Abstract
Background
Detection of glioma recurrence remains a challenge in modern neuro-oncology. Noninvasive radiographic imaging is unable to definitively differentiate true recurrence versus pseudoprogression. Even in biopsied tissue, it can be challenging to differentiate recurrent tumor and treatment effect. We hypothesized that intraoperative stimulated Raman histology (SRH) and deep neural networks can be used to improve the intraoperative detection of glioma recurrence.
Methods
We used fiber laser–based SRH, a label-free, nonconsumptive, high-resolution microscopy method (<60 sec per 1 × 1 mm2) to image a cohort of patients (n = 35) with suspected recurrent gliomas who underwent biopsy or resection. The SRH images were then used to train a convolutional neural network (CNN) and develop an inference algorithm to detect viable recurrent glioma. Following network training, the performance of the CNN was tested for diagnostic accuracy in a retrospective cohort (n = 48).
Results
Using patch-level CNN predictions, the inference algorithm returns a single Bernoulli distribution for the probability of tumor recurrence for each surgical specimen or patient. The external SRH validation dataset consisted of 48 patients (recurrent, 30; pseudoprogression, 18), and we achieved a diagnostic accuracy of 95.8%.
Conclusion
SRH with CNN-based diagnosis can be used to improve the intraoperative detection of glioma recurrence in near-real time. Our results provide insight into how optical imaging and computer vision can be combined to augment conventional diagnostic methods and improve the quality of specimen sampling at glioma recurrence.
Keywords: deep learning, frozen section, intraoperative pathology, label-free imaging, neural networks, stimulated Raman histology
Key Points.
SRH captures the diagnostic features of recurrent glioma and pseudoprogression.
Cross-validation was used to train a CNN and develop an inference algorithm.
SRH-CNN pipeline achieved a diagnostic accuracy of 95.8% (46/48).
Importance of the Study.
Diagnosing glioma tumor recurrence versus pseudoprogression is essential to determine treatment options and prognosis. Radiographic imaging does not provide a definitive diagnosis, and intraoperative neuropathologic evaluation using standard frozen sections has limitations and requires interpretation by expert neuropathologists, who are lacking at many neurosurgery centers. Additionally, optimal tissue collection at glioma recurrence is essential for patient care. We demonstrate how combining SRH, a rapid optical imaging method, and deep neural networks can be used to augment the intraoperative interpretation of surgical specimens in patients with suspected recurrent gliomas. A CNN was trained using cross-validation on an SRH training set and an inference algorithm was developed to provide patient-level diagnosis. Our trained CNN achieved a diagnostic accuracy of 95.8% on an external SRH testing set. These results demonstrate how SRH with CNN-based diagnosis can improve the intraoperative detection of glioma recurrence in near-real time.
Recurrence in gliomas is the rule rather than the exception.1 Detection of tumor recurrence, however, remains a challenge. In 2010, updated response assessment criteria were established in an attempt to formalize the radiographic definition of tumor progression.2 Even with the Response Assessment in Neuro-Oncology (RANO) criteria, there is a high incidence of false positive radiographic findings where pseudoprogression or radiation necrosis is mistaken for tumor recurrence.
Because chemotherapy and/or radiation for gliomas may result in radiographic changes that mimic tumor progression, such as an increase in tumor enhancement in up to 30% of patients, surgery is often necessary to establish definitive diagnoses.2–5 The diagnostic limitations of noninvasive radiographic methods, including structural MRI or perfusion MRI, necessitate surgical tissue sampling in select patients to identify viable recurrent tumor.
During surgery, accurate tissue diagnosis is essential given the differences in the operative goals in patients with pseudoprogression and recurrent glioma. Generally, patients with recurrent glioma benefit from safe maximal resection,6–9 whereas a more conservative surgical approach is taken in patients with pseudoprogression, since it often resolves spontaneously. We have recently observed in a prospective clinical trial that pseudoprogression and recurrent glioma are frequently confused using conventional intraoperative histologic techniques.10 Determining radiation versus tumor progression intraoperatively can be challenging for the pathologists as some features of those entities overlap, like gliosis, necrosis, and abnormal vasculature.
To improve intraoperative diagnostic accuracy and better inform surgical decision making, we hypothesized that the combination of rapid, label-free, optical imaging and deep learning–based computer vision methods can be used to differentiate recurrent glioma and pseudoprogression. Stimulated Raman histology (SRH) is a rapid, label-free optical imaging method that provides high-resolution, digital images of fresh surgical specimens. SRH generates image contrast by detecting differences in the concentration of macromolecules (lipids, proteins, nucleic acids, etc) in biomedical specimens. Recent advances in fiber laser‒based optical imaging have allowed for clinical SRH imaging in the operating room.
We recently showed that SRH combined with CNN can be used as a fully automated and rapid method for intraoperative brain tumor diagnosis.10 Here, we demonstrate the ability of an enhanced automated tissue-to-diagnosis pipeline, incorporating SRH and CNNs, to identify recurrent tumor in fresh surgical specimens.
