Abstract
The PDX-1 transcription factor plays a key role in pancreatic development and in the regulation of the insulin gene in the adult β cell. As its functions appear to be similar in humans and mice, we analyzed the functional conservation of homologous sequences important for the maintenance and the cell-specific regulation of the pdx-1 gene. Apart from the proximal promoter region, three highly homologous (PH1 to PH3) sequences were apparent in the human and mouse 5′ flanking regions of the gene. By transient transfections in β and non-β cells, we show that mainly PH1 and PH2 preferentially confer β-cell-specific activation on a heterologous promoter. DNase I footprinting and binding analyses revealed that both bind to and are transactivated by hepatocyte nuclear factor 3β (HNF-3β). Furthermore, the PH1 enhancer element also binds the PDX-1 transcription factor itself, which acts cooperatively with adjacent HNF-3β to regulate its transcriptional potency. This finding suggests a possible autoregulatory loop as a mechanism for PDX-1 to control its own expression.
The mammalian pancreas develops from the endoderm in the upper duodenal part of the foregut as dorsal and ventral buds which fuse together to form the organ. Recently, considerable progress has been made in identifying genes that control the embryonic development of the islet. Most are transcription factors such as isl-1, PDX-1, BETA2 (also called NeuroD), Nkx 2.2, Pax4, and Pax6 (for reviews, see references 8, 9, 14, 15, 26, 30, and 31). The first molecular marker identified that specifies the early pancreatic epithelium was the homeodomain-containing transcription factor PDX-1 (1, 2, 13). PDX-1 function appears to be well conserved. Its absence in both humans and mice leads to agenesis of the pancreas, while in the adult pancreatic islet, its expression is restricted to the β cell, where it acts as the glucose-sensitive transcription factor of the insulin gene (19).
Initiation of transcription appears to be the major level of regulation for many cell type-specific genes. To elucidate the regulation of pdx-1 gene expression, a 6.5-kb fragment upstream of the transcription start site of the rat pdx-1 gene (also called stf-1) (28) and a fragment extending from the region from kb −4.5 to +8.2 of the mouse pdx-1 gene (33) were shown to direct the expression of the β-galactosidase (β-Gal) reporter gene to pancreatic islet cells in transgenic mice. Using transiently transfected β cells, it was shown that the expression of the rat pdx-1 gene, apart from a proximal E box, depended on a distal enhancer element that was activated by the cooperative action of the endodermal transcription factors hepatocyte nuclear factor 3β (HNF-3β) and BETA2 (27). A study of the mouse pdx-1 promoter revealed that the region involved in regulating β-cell-specific transcription and directing appropriate developmental and adult-specific expression in transgenic animals contained an HNF-3-like element (33). Thus, these initial studies with the rat and the mouse enhancer-promoter regions suggested that HNF-3β was necessary for the transcriptional regulation of pdx-1.
As it appears that PDX-1 functions are similar in humans and rodents, we deduced that conserved sequences located within the 5′ flanking region of the gene would be of importance for its expression. To this end, we compared sequences 4.5 kb upstream of the start sites of both the mouse and human pdx-1 genes. The analysis revealed that in addition to the proximal promoter region described by Sharma et al. (28), only three relatively short sequences were highly homologous and they were designated PH1, PH2, and PH3 (for PDX-1 homology regions 1 to 3). Using transient-transfection experiments, we show that it is preferentially PH1 and PH2 that drive β-cell-specific expression of a reporter gene. Interestingly, using DNase I footprinting analyses and electrophoretic mobility shift assays (EMSAs), we demonstrate that PH1 binds the transcription factor PDX-1 itself adjacent to the endodermal factor HNF-3β, where they act cooperatively to transactivate a PH1-driven reporter construct. Furthermore, we show that these transcription factors are able to directly interact in vitro. Similar binding experiments reveal that only HNF-3β mediates the transcriptional activity of the PH2 domain. Our results therefore suggest a possible feedback mechanism whereby PDX-1 regulates its own expression.
MATERIALS AND METHODS
Cell cultures.
Hamster insulinoma HIT-T15, mouse insulinoma βTC6, and mouse glucagonoma αTC1 cells were cultured in Dulbecco's modified Eagle medium with 15% horse serum and 2.5% fetal calf serum, and with 10% fetal calf serum for CHO, HepG2, COS, and NIH 3T3 cells. One hundred units of penicillin per ml and 100 mg of streptomycin per ml were added to the media.
Cell transfections.
HIT-T15, CHO, and NIH 3T3 cells were transfected using the calcium phosphate coprecipitation method (25) with 1.5-μg quantities of human PDX-1 luciferase derivatives and 0.5 μg of the internal control cytomegalovirus β-Gal DNA plasmid. In cotransfection experiments, 1.5 μg of the reporter plasmid together with increasing amounts of expression plasmids were used as indicated in the figure legends. The cells were harvested 48 h after transfection. Approximately 100 μg of protein extracts was used to measure luciferase activity with the Luciferase Assay System (Promega, Madison, Wis.), and approximately 10 μg was used for the β-Gal assay as described previously (25). Luciferase activity was measured with a luminometer (EG&G Berthold, Bad Wildbad, Germany) and normalized to β-Gal values. COS cells were transfected with an expression plasmid for the HNF-3β and HNF-3α genes.
Plasmid constructions.
