ABSTRACT
var genes encode Plasmodium falciparum erythrocyte membrane protein-1 (PfEMP1) antigens. These highly diverse antigens are displayed on the surface of infected erythrocytes and play a critical role in immune evasion and sequestration of infected erythrocytes. Studies of var expression using non-leukocyte-depleted blood are challenging because of the predominance of host genetic material and lack of conserved var segments. Our goal was to enrich for parasite RNA, allowing de novo assembly of var genes and detection of expressed novel variants. We used two overall approaches: (i) enriching for total mRNA in the sequencing library preparations and (ii) enriching for parasite RNA with a custom capture array based on Roche’s SeqCap EZ enrichment system. The capture array was designed with probes based on the whole 3D7 reference genome and an additional >4,000 full-length var gene sequences from other P. falciparum strains. We tested each method on RNA samples from Malian children with severe or uncomplicated malaria infections. All reads mapping to the human genome were removed, the remaining reads were assembled de novo into transcripts, and from these, var-like transcripts were identified and annotated. The capture array produced the longest maximum length and largest numbers of var gene transcripts in each sample, particularly in samples with low parasitemia. Identifying the most-expressed var gene sequences in whole-blood clinical samples without the need for extensive processing or generating sample-specific reference genome data is critical for understanding the role of PfEMP1s in malaria pathogenesis.
IMPORTANCE Malaria parasites display antigens on the surface of infected red blood cells in the human host that facilitate attachment to blood vessels, contributing to the severity of infection. These antigens are highly variable, allowing the parasite to evade the immune system. Identifying these expressed antigens is critical to understanding the development of severe malarial disease. However, clinical samples contain limited amounts of parasite genetic material, a challenge for sequencing efforts further compounded by the extreme diversity of the parasite surface antigens. We present a method that enriches for these antigen sequences in clinical samples using a custom capture array, requiring minimal processing in the field. While our results are focused on the malaria parasite Plasmodium falciparum, this approach has broad applicability to other highly diverse antigens from other parasites and pathogens such as those that cause giardiasis and leishmaniasis.
KEYWORDS: P. falciparum, malaria, PfEMP1, var gene, Plasmodium falciparum, RNA-Seq
INTRODUCTION
While there has been an overall decrease in the number of malaria cases and deaths in the last decade, these numbers have plateaued since 2015 (1). More than 90% of malaria deaths occur in sub-Saharan Africa (1), primarily the result of the most severe forms of malaria caused by Plasmodium falciparum, including cerebral malaria (CM) and severe malarial anemia (SMA). Protection against malaria illness can be acquired over time in regions of endemicity, likely as a repertoire of immune responses to parasite variant surface antigens (VSAs) displayed on the surface of infected erythrocytes. Protection from clinical malaria has been associated with the presence of antibodies to these particular antigens (2–4). In P. falciparum, VSAs are encoded by three main multigene families: var, rif, and stevor genes.
The most widely studied VSAs are the P. falciparum erythrocyte membrane protein-1 (PfEMP1) antigens, which are encoded by the var gene family. Approximately 60 PfEMP1s are encoded by var genes in each P. falciparum genome (5, 6), and can be classified into different groups (A, B, C, and E) using the upstream promoter sequence (7). These are highly diverse antigens displayed on the surface of infected erythrocytes, and they play a critical role in immune evasion and sequestration. PfEMP1 antigens are composed of combinations of domains that include two to eight Duffy binding-like (DBL) domains and one to two cysteine-rich interdomain regions (CIDRs). Each infected erythrocyte displays on its surface only one particular PfEMP1 (8), which mediates binding to different host cell receptors (9, 10).
Determining PfEMP1s that are specific to different severe malaria syndromes is essential for understanding the pathogenesis of severe malaria. Studies in both adults and children have associated PfEMP1 subtypes and var groups with severe malaria syndromes, with conflicting results that suggest that some similar subtypes are associated with both cerebral malaria and severe malarial anemia. Domain cassettes (DCs), or consecutive DBL and CIDR domains found to recur across multiple PfEMP1s, have been associated with severe malarial disease. DC8 (11–14), DC13 (11, 13), and group A var genes (11, 14–17) have all been associated with severe malaria syndromes. These studies have primarily used quantitative reverse transcription PCR (qRT-PCR), targeting usually a domain or domain cassette with primers designed against a subset of known sequences and comparing the expression between groups of participants with severe malarial disease and controls with uncomplicated malarial disease. Domains or var gene groups with significantly greater transcript levels were associated with severe malaria syndromes, but these methods did not identify the most highly expressed var gene in an infection. Determining the most-expressed var gene in an infection, including identification of novel var gene sequences, requires an unbiased approach that allows detection of any independent var transcript present.
Sequencing full-length var genes from clinical samples has proven challenging. PfEMP1s often have <50% shared amino acid identity, and the full var repertoires are known for only a small number of reference genomes and clinical samples (5, 6, 18–20). The average shared amino acid identity is ∼30 to 40% within classes of DBL and CIDR domains (18), indicating that diverse sequences occur even within classes of domains. This diversity and lack of recurring motifs add to the challenge of sequencing these genes. The variable length of var genes is another challenge, ranging from ∼4,500 to ∼12,000 bp and up to 2 orders of magnitude longer than the standard sequencing read of 150 bp. Each P. falciparum genome contains multiple copies of var genes—usually ∼60—featuring an internal repeated motif structure that makes assembly difficult. Studies of var gene expression have focused on a degenerate primer approach, capturing only a few domains at a time (11–14, 21), which does not enable a comparison of the entire var gene sequence. Another challenge in sequencing var genes is the relatively small proportion of var transcripts compared to host and the total parasite transcriptome, particularly in samples collected as whole blood. Clinical samples collected as whole blood are composed of mostly host (human) RNA and DNA, with parasite genetic material composing a smaller proportion. A recent study successfully assembled var transcripts de novo from clinical malaria samples in Indonesia; however, the methods required leukocyte-depleted blood (22).
An effective method to identify and sequence highly expressed genes encoding variable antigens has the potential to provide insight into immune evasion and the pathogenesis of specific clinical syndromes for a range of pathogens, including P. falciparum, Plasmodium vivax, Giardia, and Leishmania. Here, we attempt to overcome existing challenges in generating a comprehensive profile of the expressed var genes and determining their relative expression in specific severe malaria phenotypes by applying different approaches to enrich parasite RNA in a sample and applying these methods to total RNA extracted from whole-blood clinical samples. We compared two overall approaches: (i) capture of parasite RNA, using a capture array designed from the 3D7 genome (excluding the ribosomal DNA [rDNA] loci) plus >4,000 var gene sequences from other P. falciparum strains (designated Capture), and (ii) two methods of depleting undesired RNA types, depletion of rRNA and globin mRNA (designated Depletion) and depletion of rRNA and globin mRNA followed by selection for polyadenylated transcripts [designated Depletion + poly(A)].
