Abstract
Combinatorial interaction among cardiac tissue-restricted enriched transcription factors may facilitate the expression of cardiac tissue-restricted genes. Here we show that the MADS box factor serum response factor (SRF) cooperates with the zinc finger protein GATA-4 to synergistically activate numerous myogenic and nonmyogenic serum response element (SRE)-dependent promoters in CV1 fibroblasts. In the absence of GATA binding sites, synergistic activation depends on binding of SRF to the proximal CArG box sequence in the cardiac and skeletal α-actin promoter. GATA-4's C-terminal activation domain is obligatory for synergistic coactivation with SRF, and its N-terminal domain and first zinc finger are inhibitory. SRF and GATA-4 physically associate both in vivo and in vitro through their MADS box and the second zinc finger domains as determined by protein A pullout assays and by in vivo one-hybrid transfection assays using Gal4 fusion proteins. Other cardiovascular tissue-restricted GATA factors, such as GATA-5 and GATA-6, were equivalent to GATA-4 in coactivating SRE-dependent targets. Thus, interaction between the MADS box and C4 zinc finger proteins, a novel regulatory paradigm, mediates activation of SRF-dependent gene expression.
Serum response factor (SRF) may play a leading role in the commitment of cardiac progenitors by virtue of its requirement for mesoderm formation and by its ability to activate target genes via specific protein-protein associations with other early cardiac enriched transcription factors. SRF, a member of an ancient DNA-binding protein family, shares a highly conserved DNA-binding and dimerization domain of 90 amino acids, termed the MADS box (reviewed in references 53 and 56). SRF, yeast transcription factors MCM1 and ARG80, and several plant proteins, such as Deficiens, all have a related MADS box and similar DNA sequence binding specificity. In addition, SRF-related proteins (RSRF and MEF-2) constitute a subfamily of the MADS box family of transcription factors (49, 59). SRF is especially abundant in embryonic and adult cardiac, skeletal, and smooth muscle cells (2, 12). The recent homologous recombinant knockout of the murine SRF gene locus demonstrated that SRF is absolutely required for the appearance of mesoderm and muscle lineages during mouse embryogenesis (1).
SRF interacts with other regulatory proteins and ultimately alters the regulation of specific gene programs. Studies regarding the regulation of the c-fos gene by SRF have led to the identification of several SRF accessory factors, including SAP-1, Elk-1, and Phox-1 (15, 21, 25). All of these SRF accessory factors appear to potentiate SRF's transcriptional activity on the c-fos serum response element (SRE), although the mechanisms are somewhat different. Grueneberg et al. (21) demonstrated that human SRF interacts with a novel human homeodomain protein, Phox, which enhances the exchange of SRF with its binding site in the c-fos promoter and does not require specific homeodomain DNA binding activity. Although neither Phox nor Mhox was able to activate cardiac-specified genes in the presence of SRF, our studies suggest that SRF facilitates binding of another murine homeobox transcription factor, Nkx-2.5, to SREs, resulting in the activation of the endogenous α-cardiac actin gene in fibroblasts (11). Nkx-2.5 is a potential vertebrate homologue of tinman, a factor required for Drosophila heart development (7) and the earliest known marker of vertebrate heart development.
GATA factors also play an important role in early cardiogenesis. The GATA family has been subdivided, with the GATA-1/2/3 subfamily being linked to hematopoiesis, while GATA-4/5/6 is thought to be involved with cardiac, gut, and blood vessel formation (reviewed in references 46 and 48). Each of the six GATA proteins contains a highly conserved DNA-binding domain consisting of two C4 zinc fingers of the motif Cys-X2-Cys-X17-Cys-X2-Cys. These two zinc fingers have been shown to direct binding to the DNA sequence element (A or T)GATA(A or G) (32, 40), although the carboxy zinc finger is sufficient for site-specific binding (39). Examination of the DNA-binding site specificity of all six GATA factors indicates that they are capable of binding to the same target sequence, thus suggesting their potential to substitute for one another in cells in which they are coexpressed. GATA-4 is expressed in a developmental and lineage-specific pattern within the cardiac mesoderm and gut epithelium (24, 30, 34). GATA-4 expression regulates expression of cardiac-specific genes, such as cardiac troponin C (45) and α-myosin heavy chain (41), and leads to the precocious activation of cardiac α-actin and α-myosin heavy chain gene expression when expressed in Xenopus embryos (27). GATA-4 null mice display a severe defect in formation of the cardiac tube, which is required for the migration and folding morphogenesis of the precardiogenic splanchnic mesodermal cells (33, 43). Although forced expression of antisense DNA for GATA-4 blocked expression of cardiac-specific genes in P19 cells (19), the rather normal expression of cardiac-specific genes observed in these homozygous GATA-4 knockout embryos probably reflects the redundancy of some functions in the GATA-4/5/6 subfamily. Sepulveda et al. (52) demonstrated that GATA-4 synergizes with Nkx-2.5 to activate the chicken cardiac α-actin promoter and that this activation is dependent on DNA binding by Nkx-2.5 but not by GATA-4. GATA-4 cooperates with Nkx-2.5 to activate the ANF and BNP promoters (16, 35) and synthetic promoters containing multimerized high-affinity Nkx-2.5 DNA-binding sites (NKEs) (52).
Activation of SRE-dependent muscle tissue-restricted promoters, such as α-actins, appears to be mediated through combinatorial interaction of SRF with muscle tissue-restricted transcription factors, such as Nkx-2.5 (11) and MyoD (20). We asked whether the pairing of GATA-4 and SRF activates SRE-dependent promoters. We found that SRF and GATA-4 provide robust coactivation with myogenic and smooth muscle α-actin promoters and the nonmyogenic c-fos promoter. Using isolated SREs from cardiac and skeletal α-actin promoters, we asked whether a single SRE can mediate this synergistic coactivation by SRF and GATA-4. We report here that protein-protein associations shared between SRF and GATA-4 transactivate via the SRE-laden cardiac α-actin promoter. Interactive protein regions were delineated to the SRF's MADS box and to GATA-4's second zinc finger and the C-terminal basic region. Transcriptional coactivation of the cardiac α-actin promoter depended upon the C-terminal region of GATA-4 but was inhibited by its N-terminal region and the first zinc finger. Other GATA factors expressed in the heart, such as GATA-5 and GATA-6, were equivalent to GATA-4 in coactivating cardiac α-actin promoter activity. In vivo one-hybrid assays also demonstrated coactivation of Gal4 target sequences via Gal4 fusion proteins containing either GATA-4 or SRF. Accordingly, the paired interactions of SRF with tissue-restricted cardiogenic GATA-4 confer robust levels of transcriptional activity on the cardiac α-actin promoter and tissue specificity on SRF. Thus, their coassociation in vivo might underly a primary mechanism for forming protein-protein complexes, in which each perhaps facilitates the other's recruitment to its primary DNA-binding site.
MATERIALS AND METHODS
Recombinant DNA clones.
