Abstract
Background
The online Tuberculin Skin Test/Interferon Gamma Release Assay (TST/IGRA) Interpreter V3.0 (TSTin3D), a tool for estimating the risk of active tuberculosis (TB) in individuals with latent TB infection (LTBI), has been in use for more than a decade, but its predictive performance has never been evaluated.
Methods
People with a positive TST or IGRA result from 1985 to 2015 were identified using a health data linkage that involved migrants to British Columbia, Canada. Comorbid conditions at the time of LTBI testing were identified from physician claims, hospitalizations, vital statistics, outpatient prescriptions, and kidney and HIV databases. The risk of developing active TB within 2 and 5 years was estimated using TSTin3D. The discrimination and calibration of these estimates were evaluated.
Results
A total of 37 163 individuals met study inclusion criteria; 10.4% were tested by IGRA. Generally, the TSTin3D algorithm assigned higher risks to demographic and clinical groups known to have higher active TB risks. Concordance estimates ranged from 0.66 to 0.68 in 2- and 5-year time frames. Comparing predicted to observed counts suggests that TSTin3D overestimates active TB risks and that overestimation increases over time (with relative bias of 3% and 12% in 2- and 5-year periods, respectively). Calibration plots also suggest that overestimation increases toward the upper end of the risk spectrum.
Conclusions
TSTin3D can discriminate adequately between people who developed and did not develop active TB in this linked database of migrants with predominately positive skin tests. Further work is needed to improve TSTin3D’s calibration.
Keywords: latent tuberculosis infection, TSTin3D, public health, epidemiology, validation
This external validation study that used linked administrative data found that the online Tuberculin Skin Test/Interferon Gamma Release Assay Interpreter V3.0 (TSTin3D) can distinguish adequately who among immigrants with latent tuberculosis (TB) infection will develop active TB. However, further work is needed to improve TSTin3D’s calibration in some high-risk populations.
Tuberculosis (TB) remains the leading single cause of infectious disease death globally [1]. In response to this persisting global health emergency, the World Health Organization developed the EndTB Strategy, which aims to reduce TB incidence by 90% from 2015 levels by 2035 [2]. In the absence of new technologies (ie, effective vaccination), latent TB infection (LTBI) testing and treatment in specific high-risk subpopulations will remain a core component of the EndTB strategy and will be required to hasten TB elimination in regions with low TB incidence [3, 4].
The online Tuberculin Skin Test (TST)/Interferon Gamma Release Assay (IGRA) Interpreter V3.0 (TSTin3D) is a web-based tool used for estimating the risk of developing active TB in individuals with a TST reaction of ≥5 mm and/or IGRA test results. This online tool, available at http://www.tstin3d.com, incorporates demographic, medical, radiographic, and exposure risk factors in the interpretation of positive TST or IGRA. The underlying algorithm was developed using published estimates of the impact of various risk factors on TB risk, combined with prevalence estimates of LTBI and the likelihood of a false-positive TST. It generates estimates of the annual and cumulative risk of active TB in adults with a positive TST/IGRA test result [5].
The tool was first introduced in 2008 [5] and is continually being updated with new features, such as the inclusion of IGRA and TST 5–9 mm in the latter versions, as data become available. A full list of references is available on the website, which currently has approximately 90 000 visits from 40 000 unique visitors per year. Presumably, a substantial portion of the website visits inform individual clinical decisions to initiate LTBI therapy. Also, TSTin3D is referenced in Canadian TB Guidelines as a tool to assist in TST and IGRA interpretation [6]. To date, however, TSTin3D’s predictive performance has not been formally evaluated.
In this study, we conducted an external validation of the most recent version of the TSTin3D algorithm using a population-based health administrative data linkage to determine the tool’s discrimination and calibration. Discrimination is a measure of how well a risk prediction tool can separate individuals who developed or did not develop the outcome. The concordance and D statistics are common measures of discrimination in time-to-event data [7]. Calibration, on the other hand, indicates the agreement between the risks generated by a risk prediction tool and the observed risks. Calibration can be examined quantitatively using measures such as the expected/observed (E/O) statistic or graphically by plotting observed vs predicted risks [7].
METHODS
Data Sources
We used linked data from the Immigration, Refugees, and Citizenship Canada and Population Data BC, a multiuniversity data resource based in British Columbia (BC), Canada [8]. The linked data include individual client-level, anonymized data on immigrants registered in the provincial health insurance plan of BC and include demographics, immigration information, deaths, physician visits, hospitalizations, outpatient prescription drug dispensations, and human immunodeficiency virus (HIV), kidney disease, and cancer diagnosis obtained from provincial disease registries. The data are also linked to the provincial TB Services registry, which includes TB contact status, TB screening and testing, active TB diagnosis, and active and latent TB treatment information. Detailed descriptions of the databases and linkage methods can be found in previous publications [9–11].
