Abstract
The capacity of respiratory viruses to undergo evolution within the respiratory tract raises the possibility of evolution under the selective pressure of the host environment or drug treatment. Long-term infections in immunocompromised hosts are potential drivers of viral evolution and development of infectious variants. We showed that intrahost evolution in chronic human parainfluenza virus 3 (HPIV3) infection in immunocompromised individuals elicited mutations that favored viral entry and persistence, suggesting that similar processes may operate across enveloped respiratory viruses. We profiled longitudinal HPIV3 infections from 2 immunocompromised individuals that persisted for 278 and 98 days. Mutations accrued in the HPIV3 attachment protein hemagglutinin-neuraminidase (HN), including the first in vivo mutation in HN’s receptor binding site responsible for activating the viral fusion process. Fixation of this mutation was associated with exposure to a drug that cleaves host-cell sialic acid moieties. Longitudinal adaptation of HN was associated with features that promote viral entry and persistence in cells, including greater avidity for sialic acid and more active fusion activity in vitro, but not with antibody escape. Long-term infection thus led to mutations promoting viral persistence, suggesting that host-directed therapeutics may support the evolution of viruses that alter their biophysical characteristics to persist in the face of these agents in vivo.
Keywords: Virology
Keywords: Influenza
Introduction
Human parainfluenza virus 3 (HPIV3) accounts for a large proportion of respiratory illness in humans, especially in children, the elderly, and immunocompromised individuals (1–9), yet there is currently no effective vaccine or antiviral treatment (10–12). Long-term HPIV3 infection, especially in immunocompromised patients, is a potentially important driver of viral evolution and variant emergence, both of which are major barriers to the development of therapeutics. HPIV3 virions bear 2 surface glycoproteins that cooperate to mediate viral entry into host cells: the receptor-binding protein hemagglutinin-neuraminidase (HN) and the fusion (F) protein. After HN is bound to sialic acid–containing receptors, it activates the trimeric F protein, inducing a conformational change in F that allows for insertion into the host cell membrane (13–19). The F protein then undergoes structural rearrangements that drive viral-host membrane fusion and viral entry into the target cell (13, 20–22). HN also stabilizes the F protein before the receptor is engaged to prevent viral inactivation (23). After entry and replication, HN promotes viral egress by cleaving sialic acid residues on the host cell membrane to allow the release of new viral particles (24). The individual functions of the HN/F protein complex must maintain a balance to allow for coordinated execution of these steps. This array of critical functions in the viral life cycle places the HN and F glycoproteins under significant evolutionary pressure to maintain infectivity in a given host (25–28).
Changes in the fusion machinery are likely key for viral adaptation to different environments. The propensity for HPIV3’s negative-sense, single-stranded RNA genome to rapidly mutate in the face of a viral polymerase that lacks a repair function allows adaptation to environments both in vivo and in vitro (25, 26). Growing HPIV3 in cell culture alters the dynamics of HN and F and the viral characteristics that confer viral fitness (25–29). Upon rapid adaptation to even a brief period of cell culture, HN preferentially accumulates point mutations in amino acids present in the dimer interface and across the globular domain (25), particularly at residue H552, which we have shown to be a secondary sialic acid receptor binding site and a key regulator of HN-F interaction and fusion promotion. The primary sialic acid binding site on HN, T193, also undergoes rapid adaptation in culture; the mutation T193A confers increased avidity for receptors (14, 28). Viruses bearing these mutations in HN are more fusogenic because of HN’s greater avidity to host cell receptors and F-triggering activity. Although these mutations enhance fitness in cell culture, the culture-adapted strains are no longer fit in vivo. Clinical isolates cannot be studied in a standard laboratory setting without inducing mutations through selective pressure. We have shown, using human airway epithelial (HAE) models, that clinical strains of HPIV3 grow and spread without the selective pressure that is present in monolayer culture. These models permit the study of viral growth and evolution in the human lung in the absence of a full adaptive immune response (19, 25–30). HPIV3 evolution is thus highly specific to the environment; very different variants of HN and F emerge in different experimental contexts.
The present report examines HPIV3 infection during persistent infection in an immunocompromised human host, permitting continuous monitoring of viral evolution in the living human lung. Limited but intriguing evidence has supported the concept of long-term HPIV3 replication in the lungs of healthy humans. Protracted viral shedding occurred in otherwise healthy young adults during an 8-month period of complete social isolation at the South Pole (31). The shedding of HPIV3 in these individuals several months after resolution of acute illness, in the absence of any social contact, provided evidence for the possibility of a slow, persistent infection in humans. HPIV3 illness in an immunocompromised patient is often more prolonged and severe than in an immunocompetent patient, with delayed viral clearance (2, 32). Such hosts offer a less hostile environment for viral growth, in which HPIV3 persists over an extended length of time without undergoing significant stress from the adaptive immune system that would necessitate immune evasion. We employed this unique in vivo environment to identify viral characteristics that influence propagation of HPIV3 in the human host. Samples were collected from 2 immunocompromised patients (undergoing cord blood or stem cell transplantation) infected with HPIV3 for more than 90 days, including 1 patient who consistently shed virus over a period of 9 months. Multiple samples were collected across the course of infection in each patient, and viral sequences were analyzed using metagenomic next-generation sequencing (mNGS) to assess genome-wide mutational changes in an extremely sensitive manner.
Strikingly, viral variants in the immunocompromised patients evolved to closely resemble virus grown in cell culture. Previous comparisons of viruses isolated directly from humans and those grown in cell culture revealed an inverse relationship between viral stability and efficiency in fusion and propagation (25–27). HPIV3 adapted to monolayer culture displays increased receptor avidity and fusion promotion properties of HN, along with a readily activated F protein. HPIV3 in a human host, however, displays the opposite: F is stable and resistant to activation, and the fusion complex is less receptor-avid and less fusogenic, suggesting that a more stable, less readily activated complex is suited to the host environment. In the immune-compromised patients studied here, the viruses seemed to be advantaged by properties that resembled elements of those optimal in monolayer culture, and variants emerged that were closer to those fit for growth in vitro. During infection in immunocompromised patients, which we argue provides a unique window into viral evolution during persistent infections, point mutations accumulated in the very domains of HN that alter the fusion complex’s function, affecting both fusion and cleavage activity, akin to the alterations seen during adaptation to cell culture. The variants were not driven by antibody escape, having no advantage in this respect, but rather by fusion properties. Although the particular conditions of this experiment of nature are unique, the principle of within-host evolution of respiratory viruses is likely general.
Results
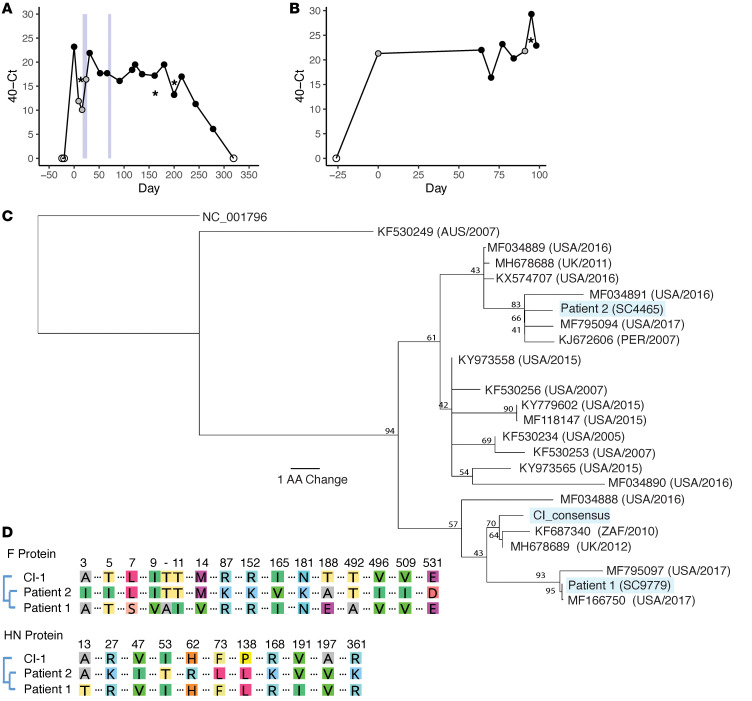
To examine the mutational changes in HPIV3 associated with a long-term persistent infection, 2 patients with HPIV3 infections of longer than 90 days duration in 2015 to 2017 were sampled throughout the course of their infection (Figure 1, A and B). Patient 1 (SC9779) was a woman in her 40s who had a double cord blood transplant to treat acute lymphoblastic leukemia that was complicated by chronic HPIV3 infection; graft versus host disease treated with mycophenolic acid, sirolimus, and extracorporeal photophoresis; and hypogammaglobulinemia, which was treated with multiple doses of i.v. Ig. Patient 1 was sampled 17 times over 278 days, with all samples being nasal swabs except for 3 bronchoalveolar lavage (BAL) specimens on days 16, 163, and 202 (see Table 1 for sample details). Patient 2 (SC4465) was a woman in her 20s who had a haploidentical peripheral blood stem cell transplant to treat acute myelogenous leukemia. Patient 2 was infected with HPIV3 for over 98 days before death; 7 specimens over 34 days (days 64–98), including a BAL specimen from day 96, were available for sequencing. Of note, patient 1 twice received the inhaled HPIV3-directed therapy (DAS181/Fludase) on days 17 to 26 and 68 to 74 (33, 34). DAS181 is an inhaled recombinant neuraminidase molecule that functions by cleaving sialic acids from the host cell surface in the lung, thereby inactivating the host cell receptor recognized by HPIV (33, 35) and preventing viral entry. The use of this compound in this individual is of interest in light of the subsequent course of infection. Both individuals were lymphopenic throughout the course of their infection (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI150506DS1). High viral loads were seen in each patient, with an average Ct across the longitudinal samples of 24.1 and 17.6, respectively.
