Abstract
Numerous studies have demonstrated that long non-coding RNAs (lncRNAs) serve an important regulatory role in ischemic injury of cardiomyocytes. lncRNA small nucleolar RNA host gene 1 (SNHG1) could effectively protect cardiomyocytes against various injuries. However, the role of SNHG1 in ischemic cardiomyocyte injury is unclear. It was hypothesized that SNHG1 may have a protective effect on cardiomyocyte injury induced by hypoxia/reoxygenation (H/R) by sponging microRNA (miRNA/miR). The purpose of the present study was to explore the role and molecular mechanism of SNHG1 in ischemic cardiomyocyte injury. A H9c2 cardiomyocyte H/R model was established. The expression levels of SNHG1 in cardiomyocytes treated with H/R were detected using reverse transcription-quantitative PCR. A luciferase reporter assay was used to analyze the associations among SNHG1, miR-16-5p and GATA binding protein 4 (GATA4). Chromatin immunoprecipitation experiments were performed to analyze the interaction between SNHG1 and GATA4. Cell Counting Kit-8, enzyme-linked immunosorbent assay, terminal deoxynucleotidyl-transferase-mediated dUTP nick end labeling and western blotting experiments were used to detect cell activity, lactate dehydrogenase release, apoptosis and apoptosis-related proteins (Bcl-2, Bax, Cleaved caspase-3 and Cleaved caspase-9), respectively. The expression levels of SNHG1 were downregulated in cardiomyocytes treated with H/R. Overexpression of SNHG1 had a protective effect on cardiomyocyte injury induced by H/R. In addition, SNHG1 could regulate the expression levels of GATA4 via sponging of miR-16-5p. Further experiments revealed that GATA4 could bind to the promoter region of SNHG1 and subsequently regulated the expression levels of SNHG1, indicating the important role of the positive feedback loop of SNHG1/miR-16-5p/GATA4 in cardiomyocyte ischemic injury. To conclude, the present study revealed the protective effect of the SNHG1/miR-16-5p/GATA4 positive feedback loop on cardiomyocyte injury induced by H/R and provided a potential therapeutic target for ischemic cardiomyocyte injury.
Keywords: small nucleolar RNA host gene 1, microRNA-16-5p, GATA binding protein 4, hypoxia/reoxygenation, cardiomyocyte
Introduction
Ischemic heart disease is the main cause of cardiovascular disease, which could induce myocardial infarction and lead to myocardial necrosis, impairing the quality of life of patients (1,2). Blood flow recovery is the most effective way to save ischemic cardiomyocytes and the lives of patients (1). However, myocardial ischemia-reperfusion (I/R) can lead to a series of adverse events, including excessive reactive oxygen species, calcium ion overload and endoplasmic reticulum stress, which could promote cardiomyocyte apoptosis and aggravate the degree of myocardial injury (3). Thus, there is an urgent need to explore the potential molecular mechanism of myocardial I/R injury to improve the quality of life of patients.
Long non-coding RNAs (lncRNAs) are a type of RNA transcripts without protein coding ability and are longer than 200 nucleotides (4). Numerous studies have demonstrated that lncRNAs are involved in numerous important processes in cell biology, including signal transduction regulation, DNA modification, transcriptional activation and protein function regulation (5). In addition, lncRNAs serve an important role in the occurrence and development of tumor cells (6). Small nucleolar RNA host gene 1 (SNHG1) is a lncRNA located on chromosome 11p2.3 (7). SNHG1 has a protective effect on cardiomyocytes under various impairments. For example, SNHG1 has been found to protect cardiomyocytes against toxic damage induced by Adriamycin (8) and inhibit cardiomyocyte apoptosis induced by H2O2 (9). A recent study revealed that SNHG1 could reduce vascular endothelial cell injury induced by hypoxia/reoxygenation (H/R) (10). In addition, SNHG1 acts as a competing endogenous RNA of microRNAs (miRNAs/miRs) to suppress the regulation of target genes (11).
Therefore, it was hypothesized that SNHG1 may have a protective effect on cardiomyocyte injury induced by H/R by sponging miRNA. However, to the best of our knowledge, there are no studies on the exact role and mechanism of SNHG1 in cardiomyocyte injury induced by H/R. Thus, the purpose of the present study was to explore the role and molecular mechanism of SNHG1 in ischemic cardiomyocyte injury.
Materials and methods
Cell culture and treatment
H9c2 cells were obtained from BeNa Culture Collection; Beijing Beina Chunglian Institute of Biotechnology Research Institute. The cells were cultured in Dulbecco's modified Eagle's medium (DMEM; HyClone; Cytiva) containing 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) and 1% penicillin/streptomycin. H9c2 cells were routinely cultured for 3 days and then subcultured. Cells in the logarithmic phase were used for subsequent experiments.
H9c2 cells were treated with H/R to establish the H/R model
Cells in the logarithmic phase were inoculated into 24-well plates at a density of 2×104 cells/well. When the cell fusion degree reached 90%, the culture medium was replaced with serum-free medium. Subsequently, cells were cultured in an incubator containing 90% N2 and 5% CO2 for 6 h at 37°C. Next, cells were returned to the normal culture conditions. Cells were then cultured in complete DMEM and maintained in a 5% CO2 incubator for 6, 12 and 24 h at 37°C.
