Abstract
Tumor-associated macrophages (TAM) are heterogeneous in nature and comprise anti-tumor M1-like (M1-TAMs) or pro-tumor M2-like (M2-TAMs) TAMs. M2-TAMs are a major component of stroma in breast tumors and enhance metastasis by reducing their phagocytic ability and increasing tumor fibrosis. However, the molecular mechanisms that regulate phenotypic plasticity of TAMs are not well known. Here we report that a novel tumor suppressor Slit2 in breast cancer by regulating TAMs in the tumor microenvironment. Slit2 reduced the in vivo growth and metastasis of spontaneous and syngeneic mammary tumor and xenograft breast tumor models. Slit2 increased recruitment of M1-TAMs to the tumor and enhanced the ability of M1-TAMs to phagocytose tumor cells in vitro and in vivo. This Slit2-mediated increase in M1-TAM phagocytosis occurred via suppression of IL6. Slit2 was also shown to diminish fibrosis in breast cancer mouse models by increasing the expression of matrix metalloproteinase 13 in M1-TAMs. Analysis of patient samples showed high Slit2 expression strongly associated with better patient survival and inversely correlated with the abundance of CD163+ TAMs. Overall, these studies define the role of Slit2 in inhibiting metastasis by activating M1-TAMs and depleting tumor fibrosis. Furthermore, these findings suggest that Slit2 can be a promising immunotherapeutic agent to redirect TAMs to serve as tumor killers for aggressive and metastatic breast cancers. In addition, Slit2 expression along with CD163+ TAMs could be used as an improved prognostic biomarker in breast cancer patients.
INTRODUCTION
Macrophages are bona fide phagocytic cells of the body and play crucial roles in tissue homeostasis by removing dead cells and immune surveillance by clearing pathogenic organisms. Depending upon the stimuli linked to tissue injury and pathogens, macrophages can polarize into different subtypes. Classically activated or M1 type macrophages are inflammatory and anti-tumor in nature, while alternatively activated or M2 type macrophages are anti-inflammatory and pro-tumor. Among various types of cells present in the tumor microenvironment, M2 type tumor-associated macrophages (TAMs) are the most abundant cell type in most of the tumors and play multifaceted roles in regulating tumor progression and metastasis. The signals from progressing tumor dampen the phagocytic ability of TAMs and polarize them towards M2-type to support migration and invasion of tumor cells and help in blood vasculature intravasation (1–3). Furthermore, abundance of TAMs correlates with fibrogenesis and reduced anti-tumor immunity, which are known contributors to metastasis (4,5). Therefore, activating M2-type TAMs to M1-type phagocytic macrophages might be a prominent strategy to inhibit tumor growth and metastasis.
Here, we analyze the role of Slit2 protein in regulating immune responses against breast cancer by activating M2-TAMs to M1-type macrophages. Slit2 is a secreted protein and preferentially binds to Roundabout receptor 1 (Robo1) (6). Initially identified as an axon guidance cue, Slit2 has also been shown to regulate mammary gland growth and development (6). Furthermore, the loss of Slit2 functions by gene deletion, epigenetic inactivation, and mutations have been reported in a variety of cancers, including breast cancer (7,8). We and others have shown that overexpression of full-length Slit2 inhibits breast tumor growth in vivo (9,10). Specifically, fibroblasts and cancer cells-secreted Slit2 has been shown to reduce tumor growth and metastasis through Robo1 in breast cancer mouse models (10,11). However, its role in regulating tumor immunity is not known. Although a recent study has shown that Slit2 regulates functions of normal macrophage (12), not much is known about its role in regulating TAMs functions, especially tumor phagocytosis.
Here, we show that Slit2 possesses strong anti-fibrotic and anti-metastatic activity. The mechanistic studies revealed that Slit2 activated macrophages are highly phagocytic, polarized towards anti-tumor M1 phenotype, and secrete fibrosis-degrading MMP13. Analysis of breast cancer patient samples also showed a positive association of Slit2 levels with improved patient survival, reduced number of TAMs, and decreased level of tumor-induced fibrosis.
MATERIALS AND METHODS
Animal studies
Animal studies were approved by institutional review board. The FVB/N and NOD-scid IL2Rgammanull (NSG) female mice of 6 weeks were used for orthotopic syngeneic and xenograft tumor growth and metastases studies. MMTV-PyMT male mice were purchased from Jackson laboratories, USA. For orthotopic tumor implantation, a total of 1 × 105 murine MVT1 cells in 100 μL of sterile saline were injected into the mammary fat pad (#4) of FVB/N mice. Similarly, 1 × 106 human MDA-MB-231 cells, or MDA-MB-231 cells overexpressing human Slit2 or vector control in NSG mice.
The tumor bearing mice were randomly divided into different groups and treated with human or mouse rSlit2-N (R&D systems, USA) in 100 μl of PBS (0.2 mg/kg body weight) or PBS alone as control, intra-peritoneally every alternate day for 3 weeks. Tumors volume was calculated according to the formula V = 0.52 × a2 × b, where a is the smallest superficial diameter and b is the largest superficial diameter. For MMTV-PyMT model, each mammary tumor was measured and total tumor volume was calculated as sum of all the tumors present in a mouse. Average tumor volume was calculated by adding total tumor volume from all the mice in a group divided by number of mice in the group. Different stages of tumor progression were analyzed in accordance with the guidelines put forth by the mouse mammary gland pathology consensus meeting in Annapolis (13).
Cell lines and culture
MVT1 cells (VEGF and c-MYC oncogenes overexpressing mouse mammary tumor cell line) (14) and MDA-MB-231 cells overexpressing Slit2 (231-Slit2) or vector control (231-Vec) were a generous gift from Dr. Ostrowski (The Ohio State University) and Dr. Hinck (University of California, Santa Cruze) respectively. RAW264.7 and MDA-MB-231 cells were purchased from ATCC, USA. All the cells cultured in Dulbecco’s modified eagle media (DMEM) (Lonza, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, USA) and 1% Penicillin/ Streptomycin (Lonza, USA).
Gene knockdown and Western blot analysis
Human Robo1 was knockdown in MDA-MB-231 cells as described earlier (15) using siRNA targeting human Robo1 (Dharmacon). Western blot analysis of lysates was done as described before (16).
Genomic analysis
Total RNA was collected from 231-Slit2 or 231-Vec or whole tumor single cells or sorted cells using RNeasy Plus mini kit (Qiagen, USA). RNA quantity and quality was analyzed using Bioanalyzer 2100 (Agilent, USA) and Qubit Fluorimeter (Invitrogen, USA). Microarray, RNASeq, and Nanostring analysis were done at the Ohio State University (Columbus, OH) core facility. nCounter® Mouse Myeloid Innate Immunity Panel v2 was used for Nanostring analysis. Microarray was performed using an Affymetrix Microarray gene U133 chip containing 40,000 human genes.
Analysis performed with in-house pipeline BISR-RNAseq (17). Gene wise counts were generated with featureCounts from the subread package v1.5.1 (18) for genes annotated by Mus_musculus.GRCm38.95, counting the primary alignment in the case of multimapped reads. Raw counts were normalized by voom and differential expression was performed with limma (19,20). Genes were tested if at least 66% of the samples had an expression of 2 CPM. Two comparisons were made to assess differential expression between groups: PBS_p vs ps_rslit2. Significant genes had FDR<0.05 and logFC>1 or <−1. Heatmaps were generated using ComplexHeatmap. Pathway analysis was performed as a Core Analysis by Ingenuity Pathway Analysis using significant genes. Expression was analyzed by GSEA (21) to identify gene sets enriched in pairwise comparison of sample groups. Permuting was performed by gene set due to small sample size. GSEA was used to compare PBS_p_CD3 vs rslit2_ps_CD3 and PBS_p vs ps_rslit2 for hallmark h.all.v6.2.symbols and GO c5.all.v6.2.symbols.
Flow cytometry
Single-cell suspension from tumors were analyzed by flow cytometry as described earlier (15). Briefly, cells were incubated with Fc receptor blocker followed by staining with anti-F4/80 PE, anti-CD11b APC, anti-CD206 Alexa Flour 488 (AF-488), anti-CD49b PE, anti-CD4 AF-488 and anti-CD8 PE (Biolegend, USA). Roundabout 1 (Robo1) receptor expression was analyzed by staining with rabbit anti-mouse Robo1 antibody (Abcam) followed by anti-rabbit AF-488 secondary antibody. For staining intracellular targets, cells were fixed and permeablized for 30 minutes at room temperature (Fixation/Permeablization Diluent, eBioscience). Cells were stained with anti-EpCAM-AF-488 (Biolegend). 10–20X106 cells were recorded on FACS fortessa (BD Biosciences) and analyzed using flowJo software.
Immunohistochemistry
Tumor sections (4μm) were analyzed using standard immunohistochemical techniques as per manufacturer’s recommendations (Vector Laboratories) using anti-mouse antibodies against Ki67 (Invitrogen, 1:100), CD31 (Santa Cruz 1:100), F4/80 (AbD Serotec, 1:150), MMP13 (Novus; 1:300), CD206 (AbCam 1:200), cleaved caspase 3 (Cell Signaling technology; 1:1000), FAP (Abcam; and Slit2 (abcam; 1:100) at 4°C for overnight. Followed by incubating with appropriate anti-rat or mouse or rabbit ImmPRESS polymer reagent tagged with horseradish peroxidase or alkaline phosphatase (Vector Laboratories). Enzyme-specific chromogen color development was performed using ImmPACT-DAB or ImmPACT-red to detection the bound primary antibodies. The cell nucleus were counterstained with Hematoxylin (blue) (Vector Laboratories). The images were acquired using light microscope (Lieca or Keyence).
Immunofluorescence
The standard immunofluorescence procedure was followed. Briefly, cells were fixed with 4% paraformaldehyde at room temperature for 20 min. Cells were washed with PBS, blocked with 5 % goat serum in TBST buffer for 60 min and incubated primary antibodies overnight at 4°C followed by incubation with secondary antibody conjugated with Alexa Flour 568 for 60 min. Cells were washed with TBST and mounted using vectashield mounting medium with DAPI and examined under Olympus FV1000 Filter confocal microscope.
In vitro MMP expression analysis
Mouse bone marrow-derived cells were obtained from femurs of FVB mice and converted to mouse bone-marrow derived macrophages (BMDMs) by culturing with M-CSF enriched L929 cell condition media for 5 days. The BMDMs were cultured with DMEM complete growth media for 24 hours to bring them in resting phase. Next, BMDMs were stimulated with rSlit2-N or PBS for 24 hours and transferred in plates containing type I collagen matrix with or without MDA-MB-231 cells. After 24 hours, the cells were harvested and total RNA was isolated. mRNA level of murine specific different MMPs were identified by RT-PCR. The co-culture of murine BMDMs with human MDA-MB-231 allowed us to detect macrophages-derived MMPs based on mmp gene sequence variations observed between species. Murine-specific primer sequences are presented in Table S1. RNA from BMDMs or MDA-MB-231 growing alone in matrix was used as species specific controls.