Figure 1. Longitudinal sampling of long-term HPIV3 infections in vivo.
Sampling time series data with associated Ct values shown for patient 1 (A) and patient 2 (B). Samples collected by nasal swabs are represented by dots; black dots are HPIV3-positive samples with associated sequencing data, gray dots are HPIV3-positive samples that were not available for sequencing, and empty dots tested negative for HPIV3. HPIV3-positive bronchoalveolar lavage (BAL) samples are represented by asterisks. Light blue boxes indicate time periods in which patient 1 was treated with DAS181. Amino acid sequences of the HN protein for patient 1 and patient 2 are placed in the context of circulating strains that were downloaded from NCBI’s GenBank (C). All sequences are labeled with GenBank accession number followed by collection location and collection date. Consensus support values are shown next to branch points. The laboratory-adapted HPIV3 reference strain (NC_001796) is used as an outgroup. Amino acid alignment of the day 0 consensus sequences of the HN and F attachment proteins for each patient were aligned with the clinical isolate used in functional assays (D). Only amino acids that differ among these isolates are depicted.
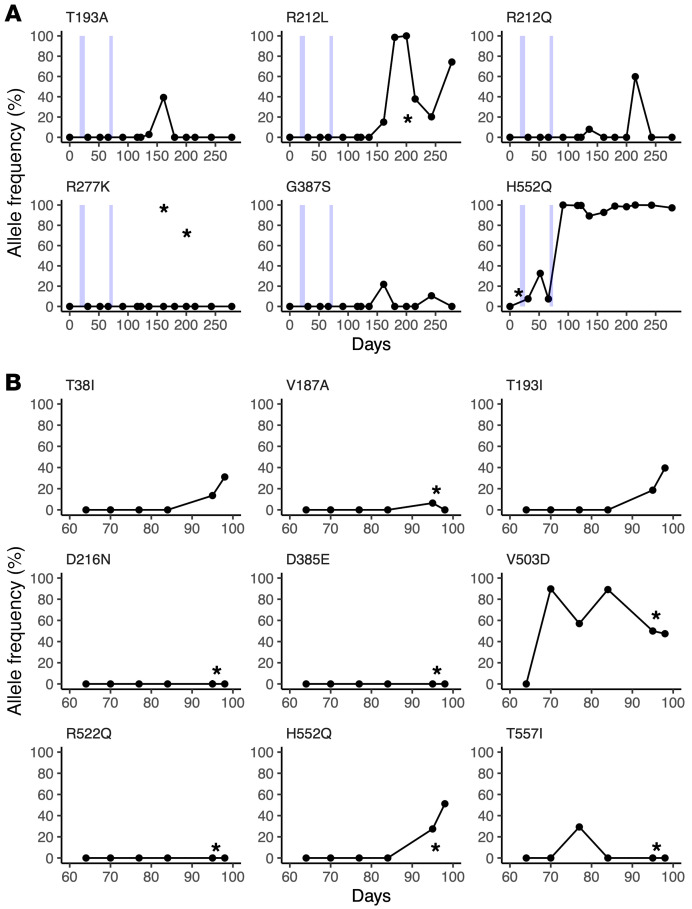
Table 1. Specimens used in this study.
Samples were prepared using a combination of metagenomic mNGS or hybridization capture sequencing. Phylogenetic analysis of the consensus HN protein sequences demonstrated that these 2 samples were representative of the circulating strains of HPIV3 from Seattle in 2015 to 2017 (Figure 1C). To examine the amino acid sequence difference between day 0 majority consensus samples and CI-1, our standard clinically isolated reference strain (27), we performed protein alignments using MAFFT (multiple alignment using fast Fourier transform) on both the HN and F protein and listed all protein coding changes across both proteins. Pairwise comparison of the attachment proteins of these 2 HPIV3 strains and CI-1 showed they differed at 11 total sites in the HN protein and 16 total sites in the F protein, of which 7 occurred in the amino terminal signal sequence (Figure 1D). We note that none of the changes between the CI-1 sample and the day 0 majority consensus sequences resided in the HN region 552 to 557, a binding/fusion promotion site on HN that is critical for viral fusion activity. Interestingly, the analysis also revealed a significant set of mutations in the M gene, which included changes in residues that are completely conserved in HPIV3 or in the entire genus (Supplemental Figure 2).
Longitudinal within-host evolution in HPIV3 localizes to the HN/F fusion complex and N-terminus of the matrix protein.
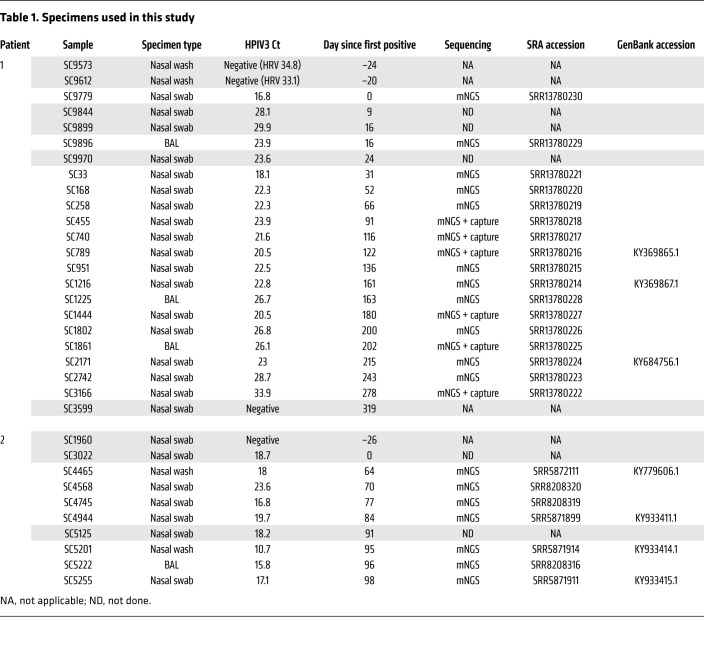
In order to assess localization and frequency of mutational changes associated with long-term persistent infection across the whole HPIV3 genome, we compared localization and maximum minor allele frequencies (>5%) for all mutational changes found across the whole genome over the course of the infection for each patient (Figure 2). Of the 38 amino acid replacements recovered longitudinally in patient 1 that occurred in at least 2 specimens, 9 were in the matrix protein (M), 11 in the F protein, and 6 in the HN protein. Of the 11 total amino acid replacement mutations that occurred during the 34 infection days profiled in patient 2, 6 were in the HN protein and 1 in the F protein.
Figure 2. Maximum allele frequency plots for nonsynonymous mutational changes across the whole genome during persistent HPIV3 infection.
Maximum allele frequency noted for nonsynonymous mutational changes across whole genome during persistent HPIV3 infection for patient 1 (A) and patient 2 (B). All changes are relative to the day 0 majority consensus for each patient. Mutational changes were determined using LAVA. Plots show nonsynonymous mutations present in at least 2 longitudinal time points that had minor allele frequency more than 5% and depth more than 10 reads and passed manual review for tagmentation artifacts. Nucleotide position is listed across the x axis and maximum allele frequency observed is plotted on the y axis. Alternating green and blue rectangles denote protein coding regions. Mutational changes observed in the HN protein and those above 50% maximum allele frequency are labeled with consensus amino acid, amino acid position relative to protein start, and amino acid change.
The mutations that affect the fusion complex during entry were pursued for study here; the M protein evolution, with parallels to other persistent paramyxovirus infections, will be explored in the future.
Nonsynonymous HN mutation allele frequencies over time.
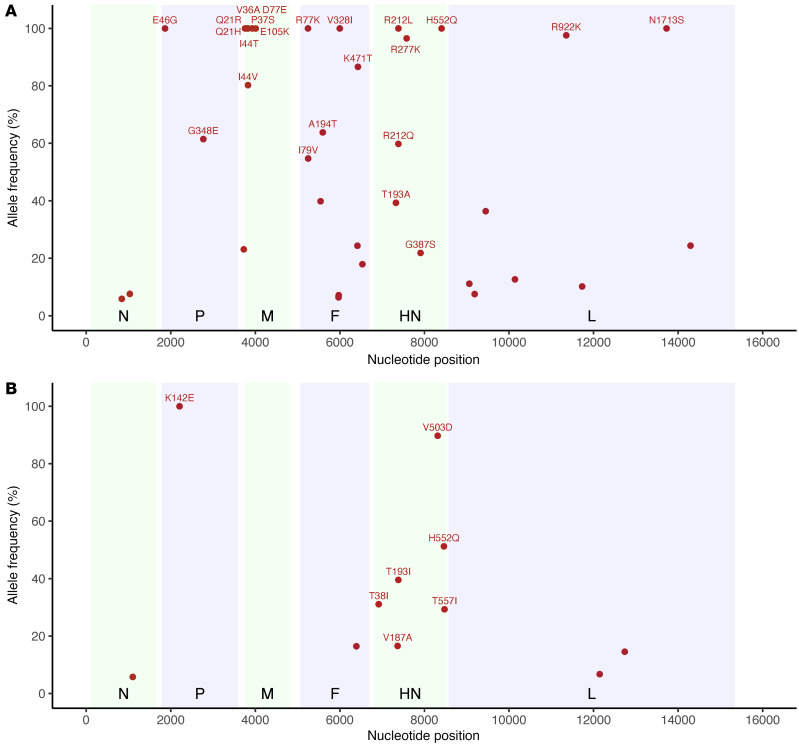
To further examine the mutational changes to the HN proteins over the course of the infection, we compared the allele frequency of nonsynonymous mutational changes in the HN protein over time (Figure 3). Intriguingly, both patients had an increase in allele frequency of the specific mutation at HN residue 552 (H552Q) that was routinely selected for during passage of HPIV3 in culture (14, 25, 29, 36). This mutation was also noted in nasal swabs and BAL samples from the same patient, albeit at significantly reduced allele frequencies in the BAL samples compared with contemporaneous nasal swab samples. Selection for H552Q was temporally associated with both DAS181 treatments in patient 1, reaching clonality that persisted over the remaining 6 months of infection. In patient 2, H552Q increased over the latter course of infection in the absence of DAS181 treatment and never reached fixation before the patient died. BAL samples from patient 1 also specifically yielded high allele frequencies of the R277K mutation in HN protein compared with contemporaneous nasal swab samples. These results suggest compartmental evolution between nasal swabs and BAL, with cell culture adaptation-like evolution of HPIV3 in BAL lagging that of virus in nasal swab samples.
