Abstract
Excessive inflammation drives the progression from sepsis to septic shock. Macrophage migration inhibitory factor (MIF) is of interest because MIF promoter polymorphisms predict mortality in different infections, and anti-MIF antibody improves survival in experimental models when administered 8 hours after infectious insult. The recent description of a second MIF superfamily member, D-dopachrome tautomerase (D-DT/MIF-2), prompted closer investigation of MIF-dependent responses. We subjected Mif–/– and Mif-2–/– mice to polymicrobial sepsis and observed a survival benefit with Mif but not Mif-2 deficiency. Survival was associated with reduced numbers of small peritoneal macrophages (SPMs) that, in contrast to large peritoneal macrophages (LPMs), were recruited into the peritoneal cavity. LPMs produced higher quantities of MIF than SPMs, but SPMs expressed higher levels of inflammatory cytokines and the MIF receptors CD74 and CXCR2. Adoptive transfer of WT SPMs into Mif–/– hosts reduced the protective effect of Mif deficiency in polymicrobial sepsis. Notably, MIF-2 lacks the pseudo-(E)LR motif present in MIF that mediates CXCR2 engagement and SPM migration, supporting a specific role for MIF in the recruitment and accumulation of inflammatory SPMs.
Keywords: Immunology, Inflammation
Keywords: Cytokines, Macrophages, Mouse models
Introduction
Sepsis is a leading cause of critical illness and mortality worldwide (1) and studies of its pathophysiology emphasize the role of a dysregulated immune response in progressive organ dysfunction and lethal shock (2). Although numerous inflammatory cytokines have been implicated in sepsis, therapeutic interventions based on glucocorticoids (3) or inhibition of particular mediators and signaling pathways (4, 5) have been uniformly disappointing. Despite improvement in supportive care, sepsis remains a major public health concern; even individuals who survive incur significant medical, social, and financial costs for health care systems (6).
Macrophage migration inhibitory factor (MIF) is a widely expressed innate cytokine with a central role in the pathophysiology of septic shock (7). MIF upregulates the expression of microbial pattern recognition receptors (8, 9), counterregulates the immunosuppressive action of glucocorticoids (10), and sustains macrophage survival in the face of activation-induced apoptosis (11). MIF is unique among cytokines in that commonly occurring functional promoter polymorphisms predict morbidity and mortality in different infectious conditions, including community-acquired pneumonia (12), pneumococcal meningitis (13), meningococcemia (14), and gram-negative and mycobacterial sepsis (9, 14, 15). Circulating MIF concentrations are high in patients with sepsis (16) and may correlate with lethal outcome (17, 18). MIF administration in murine models of septic shock, including gram-negative endotoxemia, gram-positive exotoxemia, or polymicrobial sepsis, increases systemic inflammation and mortality, whereas anti-MIF antibody reduces inflammatory cytokine production, decreases organ injury, and promotes survival (7, 19, 20). Delayed administration of anti-MIF antibody for up to 8 hours after infectious insult also protects from lethality in the robust cecal ligation and puncture (CLP) model of polymicrobial sepsis (7), which is noteworthy since sepsis diagnosis is often delayed despite intensive care unit monitoring.
MIF has long been considered to be structurally unique, but a second MIF-like protein, D-dopachrome tautomerase (D-DT or MIF-2), with 34% sequence identity and a near-identical 3D structure, was recently characterized (16). Like MIF, MIF-2 is overexpressed in patients with sepsis and circulating levels correlate with APACHE II disease severity scores (16, 21). MIF and MIF-2 are both released from activated monocytes/macrophages and signal through the cognate MIF receptor, which comprises the ligand-binding protein CD74 and the associated signal transducer CD44, suggesting that a combined MIF/MIF-2 therapeutic strategy may be therapeutically promising (16). Consideration of such an approach is made possible by ongoing clinical testing of anti-CD74 (22) and small-molecule MIF antagonists (23) in different inflammatory conditions.
This study aimed to elucidate distinguishing functions between MIF and MIF-2 in sepsis by examining Mif versus Mif-2 gene deletion in the CLP model of polymicrobial sepsis, which mimics the leading cause of sepsis in surgical patients (24). Our findings revealed an unanticipated difference in sepsis lethality between Mif–/– and Mif-2–/– mice and a unique role for MIF in the recruitment and accumulation of a highly inflammatory small peritoneal macrophage (SPM) population.
Results
Mif but not Mif-2 deficiency protects mice from septic shock.
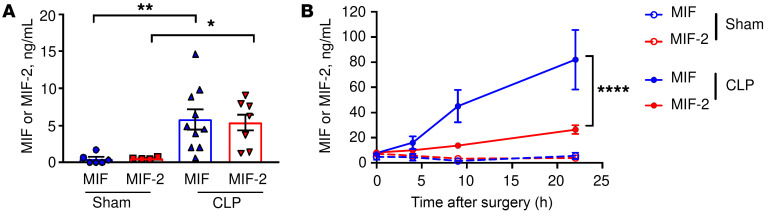
We first examined MIF and MIF-2 levels in WT mice with polymicrobial sepsis arising from peritonitis induced by CLP. Similar to MIF, MIF-2 levels were elevated in peritoneal lavage fluid after infectious insult (Figure 1A). Both MIF and MIF-2 levels increased in the systemic circulation after CLP, although MIF plasma levels were over 3 times higher at 9 and 22 hours after CLP when compared with MIF-2 levels (Figure 1B).
Figure 1. MIF and MIF-2 levels increase in sepsis.
WT mice underwent CLP or sham surgery and (A) peritoneal lavage at 22 hours and (B) peripheral blood at 0, 4, 9, and 22 hours were analyzed for MIF and MIF-2 by ELISA. (A) n = 4 to 10 animals, 1-way ANOVA with Tukey’s multiple-comparison test. (B) n = 6 to 22 animals, 2-way ANOVA with Bonferroni’s multiple-comparison test. *P < 0.05; **P < 0.01; ****P < 0.0001.
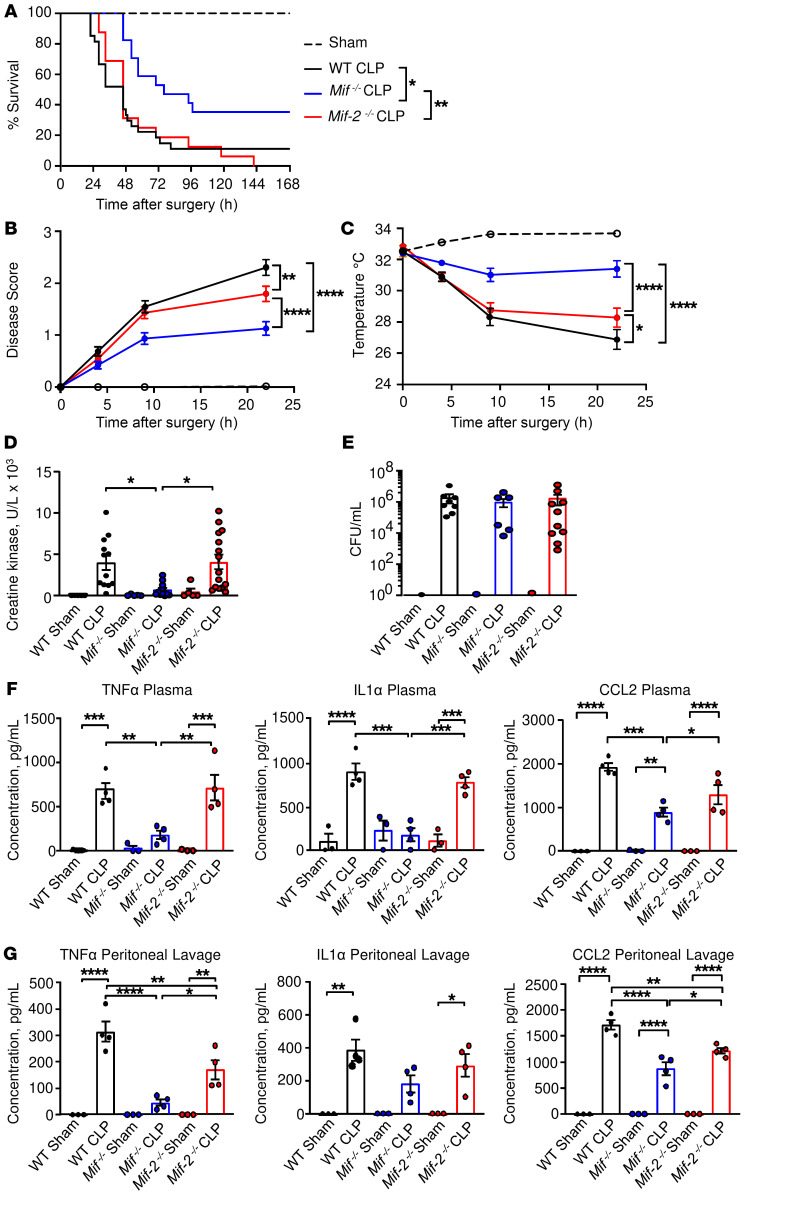
We generated Mif-2–/– mice lacking the entire Mif-2 gene (e.g., promoter and 3 exons). Notably, the genetic deletion of Mif but not Mif-2 was observed to protect mice from CLP-induced lethality (Figure 2A). The onset of lethality was delayed by 24 hours in Mif–/– mice and absolute survival was increased by 3-fold when compared with WT mice or Mif-2–/– mice. The survival benefit of Mif deficiency in polymicrobial sepsis was supported by the observations that Mif–/– mice showed a less severe disease score (Figure 2B), reduced hypothermia (Figure 2C), and lower plasma levels of creatine kinase (CK), indicating less myonecrosis, when compared with WT or Mif-2–/– mice (Figure 2D). There was no difference in the degree of bacteremia between WT, Mif–/–, and Mif-2–/– mice at 22 hours after CLP surgery (Figure 2E), and improved survival in Mif–/– mice was associated with reduced levels of inflammatory cytokines in blood plasma and within the peritoneal fluid; many of these cytokines have been reported to contribute to toxic sequelae of septic shock (ref. 25, Figure 2, F and G, and Supplemental Figures 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/JCI127171DS1). Specifically, TNF-α concentrations were significantly lower in Mif–/– mice when compared with WT or Mif-2–/– mice, both in plasma and peritoneal lavage (Figure 2, F and G), while peritoneal lavage levels for the antiinflammatory cytokine IL-10 were significantly higher in septic Mif–/– mice (Supplemental Figure 2, second row). We also observed lower circulating levels of the MIF-dependent chemokine CCL2 (26, 27) in Mif–/– mice (Figure 2, F and G), which is important for monocyte/macrophage recruitment into the peritoneum (28, 29). Collectively, these data suggest that the increased survival observed in Mif–/– mice results from a less inflammatory tissue-damaging response than in WT or Mif-2–/– mice.
