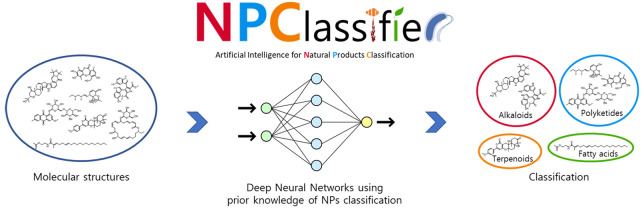
Abstract
Computational approaches such as genome and metabolome mining are becoming essential to natural products (NPs) research. Consequently, a need exists for an automated structure-type classification system to handle the massive amounts of data appearing for NP structures. An ideal semantic ontology for the classification of NPs should go beyond the simple presence/absence of chemical substructures, but also include the taxonomy of the producing organism, the nature of the biosynthetic pathway, and/or their biological properties. Thus, a holistic and automatic NP classification framework could have considerable value to comprehensively navigate the relatedness of NPs, and especially so when analyzing large numbers of NPs. Here, we introduce NPClassifier, a deep-learning tool for the automated structural classification of NPs from their counted Morgan fingerprints. NPClassifier is expected to accelerate and enhance NP discovery by linking NP structures to their underlying properties.
“Classification” is a systematic arrangement of elements into groups or categories according to established criteria to recognize, differentiate, and understand ideas or objects. In natural product (NP) research or specialized metabolite-guided drug discovery, NPs are categorized based upon their molecular structures, chemical properties, bioactivities, and biosynthetic pathways. NPs are an essential resource for drug design and discovery, as well as for pharmacological tools used in biomedical applications.1−3 Fundamentally, molecules belonging to the same class share similar properties based on the criteria used in the classification scheme; therefore, the classification of molecular structures facilitates the quick exploration of large regions of chemical space so as to derive useful information, such as new sources of drugs or bioactivity profiles.4,5
In recent years, natural product research has expanded beyond classical natural product chemistry methodologies to discover bioactive secondary metabolites by embracing new technologies such as genome mining, metabolomics, algorithms, and machine learning approaches for the rapid annotation of candidate molecular structures. Consequently, a web-based NP classification system for cheminformatic approaches is needed to leverage such structural data. For example, CANOPUS, a computational tool for systematic compound class annotation based on MS/MS data, was trained using the classification results from ClassyFire.6
Existing ontologies that provide chemical structure-based classifications include the ChEBI ontology,7 the Medical Subject Heading (MeSH) thesaurus with PubChem,8 LIPID MAPS,9 and NP-specific databases such as Super Natural II,10 MIBiG,11 the Natural Products Atlas,12 and the Dictionary of Natural Products (http://dnp.chemnetbase.com). The molecules in these databases are curated by structural classes, biological activities, or source organisms. These structures and their classification terms are used to train various tools for NP research.13−15 The ontologies and structures in these databases were manually curated from the literature. However, these databases do not have tools for automated classification of molecules. To tackle this last challenge, ClassyFire was developed to automatically classify molecular structures based on chemical properties into the ChemOnt ontology, a well-defined chemical hierarchy.16 However, ClassyFire was designed for general organic and bio-organic chemistry communities, primarily aimed at metabolomics and exposomics, and only provides partial classifications with semantic knowledge of NPs; thus, its relevance for NP research is significantly reduced. Among the 4825 classes in the ChemOnt ontology, 3514 classes describe functional groups or inorganic compounds that are not related to the semantic knowledge from NPs. Additionally, this structural motif-based ontology does not fully match with biosynthetic knowledge. For example, the ChemOnt class “lignans, neolignans, and related compounds” is outside of the phenylpropanoid and polyketide classes, even though lignans are synthesized from phenylpropanoids via the shikimate pathway. As another example, the class “alkaloids and derivatives” provides only structure-based information and does not include any insights on biological precursors such as amino acids.
Unlike structure-only based classifications, traditional classification of NPs encompasses structural information as semantically defined by NP researchers since the late 1800s. Accordingly, this classification system implies not only structural information but also various taxonomic and functional properties of the compounds. The class name “limonoids”, for example, encapsulates a variety of general information about the molecule, such as the typical source organism (Cucurbitaceae, Rutaceae, and Meliaceae), biosynthetic pathway (mevalonate pathway), bioactivities (insecticidal, antibacterial, antifungal, antimalarial, anticancer, or antiviral) and even their taste (bitter).17 These properties are commonly expected from limonoid-type NPs. Knowledge concerning the characteristics and properties of NP classes is continuously expanded and revised with new discoveries made by NP researchers. This ensures that, over time, NP classifications are semantically largely consistent and informative. Consequently, the established classification ontology for NPs allows a broader understanding of NPs.
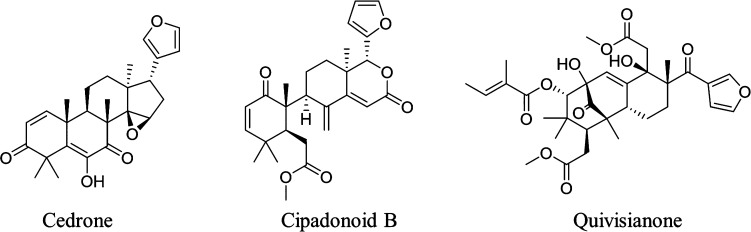
NPs exhibit a very high structural diversity that results in part from the large number of possible biosynthetic pathways, the use of multiple pathways to produce a single molecule (e.g., hybrids), and the abundance of unique tailoring reactions.18 To understand and classify NPs, various rule-based approaches such as analyzing functional groups, comparing structural similarity, and finding maximum common substructures (MCS) have been attempted.19−21 However, a rule-based system must be manually updated whenever new knowledge is gained, and any exceptions must be added by hand.22 For example, the rule-based definition of “limonoids” is any triterpenoid that is highly oxygenated and has a prototypical structure either containing or derived from a precursor with a 4,4,8-trimethyl-17-furanylsteroid skeleton such as cedrone.23 Nevertheless, as shown in Figure 1, some highly modified limonoids such as cipadonoids or quivisianones are considered in the field to be “limonoids” even though they do not fit this definition of a limonoid.24,25 As a result, current rule-based structural classification tools or ontologies have limited use for NP classification.
Figure 1.
Structures of typical (cedrone) and highly modified (cipadonoid B and quivisianone) limonoids.
To develop a new classification tool that incorporates traditional knowledge in the automated classification of NPs, we developed an NP classification system based on the traditional labels provided by the NP community. Over the last two decades, an average of 1600 new marine and microbial NPs have been reported annually,20 and most were reported with their NP classifications during the peer-review and publishing process. This provides the consistency and sustainability for classification of NPs based on the contributions of the NP community. Hence, we used standard practices in the literature concerning chemical entities and their classification in order to create the data set for NPClassifier.
As various types of data have increased for NPs in recent years, the application of deep neural networks (DNNs) to their analysis has been developed for enhancing drug discovery, genome mining, and structure elucidation.14,26−29 The power of deep learning largely derives from how the features are extracted from the data. In contrast to traditional machine learning approaches or rule-based classifications, DNNs learn features from the data in service of the task via back-propagation during training.30 Therefore, DNNs avoid issues of hand-designed features, which may be insufficient for the task. Hence, DNNs are an attractive technique to apply to the problem of NP classification.