Luciferase reporter plasmids Gal4Luc (G5Luc), c-fos, Δ56 fos, skeletal α-actin, skeletal α-actin SRE1, smooth muscle α-actin, smooth muscle γ-actin, SM22α, cardiac α-actin wild type and deletion mutants, and expression vectors for SRF, SRFpm1, SRFΔC, Gal DB, GATA-6, and the wild type and various mutants of GATA-4 have been described earlier (3, 9, 10, 16, 28, 36, 38, 52). pCGNSRFΔMADS was derived from an intermediate construct, pBSSRFΔMADS. pBSSRFΔMADS was constructed by digesting pBSKSSRF with NarI (nucleotide [nt] 565) and BglII (nt 1094) and blunt ending and religating the large fragment. The XbaI-to-BamHI fragment from pBSSRFΔMADS was subcloned into cognate sites of the pCGN vector to construct pCGNSRFΔMADS. pCDNA3GATA-5 (a gift from Mona Nemer) was constructed by subcloning the rat GATA-5 cDNA into the pCDNA3 (Clonetech) vector. pCGNGATA-4ΔZF was constructed by subcloning the XbaI-BamHI fragment containing GATA-4, with the second zinc finger deleted, from pACXVPG4ΔZF into the pCGN vector. This plasmid contains an intact nuclear localization signal sequence. pACXVPG4ΔZF will be described elsewhere (Sepulveda et al., unpublished data). Gal ZF1 + 2 was constructed by ligating the PCR-amplified fragment containing both the zinc fingers and the C-terminal basic region (nt 619 to 1051) of GATA-4 into the EcoRI- and BamHI-digested pMFH2/GAL4 vector (58). MADS Gal was constructed by subcloning the 315-bp SmaI-to-BamHI fragment of human SRF in frame with the Gal4 DNA binding domain in the vector pMFH2/GAL4.
Cell culture and transfections.
CV1 monkey kidney cells were maintained in Dulbecco modified Eagle medium containing 10% fetal bovine serum. Subconfluent cells in 60-mm plates were transfected with 1 μg of reporter, 150 ng of wild-type pCGNSRF, and 400 ng of the wild type and deletion mutants of GATA-4 expressed from the pCG vector. For the experiment comparing transcriptional coactivation by GATA-4, -5, and -6, 200 ng of reporter and 200 ng of pCGNSRF with or without 800 ng of pCDNA3-based GATA-4, -5, and -6 were transfected. For one-hybrid analysis of recruitment of GATA-4 by a MADS-Gal fusion, 200 ng of Gal4 luciferase reporter, 200 ng of Gal DB or MADS Gal, and 750 ng of GATA-4 were used. For the analysis of recruitment of SRF by Gal ZF1 + 2, 1 μg of Gal4 reporter, 1 μg of Gal DB or Gal ZF1 + 2, and 100 ng of SRF were used. Following transfection, cells were maintained in Dulbecco modified Eagle medium containing 2% horse serum and 10 μg of insulin/ml for 48 h. Cells were harvested 48 h posttransfection, and luciferase activity was measured in a luminometer.
In vitro transcription and translation and GST pull-down assays.
Full-length SRF and various deletion mutants of SRF fused to glutathione S-transferase (GST) were expressed in bacteria and purified as described earlier (11). GST pull-down experiments were performed as described by Sepulveda et al. (52). Approximately 1 μg of fusion protein immobilized on glutathione-agarose beads was incubated with 100 μg of whole-cell extract prepared from CV1 cells transfected with GATA-4, washed extensively, resolved on a sodium dodecyl sulfate (SDS)–10% polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and blotted with GATA-4 antibody. The wild type and various deletion mutants of GATA-4 hot translated in rabbit reticulocyte lysates were used to map the domains of GATA-4 required for interaction with SRF, as described previously (52).
Protein A fusion protein pullout assays.
Vectors encoding Staphylococcus aureus protein A or protein A-SRF and protein A–GATA-4 fusion were cotransfected with hemagglutinin (HA) epitope-tagged SRF and SRFPM1 into CV1 cells. Cells were lysed in EBC buffer (50 mM Tris [pH 8.0]–120 mM NaCl–0.5% Nonidet P-40 containing 2 μg of aprotinin per ml, 2 μg of leupeptin per ml, 2 μg of pepstatin per ml, and 1 mM phenylmethylsulfonyl fluoride), and the pullouts were carried out with immunoglobulin G (IgG)-Sepharose beads as described earlier (52). Proteins retained by protein A-SRF and protein A–GATA-4 were visualized by immunoblotting with anti-SRF antibody.
RESULTS
SRE-dependent promoters are coactivated by SRF and GATA-4.
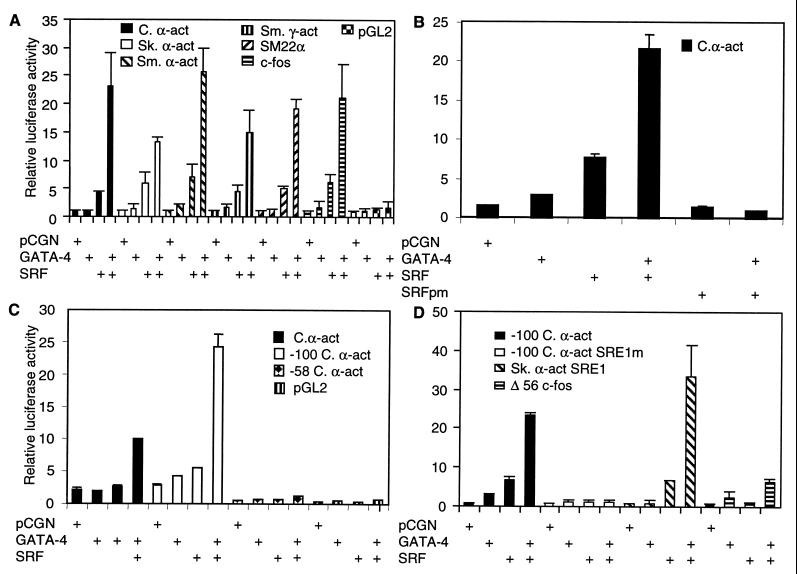
Since Nkx-2.5 interacts with GATA-4 and with SRF, we reasoned that it was also likely that SRF and GATA-4 function as coaccessory factors. To address this prospect, luciferase reporter constructs for cardiac-, skeletal-, and smooth muscle-restricted, SRE-dependent promoters, such as cardiac α-actin, skeletal α-actin, smooth muscle α-actin, smooth muscle γ-actin, and SM22α, and the ubiquitously expressed c-fos promoter were tested in cotransfection assays. Cotransfection of these reporter constructs into CV1 fibroblasts along with an expression vector encoding SRF elicited modest activation (Fig. 1A). Similarly, expression of GATA-4 with these reporters resulted in weak activation. However, coexpression of both GATA-4 and SRF, from transfected CMV-driven plasmid expression vectors, resulted in robust activation of both muscle-restricted and ubiquitous SRE-dependent promoters, as shown in Fig. 1A.
FIG. 1.
