Abstract
Several factors that mediate activation by nuclear receptors also modify the chemical and structural composition of chromatin. Prominent in this diverse group is the steroid receptor coactivator 1 (SRC-1) family, which interact with agonist-bound nuclear receptors, thereby coupling them to multifunctional transcriptional coregulators such as CREB-binding protein (CBP), p300, and PCAF, all of which have potent histone acetyltransferase activity. Additionally factors including the Brahma-related gene 1 (BRG-1) that are involved in the structural remodeling of chromatin also mediate hormone-dependent transcriptional activation by nuclear receptors. Here, we provide evidence that these two distinct mechanisms of coactivation may operate in a collaborative manner. We demonstrate that transcriptional activation by the estrogen receptor (ER) requires functional BRG-1 and that the coactivation of estrogen signaling by either SRC-1 or CBP is BRG-1 dependent. We find that in response to estrogen, ER recruits BRG-1, thereby targeting BRG-1 to the promoters of estrogen-responsive genes in a manner that occurs simultaneous to histone acetylation. Finally, we demonstrate that BRG-1-mediated coactivation of ER signaling is regulated by the state of histone acetylation within a cell. Inhibition of histone deacetylation by trichostatin A dramatically increases BRG-1-mediated coactivation of ER signaling, and this increase is reversed by overexpression of histone deacetylase 1. These studies support a critical role for BRG-1 in ER action in which estrogen stimulates an ER–BRG-1 association coupling BRG-1 to regions of chromatin at the sites of estrogen-responsive promoters and promotes the activity of other recruited factors that alter the acetylation state of chromatin.
Precise regulation of gene expression underlies the ability of a cell to control growth and to acquire and execute physiologic functions. Broad arrays of cellular signals are transduced to the nucleus, where many act on transcription factors. These diverse regulatory signals must be integrated into smaller subsets that can be transmitted to targets that modulate the basal transcription machinery. One such target is chromatin, and there exists abundant evidence that the structure and chemical composition of chromatin directly affect gene expression (35). The primary structural components of chromatin, the histones, are enzymatically acetylated, and this acetylation results in a reduced affinity for DNA and enhanced binding affinity for certain transcriptional coregulators (7). Chromatin structure is also altered via the ATP-dependent disruption of nucleosomes by large multiprotein chromatin remodeling complexes (3). One such complex, the Swi/Snf complex, is well conserved through evolution and functions as a global regulator of transcription (37). These and other mechanisms account for the link between the chemical and structural modification of chromatin and transcriptional activation by members of the nuclear receptor superfamily.
The nuclear receptor superfamily is a large family of ligand-activated transcription factors that exert control over networks of genes that regulate various aspects of cell biology. By binding to sequence-specific response elements located in the regulatory regions of target genes, they exert both positive and negative control over the rates of transcription. Their mode of activation has made nuclear receptors an attractive system in which to study the mechanisms by which transcriptional coregulation occurs. In the absence of hormone, many receptors actively repress transcription of via direct interactions with corepressors such as NCoR (22), SMRT (5), and SunCoR (48). Upon hormone binding, these corepressor complexes dissociate and the agonist-bound receptors interact with distinct multiprotein coactivator complexes that contribute to the transmission of activating signals to the general transcription machinery. While the mechanisms by which coactivation signals are transmitted are not completely understood, several studies have implicated aspects of general transcription factor function (10, 38) as well as the structure and chemical composition of chromatin (26, 32). These studies are also consistent with reports that the rate of assembly of the general transcription machinery is directly related to chromatin structure (40).
A variety of putative nuclear receptor coactivators have been identified based primarily on their ability to interact with a nuclear receptor in a hormone-dependent manner. Among these is the steroid receptor coactivator 1 (SRC-1 [also called NCoA-1]) and its related factors TIF2 (also called GRIP-1 or NCoA-2) and RAC-3 (also called AIB1, PCIP, ACTR, or TRAM) (4, 21, 33, 43, 44). These factors physically interact with members of the receptor superfamily and have been shown in functional assays to enhance their transcriptional activity. Insight into one mechanism by which the SRC-1 family potentiates nuclear receptor signaling came from the demonstration of a stable interaction between members of the SRC-1 family and the CREB-binding protein (CBP) and its homolog p300 (20, 26, 47). These multifunctional transcriptional coactivators have been proposed to modulate gene activation through direct interactions with the RNA polymerase II complex, and also via both intrinsic and associated histone acetyltransferase (HAT) activities (32). The subsequent observation of intrinsic HAT activity in SRC-1 and ACTR (4, 42) provided further evidence that one mechanism by which this complex may mediate nuclear receptor activation is through the enzymatic acetylation of histones and possibly other targets. Additionally, the demonstration that the nuclear receptor corepressors NcoR, SMRT, and SunCoR are physically associated with histone deacetylases (HDAC) provided evidence that the transcriptional repression mediated by the corepressors correlates with a reduced acetylation state. Taken together, these findings are consistent with a body of evidence that the regulation of chromatin via modulation of the acetylation state within a cell correlates with the activation and repression of nuclear receptors.
As the primary structural unit of chromatin, the nucleosome has been known to be the target of both chemical and structural modification. Several studies have shown that the acetylation of specific lysine residues within core histones results in a reduced affinity for DNA, making acetylated chromatin more accessible to transcriptional regulators (35). More recently, it has been demonstrated that the bromodomain, a domain which is well conserved in several transcriptional coactivators, exhibits high-affinity binding for acetyl-lysine (7). This finding suggests that not only does acetylation of lysine residues reduce the affinity of nucleosomes for DNA, but it also may present docking sites on the surface of the nucleosome to which bromodomain-containing factors may bind. The effects of acetylation are complemented by structural modifications of chromatin which are carried out by several distinct multiprotein chromatin remodeling complexes (29). Each of these complexes does so in a manner that is strictly ATP dependent; thus, each contains a member of the Swi-1/Snf-2 family of nuclear ATPases. The human homologs of yeast Swi-2, hBrm and hBRG-1 (hSnf-2α and hSnf-2β, respectively) (6), are crucial to the function of the Swi/Snf nucleosome remodeling complex and have been shown to interact with various nuclear receptors in a yeast-based two-hybrid assay (24). Other studies have shown that these factors can mediate transcriptional activation by several nuclear receptors (6, 30). Additionally it has been demonstrated that hBrm/BRG-1 will form a complex with the retinoblastoma gene product (Rb) and that the formation of this complex accounts for the cooperative coactivation of glucocorticoid receptor (GR) signaling by hBrm or hBRG-1 and Rb (8, 41). Consistent with their role in chromatin remodeling, two components of the hSwi/Snf complex, BRG-1 and BAF-155 (Swi-3), contribute to GR-mediated chromatin remodeling and transcriptional activation of an integrated reporter. In contrast to the stable reporter system, a transiently transfected reporter was activated by GR in a manner that was less dependent on these factors, providing functional evidence that the mechanism by which BRG-1 coactivates nuclear receptor signaling is by targeting components of chromatin (12).