Cell transfection
Short hairpin RNA (shRNA) against GATA4 (shRNA-GATA4; 5′-CTGGATGTTGGGCAGGAC −3′) (100 nM), shRNA negative control (shRNA-NC) (100 nM), miR-16-5p mimic (5′-UAGCAGCACGUAAAUAUUGGCG-3′) (50 nM), mimic negative control (miR-NC; 5′-UUCUCCGAACGUGUCACGUTT −3′, 100 nM), miR-16-5p inhibitor (5′-CGCCAAUAUUUACGUGCUGCUA −3′, 50 nM), inhibitor negative control (NC inhibitor; 5′-CAGUACUUUUGUGUAGUACAA −3′, 50 nM) pcDNA-GATA4 (100 nM) and empty control vector (pcDNA-NC; 100 nM) were synthesized by Shanghai GenePharma Co., Ltd. The SNHG1 overexpression vector (pcDNA-SNHG1; 100 nM) and empty control vector (pcDNA-NC; 100 nM) were constructed by Shanghai GenePharma Co., Ltd. Cells were seeded into 6-well plates at a density of 3×105 cells/well and cultured for 24 h at 37°C. In strict accordance with the instruction of Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) these aforementioned indicated plasmids were transfected into the cells using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.). Transfection efficiency was detected via reverse transcription-quantitative PCR (RT-qPCR) at 48 h after transfection.
RT-qPCR analysis
Total RNA was extracted from cells using TRIzol® reagent (Thermo Fisher Scientific, Inc.). RNA was synthesized into cDNA using a reverse transcription kit (Promega Corporation) according to the manufacturer's instructions. The PCR reaction was performed using SYBR Green Supermix (Applied Biosystems; Thermo Fisher Scientific, Inc.) and an ABI 7500 PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The PCR reaction conditions were as follows: 94°C for 30 sec, 55°C for 30 sec and 72°C for 30 sec (22 cycles). The relative expression levels of target genes were calculated using the 2−ΔΔCq method (12) and normalized to those of the housekeeping gene GAPDH or U6. Primers used in this study are listed in Table I.
Table I.
Primer sequences used for reverse transcription-quantitative PCR.
| Gene | Sequences (5′→3′) |
|---|---|
| SNHG1 | F: AGGCTGAAGTTACAGGTC |
| R: TTGGCTCCCAGTGTCTTA | |
| miR-16-5p | F: TAGCAGCACGTAAATATTGGCG |
| R: TGCGTGTCGTGGAGTC | |
| GATA4 | F: GAGCTGGTACCTGGCCTTC |
| R: GCTCTGCTGAAATCACTCTGA | |
| GAPDH | F: GTCAACGGATTTGGTCTGTATT |
| R: AGTCTTCTGGGTGGCAGTGAT | |
| U6 | F: CTCGCTTCGGCAGCACA |
| R: AACGCTTCACGAATTTGCGT | |
| GAPDH | F: ATCTCCTTTGTTACCGCTTCC |
| R: GAAGATGGTGATGGGATTTC |
SNHG1, small nucleolar RNA host gene 1; GATA4, GATA binding protein 4; miR, microRNA; F, forward; R, reverse.
Cell Counting Kit-8 (CCK-8) assay
Cell viability was detected using a CCK-8 assay. Cells were incubated in 96-well plates at a density of 2×103 cells/well for 24, 48 and 72 h. A total of 10 µl CCK-8 solution (cat. no. C0037; Beyotime Institute of Biotechnology) was added to each well. Then, cells were incubated for 1 h at 37°C. The culture was continued for an additional 2 h. The absorbance of each well was measured at a wavelength of 450 nm using a microplate reader (Synergy 2 Multi-Mode Microplate Reader; BioTek Instruments, Inc.).
Terminal deoxynucleotidyl-transferase-mediated dUTP nick end labeling (TUNEL) assay
Apoptosis was detected using the TUNEL Apoptosis Assay kit (cat. no. C1088; Beyotime Institute of Biotechnology). Briefly, cells were washed with PBS and fixed with 4% paraformaldehyde at room temperature for 20 min. Subsequently, cells were treated with 0.1% Triton-X-100 for 10 min. Each sample was supplemented with 50 µl TUNEL detection reagent for 40 min at 37°C in the dark. The cell nuclei were stained with 5 µg/ml DAPI at 37°C in the dark for 5 min. The morphological changes of apoptotic cells were observed in three random fields under a fluorescence microscope (Olympus FV500; Olympus Corporation; magnification ×200). Bright green fluorescence was considered to indicate apoptotic cells.
Enzyme-linked immunosorbent assay (ELISA)
Cell supernatant was collected and centrifuged at 700 × g at 4°C for 10 min. Then centrifugation was performed at 9,000 × g at 4°C for another 15 min and the supernatant collected. According to the manufacturer's instructions, an ELISA kit (cat. no. A020-1-2; Nanjing Jiancheng Bioengineering Institute) was used to analyze the level of lactate dehydrogenase (LDH) release in the supernatant. The samples of each group were analyzed using an Automatic Microplate Reader (Syngene). All the experiments in this study were repeated three times.
Luciferase reporter assay
Targets of SNHG1 were predicted using the Encyclopedia of RNA Interactomes (ENCORI) database (https://starbase.sysu.edu.cn/). The cells were inoculated into 96-well plates at a density of 2×104 cells per well. When the degree of cell fusion reached 60%, the cells were co-transfected with SNHG1 3′-untranslated region (3′-UTR) plasmid (wild-type; SNHG1 WT), SNHG11 3′-UTR plasmid (mutant; SNHG1 MUT), GATA4 3′-UTR plasmid (wild-type; GATA4 WT), GATA4 3′-UTR plasmid (mutant; GATA4 MUT) was cloned into psi-CHECK (Promega Corporation) downstream of the firefly luciferase 3′-UTR. These plasmids were amplified by Shanghai GenePharma Co., Ltd. The psi-CHECK vector also provided Renilla luciferase as the normalization signal and miR-16-5p mimics or miR-NC using Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h after transfection, the relative luciferase intensity of each group was detected using a Dual Luciferase Reporter Assay kit (Promega Corporation). Firefly luciferase activities were normalized to Renilla luciferase activities.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was performed using a ChIP assay kit (Pierce™ Agarose ChIP Kit; Pierce; Thermo Fisher Scientific, Inc.). Briefly, the chromatin fragments derived from H9c2 cells were immunoprecipitated with 10 μg antibody against GATA4 (cat. no. ab256782; Abcam) or 5 μg mouse IgG (cat. no. sc-2025; Santa Cruz Biotechnology, Inc.) were used for the immunoprecipitation. The cell lysate was incubated with a GATA antibody (1:1000) or IgG antibody (0.2 µg/ml) and 40 μl protein A/G magnetic beads (EMD Millipore) at 4°C overnight. On the second day, products of immunoprecipitation were treated with ChIP elution buffer provided by the kit. Immunoprecipitated complexes were collected by centrifugation at 10,000 × g for 10 min at 4°C. Subsequently, precipitated DNA samples were detected via RT-qPCR.
