcome the DNRE silencer activity. Therefore, this element, named 4D, has been considered an antisilencer (17). Our results show that the repressing activity of Ptx1 was abolished in the context of the native IFN-A4 promoter (Fig. 5C). In contrast, in the absence of the central antisilencer region 4D, the repressing effect of Ptx1 was observed. Thus, Ptx1 inhibits the virus-induced transcriptional activity of the IFN-A11 promoter through DNRE but not the IFN-A4 promoter containing both the distal 4DNRE and the central antisilencer region 4D. Thus, DNRE and Ptx1 appear to function as a context-dependent repressors.
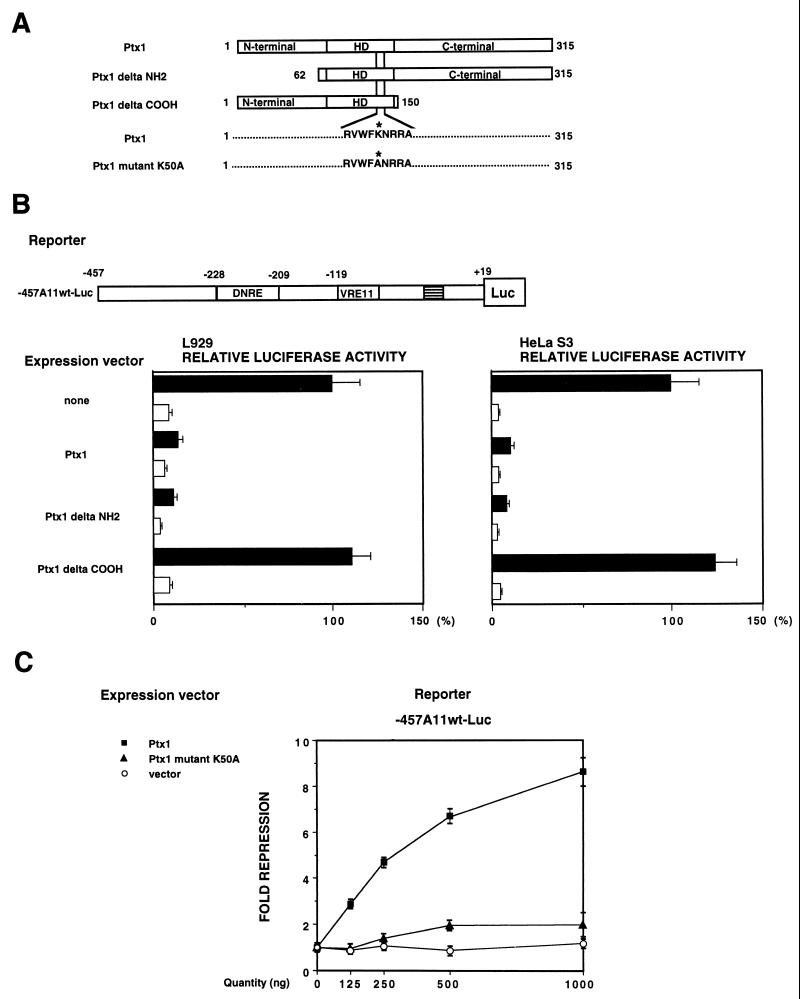
The C-terminal region and the HD of Ptx1 are required for trans repression.
To characterize the transcriptional repressive domain(s) of Ptx1, we used truncated forms of Ptx1 (38, 39) lacking either the N-terminal or the C-terminal regions but containing the HD (Fig. 6A). The bicoid-related HD is characterized by a lysine residue at position 50 of the HD. This lysine residue determines the DNA-binding specificity, and its mutagenesis (Fig. 6A) abrogates DNA binding (32, 41). The expression level and nuclear localization of Ptx1 mutant proteins have been assessed previously (38, 39). The N-terminally truncated form of Ptx1 had the same effect as the full-length Ptx1, but the C-terminally truncated and HD mutated forms of Ptx1 were unable to repress the IFN-A11 promoter after virus induction (Fig. 6B and C). These results suggest that the C-terminal region, as well as the HD of Ptx1, is required for the trans repression.
