Abstract
Multilocus sequence typing was used to characterize isolates of the major Spanish clones of penicillin-resistant and multiple-antibiotic-resistant Streptococcus pneumoniae. Isolates of the multidrug-resistant Spanish serotype 23F clone and serotype variants of this clone either had identical allelic profiles or their allelic profiles differed from this typical allelic profile at only one of the seven housekeeping loci. Similarly, isolates of the Spanish serotype 6B and 14 clones and the penicillin-resistant serotype 9V clone (and serotype variants of this clone) each had the same allelic profiles or profiles that differed at a single locus. Multilocus sequence typing therefore allows resistant pneumococci to be assigned to the Spanish clones if they have the typical allelic profile of the clone or if their profiles differ from that profile at a single locus. A few resistant isolates that had allelic profiles typical of that of a Spanish clone or whose profiles differed from that of the typical profile at only a single locus possessed penicillin-binding protein pbp1a, pbp2b, or pbp2x genes that differed from those that are characteristic of the clone. In most cases these isolates could be assigned as variant members of the clone. Since almost all serotype 9V isolates have very similar genotypes, independently emerging penicillin-resistant clones of this serotype will inevitably appear to be similar by molecular typing procedures. Analysis of the pbp genes, in addition to multilocus sequence typing (or any other molecular typing procedure), is therefore required to assign isolates unambiguously to the penicillin-resistant Spanish serotype 9V clone.
Spain was among the first countries to report a high incidence of antibiotic-resistant Streptococcus pneumoniae (3). Penicillin-resistant and multidrug-resistant isolates were first detected in the late 1970s, and their prevalence increased rapidly during the 1980s to reach a peak in 1989 of about 44% of invasive isolates with penicillin resistance (15). Since 1989, the incidence of penicillin-resistant pneumococci (MICs, ≥0.1 μg of penicillin/ml) has remained fairly constant, with penicillin resistance occurring in about 40% of all isolates (16).
Multidrug-resistant clones of serotypes 23F and 6B and a penicillin-resistant clone of serotype 9V have been common in Spain since the 1980s (10, 12, 35), and more recently, a multidrug-resistant serotype 14 clone has been identified (5). According to the newly recommended nomenclature, the latter clones are now designated Spain23F-1, Spain6B-2, France9V-3 (as this clone may have emerged within France), and Spain14-5. The first three clones have been detected in many countries and on different continents and can be considered globally distributed (2, 19, 20, 25, 26, 28, 29, 31, 34).
Studies from other countries that have a high incidence of penicillin-resistant pneumococci have identified additional distinctive clones which have presumably emerged within these countries (17, 28, 30). In addition to the increasing number of penicillin-resistant clones that are being recognized, variants of the major clones that differ in serotype are commonly observed. For example, serotype 19F variants of the Spain23F-1 clone (4) and serotype 14 variants of the France9V-3 clone (2) are now widely disseminated (7–9, 19).
Penicillin-resistant isolates are assigned to the same clone if they have identical or very similar overall genotypes and identical altered forms of the penicillin-binding protein pbp1a, pbp2b, and pbp2x genes (10). Allelic variants of these pbp genes are usually assigned by gene fingerprinting (4), but a number of different methods have been used to compare the overall relatedness of resistant pneumococci (19, 26, 29, 31, 36). As increasing numbers of penicillin-resistant clones are defined and as the genotypes of clones diversify and their serotypes change, the assignment of isolates to these clones becomes more difficult and a method is required that can unambiguously assign resistant isolates to known clones or new clones without the need for laboratories to exchange reference strains.
Multilocus sequence typing (MLST) is a procedure that fulfils these criteria as it produces unambiguous data that can be transmitted electronically via the Internet (24, 32). By this procedure the nucleotide sequences of ∼450-bp internal fragments of seven housekeeping genes are determined for each isolate. The different sequences at each locus are assigned as distinct alleles, and for each isolate, the alleles at the seven loci define the allelic profile or sequence type. MLST can resolve billions of pneumococcal genotypes, and the probability that two isolates will have the same allelic profile by chance is extremely low (11). The method is highly portable, as any laboratory can compare the sequences at the seven loci in their isolates with those in a central database on the World Wide Web (http://mlst.zoo.ox.ac.uk) and can obtain the allelic profile of each isolate. The relationship of these isolates to the known penicillin-resistant clones can then be established by comparing these allelic profiles with those in the central database, which contains the allelic profiles of the known penicillin-resistant clones (11, 28, 32).
We demonstrate that most isolates of each of the major Spanish clones have the same allelic profile or have a profile that differs from this typical allelic profile at a single locus, allowing resistant pneumococci to be assigned unambiguously to these clones via the Internet.
MATERIALS AND METHODS
Pneumococcal isolates.
The pneumococci included in this study were multiple-antibiotic-resistant pneumococci from Spain (5), penicillin-resistant pneumococci from Taiwan (28), and multiple-antibiotic-resistant serotype 6B isolates assigned to the Spain6B-2 clone by Hermans et al. (19). In addition, we studied a collection of 429 isolates from patients who recently had cases of invasive disease, including 274 isolates from eight countries described by Enright and Spratt (11) and 106 isolates from Spain (12). These isolates have the prefix M. We also searched the MLST database, which includes the allelic profiles of over 700 pneumococci.
MICs were determined according to the guidelines of the British Society for Antimicrobial Chemotherapy (1) by using the E-test (AB Biodisk, Solna, Sweden). Serotypes were kindly determined by David Griffiths, using the Quellung reaction, with sera obtained from the Statens Seruminstitut, Copenhagen, Denmark.
Genetic relatedness of isolates.
The nucleotide sequences of ∼450-bp internal regions from the aroE, ddl, gdh, gki, recP, spi, and xpt genes were amplified by PCR with the primers described previously (11). Both strands of the gene fragments were directly sequenced by using the primers that were used for the PCR and an ABI 377 Prism automated sequencer with BigDye terminators (Perkin-Elmer Applied Biosystems, Foster City, Calif.). For each isolate, the allelic profiles were obtained from the sequences at the seven loci, and the allelic profiles were compared with those in our pneumococcal MLST database by using the software at the pneumococcal MLST website (http://mlst.zoo.ox.ac.uk). The relatedness among isolates was visualized as a dendrogram, constructed from the matrix of pairwise differences between the allelic profiles by using the unweighted pair group method with arithmetic averages (UPGMA).
Analysis of pbp genes.
The pbp1a, pbp2b, and pbp2x genes were amplified from chromosomal DNA by PCR with the primers and conditions described previously (5). The pbp gene fragments were gel purified with a Qiagen QIAEX II Agarose gel extraction kit (Qiagen Ltd., Crawley, United Kingdom), and 50 ng of the purified fragments was used in a second 50-μl PCR mixture (25 cycles) that contained 0.5 μg of each primer (as described above), deoxynucleoside triphosphates (100 μM), rhodamine (R110)-labeled dUTP (1 μM), PCR buffer (Qiagen), and 2.5 U of Qiagen Taq DNA polymerase. The fluorescently labeled pbp gene fragments were ethanol precipitated, resuspended in 40 μl of distilled water, digested for 1 h with AluI (5 U; total reaction volume, 50 μl), ethanol precipitated, and resuspended in 30 μl of distilled water. One microliter of the digested fragments was mixed with 2.3 μl of loading buffer containing TAMRA-labeled GeneScan-500 internal molecular size markers (Perkin-Elmer Applied Biosystems), and the fragments were fractionated on a 4% denaturing polyacrylamide gel with an ABI Prism 377 DNA sequencer with virtual filter set C. The DNA fragment patterns were compared by using Genescan Analysis software (PE Applied Biosystems).
