Abstract
An outbreak of poliomyelitis with 20 cases occurred in Israel, Gaza, and the West Bank from October 1987 to October 1988. The wild type 1 poliovirus associated with the outbreak was most closely related to viruses found in the Nile Delta. The epidemiologic links among patients involved in the outbreak and patients with community-acquired infections during the outbreak were inferred from the evolutionary relationships among isolates of the outbreak virus. Complete VP1 sequences (906 nucleotides) were determined for 12 clinical and 4 sewage isolates. A total of 58 nucleotide differences were found among the 16 isolates; 74% of all substitutions were synonymous third-position transitions. An evolutionary tree, representing both the pathways of VP1 sequence evolution and the inferred chains of virus transmission during the outbreak, was constructed under the assumption that each substitution had occurred only once. The combined epidemiologic and molecular data suggest that a single founder strain was introduced into Israel from the vicinity of Gaza in the fall of 1987. Poliovirus circulation was apparently localized to southern communities during the winter and spread north by the following summer into the Hadera subdistrict of Israel, where it radiated via multiple chains of transmission into other communities in northern Israel and the West Bank. The close sequence matches (>99%) between clinical and sewage isolates from the same communities confirm the utility of environmental sampling as a tool for monitoring wild poliovirus circulation.
The incidence of paralytic poliomyelitis in Israel, Gaza, and the West Bank had declined sharply up to the mid-1980s (7, 25, 28). In 1986, Israel reported zero cases of polio for the first time, and the number of reported cases of polio in Gaza and the West Bank were near historic lows. However, 3 cases occurred in 1987 (2 in Israel and 1 in Gaza), followed by an additional 17 cases in 1988, 15 of which occurred in the Jewish population in Israel (25). The 20 cases in 1987 and 1988 were eventually linked as an outbreak associated with wild type 1 poliovirus.
The 1987-1988 outbreak was of particular concern because it occurred within a generally well-immunized population (>90% with neutralizing antibody titers of >1:8 to poliovirus type 1) (8) and because most of the cases occurred in people who had previously received at least three doses of oral polio vaccine (OPV) (25). In response, an immunization campaign targeting the entire population under 40 years of age in Israel, Gaza, and the West Bank was conducted in October and November 1988 (25). No cases of polio have been reported in Israel, Gaza, or the West Bank since the outbreak (15).
The last outbreak in Israel is of current interest because it occurred in a country where indigenous wild poliovirus circulation was then thought to have ceased, but circulation of wild polioviruses in neighboring countries continued (1, 7, 10). As global polio eradication progresses, a succession of newly polio-free countries are encountering similar epidemiologic conditions (1, 3). The intensive surveillance for poliomyelitis cases and wild poliovirus circulation maintained by Israel before and during the 1987-1988 outbreak (7, 25, 30) provided the opportunity for a detailed analysis of this outbreak by molecular epidemiologic methods. Specifically, we sought to determine the likely origin of the outbreak virus, to track the spread of poliovirus under various seasonal conditions, and to evaluate the use of environmental sampling as a supplementary tool for wild poliovirus surveillance during the outbreak. To approach these questions, we compared the nucleotide sequences of the outbreak isolates with each other and with the sequences of other contemporary wild type 1 polioviruses. Because significant evolution of the viral RNA genome had occurred during the 11-month outbreak, individual chains of transmission could be visualized as separate genetic lineages. A high-resolution view of the pathways of wild poliovirus transmission during the outbreak was obtained from the combined epidemiologic and sequence data.
MATERIALS AND METHODS
Cells and viruses.
Polioviruses from the 1987-1988 Israel outbreak (see Table 1 and Fig. 1) were grown in HEp-2 (human laryngeal carcinoma cell line; ATCC CCL23) and RD (human rhabdomyosarcoma cell line; ATCC CCL136) cell monolayers for isolation from clinical specimens and in BGM (buffalo green monkey kidney cell line) (5) cell monolayers for isolation from environmental samples. Wild type 1 polioviruses isolated in different regions of the world from 1977 to 1992 have been described previously (10, 24).
TABLE 1.
Poliovirus isolates from the 1987-1988 Israel outbreak sequenced in this study
| Isolatea | Date of sampling (day-mo-yr) | Subdistrict or territory |
|---|---|---|
| Rahat-A | 1 Nov 87 | Beer Sheva |
| Gaza-A | 7 Nov 87 | Gaza |
| Kfar Kasem-A | 27 Nov 87 | Petah-Tikva |
| Rahat-B | 10 Mar 88 | Beer Sheva |
| Or Akiva-3cb | 28 Aug 88 | Hadera |
| Or Akiva-S | 1 Sept 88 | Hadera |
| Akko-6 | 2 Sept 88 | Akko |
| Or Akiva-7 | 4 Sept 88 | Hadera |
| Hadera-8 | 5 Sept 88 | Hadera |
| Jenin-A | 8 Sept 88 | West Bank |
| Raanana-9 | 22 Sept 88 | Petah-Tikva |
| Hadera-14 | 7 Oct 88 | Hadera |
| Hadera-15 | 9 Oct 88 | Hadera |
| Naharia-S | 4 Oct 88 | Akko |
| Jenin-S | 4 Oct 88 | West Bank |
| Ramla-S | 4 Oct 88 | Ramla |
Cases and the associated isolates were assigned identical names and were identified by the community of residence of the patients and numbered according to the case numbers given in reference 25. Cases and isolates not described in reference 25 were assigned the suffixes “A” or “B.” Sewage isolates were identified by “S.”
Contact of polio case 3.