FIG. 6.
trans repression of IFN-A11 promoter activity by expression constructs of Ptx1. (A) Ptx1 expression constructs. The HD of Ptx1 is shown. The N-terminal truncation in mutant Ptx1 delta NH2, the C-terminal in mutant Ptx1 delta COOH, and the lysine residue changed to alanine in Ptx1 mutant K50A are shown. (B) L929 and HeLa S3 cells were cotransfected with the IFN-A11/luciferase reporter construct (-457A11wt-Luc) and with empty expression vector or vector encoding Ptx1, Ptx1 delta NH2, or Ptx1 delta COOH, as indicated. Luciferase expression in cotransfected cells is expressed relative to the induced activity of -457A11wt-Luc alone, which was set at 100%. Assay conditions were as described in the legend to Fig. 2B, except that the luciferase activity was determined. (C) L929 cells were cotransfected with the IFN-A11/luciferase reporter construct (-457A11wt-Luc) and with empty expression vector, vector encoding Ptx1, or vector encoding Ptx1 mutant K50A, as indicated. Assay conditions were as described in the legend to Fig. 2B. Luciferase activity after virus induction is reported as fold repression relative to the -457A11wt-Luc construct, which was set at 1.
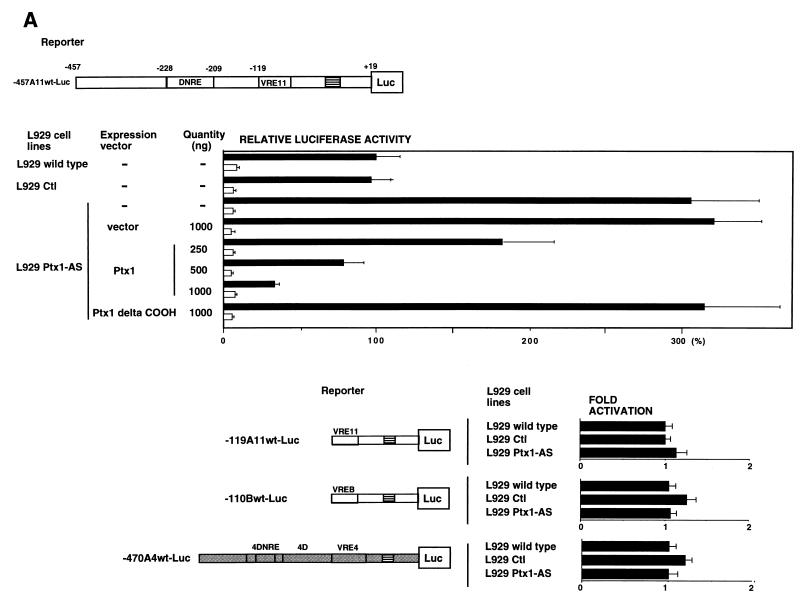
Endogenous Ptx1 participates in the repression of virus-induced IFN-A gene expression.
The importance of endogenous Ptx1 in IFN-A promoter activity was tested by stably transfecting a Ptx1 antisense RNA expression vector in L929 cells. Ptx1 antisense expression led to a significant decrease in Ptx1 DNA-binding activity without affecting other DNA-binding proteins (Fig. 4C). To test the contribution of Ptx1 to IFN-A11 repression, the native IFN-A11 promoter containing the DNRE was transfected into Ptx1 antisense clones (Fig. 7A, upper panel). The use of Ptx1 knockdown cell lines led to a significant increase in IFN-A11 promoter activity after virus induction, whereas no effect was observed in the absence of virus induction.When the Ptx1 sense expression vector was cotransfected into Ptx1 knockdown cell lines together with native IFN-A11 promoter, the enhanced activity was abolished. Cotransfection of the C-terminally truncated form of Ptx1 has no effect. Deletion of the DNRE-binding site abolished the effect of the Ptx1 antisense RNA (Fig. 7A, lower panel). Furthermore, when the IFN-B promoter or the native IFN-A4 promoter containing the antisilencer region 4D was used, the effect of Ptx1 antisense RNA was not observed.
