Abstract
We have previously described a SWI/SNF-related protein complex (PYR complex) that is restricted to definitive (adult-type) hematopoietic cells and that specifically binds DNA sequences containing long stretches of pyrimidines. Deletion of an intergenic DNA-binding site for this complex from a human β-globin locus construct results in delayed human γ- to β-globin switching in transgenic mice, suggesting that the PYR complex acts to facilitate the switch. We now show that PYR complex DNA-binding activity also copurifies with subunits of a second type of chromatin-remodeling complex, nucleosome-remodeling deacetylase (NuRD), that has been shown to have both nucleosome-remodeling and histone deacetylase activities. Gel supershift assays using antibodies to the ATPase-helicase subunit of the NuRD complex, Mi-2 (CHD4), confirm that Mi-2 is a component of the PYR complex. In addition, we show that the hematopoietic cell-restricted zinc finger protein Ikaros copurifies with PYR complex DNA-binding activity and that antibodies to Ikaros also supershift the complex. We also show that NuRD and SWI/SNF components coimmunopurify with each other as well as with Ikaros. Competition gel shift experiments using partially purified PYR complex and recombinant Ikaros protein indicate that Ikaros functions as a DNA-binding subunit of the PYR complex. Our results suggest that Ikaros targets two types of chromatin-remodeling factors—activators (SWI/SNF) and repressors (NuRD)—in a single complex (PYR complex) to the β-globin locus in adult erythroid cells. At the time of the switch from fetal to adult globin production, the PYR complex is assembled and may function to repress γ-globin gene expression and facilitate γ- to β-globin switching.
A number of different macromolecular complexes have been described in recent years that activate or repress specific mammalian gene expression by remodeling chromatin (3, 20). Among these are SWI/SNF-related complexes, also known as BAF complexes, which are very large (0.5- to 2-MDa) assemblies of up to 11 protein subunits, many of which are highly conserved from yeasts to humans (3, 20, 41). These complexes act as molecular machines, utilizing the energy of ATP to disrupt repressive chromatin structures and facilitate gene activation, presumably by permitting the binding of transcription factors to DNA regulatory elements that would otherwise be inaccessible (7, 26). Mammalian SWI/SNF complexes contain a SNF2 family helicase-ATPase subunit, either BRM or BRG1, that is probably critical for their ATP-dependent nucleosome disruption activity (21, 26, 27, 36, 54), accompanied by BRG1-associated factors (BAFs), actin-related proteins, and β-actin (4, 40, 54, 55, 59). These complexes are found in association with chromatin and are known to bind DNA structures resembling nucleosomes (42, 53, 59). By contrast, other chromatin-remodeling complexes with both nucleosome-remodeling and histone deacetylase activities (nucleosome-remodeling deacetylase [NuRD] complexes) that contain the SNF2-related helicase-ATPase Mi-2 (CHD4) (44), histone deacetylases 1 and 2, and RbAp46/48 have also been described, and they are thought to function in chromatin-mediated gene repression (24, 51, 57, 58).
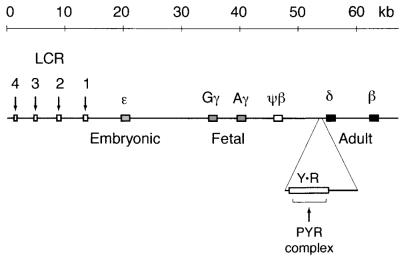
We have previously described a SWI/SNF-related complex, the PYR complex, that binds a long pyrimidine-rich sequence between the human fetal and adult β-globin-like genes (38, 39) (Fig. 1). PYR complex DNA-binding activity is specific to definitive (adult-type) hematopoietic cells and is both DNA sequence and DNA length dependent (39). Gel supershift assays and mass spectrometric sequence analysis of DNA affinity column fractions indicate that the PYR complex contains at least four known SWI/SNF subunits: INI1 (BAF47), BAF57, BAF60a, and BAF170 (39). Although the exact function of the PYR complex is unknown, we have shown that deletion of an intergenic PYR complex-binding site from a human β-globin locus construct results in delayed human fetal-to-adult globin gene switching in transgenic mice, suggesting that in erythroid cells, the PYR complex may act to facilitate this genetic switch (39) (Fig. 1).
FIG. 1.
Map of the β-globin locus on human chromosome 11. The PYR complex binds to a 250-bp pyrimidine-rich sequence (rectangle Y · R) located between the fetal (γ) and adult (δ and β) β-globin-like genes. β-Globin locus control region (LCR) DNase I-hypersensitive sites 1 through 4 are indicated by downward arrows.
We now show that, in addition to the previously described SWI/SNF proteins, PYR complex DNA-binding activity also copurifies with the hematopoietic cell-specific DNA-binding factor Ikaros and with a number of NuRD complex subunits, including the ATPase-helicase subunit of the NuRD complex, Mi-2. Antibodies to Mi-2 and Ikaros both supershift PYR complex DNA-binding activity in gel shift assays, and Mi-2 and Ikaros coimmunoprecipitate from both crude murine erythroleukemia (MEL) cell nuclear extract and chromatography fractions containing partially purified PYR complex. In addition, we show that NuRD and SWI/SNF components coimmunopurify from these fractions, indicating that they are present in a single complex.
In competition gel shift experiments using partially purified PYR complex and recombinant Ikaros protein, the PYR complex exhibits the same DNA sequence specificity as Ikaros. Furthermore, we show that DNA-dependent ATPase, histone deacetylase, and ATP-dependent nucleosome-remodeling activities also copurify with PYR complex DNA-binding activity. Taken together with our earlier observations, these data suggest that the PYR complex is a single complex containing SWI/SNF and NuRD subunits that is targeted to DNA by Ikaros and that this Ikaros-targeted chromatin-remodeling complex may function in the control of the developmental switch from fetal to adult globin synthesis.
MATERIALS AND METHODS
Purification of PYR complex and mass-spectrometric-sequence analysis.
The PYR complex was initially purified from MEL nuclear extract in five chromatographic steps as previously described (39). The final purification step utilized a sequence-specific DNA affinity column prepared from a concatenated double-stranded oligonucleotide (δ60ym) containing the PYR complex-binding site with a YY1 site mutated within it (sense strand [5′ to 3′], GATCCTCTCTCTTCCCCTCTTCCTTCCTTCCTTTCCTTATTTCTTCCTCCTCTTTCCCTC). DNA-affinity column fractions containing PYR complex activity (as determined by gel shift assay using radiolabeled δ60ym probe) were pooled, electrophoresed on sodium dodecyl sulfate (SDS)–4 to 15% gradient polyacrylamide gel, and electroblotted onto a nitrocellulose membrane. Bands identified by Ponceau S staining were eluted from the membrane and processed for internal sequence analysis as previously described (9, 29, 39). The resulting peptide pools were analyzed by matrix-assisted laser-desorption ionization (MALDI), reflectron time-of-flight (ReTOF) mass spectrometry (MS), and electrospray ionization tandem mass spectrometry (MS/MS) in conjunction with collision-induced fragmentation as previously described (39). Selected mass values from the MALDI-ReTOF experiments were taken to search a protein-nonredundant database (EBI, Hinxton, United Kingdom) using the PeptideSearch algorithm (30). MS/MS spectra were inspected for y" ion series, and the resultant information was transferred to the SequenceTag program and used as a search string (31). Any protein identification thus obtained was verified by comparing the computer-generated fragment ion series of the predicted tryptic peptide with the experimental MS/MS data. In additional experiments, the DNA-cellulose column fractions containing the peak PYR activity were pooled and run on a Superose 6 column (Amersham Pharmacia), and the fractions were analyzed.
Gel shift and gel supershift assays.
