Abstract
PCR amplifications of the 16S rRNA gene were performed on 46 specimens obtained from 43 dogs with canine leproid granuloma syndrome to help determine its etiology. Sequence capture PCR was applied to 37 paraffin-embedded specimens from 37 dogs, and nested PCR was attempted on DNA from 9 fresh tissue specimens derived from 3 of the 37 aforementioned dogs and from an additional 6 dogs. Molecular analyses of the paraffin-embedded tissues and fresh tissue specimen analyses were performed at separate institutions. PCR products with identical sequences over a 350-bp region encompassing variable regions 2 and 3 of the 16S rRNA gene were obtained from 4 of 37 paraffin-embedded specimens and from all 9 specimens of fresh tissue originating from 12 of the 43 dogs. Identical sequences were determined from amplicons obtained from paraffin-embedded and fresh specimens from one dog. The consensus DNA sequence, amplified from paraffin-embedded tissue and represented by GenBank accession no. AF144747, shared highest nucleotide identity (99.4% over 519 bp) with mycobacterial strain IWGMT 90413 but did not correspond exactly to any EMBL or GenBank database sequence. With a probe derived from the V2 region of the novel canine sequence, reverse cross blot hybridization identified an additional four paraffin-embedded specimens containing the same novel sequence. In total, molecular methodologies identified the proposed novel mycobacterial sequence in 16 of 43 dogs with canine leproid granuloma syndrome, indicating that the species represented by this sequence may be the principal etiological agent of canine leproid granuloma syndrome.
In 1998, the term canine leproid granuloma syndrome was coined to describe a nodular pyogranulomatous disease affecting the skin and subcutis of dogs (4, 25). This condition, first described in 1973 in Africa (32), is the most common mycobacterial disease of dogs in Australia and affects principally short-coated breeds (25). The lesions consist of single or multiple nodules, usually on the head and especially on the dorsal fold of the ears. Despite its prevalence, the etiology is yet to be determined, as attempts to culture acid-fast bacilli, visualized in variable numbers in the majority of lesions, have been unsuccessful despite the use of a variety of well-described methods for in vitro culturing of mycobacteria (25). PCR-based methodologies have been of considerable value for the accurate diagnosis of mycobacterial infections in animals, particularly for those mycobacteria which are nonculturable or difficult to culture (15, 27). This paper describes the use of 16S rRNA PCR-based methodologies to identify mycobacteria from both fresh biopsy material and paraffin-embedded, formalin-fixed tissue taken from lesions of dogs with canine leproid granuloma syndrome. Accurate identification and taxonomic placement of the causative mycobacterium should assist in the determination of culture requirements in the future, facilitating effective prevention, treatment, and control of the disease through tracing of its ecological niche.
MATERIALS AND METHODS
Specimens.
Thirty-seven formalin-fixed, paraffin-embedded tissues from 37 dogs with nodular granulomas of the subcutis and skin and nine fresh tissue specimens from 9 dogs were analyzed in this study. The specimens were obtained from Australian veterinary pathology laboratories between 1982 and 1999. Three of the nine fresh tissue specimens were from the aforementioned 37 dogs, and the remaining fresh specimens were from 6 additional dogs. Acid-fast bacilli had been observed in histology sections or in smears made from these tissues, but culturing, where attempted under conditions developed for mycobacterial species (6, 17, 19, 28, 33), had been unsuccessful (25).
DNA extraction. (i) Paraffin-embedded tissues.
DNA was extracted from formalin-fixed, paraffin-embedded tissue by a sequence capture methodology (26) modified as described previously (16). Briefly, a small section of each paraffin-embedded tissue was excised with a sterile blade and suspended in 500 μl of 100 mM Tris-HCl–150 mM NaCl–50 mM EDTA (pH 7.4). Each sample was dewaxed by incubation at 100°C for 10 min and agitated subsequently with 0.5 ml of zirconia beads (0.1 mm diameter; Biospec Products) for 2 min in a Mini-BeadBeater (Biospec Products). Proteinase K (50 μl, 20 mg/ml) digestion was performed at 50°C overnight, before further agitation for 2 min. 16S rRNA gene sequences were extracted from the treated tissue specimens by hybridization with 2.5 pmol of each biotinylated capture oligonucleotide, Cap pA (5′-AAAAAAGAGTTTGATCCTGGCTCAG) and Cap pH (5′-AAAAAAAGGAGGTGATCCAGCCGCA) (Gibco BRL, Life Technologies Ltd.), and subsequent capture onto streptavidin-coated magnetic beads (Dynal). Captured DNA was resuspended in 25 μl of sterile deionized water (SDW).
(ii) Fresh tissues.
DNA was recovered from fresh tissue samples by sodium dodecyl sulfate (SDS)-mediated cell lysis, phenol-chloroform extraction, and ethanol precipitation as previously described (12) after initial maceration of tissue in a mortar on ice with two sterile scalpel blades. The DNA was washed in 70% (vol/vol) ethanol, air dried, and resuspended in 200 μl of SDW.
PCR amplification. (i) Paraffin-embedded tissues.