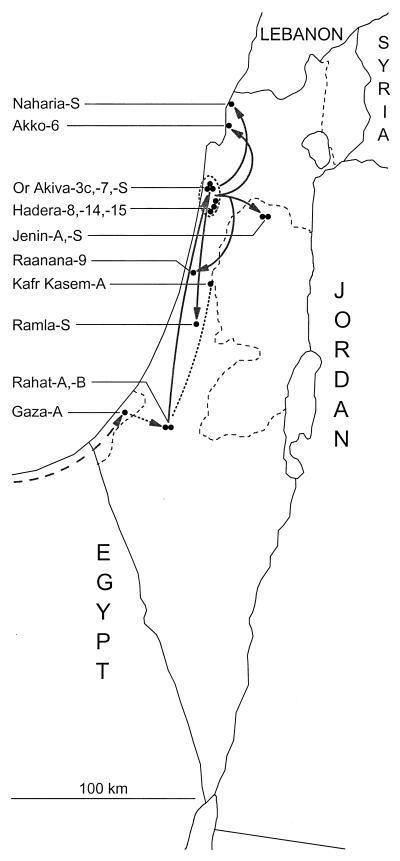
FIG. 1.
Geographic distribution of patients with clinical cases of polio and sewage samples associated with isolates of wild type 1 poliovirus (indicated by closed circles) from the 1987-1988 Israel outbreak. Gray arrows trace the main pathways of poliovirus transmission during the outbreak inferred from the epidemiologic record and the sequence relationships among poliovirus isolates. The dashed arrow indicates the proposed pathway of transmission from Egypt to Gaza and from Gaza to Rahat in the fall of 1987; the dashed line connecting Kfar Kasem and Rahat indicates linkage between the 1987 cases in these communities, but the direction of the pathway of transmission is uncertain. The dashed ellipse encloses communities in the Hadera subdistrict where the majority of polio cases occurred in 1988.
Isolation of poliovirus from sewage.
Sewage samples were either composite samples manually taken at half-hour intervals (starting 1.5 h before and ending 1.5 h after the morning peak flow) or samples taken with gauze pads submerged in the sewage stream for 24 h. Virus was obtained from the samples as described previously (14) and was plaque purified by inoculation onto BGM cell monolayers overlaid with M199 medium containing 0.9% agarose. After a 24-h incubation at 37°C, well-separated plaques were picked, diluted, and grown on BGM cell monolayers in bell tubes incubated at 40°C. Isolates that formed a cytopathic effect at 40°C by day 5 were candidate wild polioviruses. Isolates were characterized in microneutralization assays with poliovirus type-specific polyclonal antisera and with monoclonal antibodies specific to each Sabin vaccine strain (4).
Sequencing of poliovirus RNAs.
Nucleotide sequences were determined by two methods. In the first method, VP1 sequences of outbreak clinical isolates were initially determined by manual primer extension sequencing from purified virion RNA templates (24). In the second method, complete VP1 (nucleotides [nts] 2480 to 3385) and partial VP1/2A (nts 3296 to 3445) sequences of poliovirus isolates from the outbreak were determined in cycle sequencing reactions (11) containing fluorescent dye-labeled dideoxynucleotides (Applied Biosystems, Foster City, Calif.). Sequencing templates were 1,106-bp PCR products amplified from poliovirus RNAs with the primer pair Q8b (antisense [A] polarity, positions 3485 to 3504; 5′-AAGAGGTCTCT[A/G]TTCCACAT-3′) and Y7b (sense [S] polarity, positions 2399 to 2421; 5′-GGITTTGTGTCAGCITGCAAT-3′); primer positions with equimolar amounts of two different nucleotides are enclosed in brackets; deoxyinosine residues are indicated by the letter I. PCR product templates were purified for the sequencing reactions by chromatography on QiaQuick columns (Qiagen, Dusseldorf, Germany). The nucleotide sequences of both strands of each template were determined with the aid of an automated sequencer (Sequenator; model 373; Applied Biosystems). Apart from a small number of sequence ambiguities obtained with manual primer extension sequencing that were resolved by PCR cycle sequencing, the results obtained by the two methods were in complete agreement. The VP1/2A sequences of other poliovirus isolates were determined by either automated cycle sequencing (19) or manual methods (24). All primers for PCR and sequencing reactions (including, in addition to Q8b and Y7b, primers S14 [A, positions 3209 to 3228; 5′-GGGTTGTGATCATTAACCAC-3′], SR1 [A, positions 2987 to 3006; 5′-TGCCATGTGTAATCATCCCA-3′], S2Ab [S, positions 2853 to 2871; 5′-TCACCTACTCCAGATTTGA-3′], and S14F [S, positions 3209 to 3228; 5′-GTGGTTAATGATCACAACCC-3′]) were prepared and purified as described previously (32).
Analysis of VP1/2A nucleotide sequences.
Evolutionary distances between poliovirus genomes were estimated from the VP1/2A sequences (all codon positions) by using the two-parameter method of Kimura (12) to correct for multiple substitutions at a site. Calculations were performed by the program DNADIST of the PHYLIP 3.5c program package (6) by using a value of 10 for the transition/transversion ratio. The VP1/2A evolutionary distances among poliovirus isolates were summarized in a tree constructed by the neighbor-joining method with the program NEIGHBOR (6).
Reconstruction of the pathways of VP1 sequence evolution of outbreak isolates.
An evolutionary tree representing the pathways of poliovirus VP1 evolution (which represent pathways of transmission) was constructed from the combined sequence and epidemiologic data. The branches of the tree were constructed manually under the assumption that the observed substitutions were generated by the fewest number of mutational steps, and the tree was rooted to the earliest isolates. The topology of the manually constructed tree was confirmed by using phylogeny programs based upon the maximum likelihood (DNAML), maximum parsimony (DNAPARS), and neighbor-joining (NEIGHBOR) algorithms (6, 26).