Figure 2. Delayed onset of septic shock in Mif–/– but not Mif-2–/– mice.
(A) WT, Mif–/–, or Mif-2–/– mice were subjected to CLP (n = 16–19 animals) or sham surgery (n = 13 animals), and survival was monitored for at least 7 days. Log-rank (Mantel-Cox) test was performed to determine significance. (B) Disease score (see Methods) and (C) surface body temperature were recorded in the immediate postoperative period. Two-way ANOVA with Bonferroni’s multiple-comparison test. (D) Creatine kinase (CK) levels and (E) bacterial CFUs determined at 22 hours after CLP or sham surgery in plasma or heparinized whole blood, respectively. Data points are from 5 or more independent experiments and reflect n = 14 to 15 animals per group. For CFUs, each symbol represents 1 animal, n = 6–17 (Mann-Whitney test). (F and G) TNF-α, IL-1α, and CCL2 plasma (F) and peritoneal fluid (G) concentrations in WT, Mif–/–, and Mif-2–/– mice. Cytokine/chemokine levels were measured by flow cytometry using the LEGENDplex Multi Analyte Flow Assay Kit. n = 3 to 4 mice per group, 1-way ANOVA with Tukey’s multiple-comparison test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Fewer proinflammatory macrophages in the peritoneum of Mif–/– mice.
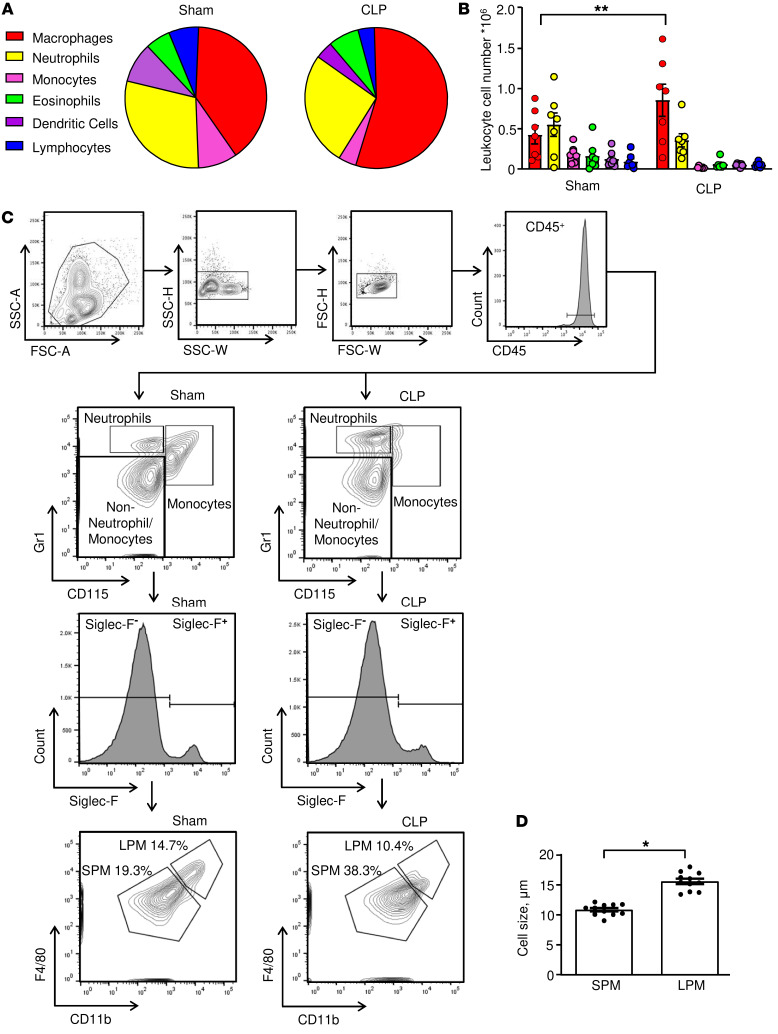
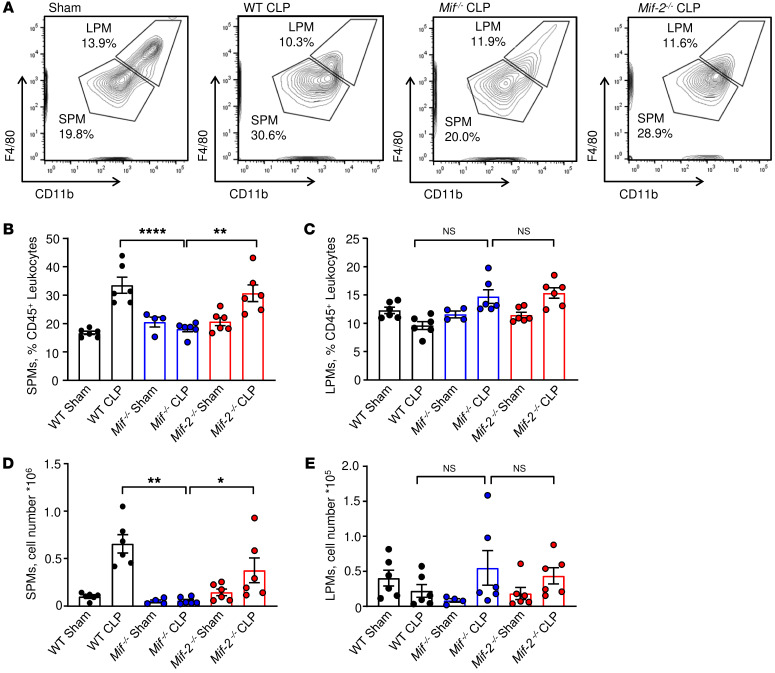
We next assessed the relative proportion of different leukocyte populations in the peritonea of septic mice. We observed septic WT mice to have a higher relative macrophage content as well as increased macrophage cell numbers in peritoneal lavage fluid when compared with sham animals (Figure 3, A and B), which is in accord with prior reports (30, 31) and consistent with the prevailing notion that macrophages drive CLP-induced septic shock. Peritoneal macrophages comprise 2 major subsets: large peritoneal macrophages (LPMs) and SPMs (32). After exclusion of monocytes, neutrophils, and eosinophils, we detected the presence of both macrophage populations with a relative increase in the SPM population from 19.3% to 38.3% after CLP induction (Figure 3C). Consistent with prior observations of SPM and LPM morphology, we also observed SPMs to be of smaller cell size than LPMs (Figure 3D and ref. 32). SPMs are derived from myeloid precursors and can be distinguished from LPMs by the expression of CX3CR1 (33). CLP and sham surgery in Cx3cr1GFP mice induced an increase in the relative and absolute numbers of peritoneal SPMs (Supplemental Figure 3, A and B). By contrast, GFP+ LPMs were barely detected, supporting the preferentially myeloid origin of the SPM population. Additionally, Ki67+GFP+ SPMs were observed after sham surgery but not after CLP (Supplemental Figure 3C), suggesting that SPMs have a low proliferation rate under inflammatory conditions and may be short-lived cells, as previously described (33, 34). Examination of SPM and LPM populations in the different genetic mouse strains after CLP revealed that Mif–/– mice contained fewer SPMs than WT or Mif-2–/– mice when analyzed as relative ratios to CD45+ leukocytes as well as absolute cell counts (Figure 4, A, B, and D). By contrast, the LPM population was not significantly altered between the different genetic mouse strains after CLP or sham surgery (Figure 4, A, C, and E).
Figure 3. Macrophages are the predominant cell type in CLP and can be classified into SPM and LPM subpopulations.
(A) Peritoneal cells were stained for flow cytometry and relative numbers of CD11b+F4/80+ macrophages, CD115–Gr1hi neutrophils, CD115+Gr1hi monocytes, Siglec F+ eosinophils, CD11c+ DCs, and CD3+/CD19+ lymphocytes; (A) was determined as percentage of CD45+ cells and (B) measured as absolute cell number. Plot of n = 7 mice from 2 separate experiments. Two-way ANOVA with Sidak’s multiple-comparison test. (C) Representative gating strategy for the identification of small (SPM) and large (LPM) peritoneal macrophages in the peritoneal lavage of WT mice 22 hours after CLP or sham surgery. (D) Cytospins of SPMs and LPMs isolated from peritoneal lavage of septic mice and stained with Wright-Giemsa stain. Macrophage size was determined by ImageJ software from n = 11 animals. SPMs measured 10.9 ± 0.3 μm; the size of LPMs was 15.6 ± 0.5 μm (unpaired, 2-tailed Student’s t test). *P < 0.05; **P < 0.01.
Figure 4. MIF deficiency is associated with reduced i.p. SPM numbers.
(A) Representative gating strategy for SPM and LPM analysis in WT, Mif–/–, and Mif-2–/– mice after CLP or sham surgery. Relative cell numbers and absolute cell count analyses of SPMs (B and D) and LPMs (C and E) in the peritoneal lavage of WT, Mif–/–, and Mif-2–/– mice after CLP or sham surgery. Flow cytometric analyses were performed on peritoneal cells collected 22 hours after CLP or sham surgery and included CountBright absolute counting beads in n = 4 to 6 animals/genotype. *P < 0.05; **P < 0.01; ****P < 0.0001 by 1-way ANOVA with Tukey’s multiple-comparison test (B) or Kruskal-Wallis with Dunn’s multiple-comparison test (C–E).
LPMs express MIF but SPMs are the more proinflammatory and MIF-responsive subset.