The fingerprints obtained by AluI digestion of the pbp genes of from isolates assigned by MLST to the major resistant Spanish clones were compared to the fingerprints obtained by AluI digestion of pbp genes from previously characterized reference isolates of each clone (5). Those with identical AluI digestion fingerprints were assigned the same allele numbers as those reported previously (5). Isolates that had pbp fingerprints which differed from those typical of the clone were assigned new allele numbers. As these variant pbp fingerprints were not compared with the AluI digestion fingerprints of each of the many alleles identified previously (5), the variant alleles were assigned roman (rather than arabic) numerals.
RESULTS
Analysis of isolates of the Spain23F-1 clone.
Previously characterized isolates of the Spain23F-1 clone (5), including serotype 19A and 19F variants of this clone (6, 7), were analyzed by MLST. All 16 of these isolates had identical allelic profiles (profile 4-4-2-4-4-1-1) except for one isolate (isolate C85) whose profile differed from this profile (Table 1). Additional isolates with the same allelic profiles and isolates whose profiles differed from this typical profile at a single locus were identified among a collection of 74 penicillin-resistant isolates from hospitals in Taiwan (28). Among these Taiwanese isolates, 16 had the typical allelic profile (11 were of serotype 23F and 5 were of serotype 19F), and a further 5 isolates (all of serotype 23F) had identical profiles (profile 4-4-2-4-6-1-1) which differed from the typical allelic profile only at the spi locus. All 21 of the Taiwanese isolates had the same pbp1a, pbp2b, and pbp2x gene fingerprints as the Spain23F-1 clone and the characteristic antibiotic resistance profile of the Spain23F-1 clone (28).
TABLE 1.
Properties of the Spain23F-1 clone
| Strain | Alleles ofa:
|
Serotype | Penicillin G MIC (μg/ml) | Country | Year | Source | Alleles of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpt | ddl | pbp1a | pbp2b | pbp2x | ||||||
| SP264 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 2 | Spain | 1984 | Blood | 1 | 1 | 1 |
| GM4 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 2 | Spain | 1988 | Eye | 1 | 1 | 1 |
| GM28 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 2 | Spain | 1988 | Eye | 1 | 1 | 1 |
| GM54 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 1 | Spain | 1988 | Eye | 1 | 1 | 1 |
| GM116 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 2 | Spain | 1989 | Eye | 1 | 1 | 1 |
| GM145 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 1 | Spain | 1989 | Lung | 1 | 1 | 1 |
| GM70 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19A | 2 | Spain | 1990 | Blood | 1 | 1 | 1 |
| VA79 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19A | 2 | Spain | 1994 | Eye | 1 | 1 | 1 |
| SJD86 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19A | 1 | Spain | 1994 | Blood | 1 | 1 | 1 |
| GA71 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | 2 | Spain | 1990 | Nose | 1 | 1 | 1 |
| SP496 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | 1 | Spain | 1985 | Lung | 1 | 1 | 1 |
| VA91 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | 2 | Spain | 1994 | Nose | 1 | 1 | 1 |
| VA95 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | 1 | Spain | 1994 | Sputum | 1 | 1 | 1 |
| UK577 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 1 | United Kingdom | 1987 | Not known | 1 | 1 | 1 |
| PO329 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | 2 | Poland | 1994 | Sputum | 1 | 1 | 1 |
| C85 | 4 | 4 | 2 | 4 | 8 | 1 | 1 | 19F | 1 | Spain | 1994 | Lung | 1 | 1 | 1 |
| TW41b | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 1 | Taiwan | 1997 | Blood | 1 | 1 | 1 |
| TW28c | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | 1 | Taiwan | 1997 | Sputum | 1 | 1 | 1 |
| TW42d | 4 | 4 | 2 | 4 | 6 | 1 | 1 | 23F | 1 | Taiwan | 1997 | Sputum | 1 | 1 | 1 |
| M41 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 1 | United Kingdom | 1997 | Sputum | 1 | 1 | 1 |
| M329 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 19F | 1 | Spain | 1997 | CSFe | 1 | 1 | 1 |
| M371 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 1 | Spain | 1998 | CSF | NDf | ND | ND |
| M382 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 2 | Spain | 1998 | CSF | ND | ND | ND |
| M414 | 4 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 2 | Spain | 1998 | CSF | ND | ND | ND |
| M170 | 4 | 4 | 2 | 4 | 5 | 1 | 1 | 23F | 2 | Denmark | 1996 | Blood | 1 | 1 | 1 |
| M390 | 8 | 4 | 2 | 4 | 4 | 1 | 1 | 23F | 1 | Spain | 1998 | CSF | ND | ND | ND |
| M405 | 4 | 4 | 2 | 4 | 4 | 1 | 47 | 23F | 2 | Spain | 1998 | CSF | ND | ND | ND |
| M417 | 4 | 4 | 2 | 4 | 4 | 1 | 47 | 23F | 1 | Spain | 1998 | CSF | ND | ND | ND |
Alleles that differ from those found in the typical allelic profile of the Spain23F-1 clone are in boldface type.
Ten additional serotype 23F isolates from Taiwan had this allelic profile and the typical pbp fingerprints of the Spain23F-1 clone.
Four additional serotype 19F isolates from Taiwan had this allelic profile and the typical pbp fingerprints of the Spain23F-1 clone.
Four additional serotype 23F isolates from Taiwan had this allelic profile and the typical pbp fingerprints of the Spain23F-1 clone.
CSF, cerebrospinal fluid.
ND, not determined.
Five isolates with the typical allelic profile and four single-locus variants were found among a collection of 429 isolates (those with an M prefix) from patients with serious invasive disease (11, 12). One isolate with the typical allelic profile was serotype 19F, but the other eight isolates were serotype 23F; all were penicillin resistant and had the characteristic antibiotic resistance profile of the Spain23F-1 clone. The pbp1a, pbp2b, and pbp2x gene fingerprints of two isolates with the typical allelic profile (isolates M41 and M329) and one of the single-locus variants (isolate M170) were examined, and all were typical of those of the Spain23F-1 clone.
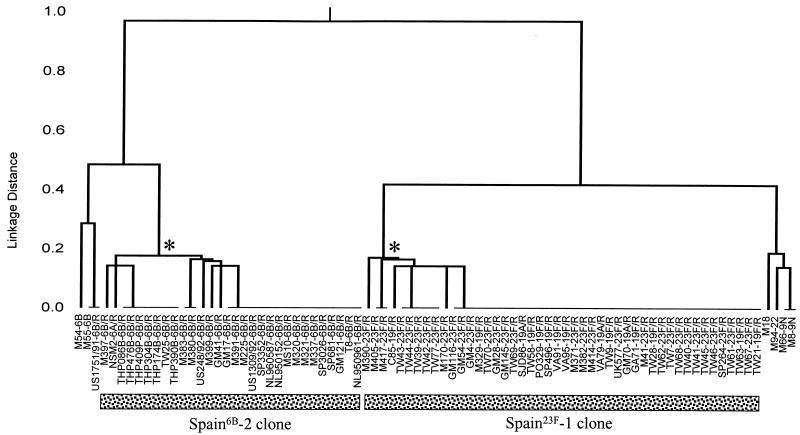
The relationships among all isolates in the MLST database that differed at less than or equal to three of the seven alleles from the typical allelic profile of the Spain23F-1 clone are shown in Fig. 1. The allelic profiles of none of the isolates in the MLST database differed from the typical allelic profile at two of seven loci. The profiles of four isolates differed at three of seven loci. The serotype of isolate M18 and the MIC of penicillin for M18 is not known since it has not survived; the other three isolates were not serotype 23F (or serotype 19A or 19F) and were penicillin susceptible. The isolates of the Spain23F-1 clone were therefore well resolved from all other isolates in the MLST database.