Nucleotide sequence accession numbers.
The sequences of the outbreak poliovirus isolates described in this article have been deposited in the EMBL/GenBank data library and have been assigned accession nos. AF139251 to AF139291 (partial VP1/2A sequences) and AJ237871 to AJ237885 (complete VP1 sequences).
RESULTS
Epidemiologic background.
Israel was one of the first countries to implement nationwide polio immunization, starting with immunization with the inactivated poliovaccine (IPV) in 1957, followed by immunization with OPV in 1961 (7, 28). In response to these initiatives, the rate of polio incidence declined sharply from >140 per 100,000 population in 1950 to <1.2 per 100,000 population after 1961. Epidemics associated with type 1 poliovirus occurred in 1958 and 1961, and sporadic cases occurred afterward from 1962 to 1986. Sporadic cases increased from 1967 to 1979, primarily among members of the non-Jewish population, presumably because of increased contact with communities in Gaza and the West Bank where polio is endemic. In 1979, immunization against polio was intensified by the introduction of a schedule that combined OPV and IPV (7, 28). However, sporadic cases continued at a reduced rate between 1983 and 1986. In October and November of 1987, three polio cases (Rahat-A, Gaza-A, and Kfar Kasem-A) were reported (Table 1; Fig. 1). In 1988, an isolated case (Rahat-B) occurred in February, followed by the occurrence of 16 additional cases from July to October (25). While 12 of the 16 cases clustered in the Hadera subdistrict (represented by Or Akiva-3c, Or Akiva-7, Hadera-8, Hadera-14, and Hadera-15), individual cases also occurred in the subdistricts of Ashkelon (isolate not analyzed), Akko (Akko-6), and Petah-Tikva (Raanana-9) in Israel and in the northern West Bank (Jenin-A). Poliovirus surveillance was intensified by the collection of sewage samples (isolates indicated by “S”) in September and October 1988 in Hadera, Or Akiva, Naharia, Jenin, and Ramla (Fig. 1; Table 1). All wild polioviruses detected during the outbreak were type 1. The appearance of polio cases in the outbreak communities of northern Israel was unexpected because of the preexisting high seroprevalence (>90%) of neutralizing antibodies (titers, >1:8) to type 1 poliovirus (7, 8, 28) and because the majority of cases occurred in people who had received at least three OPV doses (25). The last reported polio case in Israel (Hadera-15) associated with wild poliovirus had an onset date of 3 October 1988 (15, 25).
Relationships of outbreak isolates to other wild type 1 polioviruses.
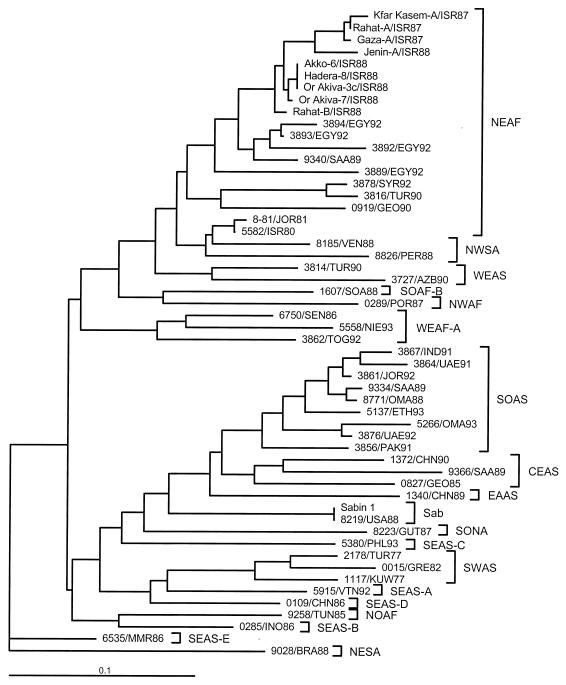
To survey the genetic relationships between the Israel outbreak isolates and wild type 1 polioviruses found elsewhere, we compared nucleotide sequences at the VP1/2A junction (positions 3296 to 3445) (10, 24). Included in the comparisons were the sequences of nine outbreak isolates, the Sabin 1 vaccine strain, a representative Sabin 1 vaccine-derived isolate, and 43 wild type 1 isolates from different parts of the world (Fig. 2). Most of the wild isolates were from patients with polio that occurred within 5 years of the Israel outbreak. However, also included were three older isolates representing a type 1 genotype (SWAS) found in the Mideast during from 1977 to 1982 and two isolates from patients with polio in Israel (1980) and Jordan (1981) (24). The 1987-1988 Israel outbreak isolates were members of the type 1 NEAF genotype, which had been widely distributed in northeastern Africa and western Asia (10, 19). Virus of this genotype was introduced into northwestern South America in about 1980, splitting off into the related type 1 genotype, NWSA (10, 24; L. De, J. Jorba, J. Boshell, R. Salas, and O. Kew, Abstr. 17th Annu. Meet. Am. Soc. Virol., abstr. W36-3, p. 123, 1998). The outbreak viruses were most closely related to viruses found in Egypt (∼95% sequence similarity) and Saudi Arabia (∼94% sequence similarity) (Fig. 2). The epidemics in Oman (1988) (1) and Jordan (1992) (19), sporadic cases in Saudi Arabia (1989) (1) and the Gulf States (1991 and 1992) (19), and endemic circulation in Ethiopia (1993) (2) were associated with another type 1 genotype (SOAS), whose largest reservoirs are in South Asia. A third type 1 genotype (CEAS) was associated with an isolated case of polio in northern Saudi Arabia in 1989 (1). The remaining wild polioviruses were representative of other genotypes not closely related to the NEAF genotype (Fig. 2).