In this report, we characterize the ability of BRG-1 to mediate estrogen receptor (ER) signaling. Consistent with previous reports, we find that ER-mediated transcriptional activation requires functional BRG-1 and that in a BRG-1-deficient background, neither SRC-1 nor CBP functions efficiently as a coactivator of estrogen signaling. Furthermore, we report that both SRC-1 and CBP can augment BRG-1-mediated coactivation of ER, suggesting a functional cooperation between the activities of the SRC-1–CBP complex and chromatin remodeling. We find that estrogen stimulates an association between ER and BRG-1 that is consistent with transcriptional activation. This association leads to the estrogen-dependent recruitment of BRG-1 to regions of chromatin which contain the estrogen-responsive elements (EREs) from promoters of genes which are known to be estrogen dependent and coincides with the histone acetylation of these promoters. The functional cooperativity between BRG-1 and factors, such as SRC-1 and CBP, that modulate the histone acetylation status within a cell is supported by the observation that inhibition of HDAC activity by trichostatin A (TSA) resulted in a dramatic increase in BRG-1-mediated coactivation and that this effect was potently reversed by overexpression of HDAC-1. These results suggest that two distinct chromatin modifying mechanisms, histone acetylation-deacetylation and ATP-dependent chromatin remodeling, are functionally linked and contribute cooperatively to the regulation of ER signaling.
MATERIALS AND METHODS
Cell lines and culture conditions.
The SW-13 adrenal carcinoma cell line (ATCC CCL-105) and the MCF-7 (ATCC HTB-22) mammary carcinoma were cultured in Dulbecco's modified Eagle medium (DMEM; Sigma) supplemented with 10 fetal calf serum (FCS; Sigma), l-glutamine (Gibco), and penicillin-streptomycin (pen-strep; Gibco) at 37°C and 5% CO2. SW-13 cells were grown to 80 to 90% confluence and passaged by standard trypsinization. MCF-7 cells were grown to 100% confluence and passaged by standard trypsinization.
Metabolic labeling.
For metabolic labeling experiments, MCF-7 cells were cultured to 70 to 80% confluence in 15-cm-diameter culture dishes. Cells were washed in phosphate-buffered saline (PBS) and starved for 1 h by incubation in a methionine-free DMEM. After starvation, 1 mCi of 35S-labeled methionine (NEN) was added to the methionine-free medium and the cells were incubated at 37°C for 3 h. Following removal of the labeling medium, cells were trypsinized, pelleted by centrifugation, and lysed in NET-N (20 mM Tris-Cl [pH 8.0], 1 mM EDTA, 100 mM NaCl, 0.05% NP-40) supplemented with 0.2 mM phenylmethylsulfonyl fluoride.
GST pulldown assay and reimmunoprecipitation.
Glutathione S-transferase (GST) fusions of the hormone binding domain of ER (amino acids 253 to 595) and the AF-2 deletion GST-Δ534 were expressed in Escherichia coli BL-21 cells, and crude bacterial lysates were prepared by sonication in TEDGN (50 mM Tris-Cl [pH 7.4] 1.5 mM EDTA, 1 mM dithiothreitol, [DTT] 10% [vol/vol] glycerol, 0.4 M NaCl) supplemented with 0.2 mM phenylmethylsulfonyl fluoride and 10 μg of leupeptin per ml. Lysates were cleared by centrifugation and stored at −80°C. These fusion proteins along with wild-type GST were bound to glutathione-Sepharose beads and incubated in the presence and absence of 1 μM 17β-estradiol. The resulting complexes were then used as affinity matrices to enrich for factors from a metabolically labeled MCF-7 whole-cell lysate. Retained fractions were washed in NET-N and eluted from the beads by boiling in a solution containing 50 mM Tris (pH 7.5), 1% sodium dodecyl sulfate (SDS), and 5 mM DTT. Eluted fractions were diluted to 1.4 ml with NET-N and subjected to immunoprecipitation with antibodies directed against hSRC-1 or hBRG-1. Retained fractions from this reimmunoprecipitation were washed in NET-N and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) on a 7.5% gel.
Far-Western assay.
BRG-1 and CBP were expressed as Flag-tagged proteins in a baculovirus expression system and purified using an anti-M2 affinity column according to the manufacturer's protocol. Three concentrations of each protein were resolved by SDS-PAGE on a 7.5% gel and transferred to nitrocellulose. The filter was incubated in blocking buffer (1× HBB [see below] plus 5% milk, 1 mM DTT, and 0.05% NP-40) and subjected to a denaturation/renaturation step by incubation in 1× HBB (25 mM HEPES-KOH [pH 7.7], 25 mM NaCl, 5 mM MgCl2) plus 6 M guanidine hydrochloride and 1 mM DTT followed by a series of twofold serial dilutions with 1× HBB–1 mM DTT. Following renaturation, filters were again incubated in blocking buffer supplemented with wild-type GST bacterial extract. The filter was probed with a 32P-labeled GST–hSRC-1(381-1360) fusion which was prepared by in vitro phosphorylation with bovine heart muscle kinase as previously described (19). Following extensive washing, filters were subjected to autoradiography.
Chromatin immunoprecipitation (CHIP).