SRF and GATA-4 synergistically activate the cardiac α-actin promoter. (A) Subconfluent CV1 cells were transfected with 1 μg of numerous myogenic and nonmyogenic promoter luciferase reporters (indicated), along with 150 ng of expression vector for SRF alone or in combination with 400 ng of GATA-4. (B) A DNA-binding mutant of SRF (SRFpm1) (150 ng) was used in addition to wild-type SRF and GATA-4. (C) The wild type and deletion mutants of the cardiac α-actin promoter and the control pGL2 basic luciferase reporters were used. (D) A deletion mutant of cardiac α-actin containing a single wild-type or mutated SRE1 and a truncated c-fos minimal promoter (Δ56 c-fos) with or without skeletal α-actin SRE1 cloned upstream was used in the cotransfection assay. The total amount of DNA was adjusted to 2 μg by balancing with the pCGN empty vector. Cells were harvested 48 h posttranscription, and the luciferase activity was measured. Results shown are means ± the standard errors of the means for three duplicate experiments (B and C) and two duplicate experiments (A and D).
Promoter-proximal SRE in cardiac α-actin and skeletal α-actin is both necessary and sufficient for SRF- and GATA-4-mediated coactivation.
Deletion mutants were used to map the 5′ regulatory borders of the cardiac α-actin promoter responsible for mediating the potent transcription activity. Deleting the cardiac α-actin promoter to −100 retained a single SRE, and this truncated cardiac α-actin promoter was sufficient for SRF- and GATA-4-mediated activation (Fig. 1C). These findings indicated that potential GATA sites located at positions −304 and −161 of the cardiac α-actin promoter were dispensable. Deletion of the proximal SRE, evaluated with the −58 bp mutant, and site-specific mutation of this SRE in the context of the −100 promoter totally abolished SRF- and GATA-4-dependent coactivation, indicating a requirement for the proximal SRE1 (Fig. 1C and D). Transfections with a dominant negative SRF mutant, SRFpm1, also blocked GATA-4-dependent activation of the cardiac α-actin promoter (Fig. 1B), thus demonstrating a dependency on intact SRE and SRF for GATA-4.
To confirm that SRE is both necessary and sufficient for mediating the SRF- and GATA-4-dependent coactivation, we used the skeletal α-actin promoter-proximal SRE cloned upstream of a heterologous c-fos minimal promoter. This construct was activated sevenfold by SRF and did not respond to GATA-4. However, coexpression of both SRF and GATA-4 very strongly activated this single SRE-containing promoter to 34-fold over background levels (Fig. 1D). In contrast, the plasmid lacking the SRE, the Δ56 fos vector, was activated sixfold.
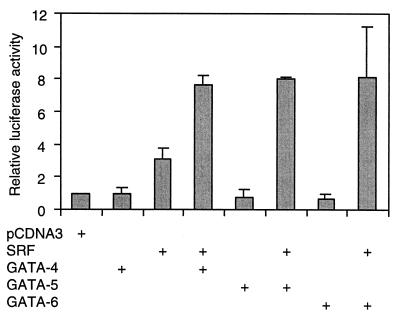
GATA-5 and GATA-6 are equivalent to GATA-4 in cotransfection assays with SRF.
Given the extensive homology between the zinc finger domains and the activation domains of GATA-4, GATA-5, and GATA-6 (45) and the fact that these GATA factors exhibited similar but nonidentical expression patterns during cardiac morphogenesis, we wanted to determine their ability to substitute for GATA-4 in transfection assays. As shown in Fig. 2, both GATA-5 and GATA-6 coactivated the cardiac α-actin promoter when cotransfected with SRF, virtually to the same extent as GATA-4 (Fig. 2). Thus, GATA factors expressed in the embryonic heart were equivalent in their abilities to drive cardiac α-actin gene activity.
FIG. 2.
GATA-5 and GATA-6 can substitute for GATA-4 in coactivation of the cardiac α-actin promoter. Subconfluent CV1 cells were transfected with 200 ng of wild-type cardiac α-actin luciferase reporter and 200 ng of an expression vector for SRF (pCGNSRF) either alone or in combination with 800 ng of pCDNA3GATA-4, -5, or -6. Cells were harvested 48 h posttranscription, and the luciferase activity was measured. Results shown are means ± standard errors of the means for two duplicate experiments.
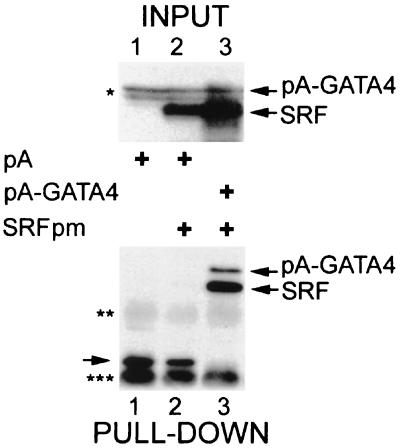
GATA-4 and SRF associate in vivo.
We asked whether GATA-4 and SRF physically associate in the cellular environment. The S. aureus protein A IgG binding domain fused to the N termini of GATA-4 and SRF was employed to immobilize protein complexes associated with these fusion proteins in transfected CV1 cells. This method allows rapid purification of associated proteins with IgG-Sepharose. When pCGN-SRFpm (which expresses the HA epitope) was cotransfected with pA-SRF in CV1 cells, a significant amount of HA-SRFpm was dimerized to pA-SRF (data not shown). Similarly, pA–GATA-4 was able to pull down HA-SRFpm, indicating that the two proteins were associated in the extracts and that the SRFpm mutation did not interfere with GATA-4 binding (Fig. 3, lane 3). In contrast with the isolated protein A, the protein A–GATA-4 fusion protein bound to coexpressed HA-SRF. These results indicated that GATA-4 and SRF physically interact when expressed in a mammalian cell. Although interaction after cell lysis cannot be ruled out, these results, together with those of the cotransfection assays, argue that the interaction between SRF and GATA-4 can occur in vivo.
FIG. 3.
GATA-4 and SRF associate in vivo. Bottom panel, CV1 cells transfected with HA-tagged SRFpm1 with either protein A (lane 2) or protein A–GATA-4 (lane 3) fusions. For lane 1, protein A vector was transfected alone. Cells were lysed, and the lysates were allowed to react with IgG-Sepharose beads. After extensive washing, proteins retained by protein A and protein A fusions were eluted by boiling in SDS sample buffer and analyzed by immunoblotting with anti-SRF antibody. The top panel shows the Western analysis of input proteins probed with HA antibody. Protein A and protein A fusions to SRF and GATA-4, as well as IgG heavy chains (double asterisk) and IgG light chains (triple asterisk), were visualized due to binding of the secondary antibody. Nonspecific anti-HA immunoreactive bands which migrate close to the pA-GATA4 band are indicated by a single asterisk.
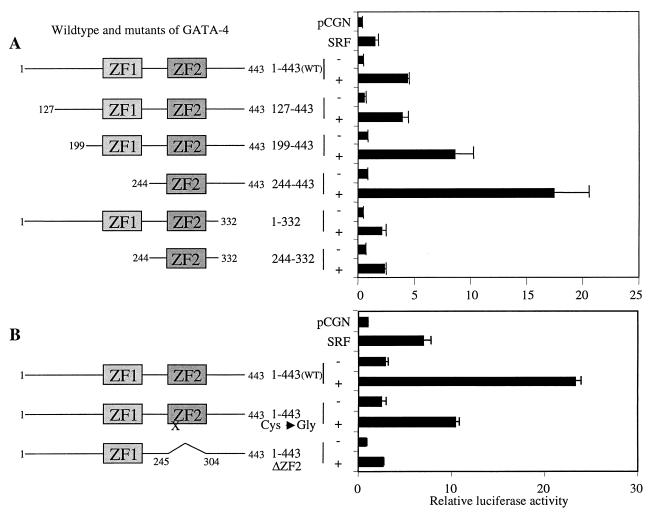
The second zinc finger and the immediate C-terminal basic region of GATA-4 are essential for synergistic activation of the cardiac α-actin promoter.