Western blot analysis
Total RNA was extracted from cells using radioimmunoprecipitation assay buffer (cat. no. P0013B; Beyotime Institute of Biotechnology). The protein concentration was detected using a bicinchoninic acid protein quantitative kit (cat. no. P0012; Beyotime Institute of Biotechnology). A total of 20 µg protein was separated via 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and subsequently transferred to a polyvinylidene fluoride membrane (cat. no. FFP36; Beyotime Institute of Biotechnology). These membranes were blocked with 5% skimmed milk for 30 min at room temperature and incubated with primary antibodies (1:1,000) against GATA4 (cat. no. ab256782; Abcam), Bcl-2 (cat. no. ab182858; Abcam), Bax (cat. no. ab182733; Abcam), caspase-3 (cat. no. ab32150; Abcam), Cleaved caspase-3 (cat. no. ab2302; Abcam), caspase-9 (cat. no. ab32539; Abcam), Cleaved caspase-9 (cat. no. ab2324; Abcam) and GAPDH (cat. no. ab9485; Abcam) overnight at 4°C. Subsequently, the membranes were incubated with the corresponding horseradish peroxidase-conjugated Goat Anti-Rabbit IgG H&L (HRP) secondary antibody (1:10,000; cat. no. ab205718; Abcam) at 37°C for 2 h at the appropriate dilutions. An ECL Plus kit (cat. no. P0018; Beyotime Institute of Biotechnology) was used to visualize the protein bands. Densitometric analysis was performed using ImageJ software (version 1.49v; National Institutes of Health).
Statistical analysis
All data were analyzed using GraphPad Prism 7 software (GraphPad Software, Inc.). The measurement data are presented as the mean ± standard deviation. Student's t-test (unpaired) was used to compare the differences between two groups. Comparisons among multiple groups were performed using one-way ANOVA followed by Tukey's post hoc test. P<0.05 was considered to indicate a statistically significant difference. All experiments were repeated at least three times.
Results
SNHG1 expression is downregulated in H9c2 cells treated with H/R
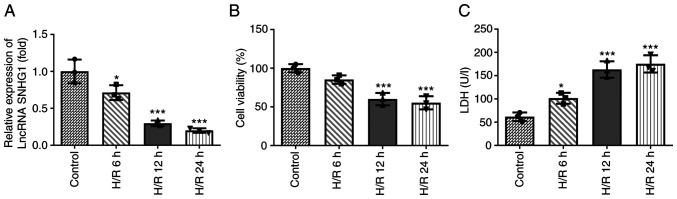
The expression levels of SNHG1 in H9c2 cells treated with H/R were assessed using RT-qPCR. As shown in Fig. 1A, the expression levels of SNHG1 in H9c2 cells treated with H/R decreased gradually with increasing treatment time. In addition, cell viability was detected using CCK-8 and LDH assays. As shown in Fig. 1B and C, compared with the control group, the viability of H9c2 cells was decreased, while the levels of LDH were increased with increasing treatment time. When the treatment time was 12 h, the change in cell viability tended to be stable. Therefore, in the follow-up experiments, the treatment time of H/R was set as 12 h.
Figure 1.
SNHG1 is downregulated in H9c2 cells treated with H/R. (A) Reverse transcription-quantitative PCR was performed to detect the expression of SNHG1 in H9c2 cells treated with H/R for 6, 12 and 24 h. (B) Cell Counting Kit-8 assay was performed to detect the viability of H9c2 cells treated with H/R for 6, 12 and 24 h. (C) ELISA was performed to detect the cytotoxicity of the H9c2 cells treated with H/R for 6, 12 and 24 h. *P<0.05 and ***P<0.001 vs. Control. SNHG1, small nucleolar RNA host gene 1; H/R, hypoxia/reoxygenation; LDH, lactate dehydrogenase; lncRNA, long non-coding RNA.
