Abstract
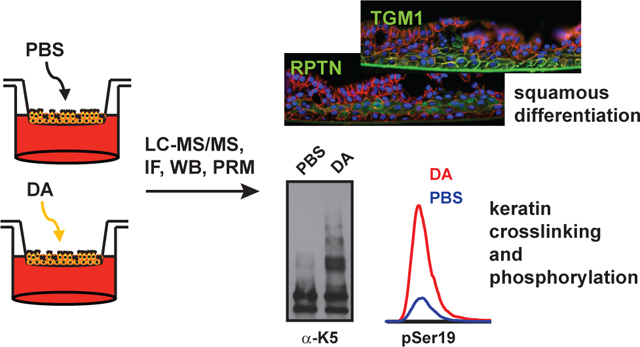
Occupational exposures to the diketone flavoring agent, diacetyl, have been associated with bronchiolitis obliterans, a rare condition of airway fibrosis. Model studies in rodents have suggested that the airway epithelium is a major site of diacetyl toxicity, but the effects of diacetyl exposure upon the human airway epithelium are poorly characterized. Here, we performed quantitative LC-MS/MS-based proteomics to study the effects of repeated diacetyl vapor exposures on three-dimensional organotypic cultures of human primary tracheobronchial epithelial cells. Using a label-free approach, we quantified approximately 3,400 proteins and 5,700 phosphopeptides in cell lysates across four independent donors. Altered expression of proteins and phosphopeptides were suggestive of loss of cilia and increased squamous differentiation in diacetyl-exposed cells. These phenomena were confirmed by immunofluorescence staining of culture cross-sections. Hyperphosphorylation, and crosslinking, of basal cell keratins were also observed in diacetyl-treated cells, and we used parallel reaction monitoring to confidently localize and quantify previously uncharacterized sites of phosphorylation in keratin 6. Collectively, these data identify numerous molecular changes in the epithelium that may be important to the pathogenesis of flavoring-induced bronchiolitis obliterans. More generally, this study highlights the utility of quantitative proteomics for the study of in vitro models of airway injury and disease.
Keywords: BOS, butter flavoring, butanedione, cornified envelope, pentanedione, repetin, RSPH4A, PRM, TGM1, squamous metaplasia
Graphical Abstract
INTRODUCTION
The inhalational exposure of workers in the microwave popcorn, baked goods and coffee manufacturing industries to the “artificial butter” flavor 2,3-butanedione (diacetyl, DA) has been linked to the development of bronchiolitis obliterans (BO), a condition of fixed obstructive lung disease and small airway fibrosis.1–4 These cases have led to strict limits on workplace DA exposure, but questions remain as to the potential toxicities of DA substitutes, including the related α-diketone 2,3,-pentanedione (PD), and of the possibility of as yet unidentified exposures to DA and PD. In addition, there is increasing awareness of the use of DA and PD in e-cigarette flavors,5, 6 raising the possibility that use of e-cigarettes may increase risk for airways disease through a similar mechanism.
Rodent models have been critical to the demonstration that respiratory exposure to DA and PD can lead to the development of BO, and suggest this process begins with epithelial injury.7–11 For example, we have shown in rats, that intratracheal instillation of DA leads to a rapid loss of acetylated tubulin (a cilia marker), followed by sloughing of the epithelium and finally airway fibrosis at these sites of severe injury.9 This is consistent with the idea that denudation of the airway epithelium may be necessary for the development of toxicant-induced BO.12 However, epithelial ablation alone is unlikely to explain the entire spectrum of flavoring-induced airway disease, as the bronchial epithelium of rats exposed to DA and PD vapor also show evidence of significant hyperplasia and squamous metaplasia.11
To better understand the effects of diacetyl on the human airway, we and others have begun to model diacetyl vapor exposure using air-liquid interface cultures of differentiated human tracheobronchial epithelial cells (TBECs). Our model utilizes three 1 h exposures on alternating days to ~1000 ppm vapor,13 a level that is near the peak (1230 ppm) that was measured at a microwave popcorn plant by Kreiss et al. in the headspace above a tank holding heated butter flavoring.1 In this model, we previously reported shedding of the pro-inflammatory and pro-fibrotic epidermal growth factor receptor ligand amphiregulin by TBECs after repeated exposures to DA.13 While these data suggest a potential mechanism for flavoring-induced fibrosis, they do not reveal the full extent of the pathological effects of DA on the airway epithelium. Recently, Fedan and coworkers also found that a 6 h exposure of differentiated normal human bronchial epithelial cells to DA vapor disrupted normal cell architecture and transepithelial electrical resistance.14 Here, we have undertaken a more comprehensive approach to characterize flavoring-induced epithelial injury, specifically, an analysis of the whole cell proteome and phosphoproteome of differentiated TBECs after three repeated short term exposures to DA vapor.
MATERIALS AND METHODS
In vitro model of diacetyl exposure.
Exposures of human tracheobronchial epithelial cells (TBECs) to DA vapor were as previously-described.13 Briefly, passage two TBECs (MatTek EpiAirway™) were isolated from donor lungs of healthy males (with no relevant disease, smoking or medication history). Tissues were obtained after death of the subject through government accredited tissue procurement agencies following IRB approval and strict adherence to all government regulations regarding informed consent and protection of human subjects. Additional characteristics were: Donor 1 (ID 9831, Caucasian, age 23); Donor 2: (ID 11257, Caucasian, age 23); Donor 3: (ID TBE-20, Caucasian, age 13); Donor 4 (ID TBE-30, Hispanic, age 50).Cells were cultured on transwells (9 mm internal diameter) and fully-differentiated at air-liquid interface.
Cells were exposed to a vapor cup containing 50 μl of 25 mM DA in PBS (or PBS vehicle control), which corresponds to a DA vapor concentration ~1000 ppm based on calculations as previously described,13 for 1 h on days 0, 2 and 4 and were harvested on day 6. Apical surfaces were washed twice with 400 μl PBS prior to each exposure, and the basolateral culture media (MatTek AIR-100) was exchanged (1 ml) each day. Cells which were harvested for proteomics were cultured in protein-free basolateral media (DMEM:F12 w/ high glucose and w/o pyruvate),1.5 mM Hepes, 0.3 mM MgCl2, 1 mM CaCl2, 80 μM ethanolamine, 0.5 mM MgSO4, 100 nM all-trans-retinoic acid) between days 5 and 6, and apical surfaces were rinsed twice with PBS on day 6 prior to freezing. Cells harvested for immunofluorescence were cultured in AIR-100 media for the entirety of the experiment, and after two apical PBS washes on day 6, were formalin-fixed and paraffin embedded (FFPE).
Sample preparation for proteomics.
Three wells of TBECs per donor per condition were lysed by scraping with 50 mM ammonium bicarbonate pH 8.0 (AmBic) containing 0.2% acid labile surfactant (ALS-1), 5 mM NaF, 1 mM Na3VO4 and 10 nM calyculin, followed by probe sonication (3 × 3 s). After centrifugation, protein recovery was quantified by Bradford assay, and 250 μg of protein per sample was denatured and reduced by addition of 10 mM DTT followed by heating at 80 °C for 10 min. Next, alkylation was performed by addition of 25 mM iodoacetamide and incubation in the dark for 30 min. TPCK-trypsin (1:50 w/w trypsin:protein) was added, and proteins were digested at 37 °C overnight. Samples were then acidified with 1% trifluoroacetic acid (TFA) and 2% acetonitrile (MeCN), followed by heating at 60 °C for 2 h, to inactivate trypsin and to degrade ALS-1. Approximately 25 μg of peptide was removed for proteome analysis, spiked with 50 fmol/μg of trypsinized yeast alcohol dehydrogenase 1 (MassPrep; Waters). QC pools were prepared by mixing equal amounts of all samples.
Phosphopeptide enrichment.
Trypsinized bovine casein was added to the remaining 225 μgs of peptide digests at 30 fmol/μg and the peptides were lyophilized to dryness. Dried peptides were reconstituted in 80% MeCN containing 1% TFA (binding buffer) and enriched using 200 μg capacity TiO2 tips (GL Sciences) using the manufacturer recommended protocol. After binding, two washes were performed with binding buffer containing 1M glycolic acid to reduce binding of non-phosphorylated peptides. After high pH elution and neutralization, peptides were lyophilized and subjected to C18 Stage tip purification. Finally, after an additional lyophilization, peptides were resuspended in 20 μl of 10 mM citrate, 2% MeCN, 1% TFA containing 10 fmol/μl of trypsinized yeast ADH1 and were transferred to Maximum Recovery LC vials (Waters). QC pools were prepared by pooling 5 μl of all samples.
Quantitative mass spectrometry analysis of the TBEC proteome.
Quantitative one-dimensional liquid chromatography, tandem mass spectrometry (1D-LC-MS/MS) was performed on 250 ng of the peptide digests per sample in singlicate, with additional QC and conditioning analyses as described in Table S1. Samples were analyzed using a nanoACQUITY UPLC system (Waters) coupled to a QExactive Plus high resolution accurate mass tandem mass spectrometer (Thermo) via a nanoelectrospray ionization source. Briefly the sample was first trapped on a Symmetry C18 180 μm × 20 mm trapping column (5 μl/min at 99.9:0.1 v/v H2O:MeCN) followed by an analytical separation using a 1.7 um ACQUITY HSS T3 C18 75 mm × 250 mm column (Waters) with a 90 min gradient of 5 to 40% MeCN with 0.1% formic acid (buffer B) at a flow rate of 400 nl/min and column temperature of 55 °C. Data collection on the QExactive Plus MS was performed in data-dependent acquisition (DDA) mode with a 70,000 resolution (@ m/z 200) full MS scan from m/z 375 to 1600 with a target AGC value of 1e6 ions followed by 10 MS/MS scans at 17,500 resolution (@ m/z 200) at a target AGC value of 5e4 ions. A 20 s dynamic exclusion was employed.