To identify the domains of GATA-4 required with SRF for coactivation of the cardiac α-actin promoter, we transfected CV1 cells with several deletion mutants of GATA-4 and full-length SRF. Deletion of the first N-terminal activation domain of GATA-4 (GATA-4Δ127) did not significantly affect transcriptional activity (Fig. 4A). A further deletion of both N-terminal activation domains of GATA-4 (GATA-4Δ199) resulted in a slight increase in promoter activity, indicating that the N-terminal activation domains of GATA-4 were dispensable for coactivation (Fig. 4). Surprisingly, deletion of both the N-terminal activation domains and the first zinc finger (GATA-4Δ244) enhanced transactivation, and these data suggest that this region overlapping a portion of the second activation domain and the entire first zinc finger (amino acids [aa] 199 to 244) revealed inhibitory sequences that might interfere with SRF. Deletion of 110 C-terminal amino acids (GATA-4 aa 1 to 332) severely reduced the ability of this GATA-4 mutant to transactivate with SRF. This loss of coactivation was not recovered by deletion of the N-terminal activation domains and the first zinc finger region (GATA-4 aa 199 to 332 and aa 244 to 332). These results indicated that the C-terminal activation domain and the second zinc finger, along with the immediate C-terminal basic domains, were vital for imparting transcriptional synergy with SRF.
FIG. 4.
The second zinc finger and the immediate C-terminal basic region of GATA-4 are essential for synergistic activation of the cardiac α-actin promoter. (A) Subconfluent CV1 cells were transfected with 1 μg of cardiac α-actin luciferase reporter and 400 ng of the wild type and various deletion mutants of GATA-4, either alone or in combination with 150 ng of SRF. The total amount of DNA was adjusted to 2 μg by balancing with the pCGN empty vector. (B) The −100 cardiac α-actin promoter containing the proximal SRE was used as the reporter. Cells were harvested 48 h posttranscription, and the luciferase activity was measured. Results shown are means ± standard errors of the means for three duplicate experiments (A) and two duplicate experiments (B). Domains of GATA-4 that are retained in each deletion mutant are diagrammatically represented on the left. ZF1 and ZF2 refer to the N- and C-terminal zinc fingers, respectively. The single amino acid mutation in ZF2 (cysteine 273 to glycine) that abolished DNA-binding activity of GATA-4 is indicated by an X.
The heterotypic cooperativity between GATA-4 and GATA-6 (10) and homotypic cooperativity with GATA-1 (40) appear to be mediated by the amino acid sequence of the second zinc finger rather than the structure of the zinc finger. To address whether the cooperativity between SRF and GATA-4 depends on the amino acid sequence or the structure of the second zinc finger, we used two additional mutants of GATA-4, one with a point mutation in the second zinc finger to alter the zinc finger structure and the other with a total deletion of the second zinc finger. As shown in Fig. 4B, the point mutation in the second zinc finger severely reduced the coactivation by more than 50%, and the deletion of the second zinc finger totally abolished coactivation between SRF and GATA-4. These results indicate that the coactivation between SRF and GATA-4 depends on the structure of the second zinc finger of GATA-4.
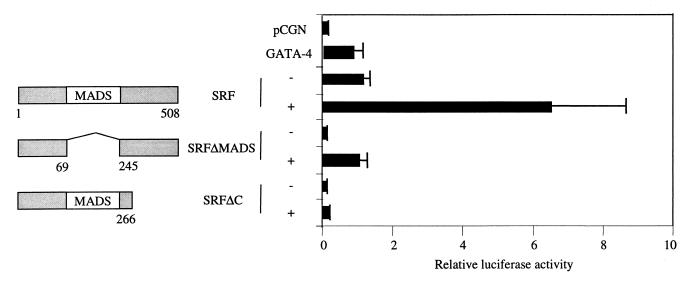
MADS box and the activation domains of SRF are necessary for coactivation of the cardiac α-actin promoter.
The inability of a DNA-binding-defective point mutant of SRF, SRFpm1, to support coactivation suggested that the DNA-binding activity of SRF was necessary for coactivation of the cardiac α-actin promoter. To confirm this result and to investigate further if the activation domain of SRF was required for coactivation, we used mutants of SRF with deletions in the conserved MADS box domain and the C-terminal activation domain. Deletion of either the MADS box domain or the activation domain abolished coactivation, suggesting that both the DNA binding and transcriptional activating activities of SRF were required for coactivation of the cardiac α-actin promoter (Fig. 5).
FIG. 5.
Subconfluent CV1 cells were transfected with 1 μg of cardiac α-actin luciferase reporter along with 400 ng of GATA-4 and 150 ng of either the wild type or deletion mutants of SRF. The total amount of DNA was adjusted to 2 μg by balancing with the pCGN empty vector. Cells were harvested 48 h posttranscription, and the luciferase activity was measured. Results shown are means ± standard errors of the means for three duplicate experiments. Domains of SRF retained in each deletion mutant are diagrammatically represented on the left.
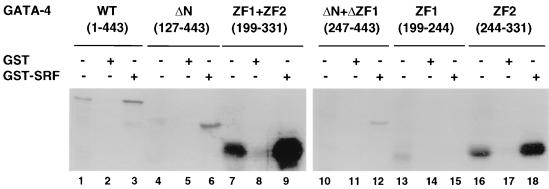
Mapping of the physical interaction domains for SRF and GATA-4.
In order to map the domains of GATA-4 interacting with SRF, various deletion mutants of in vitro-synthesized [35S]methionine-labeled GATA-4 protein were examined for their ability to bind GST-SRF. After extensive washings, the bound material was eluted and analyzed by SDS-polyacrylamide gel electrophoresis and autoradiography. As shown in Fig. 6, GATA-4 interacted with GST-SRF independently of DNA binding by SRF and GATA-4. Mutants lacking either the N terminus (aa 127 to 443) or both the N and C termini but containing both zinc fingers were bound by SRF (aa 201 to 332). The first zinc finger alone did not bind SRF (aa 305 to 332). However, the second zinc finger and the immediate C-terminal basic extension (aa 243 to 332) strongly interacted with SRF, indicating that this region of GATA-4 is both necessary and sufficient for interaction with SRF. The specificity of interaction was demonstrated by a lack of binding of wild-type and mutant GATA-4 proteins to GST and the inability of luciferase protein to associate with either GST or GST-SRF (data not shown).
FIG. 6.