Figure 3. Nonsynonymous mutational changes and allele frequencies in HN protein across long-term persistent HPIV3 infection.
Patient 1 (A); patient 2 (B). The mutational change is shown above each subplot, with all changes relative to the day 0 majority consensus for that patient. Variants above an allele frequency of 5% and a depth of 10 reads in at least 2 samples for patient 1 and 1 sample for patient 2 are depicted, given the increased number of samples. Light blue boxes indicate time periods in which patient 1 was treated with DAS181. Sample collection dates are given relative to the first sample and plotted on the x axis for each plot. Samples collected by nasal swab are represented by dots, and BAL samples are marked with an asterisk.
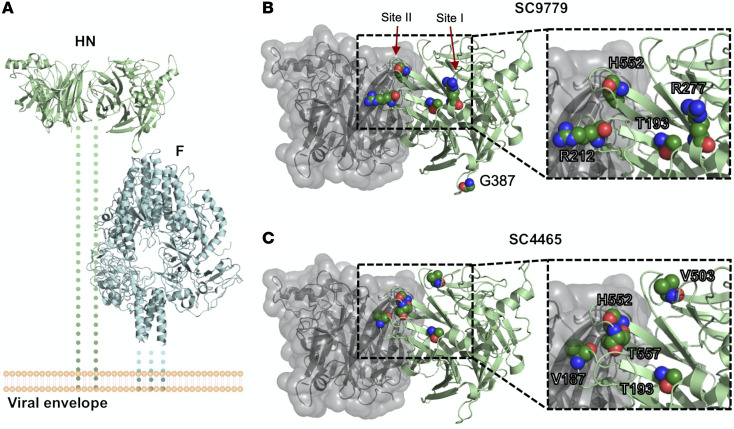
A potentially novel HN R212Q mutation appeared in patient 1, including rapid switching between R212L and R212Q. These mutations never appeared at a high level in BAL samples. We also noted that G387S appeared at a low level during the course of infection for patient 1, although never persisting. We also note the appearance of the previously described culture-adapted T557I, T193I, and T193A mutations in these individuals (14, 16, 25, 37). Figure 4 shows the locations on the HN structure of significant residues with variant changes of greater than 25% during long-term infection. The alterations were in the primary active site of HN (residue 193) and secondary binding/fusion activation site (residue 552) described above as residues that rapidly adapt to new growth environments, as well as in other regions we have previously shown to be critical for HN’s role in fusion, residues that are also under selective pressure during adaptation to culture (14, 19, 24, 28, 29, 37).
Figure 4. Location of mutated residues in HN that arose during long-term infection.
(A) Overview of HN (PDBID:4MZA) and F (PDBID:6MJZ) on the viral surface. Residues that showed variant changes of greater than 25% in the HN protein during long-term infection for (B) patient 1 and (C) patient 2 are labeled. The backbone is colored in green, and the side chains are labeled by standard atom coloring. Residues are labeled with the original reference amino acid and not the changes observed. Note the localization of minor variants to 2 of the known active sites of HPIV3 HN protein.
Patient-derived HPIV3 HN proteins have altered receptor-binding and receptor-cleaving properties.
The mutations that emerged in the patient-derived HN genes were in locations key for receptor binding and neuraminidase (receptor cleavage) activity. We assessed the receptor avidity and neuraminidase activity of these variants and quantified the release of RBCs from HN-expressing cells as a measure of the balance between avidity and receptor-cleaving properties. These properties are interrelated in their effect on virus-induced membrane fusion; enhanced avidity permits more fusion to occur during prolonged contact with the target cell, while enhanced neuraminidase activity releases virus from the target cell and leaves less opportunity for fusion.
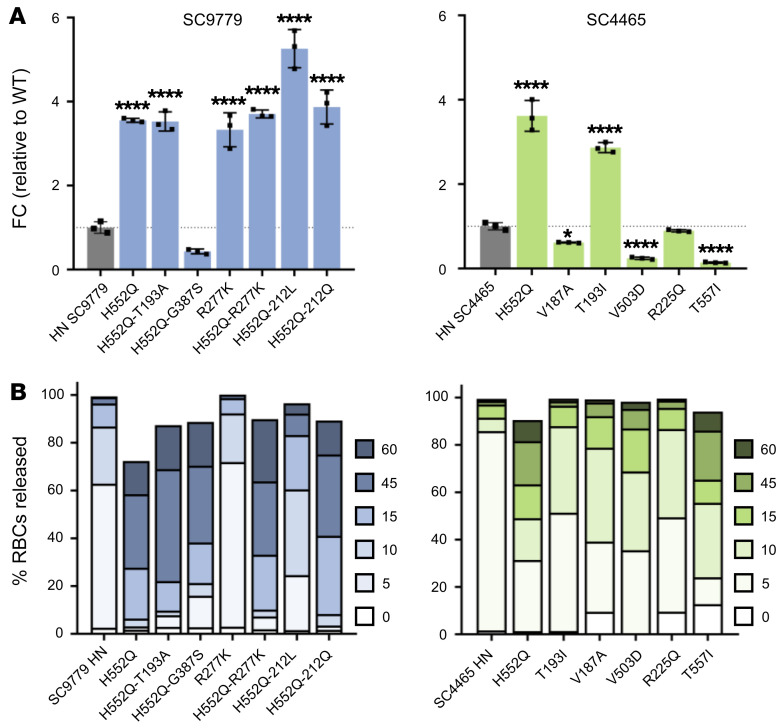
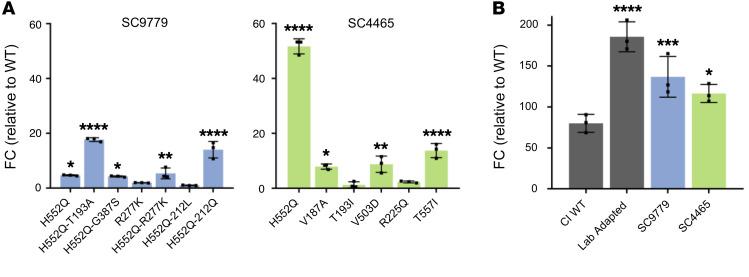
Neuraminidase activity was measured through the cleavage of 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid, sodium salt hydrate (4-MUNANA) by HN (Figure 5A). The majority of the patient-derived HNs displayed higher neuraminidase activity than WT clinical isolates, with the HNs bearing mutations in the HN dimer interface (in the background of the starting sequence of their parental patient strain) having up to 7-fold higher activity. Interestingly, the effect of individual mutations at important sites (e.g., H552Q) on neuraminidase activity was consistent between patients, and additional mutations in HN combined with H552Q led to even higher neuraminidase activity; e.g., H552Q/212L in patient 1.
Figure 5. Patient-derived HPIV3 HN proteins have altered receptor-binding and receptor-cleaving properties.
(A) Neuraminidase activity of patient-derived HPIV3 HNs. HEK293T cells were transfected with plasmids containing corresponding sequences of clinically isolated HNs. Neuraminidase activity was quantified by measuring amount of cleavage of 4-MUNANA. Results depict representative experiments from 3 biological replicates. Data indicate the mean ± SD. A 1-way ANOVA was performed to determine significance; P values are specified as follows: *P ≤ 0.05; ****P ≤ 0.0001. FC, fold change. (B) Release kinetics of patient-derived HPIV3 HNs binding sialic acid–containing RBCs at 37°C. HEK293T cells transiently expressing HNs were incubated with RBCs at 4°C for 30 minutes, washed, and transferred to 37°C. Supernatant was collected at 0, 5, 10, 15, 45, and 60 minutes and percentage of RBCs released at each time point was determined by quantification of relative absorbance at 410 nm. Results depict representative experiments from 3 biological replicates.
The HNs from both patients evolved mutations at residue T193, the primary receptor binding site of HN (see Figure 4). In patient 1, the change at T193 was to A and increased receptor avidity in this patient isolate as it did in culture (14), where this mutation arose under the selective pressure of neuraminidase treatment that decreased receptor availability (14, 36). For patient 2, the change at T193 was to I. The T193I mutation had emerged during infection under the selective pressure of treatment with zanamivir, a small molecule that is in clinical use as an influenza neuraminidase inhibitor. For HPIV3, zanamivir inhibits neuraminidase and blocks HN-receptor engagement (37, 38). The T193I HN variant was highly resistant to zanamivir.