FIG. 7.
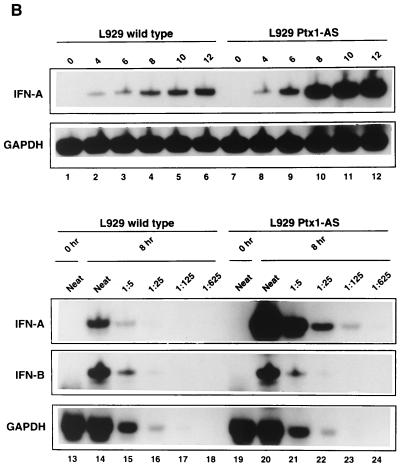
Endogenous Ptx1 participates in repression of virus-induced IFN-A gene expression. (A) Endogenous Ptx1 factor is essential for promoter-specific repression. L929 wild-type cells or L929 control clone (L929 Ctl) or L929 clones stably transfected with Ptx1 antisense RNA expression vector (L929 Ptx1-AS) were cotransfected with the IFN-A11/luciferase reporter construct (-457A11wt-Luc) and with empty expression vector or vector encoding Ptx1 sense (Ptx1-S) or Ptx1 delta COOH, as indicated. Luciferase expression in cotransfected L929 wild-type cells is expressed relative to the induced activity of -457A11wt-Luc alone, which was set at 100%. Assay conditions were as described in the legend to Fig. 6B. L929 or L929 Ptx1-AS cells were also transfected with various constructs corresponding to the IFN-A11, IFN-A4, and IFN-B promoters. Assay conditions were as described in the legend to Fig. 6B. Luciferase activity after virus induction is reported as fold repression relative to the L929 wild-type, which was set at 1. (B) Ptx1 antisense RNA experiments lead to an increase of endogenous IFN-A expression. Expression and quantification of IFN-A and IFN-B genes after virus induction in L929 and L929 Ptx1-AS cell lines (0, 4, 6, 8, 10, and 12 h postinduction) were monitored by using RT-PCR consensus conserved primers for IFN-A mRNA or specific primers for IFN-B mRNA. The level of IFN mRNA was quantified (8 h postinduction) by using serial dilution RT-PCR. As a control, expression of GAPDH mRNA is shown in the lower panels.
To further characterize the role of Ptx1 in IFN-A gene repression after virus induction, we quantitatively and qualitatively compared the endogenous IFN-A expression in L929 wild-type and L929 Ptx1 knockdown cells. In these cell lines, although no biologically active IFN protein was observed in culture media before virus induction, a different IFN activity was detected after virus induction. Indeed, at 18 h postinduction, approximately 6,400 and 102,400 IU/ml was titrated for IFN activity of L929 wild-type and L929 Ptx1 knockdown cells, respectively. The difference in IFN protein production between Ptx1 knockdown cells and wild-type cells was 16-fold. Total RNAs of both cell types were isolated before and after virus induction of IFN mRNA for quantification by RT-PCR using consensus conserved primers for IFN-A mRNA or specific primers for IFN-B mRNA. First, L929 wild-type cells expressed much lower levels of IFN-A mRNA in response to virus induction than did L929 Ptx1-AS cells (Fig. 7B, lanes 1 to 12). In both cell lines, IFN-A mRNA were undetectable in the absence of virus induction (lanes 1 and 7). At 8 h postinduction, the level of IFN mRNA was quantified by using serial dilution RT-PCR as previously described (20). IFN-B mRNA and IFN-A mRNA were both undetectable in the absence of virus induction (lanes 13 and 19). Although IFN-B gene expression level was identical in both cell lines after virus induction, IFN-A gene expression was significantly increased in L929 Ptx1 knockdown cells (lanes 14 to 18 and 20 to 24). Indeed, at 8 h postinduction, IFN-A mRNA from wild-type cells was poorly detected following fivefold dilution of cDNA (lane 15). In contrast, IFN-A mRNA from Ptx1 knockdown cells was clearly detected following 25-fold dilution (lane 22). RT-PCR products were quantified by PhosphorImager analysis, and the difference in IFN-A mRNA induction between Ptx1 knockdown cells and wild-type cells was found to be 22.8-fold. These results are in agreement with our previous results of biologically active IFN protein production. Thus, induction of IFN-A gene expression, but not of IFN-B gene expression, was significantly increased after virus induction in Ptx1 knockdown cells.