FIG. 1.
Relationships among isolates of the Spain23F-1 and Spain6B-2 clones and isolates with similar allelic profiles. A dendrogram was constructed by the UPGMA method to show the relatedness of all isolates that had allelic profiles which differed from the typical allelic profiles of the Spain23F-1 clone (profile 4-4-2-4-4-1-1) and the Spain6B-2 clone (profile 5-6-1-2-6-3-4) at less than or equal to three of the seven loci. The name of each isolate is followed (separated by a hyphen) by the serotype, and penicillin-resistant isolates are identified by the suffix /R. Isolates below each horizontal line have identical allelic profiles. The asterisks show the nodes that define the isolates of the Spain23F-1 and Spain6B-2 clones. For each clone, the profiles of all of these isolates were either identical to that of the Spain23F-1 or Spain6B-2 clones or their profiles differed from the typical profiles of the Spain23F-1 and Spain6B-2 clones at a single locus.
Analysis of isolates of the Spain6B-2 clone.
The allelic profiles of three previously characterized isolates of the Spain6B-2 clone were identical (profile 5-6-1-2-6-3-4) (Table 2). These isolates have been shown to be indistinguishable by multilocus enzyme electrophoresis (MLEE) (16 loci were examined) and by pbp1a, pbp2b, and pbp2x gene fingerprinting (5). One further isolate (isolate SP3026) that has been assigned to the Spain6B-2 clone also had the allelic profile mentioned above.
TABLE 2.
Properties of the Spain6B-2 clone and related isolates
| Strain | Alleles ofa:
|
Serotype | Penicillin G MIC (μg/ml) | Country | Year | Source | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpi | ddl | ||||||
| SP681 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1 | Spain | 1986 | Eye |
| GM17 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1 | Spain | 1988 | Abscess |
| GM121 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1.5 | Spain | 1989 | Blood |
| SP3026 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1 | Spain | NKb | Sputum |
| MS10 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1 | Spain | 1989 | Eye |
| T8 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 2 | Spain | 1989 | Ear |
| NSM2 | 5 | 6 | 39 | 2 | 6 | 3 | 4 | 6A | 1 | Spain | 1989 | Sputum |
| GM41 | 5 | 6 | 1 | 2 | 6 | 3 | 48 | 6B | 1 | Spain | 1988 | Blood |
| NL950152 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1.5 | Netherlands | 1995 | Sputum |
| NL950961 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1.5 | Netherlands | 1995 | Sputum |
| NL960587 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1.5 | Netherlands | 1996 | Nose |
| SP3352 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1.5 | Spain | 1995 | Blood |
| US1309/92 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1 | United States | 1992 | Sinus |
| US248/92 | 5 | 6 | 1 | 2 | 34 | 3 | 4 | 6B | 2 | United States | 1992 | Blood |
| ThP086B | 5 | 6 | 33 | 2 | 6 | 3 | 4 | 6B | 1.5 | Thailand | 1993-1994 | Nasopharynx |
| ThP174B | 5 | 6 | 33 | 2 | 6 | 3 | 4 | 6B | 1.5 | Thailand | 1993-1994 | Nasopharynx |
| ThP390B | 5 | 6 | 33 | 2 | 6 | 3 | 4 | 6B | 1.5 | Thailand | 1993-1994 | Nasopharynx |
| ThP304B | 5 | 6 | 33 | 2 | 6 | 3 | 4 | 6B | 1.5 | Thailand | 1993-1994 | Nasopharynx |
| ThP409P | 5 | 6 | 33 | 2 | 6 | 3 | 4 | 6B | 1.5 | Thailand | 1993-1994 | Nasopharynx |
| ThP476B | 5 | 6 | 33 | 2 | 6 | 3 | 4 | 6B | 1.5 | Thailand | 1993-1994 | Nasopharynx |
| TW25 | 5 | 6 | 33 | 2 | 6 | 3 | 4 | 6B | 1 | Taiwan | 1997 | Sputum |
| US1751/91 | 7 | 6 | 1 | 2 | 6 | 15 | 14 | 6B | 1 | United States | 1991 | Ear |
| NL951061 | 7 | 8 | 1 | 1 | 15 | 14 | 50 | 6B | 1 | The Netherlands | 1995 | NK |
| M225 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1.5 | Australia | 1997 | Blood |
| M320 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 0.5 | Spain | 1997 | CSFc |
| M321 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 0.5 | Spain | 1997 | CSF |
| M337 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1 | Spain | 1997 | CSF |
| M391 | 5 | 6 | 1 | 2 | 6 | 3 | 4 | 6B | 1 | Spain | 1998 | CSF |
| M380 | 5 | 6 | 1 | 15 | 6 | 3 | 4 | 6B | 1 | Spain | 1998 | CSF |
| M383 | 5 | 6 | 1 | 15 | 6 | 3 | 4 | 6B | 1 | Spain | 1998 | CSF |
| M397 | 5 | 6 | 41 | 2 | 6 | 3 | 4 | 6B | 0.5 | Spain | 1998 | CSF |
| M399 | 5 | 6 | 1 | 2 | 6 | 3 | 54 | 6B | 2 | Spain | 1998 | CSF |
Alleles that differ from those in the typical allelic profile of the Spain6B-2 clone are in boldface type.
NK, not known.
CSF, cerebrospinal fluid.
Three multidrug-resistant serogroup 6 isolates that have been shown (5) to be closely related in overall genotype to the Spain6B-2 clone but which differed in that they have variant forms of the pbp2b gene (isolates T8, NSM2, and MS10) were also examined. MS10 and T8 (both of which were of serotype 6B) had the typical allelic profile of the Spain6B-2 clone, whereas NSM2 (serotype 6A) differed only at gki (profile 5-6-39-2-6-3-4). These isolates were considered minor variants of the Spain6B-2 clone. A further isolate (isolate GM41, serotype 6B) was also closely related to the other isolates of the Spain6B-2 clone as determined by MLEE but possessed different forms of the pbp1a, pbp2b, and pbp2x genes. The allelic profile of this isolate (profile 5-6-1-2-6-3-48) differed from the typical allelic profile of the Spain6B-2 clone only at ddl.
We also examined 14 multidrug-resistant serotype 6B isolates assigned previously by Hermans et al. (19) to the Spain6B-2 clone on the basis of restriction fragment end labeling (Table 2). One of these isolates (isolate NL-951061) was clearly not related to the Spain6B-2 clone, as its allelic profile differed at six of seven loci from the typical allelic profile of the Spain6B-2 clone and it possessed pbp1a, pbp2b, and pbp2x genes that differed from those of the Spain6B-2 clone (19). Another of these isolates (isolate US1751/91) was also not closely related to the Spain6B-2 clone, as its profile differed from the typical allelic profile at three of seven loci and it had pbp2b and pbp2x genes that differed from those of the Spain6B-2 clone (19). Five of the other 12 isolates had the typical allelic profile (profile 5-6-1-2-6-3-4), one isolate from the United States differed from the Spain6B-2 clone only at spi (profile 5-6-1-2-35-3-4), and the other six isolates, all of which were from Thailand, were identical single-locus variants that differed from the Spain6B-2 clone at gki (profile 5-6-33-2-6-3-4). One isolate among the 74 penicillin-resistant pneumococci from Taiwan (isolate TW25) was identical to the single locus variants from Thailand, as determined by MLST, and had the characteristic pbp gene fingerprints and antibiotic resistance profile of the Spain6B-2 clone (28). All of the 12 isolates from the study of Hermans et al. (19) that were assigned to the Spain6B-2 clone and the isolate from Taiwan had the characteristic pbp1a and pbp2x genes of the Spain6B-2 clone, although, as found previously (19), 6 of the former strains had variant pbp2b genes (data not shown).
