ABSTRACT
Knowing the mechanisms that govern the persistence of infected CD4+ subpopulations could help us to design new therapies to cure HIV-1 infection. We evaluated the simultaneous distribution of the HIV-1 reservoir in 13 CD4+ subpopulations from 14 HIV-1-infected individuals on antiretroviral therapy to analyze its relationship with HIV-1 transcription, immune activation, and cell proliferation. A unique large blood donation was used to isolate CD4, CD4 resting (CD4r), CD4 activated (CD4a), T naive (TN), T stem cell memory (TSCM), T central memory (TCM), T transitional memory (TTM), T effector memory (TEM), circulating T follicular helper (cTFH), TCD20, TCD32, and resting memory TCD2high (rmTCD2high) cells. HIV-1 DNA measured by droplet digital PCR ranged from 3,636 copies/106 in TTM to 244 in peripheral blood mononuclear cells (PBMCs), with no subpopulation standing out for provirus enrichment. Importantly, all the subpopulations harbored intact provirus by intact provirus DNA assay (IPDA). TCD32, cTFH, and TTM had the highest levels of HIV-1 transcription measured by fluorescent in situ hybridization with flow cytometry (FISH/flow), but without reaching statistical differences. The subpopulations more enriched in provirus had a memory phenotype, were less activated (measured by CD38+/HLA-DR+), and expressed more programmed cell death 1 (PD-1). Conversely, subpopulations transcribing more HIV-1 RNA were not necessarily enriched in provirus and were more activated (measured by CD38+/HLA-DR+) and more proliferative (measured by Ki-67). In conclusion, the HIV reservoir is composed of a mosaic of subpopulations contributing to the HIV-1 persistence through different mechanisms such as susceptibility to infection, provirus intactness, or transcriptional status. The narrow range of reservoir differences between the different blood cell subsets tested suggests limited efficacy in targeting only specific cell subpopulations during HIV-1 cure strategies.
KEYWORDS: HIV-1, HIV-1 DNA, HIV-1 RNA, HIV-1 cure, HIV-1 reservoir, HIV-1 reservoir size
INTRODUCTION
Antiretroviral therapy (ART) effectively blocks HIV-1 replication and reduces circulating virus to undetectable levels. However, ART is not able to eradicate the virus, and viral load rebounds rapidly if it is interrupted (1). Viral rebound results from the persistence of HIV-1, mostly in long-lived CD4+ T cells infected with integrated latent virus, which is the main obstacle to eradication of HIV-1 (2).
Numerous strategies are currently being evaluated to target and kill HIV-1-infected cells, with the ultimate goal of eradicating the virus (3). However, major obstacles include the inability to distinguish between latently infected cells in vivo and to ascertain their mechanisms of persistence. Therefore, it is of great interest to define which cell subpopulations compose the HIV-1 reservoir and their specific mechanisms of persistence in order to design effective therapies to cure HIV-1 infection.
To date, various cell subpopulations and cell markers have been identified as important HIV-1 reservoir cells. Central memory T cells (TCM) were reported to be a significant source of infected cells among CD4+ T-memory cells (4). Years later, stem cell memory T cells (TSCM) (5) were also postulated as a major cell subpopulation, since their long life makes them a main target for eradication strategies (6), despite their relatively low net contribution to the total reservoir. Recent studies have postulated CD4+ T follicular helper cells (TFH) (7), resting memory TCD2high (rmTCD2high) (8), TCD32 (9, 10), and TCD20 (11) as important candidates. However, the set of cell subpopulations defined by these potential markers of viral persistence has never been studied simultaneously in the same individuals with the aim of defining more accurately their relative contribution to the structure of the viral reservoir in peripheral blood.
To address this issue, we comprehensively evaluated the distribution of the HIV-1 reservoir in the above-mentioned CD4+ T-cell subpopulations in peripheral blood and their relationship with RNA expression, immune activation, cell proliferation, and cytokine profiling.
RESULTS
Participants and study design.
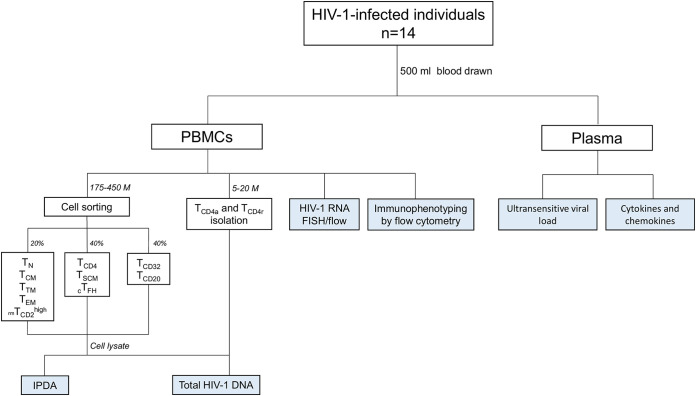
We studied 14 virologically suppressed HIV-1-infected individuals to assess the contribution of the CD4+ T-cell subpopulations to the viral reservoir. Their clinical characteristics are summarized in Table 1. All participants had been infected for more than 3 years when samples were taken, had CD4 counts over 400 cells/μL, and had undetectable viral loads for more than 2.8 years (<50 copies/mL plasma). A 500-mL blood sample was drawn to obtain peripheral blood mononuclear cells (PBMCs) and plasma. PBMCs were used for both immunomagnetic and fluorescence-activated cell sorting of the different CD4+ T-cell subpopulations of interest in order to measure the HIV-1 reservoir (total or intact), intracellular HIV-1 RNA expression, cell activation, programmed cell death 1 (PD-1) expression, and cell proliferation. Plasma was used to measure residual viremia by ultrasensitive viral load (usVL) and cytokines/chemokines in plasma (Fig. 1).
TABLE 1.
Clinical characteristics of the subjects included in the longitudinal studya
| Patient ID | Gender | Route of infection | Time since HIV-1 diagnosis (yrs) | Time with undetectable viral load (yrs) | Minimum CD4 cell count reported | Maximum viral load reported | CD4 cell count at sample (cells/μL) | usVL at sample (copies/mL) | ART at sample |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Male | MSM/BI | 16.6 | 10.3 | 382 | 640,000 | 1,200 | <0.5 | 3TC, ABC, DTG |
| 2 | Male | HTSX | 20.0 | 9.3 | 5 | 71,550 | 400 | <0.5 | 3TC, ABC, DTG |
| 3 | Male | IVDU | 21.0 | 7.5 | 310 | 450,000 | 990 | 1.7 | COB, DRV |
| 4 | Male | MSM/BI | 18.7 | 16.2 | 48 | 1,000,000 | 890 | 1.6 | COB, EVG, FTC, TAF |
| 5 | Male | MSM/BI | 9.4 | 4.3 | 536 | 830,000 | 1,110 | 2.9 | FTC, RPV, TDF |
| 6 | Male | MSM/BI | 15.1 | 14.8 | 20 | 460,000 | 760 | 14.0 | COB, FTC, TAF, EVG |
| 7 | Male | HTSX | 28.8 | 15.8 | 64 | 1,300,000 | 590 | 3.1 | COB, DRV, ETV |
| 8 | Male | HTSX | 5.3 | 3.4 | 45 | 910,000 | 610 | 2.2 | DTG, RPV |
| 9 | Male | HTSX | 11.8 | 11.7 | 129 | 19,000 | 530 | 0.6 | COB, EVG, FTC, TAF |
| 10 | Female | IVDU | 28.0 | 7.9 | 36 | 3,630,000 | 810 | 7.9 | COB, EVG, FTC, TAF |
| 11 | Male | HTSX | 28.1 | 6.4 | 302 | 170,000 | 740 | <0.5 | EFV, FTC, TDF |
| 12 | Male | MSM/BI | 3.0 | 2.8 | 540 | 287,000 | 910 | <0.5 | COB, FTC, TAF, EVG |
| 13 | Male | IVDU | 24.6 | 6.0 | 280 | 23,000 | 720 | 1.1 | TDF, COB, DRV |
| 14 | Male | MSM/BI | 12.3 | 8.0 | 400 | 550,000 | 1,310 | 2.4 | EFV, FTC, TDF |
MSM, men who have sex with men; BI, bisexual; HTSX, heterosexual; IVDU, intravenous drug users; usVL, ultrasensitive viral load; ART, antiretroviral therapy; 3TC, lamivudine; ABC, abacavir; DTG, dolutegravir; COB, cobicistat; DRV, darunavir; EVG, elvitegravir; FTC, emtricitabine; TAF, tenofovir alafenamide; RPV, rilpivirine; TDF, tenofovir disoproxil fumarate; ETV, etravirine.