To distinguish the subtypes of IFN-A gene expression from virus-induced wild-type and Ptx1 knockdown cells, cDNA were cloned and 83 randomly selected clones were analyzed by DNA sequencing. As expected, IFN-A gene expression by wild-type cells displayed a mixture of distinct subtypes (Table 1). IFN-A4 was the most abundant species detected. No cDNA clones for IFN-A11 was detected. In contrast, the IFN-A11 subtype was detected in virus-induced Ptx1 knockdown cells. Strikingly, IFN-A5 but not IFN-A4 was the most abundant species detected, suggesting that, as with the IFN-A11 gene, Ptx1 is essential for IFN-A5 repression. These results suggest that not only is endogenous IFN-A expression quantitatively affected by Ptx1 but also Ptx1 qualitatively influences the pattern of differential IFN-A gene expression.
TABLE 1.
Representation of IFN-A subtypes in virus-induced L929 wild-type and L929 Ptx1 knockdown cells
| IFN-A subtype | No. of clones with subtype/total no. in:
|
|
|---|---|---|
| L929 cells | L929 Ptx1-AS cells | |
| A2 | 4/39 | 3/44 |
| A4 | 30/39 | 8/44 |
| A5 | 3/39 | 28/44 |
| A6 | 2/39 | 2/44 |
| A11 | 0/39 | 3/44 |
DISCUSSION
The analysis of the distal silencer element DNRE, responsible for the virus-induced transcriptional repression of some IFN-A promoters, led us to clone and study the HD transcription factor, Ptx1. We show here that the promoters of IFN-A genes constitute targets for Ptx1 and that Ptx1 plays a role in the differential repression of these genes.
Ptx1, a repressor of virus-induced IFN-A gene expression.
The mechanism by which Ptx1 represses virus-induced IFN-A gene expression seems to be unrelated to other mechanisms previously described for different negative factors in the IFN-A and IFN-B gene promoters. The first factor found to repress IFN-A and IFN-B gene transcription was IRF-2 (9). IRF2 binds the VRE-B of the IFN-B gene and also recognizes the IRF-binding sites present in the VRE-A of the IFN-A genes (1, 2, 9). The role of IRF-2 as a negative regulatory factor in IFN-A and IFN-B gene expression was confirmed by the use of IRF-2-deficient mice (21). IRF-2 is described as a repressor of transcription because of its ability to antagonize activators by competing for the IRF-binding sites. In contrast to IRF-2, Ptx1 is not involved in competition for overlapping activator(s) DNRE-binding site. Furthermore, IRF-2 is not involved in the differential regulation of IFN-A genes whereas Ptx1 is implicated in this type of regulation.
Another repressor factor binding the VRE-B, PRDI-BF1, has been isolated. PRDI-BF1 is a virus-inducible gene and has been considered a postinduction repressor of the IFN-B gene (26). In contrast to PRDI-BF1, Ptx1 is present before and after virus induction.
