Abstract
The aim of this study was to evaluate the role of the regulatory small RNA (sRNA) Ern0160 in gastrointestinal tract (GIT) colonization by Enterococcus faecium. For this purpose, four strains of E. faecium were used, Aus0004 (WT), an ern0160-deleted Aus0004 mutant (Δ0160), a trans-complemented Δ0160 strain overexpressing ern0160 (Δ0160_0160), and a strain Δ0160 with an empty pAT29 vector (Δ0160_pAT29). Strains were studied both in vitro and in vivo, alone and in competitive assays. In in vitro experiments, no difference was observed between WT and Δ0160 strains cultured single while Δ0160_0160 strain grew more slowly than Δ0160_pAT29. In competitive assays, the WT strain was predominant compared to the deleted strain Δ0160 at the end of the experiment. Then, in vivo experiments were performed using a GIT colonization mouse model. Several existing models of GIT colonization were compared while a novel one, combining ceftriaxone and amoxicillin, was developed. A GIT colonization was performed with each strain alone, and no significant difference was noticed. By contrast, significant results were obtained with co-colonization experiments. With WT + Δ0160 suspension, a significant advantage for the WT strain was observed from day 5 to the end of the protocol, suggesting the involvement of ern0160 in GIT colonization. With Δ0160_0160 + Δ0160_pAT29 suspension, the strain with the empty vector took the advantage from day 3 to the end of the protocol, suggesting a deleterious effect of ern0160 overexpression. Altogether, these findings demonstrate the potential implication of Ern0160 in GIT colonization of E. faecium. Further investigations are needed for the identification of sRNA target(s) in order to decipher underlying molecular mechanisms.
Keywords: E. faecium, VRE, sRNA, pathogenicity, animal model, gut colonization
Introduction
Enterococci are ubiquitous Gram-positive cocci that are normal inhabitants of the human gut microbiota. Enterococcus faecium, which represents 15–25% of enterococcal isolates responsible for human infections, has been increasingly involved in healthcare-associated infections and hospital outbreaks (Goh et al., 2017; Weiner-Lastinger et al., 2020). Glycopeptides are usually used to treat serious E. faecium infections, but many clinical isolates have become resistant to vancomycin (VREF) especially in North America. In 2010, up to 80% of VREF were reported in the United States (O’Driscoll and Crank, 2015). According to the European Antimicrobial Resistance Surveillance Network (EARS-Net), the mean proportion of VREF among invasive isolates increased from 10.4 to 17.3% in European countries between 2014 and 2018 (European Centre for Disease Prevention and Control, 2019). These high rates of VREF prevalence could be attributable to the worldwide dissemination of a subpopulation of E. faecium hospital-adapted clones that belongs to the clonal complex 17 (CC17) and represents a public health concern (Cattoir and Giard, 2014). This CC17 actually corresponds to the phylogenetic clade A1 composed by the majority of strains responsible for infections and hospital outbreaks. Two other clades also exist, clade A2 that includes animal strains and sporadic human infection isolates and clade B that comprises human commensal fecal strains (Lebreton et al., 2013).
In recent years, bacterial regulatory RNAs, referred to as small RNAs (sRNAs), have been described as having a major role in various adaptive responses, including stress response, virulence, and antimicrobial resistance (Mraheil et al., 2010; Wagner and Romby, 2015; Klein and Raina, 2017). For example, SprX contributes to vancomycin resistance in Staphylococcus aureus (Eyraud et al., 2014) and some sRNAs are involved in stress response in Enterococcus faecalis (Michaux et al., 2014). Recently, 61 sRNA candidates were identified in E. faecium but there are currently no published data about their function (Sinel et al., 2017). Among them, 10 have been experimentally validated and their expression under sub-inhibitory concentrations (SICs) of daptomycin was observed. sRNA_0160 (renamed here Ern0160) appeared to be highly expressed and its expression decreased significantly under daptomycin exposure. Furthermore, ern0160 is conserved in all studied strains of E. faecium belonging to the three different clades (Sinel et al., 2017). These initial positive results led us to study this sRNA to evaluate its potential role in intestinal colonization.
Because the gastrointestinal tract (GIT) serves as a major reservoir from which VREF can spread to the hospital environment and GIT colonization precedes infection, understanding GIT colonization mechanisms appears to be essential to better manage those infections and to limit VREF hospital spread (Donskey et al., 2000, 2001; Ubeda et al., 2010). In vivo GIT colonization experimental models with enterococci have been described for a long time, with about one-third of these studies exploring E. faecium. Most of them used mouse models and different antibiotic protocols to eliminate the animal commensal microbiota allowing to the establishment of colonization by the studied bacteria. Several different protocols for GIT colonization with enterococci have been published, and there is no consensus on molecules used as well as routes and durations of administration. For example, Heikens et al. (2009) used a protocol with subcutaneous (SC) ceftriaxone alone, administered for 12 days. Zhang et al. (2013) associated SC ceftriaxone 2 days before inoculation, with cefoxitin added to drinking water (DW) during the protocol. Montealegre et al. (2016) published in 2016 a model of E. faecium GIT colonization using a combination of SC clindamycin and gentamicin in DW only 4 days before inoculation. As these protocols are very different, it seemed interesting to compare their performances. Moreover, as VREF intestinal colonization mostly occurs in patients treated with broad-spectrum antibiotics, it seems important to use antibiotics frequently prescribed in clinics, such as third-generation cephalosporins or ampicillin, to study the mechanisms of VREF colonization (Dubin and Pamer, 2014).