FIG 1.
Study design flow-chart.
Determination of HIV-1 DNA in CD4+ T-cell subpopulations.
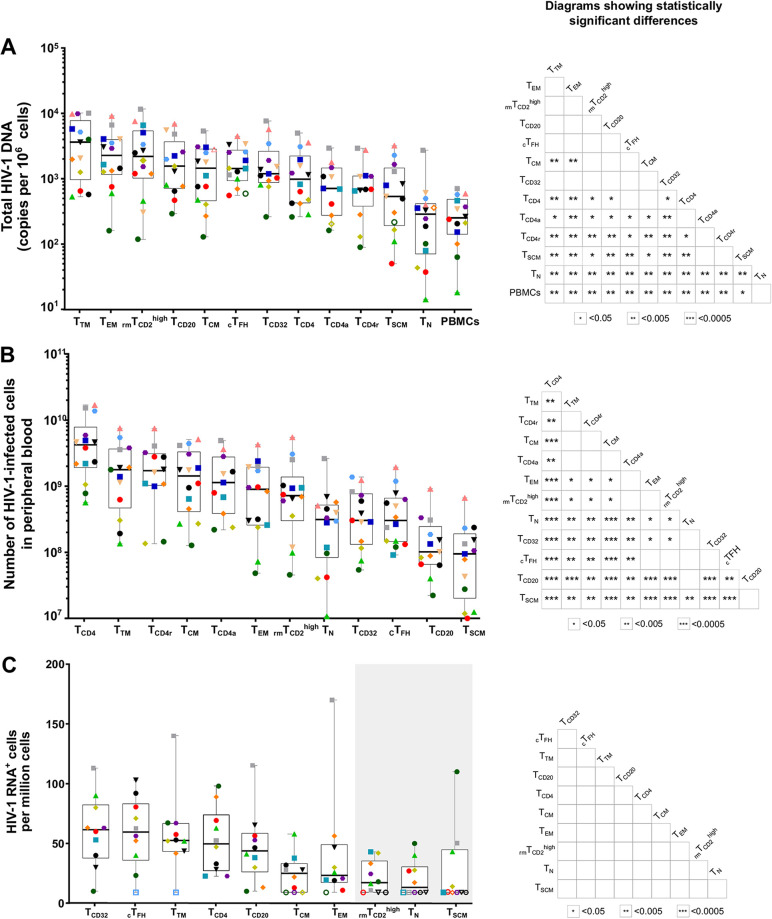
In order to determine whether the stability of infected cells could be attributed to specific CD4+ T-cell subpopulations, we sorted CD4+ T-cell subpopulations (Fig. S1; Table 2) from successfully virologically suppressed HIV-1-infected individuals based on expression of multiple surface markers and quantified total HIV-1 DNA in each subpopulation using droplet digital PCR (ddPCR). Median total HIV-1 DNA was detected in all cell subpopulations analyzed, gradually ranging from 3,636 to 244 copies per million cells of a given subset (Fig. 2A).
TABLE 2.
Number of cells sorted for each cell subpopulation
| Cell subpopulation | Mediana | Minimum | Maximum |
|---|---|---|---|
| TCD4 | 842,857 | 500,000 | 1,000,000 |
| TCD4r | 1,532,367 | 540,400 | 3,000,000 |
| TCD4a | 1,036,150 | 554,800 | 2,230,000 |
| TN | 535,246 | 126,000 | 1,450,000 |
| TCM | 475,761 | 89,000 | 835,000 |
| TTM | 55,115 | 4,400 | 141,216 |
| TEM | 454,623 | 66,000 | 753,171 |
| rmTCD2high | 271,786 | 55,000 | 500,000 |
| TSCM | 106,419 | 11,866 | 277,623 |
| cTFH | 263,779 | 19,600 | 507,000 |
| TCD32 | 113,782 | 26,000 | 318,000 |
| TCD20 | 334,635 | 118,000 | 435,000 |
Cells were equally distributed to measure total HIV-1 DNA and IPDA.
FIG 2.
Measurement of HIV-1 DNA and HIV-1 RNA in CD4+ T-cell subpopulations. (A) Total HIV-1 DNA measured in 13 sorted CD4+ T-cell subpopulations. According to the gating strategy (Fig. S1), some of these subpopulations overlap. (B) Number of HIV-1-infected cells for each CD4+ T-cell subpopulation based on the frequency of each CD4+ T-cell subpopulation and its levels of total HIV-1 DNA in peripheral blood. (C) Proportion of cells expressing HIV-1 RNA in each CD4+ T-cell subpopulation normalized to the medium control. The gray box indicates the cell subpopulations with median HIV-1+ RNA levels not different from HIV-1– individuals. (A to C) For all the panels, each colored symbol represents a single individual. On the right of each panel, matrices depict P values of the differences between paired CD4+ T-cell subpopulations. Differences were tested for statistical significance using the Wilcoxon rank sum test followed by the FDR test for multiple comparisons. Box and whisker plots indicate the median, interquartile range, and minimum and maximum values.
A representative gating strategy used for cell sorting of the cell subpopulations. Three separate cell sorters were made in order to be able to sort the overlapping subpopulations. A common gate strategy was used where PBMCs were acquired and gated according to (i) forward scatter area (FSC-A) and forward scatter height (FSC-H) to select singlets, (ii) side scatter area (SSC-A) and FSC-A to determine cell morphology, (iii) CD3 and viability staining to select living T cells, and (iv) CD4 T cells from viable cells to gate all the other cell subpopulations. In sorter 1, first, a fraction of rmTCD2high were sorted because of overlap with all the other cell subpopulations. Then, TN, TCM, TEM, and TTM were discriminated with CD45RA, CCR7, and CD27 markers as described in Materials and Methods. In sorter 2, TSCM and cTFH were sorted from CD4+ T cells. Finally, TCD32 and TCD20 were sorted in sorter 3. Download FIG S1, TIF file, 0.8 MB (844.7KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We observed that the most infected cell subpopulations had a memory phenotype, namely, T transitional memory (TTM), T effector memory (TEM), rmTCD2high, TCD20, and TCM cells (Fig. 2A). Statistically significant differences were found between multiple cell subpopulations (see P value matrices in Fig. 2A). However, although the distribution of total HIV-1 DNA gradually decreased across the CD4+ T-cell populations studied to the naive phenotype (TN) and total PBMCs, no specific subpopulation was noteworthy for enrichment in provirus. In addition, we calculated the total number of infected cells in peripheral blood for each subpopulation by considering the number of CD4+ T cells per μL in peripheral blood per individual, an assumed mean blood volume of 5 L per person, and the percentage of each subpopulation among CD4+ T cells obtained by flow cytometry (Fig. 2B). We observed a higher number of infected TTM and CD4 resting (CD4r), followed by TCM, CD4 activated (CD4a), and TEM. At the other end of the spectrum, the total number of HIV-1-infected cells in the peripheral blood of subpopulations, such as TCD20 and TSCM, was minimal.
The contribution of each subpopulation to the total HIV-1 reservoir could not be calculated, owing to the unavoidable overlap between specific cell subpopulations. Nevertheless, we calculated the contribution of the nonoverlapping TN, TSCM, TCM, TTM, and TEM cells to the HIV-1 reservoir and found the highest values for TCM cells (median contribution of 37%), followed by TTM cells (32%) and TEM cells (23%), whereas the contribution of TN cells and TSCM cells to the pool of cells harboring HIV-1 proviral DNA was marginal (8% and 0.2%, respectively) (Fig. S2A).