In this paper, we introduce a deep neural network-based NP classification tool called “NPClassifier”, which is freely available at https://npclassifier.ucsd.edu together with a web-API (see Supporting Information). NPClassifier was developed using supervised feed-forward networks with 73607 NPs collected from public databases including Pubchem, ChEBI, Chemspider, and the Universal Natural Products Database (UNPD).31−34 The distribution of molecular weights and chemical space of the data set are similar to those in the UNPD, a representative natural product database (Figure S4). NPClassifier classifies the structure of an NP at three levels into seven Pathways, 70 Superclasses, and 672 Classes, all of which are generally recognized by the NP research community (Figure 2). Already, the classification results and the ontology of NPClassifier are being used in natural products research.12,35−40
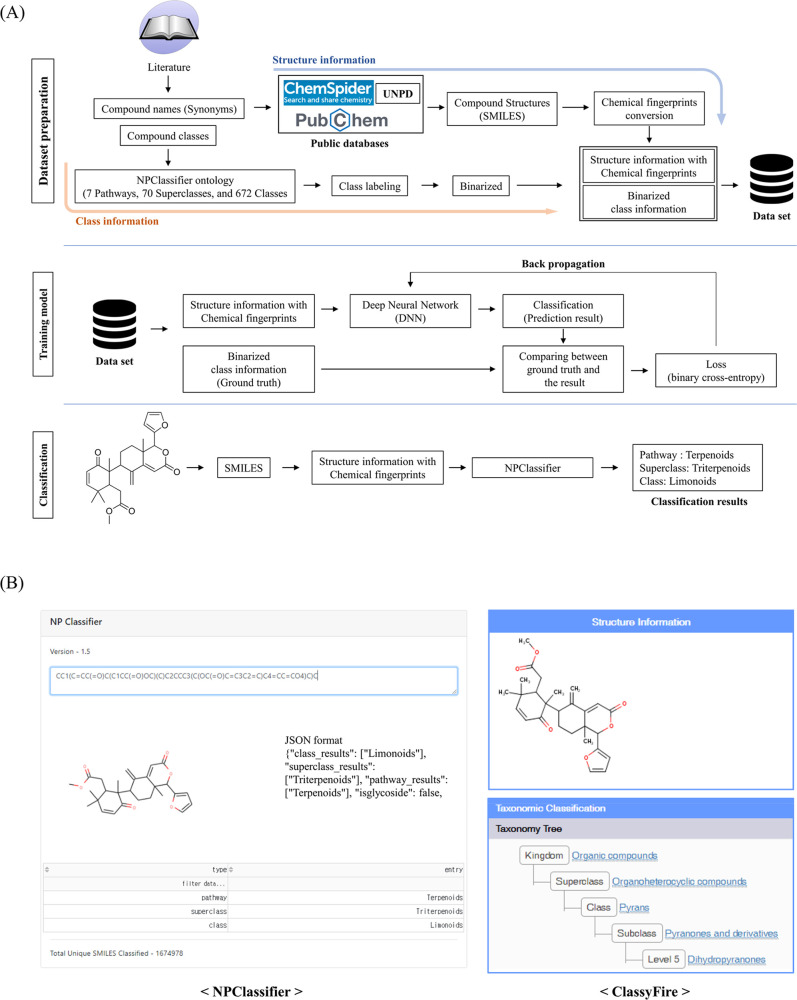
Figure 2.
Overview of NPClassifier. (A) In the data preparation stage, compound names and their class information were collected from the literature. The compound names were converted to chemical fingerprints, and class information was assigned based on the NPClassifier ontology. During the training phase, molecular fingerprints were input to a deep neural network. Binary cross-entropy loss was calculated by comparison between the prediction result from the sigmoid outputs and the ground truth and back-propagated to adjust the model parameters. In classification, a submitted chemical structure is classified by NPClassifier at three levels, including Pathway, Superclass, and Class. (B) Classification result of a highly modified limonoid, cipadonoid B, by NPClassifier and ClassyFire. NPClassifier returns the classification result with three category levels including Pathway, Superclass, and Class, which are based on the semantic knowledge of natural product research.
Results and Discussion
Training, Optimization, and Evaluation of NPClassifier Models
Classification System
A classification system was established based on the literature from the specialized metabolism of plants, marine organisms, fungi, and microorganisms17,41−43 The MIBiG database,11 which provides biosynthetic gene cluster (BGC) information on NPs, was used to ensure the correctness of the biosynthetic pathways such as NRPS-PKS hybrids or terpenoids. The categories used in NPClassifier are defined at three hierarchical levels: Pathway, Superclass, and Class.
The Pathways of NPClassifier consist of seven categories: fatty acids, polyketides, shikimates–phenylpropanoids, terpenoids, alkaloids, amino acids/peptides, and carbohydrates. The fatty acids and polyketides are major biosynthetic pathways of microorganisms and relate to the production of many antibiotics (e.g., doxycycline, erythromycin, and azithromycin) or immunosuppressants (e.g., tacrolimus, rapamycin). The shikimates–phenylpropanoids category is based on the shikimate biosynthetic pathway and includes the phenylpropanoids, which are a diverse family of organic compounds. Additionally, aromatic amino acids and many aromatic NPs are formed from phenylpropanoids via this pathway.44 It should be noted that natural products can be members of more than one pathway in our classification scheme (e.g., aromatic amino acids are both shikimates and amino acids, two of our pathways). The terpenoids are a large and diverse category of NPs derived from the mevalonate (MVA) or the 2-C-methyl-d-erythritol-4-phosphate (MEP) pathways. Terpenoids have diverse biological properties, including cytotoxicity and anti-inflammatory effects.45 Alkaloids represent nitrogenous organic compounds from NPs without obvious peptidic characteristics, although there is some diversity of opinion on this point. A number of alkaloids are part of traditional medications or have found use as single-molecule drug candidates due to their unique bioactivities.46 The amino acid/peptide category results from different biochemical mechanisms for peptide synthesis, wherein multiple amino acids are linked via amide (peptide) bonds. Ribosomal as well as nonribosomal peptide synthetase (NRPS) biosynthetic pathways are responsible for the formation of this category of NP and have been widely investigated using genome sequencing approaches. The carbohydrate category in NPClassifier includes saccharides, polyols, amino sugars, amino glycosides, and their derivatives. The glycosides of other compound classes such as flavonoid glycosides (shikimates–phenylpropanoids) or saponins (terpenoids) are omitted from the carbohydrate category.
The Superclasses represent subcategories within the Pathways, and at the present time 70 designations are proposed. The categories in the Superclass originate from the general classes of metabolites (e.g., flavonoids, meroterpenoids, or steroids), the general chemical or molecular shapes (e.g., chromanes, phloroglucinols, or macrolides), or biosynthetic information (e.g., tryptophan alkaloids, aromatic polyketides, or pseudo alkaloids). The chemical properties or taxonomic information on the chemical entities can be expected to be associated with the Superclass. For example, one of the Superclasses, steroids, is a well-known biologically active metabolite group with a specific multi-ring architecture and consistent chemical properties.47
The Superclasses are subdivided into Classes that represent specific compound families (e.g., erythromycins, penicillins, or cannabinoids), characteristic functional groups (e.g., chromones, azaphilones, indole alkaloids, or 3-spiro tetramic acids), or scaffold diversity within a Superclass (e.g., flavans, flavones, and chalcones from flavonoids). NPClassifier currently includes 672 Classes (see Supporting Information for a complete list of Pathways, Superclasses, and Classes).
Glycosides are also detected by NPClassifier. A glycoside is any molecule in which one or more sugar groups is bonded through a glycosidic bond between its anomeric carbon and a nonsugar component. Because of the numerous important roles that glycosides play in NPs, distinguishing between the sugar component and the aglycone is essential to the understanding of NPs.48 The results of glycoside detection are provided together with the three-level classification system (see Supporting Information for additional details concerning glycoside detection).