16S rRNA PCR amplifications of extracted DNA (5 μl) were performed as described previously (15) with two different primer pairs separately. Primers pA (5′-AGAGTTTGATCCTGGCTCAG; nucleotides 8 to 28 of the 16S rRNA gene of Escherichia coli) (8) and MSH-E (5′-GCGACAAACCACCTACGAG; nucleotides 557 to 539 of aligned 16S rRNA gene sequences of mycobacterial species [30]) were used to amplify variable region 2 (V2 region), corresponding to helix 10, and variable region 3 (V3 region), corresponding to helix 18, of the 16S rRNA gene (21). This amplification was attempted on all 37 paraffin-embedded specimens. Primers MSH-A (5′-CACCAACAAGCTGATAGGC; nucleotides 268 to 250 of the 16S rRNA gene of E. coli) and MSH-B (5′-GGGATAAGCCTGGGAAACT; nucleotides 147 to 165 of the 16S rRNA gene of E. coli) were used to amplify the V2 region. Amplification with primer pair MSH-A–MSH-B was attempted on specimens which did not yield PCR products with primer pair pA–MSH-E and on two specimens from which amplicons were obtained with primer pair pA–MSH-E; the latter served as positive controls. Amplification mixtures were subjected to 4 min of denaturation at 94°C; 30 cycles of 95°C for 30 s, 60°C for 1 min, and 72°C for 1 min; followed by a final extension period of 7 min at 72°C and refrigeration in a model 480 thermal cycler (Perkin-Elmer Corporation, Norwalk, Conn.). Reamplification of 5-μl aliquots of first-round PCR amplification products was performed with the same PCR conditions and cycling parameters. Negative controls for the PCR and DNA extraction process were included, and precautionary measures to prevent contamination were taken as described previously (13, 14).
(ii) Fresh tissues.
PCR amplification of a 590-bp sequence of the 16S rRNA gene including the V2 and V3 regions was performed with primer set 2 (PS2), comprised of oligonucleotide 246 (5′-AGAGTTTGATCCTGGCTCAG; nucleotide positions 8 to 28 of the 16S rRNA gene sequence of E. coli) and reverse oligonucleotide 247 (5′-TTTCACGAACAACGCGACAA; nucleotide positions 609 to 590 of the E. coli 16S rRNA gene) (2). Ten microliters of extracted DNA was added to 15 μl of the PCR mixture, with the final 25-μl volume containing 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.01% Tween 20, 0.01% gelatin, 0.01% Nonidet P-40, 100 μM deoxynucleoside triphosphates (dNTPs), 400 nM PS2 primers, and 0.5 U of DNA polymerase (DyNAZyme; Finnzymes, Espoo, Finland). Amplification was performed with an FTS960 thermal cycler (Corbett Research, Sydney, Australia) and the following profile: 95°C for 5 min; followed by 40 cycles of 96°C for 10 s, 68°C for 2 min, and 74°C for 3 min; and a final extension of 10 min at 74°C. The assay included the following controls: an extraction process negative control, a PCR mixture negative control, inhibition controls (DNA extracts spiked with 103 Mycobacterium paraffinicium bacilli), and sensitivity controls (10-fold dilutions of M. paraffinicum) to determine the end-point sensitivity of each assay for the validation of amplification performance.
To obtain sufficient DNA for sequence analysis, PCR products from fresh tissues were reamplified with nested primers internal to the PS2 primers. The internal primers M1 (5′-AGTGGCGAACGGGTGAGTAAC; nucleotide positions 105 to 126 of the 16S rRNA gene of E. coli) and R7 (5′-TTACGCCCAGTAATTCCGGACAA; nucleotide positions 573 to 551 of the 16S rRNA gene of E. coli) amplified a 469-bp sequence. Four microliters of a 1:100 dilution of the PS2 amplicon in SDW was added to the PCR mixture. The final PCR mixture volume of 50 μl contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, 0.01% Tween 20, 0.01% gelatin, 0.01% Nonidet P-40, 50 μM dNTPs, 100 nM internal primers, and 1.0 U of DNA polymerase (DyNAZyme). Thermal cycling parameters were 25 cycles of 94°C for 30 s, 68°C for 1 min, and 74°C for 15 s, with a final extension of 5 min at 74°C.
PCR product analysis. (i) Agarose gel electrophoresis.
PCR products obtained from paraffin-embedded tissues were purified directly from agarose gels with a QIAquick PCR purification kit (QIAGEN Ltd.) or, if nonspecific PCR products were a problem, from low-melting-point agarose (Gibco BRL) gels by use of a Prep-a-gene DNA purification kit (Bio-Rad Laboratories Ltd.) according to the manufacturer's instructions. For fresh tissues, M1-R7 PCR amplicons were analyzed by electrophoresis in an 0.8% (wt/vol) agarose gel. Amplicon DNA was precipitated with polyethylene glycol, leaving excess primers and nucleotides in solution (5). Purified DNA was resuspended in SDW to a concentration of approximately 100 ng/μl.
(ii) DNA sequence analysis.
Sequence analysis of purified PCR products for paraffin-embedded tissues was performed at the Advanced Biotechnology Centre, London, United Kingdom, with primers pA, MSH-E, and MSH-B. Sequences were edited with Chromas version 1.43 (Griffith University, Brisbane, Queensland, Australia) and analyzed with DNASIS version 7 software (Pharmacia Biosystems GmbH), and sequence comparisons were made with EMBL and GenBank database sequences. For fresh tissues, sequencing reactions with the M1 primer (10 pmol/μl) were performed with a model 373A automated sequencer (Applied Biosystems/Perkin Elmer, Foster City, Calif.) at the DNA sequencing facility, Institute for Clinical Pathology and Medical Research, Westmead Hospital. The M. paraffinicium positive control was sequenced with each batch of samples to provide a sequencing quality control.