The aim of this study was to evaluate the role of Ern0160 in E. faecium GIT colonization. For this purpose, in vitro experiments were performed and several existing models of GIT colonization were compared, while a novel one, as close as possible to human conditions, was developed for in vivo studies.
Materials and Methods
Bacterial Strains
Data regarding the strain characteristics are summarized in Table 1. The E. faecium Aus0004 wild-type (WT) reference strain was used (Lam et al., 2012). This vancomycin-resistant (vanB-positive) clinical isolate was recovered from a bacteremic patient and belongs to the CC17. The MIC of amoxicillin for Aus0004 strain was 256 mg/l.
TABLE 1.
Bacterial strains and plasmids used in the study.
| Strains or plasmids | Relevant characteristics | References |
| Strains | ||
| Enterococcus faecium | ||
| Aus0004 WT | vanB-positive E. faecium Aus0004 reference strain | Lam et al., 2012 |
| Δ0160 | ern0160-deleted Aus0004 | This study |
| Δ0160_pAT29 | Δ0160 carrying empty pAT29 vector | This study |
| Δ0160_Δ0160 | Δ0160 carrying recombinant plasmid pAT29Ω0160 | This study |
| Escherichia coli | ||
| EC1000 | Strain using for cloning | Leenhouts et al., 1996 |
| Plasmids | ||
| pWS3 | Temperature-sensitive pG(+)host9-derived shuttle vector used for gene disruption (Spcr) | Zhang et al., 2011 |
| pAT29 | High-copy-number shuttle vector used for cloning (Spcr) | Trieu-Cuot et al., 1990 |
| pAT29Ωern0160 | Recombinant pAT29 plasmid containing ern0160 with its native promoter and rho-independent terminator (Spcr) | This study |
Spcr, spectinomycin resistance.
Three mutants were constructed to evaluate the role of Ern0160 in GIT colonization. As described by Zhang et al. (2011), an ern0160-deleted E. faecium Aus0004 mutant (named Δ0160) was constructed by allelic exchange with a truncated copy of the gene corresponding to 5′-3′ positions of ern0160 using pWS3 and specific primers (Supplementary Table 1). The ern0160 gene with its native promoter and rho-independent terminator was cloned into the pAT29 shuttle vector using specific primers (Trieu-Cuot et al., 1990). The recombinant plasmid was introduced into Escherichia coli EC1000 and then into E. faecium Δ0160 (Leenhouts et al., 1996). The trans-complemented strain overexpressing ern0160 was named Δ0160_0160. A strain Δ0160 with an empty pAT29 vector was used as control (Δ0160_pAT29). The transformants were selected on media containing 100 mg/l (E. coli) or 300 mg/l (E. faecium) of spectinomycin.
Bacterial Growth Curves and Competitive Assays
Growth curves were performed in vitro for each strain and WT was compared to Δ0160, while Δ0160_0160 was compared to Δ0160_pAT29. Strains were cultured aerobically overnight on Trypticase Soy (TS) agar (Thermo Fisher Scientific, Waltham, MA, United States) at 35 ± 2°C. A colony was cultured in 10 ml of Brain Heart Infusion (BHI) broth (Thermo Fisher Scientific, Waltham, MA, United States) for 18 h at 35 ± 2°C under ambient air. Bacterial cultures were then adjusted to DO600 0.1, and the bacterial growth was evaluated by DO600 at each time point (2, 3, 4, 5, 6, 9, and 24 h). Two competitive assays were also performed, WT versus Δ0160 and Δ0160_0160 versus Δ0160_pAT29. Strains were initially mixed at a ratio of 1:1, and bacterial growth was measured in the same way as described above. Spectinomycin was added (300 mg/l) to BHI and TS media (BHIspec and TSspec) for all experiments with Δ0160_pAT29 and Δ0160_0160 strains to ensure the maintenance of the plasmid. For competitive assays, a sample of culture was plated on TS and TSSpec agar at each point of time, and each strain was identified by PCR, directly from colonies, under standard conditions and using specific primers (Supplementary Table 1). For each time point, 28 colonies were tested, and each experiment was performed independently three times.
Mouse Model of Gastrointestinal Tract Colonization
The same strains used for in vitro experiments were studied in in vivo protocols. For all the experiments, bacterial suspensions were calibrated as follows to inoculate mice with 108 cfu/ml. Each strain was grown aerobically overnight on TS agar at 35 ± 2°C. A colony was cultured in 10 ml of BHI broth for 18 h at 35 ± 2°C under ambient air. After centrifugation (15 min, 3,500 rpm), the pellet was resuspended in 10 ml of 0.9% saline and centrifuged again. This second pellet was resuspended in 5 ml of 0.9% saline, and the inoculum was quantified by serial dilutions plated on TS agar. For co-colonization experiment, a suspension of each strain was prepared as described above, adjusted to the same OD600, and mixed in a 1:1 ratio before being administered to mice. As previously, BHIspec and TSspec were used for all experiments with Δ0160_pAT29 and Δ0160_0160 strains. To ensure that mice GIT do not contain any E. faecium, an aerobic GIT microbiota study was performed on three cages of five mice. Ten fresh fecal pellets per cage, obtained by light abdominal massage, were suspended in 3 ml of 0.9% NaCl. This suspension was serially diluted and cultured on TS, BHI, and Bile Esculin Azide (BEA) agar plates (Sigma-Aldrich, Saint-Louis, MO, United States) aerobically at 35+/− 2°C for 24 h. The different colonies were then quantified and identified by MALDI-TOF mass spectrometry (Microflex, Bruker Daltonics, Billerica, MA, United States).