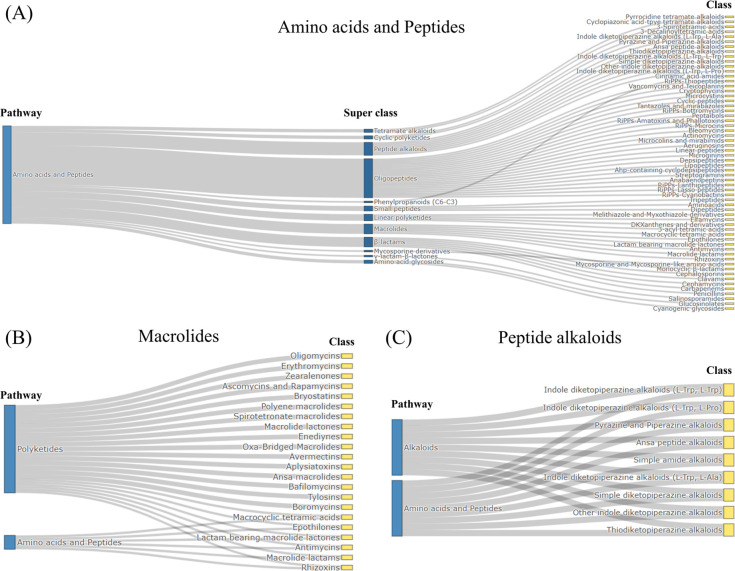
Figure 3A shows the classification system for the amino acids–peptides Pathway, in which 12 Superclasses and 51 Classes were included. Among the Superclasses of the amino acids–peptides Pathway, some Superclasses such as macrolides and peptide alkaloids are included in multiple categories. As shown in Figure 3B, the antimycin Class, which is included in the macrolides Superclass, biosynthetically belongs to hybrid NRPS-PKS synthases. Consequently, this Class is shared between both the amino acids–peptides and polyketides Pathways. The peptide alkaloids are generally considered as alkaloids but are also composed of natural amino acids linked by amide bonds.49 Thus, the peptide alkaloids Superclass is included in both the amino acid–peptides and alkaloids Pathways; hence, the classification system in NPClassifier has a directed acyclic graph structure rather than a strict hierarchy, reflecting the fact that NPs can be classified in multiple ways and may derive from more than one pathway (i.e., hybrids).
Figure 3.
Example of the classification ontology of NPClassifier. (A) Amino acids–peptides Pathway and its Superclasses and Classes in the NPClassifier classification system. This Pathway contains 12 Superclasses and 51 Classes. (B) The macrolides Superclass is involved in both polyketides and amino acids–peptides Pathways. (C) The peptide alkaloids Superclass and its Classes belong to both alkaloids and amino acids–peptides Pathways.
Chemical Descriptors
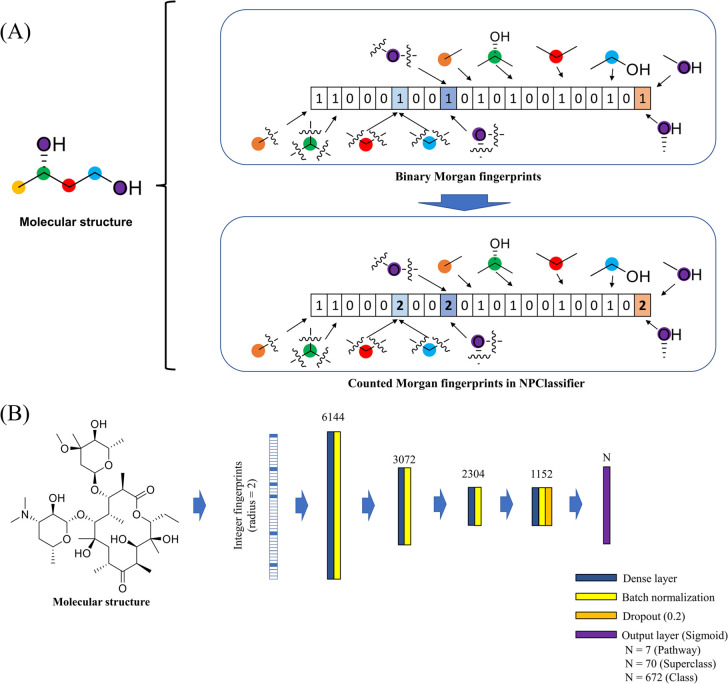
The Morgan fingerprint method was chosen as the format for inputting structural information to the neural network. Morgan fingerprints encode the structure of a molecule in a form that allows for rapid and efficient quantification of similarity between molecular structures or for finding matches to a query substructure. The molecular fingerprint method is generally used in a binary format representing the presence (indicated by a 1) or absence (indicated by a 0) of a given atomic substructure in a molecule. However, this method does not indicate the number of times that a given atomic substructure occurs in a molecule, and this is an important factor to be taken into consideration for NP classification. The count of atomic substructures is particularly important for the classification of oligomeric or structurally iterative NPs, such as proanthocyanidins, tannins, or carotenoids.
Therefore, the “counted Morgan fingerprint method” was used in NPClassifier, which uses non-negative integers to indicate the number of atomic substructures in each location of the vector. RDKit version 2019.09.3 was used to compute the counted Morgan fingerprints for training NPClassifier (Figure 4).50,51
Figure 4.
Chemical descriptor and the deep learning architecture of NPClassifier. (A) Illustration of the difference between Morgan fingerprints and counted Morgan fingerprints; the latter was used in this application. Morgan fingerprints are generally presented in a binary data format over all radii. Alternatively, the counted Morgan fingerprints have an integer format reflecting the count of atomic substructures. (B) Illustration of the structure of the neural network used for NPClassifier. Three different networks were trained: one for each level of classification in NPClassifier. The same structure was used for all three networks with just the top layers differing as a result of the number of alternatives for each level, as indicated in the legend.
Model Optimization and Evaluation
During the training experiments, the performance of the models using two kinds of chemical descriptors, Morgan fingerprints (MFs) and counted Morgan fingerprints (CMFs), was compared. As shown in Table 1, the loss of the CMF-based model was significantly lower, and the cosine similarity and mean average precision (mAP) of the CMF-based model were significantly higher than that of the MF-based model in Superclass and Class. Hence, CMFs were chosen as the input format for the DNN. These results also indicated that training deep neural networks with a semantic knowledge-based ontology resulted in excellent classification accuracy.
Table 1. Comparison of Loss, Cosine Similarity, and Mean Average Precision (mAP) from Neural Networks Trained with Different Chemical Descriptorsa.
| model | classification levels | loss (SD) | cosine similarity (SD) | mAP (SD) |
|---|---|---|---|---|
| MFs (binary) | Pathway | 0.0197 (0.0004) | 0.9863 (0.0004) | 0.9932 (0.0003)** |
| Superclass | 0.0050 (0.0000) | 0.9642 (0.0003) | 0.9423 (0.0010) | |
| Class | 0.0012 (0.0000) | 0.9314 (0.0002) | 0.8734 (0.0018) | |
| CMFs (Integer) | Pathway | 0.0211 (0.0016) | 0.9849 (0.0013) | 0.9920 (0.0004) |
| Superclass | 0.0046 (0.0001)** | 0.9682 (0.0009)** | 0.9515 (0.0030)** | |
| Class | 0.0010 (0.0000)*** | 0.9377 (0.0005)*** | 0.8951 (0.0022)*** |
Each model was optimized based on results from the validation set (n = 11777) and evaluated by using the test set (n = 14721). The results are the average values from five runs of each model. There was no significant difference in the pathway loss or cosine similarity between the two models, so neither is bolded. *Significant at p < 0.05; **significant at p < 0.005; ***significant at p < 0.001. SD = standard deviation. MFs = Morgan fingerprints. CMFs = counted Morgan fingerprints.
For the three levels of classification of NPs, two different architectures using CMFs were compared: one with three separate single-task classifiers and a multitask model. In the multitask model, the output layers were divided into three different heads to predict the three levels (Pathway, Superclass, and Class) simultaneously; this allowed for the hidden layers to receive feedback from the three output layers.52 We found that at all three levels the loss and cosine similarity were improved by using the single-task classifiers. Especially at the Class level, the results from the single-task model were significantly better in cosine similarity (0.0053 higher) and mAP (0.0238 higher) (Table 2); hence, the three single-task models were chosen for NPClassifier (see Supporting Information for additional details about the model optimization and metrics).