Study Cohort
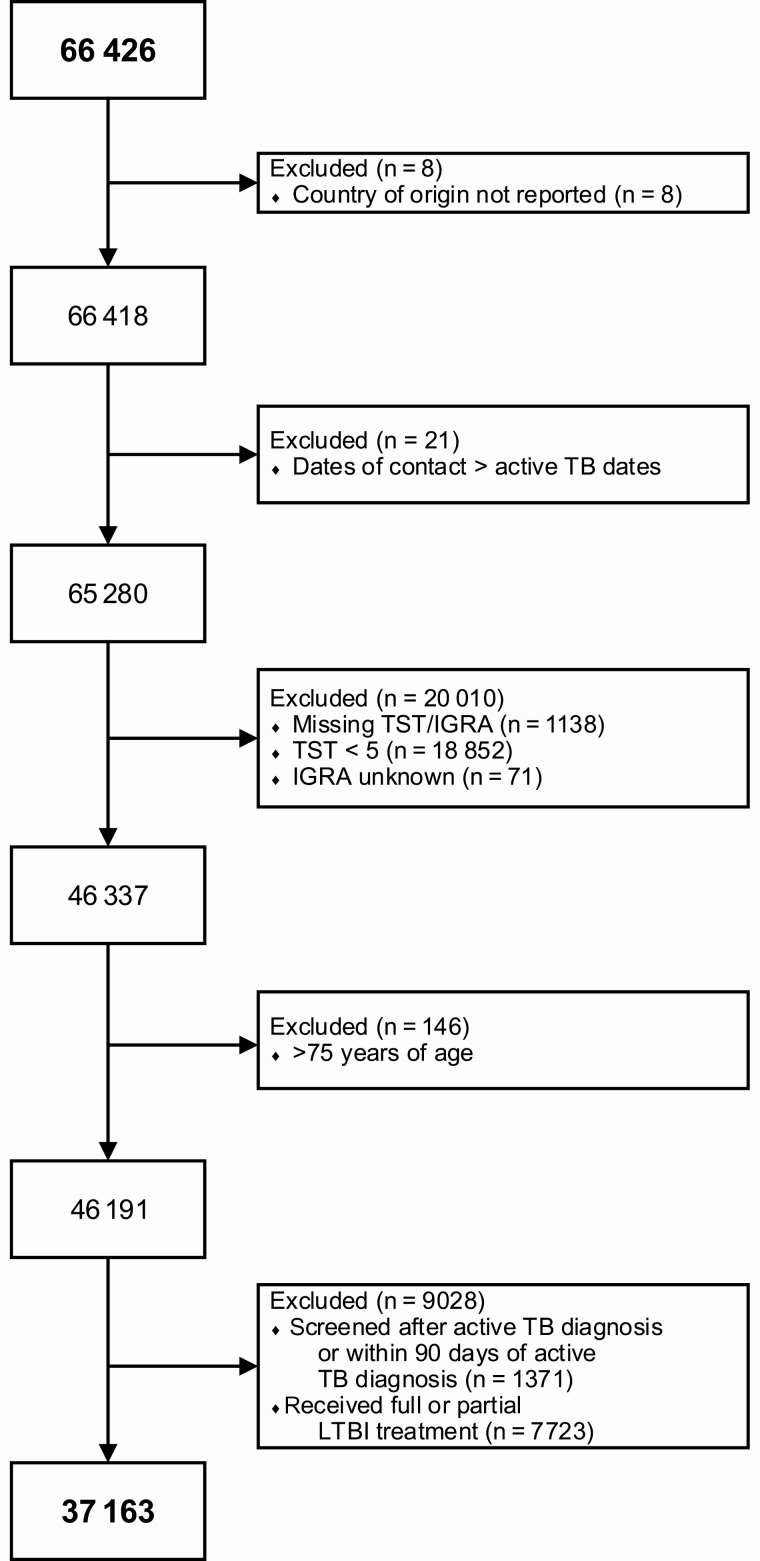
We included all individuals who established residency in and were covered by the mandatory provincial health insurance of BC at any point between 1 January 1985 and 31 December 2013 with a TST or IGRA result recorded in the provincial TB registry (n = 66 426). Individuals were excluded from the cohort if they had an unknown birth country (n = 8), missing or unknown TST/IGRA result (n = 1209), conflicting dates of TB contact (n = 21), TST induration <5 mm (n = 18 852), received LTBI treatment or TST/IGRA screening as part of an active TB diagnostic workup (defined as active TB diagnosis within 90 days of TST or IGRA testing; 9028), or were aged >75 years (TSTin3D was designed to be used for people aged <80 years; n = 146). BC does not perform systematic LTBI screening in migrants post-landing, so only a small proportion of high-risk individuals would have been referred and tested for LTBI postlanding [12]. Figure 1 shows the STrengthening the Reporting of OBservational studies in Epidemiology flow diagram for the 37 163 individuals who met study criteria.
Figure 1.
Cohort construction. Abbreviations: IGRA, interferon gamma release assay; LTBI, latent tuberculosis infection; TB, tuberculosis; TST, tuberculin skin test.
For the purposes of these analyses, time zero represents each individual’s index date, defined as the date of first TST or IGRA test. Follow-up was censored at any of the following times: active TB diagnosis, end of provincial health insurance coverage, death, or end of the study period (31 December 2015). A small proportion of the cohort (10.4%) was tested by IGRA, with 16.5% testing positive (Table 1). The TSTin3D algorithm does not account for TST size in the context of IGRA positive results, while with IGRA negative results, prior TST size was accounted for in analysis, consistent with TSTin3D algorithms.
Table 1.