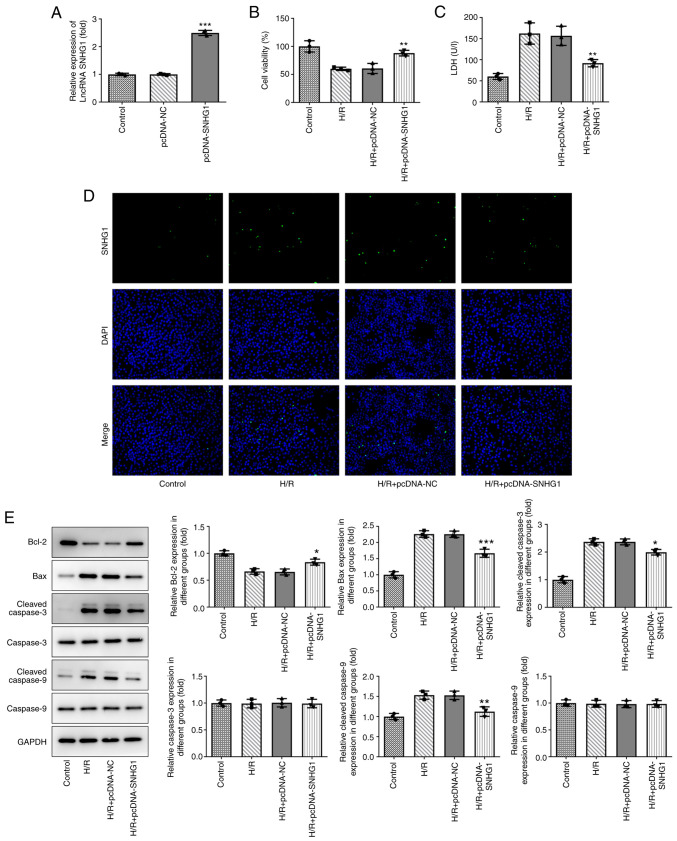
Overexpression of SNHG1 attenuates the injury of H9c2 cells induced by H/R
The expression levels of SNHG1 in H9c2 cells treated with H/R were increased by transfection with pcDNA-SNHG1. As shown in Fig. 2A, the expression levels of SNHG1 in the pcDNA-SNHG1 group were markedly higher than those in the control group. As shown in Fig. 2B and C, overexpression of SNHG1 partly alleviated the inhibition of H9c2 cell viability by H/R. In addition, a TUNEL assay and western blotting were used to detect cell apoptosis. As shown in Fig. 2D and E, compared with the control group, H/R markedly increased the number of positive apoptotic cells, decreased the expression levels of anti-apoptotic protein Bcl-2, and increased the expression levels of pro-apoptotic proteins Bax, Cleaved caspase-3 and Cleaved caspase-9. These aforementioned changes could be partially reversed by overexpression of SNHG1.
Figure 2.
Overexpression of SNHG1 attenuates H/R-induced H9c2 cell injury. (A) The efficiency of SNHG1 overexpression was examined via reverse transcription-quantitative PCR in transfected H9c2 cell lines. ***P<0.001 vs. pcDNA-NC. (B) Cell proliferation was determined using a Cell Counting Kit-8 assay. **P<0.01 vs. H/R + pcDNA-NC. (C) Cytotoxicity to the cells was measured via ELISA. **P<0.01 vs. H/R + pcDNA-NC. (D) Cell apoptosis was detected via a TUNEL assay. Bright green fluorescence was considered to indicate apoptotic cells (magnification ×200). (E) Expression of apoptosis-related proteins, cell proliferation-related proteins (Bcl-2, Bax, Cleaved caspase-3, caspase-3, Cleaved caspase-9 and caspase-9) were detected by western blotting. *P<0.05, **P<0.01 and ***P<0.001 vs. H/R + pcDNA-NC. SNHG1, small nucleolar RNA host gene 1; H/R, hypoxia/reoxygenation; LDH, lactate dehydrogenase; lncRNA, long non-coding RNA; NC, negative control.
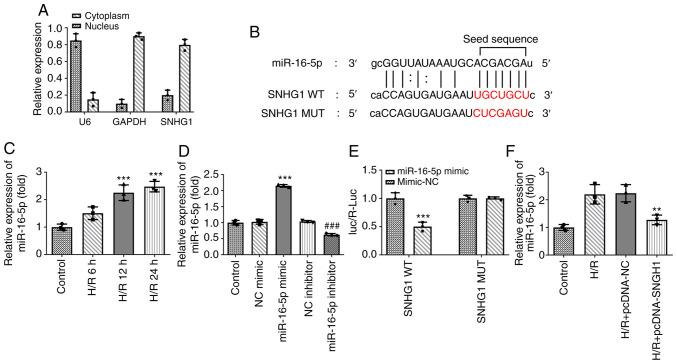
SNHG1 directly targets miR-16-5p
The present study further examined the underlying mechanism of SNHG1 in myocardial ischemic injury. miR-16-5p was predicted to be a target gene of SNHG1 using The Encyclopedia of RNA Interactomes (ENCORI) database (Fig. 3B). In addition, the results of RT-qPCR revealed that SNHG1 was localized in the cytoplasm and miR-16-5p was highly expressed in H9c2 cells treated with H/R (Fig. 3A and C). Subsequently, miR-16-5p mimic and miR-16-5p inhibitor were successfully transfected to modify the expression levels of miR-16-5p in cells (Fig. 3D). The results of the luciferase reporter assays demonstrated that compared with the miR-NC group, miR-16-5p mimic significantly decreased the luciferase activity of SNHG1 WT, while miR-116-5p mimic had no effect on the luciferase activity of SNHG1 MUT (Fig. 3E). As shown in Fig. 3F, overexpression of SNHG1 markedly inhibited miR-16-5p expression in H9c2 cells treated with H/R. In general, SNHG1 could negatively regulate miR-16-5p.
Figure 3.
SNHG1 directly targets miR-16-5p. (A) The expression of SNHG1 in the cytoplasm and nucleus was detected via RT-qPCR. (B) Sequences of miR-16-5p and the potential miRNA binding sites at the 3′ untranslated region of SNHG1 was predicted using the ENCORI database. The red characters represent the seed regions for miR-16-5p. (C) RT-qPCR was performed to detect the expression of miR-16-5p in H9c2 cells treated with H/R for 6, 12 and 24 h. ***P<0.001 vs. Control. (D) The expression of miR-16-5p after transfection of miR-16-5p mimic and miR-16-5p inhibitor was measured via RT-qPCR. ***P<0.001 vs. NC mimic; ###P<0.001 vs. NC inhibitor. (E) The relationship between miR-16-5p and SNHG1 was detected using a luciferase reporter assay. ***P<0.001 vs. mimic-NC. (F) Effect of SNHG1 overexpression on the expression of miR-16-5p in H9c2 cells was measured via RT-qPCR. **P<0.01 vs. H/R + pcDNA-NC. SNHG1, small nucleolar RNA host gene 1; miR/miRNA, microRNA; RT-qPCR, reverse transcription-quantitative PCR; H/R, hypoxia/reoxygenation; NC, negative control; WT, wild-type; MUT, mutant.
