Abstract
Light is a crucial environmental cue not only for photosynthetic energy production but also for plant growth and development. Plants employ sophisticated methods to detect and interpret information from incoming light. Five classes of photoreceptors have been discovered in the model plant Arabidopsis thaliana. These photoreceptors act either distinctly and/or redundantly in fine-tuning many aspects of plant life cycle. Unlike mobile animals, sessile plants have developed an enormous plasticity to adapt and survive in changing environment. By monitoring different information arising from ambient light, plants precisely regulate downstream signaling pathways to adapt accordingly. Given that changes in the light environment is typically synchronized with other environmental cues such as temperature, abiotic stresses, and seasonal changes, it is not surprising that light signaling pathways are interconnected with multiple pathways to regulate plant physiology and development. Indeed, recent advances in plant photobiology revealed a large network of co-regulation among different photoreceptor signaling pathways as well as other internal signaling pathways (e.g., hormone signaling). In addition, some photoreceptors are directly involved in perception of non-light stimuli (e.g., temperature). Therefore, understanding highly inter-connected signaling networks is essential to explore the photoreceptor functions in plants. Here, we summarize how plants co-ordinate multiple photoreceptors and their internal signaling pathways to regulate a myriad of downstream responses at molecular and physiological levels.
Keywords: Photomorphogenesis, Photoreceptors, Sensory proteins, Signal integration, E3 Ubiquitin ligase
One sentence summary
A battery of sensory photoreceptors with spectral specificity provides plants with multiple environmental cues including light and temperature to increase fitness, yield and biomass in agriculture.
1. Introduction
Due to sessile nature of plants, environmental factors inevitably have strong influence on plants’ physiology and development. To efficiently adapt and survive in the changing environment, plants have developed sophisticated ways of detecting external cues and translating it into internal signaling pathways. Among many environmental cues, light is one of the most crucial signals, which affects almost every step of plants lifecycle. Light not only serves as a sole energy source for CO2 fixation by photosynthesis, but also serves as a complex signaling input to modulate plant physiology and development. It is, therefore, essential for plants to correctly interpret light information through the action of multiple photoreceptors.
A series of unique photoreceptors with different wavelength absorption spectra and biochemical properties have been employed to precisely delineate plants’ light environment. To acquire the detailed information from different wavelength of the incoming light, plants have at least five classes of photoreceptors. Phytochromes (PHYA-E in model plant Arabidopsis thaliana) perceive red/far-red lights (600–750 nm); cryptochromes (CRY1, CRY2 and CRY3), phototropins (PHOT1 and PHOT2), F-box containing Flavin binding proteins (e.g., ZEITLUPE, FKF1/LKP2) for blue/UV-A light (320–500 nm); and UVR8 for UV-B light (280–320 nm) (Fig. 1) [1] Recent advances in plant photoreceptor research have identified novel roles of the receptors other than photoperception [2,3]. Thus, emerging evidences support the idea that the photoreceptors are involved in direct perception and/or modulation of responses to a wide range of environmental cues suggesting a role as a “multi-sensor”. Here, the molecular mechanism of light perception of the photoreceptors and their roles in wide range of plant responses will be discussed with an emphasis on the red/far-red light receptor phytochrome signaling pathways.
Fig. 1.
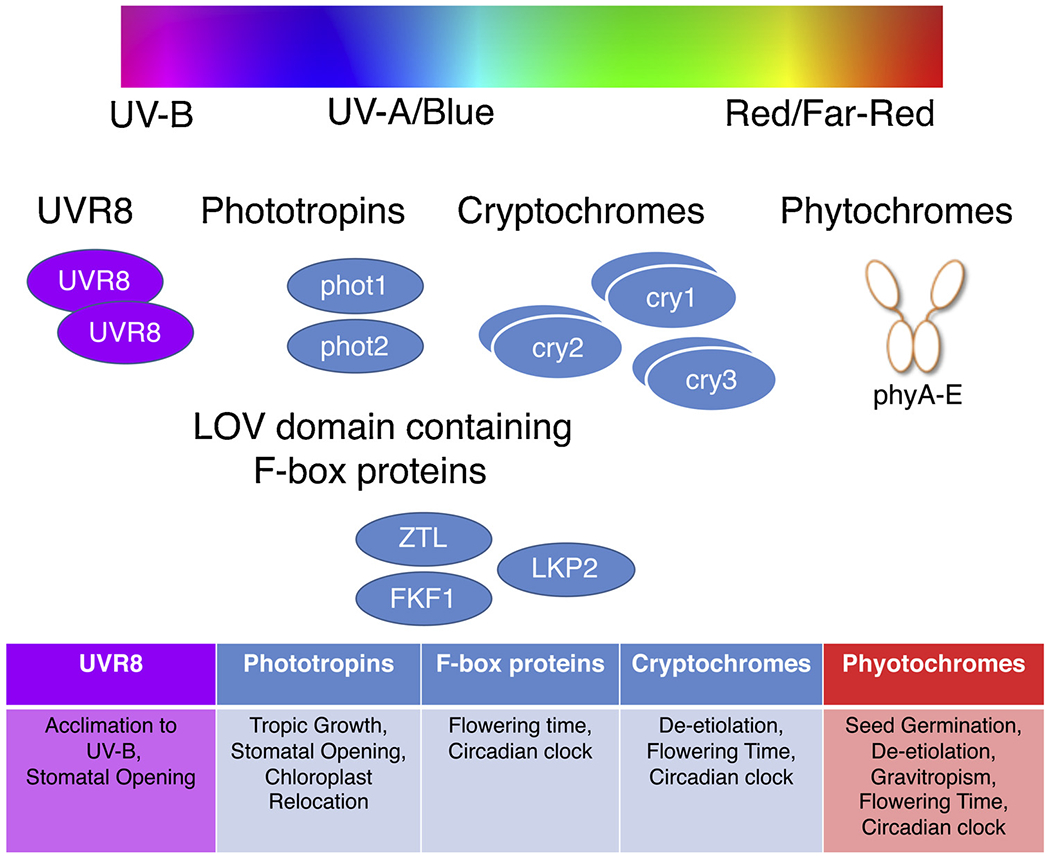
Spectral wavelength-specificity in plant photoreceptors to regulate multiple physiological pathways.
Plants utilize wavelength-specific photoreceptors to perceive and interpret incoming light signals to regulate their physiology and development. Five phytochromes (phyA-E) present in Arabidopsis perceive red (650–670 nm) and far-red (705–740 nm) lights. Three cryptochromes (cry1, cry2, and cry3), two phototropins (phot1, phot2), and three LOV domain F-box proteins (ZTL, FKF1 and LKP2) identified in Arabidopsis perceive blue light. UVR8 perceives UV-B light.
1.1. Molecular mechanisms of photoreceptors action in plants
1.1.1. Phytochrome signaling
Photoreceptors in plants utilize chromophore to detect photons in incoming light. The red/far-red light receptor phytochromes are covalently attached to a phytochromobillin tetrapyrole ring which can isomerize in response to red/far-red light to induce changes in the protein structure [1]. Phytochromes can interconvert between two isoforms: Pr and Pfr representing biologically inactive and active forms in response to far-red and red light, respectively. Although mono-chromatic red/far-red light does not exist in nature, many natural light conditions represent different red/far-red ratio [4]. Densely grown crop plants in agricultural field, for example, results in light conditions with reduced red/far-red ratio due to red light absorption by nearby vegetation. The red light can activate all five phytochromes. However, phytochrome A is the sole photoreceptor for far-red light due to its unique spectral property.