Plasmids containing fragments of the human pdx-1 promoter were kindly provided by Alan Permutt (University of Washington, St. Louis, Mo.). We subcloned and mapped about 8 kb of the 5′ flanking region of the gene and sequenced a DNA fragment from approximately kb −4.5 to +0.1. The homologous regions between the mouse and the human pdx-1 genes (PH1, PH2, and PH3) were synthesized by PCR and subcloned upstream of the minimal thymidine kinase (TK) promoter of the herpes simplex virus linked to the luciferase gene (TKLuc), creating PH1- to PH3-TKLuc. Mutations were also created by PCR, and each construct was validated by sequencing. A human PDX-1 expression plasmid was constructed as described in reference 17. Glutathione S-transferase (GST)–PDX-1 was constructed by subcloning the human PDX-1 cDNA into pGEX-2T (Amersham Pharmacia Biotech, Uppsala, Sweden) and expressed in Escherichia coli. The rat HNF-3β cDNA (kindly provided by R. H. Costa, University of Illinois, Chicago) was subcloned into pcDNA3.
Preparation of cell extracts.
Whole-cell extracts (WCE) were prepared by resuspension of the cells in high-salt extraction buffer (400 mM KCl, 20 mM Tris [pH 7.5], 20% glycerol, 2 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 20 μg of aprotinin per ml, 10 μg of leupeptin per ml). Cell lysis was performed by freezing and thawing, and the cellular debris was removed by centrifugation at 16,000 × g for 15 min at 4°C. Protein concentrations were determined by the Bradford method (4).
EMSA.
DNA-binding reactions were performed by incubating 10 μg of WCE with 0.3 ng of 32P-labeled synthetic double-stranded oligodeoxynucleotides spanning protected areas I and II of PH1 and protected area I of PH2 in ice for 20 min. The binding reaction mixtures contained 20 mM HEPES (pH 7.9), 10% glycerol, 150 mM NaCl, 1 mM dithiothreitol, and 1 μg of poly(dI-dC). Competitor oligonucleotides were incubated in 100-fold molar excess in the reaction mixtures for 10 min prior to the addition of the radiolabeled probe. Oligonucleotides were end labeled by a fill-in reaction using the Klenow fragment of DNA polymerase I. For supershift experiments, 1 μl of antibodies was added during the preincubation period. The oligonucleotides used corresponded to area I of PH1 (5′-ACACTTTAATTGGTTTACAG-3′), its mutant (5′-ACACTTcgcTGGTTTACAG-3′), area II (5′-CTTTTTTGTTTATTTATCCATA-3′), a mutant of this mutant (5′-CTTTTTTGcgTATTTATCCATA-3′), area I of the PH2 domain (5′-AGTGCAAAGTAAACACTCCGG-3′), and its corresponding mutant (5′-GAAGTGCAAcGTcggtgCTCCGGG-3′), where lowercase letters indicate the mutations.
DNase I footprint analysis.
For DNase I footprinting assays, fragments containing PH1 and PH2 (from kb −2.86 to −2.60 and from kb −2.24 to −2.05) were labeled at either end by a fill-in reaction using the Klenow fragment of DNA polymerase I and [32P]dCTP to a specific activity greater than 104 cpm/ng of DNA. Probes were incubated with 40 μg of WCE in a 50-μl reaction mixture containing 10 mM Tris (pH 7.8), 14% glycerol, 57 mM KCl, 4 mM dithiothreitol, and 0.2 μg of poly(dI-dC). Following 20 min of incubation at room temperature, 0.5 to 1 U of DNase I (Promega) diluted in 50 mM MgCl2–10 mM CaCl2 was added for 1 min. The reaction was stopped by adding 150 μl of a stop solution containing 200 mM NaCl, 20 mM EDTA, 1% sodium dodecyl sulfate, and 5 μg of Saccharomyces cerevisiae tRNA. DNA was extracted with phenol-chloroform, precipitated with ethanol, and analyzed on a denaturing 6% polyacrylamide gel. Sequencing reactions with each probe were performed using the Maxam and Gilbert procedure (18).
GST pull-down assay.
35S-labeled HNF-3β polypeptide was produced using the TNT rabbit reticulocyte lysate-coupled transcription-translation system (Promega) according to the manufacturer's protocol. The labeled HNF-3β was incubated with either GST or GST–PDX-1 proteins bound to glutathione-Sepharose beads in a binding buffer (10 mM Tris [pH 8.0], 100 mM NaCl, 20 mM EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride) for 2 h at 4°C. Binding reaction mixtures were washed three times and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The gel was dried and autoradiographed.
Nucleotide sequence accession numbers.
The following GenBank accession numbers were assigned: for human PDX-1 homologous region PH1, no. AF227990; for region PH2, no. AF227989.
RESULTS
β-Cell expression of conserved sequences in the human pdx-1 gene.
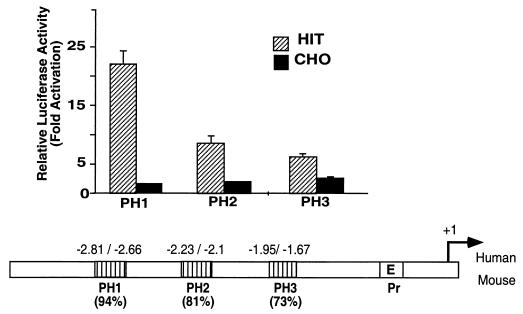
Sequence comparison upstream of the human and mouse pdx-1 promoter regions revealed three highly conserved regions, PH1, PH2, and PH3. The fragments extending from kb −2.809 to −2.655 (PH1), from −2.233 to −2.097 (PH2), and from −1.952 to −1.668 (PH3) in humans and mice present 94, 81, and 73% homology, respectively (Fig. 1). Each region was linked to the luciferase reporter gene driven by the TK promoter, and the chimeric genes were transiently transfected into HIT-T15 β cells and CHO cells. To various degrees, each fragment conferred β-cell-specific activation on a heterologous promoter. The PH1 region exhibits, in both orientations, a 13-fold preferential induction of luciferase activity in HIT-T15 compared with the level in CHO cells, whereas PH2 and PH3 exhibit approximately 4- and 2.5-fold levels of induction of luciferase, respectively (Fig. 1).