On the other hand, NRF binds the VRE-B, and expression of the NRF antisense RNA releases the constitutive endogenous IFN-B gene transcription (23). Thus, NRF is a critical component of IFN-B gene silencing prior to viral induction. In the present study, Ptx1 antisense RNA experiments show that IFN-A mRNA was undetectable in the absence of virus induction whereas IFN-A gene expression was significantly increased in L929 Ptx1 knockdown cells after virus induction. In contrast to NRF, Ptx1 is not involved in the constitutive silencing of IFN-A promoters. Thus, our data suggest a novel mechanism by which Ptx1 represses virus-induced IFN gene expression.
With regard to previously reported results concerning the overexpression of Ptx1 which led to activation of the POMC and other pituitary genes (13, 37), the effects of Ptx1 vary with promoter context, and this is the first demonstration that Ptx1 can repress gene transcription. Ptx1 now joins the class of transcription factors with dual activator-repressor functions. HD transcription factors function by positively or negatively regulating spatial and temporal patterns of gene expression. Some of these transcription factors, depending on their different promoter contexts, can positively or negatively modulate transcription. For example, the HD transcription factor Oct-1, which activates different promoters, is also involved in repression of the human PIT1 gene expression (6).
The present study shows that the HD transcription factor Ptx1 can modulate POMC or other pituitary genes and IFN-A gene expression differently. The opposite functions of Ptx1 factor may be due to the context of pituitary gene and IFN-A gene promoters. Indeed, the activity of Ptx1 as a positive regulator of transcription is synergized by cell-restricted transcription factors to confer pituitary-, lineage-, and promoter-specific expression. Several known transcriptional interaction factors act in synergy with Ptx1: basic helix-loop-helix NeuroD1 for corticotroph-specific transcription of POMC (24), Pit1 to stimulate expression of the prolactin gene (34, 37), SF-1, an orphan nuclear receptor, and Egr-1, an immediate-early response gene, to stimulate the expression of the βLH gene (37–39). The C-terminal region of Ptx1 is involved in both transcriptional activation and physical interaction with Pit1, SF1, or Egr-1. In the context of the IFN-A gene promoters, the Ptx1 factor acts as a repressor and its effect is observed only after virus induction on the VRE-A-positive activity. Ptx1 could modulate the activity of specific transcription activators involved in the regulation of the IFN-A gene expression by interaction with factors which bind to VRE-A. Different factors may be required for maximal activity of the IFN-A promoter after virus induction. Two factors of the IRF family, IRF-3 and IRF-7, have been characterized (3, 11, 20, 28, 42). Thus, Ptx1 may interact with these specific IRF factors. Furthermore, our results suggest that the transcription-repressive effects of Ptx1 are due to the C-terminal region of the protein. This last region, which has been found to interact with different factors synergizing the activity of pituitary gene promoters, may be also required for protein interactions with other factors such as IRF factors repressing the virus induction of IFN-A gene promoters.
Ptx1 and differential activation and repression of IFN-A genes.
IRF-binding sites are not the only cause of the differential IFN-A gene expression. Indeed, the repression of the murine IFN-A11 gene after virus induction is also due to the negative regulatory element DNRE and the binding of Ptx1. Furthermore, the DNRE of the IFN-A11 promoter or the similar element 4DNRE found in the IFN-A4 promoter is able to bind Ptx1, which reduces the transcriptional activity of proximal VRE-A in both the IFN-A11 and IFN-A4 promoters, but Ptx1 was unable to repress the virus induction of murine IFN-B promoter. These results demonstrate that Ptx1 functions as a promoter-specific repressor. On the other hand, the fact that the intact IFN-A4 gene promoter remains highly inducible upon virus induction whereas the intact IFN-A11 gene promoter is poorly expressed has been previously explained by the presence of a third element in the IFN-A4 promoter which is absent in the IFN-A11 promoter. This element is a central region located between the distal 4DNRE and the proximal VRE-A of the IFN-A4 promoter and is able to overcome the DNRE silencer activity. Therefore, this element has been considered an antisilencer (17). The present study shows that the central antisilencer element is able to overcome the repressive effect of Ptx1, and this result clearly demonstrates that Ptx1 functions as a context-dependent repressor. On the other hand, endogenous IFN-A expression is quantitatively affected by Ptx1. Moreover, Ptx1 qualitatively influences the pattern of differential IFN-A gene expression. Indeed, the IFN-A11 subtype was detected in virus-induced Ptx1 knockdown cells. In addition, IFN-A5, not IFN-A4, was the most abundant species detected, thus suggesting that, as with the IFN-A11 gene, Ptx1 is essential for IFN-A5 repression. For the IFN-A5 gene, a sequence (−554 to −549 [TAATCC] in the noncoding strand) within the promoter is totally homologous to the core consensus recognition DNA sequence for bicoid-related proteins. The participation of this element in the repression of the transcription of the IFN-A5 gene remains to be elucidated. In conclusion, the results of this study suggest that DNRE and Ptx1 may exert a more general modulation on the differential transcriptional strength of the promoters of different IFN-A genes. Therefore, depending on the presence or the absence of different binding sites, the modulator effects of factors such as IRF-3 and IRF-7 as activators and Ptx1 as a repressor play a role in the differential expression of the IFN-A genes after virus induction.