The active Pfr form of phytochromes translocates from cytoplasm to nucleus [5], where they can directly interact with a family of basic-helix-loop-helix transcription factors called PIFs (PHYTOCHROME INTERACTING FACTORS) to initiate light-regulated gene expression. Phytochromes can accomplish the task primarily by inducing multiple biochemical changes on the PIFs transcription factors [6]. These early responses include sequesteration [7], phosphorylation [8–10], polyubiquitylation, and subsequent degradation of the PIFs through the 26S proteasome-mediated degradation pathway [11–15].
Phytochromes also directly interact and inactivate a master negative regulator of light signaling pathway called COP1 (CONTITUTIVE PHOTOMORPHOGENIC 1)-SPA (SUPRESSOR OF PHYA-105) E3 ligase complex through multiple mechanisms. First, activated phytochromes can rearrange the COP1-SPA complex [16,17], so the complex becomes non-functional. COP1-SPA is an E3 ligase complex that prevents light induced gene expression in the dark by destabilizing multiple positively acting transcription factors of light signaling pathways such as HY5, LAF1, HFR1 and others [18]. By destabilizing the positive factors, COP1-SPA confers a strong negative regulation on the light signaling cascade in plants. COP1 activity is largely dependent on its binding partner SPA [19–21]. Photo-activated phytochromes interact with the complex to detach COP1-SPA interaction, hence inactivate COP1-SPA E3 ligase that results in stabilization of the positive factors to promote photomorphogenesis. Second, phytochromes can also inactivate COP1-SPA E3 ligase by excluding COP1 from nucleus through an unknown mechanism [22–24]. Based on these findings, it has been concluded that the active phytochromes can induce chemical/physical changes onto many of their target proteins, especially on the two major negative regulators of plant light signaling (PIFs, COP1-SPA complexes). Some of the proposed intrinsic biochemical properties of phytochrome molecule itself (e.g., kinase activity, sequestration activity) may account for some of the fore-mentioned changes on the target proteins [25,26]. However, many studies also proposed phytochrome associated enzymes (e.g., kinases, phosphatases, E3 ligases) that function to exert biochemical changes on the phytochrome target proteins [27–32]. Collectively, phytochrome action relies on the inhibition of the negative regulators and activation of the positive factors to initiate the light signaling pathways.
Under continuous activating light irradiation, phytochromes are destabilized through the 26S proteasome-mediated degradation pathway. This is a way of receptor desensitization as shown in many other receptor signaling cascades. Phytochrome A exhibits much faster degradation kinetics whereas phyB-E shows relatively slow degradation kinetics [33,34]. Thereby, these two classes of phytochromes are able to confer a distinct action mechanism on many phytochrome regulated responses in Arabidopsis.
In addition to the receptor degradation, multiple layers of regulation exist on phytochrome activity by post-translational modifications. For example, phosphorylation is one of the major modifications on both phyA and phyB [35]. Phosphorylation at different serine, threonine and even tyrosine residues on the phytochromes has been identified. The phosphorylation generally modulates phytochrome activity by affecting various aspects of the molecule. PhyB tyrosine104 phosphorylation, for example, abolishes interaction between phyB and PIF3 as well as affects the stability of phyB nuclear body [36]. Phosphorylation on oat (Avena sativa) phyA serine598 [37] also inhibits phyA-PIF3 interaction while Arabidopsis phyB serine86 phosphorylation facilitates dark reversion (will be discussed in section 3.1) of phyB to reduce the phyB activity [38]. Another post-translational modification called SUMOylation was identified on phyB lysine996 [39]. The amount of SUMOylation was increased by red light and SUMOylated phyB showed reduced activity.
One interesting molecular feature of phytochrome is that it makes nuclear bodies under light [40]. Upon activation, phyB-GFP as well as phyA-GFP makes strong nuclear speckles [41,42]. The amount of active phyB is tightly correlated with the size and number of the speckles [43]. It has been suggested that the phyB speckles are the site of protein degradation since the mutants that exhibit smaller or less phyB speckles, showed delayed degradation of phyB itself or PIF transcription factors [7,44]. However, despite numerous efforts to identify the exact molecular entity of the phyB nuclear speckle, it is not clear what components comprise the speckles and exactly how the formation of phyB speckle is achieved.
1.1.2. Cryptochrome signaling
Blue/UV-A light receptor cryptochromes (CRYs) harbor a Flavin Adenine Dinucleotide (FAD) as a chromophore. For cryptochrome activation, a series of molecular events such as phosphorylation, dimerization and photobody formation are also necessary, following photon perception by FAD [45]. Upon blue light irradiation, cryptochrome molecules undergo rapid phosphorylation. The phosphorylation of CRYs are considered as an essential modification for their function. The phosphorylated CRY2 undergoes a rapid degradation as part of receptor desensitization [45–51]. Cryptochromes are also physiologically active as homodimer. A recent study provided evidences that blue-light-induced homodimer formation is another critical step for CRY activation [52].
Similar to phytochromes, cryptochromes interact with a set of target transcription factors. Cry2, for example, interacts with CIB (CRY2-INTERACTING-BASIC-HELIX-LOOP-HELIX) to initiate downstream gene expression [53,54]. Recently, another set of cryptochrome interacting proteins named BICs were found as a potent inhibitor of cryptochrome signaling. BIC (blue-light inhibitor of cryptochromes 1) interact with cry2 under blue-light. Interestingly, BIC blocks nearly all CRY-mediated responses including early molecular events impinging on CRYs such as dimerization, phosphorylation and photobody formation. Therefore, BIC overexpressing transgenic plants exhibit phenotypes resembling cry1cry2 null mutant [52]. BIC expression itself is also under regulation of CRY signaling, suggesting a feedback inhibition of the blue light signaling cascade [55]. The presence of an active receptor desensitizing protein implies an importance of fine tuning of the photoresponse in plants.
1.1.3. Phototropin signaling
Phototropins are blue light receptors responsible for the well-known phototropic responses of plants. Phototropin molecule contains two Flavin MonoNucleotide (FMN) as a chromophore on its LOV (Light Oxygen Voltage) domains [56]. Through their c-terminal serine/threonine kinase domain, phototropins can directly phosphorylate many substrates including phot1/phot2 themselves. Phot1 can interact with NPH3 (NON-PHOTOTROPIC HYPOCOTYL 3) and PKS4 to modulate downstream signaling [57,58]. Phototropins also play an essential role in blue light mediated stomatal opening and chloroplast movement in response to light [59,60].
1.1.4. ZTL/FKF1/LKP2 family of blue light receptor signaling
ZEITLUPE (ZTL)/ FLAVIN-BINDING, KELCH REPEAT, F-BOX 1 (FKF1)/ LOV kelch protein2 (LKP2) is another class of the blue light receptor. The Flavin-binding F-box protein, ZTL and FKF1 transduce light signals primarily by altering the activity of the SCF E3 ligase complex [61]. As a consequence, targets of SCF E3 ligase alter their stability as well. ZTL mainly functions in the regulation of circadian clock as ZTL itself is positioned in interconnected clocks in plants [62]. However, FKF1 regulates the abundance of a potent photoperiodic flowering inducer, CO (CONSTANS) in Arabidopsis indirectly by controlling the level of CDF1, a Dof-domain transcription factor. This regulation results in photoperiod-dependent flowering under long-day conditions [63].