Materials and Methods
Study Design
We designed our study with the aim to (i) characterize the SRH cytologic and histoarchitectural features of recurrent gliomas, gliotic brain, and treatment effect/pseudoprogression, (ii) develop an intraoperative computer vision system to automate the detection of tumor recurrence in intraoperative specimens using SRH and deep neural networks, and (iii) perform a rigorous validation of the intraoperative system using cross-validation, external validation data (separate tertiary brain tumor center and imaging system), and an independent testing set. SRH imaging was performed at 2 tertiary medical centers: Michigan Medicine and New York-Presbyterian/Columbia University. Institutional review board approval was obtained from both centers (UM: HUM0005731, Columbia: AAAJ6163 and AAAR7365). Informed consent was obtained prior to tissue collection for all patients. The UM dataset was collected from 2 SRH imagers, a prototype SRH imager,11 and a NIO Laser Imaging System (Invenio Imaging). Inclusion criteria for the study were (i) any patient with a previous diagnosis of glioma who (ii) then underwent open biopsy or resection for suspected recurrence based on clinical or radiographic assessment. A patient could be excluded only if the collected specimen was grossly inadequate for SRH imaging due to excessive coagulation or hemorrhage. Location of tissue sampling was left to the surgeon’s discretion.
Specimens from Columbia University had been previously collected and frozen as part of a parallel, ongoing glioma recurrence study. Specimens were collected from both the core and the margin of resection cavities using stereotactic guidance. Specimens were thawed and underwent SRH imaging retrospectively. Columbia SRH images were specifically used as a large, external, independent testing dataset to evaluate model performance and generalizability. For ground truth labeling of the SRH specimens, SRH images were reviewed by one pathologist (S.C.P.) specifically for the presence of viable recurrent tumor in the imaged specimen. If recurrent tumor was identified in the SRH specimen, then the SRH image was labeled as recurrence. Otherwise, a pseudoprogression label was assigned to those specimens that lacked viable recurrent tumor. A patient with one or more specimens containing recurrent tumor was given a patient-level label of recurrence. Information on patients included in the study can be found in Supplementary Tables 1 and 2.
Intraoperative SRH Imaging
Fresh surgical tissue was loaded into a custom imaging chamber and placed in a NIO imaging system (Invenio Imaging). The tissue was excited with a dual-wavelength fiber laser with a fixed wavelength pump beam at 790 nm and a Stokes beam tunable from 1015 nm to 1050 nm. This configuration allows for spectral access to Raman shifts in the range from 2800 cm−1 to 3130 cm−1.12 Beam scanning with a spatial sampling of 450 nm/pixel, 1000 pixels per strip, and an imaging speed for 0.4 MPixel/s/Raman shift was used to acquire images. For SRH, samples were imaged sequentially at the 2 Raman shifts: 2850 cm−1 and 2950 cm−1. Lipid-rich brain regions (eg, myelinated white matter) demonstrate high stimulated Raman scattering signal at 2845 cm−1 due to CH2 (methylene) symmetric stretching in fatty acids. Cellular regions produce high 2930 cm−1 intensity and large S2930/S2845 ratios to high protein and DNA content. A virtual hematoxylin and eosin (H&E) color scheme was applied to transform the raw stimulated Raman scattering images to SRH images for intraoperative use and pathological review.11
Image Preprocessing
All images were preprocessed prior to CNN training and prediction.10 The 2845-cm−1 image was subtracted from the 2930-cm−1 image, and the resultant image was concatenated depthwise to generate a 3-channel image (2930 cm−1 –2845 cm−1, red; 2845 cm−1, green; 2930 cm−1, blue). A 300 × 300-pixel sliding window algorithm with 100-pixel step size (both horizontal and vertical direction) and valid padding was used to generate image patches for model training and inference.
This single-scale sliding window method over high-resolution, high-magnification images provides a large dataset for model training and preserves high-frequency image features (eg, chromatin structure, axonal density) without downsampling. Additionally, multiscale CNN implementations have not yielded better performance for image classification tasks involving histologic images.13 To optimize image contrast, the bottom and top 3% of pixels by intensity from each channel were clipped and images rescaled. All images were mean-zero centered by subtracting the channel-wise mean of the training set.
CNN Training, Cross-Validation, and External Testing Dataset
The Inception-ResNet-v2 CNN architecture was used for the task of image classification on 300 × 300-pixel SRH image patches.14 The CNN was initially pretrained on 3.5 million SRH images from 14 histologic subtypes.10 The pretrained convolutional layers (ie, trained SRH feature extractor) were retained and the final classification layers were modified to classify into 3 diagnostic classes: tumor recurrence, pseudoprogression, and nondiagnostic tissue.15 A total of 406 080 patches from 35 patients were used for model training and validation. We then performed 5 iterations of k-fold cross-validation with k = 7. For each fold, 30 patients were used for training and 5 patients for validation, yielding 5 predictions for each patient. During network training, we used multiple label-preserving affine transformations for data augmentation, including any uniformly distributed random combination of rotation, shift, and reflection. Models were trained for a fixed number of epochs (n = 3) for each fold without hyperparameter tuning to avoid model fitting to the validation set. After cross-validation, the best performing CNN was selected for external validation on a true testing set. Our external dataset from Columbia University contained specimens from 48 patients. This dataset was ideal for model testing, as it contained specimens sampled from both the periphery and the tumor core, resulting in less class imbalance. The model was tested for patient-level classification accuracy using our inference algorithm detailed in Supplementary Figure 1. Code repository can be found at https://github.com/toddhollon/srh_recurrence.