Physical interaction between GATA-4 and SRF is mediated by the second zinc finger and the immediate C-terminal basic region. In vitro-translated [35S]methionine-labeled wild-type (WT) GATA-4 (lanes 1 to 3), an N-terminally truncated GATA-4 (ΔN) (lanes 4 to 6), both zinc fingers of GATA-4 (ZF1 + ZF2) (lanes 7 to 9), N-terminal GATA-4 with a deletion of ZF1 (ΔN + ΔZF1) (lanes 10 to 12), the first zinc finger of GATA-4 (ZF1) (lanes 13 to 15), and the second zinc finger along with the immediate C-terminal basic region of GATA-4 (ZF2) (lanes 16 to 18) (7.5 μl each) were incubated with approximately 1 μg (lanes 2, 5, 8, 11, 14, and 17) of GST or GST-SRF fusion protein (lanes 3, 6, 9, 12, 15, and 18) immobilized on glutathione-agarose beads. The beads were washed extensively, and the bound proteins were resolved on an SDS–10% protein gel and visualized by autoradiography. For lanes 1, 4, 7, 10, and 16, 0.75-μl volumes of the lysates were run.
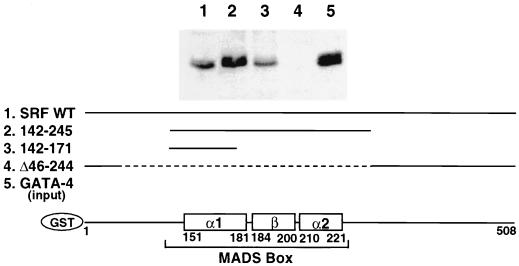
The minimal interactive regions of SRF that are required for interaction with GATA-4 were mapped by evaluating the avidity of various GST-SRF deletion mutant proteins for pull-out of the GATA-4 protein that was overexpressed in the CV1 cell extract. As shown in Fig. 7, deletions in the N and C termini of SRF (aa 142 to 245; aa 142 to 171) did not compromise the ability of SRF to interact with GATA-4. Deletion of the MADS box and dimerization domain abolished binding of SRF to GATA-4(Δ46–244). A subdomain of the MADS box containing part of the α-I helix and its N-terminal extension were necessary and sufficient for binding GATA-4 (aa 142 to 171). GST alone did not interact with GATA-4 (data not shown). Together, our mapping experiments identified the MADS box region of SRF and the second zinc finger and immediate C-terminal basic region of GATA-4 as the minimal protein-protein interaction domains.
FIG. 7.
Physical interaction between SRF and GATA-4 is mediated by the N-terminal portion of the α-I helix of the MADS box of SRF. Approximately 1 μg of wild-type GST-SRF (lane 1), N- and C-terminally truncated SRF (lane 2), the N-terminal portion of the α-I helix of the MADS box of SRF (lane 3), and the SRF with a deletion of the MADS box (lane 4) were immobilized on glutathione-Sepharose beads and incubated with 100 μg of CV1 cell extract overexpressing GATA-4. The beads were washed extensively and resolved on an SDS–10% protein gel, and Western blot analysis was done with an anti-GATA-4 antibody. Ten micrograms of the lysate was run for lane 5. A schematic diagram at the bottom of the figure shows various regions of SRF retained in the deletion mutants.
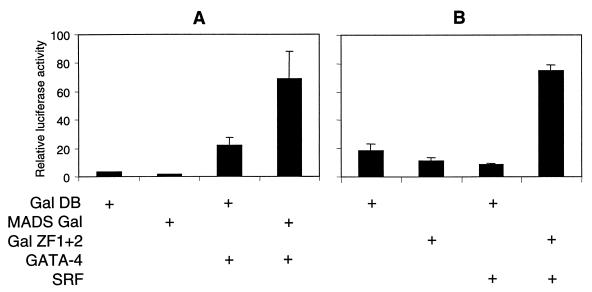
Reciprocal recruitment of SRF and GATA-4 via one-hybrid assays.
The ability of GATA-4 to activate the cardiac α-actin promoter independently of GATA binding sites indicated that GATA-4 was recruited to the promoter through its interaction with SRF. To address whether the minimal interaction domain of SRF tethered to DNA can recruit GATA-4 to the promoter and whether the activation domain of SRF is required for the transcriptional activity of the SRF–GATA-4 complex, we performed an in vivo one-hybrid analysis. CV1 cells were transfected with expression plasmids encoding the MADS box-Gal4 DNA-binding domain fusion protein (MADS-Gal) and full-length GATA-4, along with the Gal4 reporter. As shown in Fig. 8A, GATA-4 alone stimulated the reporter activity nonspecifically about sixfold. Expression of MADS-Gal repressed the basal activity of the Gal4 reporter by 50%. However, coexpression of MADS-Gal with GATA-4 relieved this repression and further enhanced the reporter activity by 45-fold. This result suggested that the MADS box domain of SRF bound to DNA was sufficient for recruitment of GATA-4 to the promoter and that the activation domain of SRF was dispensable for the transcriptional activity of the SRF–GATA-4 complex.
FIG. 8.
Reciprocal recruitment of SRF and GATA-4 via one-hybrid assays. (A) Subconfluent CV1 cells were transfected with 200 ng of Gal4 luciferase reporter and 200 ng of Gal4 DNA-binding domain (Gal DB) or Gal DB fused to the MADS box (MADS Gal) in the presence or absence of 750 ng of GATA-4. (B) One microgram of Gal4 reporter and 1 μg of Gal DB or Gal DB fused to the first and the second zinc fingers of GATA-4 (Gal ZF1 + 2) were transfected in the presence or absence of 100 ng of SRF. Cells were harvested 48 h posttranscription, and the luciferase activity was measured. Results shown are means ± standard errors of the means for two duplicate experiments.
We also asked if the zinc finger domains of GATA-4 can reciprocally recruit SRF to the promoter and if the activation domains of GATA-4 are necessary for the transcriptional activity of the GATA-4–SRF complex. In CV1 cells, coexpression of the GATA-4 zinc finger–Gal4 DNA-binding domain fusion protein (Gal ZF1 + 2) and full-length SRF resulted in a 6.5-fold increase in the transcriptional activity of the Gal4 reporter gene (Fig. 8B). These results suggest that SRF was recruited to DNA by the zinc finger domain of GATA-4 and that the activation domains of GATA-4 were not necessary for the transcriptional activity of the GATA-4–SRF complex. Together, these results suggested that the minimal DNA-binding domains of SRF (MADS box) and GATA-4 (zinc finger 2) can facilitate recruitment when either of them is tethered to DNA. It also suggested that the transcriptional activation domain of either SRF or GATA-4 can confer transcriptional activity on the SRF–GATA-4 complex.
DISCUSSION
Muscle-restricted gene expression is regulated by combinatorial interaction among various classes of transcription factors. Previously, we have shown that the Nkx-2.5 homeodomain factor interacts in pairwise combinations with the MADS box factor SRF and the zinc finger protein GATA-4 to synergistically activate cardiac α-actin gene expression (11, 52). Here we observed that GATA-4 interacts physically and functionally with SRF to drive the expression of the cardiac α-actin promoter. Deletion and point-mutational analysis of GATA-4 revealed the second zinc finger and the immediate C-terminal basic region to be essential for coactivation. In an earlier study (45), by using deletion mutants of GATA-4, two transcriptional domains were mapped to the N terminus of GATA-4. These activation domains were effective when fused to the heterologous DNA-binding domain (45). In addition to the N-terminal activation domains, the C terminus was also necessary for the transcriptional activity of GATA-4. However, this domain was transcriptionally inert in the context of a heterologous DNA-binding domain, indicating indirect participation. Recently, it was shown that acetylation of lysine residues located in the basic region C terminal to the second zinc finger and the inter-zinc finger linker region of GATA-1 results in enhanced DNA binding and transcriptional activity (8). Several of these lysine residues are conserved between GATA-1 and GATA-4. Enhancement of transcriptional activation of a mutated GATA-4 containing the second zinc finger, the inter-zinc finger linker region, and the C terminus by SRF may suggest that SRF, which binds CREB-binding protein (CBP) (also called p300) (50), may facilitate the access of GATA-4 to these transacetylating activities by interacting with and subtly altering the conformation of GATA-4 (26).