Phagocytosis analysis by immunofluorescence
CD11b+ monocytes were sorted from tumor single cell suspension using MACS Miltenyi. The sorted cells (1 X 103) were smeared on glass slides using cytospin. The cells were fixed, permeabilized and stained with F4/80 antibody (Bio-Red), Epcam antibody (Biolegend) followed by proper Alexa-floor tagged secondary antibody. The F4/80+ macrophages double positive for Epcam were identified as phagocytic macrophages using confocal microscope (Zeiss LCM 7100). The number of macrophages per smear was counted manually.
Bacterial phagocytosis assay
The BMDMs (1X105 cells) were cultured in 96 well plate overnight. The cells were pre-treated with mouse rSlit2-N (100ng/ml) or IL6 neutralization antibody (BioXCell# BE0046) for 2 hours. The culture media was replaced by live cell imaging media (Invitrogen# A14291DJ) with pH-Rhodo S. Aureus bacteria as per manufacturer’s instructions (ThermoFisher# P35367). The cells were incubated with 231-Slit2 or 231-Vec conditioned media in the presence or absence of recombinant mouse IL6 (PeproTech#216-16). The plate was incubated at 37°C in a fluorescence plate reader (Synergy 2, BioTek, USA) and the fluorescence was measured at different time points. Bacteria without BMDMs was also included as control.
TAMs-mediated Tumor cell lysis assay
The MMTV-PyMT tumors were digested and filtered to generate single cells. The F4/80+ TAMs were sorted using Biotin labelled anti-F4/80 antibody and magnetic beads conjugated anti-biotin secondary antibody and MACS cell separation system (Miltenyi Biotec, Germany). These TAMs were pretreated with rSlit2-N or PBS for 24 hour followed by co-culture with MVT1 tumor cells for additional 2 days. Tumor cells alone and TAMs alone treated with rSlit2-N were included as controls. At the end, cells were collected and stained with 7AAD for 30 minutes on ice. The 7AAD-positive lysed cells were identified using flow cytometry.
Macrophage labelling and recruitment study
The macrophages were labeled as described before (22). The tumor bearing MMTV-PyMT mice were injected with 10KD Alexa-floor 555-labelled Dextran at 10 mg/kg body weight dose. The dextran is easily engulfed by macrophages. Next day, mice were treated with rSlit2-N or PBS every day for 5 consecutive days. Next day, the animals were injected with 10KD Alexa-floor 680-labelled Dextran at 10 mg/kg body weight dose. After 2 hours, animals were euthanized, tumor harvested and digested and analyzed by flow cytometry.
Reverse transcriptase and real-time PCR
RNA was isolated from cells using TRIzol reagent. Reverse transcriptase PCR (RT-PCR) reaction was carried out using RT-PCR kits (Applied Biosystem). Expression of genes analyzed by quantitative PCR (qPCR) using specific primers and normalized to 18SRNA using the 2(−ΔCt) method (23). Primers used for RT-PCR and qPCR are listed in Table S1.
Human samples
To analyze Slit2 expression in human samples, we purchased two tissue microarrays (TMA) having normal, invasive and lymph node metastatic primary tumors from Imgenex (IMH-364 and BR1008). To study the correlation of Slit2 expression with macrophage abundance in tumors we used TMA generated by Department of Pathology at The Ohio State University. The TMA possess 318 well defined human breast cancer samples. Multiple sections of TMA were stained with Slit2 (1:300) and CD163, CD31 (1:100), CD68 (1:3000) antibodies. All the immunostained slides were analyzed and scored in a blinded manner by highly trained pathologist. The TMA cores with ≤5 macrophages were considered as none, the cores with >5 and <30 macrophages were considered as low score and ≥30 considered as high score. Slit2 mRNA expression and copy number analysis in human breast tissue samples was done using publically available UALCAN and Oncomine databases (24,25).
All the information about used reagents and resources, including, kits, proteins and antibodies have been provided in Table S2.
Statistical analysis
For the experiments using different mouse models to investigate the tumor growth, metastasis, and stromal cell populations, n=5 mice per group provides 80% power to detect a 2-fold difference between groups in endpoints (CV=0.6 and α=0.05). The percentage of macrophage distribution among various groups according to Slit2 expression was calculated by SPSS software version 15. Chi square test or student’s t-testwas used to compare the groups. Kaplan-Meier log rank t-test was used to analyze the association of Slit2 levels with patient survival.
RESULTS
Slit2 inhibits breast cancer growth and metastases.
We first analyzed if Slit2 protein expression changes during different stages of tumor progression in an autochthonous mouse model of breast cancer, mouse mammary tumor virus-polyoma middle tumor-antigen (MMTV-PyMT) (26). In this study, we found that normal mammary gland expresses high amount of Slit2, which decreased with advancing stages of tumor progression (Fig. 1A and Fig. S1A). Slit2 protein possesses an intrinsic proteolytic cleavage site which results in biologically active N-terminus of Slit2 (Slit2-N) and C-terminus Slit2 (6). Next, we tested if recombinant Slit2-N (rSlit2-N) treatment can delay tumor growth and inhibit metastases in MMTV-PyMT and MVT1 cell line-derived orthotopic syngeneic tumor models. Tumor-bearing mice were treated with rSlit2-N or PBS and over next 4 weeks, treatment significantly delayed the rate of tumor growth, which correlated with a decrease in pulmonary metastases (Fig. 1B–I and fig. S1B–D, S2A–C). A marked reduction in number of Ki67+ proliferating cancer cells and CD31+ blood vessels was observed in rSlit2-N treated MMTV-PyMT and MVT1 tumor models (fig. S3A–J). To avoid the confounding effect of tumor size on observed reduced lung metastasis in rSlit2-N treated mice, we performed another study where we allowed rSlit2-N treated tumors to grow in size similar to PBS tumors. Interestingly, the evaluation of lungs still showed reduced lung metastases in rSlit2-N treated animals, suggesting the anti-metastatic potential of rSlit2-N is not affected by tumor size (Fig. 1J–M).
Figure 1: rSlit-N treatment inhibits breast cancer growth and metastases.
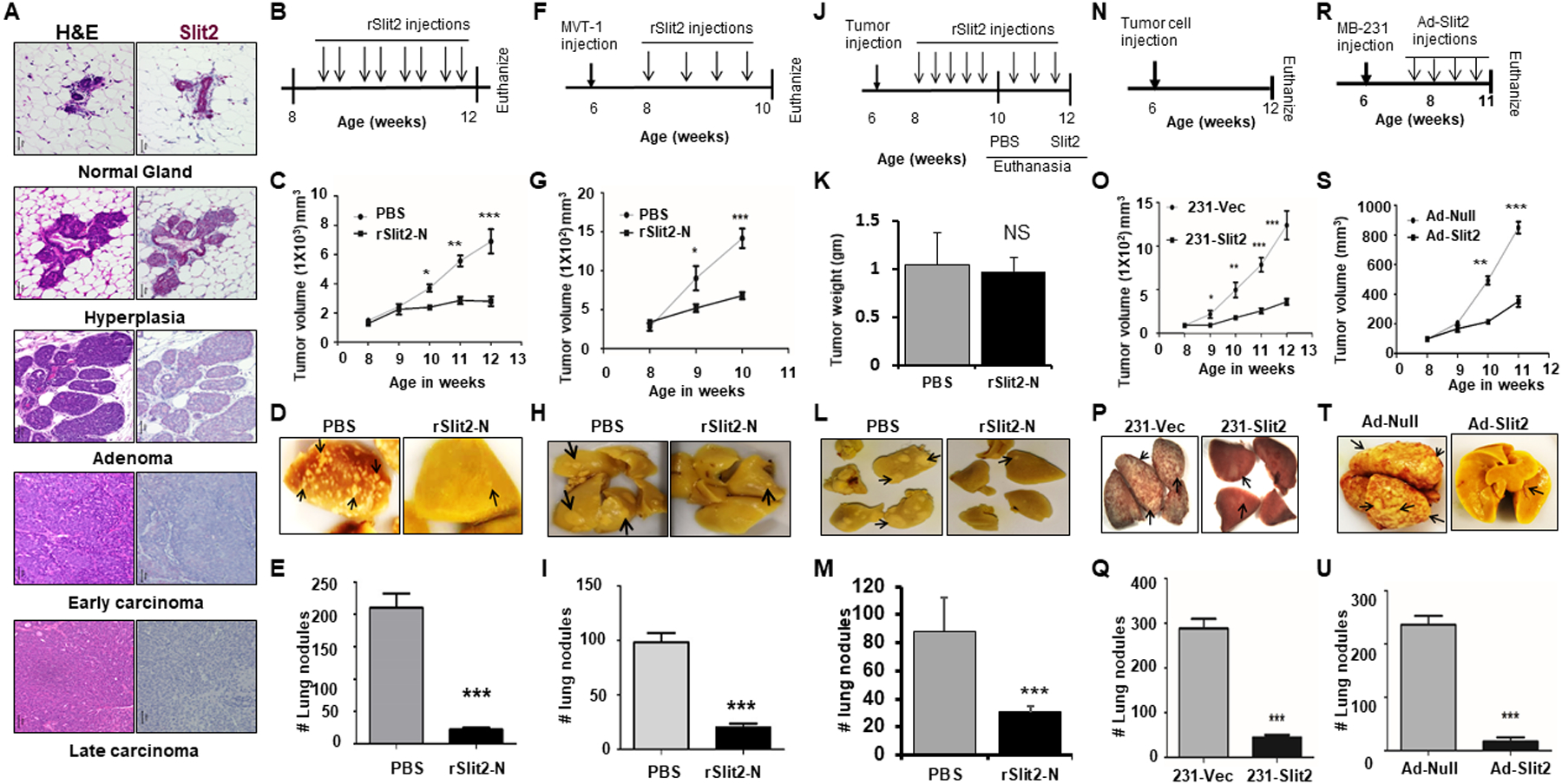
(A) Mammary glands were isolated from MMTV-PyMT at different stages of tumor progression and tissue sections were stained with H&E stain or immunostained with Slit2 antibody using the IHC technique.
B-E. (B) Eight weeks old MMTV-PyMT mice bearing spontaneous mammary tumors were treated with rSlit2-N or PBS for four weeks. (C) Tumor volume was measured every week up to 12 weeks of age. (D) Representative pictures of the lungs harvested from (B). (E) The number of metastatic nodules in the lungs.
F-M. MVT1 cells were orthotopically implanted into the six weeks old FVB wild type mice. (F) Schematics of mice treatment with rSlit2-N or PBS. N=6 mice in each group. (G) Tumor volume was measured every week. (H) Representative pictures of the lungs harvested from (F). (I) The number of metastatic nodules in the lungs. (J) Schematics of mice treatment with rSlit2-N or PBS, showing Slit2 treated tumors were allowed to grow for additional two weeks. N=5 mice in each group. (K) At the end, tumors were harvested and weight was measured. (L) Representative pictures of the lungs harvested from (J). (M) The number of metastatic nodules in the lungs.
N-Q. (N) Human breast cancer cell line MDA-MB-231 overexpressing Slit2 (231-Slit2) or vector control (231-Vec) implanted orthotopically into the NSG mice. (O) After two weeks of tumor injection, tumor volume was measured every week up to the age of 12 weeks. (P) The images of lungs harvested from (N). (Q) The harvested lungs were analyzed for the number of metastatic nodules using a dissection microscope.