Characteristics of the Study Cohort
| Characteristic | Counts | % | Developed Active TB | ||
|---|---|---|---|---|---|
| Total | 2 Years | 5 Years | |||
| All | 37 163 | 100 | 329 | 126 | 198 |
| Age at migration, y | |||||
| ≤14 | 4334 | 11.7 | 15 | 8 | 9 |
| 15–34 | 19 510 | 52.5 | 132 | 64 | 94 |
| 35–75 | 13 319 | 35.8 | 156 | 54 | 95 |
| Country of birth TB incidence (per 100 000) | |||||
| 0–9 | 455 | 1.2 | 2 | 1 | 2 |
| 10–29 | 2612 | 7.0 | 4 | 2 | 4 |
| 30–49 | 2273 | 6.1 | 5 | 4 | 5 |
| 50–99 | 6123 | 16.5 | 19 | 13 | 15 |
| 100–199 | 9805 | 26.4 | 87 | 36 | 56 |
| 200+ | 15 877 | 42.7 | 186 | 70 | 116 |
| Sex | |||||
| Male | 13 379 | 36.0 | 142 | 53 | 89 |
| Female | 23 784 | 64.0 | 161 | 73 | 109 |
| Test resultsa | |||||
| TST 5–9 mm | 4860 | 13.08 | 13 | 4 | 7 |
| TST 10–14 mm | 14 801 | 39.83 | 83 | 31 | 50 |
| TST 15+mm | 15 437 | 41.54 | 197 | 84 | 132 |
| IGRA negative | 3214 | 8.6 | 2 | 1 | 2 |
| IGRA positive | 636 | 1.7 | 9 | 6 | 8 |
| BCGb | |||||
| Never vaccinated | 15 054 | 40.5 | 184 | 56 | 105 |
| Vaccinated age <2 years | 21 188 | 57.0 | 117 | 69 | 91 |
| Vaccinated age ≥2 years | 921 | 2.5 | 2 | 1 | 2 |
| Recent contact | |||||
| None | 32 001 | 86.1 | 253 | 104 | 168 |
| Close | 1983 | 5.3 | 29 | 12 | 17 |
| Casual | 3179 | 8.6 | 21 | 10 | 13 |
| Recent TST conversion | |||||
| No | 36 357 | 97.8 | 303 | 126 | 198 |
| Yes | 806 | 2.2 | 0 | 0 | 0 |
| Carcinoma of the head and neck | |||||
| No | 37 145 | 99.9 | 303 | 126 | 198 |
| Yes | 18 | 0.1 | 0 | 0 | 0 |
| Dialysis | |||||
| No | 36 962 | 99.5 | 300 | 124 | 196 |
| Yes | 201 | 0.5 | 3 | 2 | 2 |
| Diabetes | |||||
| No | 35 134 | 94.5 | 281 | 115 | 182 |
| Yes | 2029 | 5.5 | 22 | 11 | 16 |
| Human immunodeficiency virus | |||||
| No | 37 109 | 99.9 | 300 | 124 | 195 |
| Yes | 54 | 0.2 | 3 | 2 | 3 |
| Transplantation | |||||
| No | 37 137 | 99.9 | 303 | 126 | 198 |
| Yes | 26 | 0.1 | 0 | 0 | 0 |
| Treated with glucocorticoids | |||||
| No | 36 743 | 98.9 | 298 | 124 | 196 |
| Yes | 420 | 1.1 | 5 | 2 | 2 |
| Treated with tumor necrosis factor | |||||
| No | 37 143 | 99.9 | 301 | 125 | 196 |
| Yes | 20 | 0.1 | 2 | 1 | 2 |
Abbreviations: IGRA, interferon gamma release assay; TB, tuberculosis; TST, tuberculin skin test.
aSum of counts will not be equal to the overall count (n = 36 173) as 44.5% of individuals with negative IGRA results had no previous TST results, while 8.0%, 28.1%, and 19.4% had previous TST results of 5–9 mm, 10–14 mm, and 15+ mm, respectively. TST data from those with positive IGRA results were not considered, consistent with the TSTin3D’s algorithm.
bBCG vaccination for all individuals was inferred from vaccination policies in the country and year of birth, as reported in the BCG World Atlas [13].
Variables
Our primary outcome was diagnosis of active TB, which included pulmonary or extrapulmonary TB, confirmed microbiologically or clinically. The primary analysis variables were the 2- and 5-year absolute risk estimates generated by an offline version of the TSTin3D algorithm developed based on the source code supplied by the TSTin3D developers [5].
The calculation of the 2- and 5-year risk estimates based on the TSTin3D algorithm differs slightly for different risk groups. In those with a history of close contact and/or recent TST conversion, TSTin3D calculates a 2-year risk and an annual risk that is assumed to be stable after the initial 2-year period. The 5-year risk in this group was calculated by multiplying the annual risk by 3 and adding this to the initial 2-year risk estimate. In individuals with no history of close contact and/or recent TST conversion, the TSTin3D algorithm calculates a stable annual risk. Hence, the 2- and 5-year risks for this latter group were calculated by multiplying the annual risk estimate by 2 and 5, respectively. These risk calculations are consistent with the method used by the TSTin3D to estimate the cumulative risk of active TB up to the age of 80, which assumes that cumulative risk increases by a constant rate each year (or after the first 2 years in those with close contacts and recent TST conversion histories) [5].
The TSTin3D method for estimating the 2-year and annual risks of developing active TB have previously been described in detail [5]. Essentially, the values were generated by first calculating the probability of LTBI based on the incidence of smear-positive TB in a person’s birth country, their age at immigration, and their contact history. The estimated probability of LTBI was then combined with the false-positive rate associated with BCG vaccination and skin test sensitivity to nontuberculous mycobacteria, which vary by place of birth, to calculate the positive predictive value of TST and IGRA tests. To calculate the risk of active TB, the positive predictive value of a test is combined with risk estimates for active TB based on contact history, recent TST conversion (defined as any TST ≥10 mm with a previously recorded TST <5 mm), and the presence of various comorbid conditions (eg, diagnosis of head and neck cancer, chronic kidney disease requiring dialysis, diabetes, HIV) [5].