MCF-7 cells were cultured under estrogen-free conditions for 3 days followed by treatment with 100 nM 17β-estradiol for 45 min. Following treatment, cells were fixed in 1% formaldehyde at room temperature. Cells were collected into a solution containing 100 mM Tris-HCl (pH 9.4) and 10 mM DTT and incubated for 15 min at 30°C and centrifuged for 5 min at 2,000 × g. Cell pellets were washed sequentially with 1 ml of ice-cold PBS, followed by buffer I (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 6.5]) and buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES [pH 6.5]). Cells were resuspended in 0.3 ml of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 1× protease inhibitor cocktail; Roche Molecular Biochemicals, Indianapolis, Ind.), sonicated, and then centrifuged for 10 min. Supernatants were collected and diluted in a solution containing 1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl (pH 8.1), and 1× protease inhibitor cocktail. The chromatin fragments were then immunocleared with 2 μg of sheared salmon sperm DNA, 20 μl of preimmune serum, and protein A-Sepharose (45 μl of 50% slurry in 10 mM Tris-HCl [pH 8.1], 1 mM EDTA) for 2 h at 4°C. Immunoprecipitation was performed for 6 to 12 h at 4°C with antibodies against hBRG-1 and acetylated histone H3 (Upstate Biotechnology). Following precipitation, 45 μl of protein A-Sepharose and 2 μg of salmon sperm DNA were added, and incubation was continued for 1 h. Sepharose beads were then collected and washed sequentially for 10 min each time in TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl), TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), and buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]). Beads were then washed three times with TE buffer and extracted three times with 1% SDS–0.1 M NaHCO3. Elutes were pooled and heated at 65°C for 6 h or overnight to reverse the formaldehyde cross-linking. DNA fragments were purified with a purification kit (QIAquick spin kit; Qiagen, Valencia, Calif.) and amplified with PCR.
Transient transfection.
SW-13 cells were plated at 50,000 cells per well in 24-well dishes and grown as above for 24 h to allow for attachment. Cells were washed twice with 37°C PBS and refed with phenol-free DMEM supplemented with 10% charcoal-stripped FCS, pen-strep, and l-glutamine. Four to six hours after the refeeding, cells were transfected with 10 ng of ERE2-tk-luc (see Results) and the indicated combinations of 20 ng of pcDNA 3.1 hER, 20 ng of pcDNA hER-Δ534, 150 ng of pBJ5-hBRG-1, 150 ng of pBJ5 hBRG-1 (K785R), 150 ng of pcDNA-hSRC-1, 150 ng of pRSV-mCBP, 100 ng of pcDNA-HDAC-1, and 10 ng of tk-lacZ construct. Transfections were carried out using FuGene (Roche Biochemicals) according to the manufacturer's protocol. At 20 h posttransfection, the medium was aspirated and replaced with phenol-free DMEM plus 10% charcoal-stripped FCS, pen-strep, and l-glutamine supplemented with 10 nM estradiol or ethanol (vehicle control). At 24 h after estradiol treatment, cells were lysed in NET-N and assayed for luciferase and β-galactosidase activities. All data are expressed as the fold of induction of the ratio of luciferase to β-galactosidase activity. Experiments were performed in triplicate, and error bars represent the standard error of the mean.
RESULTS
Estrogen signaling requires functional BRG-1.
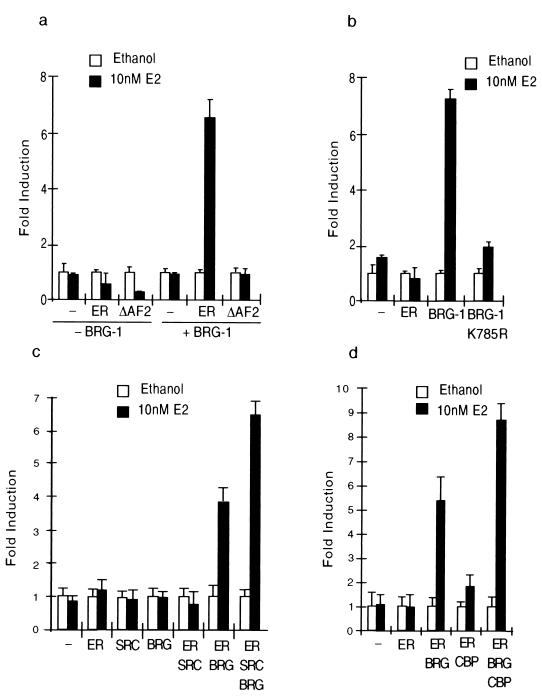
Previous studies showed that BRG-1 was capable of mediating transcriptional activation by the ER and other members of the nuclear receptor superfamily (6, 30). To further characterize this potentiation, transient transfections were carried out in the BRG-1- and Brm-deficient adrenal carcinoma cell line SW-13 (30) and an estrogen-responsive reporter system. Using this system, it was observed that in the absence of exogenous BRG-1, estrogen was incapable of stimulating a transcriptional response from a reporter gene containing tandem EREs fused to the minimal herpes simplex virus thymidine kinase promoter and luciferase (ERE2-tk-luc). Additionally we observed that overexpression of ER was insufficient to confer a transcriptional response, suggesting a deficiency in one or more components of the coactivation complex. Under these conditions, overexpression of ER and BRG-1 conferred a sevenfold transcriptional response to 10 nM 17β-estradiol (Fig. 1a). This transcriptional response requires an intact AF-2 domain, indicating that structural determinants of BRG-1-mediated coactivation overlap with those of other known coactivators (Fig. 1a). The observation that BRG-1 was required for ER-mediated transcriptional activation coupled to studies linking transcriptional activation to chromatin modifications suggested that the ATP-dependent chromatin remodeling activity of BRG-1 may contribute to the potentiation of ER activity. To test this, we transfected the point mutation BRG-1(K785R), which fails to bind to ATP, rendering it incapable of remodeling chromatin (27). This mutation resulted in the loss of BRG-1-mediated coactivation of estrogen signaling (Fig. 1b), which suggests that the chromatin remodeling activity of BRG-1 is required for efficient coactivation of estrogen signaling. Taken together, these studies demonstrate that BRG-1 potentiates hormone and AF-2-dependent transcriptional activation by ER and does so via its ATP-dependent chromatin remodeling activity.
FIG. 1.