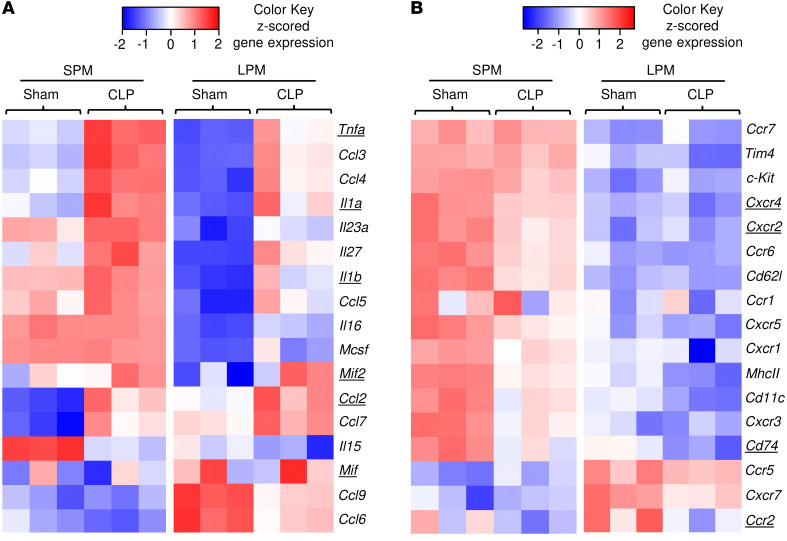
To better characterize phenotypic differences between SPM and LPM subsets that may influence sepsis lethality, we performed RNA-Seq and differential gene expression analysis of SPMs/LPMs isolated from WT mice subjected to CLP or sham surgery. Differentially expressed genes were analyzed in SPM-sham versus LPM-sham and SPM-CLP versus LPM-CLP groups, and lists were compiled for cytokines, chemokines, and their receptors, including the MIF cognate receptor CD74 and noncognate receptors CXCR2 and CXCR4 (Figure 5, A and B). Proinflammatory cytokines such as Tnfa, Il1a, and Il1b were more highly expressed in SPMs than in LPMs, with expression increasing further during CLP (Figure 5A). Additionally, transcript levels for Il15, Il16, Il23a, Il27, and Csf1 (Mcsf) were elevated in SPMs when compared with LPMs. Notably, the only cytokine that showed significantly elevated expression in LPMs under both sham and CLP conditions was Mif (P = 0.00005). By contrast, Mif-2 RNA levels were significantly increased only in the SPM-sham group compared with the LPM-sham group and showed no difference between SPMs and LPMs after CLP induction. Among chemokines, the expression of Ccl2, Ccl6, Ccl7, and Ccl9 was elevated in LPMs when compared with SPMs. These expression data support the strong proinflammatory character of SPMs with high levels of Tnfa, Il1a, and Il1b expression, which is in accord with prior reports (34). MIF can promote TNF-α and IL-1α/β production in monocytes/macrophages (11), and we confirmed prominent MIF-dependent regulation of Tnfa, Il1a, and Il1b mRNA expression by SPMs in contrast to LPMs (Figure 6, A–C).
Figure 5. SPMs exhibit a more proinflammatory phenotype than LPMs.
Visualization of differentially expressed genes in SPM-sham, LPM-sham, SPM-CLP, and LPM-CLP isolated from WT mice 22 hours after surgery showing differentially expressed cytokines and chemokines (A) and receptors (B) as a heatmap with z-scored FPKM values. Underlined genes are discussed in Results.
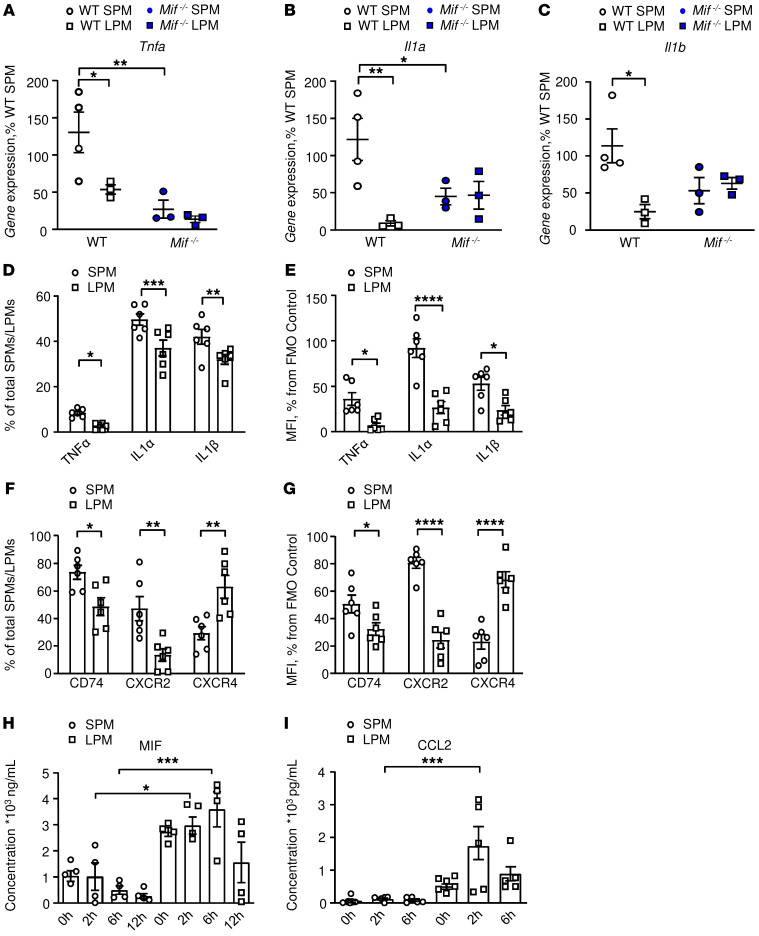
Figure 6. LPMs express higher levels of MIF and CCL2 than SPMs.
(A–C) Confirmation of MIF-dependent regulation of Tnfa, Il1a, and Il1b expression by SPMs in contrast to LPMs. Quantitative expression by qPCR of (A) Tnfa, (B) Il1a, and (C) Il1b in SPMs and LPMs isolated from WT and Mif–/– animals 22 hours after CLP. n = 4 animals per group with replication (2-way ANOVA with Sidak’s multiple-comparison test). (D–G) We confirmed cellular protein expression in SPMs and LPMs by flow cytometry. Intracellular cytokine expression assessed by flow cytometry in post-CLP SPMs and LPMs showing (D) cytokine-expressing macrophages as percentage of total SPMs/LPMs and (E) MFI as percentage of fluorescence-minus-one (FMO) control. (F and G) Flow cytometry analysis of the MIF receptors CD74, CXCR2, and CXCR4 expressed by SPMs and LPMs after CLP, (F) expressed as percentage of total SPMs/LPMs, and as (G) MFI percentage of FMO control. Results are from 3 independent experiments, n = 6 animals per group, 2-way ANOVA with Sidak’s multiple-comparison test. (H and I) In vitro confirmation of induced production of MIF and CCL2 by LPMs versus SPMs. (H) MIF and (I) CCL2 production from WT SPMs and LPMs stimulated with LPS (100 ng/mL, 1 × 105 cells/well). Cytokine concentrations in supernatants were measured by ELISA. The results are from 3 independent experiments, and data are expressed as mean ± SEM, n = 5 to 11 samples per time point. Significance determined by 1-way ANOVA with Tukey’s multiple-comparison test (H) or Kruskal-Wallis test with Dunn’s multiple-comparison test (I). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
The constitutive expression by resident LPMs of Mif and upregulation of the monocyte chemoattractant Ccl2 (Mcp1) during CLP, which is MIF dependent (35, 36), suggest an important role for these cells in the recruitment or retention of SPMs in peritoneum. This hypothesis was further supported by the expression profile of MIF receptors, which showed SPMs to express higher levels than LPMs of the MIF cognate and noncognate receptors Cd74 and Cxcr2, respectively (Figure 5B), suggesting that SPMs were preferentially responsive to MIF during CLP.
We confirmed cellular protein expression by flow cytometry and observed increased production after CLP of TNF-α, IL-1α, and IL-1β by SPMs when compared with LPMs (Figure 6, D and E). SPMs isolated after CLP also expressed increased cell surface CD74 and CXCR2 when compared with LPMs (Figure 6, F and G). We further observed the preferential induced production of MIF and CCL2 by LPMs versus SPMs in vitro (Figure 6, H and I), confirming the in vivo gene expression data. Additionally, we analyzed SPM and LPM cytokine production after LPS stimulation in vitro (Supplemental Figure 4, A–E). The cell supernatants revealed a greater inflammatory character for SPMs, with elevated production of IL-1α and TNF-α, whereas LPMs produced higher amounts of IL-6, IL-10, and in accordance with our prior observations (Figure 6I) and gene expression data of in vivo derived cells, CCL2 (Figure 5).
Taken together, these data support a model where the production of MIF and CCL2 by LPMs acts to recruit and retain SPMs within the peritoneum. SPMs in turn produce high levels of i.p. TNF-α, IL-1α, and IL-1β that contribute to systemic toxicity and lethal shock (Figure 7A).
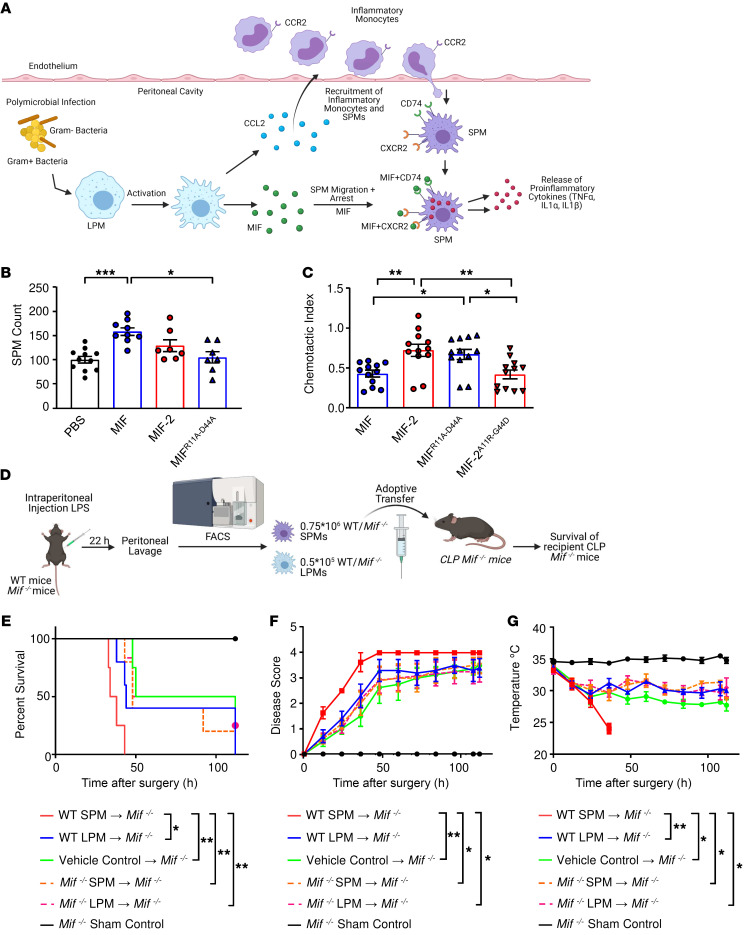
Figure 7. Adoptive transfer of WT SPMs induces premature mortality in Mif–/– CLP mice.