Deletion of N-terminal activation domains of GATA-4 located between aa 1 and 74 and aa 130 and 177 did not affect the ability of GATA-4 to coactivate with SRF, suggesting that the activation domain of SRF can compensate for the lack of activation domains on GATA-4. Interestingly, deletion of the second N-terminal activation domains and the first zinc finger of GATA-4 increased the ability of GATA-4 to synergize with SRF, suggesting that these domains interfere with the interaction of SRF and GATA-4. This interference could be mediated by binding of other proteins to these domains of GATA-4, which might preclude efficient interaction of SRF with the second zinc finger of GATA-4. This notion is supported by recent reports describing interaction of a variety of cofactors with the amino finger of GATA proteins. Multi-zinc finger coactivator proteins, such as FOG-1 and FOG-2, modulate the transcriptional activity of GATA-1 and GATA-4 by interacting with the first zinc finger (17, 37, 54, 55, 57). In a similar manner, the Drosophila GATA protein, Pannier, interacts with a zinc finger protein called U-shaped (Ush) (14, 23), which negatively regulates the transcriptional activity of Pannier toward the expression of the proneural basic HLH proteins, Achete and Scute.
The C-terminal activation domains of both SRF and GATA-4 were required for the coactivation of the cardiac α-actin promoter because deletion of the C-terminal activation domain of SRF or GATA-4 abolished coactivation. However, in the one-hybrid system wherein the activation of the GAL-4-dependent synthetic promoter was dependent on the GAL-4 DNA-binding domain in the GAL-4–SRF and GAL-4–GATA-4 hybrid proteins, the C-terminal activation domain of either SRF or GATA-4 was sufficient for activation of the GAL-4-dependent synthetic promoter. The differences in the requirement for the activation domains of SRF and GATA-4 between the two different types of assays (coactivation versus one hybrid) could be related to structural differences between the MADS box bound to DNA and the MADS box tethered to the UAS sequence via the GAL-4 DNA-binding domain. The requirement for the MADS box for both DNA binding and interaction with GATA-4 when SRF is bound to SREs may impose structural constraints that are not present when the MADS box is tethered to DNA.
The coactivation of the cardiac α-actin promoter by SRF and GATA-4 is mediated through SRE1 because deletion and specific point mutations of SRE1 reduced the basal activity of the promoter and eliminated the synergistic activation. Coactivation of the cardiac α-actin promoter was strictly dependent on SRF binding to SRE1, since deletions or point mutations that abolish DNA binding of SRF also abrogated synergistic activation. The coactivation appears to be independent of GATA-4 DNA binding, since no GATA binding site is detected in the minimal fragment of the cardiac α-actin promoter (−100) that was responsive to the SRF–GATA-4 combination. Since GATA factors are known to bind divergent GATA sites (32, 40), we performed a gel shift analysis of potential GATA sites present in the cardiac α-actin promoter to rule out direct DNA binding of GATA-4 in the context of the entire plasmid. None of these sites were bound by GATA-4 (52). Further, skeletal α-actin SRE1 that was cloned upstream of the c-fos minimal promoter was sufficient to confer synergistic activation by SRF and GATA-4. These results, and our earlier report demonstrating the absence of functional cryptic GATA sites in the luciferase vector, strongly suggest that GATA-4 is recruited to the cardiac α-actin promoter by SRF independently of binding of GATA-4 to DNA. Our claim is further supported by the ability of related GATA proteins, such as GATA-1, to activate transcription independently of DNA binding (13, 47).
Transcriptional activation of GATA binding site-dependent genes, such as those for cTnC, ANF, BNP, and troponin I, required the N-terminal activation domains of GATA-4. Further, GATA-5 and GATA-6, which share extensive homology within the N-terminal activation domains, were capable of activating these genes and substituting for GATA-4 (45). Interestingly, GATA site-independent coactivation of NKE-driven reporters by Nkx-2.5 and GATA-4 was independent of N- and C-terminal activation domains of GATA-4 (52). In contrast, synergistic activation of the ANF promoter, which contains binding sites for both GATA-4 and Nkx-2.5 by combinations of GATA-4 and Nkx-2.5, required both the N- and C-terminal activation domains of GATA-4 (16, 35). However, the C-terminal activation domain of GATA-4 was essential for the coactivation of the ANF promoter by GATA-4 and GATA-6 (10). These results indicate that differential utilization of GATA-4's activation domains may depend on the promoter context and other interactive proteins. In support of this hypothesis, transcriptional activity of GATA-1 and GATA-4 was dependent on both the target promoters and their interaction with cofactors FOG-1 and FOG-2.
GATA-4 synergistically activated various muscle-restricted promoters which are expressed in differentiated muscle types. Other cardiovascular tissue-enriched GATA factors, such as GATA-5 and GATA-6, which have an expression pattern distinct from yet overlapping that of GATA-4, were capable of interacting with SRF and substituting for GATA-4 in coactivation assays. These results suggest that the pairing of SRF with different GATA factors confers muscle subtype specificity (such as cardiac versus skeletal versus smooth). Additional degrees of muscle subtype specificity could be conferred by the interaction of the SRF-GATA complex with tissue-restricted factors, such as Nkx-2.5 and MyoD. Our unpublished results show that the cardiac tissue-restricted homeoprotein, Nkx-2.5, combinatorially interacts with both SRF and GATA-4 to strongly activate the cardiac α-actin promoter (Sepulveda et al., unpublished data). In addition to cardiac- and smooth muscle-restricted promoters, the skeletal α-actin promoter and the ubiquitous c-fos promoter, which are normally upregulated during the cardiac hypertrophic response, were also coactivated by SRF and GATA-4. Given the role of GATA-4 in mediating cardiac hypertrophy (44), the interaction of SRF with GATA-4 may have a functional role in the physiological hypertrophic response.
Pull-down assays with bacterially expressed GST-SRF and in vitro-translated GATA-4, as well as with protein A–GATA-4 and protein A-SRF, indicated that these two factors interact in solution and in mammalian cells. By analogy with the Nkx-2.5–GATA-4 interaction (16, 35, 52) and the SRF–Nkx-2.5 synergy reported by Chen and Schwartz (11), the interaction between SRF and GATA-4 required the conserved DNA-binding domains of both proteins. More specifically, the C-terminal zinc finger of GATA-4 and the 142 to 171 region (the N-terminal half of helix 1) of the MADS box were the minimum required. This region of SRF is also the minimum required for interaction with Nkx-2.5 (11) and includes the N-terminal extension of the MADS box that wraps around the DNA to interact with the minor groove of the SRE.