BRG-1 is required for ER signaling and for coactivation by SRC-1 and CBP. The BRG-1- and Brm-deficient adrenal carcinoma cell line was plated at 50,000 cells per well in 24-well dishes and transfected using FuGene according to the manufacturer's protocol. All samples were similarly processed, and data represent the fold of induction by estrogen. All data are normalized to an internal estrogen-independent reporter (tk-lacZ construct). (a) ER signaling is repressed in SW-13 cells in the absence of BRG-1. Overexpression of BRG-1 elicited a sevenfold induction in response to estrogen. This induction was dependent on hormone and an intact AF-2. (b) BRG-1-mediated coactivation of ER transcriptional activity is dependent on the ATPase activity of BRG-1. Transient transfection assays in SW-13 cells demonstrate that a point mutation in BRG-1 that abolishes ATP binding also abolishes BRG-1-mediated coactivation of ER signaling. (c) Overexpression of SRC-1 in SW-13 cells fails to coactivate ER signaling but can enhance BRG-1-mediated coactivation of estrogen signaling, suggesting that SRC-1 activity requires functional BRG-1. (c) Overexpression of CBP fails to coactivate ER signaling in SW-13 cells but can enhance BRG-1-mediated coactivation of estrogen signaling, suggesting that this activity of CBP requires functional BRG-1. All experiments were carried out in triplicate; error bars represent the standard error of the mean.
Coactivation of ER by SRC-1 and CBP requires BRG-1.
The observation that BRG-1 is required for estrogen signaling suggested that its activity may potentiate the coactivation of estrogen signaling by components of the SRC-1–CBP coactivator complex. To test this idea, transient transfection assays were performed to determine if overexpression of either SRC-1 or CBP was sufficient to activate estrogen signaling in the absence of BRG-1. In SW-13 cells, we observed that overexpression of SRC-1 was insufficient to coactivate estrogen signaling; however, in the presence of exogenous BRG-1, SRC-1 significantly augmented BRG-1-mediated coactivation of estrogen signaling (Fig. 1c). These studies suggest that BRG-1-mediated coactivation of estrogen signaling may be dramatically enhanced by SRC-1 and that the mechanisms by which SRC-1 mediates transcriptional coactivation are dependent on the actions of BRG-1. Similarly, we observed that in the absence of BRG-1, overexpression of CBP had only a modest effect on estrogen signaling and that in the presence of BRG-1, CBP enhanced coactivation to levels that were greater than those achieved by either BRG-1 or CBP alone (Fig. 1d). These studies suggest that BRG-1 activity is required for the efficient coactivation of estrogen signaling by members of the SRC-1 and CBP families of transcriptional coregulators. Additionally, it is interesting that overexpression of either SRC-1 or CBP enhanced BRG-1-mediated coactivation of ER signaling, suggesting a functional cooperativity between the contributions of SRC-1–CBP and those of BRG-1.
Recruitment of BRG-1 by ER is ligand and AF-2 dependent.
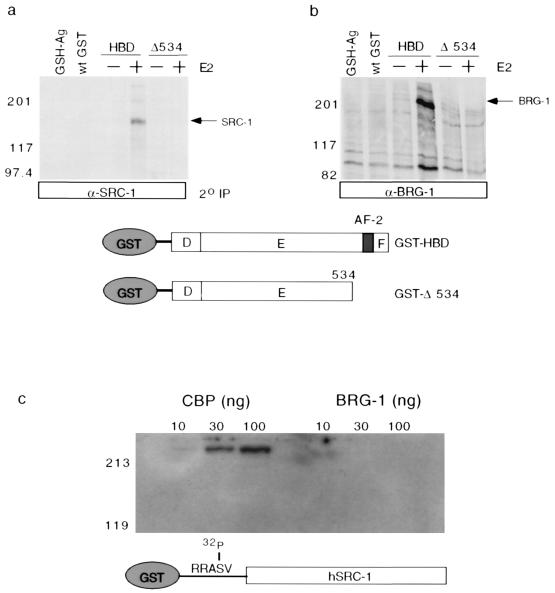
Based on the finding that BRG-1 was required for ER-mediated transcriptional activation and also on previous studies demonstrating an interaction between ER and BRG-1 in yeast-based two-hybrid assays (6), we sought to determine if BRG-1 could associate with ER in a manner consistent with transcriptional activation. To test for such an association, two versions of the ER hormone binding domain (HBD) were expressed in bacteria as GST fusion proteins. The first version represented the wild-type HBD, while the second lacked the region from amino acids 534 to 596 (ER HBD-Δ AF-2). Previously it had been shown that deletion of the ER HBD at amino acid 534 resulted in a receptor that was still capable of binding DNA, forming homodimers and binding to estrogen with high affinity, yet was transcriptionally inert. These fusion proteins were immobilized on glutathione-linked Sepharose and used as affinity matrices in the presence or absence of estrogen to enrich for interacting factors present in a metabolically labeled MCF-7. Following this enrichment, retained fractions were eluted by boiling in an SDS-containing buffer. Eluted fractions were diluted and subjected to immunoprecipitation with antibodies directed against hSRC-1 or hBRG-1. Following extensive washing, retained fractions were resolved by SDS-PAGE on a 7.5% gel and imaged by radiofluorography. Consistent with previous studies, it was observed that SRC-1 was capable of interacting with the complete HBD of ER in response to 17β-estradiol (Fig. 2a). Likewise, immunoprecipitation of BRG-1 from similarly retained fractions indicated that BRG-1 was capable of associating with the HBD of ER in a response to hormone (Fig. 2b). This association was also dependent on the AF-2, indicating that the structural requirements that support an ER–SRC-1 interaction overlap with and are sufficient to support the association between ER and BRG-1. The observation that the association between ER and BRG-1 is dependent on the presence of both hormone and an intact AF-2 domain suggests that the formation of this complex may account for BRG-1-mediated coactivation of ER signaling.
FIG. 2.
BRG-1 associates with ER in a manner that is both hormone and AF-2 dependent. (a) MCF-7 cells were metabolically labeled, and a crude whole-cell lysate was prepared. Wild-type (wt) GST and the two ER HBD fusion proteins depicted were immobilized on glutathione-linked Sepharose (GSH-Ag) and used as affinity matrices, in the presence and absence of estrogen, to enrich for factors from the MCF-7 radiolabeled lysate. Retained fractions were boiled in an SDS-containing buffer, and the eluted fraction was diluted and subjected to immunoprecipitation (IP) with a mouse monoclonal antibody raised against hSRC-1(381-1360). The retained fractions were resolved by SDS-PAGE on a 7.5% gel and detected by radiofluorography. Sizes are indicated in kilodaltons. (b) Similar GST pulldown-reimmunoprecipitation experiments were done using a rabbit polyclonal raised against hBRG-1. (c) SRC-1 interacts directly with CBP but not with BRG-1. Flag-tagged CBP and Flag-tagged BRG-1 were expressed in a baculovirus system and purified by anti-M2 affinity chromatography. Both proteins were resolved by SDS-PAGE on a 7.5% gel, transferred to a solid support, and probed with 32P-GST-SRC-1(381-1360) in a far-Western assay. RRASV, recognition sequence for heart muscle kinase.