Six-week-old non-inbred female, specific pathogen-free, Swiss mice were purchased (Janvier Labs, Le Genest-Saint-Isle, France). Five animals were housed per cage, with controlled room temperature, a 12-h light–dark cycle, and sterile standard rodent food and water ad libitum. Mice were acclimated for 1 week prior to the experiment. The experimental protocol was in keeping with French legislation on animal experimentation and approved by the Adaptive Therapeutics Animal Care and Use Committee (reference number: 2018010814547884-APAFIS#13479). To limit the bias induced by the natural murine coprophagia, mice were placed in a new cage on the day of administration of the bacterial suspension.
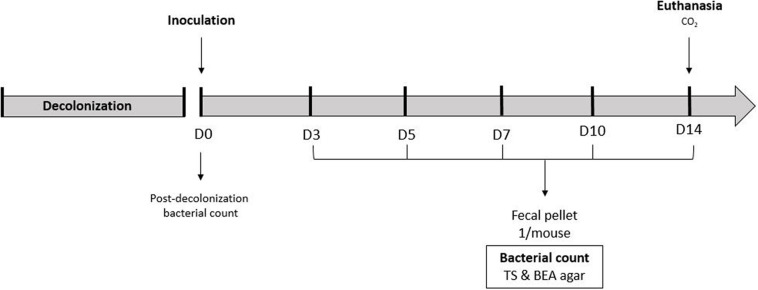
To compare antibiotic regimen efficacy, mice received antibiotics to decolonize GIT according to the different protocols described below. At day 0, mice were orally inoculated, with the bacterial suspension of E. faecium WT. To avoid standard gavage, 2 days before D0, a “water + chocolate spread” suspension was orally administered with a syringe to mice for them to get used to it. On the day of inoculation, chocolate spread was added to the bacterial suspension at the very last moment and orally administered to each mouse. GIT colonization was evaluated by quantifying E. faecium in fresh fecal pellet (one per mouse) obtained by gentle abdominal massage and collected in sterile tubes. Fecal pellets were collected before inoculation (D0) to ensure decolonization, and at days 3, 5, 7, 10, and 14 (Figure 1). Samples were weighted and homogenized in 1 ml of saline solution. Mice were euthanized with CO2 at D14. Fecal pellets were quantitatively cultured onto TS and BEA agar plates for 18 h at 35 ± 2°C under ambient air, to quantify the aerobic flora and E. faecium, respectively. Bacterial loads were expressed in log10 cfu/g of stool or tissue for each sample.
FIGURE 1.
GIT colonization model used to compare antibiotic regimen.
Before the ern0160 study, the in vivo plasmid stability was verified by performing the protocol with Δ0160_pAT29 strain on five mice. Fecal pellets were collected at D3 and D10 and plated on agar with and without spectinomycin. The stability was evaluated by the percentage of colony containing the pAT29 plasmid, calculated by the ratio.
For the ern0160 study, the same protocol was performed with 15 mice per group. Mice received antibiotics before bacterial suspension administration. The suspension contained each strain alone (WT, Δ0160, Δ0160_0160, or Δ0160_pAT29) or a two-strain combination prepared as described above (WT + Δ0160 or Δ0160_0160 + Δ0160_pAT29). A fecal pellet for each animal at D0, D3, D5, D7, and D10 were collected to be quantitatively cultured. Samples collected from mice colonized with WT or Δ0160 strains or the mixed suspension “WT + Δ0160” were cultured on TS and BEA agar. TS agar and TSspec agar were used to quantify bacterial load in samples collected from mice colonized with Δ0160_ 0160 or Δ0160_pAT29 strains and with the suspension “Δ0160_0160 + Δ0160_pAT29.” All agar plates were incubated for 18 h at 35 ± 2°C under ambient air. For co-colonization assay, each strain was identified by PCR, directly from colonies, under standard conditions and using specific primers (Supplementary Table 1). Twelve colonies were tested for each sample.