ACKNOWLEDGMENTS
S. Lopez and M.-L. Island contributed equally to this work.
We thank U. Francke for kindly providing PTX1 cDNA. We are grateful to F. Petek for help with protein purification; E. Bonnefoy for discussions and encouragement; L. Lomme for bibliographic assistance; E. Prieto for photographs; and S. Chousterman, P. Djian, and G. Vincent for critical reading of the manuscript.
The work at UPR 2228 was supported by the Centre National de la Recherche Scientifique (CNRS) and the Université René Descartes Paris V and by grants from the Association de Recherche contre le Cancer (ARC; contract 1042), Ligue Régionale contre le Cancer, and Fédération Nationale des Groupements des Entreprises Françaises et de la Lutte contre le Cancer (FEGEFLUC).
REFERENCES
- 1.Au W C, Raj N B K, Pine R, Pitha P M. Distinct activation of murine interferon-α promoter region by IRF-1/ISFG-2 and virus infection. Nucleic Acids Res. 1992;20:2877–2884. doi: 10.1093/nar/20.11.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au W C, Su Y, Raj N B K, Pitha P M. Virus mediated induction of interferon A gene requires cooperation between multiple binding factors in the interferon α promoter region. J Biol Chem. 1993;268:24032–24040. [PubMed] [Google Scholar]
- 3.Au W C, Moore P A, LaFleur D W, Tombal B, Pitha P M. Characterization of the interferon regulatory factor 7 and its potential role in the transcription activation of interferon A genes. J Biol Chem. 1998;273:29210–29217. doi: 10.1074/jbc.273.44.29210. [DOI] [PubMed] [Google Scholar]
- 4.Blaiseau P L, Isnard A D, Surdin-Kerjan Y, Thomas D. Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol Cell Biol. 1997;17:3640–3648. doi: 10.1128/mcb.17.7.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bragança J, Génin P, Bandu M T, Darracq N, Vignal M, Cassé C, Doly J, Civas A. Synergism between multiple virus-induced-factor-binding elements involved in the differential expression of IFN-A genes. J Biol Chem. 1997;272:22154–22162. doi: 10.1074/jbc.272.35.22154. [DOI] [PubMed] [Google Scholar]
- 6.Delhase M, Castrillo J L, de la Hoya M, Rajas F, Hooghe-Peters E L. AP-1 and Oct-1 transcription factors down-regulate the expression of the human PIT1/GHF1 gene. J Biol Chem. 1996;271:32349–32358. doi: 10.1074/jbc.271.50.32349. [DOI] [PubMed] [Google Scholar]
- 7.Génin P, Bragança J, Darracq N, Doly J, Civas A. A novel PRDI and TG-binding activity involved in the virus-induced transcription of the IFN-A genes. Nucleic Acids Res. 1995;23:5055–5063. doi: 10.1093/nar/23.24.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gietz D, St. Jean A, Woods R A, Schiestl R B. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425–1426. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harada H, Fujita M, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 10.Hiscott J, Cantell K, Weissmann C. Differential expression of human interferon genes. Nucleic Acids Res. 1984;12:3727–3746. doi: 10.1093/nar/12.9.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Juang Y T, Lowther W, Kellum M, Au W C, Lin R, Hiscott J, Pitha P M. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim T K, Maniatis T. The mechanism of transcriptional synergy of an in vitro assembled interferon-β enhanceosome. Mol Cell. 1997;1:119–129. doi: 10.1016/s1097-2765(00)80013-1. [DOI] [PubMed] [Google Scholar]