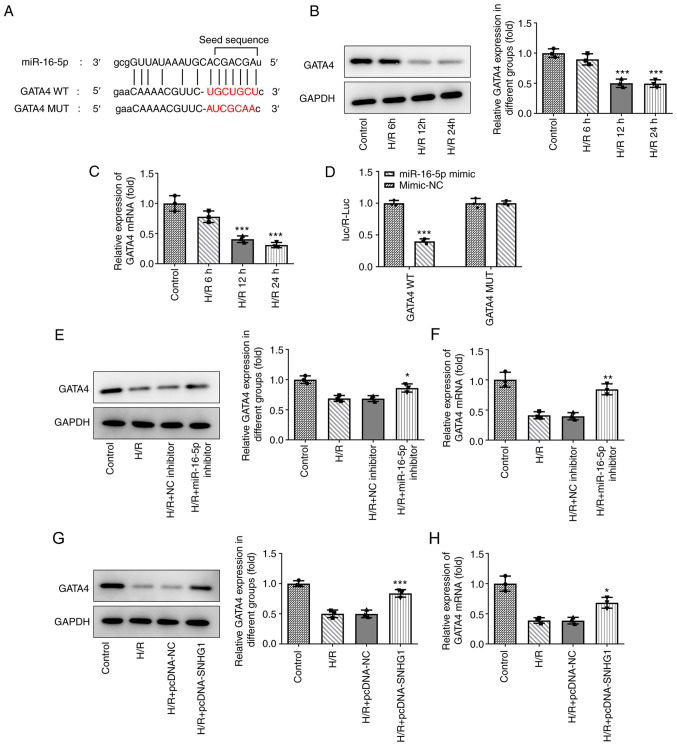
miR-16-5p directly targets GATA4
The downstream target genes of miR-16-5p were predicted using the ENCORI database. As shown in Fig. 4A, GATA4 was predicted to be a target gene of miR-16-5p. Compared with those in the control group, the expression levels of GATA4 were downregulated in H9c2 cells treated with H/R (Fig. 4B and C). The results of the luciferase reporter assays demonstrated that compared with the miR-NC group, miR-16-5p mimic markedly decreased the luciferase activity of GATA4 WT, while miR-16-5p mimic had no effect on the luciferase activity of GATA4 MUT (Fig. 4D). In addition, miR-16-5p inhibitor (Fig. 4E and F) and overexpression of SNHG1 (Fig. 4G and H) and markedly promoted GATA4 expression in H9c2 cells treated with H/R.
Figure 4.
miR-16-5p directly targets GATA4. (A) Sequences of miR-16-5p and the potential miRNA binding sites at the 3′ untranslated region of GATA4 was predicted using the ENCORI database. The red characters represent the seed regions for miR-16-5p. (B) Western blotting and (C) RT-qPCR was performed to detect the expression of GATA4 in H9c2 cells treated with H/R for 6, 12 and 24 h. ***P<0.001 vs. Control. (D) The relationship between miR-16-5p and GATA4 was detected using a luciferase reporter assay. ***P<0.001 vs. mimic-NC. Effect of miR-16-5p inhibitor on the expression of GATA4 in H9c2 cells was measured via (E) western blotting and (F) RT-qPCR. *P<0.05 and **P<0.01 vs. H/R + NC inhibitor. Effect of SNHG1 overexpression on the expression of GATA4 in H9c2 cells was measured via (G) western blotting and (H) RT-qPCR. *P<0.05 and ***P<0.001 vs. H/R + pcDNA-NC. GATA4, GATA binding protein 4; miR/miRNA, microRNA; RT-qPCR, reverse transcription-quantitative PCR; H/R, hypoxia/reoxygenation; NC, negative control; WT, wild-type; MUT, mutant.
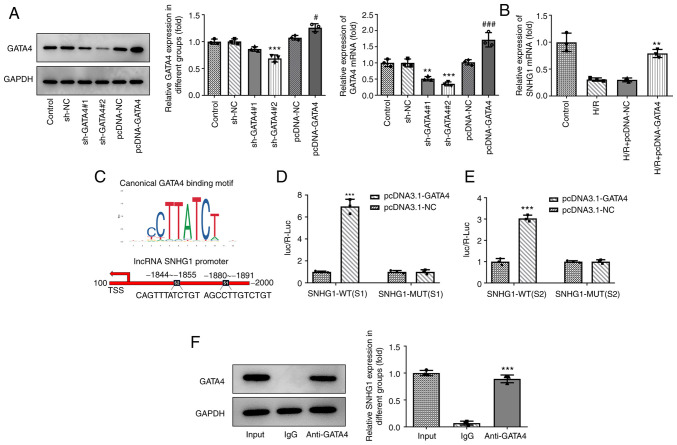
GATA4 and SNHG1 promoters combine to form the positive feedback loop
The present study aimed to examine the associations among SNHG1, miR-16-5p and GATA4. The expression levels of GATA4 in cells were altered by transfection with shRNA-GATA4 and pcDNA-GATA4. As shown in Fig. 5A, compared with those in the control group, the expression levels of GATA4 in the shRNA-GATA-2 group were the lowest and those in the pcDNA-GATA4 group were the highest. In addition, overexpression of GATA4 markedly promoted GATA4 expression (Fig. 5B). Of note, it was predicted by the JASPAR database that there are two binding sites, S1 and S2, between GATA4 and SNHG1 (Fig. 5C). The results of the luciferase reporter assay revealed that overexpression of GATA4 markedly promoted the luciferase activity of SNHG1 WT compared with the control group, while overexpression of GATA4 had no effect on the luciferase activity of SNHG1 MUT (Fig. 5D and E). The results of ChIP assay showed the enrichment of GATA4 on the promoter of SNHG1 (Fig. 5F).
Figure 5.