Antibiotic Protocols
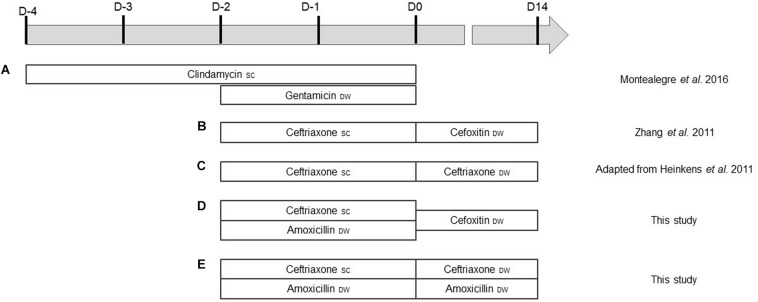
Five decolonization regimens were tested. Each protocol was administered to five mice before the inoculation at D0. Protocol A, described by Montealegre et al. (2016), combined SC clindamycin (Panpharma S.A., Luitré, France) injected every 12 h (2.4 mg/day/mouse) for 4 days and gentamicin (Panpharma) in DW (1 mg/ml) for 2 days before inoculation. Zhang et al. (2011) described protocol B, which combined SC ceftriaxone (Mylan, Canonsburg, PA, United States) and cefoxitin (Panpharma). Ceftriaxone (2.4 mg/day/mouse) was injected twice a day, 2 days before inoculation and cefoxitin (0.2 mg/ml) added to DW during the 14 days of the colonization. Protocol C was adapted from Heikens et al. (2009), who used SC ceftriaxone (2.4 mg/day/mouse) 2 days before inoculation and until the end of the study. In our adapted protocol, SC ceftriaxone was administered (2.4 mg/day/mouse) 2 days before inoculation and then in DW (0.1 mg/ml) until D14. Protocol D was the same as protocol B (ceftriaxone plus cefoxitin), with amoxicillin (Panpharma) (0.1 mg/ml) added in DW during the 2 days before inoculation. Finally, protocol E combined SC ceftriaxone (2.4 mg/day/mouse) and amoxicillin in DW (0.1 mg/mL) 2 days before inoculation; ceftriaxone (0.1 mg/ml) and amoxicillin (0.03 mg/ml) were then put in DW throughout the colonization. All regimens are summarized in Figure 2. One group received no antibiotics as a control group.
FIGURE 2.
(A–E) Antibiotic administration according to the different protocols. D0 corresponds to the day of bacterial suspension inoculation. SC, subcutaneous; DW, drinking water.
Statistical Analysis
For competitive and co-colonization assays, competitive index (CI) and normalized competitive index (nCI) were calculated for each time point as follows. CI was the ratio between “Δ0160 colonies” and “WT colonies,” or between “Δ0160_0160 colonies” and “Δ0160_pAT29 colonies.” nCI was calculated by dividing CI by CI calculated at T0 for in vitro assay, and D0 (suspension inoculated) for in vivo experiments. A normalized competitive index of 1 meant no difference, a normalized competitive index < 1 meant that WT or Δ0160_pAT29 was in higher numbers than Δ0160 or Δ0160_0160, respectively, and a competitive index > 1 meant that Δ0160 or Δ0160_0160 was in higher numbers than WT or Δ0160_pAT29, respectively. A Shapiro–Wilk test (p value >0.05) was used to evaluate normality. According to the result, CIs were analyzed using Student’s t test or Wilcoxon signed-rank test, with the null hypothesis: mean CI was not significantly different from 1 (p values of 0.05 used). Between-group cfu counts for fecal pellets were expressed by mean and standard deviation and compared with the Mann–Whitney test or Student’s t test. Antibiotic protocol results were analyzed using a Kruskal–Wallis test. p < 0.05 was considered statistically significant. Data were analyzed using GraphPad Prism 7 (GraphPad Software, Inc.).
Results
In vitro Experiments
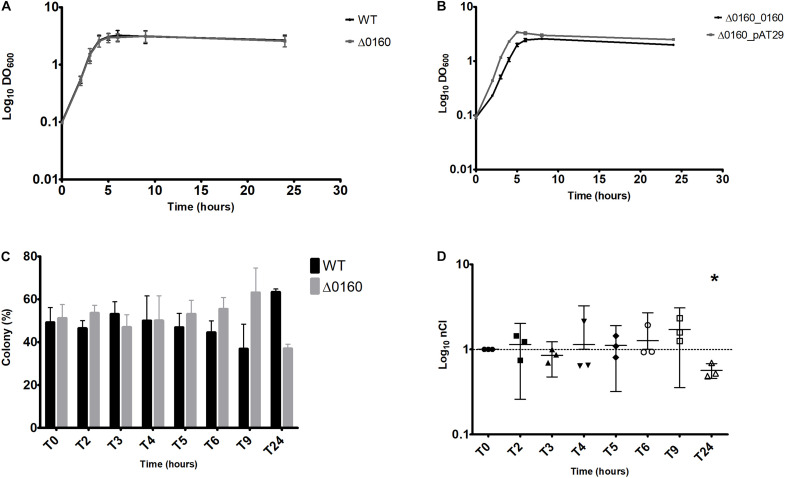
Results for all bacterial growth curves are represented in Figures 3A,B. No significant difference was observed between WT and Δ0160 growth curves. The maximum DO600 was similar and achieved simultaneously. Conversely, Δ0160_0160 and Δ0160_pAT29 showed a different growth rate. A difference of approximately 2 h to reach the growth plateau was observed between the two strains, and Δ0160_0160 grew more slowly than Δ0160_pAT29. Moreover, the maximum DO600 was lower for Δ0160_0160 bacterial growth.
FIGURE 3.
In vitro experiments. (A) Bacterial growth curves of WT and Δ0160 strains growth individually in BHI broth. (B) Bacterial growth curves of Δ0160_0160 and Δ0160_pAT29 strains growth individually in BHI broth supplemented with spectinomycin. Each mean value in panels (A) and (B) represents the mean DO600 generated from three replicates; error bars represent standard error. (C) Competitive assay of WT versus Δ0160 strains. Proportion of colonies of each strain identified by PCR colony. Each mean value represents the mean colony proportion from three replicates (28 PCR at each time point for each replicate); error bars represent standard error. (D) Competitive assay of WT versus Δ0160 strains; normalized competitive index (nCI) values of each replicate represented with means and 95% confidence interval. Asterisk indicates which results have been shown by statistical analysis to be different from 1.