Nuclear extracts were prepared from tissue culture cells as previously described (8, 38). For mouse tissues, nuclei were isolated by pelleting through a sucrose cushion (15), and nuclear extracts were then prepared using a miniextract protocol (43). Large-scale cultures of MEL cells were prepared by the National Institutes of Health (NIH) Cell Culture Center (Coon Rapids, Minn.). Recombinant glutathione S-transferase (GST)–Ikaros fusion protein, which contains the four N-terminal zinc fingers required for optimal DNA binding, has been described previously (11, 17). Gel shift and gel supershift assays using MEL crude nuclear extract were performed with 5 μg of nuclear extract and 2 μg of double-stranded poly(dI-dC) as previously described (38, 39). For gel shifts using chromatography fractions or GST-Ikaros, double-stranded poly(dG-dC), instead of poly(dI-dC), was used in empirically determined amounts as nonspecific competitor DNA. Probes and competitor DNAs (sense strand, 5′ to 3′) used were δ99, CCT CCA TCC CTT CCA TCC TCT CTC TTC CCC TCT TCC TTC CTT CCT TTC TCC ATT TCT TCC TCC TCT TTC CCT CAA TCC TTC CTT TTG GAT ATG CTC ATG; δ60ym (shown above); IKBS4, AAT TCT CAG CTT TTG GGA ATG TAT TCC CTG TCAG (34); and YY1, ACG TCG CTC CGC GGC CAT CTT GGC GGC TGGT. Antibodies were generously provided by Stephen Smale and Bradley Cobb (University of California, Los Angeles; monoclonal and polyclonal Ikaros), Gerald Crabtree (Stanford University; BAF57, BAF60a, BAF170, and BRG1), Katia Georgopoulos (Harvard University; monoclonal Mi-2), Ganjam Kalpana (Albert Einstein Medical College; INI1) and Michael Bustin (NIH; HMG-1). Antibodies to actin, HDAC2, and YY1 were obtained commercially from Santa Cruz Biotechnology (Santa Cruz, Calif.). Antibody to RbAp46/48 was obtained commercially from GeneTex (San Antonio, Tex.).
Photoactivated cross-linking.
Photoactivated UV cross-linking experiments used a 5-bromodeoxyuridine (BrdU; Life Technologies)-substituted δ99 probe. This was prepared by Klenow polymerization using a mixture of BrdU, dATP, dGTP, and [α-32P]dCTP with an M13 reverse-sequencing primer (Stratagene) on single-stranded template prepared from a pBluescript II (SK+) phagemid (Stratagene) with δ99 subcloned into its EcoRI site (pBSII-δ99). The single-stranded pBSII-δ99 template was prepared using M13 helper phage according to the manufacturer's instructions (Stratagene). After polymerization, the double-stranded DNA was digested with EcoRI, and the radiolabeled BrdU-δ99 was purified on a 5% nondenaturing polyacrylamide gel. DNA-binding assays for UV cross-linking experiments were the same as for gel shift assays except for the use of 105 to 106 cpm of BrdU-substituted δ99 as a probe. After the DNA-binding reactions, cross-linking was carried out by spotting the reactions on cling wrap and irradiating on a 302-nm UV transilluminator (Spectroline) for 4 to 8 min at room temperature. Reactions were then adjusted to 2 mM CaCl2 and 10 mM MgCl2 and digested with 1 U of micrococcal nuclease (Boehringer Mannheim Biochemicals) and 1 μg of DNase I (Worthington Biochemical) for 30 min at 37°C to remove excess probe from the DNA-protein complexes. EDTA was then added to a final concentration of 10 mM, and samples were precipitated by the addition of 4 volumes of ice-cold acetone. Pellets were resuspended in Laemmli sample buffer (Bio-Rad), heated to 95°C for 5 min, and electrophoresed on precast Tris-HCl polyacrylamide gels (Bio-Rad).
Immunoprecipitations.
Immunoprecipitations were carried out essentially as previously described (18), with a few modifications. NP-40 was added to MEL cell nuclear extract (10 μl) or extract from reactive yellow 3-agarose fraction 67 (Ikaros peak activity; 30 μl) to a final concentration of 0.01%, and the samples were then incubated with 0.5 μl of mouse monoclonal antibody at 4°C for 1 h. Immune complexes were precipitated by the addition of Protein A/G PLUS beads (Santa Cruz Biotechnology), which were then washed three times with TM-100 (20 mM Tris, 5 mM MgCl2, 20% glycerol, 0.1 M NaCl, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol [DTT], pH 7.5) with 0.1% NP-40. Proteins were then eluted from the beads by boiling in Laemmli sample buffer (Bio-Rad) and electrophoresed on sodium dodecyl sulfate (SDS)-polyacrylamide gels. Western blots were performed as previously described (18) by electroblotting onto a polyvinylidene difluoride membrane (Bio-Rad) and screening with rabbit polyclonal anti-Mi-2 antibody (1:3,000 dilution) or Ikaros antibody. Bands were visualized using a goat-anti-rabbit chemiluminescent detection kit (Amersham Pharmacia). Similar results were obtained using MEL nuclear extract both with and without preclearing with Protein A/G PLUS beads (18).
Immunoaffinity chromatography.
An anti-BAF57 immunoaffinity column was prepared using rabbit polyclonal antiserum raised against a recombinant maltose-binding protein (MBP)-BAF57 fusion protein expressed from the plasmid pMBP-BAF57, which was generously provided by Gerald Crabtree (Stanford University). The MBP-BAF57 fusion protein was expressed in Escherichia coli and purified using a pMBP protein expression kit (New England BioLabs). The anti-BAF57 antiserum was raised commercially by Cocalico Biologicals. Antibody was coupled to protein A-agarose and cross-linked using a commercial kit (protein A orientation kit; Pierce, St. Louis, Mo.) according to the manufacturer's instructions. An anti-Mi-2 immunoaffinity column was prepared using antigen-purified goat anti-Mi-2 antibody obtained commercially (Santa Cruz Biotechnology). This antibody was coupled to protein G-agarose and cross-linked using a commercial kit (Protein G PLUS orientation kit; Pierce). Immunoaffinity chromatography experiments were carried out at 4°C. DTT or protease inhibitors were not added to buffers used for immunoaffinity chromatography experiments. DNA-cellulose-purified PYR complex (0.5 ml) was diluted in 3 volumes of buffer TM 100 and recycled over a 0.5-ml immunoaffinity column for at least 1 h. The column was then washed with two column volumes of TM 100. This initial wash was pooled with the unbound material and designated flowthrough. The column was then washed twice more with two column volumes of TM 100 (washes 0.1A and 0.1B) and twice with two column volumes of TM 500 (20 mM Tris, 5 mM MgCl2, 20% glycerol, 0.5 M NaCl [pH 7.5]), designated washes 0.5A and 0.5B, and then eluted with three column volumes of 0.1 M glycine (pH 2.5). The pH 2.5 glycine eluate was mixed with 1/10 volume of 1 M Tris (pH 8.0) immediately following collection. Samples were precipitated with trichloroacetic acid, resuspended in Laemmli sample buffer, boiled for 5 min, and electrophoresed on SDS-polyacrylamide minigels (Bio-Rad). Gels were analyzed by silver stain (Plus One kit, Amersham Pharmacia) and Western blotting.
ATPase assays.
For each reaction (20-μl total volume), 4 μl of column fraction was incubated in EMSA buffer (10 mM Tris-7.5, 50 mM NaCl, 5 mM MgCl2, 20% glycerol, 1 mg of bovine serum albumin per ml, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride) containing 2 μM ATP, 1 μCi [γ-32P]ATP, and 5 μg of substrate, either calf thymus DNA (Roche), purified histones (Roche), or chicken erythrocyte chromatin (49), at 37°C for 30 min. Reactions were stopped by the addition of 1 μl of 0.5 M EDTA, and 1 μl of each reaction was spotted onto a polyethyleneimine cellulose thin-layer-chromatography plate (Sigma). Thin-layer chromatography plates were then developed for 45 min with 1 M formic acid–0.5 M LiCl, air dried, and analyzed on a Molecular Dynamics STORM scanner.
Histone deacetylase assays.
[3H]acetyllysine-labeled histones were prepared from 2 × 109 MEL cells following a previously published protocol (5), with a specific activity of approximately 2,000 cpm/μg. For each reaction (50-μl total volume), 5 μl of column fraction was incubated in EMSA buffer with 5 μl (approximately 20 μg) of labeled histones at 37°C for 30 min. The reactions were then stopped by the addition of 50 μl of stop solution (0.24 M acetic acid–1.42 M HCl) and extracted with 200 μl of ethyl acetate. A 100-μl portion of the organic phase was then assayed for released [3H]acetate using a liquid scintillation counter. For each experiment, average values were calculated from a minimum of two measurements for each fraction.