Inference Algorithm
An SRH patch-based classifier requires a method to aggregate CNN predictions from each patch in order to provide a single specimen or patient diagnosis.10 We, therefore, developed an inference algorithm to classify individual specimens and individual patients with potentially multiple specimens into either tumor recurrence or pseudoprogression. The CNN softmax output for each SRH patch is a probability distribution over the diagnostic classes. A feedforward CNN pass is performed for all patches from a given specimen, the nondiagnostic patches are removed, and the remaining output softmax vectors summed elementwise to generate an unnormalized probability distribution over the entire specimen. The residual nondiagnostic probability is set to zero, and the probability distribution is renormalized over the recurrent tumor and pseudoprogression classes, generating a valid Bernoulli probability distribution for each specimen. A diagnostic threshold can then be chosen using receiver operating characteristic (ROC) analysis or at the desired true positive or false positive rate. Patient-level diagnosis is achieved by performing above-mentioned procedure for each specimen from a patient. If any one specimen is labeled as recurrent tumor, then a patient-level diagnosis of tumor recurrence is given.
Visualization of Learned SRH Feature Representations by t-Distributed Stochastic Neighbor Embedding
To improve the interpretability of features learned by the CNN and the classification results, we used a dimensionality reduction and data visualization method, t-distributed stochastic neighbor embedding (t-SNE), to visualize how SRH images are represented in the hidden layers of the CNN. We randomly selected 500 SRH patches from the following histologic groups: normal gray matter, normal white matter, gliotic brain, necrotic tissue, dense hyalinized vasculature, recurrent low-grade glioma, and recurrent high-grade glioma. Each patch was passed through the CNN, and the 1536-dimensional feature vector from the final hidden layer was used to embed each SRH patch into a 2-dimensional graph for visualization. While this is a fully unsupervised technique, class labels were applied to demonstrate the relationship between histologic subtype and the clustering pattern.
Semantic Segmentation of SRH Images
A semantic segmentation technique was developed specifically for SRH images to segment regions of tumor recurrence, pseudoprogression, and nondiagnostic areas. Using a dense sliding window algorithm for patch generation at both training and inference times results in each pixel having an associated probability that is a function of the local overlapping patch predictions. This produces a pixel-level probability heatmap for each diagnostic class at no additional computational cost. We have previously described a similar technique for SRH images in the setting of multiclass brain tumor subtype classification.10 The proposed method for tumor recurrence is simpler and more efficient because it utilizes the trained output classes from the CNN without the need for inference classes. The probability of recurrent tumor (red), psuedoprogression (green), and nondiagnostic (blue) is uniformly mapped to an RGB lookup table and used as a semi-transparent overlay to assist surgeons and pathologists interpreting SRH images with associated CNN predictions.
Results
SRH Features of Glioma Recurrence and Treatment Effect
We first assessed the ability of SRH to effectively image the cytologic and histoarchitectural features associated with both recurrent tumors and pseudoprogression. Fig. 1 shows SRH images of recurrent lower-grade and malignant gliomas adjacent to histopathologic changes induced by chemoradiation. Regions of viable recurrent tumor imaged using SRH contain the traditionally described H&E histopathologic findings for each tumor type. Oligodendrogliomas, World Health Organization (WHO) grade II, demonstrate “fried egg”–like cells and characteristic branching, small, “chicken wire”–like blood vessels. Anaplastic astrocytomas, WHO grade III, show cellular anaplasia, hypercellularity, mitotic activity, and infiltration of normal brain parenchyma (eg, normal axons seen within tumor-infiltrated regions). Glioblastomas, WHO grade IV, have the classic atypia, mitotic figures, necrosis, and/or microvascular proliferation. Importantly, SRH imaged histopathologic features that commonly result from chemoradiation: giant cells with large nuclei and ample cytoplasm, necrosis, hyalinized vasculature, and reactive astrocytosis.
Fig. 1.
Histopathologic features of recurrent glioma and pseudoprogression in stimulated Raman histology (SRH) images. SRH images of recurrent (A) oligodendroglioma, WHO grade II, (B) anaplastic astrocytoma, WHO grade III, and (C) glioblastoma, WHO grade IV. SRH captures cytologic and histoarchitectural features not seen in conventional H&E histology, such as myelinated axons and lipid-laden macrophages. Mitotic figures are seen due to submicron spatial resolution and virtual H&E staining that highlights chromatic structure. (D) Optical sectioning allows for detection of microvascular proliferation. (E) Treatment effect can produce marked pleomorphism, giant and multinucleated cells with ample cytoplasm. (F) Necrosis seen adjacent to dense viable tumor. (G) Reactive astrocytosis, a classic finding in pseudoprogression and the peritumoral region, is well characterized in SRH images by long astrocytic processes from nonneoplastic glial cells. (H) Dense hyalinized blood vessels and (I) necrosis postradiation. Scale bars = 50 μm.