Quantitative mass spectrometry analysis of the TBEC phosphoproteome.
Quantitative one-dimensional liquid chromatography, tandem mass spectrometry (1D-LC-MS/MS) was performed on 4 μl (20%) of the enriched peptide digests per sample in singlicate, with additional QC and conditioning analyses as described in Table S4. Samples were analyzed using a nanoACQUITY UPLC system (Waters) coupled to a QExactive Plus as above except that the analytical separation used a 5 min hold at 3% MeCN/0.1% FA followed by a 90 min gradient of 3 to 30% buffer B.
Label-free quantitation.
For quantitation of each dataset, eleven raw data files (including QC analyses) were imported into Rosetta Elucidator v3.3 (Rosetta Biosoftware, Inc), and LC-MS runs were aligned based on the accurate mass and retention time of detected ions (“features”) using the PeakTeller algorithm in Elucidator. Relative peptide abundance was calculated based on area-under-the-curve (AUC) of aligned features across all runs. The proteome had 268,024 quantified features, and the phosphoproteome had 214,345 quantified features.
MS/MS data was processed using Mascot Distiller (Matrix Science), which produced 463,245 and 416,476 high collision energy (peptide fragment) spectra for the proteome and phosphophoproteome datasets, respectively. Database searches used Mascot Server (version 2.5, Matrix Sciences). Searches used a custom Swissprot database with Homo sapiens taxonomy (downloaded on 02/12/14) with additional proteins, including yeast ADH1 and bovine casein, as well as an equal number of reversed-sequence “decoys” for false discovery rate determination (40,546 total entries). Search parameters for the proteome analysis included fixed modification on Cys (carbamidomethyl) and variable modifications on Met (oxidation), Asn/Gln (deamidation). Search parameters for the phosphoproteome analysis included fixed modification on Cys (carbamidomethyl) and variable modifications on Met (oxidation), Ser/Thr (phosphorylation) and Tyr (phosphorylation). The proteome data was annotated at a 0.5% false discovery rate (FDR) after individual peptide scoring using the PeptideProphet algorithm in Elucidator. The phosphoproteome data was annotated at a ~0.5% peptide FDR using a Mascot Ion Score of 20. Finally, the phosphoproteome data was analyzed within Rosetta Elucidator using ModLoc, a probability-based phosphorylation site localization tool based on the AScore algorithm.15 For quantitative processing, the data was first curated to contain only high quality peptides with appropriate chromatographic peak shape and the dataset was intensity scaled to the robust median across all analyzed samples.
Statistical Analysis.
Statistical tests were performed in Rosetta Elucidator. Briefly, an intensity floor value of 1 was given to missing data followed by a log2 transformation. A two-sample paired t-test was performed, with or without Benjamini-Hochberg FDR correction. Unless noted, FDR-corrected p-values were used to identify differentially-expressed proteins.
Western blotting.
Western blotting was performed on up to 20 μg of the cell lystes that were used for proteomic analysis (above). Proteins were separated by SDS-PAGE using a 4–15% Tris-HCl Ready Gel (Bio-Rad). The following primary antibodies were used at 1:1000 dilution in 5% milk, including: GAPDH (Millipore MAB374), RSPH4A (Sigma-Aldrich HPA031196), repetin (Sigma-Aldrich HPA030483), TGM1 (Sigma-Aldrich HPA040171), keratin 5 (Covance PRB160P), keratin 6 (Thermo PA5–28235), keratin 14 (Novus 34270) and keratin 17 (Cell Signaling 12509). The following secondary antibodies were used at 1:3000 dilution in 5% milk: goat anti-rabbit HRP (Santa Cruz Biotech sc-2004) and goat anti-mouse HRP (Santa Cruz Biotech sc-2005).
Histology and immunofluorescence.
Sectioning and H&E staining of FFPE TBECs was performed by the NIEHS Histology Core. Immunofluorescence (IF) was performed using the following primary antibodies (sources as above unless noted): RSPH4A (1:40), TGM1 (1:3000), repetin (1:250), and mouse IgG1 anti-β catenin antibody (1:400, BD, Franklin Lakes, New Jersey). Secondary antibodies used 1:500 dilutions and: included Alexa Fluor 488 Donkey anti-Rabbit IgG, Alexa Fluor 594 Goat anti-mouse IgG1 (Invitrogen, Grand Island, NY). Sections were blocked with 5% BSA in phosphate-buffered saline (PBS) for non-specific antigen reactivity following citrate buffer retrieval. Primary and secondary antibodies were diluted in blocking solution and incubated overnight at 4°C or room temperature for 90 minutes, respectively. Slides were extensively washed in PBS and cover-slipped with DAPI (Sigma, St Louis, MO) in Fluoromont G mounting media (Southern Biotech, Birmingham, AL). Images were obtained using a Zeiss Observer.Z1 inverted fluorescent microscope (Carl Ziess, Inc., Gottingen, Germany) equipped with a digital camera and processed using AxioVision Release 4.6.3 (Carl Ziess) Software.
Targeted quantitation of phosphorylated keratin 6.
Stable isotope-labeled (SIL) peptides (SpikeTides L) were purchased from JPT (Berlin, Germany) and reconstituted in 0.1 M AmBic w/ 20% (v/v) MeCN. After reconstitution, ~100 fmol of neat peptides were analyzed by LC-MS/MS as described above for phosphopeptides except that the UPLC separation used a 30 min gradient of 3–30% buffer B. MS/MS data was searched in Mascot, and Skyline16 was used to build a spectral library and assign precursor peaks. An inclusion list (precursor mass, charge state, retention time window) was exported from Skyline and used to generate a parallel reaction monitoring (PRM) assay.17, 18 For analysis of K6 peptides by PRM, samples were spiked with ~2 fmol/μl of SIL peptides. Analysis of 2 μl of each sample used a 30 min UPLC separation as above. The MS acquisition used the Q-Exactive Plus with the following MS2 parameters: resolution, 17,500; AGC target, 5E4; Max IT, 128 ms; Isolation width, 1 m/z resolution. Data was imported into Skyline, and peptides were assigned using the spectral library, retention times of neat SIL peptides, and/or site-determining fragment ions. Intensities of native and SIL-labeled peptides were quantified by summing the area-under-the-curve of three non-interfering MS/MS fragment ions per precursor. Missing values (in controls) were replaced with the lowest detectable value in the same group, as to not overestimate the effects.
RESULTS AND DISCUSSION
Quantitative proteomic analysis of an in vitro model of diacetyl vapor exposure.
In order to better understand the cellular changes and signaling events associated with diacetyl inhalation toxicity, we treated well-differentiated tracheobronchial epithelial cells (TBECs; isolated from tissue of four healthy deceased human donors) to three exposures of DA vapor (~1000 ppm for 1 h on days 0, 2 and 4) versus control, and we performed separate quantitative analyses of the proteomes and phosphoproteomes of whole cell lysates harvested two days after the last exposure (see Methods).19, 20 The proteome analysis used 250 ng of peptide digests per sample and resulted in quantitation of 23,039 unique peptides, corresponding to 3,389 proteins (Tables S2–S3). Over 2,300 proteins were robustly quantified, as defined by having 2 or more quantified peptides and a coefficient of variation (CV) of <30% across analyses of a QC pool at the beginning, middle and end of the analysis block (Table S1). The phosphoproteome was enriched from 225 μg of peptide digests per sample, and analysis of ~20% of each enriched sample (and triplicate analyses of a QC pool; Table S4) yielded quantitative data for 5,673 unique phosphopeptides from ~2000 proteins (Table S6); at least 5,087 of the quantified phosphopeptides had at least one pSer, 663 had at least 1 pThr and 65 had at least one pTyr. Over 4,700 of these phosphopeptides were robustly quantified based on a <30% CV across triplicate analyses of a QC pool containing equal amounts of each phosphopeptide-enriched sample.
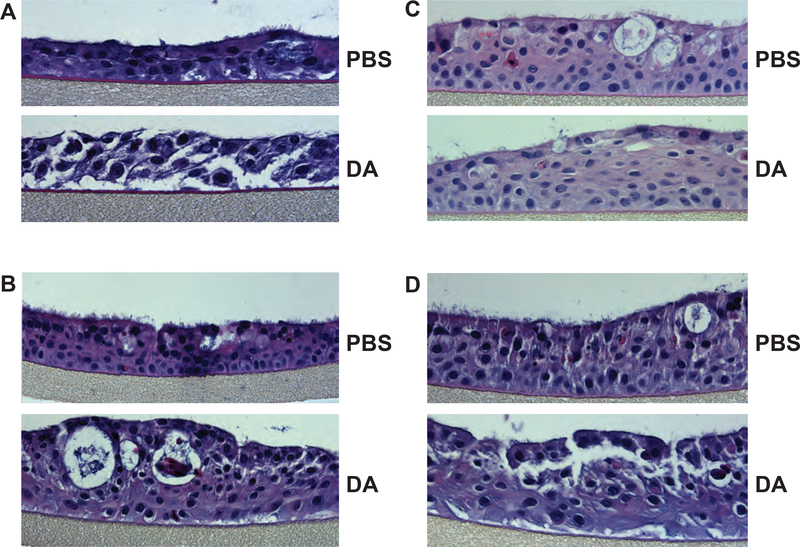
Hematoxylin and eosin (H&E) staining of TBEC cross-sections showed that DA exposure resulted in a range of pathological effects (Fig. 1). While all donors appeared to exhibit loss of cilia or cilia shortening after DA vapor exposure, there appeared to be some variability in the severity of the injury and remodeling of the epithelium. Specifically, cells from donor 1 appeared to be poorly attached to the permeable support and exhibited loss of cell-cell adhesion throughout the culture (Fig. 1A), whereas cells from donors 2–4 appeared to have more squamoid features after DA exposure but did not have a fully stratified squamous appearance that typifies squamous metaplasia (Fig. 1B–D). Nonetheless, these data showed significant evidence of injury to DA vapor across all donors and suggested that the proteome of donor 1 might be differentiated from donors 2–4 after exposure to DA vapor.