Nucleosome-remodeling assays.
Mononucleosomes were assembled by octamer transfer from donor chromatin prepared from HeLa cells as described previously (50). The beta interferon (IFN-β)-110 DNA template used is a 158-bp XbaI-ClaI restriction fragment of plasmid-110CAT and contains the enhancer region of the IFN-β gene (48). Remodeling of mononucleosomes was assayed in 25-μl DNase I footprinting reactions. For DNase I footprinting, 1 μl of DNA-cellulose column fraction, 2.5 μl of 20-mg/ml bovine serum albumin (Roche), 2.5 μl of 1-μg/μl double-stranded poly(dG-dC) competitor DNA (Amersham Pharmacia), 106 cpm of reconstituted mononucleosome probe, and 2.5 μl of 10× footprinting buffer (100 mM Tris [pH 7.5], 150 mM HEPES [pH 7.9], 0.5 M NaCl, 50 mM MgCl2, 10 mM DTT, 50% glycerol) were incubated at room temperature for 30 min in the presence or absence of 4 mM ATP. Subsequently, the reactions were digested with DNase I (Worthington) for 5 min on ice, and DNase I digestion was stopped with the addition of 2.5 M ammonium acetate and 2.5 μg of salmon sperm DNA. The reactions were then extracted with phenol-chloroform-isoamyl alcohol and precipitated with ethanol. The purified DNA was then separated on a 6% sequencing gel.
RESULTS
PYR complex DNA-binding activity is associated with SWI/SNF subunits, Mi-2, and the hematopoietic cell-restricted zinc finger protein Ikaros.
We previously reported the purification of the PYR complex from MEL nuclear extract and its identification as a SWI/SNF-related complex by MS sequence analysis and gel supershift assays (39). These experiments indicated that PYR complex contains at least four known SWI/SNF components (BAFs): BAF57, INI1 (BAF47), BAF60a, and BAF170. A histone deacetylase complex subunit, RbAp46/48, was also found to have copurified with PYR complex activity but could not be confirmed as a subunit by gel supershift (39).
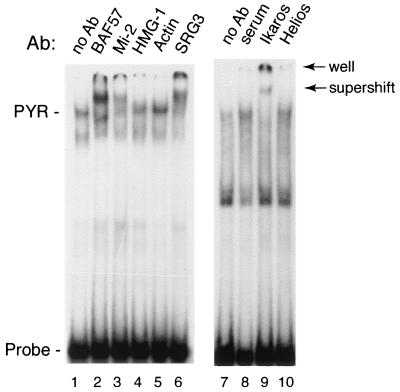
We have now identified a number of additional proteins by sequence analysis in the same DNA affinity-purified fractions. These proteins include the SNF2 family helicase-ATPase Mi-2, two additional BAFs (SRG3 and an actin-related protein similar to BAF53), an additional histone deacetylase complex subunit (HDAC2), a high-mobility group protein (HMG-1), and cytoplasmic actin (Table 1). We used gel supershift assays to test whether these proteins are components of the PYR complex or had simply copurified with it. As seen in Fig. 2, antibodies to Mi-2 and SRG3 (mouse BAF155) give clear supershifts, indicating that they are in fact PYR complex components. Antibodies to actin and HMG-1 result in negative supershifts, suggesting that they are not part of the complex. It should be noted, however, that a negative supershift does not rule out the presence of a protein in the complex, since the epitopes recognized by the antibody may be inaccessible due to the presence of other subunits of the complex.
TABLE 1.
Identification of proteins associated with PYR complex DNA-binding activity by MS sequence analysisa
| Band | Identity by MS |
|---|---|
| SWI/SNF subunits | |
| p150 | SRG3 (mouse BAF155) |
| p55a | BAF57 |
| p50 | Actin-related protein, similar to BAF53 |
| p48a | Cytoplasmic actinb |
| p48b | INI1 (BAF47) |
| NuRD subunits | |
| p220 | Mi-2 (CHD4) |
| p58 | HDAC2 |
| p55b | RbAp46 |
| Other | |
| p30 | HMG-1 |
| p22 | DR1 |
DNA-affinity column fractions containing peak PYR complex DNA-binding activity were electrophoresed on a 4- to 15%-gradient polyacrylamide gel and electroblotted onto nitrocellulose. Protein bands identified by Ponceau S staining were eluted from the membrane and analyzed by MALDI-ReTOF MS. Bands are named according to their molecular size (e.g., p220 is 220 kDa). Peptides were identified by mass analysis as described in Materials and Methods.
Six of six peptides identical to both γ and β actin.
FIG. 2.
The PYR complex contains SWI/SNF subunits, the NuRD subunit Mi-2, and the hematopoietic cell-restricted zinc finger protein Ikaros. Gel supershift assay was performed using MEL nuclear extract and labeled δ60ym DNA as a probe. Nuclear extract was preincubated for 1 h in the presence of antibody prior to the addition of probe. Supershifts are seen with anti-BAF57, anti-Mi-2, anti-SRG3, and anti-Ikaros antibodies.
It has recently been reported that in nuclear extracts from lymphocytes, both a BRG1-containing SWI/SNF-like complex and a Mi-2-containing histone deacetylase (NuRD) complex are associated with a zinc finger DNA-binding protein, Ikaros (22). To see if Ikaros is a component of the PYR complex, we tested anti-Ikaros antibody in gel supershift experiments. As seen in Fig. 2, anti-Ikaros antibody gave a very strong supershift of the PYR complex (lane 9), whereas no supershift was detected using either preimmune rabbit serum or antibody to a related lymphocyte-restricted protein, Helios (lanes 8 and 10).
The PYR complex and recombinant Ikaros protein have similar DNA-binding specificities.
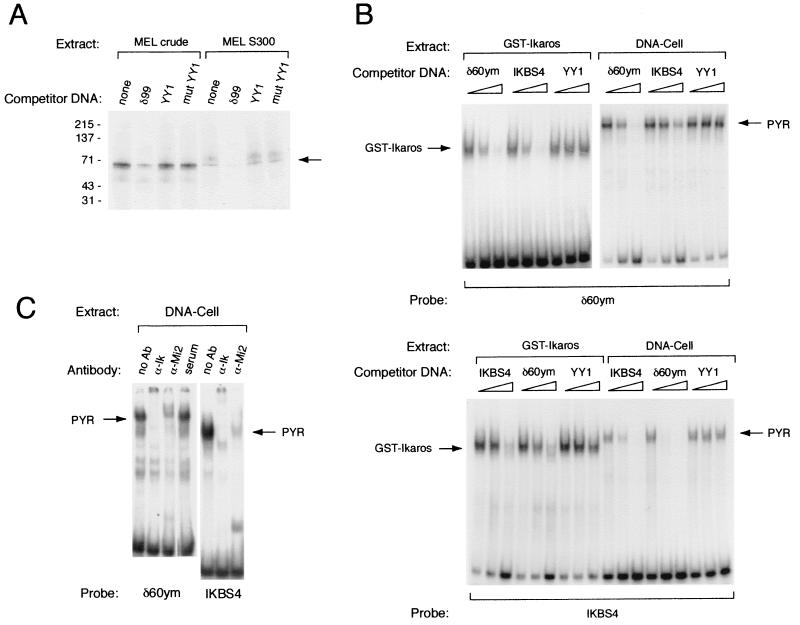
The known ability of Ikaros to bind DNA in a sequence-specific manner suggested that Ikaros might function, at least in part, as a DNA-binding subunit of the PYR complex. To determine the size of the PYR complex subunit (or subunits) that contact DNA, we performed photoactivated cross-linking experiments using a BrdU-substituted PYR DNA-binding site probe, δ99, described previously (38). These experiments indicate that the PYR complex has a 60- to 70-kDa DNA-binding subunit (Fig. 3A). This is comparable to the size of the largest Ikaros isoform, Ikaros-1, which has a molecular mass of 65 kDa.
FIG. 3.