DNA sequences obtained from both paraffin-embedded and fresh tissues were aligned with the multiple sequence alignment program CLUSTAL W, which is part of the sequence analysis package of the Genetics Computer Group (Madison, Wis.), accessed via the Australian National Genomic Information Service (ANGIS). The consensus sequence, represented by GenBank Accession Number AF144747, was compared with sequences deposited in the EMBL, GenBank, DDBJ, and PDB databases by use of the algorithm of Altschul and colleagues (1) and the BLASTN program available via the ANGIS.
(iii) RCBH. (a) Probes.
Reverse cross blot hybridization (RCBH) was performed on the 16S rRNA PCR products obtained from the paraffin-embedded tissues from 11 dogs. Amplicons were tested against a series of oligonucleotides targeted to the V2 region of 16S rRNA gene sequences of mycobacterial species and an oligonucleotide targeted to a conserved region (nucleotides 157 to 178 of the 16S rRNA gene of E. coli) of 16S rRNA gene sequences of mycobacterial species (Table 1). The oligonucleotides were synthesized with a 5′-terminal amino group (Genosys) to allow covalent binding to an activated, negatively charged Biodyne C membrane (Pall Biosupport).
TABLE 1.
Oligonucleotide sequences of probes and probe concentrations used in the RCBHa
| Probe | Target | Sequence | Probe concn (pmol/150 μl) |
|---|---|---|---|
| DPr | Potential novel mycobacterial species from dogs | 5′-ACCGGATATGACCACGAAGCGC-3′ | 100, 50, 25 |
| Y2Pr | Potential novel mycobacterial species from cattle | 5′-GGACCTCTCGGCGCATGCCTAA-3′ | 50, 25 |
| TB | M. tuberculosis complex | 5′-GGACCACGGGATGCATGTCTTGT-3′ | 50, 25 |
| 16SPr | 16S rRNA of mycobacteria | 5′-TGGGAAACTGGGTCTAATACCG-3′ | 600, 400 |
| Kans. | M. kansasii | 5′-GGACCACTTGGCGCATGCCTTGT-3′ | 100, 50 |
| Mal. | M. malmoense | 5′-GGACCCCGAGGCGCATGCCTTGG-3′ | 100, 50 |
| Intra. | M. intracellulare | 5′-GGACCTTTAGGCGCATGTCTTTA-3′ | 50, 25 |
| Lenti. | M. lentiflavum | 5′-GGACCTTTTGGCGCATGCCTTTT-3′ | 100, 25 |
| Mar. | M. marinum | 5′-GGACCACGGGATTCATGTCCTGT-3′ | 50, 25 |
| Avium | M. avium complex | 5′-GGACCTCAAGACGCATGTCTTCT-3′ | 100, 50 |
| Hiber. | M. hiberniae | 5′-GGACCGCGCGCTTCATGGTGTGT-3′ | 50, 25 |
| Nonchr. | M. nonchromogenicum | 5′-GGACCGCATGCTGCATGGTGTGT-3′ | 50, 25 |
| Fort. | M. fortuitum | 5′-TGACCACGCGCTTCATGGTGTGT-3′ | 50, 25 |
The probes were targeted to the V2 region of the 16S rRNA gene of mycobacterial species, except for 16SPr; the target for this probe, spanning nucleotide positions 157 to 178 of the 16S rRNA gene of E. coli, is a conserved region of the 16S rRNA gene for mycobacterial species but is not genus specific.
(b) Membrane preparation.
The Biodyne C membrane was activated before application of the oligonucleotides as described previously (18). Briefly, the membrane was incubated for 10 min in 16% (wt/vol) 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (Sigma Chemical Co.) and rinsed with deionized water for 2 min before being placed into a Miniblotter (MN45; Immunetics, Cambridge, Mass.). Aliquots (150 μl) of oligonucleotides in 500 mM NaHCO3 (pH 8.4) were applied to the Miniblotter slots in parallel lines, incubated for 1 min, and removed by aspiration. The membrane was inactivated in 100 mM NaOH for 10 min, washed twice in 250 ml of 2× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7])–0.1% SDS for 5 min each time at 60°C, and washed subsequently in 100 ml of 20 mM EDTA (pH 8.0) for 15 min at room temperature.
(c) Hybridization assay.
RCBH was performed by a modification of the reverse line blotting assay used for spoligotyping (18). The blot was washed in 250 ml of 2× SSPE–0.1% SDS for 5 min at 60°C and placed in the Miniblotter as described previously (20) so that the Miniblotter slots were perpendicular to the lines of applied oligonucleotides. Aliquots (20 μl) of PCR products in 150 μl of 2× SSPE–0.1% SDS were heat denatured for 10 min at 100°C and applied to the slots. Hybridization was performed at 60°C for 60 min. The membrane was washed subsequently three times in 250 ml of 2× SSPE–0.5% SDS for 10 min each time at 60°C and incubated in 10 ml of a 1:4,000 dilution of streptavidin-peroxidase conjugate (Boehringer) in 2× SSPE–0.5% SDS at 42°C for 45 min. Four washes of the membrane were performed with 250 ml of 2× SSPE–0.5% SDS for 5 min each time at 42°C, followed by two further washes with 250 ml of 2× SSPE for 5 min each time at room temperature. Chemiluminescence was detected by incubating the blot for 1 min in 20 ml of ECL detection liquid (Amersham International) and visualized by exposure to light-sensitive autoradiography film (Hyperfilm ECL; Amersham) for 1 h.