(A) Hypothesis for the role of LPM-expressed MIF and CCL2 in the recruitment and retention of SPMs into peritonea followed by SPM cytokine production. (B) SPM migration into LPS-conditioned peritonea of WT mice measured 12 hours after the i.p. injection of 1 mg/kg recombinant MIF, MIF-2, or MIF lacking the pseudo-(E)LR motif (MIFR11A–D44A). Results are normalized against control group. Data from 3 independent experiments, n = 6 to 12 mice per group (1-way ANOVA with Tukey’s multiple-comparison test). (C) Directed migration in response to CCL2 of SPMs stimulated with recombinant MIF, MIF-2, MIFR11A–D44A, or MIF-2 with recombinant amino acid insertion of the pseudo-(E)LR motif (MIF-2A11R–G44D). Shown is the relative reduction in the CCL2-triggered chemotactic index. Data from 3 independent experiments, n = 12 samples per group (1-way ANOVA with Tukey’s multiple-comparison test). (D) Experimental scheme for the adoptive transfer of WT SPMs/LPMs and Mif–/– SPMs/LPMs into Mif–/– recipient mice. SPMs/LPMs were isolated by FACS from WT and Mif–/– donor mice, 0.75 × 106 SPMs/mouse and 0.5 × 105 LPMs/mouse transferred by i.p. injection into Mif–/– mice after CLP surgery. (E) Kaplan-Meier survival plots; (F) disease score; (G) surface body temperature of Mif–/– CLP mice after adoptive transfer of WT SPM (red), WT LPM (blue), vehicle control (green), Mif–/– SPM (dotted orange), and Mif–/– LPM (dotted magenta). Sham surgery control (black). n = 4 to 6 mice per group from 4 independent experiments. P values determined by log-rank (Mantel-Cox) test (E), Kruskal-Wallis test with Dunn’s multiple-comparison test (F), or 1-way ANOVA with Tukey’s multiple-comparison test (G). Significance for F and G was determined at 36 hours after injection. Images (A and D) created with Biorender.com. *P < 0.05; **P < 0.01; ***P < 0.001.
MIF-triggered SPM migration arrest and recruitment is mediated by CXCR2.
MIF both recruits and arrests migrating leukocytes (37), although specific macrophage subsets have not been examined. We considered that the reduced i.p. accumulation of SPMs in Mif–/– versus Mif-2–/– mice and ensuing reduction in sepsis lethality could be attributable to MIF’s interaction with CXCR2. Although MIF and MIF-2 share 34% sequence identity and a near-identical 3D structure, MIF-2 lacks the pseudo-(E)LR (Arg11, Asp44) motif necessary for MIF’s noncanonical interaction with CXCR2 (37, 38). MIF binding to CXCR2 elicits a promigratory response and MIF’s eponymous migration inhibition function has been attributed to CXCR2 desensitization (37). We tested for SPM recruitment in vivo by injecting recombinant MIF or MIF-2 into endotoxin-conditioned peritoneal cavities and observed increased numbers of SPMs in MIF-injected versus MIF-2–injected (or control) mice (Figure 7B). Notably, MIF-dependent recruitment was abrogated by site-directed mutagenesis of MIF’s pseudo-(E)LR domain (MIFR11A–D44A), which yields a protein more similar in structure to MIF-2 and devoid of the site 2 binding motif for CXCR2 (38).
Using a previously established migration inhibition assay (39), we then tested for the ability of MIF and MIF-2 to arrest SPM migration in response to the monocyte chemoattractant CCL2, which is highly expressed by activated LPMs (Figure 6I). MIF but not MIF-2 reduced SPM-directed migration (Figure 7C). Moreover, migration inhibition activity was eliminated by pseudo-(E)LR mutation, and MIFR11A–D44A showed a similar lack of activity as MIF-2. We confirmed the role of the pseudo-(E)LR in migration arrest by recombinant introduction of Arg11 and Asp44 into MIF-2 to produce MIF-2A11R–G44D, yielding a protein more similar structurally to MIF by the addition of the CXCR2 site 2 binding motif. As shown in Figure 7C, the introduction of these residues into MIF-2 led to acquisition of migration inhibition activity, with MIF-2A11R–G44D reducing the CCL2-directed migration of SPMs to a similar degree as MIF. The data support the role of the pseudo-(E)LR motif in differentiating MIF versus MIF-2 action with respect to SPM accumulation at inflammatory sites of high CCL2 production.
SPMs induces premature mortality in CLP-protected Mif–/– mice.
To confirm the proinflammatory role of SPMs in CLP mortality, we isolated SPMs as well as LPMs from WT and Mif–/– mice via FACS and transferred them into recipient Mif–/– mice that had undergone CLP surgery 2 hours earlier (Figure 7D). The number of SPMs and LPMs transferred was determined according to the subset numbers in WT mice (0.75 × 106 SPMs/mouse and 0.5 × 105 LPMs/mouse; Figure 4B). Mif–/– mice that received adoptively transferred WT SPMs died sooner than the other recipient Mif–/– mice and exhibited higher disease scores and reduced body temperature (Figure 7, E–G). WT LPMs did not restore lethality under these conditions, which is consistent with their less inflammatory character compared with SPMs (Figure 5) and their more upstream (or indirect) role in progression to lethality in the adoptive transfer model. To further support the inflammatory contribution of SPMs to CLP-induced lethality in Mif–/– recipient mice, we performed an adoptive transfer of TNF-α–deficient SPMs/LPMs isolated from Tnfa–/– mice and WT SPMs/LPMs in Mif–/– recipient mice that had undergone CLP surgery 2 hours earlier (Supplemental Figure 5A). Mice receiving Tnfa–/– SPMs showed the greatest degree of protection from mortality and fall in body temperature (Supplemental Figure 5, B–D). These findings affirmed the central position of TNF-α in lethality in the CLP model (40) and support prior observations of MIF’s upstream role in fully enabling TNF action (7, 11).
These data support the conclusion that SPMs drive MIF-dependent mortality in polymicrobial sepsis (Figure 7A). LPMs produce MIF and CCL2 to recruit SPMs into peritonea, leading to an accumulation of the highly inflammatory SPM subpopulation and progression to lethal shock. SPMs express the CCL2 receptor CCR2 (33), and SPM numbers are reduced in Ccr2–/– mice (29). We confirmed the role of CCL2/CCR2 signaling in SPM migration by administering anti-CCL2 antibody to mice prior to CLP. The anti-CCL2–treated mice showed reduced SPM levels when compared with isotype-treated controls (Figure 8A). We also assessed the contribution of MIF to this pathway as CCL2 production has been reported to be reduced by Mif deficiency in unrelated models of inflammation (35, 36). We observed Ccl2 expression to be reduced in Mif–/– LPMs (Figure 8B and refs. 26, 36). Notably, our profiling data also showed reduced Ccr2 expression in CLP versus sham SPMs (Figure 5B), suggesting a downregulation of CCL2 responsiveness and migration after inflammatory activation within the peritoneum.
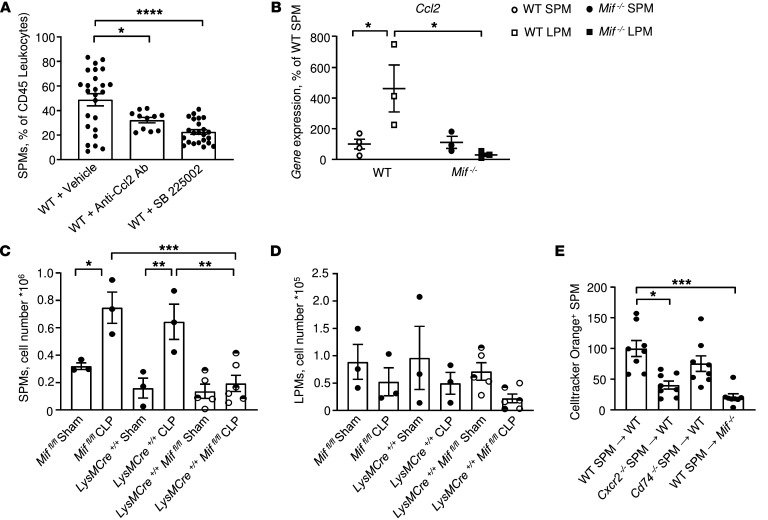
Figure 8. CCL2 and MIF regulation of LPMs and SPMs during i.p. sepsis.