1.1.5. UVR8 signaling
More recently identified UV-B receptor, UVR8, senses UV-B light in a way that is quite distinct from other photoreceptors. Rather than utilizing chromophore, a set of aromatic rings from a few tryptophan residues on the UVR8 protein, absorbs UV-B light. Upon UV-B absorption, interface between UVR8 dimer breaks, and the resulting monomer migrates into the nucleus and interact with COP1 [64–67]. This interaction results in the stabilization of HY5 to initiate UV-B mediated gene expression. Recently, two UVR8-interacting transcription factors (BES1/BIM1 and WRKY 36) have been discovered to function in the UV-B signaling pathways [68,69].
Even though the five classes of photoreceptors in Arabiodopsis have distinct biochemical mechanism to translate environmental light signals, they share some common mechanistic features. First of all, perceived photon energy is translated into changes in the molecular structure of the photoreceptors leading to the activation of the receptors. Second, activated photoreceptors convey the light information to the signaling proteins primarily by direct interaction and inducing biochemical/physical changes on the proteins. Third, the receptor desensitizing/inactivation mechanism is present to fine-tune the signaling cascades. By employing multiple photoreceptors with distinct but overlapping functions, plants can monitor their surrounding environment to assure proper physiological responses.
2. Regulation of plant development by photoreceptors
2.1. Germination
Sensing surrounding environmental conditions and determining whether to germinate or not is a crucial decision for plant’s survival. This is especially true when light availability is limited due to soil coverage over the seeds. Water imbibed seed requires red light to trigger phytochrome action to initiate the germination process in Arabidopsis. Red light activates phytochromes to promote synthesis of germination promoting hormone gibberellic acid (GA) as well as to reduce germination inhibiting hormone abscisic acid (ABA) in seeds [70]. In this particular response, a spatio-temporal regulation of phyA and phyB plays an important role in exhibiting differential activities by two phytochromes. In dry seeds, phyA expression is very limited while phyB expression level is abundant [71]. Thus, initially phyB is a major photoreceptor to perceive germination promoting red light. Upon cold stratification or about 48 h of water imbibition, phyA level increases and reaches enough amount to trigger seed germination [71]. Interestingly, only phyA can be activated under very low fluence of light, because of its unique spectral property, triggering very low fluence reponse (VLFR)-mediated seed germination [72]. It appears, in nature, seeds can respond to very weak incoming light after certain imbibition period by temporal regulation of phyA expression.
An interesting report identified a tissue specific phytochrome regulation of seed germination. By physically separating seed coat and embryo, scientists were able to dissect spatial specificity of the phytochrome action in seed germination. In canopy shade where far-red light is enriched, inactivated phyB function in seed endosperm overrides phyA-dependent germination in the embryo through the action of plant hormone ABA [73]. The spatially separated activities of phyA and phyB, together with temporal regulation of the photoreceptor expressions, allow fine modulation of seed germination in response to rapidly changing environment.
2.2. De-etiolation; transcription and post transcriptional control by photoreceptors
When germinated seeds develop into seedlings, plants display two different developmental programs in the dark and light. In nature, soil-covered seedlings need to emerge to the surface of the soil to perceive sunlight. Until then, seedlings have to protect their shoot apical meristem (SAM) by maintaining apical hook and closed cotyledons. The hypocotyls rapidly grow toward soil surface. When seedlings appear under the sunlight, they start a dramatic morphological and genetic change called photomorphogenesis; the hypocotyl cells stop rapid elongation, the cotyledons open and a massive light-induced gene expression initiates [74]. Photoreceptors in plants govern this dark-to-light transition called, de-etiolation. For example, the phyAphyB mutant that lacks two major phytochromes in Arabidopsis, is almost completely insensitive to the red light-mediated inhibition of hypocotyl elongation and cotyledon opening [75]. At molecular level, phyA plays a major role in early (~1 h after irradiation) gene expression change, whereas phyB is responsible for the prolonged light-induced gene expression changes [76].
The photoreceptors-mediated de-etiolation process can be conceptualized as the “inactivation of the negative factors and activation of the positive factors” of light signaling pathways. In the dark two major negative regulators, PIFs and COP1/SPA E3 ligase, together block the light-mediated gene expression. COP1/SPA E3 ligase complex is active in the dark and degrades the positively acting transcription factors in the light signaling pathway [18]. PIFs and COP1/SPA work synergistically to repress light signaling in the dark [77]. Upon light activation, phytochromes as well as cryptochromes interact with PIFs (phytochromes and cryptochromes) and CIBs (cryptochromes only), to regulate their function. In addition, phytcohromes and cryptochromes also interact with COP1/SPA complex to inactivate the complex, resulting in stabilization of HY5 transcription factor that can target nearly 9000 genes (Fig. 2) [78,79].
Fig. 2.
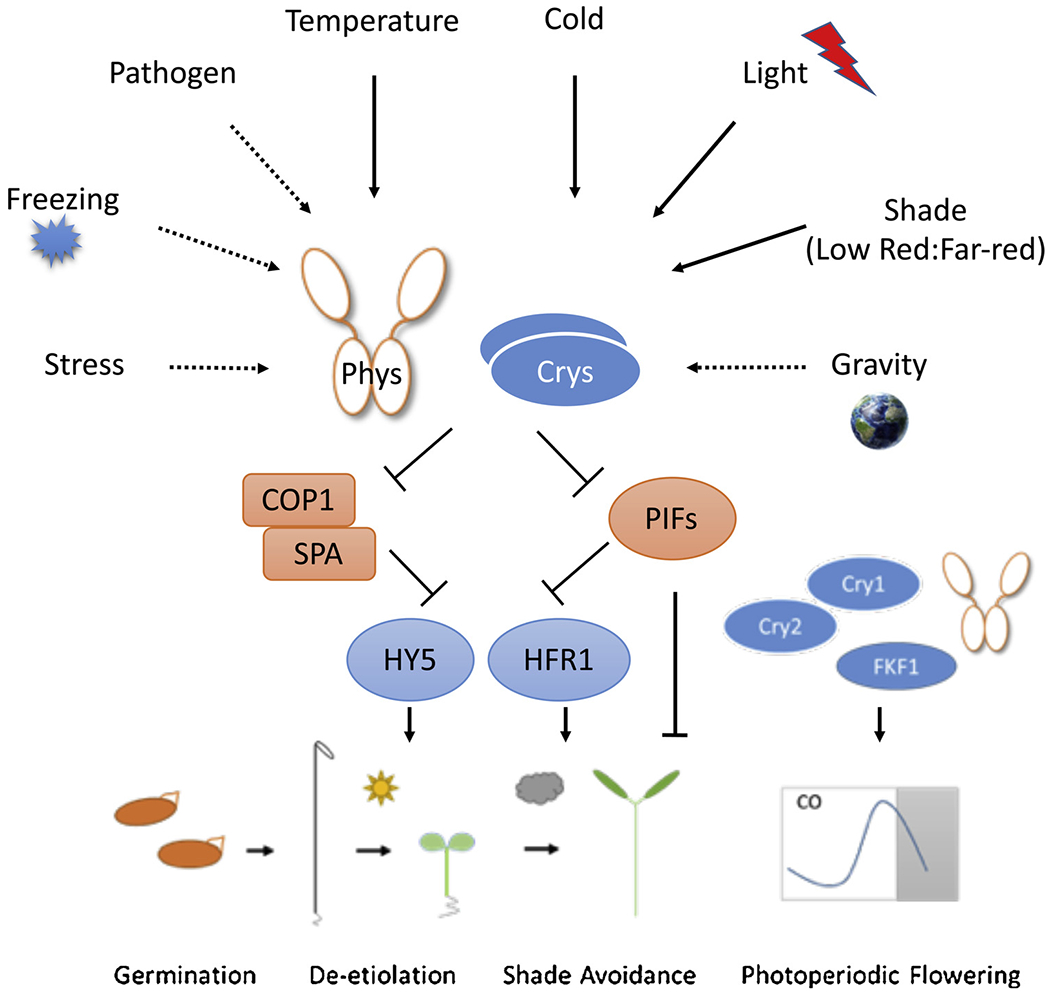
Convergence of multiple environmental cues on photoreceptors.