Nucleotide sequence accession number.
The sequence determined in this study was deposited in GenBank under accession no. AF144747.
RESULTS
PCR amplification.
PCR products (approximately 550 bp) containing the V2 and V3 regions of the 16S rRNA gene were obtained for 7 of the 37 paraffin-embedded tissue specimens with primers pA and MSH-E (Table 2). Amplification of the V2 region with primer pair MSH-A–MSH-B was carried out on all specimens which did not yield amplicons with primers pA and MSH-E and on samples 6 and 30 (from which amplicons had been obtained with primers pA and MSH-E) as positive controls. A further four specimens (2, 8, 29, and 31) yielded PCR products (approximately 120 bp) with the second set of primers (Table 2). Nested PCR products of the expected size (469 bp) were obtained from all nine fresh tissue samples. Specimens from one dog (dog 7) yielded products of 550 bp from paraffin-embedded tissue and 469 bp from fresh tissue (Table 2). In total, PCR amplicons were obtained for 20 of 46 specimens from 43 dogs.
TABLE 2.
Results of PCR amplification and species identification for formalin-fixed, paraffin-embedded tissue and fresh tissue specimens from dogs with canine leproid granuloma
| Specimen no. | Specimen typea | Acid-fast bacillib | Resultc obtained with the following:
|
Species identitye | |||||
|---|---|---|---|---|---|---|---|---|---|
| 16S rRNA primer pair
|
Identification method
|
||||||||
| pA–MSH-E | MSH-A–MSH-B | PS2-M1 | DNA sequencing | RCBH with probed
|
|||||
| 16SPr | DPr | ||||||||
| 2 | P | L | − | + | ND | ND | + | + | Novel |
| 3 | P | L | + | ND | ND | + | + | + | Novel |
| 5 | P | L | + | ND | ND | + | + | ND | Novel |
| 6 | P | M | + | + | ND | ND | + | − | U |
| 7 | P | M | + | ND | ND | + | + | + | Novel |
| F | ND | ND | + | + | ND | ND | Novel | ||
| 8 | P | L | − | + | ND | ND | + | + | Novel |
| 9 | P | S-M | − | − | ND | ||||
| 10 | P | S | + | ND | ND | ND | + | − | U |
| 12 | P | M | − | − | ND | ||||
| 13 | P | M | + | ND | ND | + | + | + | Novel |
| 14 | P | M-L | − | − | ND | ||||
| 15 | P | S | − | − | ND | ||||
| 16 | P | R | − | − | ND | ||||
| 17 | P | S | − | − | ND | ||||
| 18 | P | S | − | − | ND | ||||
| 19 | P | R | − | − | ND | ||||
| 20 | P | S | − | − | ND | ||||
| 23 | P | M | − | − | ND | ||||
| 24 | P | S | − | − | ND | ||||
| F | ND | ND | + | Novel | |||||
| 25 | P | M | − | − | ND | ||||
| 26 | P | None | − | − | ND | ||||
| 28 | P | S | − | − | ND | ||||
| 29 | P | M | − | + | ND | ND | + | + | Novel |
| 30 | P | R | + | + | ND | ND | + | − | U |
| 31 | P | R | − | + | ND | ND | + | + | Novel |
| 32 | P | M | − | − | ND | ||||
| 33 | P | S | − | − | ND | ||||
| 34 | P | R | − | − | ND | ||||
| 35 | P | S | − | − | ND | ||||
| 36 | P | L | − | − | ND | ||||
| 39 | P | S | − | − | ND | ||||
| 40 | P | S-M | − | − | ND | ||||
| 41 | P | L | − | − | ND | ||||
| 42 | P | L | − | − | ND | ||||
| 43 | P | NA | − | − | ND | ||||
| 44 | P | M-L | − | − | ND | ||||
| F | ND | ND | + | + | ND | ND | Novel | ||
| 45 | P | NA | − | − | ND | ||||
| 46 | F | S | ND | ND | + | + | ND | ND | Novel |
| 47 | F | S | ND | ND | + | + | ND | ND | Novel |
| 48 | F | M | ND | ND | + | + | ND | ND | Novel |
| 49 | F | M | ND | ND | + | + | ND | ND | Novel |
| 50 | F | M | ND | ND | + | + | ND | ND | Novel |
| 51 | F | L | ND | ND | + | + | ND | ND | Novel |
P, formalin-fixed, paraffin-embedded tissue; F, fresh tissue specimen.
L, large; M, moderate; S, small; R, rare; NA, not available.
+, positive, −, negative; ND, not done.
16SPr, general probe for 16S rRNA gene sequences; DPr, probe for novel 16S rRNA sequences from canine leproid granuloma syndrome cases.
Novel, novel mycobacterial sequence from canine leproid granuloma syndrome cases; U, species identity undetermined.
DNA sequence analysis.