R-U. NSG females were injected with MDA-MB-231 cells and treated with Adeno-Slit2 or Adeno-Null. (R) Schematics of mice treatment with Adeno-Slit2 or Adeno-Null. (S) Starting from 8 weeks of age, tumor volume was measured every week. (T) Representative pictures of the lungs harvested from (R). (U) The number of metastatic nodules in the lungs. * is p<0.05, ** is p<0.01, *** is p<0.001, NS is P value not significant using student’s t-test.
We further tested the anti-metastatic potential of Slit2 by injecting an aggressive human MDA-MB-231 cell line overexpressing human Slit2 (231-Slit2) or vector control (231-Vec) into the mammary fat pad of immunocompromised NOD-SCID IL2Rgammanull (NSG) mice. We observed a reduced rate of 231-Slit2 tumor growth and lung metastases compared to 231-Vec (Fig. 1N–Q and fig. S4A). In addition, MDA-MB-231 cells orthotopic xenograft tumors bearing NSG mice treated with adenovirus expressing human Slit2 (Ad-Slit2) showed reduced tumor growth and metastases to the lungs compared to vector control (Ad-Null) (Fig. 1R–U and fig. S4B, C). To analyze if Slit2 targets tumor cells, we performed cell apoptosis analysis by flow cytometry. The treatment of MDA-MB-231 cells with different concentrations of rSlit2-N did not induce apoptosis (Suppl. Fig. S4 D, E)
rSlit2-N treatment depletes fibrosis and activates TAMs.
To identify the rSlit2-N-mediated molecular mechanisms, we performed whole tumor RNAseq analysis on rSlit2-N or PBS treated MMTV-PyMT tumors. Ingenuity pathway analysis on differentially expressed genes identified various extracellular matrix (ECM) related genes linked to the osteoarthritis pathway (Fig. 2A and fig. S5A). We further performed a focused analysis of genes associated with ECM remodeling and observed a significantly increased expression of ECM degrading Matrix metalloproteinase 10 (MMP10) and MMP13 in rSlit2-N treated tumors (fig. S5B–D). In addition, analysis of mRNA isolated from MMTV-PyMT tumors by RNA hybridization-based Nanostring technology and a panel of genes focused on ECM and innate immunity validated the increased expression of MMP10 and MMP13 genes in rSlit2-N treated tumors compared to PBS. (Fig. 2B). We further confirmed the MMP13-mediated anti-metastatic effects of Slit2 by treating MDA-MB-231 xenografts bearing NSG mice with rSlit2-N, in the presence or absence of MMP13 inhibitor (MMP13i) (Fig. 2 C, D and fig. S5E). In support to the inhibitory effect of Slit2 on Ki67+ proliferative cells and CD31+ vessels density observed in MMTV-PyMT and MVT1 tumors, we observed that Slit2 treated MDA-MB-231 xenografts also show reduced number of Ki67+ proliferative cells and CD31+ blood vessels (fig. S6 A–D).
Figure 2. rSlit2-N treatment depletes tumor fibrosis and enhances MMP13 expression.
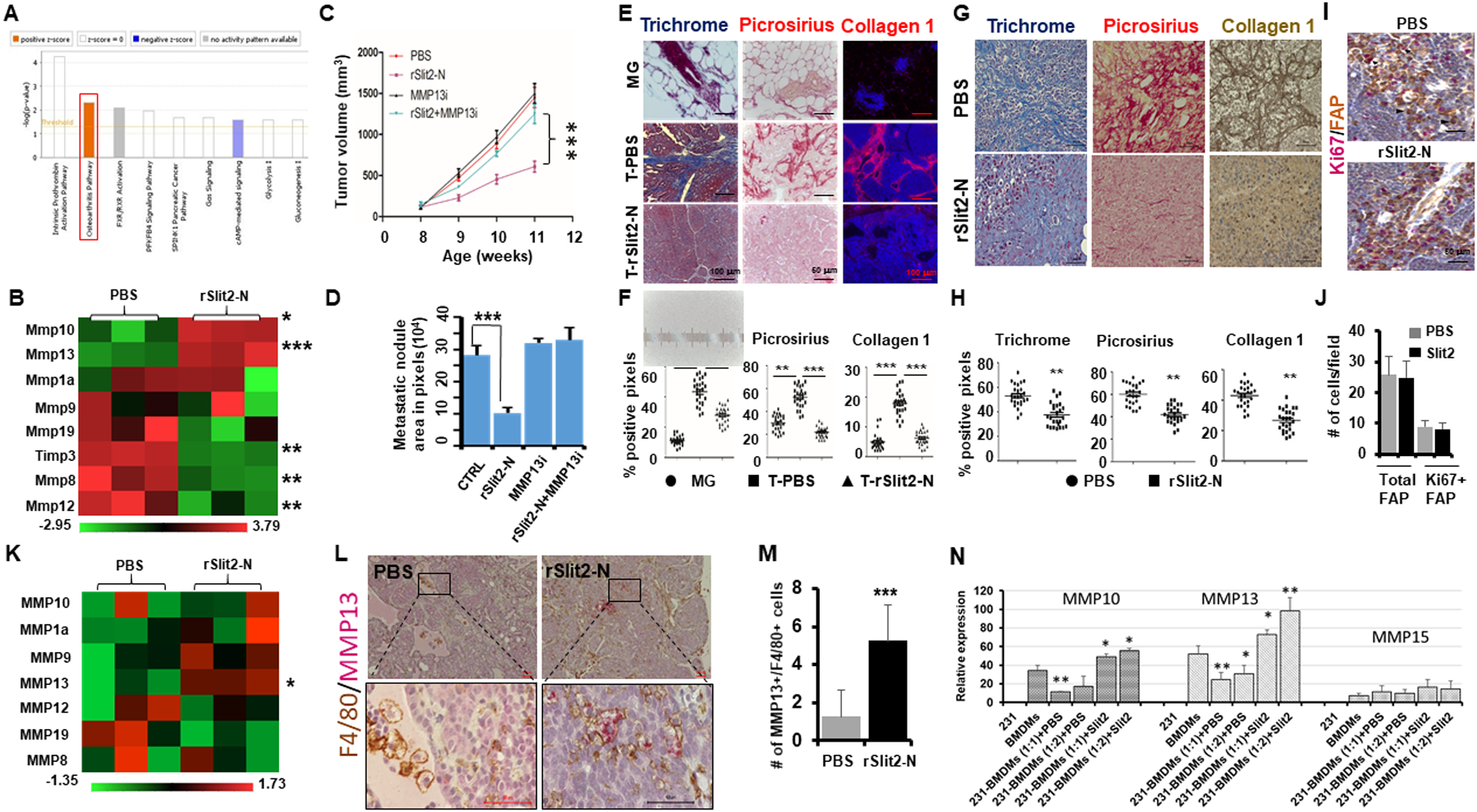
(A) The tumors isolated from MMTV-PyMT mice treated with rSlit2-N or PBS were lysed and total RNA was sequenced for gene expression levels. Ingenuity pathway analysis on differentially expressed genes showing molecular pathways altered by rSlit2-N treatment.
(B) The RNA samples from (A) were analyzed using Nanostring technology for gene expression levels. The heat map diagram shows expression of different ECM related genes.
C-D. The MDA-MB-231 xenografts bearing mice were treated with PBS or rSlit2-N in the presence or absence of MMP13 inhibitor (MMP13i) and tumor growth and metastasis were analyzed. (C) Graph showing tumor volume of different groups. (D) The area of metastasis in digital images of lungs from (C) was quantified (n=5 each group).
(E) The rSlit2-N or PBS treated MMTV-PyMT tumor or normal mammary gland tissue sections were stained with trichrome stain or picrus red stain or immunostained with collagen 1 antibody.
(F) The graphs show quantification of the staining level in tissue sections from (E) (n= 5 microscopic fields per sample, 25 fields per study group).
(G) The MDA-MB-231 xenografts treated with rSlit2-N or PBS were processed and stained with trichrome stain or picrus red stain or immunostained with collagen 1 antibody.
(H) The graphs show quantification of the staining level in tissue sections from (G), (n= 5 microscopic fields per sample, 25 fields per group).
(I) The tumor sections from (A) were immunostained with FAP and Ki67 antibodies and analyzed by dual-color IHC. The arrowhead points to only FAP+ cell and the arrow points to Ki67+/FAP+ cell.
(J) The graph depicts quantification of FAP+ total CAFs or Ki67+/FAP+ proliferative CAFs in the tumor sections from (I), (n= 5 microscopic fields per sample, 25 fields per group).
(K) Total RNA was isolated from CD11b+ cells sorted from rSlit-N or PBS treated MMTV-PyMT tumors using magnetic beads technology. The RNA was analyzed for gene expression levels using Nanostring technology. The heat map shows expression levels of different MMP genes.
(L) The rSlit2-N or PBS treated MMTV-PyMT tumors were processed and sections were immunostained with F4/80 and MMP13 antibodies. Primary antibodies were detected using peroxidase-DAB and alkaline phosphatase-Red substrate respectively.
(M) Quantification of F4/80+/MMP13+ macrophages per field as identified by IHC in (L). N= 5 mice in each group. Randomly selected 5 different areas from each tumor were analyzed.
(N) Total RNA was isolated from macrophage-tumor cell co-culture and expression of mouse-specific MMPs was analyzed by quantitative real-time PCR. * is p<0.05, ** is p<0.01, *** is p<0.001using student’s t-test.
Accumulation of ECM resulting in dense fibrosis is often observed in tumors (27,28). As we observed increased level of ECM-degrading MMPs in Slit2 treated tumors, we analyzed the level of ECM by trichrome staining, total collagens by Picrosirius Red staining, and Collagen-1 by specific antibody immunofluorescence. MMTV-PyMT tumors showed a high level of fibrosis compared to normal glands, which was significantly depleted by rSlit2-N treatment (Fig. 2E, F). We did not see a difference in the expression of other ECM proteins such as tenascin c, periostin or hyaluronan between normal gland, tumor treated with PBS or rSlit2-N (fig. S6E). Similar analysis in MDA-MB-231 xenografts treated with rSlit2-N or PBS validated the ability of Slit2 to deplete fibrosis (Fig. 2G, H). Cancer-associated fibroblasts (CAFs) are known to produce ECM in the TME (29). However, we did not see any change in the number of total fibroblasts activating protein (FAP)+ CAFs or Ki67+/FAP+ proliferative CAFs present in rSlit2-N or PBS treated tumors (Fig. 2 I, J), suggesting ECM production is not affected by rSlit2-N treatment.
Macrophage-derived MMPs have been shown to resolve fibrosis in nonmalignant disorders (30). CD11b+ TAMs harvested from rSlit2-N treated tumors showed increased expression of MMP13 mRNA (Fig. 2K). We also observed a 5 fold increase in the number of MMP13+/F4/80+ TAMs in rSlit2-N treated tumors (Fig. 2L, M, and fig. S6F). In vitro, we observed a marked reduction in the expression of BMDM-derived MMP10 and MMP13 upon co-culture with human breast cancer cells (Fig. 2N and fig. S6G). Pretreatment of BMDM with rSlit2-N attenuated the inhibitory effects of tumor cells on the expression of BMDM-derived MMP10 and MMP13 (Fig. 2N). In addition, BMDMs treated with rSlit2 showed increased expression of MMP13 protein (fig. S6H). The ability of rSlit2-N to target TAMs is further supported by the observation that the majority of Robo1-expressing cells in the MMTV-PyMT tumors are macrophages (fig. S6I). In addition, western blot analysis showed that Robo1 is expressed on human normal breast, breast cancer, and macrophage cell lines (fig. S6J)
Slit2 activated TAMs are skewed towards the anti-tumor phenotype.