Phytochromes directly perceive ambient light as well as temperature information. Through co-ordination between photoreceptors, they can modulate myriad of plants’ responses including biotic and abiotic stress responses, gravitropism, as well as multiple developmental transitions. At molecular level, photoreceptors share some of the critical signaling components such as E3 ligase COP1-SPA complex as well as transcription factors called PIFs (PHYTOCHROME INTERACTINF FACTORS), HY5, and HFR1. By inactivating negative regulators of light signaling, COP1-SPA and PIFs, photoreceptors can initiate massive gene expression changes in response to light signal. Photoreceptors also stabilize master transcription factors such as HY5 and HFR1 to target a large number of genes in Arabidopsis genome. These photoreceptor-mediated regulations eventually lead to optimized growth and fitness of plants resulting in enhanced grain and biomass yield in agriculture.
The activated phytochromes trigger massive transcriptional change primarily through destabilization/stabilization of the above mentioned transcription factors. However, apart from indirect regulation through regulation of the transcription factors, a direct role of phytochromes on transcription is not well understood. Some studies reported phytochrome association on the promoters of light and temperature regulated genes, but the biological significance of this promoter occupancy is not clear [3]. It should also be noted that only 0.7 correlation coefficient was observed in gene expression between dark-grown pifQ (pif1pif3-pif4pif5) mutant and red light grown wild type seedlings [80,81]. This suggests the existence of either additional transcription factors or non-transcriptional regulation in phytochrome-mediated signaling pathways.
In accordance with this, multiple reports suggested phytochrome regulation at the post-transcriptional level during de-etiolation. Red light and phytochrome-dependent manner, about 7% of annotated Arabidopsis genome has shown alternative splicing [82]. Specifically, a phytochrome associated splicing factor, SFPS was identified to function under red light-mediated splicing events [83]. In addition, phytochromes were shown to regulate mRNA translation efficiency of a chlorophyll biosynthetic gene, PORA [84]. Consistently, a proteomics study found phytochromes to be associated with an active translatome in etiolated Arabidopsis seedlings, suggesting a possible role of the photoreceptor in translational control [85]. Another interesting study revealed alternative promoter selections by phytochromes which ultimately resulted in production of distinct protein isoforms with differential subcellular localization [86]. Biological relevance of the alternative promoter selection in phytochrome signaling needs further study.
During early light perception, phytochromes promote the expansion of cotyledon cells while inhibit hypocotyl cell expansion, an intriguing contrasting impact of phytochromes in two different tissues. Thus, phytochromes can function in two opposite directions in cell growth in cotyledons and hypocotyl [87]. This may imply the existence of distinct signaling pathways in different tissues in the light signaling pathways, leading to a completely different physiological output in diverse tissues [88]. In support of this speculation, a recent report described a distinct tissue-specific function of phytochrome B in gravitropic response in Arabidopsis [89]. (Additional discussion in Section 3.3). Therefore, exploring tissue-specific function of photoreceptors would be of great interest.
2.3. Shade avoidance response
One of the challenges for plants in agricultural field and in nature is limitation of light sources due to neighboring vegetation. The dense canopy reduces photosynthetic activity and weakens plants’ fitness due to a low red/far-red ratio as well as low blue/green light. To survive in shade conditions, photoreceptors in plants have developed an active shade escaping strategy called shade avoidance response (SAR) that include rapid stem (hypocotyl) elongation, leaf hyponasty, and induction of flowering (Fig. 2).
Phytochrome B has a prominant role in shade avoidance response. Since the shade condition reduces active phyB in plants, phyB-mediated light signaling pathways are also inhibited under shade conditions. Consistently, phyB mutant displays constitutive shade avoidance response; elongated hypocotyl, hyponastic cotyledons and leaves, and early flowering [90]. In phyB mediated shade avoidance response, PIFs play an important role. Among all PIFs, especially PIF4, PIF5 and PIF7 were found to be essential for biosynthesis of plant growth hormone auxin, in response to shade conditions enabling a rapid cell elongation [91,92]. An inhibitory pathway also exists for phyB-PIF mediated shade avoidance response. Phytochrome A functions antagonistically to phyB under increasing far-red light [93]. In addition, PIF function is limited by an atypical bHLH protein, HFR1 to prevent excessive shade avoidance response [94]. Other photoreceptors such as cryptochrome and UVR8 also function in shade avoidance response due to a reduced level of blue and UV-B light in shade conditions. Interestingly, PIF4 and PIF5 are also involved in cryptochrome mediated shade avoidance response [95].
2.4. Flowering time
Determination of correct timing for floral induction is essential for plant reproduction in seasonal changes. For perennial and biennial plants which experience at least one winter season during lifecycle, it is critical to predict winter for spring blossom. Even for annual plants, proper transition from vegetative to reproductive phase is critical for their successful reproduction. Since the day-length is typically correlated with seasonal changes, photoreceptors play important roles in measuring day length to induce/repress flowering [96]
Three classes of photoreceptors are major positive regulators of photoperiodic flowering induction in Arabidopsis; FKF1, phytochrome A and cryptochromes. (Fig. 2.) [96]. How can plants measure the ‘day-length’? Through extensive molecular and genetic approaches, scientists have established a mechanism called the ‘external coincidence model’. Basic concept of the model is that the internal signal fluctuation and the external photoperiodic signal must coincide to induce flowering.
FT (Flowering locus T) is a florigen which can directly induce flowering in plants [97]. CO (CONSTANS) is a potent transcriptional activator of FT and is a key component in the external coincidence model [98]. CO expression level fluctuates during day and night cycles (Fig. 2). When CO expression reaches the highest in late afternoon, photoreceptors must be activated to stabilize CO protein to achieve photoperiodic flowering induction. Therefore, the changing CO expression acts as an internal factor and the photoreceptor activation in the late afternoon acts as an external factor to coincide. In long-day photoperiod (16h:8 h, light:dark), photoreceptors can be activated by light in the late afternoon. In the short-day (8h:16 h, light:dark), however, external light input is missing in late afternoon because of early sunset resulting in the lack of accumulation of CO protein.
In flowering induction, far-red and blue lights promote flowering through the action of phyA and cryptochromes, respectively. In blue light, activated cryptochrome2 can directly interact with the E3 ligase COP1/SPA complex to inactivate the complex. Through the action of cry2, COP1/SPA can no longer degrade its target CO protein resulting in an abundant CO level [99]. In addition, phyA stabilize CO in response to far-red light to promote flowering [100]. Contrary to phyA, phyB represses floral induction as phyB destabilizes CO [100]. Only recently, a novel photoperiodic flowering regulator in phyB signaling has been suggested. PHL (PHYTOCHROME-DEPENDENT LATE FLOWERING) was identified to form a complex with phyB and CO, but how PHL promotes flowering in phyB-PHL-CO complex is not clear. Phytochrome B in flowering regulation also exhibits tissue specificity. It was shown that the mesophyll-cell-expressed phyB can delay the flowering time by inhibiting FT expression in vascular bundles [101]. Further study is necessary to explore the molecular nature of phytochrome B regulation on CO level.