We recently sequenced and assembled full var repertoires from genomic sequence data from uncomplicated malaria infections in Malian children (19). As proof of principle, in the present study we applied the parasite RNA enrichment methods to 11 RNA samples from those same uncomplicated malaria infections, for which genomic data and complete, reconstructed var repertoires were available for reference. We evaluated the outcome of those methods and our protocol for de novo assembly of expressed var genes using as a reference the genomic var gene repertoires. We then applied our methods to a subset of samples from a case-control study of severe malaria, including cases with cerebral malaria or severe malarial anemia. We describe parasite RNA enrichment from whole-blood clinical samples and successful sequencing and assembly of expressed var genes using a custom capture approach, which is particularly useful for samples from low-parasitemia infections. While our results are focused on P. falciparum, this approach has broad applicability to other highly diverse antigens from a variety of parasites and pathogens such as those that cause giardiasis and leishmaniasis.
RESULTS
Sequencing data generated from each enrichment method.
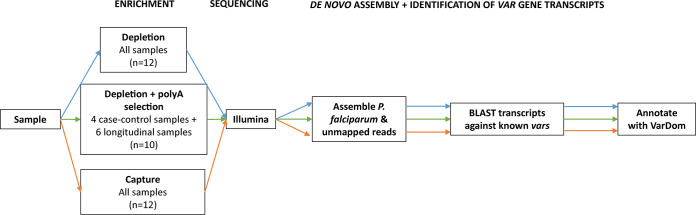
To determine the performance of different methods to enrich P. falciparum mRNA from a sample of total RNA before sequencing, we used a total of 12 samples, four from a case-control study and eight from a longitudinal study. All four samples from the case-control study were used to evaluate each method of enrichment: Depletion, Depletion + poly(A) selection, and Capture (Table 1, Fig. 1). All longitudinal study samples (n = 8) were used for Depletion and Capture, while six were used for Depletion + poly(A) selection [no Depletion + poly(A) data for samples UM 2 and UM 5 (Table 1)]. The average number of total reads sequenced was 83.9 million for the Depletion method, 72.7 million for Depletion + poly(A), and 73.2 million for Capture (Table 1; see also Table S1 and Fig. S1 in the supplemental material). Reads mapping to P. falciparum and unmapped reads were more variable, averaging 19.7 million reads for Depletion method, 15.8 million reads for Depletion + poly(A), and 72.1 million reads for Capture (Table S1 and Fig. S1).
TABLE 1.
Characteristics of samples for enrichment of parasite RNA and the enrichment methods applied to each sample
| Sample | Study source | Parasitemia (no. of parasites/μl) | Enrichment methods (total reads) |
||
|---|---|---|---|---|---|
| Depletion | Depletion + poly(A) | Capture | |||
| CM + SMA | Case-control | 1,650 | 56,722,984 | 74,499,556 | 86,791,160 |
| SMA | Case-control | 143,750 | 70,337,818 | 69,473,914 | 66,787,024 |
| UMC | Case-control | 14,700 | 63,480,128 | 52,348,806 | 82,076,018 |
| CM | Case-control | 11,680 | 73,356,546 | 68,572,664 | 75,814,008 |
| UM 1 | Longitudinal | 209,050 | 85,943,726 | 88,712,300 | 83,561,130 |
| UM 2 | Longitudinal | 15,400 | 94,326,578 | 85,030,980 | |
| UM 3 | Longitudinal | 185,400 | 94,676,230 | 67,703,494 | 42,798,838 |
| UM 5 | Longitudinal | 7,525 | 85,612,460 | 50,651,528 | |
| UM 6 | Longitudinal | 193,800 | 115,500,792 | 79,058,422 | 44,708,486 |
| UM 8 | Longitudinal | 19,275 | 91,193,310 | 70,058,484 | 48,861,952 |
| UM 9 | Longitudinal | 198,900 | 92,401,680 | 72,452,038 | 120,660,408 |
| UM 11 | Longitudinal | 14,650 | 83,334,470 | 84,384,254 | 90,965,320 |
FIG 1.
Overview of pipeline for enrichment of parasite RNA, sequencing, and assembly of transcripts. P. falciparum mRNA was enriched in all 12 samples using both globin and rRNA depletion as well as capture. Ten samples were subjected to all three methods, including globin and rRNA depletion followed by poly(A) selection. For the Depletion and Depletion + poly(A) method, enrichment took place during library preparation, and for Capture, libraries were prepared, and then Capture was applied to enrich for parasite cDNA. Reads were mapped to a concatenated reference of human and P. falciparum. We used reads that mapped to P. falciparum and unmapped reads to de novo assemble transcripts, open reading frames for the transcripts were subjected to a protein blast search to identify var transcripts, and then the protein sequences were annotated with the VarDom online database.
Comparison of the number of 20 million randomly selected reads mapping to known var genes from each sample for each library preparation. The total number of randomly selected reads from Capture (orange) that mapped to known var genes was significantly greater than Depletion (blue) and Depletion + poly(A) (green) (Wilcoxon signed-rank test). Download FIG S1, PDF file, 0.04 MB (47.8KB, pdf) .
Copyright © 2021 Stucke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sample information, total reads, Plasmodium falciparum reads, insert size, and read length. Download Table S1, XLSX file, 0.01 MB (12KB, xlsx) .
Copyright © 2021 Stucke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Capture yields a consistently greater proportion of reads from parasite transcripts.
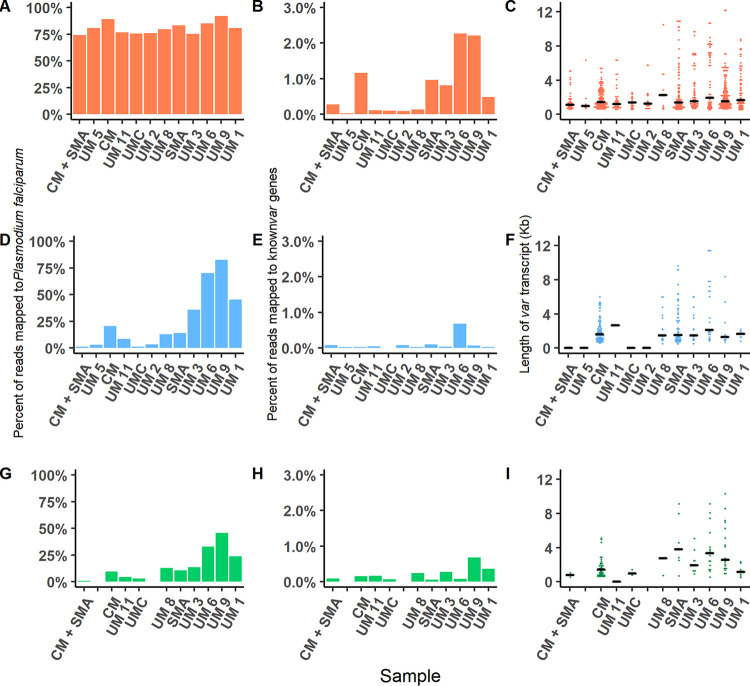
As a preliminary measure of enrichment, we first quantified the number and proportions of sequence reads that mapped to the P. falciparum reference genome to determine which library preparation most consistently recovers the greatest amount and proportion of parasite reads (Fig. 2A, D, and G). Reads mapping to the P. falciparum genome were identified from all libraries and were a greater proportion of the total reads from the Capture libraries than from the Depletion libraries. This outcome is to be expected, inasmuch as the Capture approach enriches parasite nucleic acids specifically by capturing cDNA with sequences similar to the 3D7 genome. However, we also observed a more consistent percentage of reads mapping to the P. falciparum genome from samples prepared with Capture enrichment, with a mean of 81% (range, 75 to 90%) (Fig. 2A). In contrast, for the Depletion and Depletion + poly(A) libraries, we observed a wider range of percentages mapping to the P. falciparum genome (<5% to 80%), which was positively associated with parasitemia levels. For the Depletion libraries, a mean of 23% of reads mapped to P. falciparum, with ∼1% of reads mapped to P. falciparum in the CM + SMA sample with low parasitemia (1,650 parasites/μl), versus around 80% of mapped reads in UM 6 and UM 9 with a higher parasitemia of ∼200,000 parasites/μl (Fig. 2D). For the Depletion + poly(A) libraries, we calculated a mean of 15% of reads mapped to P. falciparum, ranging from <1% in the lowest-parasitemia sample CM + SMA to ∼50% in sample UM 9 with ∼200,000 parasites/μl (Fig. 2G).