Figure 1. Hematoxylin and eosin staining of TBEC cross-sections shows evidence of DA vapor injury.
H&E staining was performed on cross sections of formalin-fixed and paraffin-imbedded PBS- and DA-exposed TBECs from (A) donor 1, (B) donor 2, (C) donor 3 and (D) donor 4. Images are representative of 3 sections per donor.
We sought to correlate changes in the proteomes and phosphoproteomes of the TBECs with these aberrant histological features. After statistical testing (paired t-test with Benjamini-Hochberg correction for false discovery rate), we visualized the data using volcano plots and by two-dimensional hierarchical clustering (Fig. S1 and Fig. 2). The volcano plots showed that ~1/3 of the robustly quantified proteins, and ~¼ of the robustly quantified phosphopeptides met a predefined significance threshold (FDR-corrected p<0.1). Hierarchical clustering showed that just over 1/3 of the differentially expressed proteome was downregulated after DA exposure (Fig. 2A), while greater than half of the differentially-expressed phosphoproteome was downregulated after DA exposure (Fig. 2B).
Figure 2. Hierarchical clustering of differentially expressed proteins and phosphoproteins.
A) Proteome data was filtered to contain proteins quantified by 2 or more peptides, with a CV of <30% for triplicate QC injections, and p<0.1 for a paired t-test w/ FDR correction. The remaining 874 proteins were normalized using Z-scoring and visualized by two-dimensional hierarchical clustering (Ward method). Cluster 7 (green) is composed predominantly of proteins expressed in multiciliated airway cells, and clusters 10–12 (yellow) are composed predominantly of proteins expressed during squamous differentiation (see Results). B) Phosphopeptides were filtered by CV <30% and p<0.1 and visualized as in (C). Data was analyzing using JMP (SAS, Cary, NC).
Decreases in protein and phosphoprotein expression largely reflect injury to multicilated cells after DA exposure.
Text mining of the proteome after hierarchical clustering identified a cluster of 89 downregulated proteins (cluster 7) that contained a large number of cilia-related proteins, including components of the cilia motor protein complex (axonemal dynein), intraflagellar transport proteins, and radial spoke head proteins (Fig. 2A and Table S3). We further queried the extensive immunohistochemical analyses of lung sections in the Human Protein Atlas (http://www.proteinatlas.org)21, 22 and found that many of these proteins are highly expressed in ciliated cells of human bronchi. We used these data to create a protein signature of DA vapor injury to the airway epithelium (Table 1 and Fig. S1A).
Table 1.
Protein signature of diacetyl injury to multiciliated cells*
| Primary Protein Name | Protein Description† | Mean fold-change | In phosho-protein signature? |
|---|---|---|---|
| DNAI1_HUMAN | Dynein intermediate chain 1, axonemal | −29.3 | |
| DTHD1_HUMAN | Death domain-containing protein 1 | −27.6 | |
| DJB13_HUMAN | DnaJ homolog subfamily B member 13 | −26.6 | |
| KAD7_HUMAN | Adenylate kinase 7 | −26.2 | |
| DYH9_HUMAN | Dynein heavy chain 9, axonemal | −19.2 | |
| DYH5_HUMAN | Dynein heavy chain 5, axonemal | −17.5 | |
| CA087_HUMAN | Uncharacterized protein C1orf87 | −16.1 | Y |
| TP53B_HUMAN | Tumor suppressor p53-binding protein 1§ | −15.6 | Y |
| CU059_HUMAN | Uncharacterized protein C21orf59 | −14.8 | |
| WDR35_HUMAN | WD repeat-containing protein 35 | −14.6 | |
| EFHC1_HUMAN | EF-hand domain-containing protein 1 | −13.0 | |
| RSH4A_HUMAN | Radial spoke head protein 4 homolog A | −11.8 | Y |
| CA173_HUMAN | Uncharacterized protein C1orf173 | −11.8 | Y |
| WDR54_HUMAN | WD repeat-containing protein 54 | −11.5 | |
| ODF3B_HUMAN | Outer dense fiber protein 3B | −11.4 | Y |
| NDK5_HUMAN | Nucleoside diphosphate kinase homolog 5 | −11.1 | |
| MAP1A_HUMAN | Microtubule-associated protein 1A | −10.5 | Y |
| CROCC_HUMAN | Rootletin | −10.4 | Y |
| RSPH9_HUMAN | Radial spoke head protein 9 homolog | −10.3 | |
| IDLC_HUMAN | Axonemal dynein light intermediate polypeptide 1 | −10.2 | Y |
| PITM1_HUMAN | Membrane-associated phosphatidylinositol transfer protein 1 | −9.3 | |
| PCM1_HUMAN | Pericentriolar material 1 protein | −8.8 | |
| HEAT2_HUMAN | HEAT repeat-containing protein 2 | −7.9 | |
| RSPH1_HUMAN | Radial spoke head 1 homolog | −7.8 | |
| FBXW9_HUMAN | F-box/WD repeat-containing protein 9 | −7.0 | Y |
| MIEAP_HUMAN | Mitochondria-eating protein | −6.8 | Y |
| LZTL1_HUMAN | Leucine zipper transcription factor-like protein 1 | −6.3 | |
| CROL3_HUMAN | Putative ciliary rootlet coiled-coil protein-like 3 protein | −6.3 | Y |
| KAD8_HUMAN | Adenylate kinase 8 | −5.9 | |
| AL3B1_HUMAN | Aldehyde dehydrogenase family 3 member B1 | −5.5 | |
| SAMH1_HUMAN | SAM domain and HD domain-containing protein 1§ | −5.2 | Y |
| IFT46_HUMAN | Intraflagellar transport protein 46 homolog | −4.9 |
Proteins ranked by average fold change of ~-5-fold or greater in DA- versus PBS-treated HBECs (quantified by 2 or more peptides, CV < 30% for replicate injections of QC pool; p<0.1, paired t-test w/FDR correction), and localization to cytoplasm/membranes of ciliated cells of human bronchi by the Human Protein Atlas (http://www.proteinatlas.org)
proteins with nuclear localization
While we could not localize the injury phenotype to a specific cluster of phosphopeptides, sorting by fold change suggested that the majority of downregulated phosphopeptides were associated with the loss of cilia or injury to multiciliated cells. We further filtered on proteins which had 2 or more associated phosphopeptides that were >5-fold lower in DA- versus PBS-treated cells, and we used these data to assemble a complementary phosphoprotein signature of injury (Table 2). While we could not exclude the possibility that dysregulated phosphorylation was due to specific alterations in kinase or phosphatase activity, the overlap in the protein and phosphoprotein signatures (12 proteins, including RSPH4A, TP53BP1 and CA087) suggested that many of the decreases in phosphorylation reflected parallel changes protein expression. Collectively, these data establish signatures of DA vapor exposure that appear to reflect extensive injury to the multiciliated cells of the airway epithelium.
Table 2.