Another blue light receptor, an F-box protein, FKF1 has an essential role in photoperiod induction of flowering [63]. FKF1 forms a complex with GI (GIGANTEA), a flowering promoting protein with a co-chaperon function [102,103]. The expression of both FKF1 and GI is under circadian clock. When FKF1 and GI expression reach the highest in late afternoon under long day, blue light-activated FKF1 can interact with GI to trigger ubiquitin-mediated degradation of CDF, a CO repressor, resulting in de-repression of CO [63]. The mechanism of FKF1 action is also an example of an external coincidence model where internal circadian clock-regulated FKF1-GI gene expression and external light activation of FKF1 by long-day can establish CO and FT expression resulting in floral induction.
3. Photoreceptor regulation of plant physiology in response to other environmental cues
3.1. Multi-functional plant photoreceptors: thermosensing
Recent advances have identified unexpected roles of the photoreceptors in plant physiology. Interestingly, other than typical red/far-red light perception, phytochrome B has been suggested as a thermosensor that can directly perceive ambient temperature [2,3]. How does this work? Thermosensors in insects and mammalian system are mostly ion channels. In mammalian system, for example, a Transient Receptor Potential (TRP) ion channel family functions as a sensor of cold and warmth to initiate ion current for downstream signaling [104]. Although, the detailed molecular mechanism of thermo-perception by the TRP channels are yet to be fully understood; however, thermodynamic structural changes of the protein under different temperature may account for the temperature sensing similar to secondary structural changes of RNA acting as thermosensors in bacteria [105].
Unlike animal system, no ion channel has been identified as a thermosensor in plants yet. However, it has been widely known that the photo-activated phytochrome B (Pfr) is thermodynamically unstable (at high energy state) and slowly converts back to the stable inactive form (Pr) in the dark through a process called “dark reversion” or “thermal reversion” [106]. Researchers found that at high ambient temperature, the active Pfr form of phyB reverts faster to the inactive Pr form compared to low temperature. As a result, at high temperature, the level of active phyB is reduced. Thus, the ambient temperature perceived by phyB is translated into Pfr/Pr ratio of phyB and ultimately into the downstream gene expression changes. Indeed, a recent study found a subset of temperature-regulated genes are also target of phyB chromatin binding [2,3]. Therefore, it was concluded that phytochromes can directly perceive at least two different environmental cues, light and temperature in Arabidopsis. Because the amount of sunlight irradiation generally correlates with the ambient temperature, it is not surprising that the photoreceptor also has developed a function as a thermosensor.
It will be intriguing to examine whether other photoreceptors (e.g., cryptochromes) can function in the thermosensory pathway. Since activated cryptochromes undergo thermal relaxation as well [52], there might be a differential inactivation rate of cryptochromes under different ambient temperatures. Although, not a direct perception of the temperature by cryptochromes, a recent study suggested that crytochrome 1 (cry1) mediates thermo-induced hypocotyl elongation through a direct interaction with the bHLH transcription factor, PIF4 [107].
3.2. Photoreceptor regulation of heat and cold stress
Photoreceptor functions are not limited to light and ambient temperature perception. Several studies suggested indirect involvement of photoreceptors in cold and heat stress responses as well as other physiological responses to abiotic challenges to plants.
Temperatures subzero or above physiologically endurable levels, result in severe damages to the plant tissues and often leads to death. Sessile plants have to cope with these extreme stresses by anticipating freezing or heating stresses. One of the early studies on molecular mechanisms of phytochrome action in cold responses revealed a light quality dependent freezing tolerance. Under low red/far-red ratio, in which phytochromes are less active, plants showed more tolerance to the freezing stress [108]. This was achieved by an induction of key transcription factors, CBF1,2,3 that play a major role in plants cold acclimation; a response where plants tolerate freezing better when they are exposed to a low temperature (above freezing temperature) prior to the freezing temperature (below 0 °C) [109]. Therefore, it seems that less phytochrome activity helps plants to anticipate freezing temperature. This is a very clever strategy adopted by plants since the lowest temperature is typically observed before the dawn when phytochrome activities are minimum during a day.
Several studies have further connected dots between phytochromes and freezing tolerance. It has been shown that phytochromes also modulate plant stress hormone signaling such as ABA and JA [110]. In addition, a direct involvement of PIF4/PIF7 to induce CBF genes were identified. These PIFs can directly bind to the CBF promoters to induce gene expression leading to freezing tolerance [111].
Unlike moderate high temperature (e.g., 28 °C for Arabidopsis) repressing phyB activity, an interesting study suggested a synergistic effect between phyB signaling and a brief heat shock. Several downstream transcription factors (e.g., PIF4, PIF5 and HY5) were shown to be involved in this synergistic effect [112]. However, it is not clear if the heat shock can directly alter phytochrome activity. In a separate study, a subset of heat shock genes were induced in response to early excess light, indicating a possible connection between the heat shock response and the light signaling pathways [113]. Furthermore, rice phyAphyB mutant showed altered expression of multiple heat shock proteins leading to a developmental defects in anther and pollen, suggesting a regulation on the abundance of heat shock proteins by phytochromes [114]. Overall, phytochromes have evolved with additional roles in sensing temperature responses.
3.3. Photoreceptor regulation of gravitropism
Sensing gravity is another critical task for photoreceptors in plants. Plants sense gravity through movement of starch-filled subcellular organelle called amyloplasts both in root and shoot tissues. Although, phytochromes do not directly perceive gravity cue, they play a prominant role in red light-induced inhibition of negative gravitropism of the shoot as well as the gravitropic curvature of the root. In dark-germinated seedlings, perceived gravity cue enables upward hypocotyl elongation to the soil surface against gravity in a process called negative gravitropism. Once the seedlings reach the surface, light becomes a major cue for plant growth, hence breaking negative-gravitropic response of the shoot through the action of photoreceptors. At molecular level, phytochromes induce a conversion of endodermal amyloplast into plastids by inactivating PIF function. By differentiating the gravity sensing organelles, amyloplasts in the endodermal cell layer of the hypocotyl, phytochromes can abolish shoot negative gravitropism [115].
Interestingly, a unique tissue-specific activity of phytochrome B was observed in this particular response. Researchers found that only epidermally expressed phyB can inhibit shoot negative gravitropism, but not the endodermal layer expressed phytochrome B. Since the amyloplasts and PIFs in the endodermal layer are required for negative gravitropism of the shoot, the phytochrome action is non-cell-autonomous [89]. This is a very intriguing phenomenon; however, the mechanistic details on how the epidermal phytochromes affect the endodermal PIFs and its downstream signaling is unknown.
Phytochromes are also required for gravitropic response in root. In response to changing gravity cue, plant roots change its growth direction. Arabidopsis phyAphyB mutant responds much less to changing gravity cue [116]. The phyAphyB mutant also exhibits shorter primary root compared to wild type suggesting an important role of the photoreceptor function in root development as well as gravitropic response.
Cryptochromes have been proposed as a magnetoreceptor in many species such as migrating bird, European robin [117]. In Arabidopsis, cryptochrome 1 has been biophysically examined and suggested to have an enhanced activity under weak external magnetic field [118]. It is still not clear whether plant cryptochromes are the bona fide magnetoreceptor and what is the physiological relevance of plant magnetoreceptor. In gravitropic response, cryptochromes seem to play a minor role.
As discussed early in this chapter, phototropism is antagonistic to gravitropism possibly due to the relative importance of phototropic responses over gravitropic responses in the presence of light. Phototropins are major photoreceptors responsible for phototropism and phytochromes indirectly assist phototropic response by inhibiting gravitropic response in light [115,119]. Therefore, the coordination between two photoreceptors can establish the fine balance between phototropism and gravitropism.