For measurement of HN receptor binding avidity, cells transiently expressing each of the patient-derived HN variants (and WT HN clinical isolates) were pretreated with neuraminidase to partially deplete receptors on the expressing cells’ surfaces. Receptor-bearing cells, in this case human erythrocytes (RBCs), with different degrees of receptor depletion were added and used to quantify HN receptor binding (17, 37). HN molecules with higher avidity bind RBCs that have lower receptor density, and the maximal level of RBC receptor depletion that permits binding provides a measure for avidity, which we express as units of neuraminidase for 50% binding, in milliunits, mU. Higher amounts of neuraminidase treatment that permit 50% binding reflect higher relative avidity. Compared with the starting avidity measurements of each isolate’s HN from the first samples taken, the mutations that arose generally increased avidity (with one minor exception, discussed later) — in most cases the increase was by several fold (Table 2). For each of the original isolates, the H552Q mutation in the secondary binding/fusion triggering site itself increased avidity; for example, for the SC4465 HN, the avidity corresponding to 50% of binding to RBCs treated with 25 mU of neuraminidase increased to 50% of binding to RBCs treated with 151 mU with this mutation. For the SC9779 (patient 1) HN, avidity increased from 50% of binding to RBCs treated with 54 mU neuraminidase to 50% binding after 205 mU neuraminidase, and even more to 50% binding after 278 mU neuraminidase in the presence of a mutation in the primary binding site of HN (T193A) in addition to H552Q. Specific alterations at HN residue 212, (adjacent to the primary binding/active site, see Figure 4) may have a tempering effect on avidity (H552Q/R212L in SC9779). However, a Q at the same site (H552Q/R212Q in SC9779) did not have the tempering effect, of note with respect to the rapid switching between R212L and R212Q in SC9779 (Figure 3).
Table 2. Relative avidity of patient-derived HPIV3 HNs binding sialic acid–containing RBCs at 4°C.
Release of bound receptor-bearing RBCs by cell-surface HNs reflects the balance of neuraminidase activity, which would cleave the bonds that tether virus to host cells, and avidity of the HNs, which mediate binding to the host cells. We quantified the amount of RBCs released over time at several time points (0, 5, 10, 15, 45, and 60 minutes) when incubated at pH 7.5 and 37°C (Figure 5B). Differences between the HNs can be readily observed by comparing the fraction of RBCs that was either released at early time points (light color) or retained until later time points (black). We observed 2 distinct groups of mutants in their ability to release RBCs. The mutants that retained the RBCs far longer than the WT clinical isolate all contained mutations in the HN dimer interface, specifically at H552 or T557, alone or in combination with other HN dimer interface/receptor binding site II mutations (e.g., H552Q/R212Q) or in combination with a mutation located in the HN primary binding site (R277K; see Figure 4). These experiments suggest that the higher receptor avidity of HN bearing H552Q can override an increase in neuraminidase activity and allow this HN to remain in prolonged contact with target cell receptors.
We addressed the possibility that the mutations in the fusion complex are escape variants arising as a result of immune pressure, and we showed that these mutations had no discernable impact on the neutralizing effect of polyclonal anti-HPIV3 serum from a healthy donor compared with neutralization of standard clinical strains (Supplemental Figure 3A). To further evaluate the potential role of selection pressure in these mutations, the neutralizing titers of the serum antibodies from these patients were evaluated; the patient sera and healthy donor sera similarly neutralized standard HPIV3 clinically isolated virus (Supplemental Figure 3B), the virus bearing the fusion complex isolated from patient 1 SC 9779 (Supplemental Figure 3C), and the virus bearing the fusion complex isolated from patient 1 SC 9779 with HN H55Q-T193A (Supplemental Figure 3D), suggesting absence of antibody-mediated selection pressure.
Patient-derived fusion complexes have enhanced fusion promotion derived from properties of both HN and F.
To assess how the emerging mutations that affected neuraminidase, avidity, or both interacted to affect HN’s key function of promoting fusion by triggering F protein, we assessed the ability of these HNs to promote fusion mediated by a lab-adapted HPIV3 F protein in a quantitative β-galactosidase complementation fusion assay (Figure 6). Cultured cells were cotransfected with HN bearing the mutation of interest on the background of the unpassaged (day 0) virus from the indicated patient and the α subunit of β-galactosidase, and then paired with a lab-adapted F to ensure that we could observe adequate levels of fusion to compare function since clinical isolate HN and F pairs result in nearly unobservable levels of fusion (25, 28). These cells were incubated with cells expressing the Ω subunit of β-galactosidase, and the resulting fusion was quantitated. In both patient-derived HN proteins, the H552Q mutation led to an increase in fusion promotion, alone or paired with R212Q, or T193A (for SC9779). For patient 1, R212L reversed the effect of H552Q, resulting in an HN similar in fusion promotion to the original (day 0) patient’s strain.
Figure 6. Fusion activity of clinically isolated HPIV3 HN and F proteins.
Fusion activity was assessed using a β-galactosidase complementation assay. HEK293T cells were cotransfected with either (A) HN sequence of the indicated patients, lab-adapted HPIV3 F, and the α subunit of β-galactosidase, or (B) F sequence of the indicated patients, lab-adapted HPIV3 HN, and the α subunit of β-galactosidase. Cells were then incubated for 6 hours with HEK293T cells expressing the Ω subunit of β-galactosidase. Fusion is depicted by luminescence relative to WT patient HN (A) or relative to mock-transfected cells (B) containing only lab-adapted HPIV3 HN and the Ω subunit of β-galactosidase. Results depict representative experiments from 3 biological replicates. Data indicate the mean ± SD. A 2-way ANOVA was performed to determine significance; P values are specified as follows: *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001.
To assess the contribution of the patient-derived strain F proteins to the overall function of the fusion complex, cultured cells were cotransfected with the F from each patient and the α subunit of β-galactosidase, and then paired with a cell culture–adapted HN, again to ensure that we could observe adequate levels of fusion to compare function (refs. 25 and 28; Figure 6B). The F from each patient was compared with the F from CI-1, our standard clinically isolated reference strain mentioned above (28), as well as to the F from our standard cell culture–adapted HPIV3 strain, relative to a mock F protein transfection. The F proteins derived from virus infecting each of the long-term patients was more fusogenic than the F from our standard clinical isolates, although not to the level of the laboratory-adapted strain. For example, the SC9779 F-mediated fusion is intermediate between the clinical isolate strain F and the laboratory-adapted F.
A number of the mutations emerging in the 2 patients that were detailed in Figures 2 and 3 have fusion complex functions similar to CI-1 and to their parent isolate (Figures 5 and 6) and were not explored further, in an effort to focus on the study of the most informative commonalities between both patients. The mutation at H552, alone or accompanied by mutation at T193, seemed to play a key role in the persistence of these isolates, and this was especially remarkable in light of the history of these mutations of emerging under selective pressure in tissue culture (14, 17–19, 25, 26, 28, 29, 36, 39).
Growth of recombinant HPIV3 bearing a patient-derived HN/F complex in HAE.
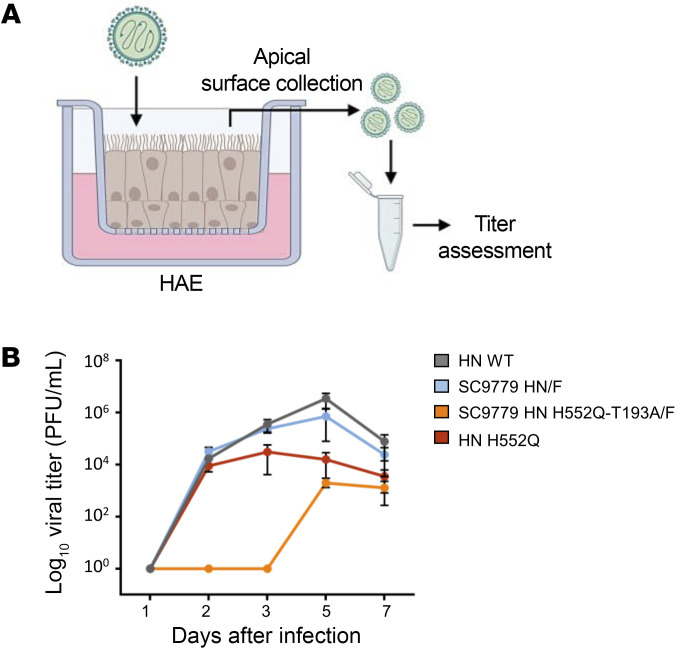
To assess the relevance and evolutionary consequences of the mutations in patient-derived fusion complexes and mutations that emerged during long-term infection in the context of authentic virus and determine the consequences of the evolution to infection in HAE, we generated recombinant viruses bearing the SC9779 patient fusion complex (original and H552Q/T193A) in the genetic background of our well-characterized HPIV-3 clinically isolated virus Cl-1 (25, 27, 28), along with an enhanced GFP (EGFP) cassette between the P and M genes (rHPIV3 Cl-1–EGFP). The SC9779 patient virus’s fusion complex was chosen for these advanced studies because that patient was sampled 15 times over an infection that persisted for 242 days, the longest of which we are aware. We have shown that HAE cultures represent an authentic model of the human lung, reflecting the cell environment and selective pressure of the natural tissue (25, 28). These airway cultures have been well-validated for evaluating features of infection and HPIV3 fitness (28, 30).