We had data for all the variables the TSTin3D algorithm required to generate risk estimates except diagnosis of AIDS, granuloma and fibronodular disease on chest X ray, infection at a young age, smoking status, and underweight status. Similar to what would be done when using the tool without this information, these conditions were assumed to be absent for everyone. BCG vaccination status was imputed based on published records of vaccination policies that were in place in an individual’s birth country at the time of birth [13].
Statistical Analyses
To evaluate TSTin3D’s discrimination and calibration, we fitted 2 separate Cox regression models that predict time to diagnosis of active TB using the 2-year and the 5-year TSTin3D risk estimates as predictors [14, 15]. As measures of discrimination, we used Somers’ D (−1 to +1) [16] and Uno’s C (0 to 1) [17]. Somers’ D values of 0.30 or greater (or −0.30 or less) [18] and Uno’s C equal to or greater than 0.64 are regarded as moderate [19].
To determine TSTin3D’s calibration, we calculated the ratio of the expected number of people with active TB (by the TSTin3D model) to the observed or actual number of people who developed active TB in the analytic cohort (expected/observed, or E/O) for a given time period. We examined E/O for the entire cohort and by subgroups [7]. We also assessed calibration graphically by plotting TSTin3D’s risk predictions against observed risks in the data linkage using a model-based calibration approach [15, 16]. All statistical analyses were performed using R (V.3.6.1), the R survival package version 2.44-1.1 [20], and SAS/STAT (V.9.4, SAS Institute, Cary, NC).
RESULTS
The analytic cohort (Figure 1) included 37 163 individuals, 329 of whom developed active TB over a 30-year period, representing an incidence rate of 80 per 100 000 person-years. The median follow-up time was 8.3 years. TB incidence rate was 179.3 per 100 000 person-years (with 126 active TB cases) in the first 2 years from the index date and 126.2 per 100 000 person-years (with 198 active TB cases) in the first 5 years from the index date.
The cohort consisted predominantly of females (N = 23 784, 64%); people migrating to Canada at ages 15–34 years (N = 19 510, 52.5%); people migrating from birth countries with TB incidence ≥100 per 100 000 population (N = 25 682, 69.1%); people with TST indurations ≥10 mm (N = 21 711, 77.3%); and people from countries with vaccination policy for children aged <2 years (N = 21 188, 57.0%; Table 1). More than 5% were close contacts and 2% had recent TST conversions. Except for diabetes (N = 2029, 5.5%) and steroid use (N = 420, 1.1%), each comorbid medical condition was present in <1% of the cohort.
Distribution of the TSTin3D Cumulative Risk
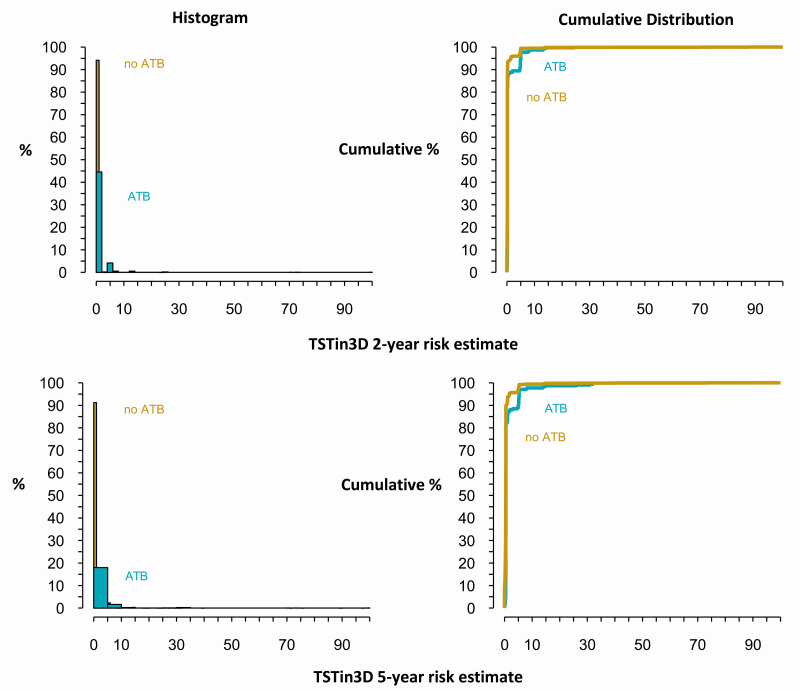
The distribution of 2- and 5-year cumulative risks generated by the TSTin3D algorithm is skewed to the right, with most risk estimates falling between 0% and 10% (Figure 2, histogram). Overall, there was a greater concentration of people with higher TSTin3D risk estimates in those who developed active TB compared with those who did not develop active TB (Figure 2).
Figure 2.
Distribution of the TSTin3D 2- and 5-year risk estimates. The histogram shows that the majority of individuals (>90%) who did not develop ATB within 2 or 5 years had lower TSTin3D risk estimates (mostly zero). On the other hand, many of those who developed ATB had nonzero risk estimates. In people who developed ATB, those in the 90th percentile of TSTin3D risk had calculated 2-year and 5-year ATB risks of 4.7 and 5.2, respectively (cumulative distribution). In contrast, of people who did not have ATB, those in the 90th percentile of TSTin3D risk had calculated 2-year and 5-year risks of 0.23 and 0.86, respectively (cumulative distribution). Abbreviations: ATB, active tuberculosis; TSTin3D, Tuberculin Skin Test/Interferon Gamma Release Assay Interpreter V3.0.