Table 2. Comparison of the Losses, Cosine Similarities, and Mean Average Precisions (mAPs) from the Multitask and Single-Task Modelsa.
| model | classification levels | loss (SD) | cosine similarity (SD) | mAP (SD) |
|---|---|---|---|---|
| multitask | Pathway | 0.0234 (0.0006) | 0.9694 (0.0009) | 0.9928 (0.0004) |
| Superclass | 0.0049 (0.0001) | 0.9571 (0.0006) | 0.9551 (0.0016) | |
| Class | 0.0011 (0.0000) | 0.9324 (0.0008) | 0.8713 (0.0009) | |
| single-task | Pathway | 0.0211 (0.0016)* | 0.9849 (0.0013)*** | 0.9920 (0.0004) |
| Superclass | 0.0046 (0.0001)* | 0.9682 (0.0009)*** | 0.9515 (0.0030) | |
| Class | 0.0010 (0.0000) | 0.9377 (0.0005)*** | 0.8951 (0.0022)*** |
Each model was optimized based on the result from the validation set (n = 11777) and evaluated by using the test set (n = 14721). The results are the average values over five runs. *Significant at p < 0.05, **significant at p < 0.005, ***significant at p < 0.001. SD = standard deviation.
Finally, we used Hyperband Tuner for TensorFlow hyperparameter tuning with keras-tuner to achieve the best performance. The performance for each category is described in the Supporting Information.
Performance of NPClassifier vs ClassyFire
To evaluate the general performance of NPClassifier, it was tested with an external test set. The test set contained representatives from three of the Pathways (amino acid–peptides, polyketides, and terpenoids) and Superclasses (flavonoids, steroids, and lignans), which were established from the Dictionary of Natural Products. These Pathways and Superclasses were chosen because they represent overlapping categories between NPClassifier and ClassyFire,16 allowing for a direct comparison. Each Pathway and Superclass had 1000 chemical entities, resulting in 6000 in total. Precision (PRE), recall (REC), and F1 score (the harmonic mean of precision and recall) are reported in Table 3. As observed in Table 3, except for amino acids and peptides, where the two models performed similarly, NPClassifier outperformed ClassyFire and generally showed excellent results. NPClassifier was especially outstanding in recognizing polyketides and lignans. The F1 scores reveal that NPClassifier returned equal or better scores in the majority of cases. We observed that in the cases where ClassyFire had better precision, its recall was much worse than NPClassifier, resulting in poorer F1 scores.
Table 3. Comparison of the Performance between NPClassifier and ClassyFire on the External Test Set from the Dictionary of Natural Products (n = 6000)a.
| NPClassifier |
ClassyFire |
||||||
|---|---|---|---|---|---|---|---|
| classification levels | name | PRE | REC | F1 score | PRE | REC | F1 score |
| Pathway | amino acids and peptides | 0.925 | 0.879 | 0.902 | 0.949 | 0.860 | 0.902 |
| polyketides | 0.781 | 0.893 | 0.834 | 0.927 | 0.331 | 0.488 | |
| terpenoids | 0.969 | 0.974 | 0.972 | 0.902 | 0.751 | 0.819 | |
| Superclass | flavonoids | 0.967 | 0.919 | 0.943 | 0.879 | 0.908 | 0.893 |
| steroids | 0.998 | 0.980 | 0.989 | 0.998 | 0.892 | 0.942 | |
| lignans | 0.992 | 0.654 | 0.788 | 0.997 | 0.353 | 0.521 | |
Each class had 1000 chemical entities.
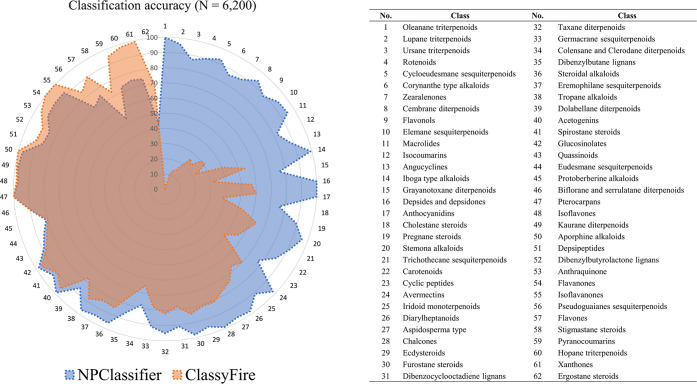
The performance of NP annotation by NPClassifier and ClassyFire was investigated in more detail at the Class level. A total of 62 classes that contained at minimum 100 chemical entities each were tested on both platforms, as shown in Figure 5. Again, NPClassifier outperformed ClassyFire for 47 Classes, often by very large margins, and performed equally or slightly worse for the remaining 15 Classes. Three classes comprising hopane triterpenoids, xanthones, and ergostane steroids classes showed significantly worse performance than ClassyFire. In the hopane triterpenoids class, 25 compounds were classified as other similar triterpenoids such as oleanane, dammarane, lupane, and cucurbitane triterpenoids. In the xanthones class, 16 compounds were unclassified, which means no output was over 0.5, four compounds were classified as flavones, three compounds were classified as anthraquinones, and two compounds were classified as catechols. In the ergostane class, 53 compounds were classified as cholestanes and four compounds were unclassified. Ergostane and cholestane steroids share similar scaffolds, differing only by a methyl group on their side chain, so that might confuse the NPClassifier network (see Supporting Information for additional details on the external test set).
Figure 5.
Comparison of the classification results from NPClassifier (blue) and ClassyFire (orange); overlap is shown in brown. Chemical entities (n = 6200, 100 chemical entities for each of 62 classes) were analyzed by NPClassifier and ClassyFire, and the classification accuracy was measured. Classes are numbered around the circumference of the circle, while the ratio of correct predictions to total predictions ranging from 0 to 100 is denoted by the scale across the radius. NPClassifier showed better results for 47 classes and equal or slightly worse results for 15 classes compared with ClassyFire.
Interpretation of Models
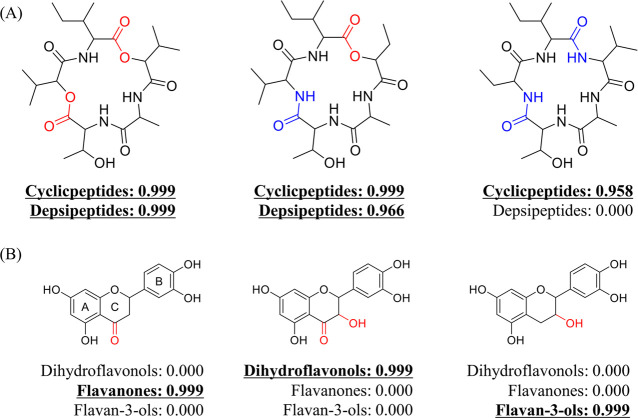
DNNs are sometimes considered to be “black boxes” because the transformations from layer to layer obscure the role of specific input variables; nevertheless, understanding how the model makes its decisions is important to improving its performance. For example, why does it fail in some cases, and how can its reliability be improved? To this end, the change in response from NPClassifier was tracked as a function of modified input structures. This allowed us to evaluate whether the model classified a molecule based on class-related structural features. As shown in Figure 6A, when the ester bonds in the cyclic depsipeptide were replaced with amide bonds, the classes of cyclic and depsipeptides were changed to cyclic peptides. In the same fashion, when the C ring of the flavonone was modified from a 4-keto to a 3-hydroxy-4-keto, the results were changed from flavonones to dihydroflavonols. Finally, when the C ring of the dihydroflavonol was modified from a 3-hydroxy-4-keto to a simple 3-hydroxy group, the results were altered from dihydroflavonols to flavan-3-ols (Figure 6B). From these experiments, the classification results from NPClassifier appear to be influenced by specific structural moieties associated with the general definition of the compound class, in this case, for flavonoids and peptides. The neural networks therefore recognize and utilize class-related features in making these designations.