Ikaros functions as the DNA-binding subunit of the PYR complex. (A) PYR complex has a 60- to 70-kDa DNA-binding subunit as detected by photoactivated cross-linking using both MEL crude nuclear extract and Sephacryl S300 chromatography fractions containing peak PYR complex DNA-binding activity. The band corresponding to this activity is competed away with excess unlabeled δ99 DNA but not by consensus (YY1) or mutant (mut YY1) YY1-binding-site DNA. (B) The PYR complex and GST-Ikaros have similar DNA-binding specificities. Upper panel: Gel shift assay using labeled δ60ym DNA probe, GST-Ikaros protein (left lanes), and partially purified PYR complex (DNA-cellulose column peak fractions [DNA-Cell], right lanes). Both GST-Ikaros and the PYR complex bind δ60ym, and these binding activities are competed away with unlabeled δ60ym DNA and DNA from a previously described high-affinity Ikaros-binding site, IKBS4, but not with YY1-binding- site DNA. The various competitor DNAs were used at 0-, 10-, and 100-fold molar excesses. Lower panel: Gel shift assay using labeled IKBS4 DNA probe, GST-Ikaros protein (left), and DNA cellulose-purified PYR complex (right). Both GST-Ikaros and the PYR complex bind labeled IKBS4, and these binding activities are competed away with unlabeled IKBS4 and δ60ym, but not with YY1-binding-site, DNA. (C) Antibodies to Ikaros and Mi-2 supershift the activity in DNA-cellulose-purified fractions that binds δ60ym (left lanes) and IKBS4 (right lanes).
We then tested whether the PYR complex bound DNA with the same sequence specificity as Ikaros. To do this, we compared the binding activity of a recombinant Ikaros fusion protein (GST-Ikaros [see Materials and Methods]) to that of partially purified PYR complex (DNA cellulose fraction with peak PYR DNA-binding activity). As seen in the upper panel of Fig. 3B, both GST-Ikaros and DNA cellulose-purified PYR complex bind to our optimal PYR complex-binding-site probe, δ60ym (39). These binding activities are competed by increasing amounts of unlabeled δ60ym DNA and DNA from a previously reported Ikaros-binding-site probe, IKBS4 (33) but not by unlabeled YY1-binding-site DNA. Unlabeled δ60ym DNA competes somewhat more strongly than IKBS4 for both the PYR complex and GST-Ikaros, suggesting that it is a higher-affinity-binding site for both the PYR complex and GST-Ikaros than IKBS4. Similar results are obtained when the identical experiment is carried out using labeled IKBS4 probe (Fig. 3B, lower panel). Both GST-Ikaros and the PYR complex bind to IKBS4, and their binding is competed away with unlabeled δ60ym and IKBS4, but not with unlabeled YY1-binding-site, DNA. In other experiments with DNA-cellulose-purified PYR complex, anti-Ikaros antibody supershifts the binding of the PYR complex to both the δ60ym and IKBS4 probes, confirming that the DNA-binding activity seen in the DNA-cellulose fraction includes Ikaros (Fig. 3C). Anti-Mi-2 antibody also supershifts the complex bound to both IKBS4 and δ60ym, showing that PYR complex DNA-binding activity in DNA cellulose-purified fractions is associated with both Ikaros and Mi-2.
PYR complex DNA-binding activity copurifies with Ikaros, NuRD, and SWI/SNF subunits from MEL nuclear extract.
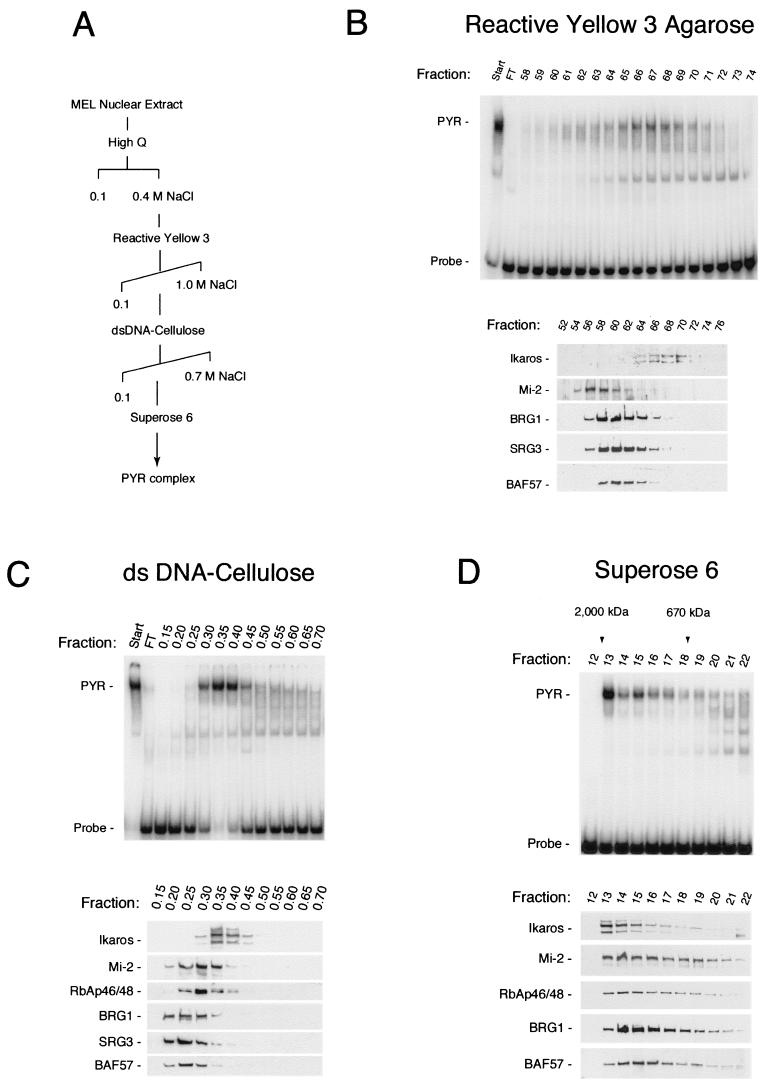
To confirm the results of the supershift experiments and to determine what proportions of the Ikaros, SWI/SNF, and NuRD proteins in MEL nuclear extract are associated with the PYR complex, we repurified the complex in four chromatographic steps (Fig. 4A). The four column steps resulted in a greater than 1,000-fold purification of the PYR complex, as determined by total protein quantitation by spectrophotometry and DNA-binding activity on a gel shift assay. Column fractions were tested for PYR complex DNA-binding activity by gel shift assay and for the presence of Ikaros, SWI/SNF, and NuRD subunits by Western blotting. By silver staining the material obtained after the final Superose 6 column step was comparable in purity to that obtained by DNA-affinity chromatography (data not shown).
FIG. 4.
PYR complex DNA-binding activity cofractionates from MEL nuclear extract with Ikaros and a subpopulation of SWI/SNF and NuRD protein. (A) Purification scheme. The PYR complex was purified from MEL nuclear extract in four chromatographic steps using High Q Macroprep (Bio-Rad; 0.1 to 0.4 M NaCl step elution), reactive yellow 3-agarose (Sigma; 0.1 to 1.0 M NaCl gradient), double-stranded (ds) DNA-cellulose (calf thymus DNA-cellulose [Sigma], 0.1 to 0.7 M NaCl step gradient), and Superose 6 (Amersham Pharmacia) columns. (B) Chromatography on reactive yellow 3-agarose shows peak PYR complex DNA-binding activity by gel shift assay (PYR) eluting in fractions 64 through 70 (upper panel), corresponding precisely with the elution pattern of Ikaros by Western blotting (lower panel). The majority of NuRD (Mi-2) and SWI/SNF (BRG1, SRG3, and BAF57) proteins elute prior to fraction 64, with some overlap into fractions 64 through 68. Fractions 65 through 70 were pooled, concentrated, adjusted to 0.1 M NaCl, and loaded onto the DNA-cellulose column (C). Peak PYR complex DNA-binding activity elutes off the DNA cellulose column in fractions 0.3 through 0.45 (upper panel), again corresponding precisely with the elution pattern of Ikaros (lower panel). Much of the NuRD (Mi-2 and RbAp46/48) and SWI/SNF (BRG1, SRG3, and BAF57) proteins elutes prior to fraction 0.3, but there is greater overlap into the PYR DNA-binding fractions than seen with reactive yellow 3-agarose. Fractions 0.3 through 0.45 were pooled, concentrated, and loaded onto the Superose 6 gel filtration column (D). Again, PYR complex DNA-binding activity, which elutes in high-molecular-mass fractions (1 to 2 MDa, upper panel), corresponds exactly with the elution pattern of Ikaros by Western blotting (lower panel). Here, however, both NuRD (Mi-2 and RbAp46/48) and SWI/SNF (BRG1 and BAF57) subunits cofractionate very closely with PYR DNA-binding activity and Ikaros. FT, failure to bind.