Contribution of memory cell subpopulations and HIV/RNA ratio. (A) Contribution of the TN, TSCM, TCM, TTM, and TEM cell subpopulations to the pool of HIV-infected cells in 14 HIV-infected individuals. Differences were tested for statistical significance using Friedman’s test followed by the false-discovery rate test for multiple comparisons. (B) Ratio between HIV-1 RNA and HIV-1 DNA. Each symbol represents a different individual. RNA/DNA ratios for TN, TSCM, and rmTCD2high were not represented due to the high number of undetectable values for intracellular HIV-1 RNA measurements. Box and whisker plots indicate the median, interquartile range, and minimum and maximum values. Download FIG S2, TIF file, 0.2 MB (235.9KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Moreover, we analyzed the correlations between total HIV-1 DNA in each CD4+ T-cell subpopulation and clinical parameters (Table S1). HIV-1 DNA was positively and significatively correlated with years with undetectable viral load in circulating T follicular helper (cTFH) (rho = 0.56, P = 0.04) and also with maximum viral load reported with TCD32 (rho = 0.58, P = 0.04). However, the significance of these correlations did not hold after adjusting for multiple comparisons.
Analysis of the correlation between total HIV-1 DNA, clinical parameters, HIV-1 expression, and cytokines/chemokines plasma levels. Download Table S1, DOCX file, 0.02 MB (22.3KB, docx) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
While subpopulations with a memory phenotype are generally richer in provirus, HIV-1 infection does not seem to occur preferentially in a very specific cell subpopulation, thus demonstrating the high heterogeneity of the HIV-1 reservoir.
Determination of intracellular HIV-1 RNA in CD4+ T-cell subpopulations.
Since most HIV-1 DNA is defective, we sought to investigate the transcriptional activity of HIV-1 in subpopulations from 10 HIV-1-infected individuals (Fig. S4). We used the novel RNA fluorescent in situ hybridization with flow cytometry (FISH/flow) protocol to measure the frequency of each subpopulation expressing intracellular HIV-1 RNA (12) and found that TCD32, cTFH, TTM, and TCD20 were the cell populations with higher HIV-1 RNA transcription (median values in HIV-1 RNA+ cells per million CD4+ T cells: TCD32, 61.5; cTFH, 59.5; TTM, 52.5; and TCD20 43.8). Subpopulations such as rmTCD2high, TN, and TSCM barely expressed HIV-1 viral transcripts, since their median levels were not different from 0 after background subtraction (Fig. 2C and Fig. S5). Nonetheless, differences between the cell subpopulations were not statistically significant, probably due to the lower number of samples assayed. We also showed that most HIV-1 DNA was not transcribed, as revealed by the HIV-1 RNA/DNA ratio, which was lower than 0.05 in all the CD4+ T-cell subpopulations analyzed (Fig. S2B).
Representative plots of HIV-1 RNA expressing cells. Levels of intracellular HIV-1 RNA in TTM cells from an uninfected donor, which is used to set up the limit of detection of the assay, and two individuals from the study. Download FIG S4, TIF file, 0.7 MB (738.7KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of levels of HIV-1 RNA expressing cells between HIV-1– and HIV-1+ individuals. Differences were tested for statistical significance using the Wilcoxon matched-pair rank test. Download FIG S5, TIF file, 0.5 MB (483.6KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Levels of intracellular HIV-1 expression were correlated with total HIV-1 DNA in TTM (rho = 0.7, P = 0.04) and TCD4 cells (rho = −0.66, P = 0.04) (Table S1). Additionally, we measured HIV-1 expression in plasma by usVL and found statistically significant positive correlations between total HIV-1 DNA and usVL in 6 subpopulations (Table S1), namely, TSCM (rho = 0.71, P = 0.01), TEM (rho = 0.7, P = 0.01), cTFH (rho = 0.7, P = 0.01), TCD32 (rho = 0.69, P = 0.01), TN (rho = 0.66, P = 0.01), and TCD20 (rho = 0.57, P = 0.04). However, after adjusting for multiple comparisons, the significance of the correlations did not hold.
Thus, these results suggest that the cells most enriched in provirus are not the most transcriptionally active ones.
Quantification of intact provirus in CD4+ T-cell subpopulations.
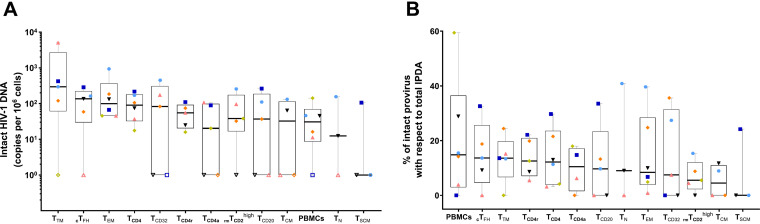
To assess the number of intact viruses in each cell subpopulation, we performed the intact provirus DNA assay (IPDA) in PBMCs from the 14 HIV-1-infected individuals. In 6 individuals, we performed IPDA in all the cell subpopulations based on sample availability. We observed that all subpopulations measured had intact provirus to a different extent, with the most transcriptionally active cells, such as TTM and cTFH, being the ones with the most intact provirus (Fig. 3A); nonetheless, there were not statistically significant differences. Of note, TTM was the subpopulation with the highest total HIV-1 DNA (Fig. 2A) and intact provirus (Fig. 3A). Moreover, the proportion of intact provirus with respect to total IPDA was generally low, with the highest median values found in PBMCs (14.8%), cTFH (13.6%), and TTM (13.6%) and the lowest in TCM (4.5%) and TSCM (0%) (Fig. 3B).
FIG 3.
Measurement of intact provirus in CD4+ T-cell subpopulations. (A) Intact proviral frequencies per 106 cells determined by IPDA for each CD4+ T-cell subpopulation from 6 HIV-1-infected individuals. (B) Percentage of intact provirus with respect to total IPDA levels. Total IPDA proviruses were determined as the sum of intact, 5′-defective, and 3′-defective proviruses from each individual. Each symbol represents 1 individual. Open symbols represent values under the limit of detection. Box and whisker plots indicate the median, interquartile range, and minimum and maximum values.
Relationship between the parameters analyzed in each CD4+ T-cell subpopulation.
We then analyzed the relative proportion of HIV-1 DNA, HIV-1 RNA, activation, proliferation, and PD-1 markers between the different subpopulations (Fig. S6). We included CD69 for early activation, HLA-DR for late activation, and double-positive cells for CD38 and HLA-DR (Fig. S7A to C). Also, we used Ki-67 as a proliferation maker, and PD-1 as a surrogate marker of HIV latency promoter (Fig. S7D and E).