FIG. 1.
Sequence comparison of the 5′ flanking regions of the human and mouse pdx-1 genes. The homologous regions are designated PH1 (94% identity), PH2 (81% identity), and PH3 (73% identity), and the promoter region is designated Pr (containing the E box) in the lower panel. The conserved PH1 to PH3 domains were linked to the herpes simplex virus TK promoter driving luciferase gene expression. The parental TKLuc and the PH1-, PH2-, and PH3-TKLuc vectors were transiently transfected into HIT-T15 (hatched bars) and CHO (black bars) cells. The activity is shown relative to that of the basic TKLuc vector. Luciferase activity was normalized to the control β-Gal values. The results represent the means of results from four experiments (± standard errors of the means [SEM]).
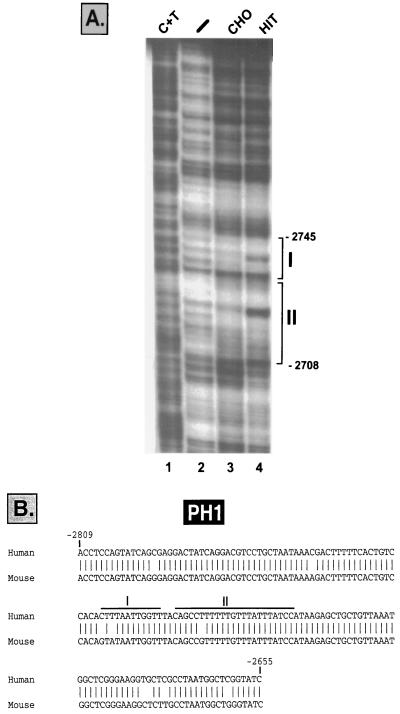
DNase I footprinting of the human PDX-1 PH1 enhancer sequence.
The transcriptional activity driven by the PH1 enhancer element of human PDX-1 suggested the presence of cis-acting regulatory elements in this region. To assess whether such putative elements interact with specific proteins, we performed a DNase I footprinting analysis using the fragment extending from kb −2.86 to −2.60 as a probe and extracts from HIT-T15 and CHO cells. As shown in Fig. 2A, two protected regions, area I (kb −2.745 to −2.734) and area II (kb −2.731 to −2.708), were obtained, each of which displayed different digestion patterns in HIT-T15 and CHO cell extracts. The homologous PH1 sequences in the human and the mouse pdx-1 genes are presented in Fig. 2B, and the footprinted areas indicated. To further characterize the trans-acting factors binding to the protected regions, double-stranded oligonucleotides spanning these sequences were synthesized and used as probes to detect HIT-T15 and CHO proteins by an EMSA.
FIG. 2.
(A) DNase I footprinting analysis was performed using the end-labeled fragment spanning the sequences between kb −2.87 and −2.61 and incubated without extract (lane 2) or with CHO cell extract (lane 3) or HIT-T15 cell extract (lane 4). A C+T sequencing reaction was run alongside as a marker (lane 1). (B) Homologous human and mouse PH1 nucleotide sequences (94% identity) of the pdx-1 gene (GenBank accession number AF227990). The footprinted areas are indicated as areas I and II.
The transcription factors PDX-1 and HNF-3β bind to the human PH1 enhancer element.
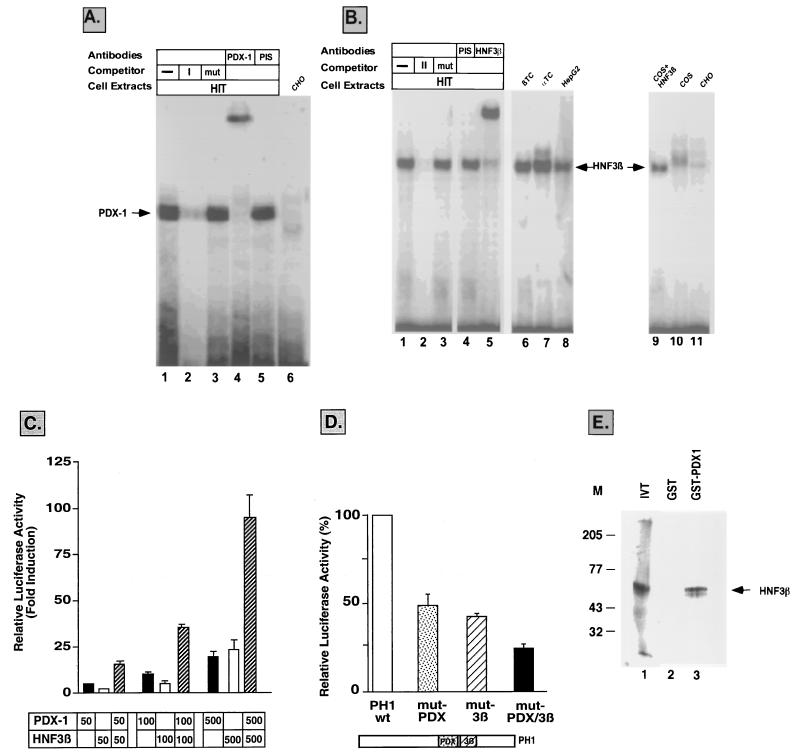
Using the area I sequence as a probe, a single β-cell-specific binding complex was obtained with β-cell extracts (Fig. 3A, lane 1, and data not shown), while no band was detected with any other cell extract tested (Fig. 3A, lane 6, and data not shown). The DNA complex was competed away by the wild-type oligonucleotide (Fig. 3A, lane 2) but not by its mutated counterpart (Fig. 3A, lane 3) or a nonspecific sequence (not shown). The binding specificity and the presence of a TAAT core sequence in area I suggested that the PDX-1 transcription factor itself may be a potential binding candidate. Indeed, anti-PDX-1-specific antibodies interacted with the protein(s) involved in the unique complex formed with the area I sequence, causing its supershift as shown in Fig. 3A (lane 4).