TAMs are skewed towards alternatively activated M2 macrophages in tumors (2). In breast, lung, and colon cancer mouse models, tumor recruited macrophages have broadly been classified into CD11b+/ MHCII+/CD206- anti-tumor M1-TAMs or CD11b+/MHCII-/CD206+ pro-tumor M2-TAMs (31–33). The number of M1-TAMs increased, and M2-TAMs decreased in rSlit2-N treated MMTV-PyMT tumors (Fig. 3A and fig. S7A, B). The decreased number of CD206+ TAMs in rSlit2-N treated MMTV-PyMT tumors was also confirmed by IHC (Fig. 3B, C). We further analyzed the M1/M2 macrophage markers in CD11b+ TAMs sorted from MMTV-PyMT tumors and identified the upregulation of M1 markers and suppression of M2 markers by rSlit2-N treatment (Fig. 3D). A longitudinal study demonstrated that the majority of new macrophages recruited after rSlit2-N treatment were M1-TAMs (Fig. 3E, F). Looking at the pre-existing macrophages, M2-TAMs were significantly low in the rSlit2-N treated tumors (fig. S8). Ex-vivo rSlit2-N treatment of CD45+ lymphocytes sorted from MMTV-PyMT tumors increased the number of MHCII+/CD80+ double positive M1-TAMs in a dose-dependent manner (Fig. 3 G–I). Similarly, rSlit2-N treatment reduced the abundance of CD206+ TAMs in MVT1 tumors (fig. S9A–D). Although we did not see any changes in the CD11b+/F4/80+ macrophages in the tumors (fig. S10A–D), the spleens of rSlit2-N treated MMTV-PyMT mice had a reduced number of CD11b+/F4/80+ macrophages (fig. S10 E&F). It is possible that Slit2 induces death in macrophages that results in reduced number of macrophages in spleen. However, we did not observe any apoptotic or cell death activity of Slit2 in primary macrophages (Fig S10G, H).
Figure 3: rSlit2-N treatment inhibits M2-TAMs polarization.
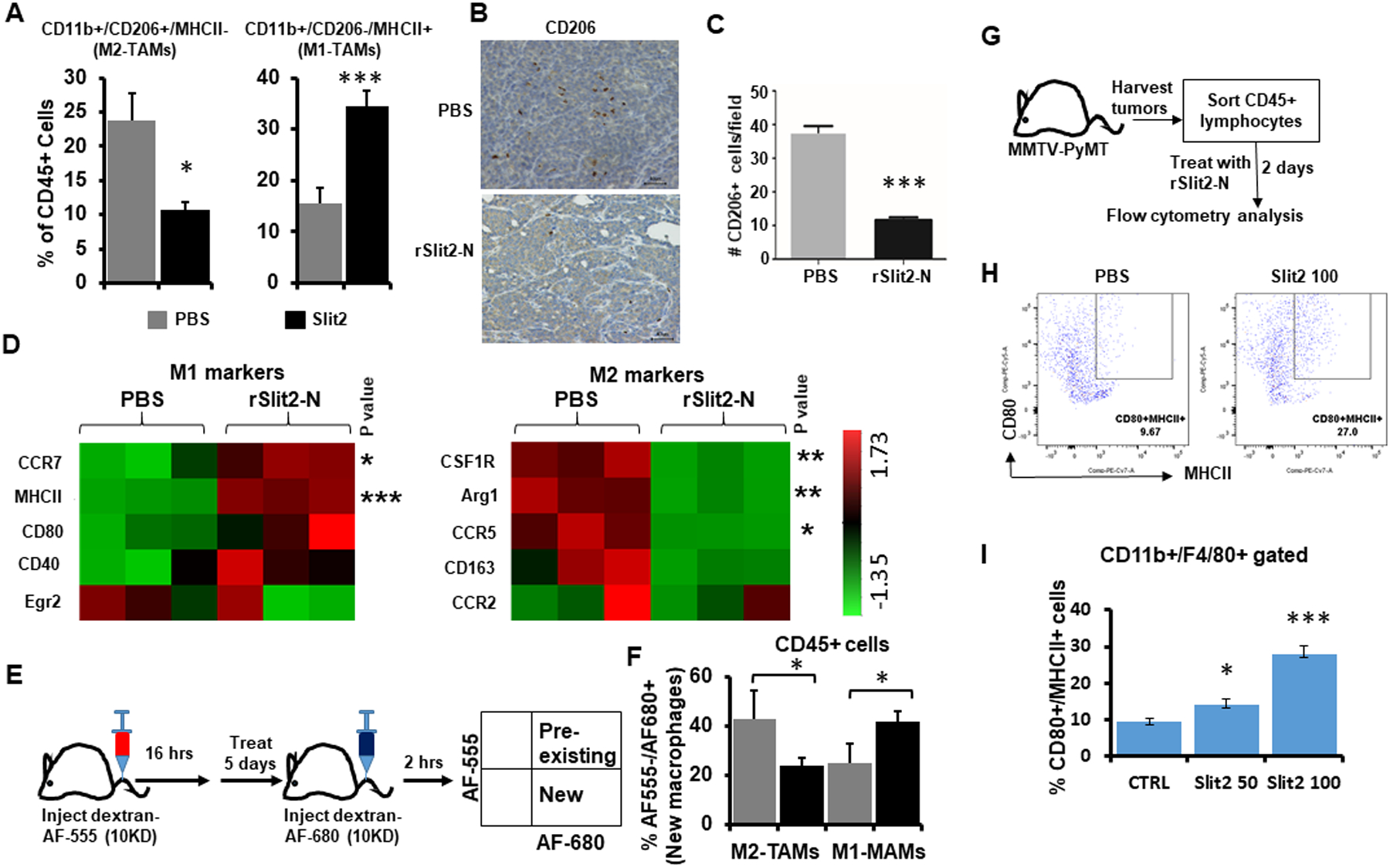
(A) The whole tumors harvested from MMTV-PyMT mice treated with rSlit2-N or PBS were processed into single cells and flow cytometry was performed. Graphs show the distribution of MHCII+ M1-TAMs and CD206+ M2-TAMs. N=5 mice per group.
(B) The representative pictures of rSlit2-N or PBS treated MMTV-PyMT tumors immunostained with CD206 antibody.
(C) The number of CD206+ cells per field was calculated.
(D) CD11b+ cells sorted from rSlit-N or PBS treated MMTV-PyMT tumors were analyzed for gene expression levels using Nanostring technology. The heat maps show expression levels of different M1 or M2 macrophage markers.
(E) To identify new and pre-existing macrophages, mice were pretreated with Alexa555 labeled dextran. The next day, mice were treated with rSlit2-N or PBS every day for 5 days. Mice were injected with Alexa680 labeled dextran two hours before euthanasia. The whole tumors were processed into single cells and flow cytometry was performed.
(F) The graph depicts the percentage of M2-TAMs and M1-TAMs present in rSlit2-N or PBS treated tumors. N=5 mice per group.
G-I. The whole tumors harvested from MMTV-PyMT mice were processed into single cells. Next, CD45+ leukocytes were sorted using magnetic beads technology. These cells were treated with 50 ng/ml rSlit2-N (Slit2 50) or 100 ng/ml rSlit2-N (Slit2 100) or PBS for 2 days. The cells were analyzed for MHCII+/CD80+ M1 type macrophages by flow cytometry. (H) Flow cytometry plots showing identification of CD80 and MHCII double positive (CD80+/MHCII+) macrophages. (I) Quantification of CD80+/MHCII+ cells in CD11b+/F4/80+ gated cells from (H), n=3 each group. * is p<0.05, ** is p<0.01, *** is p<0.001 using student’s t-test.
To verify the Slit2-mediated anti-tumor effects are macrophage dependent, we depleted macrophages in vivo using clodronate liposomes as described in our previous study (34), which showed efficient macrophage depletion (fig S10I). Macrophage depletion in MDA-MB-231 xenograft model showed that macrophage depletion and Slit2 treatment both reduces tumor growth (fig. S10J). The tumor growth was not different between Slit2 treatment and macrophage depletion in combination with Slit2 treatment groups (fig. S10J).
rSlit2-N treated TAMs are highly phagocytic.
Therapeutically reprogrammed TAMs can be detrimental to the tumor cells via enhanced phagocytic activity (31,35,36). We found a significantly increased number of phagocytic macrophages in rSlit2-N treated MMTV-PyMT tumors identified by immunofluorescence, flow cytometry, and dual-color IHC (Fig. 4 A–F and fig. S11A, B and fig. S12 A, B). We also observed increased phagocytic macrophages in MDA-MB-231 xenografts treated with rSlit2-N compared to PBS using flow cytometry analysis (fig. S13A, B). Ex-vivo, rSlit2-N pre-treated MMTV-PyMT TAMs efficiently lysed tumor cells compared to PBS (Fig. 4G and fig. S13C). In vitro analysis demonstrated that rSlit2-N treated primary mouse macrophages show increased bacterial phagocytic ability in a dose and time-dependent manner (Fig. 4H and fig. S14A, B). Altogether, these studies establish that rSlit2-N can increase the ability of TAMs to phagocytose tumor cells.
Figure 4: rSlit2-N enhances the phagocytic ability of TAMs.