Among our collection of 429 invasive isolates, 5 isolates had the typical allelic profile of the Spain6B clone (profile 5-6-1-2-6-3-4) and 4 isolates were single-locus variants (Table 2). All were multiple-antibiotic-resistant serotype 6B isolates from Spain except for which strain M225, was from Australia.
Figure 1 shows the relationships among the isolates in the MLST database whose allelic profiles differ from the typical allelic profile of the Spain6B-2 clone at less than or equal to three of the seven loci. The profiles of none of the isolates in the MLST database differed from the typical allelic profile at two of seven loci, but the profiles of three isolates (isolates M54, M55, and US1751/91) differed at three of seven loci. All three isolates were of serotype 6B, but only one was penicillin resistant. The last isolate (isolate US1751/91) was discussed above, and the difference in its allelic profile and its pbp1a and pbp2x genes from those for the Spain6B-2 clone strongly suggests it is not a member of the Spain6B-2 clone. Except for strain GM41, which appeared to be a member of the Spain6B-2 clone but which had different pbp genes, the isolates of the Spain6B-2 clone were well resolved from all other isolates in the MLST database.
Analysis of isolates of the France9V-3 clone.
A reference isolate (isolate SP665) of the France9V-3 clone (4) had allelic profile 7-11-10-1-6-8-1 (Table 3). A second reference strain from France (strain TL7/1993 [22]) had the identical allelic profile. Fourteen isolates with this allelic profile were found among the collection of 429 invasive isolates (those with an M prefix). All of these isolates were resistant to penicillin. Nine were serotype 9V and had the same pbp1a, pbp2b, and pbp2x gene fingerprints as the reference isolates of the France9V-3 clone. Two further penicillin-resistant serotype 9V isolates from Poland in the MLST database (isolates 181 and 337) also had the same allelic profile and pbp gene fingerprints as the France9V-3 clone.
TABLE 3.
Properties of the France9V-3 clone and related lineages
| Strain | Alleles ofa:
|
Serotype | Penicillin G MIC (μg/ml) | Country | Year | Source | Alleles ofb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpt | ddl | pbp1a | pbp2b | pbp2x | ||||||
| SP665 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 2 | Spain | 1988 | Ear | 1 | 1 | 1 |
| M15 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 1 | United Kingdom | 1997 | Blood | 1 | 1 | 1 |
| M40 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 2 | United Kingdom | 1997 | Sputum | 1 | 1 | 1 |
| M192 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 1 | United Kingdom | 1996 | CSFc | 1 | 1 | 1 |
| M296 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 1 | Canada | 1996 | CSF | 1 | 1 | 1 |
| M357 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 2 | Spain | 1997 | CSF | 1 | 1 | 1 |
| 181 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 1 | Poland | 1994 | Throat | 1 | 1 | 1 |
| 337 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 0.5 | Poland | 1994 | Sputum | 1 | 1 | 1 |
| M386 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 1 | Spain | 1998 | CSF | ND | ND | ND |
| M395 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 1 | Spain | 1998 | CSF | ND | ND | ND |
| M412 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 1 | Spain | 1998 | CSF | ND | ND | ND |
| M420 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 9V | 1 | United Kingdom | 1999 | CSF | ND | ND | ND |
| M134 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 14 | 2 | Denmark | 1997 | Blood | 1′ | 1 | 1 |
| M291 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 14 | 2 | Uruguay | 1996 | Blood | 1′ | 1 | 1 |
| M322 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 14 | 1 | Spain | 1997 | CSF | 1′ | 1 | 1 |
| M339 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 14 | 0.5 | Spain | 1997 | CSF | 1′ | 1 | 1 |
| M359 | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 14 | 1 | Spain | 1997 | CSF | 1′ | 1 | 1 |
| U2d | 7 | 11 | 10 | 1 | 6 | 8 | 1 | 14 | 2 | Uruguay | 1994 | Blood | 1′ | 1 | 1 |
| M144 | 7 | 11 | 10 | 1 | 1 | 8 | 1 | 9V | 1.5 | Denmark | 1997 | Blood | 1 | 1 | 1 |
| M141 | 7 | 11 | 10 | 1 | 20 | 8 | 1 | 9V | 1.5 | Denmark | 1996 | Blood | 1 | 1 | 1 |
| M143 | 7 | 11 | 10 | 1 | 21 | 8 | 1 | 9V | 2 | Denmark | 1997 | Blood | 1 | 1 | 1 |
| TW50 | 7 | 11 | 10 | 1 | 6 | 1 | 1 | 9V | 1.5 | Taiwan | 1997 | Pus | 1 | 1 | 1 |
| M288 | 7 | 11 | 10 | 1 | 6 | 34 | 1 | 14 | 1.5 | Uruguay | 1996 | CSF | 1′ | 1 | 1 |
| M325 | 7 | 11 | 10 | 1 | 4 | 8 | 1 | 14 | 0.5 | Spain | 1997 | CSF | 1′ | 1 | vii |
| M350 | 1 | 11 | 10 | 1 | 6 | 8 | 1 | 14 | 2 | Spain | 1997 | CSF | 1′ | 1 | 1 |
| M403 | 7 | 5 | 10 | 1 | 6 | 8 | 1 | 14 | 2 | Spain | 1998 | CSF | ND | ND | ND |
| M415 | 7 | 34 | 10 | 1 | 6 | 8 | 1 | 14 | 1 | Spain | 1998 | CSF | ND | ND | ND |
| U1 | 7 | 11 | 10 | 1 | 6 | 42 | 1 | 14 | 2 | Uruguay | 1993 | CSF | 1′ | 1 | 1 |
| M64 | 7 | 11 | 10 | 1 | 6 | 8 | 14 | 9V | 0.012 | United Kingdom | 1994 | CSF | S | S | S |
| M295 | 7 | 11 | 10 | 1 | 6 | 8 | 14 | 14 | 0.5 | Canada | 1995 | CSF | vi | viii | ix |
| M189 | 7 | 11 | 10 | 1 | 6 | 8 | 14 | 19F | 0.012 | United Kingdom | 1996 | CSF | S | S | S |
| M190 | 7 | 11 | 10 | 1 | 6 | 8 | 14 | 19F | 0.012 | United Kingdom | 1996 | CSF | S | S | S |
| M299 | 7 | 11 | 10 | 1 | 6 | 8 | 14 | 19F | 0.032 | Canada | 1995 | Blood | S | S | S |
| M78 | 7 | 11 | 10 | 5 | 6 | 8 | 14 | 9V | 0.012 | United Kingdom | 1996 | Blood | S | S | S |
| M142 | 7 | 11 | 10 | 1 | 1 | 8 | 14 | 9V | 0.023 | Denmark | 1997 | Blood | S | S | S |
| M180 | 7 | 11 | 10 | 1 | 10 | 8 | 14 | 9V | 0.012 | United Kingdom | 1995 | CSF | S | S | S |
| M182 | 7 | 11 | 10 | 1 | 10 | 8 | 14 | 9V | 0.006 | United Kingdom | 1995 | CSF | S | S | S |
| M187 | 7 | 11 | 10 | 1 | 10 | 8 | 14 | 19F | 0.008 | United Kingdom | 1996 | CSF | S | S | S |
| M198 | 7 | 11 | 10 | 12 | 6 | 8 | 14 | 9V | 0.016 | United Kingdom | 1995 | CSF | S | S | S |
| M14 | 7 | 11 | 10 | 6 | 6 | 8 | 14 | 19F | 0.012 | United Kingdom | NKe | Blood | S | S | S |
Alleles that differ from those found in the typical allelic profile of the France9V-3 clone are in boldface type.