Preliminary RCBH analysis of PCR products (approximately 550 bp) for specimens 3 and 5 demonstrated hybridization with the general probe for 16S rRNA mycobacterial sequences. However, the identities of these products could not be determined by RCBH, since they did not hybridize with any of the other 16S rRNA probes for mycobacterial species (Table 1). The consensus sequence obtained from specimens 3 and 5 (Fig. 1) and represented by GenBank accession no. AF144747 had the highest nucleotide identity (99.4% over 519 bp) with mycobacterial species IWGMT 90143 (35) but did not correspond exactly to any EMBL or GenBank database sequences. 16S rRNA amplicons (approximately 550 bp) from specimens 7 and 13 were sequenced subsequently and were identical to the sequence obtained from specimens 3 and 5. All nine fresh tissues yielded sequences identical to each other and to the four sequences from paraffin-embedded tissues over 350 bp of the 16S rRNA gene encompassing portions of the V2 and V3 regions. Thus, the same sequence was obtained from a total of 13 specimens from 12 dogs with canine leproid granuloma syndrome. The sequence also shared high nucleotide identity with M. tilburgii (99% over 510 bp) and with the described species M. simiae, M. interjectum, and M. genavense (98.8% over 519 bp) (differences attributed to n nucleotides are not considered). A probe (DPr) for the V2 region of this unique 16S rRNA gene sequence (Table 1) was generated to allow further amplicons to be analyzed more rapidly by RCBH for the presence of this novel mycobacterial sequence.
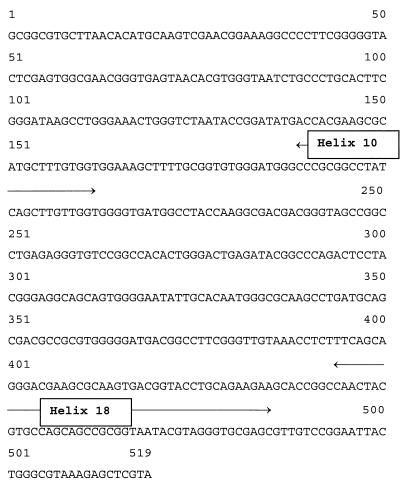
FIG. 1.
Nucleotide sequence of the 16S rRNA gene amplicons of specimens 3, 5, 7, and 13. The sequence starts at nucleotide position 2 and finishes at nucleotide position 544 of aligned mycobacterial 16S rRNA gene sequences (27). Helix 10 and helix 18 are indicated.
RCBH.
Biotinylated PCR products (approximately 550 bp and approximately 120 bp) obtained from paraffin-embedded specimens with primer pairs pA–MSH-E and MSH-A–MSH-B, respectively, were tested by RCBH against the DPr probe and several other 16S rRNA oligonucleotide probes for mycobacterial species. All of the amplicons tested hybridized to the general 16S rRNA probe, confirming their identities as 16S rRNA gene products. Amplicons from specimens 2, 3, 7, 8, 13, 29, and 31 hybridized to the DPr probe (Table 2). The efficacy of this probe and the suitability of the probe concentrations selected were confirmed by strong hybridization of DPr with amplicons from specimens 3 and 7 and from specimen 13 (data not shown). The latter served as positive controls in the RCBH assay, since sequence analysis of these amplicons had indicated previously the presence of this probe sequence. Strong hybridization signals also were obtained with amplicons from specimens 2, 8, and 29. A weak hybridization signal with the DPr probe was observed for the amplicon from specimen 31; this amplicon was associated with a weak hybridization signal with the general probe, also indicating that the quantity of amplicon applied to the blot had been overestimated. Amplicons (approximately 550 bp) from specimens 6, 10, and 30, associated with less intense hybridization, hybridized to the general probe only. The amplicon (approximately 120 bp) obtained for specimen 30 with primer pair MSH-A–MSH-B also hybridized to the general probe only but at a strong intensity.
DISCUSSION
Sequence determination of the V2 and V3 regions of the 16S rRNA gene has been recommended for the rapid identification of mycobacterial species (22, 29). This report describes PCR amplification and nucleotide sequence analysis of 16S rRNA gene sequences directly from clinical specimens of canine leproid granuloma syndrome. A unique and identical 16S rRNA gene sequence, comprising the V2 and V3 regions, was isolated from 13 lesion specimens from 12 dogs, and RCBH identified this sequence in a further 4 dogs. Importantly, also, one dog from which both paraffin-embedded and fresh materials were processed yielded the same sequence in two different laboratories, thus validating the sequence data. This result is significant, since novel sequences may be simulated by sequencing and amplification errors and chimeric molecule formation (10, 36). The sequence obtained from lesions from the 16 dogs appears to represent the predominant prokaryotic species in the tissues, as nonmycobacterial DNA was not detected, even though one laboratory used PCR primers which could amplify other bacterial species (15). Similarly, the potentially novel mycobacterial species represented by this sequence would appear to be the principal mycobacterial species in this syndrome, as only three specimens yielded amplicons which did not hybridize to the probe for the sequence. However, the samples from these dogs came from lesions on the pinnae and were grossly and histologically the same as the samples from which the novel sequence was obtained.
The sequence from the canine specimens shared the most nucleotide identity with mycobacterial isolate IWGMT 90143. Semantide and chemotaxonomy-based analyses of isolate IWGMT 90143 have resulted in its probable classification as a ribovar of M. simiae (35). The unique sequence in this study had high nucleotide identity with M. tilburgii, M. simiae, M. interjectum, and M. genavense as well. The fully described species M. simiae, M. interjectum, and M. genavense are pathogenic, slowly growing mycobacterial species which are unusual in having a short helix 18, which would otherwise be characteristic of rapidly growing mycobacteria (11). The V3 region, containing helix 18, is of particular value for higher-order taxonomic assignments (34). Analysis of the V3 region of the unique sequence indicated that the sequence is representative of the M. simiae clade of slowly growing mycobacteria (B. Springer, personal communication).