FIG. 3.
(A) Binding of PDX-1 to area I. An EMSA was performed using HIT-T15 (lanes 1–5) or CHO (lane 6) cell extracts and a 32P-labeled area I sequence. Competition for binding of HIT-T15 cell extracts to the labeled area I sequence was performed with a 100-fold excess of the unlabeled oligonucleotide (lane 2) or with its mutated counterpart (lane 3). The PDX-1 complex was identified by demonstrating the supershift of the band in the presence of antiserum against human PDX-1 (lane 4) but not preimmune serum (PIS) (lane 5). (B) HNF-3β in pancreatic cells interacts with the area II sequence of the human PH1 domain. An EMSA was performed using HIT-T15 (lanes 1 to 5), βTC6 (lane 6), αTC1 (lane 7), HepG2 (lane 8), or CHO (lane 11) cell extracts and a 32P-labeled area II sequence. Competition for binding of HIT-T15 cell extracts to the wild-type labeled area II sequence was performed with a 100-fold excess of the unlabeled oligonucleotide (lane 2) or with the mutated area II sequence (mut) (lane 3). HIT-T15 cell extracts were incubated in the presence of antiserum against HNF-3β (lane 5) or preimmune serum (lane 4). Cell extracts from HIT-T15 (lane 1) and COS cells (lane 10) or COS cells transfected with an expression vector for HNF-3β (lane 9) were incubated with the wild-type area II sequence as a probe. The HNF-3β complex is indicated by an arrow. (C) Transactivation with PDX-1 and HNF-3β in non-β cells. NIH 3T3 cells were transiently cotransfected with PDX-1 (black bars) or HNF-3β (white bars) expression plasmids or both (hatched bars) using 50, 100, and 500 ng of the plasmids and wild-type PH1-TKLuc. Luciferase activity was normalized to the control β-Gal values and is shown relative to that of the basic PH1-TKLuc vector. The results are the means of results of three to five experiments (± SEM). (D) The PH1-TKLuc wild type (PH1 wt, white bar) and the derived constructs carrying mutations in the PDX-1-binding site (mut-PDX) site (dotted bar), the HNF-3β-binding site (mut-3β) (hatched bar), or both (black bar) were transiently transfected in HIT-T15 cells. The results are the means of results of four to five experiments (± SEM), and the activity of each plasmid is represented as the percentage of that of the wild-type construct. (E) HNF-3β associates with PDX-1 in a GST pull-down assay. 35S-labeled HNF-3β polypeptide (lane 1) was incubated with bacterially expressed GST (lane 2) or GST–PDX-1 (lane 3) proteins. M, protein markers (in kilodaltons). IVT, in vitro-translated HNF-3β.
In order to elucidate the identities of the proteins binding to the area II sequences, an EMSA was performed using HIT-T15, βTC6, αTC1, CHO, COS, and HepG2 cell extracts. A strong complex was observed in pancreatic and liver cell extracts (Fig. 3B, lanes 1 and 6 to 8) but not in COS and CHO cells (Fig. 3B, lanes 10 and 11). The DNA complex was competed away by the wild-type oligonucleotide (Fig. 3B, lane 2) but not by the mutated sequences (Fig. 3B, lane 3) or the area I sequence (not shown). Sequence analysis revealed that area II is contained within an AT-rich region (Fig. 2). Computer analysis of binding sites for potential transcription factors unveiled a core motif for HNF-3β in this area. To determine whether HNF-3β was involved in the observed band, specific antibodies against this transcription factor were tested. Figure 3B demonstrates that the complex formed with the area II sequence is indeed supershifted with anti-HNF-3β (lane 5) but not with anti-HNF-3α (data not shown) antibody or preimmune serum (lane 4). Using another approach to confirm that the protein contained in the observed complex corresponds to HNF-3β, extracts from COS cells transfected with an expression plasmid for HNF-3β were analyzed for their interaction with the area II sequence. Figure 3 clearly indicates that the HNF-3β complex in COS cells migrates in a way similar to that of the complex obtained in β cells (lane 9 versus lanes 1 and 6). In a supershift assay, this complex was shown to be recognized by anti-HNF-3β antibody (data not shown). Altogether, this data establishes that endogenous HNF-3β in HIT-T15 cells specifically binds the area II sequence.
In summary, these results demonstrate that the PH1 conserved sequence in the human pdx-1 gene binds, in addition to the HNF-3β transcription factor in the protected region, area II, the product of its own gene, i.e., PDX-1 itself in the area I sequence.
Combinatorial effect of PDX-1 and HNF-3β in the activation of the PH1 sequence in the human pdx-1 gene.
To investigate the effect of the PDX-1 and HNF-3β transcription factors in controlling gene expression driven by the PH1 conserved sequence, we performed transient-transfection experiments with NIH 3T3 cells which endogenously lack both factors. To this end, the PH1-TKLuc construct was cotransfected with increasing amounts of the PDX-1 and HNF-3β expression plasmids. As presented in Fig. 3C, HNF-3β and PDX-1 separately activated the chimeric PH1 gene in a dose-dependent manner. Most significantly, cotransfection with a constant amount of PDX-1 and increasing amounts of HNF-3β strongly stimulated the expression of the gene in a synergistic manner. It appears that both sites are necessary for the expression of the PH1 chimeric gene since mutations abolishing either PDX-1 or HNF-3β binding to their respective sequences significantly impaired the transcriptional activity of the gene in β cells (Fig. 3D). Furthermore, the PH1 fragment carrying mutations in both sites showed further decreased transcriptional activity. Each mutated fragment was only partially transactivated when it was cotransfected with increasing amounts of either the PDX-1 or HNF-3β gene (data not shown). In an attempt to investigate a possible interaction between these two transcription factors, we performed a GST pull-down experiment where 35S-labeled HNF-3β polypeptide was found to bind efficiently to glutathione-Sepharose beads containing GST–PDX-1 but not GST alone (Fig. 3E). PDX-1 and HNF-3β interaction was disrupted using a buffer containing the detergent deoxycholate (not shown). In light of the results presented, we suggest that PDX-1 and HNF-3β can directly interact to activate pdx-1 gene expression.