According to these results, only the WT and Δ0160 strains were studied in competitive assays. A competitive assay was conducted during 24 h with PCR on 28 different colonies performed at each point of time. Results are reported in Figures 3C,D. Up to 9 h of culture, the nCI was not significantly different from 1, which means that the two strains were growing in a similar way. At T24, the mean nCI was 0.57 and significantly lower than 1 (p = 0.02), meaning that the WT strain was predominant compared to the deleted strain Δ0160.
In vivo Experiments
Experimental Model Development
Each protocol was tested on five mice and colonized with the E. faecium WT suspension for 14 days. Prior to beginning of our experiments, normal aerobic GIT microbiota of Swiss mice was determined on five mice. It was composed of lactobacilli (85.2%), Enterobacterales (8.5%), and E. faecalis (6.3%). For each experiment, mice were screened for E. faecium before colonization (D0) and no stool samples contained any.
Results are shown in Figure 4. In the control group, which received no antibiotic prior to WT suspension administration, no VREF were isolated from fecal pellets at D3, 5, 7, 10, and 14. With protocol A (gentamicin + clindamycin), fecal pellets contained a mean of 3.34 × 109 cfu/g at D3, 4.24 × 108 cfu/g at D5, and 3.00 × 107 cfu/g at D7. No VREF were found between D10 and D14. Whereas there were no E. faecalis at D0, they were gradually detected from D7 to D14 (3.08 × 104 cfu/g at D14). With protocols B (ceftriaxone + cefoxitin), C (ceftriaxone alone), D (ceftriaxone + cefoxitin + amoxicillin), and E (ceftriaxone + amoxicillin), a high and stable colonization was observed. Between D3 and D14, fecal pellets from mice treated with protocols B and C contained a mean of 8.86 × 109 cfu/g and 5.50 × 109 cfu/g, respectively. Some E. faecalis were detected at the end of colonization, but they remained a minority compared to VREF. The mean VREF bacterial load in fecal pellets from animals receiving protocols D and E antibiotics were 8.89 × 109 and 6.69 × 109 cfu/g, respectively. No contamination by E. faecalis was observed with protocol E. Regarding VREF colonization, no significant difference was observed between protocols B, C, D, and E (p = 0.6).
FIGURE 4.
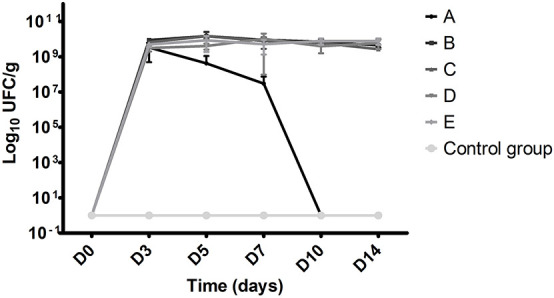
VREF bacterial load in fecal pellets according to antibiotic regimen and time. Each point represents the mean value of cfu values per gram obtained from five mice. A Clindamycin SC + gentamicin DW, B ceftriaxone SC + cefoxitin DW, C ceftriaxone SC + DW, D ceftriaxone SC + amoxicillin DW + cefoxitin DW, and E ceftriaxone + amoxicillin SC and DW (SC, subcutaneous; DW, drinking water).
Ern0160 Study
To study in vivo the role of Ern0160 in GIT colonization, the model with protocol E (ceftriaxone plus amoxicilline) was applied during 10 days, with each strain alone in a first step (WT, Δ0160, Δ0160_0160, or Δ0160_pAT29) and with a combination of two in a second step (WT + Δ0160 and Δ0160_0160 + Δ0160_pAT29). Each group was composed of 15 mice. The in vivo stability was first evaluated by determining the percentage of colony that retained the pAT29 plasmid. At D3, 94% of colonies retained the plasmid and 75% at D10, which is consistent with studies previously published (Deol et al., 2007).