(A) Impact of CCL2 antagonism (R&D Systems, AF-479-NA, 25 μg/kg, i.p.) and CXCR2 antagonism (SB225002, 10 mg/kg, i.p.) on SPM migration into LPS-conditioned peritonea (LPS 12.5 mg/kg, i.p., 2 hours previously). Data are from n = 11 to 19 animals (1-way ANOVA with Tukey’s multiple-comparison test). (B) Quantitative expression of Ccl2 by SPMs and LPMs from WT and Mif–/– mice 22 hours after CLP induction (n = 4 mice per group, 2-way ANOVA with Sidak’s multiple-comparison test). (C and D) Myeloid-derived MIF mediates i.p. SPM accumulation. Absolute cell count analyses of SPMs (C) and LPMs (D) in the peritoneal lavage of LysM-Cre+/+ Miffl/fl, LysM-Cre+/+, or Miffl/fl mice after sham or CLP surgery. LysM-Cre+/– Miffl/fl (visualized in the figure as half-filled circles) demonstrated similar SPM and LPM cell numbers as homozygous LysM-Cre+/+ Miffl/fl mice and were combined as 1 group. Flow cytometric analyses were performed on peritoneal cells collected 22 hours after CLP or sham surgery and included CountBright absolute counting beads in n = 3 animals/genotype. One-way ANOVA with Tukey’s multiple-comparison test. (E) Impact of Cxcr2 or Cd74 deficiency on SPM migration into LPS-conditioned peritonea. SPMs were isolated from WT, Cxcr2–/–, or Cd74–/– mice (after LPS injection, 12.5 mg/kg); labeled with CellTracker Orange and 1 × 106 cells transferred (i.v.) into WT mice; and followed 2 hours later by LPS challenge (12.5 mg/kg, i.p.). Labeled CellTracker Orange+ SPMs were measured in the peritoneal lavage by flow cytometry 18 hours later (Mif–/– recipients also shown as controls). Data are from n = 4 to 8 animals per group and representative of 2 replicate experiments, 1-way ANOVA with Tukey’s multiple-comparison test. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To further assess the cellular source of MIF and whether it is of myeloid nature, we performed CLP and sham surgeries on conditional knockout LysM-Cre+/+ Miffl/fl animals and analyzed the SPM and LPM cell numbers within the peritoneal cavity. Septic LysM-Cre+/+ Miffl/fl mice exhibited reduced numbers of SPMs in mice compared with septic LysM-Cre+/+ or Miffl/fl mice (Figure 8C), while the absolute cell numbers of LPMs (Figure 8D) were not significantly different between the groups after sham or CLP surgery. These findings affirmed the central position of MIF in the recruitment and migration arrest of SPMs within the peritoneal cavity as well as the role of myeloid cells as a source of MIF in septic mice.
MIF additionally can induce monocyte migration via CXCR2 (37), and we examined the potential contribution of MIF/CXCR2 signaling in SPM recruitment by treating mice with the CXCR2 antagonist SB225002. The CXCR2 antagonist reduced SPM levels by approximately 50% when compared with untreated mice (Figure 8A). CXCR2 may form a cell surface complex with the MIF cognate receptor CD74 (37), and we clarified the role of these alternate receptors by enumerating the recruitment of fluorescently labeled Cxcr2–/– or Cd74–/– SPMs into the peritonea of CLP-induced mice (Figure 8E). Cxcr2–/– but not Cd74–/– SPMs showed reduced migration into peritonea and, as expected, SPM migration was low in Mif–/– hosts. These data, taken together, support the functional role of both the CCL2/CCR2 and MIF/CXCR2 signaling pathways in SPM migration into infected peritonea.
Discussion
The MIF superfamily members MIF and MIF-2 are highly conserved in their genomic organization, amino acid sequence, and 3D structure (16). MIF has been well studied in different infectious conditions and as a genetic modifier of clinical outcome (7, 9, 12–15, 41–43), but much less is known about MIF-2. The 2 proteins are coordinately expressed during inflammation and show evidence of functional overlap by activation of the common MIF family receptor CD74. An analysis of circulating levels indicated that the 2 proteins also correlate similarly with APACHE II disease severity scores (16, 21). In contrast to MIF, the human MIF-2 gene (DDT) lacks common polymorphisms, and recent studies suggest that MIF-2 may exert a more constitutive role in CD74-dependent cytoprotection and tissue repair (44). To our knowledge, the present study is the first to investigate Mif-2–/– mice in a comparative survival analysis and revealed a critical and distinct role for MIF in the recruitment of SPMs during polymicrobial sepsis. Although MIF and MIF-2 were produced in similar levels in peritonea after CLP, Mif-2–/– mice showed similar survival and disease severity indices as WT mice. This result contrasts with prior data showing protection by immunoneutralization of MIF or MIF-2 in endotoxemia (16) and affirms the limitations of the endotoxic shock model in investigating host responses to life-threatening infection (45).
This study also illuminates the role of discrete macrophage subpopulations and their network of interactions in the host response to sepsis. Our data indicate that CLP led to an i.p. enrichment of highly inflammatory SPMs, which were reduced in Mif–/– but not Mif-2–/– mice. Transcriptional profiling confirmed by cytokine production analysis indicated that although resident LPMs had a less inflammatory character than SPMs during peritoneal infection, with reduced expression of Tnfa, Il1a, and Il1b, LPMs expressed higher levels of Ccl2 and Mif than SPMs. CCL2 and MIF were also preferentially induced by LPMs compared with SPMs in response to inflammatory stimulation, which is consistent with their role in promoting SPM migration and retention within infected peritoneum (Figure 7A). Mif–/– mice with polymicrobial sepsis also showed reduced circulating levels of CCL2 and decreased expression of Ccl2 by LPMs, which is in accord with MIF’s known role in upregulating CCL2 production in other contexts (35, 36). SPMs also expressed higher levels of the MIF receptors CD74 and CXCR4 than LPMs, further supporting the notion that SPMs are a critical MIF-responsive effector population in peritoneal infection. Finally, MIF deficiency was associated with a reduction in the intrinsic proinflammatory character of SPMs, with decreased Tnfa, Il1a, and Il1b expression. Experimentally, the delayed onset of lethality from sepsis associated with reduced accumulation of i.p. SPMs in Mif–/– mice was diminished by the adoptive transfer of WT SPMs.
We provide mechanistic evidence for differential MIF versus MIF-2 action in polymicrobial sepsis based on MIF’s noncanonical pseudo-(E)LR domain, which mediates CXCR2 interaction and is absent in MIF-2 (38). MIF inhibits the directed migration of peripheral blood monocytes by a mechanism that involves CXCR2 receptor desensitization (37–39), and we showed that MIF but not MIF-2 arrested the CCL2-directed migration of SPMs. Site-directed mutagenesis of the 2 residues that comprise the MIF pseudo-(E)LR domain, which represents the site 2 binding motif for CXCR2, eliminated both migration and migration inhibition activity, while the recombinant substitution of these 2 residues into homologous positions into MIF-2 produced de novo migration inhibition activity. The role of CXCR2 signaling was confirmed with Cxcr2–/– SPMs and CXCR2 antagonist–treated mice, which showed reduced SPM migration and lower i.p. SPM counts.
Current paradigms of sepsis pathophysiology emphasize the tightly regulated balance between proinflammatory and antiinflammatory responses for eliminating invasive pathogens while restricting excessive tissue-damaging inflammation (2, 46). MIF acting through SPMs regulates this network of interactions during i.p. infection. Resident LPMs produce MIF, along with chemokines such as CCL2, to promote SPM entry into infected peritoneum. Transfer of WT LPMs did not lead to a higher mortality rate in CLP-challenged Mif–/– mice, suggesting that other cell types may be important contributory sources of MIF for the recruitment or activation of i.p. SPMs. The present findings nevertheless indicate that high i.p. MIF levels promote the migration arrest of inflammatory SPMs, resulting in the high expression of tissue-damaging cytokines. The enhanced survival of LysM-Cre+/+ Miffl/fl mice after CLP additionally affirmed the critical role of the myeloid compartment in producing the MIF necessary for sepsis lethality.
In summary, these data reinforce MIF’s central role in the pathophysiology of polymicrobial sepsis, as first revealed when anti-MIF was found to protect mice from lethal CLP or E. coli injection even when administered 8 hours after insult (7). High-expression MIF alleles, which occur commonly in the population, have since been described to be associated with adverse outcomes in infections, such as invasive pneumococcus (13) or meningococcemia (14), where excessive inflammation dominates clinical manifestations. The present study identified an important distinguishing feature between MIF and MIF-2 in the immune response to sepsis, but a specific role of MIF-2 in sepsis physiology remains to be established. We note accruing literature supporting roles for MIF-2 in tissue protection and the homeostatic response to cellular stress, which involve CD74-dependent activation of the energy sensor AMPK and the transcription factors eIF2α and ATF (44, 47). Such reparative pathways are likely to be coordinately activated during sepsis, and a closer examination of MIF-2 function in sepsis-induced tissue injury, for instance in the lung, kidney, and heart, is warranted (47–50).
The present work also prompts closer attention to the specific signaling pathways that govern the maintenance and function of specialized macrophage subpopulations. More selective interventions directed at the LPM/SPM signaling network and potentially at MIF in genetically at-risk hosts (22, 23) could offer new therapeutic approaches to reducing mortality in discrete sepsis syndromes.
Methods
Study design.
The primary objective of this study was to identify functional differences between the cytokine MIF and its structural homologue MIF-2 and their regulatory influence on 2 macrophage subsets in septic mice. We developed Mif-2–/– mice and elucidated the functional characteristics of MIF-2 and MIF in comparative studies using Mif–/– and WT mice. All mice were generated in a C57BL/6J background and all parameters of genetically modified mice were compared with littermate controls. Mice were age-matched within 2 weeks for each experiment. For surgical experiments, at least 4 animals were included in each group and sample sizes were predetermined based on previous experience using a minimum of 3 mice per group. After inducing sepsis in mice, the animals were randomized into separate groups for cell or protein administration, and where possible, treatment groups were blinded until statistical analysis.
Mice.
Mif–/– mice lacking the entire Mif gene (promoter and 3 exons) and maintained in the pure C57BL/6J background have been described previously (51). We generated a constitutive Mif-2–/– mouse strain as previously described (44). Cxcr2tm1Mwm mice (37) (also known as mIL-8Rh-, stock number 002724) were obtained from the Jackson Laboratory. C57BL/6 WT controls and breeding pairs of Cx3cr1GFP and Tnfa–/– mice were purchased from Charles River Laboratories. To generate LysM-Cre Miffl/fl mice, mice expressing Cre recombinase under the LysM promoter were intercrossed with Miffl/fl mice (51). LysM-Cre+/– Miffl/fl, LysM-Cre+/+, and Miffl/fl mice were bred and included as experimental controls.
All mouse strains were bred and maintained in the Yale School of Medicine animal facility. All experiments were performed in 6- to 12-week-old animals that were age matched (within 2 weeks) for each experiment.
CLP model of polymicrobial sepsis.
CLP surgery was performed as described previously (52). Briefly, a midline laparotomy incision was made, the cecum exteriorized, and 80% of the cecum was ligated and through-and-through punctured with a 21G needle followed by extrusion of a small drop of fecal contents. The cecum was then returned to the peritoneal cavity, and the incision was closed in 2 layers (muscle and skin). For sham surgeries, the cecum was exteriorized and returned to the peritoneum without ligation or puncture. Mice received 1 mL warm PBS solution (s.c.) immediately after surgery and buprenorphine analgesia at 4- to 12-hour intervals. Postoperatively, mice were monitored for survival, disease score (0, bright, alert, responsive; 1, slightly lethargic; 2, lethargic and hunched; 3, very lethargic and shaky; 4, dead), and surface body temperature, which was measured with an infrared thermometer.