SRF increases the rate of assembly of the preinitiation complex at the target promoter (60), in part by interacting with the Rap74 subunit of TFIIF (29). Little is currently understood about the molecular mechanisms by which GATA-4 activates transcription. One possible mechanism by which SRF and GATA-4 interaction results in increased transcriptional activity relates to the ability of SRF to recruit the coactivator and protein acetylases CBP (50) and SRC-1 (31). GATA-1 also binds CBP (6) and undergoes a conformational change after acetylation by CBP that correlates with activation (26). It is possible that synergistic activation results from a cooperative recruitment of the holoenzyme by SRF (through TFIIF) and of CBP by the SRF–GATA-4 complex.
GATA proteins have been reported to interact with a multitude of transcription factors, but this is the first demonstration of interaction between a GATA protein and SRF. Several functional interactions of SRF with the zinc finger protein Sp1 have been described (4, 51), but physical association of the two proteins has not been demonstrated, while MEF2 has been shown to associate with Sp1 (18). MEF2C, a member of the MADS box family, activates the expression of several muscle-specific genes, either directly by binding to the regulatory regions of the target genes or indirectly by interacting with other muscle-restricted factors, such as MyoD and myogenin (reviewed in references 5 and 42). Reciprocal recruitment of SRF and GATA-4 to the promoter independently of DNA binding by either SRF or GATA-4 is analogous to the cross-recruitment between MEF2C and myogenic bHLH proteins. The reciprocal recruitment between SRF and GATA-4 would expand the spectrum of genes regulated by either of these factors while conferring an additional level of specificity. Our results demonstrating interaction between MADS box proteins, such as SRF and MEF2C, and the zinc finger protein GATA-4 underscore the ability of these proteins to interact combinatorially to drive the myogenic program of gene expression.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grant P01-HL49953, USDA-/ARS6250-6100, and NSBRI (NASA).
REFERENCES
- 1.Arsenian S, Weinhold B, Oelgeschlager M, Ruther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belaguli N S, Schildmeyer L A, Schwartz R J. Organization and myogenic restricted expression of the murine serum response factor gene. J Biol Chem. 1997;272:18222–18231. doi: 10.1074/jbc.272.29.18222. [DOI] [PubMed] [Google Scholar]
- 3.Belaguli N S, Zhou W, Trinh T-H T, Majesky M W, Schwartz R J. Dominant negative murine serum response factor: alternative splicing within the activation domain inhibits transactivation of serum response factor binding targets. Mol Cell Biol. 1999;19:4582–4591. doi: 10.1128/mcb.19.7.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biesiada E, Hamamori Y, Kedes L, Sartorelli V. Myogenic basic helix-loop-helix proteins and Sp1 interact as components of a multiprotein transcriptional complex required for activity of the human cardiac α-actin promoter. Mol Cell Biol. 1999;19:2577–2584. doi: 10.1128/mcb.19.4.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black B L, Olson E N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 6.Blobel G A, Nakajima T, Eckner R, Montminy M, Orkin S H. CREB-binding protein (CBP) cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc Natl Acad Sci USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bodmer R. The gene tinman is required for specification of the heart and visceral muscle in Drosophila. Development. 1993;118:719–729. doi: 10.1242/dev.118.3.719. [DOI] [PubMed] [Google Scholar]
- 8.Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- 9.Browning C L, Culberson D E, Argon I V, Fillmore R A, Croissant J D, Schwartz R J, Zimmer W E. The developmentally regulated expression of serum response factor plays a key role in the control of smooth muscle-specific genes. Dev Biol. 1998;194:18–37. doi: 10.1006/dbio.1997.8808. [DOI] [PubMed] [Google Scholar]
- 10.Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Mol Cell Biol. 1999;19:4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C-Y, Schwartz R J. Recruitment of the tinman homolog Nkx-2.5 by serum response factor activates cardiac α-actin gene transcription. Mol Cell Biol. 1996;16:6372–6384. doi: 10.1128/mcb.16.11.6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croissant J D, Kim J H, Eichele G, Goering L, Lough J, Prywes R, Schwartz R J. Avian serum response factor expression restricted primarily to muscle cell lineages is required for alpha-actin gene transcription. Dev Biol. 1996;177:250–264. doi: 10.1006/dbio.1996.0160. [DOI] [PubMed] [Google Scholar]
- 13.Crossley M, Merika M, Orkin S H. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15:2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cubadda Y, Heitzler P, Ray R P, Bourouis M, Ramain P, Gelbart W, Simpson P, Haenlin M. u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes Dev. 1997;11:3083–3095. doi: 10.1101/gad.11.22.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- 16.Durocher D, Charron F, Warren R, Schwartz R J, Nemer M. The cardiac transcription factors Nkx-2.5 and GATA-4 are mutual cofactors. EMBO J. 1997;16:5687–5696. doi: 10.1093/emboj/16.18.5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox A H, Liew C, Holmes M, Kowalski K, Mackay J, Crossley M. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 1999;18:2812–2822. doi: 10.1093/emboj/18.10.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grayson J, Bassel-Duby R, Williams S R. Collaborative interactions between MEF-2 and Sp1 in muscle-specific gene regulation. J Cell Biochem. 1998;70:366–375. [PubMed] [Google Scholar]
- 19.Grepin C, Robitaille L, Antakly T, Nemer M. Inhibition of transcription factor GATA-4 expression blocks in vitro cardiac muscle differentiation. Mol Cell Biol. 1995;15:4095–4102. doi: 10.1128/mcb.15.8.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Groisman R, Masutani H, Leibovitch M-P, Robin P, Soudant I, Trouche D, Harel-Bellan A. Physical interaction between the mitogen-responsive serum response factor and myogenic basic-helix-loop-helix proteins. J Biol Chem. 1996;271:5258–5264. doi: 10.1074/jbc.271.9.5258. [DOI] [PubMed] [Google Scholar]
- 21.Grueneberg D A, Natesan S, Alexandre C, Gilman M Z. Human and Drosophila homeodomain proteins that enhance the DNA-binding activity of serum response factor. Science. 1992;257:1089–1095. doi: 10.1126/science.257.5073.1089. [DOI] [PubMed] [Google Scholar]
- 22.Gulick J, Subramaniam A, Neumann J, Robbins J. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J Biol Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- 23.Haenlin M, Cubadda Y, Blondeau P, Heitzler P, Lutz P, Simpson P, Ramain P. Transcriptional activity of pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev. 1997;11:3096–3108. doi: 10.1101/gad.11.22.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heikinheimo M, Scandrett J M, Wilson D B. Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev Biol. 1994;164:361–373. doi: 10.1006/dbio.1994.1206. [DOI] [PubMed] [Google Scholar]
- 25.Hipskind R A, Rao V N, Mueller C G, Reddy E S, Nordheim A. Ets-related protein Elk-1 is homologous to the c-fos regulatory factor p62TCF. Nature. 1991;354:531–534. doi: 10.1038/354531a0. [DOI] [PubMed] [Google Scholar]