Figure 6.
Examples of the correlations between structural modifications and classification results. (A) Ester bonds of a cyclic depsipeptide were sequentially replaced with amide bonds, and the classification result changed from cyclic peptide and depsipeptides to cyclic peptides. (B) Correlations between the modification of the C-ring substituents in flavonoids and the resulting classifications.
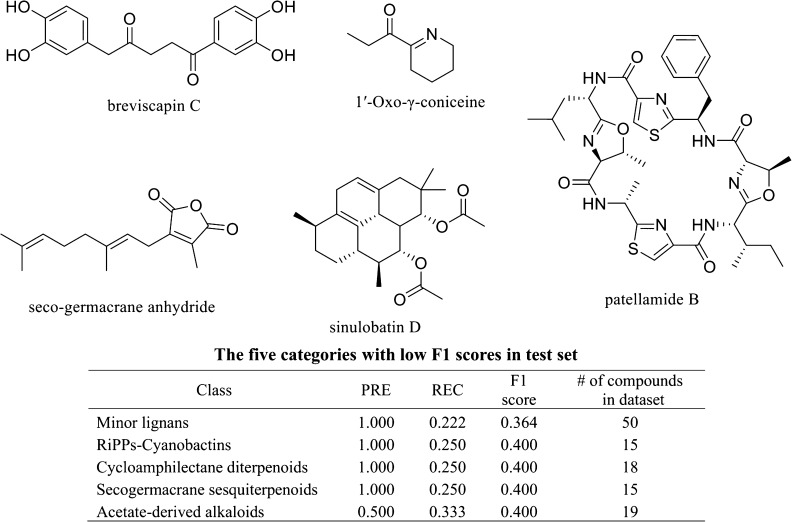
Additionally, the cases where NPClassifier failed were investigated to understand the conditions underlying these incorrect classifications. Incorrectly classified examples were chosen from five categories with the lowest F1 scores in the test set (Figure 7). In the minor lignan class, the norlignan named breviscapin C was classified correctly at the pathway level as shikimates–phenylpropanoids, but unclassified, which means no output was over 0.5 in the Superclass and Class levels. In the acetate-derived alkaloids class, the hemlock alkaloid 1′-oxo-γ-coniceine was classified correctly in the alkaloids at the Pathway level, but was left unclassified at the Superclass and Class levels. The seco-germacrane anhydride shown in Figure 7, which is a ring-opened germacrane sesquiterpenoid, was recognized as a terpenoid, but incorrectly classified as a prenylquinone meroterpenoid at the Superclass level and unclassified at the Class level. Sinulobatin D, a cycloamphilectane diterpenoid, was correctly classified as a terpenoid at the Pathway level and diterpenoid at the Superclass level, but unclassified in the Class level. Patellamide B, which is a cyclic peptide and also a kind of cyanobactin, which is a member of the ribosomally synthesized and post-translationally modified peptides (RiPPs), was expected to be classified as an amino acid and peptide at the Pathway level, an oligopeptide at the Superclass level, and cyclic peptides and RiPP–cyanobactin at the Class level. It was classified correctly as an amino acid and peptide at the Pathway level and an oligopeptide at the Superclass level. However, it was just classified as a cyclic peptide at the Class level and was not classified as a RiPP–cyanobactin.
Figure 7.
Incorrectly classified structures and five categories with low F1 scores in the test set.
The common feature in these unclassified or misclassified metabolites is that the number of representatives in the training set is smaller than the chemical diversity present within their respective Superclass or Class. For example, the minor lignan class represents a group of metabolites with a rarely reported lignan scaffold. Rearranged structures, such as seco- or cyclo-scaffolds, are less often reported than the general parent scaffolds.
In summary, incorrect outputs of NPClassifier can be traced to deficiencies in the training data set or limitations imposed by a limiting number of different classes. The addition of more entries to the reference data set or expanding the number of metabolite classes is expected to reduce these deficiencies. Therefore, user feedback and evaluation forms have been created on the NPClassifier website to collect the community’s contributions and suggestions.
Application of NPClassifier to Natural Products Research and Drug Discovery
NPClassifier was designed for natural product classification and is expected to assist natural product research in a variety of ways. The possible applications where NPClassifier would be useful include
providing a quick chemical overview of a set of natural products, highlighting possible novel chemical architectures
accelerating large-scale genome–metabolome-based natural products discovery studies through compound class-based selection of possible BGC–molecular links; BGCs are better aligned with NPClassifier natural product ontologies than with ClassyFire53,54
analyzing the distribution of secondary metabolite chemical pathways among different environments55
Additionally, we demonstrate here the application of NPClassifier in conjunction with the Natural Products Atlas (NP Atlas) database.
Analysis and Interpretation of Databases
The NP Atlas is an open source database that provides compound names, chemical structures, organism sources, and a structure similarity-based chemical space for natural products from fungi and bacteria along with their literature references.12 In a previous study of NP Atlas, the metabolites from fungi and bacteria were well distinguished using a MAP4-based support vector machine, even though the biosynthetic pathways in bacteria and fungi are generally quite similar.56 However, this study did not explain what factors were responsible for distinguishing between fungal and bacterial metabolites, but suggested that the difference of molecular weight range, the fraction of sp3 carbons, and the presence of glycoside moieties might be responsible.
To further explore the differences between secondary metabolites produced by bacteria and fungi from a biosynthetic perspective, we classified all of the molecules in the NP Atlas using NPClassifier. These results provided some interesting insights into the basis of this separation. At the pathway level, the major difference between the two types of organisms was the number of reported terpenoids (Figure 8A). Over 3000 terpenoids of various sizes were reported from fungi, whereas only around 300 were obtained from bacteria. This is similar to the ratio of BGCs reported from the two types of organisms in the MIBiG database (Figure 8A). We also compared the polyketide metabolites in these two groups; this is a biosynthetic class that is abundantly produced by both fungi and bacteria, and many polyketide synthase (PKS) biosynthetic gene clusters are reported in the MIBiG database for these organisms.
Figure 8.
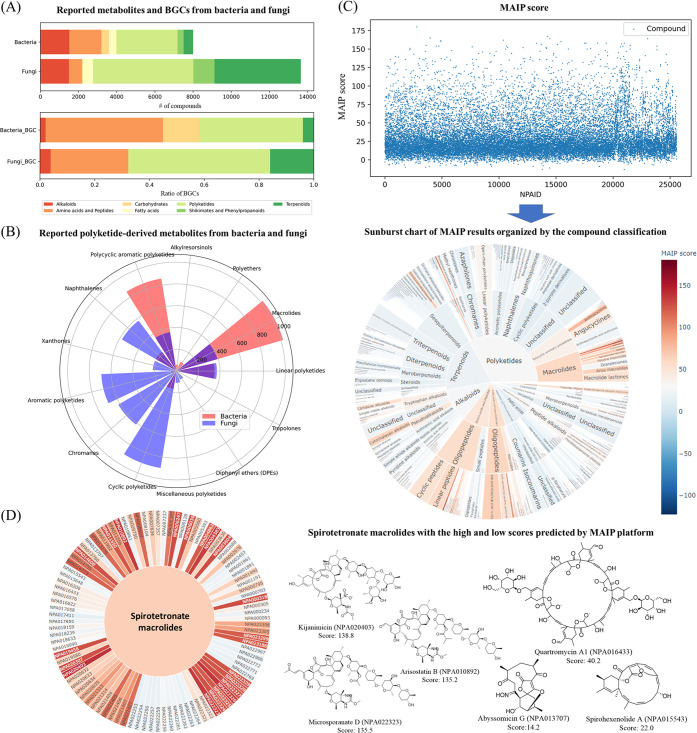
Application of NPClassifier to natural products research and drug discovery. (A) NPClassifier analysis of the diversity of metabolites and BGCs from bacteria and fungi (see text for more details). (B) Distribution of PKS-derived metabolites from bacteria and fungi. (C) The results of in silico antimalarial screening of NP Atlas using the MAIP tool (upper) and the analysis of these results using NPClassifier (lower). The level of predicted antimalarial activity is colored red for active and blue for inactive. (D) Spirotetronate macrolides with high (decalin containing) and low (non-decalin containing) MAIP scores present in the NP Atlas database.