FIG 2.
Reads mapping to P. falciparum, known var genes, and number and length of assembled var gene transcripts. Results were quantified for Capture (orange; first row), Depletion (blue; second row), and Depletion + poly(A) (green; third row). Reads mapping to P. falciparum were quantified for the three methods (A, D, and G); percentages of the random selection of 20 million reads mapping to known var genes were quantified for the three methods (B, E, and H); and the number and length of unique transcripts were quantified for the three methods (C, F, and I). Capture retained the greatest percentages of P. falciparum and var reads, and more unique var transcripts were assembled from the Capture-prepared samples. Samples are arranged from least (left) to greatest parasitemia. CM, cerebral malaria; UM, uncomplicated malaria; UMC, uncomplicated malaria control; SMA, severe malarial anemia.
Capture produces greater percentages of var reads and assembled var transcripts.
To determine which preparation produced the greatest proportion of reads mapping to var genes, we selected a random sample of 20 million paired reads from each library to normalize by data set size to the smallest data set sequenced. These read subsets were mapped to an index of >4,000 var gene sequences from lab strains, reference genomes, and clinical samples (described in Materials and Methods) to determine the percentage of reads mapping to known var genes (Fig. 2B, E, and H). Fewer than 1% of reads mapped to the index of known var genes from the Depletion and Depletion + poly(A) libraries, which contrasts with Capture libraries in which reads mapping to var genes were identified from every sample. Capture produced greater percentages of var reads, which surpassed 1% for three samples (CM, UM 6, and UM 9) (Fig. 2B). Using the 20 million randomly selected reads, Capture libraries had a significantly greater median number of reads mapping to known var genes than Depletion libraries (Wilcoxon signed-rank test, P < 0.001 [Fig. S2]) and Depletion + poly(A) libraries (Wilcoxon signed-rank test, P < 0.05). The Capture approach thus produced more reads, and reads from the total library that were then assembled into more transcripts for each sample than the other library preparation methods (Fig. 2C).
Total number of reads mapping to the P. falciparum reference genome or unmapped (red) or the human reference genome (blue) for each library preparation [Capture, left; Depletion, center; Depletion + poly(A), right]. Reads mapping to P. falciparum or unmapped are a larger proportion of total reads in the Capture (left) library preparation than in those of Depletion (center) and Depletion + poly(A) (right). Samples are arranged from least to greatest parasitemia, left to right. Download FIG S2, PDF file, 0.03 MB (35.6KB, pdf) .
Copyright © 2021 Stucke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We compared the use of two different references to identify var transcripts from the reconstructed transcriptome, by using (i) as reference var sequences from the 3D7 reference genome versus (ii) an extended set of known var sequences from several reference genomes and clinical isolates included in the design of the capture array. For Capture, we were able to identify all transcripts with PfEMP1 domains using either reference set. For Depletion + poly(A), we identified 95% of transcripts with PfEMP1 domains using only the 3D7 reference. While the larger database strategy matched sequences with greater amino acid identity, the transcripts with var domains matched 3D7 var sequences with sufficiently high amino acid identity to be recognized.
To compare the assemblies of expressed var genes from each library, we quantified the number of de novo-assembled var transcripts and determined their length. We were able to assemble transcripts with one or more domains from all libraries prepared with Capture (Fig. 2C); however, for Depletion and Depletion + poly(A) libraries for samples with lower parasitemias (parasitemia of <15,000/μl) we assembled very few or no transcripts (Fig. 2F and I). This included no transcripts for four samples using the Depletion library preparation (CM + SMA, UM 5, UMC [uncomplicated malaria control], and UM 2) and for one sample using the Depletion + poly(A) preparation (UM 11) (Fig. 2F). We were able to assemble multiple transcripts from sample CM using each of the enrichment methods, but the percentage of reads and the number of transcripts were improved with Capture. Overall, Capture produced greater percentages of reads mapping to var genes and more de novo-assembled var transcripts.
Enrichment methods yield similar results for most-expressed var genes.
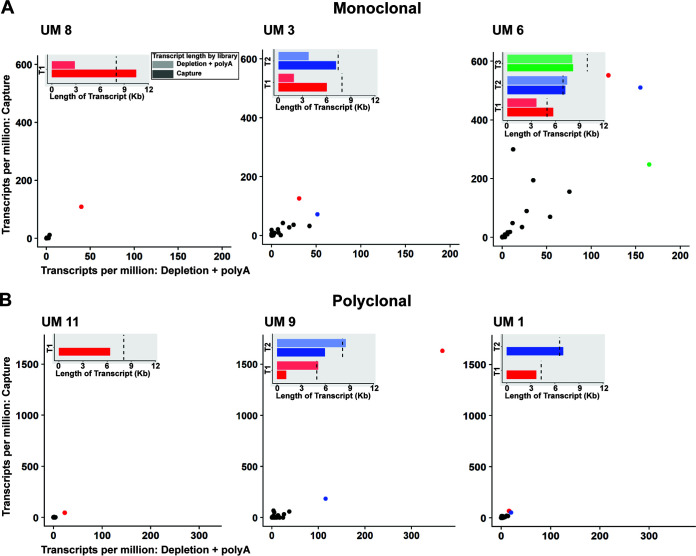
As proof of principle for our assembly and quantification methods, namely, to confirm that we are reconstructing var genes of the correct length, not chimeras, and that reconstructed transcripts can be used to quantify expression, we focused on the longitudinal study samples with corresponding genomic data (UM 1, UM 2, UM 3, UM 8, UM 9, and UM 11). To determine which library preparation yields the most reliable relative quantification of var gene expression and if expression levels were similar between the Depletion + poly(A) and Capture enrichment methods, we mapped reads to the genomic var sequences from each sample (Fig. 3). For each uncomplicated malaria sample with a genomic var reference, the same var genes were detected in both libraries, but with greater transcripts per million (TPMs) in those from Capture. For samples UM 8, UM 11, and UM 9, there was a single most-expressed var gene identified by both enrichment methods. In samples UM 3 and UM 1, the top two most-expressed var genes were the same but the order of expression magnitude differed. For sample UM 6, the most-expressed var gene was the same, but the order of the second and third most-expressed differed between Capture and Depletion + poly(A). For the case-control samples with no genomic reference, we compared var gene expression levels by mapping the reads from each library separately to the reference using a common de novo var reference assembled with reads from both Capture and Depletion + poly(A) libraries (Fig. S3). We detected the same var genes from both libraries, and no var genes detected by Depletion + poly(A) were not detected by Capture, indicating that the use of Capture probes did not result in a failure to detect particular transcripts.
