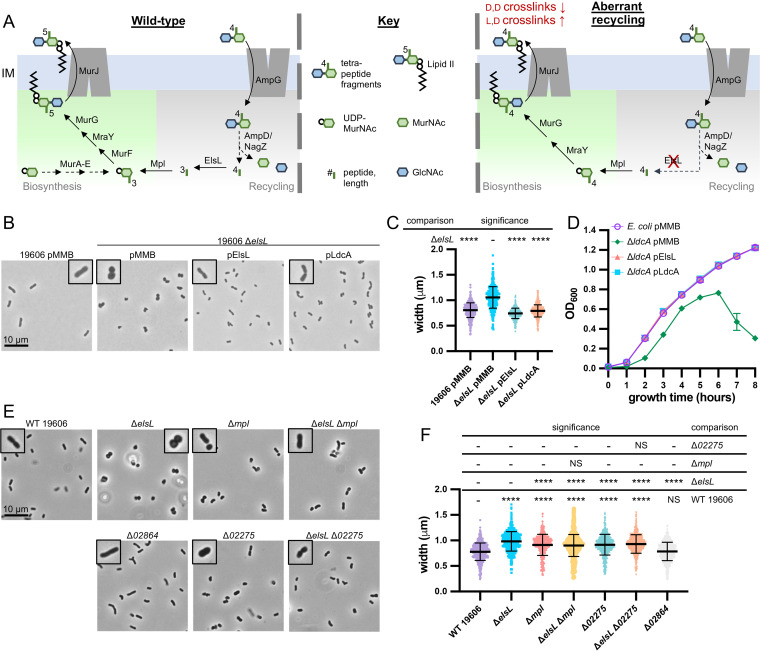
FIG 3.
Genetic evidence supports that A. baumannii ElsL is an l,d-carboxypeptidase. (A) Depiction of peptidoglycan recycling pathway for tetrapeptide fragments released during peptidoglycan remodeling (left) and the aberrant recycling when an l,d-carboxypeptidase, like ElsL, is disrupted (right). Briefly, the permease AmpG imports tetrapeptide peptidoglycan fragments into the cytoplasm. Fragments are then degraded by a β-N-acetylglucosaminidase, NagZ; amidase, AmpD; and l,d-carboxypeptidase, ElsL, into individual sugars N-acetylglucosamine (GlcNAc; blue hexagon) and N-acetylmuramic acid (MurNAc; green hexagons), tripeptides, and d-alanine. Tripeptides can feed directly back into peptidoglycan synthesis through Mpl which ligates them to a fresh UDP-activated MurNAc, and they enter the biosynthesis pathway upstream of the d-ala-d-ala ligase MurF. In the absence of an l,d-carboxypeptidase (right), a tetrapeptide is instead released and ligated to UDP-MurNAc. MurF is bypassed because the fourth amino acid d-ala is already present and a tetrapeptide peptidoglycan precursor is produced. Tetrapeptide peptidoglycan precursors are unable to be utilized by d,d-transpeptidases but can be utilized by l,d-transpeptidases. (B and C) Phase-contrast microscopy of 19606 ΔelsL mutants with empty (pMMB) or complementation plasmids encoding ElsL from A. baumannii or the known l,d-carboxypeptidase LdcA from E. coli. (D) Averages of quadruplicate growth curves of E. coli ΔldcA mutants with empty (pMMB) or complementation plasmids encoding ElsL from A. baumannii or LdcA, from E. coli. (E and F) Phase-contrast microscopy of 19606 mutants that disrupt peptidoglycan recycling genes elsL and mpl and two possible homologs of ampG, namely, 02275 and 02864. Δmpl and Δ02275 mutants have the same morphological changes that are dominant to the cell rounding of the ΔelsL mutant, indicating their shared role in peptidoglycan recycling. Microscopy (B and E) imaged at ×100 magnification and all fields of view were resized identically with a 10-μm scale bar on the first image of each panel. A single cell from the field of view is highlighted with a 2× magnified inset. Cell measurements (C and F) were performed on ≥400 cells with MicrobeJ and assessed for significant differences as indicated in the Materials and Methods; NS indicates not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.