HNF-3β binding to the PH2 conserved sequence in the human pdx-1 gene.
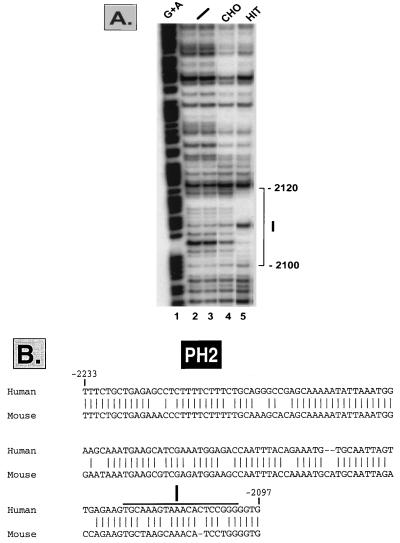
The PH2 conserved sequence also showed preferential β-cell-specific expression, albeit to a lesser degree than the PH1 sequence (Fig. 1). In order to analyze the potential regulatory sequences within this region which interact with extracts from β cells and non-β cells, DNase I footprinting analysis was performed. A fragment extending from kb −2.24 to −2.05 was used as a probe together with extracts from HIT-T15 and CHO cells. The protected sequence (kb −2.120 to −2.1) shows a hypersensitive site in the presence of HIT-T15 cell extracts (Fig. 4A, lane 5) as well as with extracts from αTC1 and HepG2 cells (data not shown). The homologous PH2 sequences of the human and the mouse pdx-1 genes are presented in Fig. 4B, and protected area I is indicated.
FIG. 4.
(A) A DNase I footprinting analysis was performed using the end-labeled fragment spanning the sequences between kb −2.27 and −2.05 and incubated without extract (lanes 2 and 3) or with extracts from CHO (lane 4) and HIT-T15 (lane 5) cells. A G+A sequencing reaction was run alongside as a marker (lane 1). (B) Homologous human and mouse PH2 nucleotide sequences (81% identity) of the pdx-1 gene (GenBank accession number AF227989). The area I footprinted region is indicated.
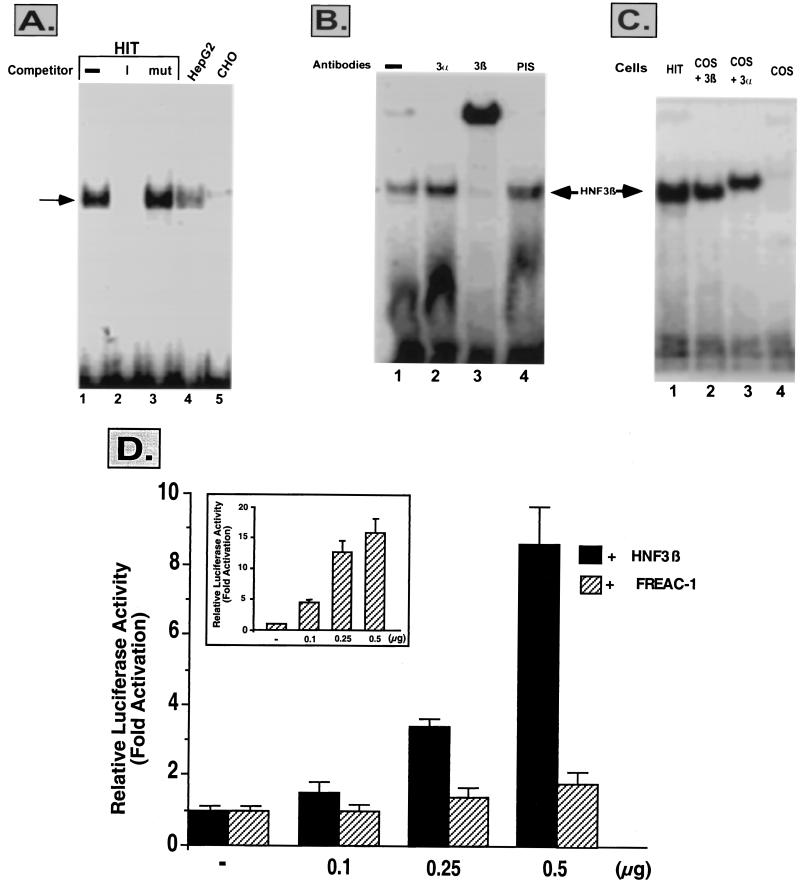
To further characterize the trans-acting factors binding to the footprinted region, a double-stranded oligonucleotide spanning this sequence was synthesized and used as a probe to detect HIT-T15, HepG2, and CHO proteins by an EMSA. Figure 5A shows a single binding complex in HIT-T15 (lane 1) and in HepG2 (lane 4) but not in CHO (lane 5) cell extracts with the PH2 protected sequence. The complex was competed away with the wild-type oligonucleotide (lane 2) but not with a mutated one (lane 3). The binding pattern in HIT and HepG2 extracts as well as a core binding sequence for the HNF-3β transcription factor prompted us to test the interaction with antibodies against this transcription factor. As shown in Fig. 5B, anti-HNF-3β (lane 3) but not anti-HNF-3α (lane 2) antibody supershifted the single complex. In confirmation of these results, cell extracts from COS cells transfected with an expression plasmid for HNF-3β (Fig. 5C, lane 2) but not for HNF-3α (Fig. 5C, lane 3) formed a complex with the PH2 protected sequence which migrates in a way similar to that of the complex obtained in HIT-T15 cells (lane 1). The protected sequence also showed a core-binding site (RTAAAYA) for the forkhead-related family of transcription factors, the fork-head-related activators (FREACs) (16, 24). However, unlike with HNF-3β, increasing amounts of FREAC-1 (or FREAC-2 and FREAC-4, data not shown) had no effect on the transactivation of the PH2-TKLuc construct (Fig. 5D).