Gradient elution from the reactive yellow 3-agarose column shows that PYR complex DNA-binding activity on gel shift assays (Fig. 4B, upper panel) copurifies precisely with Ikaros as assayed by Western blotting (Fig. 4B, lower panel). The bulk of SWI/SNF and NuRD proteins in MEL nuclear extract elutes in earlier fractions than PYR complex DNA-binding activity and Ikaros, although a small amount of SWI/SNF and NuRD also coelutes with PYR complex-binding activity. This would suggest the presence in MEL cells of many different complexes containing SWI/SNF or NuRD subunits in addition to the PYR complex. Elution off the next column, DNA cellulose (Fig. 4C), again shows peak PYR complex DNA-binding activity on gel shift assays corresponding precisely to the elution pattern of Ikaros on Western blotting. Much of the NuRD (Mi-2 and RbAp46/48) and SWI/SNF (BRG1, SRG3, and BAF57) proteins elutes prior to PYR complex DNA-binding activity and Ikaros, but there is greater overlap into the PYR DNA-binding fractions than seen off reactive yellow 3-agarose. Analysis of fractions off the Superose 6 gel filtration column (Fig. 4D) continues to show PYR complex DNA-binding activity, which elutes in high-molecular-mass fractions (1 to 2 MDa), corresponding exactly with the elution pattern of Ikaros. Here, however, both NuRD (Mi-2 and RbAp46/48) and SWI/SNF (BRG1 and BAF57) subunits cofractionate very closely with PYR DNA-binding activity and Ikaros. Thus, with continued purification through DNA-cellulose and Superose 6, SWI/SNF and NuRD subunits begin to coelute with PYR DNA-binding activity and Ikaros. Taken together, these results indicate that PYR complex DNA-binding activity on gel shift assays represents Ikaros in close association with a subpopulation of the NuRD and SWI/SNF proteins in MEL cells. Our results also suggest that Ikaros in erythroid cells is found only associated in chromatin-remodeling complexes and is not detectable as free protein.
Ikaros, Mi-2, and SWI/SNF proteins coimmunopurify from MEL chromatography fractions.
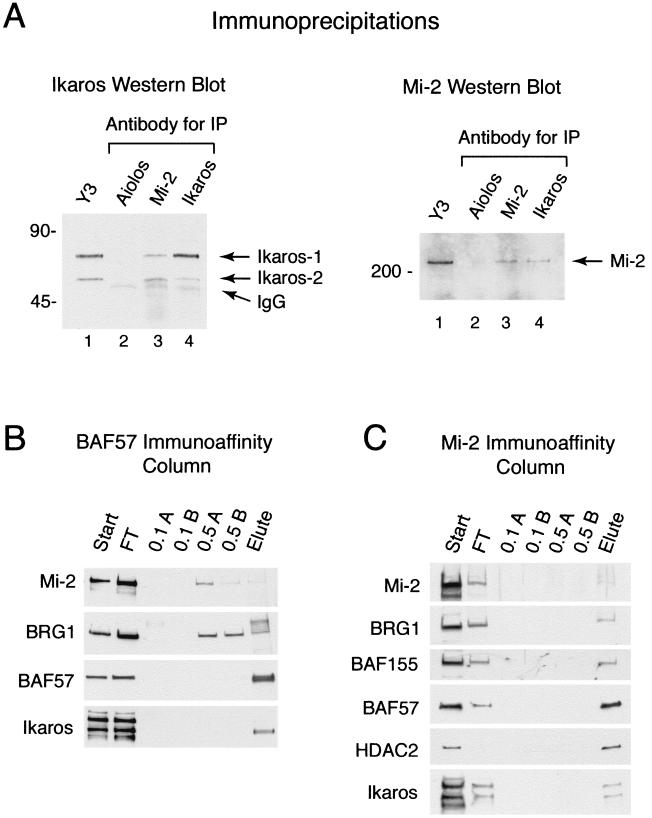
To determine whether Ikaros, Mi-2, and SWI/SNF proteins are physically associated in MEL nuclear extract by using a method other than gel supershift assays and conventional chromatography, we performed coimmunoprecipitations on both MEL crude nuclear extract (data not shown) and chromatography fractions containing partially purified PYR complex, using antibodies to Mi-2 and Ikaros (Fig. 5A). As seen in the left panel of Fig. 5, monoclonal antibodies to Mi-2 and Ikaros both precipitate Ikaros isoforms 1 and 2 (47), which are detected as 65- and 55-kDa bands, respectively, by Western blotting with polyclonal anti-Ikaros antiserum. As a control, antibody to the lymphocyte-restricted Ikaros family member Aiolos does not precipitate Ikaros from DNA cellulose-purified PYR complex. Similar results are seen in the reverse experiment, in which monoclonal antibodies to Mi-2 and Ikaros both precipitate Mi-2, detected as a 220-kDa band by Western blotting with anti-Mi-2 antiserum (Fig. 5A, right). In the control lane, antibody to Aiolos does not precipitate Mi-2. The results indicate that Ikaros and Mi-2 are in physical association in both crude MEL nuclear extract and in chromatography fractions containing PYR complex DNA-binding activity, in support of our observations with gel supershift experiments.
FIG. 5.
Ikaros, NuRD, and SWI/SNF proteins coimmunopurify from chromatography fractions containing peak PYR complex DNA-binding activity. (A) Ikaros and Mi-2 coimmunoprecipitate from reactive yellow 3-agarose-fractionated MEL extract. (Left panel) Western blot analysis with rabbit polyclonal anti-Ikaros antibody. Mouse monoclonal antibodies used for immunoprecipitation (IP) are indicated above lanes 2 through 4. The sample for lane 1 was 1/10 the volume of reactive yellow 3-agarose-purified PYR complex (yellow 3 fraction 67 [Y3]) before precipitation as a positive control for the Ikaros signal (Ik-1, 65 kDa; Ik-2, 55 kDa). Weak cross-reaction of the anti-rabbit IgG secondary antibody with the mouse IgG heavy chain results in a faint band of approximately 50 kDa in lanes 2 through 4. (Right panel) Western blot analysis with an anti-Mi-2 rabbit polyclonal antibody. Lane 1, reactive yellow 3-agarose fraction 67; lane 2, anti-Aiolos precipitate; lane 3, anti-Mi-2 precipitate; lane 4, anti-Ikaros precipitate. (B) Ikaros, BRG1, and Mi-2 copurify with BAF57 off an anti-BAF57 immunoaffinity column. The start material (Start) was DNA-cellulose-purified PYR complex. The vast bulk of the protein loaded onto the column does not bind (FT). This was confirmed by silver staining (data not shown). The column was washed twice with 0.1 M NaCl buffer (0.1 A and B) and twice with 0.5 M NaCl buffer (0.5 A and B) and then eluted with 0.1 M glycine (pH 2.5) (Elute). BAF57 and Ikaros, particularly Ikaros isoform 2, bind very tightly to the column, withstanding the 0.5 M salt washes. Both BRG1 and Mi-2 bind to the column with somewhat lower affinity, eluting in the 0.5 M salt and pH 2.5 glycine fractions. (C) Ikaros, NuRD, and SWI/SNF proteins copurify with Mi-2 off an anti-Mi-2 immunoaffinity column. The start material (Start) was DNA-cellulose-purified PYR complex. Silver-stained gels showed that the vast bulk of the protein loaded onto the column does not bind (data not shown). The column was washed and eluted as described for panel B. Mi-2 binds very tightly to the column, with much of the Mi-2 remaining on the column after the pH 2.5 elution step. HDAC2, Ikaros isoforms 1 and 2, and SWI/SNF proteins (BAF57, BRG1, and BAF155) bind to the column with high affinity, eluting only in the pH 2.5 glycine fraction.