FIG 3.
Comparison of expression and assembly results from Capture and Depletion + poly(A) mapping to genomic sequences. Each scatterplot compares transcripts per million (TPMs) in Depletion + poly(A) on the x axis to Capture on the y axis by mapping the reads from each library to the genomic reference. The bar graph in the inset of each panel shows the length of the most-expressed transcripts de novo assembled from the RNA-Seq data [Capture, darker shade; Depletion + poly(A), lighter shade] and the length of the corresponding genomic sequence (dotted black line). For each uncomplicated malaria sample, the same var genes were identified in the two libraries, but Capture yielded greater transcripts per million (scatterplots). Predominantly expressed var genes de novo assembled from RNA-Seq data were similar in length (bar graphs) to the genome reference var genes (dotted black line), with de novo-assembled sequences from capture typically longer than those from Depletion + poly(A). Panel A (UM 8, UM 3, and UM 6) includes monoclonal (one infecting parasite clone) samples, and panel B includes polyclonal (more than one infecting parasite clone) samples (UM 11, UM 9, and UM 1). Samples are arranged from least to greatest parasitemia within each row, from left to right. The color of the dots in the scatterplot (red, blue, or green) corresponds to the same transcript (T1, T2, or T3) shown assembled in the inset bar graph. UM, uncomplicated malaria; T1, transcript 1; T2, transcript 2; T3, transcript 3.
Comparison of transcripts per million for expressed var genes in Depletion + poly(A) and Capture. TPMs shown in scatterplots are for the var genes expressed in the four case-control study samples. TPMs are calculated from mapping to a common reference de novo assembled by combining reads from both Depletion + poly(A) and Capture libraries. Capture identified the same expressed var genes in the case-control samples as did Depletion + poly(A), but with more transcripts per million. CM, cerebral malaria; UMC, uncomplicated malaria control; SMA, severe malarial anemia. Download FIG S3, PDF file, 0.02 MB (17.1KB, pdf) .
Copyright © 2021 Stucke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We next determined if we were able to assemble the sequences of the most-expressed var genes from the RNA sequencing (RNA-Seq) data. For each sample, we identified sequences from the transcriptome sequencing de novo assembly matching with >99% identity to the genomic var sequences (Fig. 3, inset). Predominantly expressed var genes were similar in length to the genomic sequences (Fig. 3, dotted black lines in insets) and most commonly assembled with Capture. However, we were not able to assemble sequences matching the most-expressed var genes from two samples—UM 11 and UM 1—with the Depletion + poly(A) method. Depletion + poly(A) worked particularly well for sample UM 9, and while we identified matching sequences with Capture, the sequences were not the same length as the genomic sequence.
Most-expressed var genes were similar whether using genomic or de novo-assembled sequences as reference.
Unlike the present study, most var transcription studies using field samples lack reference whole-genome sequences, and quantification of expression will need to be done using de novo-assembled transcripts as reference. To determine the reliability of this approach, we identified the most-expressed var genes from mapping the RNA-Seq reads to both the genomic reference as well as to the var transcripts assembled de novo from the RNA-Seq data. Using data generated from Capture, we found that 5/8 samples had the same most-expressed var gene with mapping to either the genomic or de novo transcripts (UM 3, UM 5, UM 8, UM 9, and UM 11 [Fig. S4]). In an additional two samples, the same var genes were the top two most expressed; however, the order of expression varied for each reference (UM 1 and UM 6 [Fig. S4]).
Proportions of var gene expression for Capture mapping to de novo-assembled sequences and to genomic sequences. The most-expressed var gene was the same mapping to the de novo and to the genomic references for samples UM 3, UM 5, UM 8, UM 9, and UM 11. Each color represents a distinct var sequence within each sample, except for the checkered wedges, which represent var sequences with expression of <5% of the total var expression. Wedges that are different pieces but the same color within a de novo reference pie correspond to two different parts of the same genomic sequence (only within the same sample); however, the sequences assembled as two different distinct sequences in the de novo assembly with rnaSPAdes. UM, uncomplicated malaria. Download FIG S4, PDF file, 0.2 MB (193.8KB, pdf) .
Copyright © 2021 Stucke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
To understand the impact of PfEMP1 in malaria pathogenesis, it is essential to determine which var genes are expressed in an infection and which transcript(s) is the most abundant. Sequencing expressed var genes from clinical samples has proven difficult owing to the diversity and motif repetition of the gene family. Achieving those goals with a single conserved set of primers to amplify all var genes has not been possible because of the extreme variation and the large numbers of var genes per genome. Another difficulty in identifying var transcripts from clinical samples is obtaining adequate parasite RNA for sequencing as almost all RNA in these specimens belongs to the human host. These obstacles pose significant challenges to understanding expression of var genes in clinical samples. Therefore, the goal of this work was to determine the viability of mRNA and parasite RNA enrichment methods to recover this information.
One approach, enriching for mRNA during library preparation [Depletion and Depletion + poly(A)], was intended to enrich for parasite mRNA in the sample by eliminating the two most abundant transcripts in blood samples, namely, rRNA, which altogether constitutes over ∼90% of all host RNA, and globin RNA, the next most abundant transcript in human blood (23). The potential pitfall of this method is that it also enriches for human mRNA. An alternative approach available to us was to enrich specifically for all parasite RNA using capture probes based on the 3D7 reference genome supplemented with additional sequences of the variant surface antigens PfEMP1s, RIFINs, and STEVORs. Potential pitfalls of the capture approach were that our probe set included probes for the entire genome sequence (albeit enriched for variant surface antigens) and that var transcript detection depended on hybridization to the (potentially quite different) reference var sequences used in probe design. To evaluate our assembly methods and the Capture approach, we used two sets of samples. One set was used for proof of principle, since the corresponding genome assembly was available and allowed us to determine the accuracy of the reconstructed var transcripts, as well as to determine if their relative abundance matches would be expected from mapping the reads to the genome. With the second set of samples, we aimed to determine the feasibility of this overall study with samples from a case-control study for which no reference genome data were available. The proof-of-principle approach showed that the var transcripts were reconstructed accurately from the RNA-Seq reads obtained. This is supported by a study that reconstructed genomic var gene sequences from whole-genome sequencing data for >2,000 samples (5). In addition, the proof-of-principle samples showed that the Capture approach resulted in an increased proportion of var reads relative to the Depletion methods and that the reconstructed transcriptome can serve as a reference to identify the most abundantly transcribed var gene. The application of these approaches to the case-control study suggests that, at least with this limited number of samples, the Capture-based method did not result in missed var transcripts, since the Depletion methods did not reveal novel var transcripts that were not also identified with Capture.