It was thought that the high nucleotide identity between the unique sequence and the described species with fastidious growth requirements may have facilitated determination of the growth conditions of this potentially novel mycobacterial species. The culture requirements of M. tilburgii isolated from a disseminated infection in an immunocompetent person have not yet been reported. Of the three other mycobacterial species sharing high nucleotide identity with the unique 16S rRNA gene sequence from canines, M. genavense has fastidious growth requirements (3, 28). However, preliminary attempts that have used atmospheric and nutritional conditions known to favor the growth of M. genavense have failed to allow the isolation of acid-fast bacilli.
Full 16S rRNA analysis may provide more insight into the taxonomic status of the sequence identified from canine leproid granuloma syndrome samples. Determination of DNA-DNA hybridization values also may be useful in clarifying its taxonomy. Different mycobacterial species may have identical 16S rRNA gene sequences (11), and recently diverged species may not be differentiated by 16S rRNA gene analysis (9). DNA-DNA hybridization studies were not considered here because the inability to culture the mycobacterial species limited the amount of DNA available for such studies. Discrepancies in DNA-DNA hybridization values may occur due to experimental conditions (24), and although DNA-DNA hybridization is considered the gold standard, a polyphasic taxonomy approach has been recommended (35).
In this study, the value of molecular methods for the identification of an as-yet-uncultured mycobacterial species from canine leproid granuloma syndrome samples is evident. Two different molecular methods, 16S rRNA sequence analysis and RCBH, were used to analyze PCR products obtained from canine leproid granuloma syndrome specimens. The latter type of assay, involving hybridization of PCR products to 16S rRNA probes attached to a solid support, has been used previously with success for the identification of mycobacteria from clinical specimens and cultures (7, 23, 31). Nucleotide sequence determination of PCR products from canine leproid granuloma syndrome specimens was necessary since the amplicons did not hybridize to any of the probes used in the initial RCBH assay. In agreement with earlier studies, RCBH allowed more rapid diagnosis of a larger number of specimens than 16S rRNA sequencing. The former technique also has the advantage of being able to identify mixed populations of mycobacterial species, which have been previously associated with veterinary specimens (13, 15). A potential problem associated with RCBH is the possible need to standardize the concentration of the PCR product used for hybridization. It has been reported that different amounts of PCR products did not influence the intensity of the hybridization signal (31), but in this study some variation in intensity was noted. In contrast to what was done in previous studies (7, 23, 31), the probe concentrations in this study were varied, as for spoligotyping, to enhance the hybridization of individual probes. Such assays may require optimization before use as diagnostic tests.
A difference in the efficacy of PCR amplification of DNA extracted from fresh tissues versus paraffin-embedded tissues was apparent. The former was more successful and sensitive; products were obtained for all nine specimens which were associated with variable (small to large) numbers of acid-fast bacilli. Different extraction procedures and PCR conditions were used for the fresh tissues and the formalin-fixed, paraffin-embedded tissues, since analyses of the two types of specimens were performed separately in two different laboratories. However, the greater success of the PCR with fresh tissue specimens was probably more attributable to a larger specimen size with a correspondingly larger total input of bacteria available for amplification. DNA degradation due to formalin fixation may have contributed to the failure to obtain amplicons from some of the specimens which underwent various formalin fixation treatments. In some cases, a product of approximately 120 bp was obtained with primer pair MSH-A–MSH-B, whereas a product of approximately 550 bp was not obtained with primer pair pA–MSH-E, although the higher efficiency of amplification of smaller targets also may have influenced these results.
This study has provided an initial step in identifying and characterizing a probable etiological agent of canine leproid granuloma syndrome. A specific PCR-based diagnostic test could be developed for more rapid identification of this species from further cases of the disease. Further molecular characterization of the sequence should help clarify the taxonomic position of the species. The inability at present to culture this mycobacterial species limits experimental reproduction of the disease and also prevents full semantide and chemotaxonomy-based analyses to be performed. With increasing use of molecular diagnostic methods, it is envisaged that this problem will become more prevalent. Validly described culturable prokaryotes do not represent the diversity of actual prokaryotic species (36). The possibility of other causative agents of canine leproid granuloma syndrome requires further study as well. Amplicons obtained from three canine leproid granuloma syndrome specimens did not hybridize to the probe for the novel sequence or to any of the other mycobacterial species-specific probes used in this study. The amplicons (approximately 550 bp) obtained for these specimens with primer pair pA–MSH-E hybridized with less intensity to the general probe, suggesting that lower concentrations of DNA were used in the assay for these amplicons than for most of the other amplicons; this situation could have resulted in false-negative hybridization results with the DPr probe. However, the amplicon (approximately 120 bp) obtained for specimen 30 with primer pair MSH-A–MSH-B hybridized to the general probe only with a strong intensity, suggesting the possible involvement of another species in this syndrome. The identities of these amplicons can be established by nucleotide sequence analysis to determine their involvement in this disease.
Thus, while it is possible that other mycobacterial species may be associated with this syndrome, the results presented here implicate a potentially novel mycobacterium as a probable important etiological agent in canine leproid granuloma syndrome. The challenges for further investigations include finding the ecological niche of the species, growing the microorganisms, and clarifying whether they are a single species. The methodologies described here may well assist in these investigations, and the taxonomic placement of the species should aid in the development of conditions for its cultivation in vitro.