To determine whether PYR DNA-binding activity represents either one or two protein complexes, we then performed similar experiments with anti-BAF57 and anti-Mi-2 immunoaffinity columns, using DNA cellulose-purified PYR complex as start material. As seen in Fig. 5B, both BAF57 and Ikaros—in particular Ikaros isoform 2—bind strongly to the anti-BAF57 column after extensive washes to eliminate nonspecific binding. Both BRG1 and Mi-2 also bind specifically to the column with somewhat lower affinity. Using the anti-Mi-2 column, we see strong specific binding of Mi-2 (some of which remains bound to the column), Ikaros, HDAC2, and three SWI/SNF proteins (BRG1, BAF155, and BAF57 [Fig. 5C]). Taken together with the results of the supershift, conventional chromatography, and immunoprecipitation experiments, these results indicate that in MEL cells Ikaros is closely associated with SWI/SNF and NuRD proteins in a single complex.
Chromatography fractions with PYR complex DNA-binding activity have chromatin-remodeling and histone deacetylase activities.
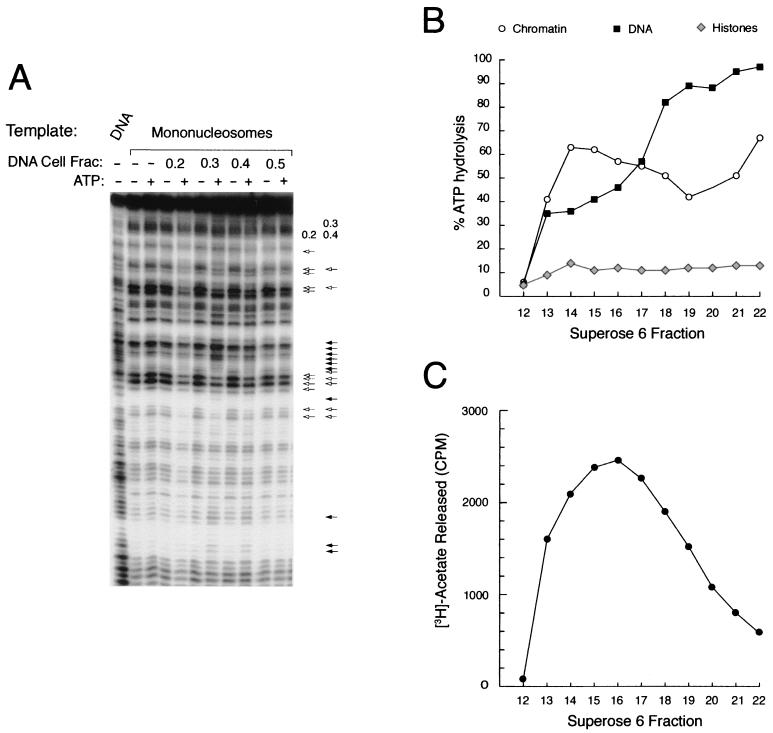
The presence of SWI/SNF and NuRD components in the PYR complex indicates that the purified complex should exhibit a number of activities associated with chromatin-remodeling complexes. SWI/SNF complexes are known to have DNA-dependent ATPase activity and have been shown to remodel nucleosome templates in the presence of ATP (46). NuRD complexes display both of these activities as well as a histone deacetylase activity (58). We tested fractions from the DNA-cellulose column for their ability to remodel mononucleosome templates in the presence or absence of ATP. Remodeling activity, characterized by a combination of enhanced and diminished DNase I hypersensitivity with the addition of ATP, is detected in the 0.3 and 0.4 M NaCl fractions (Fig. 6A), corresponding to the pattern of PYR complex DNA-binding activity off that column (Fig. 4C).
FIG. 6.
The PYR complex is associated with nucleosome remodeling and histone deacetylase activities. (A) Calf thymus DNA cellulose column fractions with peak PYR complex DNA-binding activity remodel mononucleosomes in an ATP-dependent manner. DNA cellulose column fractions (Fig. 4C) were tested for the ability to remodel a radiolabeled probe (IFNβ-110) packaged into a mononucleosome template in the presence (+) or absence (−) of ATP. Remodeling activity, indicated by the combination of increased (filled arrows) and decreased (open arrows) DNase I sensitivity in the presence of ATP, is found in the 0.3 and 0.4 M NaCl fractions. A different pattern of activity seen in the 0.2 M fraction, suggestive of DNase I protection, is also indicated. (B) Superose 6-purified PYR complex is associated with DNA-dependent ATPase activity. DNA-dependent ATPase activity, as measured by percent ATP hydrolysis, is detected in all of the fractions off the Superose 6 column (Fig. 4D) except fraction 12. Two different activities elute off the column, a high-molecular-weight activity that prefers chromatin as its substrate (fractions 13 through 17) and a lower-molecular-weight activity that prefers naked DNA (fractions 18 through 22). No appreciable ATPase activity is seen in control reactions that use purified histones as the substrate. (C) Superose 6-purified PYR complex is associated with histone deacetylase activity. Histone deacetylase activity, as measured by counts per minute of released [3H]acetate, is detected in the high-molecular-weight fractions (peak activity in fractions 13 through 18).
From the Superose 6 gel filtration column, PYR complex DNA-binding activity by gel shift assay elutes in fractions close to the void volume, corresponding to a molecular size of 1 to 2 MDa (Fig. 4D, fractions 13 through 17). Western blot analysis shows that Ikaros, SWI/SNF, and NuRD subunits also elute in these fractions (Fig. 4D). Two different ATPase activities elute from the Superose 6 column. An activity that is higher in the presence of chicken erythrocyte chromatin elutes in fractions 13 through 18, corresponding to PYR complex DNA-binding activity, Ikaros, SWI/SNF, and NuRD subunits (Fig. 6B). A second activity that prefers naked DNA as a substrate peaks in lower-molecular-weight fractions (fractions 18 through 22), unrelated to the PYR complex (Fig. 6B). Histone deacetylase activity peaks in fractions 13 through 18, associated with the PYR complex (Fig. 6C). These results indicate that the PYR complex is associated with both DNA-dependent ATPase and histone deacetylase activities.
DISCUSSION
We report here the further characterization of the PYR complex, both structurally and functionally, in adult erythroid cells. Most importantly, we show that the PYR complex contains the hematopoietic cell-specific zinc finger DNA-binding protein Ikaros and that the PYR complex and recombinant Ikaros protein bind DNA with similar sequence specificity. These data indicate that Ikaros is a DNA-binding subunit of the PYR complex.
Ikaros gene function has been shown to be required for normal T- and B-cell development (12, 52). Ikaros has four N-terminal DNA-binding zinc fingers and two C-terminal zinc fingers; the C-terminal fingers permit homo- and heterodimerization of Ikaros isoforms (13, 28, 47). A number of different Ikaros isoforms have been identified, all of which are products of alternative RNA splicing, and different Ikaros isoforms are found in different hematopoietic cell lineages (23). Ikaros has recently been shown to be associated with two chromatin-remodeling complexes in lymphoid cells, a SWI/SNF-like complex and a NuRD complex containing Mi-2 (22). Ikaros has not previously been shown to be present in chromatin-remodeling complexes in erythroid cells. The complexes in lymphocytes are thought to function via the promoters and enhancers of lymphoid cell-specific genes (10, 11, 28). To our knowledge, ours is the first report to show that an Ikaros-containing chromatin-remodeling complex interacts with intergenic DNA sequences.
Ikaros in lymphocytes has been shown to occur in complexes with related lymphocyte-restricted proteins, Helios (16) and Aiolos (22, 35). It is possible that in erythroid cells Ikaros is in a complex with a similar Ikaros-like protein and/or perhaps with erythroid-specific factors such as GATA-1. GATA-1 has a strong binding site near the PYR complex-binding site between the fetal and adult β-globin-like genes (38), but it is not clear whether GATA-1 cooperatively binds with PYR complex to this region.
More recently, defective and/or deficient Ikaros protein production has been shown to cause erythroid as well as lymphoid cell defects in Ikaros-knockout mice (37). In these studies, it has been shown that Ikaros-null mice have aberrant erythroid colony activity and that other knockout mice with a dominant negative phenotype have severe anemia (12, 37, 52). Hemoglobin switching in these mice has not been investigated. Hematopoietic stem cell activity in Ikaros-knockout mice is also significantly decreased (37). These results are indicative of pleiotropic effects of Ikaros in hematopoietic cells, consistent with our data presented here and with other observations indicating the presence of high-molecular-weight Ikaros-containing complexes in megakaryocytic and granulocytic cells as well (unpublished observations).