Using the SeqCap enrichment system with custom capture probes, we were able to sequence mostly P. falciparum RNA and retain a greater proportion of var-like reads compared with depletion of globin mRNA and rRNA transcripts or with depletion of globin mRNA and rRNA transcripts followed by selection of polyadenylated transcripts. In addition, this method produced more unique transcripts, including transcripts from low-parasitemia samples that were not recovered either with depletion of globin mRNA and rRNA transcripts or with depletion of globin mRNA and rRNA transcripts followed by selection of polyadenylated transcripts. Interestingly, in the case-control samples, we observed greater percentages of reads mapping to var genes in the severe malaria samples from Capture than from the other enrichment methods, even for samples with lower parasitemia. This could indicate that parasites from severe malaria infections are producing more var transcripts than those from uncomplicated malaria infections.
Assembly and reconstruction of var genes from clinical samples have been successful using genomic sequencing data (5, 6, 19), and one recent study has successfully assembled var genes from RNA sequencing of clinical samples (22). A published study presented var gene repertoires assembled from >2,000 clinical isolates from 12 countries (5), de novo assembling var genes from Illumina sequence data from reads mapping to the 3D7 reference genome. The completeness of each var complement was assessed by quantifying a conserved motif in the first domain of most var genes, DBLα, and by limiting the quantified var genes to those of >3 kb. The de novo assemblies of var genes were confirmed by comparison to whole-genome assemblies of 15 field isolates with PacBio sequencing (6). While PacBio sequencing has the advantage of longer reads that will sequence through the entire length of a variant surface antigen, this technology requires a large amount of genetic material that is usually generated from short-term parasite cultures. One study thus far has assembled var genes to quantify var gene expression in clinical samples using Illumina sequencing data. Similar to our approach, a study of severe and uncomplicated malaria with Indonesian samples used de novo assembly methods to assemble var gene transcripts from RNA sequencing data (22). However, this method for RNA sequencing was limited to leukocyte-depleted blood, which requires a time-consuming process that must be conducted in the field at sample collection. In contrast, we employ a Capture method that can be applied on a large scale without extensive sample processing in the field to understand how var expression, defined in terms of transcripts with multiple domains, is associated with severe malaria. Furthermore, we were able to evaluate and validate our assembly methods by comparison with whole-genome sequencing data generated from samples collected at the same time.
Our study did have some limitations. We included only a small subset of samples from a case-control study of severe malaria and thus cannot draw any definitive conclusions about the nature of transcription in severe malaria samples. We were also limited in testing different enrichment methods on all included samples by the amount of RNA that we were able to isolate. The amount of RNA was sufficient to employ all three of our enrichment methods on the subset of case-control samples. However, we had adequate RNA to test only two enrichment methods on six samples of the 11 uncomplicated malaria samples from the longitudinal study and only one method for the remaining five.
The Capture method may also have some limitations. It is possible that enrichment with Capture may produce some bias toward the expression of particular var genes, but we did not find evidence of this in comparing expression methods. Expression using different methods for the same sample was generally in agreement in terms of the most-expressed var sequences. We did not find any var genes highly expressed in the Depletion + poly(A) preparation that were not observed with Capture. The broad concordance between the two methods indicates that we are able to identity the most-expressed var gene. However, the rank order of var expression varied in the situation when the expression levels of particular var genes were close in magnitude. The results from Capture would benefit from confirmation with other methods to study expression, such as qRT-PCR to confirm the most-expressed var gene. Finally, capture approaches can be expensive relative to leukocyte depletion, representing a tradeoff between cost and sample processing time in the field. Capture, costing an additional ∼$1,000 per sample, produced on average 3 to 4 times more reads mapping to P. falciparum than did Depletion and Depletion + poly(A) at an additional $125 per sample. To generate the same number of P. falciparum reads, the libraries from samples prepared using Depletion or Depletion + poly(A) would have to be sequenced 3 to 4 times more, thereby tripling or quadrupling the sequencing cost of these enrichment methods, an increase of several hundreds of dollars. In addition, the cost of Capture for these results was estimated based on a singleplex reaction (single capture per reaction), and multiplexing 3 to 4 samples in the Capture reaction could reduce cost by an added ∼$250 to 300 per sample. While we have established the feasibility of multiplexing genome samples, we are now assessing whether multiplexing RNA samples significantly impacts the dynamic range of transcript abundance estimation.
Enrichment of parasite RNA from whole-blood clinical samples via Capture allowed the successful application of a de novo transcript assembly approach for the discovery of novel var transcripts, ideal for a diverse gene family. We identified a method to enrich for parasite RNA using Roche’s SeqCap EZ enrichment system with custom capture probes (Capture) to sequence constitutive domains of the expressed var genes. This efficient method does not require leukocyte depletion at collection. This comprehensive probe set targets the whole-parasite genome, hence enabling a multitude of applications. However, given the success of this approach, studies that focus exclusively on var gene expression could contemplate using a probe set that targets only var genes. This method produces a wealth of information from the sequence data of the expressed var genes from clinical samples, making possible an analysis of var gene expression at multiple levels, including transcript, constitutive domains, domain cassettes, and motifs. Generating these data on a large scale from an observational study without the need to generate whole-genome sequence data facilitates understanding the role of PfEMP1s in malaria pathogenesis. There are aspects of PfEMP1 biology that have not been addressed due to feasibility. Before we can make biological inferences, we must conduct studies on a larger scale that also would allow for the detection of novel variants. This requires an investment in methods that allows for the collection of samples from a large-scale study that can be subjected to RNA sequencing. Additional applications beyond var genes in P. falciparum include antigen families in other malaria parasite species such as vir genes in P. vivax, as well as highly diverse antigens in other parasites such as those that cause giardiasis and leishmaniasis.
MATERIALS AND METHODS
Sample collection.
A total of 2.5 ml of whole blood was obtained from subjects in a case-control study of severe malaria in children in Mali, West Africa, including three children with severe malaria and one uncomplicated malaria control (UMC). The three severe malaria cases included one case of cerebral malaria (CM), one case of severe malarial anemia (SMA), and one case of both cerebral malaria and severe malarial anemia (CM + SMA). Cerebral malaria was defined using the following criteria: Blantyre coma score of ≤2, no other apparent cause of coma, and confirmation of cerebral malaria by malarial retinopathy, an indicator of infected erythrocytes sequestered in the microvasculature of the brain (24). Severe malarial anemia was defined as malarial illness with a hemoglobin level of ≤5 g/dl. A total of 2.5 ml of whole blood was also collected from 11 uncomplicated, symptomatic malaria (UM 1 to 3, 5, 6, 8, 9, and 11 [Table 1]) episodes as part of a longitudinal study of malaria incidence at a vaccine testing site in Bandiagara, Mali (25). For these same 11 samples, both Illumina and Pacific Biosciences (PacBio) whole-genome sequence data were used to reconstruct var gene repertoires for each sample (19).