Not surprisingly, recent studies also suggested photoreceptors function in biotic stress response (e.g., pathogen response) in plants. Generally, mutants with reduced light signaling (e.g., phyB) showed reduced fitness. As a result, the high susceptibility to pathogens observed in these mutants were considered as an indirect effect of the unhealthy plants. Instead, recent genetic studies identified direct regulation of the photoreceptors and their signaling components in plant-pathogen responses especially through a defense hormone jasmonic acid (JA) [120]. Readers are referred to recent reviews covering this topic in detail [6,121].
4. Conclusion
Sessile plants have the most sophisticated photoreceptor system among all living organisms. Recent advances in photoreceptor study revealed novel roles of plant photoreceptors. As discussed in this review, phytochromes can directly perceive at least two environmental cues; red/far-red light and ambient temperature. Cryptochrome, Phototrpin, ZTL and UVR8 photoreceptors are functioning to orchestrate plant development and responses to dynamic environmental cues. Importantly, plants responses to the multiple non-light environmental cues are directly or indirectly regulated by these photoreceptors.
It should be noted that the striking similarity found in plant responses to the shade (low red: far-red ratio), the high ambient temperature and the stress responses implies a shared perception or signaling pathways among these responses. Thus, it is not surprising that the red/far-red light receptor phytochrome can function as a ‘multi-sensor’. Many of the critical light-signaling components are shared by different photoreceptors, again suggesting that photoreceptors are co-operatively working as multi-functional receptors.
Although, many of the downstream signaling factors are commonly shared by photoreceptors, plants still can generate delicate and distinct responses to different external cues. This has been achieved through a spatio-temporal regulation of photoreceptors and their signaling molecules, a distinct biochemical and spectral properties of the photoreceptors themselves, and a combination with specific signaling factors. Although not proven yet, it might be also possible that plant photoreceptors can utilize unknown mobile signals (e.g., peptide hormones) as well as second messenger molecules such as Ca2+ ion flux for their signaling cascades as proposed earlier [122].
Plants photoreceptors govern plant-environment interactions. It would be of extreme interest to explore if photoreceptors involved in other non-light related responses and the molecular mechanism of how that is achieved. The emerging roles of the plant photoreceptors are opening doors to new arena in plant photobiology.
Acknowledgments
We thank Dr. Praveen Kathare and Vinh Pham for critical reading of this manuscript. Due to space constraints, we apologize that many recent articles from other colleagues could not be discussed. No conflict of interest is declared.
Funding
We acknowledge support by grants from the National Institute of Health (NIH) (1R01 GM-114297) and National Science Foundation (MCB-1543813) to E.H.
References
- [1].Bae G, Choi G, Decoding of light signals by plant phytochromes and their interacting proteins, Annu. Rev. Plant Biol 59 (2008) 281–311. [DOI] [PubMed] [Google Scholar]
- [2].Legris M, et al. , Phytochrome B integrates light and temperature signals in Arabidopsis, Science 354 (6314) (2016) 897–900. [DOI] [PubMed] [Google Scholar]
- [3].Jung JH, et al. , Phytochromes function as thermosensors in Arabidopsis, Science 354 (6314) (2016) 886–889. [DOI] [PubMed] [Google Scholar]
- [4].Casal JJ, Shade avoidance, Arabidopsis Book 10 (2012) p. e0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Sakamoto K, Nagatani A, Nuclear localization activity of phytochrome B, Plant J. 10 (5) (1996) 859–868. [DOI] [PubMed] [Google Scholar]
- [6].Paik I, et al. , Expanding roles of PIFs in signal integration from multiple processes, Mol. Plant 10 (8) (2017) 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Park E, et al. , Phytochrome B inhibits binding of phytochrome-interacting factors to their target promoters, Plant J. 72 (4) (2012) 537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Al-Sady B, et al. , Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation, Mol. Cell 23 (3) (2006) 439–446. [DOI] [PubMed] [Google Scholar]
- [9].Shen H, et al. , Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes, Plant Cell 20 (6) (2008) 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Shen Y, et al. , Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation, Plant Physiol. 145 (3) (2007) 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Oh E, et al. , Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis, Plant J. 47 (1) (2006) 124–139. [DOI] [PubMed] [Google Scholar]
- [12].Bauer D, et al. , Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis, Plant Cell 16 (6) (2004) 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Park E, et al. , Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling, Plant Cell Physiol. 45 (8) (2004) 968–975. [DOI] [PubMed] [Google Scholar]
- [14].Huq E, et al. , Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis, Science 305 (5692) (2004) 1937–1941. [DOI] [PubMed] [Google Scholar]
- [15].Shen H, Moon J, Huq E, PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis, Plant J. 44 (6) (2005) 1023–1035. [DOI] [PubMed] [Google Scholar]
- [16].Lu XD, et al. , Red-light-dependent interaction of phyB with SPA1 promotes COP1-SPA1 dissociation and photomorphogenic development in Arabidopsis, Mol. Plant 8 (3) (2015) 467–478. [DOI] [PubMed] [Google Scholar]
- [17].Sheerin DJ, et al. , Light-activated phytochrome A and B interact with members of the SPA family to promote photomorphogenesis in Arabidopsis by reorganizing the COP1/SPA complex, Plant Cell 27 (1) (2015) 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lau OS, Deng XW, The photomorphogenic repressors COP1 and DET1: 20 years later, Trends Plant Sci. 17 (10) (2012) 584–593. [DOI] [PubMed] [Google Scholar]
- [19].Hoecker U, Quail PH, The phytochrome A-specific signaling intermediate SPA1 interacts directly with COP1, a constitutive repressor of light signaling in Arabidopsis, J. Biol. Chem 276 (41) (2001) 38173–38178. [DOI] [PubMed] [Google Scholar]
- [20].Menon C, Sheerin DJ, Hiltbrunner A, SPA proteins: SPAnning the gap between visible light and gene expression, Planta 244 (2) (2016) 297–312. [DOI] [PubMed] [Google Scholar]
- [21].Seo HS, et al. , LAF1 ubiquitination by COP1 controls photomorphogenesis and is stimulated by SPA1, Nature 423 (6943) (2003) 995–999. [DOI] [PubMed] [Google Scholar]
- [22].von Arnim AG, et al. , Genetic and developmental control of nuclear accumulation of COP1, a repressor of photomorphogenesis in Arabidopsis, Plant Physiol. 114 (3) (1997) 779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Subramanian C, et al. , The Arabidopsis repressor of light signaling, COP1, is regulated by nuclear exclusion: mutational analysis by bioluminescence resonance energy transfer, Proc. Natl. Acad. Sci. U. S. A 101 (17) (2004) 6798–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stacey MG, Hicks SN, von Arnim AG, Discrete domains mediate the light-responsive nuclear and cytoplasmic localization of Arabidopsis COP1, Plant Cell 11 (3) (1999) 349–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Shin AY, et al. , Evidence that phytochrome functions as a protein kinase in plant light signalling, Nat. Commun 7 (2016) 11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Yeh KC, Lagarias JC, Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry, Proc. Natl. Acad. Sci. U. S. A 95 (23) (1998) 13976–13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Bu Q, et al. , Phosphorylation by CK2 enhances the rapid light-induced degradation of phytochrome interacting factor 1 in Arabidopsis, J. Biol. Chem 286 (14) (2011) 12066–12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bu Q, Zhu L, Huq E, Multiple kinases promote light-induced degradation of PIF1, Plant Signal. Behav 6 (8) (2011) 1119–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ni W, et al. , PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3, Nat. Commun 8 (2017) 15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ryu JS, et al. , Phytochrome-specific type 5 phosphatase controls light signal flux by enhancing phytochrome stability and affinity for a signal transducer, Cell 120 (3) (2005) 395–406. [DOI] [PubMed] [Google Scholar]