In this paper, we have also shown that the ATPase-helicase subunit of the NuRD complex, Mi-2, is a component of the PYR complex. Mi-2 has a variety of interesting protein motifs in addition to its SNF2 family helicase-ATPase domain. These include paired chromo (chromatin organization modifier) domains, two plant homeodomain fingers, and Myb- and HMG-like motifs (56). Many chromo domain-containing proteins, such as Polycomb and heterochromatin-associated protein 1 (HP1) in Drosophila spp., function in heterochromatin-mediated gene silencing (6). Plant homeodomain fingers have been described in both positive and negative regulators of chromatin-mediated transcriptional regulation, and they are believed to be involved in protein-protein interactions (1). Mi-2 has been identified in protein complexes in human cells that have both ATP-dependent nucleosome remodeling and histone deacetylase activities, and they are thought to function in chromatin-mediated transcriptional repression (22, 25, 51, 57, 58). Our previous observations in transgenic mice (39) that deletion of the PYR complex-binding site delays fetal-to-adult globin gene switching could be explained by the targeting of a repressive complex to the region of the fetal genes late in development. This could allow the locus control region to interact with the adult β-globin genes by default, facilitating β-globin gene expression. The possibility that a complex with histone deacetylase activity might inhibit human γ-globin gene expression is also consistent with the observation that sodium butyrate, a potent inhibitor of histone deacetylase, is known to enhance γ-globin gene expression in adult erythroid cells (19, 32).
In addition to the presence of Ikaros, Mi-2, and several BAF proteins (INI1, BAF57, BAF60a, SRG3, and BAF170) in the PYR complex, we show that a number of other NuRD and SWI/SNF subunits copurify with PYR complex DNA-binding activity. These include NuRD subunits HDAC2 and RbAp46/48 and the SWI/SNF subunit BRG1, which, like Mi-2, is a SNF2 family ATPase-helicase. The gel supershift, conventional and immunoaffinity chromatography, and immunoprecipitation results are most consistent with the presence of Ikaros, SWI/SNF, and NuRD components in a single molecular complex. In particular, the supershift data and the coimmunopurification of SWI/SNF (BAF57, BAF155, and BRG1) and NuRD (Mi-2 and HDAC2) subunits from column fractions containing the PYR complex by gel shift assay support this conclusion. It is, of course, possible that, in vivo, the NuRD and SWI/SNF subunits are each associated with Ikaros in two closely related complexes, one containing SWI/SNF and the other containing NuRD subunits.
It is likely that the control of chromatin structure plays an important role in the developmental regulation of globin gene expression. A SNF2 family helicase-ATPase, ATRX, is thought to be a positive regulator of α-globin gene expression, since naturally occurring mutations in this gene in humans are associated with α-thalassemia (14). As a SNF2 homologue, it seems likely that ATRX functions as part of a complex similar to the one we describe. In addition, a SWI/SNF-like complex has previously been described in MEL cells that permits the erythroid transcription factor EKLF to activate globin gene transcription from a β-globin promoter packaged into chromatin (2). Also, a heritable state of β-globin silencing similar to Polycomb-mediated repression has been described in hybrids of MEL cells and mouse embryonic erythroblasts and suggests that chromatin-mediated gene silencing also occurs at the β-globin locus (45). Our data suggest that Ikaros may target both activator (SWI/SNF) and repressor (NuRD) complexes to the β-globin locus in adult erythroid cells at the time of the switch from fetal to adult globin production. Taken together with our previous findings in transgenic mice (39), we propose that Ikaros-targeted chromatin-remodeling complexes appear late in erythroid development and help to regulate this switch. The further biochemical characterization of the PYR complex, together with experiments studying the erythroid phenotype of Ikaros-knockout mice in more detail, should help to clarify the specific role of Ikaros in erythroid gene regulation.
ACKNOWLEDGMENTS
This work was supported by PHS grants DK25274 and HL28381 from the National Institutes of Health, a grant from the Ahepa Cooley's Anemia Foundation, and NCI grant P30 CA08748. D.W.O. was supported by NIH Clinical Investigator Award DK02260.
We thank the National Cell Culture Center for large-scale MEL cell cultures; Eugene Leung for help with the large-scale preparation of MEL nuclear extracts; Lynne Lacomis, Mary Lui, Anita Grewal, and Scott Geromanos for help with MS analysis; Matthias Mann for the PeptideSearch and SequenceTag programs; Christina Starke and Jason Garyu for technical assistance; Una Terrie Collins for assistance in the preparation of the manuscript; and Stephen Smale, Bradley Cobb, Gerald Crabtree, Katia Georgopoulos, Ganjam Kalpana, and Michael Bustin for generously supplying antibodies.
REFERENCES
- 1.Aasland R, Gibson T J, Stewart A F. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong J A, Bieker J J, Emerson B M. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell. 1998;95:93–104. doi: 10.1016/s0092-8674(00)81785-7. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong J A, Emerson B M. Transcription of chromatin: these are complex times. Curr Opin Genet Dev. 1998;8:165–172. doi: 10.1016/s0959-437x(98)80137-8. [DOI] [PubMed] [Google Scholar]
- 4.Cairns B R, Erdjument-Bromage H, Tempst P, Winston F, Kornberg R D. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 1998;2:639–651. doi: 10.1016/s1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 5.Carmen A A, Rundlett S E, Grunstein M. HDA1 and HDA3 are components of a yeast histone deacetylase (HDA) complex. J Biol Chem. 1996;271:15837–15844. doi: 10.1074/jbc.271.26.15837. [DOI] [PubMed] [Google Scholar]
- 6.Cavalli G, Paro R. Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Curr Opin Cell Biol. 1998;10:354–360. doi: 10.1016/s0955-0674(98)80011-2. [DOI] [PubMed] [Google Scholar]
- 7.Cote J, Peterson C L, Workman J L. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc Natl Acad Sci USA. 1998;95:4947–4952. doi: 10.1073/pnas.95.9.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 9.Erdjument-Bromage H, Liu M, Lacomis L, Grewal A, Annan R S, McNulty D E, Carr S A, Tempst P. Examination of micro-tip reversed-phase liquid chromatographic extraction of peptide pools for mass spectrometric analysis. J Chromatogr A. 1998;826:167–181. doi: 10.1016/s0021-9673(98)00705-5. [DOI] [PubMed] [Google Scholar]
- 10.Ernst P, Hahm K, Smale S T. Both Lyf-1 and an Ets protein interact with a critical promoter element in the murine terminal transferase gene. Mol Cell Biol. 1993;13:2982–2992. doi: 10.1128/mcb.13.5.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst P, Hahm K, Trinh L, Davis J N, Roussel M F, Turck C W, Smale S T. A potential role for Elf-1 in terminal transferase gene regulation. Mol Cell Biol. 1996;16:6121–6131. doi: 10.1128/mcb.16.11.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgopoulos K, Bigby M, Wang J H, Molnar A, Wu P, Winandy S, Sharpe A. The Ikaros gene is required for the development of all lymphoid lineages. Cell. 1994;79:143–156. doi: 10.1016/0092-8674(94)90407-3. [DOI] [PubMed] [Google Scholar]
- 13.Georgopoulos K, Moore D D, Derfler B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science. 1992;258:808–812. doi: 10.1126/science.1439790. [DOI] [PubMed] [Google Scholar]
- 14.Gibbons R J, Picketts D J, Villard L, Higgs D R. Mutations in a putative global transcriptional regulator cause X-linked mental retardation with alpha-thalassemia (ATR-X syndrome) Cell. 1995;80:837–845. doi: 10.1016/0092-8674(95)90287-2. [DOI] [PubMed] [Google Scholar]