The protocols for the case-control study (HP-00075140) and the longitudinal study of malaria incidence (HP-00041382) were approved by the institutional review boards of the Faculty of Medicine, Pharmacy and Dentistry, Bamako, Mali (FWA00001769), and the University of Maryland School of Medicine (FWA00007145). The consent form was approved by the Institutional Review Board of the Faculty of Medicine, Pharmacy and Dentistry, Bamako, Mali. Written informed consent was obtained from parents or guardians of all study participants, and the study was performed in accordance with the Declaration of Helsinki. All methods were performed in accordance with the relevant guidelines and regulations. Participants consented to publication of study results provided that they were identified only by participant number.
Whole venous blood was preserved in PreAnalytiX PAXgene blood RNA tubes (Qiagen/BD), which were inverted 10 times, stored at room temperature for 2 to 72 h, and then frozen for storage at −80°C according to the manufacturer’s instructions. RNA was extracted with the PreAnalytiX PAXgene blood RNA kit according to the manufacturer’s instructions after thawing the tubes and bringing the tubes to room temperature before extraction. Before processing the RNA for sequencing, samples were evaluated for quality with an Agilent Bioanalyzer to determine the RNA integrity number (RIN), a ratio of the 28S to 18S rRNA, which is an indicator of the sample quality.
Enrichment methods.
Our first approach was to enrich for parasite RNA with Roche’s SeqCap EZ enrichment system (“Capture”). Biotinylated probes were designed in conjunction with Roche by providing sequences consisting of the 3D7 P. falciparum genome (PlasmoDB v24) with rRNA-encoding genes removed, as well as 2,885 rif and stevor sequences extracted from clinical isolates, and 4,695 var sequences from additional lab strains (DD2, RAJ116, IGH-CR14, 7G8, NF135, and NF166) (7, 18, 20) and clinical isolates from West Africa, East Africa, and Southeast Asia. Probes with lengths between 50 and 150 bp were designed by Roche, using a proprietary algorithm. Probes bound to SeqCap EZ capture beads were then hybridized in solution with the RNA-Seq library, resulting in enrichment of parasite RNA sequences. Unbound sequences were washed away, and the remaining cDNA was amplified and then sequenced using an Illumina HiSeq 4000 platform.
Two methods were used for the second approach of enriching for mRNA: depletion of human globin mRNA and depletion of host and parasite rRNA ((which we refer to, here, as “Depletion”) (Globin-Zero Gold kit; Illumina) and globin mRNA and rRNA depletion followed by poly(A) selection [here “Depletion + poly(A)”] [Ribo-Zero Gold rRNA removal kit (Epidemiology) (Illumina) and NEBNext poly(A) mRNA magnetic isolation module (New England BioLabs)].
Globin mRNA and rRNA were removed with probes that hybridize to rRNA and globin mRNA, leaving mRNA and regulatory RNAs, which are included in the RNA preparation. After enrichment using globin and rRNA depletion, cDNA was generated and libraries with an insert size of 300 to 500 bp were prepared for sequencing with the TruSeq stranded mRNA library prep kit (Illumina) and then sequenced using the Illumina HiSeq 4000.
RNA-Seq pipeline.
After sequencing, raw data were evaluated for quality by examining the FastQC files, and sequence reads were trimmed if necessary. Using the Eukaryotic RNASeq pipeline at the Institute for Genome Sciences at the University of Maryland School of Medicine, the reads were mapped to a concatenated reference composed of the reference genomes for human (GRCh37) and P. falciparum (PlasmoDB v24) using HISAT2 (v2.0.4) (26). At this step, we determined the percentage of reads mapping to either the human genome or the P. falciparum genome. Using the BAM files converted from SAM files, the files were sorted using SAMtools (v0.1.19) (27), and then seqtk (v1.0) (https://github.com/lh3/seqtk) was used to subset unmapped reads and reads mapping to P. falciparum from the fastq file.
To estimate the number of reads mapping to var genes, we constructed an index of 4,695 available var gene sequences. From each library, we used seqtk (v1.0) to select a random sample of 20 million paired reads from each library to normalize to the smallest library size and mapped the sample of reads to the known var genes to determine the overall alignment rate to known var genes. Using a random selection of reads allowed us to compare reads from each library without bias from larger libraries that may have more var reads just because more of the sample was sequenced. We used the Wilcoxon signed-rank test to compare the absolute numbers of randomly selected reads mapping from paired samples.
De novo assembly.
Any reads mapping to only the human genome were removed, and the remaining reads (unmapped reads and reads mapping to P. falciparum) were then de novo assembled into transcripts using rnaSPAdes with default parameters (v3.10.1) (28). Redundant transcripts were clustered and condensed using CD-HIT (v4.6) and the default sequence identity parameter of 90% (29). The sequence of the longest open reading frame (ORF) from each assembled transcript and its translation were obtained using EMBOSS’s getorf utility (EMBOSS v6.6.0.0), with minimum ORF size set at 500 nucleotides. The resulting peptide sequences were searched using BLASTp (BLAST 2.2.26+) against all ∼4,700 PfEMP1 amino acid sequences from reference genomes and clinical isolates included in the design of the capture array. The protein blast search was repeated against a database of only var sequences from the 3D7 reference genome for comparison. Any transcript matching a PfEMP1 amino acid sequence was evaluated for the presence of var domains using the VarDom database (18). Sequences for which var domains could be annotated with the VarDom database were considered var transcripts. The number and length of these transcripts were then evaluated for each sample, from each enrichment method.
Comparing de novo-assembled var genes to genome assembly sequences.
Full var repertoires are known for a few reference genomes and clinical samples (6, 18). Recently, we sequenced and assembled var genes from 12 uncomplicated malaria infections in Malian children with a novel algorithm, providing full genomic var repertoires for these clinical infections from a longitudinal study of malaria incidence (19). Samples for RNA extraction were collected and preserved at the same time as those from which DNA was obtained to generate the whole-genome sequence data and full var repertoires. As a proof of principle, we compared the de novo-assembled transcripts identified as var genes with the var sequences from the genomic data, which were reconstructed using both PacBio and Illumina reads. Assembled transcripts were compared with the genomic sequences for both coverage (length) and nucleotide sequence identity.
Quantification of var expression.
To compare expression levels estimated from different enrichment methods, and to determine whether we would identify similar predominantly expressed var genes, we used the genomic var repertoires reconstructed from the genomic data as a reference. An index was created for each sample using the genomic var sequences. Using HISAT2, we mapped the reads from each library preparation method using the same index (26) and then determined gene counts with htseq-count (30). Gene counts were converted to transcripts per million (TPMs) to normalize for gene length and library size. TPMs were used to create scatterplots comparing the TPMs from Depletion + poly(A) to Capture. To compare expression levels in the case-control samples with no corresponding genomic reference, for each sample, we generated a reference by de novo assembling transcripts using fastq files of combined reads from the Capture and Depletion + poly(A) libraries.
Data availability.
Sequencing data are available at NCBI through the following accession numbers: SAMN08815609 to SAMN08815630 (longitudinal study samples) and SAMN20286136 to SAMN20286147 (case-control samples).