- [31].Dong J, et al. , Light-dependent degradation of PIF3 by SCFEBF1/2 promotes a photomorphogenic response in Arabidopsis, Curr. Biol 27 (16) (2017) 2420–2430 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ni W, et al. , A mutually assured destruction mechanism attenuates light signaling in Arabidopsis, Science 344 (6188) (2014) 1160–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Seo HS, et al. , Photoreceptor ubiquitination by COP1 E3 ligase desensitizes phytochrome A signaling, Genes Dev. 18 (6) (2004) 617–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Furuya M, Molecular properties and biogenesis of phytochrome I and II, Adv. Biophys 25 (1989) 133–167. [DOI] [PubMed] [Google Scholar]
- [35].Kim JI, et al. , Phytochrome phosphorylation in plant light signaling, Photochem. Photobiol. Sci 4 (9) (2005) 681–687. [DOI] [PubMed] [Google Scholar]
- [36].Nito K, et al. , Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors, Cell Rep. 3 (6) (2013) 1970–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kim JI, et al. , Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction, Plant Cell 16 (10) (2004) 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Medzihradszky M, et al. , Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis, Plant Cell 25 (2) (2013) 535–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sadanandom A, et al. , SUMOylation of phytochrome-B negatively regulates light-induced signaling in Arabidopsis thaliana, Proc. Natl. Acad. Sci. U. S. A 112 (35) (2015) 11108–11113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Chen M, Phytochrome nuclear body: an emerging model to study interphase nuclear dynamics and signaling, Curr. Opin. Plant Biol 11 (5) (2008) 503–508. [DOI] [PubMed] [Google Scholar]
- [41].Kircher S, et al. , Nucleocytoplasmic partitioning of the plant photoreceptors phytochrome A, B, C, D, and E is regulated differentially by light and exhibits a diurnal rhythm, Plant Cell 14 (7) (2002) 1541–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kircher S, et al. , Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B, Plant Cell 11 (8) (1999) 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen M, Schwab R, Chory J, Characterization of the requirements for localization of phytochrome B to nuclear bodies, Proc. Natl. Acad. Sci. U. S. A 100 (24) (2003) 14493–14498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Chen M, et al. , Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes, Cell 141 (7) (2010) 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yu X, et al. , Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation, Plant Cell 21 (1) (2009) 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shalitin D, et al. , Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation, Nature 417 (6890) (2002) 763–767. [DOI] [PubMed] [Google Scholar]
- [47].Shalitin D, et al. , Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1, Plant Cell 15 (10) (2003) 2421–2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Yu X, et al. , Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus, Plant Cell 19 (10) (2007) 3146–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tan ST, et al. , Arabidopsis casein kinase1 proteins CK1.3 and CK1.4 phosphorylate cryptochrome2 to regulate blue light signaling, Plant Cell 25 (7) (2013) 2618–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Liu Q, et al. , Molecular basis for blue light-dependent phosphorylation of Arabidopsis cryptochrome 2, Nat. Commun 8 (2017) 15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Wang Q, et al. , The blue light-dependent phosphorylation of the CCE domain determines the photosensitivity of Arabidopsis CRY2, Mol. Plant 8 (4) (2015) 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wang Q, et al. , Photoactivation and inactivation of Arabidopsis cryptochrome 2, Science 354 (6310) (2016) 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Liu H, et al. , The action mechanisms of plant cryptochromes, Trends Plant Sci. 16 (12) (2011) 684–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Liu H, et al. , Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis, Science 322 (5907) (2008) 1535–1539. [DOI] [PubMed] [Google Scholar]
- [55].Wang X, et al A CRY-BIC negative-feedback circuitry regulating blue light sensitivity of Arabidopsis, Plant J. 92 (3) (2017) 426–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Okajima K, Molecular mechanism of phototropin light signaling, J. Plant Res 129 (2) (2016) 149–157. [DOI] [PubMed] [Google Scholar]
- [57].Pedmale UV, Liscum E, Regulation of phototropic signaling in Arabidopsis via phosphorylation state changes in the phototropin 1-interacting protein NPH3, J. Biol. Chem 282 (27) (2007) 19992–20001. [DOI] [PubMed] [Google Scholar]
- [58].Demarsy E, et al. , Phytochrome Kinase Substrate 4 is phosphorylated by the phototropin 1 photoreceptor, EMBO J. 31 (16) (2012) 3457–3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kinoshita T, et al. , Phot1 and phot2 mediate blue light regulation of stomatal opening, Nature 414 (6864) (2001) 656–660. [DOI] [PubMed] [Google Scholar]
- [60].Kong SG, Wada M, Molecular basis of chloroplast photorelocation movement, J. Plant Res 129 (2) (2016) 159–166. [DOI] [PubMed] [Google Scholar]
- [61].Song YH, et al. , Distinct roles of FKF1, Gigantea, and Zeitlupe proteins in the regulation of Constans stability in Arabidopsis photoperiodic flowering, Proc. Natl. Acad. Sci. U. S. A Ill (49) (2014) 17672–17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Zoltowski BD, Imaizumi T, Structure and function of the ZTL/FKF1/LKP2 group proteins in Arabidopsis, Enzymes 35 (2014) 213–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Song YH, et al. , FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering, Science 336 (6084) (2012) 1045–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Rizzini L, et al. , Perception of UV-B by the Arabidopsis uvr8 protein, Science 332 (6025) (2011) 103–106. [DOI] [PubMed] [Google Scholar]
- [65].Cloix C, et al. , C-terminal region of the UV-B photoreceptor UVR8 initiates signaling through interaction with the COP1 protein, Proc. Natl. Acad. Sci. U. S. A 109 (40) (2012) 16366–16370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Huang X, et al. , Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B, Proc. Natl. Acad. Sci. U. S. A 110 (41) (2013) 16669–16674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Wu D, et al. , Structural basis of ultraviolet-B perception by UVR8, Nature 484 (7393) (2012) 214–219. [DOI] [PubMed] [Google Scholar]
- [68].Liang T, et al. , UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis, Dev. Cell 44 (4) (2018) 512–523 e5. [DOI] [PubMed] [Google Scholar]
- [69].Yang Y, et al. , UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis, Nat. Plants 4 (2) (2018) 98–107. [DOI] [PubMed] [Google Scholar]
- [70].Yamaguchi S, et al. , Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds, Plant Cell 10 (12) (1998) 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Shinomura T, et al. , Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana, Proc. Natl. Acad. Sci. U. S. A 93 (15) (1996) 8129–8133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Botto JF, et al. , Phytochrome a mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis, Plant Physiol. 109 (2) (1996) 439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Lee KP, et al. , Spatially and genetically distinct control of seed germination by phytochromes A and B, Genes Dev. 26 (17) (2012) 1984–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Xu X, et al. , Illuminating progress in phytochrome-mediated light signaling pathways, Trends Plant Sci. 20 (10) (2015) 641–650. [DOI] [PubMed] [Google Scholar]
- [75].Reed JW, et al. , Phytochrome a and phytochrome B have overlapping but distinct functions in Arabidopsis development, Plant Physiol. 104 (4) (1994) 1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Tepperman JM, Hwang YS, Quail PH, phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation, Plant J. 48 (5) (2006) 728–742. [DOI] [PubMed] [Google Scholar]