Phosphoprotein signature of diacetyl injury to multiciliated cells*
| Primary Protein Name | Protein Description | # of unique phospho-peptides | In protein signature? |
|---|---|---|---|
| MAP1A_HUMAN | Microtubule-associated protein 1A | 18 | Y |
| RSH4A_HUMAN | Radial spoke head protein 4 homolog A | 13 | Y |
| K121L_HUMAN | Uncharacterized protein KIAA1211-like | 9 | |
| CA173_HUMAN | Uncharacterized protein C1orf173 | 8 | Y |
| MAP1B_HUMAN | Microtubule-associated protein 1B | 7 | |
| CA087_HUMAN | Uncharacterized protein C1orf87 | 5 | Y |
| CG057_HUMAN | Uncharacterized protein C7orf57 | 5 | |
| CROCC_HUMAN | Rootletin | 5 | Y |
| MIEAP_HUMAN | Mitochondria-eating protein | 4 | Y |
| TP53B_HUMAN | Tumor suppressor p53-binding protein 1§ | 4 | Y |
| CC151_HUMAN | Coiled-coil domain-containing protein 151 | 3 | |
| CCD13_HUMAN | Coiled-coil domain-containing protein 13 | 3 | |
| CCD19_HUMAN | Coiled-coil domain-containing protein 19, mitochondrial | 3 | |
| CROL3_HUMAN | Putative ciliary rootlet coiled-coil protein-like 3 protein | 3 | Y |
| GA2L2_HUMAN | GAS2-like protein 2 | 3 | |
| IDLC_HUMAN | Axonemal dynein light intermediate polypeptide 1 | 3 | Y |
| MLF1_HUMAN | Myeloid leukemia factor 1 | 3 | |
| ODF3B_HUMAN | Outer dense fiber protein 3B | 3 | Y |
| SPAT6_HUMAN | Spermatogenesis-associated protein 6 | 3 | |
| SYNE1_HUMAN | Nesprin-1 | 3 | |
| CE164_HUMAN | Centrosomal protein of 164 kDa | 2 | |
| CH047_HUMAN | Uncharacterized protein C8orf47 | 2 | |
| CP110_HUMAN | Centriolar coiled-coil protein of 110 kDa | 2 | |
| FBXW9_HUMAN | F-box/WD repeat-containing protein 9 | 2 | Y |
| INP5E_HUMAN | 72 kDa inositol polyphosphate 5-phosphatase | 2 | |
| KI21A_HUMAN | Kinesin-like protein KIF21A | 2 | |
| KIF19_HUMAN | Kinesin-like protein KIF19 | 2 | |
| KTU_HUMAN | Protein kintoun | 2 | |
| LCA5_HUMAN | Lebercilin | 2 | |
| MARE3_HUMAN | Microtubule-associated protein RP/EB family member 3 | 2 | |
| SAMH1_HUMAN | SAM domain and HD domain-containing protein 1§ | 2 | Y |
| TMC5_HUMAN | Transmembrane channel-like protein 5 | 2 | |
| TPPP_HUMAN | Tubulin polymerization-promoting protein | 2 |
Phosphoproteins ranked by number of unique phosphopeptides with ~-5-fold or greater in DA- versus PBS-treated HBECs (CV < 30% for replicate injections of QC pool; p<0.1, paired t-test w/FDR correction), and localization to cytoplasm/membranes of ciliated cells of human bronchi by the Human Protein Atlas (http://www.proteinatlas.org)
proteins with nuclear localization
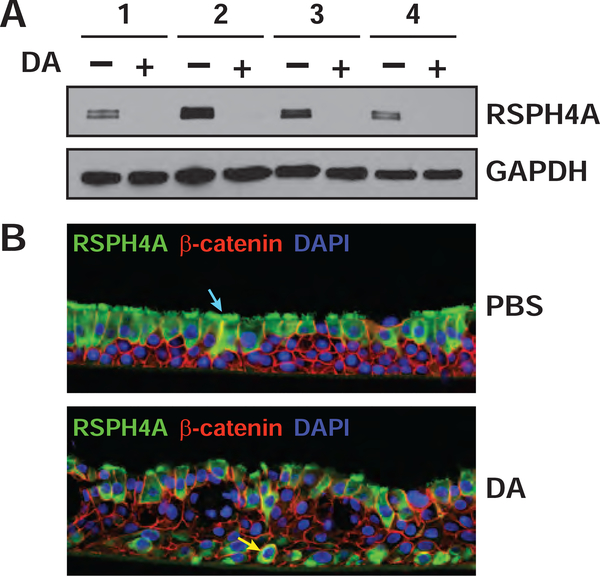
Radial spokehead homolog 4A (RSPH4A) was found in both the protein and phosphoprotein signatures, and we hypothesized that the downregulation of RSHPH4A phosphorylation (Table 2 and Fig. S1B) was due to the concomitant decrease in protein expression (as visualized for a representative RSPH4A peptide in Fig. S3). RSHP4A is highly expressed in the cytoplasm and membranes of multiciliated cells of the human airway22 and is the core protein of the radial spoke head, a complex which has a role in mechanical transduction of cilia.23 By western blotting, RSPH4A expression was essentially undetectable in total lysates of DA-treated cells versus controls, which appeared to validate the results of the proteomic analysis (Fig. 3A and Fig. S2). Immunofluorescence analysis of TBEC cross sections also showed a reduction in RSPH4A expression in the ciliated cells lining the apical surface of the DA-exposed epithelium. Interestingly, RSPH4A expression was increased in the basal layer of injured cells (Fig. 3B and Fig. S4), which could suggest some capacity of these cultures for re-differentiation to a ciliated phenotype. It should be noted that the RSPH4A polyclonal antibody used for these analyses (HPA031196) has been shown by protein array to bind only to its antigen (http://www.proteinatlas.org). Nonetheless, further studies are necessary to determine whether the IF staining represents detection of an authentic RSHP4A epitope in basal cells or cross reactivity with an unrelated antigen that is expressed in the airway epithelium after DA exposure.
Figure 3. The cilia-specific marker, RSPH4A was markedly reduced after DA treatment.
A) The levels of RSPH4A and GAPDH in cell lysates were visualized by western blotting. Western blotting performed on a replicate across the entire MW range (Figure S2) showed that only a single antigen was recognized by this antibody. B) Immunofluorescence was performed on cross sections of formalin-fixed and paraffin-imbedded TBECs to visualize RSPH4A (green, Alexa Fluor 488), β-catenin (red, Alexa Fluor 594) and nuclei (blue, DAPI) in donor 3. Localization of RSPH4A in ciliated cells is shown by blue arrow (upper image) and localization to basal cells is shown by yellow arrow (lower image). Images in (B) are representative of three different sections per condition (Fig. S3, A–B), and results were replicated in single sections from donors 2 and 4 (Fig. S3, C–F).
Markers of squamous differentiation are upregulated after DA exposure.
We next analyzed the proteins that were increased in DA-treated cells to determine whether they might reveal a molecular basis for repair or regeneration processes. Among robustly quantified and differentially expressed proteins, the most highly overexpressed proteins included: ECM1 (+8-fold), a extracellular matrix protein that localizes to the basement membrane of skin and to squamous epithelium;21 GDF15 (+7.5-fold), a member of the TGF-β superfamily that is thought to play an early role in response to lung injury24 and is upregulated in airway epithelial cells after cigarette smoke-exposure;25 and sequestosome-1 (+5-fold), a negative regulator of autophagy that has been shown to be upregulated in the epithelium of DA-exposed mice.26 These proteins did not appear to belong to a single common pathway. However, squamous differentiation was suggested by the localization of numerous small proline-rich proteins (SPR1B, SPR2A, SPRR3) and protein-glutamine gamma-glutamyltransferase K (TGM1) to a branch of the cluster dendrogram (clusters 10–12; Fig. 2B). Based on fold-change these proteins were also among the mostly highly upregulated in DA- versus PBS-treated cells (Table S3). We noted that a number of other putative markers of squamous differentiation—including involucrin27, 28 and repetin29—were highly expressed in donors 2–4, and available data in the Human Protein Atlas also showed that most of these proteins are more highly expressed in squamous (versus respiratory) epithelial cells. These data formed the basis for a protein signature of squamous differentiation (Table 3 and Fig. S1A) and appeared to be consistent with the H&E staining of donors 2–4 (Figs. 1B–D) versus donor 1 (Fig. 1A).
Table 3.
Protein Signature of Squamous Differentiation*
| Primary Protein Name | Protein Description | Fold-change donor 1 DA vs. PBS | Average Fold-change donors 2–4 DA vs. PBS | paired t-test | paired t-test FDR |
|---|---|---|---|---|---|
| ISK7_HUMAN | Serine protease inhibitor Kazal-type 7 | −1.1 | 25.1 | 0.17 | 0.29 |
| RPTN_HUMAN | Repetin | 1.1 | 21.3 | 0.06 | 0.14 |
| ECM1_HUMAN | Extracellular matrix protein 1 | 6.7 | 12.1 | 0.01 | 0.05 |
| SPR2B_HUMAN | Small proline-rich protein 2B | −1.5 | 11.6 | 0.12 | 0.23 |
| SPR2A_HUMAN | Small proline-rich protein 2A | 2.8 | 8.6 | 0.01 | 0.06 |
| IL36A_HUMAN | Interleukin-36 alpha | −1.5 | 8.5 | 0.12 | 0.23 |
| CRNN_HUMAN | Cornulin | 1.2 | 6.5 | 0.06 | 0.15 |
| TGM1_HUMAN | Protein-glutamine gamma-glutamyltransferase K | 2.6 | 6.3 | 0.01 | 0.05 |
| SPRR3_HUMAN | Small proline-rich protein 3 | 2.4 | 6.2 | 0.01 | 0.07 |
| INVO_HUMAN | Involucrin | −1.2 | 5.5 | 0.09 | 0.19 |
| CI169_HUMAN | UPF0574 protein C9orf169 | 3.2 | 4.5 | 0.00 | 0.03 |
| HSPB8_HUMAN | Heat shock protein beta-8 | 1.2 | 4.3 | 0.04 | 0.12 |
| SPR1A_HUMAN | Cornifin-A | 1.2 | 3.2 | 0.04 | 0.11 |
| ABHD5_HUMAN | 1-acylglycerol-3-phosphate O-acyltransferase 5 | 2.0 | 3.1 | 0.00 | 0.04 |
| S10AG_HUMAN | Protein S100-A16 | 1.8 | 3.0 | 0.01 | 0.05 |
| SCEL_HUMAN | Sciellin | 1.1 | 3.0 | 0.05 | 0.14 |
| SPR1B_HUMAN | Cornifin-B | 1.2 | 2.8 | 0.03 | 0.10 |
| ST2B1_HUMAN | Sulfotransferase family cytosolic 2B member 1 | −1.3 | 2.5 | 0.13 | 0.24 |
Proteins ranked by average fold change in donors 2–4 in DA- versus PBS-treated HBECs (quantified by 2 or more peptides, CV < 30% for replicate injections of QC pool; required >2-fold change for donors 2–4)
Although most of the proteins in the signature of squamous differentiation serve a structural role, there were several enzymes of note, including TGM1, 1-acylglycerol-3-phosphate O-acyltransferase (ABHD5) and sulfotransferase family cytosolic 2B member 1 (ST2B1). TGM1 crosslinks SPRs, keratins and other proteins,30–32 as well as ω-OH-(glucosyl)acylceramide (GlyAcylCer)33 within the cornified envelope, a protective layer produced below the plasma membrane of terminally differentiating stratified squamous epithelia.30 ABHD5 has a role in triacylglycerol metabolism upstream of the aforementioned GlyAcylCer.33 Finally, ST2B1 possesses cholesterol sulfotransferase activity and is expressed in epidermis with fillagrin, a marker of late squamous differentiation;34 the product of this enzyme, cholesterol sulfate, is incorporated into the cornified envelope and its expression parallels that of TGM1.35.