FIG. 2.
Tree summarizing sequence relatedness across the interval of nts 3296 to 3445 (VP1/2A region) among 9 clinical isolates from the 1987-1988 Israel outbreak, 43 wild type 1 poliovirus isolates from the Middle East and other regions of the world (usually from cases that had occurred within 5 years of the Israel outbreak), the Sabin 1 vaccine strain, and a representative Sabin 1 vaccine-derived clinical isolate (8219/USA88). Isolates are identified by laboratory number, three-letter country code, and year of isolation. Country abbreviations: AZB, Azerbaijan; BRA, Brazil; CHN, China; EGY, Egypt; ETH, Ethiopia; GEO, Georgia; GRE, Greece; GUT; Guatemala; IND, India; INO, Indonesia; ISR, Israel; JOR, Jordan; KUW, Kuwait; MMR, Myanmar; NIE, Nigeria; OMA, Oman; PAK, Pakistan; PER: Peru; PHL, Philippines; POR, Portugal; SAA, Saudi Arabia; SEN, Senegal; SOA, South Africa; TOG, Togo; TUN, Tunisia; TUR, Turkey; UAE, United Arab Emirates; USA, United States; VEN, Venezuela; VTN, Vietnam (31). Wild poliovirus genotypes within each serotype are identified by a four-letter code; the first two letters indicate eight compass points (NO, NE, EA, SE, SO, SW, WE, NW) or central (CE), and the last two letters indicate continents (AF [Africa], AS [Asia], EU [Europe], NA [North America], SA [South America]). When more than one type 1 genotype has been endemic to a region, the individual genotypes are distinguished by a letter suffix (e.g., SEAS-A). Sabin vaccine strain-related isolates are identified as Sab.
Comparison of VP1 sequences of outbreak isolates.
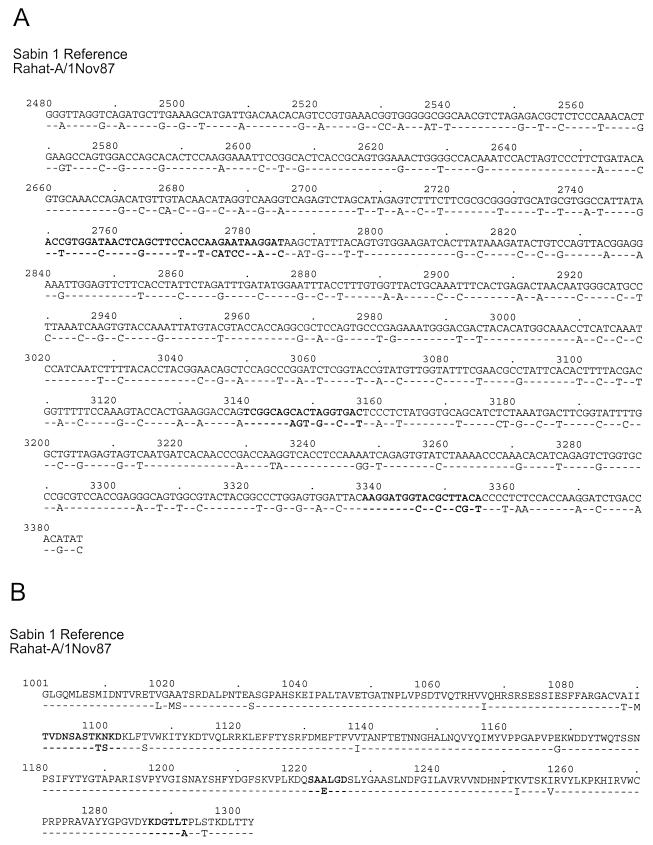
VP1 sequences (906 nts) were determined for 12 isolates from stools taken from 11 case patients and 1 contact of a patient with polio during the outbreak (October 1987 to September 1988) and for 4 isolates from sewage samples taken during September and October 1988 in communities with (Or Akiva, Jenin) or without (Naharia, Ramla) cases of polio. The VP1 sequence of the earliest outbreak isolate, Rahat-A (1 November 1987), differed from the sequence of the Sabin 1 strain at 21% (192 of 906) of the nucleotide positions (Fig. 3A) and at 5.6% (17 of 302) of the amino acid residues (Fig. 3B). All of the amino acid differences occurred at sites known to vary among type 1 polioviruses (C.-F. Yang and S.-J. Yang, unpublished data), with the Rahat-A residues generally corresponding to the consensus for wild type 1 polioviruses at the variable positions. Four of the amino acid differences clustered in the three neutralizing antigenic (NAg) sites of VP1. The NAg I and III amino acid sequences of Rahat-A match the consensus amino acid sequence for wild type 1 polioviruses (17; Yang and Yang, unpublished data), and the NAg II sequence SAELGD is frequently found among isolates of other type 1 genotypes (Yang and Yang, unpublished data). It appears likely that these differences in virion surface residues contribute to the observed antigenic differences between the outbreak isolates and the Sabin 1 vaccine strain (8, 25).
FIG. 3.
VP1 nucleotide (A) and amino acid (B) sequence alignment of the Sabin 1 reference strain and the 1987 isolate, Rahat-A. Sabin 1 strain nucleotide positions are numbered as described by Nomoto et al. (21); those of the Rahat-A isolate are numbered similarly for comparability. Capsid amino acid positions are indicated by a four-digit number: the first digit identifies the virion protein, and the next three digits specify residue position (e.g., 1001 indicates residue 1 of VP1). Boldface letters identify residues that encode or form NAg I (nts 2750 to 2785; amino acids 1091 to 1102), NAg II (nts 3140 to 3157; amino acids 1221 to 1226), and NAg III (nts 3338 to 3355; amino acids 1287 to 1292) (16).