Based on the observation of an association between ER and BRG-1 that was both ligand and AF-2 dependent, we sought to determine if these factors may be involved in a direct interaction. To test this, 35S-labeled BRG-1 was generated by coupled in vitro transcription-translation and incubated with GST-ER HBD in the presence and absence of 17β-estradiol. These complexes were captured on glutathione-linked Sepharose, and retained fractions were resolved by SDS-PAGE. Under conditions in which 35S-labeled SRC-1 would interact in a hormone-dependent manner, we observed no interaction between ER and BRG-1 (data not shown). These results indicate that the association between ER and BRG-1 is unlikely to be mediated by a direct interaction but rather through additional factors present in MCF-7 whole-cell extracts and not in the rabbit reticulocyte lysate used to generate the 35S-labeled BRG-1. Since the chemical and structural requirements for the observed ER–BRG-1 association overlapped with those of the ER–SRC-1 direct association, we hypothesized that SRC-1 may be capable of mediating the association between ER and BRG-1 via an interaction between SRC-1 and BRG-1. To test this hypothesis, far-Western studies were carried out to determine if SRC-1 and BRG-1 physically interact. Our data demonstrate that under conditions in which 32P-GST-SRC-1 binds to baculovirus-produced and affinity-purified CBP, there is no detectable interaction between SRC-1 and baculovirus-produced and affinity-purified BRG-1 (Fig. 2c). These data suggest that SRC-1 is unlikely to be the only factor which mediates the association between ER and BRG-1 and are supported by GST pulldown assays in which SRC-1 is insufficient to reconstitute the association between the ER and BRG-1 (data not shown). Additionally, we tested a panel of monoclonal antibodies raised against SRC-1 for the ability to coimmunoprecipitate BRG-1. None of the antibodies was capable of coimmunoprecipitating BRG-1. Taken together, these studies suggest that the association between ER and BRG-1 requires additional factors present in an MCF-7 nuclear extract that are distinct from SRC-1.
BRG-1 binds to estrogen-responsive promoters in response to estrogen.
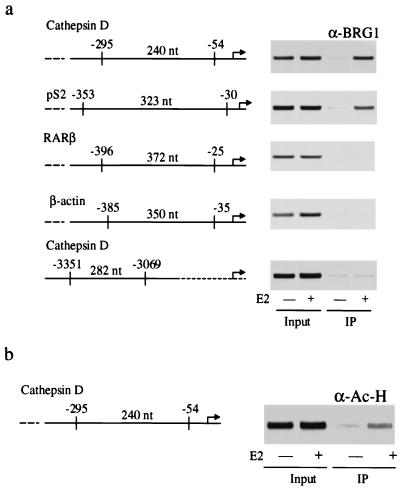
The observed association between BRG-1 and ER coupled to the known role of BRG-1 in chromatin remodeling suggested that BRG-1 might be recruited in a hormone-dependent manner to regions of chromatin that are proximal to the EREs of known target genes. To test this hypothesis, we developed a CHIP assay that would allow us to detect the presence of various factors in association with the chromatin of known target genes in vivo. Briefly, MCF-7 cells were treated with either 100 nM estrogen or a vehicle control. Following this treatment, the chromatin-associated proteins were cross-linked by fixation in formaldehyde and chromatin was extracted from the cells. These fixed chromatin fractions were sheared by sonication and subjected to immunoprecipitation. After extensive washing, the cross-linking was reversed and the retained DNA fragments were purified. These fractions were amplified by PCR using primers that were targeted to two distinct estrogen-responsive genes encoding cathepsin D and pS2. Other non-estrogen-responsive fragments were also targeted as controls for the estrogen dependence of the observed interactions. In these studies, it was observed that antibodies directed against BRG-1 efficiently precipitated the estrogen-responsive regions of cathepsin D and pS2 (1, 15) in a manner that was dependent on treatment of MCF-7 cells with estrogen (Fig. 3a). These studies indicate that estrogen treatment of MCF-7 cells results in the recruitment of BRG-1 to regions of chromatin which contain the EREs of the cathepsin D and pS2 genes. In similar studies, BRG-1 antibodies failed to precipitate regions of chromatin that represented promoters of two non-estrogen-dependent genes, the retinoic acid receptor β and β-actin genes. Importantly, a region of the cathepsin D promoter that does not contain estrogen-responsive sequences was not precipitated by BRG-1 antibodies, suggesting that the estrogen-stimulated recruitment BRG-1 required an ERE. These studies suggest that the mechanism by which BRG-1 mediates coactivation of ER signaling is by being recruited to estrogen-responsive regions of target genes. These findings are consistent with the observed association between ER and BRG-1 and suggest a complex interaction between ER, BRG-1, and estrogen-responsive promoters.
FIG. 3.
BRG-1 binds to ERE-containing chromatin fragments in a hormone-dependent manner. (a) MCF-7 cells were deprived of estrogen for 72 h and then treated with either 100 nM 17β-estradiol (E2) or a vehicle control. At 45 min posttreatment, cells were harvested and fixed in 1% formaldehyde, resulting in the cross-linking of chromatin-associated factors to DNA. The fixed cells were sonicated, and the fragmented chromatin was subjected to immunoprecipitation with an antibody directed against BRG-1. Antibody-antigen complexes were captured on protein A-linked Sepharose and washed extensively. The retained fraction were incubated at 60°C overnight, resulting in the liberation of retained DNA fragments. These fragments were purified and subjected to PCR analysis to test for the presence or absence of the indicated promoter regions. nt, nucleotides. (b) Immunoprecipitation of chromatin with antibodies directed against acetylhistone indicates that estrogen-responsive genes undergo enhanced acetylation in response to estrogen treatment. CHIP assays were performed as above, using an antibody directed acetylated histone H3 (α-Ac-H; Upstate Biotechnology). RARβ, retinoic acid receptor β.