The engineered viruses permit us to evaluate the outcome of evolution within the immunocompromised host and its impact on viral fitness. HAE cultures at an air-liquid interface, as diagrammed in Figure 7A, were infected with viruses bearing WT Cl-1 HN/Cl-1 F, Cl-1 HN H552Q/Cl-1 F, SC9779 HN/SC9779 F, or SC9779 HN H552Q/T193A/SC9779 F and expressing EGFP. Compared with the WT Cl-1, the virus bearing the SC9779 HN and F had similar growth in HAE (Figure 7B, gray and blue curves). Introduction of the H552Q mutation into the clinical isolate HN reduced the production of new viral particles (red curve). Introduction of the H552Q/T193A mutations into the SC9779 HN drastically reduced growth of the virus bearing the SC9779 fusion complex in HAE (orange curve), reflecting marked growth reduction for the SC9779 bearing H552Q/T193A compared with the starting SC9779 virus from patient 1.
Figure 7. Growth of recombinant CI-1 HPIV3 EGFP containing SC9779 HN/F in HAE cells.
(A) HAE cells were infected with 200 PFU of CI-1 HPIV3 EGFP containing SC9779 HN/F, SC9779 HN T193A-H552Q/F, or HN H552Q/F at the apical surface. Viruses were collected from the apical surface of the HAE cells on days 1, 2, 3, 5, and 7 after initial infection. (B) Titration of HPIV-3 CI-1 EGFP viruses (WT CI-1 or HN mutants) of virus collected from HAE cells 1, 2, 3, 5, and 7 days after initial infection (PFU/mL). Results depict a representative experiment from 3 biological replicates; data shown as mean ± SEM.
Infectivity of patient-derived viruses in vivo.
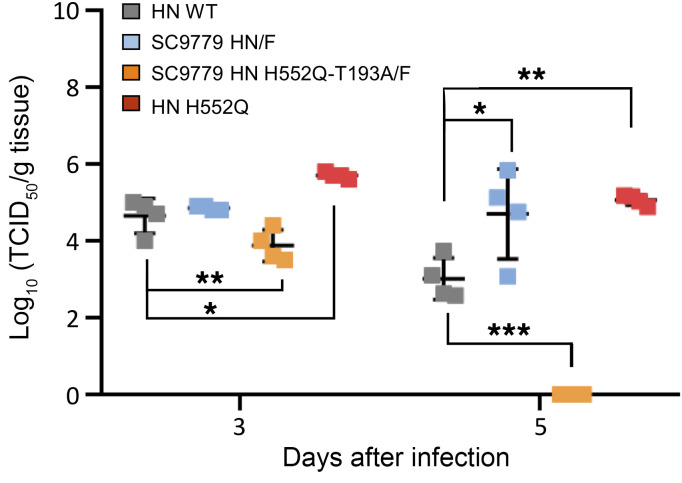
HPIV3 clinical isolates grow efficiently in cotton rats, a model that has been well-validated for viral respiratory disease (27, 33, 40). Cotton rats (4 per group) were infected with CI-1, CI-1 bearing H552Q HN, CI-1 bearing the SC9779 HN/F glycoproteins, and CI-1 bearing the SC9779 HN/F glycoproteins with HN H552Q-T193A/F (Figure 8). The animals were euthanized on 3 and 5 days after infection (HPIV3 titers peak in cotton rats in this range; ref. 40), and the viral titers from the left lung were measured by plaque assay. The viruses attained similar titers of approximately 105 PFU/g of lung and nasal turbinate tissue at day 3, showing that the day 0 virus infecting the SC9779 patient had no apparent deficit in fitness in vivo owing to its fusion complex, and the CI-1 bearing the SC9779 HN/F glycoproteins with HN H552Q-T193A/F was somewhat impaired. This finding is in line with the severe reduction in HAE growth resulting from the H552Q/T193A mutations shown in Figure 7. This difference is even more striking at day 5.
Figure 8. In vivo infectivity of patient-derived viruses.
Cotton rats were infected intranasally with CI-1, CI-1 HN H552Q, CI-1 HN/F SC9779, and CI-1 HN/F SC9779 H552Q-T193A, and the left lungs were collected after 3 and 5 days. A 1-way ANOVA was performed to determine significance; P values are specified as follows: *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001. TCID50, 50% tissue culture infectious dose.
Discussion
The sequences of a diverse assortment of clinical isolates that have been collected during acute infections have shown significant differences from the monolayer culture–adapted strains. In fact, none of the mutations that emerged in monolayer culture, specifically those in the primary and secondary binding sites of HN, were identified in any previous patient isolates that we sequenced directly from humans with acute HPIV3 infections. Thus, it was remarkable to find that during persistent infection in immunocompromised patients, the very residues important for culture adaptation were altered, especially in the HN dimer interface that modulates fusion during entry.
HPIV3 variant viruses with HN mutations emerged under the pressure of neuraminidase-mediated depletion of receptors in cell monolayers (14, 18, 41, 42). The variants escaped the constraint of neuraminidase-induced receptor depletion, either by enhanced receptor avidity or enhanced F triggering, and grew effectively in cell culture. The H552Q mutation in HN emerged during HPIV3 adaptation to cell culture and pointed to the existence of a key second functional site distinct from the primary binding site. This second site, at the HN dimer interface, is important for receptor binding and for activating F (28, 29, 43). The dimer interface mutations were associated with much higher fusogenicity in culture, by virtue of both increased avidity for sialic acid and increased inherent F-triggering activity, by enhancing HN dimer interface interactions (23, 43).
During growth of clinical isolates in culture, the H552Q mutation in HN emerges and confers increased neuraminidase activity as well as higher receptor avidity and intrinsically enhanced triggering of F (26). In culture-adapted variants, the binding and fusion features conferred by H552Q HN dominate the phenotype (18, 25), impairing growth in HAE and in vivo. However, when HN H552Q was on the background of the clinical strain CI-1, the increased neuraminidase activity balanced the other features, and the clinical isolates with HN H552Q retained viability in HAE and in vivo. It is therefore of interest that patient 1 was treated with DAS181 (Fludase), which cleaves sialic acids from the host cell surface in the lung, inactivating the HPIV3 host cell receptor (33, 35) and preventing viral entry, similarly to the selection process that led to the emergence of HN H552Q in cell culture.
DAS181 has been used in a number of clinical trials, including a larger multicenter trial of immunocompromised individuals with lower respiratory tract HPIV3 infection that showed promise in those requiring supplemental oxygen (44). In preclinical proof-of-concept studies of DAS181 for HPIV3, we assessed DAS181 activity in vitro and in HAE (33). Treating cultures with DAS181 also enhanced the production of infectious variant HPIV3 HN H552Q particles in HAE (data not shown). The fact that such variants emerged under the pressure of neuraminidase treatment and that their growth in HAE was enhanced by the presence of DAS181 raised the concern that such resistant variants might emerge under selective pressure of DAS181 treatment in vivo. DAS181 treatment in effect replicates the selective pressure that produced such resistant variants by depleting cell-surface receptors for HPIV3.
We had previously conjectured that some variants with HN mutations at the dimer interface fusion promotion site might not be fit in vivo, and the present study showed that such variants can emerge in the face of immunosuppression and persist in the human lung. A recent report describes an immunosuppressed person infected with SARS-CoV-2 who, like the HPIV3 patients in this study, remained viremic for multiple months (45). The prolonged replication in that individual resulted in evolution of SARS-CoV-2 within the patient with multiple mutations over time, including the spike (S) gene mutation E484K currently found in highly transmissible variant viruses globally. However, in that report, in contrast to our findings for HPIV3, the mutations conferred resistance to antibodies found in convalescent sera as well as to monoclonal antibodies in clinical use (46).
The HN mutation T193A alters a residue that contributes to the primary sialic acid binding site on HN (14, 28). The substitution of threonine for alanine opens up the binding cleft slightly as a result of the loss of threonine’s methyl and hydroxyl groups, reducing steric clash. R277K is a mutation in a surface site near the sialic acid binding pocket. This mutation only appears in the absence of H552Q (the presence of both mutations severely impairs growth; see Figure 7) and appeared only in the BAL samples. Mutations in residues 277 and 193, in proximity to the sialic acid binding site, have been shown to functionally alter fusion activity, consistent with the direct link between HN’s receptor engagement and fusion activation.
In light of the intricate balance of entry and egress for HPIV3 attachment proteins, long-term evolution of HPIV3 in the immunocompromised patients described here led to tipping the balance in favor of cellular entry rather than exit. These viruses are receptor-avid and primed for entry and replication within the tissue. After infection has been established in the lung epithelium, perhaps continuous infection of new cells occurs via spread from cell to cell and low-level replication, permitting a carrier state. Enhanced receptor binding and fusion activation, features that could impede transmission to a new host, are well-suited to this local cycle. Although it seems that this ongoing infection might cause lung damage, we cannot assess the etiology of lung disease for the patients in this report, who had several simultaneous conditions that could account for lung damage. Future studies in human lung models (26, 27, 30) will address this question.
We speculate that after lung infection has been established and viruses need only spread from cell to cell within a limited space, the requirements for fitness are altered. A virus that is best able to bind and enter cells that are directly adjacent to its current host cell would have an advantage, similar to the situation in cultured cells. However, different features are needed for transmission, which requires production of virions equipped to transverse the upper airway and to resist activation until they reach the lower airway. We hypothesize that such viruses, emerging during prolonged localized infection in a human, would be poorly transmissible from human to human. The balance of features of the fusion complex that favors persistent infection, leading to a high-affinity, fusogenic virus, is likely to exact a fitness cost.