ACKNOWLEDGMENTS
We thank the team of the Bandiagara Malaria Project for their dedication; the community of Bandiagara, Mali; and the University of Maryland School of Medicine Malaria Research Program. We also thank Matthew Chung for bioinformatics assistance; Christopher Stucke for assistance with Adobe InDesign; and Ana Raquel da Costa, Tina Williams, James Munro, Joanne Morrison, and Nicole Eddington Johnson for administrative support.
We declare no competing interests.
This project was funded in part by federal funds from the National Institute of Allergy and Infectious Diseases and National Institute of Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services under NIH grants U19AI110820 and R01AI141900; cooperative agreement (U19AI065683) from the NIAID; NIAID GSCID contract no. HHSN272200900009C; NIH grant D43TW001589 from the Fogarty International Center; NIH IRID grant R01AI099628; NIH R01HL130750 R01HL146377; a Burroughs Wellcome Fund/American Society of Tropical Medicine and Hygiene Postdoctoral Fellowship to M. A Travassos; and an award to C. V. Plowe from the Howard Hughes Medical Institute. Student support for E. M. Stucke provided by an NIGMS Initiative for Maximizing Student Development Grant (2 R25-GM55036) as part of the Meyerhoff Graduate Fellows Program and NIH grant 1F31HL147471-01. Partial funding for open access was provided by the University of Maryland Health Sciences and Human Services Library’s Open Access Fund.
E. M. Stucke, M. A. Travassos, and J. C. Silva designed and analyzed the study and drafted the manuscript. C. V. Plowe, M. A. Thera, D. Coulibaly, O. K. Doumbo, M. B. Laurens, A. Ouattara, and M. A. Travassos developed, funded, and coordinated the clinical studies. S. Takala-Harrison and J. C. Silva funded and contributed to the analyses. A. Dara, A. Dwivedi, T. K. Hodges, A. E. Zhou, and D. Serre contributed bioinformatics assistance and to the analyses. S. Ott, L. J. Tallon, and L. Sadzewicz provided scientific guidance on sequencing and enrichment methods, prepared samples for sequencing, and managed sequencing data. D. Coulibaly, A. K. Koné, K. Traoré, B. Guindo, B. M. Tangara, A. Niangaly, M. Daou, I. Diarra, Y. Tolo, M. Sissoko, B. Kouriba, O. K. Doumbo, and M. A. Thera enrolled participants, provided participant care, and collected and processed participant data and samples. With the exception of the deceased authors, all authors edited the manuscript.
Contributor Information
Mark A. Travassos, Email: mtravass@som.umaryland.edu.
Joana C. Silva, Email: jcsilva@som.umaryland.edu.
Marta M. Gaglia, Tufts University
REFERENCES
- 1.World Health Organization. 2020. World malaria report 2020: 20 years of global progress and challenges. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Crompton PD, Kayala MA, Traore B, Kayentao K, Ongoiba A, Weiss GE, Molina DM, Burk CR, Waisberg M, Jasinskas A, Tan X, Doumbo S, Doumtabe D, Kone Y, Narum DL, Liang X, Doumbo OK, Miller LH, Doolan DL, Baldi P, Felgner PL, Pierce SK. 2010. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc Natl Acad Sci USA 107:6958–6963. doi: 10.1073/pnas.1001323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JA, Howell KB, Reiling L, Ataide R, Mackintosh CL, Fowkes FJ, Petter M, Chesson JM, Langer C, Warimwe GM, Duffy MF, Rogerson SJ, Bull PC, Cowman AF, Marsh K, Beeson JG. 2012. Targets of antibodies against Plasmodium falciparum-infected erythrocytes in malaria immunity. J Clin Invest 122:3227–3238. doi: 10.1172/JCI62182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J-A, Fowkes FJI, Beeson JG. 2014. Surface antigens of Plasmodium falciparum-infected erythrocytes as immune targets and malaria vaccine candidates. Cell Mol Life Sci 71:3633–3657. doi: 10.1007/s00018-014-1614-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otto TD, Assefa SA, Bohme U, Sanders MJ, Kwiatkowski D, Pf3k consortium, Berriman M, Newbold C. 2019. Evolutionary analysis of the most polymorphic gene family in falciparum malaria. Wellcome Open Res 4:193. doi: 10.12688/wellcomeopenres.15590.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otto TD, Bohme U, Sanders M, Reid A, Bruske EI, Duffy CW, Bull PC, Pearson RD, Abdi A, Dimonte S, Stewart LB, Campino S, Kekre M, Hamilton WL, Claessens A, Volkman SK, Ndiaye D, Amambua-Ngwa A, Diakite M, Fairhurst RM, Conway DJ, Franck M, Newbold CI, Berriman M. 2018. Long read assemblies of geographically dispersed Plasmodium falciparum isolates reveal highly structured subtelomeres. Wellcome Open Res 3:52. doi: 10.12688/wellcomeopenres.14571.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, Paulsen IT, James K, Eisen JA, Rutherford K, Salzberg SL, Craig A, Kyes S, Chan MS, Nene V, Shallom SJ, Suh B, Peterson J, Angiuoli S, Pertea M, Allen J, Selengut J, Haft D, Mather MW, Vaidya AB, Martin DM, Fairlamb AH, Fraunholz MJ, Roos DS, Ralph SA, McFadden GI, Cummings LM, Subramanian GM, Mungall C, Venter JC, Carucci DJ, Hoffman SL, Newbold C, Davis RW, Fraser CM, Barrell B. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, Gysin J, Lanzer M. 1998. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J 17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 10.Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, Taraschi TF, Howard RJ. 1995. Cloning the P falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 11.Lavstsen T, Turner L, Saguti F, Magistrado P, Rask TS, Jespersen JS, Wang CW, Berger SS, Baraka V, Marquard AM, Seguin-Orlando A, Willerslev E, Gilbert MT, Lusingu J, Theander TG. 2012. Plasmodium falciparum erythrocyte membrane protein 1 domain cassettes 8 and 13 are associated with severe malaria in children. Proc Natl Acad Sci USA 109:E1791–E1800. doi: 10.1073/pnas.1120455109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertin GI, Lavstsen T, Guillonneau F, Doritchamou J, Wang CW, Jespersen JS, Ezimegnon S, Fievet N, Alao MJ, Lalya F, Massougbodji A, Ndam NT, Theander TG, Deloron P. 2013. Expression of the domain cassette 8 Plasmodium falciparum erythrocyte membrane protein 1 is associated with cerebral malaria in Benin. PLoS One 8:e68368. doi: 10.1371/journal.pone.0068368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernabeu M, Danziger SA, Avril M, Vaz M, Babar PH, Brazier AJ, Herricks T, Maki JN, Pereira L, Mascarenhas A, Gomes E, Chery L, Aitchison JD, Rathod PK, Smith JD. 2016. Severe adult malaria is associated with specific PfEMP1 adhesion types and high parasite biomass. Proc Natl Acad Sci USA 113:E3270–E3279. doi: 10.1073/pnas.1524294113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mkumbaye SI, Wang CW, Lyimo E, Jespersen JS, Manjurano A, Mosha J, Kavishe RA, Mwakalinga SB, Minja DTR, Lusingu JP, Theander TG, Lavstsen T. 2017. The severity of Plasmodium falciparum infection is associated with transcript levels of var genes encoding endothelial protein C receptor-binding P. falciparum erythrocyte membrane protein 1. Infect Immun 85:e00841-16. doi: 10.1128/IAI.00841-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kyriacou HM, Stone GN, Challis RJ, Raza A, Lyke KE, Thera MA, Kone AK, Doumbo OK, Plowe CV, Rowe JA. 2006. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol Biochem Parasitol 150:211–218. doi: 10.1016/j.molbiopara.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rottmann M, Lavstsen T, Mugasa JP, Kaestli M, Jensen AT, Muller D, Theander T, Beck HP. 2006. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect Immun 74:3904–3911. doi: 10.1128/IAI.02073-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jespersen JS, Wang CW, Mkumbaye SI, Minja DT, Petersen B, Turner L, Petersen JE, Lusingu JP, Theander TG, Lavstsen T. 2016. Plasmodium falciparum var genes expressed in children with severe malaria encode CIDRalpha1 domains. EMBO Mol Med 8:839–850. doi: 10.15252/emmm.201606188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rask TS, Hansen DA, Theander TG, Gorm PA, Lavstsen T. 2010. Plasmodium falciparum erythrocyte membrane protein 1 diversity in seven genomes–divide and conquer. PLoS Comput Biol 6:e1000933. doi: 10.1371/journal.pcbi.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dara A, Drabek EF, Travassos MA, Moser KA, Delcher AL, Su Q, Hostelley T, Coulibaly D, Daou M, Dembele A, Diarra I, Kone AK, Kouriba B, Laurens MB, Niangaly A, Traore K, Tolo Y, Fraser CM, Thera MA, Djimde AA, Doumbo OK, Plowe CV, Silva JC. 2017. New var reconstruction algorithm exposes high var sequence diversity in a single geographic location in Mali. Genome Med 9:30. doi: 10.1186/s13073-017-0422-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moser KA, Drabek EF, Dwivedi A, Stucke EM, Crabtree J, Dara A, Shah Z, Adams M, Li T, Rodrigues PT, Koren S, Phillippy AM, Munro JB, Ouattara A, Sparklin BC, Dunning Hotopp JC, Lyke KE, Sadzewicz L, Tallon LJ, Spring MD, Jongsakul K, Lon C, Saunders DL, Ferreira MU, Nyunt MM, Laufer MK, Travassos MA, Sauerwein RW, Takala-Harrison S, Fraser CM, Sim BKL, Hoffman SL, Plowe CV, Silva JC. 2020. Strains used in whole organism Plasmodium falciparum vaccine trials differ in genome structure, sequence, and immunogenic potential. Genome Med 12:6. doi: 10.1186/s13073-019-0708-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdi AI, Kariuki SM, Muthui MK, Kivisi CA, Fegan G, Gitau E, Newton CR, Bull PC. 2015. Differential Plasmodium falciparum surface antigen expression among children with malarial retinopathy. Sci Rep 5:18034. doi: 10.1038/srep18034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tonkin-Hill GQ, Trianty L, Noviyanti R, Nguyen HHT, Sebayang BF, Lampah DA, Marfurt J, Cobbold SA, Rambhatla JS, McConville MJ, Rogerson SJ, Brown GV, Day KP, Price RN, Anstey NM, Papenfuss AT, Duffy MF. 2018. The Plasmodium falciparum transcriptome in severe malaria reveals altered expression of genes involved in important processes including surface antigen-encoding var genes. PLoS Biol 16:e2004328. doi: 10.1371/journal.pbio.2004328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, Johnson R, Segre AV, Djebali S, Niarchou A, GTEx Consortium, Wright FA, Lappalainen T, Calvo M, Getz G, Dermitzakis ET, Ardlie KG, Guigo R. 2015. Human genomics. The human transcriptome across tissues and individuals. Science 348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beare NA, Taylor TE, Harding SP, Lewallen S, Molyneux ME. 2006. Malarial retinopathy: a newly established diagnostic sign in severe malaria. Am J Trop Med Hyg 75:790–797. doi: 10.4269/ajtmh.2006.75.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coulibaly D, Travassos MA, Kone AK, Tolo Y, Laurens MB, Traore K, Diarra I, Niangaly A, Daou M, Dembele A, Sissoko M, Guindo B, Douyon R, Guindo A, Kouriba B, Sissoko MS, Sagara I, Plowe CV, Doumbo OK, Thera MA. 2014. Stable malaria incidence despite scaling up control strategies in a malaria vaccine-testing site in Mali. Malar J 13:374. doi: 10.1186/1475-2875-13-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li W, Godzik A. 2006. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 30.Anders S, Pyl PT, Huber W. 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the number of 20 million randomly selected reads mapping to known var genes from each sample for each library preparation. The total number of randomly selected reads from Capture (orange) that mapped to known var genes was significantly greater than Depletion (blue) and Depletion + poly(A) (green) (Wilcoxon signed-rank test). Download FIG S1, PDF file, 0.04 MB (47.8KB, pdf) .
Copyright © 2021 Stucke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Sample information, total reads, Plasmodium falciparum reads, insert size, and read length. Download Table S1, XLSX file, 0.01 MB (12KB, xlsx) .
Copyright © 2021 Stucke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Total number of reads mapping to the P. falciparum reference genome or unmapped (red) or the human reference genome (blue) for each library preparation [Capture, left; Depletion, center; Depletion + poly(A), right]. Reads mapping to P. falciparum or unmapped are a larger proportion of total reads in the Capture (left) library preparation than in those of Depletion (center) and Depletion + poly(A) (right). Samples are arranged from least to greatest parasitemia, left to right. Download FIG S2, PDF file, 0.03 MB (35.6KB, pdf) .
Copyright © 2021 Stucke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Comparison of transcripts per million for expressed var genes in Depletion + poly(A) and Capture. TPMs shown in scatterplots are for the var genes expressed in the four case-control study samples. TPMs are calculated from mapping to a common reference de novo assembled by combining reads from both Depletion + poly(A) and Capture libraries. Capture identified the same expressed var genes in the case-control samples as did Depletion + poly(A), but with more transcripts per million. CM, cerebral malaria; UMC, uncomplicated malaria control; SMA, severe malarial anemia. Download FIG S3, PDF file, 0.02 MB (17.1KB, pdf) .
Copyright © 2021 Stucke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Proportions of var gene expression for Capture mapping to de novo-assembled sequences and to genomic sequences. The most-expressed var gene was the same mapping to the de novo and to the genomic references for samples UM 3, UM 5, UM 8, UM 9, and UM 11. Each color represents a distinct var sequence within each sample, except for the checkered wedges, which represent var sequences with expression of <5% of the total var expression. Wedges that are different pieces but the same color within a de novo reference pie correspond to two different parts of the same genomic sequence (only within the same sample); however, the sequences assembled as two different distinct sequences in the de novo assembly with rnaSPAdes. UM, uncomplicated malaria. Download FIG S4, PDF file, 0.2 MB (193.8KB, pdf) .
Copyright © 2021 Stucke et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Sequencing data are available at NCBI through the following accession numbers: SAMN08815609 to SAMN08815630 (longitudinal study samples) and SAMN20286136 to SAMN20286147 (case-control samples).