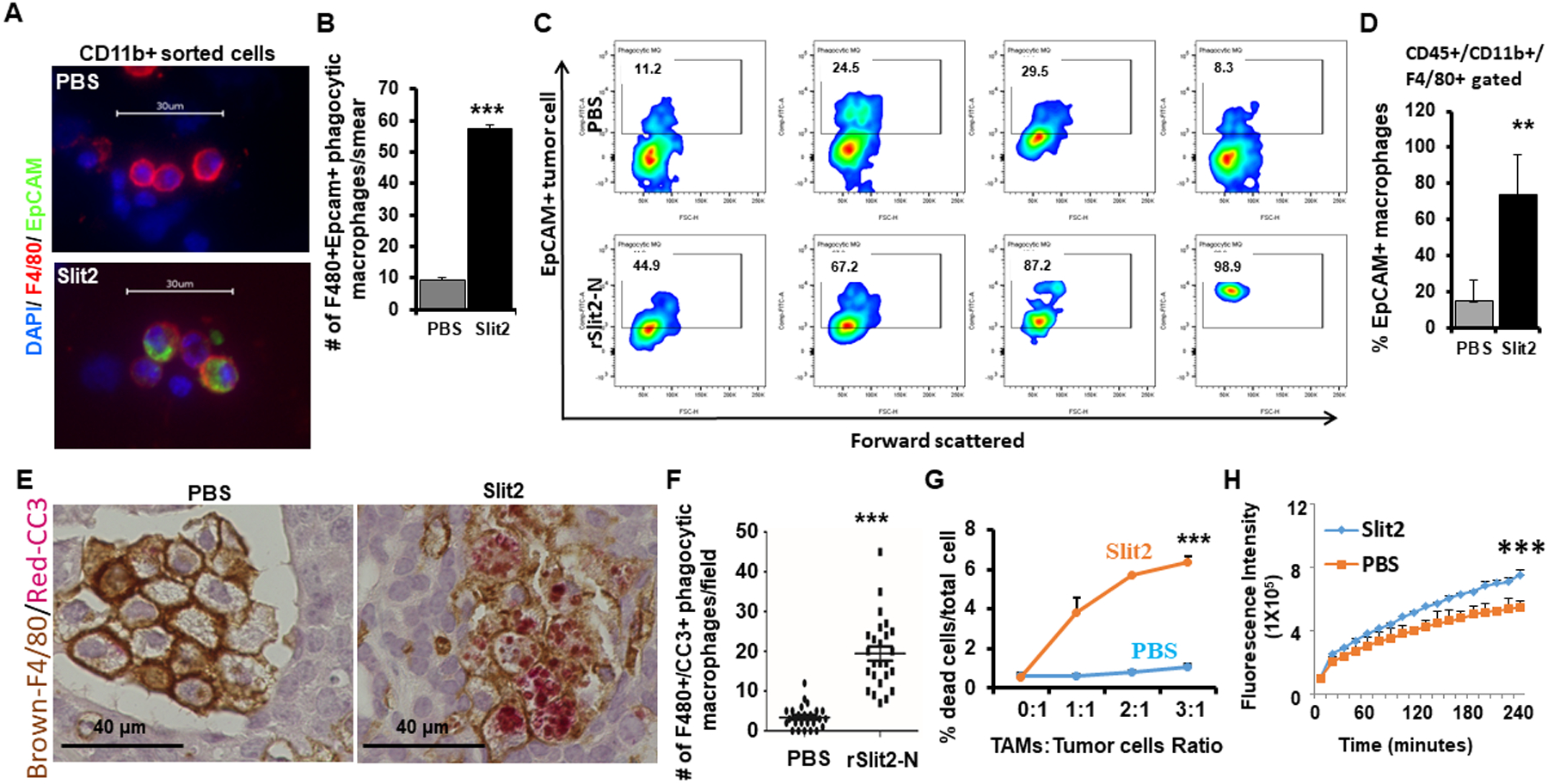
(A) Whole tumors obtained from MMTV-PyMT mice treated with rSlit2-N or PBS were digested into single cells and CD11b+ cells were sorted. The cell aliquots were smeared onto a glass slide using cytospin, permeabilized, and stained with F4/80 and Epcam specific antibodies. The representative images showing phagocytic macrophages as F4/80+ cells that contain intracellular EpCAM, a marker of tumor cell.
(B) Quantification of phagocytic macrophages per smear as identified in (A).
(C) Single-cell preparations from rSlit2-N or PBS treated MMTV-PyMT tumors were also analyzed for phagocytic macrophages using intracellular flow cytometry. The cytometry plots are showing intracellular EpCAM signal inside CD45+/CD11b+/F4/80+ macrophages.
(D) The graph shows the percentage of EpCAM+ macrophages out of total macrophages in PBS or rSlit2-N treated tumors as identified in (C).
(E) Representative image of PBS or rSlit2-N treated MMTV-PyMT tumor sections immunostained with F4/80 and cleaved caspase 3 (CC3) antibodies and detected using peroxidase-DAB and alkaline phosphatase-Red substrate respectively to identify CC3-positive apoptotic bodies within macrophages.
(F) Quantification of F4/80+/CC3+ double-positive phagocytic macrophages per field as identified by IHC in (E). N= 5 mice in each group. Randomly selected 5 different areas from each tumor were analyzed.
(G) F4/80+ TAMs were sorted from tumors harvested from MMTV-PyMT mice. The TAMs were treated with rSlit2 or PBS overnight, followed by co-culture with tumor cells for 24 hours. Tumor cell death in vitro was analyzed by 7-aminoactinomycin D (7-AAD) labeling and flow cytometry. The graph shows a representative assay from two independent experiments, each performed in triplicate.
(H) The rSlit2-N or PBS pre-treated BMDMs were incubated with pHRhodo tagged S. Aureus bacteria at 37° C in a fluorescence plate reader and kinetics of bacterial phagocytosis was analyzed up to 4 hours. The graph represents fluorescence intensity generated by phagocytosis of bacterial particles. ** is p<0.01, *** is p<0.001 using student’s t-test.
Slit2 suppresses IL6 expression to inhibit pro-tumor M2-TAMs activities.
To decipher the underlying molecular mechanism of Slit2 anti-tumor activity, we reanalyzed mRNA expression data from CD11b+ macrophages sorted from rSlit2-N or PBS treated MMTV-PyMT tumors and 231-Slit2 and 231-Vec cells. Differentially expressed genes analysis identified IL6 as the top gene suppressed by rSlit2-N treatment (Fig. 5A, B, and Table S3). We further confirm the reduced expression of IL6 protein in 231-Slit2 cells by cytokine array and ELISA (Fig. 5C, D). Next, we analyzed Slit2-mediated transcriptional regulation of IL6 expression. Transcription factor NF-κB is known to regulate IL6 gene expression (37). We found that Slit2 inhibits IL6 in MDA-MB-231 cells by targeting the activation of NF-κB in Robo1 receptor dependent manner (Fig. 5 E–G).
Figure 5. Slit2 reduces the expression of interleukin-6, thereby inhibiting TAMs activity.
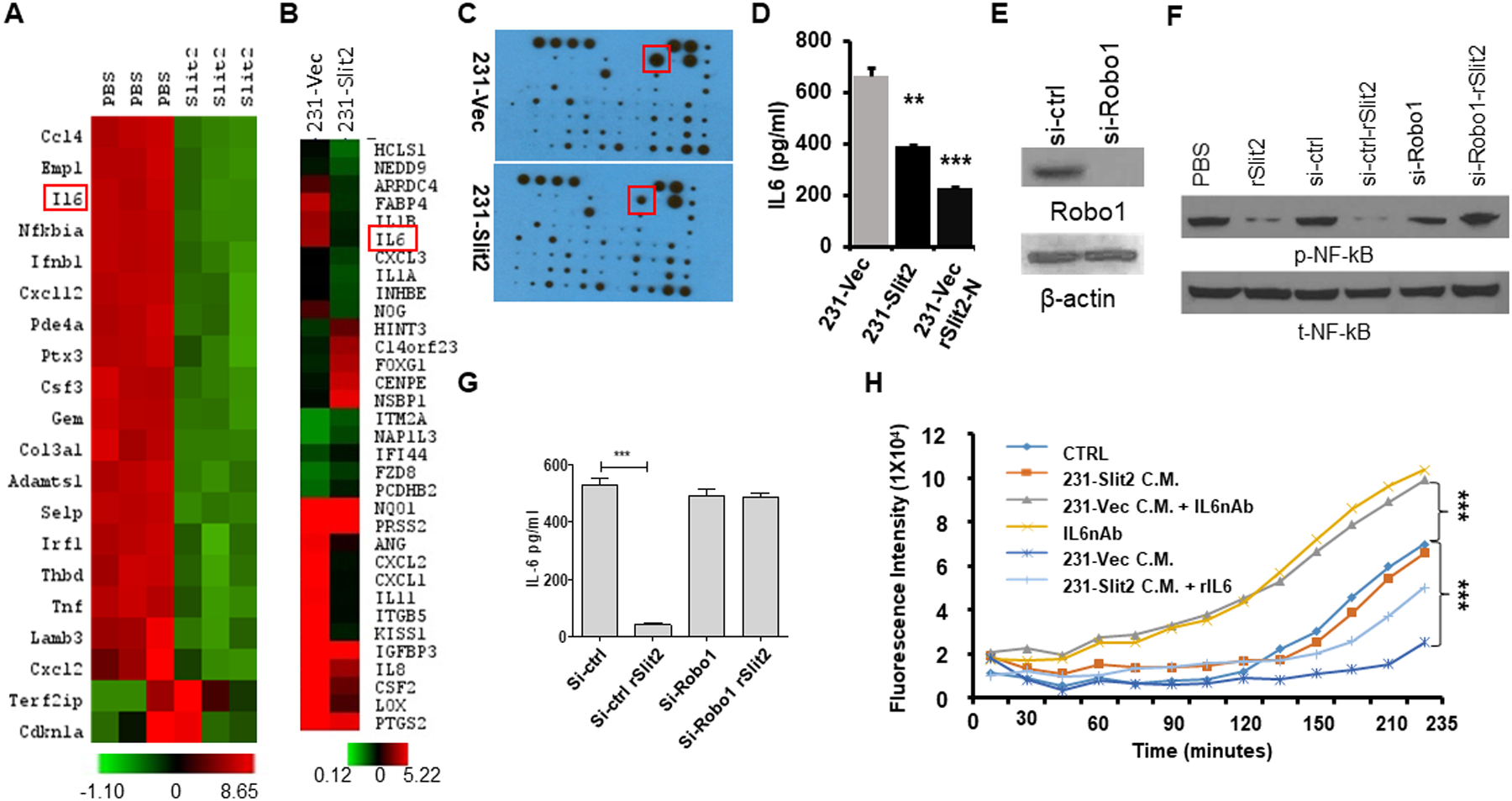
(A) Total RNA of CD11b+ cells sorted from rSlit2-N PBS treated MMTV-PyMT tumors was subjected to gene expression analysis using Nanostring technology. The heat map shows differentially expressed top genes.
(B) Total RNA was isolated from MDA-MB-231 cells overexpressing Slit2 (231-Slit2) or vector control (231-vec) and analyzed for gene expression using microarray technology. The heat map shows differentially expressed top genes.
(C) 231-Slit2 or 231-Vec cells conditioned media (CM) was subjected to cytokine array analysis. The representative image shows differentially expressed molecules.
(D) The graph shows levels of IL6 in CM derived from 231-Sli2 or Vec CM detected by ELISA.
(E) Expression of Robo1 in MDA-MB-231 cells transduced with lentivirus expressing siRNA specific to Robo1 (si-Robo1) or control (si-ctrl) by western blot.
(F) β-actin was used as loading control. MDA-MB-231 parental control, si-Robo1 or si-ctrl from (E) were treated with rSlit2-N or PBS and phosphorylation at p65 of NF-κB (p-NF-κB) or total NF-κB (t-NF-κB) was analyzed.
(G) Graph showing levels of IL6 in si-Robo1 or si-ctrl MDA-MB-231 cells treated with rSlit2-N or PBS detected by ELISA (n=3 each group).
(H) BMDMs were pre-treated with serum-free Conditioned Media (CM) (CTRL), or 231-Slit2 CM or 231-Vec CM or 231-Vec CM pre-incubated with IL6 nAb (231-Vec + IL6nAb) or IL6 nAb. After 2 hours, pH-Rhodo labeled S. Aureus particles were added to the cells and recombinant IL6 was added to the 231-Slit2 CM cells (231-Slit2-rIL6). The kinetics of S. Aureus phagocytosis was analyzed using a fluorescence plate reader at every 15 minutes up to 4 hours.
* is p<0.05; ** is p <0.01; *** is p <0.001 using Student’s t-test.