On average, TSTin3D generated higher risk estimates in groups known to have higher rates of active TB (Table 2). For example, the 5-year risk of active TB calculated by TSTin3D in people aged 35–75 years was 0.96% (95% confidence interval [CI], .92%–1.00%) compared with 0.56% (95% CI, .53%–.59%) in people aged ≤14 years. Similarly, people from birth countries with a TB incidence of >200 per 100 000 population were calculated by TSTin3D to have a 5-year risk of 0.96% (95% CI, .92%–1.00%), whereas people born in countries with TB incidence <10 per 100 000 had a 5-year risk of 0.29% (95% CI, .28%–.30%). TSTin3D also correctly assigned higher TB risks to males (5-year risk of 0.97%) compared with females (5-year risk of 0.80%), despite sex not being explicitly used by the algorithm as a factor to estimate risk.
Table 2.
Average Tuberculin Skin Test/Interferon Gamma Release Assay Interpreter V3.0–Generated Cumulative Risk by Cohort Characteristics
| Characteristic | Cumulative Risk, % | |||
|---|---|---|---|---|
| 2 Year | 95% CI | 5 Year | 95% CI | |
| Age at immigration, y | ||||
| ≤14 | 0.51 | .40–.61 | 0.70 | .60–.80 |
| 15–34 | 0.53 | .49–.58 | 0.82 | .78–.87 |
| 35–75 | 0.70 | .64–.77 | 1.05 | .99–1.12 |
| Tuberculosis incidence in country of birth (per 100 000) | ||||
| 0–9 | 0.19 | .11–.27 | 0.40 | .25–.56 |
| 10–29 | 0.25 | .17–.34 | 0.45 | .37–.54 |
| 30–49 | 0.27 | .20–.34 | 0.51 | .43–.58 |
| 50–99 | 0.44 | .36–.52 | 0.69 | .61–.77 |
| 100–199 | 0.63 | .57–.70 | 0.94 | .87–1.01 |
| 200+ | 0.73 | .67–.80 | 1.08 | 1.01–1.14 |
| Sex | ||||
| Male | 0.66 | .60–.72 | 0.98 | .92–1.05 |
| Female | 0.55 | .51–.60 | 0.84 | .80–.88 |
| Test results | ||||
| TST 5–9 mm | 0.67 | .58–.76 | 0.95 | .85–1.05 |
| TST 10–14 mm | 0.61 | .55–.67 | 0.90 | .84–.96 |
| TST 15+ mm | 0.55 | .50–.60 | 0.89 | .84–.94 |
| IGRA negative | 0.91 | .74–1.07 | 1.14 | .97–1.31 |
| IGRA positive | 1.78 | 1.23–2.32 | 2.31 | 1.74–2.87 |
| BCG | ||||
| Never vaccinated | 0.65 | .59–.70 | 1.01 | .95–1.07 |
| Vaccinated age <2 years | 0.57 | .52–.61 | 0.84 | .79–.88 |
| Vaccinated age ≥2 years | 0.15 | .10–.20 | 0.26 | .20–.32 |
| Recent contact | ||||
| None | 0.19 | .19–.20 | 0.49 | .48–.49 |
| Close | 7.57 | 7.00–8.14 | 7.92 | 7.34–8.50 |
| Casual | 0.23 | .22–.25 | 0.60 | .55–.64 |
| Recent TST conversion | ||||
| No | 0.49 | .47–.51 | 0.79 | .76–.82 |
| Yes | 5.11 | 3.98–6.25 | 5.46 | 4.33–6.60 |
| Carcinoma of the head and neck | ||||
| No | 0.59 | .56–.62 | 0.89 | .85–.93 |
| Yes | 1.26 | .96–1.56 | 5.05 | 3.84–6.25 |
| Dialysis | ||||
| No | 0.56 | .53–.59 | 0.84 | .81–.87 |
| Yes | 6.78 | 3.72–9.84 | 9.84 | 6.74–12.95 |
| Diabetes | ||||
| No | 0.52 | .48–.55 | 0.79 | .75–.82 |
| Yes | 1.88 | 1.57–2.19 | 2.68 | 2.36–3.00 |
| Human immunodeficiency virus | ||||
| No | 0.58 | .54–.61 | 0.87 | .83–.90 |
| Yes | 8.85 | 2.68–15.01 | 18.64 | 11.98–25.31 |
| Transplantation | ||||
| No | 0.59 | .55–.62 | 0.89 | .85–.92 |
| Yes | 1.33 | .42–2.23 | 5.17 | 1.66–8.69 |
| Treated with glucocorticoids | ||||
| No | 0.57 | .54–.61 | 0.87 | .83–.90 |
| Yes | 2.02 | 1.29–2.74 | 2.92 | 2.15–3.68 |
| Treated with tumor necrosis factor | ||||
| No | 0.59 | .55–.62 | 0.89 | .85–.93 |
| Yes | 1.53 | .00–4.26 | 2.28 | .00–5.14 |
The numbers represent the average cumulative risk (in percentage units) of developing active tuberculosis (TB) within 2 and 5 years in each demographic/clinical group, as calculated by TSTin3D. For example, an individual with human immunodeficiency virus, on average, has an 8.71% and 18.9% risk of developing active TB within 2 and 5 years, respectively.
Abbreviations: CI, confidence interval; IGRA, interferon gamma release assay; TST, tuberculin skin test.
Discrimination of TSTin3D
Overall, TSTin3D’s discrimination was moderate and tended to be slightly higher over longer time frames (Table 3). For 2-year cumulative risks, Somers’ D and Uno’s C were 0.31 (95% CI, .28–.41) and 0.66 (95% CI, .62–.70), respectively, whereas for the 5-year cumulative risks, the same indices were 0.34 (95% CI, .23–.40) and 0.68 (95% CI, .65–.71).
Table 3.