The ability to establish persistent infections in vitro and in vivo is a hallmark of paramyxoviruses (47–51) and may have significant consequences to the hosts. For example, in the case of measles virus, persistence can lead to fatal complications in the CNS (52, 53). Evidence of persistence in humans includes the prolonged shedding of HPIV3 without opportunity for having acquired new infection (31), as well as evidence of long-term presence of HPIV3 in bone marrow (54, 55). However, molecular mechanisms of persistence for RNA viruses have been difficult to identify. For measles virus, several molecular mechanisms have been uncovered in the pathogenesis of long-term infection of the CNS, including biased hypermutation of the H (receptor binding) and M protein genes (56). Recently, we and others have studied viruses from brains of individuals who died of measles inclusion body encephalitis (MIBE) and subacute sclerosing panencephalitis (SSPE) and found that these viruses bear H/F fusion complexes that permit them to enter cells of the CNS and spread from cell to cell without using the usual complement of host receptors (53, 57–59). Interestingly, like the HPIV3 viruses in this study that evolved in a setting of reduced immune system activity, these MIBE/SSPE viruses emerged in the context of the CNS, a site that is relatively protected from the immune system compared with the other sites of measles virus replication. The phenotypes of our HPIV3 patient variants seem analogous to these measles variants arising in the CNS, advantaged at spread within the host tissue, pointing to the theme of persistence accomplished by favoring entering and cell-cell spread over exit of infectious virus.
In the case of another parainfluenza virus, PIV5, persistent infection can result if replication is suppressed by phosphorylation of the P protein, a component of the viral RNA-dependent RNA polymerase complex, in turn downregulating RNA polymerase activity and viral growth (47). This suppression of replication is thought to permit persistence, and such variants might be at a selective advantage in the presence of innate immunity, whereas switching to productive infection would promote transmission (48). In the case of the HPIV3 isolates we describe, the viral replication machinery is unaffected and growth in HAE is unaltered, suggesting that the mechanism identified for PIV5 is different from that in operation here.
Defective viral genomes or defective interfering particles may play a role in the establishment of persistence, a mechanism well-established in culture and in experimental infections, and more recently shown to be present in natural infections with negative-sense RNA viruses, potentially affecting the course of disease (50, 60). It is as of yet not clear whether defective viral genomes contribute to the establishment of persistence in humans, although there are intriguing examples that suggest this may be the case. Defective interfering particles of measles virus have been found in SSPE, and it has been suggested that they may play a role in establishment of this disease by helping to establish persistence (51). We did not detect evidence of high levels of defective genomes in the isolates from the patients in this study, although the number of patients and use of short read sequencing and tagmentation may affect defective viral genome detection.
In the viruses isolated during long-term infection in the immunocompromised patients described here, the host environment favored evolution of the surface glycoproteins that constitute the viral entry complex, altering the regulation of entry. This theme is similar to the MIBE/SSPE variants arising in the CNS, similarly protected from immune activity, that are skewed toward fusing and entering, even in cells that do not bear any known receptor, and have an advantage at spread within the host tissue. Arriving at this result via a very different set of specific mutations than those in the measles variants, these HPIV3 viruses similarly prioritize setting up residence within the tissue, spreading from cell to cell, adhering tightly to their host cells, and resisting release into the environment.
Methods
Viruses.
Recombinant viruses were generated by reverse genetics as previously described using an HPIV3 CI-1-EGFP background and a recombinant, clinically isolated (CI-1) HPIV3 virus sequence (28) containing an EGFP cassette between genes P and M. HN and F sequences isolated from day 1 collection from patient samples of the current study replaced the corresponding CI-1 genes, with point mutations made in the corresponding consensus sequences. Resulting viruses were propagated using the HAE EpiAirway AIR-100 system (MatTek Corporation; see below), and harvested in 1× PBS with magnesium and calcium (MatTek Corporation). All recombinant viruses were sequenced using described mNGS prior to experimental use. Viruses were titered by limiting dilution infection of Vero cells (ATCC), with infected cells quantified using InCell Analyzer 2000.
Constructs.
Plasmids encoding HPIV3 HN and F from clinically isolated and laboratory-adapted variants were generated through site-directed mutagenesis of a previously constructed pCAGGS mammalian expression vector (27) and sequenced via Sanger sequencing prior to experimental use. Transfections with plasmids were performed in HEK293T cells (ATCC) using Lipofectamine 2000 per the manufacturer’s specifications (Invitrogen). Consensus sequence of the laboratory-adapted strain of HPIV3 (Wash/47885/57) used throughout the study was obtained from the NIH (HA-1, NIH no. 47885, catalog no. V323-002-020; ref. 61).
Cells.
HEK293T cells (ATCC) for transfections were grown in DMEM (Gibco) supplemented with 10% FBS and 1% penicillin-streptomycin (Gibco) at 37°C and 5% CO2. Vero cells (ATCC) were grown in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin (Gibco) at 37°C and 5% CO2. All cells tested negative for mycoplasma presence using MycoAlert Mycoplasma Detection kit per the manufacturer’s specifications (Lonza).
HAE culture.
The HAE EpiAirway AIR-100 system (MatTek Corporation) comprises a cultured human-derived tracheo/bronchial epithelium that enables the formation of pseudostratified, differentiated mucociliary epithelium recapitulating in vivo human tissue. Upon receipt from the manufacturer, HAE cultures were transferred to provided 6-well plates containing 2 mL of AIR-100-ASY assay medium (MatTek Corporation) per well with the apical surface remaining exposed to air and incubated at 37°C in 5% CO2 overnight prior to infection.
HAE cultures were infected with the listed recombinant viruses at the apical surface for 90 minutes at 37°C, followed by inoculum removal and incubation at 37°C for the remainder of the experiment. Provided maintenance medium (MatTek Corporation) was changed every other day with 2 mL medium via the basolateral surface. Viruses were harvested by adding 200 μL of provided 1× PBS containing magnesium and calcium (MatTek Corporation) per well via the apical surface and incubated for 30 minutes at 37°C. Supernatant was subsequently collected and viral titers determined by fluorescent quantification as described above.
HN neuraminidase assay.
Neuraminidase activity of patient-derived HNs were measured as previously described (26, 38). Briefly, HEK293T cells were transfected with plasmids encoding specified HNs. After overnight incubation, media was aspirated and replaced with CO2-independent medium (pH 4.7; Gibco) and incubated for 10 minutes at 37°C. Then, 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid, sodium salt hydrate (4-MUNANA) was added to a final concentration of 7.7 mM and rate of cleavage was quantified via fluorescence readings every 2 minutes for 1 hour at 37°C with a microplate reader; excitation 365 nm, emission 450 nm (Tecan M1000 Pro).
β-Galactosidase complementation assay.
The ability of patient-derived HNs to promote fusion, as well as fusion capability of patient-derived Fs, were measured through a β-galactosidase complementation assay as previously described (17). To assess individual patient-derived HNs and Fs, assays were conducted using paired HPIV3 F or HNs from a laboratory-adapted reference strain (see Viruses). Extent of fusion was quantified by luminescence readings (Tecan M1000 Pro).
Release.
The ability of cells transiently expressing different HPIV3 HNs to bind and release human RBCs was measured as described previously (15, 18). Briefly, 6-well plates containing HEK293T cells transiently transfected with specified HNs were treated with CO2-independent medium (Gibco) containing 240 mU of neuraminidase (Sigma-Aldrich) for 90 minutes at 37°C. Cells were then placed on ice to equilibrate and media was aspirated and replaced with CO2-independent medium containing 1% RBCs. RBCs were left to bind for 30 minutes at 4°C. After incubation, cells were washed with CO2-independent medium and replaced with 1× PBS. Cells were then incubated at 37°C for the indicated times prior to collection and 1× PBS replacement for the next time point. After the final time point, RBCs that remained bound to the cells were lysed with ACK lysis buffer (Gibco). RBC release was quantified by measurement of absorbance at 410 nm.
Avidity.
The ability of cells transiently expressing different HPIV3 HNs to bind sialic acid–bearing receptors on RBCs was measured as described previously (18, 37). Briefly, 48-well plates containing HEK293T cells transiently transfected with specified HNs were treated with CO2-independent medium (Gibco) containing 15 mU of neuraminidase (Sigma-Aldrich) for 90 minutes at 37°C. Cells were then placed on ice to equilibrate and media was aspirated and replaced with CO2-independent medium containing 1% RBCs treated with varying levels of receptor depletion by virtue of the RBCs being preincubated with increasing concentrations of neuraminidase (Sigma-Aldrich). RBCs were then left to bind for 30 minutes at 4°C. After incubation, cells were washed with CO2-independent medium and treated with ACK lysis buffer (Gibco). The amount of bound RBCs was quantified by measurement of absorbance at 410 nm.
Animals.
Inbred Hsd cotton rats were obtained from Envigo. Female and male animals, 6 to 8 weeks of age, were used. The animals were purchased in a specific pathogen–free condition according to the breeder’s specifications and were maintained in a barrier system. Animals were kept under controlled environmental conditions of 22°C ± 1°C and a 12-hour light/12-hour dark cycle.
Cotton rats were isoflurane-anesthetized and infected intranasally with 105 TCID50 of virus in 100 μL of PBS. Animals were euthanized by CO2 inhalation 3 days after infection and left lungs were collected and homogenized with a glass homogenizer. Homogenates were frozen until titration at −80°C.
Neutralization assay.
The effects of the patient antibodies on standard clinically isolated viruses and clinically isolated viruses from the patients, as well as the effects of control human antibodies on patient clinically isolated viruses, were assessed using a plaque reduction assay as described previously (62). The assay measures the effect of each treatment on viral entry measured as percentage inhibition of entry and was performed using 3 HPIV3 viruses: CI-1 (2000 PFU/well); CI-1 with patient 1 (SC9779) HN and F (2000 PFU/well); CI-1 with patient 1 (SC9779) and T193A H552Q HN and F (200 PFU/well). Briefly, Vero cell monolayers were infected with the indicated virus in the presence of serial dilutions of the indicated sera. After 90 minutes, the plates were overlaid with carboxymethylcellulose; 24 hours later the cellulose was removed, the cells were fixed, and the infected cells were counted. Serum samples were obtained from blood draws at indicated points during the patient’s infection; immune serum was obtained from a normal donor.