GATA4 and SNHG1 promoters combine to form the positive loop feedback. (A) The expression of GATA4 after transfection of sh-GATA4 and pcDNA-GATA4 was measured via RT-qPCR and western blotting. **P<0.01 and ***P<0.001 vs. sh-NC; #P<0.05 and ###P<0.001 vs. pcDNA-NC. (B) Effect of GATA4 overexpression on the expression of SNHG1 in H9c2 cells was measured via RT-qPCR. **P<0.01 vs. H/R + pcDNA-NC. (C) JASPAR analysis predicted two GATA4 binding sites, S1 and S2, on the SNHG1 promoter. (D and E) The relationship between GATA4 and SNHG1 was detected using a luciferase reporter assay. ***P<0.001 vs. pcDNA-NC. (F) Chromatin immunoprecipitation assay-coupled to RT-qPCR to verify GATA4 binding to the promoter of SNHG1. ***P<0.001 vs. IgG. GATA4, GATA binding protein 4; SNHG1, small nucleolar RNA host gene 1; sh-, short hairpin RNA; RT-qPCR, reverse transcription-quantitative PCR; NC, negative control; H/R, hypoxia/reoxygenation; WT, wild-type; MUT, mutant.
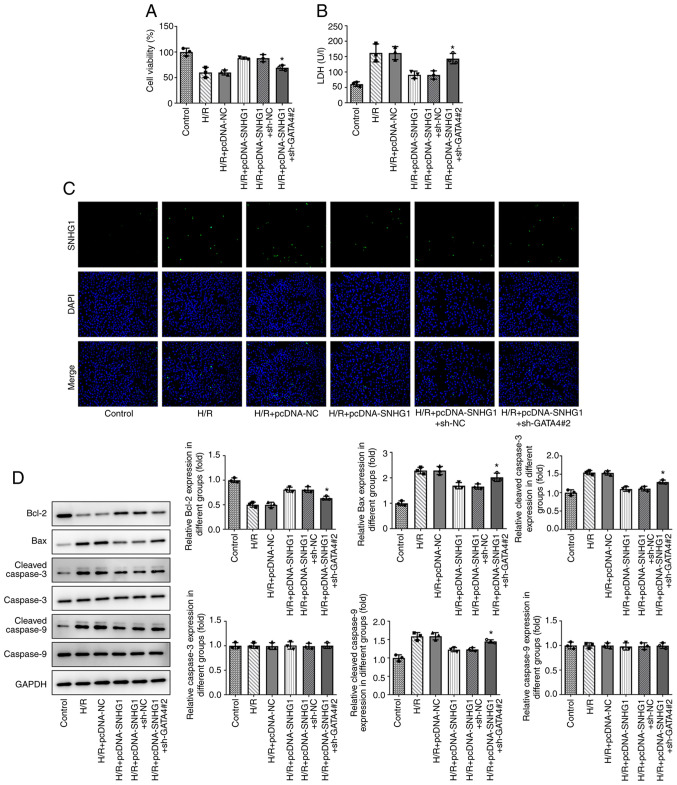
Overexpression of SNHG1 attenuates the injury of H9c2 cells treated with H/R by upregulating GATA4
As shown in Fig. 6A, CCK-8 revealed that GATA4 knockdown reduced the inhibitory effect of SNHG1 overexpression on the viability of H9c2 cells treated with H/R. Furthermore, the results of LDH showed that GATA4 knockdown reduced the inhibitory effect of SNHG1 overexpression on the expression of LDH in H9c2 cells treated with H/R (Fig. 6B). As shown in Fig. 6C and D, TUNEL analysis and western blotting demonstrated that GATA4 knockdown partially reversed the effect of SNHG1 overexpression on the apoptosis of H9c2 cells treated with H/R.
Figure 6.
Overexpression of SNHG1 attenuates H/R-induced H9c2 cell injury by upregulating GATA4. (A) Effect of SNHG1 on the viability of H/R-induced H9c2 cells via regulation of GATA4 was detected using a Cell Counting Kit-8 assay. (B) Effect of SNHG1 on the cytotoxicity of H/R-induced H9c2 cells via regulation of GATA4 was detected via ELISA. (C) Effect of SNHG1 on the apoptosis of H/R-induced H9c2 cells by regulating GATA4 was detected by a TUNEL assay (magnification ×200). (D) Effect of SNHG1 on the expression levels of apoptosis-related proteins and cell proliferation-related proteins (Bcl-2, Bax, Cleaved caspase-3, caspase-3, Cleaved caspase-9 and caspase-9) in H9c2 cells treated with H/R by regulating GATA4 were detected via western blotting. *P<0.05 vs. H/R + pcDNA-SNHG1 + sh-NC. SNHG1, small nucleolar RNA host gene 1; H/R, hypoxia/reoxygenation; GATA4, GATA binding protein 4; NC, negative control; sh-, short hairpin RNA.
Discussion
To the best of our knowledge, the present study was the first to demonstrate the protective effect of SNHG1 on ischemic cardiomyocyte injury. Specifically, SNHG1 inhibited ischemic cardiomyocyte injury via regulation of the miR-16-5p/GATA4 axis. Notably, GATA4 could bind to the promoter region of SNHG1. These results demonstrated the existence of the SNHG1/miR-16-5p/GATA4 positive feedback loop and provided a novel therapeutic target for the treatment of ischemic cardiomyocyte injury.