Pipeline for Automated Intraoperative Detection of Tumor Recurrence
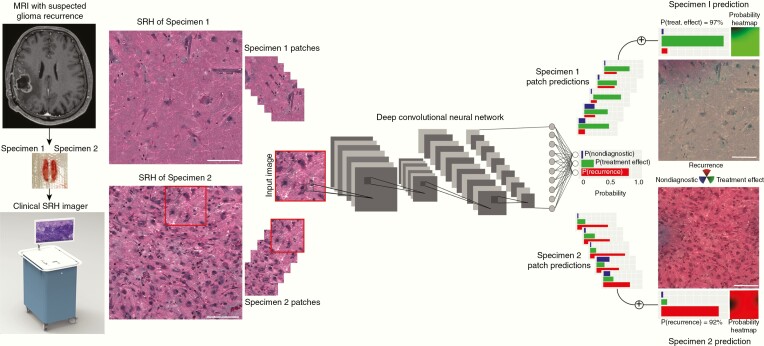
After demonstrating that SRH can image the histologic features of recurrence and pseudoprogression, we developed an intraoperative pipeline, called the SRH-CNN pipeline, to automate the detection of tumor recurrence using SRH and CNN (Fig. 2). Fresh, unprocessed surgical specimens are passed off the operative field and a small sample (eg, 3 mm3) is compressed into a custom microscope slide. After inserting the slide into the SRH imager, images are acquired at 2 Raman shifts, 2845 cm−1 and 2930 cm−1, to generate high-resolution digital images. SRH whole-slide images are then processed via a dense sliding window algorithm to generate overlapping, single-scale, and high-magnification patches used for CNN training and inference. In the prediction stage, individual patches are passed through the trained Inception-ResNet-v2 network for image classification.14 Following feedforward CNN pass on each SRH patch, the patch-level predictions are used as input for an inference algorithm designed to return a single whole-slide or patient-level diagnosis. The inference algorithm optimized for prediction on SRH images is shown in Supplementary Figure 1.
Fig. 2.
SRH and CNN pipeline for automated detection of recurrent glioma. Rapid, bedside, label-free optical images are generating using clinical SRH imager. A 1 × 1-mm SRH image is captured in approximately 60 seconds. SRH patches (300 × 300-pixel) are produced using a dense sliding window algorithm and each SRH patch undergoes a feedforward CNN pass. The final softmax layer outputs a categorical probability distribution over three classes: recurrence, pseudoprogression/treatment effect, and nondiagnostic. An inference algorithm is then used to aggregate the patch-level prediction probabilities and output a single probability of recurrence for each specimen and/or each patient. We also implemented a semantic segmentation method that overlays SRH-CNN probability heatmaps to identify spatial regions of tumor recurrence or pseudoprogression. An RGB transparency (red, recurrence; green, pseudoprogression; blue; nondiagnostic) is overlaid on the SRH whole-slide image to provide both surgeons and pathologists spatial information in addition to prediction probabilities. Scale bars = 50 μm.
CNN Training and Cross-Validation Results
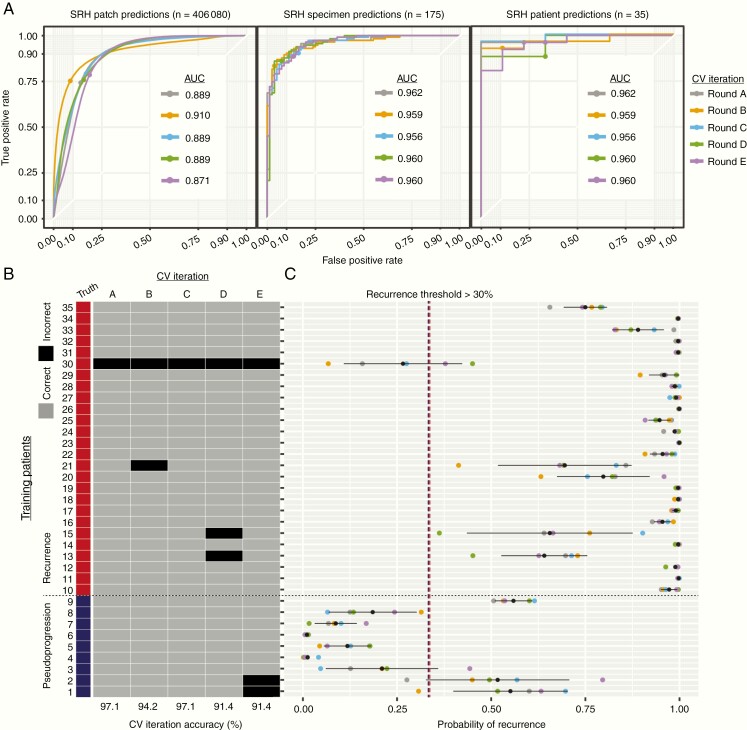
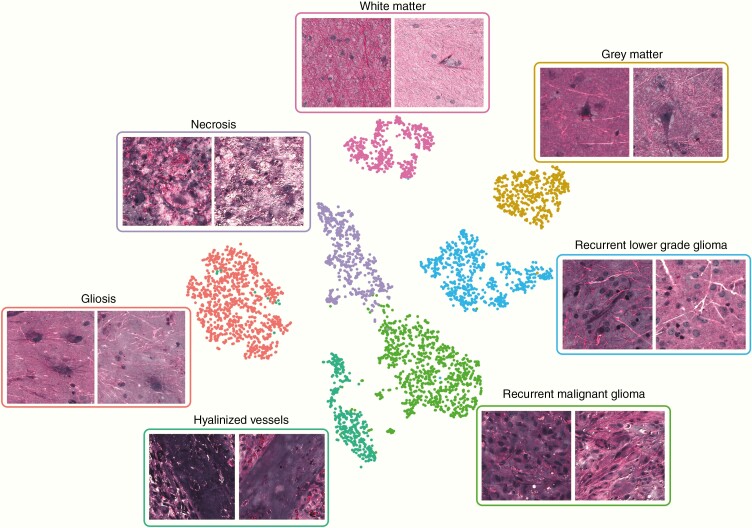
Using our 35-patient training set (Supplementary Table 1), we performed 5 iterations of cross-validation to confirm optimal CNN training and identify an ideal probability threshold for diagnosis of glioma recurrence (Fig. 3). Our dataset contained 26 patients with glioma recurrence and 9 with pseudoprogression. Cross-validation results showed stable training and validation results over each iteration, with area under the curve (AUC) values ranging 95.6–96.2% for specimen-level diagnosis and 96.2–98.7% for patient-level diagnosis. Fig. 3B plots the probability of recurrence for each patient and each cross-validation iteration as determined by our inference algorithm. Patient 30 was excluded from the final thresholding analysis due to having a consistently low probability of recurrence and was an outlier in the recurrence group. Error analysis demonstrated that the final histopathologic diagnosis was recurrent low-grade glioneuronal tumor, WHO grade I, and the patient had not received chemotherapy or radiation. We chose a recurrence probability threshold of 30%, corresponding to a sensitivity of 100.0%, specificity of 82.2%, and accuracy of 92.4% on the validation set. This threshold was tuned to maximize sensitivity to avoid false negatives intraoperatively and potentially miss a diagnosis of glioma recurrence. To improve model interpretability, we used t-SNE to embed the most common histopathologies seen in pseudoprogression and glioma recurrence in a 2D graph based on the learned internal CNN representations (Fig. 4). Patches within each histopathologic subtype form discrete clusters based on the internal CNN 1536-dimensional feature vector. The t-SNE results indicate that similar histopathologies share common feature representations in the final CNN hidden layer. Using our cross-validation SRH dataset, our semantic segmentation method was able to identify regions of diagnostic, dense tumor recurrence within whole-slide SRH images (Fig. 5). Glioma recurrence adjacent to gliotic peritumoral regions was segmented with high confidence, improving the clinician’s review of CNN predictions.