Measurements of MIF, MIF-2, CCL2, CK levels, and CFUs.
At 0, 4, 9, and 22 hours after surgery, blood was drawn from mice by retroorbital bleeding and prepared in separator tubes (BD Pharmingen). MIF and MIF-2 levels were measured in plasma or cell-free peritoneal lavage by ELISA (16).
CK levels in plasma collected at 22 hours after surgery were measured with the CK reagent set (Pointe Scientific) according to the manufacturer’s recommendations. For determination of CFUs, heparinized whole blood was collected 22 hours after surgery, serially diluted with sterile water, plated on BBL agar plates (BD Pharmingen), and incubated at 37°C for 24 hours. Bacterial colonies were then counted.
MIF and CCL2 levels were measured in SPM/LPM cell culture supernatants after 0, 2, 6, or 12 hours with LPS (Sigma-Aldrich L2630, 100 ng/mL) using CCL2 ELISA (eBioscience) or MIF ELISA (Abcam), respectively, according to the manufacturer’s instructions.
Flow cytometry and cytometric bead array.
Mice were euthanized at 22 hours after surgery. Abdominal fur was removed and 3 mL sterile ice-cold PBS injected twice into the peritoneal cavity. The abdomen was gently massaged and the fluid aspirated. After red blood cell lysis by ACK lysing buffer (Lonza), cells were labeled at 4°C for 20 to 30 minutes in FACS buffer in the presence of mouse FC block (clone 2.4G2, 553141, BD Pharmingen) with the following fluorophore-conjugated antibodies: CD11b–Alexa Fluor 700 (clone M1/70, 56-0112-82), F4/80–eFluor 450 (clone BM8, 48-4801-82), CD19-APC (clone eBio1D3, 17-0193-82), CD44-APC (clone IM7, 17-0441-82), CXCR4-APC (clone 2B11, 17-9991-82) (prior antibodies from eBioscience); Gr1-FITC (clone RB6-8C5, 108405), CD3-PECy7 (clone 17A2, 100219), CXCR2-PerCP (clone SA045E1, 149605), CD11c-PeCy7 (clone N418, 117317), CD3-APCCy7 (clone 17A2, 100221), CD19-APCCy7 (clone 6D5, 115529), CD45-APC (clone 30-F11, 103111), CD45-PE (clone 30-F11, 103105), CD45-PerCP (clone 30-F11, 103129) (prior antibodies from BioLegend); SiglecF-PE (clone E50-2440, 562068, BD Pharmingen), or CD74-PE (clone ln-2, sc-6262 PE, Santa Cruz Biotechnology). For intracellular cytokine staining, the cells were then fixed, permeabilized using Foxp3 fix/perm buffer (eBioscience) for 30 minutes, and labeled with the following fluorophore-conjugated antibodies: TNF-α–PE (clone MP6-XT22, 12-7321-41, eBioscience), IL-1α–PE (clone ALF-161, 503203, BioLegend), IL-1β–PE (clone 166931, IC4013P, R&D Systems), and Ki67–PE-Cy7 (clone SolA15, 25-5698-82, Invitrogen/Thermo Fisher Scientific). Cell proliferation was assessed by intracellular staining of proliferation marker Ki67. Flow cytometry was performed on an LSRII cytometer (BD Pharmingen) and analyzed with Flow Jo software (Tree Star).
To determine absolute peritoneal cell counts, peritoneal cells were isolated from mice after CLP or sham surgery by peritoneal lavage as described in the prior paragraph. After red blood cell lysis, cells were surface stained at 4°C for 20 to 30 minutes in FACS buffer in the presence of mouse FC block with fluorophore-conjugated antibodies. After surface staining, CountBright absolute counting beads (Invitrogen/Thermo Fisher Scientific) were added to every sample and the samples analyzed by flow cytometry on an LSRII cytometer.
Cytokine levels in the blood plasma and peritoneal lavage of WT, Mif–/–, and Mif-2–/– mice were measured by flow cytometry using the mouse inflammation LEGENDplex Multi Analyte Flow Assay Kit (BioLegend).
Cytospin.
Peritoneal lavage was obtained from mice 22 hours after CLP surgery and cells stained for flow cytometry as described above. SPMs and LPMs were sorted by FACS and spun onto slide chambers with a cytocentrifuge (Thermo Fisher Scientific). Subsequently, slides were stained with Wright-Giemsa stain (Merck Millipore) according to the manufacturer’s recommendations. ImageJ software (NIH) was used to analyze the cell size of SPMs and LPMs.
RNA-Seq and differential gene expression analysis.
Sequencing and expression analysis were performed individually on SPMs and LPMs from triplicate samples. SPMs and LPMs were isolated by FACS of peritoneal lavage from 6 independent donor mice after sham or CLP surgery constituting 1 sample. Total RNA was extracted using the RNeasy Plus Mini MinElute Cleanup Kit (Qiagen). Total RNA quality was determined by estimating the A260/A280 and A260/A230 ratios using a NanoDrop spectrophotometer (Thermo Fisher Scientific) and RNA integrity was verified by Agilent 2100 Bioanalyzer based on the relative abundance of 18S and 28S rRNA. Twelve sequencing libraries were produced by the Illumina TruSeq stranded protocol for 76-bp paired-end sequencing using Illumina HiSeq 2500. Adapter sequences, empty reads, and low-quality sequences were removed. The nucleotides at the 5′ and 3′ end with a quality score below 20 for each read were trimmed using in-house scripts, and read pairs with either end shorter than 45 bp after trimming were discarded. Reads passing quality control were aligned using Tophat v.2.0.13 (53) to perform spliced alignment of the reads against to the reference UCSC mouse genome and transcript annotation. Only the reads that mapped to a single unique location within the genome, with a maximum of 2 mismatches in the anchor region of the spliced alignment, were reported in these results. We used the default settings for all other Tophat options. Tophat alignments were then processed by Cufflinks v2.2.1 (54) to quantify the abundance of each transcript. The transcript abundance was measured in fragments per kb of exon per million mapped fragments (FPKM) to normalize the read count of a transcript by both its length and library size. These normalized transcript abundances then were analyzed to identify differential gene expression between conditions using Cuffdiff (55) with default options. After differential gene expression analysis, the significantly differentially expressed genes in the SPM-sham, LPM-sham, SPM-CLP, and LPM-CLP groups were chosen using a cutoff of FDR-controlled P value less than 0.05. The significantly differentially expressed identified from SPM-sham versus LPM-sham were compared with the those identified from SPM-CLP versus LPM-CLP. These comparisons revealed 2629 genes to be differentially regulated between SPM and LPM under the sham condition and 1653 genes to be differentially regulated between SPM and LPM only under the CLP condition (Supplemental Figure 6). Next, 223 genes were found to be differentially regulated between SPMs and LPMs and showed opposite regulation in sham versus CLP conditions (Supplemental Figure 6). Overlapping genes (3991 genes) were then analyzed for the differential expression of cytokines, chemokines, chemokine receptors, and MIF/MIF-2–related receptors between SPM and LPM in both sham and CLP conditions with a cutoff value of fold-change greater than 0.7. FPKM values of differentially expressed genes were visualized in heatmaps and z score normalized. Gene expression data files are available upon publication in the International MIF Consortium database (http://www.biochemmcb.rwth-aachen.de/mif_consortium_ public/index.php).
Quantitative real-time PCR.
Messenger RNA from WT and Mif–/– SPMs and LPMs was extracted by the RNeasy Mini Kit (Qiagen) and cDNA synthesis was performed with the QuantiTect Reverse Transcription Kit (Qiagen) following the manufacturer’s instructions. Complementary DNA was subject to quantitative real-time PCR (qPCR) using SYBR Green Master Mix (Bio-Rad Laboratories, Inc.). Using the following specific primer pairs, the expression of Ccl2, Il1a, Il1b, Tnfa, and Gapdh was determined: 5′-TTCTTCGATTTGGGTCTCCTTG and 5′-GTGCAGCTCTTGTCGGTGAA (for Ccl2); 5′-TCTATGATGCAAGCTATGGCTCA and 5′-CGGCTCTCCTTGAAGGTGA (for Il1a), 5′-GAAATGCCACCTTTTGACAGTG and 5′-TGGATGCTCTCATCAGGACAG (for Il1b), 5′-GCGGCCACAGAAAACACTC and 5′-CTCCCAATGGTCAAGGCATC (for Tnfa), 5′-AATGGATTTGGACGCATTGGT and 5′-TTTGCACTGGTACGTGTTGAT (for Gapdh). GAPDH was used as a reference gene. Data were analyzed with the comparative cycle time (CT) method.
In vitro confirmation of SPM and LPM cytokine phenotype.
To assess cytokine production, SPMs and LPMs were isolated from the peritoneal cavities of WT mice after 20 hours of LPS challenge (i.p., 6 mg/kg, Sigma-Aldrich, L2630). SPMs and LPMs then were sorted via FACS as previously described and seeded (1 × 105 cells/well) into 96-well plates in culture media (RPMI + L-glutamine, 10% FBS, 1% penicillin-streptomycin) followed by stimulation with 100 ng/mL LPS (Sigma-Aldrich, L2630). We measured the cell supernatant cytokine levels at different time points using the mouse inflammation LEGENDplex Multi Analyte Flow Assay Kit.
Recombinant MIF and MIF-2 and pseudo-(E)LR mutagenesis.
Recombinant mouse MIF, MIF-2, and mutant proteins were produced and purified as previously described (19), including modifications described in a previous study (56). Briefly, recombinant proteins were expressed in E. coli and purified by anion exchange chromatography (Q-Sepharose) followed by reversed-phase chromatography (C18 column) and acetonitrile gradient elution. The eluted proteins were lyophilized, refolded, and confirmed to have very low endotoxin content (<1.9 EU/μg protein, reference E. coli 055:B5). Mouse pseudo-(E)LR mutant MIFR11A–D44A was generated by site-directed mutagenesis, expressed, and purified as previously described (38). Mouse pseudo-(E)LR mutant MIF-2A11R–G44D was produced using a synthetic oligonucleotide optimized for E.coli expression. The MIF-2A11R–G44D coding DNA was cloned into pUC57, subcloned into the pET-11b expression vector, and expressed in BL21DE3 Gold cells (16).