IL6 has been shown to induce alternatively activated M2-TAMs, increase fibrosis and reduce phagocytosis (38,39). Next, we examined the effect of Slit2 and IL6 crosstalk on macrophage phagocytic ability. The ability of BMDMs to phagocytose pH-Rhodo tagged S. Aureus particles was reduced significantly in the presence of 231-Vec conditioned medium (CM) compared to control media, which was restored by neutralizing IL6 (Fig. 5H). Furthermore, 231-Slit2 CM did not reduce the phagocytic ability and the addition of recombinant mouse IL6 (rIL6) in 231-Slit2 CM significantly decreased the phagocytic ability of BMDMs (Fig. 5H). Recombinant IL6 also induced expression of M2 macrophage markers in human THP1 monocyte cell line differentiated to macrophages and BMDMs, (fig. S15 A–C). In addition, 231-Vec CM enhanced the expression of M2 macrophage marker CD206 in THP1-macrophages, while 231-Slit2 CM did not induce the expression of CD206 (fig. S15A, B). Furthermore, addition of IL6 to 231-Vec CM enhanced CD206 expression marginally, but had no effect on 231-Slit2 CM (fig. S15A, B). These observations suggest that suppression of IL6 by Slit2 prevents tumor-induced polarization of macrophages towards M2 TAMs phenotype and enhances their phagocytic activity.
Slit2 positively correlates with improved overall patient survival.
To identify the clinical relevance of Slit2, we used human tissue microarrays containing normal breast and different types of breast cancer tumors. We observed lower Slit2 expression levels in invasive and metastatic tissue samples compared to normal breast (Fig. 6A, B). Using publicly available ULCAN and Oncomine databases, we validated reduced levels of Slit2 mRNA and gene copy number in a large cohort of breast cancer patients as compared to healthy individuals (Fig. 6C, D). We further evaluated the association of Slit2 mRNA and protein expression with various prognostic markers and observed a significant reduction of Slit2 in high grade, Estrogen Receptor (ER) negative, Progesterone Receptor (PR) negative, and triple-negative breast cancer (TNBC) patients (Fig. 6C and S16A and Table S4). The selected patient cohort showed a negative association of patient survival with high tumor stage and high blood vessel density, suggesting an unbiased cohort of the patient (fig. S16 B, C).
Figure 6. Slit2 expression levels predict breast cancer patient survival.
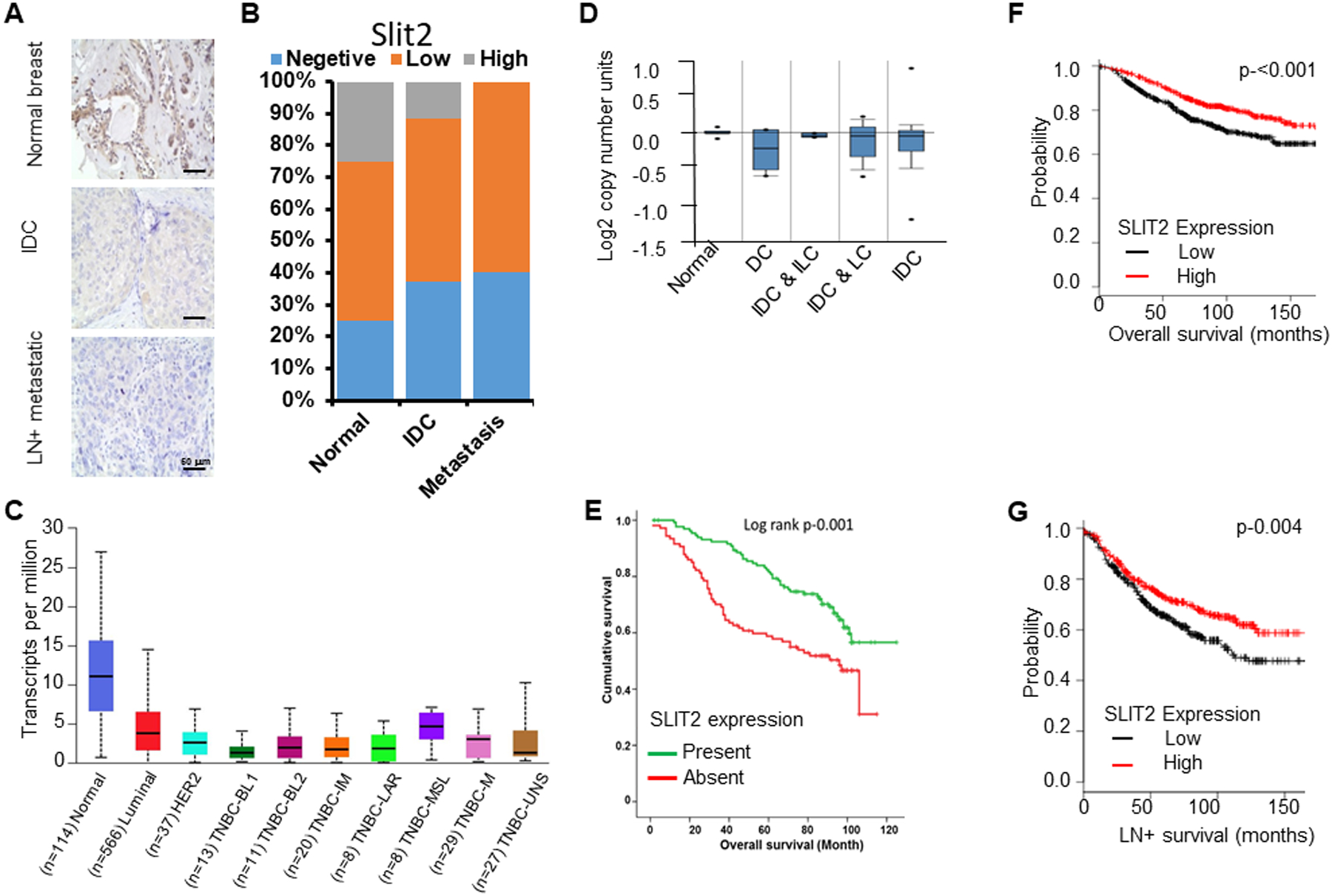
(A) Human breast tissue microarray (pilot TMA) was immunostained with Slit2 antibody using IHC. Representative images showing Slit2 expression levels in normal (n=10), infiltrative ductal carcinoma (IDC) (n=50) and lymph node-positive metastatic (LN+ metastatic) (n=40) breast tissues.
(B) The graph depicts the percentage of human breast samples expressing different levels of Slit2.
(C) The expression level of Slit2 in human breast tissues using UALCAN database. BL1, basal-like 1; BL2, basal-like 2; IM, immunomodulatory; M, mesenchymal; MSL, mesenchymal stem-like; LAR, luminal androgen receptor; UNS, unspecified.
(D) Analysis of Slit2 gene copy number in TCGA dataset using Oncomine. N, normal breast (n-111); DC, ductal carcinoma (n-5); IDC & ILC, invasive ductal and invasive lobular carcinoma (n-5); IDC & LC, invasive ductal and lobular carcinoma (n-14); IDC, invasive ductal carcinoma (n-639).
(E) Using experimental TMA, the association of Slit2 protein expression levels with patient survival was analyzed. The Kaplan-Meier graph demonstrating the association of Slit2 expression level with patient survival.
(F) Analysis of breast cancer patient overall survival based on expression levels of Slit2 mRNA in the KM-plotter database (n=1115).
(G) Analysis of breast cancer lymph node-positive (LN+) metastases patient survival based on expression levels of Slit2 in the KM-plotter database (n=936).
We next explored if Slit2 protein expression also correlates with patient overall survival and observed that patients expressing Slit2 protein showed a significantly better survival as compared to patients not expressing Slit2 (n=242) (Fig. 6E). Furthermore, the mining of publically available datasets confirmed the positive association of Slit2 expression with better overall and lymph node metastases positive survival (Fig. 6F, G and fig. S16 D–F).
Based on our results showing the role of Slit2 in inhibiting fibrosis and M2-TAMs, an inverse correlation of Slit2 expression with the level of fibrosis and number of CD163+ M2-TAMs was observed in these patient samples (Fig. 7A, B and Table S5). In addition, we stained experimental TMA with CD68 antibody to detect total macrophages. Although no association between Slit2 and macrophages was observed (Table S5), the patients with tumors positive for macrophages and Slit2 showed better survival as compared to patients devoid of macrophages and expressing Slit2 or patients with macrophages but not expressing Slit2 (Fig. 7C, D). These observations suggest a strong association of Slit2 levels with better survival in breast cancer patients, especially those who harbor macrophages.
Figure 7. Slit2 expression inversely correlates with TAMs and fibrosis in the breast cancer patient.
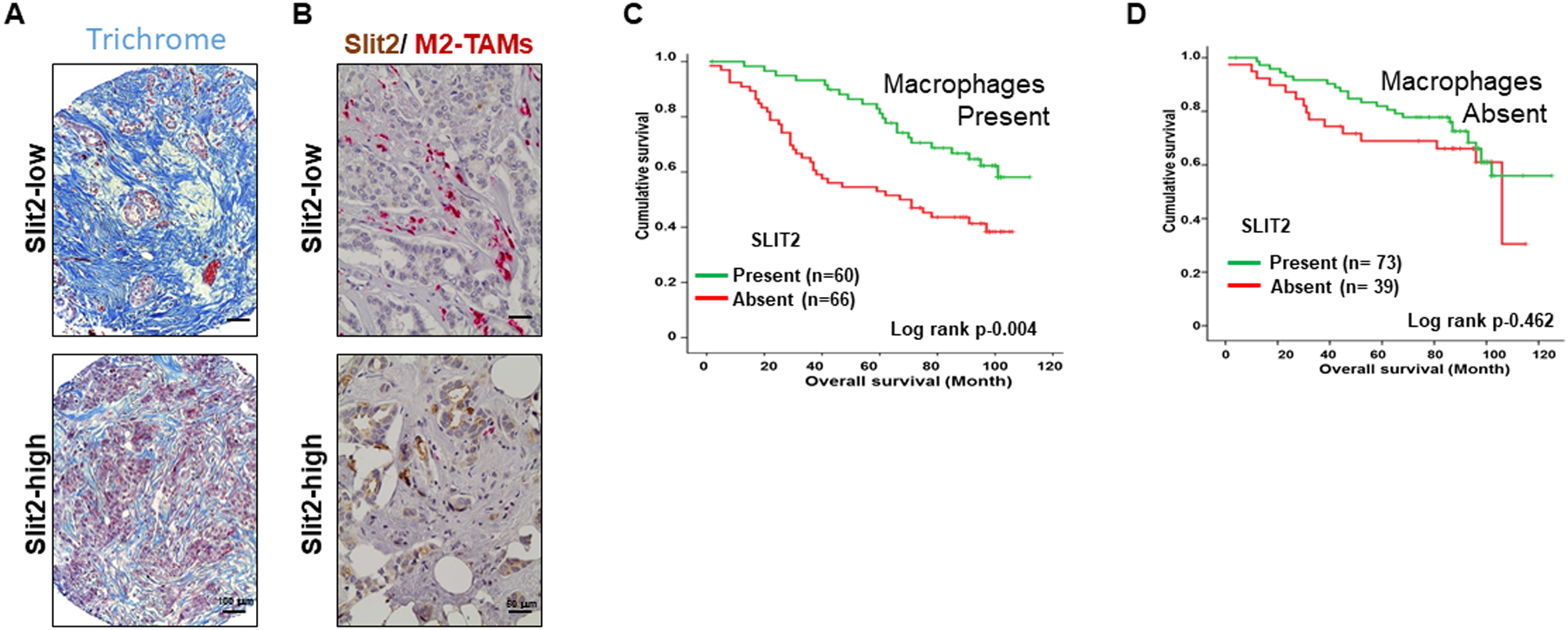
(A) The images showing the detection of fibrosis in patient samples by Trichrome staining.