Fig. 3.
CNN training and cross-validation results. Five iterations of cross validation were performed. In our implementation, k was 7, which yields 30 training cases and 5 validation cases for each fold. CNN was trained for 3 epochs without additional training callbacks or hyperparameter tuning to avoid overfitting the held-out data sets. (A) Patch-, specimen-, and patient-level ROC curves are shown for each cross-validation iteration. ROC-AUC was similar across each iteration and expectedly increases as the predictions scale up from patch- to patient-level predictions, with a maximum patient-level AUC of 98.7%. (B) Accuracy results for each patient and each cross-validation (CV) iteration. Pseudoprogression patients (blue, patients 1–9) and recurrence patients (red, patients 10–35) are presented as rows and each CV iteration is a column. Whether the patient was correctly classified for each iteration is color coded (gray, correct; black, incorrect). CV iterations A and C had the highest accuracy of 97.1% (34/35 patients). (C) Patient-level prediction probability of recurrence is plotted for each CV iteration with the mean ± standard deviation (black point and line). Patient 30 was consistently predicted to have a low probability of recurrence, while the remaining recurrence patients had a high prediction probability. Based on the results displayed in (C), we choose a recurrence probability threshold of 30% to be used in the external testing set.
Fig. 4.
Visualization of final hidden layer CNN representations of SRH patches.
The t-SNE plot of sampled patches from 7 histologic subtypes encountered in glioma recurrence and pseudoprogression. Discrete clusters based on the 1536-dimensional feature vector from the final hidden layer. Each of the 7 subtypes are color coded for easier interpretation of the t-SNE plot. This graph indicates that the trained CNN learned to represent common histologies with specific feature vectors, thereby improving the diagnostic accuracy of the final softmax classification layer.
Fig. 5.
Semantic segmentation of SRH images for regions of glioma recurrence. (A) Probability heatmaps are shown for each of the 3 output classes. Because patch-level predictions are generated from a dense, overlapping sliding window algorithm, every pixel in the image has a probability distribution over the output classes that is a function of the local neighborhood of overlapping patch predictions. This method produces a higher resolution and smoother heatmap. Each probability heatmap is mapped to a specific RBG channel to produce a prediction overlay for the SRH whole-slide image. (B) An example of an SRH image taken from a patient with recurrent glioblastoma, WHO IV. A dense region of recurrent tumor (red) was identified adjacent to nondiagnostic hemorrhagic and necrotic tissue (blue) and gliotic brain (green). Our semantic segmentation method allows for the interpretation of SRH images in the context of CNN predictions with spatial information regarding regions of recurrent tumor. Scale bars = 50 μm.