A total of 58 nucleotide differences in VP1 were found among the 16 isolates (Fig. 4). The maximum difference between any pair of isolates was 2.5% (23 of 906). Each isolate had a unique VP1 sequence. Most (47 of 58; 81%) of the substitutions occurred in the third codon position, and 93% (54 of 58) of all substitutions were transitions. Only 11 of the 58 nucleotide substitutions encoded amino acid changes, three of which (S1221L, A1222V, and L1224S) mapped to NAg II (Fig. 4).
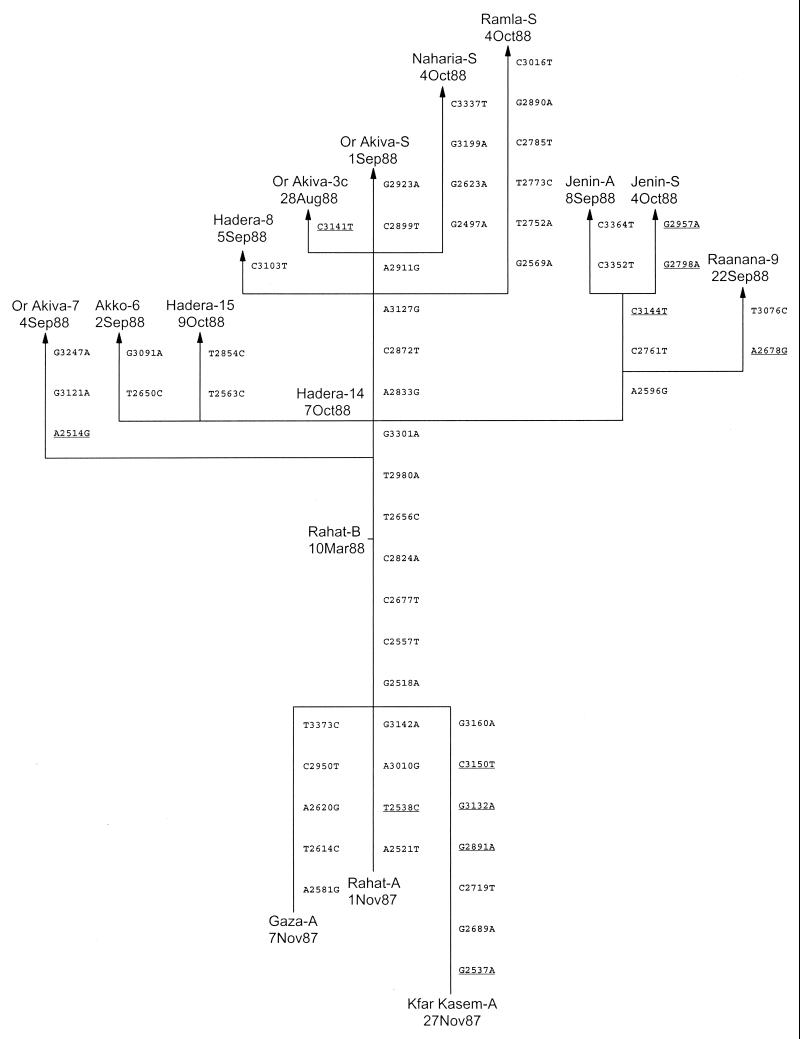
FIG. 4.
Tree summarizing evolutionary relationships among outbreak isolates based upon differences in VP1 nucleotide sequences. The branch structure of the tree was constructed under the assumption that each substitution occurred only once, and the tree was rooted to three 1987 isolates, Rahat-A, Gaza-A, and Kfar Kasem-A. Vertical branches are scaled to the number of nucleotide differences between VP1 sequences. The VP1 nucleotide substitutions that encode amino acid changes (A2514G, N1012S; G2537A, A1020T; T2538C, M1020T; A2678G, I1067V; G2798A, V1107M; G2891A, V1138I; G2957A, V1160I; G3132A, R1218K; C3141T, S1221L; C3144T, A1222V; C3150T, S1224L) are underlined. Dates shown are the dates on which the samples were taken. The direction of the substitutions above the node between Rahat-A and Rahat-B, representing the sequence of the hypothetical founder virus, is unambiguous. The direction below the node considers three alternative lineages connecting each 1987 isolate to the hypothetical founder.
Pathways of wild poliovirus evolution and transmission during the outbreak.
The rate, extent, and pattern of VP1 sequence evolution among the outbreak isolates enabled us to reconstruct from the genetic data the main pathways of poliovirus transmission. The observed VP1 sequence differences among isolates from the Israel outbreak are represented in a tree (Fig. 4) constructed under the assumption that each nucleotide substitution had occurred only once (i.e., under a minimum evolution model). The dates shown on the tree are the dates on which the samples were taken. Branch points (nodes) represent the inferred VP1 sequences of intermediate viruses that were usually not observed as isolates. The vertical length of each segment (connecting an observed sequence to a node or connecting two interior nodes) is proportional to the number of nucleotide substitutions that define that segment. When a segment is defined by more than one substitution, the temporal order of substitutions is indeterminate.