Interestingly, classifications of the PKS products present in bacteria and fungi showed little overlap with each other (Figure 8B). Macrolides and polycyclic aromatic polyketides represent the majority of the bacteria-produced polyketides. In contrast, cyclic polyketides, chromones, aromatic polyketides, and naphthalene derivatives were mainly reported from fungi. These results are similar to those obtained from a large-scale comparison study between bacterial and fungal biosynthetic gene cluster families.57 This study revealed dramatic differences in the biosynthetic logic and chemical space between the two types of organisms. Therefore, even though the biosynthetic pathways used by these two classes of organisms are similar, the products are notably different, and this possibly explains why the previous classification using a machine learning algorithm was successful in this regard (see Supporting Information for all classified results of the NP Atlas database using NPClassifier).
Natural Product Scaffolds-Based in Silico Screening
Over the past few decades, improvement in virtual screening has resulted from the increased size of real as well as virtual compound databases, as well as improvements in the applied algorithms.58 Natural products possess high structural diversity, although this diversity is readily mapped to the outputs of specific biosynthetic pathways. If a specific scaffold is chosen as a candidate structure type from in silico screening, NPClassifier can help identify related source organisms and their compounds for further investigation. This can provide insights into structure–activity relationships (SAR) by integrating NPClassifier with other target prioritization strategies and databases. Such an approach can also be integrated with engineered biochemical pathways and synthetic biology.
For example, using the malaria inhibitor prediction (MAIP) platform from EMBL-EBI (https://www.ebi.ac.uk/chembl/maip/), 25523 chemical entities from the NP Atlas were screened to find potential natural product-derived antimalarial agents.59 The MAIP platform is a machine learning based web service for predicting blood-stage malaria inhibitors and was trained with the results of 4 million screening results. The predicted antimalarial compounds from this MAIP analysis were classified by NPClassifier and labeled with their MAIP score (Figure 8C). The predicted active compounds could then be organized by Pathway, Superclass, and Class. Interestingly, a number of scaffolds were predicted as having highly potent antimalarial activity among the chemical entities in the NP Atlas database (a high-resolution interactive sunburst chart described in Figure 8C is available on https://zenodo.org/record/5068687#.YPm57ehKiUl).
For example, spirotetronate macrolides from the polyketide pathway in the NP Atlas showed a mild MAIP score (65.8), but the range of the scores was quite wide, from 14.2 to 138.8. Among the 100 spirotetronate macrolides in the NP Atlas database, we found that the presence of a decalin moiety as part of the macrolide structure was highly correlated with better MAIP potency. The average MAIP score of decalin-containing macrolides (n = 55) was 97.2, whereas it was only 25.1 when this structural motif was absent (Figure 8D). Interestingly, these prediction results are consistent with previous antimalarial studies where kijanimicin was quite active but the abyssomicins were not.60,61 In this regard, scaffold-based analysis as enabled by NPClassifier could allow for scaffold prioritization in NP drug discovery efforts or could be used to initiate SAR studies of synthetically modified NPs.
Conclusion
In this study, we introduce NPClassifier, a tool that is designed for the classification of natural products using deep learning, including a specific training strategy, optimized parameters, and its evaluation and application. The classification ontology used in NPClassifier is categorized into three hierarchical levels based on expert knowledge. The specialized metabolism (Pathway), chemical properties or chemotaxonomic information (Superclass), and structural details (Class) of the analyzed molecules can be deduced from the classification results of NPClassifier. We anticipate that by supporting large-scale computationally driven NP discovery studies, such as linking the results from genomic and metabolomic mining, NPClassifier can assist with natural products drug discovery as well as understanding the molecular basis for ecological interactions, including human health and the microbiome.
Experimental Section
Data Set Preparation
The data set for training the neural network was prepared using Class, Superclass, and Pathway categories. Compounds with these same category descriptions were manually collected from hundreds of research papers, review papers, titles, books, and abstracts. These collected keywords were converted to structures using the PubChem identifier exchange service (https://pubchem.ncbi.nlm.nih.gov/idexchange) and Chemspider (http://www.chemspider.com). Additional compounds from the ChEBI database7 were added to improve the data set balance between the different classes of NPs. The structures of unconverted keywords via PubChem (https://pubchem.ncbi.nlm.nih.gov) or Chemspider were manually curated by searching the UNPD database34 or by drawing the structures from primary literature sources. Duplicates were removed by comparing InChIKey representations; these in turn were produced by conversion from their SMILES strings. In total, 73607 natural products were labeled and established as a data set for training NPClassifier. This data set was split in a stratified fashion using the Class labels; 64% were assigned to the training set, 16% to the validation set, and 20% to the test set. After hyperparameter tuning, the training and validation sets were merged together and subjected to the final model training.
Preparing an External Evaluation Test Set
To evaluate and compare the performance between different platforms, compound Classes that were included in both the NPClassifier and ClassyFire platforms were chosen from the Dictionary of Natural Products (http://dnp.chemnetbase.com/). In the external test set, 3000 chemical entities for three Pathways (amino acid–peptides, polyketides, and terpenoids), 3000 compounds for three Superclasses (flavonoids, lignans, and steroids), and 6200 compounds for 62 Classes were included. As these structures were used from a commercial library, these data were only used for testing and were not included in the training set of NPClassifier.
Data Labeling and Evaluation Metric
Each unique category from the three classification levels was encoded by the binary encoding method. Cosine scoring was used to measure the similarity between these binarized vectors, and this allowed comparison between the predicted results and the ground truth results during training.
To compare the performance between each model, (1) average precision, (2) mean average precision, and (3) F1 scores were computed from the results. In the precision recall curves, the trade-off between precision and recall was shown for different thresholds. A high area under the curve represents both high recall and high precision, where high precision relates to a low false positive rate, and high recall relates to a low false negative rate. Thus, high scores for both demonstrate that NPClassifier is returning correct results (high precision) as well as a majority of all correct results (high recall). Average precision (AP) summarizes a precision–recall curve as the weighted mean of precisions achieved at each threshold, with the increase in recall from the previous threshold used as the weight:
| 1 |
where Pk and Rk are the precision and recall at the kth threshold.
Mean average precision is the average AP from k classes in order to measure the performance of multilabel classification problems.
| 2 |
F1 score is defined as the harmonic mean of precision and recall. This score is often used in the field of information retrieval for measuring search, document classification, and query classification performance.
| 3 |
Deep Neural Network Architectures
The training of NPClassifier was performed on a server with an Intel Core i7-6850K CPU, NVIDIA GeForce GTX 1080 with 8GB video memory GPU, and 64 GB RAM. For the purpose of this research, the Python programming language was used and the TensorFlow 2.3.0 deep learning framework was used. The DNN for the NPClassifier was composed of three different networks that classified the molecular structure at the three levels of hierarchy with feed-forward neural network architecture. For each network, there was an input layer, representing the counted fingerprints, followed by three hidden layers and a fully connected layer to the output. Dropout was applied to the fully connected layers to improve generalization. The activation function for the hidden layers used the ReLu function and all hidden layers were normalized by batch normalization. Hyperparameters including the number of hidden layer units, learning rates, regularization, and dropout rate were optimized by the Hyperband algorithm in Keras Tuner.