Performance Indices of the Tuberculin Skin Test/Interferon Gamma Release Assay Interpreter V3.0 Algorithm
| Performance Index | 2 Year | 95% CI | 5 Year | 95% CI |
|---|---|---|---|---|
| Somers’ D | 0.27 | .25–.38 | 0.32 | .18–.36 |
| Uno’s C | 0.64 | .60–.68 | 0.67 | .63–.70 |
| Harrel’s C | 0.64 | .59–.68 | 0.66 | .62–.69 |
| Ratio of expected to observed | 1.03 | 1.02–1.03 | 1.12 | 1.10–1.14 |
Uno’s C is an estimate of concordance that is weighted by the censoring distribution. Harrel’s C is the regular concordance statistic and can be derived from Somers’ D using the formula: D/2 + .5. Higher values of Somers’ D and the concordance statistics indicate better performance (TSTin3D risk estimates are associated with higher predicted risks). E/O (ratio of expected to observed) values that are closer to 1.0 are better, suggesting that the expected number of people who will develop active tuberculosis (TB) is equal to the actual number of people who developed active TB.
Abbreviations: CI, confidence interval.
Calibration of TSTin3D
When the TSTin3D risk calculations were used as a predictor in a Cox regression for time to diagnosis of active TB, the raw estimates generated by TSTin3D were substantially higher than the risks modeled from the cohort (Supplementary Figure 1). For example, in the 3472 people with a TSTin3D risk ≥1%, the mean TSTin3D 5-year raw estimate was 5.27%, whereas the mean 5-year risk modeled from the data was 0.71% (Supplementary Table 1). Meanwhile, in the 244 people with a TSTin3D predicted risk ≥10%, the mean 5-year risk generated by TSTin3D was 31.6% compared with the modeled 5-year mean risk of 1.8%.
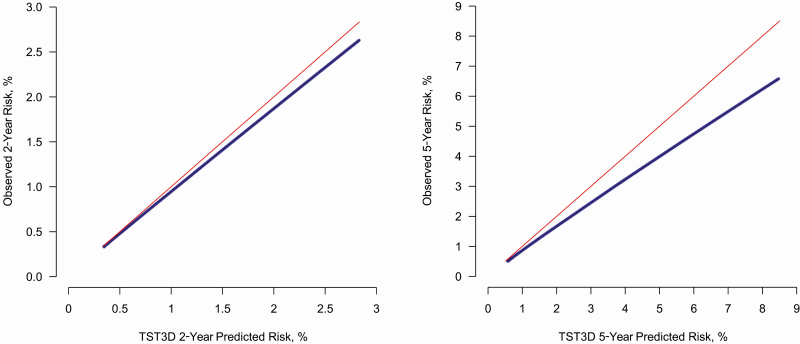
Calibration as measured by E/O suggests that TSTin3D was better or more accurate in the shorter time frame, overpredicting risks by 3.4% (E/O = 1.03, 95% CI, 1.03–1.04) in a 2-year period vs 12.0% (E/O = 1.12, 95% CI, 1.10–1.14) in a 5-year period (Table 3). Calibration plots also indicate that the discrepancy between TSTin3D-predicted and observed risks increased toward the upper end of the risk spectrum (Figure 3).
Figure 3.
Calibration plot. These plots represent the correspondence between the risk predictions generated by the TSTin3D and the observed risks. The data points were generated by regressing the observed risk on the predicted risks following the previously described approach. The upper diagonal line represents perfect calibration. Abbreviation: TSTin3D, Tuberculin Skin Test/Interferon Gamma Release Assay Interpreter V3.0.
TSTin3D’s Calibration by Demographic and Clinical Subgroups
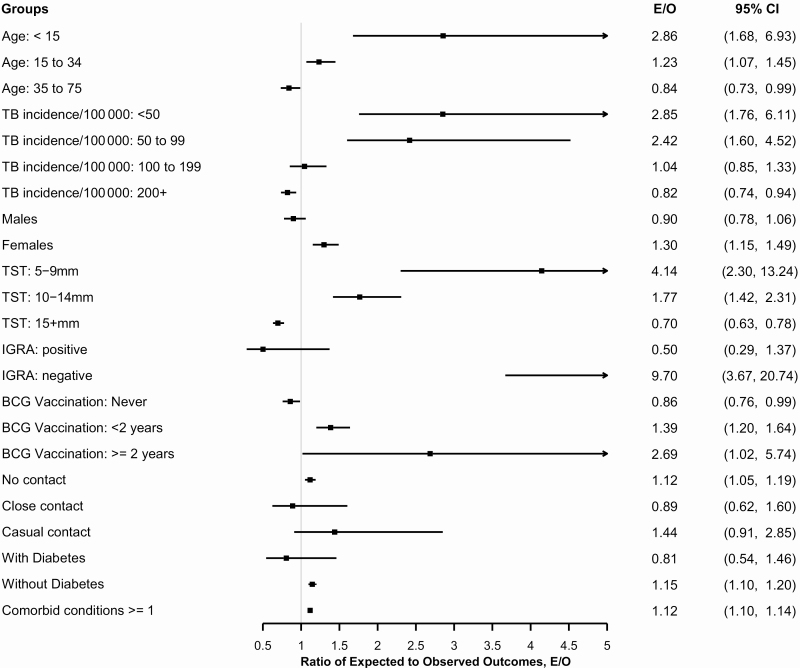
Across most demographic and clinical subgroups, TSTin3D appeared to overpredict active TB risk. However, observed risk was underpredicted in a few subgroups, with the strongest evidence for underprediction found in individuals with TST indurations ≥15 mm (E/O = 0.67, 95% CI, .60–.75). TSTin3D also underpredicted observed risks in people who originated from countries with TB incidence >200 per 100 000 population or from countries with no vaccination policy for children aged <2 years (Figure 4).
