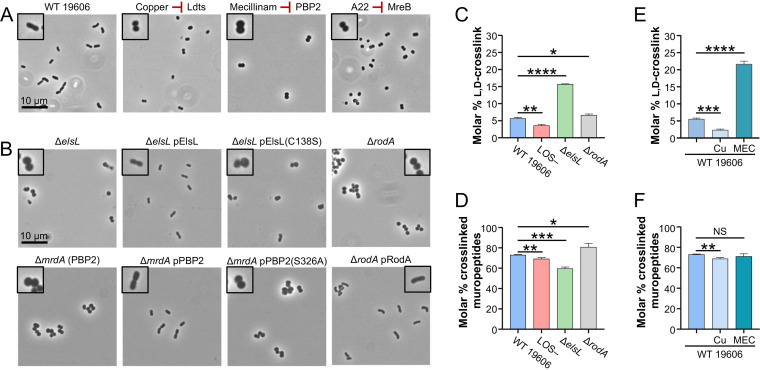
FIG 2.
A. baumannii elongasome depends on the recycling subunit ElsL. (A) Inhibitors of ElsL [copper (II), 3.12 mM], PBP2 (mecillinam, 250 μg/mL), and MreB (A22, 15.6 μg/mL) disrupt activity of the A. baumannii 19606 elongasome and result in coccoid cells. (B) Disruption of ElsL and PBP2 (encoded by mrdA) by deletion or catalytic mutations disrupt activity of the elongasome, indicating that catalytic activity of both proteins is required for elongasome activity. Deletion and complementation of rodA were performed for comparison. IPTG was supplemented at 100 μM for pElsL, 25 μM for pPBP2, and 100 μM for pRodA. Phase-contrast microscopy was imaged at ×100 magnification, and all field of views were resized identically with a 10-μm scale bar on the first image of each panel. A single cell from each field of view is highlighted with a 2× magnified inset. (C to E) Assessment of l,d-cross-links (C and E) and overall cross-linking (D and F) detected by muropeptide analysis of 19606 mutants (C and D) or wild-type 19606 treated with chemical inhibitors (E and F), copper (II) (indicated as Cu) and mecillinam (indicated as MEC). Data represent mean values with the standard deviation from three independent cultures. Significance was calculated using Student’s unpaired t test. NS indicates not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.