The observation that BRG-1 becomes associated with regions of chromatin that contain estrogen-responsive regulatory sequences coupled to the observed association between ER and BRG-1 suggested that BRG-1 may be involved in a bipartite association that allows for hormone-dependent recruitment to ER target genes. Further, it suggests that recruitment of the HAT activities of the SRC-1–CBP complex might also become associated with chromatin in a manner that is targeted by hormone-bound ER. If true, then it may be possible to detect an enhanced state of histone acetylation on ER target genes in response to estrogen. To test this idea, CHIP assays were performed using antibodies directed against acetylated histones (Fig. 3b). In these assays, we observed increased histone acetylation of the estrogen-responsive region of the cathepsin D promoter in response to estrogen. Interestingly, the CHIP assays in these studies and in the BRG-1 studies described in Fig. 3a represent the state of the chromatin after 45 min of estrogen treatment. This indicates that there is a temporal overlap between two distinct hormone-regulated events, the recruitment of BRG-1 and the acetylation of histones. Taken together, these results suggest that multiple mechanisms of coactivation contribute to the activation of estrogen-responsive genes and that they may be targeted to these genes by the hormone-bound ER.
Modulation of BRG-1 activity by acetylation and deacetylation.
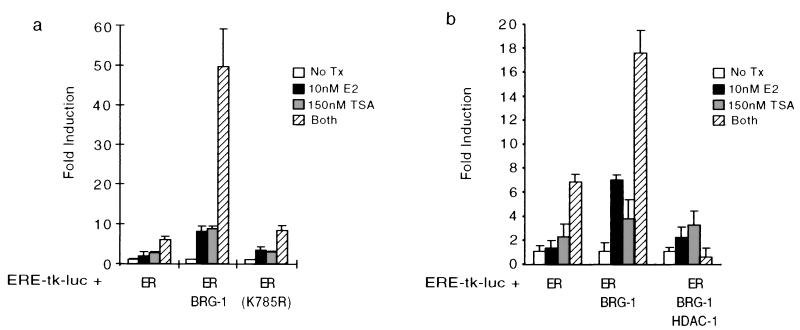
Several studies have demonstrated that factors which regulate the acetylation state of histones contribute to the coactivation of nuclear receptors and other transcription factors. Several coactivators which are known to contribute to nuclear receptor function, including CBP-p300, PCAF, SRC-1, and ACTR, have been shown to have measurable HAT activity (2, 4, 11, 32, 42). Additionally, the observed enhancement of BRG-1-mediated coactivation by CBP and SRC-1 and the finding that estrogen stimulates the recruitment of BRG-1 to and enhanced histone acetylation of ER target genes raised the possibility that the acetylation state of histones and possibly other factors may modulate BRG-1 activity. To test this hypothesis, we used the HDAC inhibitor TSA to enhance acetylation within a cell and measured the effects of this treatment on BRG-1-mediated coactivation in transient transfection assays. In the absence of exogenous BRG-1, neither estrogen nor TSA could stimulate ER-mediated transcription in SW-13 cells; however, together they elicited a sevenfold induction relative to untreated controls (Fig. 4a). The observation that TSA can potentiate estrogen-dependent transcription suggested that one mechanism of ER activation involves the modulation of histone acetylation. Under these conditions, overexpression of BRG-1, but not the ATPase-deficient mutant K785R, elicited an 8-fold coactivation of estrogen signaling alone and a 48-fold coactivation in the presence of TSA (Fig. 4a). These results suggest a positive correlation between the acetylation state of a cell and the ability of BRG-1 to coactivate estrogen signaling. Taken together, these results demonstrate that factors involved in two distinct mechanisms of transcriptional coactivation, histone acetylation and nucleosome remodeling, contribute to nuclear receptor signaling and may be functionally linked in such a way that they contribute to the maximal activity of nuclear receptors.
FIG. 4.
BRG-1-mediated coactivation of estrogen signaling is positively and negatively regulated by the acetylation state of a cell. Transient transfection assays were carried out essentially as for Fig. 1. (a) Inhibition of HDAC activity dramatically augments BRG-1-mediated coactivation of estrogen signaling. Transient transfection of SW-13 cells was performed as described in the text, and cells were treated with a vehicle control (No Tx), 10 nM 17β-estradiol (E2), 150 nM TSA, or 10 nM 17β-estradiol plus 150 nM TSA (Both). (b) Overexpression of HDAC-1 diminishes BRG-1-mediated activation of ER signaling and abolishes the cooperative enhancement of BRG-1 activity observed with TSA. All experiments were carried out in triplicate; error bars represent the standard error of the mean.
The observation that either overexpression of SRC-1–CBP or treatment with TSA was capable of enhancing BRG-1-mediated coactivation of estrogen signaling suggests that BRG-1 activity is regulated via the modulation of the acetylation state of a cell. Furthermore, the observed enhancement of BRG-1 activity by TSA also implies that factors that decrease the acetylation state within a cell may negatively regulate the ability of BRG-1 to function as a coactivator of ER signaling. To test this, we measured the effects of overexpression of HDAC-1 on the ability of BRG-1 and TSA to function cooperatively in the coactivation of estrogen signaling. In these studies, we observed that overexpression of HDAC-1 significantly reduced BRG-1-mediated coactivation of estrogen signaling (Fig. 4b). Additionally, the TSA-mediated enhancement of BRG-1 activity was completely abolished by overexpression of HDAC-1. These studies suggest that estrogen signaling may be negatively regulated by increased HDAC activity, which is consistent with studies correlating the activity of HATs and HDACs with positive and negative regulation of nuclear receptor function. These studies also support the conclusion that the ability of BRG-1 to potentiate estrogen signaling is modulated by factors that regulate the acetylation state within a cell. Taken together, these studies support a model in which two distinct mechanisms by which the chemical and structural integrity of chromatin work cooperatively to support nuclear receptor activation.