The pathways of VP1 evolution shown in the tree also trace the major chains of poliovirus transmission during the outbreak (Fig. 1). The true root of the tree (i.e., the VP1 sequence corresponding to the actual 1987 ancestral virus) cannot be unambiguously determined from the sequence data. Consequently, the tree is shown as having three alternative roots, representing the sequences of the 1987 isolates Rahat-A, Gaza-A, and Kfar Kasem-A (Fig. 4). A potential fourth root, defined by the node connecting the sequences of the three 1987 isolates, represents the sequence of a hypothetical genetic founder (i.e., direct ancestral virus) for all 1988 isolates. The sequences of all 1988 outbreak viruses differed from those of the hypothetical founder (and all three 1987 isolates) by the substitutions G2518A, C2557T, C2677A, and C2824A (Fig. 4). The 1987 isolates differed from the hypothetical founder at four (Rahat-A), five (Gaza-A), or seven (Kfar Kasem-A) VP1 positions. We cannot ascertain from the sequence data whether the 1987 isolates were parental to or were derived from the hypothetical founder virus.
The earliest 1988 isolate, Rahat-B (isolated on 10 March 1988), appears to be a genetic intermediate between the 1987 ancestral virus and all later 1988 outbreak viruses. Infection with this virus occurred during the seasonal low point for poliovirus circulation in this region (7). Only one chain of transmission, represented by Rahat-B, was observed to have continued through the winter of 1987-1988. All subsequent 1988 isolates appear to be descendents of Rahat-B that further evolved via a common pathway (chain of transmission) identified by the VP1 substitutions T2656C and T2980A. By the summer of 1988, the common chain of transmission had diverged into multiple independent lineages (Fig. 4). The first lineage (identified by substitutions A2514G, G3121A, and G3247A) observed to have split from the main transmission pathway terminates with the isolate Or Akiva-7. Several more lineages branch off from the main pathway at the next node (identified by the sequence of isolate Hadera-14), with two separate branches leading to isolates Akko-6 and Hadera-15 and a third branch splitting further into three sublineages that terminate with isolates Raanana-9, Jenin-A, and Jenin-S. Above the node defined by Hadera-14, the main transmission pathway is identified by the substitutions A2833G, C2872T, and A3127G. Isolates Or Akiva-3c, Hadera-8, and Or Akiva-S are derived from the main transmission pathway and form a close genetic cluster (three to four nucleotide differences) near the top of the tree. These isolates differ from the Or Akiva-7 isolate at 8 to 10 nucleotide positions. The sewage isolates Ramla-S and Naharia-S appear to be derived from the Or Akiva-3c and Hadera-8 cluster of viruses, but apparently, each represents a separate pathway of transmission, as these isolates differ from each other at 11 VP1 positions. The other two sewage isolates, Or Akiva-S and Jenin-S, were most closely related to case isolates from their respective communities (Fig. 4).
DISCUSSION
This study demonstrates that the high rate of poliovirus genomic evolution permits the use of comparative nucleotide sequencing to resolve the fine structure of a poliomyelitis outbreak. Individual chains of transmission were visualized as separate genetic lineages, and lineages not observable through the appearance of poliomyelitis cases were detected by environmental sampling. Infections that were temporally and geographically coincident (such as those represented by isolates Or Akiva-3c and Or Akiva-7) were sometimes found to be associated with different lineages. Molecular epidemiologic methods have frequently been used to distinguish between alternate models for poliovirus transmission (10); however, most studies have distinguished between isolates of different genotypes, which had diverged many years earlier (9, 18, 24), instead of between individual lineages derived from a recent common ancestral infection (13, 22).
The relationships among poliovirus infections, determined from the combined sequence and epidemiologic data, reveal the underlying seasonal dynamics of poliovirus circulation in a subtropical region. At least three wild poliovirus lineages, represented by the isolates Rahat-A, Gaza-A, and Kfar Kasem-A, were circulating in the vicinity of Israel in the fall of 1987. These lineages had probably diverged during the 1987 summer-fall peak transmission season (7). Only one lineage, possibly localized near Rahat, was found to have passed through the bottleneck of the 1987-1988 winter low-transmission season. The other two lineages, represented by isolates Gaza-A and Kfar Kasem-A, apparently terminated during that low-transmission season. Only the second case of polio in Rahat (onset date, 24 February 1988) signaled the presence of continuing poliovirus transmission during the 9 months between late November 1987 and late July 1988 (onset date of first Or Akiva case, 31 July 1988). By the time cases appeared in the Hadera subdistrict in late July and August, poliovirus was circulating by multiple chains of transmission. Virus radiated by independent pathways from the Hadera subdistrict where the epidemic was occurring to other communities in northern Israel and the West Bank (Fig. 1). In view of the widespread circulation of poliovirus in Israel by the summer of 1988, the nationwide OPV immunization campaign launched in October and November was probably essential to ensure the cessation of further poliovirus transmission (25). Circulation of the outbreak lineages apparently stopped in the fall of 1988, as no direct descendents of these lineages have been isolated since that time (15). The occurrence of the highest number of polio cases in the Hadera subdistrict is most likely attributable to the immune status of the local population rather than to any unusual exposure event in that community (25).
The immediate source of the outbreak virus was probably Gaza (Fig. 1 and 4), as the patient with the first case in Rahat had direct contact with Gaza at the time of infection. However, the original reservoir of endemicity appears to have been communities in northern Egypt, whose populations are in regular contact with the population in Gaza. Over the past decade, Egypt has reduced endemic poliovirus circulation to low levels (3; S.-J. Yang, T. Naguib, C.-F. Yang, and O. Kew, Abstr. 17th Annu. Meet. Am. Soc. Virol., abstr. P26-4, p. 193, 1998), so that the risks of importation into Israel of wild polioviruses from this source have declined sharply. At the same time, Israel has improved its own national program for polio immunization (7, 28), further reducing the risk of spread of any imported polioviruses. Possibly as a result of the higher vaccine coverage rates, no cases occurred in the West Bank or Israel during or after the 1992 polio outbreak in the eastern Jordan valley (28).