(B) The representative images of breast cancer patient sample immunostained using antibodies specific for Slit2 and CD163 and double color IHC.
C-D. The TMA was stained with macrophage-specific CD68 antibody and patients were divided into macrophage present or macrophage absent groups. (C) The graph showing the survival analysis of patients with macrophages based on Slit2 expression present or absent. (D) The graph showing the survival analysis of patients without macrophages based on Slit2 expression present or absent.
DISCUSSION
We and others have shown that ectopic expression of Slit2 in human breast cancer cells reduces the rate of tumor growth(9,10). Full-length Slit2 is a large protein of ~200KD size. Slit2-N is a shorter and biologically active ~120KD fragment of Slit2 and therefore, the recombinant form of Slit2-N is a more suitable candidate to test for therapeutic implications. We have also developed novel Slit2-N expressing Adenovirus to evaluate the pre-clinical potential of Slit2-N as a gene therapy. Here, we show that rSlit2-N or Ad-Slit2 treatment inhibits the growth of established tumors, blocks tumor progression, and regress fibrosis in various mouse models of breast cancer. In addition, for the first time, we show that rSlit2-N or Ad-Slit2 treatment markedly reduces metastases to the lungs in various pre-clinical breast cancer mouse models, including spontaneous transgenic MMTV-PyMT, orthotopic and human cell line-derived xenograft tumor mouse models.
The whole tumor RNASeq performed to identify the rSlit2-N-mediated mechanisms revealed various ECM related genes. Increased deposition of fibroblasts-secreted ECM leads to the fibrotic stroma, which negatively correlates with patient survival (40). We found that rSlit2-N treatment can regress tumor-associated fibrosis and these anti-fibrotic mechanisms are linked with the upregulation of macrophage-secreted MMP13. The infiltrative macrophages upregulate MMP13 to degrade ECM and resolve kidney fibrosis (41). Macrophages-derived MMP13 has been recently reported to increase during the resolution of liver and pancreatic cancer fibrosis (42,43). In addition, Slit2 has been reported to resolve fibrosis in non-malignant kidney and lungs (44,45). These studies suggest that rSlit2-N resolves fibrosis in malignant tissues by educating TAMs to degrade ECM.
We did not see a reduction in the number of macrophages recruited to the tumors treated with rSlit2-N. However, macrophages were reduced in the spleen of rSlit2-N treated tumor-bearing mice. These results suggest that tumor-derived factors overcome the macrophage migration inhibitory effects of rSlit2-N, whereas spleen, that is not exposed to these signals and is thus depleted for macrophages. Given that the rate of macrophage infiltration in the tumors was not changed, we asked whether TAMs functions were altered by Slit2. Indeed, macrophages present in the rSlit2-N treated tumors were polarized towards anti-tumor M1-type and possessed enhanced phagocytic activity against tumor cells. Cell-based assays established the ability of rSlit2-N to decrease alternatively activated M2 macrophage markers and enhance the phagocytic activity, which is consistent with blunted tumor-promoting functions. CD206+ TAMs have been shown to escort tumors cells to intravasate into the blood vessels (46). A recent study suggests that CD206+ M2-type TAMs orchestrate early dissemination and metastases of Her2+ mammary tumor cells (47). Inhibition of CD206+ TAMs may explain the reduced metastases observed in Slit2-N treated animals. Molecular targeting strategies against M2-type TAMs show therapeutic benefits (35,48). In addition, the M2-type TAMs are less phagocytic and therapeutically enhancing the phagocytic activity of these TAMs can regress colorectal tumors (31). Depletion of CD206+ TAMs in mammary tumors by rSlit2-N suggests that it can be used as an agent to specifically deplete harmful subtype of TAMs without depleting anti-tumor macrophages.
Our genomics and protein array analysis identified IL6 as a top molecule suppressed by Slit2 in TAMs, as well as in tumor cells. Cell-based in vitro assays allowed us to establish the role of tumor-derived IL6 in promoting M2 polarization and decreasing the phagocytic ability of normal macrophages. Previous reports have also suggested a crucial role of IL6 in polarizing macrophages towards M2 phenotype (38,39). The ability of rSlit2-N to inhibit M2-TAMs polarization and increase phagocytic activity by suppressing IL6 suggests that Slit2 could successfully be used as an agent to activate macrophages against breast cancer. We have also shown that Slit2-N treatment reduces tumor angiogenesis. These results suggest that Slit2 may regulate tumor growth and metastasis by blocking the expansion of tumor vasculature.
We have also analyzed the expression of Slit2 in normal and malignant breast cancer samples and correlated its expression with various prognostic factors. Using publically available datasets, we found that higher Slit2 mRNA expression correlates with better overall survival, lymph node-positive, and distant metastases free survival. We further confirmed the reduced expression of Slit2 protein in invasive and lymph node metastatic primary tumor tissue compared to normal tissue using tissue microarrays. We also showed that patients expressing high Slit2 contain a reduced number of CD163+ M2-TAMs and less tumor fibrosis. It has been shown that fibroblasts-derived Slit2 mRNA in metastatic breast cancer samples is significantly reduced as compared to non-metastatic patient samples (10). In addition, Yu et. al. observed better overall survival with higher Slit2 mRNA expression in prostate cancer patients (49).
Our present study provides strong evidences for the anti-metastatic activity of the N-terminal Slit2 fragment. The mechanistic studies revealed novel functions of Slit2 in activating M2-TAMs into M1-type phagocytic macrophages and thus, exhibit antitumoral activity. We have also shown that Slit2 inhibits tumor fibrosis by activating ECM degrading MMP13 secretion from macrophages. The analysis of clinical samples demonstrates that Slit2 expression is significantly reduced in breast cancer, including high-grade tumors and TNBC. Furthermore, the presence of Slit2 is positively associated with improved patient survival. In addition, we showed Slit2 expression negatively correlates with fibrosis and CD163+ TAMs. These studies suggest that Slit2 possesses several clinical implications. The ability of Slit2-N to resolve tumor fibrosis is novel and can be utilized to prime the fibrotic tumors for chemotherapies and immunotherapies. Furthermore, Slit2 has the ability to re-educating M2-type TAMs to M1 type macrophages. These M1 macrophages were shown to possess phagocytic capabilities against tumor cells. Slit2-mediated effects against TAMs may represent a better approach to improve the anti-tumor functions of macrophages rather deleting them. In addition, Slit2 expression along with CD163+ TAMs could be used as better prognostic biomarkers in breast cancer patients. Overall, our comprehensive studies using in vitro assays, in vivo mouse models and patient samples suggests novel roles of Slit2 in regulating anti-tumor immune responses, especially macrophage-mediated innate immune responses (Fig. 8). Furthermore, soluble Slit2-N could be used as a novel therapeutic strategy to inhibit aggressive and metastatic breast cancers.
Figure 8. Schematic diagram of anti-tumor effects of rSlit2-N.
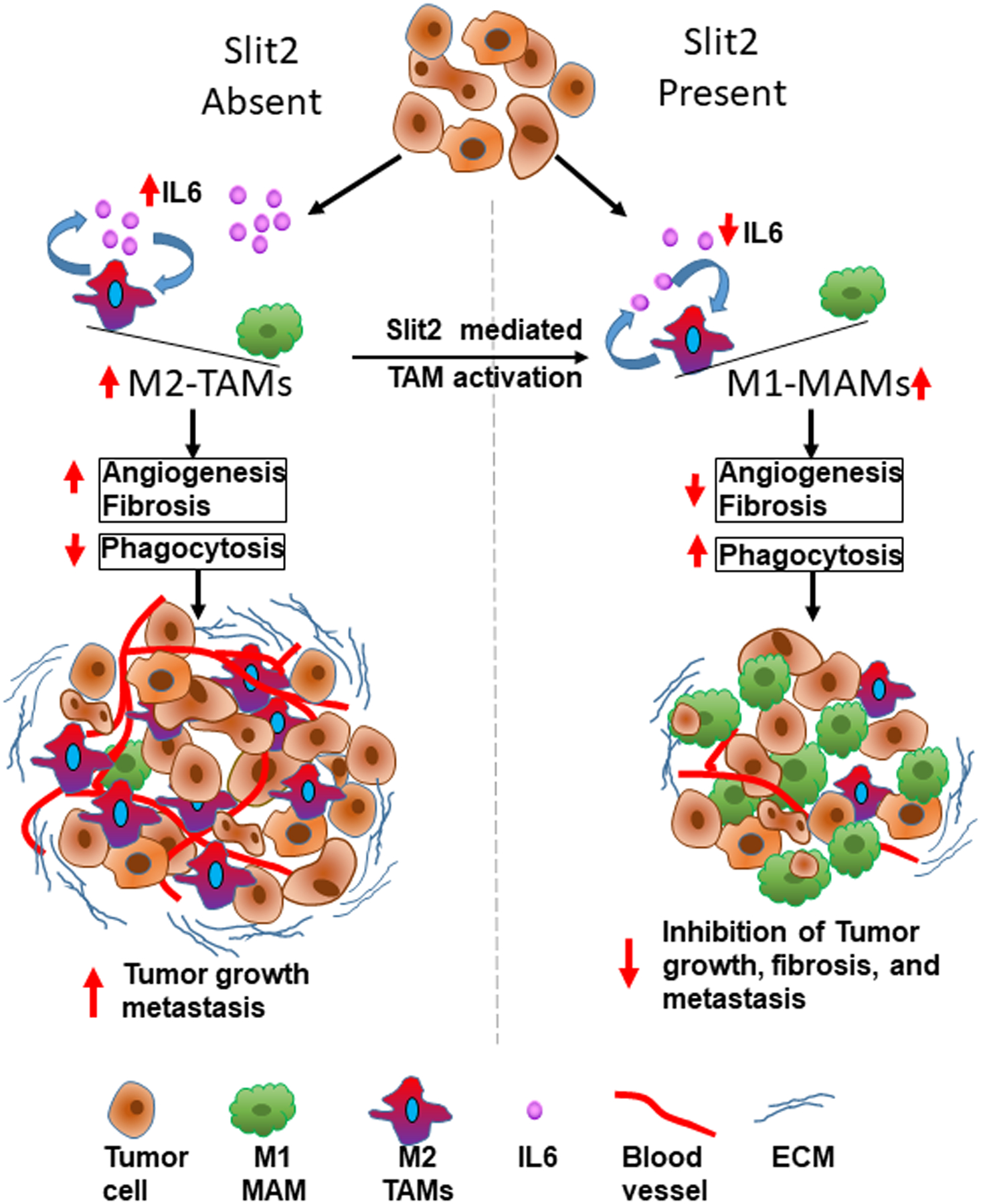
The diagram showing different roles of Slit2 in depleting fibrosis, suppressing TAMs, and enhancing phagocytic activity of macrophages that leads to inhibition of tumor growth and metastasis.