DISCUSSION
The mechanisms by which nuclear receptors transmit a hormone binding signal to core transcription machinery have been the focus of intensive research. These efforts have lead to the identification and characterization of several distinct multiprotein complexes which directly interact with agonist-bound nuclear receptors (9, 16, 28, 45, 46). The SRC-1 family of nuclear receptor coactivators appear to play a critical role in mediating the association of one of these complexes to nuclear receptors. This complex has been shown to contain potent HAT activities in p300-CBP and also the p300/CBP-associated factor PCAF. Additionally, these factors have been identified in complexes that contain intrinsic chromatin remodeling activity. These findings suggest that the SRC-1-containing complex may have as its primary purpose the chemical modification of the chromatin surrounding a target gene to which it is recruited. A second complex, identified on the basis of its ability to interact with activated nuclear receptors, is the vitamin D receptor-interacting protein complex (DRIP) (38), also known as the thyroid receptor-associated protein complex (10). This complex has also been shown to play a critical role in the coactivation of several classes of transcription factors other than nuclear receptors (31). Detailed biochemical analyses of the subunits of this complex have provided strong evidence that this complex is an advanced homolog of the yeast mediator complex, and there is evidence of direct interactions between subunits of DRIP and subunits of the TFIID complex. The identification of these two unique complexes supports a two-step model for the coactivation of nuclear receptor signaling in which the modification of chromatin structure and the direct transmission of a hormone binding signal to basal transcription machinery both contribute to the regulation of nuclear receptor function (25). While it remains unproven that these two complexes work cooperatively, there is abundant evidence that activities which relax the structure of chromatin have been associated with an increased rate of preinitiation complex formation upon a basal promoter (34). The close association between the preinitiation complex and the mediator complex might also imply that the modification of chromatin structure can enhance the rate at which complexes like DRIP engage both the basal transcription machinery and activated transcription factors.
There is substantial evidence that while they are biochemically separable, there is a functional link between histone acetylation and chromatin remodeling. Chromatin structure is altered by large multiprotein nucleosome remodeling complexes such as Swi/Snf, RSC, ACF, CHRAC, and NURF (3). Each of these complexes was purified as a distinct ATP-dependent chromatin remodeling activity, and each appears to be biochemically distinct. One element common to each of these complexes is a member of the Swi-2/Snf-2 family of nuclear ATPases which includes yeast Swi-2/Snf-2, Drosophila Brahma and I-Swi, and human Brm and BRG-1 among others. These factors have been proposed to function as molecular motors that use the catalysis of ATP to drive a variety of remodeling activities. Studies of mutations in a yeast HAT complex have suggested a link to between histone acetylation and Swi/Snf function, while other studies have reported the identification of histone acetylases (17, 36, 39) closely associated with chromatin remodeling machines. Consistent with the role of acetylation status are studies in which diverse members of the HDAC family are implicated in the corepression of nuclear receptor activity (14, 18, 23). Taken together, these studies along with data presented here support a model in which an acetylated nucleosome may be a better substrate for the ATP-dependent chromatin remodeling machines, such as Swi/Snf. The loss of a positively charged amino acid residue that is the result of the acetylation of lysine reduces the affinity of the nucleosome for DNA, which, in turn, may contribute to the nucleosome being a more labile substrate for remodeling machines. Additionally, a domain which is well conserved in several transcriptional coregulators, the bromodomain, has recently been shown to bind to acetyl-lysine with high affinity (7). This observation may suggest that in addition to reducing the affinity of the nucleosome core for DNA, acetylation of the external arms of the histones may actually provide high-affinity docking sites upon which bromodomain-containing proteins can bind. In this way, acetylation may enhance chromatin remodeling activity by two distinct mechanisms: the reduction of the DNA binding affinity of the nucleosome, and the presentation of docking sites upon which bromodomain containing proteins can bind.
In this paper we report an association between ER and BRG-1 that we believe is mediated by additional factors. This association was observed in crude whole-cell extracts and could not be reconstituted using recombinant proteins. These observations are likely to support a model in which BRG-1 is recruited to the activated ER as a member of a large multisubunit complex of proteins. Several reports have identified factors that interact with nuclear receptors in a manner that is hormone and AF-2 dependent and that are distinct from the SRC-1 family. Given that the association between ER and BRG-1 is dependent on the presence of both hormone and AF-2, it is plausible that one or more of these factors may mediate the association between ER and BRG-1. Previous studies have demonstrated interactions between BRG-1 and members of the nuclear receptor superfamily, including ER, retinoic acid receptor, and GR in yeast-based two-hybrid systems (6, 30). There are several possible explanations for this apparent conflict. While the two-hybrid systems are designed to measure direct interactions between two chimeric proteins, it is difficult to control for the interactions of additional endogenous factors. Equally possible is that the interaction between ER and BRG-1 in yeast is direct and may be the result of distinct mechanisms by which the heterologous transcriptional activation by nuclear receptors is mediated in yeast (13). This possibility is particularly intriguing given that there appear to be no obvious SRC-1 family homologs encoded in the yeast genome and may suggest that the SRC-1 family members evolved as a more dynamic platform upon which coactivator complexes are assembled. Thus, it may be possible that the observed interaction in yeast represents a more global mechanism of transcriptional activation that has been replaced by factors such as the SRC-1 family in higher eukaryotes.
The findings presented in this report are consistent with a model in which BRG-1 is required for transcriptional activation by the ER. Our data suggest that upon hormone binding, the ER associates with BRG-1, thereby recruiting BRG-1 to the sites of estrogen-responsive chromatin. Similar CHIP assays using antibodies directed against SRC-1 and acetyl-lysine have suggested that the association between BRG-1 and estrogen-responsive promoter regions is accompanied by the interaction of SRC-1 and CBP (Y. Shang et al., submitted for publication). Coactivation of estrogen signaling by members of the SRC-1 family or members of the CBP family is also dependent on the actions of BRG-1. These studies also suggest that factors which enhance the acetylation state within a cell elicit a corresponding enhancement of BRG-1-mediated coactivation of estrogen signaling. This enhancement may be accounted for by increased access of BRG-1 to acetylated chromatin, which in turn might render an acetylated nucleosome a better substrate for the remodeling activity of BRG-1. Additionally, the presence of a bromodomain at the C terminus of BRG-1 may suggest that under conditions of enhanced acetylation, BRG-1 can associate via the recently identified interaction between a bromodomain and acetyl-lysine. Such mechanisms may account for the collaborative effect of BRG-1 and factors that promote histone acetylation upon the coactivation of estrogen signaling.
ACKNOWLEDGMENTS
We thank David Livingston and Mark Ewen for helpful discussions regarding the manuscript and James DeCaprio and Jenny Gan for assistance in developing the hSRC-1 monoclonal antibodies.
J.D. is supported by U.S. Department of Defense Career Development Award DAMD17-99-1-9163. This work was supported by NIH grant CA57374 and Department of Defense Academic Award DAMD17-99-1-9161 to M.B.