Representative example of the gating strategy used for analyses in Fig. S7A to E. PBMCs were acquired and gated according to (i) time to ensure homogeneous acquisition, (ii) SSC-A and FSC-A to determine cell morphology, (iii) FSC-A and FSC-H to select singlets, (iv) CD3 and viability staining to select living T cells, and (v) CD4 T cells from viable cells to gate all the other cell subpopulations. Then CD69, CD38, HLA-DR, PD-1, and CD38+HLADR+ were gated on every cell subpopulation based on FMOs. The graph shows a representative gate strategy for CD4+ T cells, but the same was made for TN, TSCM, TCM, TTM, TEM, cTFH, TCD20, TCD32, and rmTCD2high that were gated as in Fig. S1. Download FIG S6, TIF file, 0.7 MB (745.7KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of activation, proliferation, and exhaustion in CD4+ T-cell subpopulations. (A to C) Percentage of activation levels based on (A) CD69+, (B) HLA-DR+, and (C) double positive CD38+ and HLA-DR+ cells for each CD4+ T-cell subpopulation. (D) Percentage of cell proliferation measured using Ki-67+ cells and (E) percentage of PD-1+ cells. Each symbol represents 1 individual. (A to E) Matrices depict P values of the differences between paired CD4+ T-cell subpopulations. Differences were tested for statistical significance using the Wilcoxon rank sum test followed by the false-discovery rate test for multiple comparisons. Box and whisker plots indicate the median, interquartile range, and minimum and maximum values. Download FIG S7, TIF file, 0.6 MB (621KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
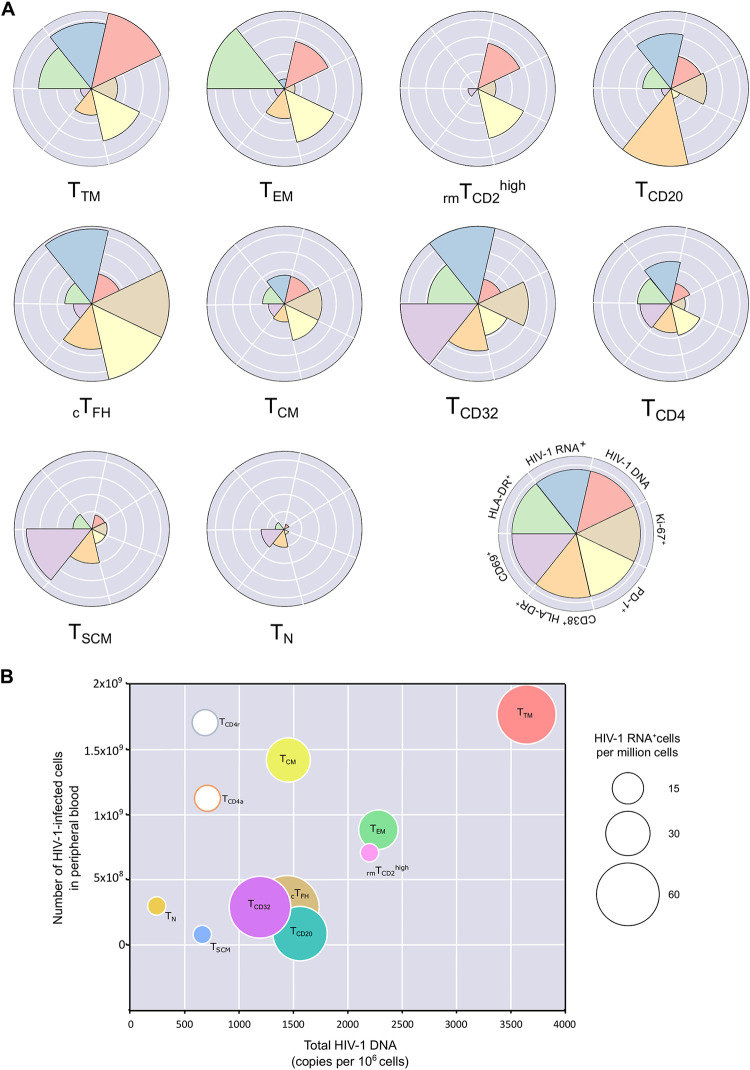
We observed that the subpopulations with the highest total HIV-1 DNA value, with the exception of TCD20, had higher expression of PD-1, which has been described as actively promoting HIV latency (13, 14) (Fig. 4A and Fig. S7E). It is noteworthy that these subpopulations were those with a more differentiated phenotype, such as TEM and TTM, while the less differentiated cells and those with less provirus (TN and TSCM) generally had very low frequencies of PD-1 (Fig. 4A and Fig. S7E). Similarly, the activation profile based on the double expression of CD38/HLA-DR (Fig. S7C) showed that cell subpopulations with a larger reservoir were not highly activated (Fig. 4A). Regarding proliferation, the cell subpopulations with the highest levels of Ki-67 were those that also had the highest levels of HIV-1 RNA expression, such as cTFH and TCD32 (Fig. S7D). Interestingly, TCD32 cells (and to a lesser extent total CD4+ T cells) had average total HIV-1 DNA but high HIV-1 expression, activation, and proliferation, thus indicating a nonquiescent state (Fig. 4A). In contrast, subpopulations with the lowest levels of total HIV-1 DNA (TN and TSCM) expressed less PD-1 and were nonproliferating and did not express HIV-1 RNA (Fig. 4A).
FIG 4.
Relationship between HIV-1 DNA, HIV-1 RNA, and parameters of activation, exhaustion, and proliferation for each CD4+ T-cell subpopulation. (A) The Nightingale rose plots show the patterns of 7 parameters (total HIV-1 DNA, HIV-1 RNA+, Ki-67+, PD1+, CD38+HLA-DR+, CD69+, HLA-DR+) for 10 CD4+ T-cell subpopulations. Each wedge represents a parameter studied and is depicted in a different color. The size of the wedge for each parameter depicts a proportional magnitude of the parameter relative to the maximum magnitude across the different CD4+ T-cell subpopulations. (B) Three-dimensional graph showing the correlation between total HIV-1 DNA and the number of HIV-1-infected cells in peripheral blood; the bubble size represents the levels of intracellular HIV-1 RNA in each cell subpopulation. TCD4r and TCD4a bubbles are not sized according to their intracellular HIV-1 RNA. TN, TSCM, and rmTCD2high are given a small bubble size since their intracellular HIV-1 RNA levels are not different from those of HIV-1– individuals.
We performed a more in-depth analysis of the relationship between total HIV-1 DNA per CD4 T-cell subpopulation, the number of HIV-1-infected cells in total peripheral blood, and the expression of intracellular HIV-1 RNA measured by FISH/flow in each cell subpopulation (Fig. 4B). With the exception of TCD4r and TCD4a cell populations, most of the CD4+ T-cell subpopulations showed a certain degree of correlation between total HIV-1 DNA per CD4+ T-cell subpopulation and the number of HIV-1-infected cells in total peripheral blood. However, expression of intracellular HIV-1 RNA varied more widely between cell subpopulations. Thus, TTM was the most infected CD4+ T-cell subpopulation, the most abundant in peripheral blood, and one of the most highly transcriptionally active (Fig. 4B). The most transcriptionally active remaining cell subpopulations (TCD32, cTFH, TCD20) clustered together with intermediate levels of HIV-1 DNA and an average number of infected cells in total peripheral blood (Fig. 4B).
Overall, these results indicate that the transcriptionally inactive HIV-1 reservoir is composed mainly of cells with low levels of activation, high levels of PD-1, and low proliferation, potentially indicating the quiescent nonactivated state of the cells that constitute the HIV-1 reservoir in individuals receiving ART.
Cytokines and chemokines in plasma.
To further characterize how residual inflammation might affect the composition of the viral CD4+ T-cell reservoir, we measured 10 cytokines/chemokines using Luminex in the plasma of 14 ART-treated individuals. We found statistically significant negative correlations between interferon gamma (IFN-γ) and total HIV-1 DNA in TCD32, cTFH, and TSCM (Table S1), as well as between tumor necrosis factor (TNF) and TCM, TCD32, and TCD4. Moreover, statistically significant negative correlations were observed for the cytokine interleukin 17A (IL-17A)/cytotoxic-T-lymphocyte-associated antigen 8 (CTLA8) and total HIV-1 DNA in 4 out of 10 subpopulations studied, including TTM, rmTCD2high, TCM, and TSCM (Table S1). Nonetheless, correlations did not hold after false-discovery rate (FDR) adjustment. These results suggest a subtle link between levels of HIV-1 reservoir in the different cell subpopulations and the inflammation profile.
DISCUSSION
Elimination of HIV-1-infected cells is one of the current priorities in HIV-1 research (15). However, we are still unable to identify infected cells in vivo that can be specifically targeted in therapeutic strategies. In this study, we simultaneously and comprehensively analyzed proviral DNA levels, transcription, activation status, and inflammatory profile in up to 13 relevant CD4 T-cell subsets from the same blood samples of a unique cohort of people with HIV who were on suppressive antiretroviral therapy. Therefore, this study helps to establish a more precise comparison among previously described viral reservoirs in blood cells, diminishing the effect of patient, sample, and technique variability.
First, we observed that total HIV-1 DNA content was always present and gradually distributed in all the CD4+ T-cell subpopulations analyzed, with a maximum variation of the medians of about 1 log. No specific subpopulation stood out from the rest regarding enrichment in provirus, and therefore targeting a specific subpopulation in HIV eradication strategies will result in limited changes of the total reservoir size. However, as previously described, cell subpopulations with a memory phenotype, such as TTM, TEM, and rmTCD2high, were more enriched in provirus, while TN cells harbored the smallest reservoir (4). Thus, driving the HIV-1-infected cells toward terminal effector cell subpopulations that have shorter half-lives might help to diminish the viral reservoir, as recently suggested by Grossman et al. (16).
Nevertheless, to fully estimate the final contribution of the viral reservoir in peripheral blood, the size of each cell subpopulation must also be taken into account. In this regard, TTM, TCD4r, and TCM subpopulations were the main contributors to the HIV reservoir in peripheral blood. This is probably the result of the long half-life and self-renewal capacities that characterize the memory subsets (17, 18).