Figure 4.
Number of individuals with active TB as predicted by Tuberculin Skin Test/Interferon Gamma Release Assay Interpreter V3.0 (TSTin3D) vs the actual number of individuals who developed active TB within 5 years. Note: E/O is the ratio of the number of individuals with active TB predicted by TSTin3D to the actual number of observed outcomes within a 5-year period. Confidence intervals were estimated using the percentile bootstrap method (B = 10 000). Abbreviations: CI, confidence interval; E/O, ratio of expected to observed; IGRA, interferon gamma release assay; TB, tuberculosis; TST, tuberculin skin test.
Sensitivity Analyses
Results of sensitivity analyses (Supplementary Table 2) indicated that the exclusion of people who received partial or full LTBI treatment (n = 7723) from the primary analyses led to only small changes in measures of discrimination (concordance statistic of 0.68 vs 0.65 without the exclusion; and Somers’ D of 0.34 vs 0.28 without the exclusion). The exclusion did not affect calibration (Supplementary Figure 2).
DISCUSSION
To our knowledge, this is the first study to use administrative data to externally validate TSTin3D, a widely used tool for understanding active TB risk in people with positive LTBI testing. Our results demonstrate that TSTin3D has adequate discrimination when used over a 2- and 5-year time frame to assess risk of active TB. However, a number of caveats are in order when using the tool for individual-level prognostication.
First, our results suggest that in this population, TSTin3D generally overestimated risk, particularly in those deemed at highest risk by the TSTin3D algorithm. Given this observation and lack of other external validations for this tool, we believe that TSTin3D outputs should be interpreted with some degree of caution in the clinical context. Specifically, we recommend that TSTin3D be used as a clinical decision support tool that can be used in conjunction with other clinical information to help understand and communicate an individual’s risk of active TB, and not as a precise estimator of individual risk of active TB.
Second, the tool tended to overestimate actual risks in longer time frames. This may be due to TSTin3D’s assumption of stable annual risks after the first or second year of immigration. Clinicians may, therefore, wish to avoid using TSTin3D to assess whether patients are at high risk of developing active TB beyond 2 or 5 years.
Third, while TSTin3D overestimated TB risk on average in this population, the tool also underestimated TB risk in certain subgroups, such as those with TST indurations ≥15 mm and those who were born in countries with no BCG vaccination policy for children aged <2 years. Risk assessments for these groups may need to be adjusted upward.
We note that this study is an external validation using administrative data and not prospectively collected clinical data. Also, models generally perform differently or poorly when externally validated or applied to different populations [16, 21]. Given these contexts and the consideration that predictive models with concordance values of 0.64 are moderately useful [19], TSTin3D’s discrimination or 5-year concordance statistic of 0.68 appears acceptable. Unfortunately, the concordance values obtained in this study could not be compared with results from previous studies, but a comparison can be made between the concordance values obtained in this study with results from a Cox regression model fitted with the same set of variables that TSTin3D uses as inputs. When we ran this analysis, we obtained a concordance value of 0.73 (95% CI, .70–.76) or .70 when corrected for optimism via 20 × 10-fold cross-validation. This suggests that the concordance values obtained in this external validation study are likely to be as high as could be expected from models that use the same set of data or information as TSTin3D.
It may be possible to address TSTin3D’s tendency to overpredict risks by adjusting the algorithm’s risk estimates downward. One way this could be done is by using more recent data on annual risk of infection, which has declined from the 2009 figures used in the most recent version of the TSTin3D [5]. Another approach is to use the coefficients and the baseline cumulative risks obtained from the regression models fitted in this study to shrink the risk estimates. This adjustment could be useful in other settings, particularly in other low-incidence regions with similar baseline cumulative risks. This adjustment, however, may not fix the underprediction issue in some high-risk groups. Resolving this particular issue requires additional work in terms of reevaluating or updating the weights used in the TSTin3D for age, BCG vaccination history, TST induration size, and medical comorbidities. Members of our team are actively reviewing literature and data, including those from this study, to prepare the next version of the TSTin3D.
This external validation study has some limitations. First, only one-tenth of the cohort was tested with IGRA, and this may have affected our assessment of TSTin3D’s discrimination and calibration. Second, we lack individual-level data on BCG vaccination status and data for some of the comorbid conditions. Given that all missing comorbidities and risk factors increased TSTin3D calculated risk and that they were assumed to be absent in our cohort, we expect that TSTin3D may further overestimate risk in some populations. The impact of BCG imputation, however, is less clear given variance in BCG uptake.
Third, the individuals who were included in the cohort were from the provincial TB registry and represent a segment of the population with positive TST/IGRA results not treated for LTBI. This untreated population may be at lower risk for active TB compared with the population that received a course of LTBI therapy. Last, this external validation study is based on a retrospective analysis of linked administrative data, which carries some risk of misclassification and missing data.
CONCLUSIONS
The TSTin3D tool appears to have adequate ability to distinguish individuals who have high and low risk of being diagnosed with active TB. This tool, however, requires further refinement to ensure that its risk predictions are well calibrated to outcomes seen in low-incidence regions such as BC. We suggest further investigation into calibration and perhaps assessment of opportunities to facilitate more meaningful interpretation of the risk estimates the tool generates. These data could be provided by qualitative work in understanding interpretation of risk estimates in people at risk for active TB.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. Access to the linked health data was granted by Immigration, Refugees, and Citizenship Canada; British Columbia (BC) Ministry of Health; BC Cancer Agency; and BC Renal Agency. Data access was facilitated by Population Data BC.