Several phosphoproteins that were represented by multiple upregulated phosphopeptides (Table S6). These included: TGM1; sequestosome-1 (SQSTM1/p62) (Fig. S1B), which was also upregulated at the protein level; the basal cell cytokeratins 6 and 14; SLC6A14, an amino acid transporter expressed in the airway epithelium; and tristetraprolin (TTP), an mRNA-binding protein that can destabilize interleukin 8 mRNA in the airway epithelium.36 Of the phosphoproteins that met strict filtering critera, only TGM1 was also found in the protein signature of squamous differentiation (Table 3); and only two other phosphopeptides with an obvious role in squamous differentiation, one each from SPR2A and late cornified envelope protein 3D, were highly upregulated after DA treatment (Table S6). When we relaxed the p-value or significance cutoffs, we were able to identify phosphopeptides to additional markers of squamous differentiation, specifically fillagrin, repetin and small proline-rich protein 3, which had high expression in donors 2–4 (but not donor 1; Table S6).
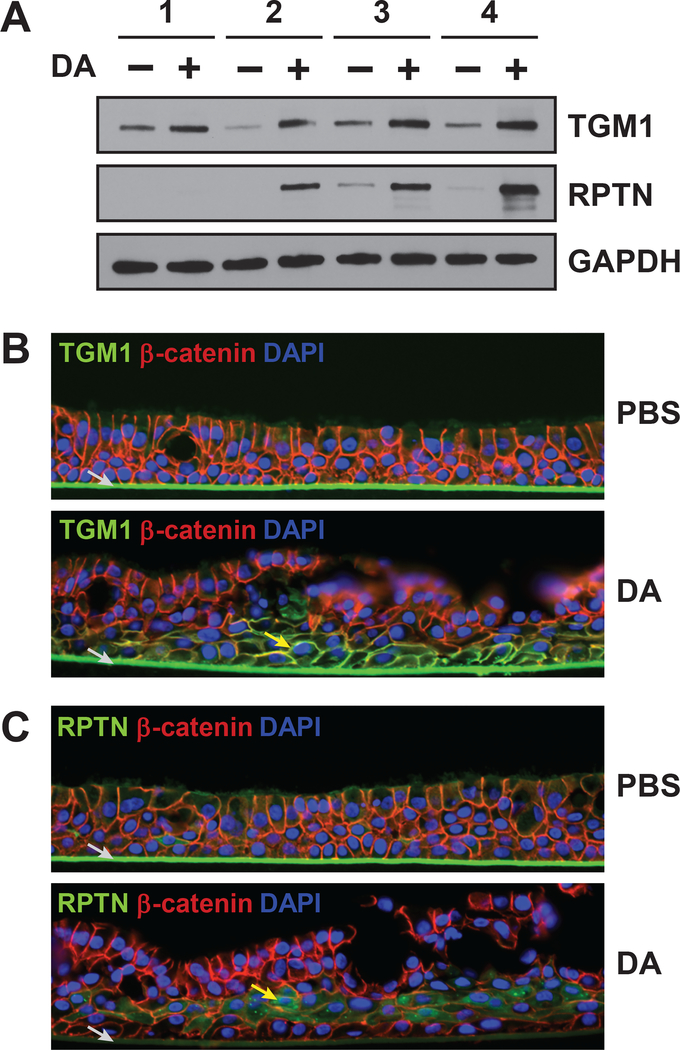
We used western blotting and immunofluorescence to confirm the increased expression of TGM1 and repetin. Consistent with the proteomic analysis, TGM1 levels were higher in all four donors after DA treatment, while levels of repetin were only elevated in donors 2–4 (Fig. 4A). Immunofluorescence analysis of cross-sections further showed that TGM1 localized to the plasma membrane in DA-treated TBECs with qualitatively highest expression in the basal-most layer of cells (Fig. 4B and Fig. S5). Repetin, which is expressed very late in squamous differentiation,29 was highly expressed in the suprabasal (but not basal) layer of DA-treated cells (Fig. 4C and Fig. S6). This is consistent with models of keratinocyte differentiation, where stem cells remain at the basal layer and differentiating cells move upwards.37 Collectively, these data suggests that in our model, DA-exposed cells largely undergo aberrant repair (i.e. squamous differentiation) rather than regeneration of a normal airway epithelium.
Figure 4. Markers of squamous differentiation are increased in DA-exposed cells.
A) The levels of TGM1, repetin and GAPDH in cell lysates were visualized by western blotting. B-C) Immunofluorescence was performed on cross sections of formalin-fixed and paraffin-imbedded TBECs to visualize (B) repetin (green, Alexa Fluor 488), β-catenin (red, Alexa Fluor 594) and nuclei (blue, DAPI) in donor 3, and (C) TGM1 (green, Alexa Fluor 488), β-catenin (red, Alexa Fluor 594) and nuclei (blue, DAPI) in donor 3. Localization of repetin and TGM1 immunostaining is indicated by yellow arrows in (B) and (C), respectively. Non-specific staining of transwell membrane by secondary antibody is indicated by grey arrow. Images in (B) are representative of three different sections per condition (Fig. S4, A–B), and results were replicated in a single section from donor 2 (Fig. S4, C–D). Images in (C) are representative of three different sections per condition (Fig. S5, A–B), and results were replicated in single sections from donors 2 and 4 (Fig. S5, C–F).
Basal cell cytokeratins are crosslinked and hyperphosphorylated following diacetyl-induced injury.
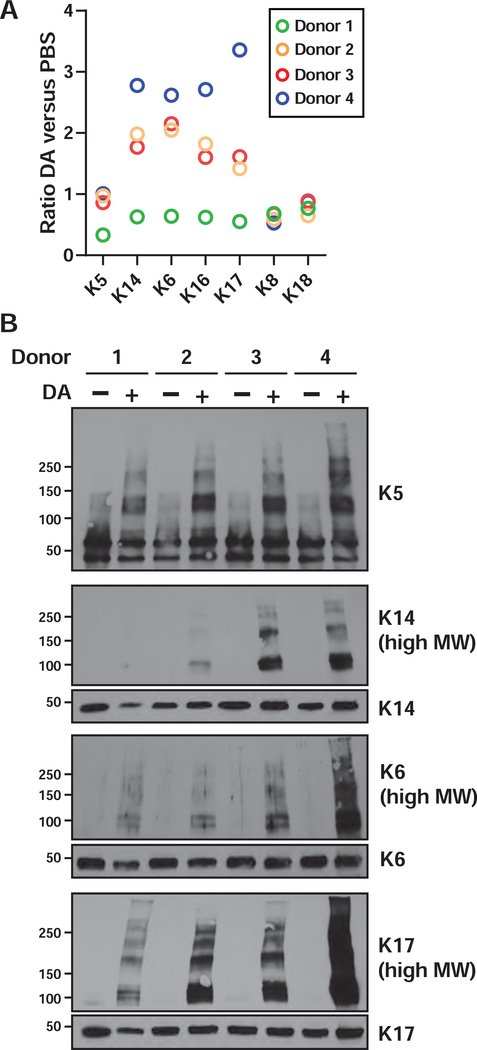
The squamous phenotype suggested a role for airway “progenitor” cell-types in the remodeling of the epithelium after DA injury. Among airway stem cells, basal cells, which are often characterized based on expression of cytokeratins,38 constitute as much as 30% of the pseudostratified human airway epithelium,39, 40 and basal cells can also express many of the proteins that we have associated with the squamous phenotype.28, 41, 42 Our proteomic analysis quantified numerous “basal cell” keratins, including type II keratins 5 and 6 (K5 and K6) and type I keratins 14, 16 and 17 (K14, K16 and K17). We wondered whether our proteomic data might provide better quantitation of the overall fate of airway progenitor cells in this model. In donor 1, which had low levels of squamous differentiation markers, all basal cell keratins were unchanged or lower after DA treatment (Fig. 5A), suggesting that basal cells might be lower in this donor after injury. In the other three donors, K6, K14, K16 and K17 were all increased after DA treatment (Fig. 5A), which could be a consequence of basal cell proliferation and/or squamous differentiation. In contrast, levels of the type I keratin 18 (K18) and its partner type II keratin 8 (K8), which are expressed in simple, non-stratified, epithelia,43 were essentially unchanged across all donors.
Figure 5. DA exposure induces crosslinking of basal cell cytokeratins.
A) Fold changes were plotted for the basal cell keratins, and clara cell marker CCSP, as determined by proteomic analysis. Note that keratin expression values were re-calculated from isoform-specific peptides (Tables S7–S8). B) The expression and “crosslinking” of cytokeratins was visualized by western blotting of whole cell lysates. For K14, K6 and K17, panels labeled as (high MW) are longer exposures of the identical membranes from which the ~50 kDa keratin monomers were visualized.