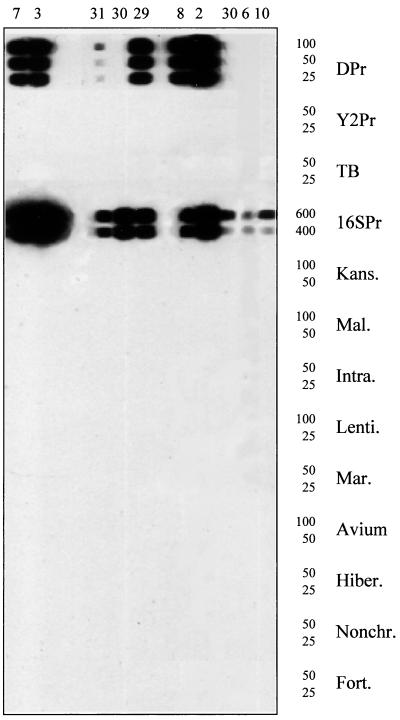
FIG. 2.
RCBH of 16S rRNA amplicons from formalin-fixed, paraffin-embedded tissue specimens from dogs with canine leproid granuloma syndrome by use of mycobacterial probes and the general probe 16SPr. Probe names (Table 1) and concentrations (picomoles/150 μl) are shown. Hybridization patterns of PCR amplicons for specimens 7 (∼550 bp), 3 (∼550 bp), 31 (∼120 bp), 30 (∼120 bp), 29 (∼120 bp), 8 (∼120 bp), 2 (∼120 bp), 30 (∼550 bp), 6 (∼550 bp), and 10 (∼550 bp) are shown.
ACKNOWLEDGMENTS
We thank R. A. Skuce, M. J. Taylor, and D. Brittain, Veterinary Sciences Division, for their assistance and B. Springer and E. C. Böttger, Medizinische Hochschule Hannover, for helpful information regarding the sequence isolated from the canine leproid granuloma specimens. This work benefited from the use of the HGMP-RC Bioinformatics Service.
R. Malik is supported by the Valentine Charlton Bequest of the Post Graduate Foundation in Veterinary Science of the University of Sydney.
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programmes. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Böddinghaus B, Rogall T, Flohr T, Blöcker H, Böttger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Böttger E C, Hirschel B, Coyle M B. Mycobacterium genavense sp. nov. Int J Syst Bacteriol. 1993;43:841–843. doi: 10.1099/00207713-43-4-841. [DOI] [PubMed] [Google Scholar]
- 4.Charles J, Martin P, Wigney D I, Malik R, Love D N. Histopathology of canine leproid granuloma syndrome. Aust Vet J. 1999;77:799–803. doi: 10.1111/j.1751-0813.1999.tb12948.x. [DOI] [PubMed] [Google Scholar]
- 5.Craxton M. Cosmid sequencing. Methods Mol Biol. 1993;23:149–67. doi: 10.1385/0-89603-248-5:149. [DOI] [PubMed] [Google Scholar]
- 6.Dawson D J. A simple identification scheme for mycobacteria. Aust J Med Technol. 1971;2:7–15. [Google Scholar]
- 7.De Beenhouwer H, Liang Z, de Rijk P, van Eekeren C, Portaels F. Detection and identification of mycobacteria by DNA amplification and oligonucleotide-specific capture plate hybridization. J Clin Microbiol. 1995;33:2994–2998. doi: 10.1128/jcm.33.11.2994-2998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards U, Rogall T, Blöcker H, Emde M, Böttger E C. Isolation and direct sequencing of entire genes. Characterization of a gene coding for 16S ribosomal RNA. Nucleic Acids Res. 1989;17:7843–7853. doi: 10.1093/nar/17.19.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 10.Gest H. Bacterial classification and taxonomy: a ‘primer’ for the new millennium. Microbiol Today. 1999;26:70–72. [Google Scholar]
- 11.Goodfellow M, Magee J G. Taxonomy of mycobacteria. In: Gangadharam P R J, Jenkins P A, editors. Mycobacteria. I. Basic aspects. Boston, Mass: Kluwer Academic Publishers; 1997. pp. 1–50. [Google Scholar]
- 12.Greenfield L, White T J. Sample preparation methods. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C.: American Society for Microbiology; 1993. pp. 122–137. [Google Scholar]
- 13.Hughes M S, Skuce R A, Beck L-A, Neill S D. Identification of mycobacteria from animals by restriction enzyme analysis and direct DNA cycle sequencing of polymerase chain reaction-amplified 16S rRNA gene sequences. J Clin Microbiol. 1993;31:3216–3222. doi: 10.1128/jcm.31.12.3216-3222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes M S, Beck L-A, Skuce R A. Identification and elimination of DNA sequences in Taq DNA polymerase. J Clin Microbiol. 1994;32:2007–2008. doi: 10.1128/jcm.32.8.2007-2008.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes M S, Ball N W, Beck L-A, deLisle G W, Skuce R A, Neill S D. Determination of the etiology of presumptive feline leprosy by 16S rRNA gene analysis. J Clin Microbiol. 1997;35:2464–2471. doi: 10.1128/jcm.35.10.2464-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes M S, Ball N W, Love D N, Canfield P J, Wigney D I, Dawson D, Davis P E, Malik R. Disseminated Mycobacterium genavense infection in a FIV-positive cat. J Feline Med Surg. 1999;1:23–29. doi: 10.1016/S1098-612X(99)90006-2. [DOI] [PubMed] [Google Scholar]
- 17.Jackson K, Sievers A, Ross B C, Dwyer B. Isolation of a fastidious Mycobacterium species from two AIDS patients. J Clin Microbiol. 1992;30:2934–2937. doi: 10.1128/jcm.30.11.2934-2937.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katila M L, Mattila J, Brander E. Enhancement of growth of Mycobacterium malmoense by acidic pH and pyruvate. Eur J Clin Microbiol Infect Dis. 1989;8:998–1000. doi: 10.1007/BF01967574. [DOI] [PubMed] [Google Scholar]