- 13.Lamonerie T, Tremblay J J, Lanctôt C, Therrien M, Gauthier Y, Drouin J. Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 1996;10:1284–1295. doi: 10.1101/gad.10.10.1284. [DOI] [PubMed] [Google Scholar]
- 14.Lanctôt C, Lamolet B, Drouin J. The bicoid-related homeoprotein Ptx1 defines the most anterior domain of the embryo and differentiates posterior from anterior lateral mesoderm. Development. 1997;124:2807–2817. doi: 10.1242/dev.124.14.2807. [DOI] [PubMed] [Google Scholar]
- 15.Lanctôt C, Gauthier Y, Drouin J. Pituitary homeobox 1 (Ptx1) is differencially expressed during pituitary development. Endocrinology. 1999;140:1416–1422. doi: 10.1210/endo.140.3.6549. [DOI] [PubMed] [Google Scholar]
- 16.Lanctôt C, Gauthier Y, Drouin J. Hindlimb patterning and mandible development require the Ptx1 gene. Development. 1999;126:1805–1810. doi: 10.1242/dev.126.9.1805. [DOI] [PubMed] [Google Scholar]
- 17.Lopez S, Reeves R, Island M L, Bandu M T, Christeff N, Doly J, Navarro S. Silencer activity in the interferon A gene promoters. J Biol Chem. 1997;272:22788–22800. doi: 10.1074/jbc.272.36.22788. [DOI] [PubMed] [Google Scholar]
- 18.MacDonald N J, Kuhl D, Maguire D, Näf D, Gallant P, Goswamy A, Hug H, Büeler H, Chaturvedi M, De la Fuente J, Ruffner H, Meyer F, Weissmann C. Different pathways mediate virus inducibility of the human IFN-α1 and IFN-β genes. Cell. 1990;60:767–779. doi: 10.1016/0092-8674(90)90091-r. [DOI] [PubMed] [Google Scholar]
- 19.Maniatis T, Whittemore L A, Du W, Fan C M, Keller A D, Palombella V J, Thanos D N. Positive and negative control of human Interferon-β gene expression. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 1193–1220. [Google Scholar]
- 20.Marié I, Durbin J E, Levy D E. Differential viral induction of distinct interferon-α genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kündig T, Amakawa R, Kishihara K, Wakeman A, Potter J, Furlonger C L, Narendran A, Suzuki H, Ohashi P S, Paige C J, Taniguchi T, Mak T W. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 22.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 23.Nourbakhsh M, Hauser H. Constitutive silencing of IFN-β promoter is mediated by NRF (NF-κB-repressing factor), a nuclear inhibitor of NF-κB. EMBO J. 1999;18:6415–6425. doi: 10.1093/emboj/18.22.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poulin G, Turgeon B, Drouin J. NeuroDl/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol Cell Biol. 1997;17:6673–6682. doi: 10.1128/mcb.17.11.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raj N B K, Au W C, Pitha P M. Identification of a novel virus-responsive sequence in the promoter of murine interferon-α genes. J Biol Chem. 1991;266:11360–11365. [PubMed] [Google Scholar]
- 26.Ren B, Chee K J, Kim T H, Maniatis T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999;13:125–137. doi: 10.1101/gad.13.1.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roffet P, Lopez S, Navarro S, Bandu M T, Coulombel C, Vignal M, Doly J, Vodjdani G. Identification of distal silencing elements in the murine interferon-A11 gene promoter. Biochem J. 1996;317:697–706. doi: 10.1042/bj3170697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka M. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 29.Semina E V, Reiter R, Leysens N J, Alward W L, Small K W, Datson N A, Siegel-Bartelt J, Bierke-Nelson D, Bitoun P, Zabel B U, Carey J C, Murray J C. Cloning and characterization of a novel bicoid-related homeobox transcription factor gene, RIEG, involved in Rieger syndrome. Nat Genet. 1996;14:392–399. doi: 10.1038/ng1296-392. [DOI] [PubMed] [Google Scholar]