Another important factor to consider is the intactness of the provirus, since only intact proviruses are likely to be infectious and therefore contribute to viral rebound if the pressure of ART is interrupted (19). Importantly, we detected intact provirus measured by IPDA in all the cell subpopulations analyzed in the subset of individuals whose samples were available, suggesting that all the infected cell subpopulations might be a source of viral rebound, which would be in line with previous reports (20) In all cases, however, intact provirus represented less than 15% of the total HIV-1 DNA, as previously reported (21). Although there were not statistically significant differences, likely due to the small sample size, the results suggested that TTM, cTFH, TEM, and TCD32 were slightly more enriched in intact provirus.
Latently infected cells in individuals on ART are generally not transcriptionally active (22), although this observation might differ depending on the cell type, owing to differences in activation state or the proportion of defective provirus. We found that cell subpopulations with higher intact provirus, in general, expressed more intracellular HIV-1 RNA as measured by FISH/flow, including TCD32, cTFH, and TTM, indicating that it would be important to tackle not only cells in a resting state, but also the active viral reservoir, since these might be sources of residual viremia during ART and viral rebound if ART is discontinued. In fact, a recent study showed that viral RNA expression after latency reversal was detected in all CD4+ T-cell subpopulations assayed (23), indicating that when properly stimulated, most cells are able to sustain viral transcription.
We also considered whether there were differences in the activation, proliferation, and PD-1 levels between the different cell subpopulations. It is known that a larger fraction of the viral reservoir is mostly composed of resting and exhausted cells with low rates of proliferation (2, 24), a finding that is consistent with our results. Specifically, highly activated cell subpopulations measured by CD38+HLA-DR+, such as cTFH and TCD32, were, as expected, highly transcriptionally active but not extraordinarily enriched in provirus, confirming previous findings on TCD32 cells (25, 26). On the other hand, the TTM, TEM, and rmTCD2high subpopulations that harbored more provirus, were also the ones expressing more PD-1, probably due to the persistent exposure to HIV-1 antigens. The frequencies and distribution of CD4+ T cells expressing PD-1 were similar to those reported in the study from Fromentin et al. (24). Importantly, clonal proliferation of HIV-1-infected cells has been identified as a key mechanism of viral persistence in vivo (27, 28). In this regard, we found that some of the most transcriptionally active subsets (TCD32 and cTFH cells) also have a higher cell proliferation capacity, as measured according to expression of Ki-67. The way in which these subpopulations might be associated with the clonal expansion of specific HIV-1-infected cells in vivo warrants further investigation.
Surprisingly, we found a negative correlation between levels of HIV-1 DNA in several cell subpopulations and the plasma cytokine IL-17. An association has been observed between high levels of plasma viremia and depletion of Th17 cells, which produce IL-17 (29). Although we did not study Th17 cells, the positive correlation found between usVL and levels of total HIV-1 DNA leads us to speculate that individuals with a larger HIV-1 reservoir might have a lower frequency of Th17 cells and, therefore, lower levels of IL-17 in plasma. Given the recent finding implicating Th17 cells as sanctuaries for HIV-1 reservoirs (30), viral persistence may modulate cell epigenetic states and signaling pathways to allow viral latency that can impair IL-17 release, thus rendering Th17 cells dysfunctional (31). Finally, as IL-17 contributes to the integrity of the gut barrier, low levels of Th17 may be associated with greater microbial translocation, which may increase residual inflammation and favor HIV-1 persistence. Nonetheless, these explanations are speculative, and further investigations should be performed to prove them.
Our findings indicate that the HIV-1 reservoir in peripheral blood is composed of a mosaic of HIV-1-infected cell subpopulations contributing to the persistence of HIV-1 through a series of mechanisms, including susceptibility to infection (TTM, TEM, rmTCD2high), higher rates of intact provirus (TTM, cTFH, TEM), transcriptionally active subpopulations (cTFH, TCD32), and subpopulations with a long half-life (TSCM, TCM) (5, 18). Therefore, these results suggest that eradication strategies relying solely upon targeting unique cell subpopulations would hardly be effective. In fact, a recent study where they did a phenotypic analysis of the CD4 T-cell reservoir using cytometry by time of flight (CyTOF), reached a similar conclusion that a single surface marker/cell subpopulation does not exist to target the entire HIV-1 reservoir at once (32). A combination of strategies targeting their different characteristics would be more desirable.
Our study was subject to a series of limitations. First, it was only performed in peripheral blood, while it is known that most infected cells are found in lymph nodes and gut-associated lymphoid tissue and that the frequencies of cell subpopulations in tissues might differ. However, given the low frequencies of some of the cell subpopulations in the study, it would have been extremely challenging to perform the same comparative analysis in other tissues. These same low frequencies can also limit, in some cases, the precision of quantification of total HIV-1 DNA and measurements of intact proviruses in blood. Also, the technique used for measuring HIV-1-expressing cells has a relatively high background in HIV-1– individuals. Finally, we did not analyze the functionality of the different cell subpopulations, for example, by measuring the replication competence of the provirus.
In conclusion, we found that the HIV-1 reservoir under ART is distributed throughout all CD4 T-cell subpopulations and has heterogeneous levels of HIV-1 expression, activation, and proliferation and, thus, contributes to HIV-1 persistence through different mechanisms, such as susceptibility to infection, rates of provirus intactness, transcriptional status, and half-life. This research provides new knowledge on the composition of the HIV-1 reservoir suggesting that eradication strategies relying solely upon targeting unique cell subpopulations will result in limited changes of the total reservoir size.
MATERIALS AND METHODS
Study participants.
Samples from 14 HIV-1-infected individuals on suppressive ART were obtained from the HIV unit of Hospital Universitari Vall d’Hebron in Barcelona, Spain. A 500-mL sample of blood was extracted to collect large numbers of peripheral blood mononuclear cells (PBMCs) and plasma. The local ethics committee approved the protocol [PR(AG)192/2018], and the study was performed in accordance with the principles of the Declaration of Helsinki. All participants provided written informed consent before initiation. Samples were obtained only from adults and were totally anonymous and untraceable.
Cell sorting.
PBMCs were isolated from fresh blood (14 individuals) using Ficoll-Paque centrifugation. Resting and activated CD4+ T cells (CD4r and CD4a, respectively) were isolated from PBMCs using magnetic beads (MACS; Miltenyi Biotec) based on CD69, CD25, and HLA-DR activation markers. Then, starting from 175 × 106 to 450 × 106 PBMCs, other CD4+ T-cell subpopulations were sorted on a FACSAria II device (BD Biosciences) using 3 different sorting panels because of the overlap between the cell subpopulations (see sorting strategy in Fig. S1). PBMCs were incubated with the following series of antibodies: live/dead (allophycocyanin Cy7 [APC-Cy7]; Invitrogen), CD3 (BV510; BD Biosciences), CD4 (AF700; BD Biosciences), CD45RA (APC; BioLegend), CCR7 (PE Dazzle; BioLegend), CD27 (fluorescein isothiocyanate [FITC]; BD Biosciences), CD95 (phycoerythrin Cy5 [PE-cy5]; BD Biosciences), CXCR5 (PE-Cy7; BD Biosciences), PD-1 (BV421; BioLegend), HLA-DR (BV421; BD Biosciences), CD2 (BV605; BD Biosciences), CD32 (phycoerythrin [PE]; BioLegend), and CD20 (PerCPCy5.5; BD Biosciences). All sorted CD4+ T-cell subpopulations were defined as live CD3+CD4+ T cells. T-cell maturation was based on the expression of the surface markers CD45RA, CCR7, CD27, and CD95 in order to define the following subpopulations: T naive (TN, CD45RA+CCR7+CD27+), T stem cell memory (TSCM, CD45RA+CCR7+CD27+CD95+), T central memory (TCM, CD45RA-CCR7+CD27+), T transitional memory (TTM, CD45RA–CCR7–CD27+), and T effector memory (TEM, CD45RA–CCR7–CD27–). The remaining CD4+ T-cell subpopulations were circulating T follicular helper (cTFH; CXCR5+, PD1+), TCD32 (CD32+), TCD20 (CD20+), and rmTCD2high (CD45RA–, HLA-DR–, CD2high) cells.
Flow cytometry.