Alleles in arabic numerals correspond to those in reference 5 allele 1′ is the variant of pbp1a allele 1 found in the serotype 14 isolates of the France9V-3 clone; alleles in lowercase roman numerals differ from those characteristic of the France9V-3 clone, but they were not compared with those in reference 5. S, penicillin-susceptible isolates; ND, not determined.
CSF, cerebrospinal fluid.
Five other penicillin-resistant serotype 14 isolates from Uruguay had the same allelic profile.
NK, not known.
The other five invasive isolates with the allelic profile 7-11-10-1-6-8-1 and six further isolates from Uruguay with this profile were serotype 14 and had the pbp2x and pbp2b gene fingerprints characteristic of this clone but had a slightly different pbp1a gene fingerprint (Table 3). These serotype 14 isolates have been shown to be variants of the France9V-3 clone (2) that have arisen by a recombinational exchange which replaces the capsular biosynthetic region and part of the downstream pbp1a gene with the corresponding region from a serotype 14 donor strain (8).
The allelic profiles of one penicillin-resistant isolate from Taiwan (strain TW50) and one from Uruguay (strain U1) differed from the typical allelic profile of the France9V-3 clone at a single locus, and there were an additional 9 penicillin-resistant single-locus variants among the 429 invasive isolates. All of the penicillin-resistant serotype 9V single-locus variants produced pbp1a fingerprints identical to that of the France9V-3 clone, whereas (except for strain M295) the resistant serotype 14 single-locus variants had the variant pbp1a fingerprint characteristic of the serotype 14 variants of this clone (8). The pbp1a, pbp2b, and pbp2x fingerprints of resistant serotype 14 isolate M295 were each different from those of the France9V-3 clone.
There were four penicillin-susceptible single-locus variants of the France9V-3 clone among the collection of invasive isolates. One of these was serotype 9V, and the others were serotype 19F; all had the same allelic profile (profile 7-11-10-1-6-8-14). The profiles of a further seven invasive isolates (five serotype 9V isolates and two serotype 19F isolates differed from the profile of the France9V-3 clone at two loci. All of these were penicillin susceptible (Table 3).
One striking feature of the isolates whose allelic profiles differed from the allelic profile of the France9V-3 clone at one or two loci was that, except for strain M295, those that had allele 14 at the ddl locus were penicillin susceptible, whereas those with allele 1 were invariably penicillin resistant. A possible explanation for this finding is given below.
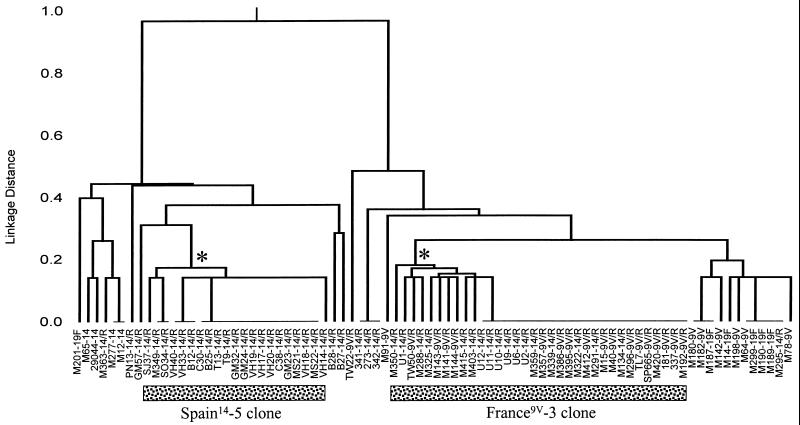
Figure 2 shows the relatedness of all isolates in the MLST database whose allelic profiles differ from the typical allelic profile of the France9V-3 clone at less than or equal to three of the seven loci.
FIG. 2.
Relationships among isolates of the France9V-3 and Spain14-5 clones and isolates with similar allelic profiles. The dendrogram includes all isolates with allelic profiles that differ from the typical allelic profiles of the France9V-3 clone (profile 7-11-10-1-6-8-1) and Spain6B-2 clone (profile 5-6-1-2-6-3-4) at less than or equal to three of the seven loci. The isolate designations are as described in the legend to Fig. 1. The asterisks show the nodes that define the isolates of the France9V-3 and Spain6B-2 clones. For each clone, all of these isolates were either identical to that of the France9V-3 or Spain6B-2 clones or differed from the typical profiles of the France9V-3 and Spain6B-2 clones at a single locus.
Analysis of isolates of the Spain14-5 clone.
Eighteen previously characterized isolates of the Spain14-5 clone were analyzed. All of these isolates have been shown to be closely related in terms of their overall genotypes and to have identical pbp1a, pbp2b, and pbp2x gene fingerprints (5). Fourteen had the same allelic profile (profile 1-5-4-11-9-3-16), three were identical single-locus variants (profile 1-5-4-18-9-3-16), and one was a different single-locus variant (profile 1-5-4-4-9-3-16) (Table 4).
TABLE 4.
Properties of the Spain14-5 clone and related isolates
| Strain | Alleles ofa:
|
Serotype | Penicillin G MIC (μg/ml) | Country | Year | Source | Alleles of:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpt | ddl | pbp1a | pbp2b | pbp2x | ||||||
| VH18 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 1 | Spain | 1990 | Blood | 25 | 19 | 46 |
| VH20 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 1 | Spain | 1990 | Blood | 25 | 19 | 46 |
| MS21 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 1 | Spain | 1990 | Nose | 25 | 19 | 46 |
| MS22 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 2 | Spain | 1990 | Lung | 25 | 19 | 46 |
| GM23 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 1 | Spain | 1993 | Blood | 25 | 19 | 46 |
| C38 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 2 | Spain | 1994 | Blood | 25 | 19 | 46 |
| VH17 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 1 | Spain | 1990 | Blood | 25 | 19 | 46 |
| VH19 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 1 | Spain | 1990 | Blood | 25 | 19 | 46 |
| GM24 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 1 | Spain | 1993 | Blood | 25 | 19 | 46 |
| GM32 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 2 | Spain | 1990 | Pus | 25 | 19 | 46 |
| T9 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 0.5 | Spain | 1991 | Eye | 25 | 19 | 46 |
| T13 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 2 | Spain | 1990 | Ear | 25 | 19 | 46 |
| B25 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 2 | Spain | 1993 | Sputum | 25 | 19 | 46 |
| C30 | 1 | 5 | 4 | 11 | 9 | 3 | 16 | 14 | 1 | Spain | 1994 | Eye | 25 | 19 | 46 |
| VH14 | 1 | 5 | 4 | 4 | 9 | 3 | 16 | 14 | 2 | Spain | 1990 | Blood | 25 | 19 | 46 |
| B12 | 1 | 5 | 4 | 18 | 9 | 3 | 16 | 14 | 2 | Spain | 1992 | Blood | 25 | 19 | 46 |
| VH33 | 1 | 5 | 4 | 18 | 9 | 3 | 16 | 14 | 2 | Spain | 1990 | Blood | 25 | 19 | 46 |
| VH40 | 1 | 5 | 4 | 18 | 9 | 3 | 16 | 14 | 1 | Spain | 1994 | Blood | 25 | 19 | 46 |
| SJ37 | 1 | 5 | 4 | 11 | 9 | 3 | 4 | 14 | 1 | Spain | 1994 | Blood | 25 | 28 | 46 |
| SO34 | 1 | 5 | 4 | 11 | 9 | 3 | 47 | 14 | 1 | Spain | 1990 | Ear | 25 | 34 | 51 |
| M349 | 1 | 5 | 4 | 11 | 9 | 3 | 47 | 14 | 1 | Spain | 1997 | CSFb | NDc | ND | ND |
| B27 | 1 | 5 | 2 | 11 | 9 | 3 | 51 | 14 | 1 | Spain | 1994 | Pus | 25 | 32 | 45 |
| B28 | 1 | 5 | 2 | 1 | 9 | 3 | 49 | 14 | 1 | Spain | 1994 | Blood | 25 | 33 | 50 |
| GM57 | 1 | 5 | 4 | 11 | 9 | 43 | 52 | 14 | 0.5 | Spain | 1988 | Blood | 28 | 21 | 1 |
Alleles that differ from those in the typical allelic profile of the Spain14-5 clone are in boldface type.