- 15.Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- 16.Hahm K, Cobb B S, McCarty A S, Brown K E, Klug C A, Lee R, Akashi K, Weissman I L, Fisher A G, Smale S T. Helios, a T cell-restricted Ikaros family member that quantitatively associates with Ikaros at centromeric heterochromatin. Genes Dev. 1998;12:782–796. doi: 10.1101/gad.12.6.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hahm K, Ernst P, Lo K, Kim G S, Turck C, Smale S T. The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol. 1994;14:7111–7123. doi: 10.1128/mcb.14.11.7111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harlow E, Lane D. Using antibodies: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 19.Ishiguro K, Sartorelli A C. Coinduction of embryonic and adult-type globin mRNAs by sodium butyrate and trichostatin A in two murine interleukin-3-dependent bone marrow-derived cell lines. Blood. 1998;92:4383–4393. [PubMed] [Google Scholar]
- 20.Kadonaga J. Eukaryotic transcription: an interlaced network of transcription factors and chromatin-modifying machines. Cell. 1998;92:307–313. doi: 10.1016/s0092-8674(00)80924-1. [DOI] [PubMed] [Google Scholar]
- 21.Khavari P A, Peterson C L, Tamkun J W, Mendel D B, Crabtree G R. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 22.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 23.Klug C A, Morrison S J, Masek M, Hahm K, Smale S T, Weissman I L. Hematopoietic stem cells and lymphoid progenitors express different Ikaros isoforms, and Ikaros is localized to heterochromatin in immature lymphocytes. Proc Natl Acad Sci USA. 1998;95:657–662. doi: 10.1073/pnas.95.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knoepfler P S, Eisenman R N. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- 25.Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 27.Laurent B C, Treich I, Carlson M. The yeast SNF2/SWI2 protein has DNA-stimulated ATPase activity required for transcriptional activation. Genes Dev. 1993;7:583–591. doi: 10.1101/gad.7.4.583. [DOI] [PubMed] [Google Scholar]
- 28.Lo K, Landau N R, Smale S T. LyF-1, a transcriptional regulator that interacts with a novel class of promoters for lymphocyte-specific genes. Mol Cell Biol. 1991;11:5229–5243. doi: 10.1128/mcb.11.10.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lui M, Tempst P, Erdjument-Bromage H. Methodical analysis of protein-nitrocellulose interactions to design a refined digestion protocol. Anal Biochem. 1996;241:156–166. doi: 10.1006/abio.1996.0393. [DOI] [PubMed] [Google Scholar]
- 30.Mann M, Hojrup P, Roepstorff P. Use of mass spectrometric molecular weight information to identify proteins in sequence databases. Biol Mass Spectrom. 1993;22:338–345. doi: 10.1002/bms.1200220605. [DOI] [PubMed] [Google Scholar]
- 31.Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66:4390–4399. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 32.McCaffrey P G, Newsome D A, Fibach E, Yoshida M, Su M S. Induction of gamma-globin by histone deacetylase inhibitors. Blood. 1997;90:2075–2083. [PubMed] [Google Scholar]
- 33.Molnar A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14:8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molnar A, Wu P, Largespada D A, Vortkamp A, Scherer S, Copeland N G, Jenkins N A, Bruns G, Georgopoulos K. The Ikaros gene encodes a family of lymphocyte-restricted zinc finger DNA binding proteins, highly conserved in human and mouse. J Immunol. 1996;156:585–592. [PubMed] [Google Scholar]
- 35.Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, Wu P, Neben S, Georgopoulos K. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nichogiannopoulou A, Trevisan M, Neben S, Friedrich C, Georgopoulos K. Defects in hemopoietic stem cell activity in Ikaros mutant mice. J Exp Med. 1999;190:1201–1214. doi: 10.1084/jem.190.9.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Neill D, Bornschlegel K, Flamm M, Castle M, Bank A. A DNA-binding factor in adult hematopoietic cells interacts with a pyrimidine-rich domain upstream from the human delta-globin gene. Proc Natl Acad Sci USA. 1991;88:8953–8957. doi: 10.1073/pnas.88.20.8953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Neill D, Yang J, Erdjument-Bromage H, Bornschlegel K, Tempst P, Bank A. Tissue-specific and developmental stage-specific DNA binding by a mammalian SWI/SNF complex associated with human fetal-to-adult globin gene switching. Proc Natl Acad Sci USA. 1999;96:349–354. doi: 10.1073/pnas.96.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peterson C L, Zhao Y, Chait B T. Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J Biol Chem. 1998;273:23641–23644. doi: 10.1074/jbc.273.37.23641. [DOI] [PubMed] [Google Scholar]
- 41.Pollard K J, Peterson C L. Chromatin remodeling: a marriage between two families? BioEssays. 1998;20:771–780. doi: 10.1002/(SICI)1521-1878(199809)20:9<771::AID-BIES10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 42.Quinn J, Fyrberg A M, Ganster R W, Schmidt M C, Peterson C L. DNA-binding properties of the yeast SWI/SNF complex. Nature. 1996;379:844–847. doi: 10.1038/379844a0. [DOI] [PubMed] [Google Scholar]
- 43.Schreiber E, Matthias P, Muller M M, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts,’ prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seelig H P, Moosbrugger I, Ehrfeld H, Fink T, Renz M, Genth E. The major dermatomyositis-specific Mi-2 autoantigen is a presumed helicase involved in transcriptional activation. Arth Rheum. 1995;38:1389–1399. doi: 10.1002/art.1780381006. [DOI] [PubMed] [Google Scholar]
- 45.Stanworth S J, Roberts N A, Sharpe J A, Sloane-Stanley J A, Wood W G. Established epigenetic modifications determine the expression of developmentally regulated globin genes in somatic cell hybrids. Mol Cell Biol. 1995;15:3969–3978. doi: 10.1128/mcb.15.8.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steger D J, Utley R T, Grant P A, John S, Eberharter A, Cote J, Owen-Hughes T, Ikeda K, Workman J L. Regulation of transcription by multisubunit complexes that alter nucleosome structure. Cold Spring Harb Symp Quant Biol. 1998;63:483–491. doi: 10.1101/sqb.1998.63.483. [DOI] [PubMed] [Google Scholar]
- 47.Sun L, Liu A, Georgopoulos K. Zinc finger-mediated protein interactions modulate Ikaros activity, a molecular control of lymphocyte development. EMBO J. 1996;15:5358–5369. [PMC free article] [PubMed] [Google Scholar]
- 48.Thanos D, Maniatis T. The high mobility group protein HMG I(Y) is required for NF-kappa B-dependent virus induction of the human IFN-beta gene. Cell. 1992;71:777–789. doi: 10.1016/0092-8674(92)90554-p. [DOI] [PubMed] [Google Scholar]
- 49.Thomas J O. Isolation and fractionation of chromatin and linker histones. In: Gould H, editor. Chromatin: a practical approach. New York, N.Y: Oxford University Press; 1998. pp. 1–34. [Google Scholar]
- 50.Utley R T, Owen-Hughes T A, Juan L J, Cote J, Adams C C, Workman J L. In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Methods Enzymol. 1996;274:276–291. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 51.Wade P A, Jones P L, Vermaak D, Wolffe A P. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- 52.Wang J H, Nichogiannopoulou A, Wu L, Sun L, Sharpe A H, Bigby M, Georgopoulos K. Selective defects in the development of the fetal and adult lymphoid system in mice with an Ikaros null mutation. Immunity. 1996;5:537–549. doi: 10.1016/s1074-7613(00)80269-1. [DOI] [PubMed] [Google Scholar]
- 53.Wang W, Chi T, Xue Y, Zhou S, Kuo A, Crabtree G R. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci USA. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W, Cote J, Xue Y, Zhou S, Khavari P A, Biggar S R, Muchardt C, Kalpana G V, Goff S P, Yaniv M, Workman J L, Crabtree G R. Purification and biochemical heterogeneity of the mammalian SWI/SNF complex. EMBO J. 1996;15:5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 55.Wang W, Xue Y, Zhou S, Kuo A, Cairns B R, Crabtree G R. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 56.Woodage T, Basrai M A, Baxevanis A D, Hieter P, Collins F S. Characterization of the CHD family of proteins. Proc Natl Acad Sci USA. 1997;94:11472–11477. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xue Y, Wong J, Moreno G T, Young M K, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, LeRoy G, Seelig H P, Lane W S, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- 59.Zhao K, Wang W, Rando O J, Xue Y, Swiderek K, Kuo A, Crabtree G R. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]