Supplementary Material
SIGNIFICANCE.
This study provides evidence that the antitumor effect of Slit2 in breast cancer occurs by activating the phagocytic activity of M1-like tumor-associated macrophages against tumor cells and diminishing fibrosis.
ACKNOWLEDGMENTS
We thank Dr. Ostrowski, M. for providing MVT1 and Dr. Hinck, L. for providing MDA-MB-231 cells overexpressing Slit2 or vector control. We thank Dr. Sundar, G.V. for assisting with data analysis. We Thank Varun Kavvori, Areeb Narvel and Kunj Vyas for their technical assistance.
Funding:
This work was supported by National Cancer Institute, NIH, USA, grant # R01CA109527, DoD breast cancer breakthrough awards (W81XWH-19-1-0088, W81XWH-17-1-0025, W81XWH-16-1-0037), and Pelotonia grant to RKG and Pelotonia Postdoctoral award to DKA.
Financial support:
This work was supported by National Cancer Institute, NIH, USA, grant # R01CA109527, DoD breast cancer breakthrough awards (W81XW-19-1-0088), and Pelotonia grant to RKG and Pelotonia Postdoctoral award to DKA and SM.
Footnotes
COI: The authors have declared that no conflict of interest exists.
REFERENCES
- 1.Cassetta L, Fragkogianni S, Sims AH, Swierczak A, Forrester LM, Zhang H, et al. Human Tumor-Associated Macrophage and Monocyte Transcriptional Landscapes Reveal Cancer-Specific Reprogramming, Biomarkers, and Therapeutic Targets. Cancer Cell 2019;35:588–602 e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell 2010;141:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res 2007;67:2649–56 [DOI] [PubMed] [Google Scholar]
- 4.Zhu Y, Herndon JM, Sojka DK, Kim KW, Knolhoff BL, Zuo C, et al. Tissue-Resident Macrophages in Pancreatic Ductal Adenocarcinoma Originate from Embryonic Hematopoiesis and Promote Tumor Progression. Immunity 2017;47:597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen IX, Chauhan VP, Posada J, Ng MR, Wu MW, Adstamongkonkul P, et al. Blocking CXCR4 alleviates desmoplasia, increases T-lymphocyte infiltration, and improves immunotherapy in metastatic breast cancer. Proc Natl Acad Sci U S A 2019;116:4558–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macias H, Moran A, Samara Y, Moreno M, Compton JE, Harburg G, et al. SLIT/ROBO1 signaling suppresses mammary branching morphogenesis by limiting basal cell number. Dev Cell 2011;20:827–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nones K, Waddell N, Song S, Patch AM, Miller D, Johns A, et al. Genome-wide DNA methylation patterns in pancreatic ductal adenocarcinoma reveal epigenetic deregulation of SLIT-ROBO, ITGA2 and MET signaling. Int J Cancer 2014;135:1110–8 [DOI] [PubMed] [Google Scholar]
- 8.Dallol A, Da Silva NF, Viacava P, Minna JD, Bieche I, Maher ER, et al. SLIT2, a human homologue of the Drosophila Slit2 gene, has tumor suppressor activity and is frequently inactivated in lung and breast cancers. Cancer Res 2002;62:5874–80 [PubMed] [Google Scholar]
- 9.Prasad A, Paruchuri V, Preet A, Latif F, Ganju RK. Slit-2 induces a tumor-suppressive effect by regulating beta-catenin in breast cancer cells. J Biol Chem 2008;283:26624–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang PH, Hwang-Verslues WW, Chang YC, Chen CC, Hsiao M, Jeng YM, et al. Activation of Robo1 signaling of breast cancer cells by Slit2 from stromal fibroblast restrains tumorigenesis via blocking PI3K/Akt/beta-catenin pathway. Cancer Res 2012;72:4652–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tavora B, Mederer T, Wessel KJ, Ruffing S, Sadjadi M, Missmahl M, et al. Tumoural activation of TLR3-SLIT2 axis in endothelium drives metastasis. Nature 2020;586:299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhosle VK, Mukherjee T, Huang YW, Patel S, Pang BWF, Liu GY, et al. SLIT2/ROBO1-signaling inhibits macropinocytosis by opposing cortical cytoskeletal remodeling. Nat Commun 2020;11:4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cardiff RD, Anver MR, Gusterson BA, Hennighausen L, Jensen RA, Merino MJ, et al. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene 2000;19:968–88 [DOI] [PubMed] [Google Scholar]
- 14.Pei XF, Noble MS, Davoli MA, Rosfjord E, Tilli MT, Furth PA, et al. Explant-cell culture of primary mammary tumors from MMTV-c-Myc transgenic mice. In Vitro Cell Dev Biol Anim 2004;40:14–21 [DOI] [PubMed] [Google Scholar]
- 15.Charan M, Das S, Mishra S, Chatterjee N, Varikuti S, Kaul K, et al. Macrophage migration inhibitory factor inhibition as a novel therapeutic approach against triple-negative breast cancer. Cell Death Dis 2020;11:774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wani N, Nasser MW, Ahirwar DK, Zhao H, Miao Z, Shilo K, et al. C-X-C motif chemokine 12/C-X-C chemokine receptor type 7 signaling regulates breast cancer growth and metastasis by modulating the tumor microenvironment. Breast Cancer Res 2014;16:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadepalli VS, Ozer HG, Yilmaz AS, Pietrzak M, Webb A. BISR-RNAseq: an efficient and scalable RNAseq analysis workflow with interactive report generation. BMC Bioinformatics 2019;20:670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30 [DOI] [PubMed] [Google Scholar]
- 19.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohela M, Casbon AJ, Olow A, Bonham L, Branstetter D, Weng N, et al. Intravital imaging reveals distinct responses of depleting dynamic tumor-associated macrophage and dendritic cell subpopulations. Proc Natl Acad Sci U S A 2014;111:E5086–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–8 [DOI] [PubMed] [Google Scholar]
- 24.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017;19:649–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, et al. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am J Pathol 2003;163:2113–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeFilippis RA, Chang H, Dumont N, Rabban JT, Chen YY, Fontenay GV, et al. CD36 repression activates a multicellular stromal program shared by high mammographic density and tumor tissues. Cancer Discov 2012;2:826–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandler C, Liu T, Buckanovich R, Coffman LG. The double edge sword of fibrosis in cancer. Transl Res 2019;209:55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer 2020;20:174–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duffield JS, Forbes SJ, Constandinou CM, Clay S, Partolina M, Vuthoori S, et al. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J Clin Invest 2005;115:56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon SR, Maute RL, Dulken BW, Hutter G, George BM, McCracken MN, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017;545:495–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Movahedi K, Laoui D, Gysemans C, Baeten M, Stange G, Van den Bossche J, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res 2010;70:5728–39 [DOI] [PubMed] [Google Scholar]
- 33.Buchta Rosean C, Bostic RR, Ferey JCM, Feng TY, Azar FN, Tung KS, et al. Preexisting Commensal Dysbiosis Is a Host-Intrinsic Regulator of Tissue Inflammation and Tumor Cell Dissemination in Hormone Receptor-Positive Breast Cancer. Cancer Res 2019;79:3662–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nasser MW, Qamri Z, Deol YS, Ravi J, Powell CA, Trikha P, et al. S100A7 enhances mammary tumorigenesis through upregulation of inflammatory pathways. Cancer Res 2012;72:604–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beatty GL, Chiorean EG, Fishman MP, Saboury B, Teitelbaum UR, Sun W, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science 2011;331:1612–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buhtoiarov IN, Sondel PM, Wigginton JM, Buhtoiarova TN, Yanke EM, Mahvi DA, et al. Anti-tumour synergy of cytotoxic chemotherapy and anti-CD40 plus CpG-ODN immunotherapy through repolarization of tumour-associated macrophages. Immunology 2011;132:226–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanaka T, Narazaki M, Kishimoto T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb Perspect Biol 2014;6:a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin Z, Ma T, Lin Y, Lu X, Zhang C, Chen S, et al. IL-6/STAT3 pathway intermediates M1/M2 macrophage polarization during the development of hepatocellular carcinoma. J Cell Biochem 2018;119:9419–32 [DOI] [PubMed] [Google Scholar]
- 39.Mauer J, Chaurasia B, Goldau J, Vogt MC, Ruud J, Nguyen KD, et al. Signaling by IL-6 promotes alternative activation of macrophages to limit endotoxemia and obesity-associated resistance to insulin. Nat Immunol 2014;15:423–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whatcott CJ, Diep CH, Jiang P, Watanabe A, LoBello J, Sima C, et al. Desmoplasia in Primary Tumors and Metastatic Lesions of Pancreatic Cancer. Clin Cancer Res 2015;21:3561–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren J, Zhang J, Rudemiller NP, Griffiths R, Wen Y, Lu X, et al. Twist1 in Infiltrating Macrophages Attenuates Kidney Fibrosis via Matrix Metallopeptidase 13-Mediated Matrix Degradation. J Am Soc Nephrol 2019;30:1674–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abe H, Kamimura K, Kobayashi Y, Ohtsuka M, Miura H, Ohashi R, et al. Effective Prevention of Liver Fibrosis by Liver-targeted Hydrodynamic Gene Delivery of Matrix Metalloproteinase-13 in a Rat Liver Fibrosis Model. Mol Ther Nucleic Acids 2016;5:e276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Long KB, Gladney WL, Tooker GM, Graham K, Fraietta JA, Beatty GL. IFNgamma and CCL2 Cooperate to Redirect Tumor-Infiltrating Monocytes to Degrade Fibrosis and Enhance Chemotherapy Efficacy in Pancreatic Carcinoma. Cancer Discov 2016;6:400–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yuen DA, Huang YW, Liu GY, Patel S, Fang F, Zhou J, et al. Recombinant N-Terminal Slit2 Inhibits TGF-beta-Induced Fibroblast Activation and Renal Fibrosis. J Am Soc Nephrol 2016;27:2609–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilling D, Zheng Z, Vakil V, Gomer RH. Fibroblasts secrete Slit2 to inhibit fibrocyte differentiation and fibrosis. Proc Natl Acad Sci U S A 2014;111:18291–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Egeblad M, Ewald AJ, Askautrud HA, Truitt ML, Welm BE, Bainbridge E, et al. Visualizing stromal cell dynamics in different tumor microenvironments by spinning disk confocal microscopy. Dis Model Mech 2008;1:155–67; discussion 65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Linde N, Casanova-Acebes M, Sosa MS, Mortha A, Rahman A, Farias E, et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat Commun 2018;9:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Papadopoulos KP, Gluck L, Martin LP, Olszanski AJ, Tolcher AW, Ngarmchamnanrith G, et al. First-in-Human Study of AMG 820, a Monoclonal Anti-Colony-Stimulating Factor 1 Receptor Antibody, in Patients with Advanced Solid Tumors. Clin Cancer Res 2017;23:5703–10 [DOI] [PubMed] [Google Scholar]
- 49.Yu J, Cao Q, Wu L, Dallol A, Li J, Chen G, et al. The neuronal repellent SLIT2 is a target for repression by EZH2 in prostate cancer. Oncogene 2010;29:5370–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