HPIV3 genome recovery.
Two individuals with HPIV3 shedding of more than 90 days were identified and BAL, nasal swab, and nasal wash specimens were subjected to deep sequencing. HPIV3 Ct values were obtained from the diagnostic respiratory RT-PCR assay (16825353). Metagenomic sequencing libraries were prepared as previously described (25, 63) for all of the primary clinical samples. RNA was extracted from the primary samples using Zymo Viral RNA kit and DNased using Turbo DNase I kit (Invitrogen). DNased RNA was synthesized to double-stranded cDNA using SSIII reverse transcriptase (Invitrogen)and Sequenase V2.0 (Life Technologies). Shotgun sequencing libraries were made using 0.3× of reaction volume of Nextera XT (Illumina) with 22 cycles and cleaned up using 0.8× Ampure XP beads and sequenced on Illumina MiSeq and NovaSeq 6000. Libraries that failed to give sufficient coverage by shotgun metagenomics were enriched for HPIV3 reads using custom biotinylated 120 bp xGen lockdown probes (IDTDNA) based on the HPIV3 reference genome (GenBank, NC_001796). Double-stranded cDNA was created as above and tagmented using half-reactions of Nextera Flex prior to amplification and overnight hybridization capture and reamplification following the manufacturer’s protocol. These libraries were also sequenced on a NovaSeq 6000. Sequencing reads are available at NCBI’s BioProject (PRJNA704707).
Sequencing library analysis.
Metagenomic and capture sequencing reads were adapter and Q20 quality-trimmed using Trimmomatic v0.39. Variants for all samples were called with LAVA (64). Briefly, sequencing reads for the first sample for each patient were aligned to the HPIV3 GenBank reference sequence (NC_001796.2) using bwa-mem v0.7.17-r1188, and the majority consensus for each patient was then extracted using bcftools v1.9 and annotated via VarScan v2.3. Subsequent samples for that patient were then aligned to this majority consensus to call relative mutational changes. Variant alleles that were determined to be sequencing artifacts based on position in read were removed. We then recorded variants that were present in at least 2 longitudinal time points and arose with a minimum allelic depth of 3 or higher in all samples from all patients. Figure generation code is publicly available on GitHub (https://github.com/greninger-lab/hpiv3-iterative).
Statistics.
Statistical analysis was done using 1-way ANOVA (Figures 5 and 8) or 2-way ANOVA (Figure 6) where appropriate using GraphPad Prism 7. Results are mean ± SD unless otherwise stated. P values less than 0.05 were considered statistically significant.
Study approval.
This study was approved by the University of Washington IRB (Seattle, WA) and by the Fred Hutchinson Cancer Research Center IRB (Seattle, WA), and written informed consent was obtained from study participants.
Author contributions
ALG, KR, SN, BKR, MP, and AM designed research studies; ALG, KR, MJL, RCS, JDB, NM, OH, FB, SRB, and MP conducted experiments and acquired data; ALG, KR, TCM, FB, SRB, SN, BKR, MP, and AM analyzed data; MB provided serum specimens; MP and AM provided reagents; ALG, KR, TCM, SRB, SN, BKR, MP, and AM contributed to writing the manuscript and/or preparing figures.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01AI114736 to AM and by the Sharon Golub fund.
Version 1. 10/05/2021
In-Press Preview
Version 2. 12/01/2021
Electronic publication
Footnotes
Conflict of interest: MB received research support from Ansun BioPharma.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(23):e150506.https://doi.org/10.1172/JCI150506.
Contributor Information
Alexander L. Greninger, Email: agrening@uw.edu.
Ksenia Rybkina, Email: xr2141@cumc.columbia.edu.
Michelle J. Lin, Email: mjlin@uw.edu.
Jennifer Drew-Bear, Email: jd3730@cumc.columbia.edu.
Tara C. Marcink, Email: tm2996@cumc.columbia.edu.
Ryan C. Shean, Email: ryanshean1@gmail.com.
Negar Makhsous, Email: nmakhsous@gmail.com.
Michael Boeckh, Email: mboeckh@fredhutch.org.
Olivia Harder, Email: harder.33@osu.edu.
Francesca Bovier, Email: francescabovier@gmail.com.
Shana R. Burstein, Email: shana.burstein@gmail.com.
Stefan Niewiesk, Email: Stefan.Niewiesk@cvm.osu.edu.
Bert K. Rima, Email: b.rima@qub.ac.uk.
Matteo Porotto, Email: mprotto@gmail.com.
Anne Moscona, Email: am939@cumc.columbia.edu.
References
- 1.Nair H, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375(9725):1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maziarz RT, et al. Control of an outbreak of human parainfluenza virus 3 in hematopoietic stem cell transplant recipients. Biol Blood Marrow Transplant. 2010;16(2):192–198. doi: 10.1016/j.bbmt.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah DP, et al. Parainfluenza virus infections in hematopoietic cell transplant recipients and hematologic malignancy patients: a systematic review. Cancer Lett. 2016;370(2):358–364. doi: 10.1016/j.canlet.2015.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lo MS, et al. The impact of RSV, adenovirus, influenza, and parainfluenza infection in pediatric patients receiving stem cell transplant, solid organ transplant, or cancer chemotherapy. Pediatr Transplant. 2012;17(2):133–143. doi: 10.1111/petr.12022. [DOI] [PubMed] [Google Scholar]
- 5.Weinberg GA, et al. Parainfluenza virus infection of young children: estimates of the population-based burden of hospitalization. J Pediatr. 2009;154(5):694–699. doi: 10.1016/j.jpeds.2008.11.034. [DOI] [PubMed] [Google Scholar]
- 6.Seo S, et al. Parainfluenza virus lower respiratory tract disease after hematopoietic cell transplant: viral detection in the lung predicts outcome. Clin Infect Dis. 2014;58(10):1357–1368. doi: 10.1093/cid/ciu134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henrickson KJ, et al. National disease burden of respiratory viruses detected in children by polymerase chain reaction. Pediatr Infect Dis J. 2004;23(1 suppl):S11–S18. doi: 10.1097/01.inf.0000108188.37237.48. [DOI] [PubMed] [Google Scholar]
- 8.Chemaly RF, et al. The characteristics and outcomes of parainfluenza virus infections in 200 patients with leukemia or recipients of hematopoietic stem cell transplantation. Blood. 2012;119(12):2738–2745. doi: 10.1182/blood-2011-08-371112. [DOI] [PubMed] [Google Scholar]
- 9.Roghmann M, et al. Active surveillance for respiratory virus infections in adults who have undergone bone marrow and peripheral blood stem cell transplantation. Bone Marrow Transplant. 2003;32(11):1085–1088. doi: 10.1038/sj.bmt.1704257. [DOI] [PubMed] [Google Scholar]
- 10.Schmidt AC, et al. Progress in the development of human parainfluenza virus vaccines. Expert Rev Respir Med. 2011;5(4):515–526. doi: 10.1586/ers.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Englund JA, et al. Safety and infectivity of two doses of live-attenuated recombinant cold-passaged human parainfluenza type 3 virus vaccine rHPIV3cp45 in HPIV3-seronegative young children. Vaccine. 2013;31(48):5706–5712. doi: 10.1016/j.vaccine.2013.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Asten L, et al. Mortality attributable to 9 common infections: significant effect of influenza A, respiratory syncytial virus, influenza B, norovirus, and parainfluenza in elderly persons. J Infect Dis. 2012;206(5):628–639. doi: 10.1093/infdis/jis415. [DOI] [PubMed] [Google Scholar]
- 13.Chang A, Dutch RE. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses. 2012;4(4):613–636. doi: 10.3390/v4040613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moscona A, Peluso RW. Relative affinity of the human parainfluenza virus type 3 hemagglutinin-neuraminidase for sialic acid correlates with virus-induced fusion activity. J Virol. 1993;67(11):6463–6468. doi: 10.1128/jvi.67.11.6463-6468.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Porotto M, et al. Triggering of human parainfluenza virus 3 fusion protein(F) by the hemagglutinin-neuraminidase (HN): an HN mutation diminishing the rate of F activation and fusion. J Virol. 2003;77(6):3647–3654. doi: 10.1128/JVI.77.6.3647-3654.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moscona A. Entry of parainfluenza virus into cells as a target for interrupting childhood respiratory disease. J Clin Invest. 2005;115(7):1688–1698. doi: 10.1172/JCI25669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palermo LM, et al. Fusion promotion by a paramyxovirus hemagglutinin-neuraminidase protein: pH modulation of receptor avidity of binding sites I and II. J Virol. 2007;81(17):9152–9161. doi: 10.1128/JVI.00888-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Porotto M, et al. A second receptor binding site on human parainfluenza virus type 3 hemagglutinin-neuraminidase contributes to activation of the fusion mechanism. J Virol. 2007;81(7):3216–3228. doi: 10.1128/JVI.02617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palermo L, et al. Human parainfluenza virus infection of the airway epithelium: viral hemagglutinin-neuraminidase regulates fusion protein activation and modulates infectivity. J Virol. 2009;83(13):6900–6908. doi: 10.1128/JVI.00475-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15(7):690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White JM, et al. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43(3):189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Plattet P, Plemper RK. Envelope protein dynamics in paramyxovirus entry. mBio. 2013;4(4):e00413-13. doi: 10.1128/mBio.00413-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porotto M, et al. Regulation of paramyxovirus fusion activation: the hemagglutinin-neuraminidase protein stabilizes the fusion protein in a pretriggered state. J Virol. 2012;86(23):12838–12848. doi: 10.1128/JVI.01965-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huberman K, et al. Hemagglutinin-neuraminidase of human parainfluenza 3: role of the neuraminidase in the viral life cycle. Virology. 1995;214(1):294–300. doi: 10.1006/viro.1995.9925. [DOI] [PubMed] [Google Scholar]