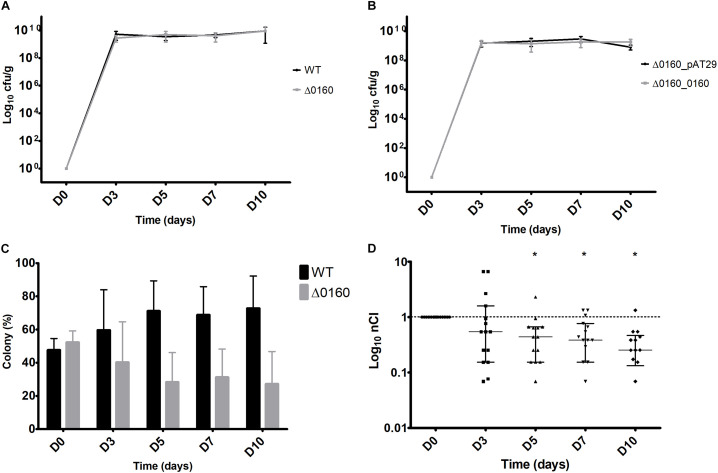
Results obtained with GIT colonization with single strains are shown in Figures 5A,B. At D3, the mean bacterial load was significantly higher with the WT strain compared to Δ0160 (5.2 × 109 cfu/g and 2.8 × 109 cfu/g, respectively, p = 0.008). Then, from D5 to D10, GIT colonization remained high and stable with WT and Δ0160 strains, with a mean bacterial load of 5.78 × 109 and 6.03 × 109 cfu/g, respectively, which was not significantly different (Figure 5A). A high and stable GIT colonization was also obtained from D3 with mutant strains Δ0160_0160 and Δ0160_pAT29. At D3, the mean bacterial load was 1.49 × 109 and 1.61 × 109 cfu/g with Δ0160_0160 and Δ0160_pAT29, respectively, which was not significantly different. From D5 to D10, GIT colonization remained stably high with Δ0160_0160 and Δ0160_pAT29 strains, with a mean bacterial load of 5.93 × 109 and 1.70 × 109 cfu/g, respectively, which was not significantly different (Figure 5B).
FIGURE 5.
GIT colonization with each strain individually and co-colonization with WT and Δ0160 strains mixed. (A) Mean bacterial load per gram in fecal pellets from 15 mice inoculated with the WT strain, and 15 mice with the Δ0160 strain. (B) Mean bacterial load per gram in fecal pellets from mice inoculated with mutant strains Δ0160_0160 and Δ0160_pAT29. Error bars represent standard deviation. (C) Proportion of colonies of each strain identified by PCR colony. Each mean value represents the mean colony proportion from 15 mice (12 PCR at each time point for each sample). (D) Normalized competitive index (nCI) values calculated for each mouse represented with means and 95% confidence interval. Asterisk indicates which results have been shown by statistical analysis to be different from 1.
The co-colonization model was performed with the two combinations of strains. With WT + Δ0160 suspension, the nCI was not significantly different from 1 at D3; the two strains colonized GIT in the same way. From D5 to D10, nCI significantly decreased (p = 0.01 at D5, and p < 0.01 at D7 and D10) with a predominance of the WT strain (Figures 5C,D). With Δ0160_0160 + Δ0160_pAT29 suspension, an nCI under 0.1 was observed from D3 to the end of the protocol for all mice. At D10, all colonies tested belonged to the Δ0160_pAT29 strain and no Δ0160_0160 colony was isolated.
Discussion
In the last decade, bacterial sRNAs have been more and more investigated, and even if functions of many of these riboregulators are still unknown, several of them are involved in stress response or virulence. Few data are available on sRNAs in E. faecium; indeed, they have been identified only recently (Sinel et al., 2017). In their study, Sinel et al. showed that the expression level of one sRNA, Ern0160, was significantly downregulated under SIC daptomycin exposure. More recently, Dejoies et al. (2021) described a significant reduction in ern0160 expression level under SICs of biocides. These two studies suggest that Ern0160 could be involved in antibiotic and biocide stress responses in E. faecium, and for these reasons, it appeared to be an interesting candidate potentially involved in its pathogenicity. GIT colonization is the main starting point of VREF infection and hospital outbreaks. Colonization mechanisms remain unclear, so this study sought to highlight whether Ern0160 could be implicated.
GIT colonization studies are difficult to reproduce in vitro because many parameters are involved, such as pH, cell diversity, or enzymatic activity. Experimental models have been developed to study enterococcus infectious diseases since 1899 (Maccallum and Hastings, 1899). These models enabled researchers to study enterococcal pathogenesis in the context of the innate and adaptive immune responses, as closely as possible to human conditions. Moreover, experimental models are reproducible and reliable statistical analysis on data could be done. Mice are the most frequently used animal to study enterococci in GIT (Goh et al., 2017). In our study, we evaluated for the first time sRNA of E. faecium in a GIT colonization model optimized for this purpose. Several models of GIT colonization have been published, based on the administration of a suspension of enterococci via oral gavage or in the drinking water. Oral gavage is an invasive procedure that can damage the esophagus, but a calibrated quantity of bacteria could be administered. Because mice are housed by five, if the bacterial suspension is administered in the drinking water, the number of bacteria taken up by each mouse is not precisely quantifiable. Because of these disadvantages, an alternative method has been chosen for our study (Scarborough et al., 2020). A “water+chocolate spread” solution was orally administered to mice a few days before starting colonization and then added to bacterial suspension. Thus, a calibrated suspension of bacteria could be administered without invasive procedure and thus avoid the possible associated complications. Prior to inoculation, mice received antibiotics to eliminate GIT normal flora. Several antibiotic regimens have been published and compared in our study. All tested protocols led to an important colonization but had some limitations. With the one associating clindamycin and gentamicin, the VREF colonization did not persist long enough and with protocols composed of β-lactams without amoxicillin, a significant growth of E. faecalis was observed. A high and stable colonization without E. faecalis contamination was obtained with two regimens, ceftriaxone associated with amoxicillin and ceftriaxone associated with amoxicillin and cefoxitin. As cefoxitin is not frequently used in human medicine unlike ceftriaxone, the protocol without cefoxitin was preferred, to be as close as possible to human conditions.