CSF, cerebrospinal fluid.
ND, not determined.
The allelic profiles of two additional multidrug-resistant serotype 14 isolates that had been shown to be closely related to the Spain14-5 clone by MLEE also differed from the typical allelic profile at only a single locus. However, these isolates had pbp2b genes (strain SJ37) or pbp2b and pbp2x genes (strain SO34) that differed from those of the Spain14-5 clone. No pneumococci that had the typical allelic profile of the Spain14-5 clone were found among the collection of 429 invasive isolates, but one invasive isolate from Spain (isolate M349) was a single-locus variant that had the same allelic profile as SO34. This isolate was resistant to penicillin and had the typical antibiotic resistance profile of the Spain14-5 clone. These isolates were tentatively assigned as variant members of the Spain14-5 clone.
Figure 2 shows the relatedness of all isolates in the MLST database whose allelic profiles differ from the allelic profile of the Spain14-5 clone at less than or equal to three of the seven loci. The allelic profile of one resistant isolate (isolate GM57) that had pbp1a, pbp2b, and pbp2x genes which differed from those of the Spain14-5 clone and which has been shown to be only distantly related to this clone by MLEE (5) differed at two of seven loci as determined by MLST. This isolate also differed from the Spain14-5 clone in that it was susceptible to tetracycline and was not considered a member of the clone. The profiles of two further well-characterized resistant isolates (isolates B27 and B28) differed at two of seven and three of seven loci, respectively, and they also had different pbp2b and pbp2x genes (5) and were not considered members of the Spain14-5 clone. Besides the isolates described above, the profiles of none of the penicillin-susceptible or -resistant isolates in the MLST database differed at two of seven loci. The profiles of seven isolates (six serotype 14 and one serotype 19F) differed at three of seven loci (Fig. 2). Three of these serotype 14 isolates were resistant to penicillin (MICs, ≥1 μg/ml), and although their pbp genes were not examined, they are very unlikely to be members of the Spain14-5 clone.
DISCUSSION
The identification of the major clones, minor clones, and unique isolates among a collection of penicillin-resistant S. pneumoniae isolates that have been recovered within a community over a short time period is usually relatively straightforward. Many molecular typing procedures can successfully identify the clusters of closely related isolates that indicate the predominant penicillin-resistant clones in such populations (10, 21, 35). However, determination of whether these clones are novel or correspond to penicillin-resistant clones that have been described previously is more problematic. The difficulty arises from the fact that several of the major penicillin-resistant clones probably arose in the 1970s, and as they have spread their genotypes have diversified and their serotypes have changed so that they are no longer uniquely defined by any of the molecular typing procedures that are commonly used.
A second problem arises because penicillin resistance has emerged predominantly in a small number of serotypes, typically those that are commonly carried by children (serotypes 6B, 9V, 14, 19F, 23F [27]), and although isolates of the same serotype are often only distantly related in terms of overall genotype (11), most isolates of a few serotypes (e.g., serotypes 9V and 7F) appear to have very similar genotypes (11, 18, 23, 33). The genotypes of two penicillin-resistant clones of one of the latter serotypes that emerge independently will therefore almost inevitably appear to be closely related. The similarity of genotypes therefore does not always imply that two penicillin-resistant isolates are descended from the same ancestral resistant isolate.
MLST provides a new way of defining the genotype of a penicillin-resistant isolate as a string of integers, the alleles at each of the seven housekeeping loci. Comparison of allelic profiles is less ambiguous than comparison of patterns of DNA fragments on agarose gels, but as with all molecular typing procedures, there can be some ambiguity in assigning penicillin-resistant isolates as members of a clone, as variants of the clone, or as genetically similar isolates that have emerged independently. This problem can largely be circumvented by analyzing the pbp1a, pbp2b, and pbp2x genes, as well as the overall genotype, as it is extremely unlikely that independently arising penicillin-resistant clones will have acquired indistinguishable mosaic pbp genes.
A major advantage of MLST is that the allelic profiles of pneumococcal isolates, including reference isolates of all of the known penicillin-resistant and multidrug-resistant clones, together with epidemiological data, can be stored in a single central database which can be interrogated remotely via the Internet (32). MLST can be used to identify the major clones among large collections of resistant pneumococci from a geographic region (28), but for many laboratories this may not be practical, and in this case MLST can be applied to a single example of each clone identified by other methods (e.g., pulsed-field gel electrophoresis). By using software available on the pneumococcal MLST website (http://mlst.zoo.ox.ac.uk), the sequences at the seven loci can be used to identify whether each of the identified penicillin-resistant clones is already known or whether the sequences define new clones. The allelic profiles of the isolates and the associated epidemiological data can be deposited in the pneumococcal database on the MLST website to provide a tool for global epidemiology and surveillance.
This paper addresses the variation in allelic profiles that is found among the Spanish clones of penicillin-resistant pneumococci (which are probably among the oldest and which are certainly the most widely disseminated of such clones), as this determines the ease with which they can be identified unambiguously by MLST.
Previously characterized isolates of each penicillin-resistant clone had the same allelic profile (the typical profile) or their profiles differed from this profile at only a single locus. For example, all isolates with the typical allelic profile of the Spain23F-1 clone and all single-locus variants expressed high-level resistance to penicillin and had the antibiotic resistance profile and the typical pbp gene fingerprints of this clone. The allelic profiles of none of the isolates in the database differed from the typical allelic profile of the Spain23F-1 clone at two loci, and all the isolates whose profiles differed at three loci were penicillin susceptible. The absence from the MLST database of penicillin-susceptible isolates that are closely related in genotype to the Spain23F-1 clone suggests that the parental serotype 23F isolate from which the Spain23F-1 clone emerged may have had a rare genotype. The rarity of closely related genotypes allows an unambiguous identification of this clone by MLST, and a multidrug-resistant pneumococcus that has the typical allelic profile of the Spain23F-1 clone or whose profile differs from this profile at a single locus can confidently be assigned as a member of the Spain23F-1 clone.
Isolates of the Spain6B-2 clone that had the characteristic pbp gene fingerprints of this clone also had identical allelic profiles or profiles that differed from this profile at a single locus. However, in contrast to the Spain23F-1 clone, which had uniform pbp gene fingerprints, there was some variation among isolates of the Spain6B-2 clone in terms of their pbp2b fingerprints. Furthermore, one isolate (isolate GM41) whose allelic profile differed from the typical allelic profile of the Spain6B-2 clone at only a single locus had pbp1a, pbp2b, and pbp2x genes that were distinct from those found in this clone. This isolate was also very closely related to the Spain6B-2 clone by MLEE (5) and, on the basis of the criterion of genetic relatedness, would be assigned to the Spain6B-2 clone, although the differences in all three pbp genes suggest that it is not a member of the clone. This is probably an example of the independent emergence of penicillin resistance in a serotype 6B isolate that had a genotype similar to that of the Spain6B-2 clone.