FIG. 5.
HNF-3β in pancreatic cells interacts with the area I sequence of the human PH2 domain. (A) An EMSA was performed using HIT-T15 (lanes 1 to 3), HepG2 (lane 4), or CHO (lane 5) cell extracts and a 32P-labeled area I sequence of the PH2 domain. Competition for binding of HIT-T15 cell extracts to the labeled area I sequence was performed with a 100-fold excess of the unlabeled oligonucleotide (lane 2) or with its mutated counterpart (mut) (lane 3). (B) An EMSA was performed with HIT-T15 cell extracts incubated with the labeled probe (lane 1) in the presence of antiserum against HNF-3α (lane 2), HNF-3β (lane 3), or preimmune serum (PIS) (lane 4). (C) Cell extracts from HIT-T15 (lane 1) and COS (lane 4) cells or COS cells transfected with an expression vector for HNF-3β (lane 2) or for HNF-3α (lane 3) were incubated with the wild-type area I sequence as a probe. The HNF-3β complex is indicated by an arrow. (D) Transactivation with HNF-3β or FREAC-1 in non-β cells. NIH 3T3 cells were transiently cotransfected with HNF-3β (black bars) or FREAC-1 (hatched bars) expression plasmids and wild-type PH2-TKLuc. The insert represents the activity of a luciferase gene driven by a multimerized FREAC site cotransfected by increasing amounts of the FREAC-1 expression plasmid. Luciferase activity was normalized to the control β-Gal values and is shown relative to that of the basic PH2-TKLuc vector. The results are the means of results of three to six experiments (± SEM).
DISCUSSION
PDX-1 has been shown to be expressed early during development in cells of both exocrine and endocrine origins. Later it becomes restricted primarily to β cells, where it regulates the expression of β-cell-specific genes and, most importantly, mediates the glucose effect on insulin gene transcription. It was therefore important to identify the molecular mechanisms that specifically govern the expression of PDX-1 in the mature β cell. It was known from previous studies that fragments carrying, respectively, upstream sequences extending to kb −6.5 and −4.5 of the rat (28) and the mouse (33) pdx-1 genes contain the information necessary to target PDX-1 expression to islet cells. Therefore, in order to analyze the potential regulatory elements of the human pdx-1 gene, we sequenced about 4.5 kb in the 5′ flanking region of the gene and compared it to that of the mouse pdx-1 gene. A striking divergence at the nucleotide level was observed between the two species. In addition to the promoter region previously described by Sharma et al. (28), only three short conserved regions, designated PH1, PH2, and PH3, were revealed. Each of these regions confers β-cell-specific expression with various levels of potency on a reporter gene driven by the TK promoter. Since the β-cell specificity of PH3 was relatively weak (2.5-fold), we concentrated our efforts on investigating the cis-acting regulatory elements of the PH1 and PH2 conserved regions and the binding factors which are responsible for their transcriptional activity.
DNase I footprint analysis of the PH1 enhancer element, using HIT-T15 and CHO cell extracts, led to the identification of two protected AT-rich sequences with different digestion patterns, designated areas I and II. We analyzed various cell extracts for their capacity to bind to the area I sequence by EMSA. Surprisingly, a single complex was obtained exclusively with β-cell extracts. The β-cell specificity and core TAAT sequences raised the possibility that the PDX-1 transcription factor itself might bind to area I. Indeed, antibodies raised against the PDX-1 protein interacted with the β-cell-specific complex, confirming its binding to the PH1 element. Furthermore, we demonstrated that in the non-β-cell NIH 3T3 cell line, PDX-1 strongly stimulated transcriptional activity driven by PH1.
We have previously demonstrated that in the normal adult islet, PDX-1 (previously named GSF) acts as a glucose sensitive factor controlling the physiological expression of the insulin gene (19). Similar to the results obtained here, we showed that in non-insulin-producing cell lines, PDX-1 strongly transactivates the human insulin promoter. However, in β cells, high levels of ectopic PDX-1 has an inhibitory effect on the expression of the human insulin gene (17). We suggested that this repression might occur by a protein-protein interaction; e.g., PDX-1 might compete for a coactivator present only in limited amounts in normal adult islets. Cooperative interactions between PDX-1 and other transcription factors have been previously described by several groups. Peers et al. (22) have reported the involvement of the helix-loop-helix protein E47 in inducing insulin gene expression. Recently, Ohneda et al. (20) have shown that the transcription factor BETA2 and the nuclear high-mobility group protein I (Y) contribute to the PDX-1–E47 synergy, through direct interaction with the homeodomain of PDX-1.
It was also demonstrated that PDX-1 forms a heterodimeric complex with Pbx, the mammalian homologue of the drosophila extradenticle. This heterodimer bound the TAAT sequence of the somatostatin promoter but not the same sequence (A3 motif) of the insulin promoter, suggesting that this preference may form the basis for target site selection in developing islet cells (23) and that the transcriptional function of the gene may thus be highly context dependent.