It has been demonstrated that lncRNAs serve an important role in the occurrence and development of tumors, and abnormal lncRNA expression has been used as a prognostic marker for numerous tumors, such as hepatocellular carcinoma, colorectal cancer and breast cancer (4–6). lncRNA SNHG1, located on chromosome 11, is abnormally highly expressed in a variety of tumor tissues and can promote the proliferation of lung, liver, gastric and prostate cancer, and is associated with poor prognosis in tumors (13–16). Beyond this tumor-suppressive role, SNHG1 has a protective effect on cardiomyocyte injury induced by various factors. For example, SNHG1 protects cardiomyocytes from toxic damage induced by doxorubicin by inhibiting miR-195 (8). SNHG1 has also been found to inhibit cardiomyocyte injury induced by H2O2 by regulating the miR-195/Bcl-2 axis (9). In addition, a recent study revealed that SNHG1 attenuates vascular endothelial cell injury induced by sponging miR-140-3p (10). Therefore, it was suggested that SNHG1 may have a protective effect on cardiomyocyte injury induced by H/R. In the present study, SNHG1 expression was downregulated in H9c2 cells induced by H/R. Overexpression of SNHG1 markedly promoted the viability of H9c2 cells treated with H/R and inhibited apoptosis. These results suggested that SNHG1 serves an important protective role in cardiomyocyte injury induced by H/R.
A recent study revealed that lncRNAs contain miRNA response elements, which can bind to their target miRNAs to inhibit their biological functions (17). This mechanism has been repeatedly demonstrated in multiple tumor models (18–20). In addition, miRNAs serve an important role in the occurrence and development of various cardiovascular diseases (21). In the present study, bioinformatics analysis revealed that miR-16-5p was a potential target gene of SNHG1. The results of the luciferase reporter gene assay revealed that SNHG1 could directly target miR-16-5p. In addition, miR-16-5p expression is downregulated in a variety of tumor tissues and exerts an antitumor effect via induction of apoptosis (22,23). It is worth noting that miR-16-5p has been found to be highly expressed in AC16 cardiomyocytes treated with I/R, and miR-16-5p knockdown could markedly promote cell viability and angiogenesis via inhibition of apoptosis (24). In the present study, miR-16-5p expression was upregulated in H9c2 cells treated with H/R, and the overexpression of SNHG1 markedly decreased the expression levels of miR-16-5p.
The present study further explored the mechanism of SNHG1 in cardiomyocyte injury induced by H/R. GATA4 was predicted as a potential target gene of miR-16-5p using bioinformatics analysis. GATA4 is an important regulator in the early stage of development and serves an important role in the development of the heart and intestines (25). A previous study revealed that overexpression of GATA4 promoted P19 cells to differentiate into cardiomyocytes (26). It is worth noting that downregulation of miR-122 has been shown to reduce cardiomyocyte apoptosis induced by H/R by regulating GATA4 (27). In addition, Astragaloside IV can enhance the activity of H9c2 cells treated with H/R by upregulating GATA4 (28). Importantly, using the JASPAR database in the present study, it was identified that there may be two binding sites between GATA4 and SNHG1, indicating the existence of the SNHG1/miR-16-5p/GATA4 positive feedback loop. In the present study, miR-16-5p could negatively target GATA4. The results of the luciferase reporter gene and ChIP assays demonstrated that GATA4 could bind to the SNHG1 promoter. SNHG1 knockdown partially reversed the effects of SNHG1 overexpression on cardiomyocyte viability and apoptosis induced by H/R.
In conclusion, the present results demonstrated that the positive feedback loop of SNHG1/miR-16-5p/GATA4 could potentially serve an important role in cardiomyocyte injury induced by H/R. SNHG1 could improve cardiomyocyte injury via upregulation of GATA4 by targeting miR-16-5p. However, due to the limitation of time and funds, only H9c2 cells were used to explore the regulatory effects of SNHG1 in H/R-induced cardiomyocyte injury via upregulation of GATA4 by targeting miR-16-5p. Further studies are needed to focus on nuclear factor E2-associated factor 2/heme oxygenase 1 signaling pathways, and future experiments should include animal models to further support the findings of the present study.
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JG and LD carried out the data collection and data analysis. JG and YZ conceived and designed the study, and wrote and revised the manuscript. JG and YZ confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hall TM, Gordon C, Roy R, Schwenke DO. Delayed coronary reperfusion is ineffective at impeding the dynamic increase in cardiac efferent sympathetic nerve activity following myocardial ischemia. Basic Res Cardiol. 2016;111:35. doi: 10.1007/s00395-016-0556-3. [DOI] [PubMed] [Google Scholar]
- 2.Hao PP, Jiang F, Chen YG, Yang J, Zhang K, Zhang MX, Zhang C, Zhao YX, Zhang Y. Traditional Chinese medication for cardiovascular disease. Nat Rev Cardiol. 2015;12:115–122. doi: 10.1038/nrcardio.2014.177. [DOI] [PubMed] [Google Scholar]
- 3.Writing Group Members, corp-author. Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Després JP, et al. Heart disease and stroke statistics-2016 update: A report from the American heart association. Circulation. 2016;133:e38–e360. doi: 10.1161/CIR.0000000000000366. [DOI] [PubMed] [Google Scholar]
- 4.Ou F, Su K, Sun J, Liao W, Yao Y, Zheng Y, Zhang Z. The LncRNA ZBED3-AS1 induces chondrogenesis of human synovial fluid mesenchymal stem cells. Biochem Biophys Res Commun. 2017;487:457–463. doi: 10.1016/j.bbrc.2017.04.090. [DOI] [PubMed] [Google Scholar]