- 30.Shang J, Luo Y, Clayton D A. Backfoot is a novel homeo box gene expressed in the mesenchyme of developing hind limb. Dev Dyn. 1997;209:242–253. doi: 10.1002/(SICI)1097-0177(199706)209:2<242::AID-AJA10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 31.Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simeone A, Acampora D, Mallamaci A, Stornaiuolo A, D'Apice M R, Nigro V, Boncinelli E. A vertebrate gene related to orthodenticle contains a homeo domain of the bicoid class and demarcates anterior neuroectoderm in the gastrulating mouse embryo. EMBO J. 1993;12:2735–2747. doi: 10.1002/j.1460-2075.1993.tb05935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smidt M P, van Schaick H S, Lanctôt C, Tremblay J J, Cox J J, van der Kleij A, Wolterink G, Drouin J, Burbach J P. A homeodomain gene Ptx3 has highly restricted brain expression in mesencephalic dopaminergic neurons. Proc Natl Acad Sci USA. 1997;94:13305–13310. doi: 10.1073/pnas.94.24.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szeto D P, Ryan A K, O'Connell S M, Rosenfeld M G. P-OTX: a PIT-1-interacting homeodomain factor expressed during anterior pituitary gland development. Proc Natl Acad Sci USA. 1996;93:7706–7710. doi: 10.1073/pnas.93.15.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szeto D P, Rodriguez-Esteban C, Ryan A K, O'Connell S M, Liu F, Kioussi C, Gleiberman A S, Izpisua-Belmonte J C, Rosenfeld M G. Role of the bicoid-related homeodomain factor Pitx1 in specifying hindlimb morphogenesis and pituitary development. Genes Dev. 1999;13:484–494. doi: 10.1101/gad.13.4.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thanos D, Maniatis T. Virus induction of IFN β gene expression requires of the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 37.Tremblay J J, Lanctôt C, Drouin J. The pan-pituitary activator of transcription, Ptx1 (pituitary homeobox 1), acts in synergy with SF-1 and Pit1 and is an upstream regulator of the Lim-homeodomain gene Lim3/Lhx3. Mol Endocrinol. 1998;12:428–441. doi: 10.1210/mend.12.3.0073. [DOI] [PubMed] [Google Scholar]
- 38.Tremblay J J, Drouin J. Egr-1 is a downstream effector of GnRH and synergizes by direct interaction with Ptx1 and SF-1 to enhance luteinizing hormone β gene transcription. Mol Cell Biol. 1999;19:2567–2576. doi: 10.1128/mcb.19.4.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tremblay J J, Marcil A, Gauthier Y, Drouin J. Ptx1 regulates SF-1 activity by an interaction that mimics the role of the ligand-binding domain. EMBO J. 1999;18:3431–3441. doi: 10.1093/emboj/18.12.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors of the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 41.Wilson D S, Sheng G, Jun S, Desplan C. Conservation and diversification in homeo domain-DNA interactions: a comparative genetic analysis. Proc Natl Acad Sci USA. 1996;93:6886–6891. doi: 10.1073/pnas.93.14.6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoneyama M, Suhara W, Fujuhara Y, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing of IRF-3 and CBP/P300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]