The footprinting sequences described as region II in PH1 and region I in PH2 showed similarity to the consensus binding site for HNF-3β (21, 24). We were able to show that it is indeed HNF-3β that binds and stimulates the activity of the human PH1 and PH2 enhancer elements in non-β cells. While this paper was under review, Gerrish et al. (11) also reported that conserved sequences between the human and mouse pdx-1 genes (termed areas I and II) confer β-cell-selective gene expression. Their data supported the fact that HNF-3β associates with these regions and is necessary for area I transcriptional activity. In addition, the levels of PDX-1 mRNA were markedly impaired in embryonic stem cells in which the HNF-3β gene was inactivated and upon differentiation to embryoid bodies. In the rat pdx-1 gene, Sharma et al. (27) characterized a distal enhancer element to which the nuclear factors HNF-3β and BETA2 bind and cooperatively induce PDX-1 expression in islets. Thus, it can be stated that at least some aspects of PDX-1 expression rely on the transcription factor HNF-3β.
HNF-3β, a member of the forkhead and winged-helix family of transcription factors, is essential for endodermal cell lineages (12, 34). It is structurally related to histone H5, which can alter the nucleosomal structure and thus prime target genes for expression by opening the chromatin structure to provide promoter access to other transcription factors (6, 29). Since HNF-3β is not restricted to β cells, the selective transcription of pdx-1 is likely to rely on an additional factor(s). Our findings that the PH1 enhancer element binds both HNF-3β and PDX-1, that mutations in each individual site dramatically impair its transcriptional activity, and in addition that HNF-3β and PDX-1 directly interact in vitro suggest cooperativity between these factors. We therefore propose that a possible feedback mechanism might control the expression of PDX-1 at different stages during development. It is conceivable that the functional conservation of different enhancer elements in the 5′ flanking region of the pdx-1 gene is crucial for its maintenance and cell specificity. Indeed, multiple enhancer elements were shown to regulate CD8 expression in diverse subsets of cells during development as was shown in vivo for T cells by Ellmeier et al. (10).
The control of pdx-1 gene expression in many aspects resembles that of the tissue-specific POU domain factor Pit-1 (also called GHF-1), which is required for the formation of three cell phenotypes in the anterior pituitary gland (3, 32). The Pit-1 gene utilizes distinct enhancers for initial gene activation and for subsequent autoregulation required for the maintenance of its own expression (7). Like PDX-1, Pit-1 also shows a biphasic pattern of expression during ontogeny. Its transcripts are observed in the rat neural tube and neural plate (embryonic days 10 to 11) and disappear thereafter (day 13), only to reappear exclusively in the anterior pituitary gland (day 15) just before activation of prolactin and growth hormone. Interestingly, it was shown that Pit-1 can positively autoregulate the expression of its own gene by binding to two Pit-1-binding elements (5). The results presented here suggest that PDX-1 may well be regulated in a similar way. Distinct regulatory elements may function at specific stages during development, and an autoregulatory loop may be necessary for its maintenance in the adult β cell. To fully understand the functional relevance of these distinct regulatory elements, in vivo testing with transgenic models will be required.
ACKNOWLEDGMENTS
We thank Roland Stein (Vanderbilt University, Nashville, Tenn.) for the mouse pdx-1 sequence and Alan M. Permutt (Washington University) for the human pdx-1 gene. We are infinitely grateful to Robert Costa (University of Illinois at Chicago) for the gifts of HNF-3α and HNF-3β expression vectors and antibodies and to Peter Carlsson (Göteborg University, Göteborg, Sweden) for FREAC-1, FREAC-2, and FREAC-4 expression vectors. Our sincere thanks go to Rahel Oron for excellent technical help.
This work was supported by grants from the Juvenile Diabetes Foundation International, BIOMED 2 (BMH4-CT98-3448), and the Israel Science Foundation.
REFERENCES
- 1.Ahlgren U, Jonsson J, Edlund H. The morphogenesis of the pancreatic mesenchyme is uncoupled from that of the pancreatic epithelium in IPF1/PDX1-deficient mice. Development. 1996;122:1409–1416. doi: 10.1242/dev.122.5.1409. [DOI] [PubMed] [Google Scholar]
- 2.Ahlgren U, Jonsson J, Jonsson L, Simu K, Edlund H. Beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 1998;12:1763–1768. doi: 10.1101/gad.12.12.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andersen B, Rosenfeld M G. Pit-1 determines cell types during development of the anterior pituitary gland. A model for transcriptional regulation of cell phenotypes in mammalian organogenesis. J Biol Chem. 1994;269:29335–29338. [PubMed] [Google Scholar]
- 4.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen R P, Ingraham H A, Treacy M N, Albert V R, Wilson L, Rosenfeld M G. Autoregulation of pit-1 gene expression mediated by two cis-active promoter elements. Nature. 1990;346:583–586. doi: 10.1038/346583a0. [DOI] [PubMed] [Google Scholar]
- 6.Clark K L, Halay E D, Lai E, Burley S K. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 7.DiMattia G E, Rhodes S J, Krones A, Carriere C, O'Connell S, Kalla K, Arias C, Sawchenko P, Rosenfeld M G. The Pit-1 gene is regulated by distinct early and late pituitary-specific enhancers. Dev Biol. 1997;182:180–190. doi: 10.1006/dbio.1996.8472. [DOI] [PubMed] [Google Scholar]