Disclaimer. All inferences, opinions, and conclusions drawn in this article are those of the authors and do not reflect the opinions or policies of the data stewards(s) and funding agencies.
Financial support. This work is funded through grants provided by the Michael Smith Foundation for Health Research, the BC Lung Association, and the Canadian Institutes of Health Research.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1. MacNeil A, Glaziou P, Sismanidis C, Maloney S, Floyd K. Global epidemiology of tuberculosis and progress toward achieving global targets—2017. MMWR Morb Mortal Wkly Rep 2019; 68:263–6. Available at: http://www.cdc.gov/mmwr/volumes/68/wr/mm6811a3.htm?s_cid=mm6811a3_w. Accessed 4 November 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. World Health Organization. The end TB strategy. Geneva, Switzerland: WHO, 2014: 20. Available at: http://www.who.int/tb/strategy/en/. Accessed 4 November 2019. [Google Scholar]
- 3. Pareek M, Greenaway C, Noori T, Munoz J, Zenner D. The impact of migration on tuberculosis epidemiology and control in high-income countries: a review. BMC Med 2016; 14:48. Available at: http://bmcmedicine.biomedcentral.com/articles/10.1186/s12916-016-0595-5. Accessed 21 November 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Greenaway C, Sandoe A, Vissandjee B, et al. Tuberculosis: evidence review for newly arriving immigrants and refugees. CMAJ 2011; 183:E939–51. Available at: https://www.cmaj.ca/content/183/12/E939. Accessed 4 November 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Menzies D, Gardiner G, Farhat M, Greenaway C, Pai M. Thinking in three dimensions: a web-based algorithm to aid the interpretation of tuberculin skin test results. Int J Tuberc Lung Dis 2008; 12:498–505. [PubMed] [Google Scholar]
- 6. Public Health Agency of Canada and Canadian Lung Association/Canadian Thoracic Society. Canadian Tuberculosis Standards. 2014. Available at: https://www.canada.ca/en/public-health/services/infectious-diseases/canadian-tuberculosis-standards-7th-edition.html. Accessed 4 July 2019.
- 7. Riley RD, Ensor J, Snell KIE, et al. External validation of clinical prediction models using big datasets from e-health records or IPD meta-analysis: opportunities and challenges. BMJ 2016; 353:i3140. Available at: http://www.bmj.com/lookup/doi/10.1136/bmj.i3140. Accessed 28 June 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Population Data BC. Pop Data BC. Available at: http://www.popdata.bc.ca/data. Accessed 14 October 2010.
- 9. Ronald LA, Campbell JR, Balshaw RF, et al. Predicting tuberculosis risk in the foreign-born population of British Columbia, Canada: study protocol for a retrospective population-based cohort study. BMJ Open 2016; 6:e013488. Available at: http://bmjopen.bmj.com/lookup/doi/10.1136/bmjopen-2016–013488. Accessed 21 November 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ronald LA, Campbell JR, Rose C, et al. Estimated impact of World Health Organization latent tuberculosis screening guidelines in a region with a low tuberculosis incidence: retrospective cohort study. Clin Infect Dis 2019; 69:2101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ronald LA, Campbell JR, Balshaw RF, et al. Demographic predictors of active tuberculosis in people migrating to British Columbia, Canada: a retrospective cohort study. CMAJ 2018; 190:E209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roth D, Johnston J, Cook V. TB in foreign-born patients. BC Med J 2012; 54:387–388. Available at: https://www.bcmj.org/bccdc/tb-foreign-born-patients. Accessed 16 April 2020. [Google Scholar]
- 13. BCG World Atlas: A Database of Global Vaccination Policies and Practices. Available at: http://bcgatlas.org/about.php. Accessed 8 August 2019. [DOI] [PMC free article] [PubMed]
- 14. Royston P, Altman DG. External validation of a Cox prognostic model: principles and methods. BMC Med Res Methodol 2013; 13:33. Available at: https://bmcmedresmethodol.biomedcentral.com/articles/10.1186/1471-2288-13-33. Accessed 25 February 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crowson CS, Atkinson EJ, Therneau TM. Assessing calibration of prognostic risk scores. Stat Methods Med Res 2016; 25:1692–1706. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3933449/. Accessed 16 July 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harrell F. Regression modeling strategies: with applications to linear models, logistic and ordinal regression, and survival analysis. 2nd ed. New York: Springer International Publishing, 2015. Available at: https://www.springer.com/gp/book/9783319194240. Accessed 16 June 2019. [Google Scholar]
- 17. Uno H, Cai T, Pencina MJ, D’Agostino RB, Wei LJ. On the C-statistics for evaluating overall adequacy of risk prediction procedures with censored survival data. Stat Med 2011; 30:1105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cohen J. A power primer. Psychol Bull 1992; 112:155. [DOI] [PubMed] [Google Scholar]
- 19. Helmus LM, Babchishin KM. Primer on risk assessment and the statistics used to evaluate its accuracy. Crim Justice Behav 2017; 44:8–25. Available at: http://journals.sagepub.com/ doi/10.1177/0093854816678898. Accessed 5 September 2019. [Google Scholar]
- 20. Therneau TM. A Package for Survival Analysis in R. R package version 3.2-3. 2020. Available at: CRAN.R-project.org/package=survival. Accessed 26 June 2020.
- 21. Bleeker SE, Moll HA, Steyerberg EW, et al. External validation is necessary in prediction research: a clinical example. J Clin Epidemiol 2003; 56: 826–32. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.