Some of the penicillin-resistant serotype 6B isolates that we examined by MLST had been identified as members of the Spain6B-2 clone by using restriction fragment end labeling (19). Although most of these isolates were confirmed to be members of the Spain6B-2 clone by MLST, two isolates were clearly not members of this clone. These isolates differed from the Spain6B-2 clone at three of seven and six of seven loci, respectively, as determined by MLST and had completely different alleles at the pbp1a, pbp2b, and pbp2x genes (19). The six resistant serotype 6B isolates from Thailand were identical by MLST and were single-locus variants of the Spain6B-2 clone. These isolates were previously correctly identified by Hermans et al. (19) as members of the Spain6B-2 clone, but by restriction fragment end labeling, they were scattered across the part of the dendrogram which included the members of this clone. MLST was therefore able to identify in Thailand a distinctive variant of the Spain6B-2 clone which was not apparent by a method that depends on computer-assisted comparisons of DNA fragment sizes on gels. The identification of one multidrug-resistant serotype 6B isolate from Taiwan with the same allelic profile as the isolates from Thailand suggests that this distinctive variant of the Spain6B-2 clone has spread within Southeast Asia and also demonstrates the power of MLST to identify unambiguously a single isolate whose allelic profile matches a particular allelic profile within a database of any size.
The definition of the France9V-3 clone is likely to be slightly problematic by any typing procedure since the genotypes of all serotype 9V isolates (penicillin susceptible or penicillin resistant) appear to be closely related (11, 18), and this was the only Spanish resistant clone where there were penicillin-susceptible isolates which had allelic profiles that differed from those of the resistant clones at a single locus. Furthermore, three serotypes were represented and there was a poor congruence between serotype and genotype among the cluster of isolates that are most similar to the France9V-3 clone (Fig. 2), suggesting that changes of serotype may been more common among these isolates than among those of most other pneumococcal lineages (11). Examination of the pbp gene fingerprints is therefore essential for distinguishing isolates of the France9V-3 clone from genotypically similar penicillin-resistant isolates (e.g., isolate M295) that have emerged independently.
With one exception, all of the allelic variants of the France9V-3 clone that possessed allele 14 at the ddl locus were penicillin susceptible, whereas those with allele 1 were penicillin resistant. The ddl locus is known to be 783 bp downstream of pbp2b, and recombinational exchanges that replace the normal pbp2b gene of penicillin-susceptible isolates with that from related streptococcal species or from another penicillin-resistant pneumococcus often extend into or through ddl (13). A likely scenario is that a penicillin-susceptible serotype 9V clone (allelic profile 7-11-10-1-6-8-14) was the progenitor of the France9V-3 clone (allelic profile 7-11-10-1-6-8-1) and that, during the emergence of penicillin resistance, the recombinational exchange that introduced the mosaic pbp2b gene (probably from an isolate of the Spain23F-1 clone [4]) extended into ddl to result in a change from allele 14 to allele 1. The one exceptional isolate, which possessed ddl allele 14 but which was penicillin resistant, almost certainly emerged independently, as it possessed pbp1a, pbp2b, and pbp2x genes that differed from those of the France9V-3 clone.
Penicillin-resistant isolates that have the allelic profile 7-11-10-1-6-8-1 are clearly assigned as members of the France9V-3 clone. Penicillin-resistant isolates that differ from the typical allelic profile at a single locus are also likely to be members of the clone (although caution is required if they have a different ddl allele), especially if they are susceptible to chloramphenicol and tetracycline and resistant to trimethoprim-sulfamethoxazole. Comparison of the pbp gene fingerprints to those of a reference isolate of the France9V-3 clone is required for a definitive assignment to the clone.
All previously characterized isolates of the Spain14-5 clone that had the characteristic pbp gene fingerprints of this clone were identical by MLST or differed from the typical allelic profile at a single locus. Penicillin-resistant isolates can therefore be assigned as members of the Spain14-5 clone if they have the typical allelic profile of this clone or have a profile that differs from the typical profile at a single locus.
It is interesting that the allelic profiles of the penicillin-resistant and multidrug-resistant Spanish clones are not uniform and that the profiles of about 25% of the members of the Spanish clones differ from those of typical isolates of the Spanish clones at one of the seven housekeeping genes. The common ancestors of these clones must have emerged very recently on an evolutionary time scale and have certainly emerged since the introduction of penicillin in the 1940s, and it might be expected that the profiles of members of these very young clones would be identical at these neutral loci. The relatively rapid rate of clonal diversification in the resistant clones, which will make it increasingly difficult to define them unambiguously by MLST or any other molecular typing procedure, has been shown to be due to high rates of recombinational exchanges in the naturally transformable pneumococcus (14).
In conclusion, penicillin-resistant and multidrug-resistant isolates can be assigned to the major penicillin-resistant and multidrug-resistant Spanish clones by MLST if they have the typical allelic profile of the clone or if their profiles differ from the typical profile at a single locus. However, definitive assignment to a clone by MLST (or any other typing procedure) may require an analysis of their pbp1a, pbp2b, and pbp2x genes.
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust. B.G.S. is a Wellcome Trust Principal Research Fellow.
We thank Tracey Coffey for allowing the use of unpublished data, David Griffiths and the Oxford Vaccine Group for serotyping, and Marcel Sluijter for providing pneumococcal isolates.
REFERENCES
- 1.Anonymous. A guide to sensitivity testing. Report of the Working Party on Antibiotic Sensitivity Testing of the British Society for Antimicrobial Chemotherapy. J Antimicrob Chemother. 1991;27(Suppl. D):1–50. [PubMed] [Google Scholar]
- 2.Camou T, Hortal M, Tomasz A. The apparent importation of penicillin-resistant capsular type 14 Spanish/French clone of Streptococcus pneumoniae into Uruguay in the early 1990's. Microb Drug Resist. 1998;4:219–224. doi: 10.1089/mdr.1998.4.219. [DOI] [PubMed] [Google Scholar]
- 3.Casal J. Antimicrobial susceptibility of Streptococcus pneumoniae: serotype distribution of penicillin-resistant strains in Spain. Antimicrob Agents Chemother. 1982;22:222–225. doi: 10.1128/aac.22.2.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 5.Coffey T J, Berrón S, Daniels M, Garcia-Leoni M E, Cercenado E, Bouza E, Fenoll A, Spratt B G. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988–1994): novel major clones of serotypes 14, 19F and 15F. Microbiology. 1996;142:2747–2757. doi: 10.1099/13500872-142-10-2747. [DOI] [PubMed] [Google Scholar]
- 6.Coffey T J, Enright M C, Daniels M, Wilkinson P, Berrón S, Fenoll A, Spratt B G. Serotype 19A variants of the Spanish serotype 23F multiresistant clone of Streptococcus pneumoniae. Microb Drug Resist. 1998;4:51–55. doi: 10.1089/mdr.1998.4.51. [DOI] [PubMed] [Google Scholar]