In our in vitro experiments, the deleted mutant strain Δ0160 grew like the wild-type strain. These results have been confirmed in in vivo experiments. Indeed, a similar colonization profile was obtained with the two strains tested individually. Competitive assays have also been performed in vitro and in vivo. In vitro, neither of the two strains grew more than the other up to 24 h of culture. At 24 h, the wild-type strain was slightly predominant. As in vitro experiments were performed in BHI broth, which is far from the composition of the digestive tract, it was important to perform competition assays in mice. In the co-colonization model, the wild-type strain was significantly predominant from day 3 until the end of the protocol. These results suggested that Ern0160 could be involved in the GIT colonization process. To complete the results obtained with the deleted strain, identical in vitro and in vivo experiments were performed with the trans-complemented strain Δ0160_0160, which overexpressed ern0160, knowing that overexpression experiments are classically used to study sRNA functions (Brantl and Jahn, 2015). The strain Δ0160_pAT29, with an empty pAT29 vector, was used as control to compare strains with supposed similar fitness costs. In individual bacterial growth curve assays, the Δ0160_0160 strain appeared to grow more slowly than the Δ0160_pAT29 strain. The overexpression of ern0160 by the strain Δ0160_0160 may induce an energetic cost for the bacterium and could therefore explain the shift observed in growth kinetics. In the colonization model with only a strain, the two strains colonized mice in the same way during all protocol. In the co-colonization model, the Δ0160_pAT29 strain was largely predominant by day 3 and until the end of the protocol. Based on our first results, it would have been expected that the complemented strain Δ0160_0160 would colonize GIT more than the Δ0160_pAT29 strain. As the Δ0160_0160 strain overexpressed ern0160 compared to the WT strain, these results suggested that an overexpression of ern0160 could be deleterious to the strain in GIT colonization. Altogether, these findings show the involvement of Ern0160 in GIT colonization with a probable very tight regulation. Further studies, in particular with a cis-complemented strain, may provide additional information. As Ern0160 has probably pleiotropic effects, it will also be important to determine which targets are regulated by Ern0160 to understand precisely molecular mechanisms. As the different pathways of adaptation or virulence often involve several RNAs, it will also be interesting to determine if other sRNAs are involved in GIT colonization and if there is a cross-regulation with Ern0160. This study was the first to explore in vivo E. faecium regulatory RNAs and its potential function in GIT colonization. Further studies are needed to determine the targets of this sRNA in order to decipher its role and regulatory circuits.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by Adaptive Therapeutics Animal Care and Use Committee.
Author Contributions
SR, BF, VC, and MR conceived and designed the experiments, analyzed the data, and wrote the manuscript. SR, KL, VB, LD, and AL performed the experiments. VB, BF, VC, and MR contributed with reagents, materials, and analysis tools. All the authors discussed the results and commented on the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Biosit Arche platform (Rennes 1 University, France), for animal house facilities.
Funding
This work was supported by the “Fondation pour la Recherche Médicale” (to SR, grant number FDM 20170637920), the “Institut National de la Santé et de la Recherche Médicale,” and Rennes 1 University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.757227/full#supplementary-material
References
- Brantl S., Jahn N. (2015). sRNAs in bacterial type I and type III toxin-antitoxin systems. FEMS Microbiol. Rev. 39 413–427. 10.1093/femsre/fuv003 [DOI] [PubMed] [Google Scholar]
- Cattoir V., Giard J.-C. (2014). Antibiotic resistance in Enterococcus faecium clinical isolates. Expert Rev. Anti Infect. Ther. 12 239–248. 10.1586/14787210.2014.870886 [DOI] [PubMed] [Google Scholar]
- Dejoies L., Le Neindre K., Reissier S., Felden B., Cattoir V. (2021). Distinct expression profiles of regulatory RNAs in the response to biocides in Staphylococcus aureus and Enterococcus faecium. Sci. Rep. 11:6892. 10.1038/s41598-021-86376-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deol P., Zaiss D. M. W., Monaco J. J., Sijts A. J. A. M. (2007). Rates of processing determine the immunogenicity of immunoproteasome-generated epitopes. J. Immunol. 178 7557–7562. 10.4049/jimmunol.178.12.7557 [DOI] [PubMed] [Google Scholar]
- Donskey C. J., Hanrahan J. A., Hutton R. A., Rice L. B. (2000). Effect of parenteral antibiotic administration on the establishment of colonization with vancomycin−resistant Enterococcus faecium in the mouse gastrointestinal tract. J. Infect. Dis. 181 1830–1833. 10.1086/315428 [DOI] [PubMed] [Google Scholar]
- Donskey C. J., Hoyen C. K., Das S. M., Farmer S., Dery M., Bonomo R. A. (2001). Effect of oral Bacillus coagulans administration on the density of vancomycin-resistant enterococci in the stool of colonized mice. Lett. Appl. Microbiol. 33 84–88. 10.1046/j.1472-765X.2001.00948.x [DOI] [PubMed] [Google Scholar]
- Dubin K., Pamer E. G. (2014). Enterococci and their interactions with the intestinal microbiome. Microbiol. Spectr. 5. 10.1128/microbiolspec.BAD-0014-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control (2019). Surveillance of Antimicrobial Resistance in Europe 2018. Stockholm: ECDC, 110. [Google Scholar]