In vitro and in vivo migration assays.
In vitro macrophage migration studies were conducted in Transwell migration chambers (8 μm pore size, Corningen) with thioglycollate-elicited peritoneal macrophages. Cells were stimulated with recombinant MIF, MIF-2, MIFR11A–D44A, MIF-2A11R–G44D (200 ng/mL) and allowed to migrate toward CCL2 (10 ng/mL) in the presence of different MIF proteins for 22 hours. Macrophages migrated through the insert pores to the opposite side of the insert membrane and remained adherent there. After 22 hours, the inserts were washed with PBS and nonmigrating cells remaining on the upper surface of the insert were removed with a cotton swab. Migrated cells adherent to insert were fixed and stained with Wright-Giemsa stain according to the manufacturer’s recommendations, counted, and the chemotactic index calculated by dividing the number of migrated macrophages in the presence of the cytokines by the number of migrated macrophages of the chemokinesis control (no cytokines).
In vivo macrophage migration was studied in WT mice after 12 mg/kg LPS (Sigma-Aldrich, L2630) injection (i.p.). Recombinant MIF, MIF-2, and MIFR11A–D44A proteins (1 mg/kg) were i.p. administered 6 hours after LPS injection (Sigma-Aldrich, L2630). Mice were euthanized 12 hours after protein administration and peritoneal cells were collected by peritoneal lavage. Cells then were stained with CD11b–Alexa Fluor 700 (clone M1/70, 56-0112-82, eBioscience), F4/80–eFluor 450 (clone BM8, 48-4801-82, eBioscience), and CD45-PerCP (clone 30-F11, 103129, BioLegend). The number of migrated SPMs after protein injection was quantified by flow cytometry. As the vehicle control, WT mice were injected with PBS instead of recombinant proteins.
Antibodies and antagonists.
Anti-CCL2 studies were conducted by injecting mice with anti-CCL2 antibody (R&D Systems, AF-479-NA, i.p., 25 μg/kg) 2 hours before LPS challenge (Sigma-Aldrich, L2630, i.p., 12.5 mg/kg). CXCR2 blockade was with the CXCR2 antagonist (Santa Cruz Biotechnology, SB225002, i.p., 10 mg/kg; ref. 57) 2 hours before LPS injection (i.p., 12.5 mg/kg). Vehicle controls received PBS or DMSO. Peritoneal lavage SPM content was evaluated by flow cytometry 18 hours after LPS injection.
Adoptive transfer of SPMs.
Peritoneal lavage cells were collected from WT and Mif–/– mice 22 hours after i.p. LPS injection (12.5 mg/kg) and SPMs and LPMs isolated by FACS (FACSAria II cytometer, BD Pharmingen) employing the antibodies CD11b–Alexa Fluor 700 (clone M1/70, 56-0112-82, eBioscience), F4/80–eFluor 450 (clone BM8, 48-4801-82, eBioscience), and CD45-PerCP (clone 30-F11, 103129, BioLegend). Approximately 0.75 × 106 SPMs/mouse and 0.5 × 105 LPMs/mouse then were i.p. injected into recipient Mif–/– mice that had undergone CLP surgery 2 hours prior to injection. Mif–/– mice injected with vehicle (PBS) served as controls. Subsequently, mice were monitored for survival, disease score, and surface body temperature as described above. Additional adoptive transfer studies were performed in which SPMs/LPMs of Tnfa–/– mice and WT SPMs/LPMs were isolated via FACS and injected into Mif–/– recipient mice that had undergone CLP surgery 2 hours earlier.
Adoptive transfer of CellTracker Orange–labeled SPMs.
SPMs isolated by FACS from the LPS-conditioned peritoneal cavities (12.5 mg/kg, i.p.) of Cxcr2–/–, Cd74–/–, or WT mice were labeled with CellTracker Orange CMFDA dye (Invitrogen) according to the manufacturer’s instructions. Briefly, 1 × 106 cells were incubated with 1 μM dye in warm PBS for 30 minutes at 30°C. After staining, the cells were washed and resuspended in 1 × PBS. Next, 1 × 106 cells then were injected into WT or Mif–/– recipient mice through the retroorbital plexus. Two hours later, the recipient mice were challenged with LPS (12.5 mg/kg, i.p.), and peritoneal lavage fluid was obtained 18 hours later. After red blood cell lysis (ACK lysis buffer, Sigma-Aldrich), the cells were surface stained with CD45-PerCP (clone 30-F11, 103129, BioLegend), CD11b–Alexa Fluor 700 (clone M1/70, 56-0112-82, eBioscience), CD115-APC (clone AFS98, 135510, BioLegend), Gr1-FITC (clone RB6-8C5, 108405, BioLegend), and F4/80–eFluor 450 (clone BM8, 48-4801-82, eBioscience) at 4°C for 30 minutes and analyzed by flow cytometry on an LSRII cytometer (BD Pharmingen) and analyzed with Flow Jo software (Tree Star).
Statistics.
Data are representative of at least 3 independent experiments (unless stated otherwise), and statistical analyses were conducted using GraphPad Prism software. Results are expressed as mean ± SEM. Statistical tests for each graph are described in figure legends. P values of less than 0.05 were considered significant.
Study approval.
All experimental procedures were approved by the Yale University IACUC and conducted in accordance with the IACUC’s guidelines.
Author contributions
PVT designed the study; conducted plasma cytokine measurements; performed FACS experiments for intracellular cytokine measurements, cell surface receptor expression and confirmation of SPM origin; conducted in vivo and in vitro migration assays; conducted qPCR experiments; performed MIF/CCL2 ELISA of SPMs and LPMs; performed cytokine bead arrays; performed adoptive transfer experiments and survival studies; conducted antibody/antagonist treatment studies; illustrated the SPM/LPM network; generated and established mouse strains; conducted statistical analysis; and wrote the manuscript. WS designed pilot experiments, performed pilot FACS experiments to identify macrophage populations, established the Mif-2–/– mouse strain, designed and conducted survival studies, performed MIF/MIF-2 ELISA, measured CK levels and CFU, performed RNA-Seq experiments, and assisted with manuscript writing. TH designed and conducted initial flow cytometry analysis. BSK assisted with RNA-Seq data analysis. JN and MS assisted with data acquisition and analysis. MP assisted with mouse strain generation and provided technical assistance. GP, EL, and JB contributed new reagents and analytic tools. LL assisted with general experimental design and provided thoughtful discussions. GFR designed the mouse strain. RB supervised the project, provided grant support, designed the study, analyzed data, and finalized the manuscript.
Supplementary Material
Acknowledgments
We thank the Yale Center for Genome Analysis (YCGA) for performing the RNA-Seq analysis. We thank Francesco Lopez and Xiaoqing Yu for their assistance with the gene expression profile analysis. We also thank Christine Wong and the Goldstein lab for providing us with Cx3Cr1GFP mice. We thank Wendy Walker for her assistance with the CLP model and Scott Roberts for the technical assistance. The authors were supported by research fellowships of the Deutsche Forschungsgemeinschaft (DFG) SCHU2851/1-1 (to WS) and KI1973/1-1 (to BSK); the START program of RWTH Aachen University 691346, START 2013-2 (to BSK); DFG grants SFB 1123/A03, BE1977/7-1, and BE1977/11-1 (to JB); NIH grants AR049610, AR078334, AI 5R01-51306, and HL130669 and the Rheumatology Research Foundation (to RB); NIH grants R01 HL155948 and K08HL135402 (to MS); and an Arthritis Foundation Award 548970 (to PVT). WS is a participant in the BIH Charité Clinician Scientist Program funded by the Charité — Universitätsmedizin Berlin and the Berlin Institute of Health.
Version 1. 12/01/2021
Electronic publication
Footnotes
Conflict of interest: RB and JB are listed as co-inventors on issued patents (US20120149044A1 and US20100267714A1) for MIF antagonists.
Copyright: © 2021, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2021;131(23):e127171.https://doi.org/10.1172/JCI127171.
Contributor Information
Pathricia Veronica Tilstam, Email: pathricia_tilstam@hms.harvard.edu.
Wibke Schulte, Email: wibke.schulte@charite.de.
Thomas Holowka, Email: tomholowka@gmail.com.
Bong-Sung Kim, Email: Bong-Sung.Kim@usz.ch.
Jessica Nouws, Email: jessica.nouws@yale.edu.
Maor Sauler, Email: maor.sauler@yale.edu.
Marta Piecychna, Email: Marta.piecychna@yale.edu.
Georgios Pantouris, Email: gpantouris@PACIFIC.EDU.
Elias Lolis, Email: elias.lolis@yale.edu.
Lin Leng, Email: lin.leng@yale.edu.
Günter Fingerle-Rowson, Email: g.fingerle-rowson@gmx.de.
Richard Bucala, Email: richard.bucala@yale.edu.