- 5.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 6.Xu Y, Zheng Y, Liu H, Li T. Modulation of IGF2BP1 by long non-coding RNA HCG11 suppresses apoptosis of hepatocellular carcinoma cells via MAPK signaling transduction. Int J Oncol. 2017;51:791–800. doi: 10.3892/ijo.2017.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tycowski KT, Shu MD, Steitz JA. Requirement for intron-encoded U22 small nucleolar RNA in 18S ribosomal RNA maturation. Science. 1994;266:1558–1561. doi: 10.1126/science.7985025. [DOI] [PubMed] [Google Scholar]
- 8.Chen S, Wang J, Zhou Y. Long non-coding RNA SNHG1 protects human AC16 cardiomyocytes from doxorubicin toxicity by regulating miR-195/Bcl-2 axis. Biosci Rep. 2019;39:BSR20191050. doi: 10.1042/BSR20191050. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Zhang N, Meng X, Mei L, Hu J, Zhao C, Chen W. The long non-coding RNA SNHG1 attenuates cell apoptosis by regulating miR-195 and BCL2-like protein 2 in human cardiomyocytes. Cell Physiol Biochem. 2018;50:1029–1040. doi: 10.1159/000494514. [DOI] [PubMed] [Google Scholar]
- 10.Liang S, Ren K, Li B, Li F, Liang Z, Hu J, Xu B, Zhang A. LncRNA SNHG1 alleviates hypoxia-reoxygenation-induced vascular endothelial cell injury as a competing endogenous RNA through the HIF-1α/VEGF signal pathway. Mol Cell Biochem. 2020;465:1–11. doi: 10.1007/s11010-019-03662-0. [DOI] [PubMed] [Google Scholar]
- 11.Huang L, Jiang X, Wang Z, Zhong X, Tai S, Cui Y. Small nucleolar RNA host gene 1: A new biomarker and therapeutic target for cancers. Pathol Res Pract. 2018;214:1247–1252. doi: 10.1016/j.prp.2018.07.033. [DOI] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative Pcr and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated lncRNA SNHG1 contributes to progression of non-small cell lung cancer through inhibition of miR-101-3p and activation of Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:17785–17794. doi: 10.18632/oncotarget.14854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, Zhou D, Ying M, Chen M, Chen P, Chen Z, Zhang F. Expression of long non-coding RNA (lncRNA) small nucleolar RNA host gene 1 (SNHG1) exacerbates hepatocellular carcinoma through suppressing miR-195. Med Sci Monit. 2016;22:4820–4829. doi: 10.12659/MSM.898574. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Hu Y, Ma Z, He Y, Liu W, Su Y, Tang Z. LncRNA-SNHG1 contributes to gastric cancer cell proliferation by regulating DNMT1. Biochem Biophys Res Commun. 2017;491:926–931. doi: 10.1016/j.bbrc.2017.07.137. [DOI] [PubMed] [Google Scholar]
- 16.Li J, Zhang Z, Xiong L, Guo C, Jiang T, Zeng L, Li G, Wang J. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;487:146–152. doi: 10.1016/j.bbrc.2017.03.169. [DOI] [PubMed] [Google Scholar]
- 17.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden. RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Q, Shan S, Li Y, Zhu D, Jin W, Ren T. Long noncoding RNA SNHG1 promotes non-small cell lung cancer progression by up-regulating MTDH via sponging miR-145-5p. FASEB J. 2018;32:3957–3967. doi: 10.1096/fj.201701237RR. [DOI] [PubMed] [Google Scholar]
- 19.Zhao L, Kong H, Sun H, Chen Z, Chen B, Zhou M. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J Cell Physiol. 2018;233:4044–4055. doi: 10.1002/jcp.26072. [DOI] [PubMed] [Google Scholar]
- 20.Mou K, Liu B, Ding M, Mu X, Han D, Zhou Y, Wang LJ. lncRNA-ATB functions as a competing endogenous RNA to promote YAP1 by sponging miR-590-5p in malignant melanoma. Int J Oncol. 2018;53:1094–1104. doi: 10.3892/ijo.2018.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meng Q, Liu Y, Huo X, Sun H, Wang Y, Bu F. MicroRNA-221-3p contributes to cardiomyocyte injury in H2O2-treated H9c2 cells and a rat model of myocardial ischemia-reperfusion by targeting p57. Int J Mol Med. 2018;42:589–596. doi: 10.3892/ijmm.2018.3628. [DOI] [PubMed] [Google Scholar]
- 22.Xu F, Zhang X, Lei Y, Liu X, Liu Z, Tong T, Wang W. Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma. J Cell Biochem. 2010;111:727–734. doi: 10.1002/jcb.22762. [DOI] [PubMed] [Google Scholar]
- 23.Jang JY, Lee JK, Jeon YK, Kim CW. Exosome derived from epigallocatechin gallate treated breast cancer cells suppresses tumor growth by inhibiting tumor-associated macrophage infiltration and M2 polarization. BMC Cancer. 2013;13:421. doi: 10.1186/1471-2407-13-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Shang Y, Dai S, Wu W, Yi F, Cheng L. MicroRNA-16-5p aggravates myocardial infarction injury by targeting the expression of insulin receptor substrates 1 and mediating myocardial apoptosis and angiogenesis. Curr Neurovasc Res. 2020;17:11–17. doi: 10.2174/1567202617666191223142743. [DOI] [PubMed] [Google Scholar]
- 25.Molkentin JD. The zinc finger-containing transcription factors GATA-4,-5, and-6 ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem. 2000;275:38949–38952. doi: 10.1074/jbc.R000029200. [DOI] [PubMed] [Google Scholar]
- 26.Ang YS, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu JD, Spencer CI, et al. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. 2016;167:1734–1749. e22. doi: 10.1016/j.cell.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang W, Guo J, Li J, Bai C, Dong Y. Downregulation of miR-122 attenuates hypoxia/reoxygenation (H/R)-induced myocardial cell apoptosis by upregulating GATA-4. Biochem Biophys Res Commun. 2016;478:1416–1422. doi: 10.1016/j.bbrc.2016.08.139. [DOI] [PubMed] [Google Scholar]
- 28.Yang JJ, Zhang XH, Ma XH, Duan WJ, Xu NG, Chen YJ, Liang L. Astragaloside IV enhances GATA-4 mediated myocardial protection effect in hypoxia/reoxygenation injured H9c2 cells. Nutr Metab Cardiovasc Dis. 2020;30:829–842. doi: 10.1016/j.numecd.2020.01.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.