- Eyraud A., Tattevin P., Chabelskaya S., Felden B. (2014). A small RNA controls a protein regulator involved in antibiotic resistance in Staphylococcus aureus. Nucleic Acids Res. 42 4892–4905. 10.1093/nar/gku149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh H. M. S., Yong M. H. A., Chong K. K. L., Kline K. A. (2017). Model systems for the study of enterococcal colonization and infection. Virulence 8 1525–1562. 10.1080/21505594.2017.1279766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikens E., Leendertse M., Wijnands L. M., van Luit-Asbroek M., Bonten M. J., Poll T., et al. (2009). Enterococcal surface protein Esp is not essential for cell adhesion and intestinal colonization of Enterococcus faecium in mice. BMC Microbiol. 9:19. 10.1186/1471-2180-9-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G., Raina S. (2017). Small regulatory bacterial RNAs regulating the envelope stress response. Biochem. Soc. Trans. 45 417–425. 10.1042/BST20160367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam M. M. C., Seemann T., Bulach D. M., Gladman S. L., Chen H., Haring V., et al. (2012). Comparative analysis of the first complete Enterococcus faecium genome. J. Bacteriol. 194 2334–2341. 10.1128/JB.00259-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebreton F., van Schaik W., McGuire A. M., Godfrey P., Griggs A., Mazumdar V., et al. (2013). Emergence of epidemic multidrug-resistant Enterococcus faecium from animal and commensal strains. mBio 4 e00534–13. 10.1128/mBio.00534-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts K., Buist G., Bolhuis A., Ten Berge A., Kiel J., Mierau I., et al. (1996). A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253 217–224. 10.1007/s004380050315 [DOI] [PubMed] [Google Scholar]
- Maccallum W. G., Hastings T. W. (1899). A case of acute endocarditis caused by Micrococcus zymogenes (nov. spec.), with a description of the microorganism. J. Exp. Med. 4 521–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaux C., Hartke A., Martini C., Reiss S., Albrecht D., Budin-Verneuil A., et al. (2014). Involvement of Enterococcus faecalis small RNAs in stress response and virulence. Infect. Immun. 82 3599–3611. 10.1128/IAI.01900-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montealegre M. C., Singh K. V., Murray B. E. (2016). Gastrointestinal tract colonization dynamics by different Enterococcus faecium clades. J. Infect. Dis. 213 1914–1922. 10.1093/infdis/jiv597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mraheil M. A., Billion A., Kuenne C., Pischimarov J., Kreikemeyer B., Engelmann S., et al. (2010). Comparative genome-wide analysis of small RNAs of major Gram-positive pathogens: from identification to application. Microb. Biotechnol. 3 658–676. 10.1111/j.1751-7915.2010.00171.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Driscoll T., Crank C. W. (2015). Vancomycin-resistant enterococcal infections: epidemiology, clinical manifestations, and optimal management. Infect. Drug Resist. 8 217–230. 10.2147/IDR.S54125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarborough J., Mueller F., Arban R., Dorner-Ciossek C., Weber-Stadlbauer U., Rosenbrock H., et al. (2020). Preclinical validation of the micropipette-guided drug administration (MDA) method in the maternal immune activation model of neurodevelopmental disorders. Brain Behav. Immun. 88 461–470. 10.1016/j.bbi.2020.04.015 [DOI] [PubMed] [Google Scholar]
- Sinel C., Augagneur Y., Sassi M., Bronsard J., Cacaci M., Guérin F., et al. (2017). Small RNAs in vancomycin-resistant Enterococcus faecium involved in daptomycin response and resistance. Sci. Rep. 7:11067. 10.1038/s41598-017-11265-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Carlier C., Poyart-Salmeron C., Courvalin P. (1990). A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 18:4296. 10.1093/nar/18.14.4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C., Taur Y., Jenq R. R., Equinda M. J., Son T., Samstein M., et al. (2010). Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120 4332–4341. 10.1172/JCI43918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E. G. H., Romby P. (2015). “Chapter three–small RNAs in bacteria and archaea: who they are, what they do, and how they do it,” in Advances in Genetics, eds Friedmann T., Dunlap J. C., Goodwin S. F. (Cambridge, MA: Academic Press; ), 133–208. 10.1016/bs.adgen.2015.05.001 [DOI] [PubMed] [Google Scholar]
- Weiner-Lastinger L. M., Abner S., Edwards J. R., Kallen A. J., Karlsson M., Magill S. S., et al. (2020). Antimicrobial-resistant pathogens associated with adult healthcare-associated infections: summary of data reported to the National Healthcare Safety Network, 2015–2017. Infect. Control Hosp. Epidemiol. 41 1–18. 10.1017/ice.2019.296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Top J., de Been M., Bierschenk D., Rogers M., Leendertse M., et al. (2013). Identification of a genetic determinant in clinical Enterococcus faecium strains that contributes to intestinal colonization during antibiotic treatment. J. Infect. Dis. 207 1780–1786. 10.1093/infdis/jit076 [DOI] [PubMed] [Google Scholar]
- Zhang X., Vrijenhoek J. E. P., Bonten M. J. M., Willems R. J. L., van Schaik W. (2011). A genetic element present on megaplasmids allows Enterococcus faecium to use raffinose as carbon source. Environ. Microbiol. 13 518–528. 10.1111/j.1462-2920.2010.02355.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.