For phenotypic analysis, we stained PBMCs with the following series of antibodies: live/dead (APC-Cy7; Invitrogen), CD3 (APC-CY7; BioLegend), CD4 (AF700; BD Biosciences), CD2 (BV605; BD Biosciences), CD32 (PE; BioLegend), CD45RA (BV510; BioLegend), CCR7 (PE Dazzle; BioLegend), CD27 (FITC; BD Biosciences), CD95 (PE-Cy5; BD Biosciences), CXCR5 (PE Dazzle; BioLegend), PD-1 (BV421; BioLegend), HLA-DR (BV786; BD Biosciences), CD38 (BV711; BioLegend), CD20 (BV570; BioLegend), and CD69 APC; BD Biosciences). We ran the assays on an LSR Fortessa device (BD Biosciences), and data were analyzed using FlowJo software v10. Cutoffs for continuous markers were based on fluorescence-minus-one (FMO) controls (Fig. S6).
Total HIV-1 DNA quantification.
Total HIV-1 DNA was measured in cell lysates of each cell subpopulation isolated using droplet digital PCR (ddPCR), as previously described (33). Briefly, the 5′ long terminal repeat (5′LTR) region or Gag was amplified, and the RPP30 housekeeping gene was quantified in parallel to normalize sample input. Raw ddPCR data were analyzed using the QX100 droplet reader and QuantaSoft v1.6 software (Bio-Rad).
Undetectable samples are expressed as the limit of detection, which varies between samples depending on cell input. PBMCs from HIV-negative donors were used as negative controls and assayed in each plate to set the positive/negative threshold for ddPCR analysis.
RNA FISH/flow assay of single cells expressing HIV-1 RNA transcripts.
PBMCs from 10 ART-treated HIV-infected patients were obtained using Ficoll Hypaque, and CD4+ T cells were isolated by negative selection using magnetic beads (MagniSort human CD4+ T cell enrichment; eBioscience). Then, cells underwent the RNA FISH/flow protocol for the detection of HIV-1 mRNA transcripts following the manufacturer’s instructions (Human PrimeFlow RNA assay; eBioscience), with some modifications, as previously described (12). Briefly, freshly isolated CD4+ T cells were stained with antibodies against cell surface markers and viability dye. Cells were then fixed, permeabilized, and intracellularly stained. After an additional fixation step, cells were prepared for 3 h with hybridization probes at 40 ± 1°C using a high-sensitivity target-specific set of 50 probes spanning the whole Gag-Pol HIV mRNA sequence (bases 1165 to 4402 of the HIV-1HXB2 consensus genome). Subsequently, cells underwent various amplification steps (sequential 2-h incubations at 40°C). Finally, multiple-label probes were hybridized with the specific amplifiers (1 h at 40°C), and samples were run on an LSR Fortessa 4-laser flow cytometer (Becton, Dickinson).
In these experiments, isolated CD4+ T cells were stained with 2 different antibody panels (Fig. S3). The first panel comprised the following antibodies for cell surface staining: CD3 (AF700; BioLegend), CCR7 (PE-CF594; BD Biosciences), CD45RO (BV605; BioLegend), CD27 (FITC; BD Biosciences), CD95 (PE-Cy5; BD Biosciences), CD20 (BV786; BD Biosciences), CD32 (PE-Cy7; BioLegend), CD69 (PE; BD Biosciences), HLA-DR (BV711; BD Biosciences). Ki-67 (BV510; BD Biosciences) was used for intracellular staining. The second panel comprised the following antibodies: CD3 (AF700; BioLegend), CD45RO (BV605; BioLegend), CD2 (PE-Cy5; BD Biosciences), PD-1 (PE-Cy7; BD Biosciences), CXCR5 (AF488; BD Biosciences), CD69 (PE; BD Biosciences), HLA-DR (BV711; BD Biosciences), and Ki-67 (BV510; BD Biosciences). The expression of HIV-1 RNA transcripts was analyzed with target-specific AF647-labeled probes, and cell viability was determined using a violet viability dye for flow cytometry (live/dead fixable violet dead cell stain kit; Invitrogen). Negative controls were included in all experiments with cells from non-HIV-1-infected donors. The normalized percentage of HIV-1 RNA expression was calculated for each subpopulation by subtracting the mean value of the negative control from the signal obtained with the analyzed sample.
A representative example of the gating strategy used for analyses in Fig. 2C. Download FIG S3, TIF file, 0.3 MB (315.2KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Intact proviral DNA assay.
Intact provirus in each subpopulation was assessed by intact provirus DNA assay (IPDA) when sample was available. IPDA is based on a duplex ddPCR targeting 2 regions in the viral genome that are present in most intact proviruses, namely, the HIV-1 packaging signal (Ψ) and the Rev responsive element in envelope (Env) (21). RPP30 was targeted to determine the cell number and DNA shearing. The number of intact proviruses was determined based on double positives for Ψ and Env.
Residual viremia by ultrasensitive viral load assay.
Residual viremia (HIV-1 RNA) was measured by ultracentrifugation in 9 ml of plasma at 170,000 × g at 4°C for 30 min, followed by viral RNA extraction using the m2000sp Abbot RealTime HIV-1 (34) assay device and laboratory-defined software applications from the instrument. HIV-1 RNA copies in the low range were determined based on an in-house calibration curve set of standards (range, 10 to 103 absolute copies) (35), which had previously been validated using a standard HIV-1 DNA control from the WHO in the range of 0.5 to 128 copies per mL. The limit of detection was calculated relative to the plasma volume used in each sample (down to 0.5 HIV-1 RNA copies/mL when using 9 mL of plasma).
Cytokine and chemokine plasma levels.
Concentrations of the cytokines and chemokines IFN-γ, IL-12p70, IL-17a/CTLA8, IL-1b, IL-2, IL-7, IP10/CXCL10, MCP1/CCL2, MIP1β/CCL4, and TNF were quantified in 25 μL of plasma using a bead-based multiplex immunoassay (Milliplex; Merck Millipore) according to the manufacturer’s recommendations. Measurements were performed using a Luminex 100 instrument (Luminex Corp.) and analyzed using a standard curve for each cytokine.
Statistical analysis.
Data are summarized as box and whisker plots indicating the median, interquartile range, and minimum and maximum values. Spearman correlation coefficients were calculated to analyze correlations between study variables. Differences were tested for statistical significance using the Wilcoxon rank sum test or Friedman’s test followed by the Benjamini-Hochberg false-discovery rate adjustment for multiple comparisons. The analyses were performed with R (v3.4) and GraphPad (v8.4).
ACKNOWLEDGMENTS
We thank the IGTP Cytometry Core Facility and staff (M.A. Fernández and G. Requena) for their contribution to this publication. We also thank the CERCA Programme/Generalitat de Catalunya 2017 SGR 252.
This work was sponsored in part by Grifols and by Gilead (GLD17/00204). The funding organizations had no input in the design of the study or in the collection, analyses, and interpretation of the data. Similarly, they played no role in the writing of the manuscript or in the decision to submit the study for publication. J.M.-P. is supported by grant PID2019-109870RB-I00 from the Spanish Ministry of Science and Innovation. J.M.-P. and M.J.B. are supported by the RETIC RD16/0025/0041 and RD16/0025/0007 (cofunded by the European Regional Development Fund/European Social Fund). M.J.B. is supported by grant RTI2018-101082-B-I00 (MINECO/FEDER) and by the Miguel Servet program funded by the Spanish Health Institute Carlos III (CP17/00179). C.G. was supported by the Ph.D. fellowship of the Spanish Ministry of Education, Culture, and Sport (FPU15/03698).
We declare no competing interests related to this work.
J.M.-P. and M.J.B. conceived and designed the study; C.G. and J.G.-E. performed the experiments; C.G., V.U., and J.G.-E. analyzed the data; C.G., J.G.-E., V.U., B.C., V.F., J.M.-P., and M.J.B. interpreted the results. C.G. and J.M.-P. wrote the paper. All the authors read, reviewed, and approved the final version of the paper.
Contributor Information
Maria José Buzón, Email: mariajose.buzon@vhir.org.
Javier Martinez-Picado, Email: jmpicado@irsicaixa.es.