- [77].Xu X, et al. , PHYTOCHROME INTERACTING FACTOR1 enhances the E3 ligase activity of CONSTITUTIVE PHOTOMORPHOGENIC1 to synergistically repress photomorphogenesis in Arabidopsis, Plant Cell 26 (5) (2014) 1992–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zhang H, et al. , Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation, Plant J. 65 (3) (2011) 346–358. [DOI] [PubMed] [Google Scholar]
- [79].Lee J, et al. , Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development, Plant Cell 19 (3) (2007) 731–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Leivar P, et al. , Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness, Curr. Biol 18 (23) (2008) 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Shin J, et al. , Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors, Proc. Natl. Acad. Sci. U. S. A 106 (18) (2009) 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Shikata H, et al. , Phytochrome controls alternative splicing to mediate light responses in Arabidopsis, Proc. Natl. Acad. Sci. U. S. A 111 (52) (2014) 18781–18786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Xin R, et al. , SPF45-related splicing factor for phytochrome signaling promotes photomorphogenesis by regulating pre-mRNA splicing in Arabidopsis, Proc. Natl. Acad. Sci. U. S. A 114 (33) (2017) E7018–E7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Paik I, Yang S, Choi G, Phytochrome regulates translation of mRNA in the cytosol, Proc. Natl. Acad. Sci. U. S. A 109 (4) (2012) 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Reichel M, et al. , In planta determination of the mRNA-binding proteome of Arabidopsis etiolated seedlings, Plant Cell 28 (10) (2016) 2435–2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Ushijima T, et al. , Light controls protein localization through phytochrome-mediated alternative promoter selection, Cell 171 (6) (2017) 1316–1325 e12. [DOI] [PubMed] [Google Scholar]
- [87].Oh S, Warnasooriya SN, Montgomery BL, Downstream effectors of light- and phytochrome-dependent regulation of hypocotyl elongation in Arabidopsis thaliana, Plant Mol. Biol 81 (6) (2013) 627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Montgomery BL, Spatial-specific phytochrome responses during de-etiolation in Arabidopsis thaliana, Plant Signal. Behav 4 (1) (2009) 47–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Kim J, et al. , Epidermal phytochrome B inhibits hypocotyl negative gravitropism non-cell-Autonomously, Plant Cell 28 (11) (2016) 2770–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Reed JW, et al. , Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development, Plant Cell 5 (2) (1993) 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Hornitschek P, et al. , Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling, Plant J. 71 (5) (2012) 699–711. [DOI] [PubMed] [Google Scholar]
- [92].Li L, et al. , Linking photoreceptor excitation to changes in plant architecture, Genes Dev. 26 (8) (2012) 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Martinez-Garcia JF, et al. , The shade avoidance syndrome in Arabidopsis: the antagonistic role of phytochrome a and B differentiates vegetation proximity and canopy shade, PLoS One 9 (10) (2014) e109275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hornitschek P, et al. , Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers, EMBO J. 28 (24) (2009) 3893–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Pedmale UV, et al. , Cryptochromes interact directly with PIFs to control plant growth in limiting blue light, Cell 164 (1-2) (2016) 233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Song YH, et al. , Photoperiodic flowering: time measurement mechanisms in leaves, Annu. Rev. Plant Biol 66 (2015) 441–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Corbesier L, et al. , FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis, Science 316 (5827) (2007) 1030–1033. [DOI] [PubMed] [Google Scholar]
- [98].Putterill J, Laurie R, Macknight R, It’s time to flower: the genetic control of flowering time, Bioessays 26 (4) (2004) 363–373. [DOI] [PubMed] [Google Scholar]
- [99].Zuo Z, et al. , Blue light-dependent interaction of CRY2 with SPA1 regulates COP1 activity and floral initiation in Arabidopsis, Curr. Biol 21 (10) (2011) 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Valverde F, et al. , Photoreceptor regulation of CONSTANS protein in photoperiodic flowering, Science 303 (5660) (2004) 1003–1006. [DOI] [PubMed] [Google Scholar]
- [101].Endo M, et al. , Phytochrome B in the mesophyll delays flowering by suppressing FLOWERING LOCUS T expression in Arabidopsis vascular bundles, Plant Cell 17 (7) (2005) 1941–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Cha JY, et al. , GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock, Nat. Commun 8 (1) (2017) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Sawa M, et al. , FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis, Science 318 (5848) (2007) 261–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Wang H, Siemens J, TRP ion channels in thermosensation, thermoregulation and metabolism, Temperature (Austin) 2 (2) (2015) 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Johansson J, et al. , An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes, Cell 110 (5) (2002) 551–561. [DOI] [PubMed] [Google Scholar]
- [106].Hofmann NR, Phosphorylation and dark reversion of phytochrome B, Plant Cell 25 (2) (2013) 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Ma D, et al. , Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light, Proc. Natl. Acad. Sci. U. S. A 113 (1) (2016) 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Franklin KA, Whitelam GC, Light-quality regulation of freezing tolerance in Arabidopsis thaliana, Nat. Genet 39 (11) (2007) 1410–1413. [DOI] [PubMed] [Google Scholar]
- [109].Kim HJ, et al. , Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana, Plant J. 29 (6) (2002) 693–704. [DOI] [PubMed] [Google Scholar]
- [110].Carvalho RF, Campos ML, Azevedo RA, The role of phytochrome in stress tolerance, J. Integr. Plant Biol 53 (12) (2011) 920–929. [DOI] [PubMed] [Google Scholar]
- [111].Lee CM, Thomashow MF, Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana, Proc. Natl. Acad. Sci. U. S. A 109 (37) (2012) 15054–15059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Karayekov E, et al. , Heat shock-induced fluctuations in clock and light signaling enhance phytochrome B-mediated Arabidopsis deetiolation, Plant Cell 25 (8) (2013) 2892–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Jung HS, et al. , Subset of heat-shock transcription factors required for the early response of Arabidopsis to excess light, Proc. Natl. Acad. Sci. U. S. A 110 (35) (2013) 14474–14479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Sun W, et al. , The rice phytochrome genes, PHYA and PHYB, have synergistic effects on anther development and pollen viability, Sci. Rep 7 (1) (2017) 6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kim K, et al. , Phytochromes inhibit hypocotyl negative gravitropism by regulating the development of endodermal amyloplasts through phytochrome-interacting factors, Proc. Natl. Acad. Sci. U. S. A 108 (4) (2011) 1729–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Correll MJ, Kiss JZ, The roles of phytochromes in elongation and gravitropism of roots, Plant Cell Physiol. 46 (2) (2005) 317–323. [DOI] [PubMed] [Google Scholar]
- [117].Liedvogel M, et al. , Chemical magnetoreception: bird cryptochrome 1a is excited by blue light and forms long-lived radical-pairs, PLoS One 2 (10) (2007) e1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Solov’yov IA, Chandler DE, Schulten K, Magnetic field effects in Arabidopsis thaliana cryptochrome-1, Biophys. J 92 (8) (2007) 2711–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Christie JM, et al. , Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism, Science 282 (5394) (1998) 1698–1701. [DOI] [PubMed] [Google Scholar]
- [120].Campos ML, et al. , Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs, Nat. Commun 7 (2016) 12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Leivar P, Monte E, PIFs: systems integrators in plant development, Plant Cell 26 (1) (2014) 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Bowler C, et al. , Cyclic GMP and calcium mediate phytochrome phototransduction, Cell 77 (1) (1994) 73–81. [DOI] [PubMed] [Google Scholar]