- 20.Kaufold A, Podbielski A, Baumgarten G, Blokpoel M, Top J, Schouls L. Rapid typing of group A streptococci by the use of DNA amplification and non-radioactive allele-specific oligonucleotide probes. FEMS Microbiol Lett. 1994;119:19–26. doi: 10.1111/j.1574-6968.1994.tb06861.x. [DOI] [PubMed] [Google Scholar]
- 21.Kempsell K E, Ji Y-E, Estrada I C, Colston M J, Cox R A. The nucleotide sequence of the promoter, 16S rRNA and spacer region of the ribosomal RNA operon of Mycobacterium tuberculosis and comparison with Mycobacterium leprae precursor rRNA. J Gen Microbiol. 1992;138:1717–1727. doi: 10.1099/00221287-138-8-1717. [DOI] [PubMed] [Google Scholar]
- 22.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F-C, Böttger E C. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kox L F F, van Leeuwen J, Knijper S, Jansen H M, Kolk A H J. PCR assay based on DNA coding for 16S rRNA for detection and identification of mycobacteria in clinical samples. J Clin Microbiol. 1995;33:3225–3233. doi: 10.1128/jcm.33.12.3225-3233.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lévy-Frébault V V, Portaels F. Proposed minimal standards for the genus Mycobacterium and for description of new slowly growing Mycobacterium species. Int J Syst Bacteriol. 1992;42:315–323. doi: 10.1099/00207713-42-2-315. [DOI] [PubMed] [Google Scholar]
- 25.Malik R, Love D N, Wigney D I, Martin P. Mycobacterial nodular granulomas affecting the subcutis and skin of dogs (canine leproid granuloma syndrome) Aust Vet J. 1998;76:403–407. doi: 10.1111/j.1751-0813.1998.tb12388.x. [DOI] [PubMed] [Google Scholar]
- 26.Mangiapan G, Vokurka M, Schouls L, Cadranel J, Lecossier D, van Embden J, Hance A J. Sequence capture-PCR improves detection of mycobacterial DNA in clinical specimens. J Clin Microbiol. 1996;34:1209–1215. doi: 10.1128/jcm.34.5.1209-1215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramis A, Ferrer L, Aranaz A, Liébana E, Mateos A, Domínguez L, Pascual C, Fdez-Garayazabal J, Collins M D. Mycobacterium genavense infection in canaries. Avian Dis. 1996;40:246–251. [PubMed] [Google Scholar]
- 28.Realini L, de Ridder K, Palomino J-C, Hirschel B, Portaels F. Microaerophilic conditions promote growth of Mycobacterium genavense. J Clin Microbiol. 1998;36:2565–2570. doi: 10.1128/jcm.36.9.2565-2570.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogall T, Flohr T, Böttger E C. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol. 1990;136:1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- 30.Rogall T, Wolters J, Flohr T, Böttger E C. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int J Syst Bacteriol. 1990;40:323–330. doi: 10.1099/00207713-40-4-323. [DOI] [PubMed] [Google Scholar]
- 31.Sanguinetti M, Posteraro B, Ardito F, Zanetti S, Cingolani A, Sechi L, De Luca A, Ortona L, Fadda G. Routine use of PCR-reverse cross-blot hybridization assay for rapid identification of Mycobacterium species growing in liquid media. J Clin Microbiol. 1998;36:1530–1533. doi: 10.1128/jcm.36.6.1530-1533.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith R I E. Canine skin tuberculosis. Rhod Vet J. 1973;3:63–64. [Google Scholar]
- 33.Sompolinsky D, Lagziel A, Naveh D, Yankilewitz T. Mycobacterium haemophilum sp. nov., a new pathogen of humans. Int J Syst Bacteriol. 1978;28:67–75. [Google Scholar]
- 34.Springer B, Stockman L, Teschner K, Roberts G D, Böttger E C. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996;34:296–303. doi: 10.1128/jcm.34.2.296-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wayne L G, Good R C, Böttger E C, Butler R, Dorsch M, Ezaki T, Gross W, Jonas V, Kilburn J, Kirschner P, Krichevsky M I, Ridell M, Shinnick T M, Springer B, Stackebrandt E, Tarnok I, Tarnok Z, Tasaka H, Vincent V, Warren N G, Knott C A, Johnson R. Semantide- and chemotaxonomy-based analyses of some problematic phenotypic clusters of slowly growing mycobacteria, a cooperative study of the international working group on mycobacterial taxonomy. Int J Syst Bacteriol. 1996;46:280–297. doi: 10.1099/00207713-46-1-280. [DOI] [PubMed] [Google Scholar]
- 36.Winztingerode F, Göbel U B, Stackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]