Acknowledgments
We gratefully acknowledge financial support by the U.S. National Institutes of Health (NIH) (R01 GM107550) to P.C.D., W.H.G., and G.W.C., the Gordon and Betty Moore Foundation grant (GBMF7622) to P.C.D., W.H.G., and G.W.C., and The Netherlands eScience Center (NLeSC) ASDI eScience grant (ASDI.2017.030) to J.J.J.v.d.H. K.B.K was supported by a National Research Foundation of Korea (NRF) grant funded by the Ministry of Science, ICT, and Future Planning (NRF-2020R1C1C1004046). We thank anonymous reviewers for highly insightful comments on the manuscript.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jnatprod.1c00399.
The authors declare the following competing financial interest(s): Garrison W. Cottrell, and William H. Gerwick are the cofounders of NMR Finder LLC. Mingxun Wang is the founder of Ometa Laboratories LLC.
Notes
The web application is available at https://npclassifier.ucsd.edu/. The source code for the NPClassifier is available on GitHub at https://github.com/mwang87/NP-Classifier. The data set used in the NPClassifier is available on https://zenodo.org/record/5068687#.YOKJQOgzaUl.
Supplementary Material
References
- Lachance H.; Wetzel S.; Kumar K.; Waldmann H. J. Med. Chem. 2012, 55, 5989–6001. 10.1021/jm300288g. [DOI] [PubMed] [Google Scholar]
- Grisoni F.; Merk D.; Consonni V.; Hiss J. A.; Tagliabue S. G.; Todeschini R.; Schneider G. Commun. Chem. 2018, 1, 44. 10.1038/s42004-018-0043-x. [DOI] [Google Scholar]
- Wu M. C.; Law B.; Wilkinson B.; Micklefield J. Curr. Opin. Biotechnol. 2012, 23, 931–40. 10.1016/j.copbio.2012.03.008. [DOI] [PubMed] [Google Scholar]
- Reymond J. L.; Awale M. ACS Chem. Neurosci. 2012, 3, 649–657. 10.1021/cn3000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldivar-Gonzalez F. I.; Lenci E.; Trabocchi A.; Medina-Franco J. L. RSC Adv. 2019, 9, 27105–27116. 10.1039/C9RA04841C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dührkop K.; Nothias L.-F.; Fleischauer M.; Reher R.; Ludwig M.; Hoffmann M. A.; Petras D.; Gerwick W. H.; Rousu J.; Dorrestein P. C.; Böcker S. Nat. Biotechnol. 2021, 39, 462–471. 10.1038/s41587-020-0740-8. [DOI] [PubMed] [Google Scholar]
- Hastings J.; Owen G.; Dekker A.; Ennis M.; Kale N.; Muthukrishnan V.; Turner S.; Swainston N.; Mendes P.; Steinbeck C. Nucleic Acids Res. 2016, 44, 1214–1219. 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers F. B. Bull. Med. Libr. Assoc. 1963, 51, 114–116. [PMC free article] [PubMed] [Google Scholar]
- Fahy E.; Subramaniam S.; Murphy R. C.; Nishijima M.; Raetz C. R. H.; Shimizu T.; Spener F.; van Meer G.; Wakelam M. J. O.; Dennis E. A. J. Lipid Res. 2009, 50, 9–14. 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee P.; Erehman J.; Gohlke B. O.; Wilhelm T.; Preissner R.; Dunkel M. Nucleic Acids Res. 2015, 43, 935–939. 10.1093/nar/gku886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautsar S. A.; Blin K.; Shaw S.; Navarro-Munoz J. C.; Terlouw B. R.; van der Hooft J. J. J.; van Santen J. A.; Tracanna V.; Duran H. G. S.; Andreu V. P.; Selem-Mojica N.; Alanjary M.; Robinson S. L.; Lund G.; Epstein S. C.; Sisto A. C.; Charkoudian L.; Collemare J.; Linington R. G.; Weber T.; Medema M. H. Nucleic Acids Res. 2019, 48, 454–458. 10.1093/nar/gkz882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Santen J. A.; Jacob G.; Singh A. L.; Aniebok V.; Balunas M. J.; Bunsko D.; Neto F. C.; Castano-Espriu L.; Chang C.; Clark T. N.; Little J. L. C.; Delgadillo D. A.; Dorrestein P. C.; Duncan K. R.; Egan J. M.; Galey M. M.; Haeckl F. P. J.; Hua A.; Hughes A. H.; Iskakova D.; Khadilkar A.; Lee J. H.; Lee S.; LeGrow N.; Liu D. Y.; Macho J. M.; McCaughey C. S.; Medema M. H.; Neupane R. P.; O’Donnell T. J.; Paula J. S.; Sanchez L. M.; Shaikh A. F.; Soldatou S.; Terlouw B. R.; Tran T. A.; Valentine M.; van der Hooft J. J. J.; Vo D. A.; Wang M. X.; Wilson D.; Zink K. E.; Linington R. G. ACS Cent. Sci. 2019, 5, 1824–1833. 10.1021/acscentsci.9b00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M.; Shin H. K.; Myung Y.; Hwang S.; No K. T. J. Cheminf. 2020, 12, 6. 10.1186/s13321-020-0410-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Trevino S. H.; Uc-Cetina V.; Fernandez-Herrera M. A.; Merino G. J. Chem. Inf. Model. 2020, 60, 3376–3386. 10.1021/acs.jcim.0c00293. [DOI] [PubMed] [Google Scholar]
- Korotcov A.; Tkachenko V.; Russo D. P.; Ekins S. Mol. Pharmaceutics 2017, 14, 4462–4475. 10.1021/acs.molpharmaceut.7b00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feunang Y. D.; Eisner R.; Knox C.; Chepelev L.; Hastings J.; Owen G.; Fahy E.; Steinbeck C.; Subramanian S.; Bolton E.; Greiner R.; Wishart D. S. J. Cheminf. 2016, 8, 61. 10.1186/s13321-016-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S.; Liu B. Org. Chem. Front. 2020, 7, 1903–1947. 10.1039/D0QO00203H. [DOI] [Google Scholar]
- Morrison K. C.; Hergenrother P. J. Nat. Prod. Rep. 2014, 31, 6–14. 10.1039/C3NP70063A. [DOI] [PubMed] [Google Scholar]
- Koch M. A.; Schuffenhauer A.; Scheck M.; Wetzel S.; Casaulta M.; Odermatt A.; Ertl P.; Waldmann H. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 17272–17277. 10.1073/pnas.0503647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye C. R.; Bertin M. J.; Lokey R. S.; Gerwick W. H.; Linington R. G. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 5601–5606. 10.1073/pnas.1614680114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y. Q.; Jiang T.; Girke T. Bioinformatics 2008, 24, 366–374. 10.1093/bioinformatics/btn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A.; Bi L.; Kim J.; Feng D. D. In Biomedical Information Technology, 2nd ed; Feng D. D., Ed.; Academic Press, 2020; Chapter 5, pp 167–196. [Google Scholar]
- Roy A.; Saraf S. Biol. Pharm. Bull. 2006, 29, 191–201. 10.1248/bpb.29.191. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Y.; Xu H. RSC Adv. 2017, 7, 35191–35220. 10.1039/C7RA04715K. [DOI] [Google Scholar]
- Tian X.; Li H.; An F.; Li R.; Zhou M.; Yang M.; Kong L.; Luo J. Planta Med. 2017, 83, 341–350. 10.1055/s-0042-117114. [DOI] [PubMed] [Google Scholar]