We used western blotting to independently validate the relative levels of a number of basal cell keratins. These data showed mostly lower levels of keratin monomers in DA-exposed donor 1 cells, but interestingly, DA treatment appeared to result in extensive non-reducible crosslinking of keratins, as reflected in shifts of K5, K14, K6 and K17 to higher molecular weights (of ~2 times or greater mass) with small overall changes in the levels of keratin monomers (Fig. 5B). The cross-linked species were detectable, but overall lower in TBECs in cells from donor 1, which correlated with the lower expression of basal keratins and squamous differentiation markers (including repetin). These data suggest that the increased levels of basal cell keratins, as measured by mass spectrometry, reflect an overall increase in total (monomer and crosslinked) keratins. This phenomenon could be due to polyubiquitinylation; however, because of the specific mass shifts, and the concomitant increase in expression of TGM1 throughout the basolateral layer of cells exposed to DA vapor (Fig. 4B), we believe that these high MW species are formed by TGM1-mediated Nε-(γ-glutamyl) lysine crosslinking. Transglutaminase mediated crosslinking of K5 and K6 to the cornified envelope has been previously demonstrated32 although to our knowledge, transglutaminase-dependent crosslinking of type I keratins (i.e. K14 and K17) has not been previously shown.
Keratin phosphorylation has been shown to be important for the structure/function of the intermediate filament (IF) and to occur under conditions of mitosis and cellular stress.44 Within the phosphoproteome dataset, ~100 unique phosphopeptides were assigned to basal cell keratins (Table S6). Twenty-five keratin peptides, mostly from K6 and K14, were significantly upregulated in the DA and control groups. An overlay of these peptides onto the keratin protein sequences showed that a majority of the quantified phosphorylation sites were within the head and tail domains (Fig. S7), whereas we obtained high sequence coverage of non-phosphorylated peptides across the entire protein sequences (except for the Gly-rich region of the keratin head domain; Fig. S6). These data suggest that the disordered head and tail domains may be more accessible to kinases than the central alpha-helical rod domain and are also consistent with previously characterized phosphorylation sites in keratins and other IF proteins.44, 45 However, it should be also note that the most well-characterized site of type II keratin phosphorylation, a target of p38 MAPK that is conserved in K6 (Thr146),46 does not appear to be accessible by trypsin digestion of the protein (Fig. S7).
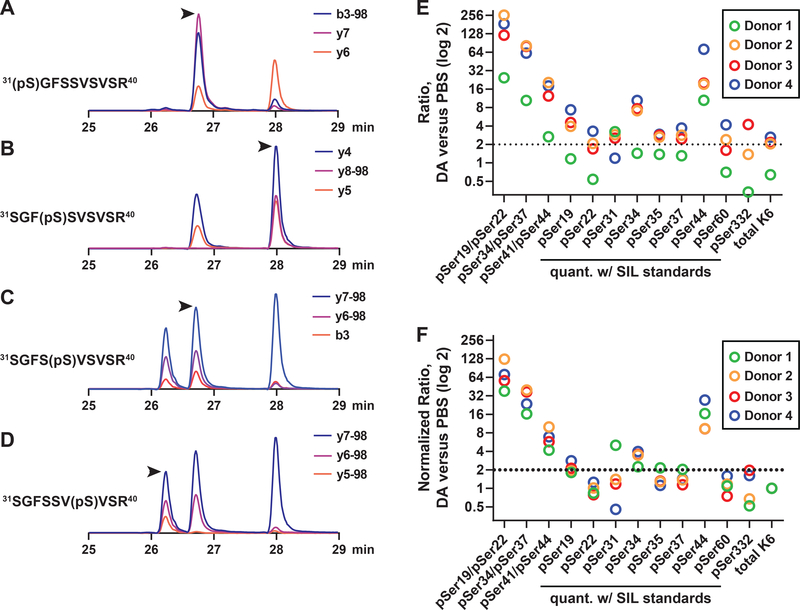
Based on their high differential expression in DA-treated versus control cells, we selected a majority of the K6 phosphopeptides for validation using a targeted proteomic approach (parallel reaction monitoring; PRM).17, 18 We focused primarily on a region in K6 between amino acids 18–60 that contained a total of 12 Ser residues, 9 of which were putatively phosphorylated in our dataset (Table S6 and Fig. S7). For peptides that contained multiple potential acceptor sites, we used authentic stable isotope-labeled (SIL) standards to confidently assign site localization of phosphorylation based on specific MS/MS fragment ions and retention times. This allowed us to develop a PRM assay to quantify four unique phosphopeptides (pSer31, 31, 35 and 37) which had an identical precursor m/z (Fig. 6), including the nearly co-eluting pSer31 and pSer35 peptides (Fig. S8).
Figure 6. DA exposure results in hyperphosphorylation of keratin 6.
A-D) Parallel reaction monitoring was used to analyze a mixture of mono-phosphorylated, stable isotope-labeled peptide standards with sequence 31SGFSSVSVSR40 (precursor m/z 551.7438). Isoform-specific product ions are shown for the peptides assigned to (A) 31(pSer)GFSSVSVSR40, (B) 31SGF(pSer)SVSVSR40, (C) 31SGFS(pSer)VSVSR40 and (D) 31SGFSSV(pSer)VSR40. The position of the properly assigned peak is indicated by an arrow. Note that the peptides containing pSer31 and pSer35 are virtually overlapping but can be distinguished based on unique product ions and retention times (as shown in Fig. S6). E) K6 phosphopeptides were quantified by PRM. For each donor, phosphopeptide ratios were determined for DA versus PBS samples. Multiply-phosphorylated peptides used label-free quantitation, and mono-phosphorylated peptides were normalized to SIL internal standards. Keratin 6 protein ratios are also shown (from Fig. 5). F) Data in (A) was normalized to ratio of K6 protein.
The PRM assay was used to unambiguously quantify K6 phosphorylation in response to DA-induced injury. Multiply phosphorylated peptides, which could not be readily synthesized in small scale, were quantified using label-free approach, whereas singly phosphorylated peptides were normalized to the SIL internal standards. Finally, ratios were used to visualize changes in phosphorylation in DA- versus PBS-treated cells (Fig. 6E). Numerous phosphopeptides were highly upregulated in all donors; however, relative changes did appear to reflect K6 protein expression, with a trend toward lower expression in donor 1. To correct for changes in K6 protein expression, the data was further normalized to total K6 ratios in each of the four donors (Fig. 6F). The normalized data showed that singly phosphophorylated peptides with pSer19, pSer34 and pSer44 were ~2-fold or more upregulated in DA-treated cells, with pSer44 increasing an average of ~15-fold in DA-treated cells. Increases in two of the doubly-phosphorylated peptides (pSer19/pSer22 and pSer34/pSer37) in DA-treated cells were much greater than the corresponding mono-phosphorylated peptides, suggesting that there may be cooperative phosphorylation of these sites by one or more kinases.
The identification of the kinase(s) that phosphorylate the N-terminus of K6 will be critical for determining the pathophysiological significance of these PTMs. To date, only a few kinases have been implicated in keratin phosphorylation, principally p38 MAPK,46 and p90RSK/RSK1.45 Of the newly validated K6 sites, none are proline-directed (i.e. pSer-Pro or pThr-Pro), which excludes MAPKs such as p38 as direct effector kinases. This is in contrast to the dataset as a whole, where ~50% of quantified phosphorylation sites have pSerPro or pThrPro motifs (data not shown). One of the upregulated sites (Ser 19) has a RXXpS motif that is required for RSK1 phosphorylation, and it is intriguing that this site in K6 shares the same RGFpS sequence as pSer380 of RSK1, a site of autoactivation.47 The other highly induced phosphopeptides are contained within Arg-rich motifs, including R-R-X-pS-X-pS, R-pS-R-X-pS that are preferred by AGC kinases,48 but the specific modulation of kinases, or kinase cascades, will be needed to determine the identity of the kinases that respond to DA injury by phosphorylating K6..
CONCLUSIONS
Exposure to diacetyl is implicated both clinically and experimentally in the development of bronchiolitis obliterans, and in vivo models have suggested that epithelial injury is a critical step in this process. Our proteomic analysis has shown that this in vitro model recapitulates many key pathological and molecular changes to the epithelium in rodent models of flavoring-induced BO, including reduced expression of cilia-related proteins,49 squamous metaplasia and keratinization10, 11 and upregulation of TGM149 and SQSTM1/p6250 Collectively, these data suggest that parallel studies in established in vitro and in vivo models should provide complementary insights into the pathogenesis of flavoring-induced BO. More generally, this study highlights the utility of primary differentiated TBECs for modeling airway injury and repair. While most proteomic studies have previous focused on monolayers of isogenic airway epithelial cells, these studies belie both the complexity of the airway—which is composed of multiple distinct cell-types (e.g. basal, goblet, multiciliated)—as well as the biological variability across cells isolated from unique individuals.
Squamous differentiation has been previously linked to airway inflammation and fibrosis, and this phenotype may be important to the pathogenesis of flavoring-induced BO. It is notable that airways of smokers, and patients with chronic obstructive pulmonary disease, express common markers of squamous differentiation, including involucrin;27, 28 and exposure of differentiated TBECs to cigarette smoke extract, or differentiation of basal cells under conditions of serum starvation or epidermal growth factor receptor stimulation can replicate these phenomena.28, 42 Under these conditions, the airway epithelium expresses pro-inflammatory and pro-fibrotic mediators, including interleukin-1β and transforming growth factor beta.27, 51 The secreted epidermal growth factor receptor ligand amphiregulin (AREG), a potent modulator of TGF-beta dependent pulmonary fibrosis,52 is also highly expressed in airway basal cells41 and upregulated under conditions of squamous differentiation.28 Importantly, we have previously shown that AREG expression, and shedding, are increased in both this in vitro model and in rats exposed to DA.13 Collectively, these data suggest that aberrant epithelial repair (i.e. squamous differentiation of basal cells) following DA exposure may promote the secretion of pro-inflammatory and pro-fibrotic mediators. To better connect flavoring-induced epithelial injury to BO pathogenesis, we are also pursuing a complementary proteomic analysis of TBEC secretions in this in vitro model.