External Validation of SRH with CNN-Based Diagnosis of Glioma Recurrence
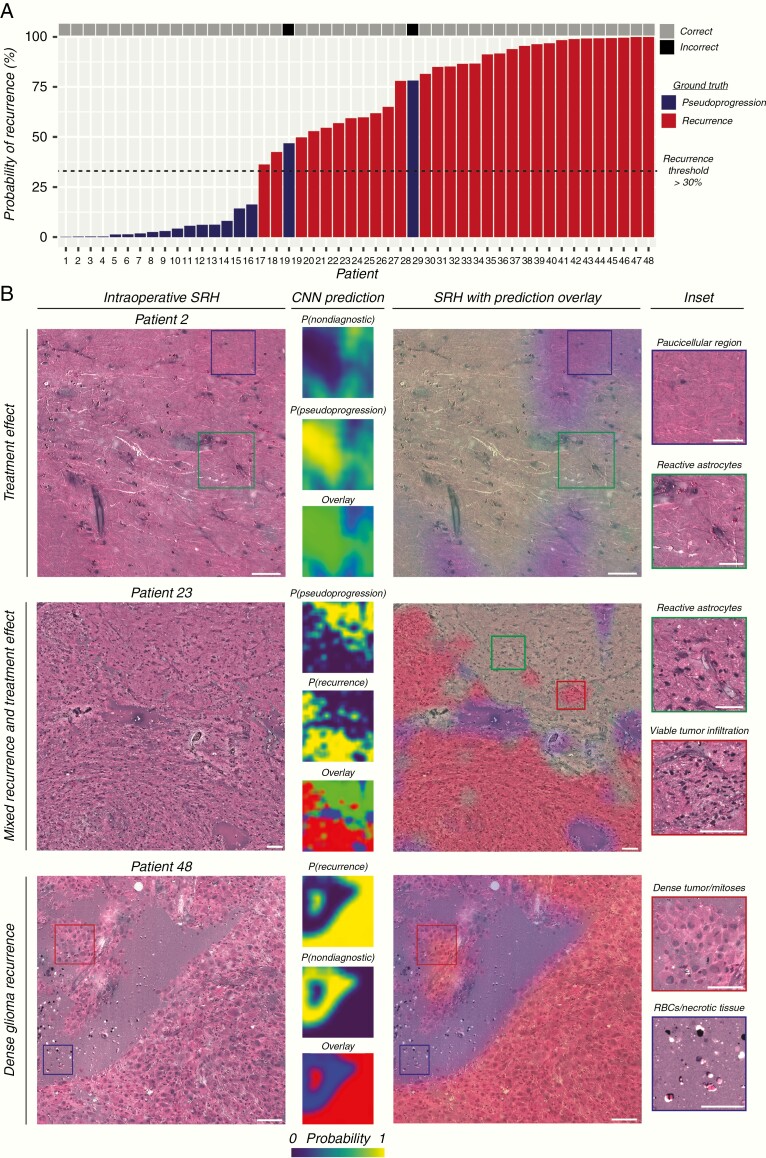
Following CNN training and cross-validation, we aimed to test our SRH-CNN pipeline on an external SRH dataset from New York-Presbyterian/Columbia University. Moreover, a semantic segmentation technique was implemented on the external dataset to improve surgeon and pathologist ability to rapidly identify regions of tumor recurrence within SRH whole-slide images (Fig. 6). Probability heatmaps for recurrence, pseudoprogression, and nondiagnostic regions are overlaid as a RGB transparency onto the whole-slide SRH image. Regions highlighted red correspond to areas of high probability of tumor recurrence. Forty-eight patients (30 labeled recurrent, 18 labeled pseudoprogression) were included in the external testing set. A recurrence probability threshold of 30%, as determined by our cross-validation experiments, was used to provide a final intraoperative diagnosis of glioma recurrence. Our SRH-CNN pipeline resulted in a sensitivity of 100% (30/30), specificity of 88.9% (16/18), and overall accuracy of 95.8% (46/48). Regions of viable tumor were segmented in patients with both lower-grade and malignant glioma recurrence. Gliotic brain regions with reactive astrocytes were identified in pseudoprogression patients without evidence of tumor recurrence. Importantly, regions of mixed glioma recurrence and treatment effect were effectively segmented in the same specimen and expectedly resulted in intermediate diagnostic probabilities.
Fig. 6.
External validation of SRH with CNN-based diagnosis of glioma recurrence. (A) SRH-CNN prediction for each of the patients in the external validation set. The probability of recurrence is plotted on the vertical axis. Ground truth label is color coded (red, recurrence; blue, pseudoprogression). Only 2 of 18 patients labeled as pseudoprogression were incorrectly classified as having tumor recurrence (88.9% specificity); all patients with tumor recurrence were correctly diagnosed (100.0% sensitivity). Overall diagnostic accuracy of glioma recurrence using SRH-CNN was 95.8%. (B) Example specimens from the external validation set with SRH images and CNN probability heatmaps. SRH images from patient 2 did not show evidence of tumor recurrence, and our semantic segmentation method correctly identified regions of reactive astrocytosis as pseudoprogression. Patient 34 was found to have recurrent oligodendroglioma, WHO grade II, and dense regions of infiltrative low-grade glioma were correctly identified. Patient 48 was found to have dense, viable recurrent glioblastoma, WHO grade IV. These findings indicate that our segmentation method correctly segmented regions of both lower-grade and malignant gliomas with similar accuracy. Scale bars = 50 μm.
Discussion
Here, we demonstrated how SRH and deep neural networks combine to form a complementary intraoperative pathway for the diagnosis of glioma recurrence that is independent of a traditional pathology laboratory. Clinical SRH was able to image the most diagnostic features associated with recurrent glial tumors and treatment-induced histologic findings commonly seen in pseudoprogression. Cross-validation was performed for CNN training and to identify a clinically optimal patient-level probability threshold for diagnosing glioma recurrence. Using SRH image data collected from an external validation medical center, we showed that our SRH-CNN pipeline was able to achieve a testing set diagnostic accuracy of 95.8%. Regions of dense tumor, in both lower-grade and malignant glioma, were segmented to highlight regions of high probability of recurrence and improve the clinician’s interpretation of automated diagnostic results.
A major challenge in glioma surveillance is that conventional structural MRI does not provide sufficient molecular or metabolic data for diagnostic purposes, with sensitivity and specificity of 68% (95% CI: 51–81) and 77% (45–93), respectively.16 As a result, advanced MRI methods have been used for glioma surveillance and produce better diagnostic performance compared with structural MRI. In a survey of 220 medical centers, perfusion MRI was regarded as the advanced modality of choice to distinguish radiation effects from tumor progression by the majority of centers, with sensitivity of 87% (82–91) and specificity of 86% (77–91).16,17 Magnetic resonance spectroscopy can produce higher diagnostic accuracy than structural MRI but is less suitable for routine and universal application in glioma surveillance.