- Reher R.; Kim H. W.; Zhang C.; Mao H. H.; Wang M. X.; Nothias L. F.; Caraballo-Rodriguez A. M.; Glukhov E.; Teke B.; Leao T.; Alexander K. L.; Duggan B. M.; Van Everbroeck E. L.; Dorrestein P. C.; Cottrell G. W.; Gerwick W. H. J. Am. Chem. Soc. 2020, 142, 4114–4120. 10.1021/jacs.9b13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merwin N. J.; Mousa W. K.; Dejong C. A.; Skinnider M. A.; Cannon M. J.; Li H. X.; Dial K.; Gunabalasingam M.; Johnston C.; Magarvey N. A. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 371–380. 10.1073/pnas.1901493116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.; Idelbayev Y.; Roberts N.; Tao Y. W.; Nannapaneni Y.; Duggan B. M.; Min J.; Lin E. C.; Gerwick E. C.; Cottrell G. W.; Gerwick W. H. Sci. Rep. 2017, 7, 14243. 10.1038/s41598-017-13923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S. J.; Yan X.; Gu Q.; Yang Y. D.; Du Y. F.; Lu Y. T.; Xu J. J. Cheminf. 2019, 11, 5. 10.1186/s13321-019-0328-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y.; Bengio Y.; Hinton G. Nature 2015, 521, 436–444. 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- de Matos P.; Dekker A.; Ennis M.; Hastings J.; Haug K.; Turner S.; Steinbeck C. J. Cheminf. 2010, 2, 6. 10.1186/1758-2946-2-S1-P6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.; Thiessen P. A.; Bolton E. E.; Chen J.; Fu G.; Gindulyte A.; Han L. Y.; He J. E.; He S. Q.; Shoemaker B. A.; Wang J. Y.; Yu B.; Zhang J.; Bryant S. H. Nucleic Acids Res. 2016, 44, 1202–1213. 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence H. E.; Williams A. J. Chem. Educ. 2010, 87, 1123–1124. 10.1021/ed100697w. [DOI] [Google Scholar]
- Gu J. Y.; Gui Y. S.; Chen L. R.; Yuan G.; Lu H. Z.; Xu X. J. PLoS One 2013, 8, 62839. 10.1371/journal.pone.0062839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz A.; Sorokina M.; Galgonek J.; Mietchen D.; Willighagen E.; Graham J.; Stephan R.; Page R.; Vondrášek J.; Steinbeck C.; Pauli G. F.; Wolfender J.-L.; Bisson J.; Allard P.-M.. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianza M.; Leroy R.; Machado Rodrigues C.; Borie N.; Sayagh C.; Remy S.; Kuhn S.; Renault J.-H.; Nuzillard J.-M. Molecules 2021, 26, 637. 10.3390/molecules26030637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings J.; Glauer M.; Memariani A.; Neuhaus F.; Mossakowski T. J. Cheminf. 2021, 13, 23. 10.1186/s13321-021-00500-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokina M.; Merseburger P.; Rajan K.; Yirik M. A.; Steinbeck C. J. Cheminf. 2021, 13, 2. 10.1186/s13321-020-00478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M. A.; Nothias L.-F.; Ludwig M.; Fleischauer M.; Gentry E. C.; Witting M.; Dorrestein P. C.; Dührkop K.; Böcker S.. bioRxiv 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewick P. M.Medicinal Natural Products: A Biosynthetic Approach, 3rd ed.; John Wiley & Sons: Chichester, 2009. [Google Scholar]
- Fattorusso E.; Gerwick W. H.; Taglialatela-Scafati O.. Handbook of Marine Natural Products; Springer: Dordrecht, 2012. [Google Scholar]
- Kinghorn A. D.; Falk H.; Gibbons S.; Kobayashi J.. Progress in the Chemistry of Organic Natural Products 106; Springer International Publishing: Cham, 2017. [Google Scholar]
- Averesch N. J. H.; Krömer J. O. Front. Bioeng. Biotechnol. 2018, 6, 32. 10.3389/fbioe.2018.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downer E. J. ACS Chem. Neurosci. 2020, 11, 659–662. 10.1021/acschemneuro.0c00075. [DOI] [PubMed] [Google Scholar]
- Kittakoop P.; Mahidol C.; Ruchirawat S. Curr. Top. Med. Chem. 2013, 14, 239–252. 10.2174/1568026613666131216105049. [DOI] [PubMed] [Google Scholar]
- Rahman S. U.; Ismail M.; Khurram M.; Ullah I.; Rabbi F.; Iriti M. Molecules 2017, 22, 2156. 10.3390/molecules22122156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kytidou K.; Artola M.; Overkleeft H. S.; Aerts Front. Plant Sci. 2020, 11, 357. 10.3389/fpls.2020.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat K. L.; Joullie M. M. J. Chem. Educ. 1987, 64, 21–27. 10.1021/ed064p21. [DOI] [Google Scholar]
- Landrum G.Abstr. Pap. Am. Chem. Soc. 2019, 258. [Google Scholar]
- Rogers D.; Hahn M. J. Chem. Inf. Model. 2010, 50, 742–754. 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Wenzel J.; Matter H.; Schmidt F. J. Chem. Inf. Model. 2019, 59, 1253–1268. 10.1021/acs.jcim.8b00785. [DOI] [PubMed] [Google Scholar]
- van der Hooft J. J. J.; Mohimani H.; Bauermeister A.; Dorrestein P. C.; Duncan K. R.; Medema M. H. Chem. Soc. Rev. 2020, 49, 3297–3314. 10.1039/D0CS00162G. [DOI] [PubMed] [Google Scholar]
- Beniddir M. A.; Kang K. B.; Genta-Jouve G.; Huber F.; Rogers S.; van der Hooft J. J. J.. Nat. Prod. Rep. 2021, 10.1039/D1NP00023C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer J. P.; Nothias L.-F.; Thompson L. R.; Sanders J. G.; Salido R. A.; Couvillion S. P.; Brejnrod A. D.; Huang S.; Lejzerowicz F.; Lutz H. L.; Zhu Q.; Martino C.; Morton J. T.; Karthikeyan S.; Nothias-Esposito M.; Dührkop K.; Böcker S.; Kim H.; Aksenov A. A.; Bittremieux W.; Minich J. J.; Marotz C.; Bryant M. M.; Sanders K.; Schwartz T.; Humphrey G.; Vásquez-Baeza Y.; Tripathi A.; Parida L.; Carrieri A. P.; Haiminen N.; Beck K. L.; Das P.; González A.; McDonald D.; Karst S. M.; Albertsen M.; Ackermann G.; DeReus J.; Thomas T.; Petras D.; Shade A.; Stegen J.; Song S. J.; Metz T. O.; Swafford A. D.; Dorrestein P. C.; Jansson J. K.; Gilbert J. A.; Knight R.. bioRxiv 2021. [Google Scholar]
- Capecchi A.; Reymond J. L. Biomolecules 2020, 10, 1385. 10.3390/biom10101385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robey M. T.; Caesar L. K.; Drott M. T.; Keller N. P.; Kelleher N. L. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, 118. 10.1073/pnas.2020230118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. E. J. Chem. Inf. Model. 2020, 60, 4120–4123. 10.1021/acs.jcim.0c00101. [DOI] [PubMed] [Google Scholar]
- Bosc N.; Felix E.; Arcila R.; Mendez D.; Saunders M. R.; Green D. V. S.; Ochoada J.; Shelat A. A.; Martin E. J.; Iyer P.; Engkvist O.; Verras A.; Duffy J.; Burrows J.; Gardner J. M. F.; Leach A. R. J. Cheminf. 2021, 13, 13. 10.1186/s13321-021-00487-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waitz J. A.; Horan A. C.; Kalyanpur M.; Lee B. K.; Loebenberg D.; Marquez J. A.; Miller G.; Patel M. G. J. Antibiot. 1981, 34, 1101–1106. 10.7164/antibiotics.34.1101. [DOI] [PubMed] [Google Scholar]
- Sadaka C.; Ellsworth E.; Hansen P. R.; Ewin R.; Damborg P.; Watts J. L. Molecules 2018, 23, 1371. 10.3390/molecules23061371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.