- 8.Edlund H. Pancreas: how to get there from the gut? Curr Opin Cell Biol. 1999;11:663–668. doi: 10.1016/s0955-0674(99)00033-2. [DOI] [PubMed] [Google Scholar]
- 9.Edlund H. Transcribing pancreas. Diabetes. 1998;47:1817–1823. doi: 10.2337/diabetes.47.12.1817. [DOI] [PubMed] [Google Scholar]
- 10.Ellmeier W, Sunshine M J, Losos K, Littman D R. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity. 1998;9:485–496. doi: 10.1016/s1074-7613(00)80632-9. [DOI] [PubMed] [Google Scholar]
- 11.Gerrish K, Gannon M, Shih D, Henderson E, Stoffel M, Wright C V, Stein R. Pancreatic beta cell-specific transcription of the pdx-1 gene. The role of conserved upstream control regions and their hepatic nuclear factor 3beta sites. J Biol Chem. 2000;275:3485–3492. doi: 10.1074/jbc.275.5.3485. [DOI] [PubMed] [Google Scholar]
- 12.Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman J R, Zaret K S. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- 13.Guz Y, Montminy M R, Stein R, Leonard J, Gamer L W, Wright C V, Teitelman G. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- 14.Larsson L I, St.-Onge L, Hougaard D M, Sosa-Pineda B, Gruss P. Pax 4 and 6 regulate gastrointestinal endocrine cell development. Mech Dev. 1998;79:153–159. doi: 10.1016/s0925-4773(98)00182-8. [DOI] [PubMed] [Google Scholar]
- 15.Madsen O D, Jensen J, Petersen H V, Pedersen E E, Oster A, Andersen F G, Jorgensen M C, Jensen P B, Larsson L I, Serup P. Transcription factors contributing to the pancreatic beta-cell phenotype. Horm Metab Res. 1997;29:265–270. doi: 10.1055/s-2007-979035. [DOI] [PubMed] [Google Scholar]
- 16.Mahlapuu M, Pelto-Huikko M, Aitola M, Enerback S, Carlsson P. FREAC-1 contains a cell-type-specific transcriptional activation domain and is expressed in epithelial-mesenchymal interfaces. Dev Biol. 1998;202:183–195. doi: 10.1006/dbio.1998.9010. . (Erratum, 207:476, 1999.) [DOI] [PubMed] [Google Scholar]
- 17.Marshak S, Totary H, Cerasi E, Melloul D. Purification of the beta-cell glucose-sensitive factor that transactivates the insulin gene differentially in normal and transformed islet cells. Proc Natl Acad Sci USA. 1996;93:15057–15062. doi: 10.1073/pnas.93.26.15057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 19.Melloul D, Ben-Neriah Y, Cerasi E. Glucose modulates the binding of an islet-specific factor to a conserved sequence within the rat I and the human insulin promoters. Proc Natl Acad Sci USA. 1993;90:3865–3869. doi: 10.1073/pnas.90.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohneda K, Mirmira R G, Wang J, Johnson J D, German M S. The homeodomain of PDX-1 mediates multiple protein-protein interactions in the formation of a transcriptional activation complex on the insulin promoter. Mol Cell Biol. 2000;20:900–911. doi: 10.1128/mcb.20.3.900-911.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Overdier D G, Porcella A, Costa R H. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino acid residues adjacent to the recognition helix. Mol Cell Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peers B, Leonard J, Sharma S, Teitelman G, Montminy M R. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol Endocrinol. 1994;8:1798–1806. doi: 10.1210/mend.8.12.7708065. [DOI] [PubMed] [Google Scholar]
- 23.Peers B, Sharma S, Johnson T, Kamps M, Monteminy M. The pancreatic islet factor STF-1 binds cooperatively with Pbx to a regulatory element in the somatostatin promoter: importance of the FPWMK motif and of the homeodomain. Mol Cell Biol. 1995;15:7091–7097. doi: 10.1128/mcb.15.12.7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 1994;13:5002–5012. doi: 10.1002/j.1460-2075.1994.tb06827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 26.Sander M, German M S. The beta cell transcription factors and development of the pancreas. J Mol Med. 1997;75:327–340. doi: 10.1007/s001090050118. [DOI] [PubMed] [Google Scholar]
- 27.Sharma S, Jhala U S, Johnson T, Ferreri K, Leonard J, Montminy M. Hormonal regulation of an islet-specific enhancer in the pancreatic homeobox gene STF-1. Mol Cell Biol. 1997;17:2598–2604. doi: 10.1128/mcb.17.5.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma S, Leonard J, Lee S, Chapman H D, Leiter E H, Montminy M R. Pancreatic islet expression of the homeobox factor STF-1 relies on an E-box motif that binds USF. J Biol Chem. 1996;271:2294–2299. doi: 10.1074/jbc.271.4.2294. [DOI] [PubMed] [Google Scholar]
- 29.Shim E Y, Woodcock C, Zaret K S. Nucleosome positioning by the winged helix transcription factor HNF3. Genes Dev. 1998;12:5–10. doi: 10.1101/gad.12.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slack J M. Developmental biology of the pancreas. Development. 1995;121:1569–1580. doi: 10.1242/dev.121.6.1569. [DOI] [PubMed] [Google Scholar]
- 31.Sussel L, Kalamaras J, Hartigan-O'Connor D J, Meneses J J, Pedersen R A, Rubenstein J L, German M S. Mice lacking the homeodomain transcription factor Nkx2.2 have diabetes due to arrested differentiation of pancreatic beta cells. Development. 1998;125:2213–2221. doi: 10.1242/dev.125.12.2213. [DOI] [PubMed] [Google Scholar]
- 32.Theill L E, Karin M. Transcriptional control of GH expression and anterior pituitary development. Endocr Rev. 1993;14:670–689. doi: 10.1210/edrv-14-6-670. [DOI] [PubMed] [Google Scholar]
- 33.Wu K L, Gannon M, Peshavaria M, Offield M F, Henderson E, Ray M, Marks A, Gamer L W, Wright C V, Stein R. Hepatocyte nuclear factor 3β is involved in pancreatic β-cell-specific transcription of the pdx-1 gene. Mol Cell Biol. 1997;17:6002–6013. doi: 10.1128/mcb.17.10.6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaret K S. Molecular genetics of early liver development. Annu Rev Physiol. 1996;58:231–251. doi: 10.1146/annurev.ph.58.030196.001311. [DOI] [PubMed] [Google Scholar]