The findings of this study also highlight the utility of environmental sampling as a supplementary tool for wild poliovirus surveillance. Detection of wild polioviruses in the sewage of two separate communities with no cases of polio confirmed the widespread distribution of the outbreak virus and revealed the existence of additional lineages not associated with cases. When clinical and sewage isolates were available from the same community, they were found to be very closely related. Virologic analysis of environmental samples is a powerful method for the detection of poliovirus importation into countries where a virus is not endemic (15, 23, 29). It is especially valuable in countries such as Israel that have close contact with areas where polio is endemic but where a very high proportion of any wild poliovirus infections would be inapparent because of high vaccine coverage rates. Environmental sampling can also increase the sensitivities of wild poliovirus detection in areas of endemicity (27), particularly during the low-transmission season, when circulation in reservoir communities is only infrequently signaled by poliomyelitis cases (20). Finally, as shown here and by others (13, 23), sampling of sewage can improve the resolution of molecular epidemiologic studies into the patterns of poliovirus circulation during outbreaks.
ACKNOWLEDGMENTS
We thank Tova Halmut, Mina Neuman, Batya Abramovitch, and Hagit Rudich (Central Virology Laboratory) for assistance with the isolation of virus from clinical specimens and environmental samples and Baldev Nottay, Lina De, Jaume Jorba, David R. Kilpatrick, and Yvonne Stone (Centers for Disease Control and Prevention) for assistance in the further characterization of the isolates. We thank virologists from the World Health Organization Global Polio Laboratory Network for contributing poliovirus isolates. We thank Larry Anderson for critical review of the manuscript.
This study was funded in part by a grant from the United States-Israel Binational Science Foundation.
Footnotes
This paper is dedicated to the memory of our friend and colleague, Ami Vonsover.
REFERENCES
- 1.Afif H, Sutter R W, Fontaine R E, Kew O M, Pallansch M A, Goyal M K, Cochi S L. Outbreak of poliomyelitis in Gizan, Saudi Arabia: co-circulation of wild type 1 polioviruses from three separate origins. J Infect Dis. 1997;175(Suppl. 1):S71–S75. doi: 10.1093/infdis/175.supplement_1.s71. [DOI] [PubMed] [Google Scholar]
- 2.Alexander J P, Jr, Alemu W, Kilpatrick D, Kebede S, Yang C-F, Beyene H, Kew O M. Poliomyelitis in Ethiopia: virologic links to poliomyelitis cases in the Indian subcontinent. Pediatr Infect Dis J. 1996;15:629–631. doi: 10.1097/00006454-199607000-00015. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Virologic surveillance and progress toward poliomyelitis eradication—Eastern Mediterranean Region, 1995–September 1998. Morbid Mortal Weekly Rep. 1998;47:1001–1005. [PubMed] [Google Scholar]
- 4.Crainic R, Couillin P, Blondel B, Cabau N, Boue A, Horodniceanu F. Natural variation of poliovirus neutralization epitopes. Infect Immun. 1983;41:1217–1225. doi: 10.1128/iai.41.3.1217-1225.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahling D R, Berg G, Berman D. BGM, a continuous cell line more sensitive than primary rhesus and African green kidney cells for the recovery of viruses from water. Health Lab Sci. 1974;11:275–282. [PubMed] [Google Scholar]
- 6.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 7.Goldblum N, Gerichter C B, Tulchinsky T H, Melnick J L. Poliomyelitis control in Israel, the West Bank and Gaza Strip: changing strategies with the goal of eradication in an endemic area. Bull W H O. 1994;72:783–796. [PMC free article] [PubMed] [Google Scholar]
- 8.Green M S, Handsher R, Cohen D, Melnick J L, Slepon R, Mendelson E, Danon Y L. Age differences in immunity against wild and vaccine strains of poliovirus prior to the 1988 outbreak in Israel and response to booster immunization. Vaccine. 1993;11:75–81. doi: 10.1016/0264-410x(93)90342-u. [DOI] [PubMed] [Google Scholar]
- 9.Huovilainen A, Mulders M N, Agboatwalla M, Pöyry T, Stenvik M, Hovi T. Genetic divergence of poliovirus strains isolated in the Karachi region of Pakistan. J Gen Virol. 1995;76:3079–3088. doi: 10.1099/0022-1317-76-12-3079. [DOI] [PubMed] [Google Scholar]
- 10.Kew O M, Mulders M N, Lipskaya G Y, da Silva E E, Pallansch M A. Molecular epidemiology of polioviruses. Semin Virol. 1995;6:401–414. [Google Scholar]
- 11.Kilpatrick D R, Nottay B, Yang C-F, Yang S-J, Mulders M N, Holloway B P, Pallansch M A, Kew O M. Group-specific identification of polioviruses by PCR using primers containing mixed-base or deoxyinosine residues at positions of codon degeneracy. J Clin Microbiol. 1996;34:2990–2996. doi: 10.1128/jcm.34.12.2990-2996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 13.Kinnunen L, Pöyry T, Hovi T. Generation of virus genetic lineages during an outbreak of poliomyelitis. J Gen Virol. 1991;72:2483–2489. doi: 10.1099/0022-1317-72-10-2483. [DOI] [PubMed] [Google Scholar]