Supplementary Material
Table S1. Run order and raw filename associations for proteome analysis
Table S2. Peptide expression table (proteome)
Table S3. Proteins expression table (proteome)
Table S4. Run order and raw filename associations for phosphoproteome analysis
Table S5. Phosphooeptide expression table (all quantified peptides) from TiO2-enriched proteome
Table S6. Phosphopeptide expression table (quantified phosphopeptides only) from TiO2-enriched
Table S7. Keratin isoform-specific peptide expression
Table S8. Recalculated keratin protein expression.
Figure S1. Visualization of proteomic data by volcano plots.
Figure S2. Extacted ion chromatograms the total and phosphorylated forms of a representative RSPH4A peptide
Figure S3. Western blotting of RSPH4A
Figure S4. Immunofluorescence staining of RSPH4A in donors 2–4
Figure S5. Immunofluorescence staining of TGM1 in donors 2 and 3
Figure S6. Immunofluorescence staining of repetin in donors 2–4.
Figure S7. Summary of identified and quantified phosphorylation sites in K6 and K14
Figure S8. Overlay of quantified transitions for pSer31 and pSer35 peptides of K6.
ACKNOWLEDGEMENTS
This work was supported in part by R21 OH010490 (M.W.F. and S.M.P.)
Footnotes
ASSOCIATED CONTENT.
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
Supplemental Electronic Data:
The raw data for the label-free quantitative proteomic analyses was deposited at Chorus Project (www.chorusproject.org) under the project “EpiAir Diacetyl Proteomics” and experiment “EpiAir Diacetyl Proteome”. Skyline files containing results of PRM analysis have been made public as part of the Panorama Targeted Proteomics data repository (panoramaweb.org).65 Data can be viewed at https://goo.gl/e5gJRk and downloaded at https://goo.gl/3Z905f.
REFERENCES
- 1.Kreiss K; Gomaa A; Kullman G; Fedan K; Simoes EJ; Enright PL, Clinical bronchiolitis obliterans in workers at a microwave-popcorn plant. N Engl J Med 2002, 347, (5), 330–8. [DOI] [PubMed] [Google Scholar]
- 2.Kreiss K, Flavoring-related bronchiolitis obliterans. Curr Opin Allergy Clin Immunol 2007, 7, (2), 162–7. [DOI] [PubMed] [Google Scholar]
- 3.Duling MG; LeBouf RF; Cox-Ganser JM; Kreiss K; Martin SB Jr.; Bailey RL, Environmental characterization of a coffee processing workplace with obliterative bronchiolitis in former workers. J Occup Environ Hyg 2016, 13, (10), 770–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holden VK; Hines SE, Update on flavoring-induced lung disease. Curr Opin Pulm Med 2016, 22, (2), 158–64. [DOI] [PubMed] [Google Scholar]
- 5.Farsalinos KE; Kistler KA; Gillman G; Voudris V, Evaluation of electronic cigarette liquids and aerosol for the presence of selected inhalation toxins. Nicotine Tob Res 2015, 17, (2), 168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen JG; Flanigan SS; LeBlanc M; Vallarino J; MacNaughton P; Stewart JH; Christiani DC, Flavoring Chemicals in E-Cigarettes: Diacetyl, 2,3-Pentanedione, and Acetoin in a Sample of 51 Products, Including Fruit-, Candy-, and Cocktail-Flavored E-Cigarettes. Environ Health Perspect 2016, 124, (6), 733–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbs AF; Battelli LA; Goldsmith WT; Porter DW; Frazer D; Friend S; Schwegler-Berry D; Mercer RR; Reynolds JS; Grote A; Castranova V; Kullman G; Fedan JS; Dowdy J; Jones WG, Necrosis of nasal and airway epithelium in rats inhaling vapors of artificial butter flavoring. Toxicol Appl Pharmacol 2002, 185, (2), 128–35. [DOI] [PubMed] [Google Scholar]
- 8.Hubbs AF; Goldsmith WT; Kashon ML; Frazer D; Mercer RR; Battelli LA; Kullman GJ; Schwegler-Berry D; Friend S; Castranova V, Respiratory toxicologic pathology of inhaled diacetyl in sprague-dawley rats. Toxicol Pathol 2008, 36, (2), 330–44. [DOI] [PubMed] [Google Scholar]
- 9.Palmer SM; Flake GP; Kelly FL; Zhang HL; Nugent JL; Kirby PJ; Foley JF; Gwinn WM; Morgan DL, Severe airway epithelial injury, aberrant repair and bronchiolitis obliterans develops after diacetyl instillation in rats. PLoS One 2011, 6, (3), e17644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan DL; Jokinen MP; Price HC; Gwinn WM; Palmer SM; Flake GP, Bronchial and bronchiolar fibrosis in rats exposed to 2,3-pentanedione vapors: implications for bronchiolitis obliterans in humans. Toxicol Pathol 2012, 40, (3), 448–65. [DOI] [PubMed] [Google Scholar]
- 11.Morgan DL; Jokinen MP; Johnson CL; Price HC; Gwinn WM; Bousquet RW; Flake GP, Chemical Reactivity and Respiratory Toxicity of the alpha-Diketone Flavoring Agents: 2,3-Butanedione, 2,3-Pentanedione, and 2,3-Hexanedione. Toxicol Pathol 2016, 44, (5), 763–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Koren EG; Hogan BL; Gunn MD, Loss of basal cells precedes bronchiolitis obliterans-like pathological changes in a murine model of chlorine gas inhalation. Am J Respir Cell Mol Biol 2013, 49, (5), 788–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelly FL; Sun J; Fischer BM; Voynow JA; Kummarapurugu AB; Zhang HL; Nugent JL; Beasley RF; Martinu T; Gwinn WM; Morgan DL; Palmer SM, Diacetyl induces amphiregulin shedding in pulmonary epithelial cells and in experimental bronchiolitis obliterans. Am J Respir Cell Mol Biol 2014, 51, (4), 568–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaccone EJ; Goldsmith WT; Shimko MJ; Wells JR; Schwegler-Berry D; Willard PA; Case SL; Thompson JA; Fedan JS, Diacetyl and 2,3-pentanedione exposure of human cultured airway epithelial cells: Ion transport effects and metabolism of butter flavoring agents. Toxicol Appl Pharmacol 2015, 289, (3), 542–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beausoleil SA; Villen J; Gerber SA; Rush J; Gygi SP, A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 2006, 24, (10), 1285–92. [DOI] [PubMed] [Google Scholar]
- 16.MacLean B; Tomazela DM; Shulman N; Chambers M; Finney GL; Frewen B; Kern R; Tabb DL; Liebler DC; MacCoss MJ, Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010, 26, (7), 966–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gallien S; Duriez E; Crone C; Kellmann M; Moehring T; Domon B, Targeted proteomic quantification on quadrupole-orbitrap mass spectrometer. Mol Cell Proteomics 2012, 11, (12), 1709–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterson AC; Russell JD; Bailey DJ; Westphall MS; Coon JJ, Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics 2012, 11, (11), 1475–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoos MD; Richardson BM; Foster MW; Everhart A; Thompson JW; Moseley MA; Colton CA, Longitudinal study of differential protein expression in an Alzheimer’s mouse model lacking inducible nitric oxide synthase. J Proteome Res 2013, 12, (10), 4462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schechter MA; Hsieh MK; Njoroge LW; Thompson JW; Soderblom EJ; Feger BJ; Troupes CD; Hershberger KA; Ilkayeva OR; Nagel WL; Landinez GP; Shah KM; Burns VA; Santacruz L; Hirschey MD; Foster MW; Milano CA; Moseley MA; Piacentino V 3rd; Bowles DE, Phosphoproteomic profiling of human myocardial tissues distinguishes ischemic from non-ischemic end stage heart failure. PLoS One 2014, 9, (8), e104157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uhlen M; Fagerberg L; Hallstrom BM; Lindskog C; Oksvold P; Mardinoglu A; Sivertsson A; Kampf C; Sjostedt E; Asplund A; Olsson I; Edlund K; Lundberg E; Navani S; Szigyarto CA; Odeberg J; Djureinovic D; Takanen JO; Hober S; Alm T; Edqvist PH; Berling H; Tegel H; Mulder J; Rockberg J; Nilsson P; Schwenk JM; Hamsten M; von Feilitzen K; Forsberg M; Persson L; Johansson F; Zwahlen M; von Heijne G; Nielsen J; Ponten F, Proteomics. Tissue-based map of the human proteome. Science 2015, 347, (6220), 1260419. [DOI] [PubMed] [Google Scholar]
- 22.Lindskog C; Fagerberg L; Hallstrom B; Edlund K; Hellwig B; Rahnenfuhrer J; Kampf C; Uhlen M; Ponten F; Micke P, The lung-specific proteome defined by integration of transcriptomics and antibody-based profiling. FASEB J 2014, 28, (12), 5184–96. [DOI] [PubMed] [Google Scholar]
- 23.Frommer A; Hjeij R; Loges NT; Edelbusch C; Jahnke C; Raidt J; Werner C; Wallmeier J; Grosse-Onnebrink J; Olbrich H; Cindric S; Jaspers M; Boon M; Memari Y; Durbin R; Kolb-Kokocinski A; Sauer S; Marthin JK; Nielsen KG; Amirav I; Elias N; Kerem E; Shoseyov D; Haeffner K; Omran H, Immunofluorescence Analysis and Diagnosis of Primary Ciliary Dyskinesia with Radial Spoke Defects. Am J Respir Cell Mol Biol 2015, 53, (4), 563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zimmers TA; Jin X; Hsiao EC; McGrath SA; Esquela AF; Koniaris LG, Growth differentiation factor-15/macrophage inhibitory cytokine-1 induction after kidney and lung injury. Shock 2005, 23, (6), 543–8. [PubMed] [Google Scholar]