References
- 1.Vincent JL, et al. Assessment of the worldwide burden of critical illness: the intensive care over nations (ICON) audit. Lancet Respir Med. 2014;2(5):380–386. doi: 10.1016/S2213-2600(14)70061-X. [DOI] [PubMed] [Google Scholar]
- 2.Hotchkiss RS, et al. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. 2013;13(12):862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volbeda M, et al. Glucocorticosteroids for sepsis: systematic review with meta-analysis and trial sequential analysis. Intensive Care Med. 2015;41(7):1220–1234. doi: 10.1007/s00134-015-3899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown KA, et al. Targeting cytokines as a treatment for patients with sepsis: a lost cause or a strategy still worthy of pursuit? Int Immunopharmacol. 2016;36:291–299. doi: 10.1016/j.intimp.2016.04.041. [DOI] [PubMed] [Google Scholar]
- 5.Marti-Carvajal AJ, et al. Human recombinant protein C for severe sepsis and septic shock in adult and paediatric patients. Cochrane Database Syst Rev. 2012;12:CD004388. doi: 10.1002/14651858.CD004388.pub6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwashyna TJ, et al. Long-term cognitive impairment and functional disability among survivors of severe sepsis. JAMA. 2010;304(16):1787–1794. doi: 10.1001/jama.2010.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calandra T, et al. Protection from septic shock by neutralization of macrophage migration inhibitory factor. Nat Med. 2000;6(2):164–170. doi: 10.1038/72262. [DOI] [PubMed] [Google Scholar]
- 8.Roger T, et al. Macrophage migration inhibitory factor promotes innate immune responses by suppressing glucocorticoid-induced expression of mitogen-activated protein kinase phosphatase-1. Eur J Immunol. 2005;35(12):3405–3413. doi: 10.1002/eji.200535413. [DOI] [PubMed] [Google Scholar]
- 9.Das R, et al. Macrophage migration inhibitory factor (MIF) is a critical mediator of the innate immune response to Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2013;110(32):E2997–E3006. doi: 10.1073/pnas.1301128110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calandra T, et al. MIF as a glucocorticoid-induced modulator of cytokine production. Nature. 1995;377(6544):68–71. doi: 10.1038/377068a0. [DOI] [PubMed] [Google Scholar]
- 11.Mitchell RA, et al. Macrophage migration inhibitory factor (MIF) sustains macrophage proinflammatory function by inhibiting p53: regulatory role in the innate immune response. Proc Natl Acad Sci U S A. 2002;99(1):345–350. doi: 10.1073/pnas.012511599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yende S, et al. The influence of macrophage migration inhibitory factor gene polymorphisms on outcome from community-acquired pneumonia. FASEB J. 2009;23(8):2403–2411. doi: 10.1096/fj.09-129445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Savva A, et al. Functional polymorphisms of macrophage migration inhibitory factor as predictors of morbidity and mortality of pneumococcal meningitis. Proc Natl Acad Sci U S A. 2016;113(13):3597–3602. doi: 10.1073/pnas.1520727113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Renner P, et al. A functional microsatellite of the macrophage migration inhibitory factor gene associated with meningococcal disease. FASEB J. 2012;26(2):907–916. doi: 10.1096/fj.11-195065. [DOI] [PubMed] [Google Scholar]
- 15.Das R, et al. Functional polymorphisms in the gene encoding macrophage migration inhibitory factor are associated with Gram-negative bacteremia in older adults. J Infect Dis. 2014;209(5):764–768. doi: 10.1093/infdis/jit571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Merk M, et al. The D-dopachrome tautomerase (DDT) gene product is a cytokine and functional homolog of macrophage migration inhibitory factor (MIF) Proc Natl Acad Sci U S A. 2011;108(34):E577–E585. doi: 10.1073/pnas.1102941108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beishuizen A, et al. Macrophage migration inhibitory factor and hypothalamo-pituitary-adrenal function during critical illness. J Clin Endocrinol Metab. 2001;86(6):2811–2816. doi: 10.1210/jcem.86.6.7570. [DOI] [PubMed] [Google Scholar]
- 18.Brenner T, et al. Macrophage migration inhibitory factor (MIF) and manganese superoxide dismutase (MnSOD) as early predictors for survival in patients with severe sepsis or septic shock. J Surg Res. 2010;164(1):e163–171. doi: 10.1016/j.jss.2010.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Bernhagen J, et al. MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature. 1993;365(6448):756–759. doi: 10.1038/365756a0. [DOI] [PubMed] [Google Scholar]
- 20.Calandra T, et al. Macrophage migration inhibitory factor is a critical mediator of the activation of immune cells by exotoxins of Gram-positive bacteria. Proc Natl Acad Sci U S A. 1998;95(19):11383–11388. doi: 10.1073/pnas.95.19.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pohl J, et al. Elevated MIF-2 levels predict mortality in critically ill patients. J Crit Care. 2017;40:52–57. doi: 10.1016/j.jcrc.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 22.Wallace D, et al. IMMU-115 (humanized anti-CD74 antibody) for subcutaneous administration: a phase Ib study in patients with systemic lupus erythematosus (SLE) Ann Rheum Dis. 2016;75:291. doi: 10.1136/annrheumdis-2016-eular.6397. [DOI] [Google Scholar]
- 23.Fox RJ, et al. Phase 2 trial of ibudilast in progressive multiple sclerosis. N Engl J Med. 2018;379(9):846–855. doi: 10.1056/NEJMoa1803583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore LJ, et al. The epidemiology of sepsis in general surgery patients. J Trauma. 2011;70(3):672–680. doi: 10.1097/TA.0b013e31820e7803. [DOI] [PubMed] [Google Scholar]
- 25.Cannon JG, et al. Circulating interleukin-1 and tumor necrosis factor in septic shock and experimental endotoxin fever. J Infect Dis. 1990;161(1):79–84. doi: 10.1093/infdis/161.1.79. [DOI] [PubMed] [Google Scholar]
- 26.Xie J, et al. Macrophage migration inhibitor factor upregulates MCP-1 expression in an autocrine manner in hepatocytes during acute mouse liver injury. Sci Rep. 2016;6:27665. doi: 10.1038/srep27665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leng L, et al. A small-molecule macrophage migration inhibitory factor antagonist protects against glomerulonephritis in lupus-prone NZB/NZW F1 and MRL/lpr mice. J Immunol. 2011;186(1):527–538. doi: 10.4049/jimmunol.1001767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deshmane SL, et al. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res. 2009;29(6):313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ajuebor MN, et al. Endogenous monocyte chemoattractant protein-1 recruits monocytes in the zymosan peritonitis model. J Leukoc Biol. 1998;63(1):108–116. doi: 10.1002/jlb.63.1.108. [DOI] [PubMed] [Google Scholar]
- 30.Walker W, et al. Changes in peritoneal macrophage composition during sepsis. J Immunol. 2018;200(1 suppl):42.4 [Google Scholar]
- 31.Ayala A, et al. Sepsis-induced potentiation of peritoneal macrophage migration is mitigated by programmed cell death receptor-1 gene deficiency. J Innate Immun. 2014;6(3):325–338. doi: 10.1159/000355888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosn EE, et al. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc Natl Acad Sci U S A. 2010;107(6):2568–2573. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cain DW, et al. Identification of a tissue-specific, C/EBPβ-dependent pathway of differentiation for murine peritoneal macrophages. J Immunol. 2013;191(9):4665–4675. doi: 10.4049/jimmunol.1300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cassado Ados A, et al. Revisiting mouse peritoneal macrophages: heterogeneity, development, and function. Front Immunol. 2015;6:225. doi: 10.3389/fimmu.2015.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gregory JL, et al. Macrophage migration inhibitory factor induces macrophage recruitment via CC chemokine ligand 2. J Immunol. 2006;177(11):8072–8079. doi: 10.4049/jimmunol.177.11.8072. [DOI] [PubMed] [Google Scholar]
- 36.Das R, et al. Macrophage migration inhibitory factor promotes clearance of pneumococcal colonization. J Immunol. 2014;193(2):764–772. doi: 10.4049/jimmunol.1400133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bernhagen J, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13(5):587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 38.Weber C, et al. Structural determinants of MIF functions in CXCR2-mediated inflammatory and atherogenic leukocyte recruitment. Proc Natl Acad Sci U S A. 2008;105(42):16278–16283. doi: 10.1073/pnas.0804017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hermanowski-Vosatka A, et al. Enzymatically inactive macrophage migration inhibitory factor inhibits monocyte chemotaxis and random migration. Biochemistry. 1999;38(39):12841–12849. doi: 10.1021/bi991352p. [DOI] [PubMed] [Google Scholar]
- 40.Schulte W, et al. Cytokines in sepsis: potent immunoregulators and potential therapeutic targets--an updated view. Mediators Inflamm. 2013;2013:165974. doi: 10.1155/2013/165974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakuragi T, et al. Lung-derived macrophage migration inhibitory factor in sepsis induces cardio-circulatory depression. Surg Infect (Larchmt) 2007;8(1):29–40. doi: 10.1089/sur.2006.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sprong T, et al. Macrophage migration inhibitory factor (MIF) in meningococcal septic shock and experimental human endotoxemia. Shock. 2007;27(5):482–487. doi: 10.1097/01.shk.0000246898.65692.34. [DOI] [PubMed] [Google Scholar]
- 43.Awandare GA, et al. MIF (macrophage migration inhibitory factor) promoter polymorphisms and susceptibility to severe malarial anemia. J Infect Dis. 2009;200(4):629–637. doi: 10.1086/600894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qi D, et al. The vestigial enzyme D-dopachrome tautomerase protects the heart against ischemic injury. J Clin Invest. 2014;124(8):3540–3550. doi: 10.1172/JCI73061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Echtenacher B, et al. Differences in innate defense mechanisms in endotoxemia and polymicrobial septic peritonitis. Infect Immun. 2001;69(12):7271–7276. doi: 10.1128/IAI.69.12.7172-7276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamayo E, et al. Pro- and anti-inflammatory responses are regulated simultaneously from the first moments of septic shock. Eur Cytokine Netw. 2011;22(2):82–87. doi: 10.1684/ecn.2011.0281. [DOI] [PubMed] [Google Scholar]
- 47.Ochi A, et al. MIF-2/D-DT enhances proximal tubular cell regeneration through SLPI- and ATF4-dependent mechanisms. Am J Physiol Renal Physiol. 2017;313(3):F767–F780. doi: 10.1152/ajprenal.00683.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donnelly SC, et al. Regulatory role for macrophage migration inhibitory factor in acute respiratory distress syndrome. Nat Med. 1997;3(3):320–323. doi: 10.1038/nm0397-320. [DOI] [PubMed] [Google Scholar]
- 49.Ma Y, et al. Cardiomyocyte d-dopachrome tautomerase protects against heart failure. JCI Insight. 2019;4(17):e128900. doi: 10.1172/jci.insight.128900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song S, et al. D-dopachrome tautomerase contributes to lung epithelial repair via atypical chemokine receptor 3-dependent Akt signaling. EBioMedicine. 2021;68:103412. doi: 10.1016/j.ebiom.2021.103412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fingerle-Rowson G, et al. The p53-dependent effects of macrophage migration inhibitory factor revealed by gene targeting. Proc Natl Acad Sci U S A. 2003;100(16):9354–9359. doi: 10.1073/pnas.1533295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rittirsch D, et al. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat Protoc. 2009;4(1):31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C, et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31(1):46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pantouris G, et al. Structural plasticity in the c-terminal region of macrophage migration inhibitory factor-2 is associated with an induced fit mechanism for a selective inhibitor. Biochemistry. 2018;57(26):3599–3605. doi: 10.1021/acs.biochem.8b00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cao Q, et al. Therapeutic inhibition of CXC chemokine receptor 2 by SB225002 attenuates LPS-induced acute lung injury in mice. Arch Med Sci. 2018;14(3):635–644. doi: 10.5114/aoms.2017.64980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.