- 14.Manor Y, Handsher R, Halmut T, Neuman M, Abramovitz B, Mates A, Mendelson E. A double-selective tissue culture system for isolation of wild-type poliovirus from sewage applied in a long-term environmental surveillance. Appl Environ Microbiol. 1999;65:1794–1797. doi: 10.1128/aem.65.4.1794-1797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manor Y, Handsher R, Halmut T, Neuman M, Bobrov A, Rudich H, Vonsover A, Shulman L, Kew O, Mendelson E. Detection of poliovirus circulation by environmental surveillance in the absence of clinical cases in Israel and the Palestinian Authority. J Clin Microbiol. 1999;37:1670–1675. doi: 10.1128/jcm.37.6.1670-1675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minor P D. Antigenic structure of picornaviruses. Curr Top Microbiol Immunol. 1990;161:121–154. doi: 10.1007/978-3-642-75602-3_5. [DOI] [PubMed] [Google Scholar]
- 17.Minor P D, Ferguson M, Phillips A, Magrath D I, Houvalainen A, Hovi T. Conservation in vivo of protease cleavage sites in antigenic sites of poliovirus. J Gen Virol. 1987;68:1857–1865. doi: 10.1099/0022-1317-68-7-1857. [DOI] [PubMed] [Google Scholar]
- 18.Morvan J M, Chezzi C, Gouandjika I, Reimerink J H, van der Avoort H G. The molecular epidemiology of type 1 poliovirus in Central African Republic. J Gen Virol. 1997;78:591–599. doi: 10.1099/0022-1317-78-3-591. [DOI] [PubMed] [Google Scholar]
- 19.Mulders M N, Lipskaya G Y, van der Avoort H G A M, Koopmans M P G, Kew O M, van Loon A M. Molecular epidemiology of wild poliovirus type 1 in Europe, the Middle East, and the Indian Subcontinent. J Infect Dis. 1995;171:1399–1405. doi: 10.1093/infdis/171.6.1399. [DOI] [PubMed] [Google Scholar]
- 20.Nathanson N, Martin J R. The epidemiology of poliomyelitis: enigmas surrounding its appearance, epidemicity, and disappearance. Am J Epidemiol. 1979;110:672–692. doi: 10.1093/oxfordjournals.aje.a112848. [DOI] [PubMed] [Google Scholar]
- 21.Nomoto A, Omata T, Toyoda H, Kuge S, Horie H, Kataoka Y, Genba Y, Nakano Y, Imura N. Complete nucleotide sequence of the attenuated poliovirus Sabin 1 strain genome. Proc Natl Acad Sci USA. 1982;79:5793–5797. doi: 10.1073/pnas.79.19.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nottay B K, Kew O M, Hatch M H, Heyward J T, Obijeski J F. Molecular variation of type 1 vaccine-related and wild polioviruses during replication in humans. Virology. 1981;108:405–423. doi: 10.1016/0042-6822(81)90448-7. [DOI] [PubMed] [Google Scholar]
- 23.Pöyry T, Stenvik M, Hovi T. Viruses in sewage waters during and after a poliomyelitis outbreak and subsequent nationwide oral poliovirus vaccination campaign in Finland. Appl Environ Microbiol. 1988;54:371–374. doi: 10.1128/aem.54.2.371-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rico-Hesse R, Pallansch M A, Nottay B K, Kew O M. Geographic distribution of wild poliovirus type 1 genotypes. Virology. 1987;160:311–322. doi: 10.1016/0042-6822(87)90001-8. [DOI] [PubMed] [Google Scholar]
- 25.Slater P E, Orenstein W A, Morag A, Avni A, Handsher R, Green M S, Costin C, Yarrow A, Rishpon S, Havkin O, Ben-Zvi T, Kew O M, Rey M, Epstein I, Swartz T A, Melnick J L. Poliomyelitis outbreak in Israel in 1988: a report with two commentaries. Lancet. 1990;335:1192–1198. doi: 10.1016/0140-6736(90)92705-m. [DOI] [PubMed] [Google Scholar]
- 26.Swofford D L, Olsen G J, Wadell D J, Hillis D M. Phylogenetic inference. In: Hillis D M, Moritz C, Mable B K, editors. Molecular systematics. 2nd ed. Sunderland, Mass: Sinauer Associates; 1996. pp. 407–514. [Google Scholar]
- 27.Tambini G, Andrus J K, Marques E, Boshell J, Pallansch M, de Quadros C A, Kew O M. Direct detection of wild poliovirus transmission by stool surveys of healthy children and analysis of community wastewater. J Infect Dis. 1993;168:1510–1514. doi: 10.1093/infdis/168.6.1510. [DOI] [PubMed] [Google Scholar]
- 28.Tulchinsky T, Abed Y, Handsher R, Toubassi N, Acker C, Melnick J. Successful control of poliomyelitis by a combined OPV/IPV polio vaccine program in the West Bank and Gaza, 1978-93. Isr J Med Sci. 1994;30:489–494. [PubMed] [Google Scholar]
- 29.van der Avoort H G, Reimerink J H, Ras A, Mulders M N, van Loon A M. Isolation of epidemic poliovirus from sewage during the 1992-3 type 3 outbreak in The Netherlands. Epidemiol Infect. 1995;114:481–491. doi: 10.1017/s0950268800052195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vonsover A, Handsher R, Neuman M, Guillot S, Balanant J, Rudich H, Mendelson E, Swartz T, Crainic R. Molecular epidemiology of type 1 polioviruses isolated in Israel and defined by restriction fragment length polymorphism assay. J Infect Dis. 1993;167:199–203. doi: 10.1093/infdis/167.1.199. [DOI] [PubMed] [Google Scholar]
- 31.World Health Organization. Manual for the virologic investigation of poliomyelitis WHO/EPI/GEN/97.1. Geneva, Switzerland: World Health Organization; 1997. [Google Scholar]
- 32.Yang C-F, De L, Holloway B P, Pallansch M A, Kew O M. Detection and identification of vaccine-related polioviruses by the polymerase chain reaction. Virus Res. 1991;20:159–179. doi: 10.1016/0168-1702(91)90107-7. [DOI] [PubMed] [Google Scholar]