Without a definitive noninvasive method to detect glioma recurrence, tissue diagnosis is needed in patients with an equivocal radiographic and clinical course. Moreover, the frequency of tissue sampling at time of recurrence is likely to increase in the era of personalized medicine. RANO investigators state that “the suboptimal employment of histological and molecular assessment at recurrence represents a missed opportunity to proactively guide patient management and increase knowledge. . . . Progress in this area will almost certainly require improved specimen sampling.” 1 They claim that neuro-oncology is lagging behind other oncology specialties that routinely use tissue-based analyses to document the molecular and cellular features at disease recurrence. In addition to the increase in the number of patients who undergo tissue sampling for recurrent gliomas, the number of specimens per patient may increase due to the known spatial histologic and molecular heterogeneity of gliomas.18–21 Determining the diagnostic yield for each specimen at the time of surgery is essential to ensure that the optimal amount of viable tissue is available for subsequent tissue analyses. Consistently sending multiple intraoperative specimens is not feasible using our current pathology workflow. Additionally, our labor, time, and resource limitations are unlikely to improve in the foreseeable future. Both globally and within the US, there is an uneven distribution of expert pathologists available to provide intraoperative diagnosis. For example, many centers performing brain tumor surgery currently have no neuropathologist on staff, and further shortages are expected as there is a 42% vacancy rate in neuropathology fellowship programs.22,23
Recent advances in deep learning and computer vision have resulted in human-level performance on diagnostic tasks in ophthalmology,24 radiology,25 dermatology,26 and pathology.10,27–29 These advances provide insight into how artificial intelligence can ameliorate the effect of labor shortages in health care and augment standard-of-care diagnostic techniques. Here, we show that fiber laser–based clinical optical imaging combined with state-of-the-art deep learning methods can augment the diagnostic accuracy of the detection of recurrent gliomas intraoperatively in a fully automated fashion. Multiple specimens can be imaged in an order of magnitude in less time than conventional methods, decreasing the likelihood of intraoperative sampling error. Rapid SRH imaging allows for multiple specimens to be collected with confirmed diagnostic tissue for subsequent molecular analysis and assessment for spatial heterogeneity.21
A limitation of our study is the challenge of determining a definitive labeling strategy for tumor recurrence. This is a challenge inherent to a study investigating glioma recurrence, which is a complex and heterogeneous diagnosis. We aimed to keep the labeling task as simple as possible (ie, binary labeling) to avoid introducing additional levels of complexity and opted to have a single reviewer label the SRH images to maintain a consistent ground truth. As has been the case in our previous SRH studies, we believe that the detection of tumor is easier in SRH images compared with conventional H&E histology for both clinicians and computer-aided diagnostic systems because specimens are imaged fresh without tissue processing that has the potential to induce artifacts. Additional studies will be needed to confirm that our results generalize to other medical centers.
Future applications of SRH and deep learning include moving beyond histopathologic diagnosis toward molecular classification. The WHO classification integrates both histopathology and molecular diagnostics to stratify patients into the most informative subgroups. Intraoperative, real-time identification of the known spatial and temporal molecular heterogeneity of gliomas could provide a much richer understanding of clonal evolution.21,30 Preliminary studies using Raman spectroscopy have shown promising results for detecting some molecular subtypes of gliomas.31,32 We hope that the nonlinear excitation induced by SRH will generate a rapid and accurate method to detect glioma driver mutations in near-real time.
In conclusion, rapid, label-free detection of glioma recurrence can be achieved using intraoperative SRH and deep neural networks. A parallel, automated, intraoperative pathology pipeline for differentiating true progression versus pseudoprogression will be indispensable in the era of personalized medicine. Accurate tumor sampling at the time of glioma recurrence will provide invaluable diagnostic information to proactively guide patient management and increase knowledge of glioma biology.
Supplementary Material
Funding
This work was supported by the NIH National Cancer Institute (R01CA226527, D.A.O), and The Cook Family Foundation. The collection of MRI-localized samples from Columbia University was funded by R01 NS103473 (P.C., J.N.B.).
Conflict of interest statement. D.A.O. is an adviser and shareholder of Invenio Imaging, Inc, a company developing SRH microscopy systems. C.W.F. is an employee and shareholder of Invenio Imaging, Inc.
Authorship statement. Conception/design: TCH, SSK, PC, SC-P, HL, DAO. Data acquisition: TCH, BP, EU, AVS, ARA, SS, ZF, TM, JNB, WNA, NKJ, KE, JAH, SSK, CWF. Data analysis/interpretation: TCH, BP, EU, AVS, ARA, SS, NKJ, KE, SSK, KC, PC, CWF, SC-P, HL, DAO. Experimental work: TCH, BP, EU, AVS, ARA, SS, NKJ, KE, JAH, SSK, SC-P. Drafting of article: TCH, HL, PC, DAO. Revision of article: all authors. Statistical expertise: TCH, SSK, HL. Admin., technical, or other support: TCH, KE, SSK.