Nancie M. Archin, University of North Carolina at Chapel Hill
Vinayaka R. Prasad, Albert Einstein College of Medicine
REFERENCES
- 1.Chun TW, Davey RT, Engel D, Lane HC, Fauci AS. 1999. Re-emergence of HIV after stopping therapy. Nature 401:874–875. doi: 10.1038/44755. [DOI] [PubMed] [Google Scholar]
- 2.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med 5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 3.Margolis DM. 2017. Towards an HIV cure: a view of a developing field. J Infect Dis 215:S109–S110. doi: 10.1093/infdis/jiw633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sékaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buzon MJ, Sun H, Li C, Shaw A, Seiss K, Ouyang Z, Martin-Gayo E, Leng J, Henrich TJ, Li JZ, Pereyra F, Zurakowski R, Walker BD, Rosenberg ES, Yu XG, Lichterfeld M. 2014. HIV-1 persistence in CD4+ T cells with stem cell-like properties. Nat Med 20:139–142. doi: 10.1038/nm.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaafoura S, de de Herve MGG, Hernandez-Vargas EA, Hendel-Chavez H, Abdoh M, Mateo MC, Krzysiek R, Merad M, Seng R, Tardieu M, Delfraissy JF, Goujard C, Taoufik Y. 2014. Progressive contraction of the latent HIV reservoir around a core of less-differentiated CD4+ memory T cells. Nat Commun 5:1–8. doi: 10.1038/ncomms6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perreau M, Savoye AL, De Crignis E, Corpataux J-M, Cubas R, Haddad EK, De Leval L, Graziosi C, Pantaleo G. 2013. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 210:143–156. doi: 10.1084/jem.20121932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iglesias-Ussel M, Vandergeeten C, Marchionni L, Chomont N, Romerio F. 2013. High levels of CD2 expression identify HIV-1 latently infected resting memory CD4+ T cells in virally suppressed subjects. J Virol 87:9148–9158. doi: 10.1128/JVI.01297-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Descours B, Petitjean G, López-Zaragoza JL, Bruel T, Raffel R, Psomas C, Reynes J, Lacabaratz C, Levy Y, Schwartz O, Lelievre JD, Benkirane M. 2017. CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature 543:564–567. doi: 10.1038/nature21710. [DOI] [PubMed] [Google Scholar]
- 10.Darcis G, Kootstra NA, Hooibrink B, van Montfort T, Maurer I, Groen K, Jurriaans S, Bakker M, van Lint C, Berkhout B, Pasternak AO. 2020. CD32+CD4+ T cells are highly enriched for HIV DNA and can support transcriptional latency. Cell Rep 30:2284–2296.e3. doi: 10.1016/j.celrep.2020.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Serra-Peinado C, Grau-Expósito J, Luque-Ballesteros L, Astorga-Gamaza A, Navarro J, Gallego-Rodriguez J, Martin M, Curran A, Burgos J, Ribera E, Raventós B, Willekens R, Torrella A, Planas B, Badía R, Garcia F, Castellví J, Genescà M, Falcó V, Buzon MJ. 2019. Expression of CD20 after viral reactivation renders HIV-reservoir cells susceptible to rituximab. Nat Commun 10:1–15. doi: 10.1038/s41467-019-11556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grau-Expósito J, Serra-Peinado C, Miguel L, Navarro J, Curran A, Burgos J, Ocaña I, Ribera E, Torrella A, Planas B, Badía R, Castellví J, Falcó V, Crespo M, Buzon MJ. 2017. A novel single-cell FISH-flow assay identifies effector memory CD4+ T cells as a major niche for HIV-1 transcription in HIV-infected patients. mBio 8:e00876-17. doi: 10.1128/mBio.00876-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans VA, van der Sluis RM, Solomon A, Dantanarayana A, McNeil C, Garsia R, Palmer S, Fromentin R, Chomont N, Sékaly R-P, Cameron PU, Lewin SR. 2018. Programmed cell death-1 contributes to the establishment and maintenance of HIV-1 latency. AIDS 32:1491–1497. doi: 10.1097/QAD.0000000000001849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fromentin R, DaFonseca S, Costiniuk CT, El-Far M, Procopio FA, Hecht FM, Hoh R, Deeks SG, Hazuda DJ, Lewin SR, Routy JP, Sékaly RP, Chomont N. 2019. PD-1 blockade potentiates HIV latency reversal ex vivo in CD4 + T cells from ART-suppressed individuals. Nat Commun 10:814. doi: 10.1038/s41467-019-08798-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deeks SG, Autran B, Berkhout B, Benkirane M, Cairns S, Chomont N, Chun T-W, Churchill M, Di Mascio M, Katlama C, Lafeuillade A, Landay A, Lederman M, Lewin SR, Maldarelli F, Margolis D, Markowitz M, Martinez-Picado J, Mullins JI, Mellors J, Moreno S, O’Doherty U, Palmer S, Penicaud M-C, Peterlin M, Poli G, Routy J-P, Rouzioux C, Silvestri G, Stevenson M, Telenti A, Van Lint C, Verdin E, Woolfrey A, Zaia J, Barré-Sinoussi F, International AIDS Society Scientific Working Group on HIV Cure . 2012. Towards an HIV cure: a global scientific strategy. Nat Rev Immunol 12:607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossman Z, Singh NJ, Simonetti FR, Lederman MM, Douek DC, Deeks SG, Kawabe T, Bocharov G, Meier-Schellersheim M, Alon H, Chomont N, Grossman Z, Sousa AE, Margolis L, Maldarelli F. 2020. ‘Rinse and replace’: boosting T cell turnover to reduce HIV-1 reservoirs. Trends Immunol 41:466–480. doi: 10.1016/j.it.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Michie CA, McLean A, Alcock C, Beverley PCL. 1992. Lifespan of human lymphocyte subsets defined by CD45 isoforms. Nature 360:264–265. doi: 10.1038/360264a0. [DOI] [PubMed] [Google Scholar]
- 18.Hellerstein MK, Hoh RA, Hanley MB, Cesar D, Lee D, Neese RA, McCune JM. 2003. Subpopulations of long-lived and short-lived T cells in advanced HIV-1 infection. J Clin Invest 112:956–966. doi: 10.1172/JCI17533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu CL, Pai JA, Nogueira L, Mendoza P, Gruell H, Oliveira TY, Barton J, Lorenzi JCC, Cohen YZ, Cohn LB, Klein F, Caskey M, Nussenzweig MC, Jankovic M. 2018. Relationship between intact HIV-1 proviruses in circulating CD4+ T cells and rebound viruses emerging during treatment interruption. Proc Natl Acad Sci USA 115:E11341–E11348. doi: 10.1073/pnas.1813512115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon KJ, Timmons AE, Sengupta S, Simonetti FR, Zhang H, Hoh R, Deeks SG, Siliciano JD, Siliciano RF. 2020. Different human resting memory CD4+ T cell subsets show similar low inducibility of latent HIV-1 proviruses. Sci Transl Med 12:6795. doi: 10.1126/scitranslmed.aax6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, Bertagnolli LN, Capoferri AA, Kufera JT, Timmons A, Nobles C, Gregg J, Wada N, Ho YC, Zhang H, Margolick JB, Blankson JN, Deeks SG, Bushman FD, Siliciano JD, Laird GM, Siliciano RF. 2019. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566:120–125. doi: 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Lint C, Bouchat S, Marcello A. 2013. HIV-1 transcription and latency: an update. Retrovirology 10:67. doi: 10.1186/1742-4690-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grau-Expósito J, Luque-Ballesteros L, Navarro J, Curran A, Burgos J, Ribera E, Torrella A, Planas B, Badía R, Martin-Castillo M, Fernández-Sojo J, Genescà M, Falcó V, Buzon MJ. 2019. Latency reversal agents affect differently the latent reservoir present in distinct CD4+ T subpopulations. PLoS Pathog 15:e1007991. doi: 10.1371/journal.ppat.1007991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fromentin R, Bakeman W, Lawani MB, Khoury G, Hartogensis W, DaFonseca S, Killian M, Epling L, Hoh R, Sinclair E, Hecht FM, Bacchetti P, Deeks SG, Lewin SR, Sékaly RP, Chomont N. 2016. CD4+ T cells expressing PD-1, TIGIT and LAG-3 contribute to HIV persistence during ART. PLoS Pathog 12:e1005761. doi: 10.1371/journal.ppat.1005761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abdel-Mohsen M, Kuri-Cervantes L, Grau-Exposito J, Spivak AM, Nell RA, Tomescu C, Vadrevu SK, Giron LB, Serra-Peinado C, Genescà M, Castellví J, Wu G, Del Rio Estrada PM, González-Navarro M, Lynn K, King CT, Vemula S, Cox K, Wan Y, Li Q, Mounzer K, Kostman J, Frank I, Paiardini M, Hazuda D, Reyes-Terán G, Richman D, Howell B, Tebas P, Martinez-Picado J, Planelles V, Buzon MJ, Betts MR, Montaner LJ. 2018. CD32 is expressed on cells with transcriptionally active HIV but does not enrich for HIV DNA in resting T cells. Sci Transl Med 10:6759. doi: 10.1126/scitranslmed.aar6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badia R, Ballana E, Castellví M, García-Vidal E, Pujantell M, Clotet B, Prado JG, Puig J, Martínez MA, Riveira-Muñoz E, Esté JA. 2018. CD32 expression is associated to T-cell activation and is not a marker of the HIV-1 reservoir. Nat Commun 9:1–10. doi: 10.1038/s41467-018-05157-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maldarelli F, Wu X, Su L, Simonetti FR, Shao W, Hill S, Spindler J, Ferris AL, Mellors JW, Kearney MF, Coffin JM, Hughes SH. 2014. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345:179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, Wells D, Su L, Luke BT, Halvas EK, Besson G, Penrose KJ, Yang Z, Kwan RW, Van Waes C, Uldrick T, Citrin DE, Kovacs J, Polis MA, Rehm CA, Gorelick R, Piatak M, Keele BF, Kearney MF, Coffin JM, Hughes SH, Mellors JW, Maldarelli F. 2016. Clonally expanded CD4 + T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci USA 113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ndhlovu LC, Chapman JM, Jha AR, Snyder-Cappione JE, Pagán M, Leal FE, Boland BS, Norris PJ, Rosenberg MG, Nixon DF. 2008. Suppression of HIV-1 plasma viral load below detection preserves IL-17 producing T cells in HIV-1 infection. AIDS 22:990–992. doi: 10.1097/QAD.0b013e3282ff884e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Planas D, Routy JP, Ancuta P. 2019. New Th17-specific therapeutic strategies for HIV remission. Curr Opin HIV AIDS 14:85–92. doi: 10.1097/COH.0000000000000522. [DOI] [PubMed] [Google Scholar]
- 31.Leal FE, Ndhlovu LC, Hasenkrug AM, Bruno FR, Carvalho KI, Wynn-Williams H, Neto WK, Sanabani SS, Segurado AC, Nixon DF, Kallas EG. 2013. Expansion in CD39+ CD4+ immunoregulatory T cells and rarity of Th17 cells in HTLV-1 infected patients is associated with neurological complications. PLoS Negl Trop Dis 7:e2028. doi: 10.1371/journal.pntd.0002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neidleman J, Luo X, Frouard J, Xie G, Hsiao F, Ma T, Morcilla V, Lee A, Telwatte S, Thomas R, Tamaki W, Wheeler B, Hoh R, Somsouk M, Vohra P, Milush J, James KS, Archin NM, Hunt PW, Deeks SG, Yukl SA, Palmer S, Greene WC, Roan NR. 2020. Phenotypic analysis of the unstimulated in vivo HIV CD4 T cell reservoir. Elife 9:e60933. doi: 10.7554/eLife.60933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morón-López S, Puertas MC, Gálvez C, Navarro J, Carrasco A, Esteve M, Manyé J, Crespo M, Salgado M, Martinez-Picado J. 2017. Sensitive quantification of the HIV-1 reservoir in gut-associated lymphoid tissue. PLoS One 12:e0175899. doi: 10.1371/journal.pone.0175899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puertas MC, Salgado M, Morón-López S, Ouchi D, Muñoz-Moreno JA, Moltó J, Clotet B, Martinez-Picado J. 2014. Effect of lithium on HIV-1 expression and proviral reservoir size in the CD4+ T cells of antiretroviral therapy suppressed patients. AIDS 28:2157–2159. doi: 10.1097/QAD.0000000000000374. [DOI] [PubMed] [Google Scholar]
- 35.Amendola A, Bloisi M, Marsella P, Sabatini R, Bibbò A, Angeletti C, Capobianchi MR. 2011. Standardization and performance evaluation of “modified” and “ultrasensitive” versions of the Abbott RealTime HIV-1 assay, adapted to quantify minimal residual viremia. J Clin Virol 52:17–22. doi: 10.1016/j.jcv.2011.04.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A representative gating strategy used for cell sorting of the cell subpopulations. Three separate cell sorters were made in order to be able to sort the overlapping subpopulations. A common gate strategy was used where PBMCs were acquired and gated according to (i) forward scatter area (FSC-A) and forward scatter height (FSC-H) to select singlets, (ii) side scatter area (SSC-A) and FSC-A to determine cell morphology, (iii) CD3 and viability staining to select living T cells, and (iv) CD4 T cells from viable cells to gate all the other cell subpopulations. In sorter 1, first, a fraction of rmTCD2high were sorted because of overlap with all the other cell subpopulations. Then, TN, TCM, TEM, and TTM were discriminated with CD45RA, CCR7, and CD27 markers as described in Materials and Methods. In sorter 2, TSCM and cTFH were sorted from CD4+ T cells. Finally, TCD32 and TCD20 were sorted in sorter 3. Download FIG S1, TIF file, 0.8 MB (844.7KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Contribution of memory cell subpopulations and HIV/RNA ratio. (A) Contribution of the TN, TSCM, TCM, TTM, and TEM cell subpopulations to the pool of HIV-infected cells in 14 HIV-infected individuals. Differences were tested for statistical significance using Friedman’s test followed by the false-discovery rate test for multiple comparisons. (B) Ratio between HIV-1 RNA and HIV-1 DNA. Each symbol represents a different individual. RNA/DNA ratios for TN, TSCM, and rmTCD2high were not represented due to the high number of undetectable values for intracellular HIV-1 RNA measurements. Box and whisker plots indicate the median, interquartile range, and minimum and maximum values. Download FIG S2, TIF file, 0.2 MB (235.9KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of the correlation between total HIV-1 DNA, clinical parameters, HIV-1 expression, and cytokines/chemokines plasma levels. Download Table S1, DOCX file, 0.02 MB (22.3KB, docx) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Representative plots of HIV-1 RNA expressing cells. Levels of intracellular HIV-1 RNA in TTM cells from an uninfected donor, which is used to set up the limit of detection of the assay, and two individuals from the study. Download FIG S4, TIF file, 0.7 MB (738.7KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of levels of HIV-1 RNA expressing cells between HIV-1– and HIV-1+ individuals. Differences were tested for statistical significance using the Wilcoxon matched-pair rank test. Download FIG S5, TIF file, 0.5 MB (483.6KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Representative example of the gating strategy used for analyses in Fig. S7A to E. PBMCs were acquired and gated according to (i) time to ensure homogeneous acquisition, (ii) SSC-A and FSC-A to determine cell morphology, (iii) FSC-A and FSC-H to select singlets, (iv) CD3 and viability staining to select living T cells, and (v) CD4 T cells from viable cells to gate all the other cell subpopulations. Then CD69, CD38, HLA-DR, PD-1, and CD38+HLADR+ were gated on every cell subpopulation based on FMOs. The graph shows a representative gate strategy for CD4+ T cells, but the same was made for TN, TSCM, TCM, TTM, TEM, cTFH, TCD20, TCD32, and rmTCD2high that were gated as in Fig. S1. Download FIG S6, TIF file, 0.7 MB (745.7KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Analysis of activation, proliferation, and exhaustion in CD4+ T-cell subpopulations. (A to C) Percentage of activation levels based on (A) CD69+, (B) HLA-DR+, and (C) double positive CD38+ and HLA-DR+ cells for each CD4+ T-cell subpopulation. (D) Percentage of cell proliferation measured using Ki-67+ cells and (E) percentage of PD-1+ cells. Each symbol represents 1 individual. (A to E) Matrices depict P values of the differences between paired CD4+ T-cell subpopulations. Differences were tested for statistical significance using the Wilcoxon rank sum test followed by the false-discovery rate test for multiple comparisons. Box and whisker plots indicate the median, interquartile range, and minimum and maximum values. Download FIG S7, TIF file, 0.6 MB (621KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A representative example of the gating strategy used for analyses in Fig. 2C. Download FIG S3, TIF file, 0.3 MB (315.2KB, tif) .
Copyright © 2021 Gálvez et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.