- 7.Coffey T J, Enright M C, Daniels M, Morona J K, Morona R, Hryniewicz W, Paton J C, Spratt B G. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27:73–83. doi: 10.1046/j.1365-2958.1998.00658.x. [DOI] [PubMed] [Google Scholar]
- 8.Coffey T J, Daniels M, Enright M C, Spratt B G. Serotype 14 variants of the Spanish penicillin-resistant serotype 9V clone of Streptococcus pneumoniae arose by large recombinational replacements of the cpsA-pbp1a region. Microbiology. 1999;145:2023–2031. doi: 10.1099/13500872-145-8-2023. [DOI] [PubMed] [Google Scholar]
- 9.Corso A, Severina E P, Petruk V F, Mauriz Y R, Tomasz A. Molecular characterization of penicillin-resistant Streptococcus pneumoniae isolates causing respiratory disease in the United States. Microb Drug Resist. 1998;4:325–337. doi: 10.1089/mdr.1998.4.325. [DOI] [PubMed] [Google Scholar]
- 10.Crook D W M, Spratt B G. Multidrug resistance in Streptococcus pneumoniae. Br Med Bull. 1998;54:593–608. doi: 10.1093/oxfordjournals.bmb.a011713. [DOI] [PubMed] [Google Scholar]
- 11.Enright M C, Spratt B G. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology. 1998;144:3049–3060. doi: 10.1099/00221287-144-11-3049. [DOI] [PubMed] [Google Scholar]
- 12.Enright M C, Fenoll A, Griffiths D, Spratt B G. The three major Spanish clones of penicillin-resistant Streptococcus pneumoniae are the most common clones recovered from recent cases of meningitis in Spain. J Clin Microbiol. 1999;37:3210–3216. doi: 10.1128/jcm.37.10.3210-3216.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright M C, Spratt B G. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol Biol Evol. 1999;16:1687–1695. doi: 10.1093/oxfordjournals.molbev.a026082. [DOI] [PubMed] [Google Scholar]
- 14.Feil, E. J., J. Maynard Smith, M. C. Enright, and B. G. Spratt. Estimating recombinational parameters in Streptococcus pneumoniae from multilocus sequence typing data. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 15.Fenoll A, Martín Bourgon C, Muñoz R, Vicioso D, Casal J. Serotype distribution and antimicrobial resistance of Streptococcus pneumoniae isolates causing systemic infections in Spain, 1979–1989. Rev Infect Dis. 1991;13:56–60. doi: 10.1093/clinids/13.1.56. [DOI] [PubMed] [Google Scholar]
- 16.Fenoll A, Jado I, Vicioso D, Pérez A, Casal J. Evolution of Streptococcus pneumoniae serotypes and antibiotic resistance in Spain: update (1990–1996) J Clin Microbiol. 1998;36:3447–3454. doi: 10.1128/jcm.36.12.3447-3454.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figueiredo A M, Austrian R, Urbaskova P, Teixeira L A, Tomasz A. Novel penicillin-resistant clones of Streptococcus pneumoniae in the Czech Republic and in Slovakia. Microb Drug Resist. 1995;1:71–78. doi: 10.1089/mdr.1995.1.71. [DOI] [PubMed] [Google Scholar]
- 18.Gasc A M, Giammarinaro P, Ton-Hoang B, Geslin P, van der Giezen M, Sicard M. Structural organization of the Streptococcus pneumoniae chromosome and relatedness of penicillin-sensitive and -resistant strains in type 9V. Microb Drug Resist. 1997;3:65–72. doi: 10.1089/mdr.1997.3.65. [DOI] [PubMed] [Google Scholar]
- 19.Hermans P W, Sluijter M, Dejsirilert S, Lemmens N, Elzenaar K, van Veen A, Goessens W H, de Groot R. Molecular epidemiology of drug-resistant pneumococci: toward an international approach. Microb Drug Resist. 1997;3:243–251. doi: 10.1089/mdr.1997.3.243. [DOI] [PubMed] [Google Scholar]
- 20.Klugman K P, Coffey T J, Smith A, Wasas A, Myers M, Spratt B G. Cluster of an erythromycin-resistant variant of the Spanish multiply resistant 23F clone of Streptococcus pneumoniae in South Africa. Eur J Clin Microbiol. 1994;13:171–174. doi: 10.1007/BF01982193. [DOI] [PubMed] [Google Scholar]
- 21.Klugman K P. Epidemiology, control, and treatment of multiresistant pneumococci. Drugs. 1996;52:S2–S6. doi: 10.2165/00003495-199600522-00009. [DOI] [PubMed] [Google Scholar]
- 22.Lefevre J C, Bertrand M A, Faucon G. Molecular analysis by pulsed-field gel electrophoresis of penicillin-resistant Streptococcus pneumoniae from Toulouse, France. Eur J Clin Microbiol Infect Dis. 1995;14:491–497. doi: 10.1007/BF02113426. [DOI] [PubMed] [Google Scholar]
- 23.Louie M, Louie L, Papia G, Talbot J, Lovgren M, Simor A E. Molecular analysis of the genetic variation among penicillin-susceptible and penicillin-resistant Streptococcus pneumoniae serotypes in Canada. J Infect Dis. 1999;179:892–900. doi: 10.1086/314664. [DOI] [PubMed] [Google Scholar]
- 24.Maiden M C J, Bygraves J A, Feil E, Morelli G, Russell J E, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant D A, Feavers I M, Achtman M, Spratt B G. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGee L, Klugman K P, Friedland D, Lee H J. Spread of the Spanish multi-resistant serotype 23F clone of Streptococcus pneumoniae to Seoul, Korea. Microb Drug Resist. 1997;3:253–257. doi: 10.1089/mdr.1997.3.253. [DOI] [PubMed] [Google Scholar]
- 26.Muñoz R, Coffey T J, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, Tomasz A. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 27.Scott J A G, Hall A J, Dagan R, Dixon J M S, Eykyn S J, Fenoll A, Hortal M, Jette L P, Jorgensen J H, Lamothe F, Latorre C, MacFarlane J T, Shlaes D M, Smart L E, Tauney A. Serogroup-specific epidemiology of Streptococcus pneumoniae—associations with age, sex, and geography in 7,000 episodes of invasive disease. Clin Infect Dis. 1996;22:973–981. doi: 10.1093/clinids/22.6.973. [DOI] [PubMed] [Google Scholar]
- 28.Shi Z-Y, Enright M C, Wilkinson P, Griffiths D, Spratt B G. Identification of three major clones of multiply antibiotic-resistant Streptococcus pneumoniae in Taiwanese hospitals using multilocus sequence typing. J Clin Microbiol. 1998;36:3514–3519. doi: 10.1128/jcm.36.12.3514-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sibold C, Wang J, Henrichsen J, Hakenbeck R. Genetic relationships of penicillin-susceptible and -resistant Streptococcus pneumoniae strains isolated on different continents. Infect Immun. 1992;60:4119–4126. doi: 10.1128/iai.60.10.4119-4126.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith A M, Klugman K P. Three predominant clones identified within penicillin-resistant South African isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:385–389. doi: 10.1089/mdr.1997.3.385. [DOI] [PubMed] [Google Scholar]
- 31.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980's. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 32.Spratt B G. Multilocus sequence typing: molecular typing of bacterial pathogens in an era of rapid DNA sequencing and the Internet. Curr Opin Microbiol. 1999;2:312–316. doi: 10.1016/S1369-5274(99)80054-X. [DOI] [PubMed] [Google Scholar]
- 33.Takala A K, Vuopio-Varkila J, Tarkka E, Leinonen M, Musser J M. Subtyping of common pediatric pneumococcal serotypes from invasive disease and pharyngeal carriage in Finland. J Infect Dis. 1996;173:128–135. doi: 10.1093/infdis/173.1.128. [DOI] [PubMed] [Google Scholar]
- 34.Tarasi A, Chong Y, Lee K, Tomasz A. Spread of the serotype 23F multidrug-resistant Streptococcus pneumoniae clone to South Korea. Microb Drug Resist. 1997;3:105–109. doi: 10.1089/mdr.1997.3.105. [DOI] [PubMed] [Google Scholar]
- 35.Tomasz A. Antibiotic resistance in Streptococcus pneumoniae. Clin Infect Dis. 1997;24:S85–S88. doi: 10.1093/clinids/24.supplement_1.s85. [DOI] [PubMed] [Google Scholar]
- 36.Versalovic J, Kapur V, Mason E O, Shah U, Koeuth T, Lupski J R, Musser J M. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J Infect Dis. 1993;167:850–856. doi: 10.1093/infdis/167.4.850. [DOI] [PubMed] [Google Scholar]