References
- 1. Haider AS, van den Bent M, Wen PY, et al. Towards a standard pathological and molecular characterization of recurrent glioma in adults: a RANO effort. Neuro Oncol. 2020;22(4):450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 3. van West SE, de Bruin HG, van de Langerijt B, Swaak-Kragten AT, van den Bent MJ, Taal W. Incidence of pseudoprogression in low-grade gliomas treated with radiotherapy. Neuro Oncol. 2017;19(5):719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abbasi AW, Westerlaan HE, Holtman GA, Aden KM, van Laar PJ, van der Hoorn A. Incidence of tumour progression and pseudoprogression in high-grade gliomas: a systematic review and meta-analysis. Clin Neuroradiol. 2018;28(3):401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thust SC, van den Bent MJ, Smits M. Pseudoprogression of brain tumors. J Magn Reson Imaging. 2018. doi: 10.1002/jmri.26171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hervey-Jumper SL, Berger MS. Reoperation for recurrent high-grade glioma: a current perspective of the literature. Neurosurgery. 2014;75(5):491–499; discussion 498–499. [DOI] [PubMed] [Google Scholar]
- 7. Ramakrishna R, Hebb A, Barber J, Rostomily R, Silbergeld D. Outcomes in reoperated low-grade gliomas. Neurosurgery. 2015;77(2):175–84; discussion 184. [DOI] [PubMed] [Google Scholar]
- 8. Oppenlander ME, Wolf AB, Snyder LA, et al. An extent of resection threshold for recurrent glioblastoma and its risk for neurological morbidity. J Neurosurg. 2014;120(4):846–853. [DOI] [PubMed] [Google Scholar]
- 9. Bloch O, Han SJ, Cha S, et al. Impact of extent of resection for recurrent glioblastoma on overall survival: clinical article. J Neurosurg. 2012;117(6):1032–1038. [DOI] [PubMed] [Google Scholar]
- 10. Hollon TC, Pandian B, Adapa AR, et al. Near real-time intraoperative brain tumor diagnosis using stimulated Raman histology and deep neural networks. Nat Med. 2020. doi: 10.1038/s41591-019-0715-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Orringer DA, Pandian B, Niknafs YS, et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat Biomed Eng. 2017;1. doi: 10.1038/s41551-016-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Freudiger CW, Yang W, Holtom GR, Peyghambarian N, Xie XS, Kieu KQ. Stimulated Raman scattering microscopy with a robust fibre laser source. Nat Photonics. 2014;8(2):153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu Y, Gadepalli K, Norouzi M, et al. Detecting cancer metastases on gigapixel pathology images. arXiv [csCV]. 2017. doi:1703.02442. [Google Scholar]
- 14. Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Inception-v4, Inception-ResNet and the impact of residual connections on learning. AAAI. Vol 4. 2017:12. [Google Scholar]
- 15. Razavian AS, Azizpour H, Sullivan J, Carlsson S. CNN features off-the-shelf: an astounding baseline for recognition. arXiv [csCV]. 2014. doi:1403.6382. [Google Scholar]
- 16. van Dijken BRJ, van Laar PJ, Holtman GA, van der Hoorn A. Diagnostic accuracy of magnetic resonance imaging techniques for treatment response evaluation in patients with high-grade glioma, a systematic review and meta-analysis. Eur Radiol. 2017;27(10):4129–4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thust SC, Heiland S, Falini A, et al. Glioma imaging in Europe: a survey of 220 centres and recommendations for best clinical practice. Eur Radiol. 2018;28(8):3306–3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Szerlip NJ, Pedraza A, Chakravarty D, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci U S A. 2012;109(8):3041–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aum DJ, Kim DH, Beaumont TL, Leuthardt EC, Dunn GP, Kim AH. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg Focus. 2014;37(6):E11. [DOI] [PubMed] [Google Scholar]
- 21. Suzuki H, Aoki K, Chiba K, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47(5): 458–468. [DOI] [PubMed] [Google Scholar]
- 22. Robboy SJ, Weintraub S, Horvath AE, et al. Pathologist workforce in the United States: I. Development of a predictive model to examine factors influencing supply. Arch Pathol Lab Med. 2013;137(12): 1723–1732. [DOI] [PubMed] [Google Scholar]
- 23. Metter DM, Colgan TJ, Leung ST, Timmons CF, Park JY. Trends in the US and Canadian Pathologist Workforces From 2007 to 2017. JAMA Netw Open. 2019;2(5):e194337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316(22):2402–2410. [DOI] [PubMed] [Google Scholar]
- 25. Titano JJ, Badgeley M, Schefflein J, et al. Automated deep-neural-network surveillance of cranial images for acute neurologic events. Nat Med. 2018;24(9):1337–1341. [DOI] [PubMed] [Google Scholar]
- 26. Esteva A, Kuprel B, Novoa RA, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Litjens G, Sánchez CI, Timofeeva N, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016;6:26286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coudray N, Ocampo PS, Sakellaropoulos T, et al. Classification and mutation prediction from non-small cell lung cancer histopathology images using deep learning. Nat Med. 2018;24(10):1559–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He K, Zhang X, Ren S, Sun J. Delving deep into rectifiers: surpassing human-level performance on ImageNet classification. arXiv [csCV]. 2015. doi:1502.01852. [Google Scholar]
- 30. Johnson BE, Mazor T, Hong C, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Livermore LJ, Isabelle M, Bell IM, et al. Rapid intraoperative molecular genetic classification of gliomas using Raman spectroscopy. Neurooncol Adv. 2019;1(1):vdz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Uckermann O, Yao W, Juratli TA, et al. IDH1 mutation in human glioma induces chemical alterations that are amenable to optical Raman spectroscopy. J Neurooncol. 2018;139(2):261–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.