- 25.Iketani S, et al. Viral entry properties required for fitness in humans are lost through rapid genomic change during viral isolation. mBio. 2018;9(4):e00898-18. doi: 10.1128/mBio.00898-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmer SG, et al. Adaptation of human parainfluenza virus to airway epithelium reveals fusion properties required for growth in host tissue. mBio. 2012;3(3):e00137-12. doi: 10.1128/mBio.00137-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palermo LM, et al. Features of circulating parainfluenza virus required for growth in human airway. mBio. 2016;7(2):e00235. doi: 10.1128/mBio.00235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer SG, et al. Circulating clinical strains of human parainfluenza virus reveal viral entry requirements for in vivo infection. J Virol. 2014;88(22):13495–13502. doi: 10.1128/JVI.01965-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu R, et al. Interaction between the hemagglutinin-neuraminidase and fusion glycoproteins of human parainfluenza virus type III regulates viral growth in vivo. mBio. 2013;4(5):e00803–e00813. doi: 10.1128/mBio.00803-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porotto M, et al. Authentic modeling of human respiratory virus infection in human pluripotent stem cell-derived lung organoids. mBio. 2019;10(3):e00723-19. doi: 10.1128/mBio.00723-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muchmore H, et al. Persistent parainfluenza virus shedding during isolation at the South Pole. Nature. 1981;289(5794):187–189. doi: 10.1038/289187a0. [DOI] [PubMed] [Google Scholar]
- 32.Srinivasan A, et al. Symptomatic parainfluenza virus infections in children undergoing hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2011;17(10):1520–1527. doi: 10.1016/j.bbmt.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moscona A, et al. A recombinant sialidase fusion protein effectively inhibits human parainfluenza viral infection in vitro and in vivo. J Infect Dis. 2010;202(2):234–241. doi: 10.1086/653621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waghmare A, et al. Successful treatment of parainfluenza virus respiratory tract infection with DAS181 in 4 immunocompromised children. J Pediatric Infect Dis Soc. 2015;4(2):114–118. doi: 10.1093/jpids/piu039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Triana-Baltzer GB, et al. DAS181, a sialidase fusion protein, protects human airway epithelium against influenza virus infection: an in vitro pharmacodynamic analysis. J Antimicrob Chemother. 2010;65(2):275–284. doi: 10.1093/jac/dkp421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moscona A, Peluso RW. Analysis of human parainfluenza virus 3 receptor binding variants: evidence for the use of a specific sialic acid-containing receptor. Microb Pathog. 1996;20(3):179–184. doi: 10.1006/mpat.1996.0016. [DOI] [PubMed] [Google Scholar]
- 37.Murrell M, et al. Mutations in human parainfluenza virus type 3 HN causing increased receptor binding activity and resistance to the transition state sialic acid analog 4-GU-DANA (zanamivir) J Virol. 2003;77:309–317. doi: 10.1128/JVI.77.1.309-317.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Greengard O, et al. The anti-influenza virus agent 4-GU-DANA (zanamivir) inhibits cell fusion mediated by human parainfluenza virus and influenza virus HA. J Virol. 2000;74(23):11108–11114. doi: 10.1128/JVI.74.23.11108-11114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Porotto M, et al. Paramyxovirus receptor-binding molecules: engagement of one site on the hemagglutinin-neuraminidase protein modulates activity at the second site. J Virol. 2006;80(3):1204–1213. doi: 10.1128/JVI.80.3.1204-1213.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niewiesk S. Current animal models: cotton rat animal model. Curr Top Microbiol Immunol. 2009;330:89–110. doi: 10.1007/978-3-540-70617-5_5. [DOI] [PubMed] [Google Scholar]
- 41.Porotto M, et al. Human parainfluenza virus type 3 HN-receptor interaction: the effect of 4-GU-DANA on a neuraminidase-deficient variant. J Virol. 2001;76:7481–7488. doi: 10.1128/JVI.75.16.7481-7488.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porotto M, et al. Influence of the human parainfluenza virus 3 attachment protein’s neuraminidase activity on its capacity to activate the fusion protein. J Virol. 2005;79(4):2383–2392. doi: 10.1128/JVI.79.4.2383-2392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Porotto M, et al. The second receptor binding site of the globular head of the Newcastle disease virus (NDV) hemagglutinin-neuraminidase activates the stalk of multiple paramyxovirus receptor binding proteins to trigger fusion. J Virol. 2012;86(10):5730–5741. doi: 10.1128/JVI.06793-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chemaly RF, et al. DAS181 treatment of severe lower respiratory tract parainfluenza virus infection in immunocompromised patients: a phase 2 randomized, placebo-controlled study. Clin Infect Dis. 2021;73(3):e773–e781. doi: 10.1093/cid/ciab113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi B, et al. Persistence and evolution of SARS-CoV-2 in an immunocompromised host. N Engl J Med. 2020;383(23):2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark SA, et al. ISSCR guidelines for stem cell research and clinical translation: the 2021 update. Stem Cell Reports. 2021;16(6):1398–1408:S2213-6711(21)00263-0. doi: 10.1016/j.stemcr.2021.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young DF, et al. The switch between acute and persistent paramyxovirus infection caused by single amino acid substitutions in the RNA polymerase P subunit. PLoS Pathog. 2019;15(2):e1007561. doi: 10.1371/journal.ppat.1007561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lidsky PV, et al. Variability in viral pathogenesis: modeling the dynamic of acute and persistent infections. Curr Opin Virol. 2017;23:120–124. doi: 10.1016/j.coviro.2017.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fearns R, et al. Evidence that the paramyxovirus simian virus 5 can establish quiescent infections by remaining inactive in cytoplasmic inclusion bodies. J Gen Virol. 1994;75(pt 12):3525–3539. doi: 10.1099/0022-1317-75-12-3525. [DOI] [PubMed] [Google Scholar]
- 50.Manzoni TB, Lopez CB. Defective (interfering) viral genomes re-explored: impact on antiviral immunity and virus persistence. Future Virol. 2018;13(7):493–503. doi: 10.2217/fvl-2018-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Randall RE, Griffin DE. Within host RNA virus persistence: mechanisms and consequences. Curr Opin Virol. 2017;23:35–42. doi: 10.1016/j.coviro.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norrby E, Kristensson K. Measles virus in the brain. Brain Res Bull. 1997;44(3):213–220. doi: 10.1016/S0361-9230(97)00139-1. [DOI] [PubMed] [Google Scholar]
- 53.Angius F, et al. Analysis of a subacute sclerosing panencephalitis genotype B3 virus from the 2009–2010 South African measles epidemic shows that hyperfusogenic F proteins contribute to measles virus infection in the brain. J Virol. 2019;93(4):e01700-18. doi: 10.1128/JVI.01700-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goswami K, et al. Evidence for the persistence of paramyxoviruses in human bone marrows. J Gen Virol. 1984;65(pt 11):1881–1888. doi: 10.1099/0022-1317-65-11-1881. [DOI] [PubMed] [Google Scholar]
- 55.Basle M, et al. Paramyxovirus antigens in osteoclasts from Paget’s bone tissue detected by monoclonal antibodies. J Gen Virol. 1985;66(pt 10):2103–2110. doi: 10.1099/0022-1317-66-10-2103. [DOI] [PubMed] [Google Scholar]
- 56.Rima BK, Duprex WP. Molecular mechanisms of measles virus persistence. Virus Res. 2005;111(2):132–147. doi: 10.1016/j.virusres.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 57.Jurgens EM, et al. Measles fusion machinery is dysregulated in neuropathogenic variants. mBio. 2015;6(1):e02528-14. doi: 10.1128/mBio.02528-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ayata M, et al. The F gene of the Osaka-2 strain of measles virus derived from a case of subacute sclerosing panencephalitis is a major determinant of neurovirulence. J Virol. 2010;84(21):11189–11199. doi: 10.1128/JVI.01075-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ayata M, et al. Amino acid substitutions in the heptad repeat A and C regions of the F protein responsible for neurovirulence of measles virus Osaka-1 strain from a patient with subacute sclerosing panencephalitis. Virology. 2016;487:141–149. doi: 10.1016/j.virol.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 60.Ziegler CM, Botten JW. Defective interfering particles of negative-strand RNA viruses. Trends Microbiol. 2020;28(7):554–565. doi: 10.1016/j.tim.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moscona A, Galinski MS. Characterization of human parainfluenza virus type 3 persistent infection in cell culture. J Virol. 1990;64(7):3212–3218. doi: 10.1128/jvi.64.7.3212-3218.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marcink TC, et al. Hijacking the fusion complex of human parainfluenza virus as an antiviral strategy. mBio. 2020;11(1):e03203-19. doi: 10.1128/mBio.03203-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Greninger AL, et al. Rapid metagenomic next-generation sequencing during an investigation of hospital-acquired human parainfluenza virus 3 infections. J Clin Microbiol. 2017;55(1):177–182. doi: 10.1128/JCM.01881-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lin MJ, et al. LAVA: a streamlined visualization tool for longitudinal analysis of viral alleles [preprint]. Posted on bioRxiv December 18, 2019. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.