- 25.Wu Q; Jiang D; Matsuda JL; Ternyak K; Zhang B; Chu HW, Cigarette Smoke Induces Human Airway Epithelial Senescence via GDF15 Production. Am J Respir Cell Mol Biol 2016. [DOI] [PubMed] [Google Scholar]
- 26.Hubbs AF; Fluharty KL; Edwards RJ; Barnabei JL; Grantham JT; Palmer SM; Kelly F; Sargent LM; Reynolds SH; Mercer RR; Goravanahally MP; Kashon ML; Honaker JC; Jackson MC; Cumpston AM; Goldsmith WT; McKinney W; Fedan JS; Battelli LA; Munro T; Bucklew-Moyers W; McKinstry K; Schwegler-Berry D; Friend S; Knepp AK; Smith SL; Sriram K, Accumulation of Ubiquitin and Sequestosome-1 Implicate Protein Damage in Diacetyl-Induced Cytotoxicity. Am J Pathol 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herfs M; Hubert P; Poirrier AL; Vandevenne P; Renoux V; Habraken Y; Cataldo D; Boniver J; Delvenne P, Proinflammatory cytokines induce bronchial hyperplasia and squamous metaplasia in smokers: implications for chronic obstructive pulmonary disease therapy. Am J Respir Cell Mol Biol 2012, 47, (1), 67–79. [DOI] [PubMed] [Google Scholar]
- 28.Araya J; Cambier S; Markovics JA; Wolters P; Jablons D; Hill A; Finkbeiner W; Jones K; Broaddus VC; Sheppard D; Barzcak A; Xiao Y; Erle DJ; Nishimura SL, Squamous metaplasia amplifies pathologic epithelial-mesenchymal interactions in COPD patients. J Clin Invest 2007, 117, (11), 3551–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krieg P; Schuppler M; Koesters R; Mincheva A; Lichter P; Marks F, Repetin (Rptn), a new member of the “fused gene” subgroup within the S100 gene family encoding a murine epidermal differentiation protein. Genomics 1997, 43, (3), 339–48. [DOI] [PubMed] [Google Scholar]
- 30.Candi E; Schmidt R; Melino G, The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 2005, 6, (4), 328–40. [DOI] [PubMed] [Google Scholar]
- 31.Jetten AM, Multistep process of squamous differentiation in tracheobronchial epithelial cells in vitro: analogy with epidermal differentiation. Environ Health Perspect 1989, 80, 149–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Candi E; Tarcsa E; Digiovanna JJ; Compton JG; Elias PM; Marekov LN; Steinert PM, A highly conserved lysine residue on the head domain of type II keratins is essential for the attachment of keratin intermediate filaments to the cornified cell envelope through isopeptide crosslinking by transglutaminases. Proc Natl Acad Sci U S A 1998, 95, (5), 2067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radner FP; Fischer J, The important role of epidermal triacylglycerol metabolism for maintenance of the skin permeability barrier function. Biochim Biophys Acta 2014, 1841, (3), 409–15. [DOI] [PubMed] [Google Scholar]
- 34.Higashi Y; Fuda H; Yanai H; Lee Y; Fukushige T; Kanzaki T; Strott CA, Expression of cholesterol sulfotransferase (SULT2B1b) in human skin and primary cultures of human epidermal keratinocytes. J Invest Dermatol 2004, 122, (5), 1207–13. [DOI] [PubMed] [Google Scholar]
- 35.Rearick JI; Stoner GD; George MA; Jetten AM, Cholesterol sulfate accumulation in tumorigenic and nontumorigenic rat esophageal epithelial cells: evidence for defective differentiation control in tumorigenic cells. Cancer Res 1988, 48, (18), 5289–95. [PubMed] [Google Scholar]
- 36.Prabhala P; Ammit AJ, Tristetraprolin and its role in regulation of airway inflammation. Mol Pharmacol 2015, 87, (4), 629–38. [DOI] [PubMed] [Google Scholar]
- 37.Eckert RL; Rorke EA, Molecular biology of keratinocyte differentiation. Environ Health Perspect 1989, 80, 109–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rackley CR; Stripp BR, Building and maintaining the epithelium of the lung. J Clin Invest 2012, 122, (8), 2724–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wansleeben C; Barkauskas CE; Rock JR; Hogan BL, Stem cells of the adult lung: their development and role in homeostasis, regeneration, and disease. Wiley Interdiscip Rev Dev Biol 2013, 2, (1), 131–48. [DOI] [PubMed] [Google Scholar]
- 40.Rock JR; Randell SH; Hogan BL, Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech 2010, 3, (9–10), 545–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hackett NR; Shaykhiev R; Walters MS; Wang R; Zwick RK; Ferris B; Witover B; Salit J; Crystal RG, The human airway epithelial basal cell transcriptome. PLoS One 2011, 6, (5), e18378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaykhiev R; Zuo WL; Chao I; Fukui T; Witover B; Brekman A; Crystal RG, EGF shifts human airway basal cell fate toward a smoking-associated airway epithelial phenotype. Proc Natl Acad Sci U S A 2013, 110, (29), 12102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutton E; Paladini RD; Yu QC; Yen M; Coulombe PA; Fuchs E, Functional differences between keratins of stratified and simple epithelia. J Cell Biol 1998, 143, (2), 487–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hyder CL; Pallari HM; Kochin V; Eriksson JE, Providing cellular signposts--post-translational modifications of intermediate filaments. FEBS Lett 2008, 582, (14), 2140–8. [DOI] [PubMed] [Google Scholar]
- 45.Pan X; Kane LA; Van Eyk JE; Coulombe PA, Type I keratin 17 protein is phosphorylated on serine 44 by p90 ribosomal protein S6 kinase 1 (RSK1) in a growth- and stress-dependent fashion. J Biol Chem 2011, 286, (49), 42403–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toivola DM; Zhou Q; English LS; Omary MB, Type II keratins are phosphorylated on a unique motif during stress and mitosis in tissues and cultured cells. Mol Biol Cell 2002, 13, (6), 1857–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dalby KN; Morrice N; Caudwell FB; Avruch J; Cohen P, Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J Biol Chem 1998, 273, (3), 1496–505. [DOI] [PubMed] [Google Scholar]
- 48.Pearce LR; Komander D; Alessi DR, The nuts and bolts of AGC protein kinases. Nat Rev Mol Cell Biol 2010, 11, (1), 9–22. [DOI] [PubMed] [Google Scholar]
- 49.Morgan DL; Merrick BA; Gerrish KE; Stockton PS; Wang Y; Foley JF; Gwinn WM; Kelly FL; Palmer SM; Ton TV; Flake GP, Gene expression in obliterative bronchiolitis-like lesions in 2,3-pentanedione-exposed rats. PLoS One 2015, 10, (2), e0118459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hubbs AF; Fluharty KL; Edwards RJ; Barnabei JL; Grantham JT; Palmer SM; Kelly F; Sargent LM; Reynolds SH; Mercer RR; Goravanahally MP; Kashon ML; Honaker JC; Jackson MC; Cumpston AM; Goldsmith WT; McKinney W; Fedan JS; Battelli LA; Munro T; Bucklew-Moyers W; McKinstry K; Schwegler-Berry D; Friend S; Knepp AK; Smith SL; Sriram K, Accumulation of Ubiquitin and Sequestosome-1 Implicate Protein Damage in Diacetyl-Induced Cytotoxicity. Am J Pathol 2016, 186, (11), 2887–2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen YT; Nikulina K; Lazarev S; Bahrami AF; Noble LB; Gallup M; McNamara NA, Interleukin-1 as a phenotypic immunomodulator in keratinizing squamous metaplasia of the ocular surface in Sjogren’s syndrome. Am J Pathol 2010, 177, (3), 1333–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Y; Lee JY; Lee CM; Cho WK; Kang MJ; Koff JL; Yoon PO; Chae J; Park HO; Elias JA; Lee CG, Amphiregulin, an epidermal growth factor receptor ligand, plays an essential role in the pathogenesis of transforming growth factor-beta-induced pulmonary fibrosis. J Biol Chem 2012, 287, (50), 41991–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Run order and raw filename associations for proteome analysis
Table S2. Peptide expression table (proteome)
Table S3. Proteins expression table (proteome)
Table S4. Run order and raw filename associations for phosphoproteome analysis
Table S5. Phosphooeptide expression table (all quantified peptides) from TiO2-enriched proteome
Table S6. Phosphopeptide expression table (quantified phosphopeptides only) from TiO2-enriched
Table S7. Keratin isoform-specific peptide expression
Table S8. Recalculated keratin protein expression.
Figure S1. Visualization of proteomic data by volcano plots.
Figure S2. Extacted ion chromatograms the total and phosphorylated forms of a representative RSPH4A peptide
Figure S3. Western blotting of RSPH4A
Figure S4. Immunofluorescence staining of RSPH4A in donors 2–4
Figure S5. Immunofluorescence staining of TGM1 in donors 2 and 3
Figure S6. Immunofluorescence staining of repetin in donors 2–4.
Figure S7. Summary of identified and quantified phosphorylation sites in K6 and K14
Figure S8. Overlay of quantified transitions for pSer31 and pSer35 peptides of K6.