- 26.Hung H-L, Lau J, Kim A Y, Weiss M J, Blobel G A. CREB-binding protein acetylates hematopoietic transcription factor GATA-1 at functionally important sites. Mol Cell Biol. 1999;19:3496–3505. doi: 10.1128/mcb.19.5.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang Y, Evans T. The Xenopus GATA-4/5/6/genes are associated with cardiac specification and can regulate cardiac-specific transcription during embryogenesis. Dev Biol. 1996;174:258–270. doi: 10.1006/dbio.1996.0071. [DOI] [PubMed] [Google Scholar]
- 28.Johansen F-E, Prywes R. Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by using GAL4-SRF constructs. Mol Cell Biol. 1993;13:4640–4647. doi: 10.1128/mcb.13.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jollot V, Demma M, Prywes R. Interaction with RAP74 subunit of TFIIF is required for transcriptional activation by serum response factor. Nature. 1995;373:632–635. doi: 10.1038/373632a0. [DOI] [PubMed] [Google Scholar]
- 30.Kelley C, Blumberg H, Zon L I, Evans T. GATA-4 is a novel transcription factor expressed in endocardium of the developing heart. Development. 1993;118:817–827. doi: 10.1242/dev.118.3.817. [DOI] [PubMed] [Google Scholar]
- 31.Kim J-H, Kim H-J, Lee W J. Steroid receptor coactivator-1 interacts with serum response factor and coactivates serum response element-mediated transactivations. J Biol Chem. 1998;273:28564–28567. doi: 10.1074/jbc.273.44.28564. [DOI] [PubMed] [Google Scholar]
- 32.Ko L J, Engel J D. DNA-binding specificities of the GATA transcription factor family. Mol Cell Biol. 1993;13:4011–4022. doi: 10.1128/mcb.13.7.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuo C T, Morrisey E E, Anandappa R, Sigrist K, Lu M M, Parmacek M S, Soudais C, Leiden J M. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 34.Laverriere A C, MacNeill C, Mueller C, Poelmann R E, Burch J B E, Evans T. GATA-4/5/6: a subfamily of three transcription factors expressed in developing heart and gut. J Biol Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- 35.Lee Y, Shioi T, Kasahara H, Jobe S M, Wiese R J, Markham B E, Izumo S. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol Cell Biol. 1998;18:3120–3129. doi: 10.1128/mcb.18.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li L, Liu Z-C, Mercer B, Overbeek P, Olson E N. Evidence for serum response factor-mediated regulatory networks governing SM22α transcription in smooth, skeletal, and cardiac muscle cells. Dev Biol. 1997;187:311–321. doi: 10.1006/dbio.1997.8621. [DOI] [PubMed] [Google Scholar]
- 37.Lu J-R, McKinsey T A, Xu H, Wang D-Z, Richardson J A, Olson E N. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol Cell Biol. 1999;19:4495–4502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacLellan W R, Lee T C, Schwartz R J, Schneider M D. Transforming growth factor-beta response elements of the skeletal alpha-actin gene. Combinatorial action of serum response factor, YY1, and the SV40 enhancer-binding protein, TEF-1. J Biol Chem. 1994;269:16754–16760. [PubMed] [Google Scholar]
- 39.Martin D I K, Orkin S H. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf1. Genes Dev. 1990;4:1886–1898. doi: 10.1101/gad.4.11.1886. [DOI] [PubMed] [Google Scholar]
- 40.Merika M, Orkin S H. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999–4010. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molkentin J D, Kalvakolanu D V, Markham B E. Transcription factor GATA-4 regulates cardiac muscle-specific expression of the α-myosin heavy-chain gene. Mol Cell Biol. 1994;14:4947–4957. doi: 10.1128/mcb.14.7.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molkentin J D, Black B L, Martin J F, Olson E N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 43.Molkentin J D, Lin Q, Duncan S A, Olson E N. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 44.Molkentin J D, Lu J, Antons C L, Markham B, Richardson J, Robbins J, Grant S R, Olson E N. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1999;93:1–20. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morrisey E E, Ip H S, Tang Z, Parmacek M S. GATA-4 activates transcription via two novel domains that are conserved within the GATA-4/5/6 subfamily. J Biol Chem. 1997;272:8515–8524. doi: 10.1074/jbc.272.13.8515. [DOI] [PubMed] [Google Scholar]
- 46.Orkin S H. Erythropoiesis: how does it happen? Curr Opin Cell Biol. 1995;7:870–877. doi: 10.1016/0955-0674(95)80072-7. [DOI] [PubMed] [Google Scholar]
- 47.Osada H, Grutz G, Axelson H, Forster A, Rabbitts T H. Association of erythroid transcription factors: complexes involving the LIM protein RBTN2 and the zinc finger protein GATA1. Proc Natl Acad Sci USA. 1995;92:9585–9589. doi: 10.1073/pnas.92.21.9585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parmacek M S, Leiden J M. GATA transcription factors and cardiac development. In: Harvey R, Rosenthal N, editors. Heart development. San Diego, Calif: Academic Press; 1999. pp. 291–306. [Google Scholar]
- 49.Pollock R, Treisman R. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 1991;5:2327–2341. doi: 10.1101/gad.5.12a.2327. [DOI] [PubMed] [Google Scholar]
- 50.Ramirez S, Ali S A S, Robin P, Trouche D, Harel-Bellan A. The CREB-binding protein (CBP) cooperates with the serum response factor for transactivation of the c-fos serum response element. J Biol Chem. 1997;272:31016–31021. doi: 10.1074/jbc.272.49.31016. [DOI] [PubMed] [Google Scholar]
- 51.Sartorelli V, Webster K, Kedes L. Muscle-specific expression of the cardiac α-actin gene requires MyoD1, CArG-box binding factor, and SP1. Genes Dev. 1990;4:1811–1822. doi: 10.1101/gad.4.10.1811. [DOI] [PubMed] [Google Scholar]
- 52.Sepulveda J L, Belaguli N S, Nigam V, Chen C-Y, Nemer M, Schwartz R J. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol Cell Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shore P, Sharrocks A D. The MADS-box family of transcription factors. Eur J Biochem. 1995;229:1–13. doi: 10.1111/j.1432-1033.1995.tb20430.x. [DOI] [PubMed] [Google Scholar]
- 54.Svensson E C, Tufts R L, Polk C E, Leiden J M. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci USA. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tevosian S G, Deconinck A E, Cantor A B, Rieff H I, Fujiwara Y, Corfas G, Orkin S H. FOG-2: a novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc Natl Acad Sci USA. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Treisman R. Journey to the surface of the cell: Fos regulation and the SRE. EMBO J. 1995;14:4905–4913. doi: 10.1002/j.1460-2075.1995.tb00173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tsang A P, Visvader J E, Turner C A, Fujiwara Y, Yu C, Weiss M, Crossley M, Orkin S H. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- 58.Witzgall R, O'Leary E, Bonventre J V. A mammalian expression vector for the expression of GAL4 fusion proteins with an epitope tag and histidine tail. Anal Biochem. 1994;223:291–298. doi: 10.1006/abio.1994.1587. [DOI] [PubMed] [Google Scholar]
- 59.Yu Y-T, Breitbart R E, Smoot L B, Lee Y, Mahdavi V, Nadal-Ginard B. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 1992;6:1783–1798. doi: 10.1101/gad.6.9.1783. [DOI] [PubMed] [Google Scholar]
- 60.Zhu H, Roy A L, Roeder R G, Prywes R. Serum response factor affects preinitiation complex formation by TFIID in vitro. New Biol. 1991;3:455–464. [PubMed] [Google Scholar]