REFERENCES
- 1.Augereau P, Miralles F, Cavailles V, Gaudelet C, Parker M, Rochefort H. Characterization of the proximal estrogen-responsive element of human cathepsin D gene. Mol Endocrinol. 1994;8:693–703. doi: 10.1210/mend.8.6.7935485. [DOI] [PubMed] [Google Scholar]
- 2.Blanco J C, Minucci S, Lu J, Yang X J, Walker K K, Chen H, Evans R M, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cairns B R. Chromatin remodeling machines: similar motors, ulterior motives. Trends Biochem Sci. 1998;23:20–25. doi: 10.1016/s0968-0004(97)01160-2. [DOI] [PubMed] [Google Scholar]
- 4.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 5.Chen J D, Evans R M. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 6.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhalluin C, Carlson J E, Zeng L, He C, Aggarwal A K, Zhou M M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 8.Dunaief J L, Strober B E, Guha S, Khavari P A, Alin K, Luban J, Begemann M, Crabtree G R, Goff S P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 9.Edwards D P. Coregulatory proteins in nuclear hormone receptor action. Vitam Horm. 1999;55:165–218. doi: 10.1016/s0083-6729(08)60936-x. [DOI] [PubMed] [Google Scholar]
- 10.Fondell J D, Roy A L, Roeder R G. Unliganded thyroid hormone receptor inhibits formation of a functional preinitiation complex: implications for active repression. Genes Dev. 1993;7:1400–1410. doi: 10.1101/gad.7.7b.1400. [DOI] [PubMed] [Google Scholar]
- 11.Forsberg E C, Lam L T, Yang X J, Nakatani Y, Bresnick E H. Human histone acetyltransferase GCN5 exists in a stable macromolecular complex lacking the adapter ADA2. Biochemistry. 1997;36:15918–15924. doi: 10.1021/bi971664x. [DOI] [PubMed] [Google Scholar]
- 12.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 13.Garabedian M J, Yamamoto K R. Genetic dissection of the signaling domain of a mammalian steroid receptor in yeast. Mol Biol Cell. 1992;3:1245–1257. doi: 10.1091/mbc.3.11.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci P G, Lazar M A. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giamarchi C, Solanas M, Chailleux C, Augereau P, Vignon F, Rochefort H, Richard-Foy H. Chromatin structure of the regulatory regions of pS2 and cathepsin D genes in hormone-dependent and -independent breast cancer cell lines. Oncogene. 1999;18:533–541. doi: 10.1038/sj.onc.1202317. [DOI] [PubMed] [Google Scholar]
- 16.Glass C K, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 17.Grant P A, Duggan L, Cote J, Roberts S M, Brownell J E, Candau R, Ohba R, Owen-Hughes T, Allis C D, Winston F, Berger S L, Workman J L. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 18.Guenther M G, Lane W S, Fischle W, Verdin E, Lazar M A, Shiekhattar R. A core SMRT corepressor complex containing HDAC3 and TBL1, a WD40-repeat protein linked to deafness. Genes Dev. 2000;14:1048–1057. [PMC free article] [PubMed] [Google Scholar]
- 19.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 20.Hanstein B, Eckner R, DiRenzo J, Halachmi S, Liu H, Searcy B, Kurokawa R, Brown M. p300 is a component of an estrogen receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horlein A J, Naar A M, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass C K, Rosenfeld M G. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- 23.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 24.Ichinose H, Garnier J M, Chambon P, Losson R. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene. 1997;188:95–100. doi: 10.1016/s0378-1119(96)00785-8. [DOI] [PubMed] [Google Scholar]
- 25.Jenster G, Spencer T E, Burcin M M, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor induction of gene transcription: a two-step model. Proc Natl Acad Sci USA. 1997;94:7879–7884. doi: 10.1073/pnas.94.15.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 27.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 28.Manteuffel-Cymborowska M. Nuclear receptors, their coactivators and modulation of transcription. Acta Biochim Pol. 1999;46:77–89. [PubMed] [Google Scholar]
- 29.Muchardt C, Yaniv M. ATP-dependent chromatin remodelling: SWI/SNF and Co. are on the job. J Mol Biol. 1999;293:187–198. doi: 10.1006/jmbi.1999.2999. [DOI] [PubMed] [Google Scholar]
- 30.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naar A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 32.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 33.Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 34.Paranjape S M, Kamakaka R T, Kadonaga J T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 35.Paranjape S M, Krumm A, Kadonaga J T. HMG17 is a chromatin-specific transcriptional coactivator that increases the efficiency of transcription initiation. Genes Dev. 1978;9:1978–1991. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Martin J, Johnson A D. Mutations in chromatin components suppress a defect of Gcn5 protein in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:1049–1054. doi: 10.1128/mcb.18.2.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peterson C L, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 38.Rachez C, Suldan Z, Ward J, Chang C P, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts S M, Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan M P, Jones R, Morse R H. SWI-SNF complex participation in transcriptional activation at a step subsequent to activator binding. Mol Cell Biol. 1998;18:1774–1782. doi: 10.1128/mcb.18.4.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh P, Coe J, Hong W. A role for retinoblastoma protein in potentiating transcriptional activation by the glucocorticoid receptor. Nature. 1995;374:562–565. doi: 10.1038/374562a0. [DOI] [PubMed] [Google Scholar]
- 42.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 43.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits distinct properties from steroid receptor coactivator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 44.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 45.Westin S, Rosenfeld M G, Glass C K. Nuclear receptor coactivators. Adv Pharmacol. 2000;47:89–112. doi: 10.1016/s1054-3589(08)60110-6. [DOI] [PubMed] [Google Scholar]
- 46.Xu L, Glass C K, Rosenfeld M G. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–147. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- 47.Yao T-P, Ku G, Zhou N, Scully R, Livingston D M. The nuclear hormone receptor co-activator SRC-1 is a specific target of p300. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zamir I, Dawson J, Lavinsky R M, Glass C K, Rosenfeld M G, Lazar M A. Cloning and characterization of a corepressor and potential component of the nuclear hormone receptor repression complex. Proc Natl Acad Sci USA. 1997;94:14400–14405. doi: 10.1073/pnas.94.26.14400. [DOI] [PMC free article] [PubMed] [Google Scholar]